Summary
A common strategy for multi-protein expression is to link genes by self-cleaving 2A peptide sequences. Yet, little is known how the 2A peptide-derived N-terminal proline or adjacent non-native residues introduced during cDNA cloning affects protein stoichiometry. Polycistronic reprogramming constructs with altered KLF4 protein stoichiometry can influence induced pluripotent stem cell (iPSC) generation. We studied the impact of N-terminal 2A peptide-adjacent residues on the protein stability of two KLF4 isoforms, and assayed their capacity to generate iPSCs. Here, we show that the N-terminal proline remnant of the 2A peptide, alone or in combination with leucine, introduced during polycistronic cloning, destabilizes KLF4 resulting in increased protein degradation, which hinders reprogramming. Interestingly, the addition of charged and hydrophilic amino acids, such as glutamate or lysine stabilizes KLF4, enhancing reprogramming phenotypes. These findings raise awareness that N-terminal modification with 2A peptide-derived proline or additional cloning conventions may affect protein stability within polycistronic constructs.
Keywords: 2A peptide, stoichiometry, KLF4, protein stability, proline/N-end rule, polycistronic cloning, MG132, cycloheximide, iPSCs, reprogramming
Graphical Abstract

Highlights
-
•
2A peptide-derived N-terminal adjacent non-native residues affect KLF4 stability
-
•
KLF4 stability is related with amino acid charge and hydrophobicity at the N-terminus
-
•
Reprogramming phenotypes are highly associated with KLF4 stability
In this article, Woltjen and colleagues show that protein expression levels in polycistronic reprogramming constructs depend upon the method of 2A peptide linkage. Specifically, the 2A peptide-derived N-terminal proline along with non-native amino acids derived during cloning affect the stability of KLF4. Their results correlate with discrepancies in reprogramming outcomes and reports of cDNA order-effects in other 2A-linked constructs.
Introduction
Multigene expression is commonly employed to introduce reporter genes (Zheng et al., 2018), investigate multi-protein complexes (Momose and Morikawa, 2016), conduct cellular reprogramming (Okita et al., 2008, Kaji et al., 2009, Okita et al., 2013), or induce differentiation (Wang et al., 2015, Imamura et al., 2017). A widely used strategy to achieve fixed stoichiometric multi-protein expression is viral-derived 2A peptides that link cDNAs to form polycistronic constructs that produce independent proteins. 2A peptides, also known as CHYSEL (cis-acting hydrolase element) (de Felipe, 2004), are 18–22 amino acids long (Szymczak-Workman et al., 2012) and share the functional motif DxExNPGˆP (Donnelly et al., 2001). This motif appears to interrupt translation such that the formation of the last glycine-proline linkage is “skipped” by the ribosome, releasing the upstream protein which is appended by the C-terminal 2A peptide sequence. Translation continues using proline as the first amino acid in the downstream protein (Donnelly et al., 2001, Doronina et al., 2008).
It has been shown that protein stoichiometry can be influenced by gene order in polycistronic constructs (Wang et al., 2015). The appended C-terminal 2A peptides can impact stability of the upstream protein (Hasegawa et al., 2007, Lengler et al., 2005), while less is known about the destabilizing effect of the N-terminal proline modification (Momose and Morikawa, 2016). Furthermore, additional non-native amino acid residues can modify the N terminus, as unique restriction sites are often introduced downstream of the 2A peptide during polycistronic cDNA cloning (Liu et al., 2017, Szymczak-Workman et al., 2012). N-Terminal proline [P] and up to five adjacent residues can destabilize proteins by recruiting proline/N-end rule pathway triggering proteasomal degradation (Chen et al., 2017), implicating N-terminal sequence identity in determining protein stability.
The transcription factor Krüppel-like factor 4 (KLF4) has been predicted to exist as one of two possible isoforms: a 483-amino acid long isoform (KLF4L) and a shorter 474-amino acid isoform truncated by nine N-terminal amino acids (KLF4S) (Shields et al., 1996, Garrett-Sinha et al., 1996). A comparison of polycistronic vectors expressing either isoform together with OCT3/4, SOX2, and c-MYC revealed distinct KLF4 protein stoichiometry and induced pluripotent stem cell (iPSC) phenotypes (Kim et al., 2015, Kagawa et al., 2018). Polycistronic vectors with KLF4S had low expression resulting in partially reprogrammed cells, while those with KLF4L achieved high expression leading to improved reprogramming efficiencies. Because monocistronic expression of either KLF4 isoform results in similar expression levels (Kim et al., 2015), it seems likely that the cause of altered KLF4 stoichiometry originates from the 2A peptide polycistronic system rather than the native KLF4 N-terminal residues alone.
Based on this model, we investigated if 2A peptide-derived proline [P] and adjacent residues affect KLF4 stability. Here, we show that the 2A peptide-derived proline and non-native amino acids, such as leucine introduced by cloning impairs protein stability of the KLF4S isoform. Addition of glutamate increases KLF4S expression and rescues reprogramming outcomes. Our findings reveal that additional N-terminal residues introduced by 2A peptides or cloning conventions can impact protein stability and their effect should be considered when using polycistronic expression systems.
Results
Additional Non-native N-Terminal Amino Acid Residues Impairs Expression of KLF4S in a Polycistronic System
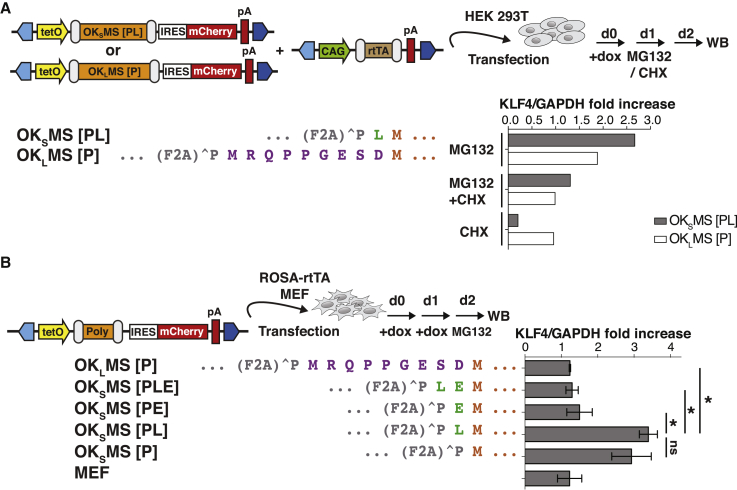
To establish if additional N-terminal amino acids from the 2A peptide or from polycistronic cloning are correlated with reduced KLF4 protein expression we overexpressed various mono- and polycistronic constructs in HEK293T cells (Figure 1A). First, we compared both KLF4 isoforms (GenBank: U70662.1 and U20344.1) without N-terminal modification in doxycycline (dox)-inducible monocistronic (KLF4S and KLF4L) or polycistronic (KSMS and KLMS) systems (Figures 1A, S1A, and S2A). Overexpression resulted in comparable KLF4 protein levels between monocistronic vectors or between KSMS and KLMS polycistronic vectors, although expression in a polycistronic context was approximately half that of the monocistronic. Furthermore, we verified our previous report (Kim et al., 2015) showing that significantly reduced KLF4 expression is observed for the polycistronic reprogramming vector OKSMS [PL] compared with the elongated OKLMS [P] (Figure 1A). These data suggest that comparable expression of KLF4 isoforms can be achieved in a polycistronic context when KLF4 is placed in the first position without 2A peptide modification, suggesting that the non-native amino acids proline-leucine [PL] or proline [P] preceding methionine also impact KLF4S levels in OKSMS [PL].
Figure 1.
N-Terminal Residues Impact KLF4S Expression in Polycistronic Vectors
(A) The top panel depicts the schedule of transient transfection of HEK293T cells. The left side shows monocistronic KLF4S and KLF4L, the cassette structure of polycistronic constructs and the sequences of their 2A peptide-KLF4 junctions. The two KLF4 isoforms used to create the original (underlined) OKMS (KLF4S) and OSKM (KLF4L) were based on the predicted open-reading frames of murine KLF4 (GenBank: U70662.1 and U20344.1, respectively). OKSMS [PE] and OSKLM [PLE] were engineered to express the OKLMS [P] and OSKSM [PLE] isoform. The right side depicts western blot analysis of KLF4 expression that was normalized to GADPH. Additional N-terminal amino acids generated during polycistronic cloning by the Xho1 restriction site are highlighted in green. E2A (blue dot), equine rhinitis A virus; P2A (gray dot), porcine teschovirus-1 2A; T2A (scarlet dot), thosea asigna virus 2A; F2A (green dot), foot-and-mouth disease virus. Means ± SEM for three independent experiments. ∗p < 0.05 of two-sided unpaired t tests against OKSMS [PL].
(B) The top panel depicts schedule of transient transfection of MEF cells. Left visualizes polycistronic constructs and sequences of the 2A peptide-KLF4 junctions. Right shows western blot analysis on reprogramming day 2 (d2). KLF4 expression levels were normalized to GAPDH. Means ± SEM for three independent experiments. ∗p < 0.05 of two-sided unpaired t tests against OKSMS [PL].
To investigate this effect further, we employed a second reprogramming vector OSKLM (Carey et al., 2009) that includes a “CTCGAG” XhoI restriction endonuclease site resulting in [PLE] preceding the KLF4 methionine (Figure S2A). OSKLM [PLE] has been shown to produce higher levels of KLF4 than OKSMS [PL] (Kim et al., 2015), which we verified by quantitative densitometry (Figure 1A). Since OSKLM [PLE] originally expressed KLF4L, we eliminated the nine N-terminal amino acids to specifically test the effect of the preceding [PLE] on KLF4S. Surprisingly, the expression of OSKSM [PLE] was similar to OSKLM (Figure 1A). These data suggest that the identity of the preceding non-native amino acids affects KLF4S expression levels in polycistronic overexpression.
To understand how individual non-native N-terminal amino acids impact KLF4S expression in a relevant cell system, we systematically generated all preceding [PLE] variants and measured their expression at the onset of mouse embryonic fibroblast (MEF) reprogramming (Figure 1B). Expression of transgenes was induced with dox 24 h after transfection of MEFs and protein expression was detected after 48 h by western blot analysis. Consistent with expression in HEK293T cells (Figure 1A), removal of the nine N-terminal amino acids in OSKLM [PLE] to generate OSKSM [PLE] did not have a major impact on KLF4 levels (Figure 1B). Similarly, preceding KLF4S with [PLE] significantly increased expression in OKSMS [PLE], an effect nearly as dramatic as elongating KLF4S with the nine amino acids of the KLF4L isoform (OKLMS [P]). Interestingly, we found N-terminal PE sufficient to maintain high KLF4S expression in OKSMS [PE] and OSKSM [PE], and its absence in both OKSMS and OSKSM [PL] or [P] variants led to the lowest KLF4 protein levels. Modifying the preceding N-terminal amino acids of KLF4S did not impact expression of other reprogramming factors, as shown for OCT3/4 (Figures S1B and S1C).
In summary, preceding KLF4S with proline [P] or proline plus leucine [PL] diminished protein expression in polycistronic OSKSM and OKSMS reprogramming vectors, while addition of a glutamate residue [E] enhances KLF4S expression.
Hydrophobic N-Terminal Non-native Amino Acids Cause Proteasomal Degradation of KLF4S
Because a hydrophobic N-terminus can impair protein stability (Abe et al., 2014), we evaluated the hydrophobicity of the N-terminus for each KLF4 variant (Figures S2A and S2B). The first nine N-terminal amino acids of KLF4L encode mostly hydrophilic residues, while the N-terminus of KLF4S was found to be mainly hydrophobic (Figure S2A, right). The hydrophilic N-terminal amino acids of KLF4L are retained in the high-KLF4 expression vector OKLMS [P], suggesting improved protein stability as the mechanism. Similarly, we found previously that unrelated amino acids encoded by the hydrophilic HA tag in OKS+HAMS [P + HA] also give rise to increased KLF4 levels, and characteristic gene expression in reprogramming (Kim et al., 2015). Interestingly, [P] and [PL] preceding the hydrophobic N-terminus of KLF4S resulted in further raised hydrophobicity scores suggesting that OKSMS [P] and [PL] are subject to protein instability. In contrast, inclusion of glutamate in [PLE] and [PE], which is correlated with increased KLF4S protein levels, reduced hydrophobicity scores for OKSMS [PLE] and [PE].
To elucidate whether protein degradation or aberrant synthesis causes reduced KLF4S expression, we transfected HEK293T cells with polycistronic KLF4 constructs and treated them with the proteasome inhibitor MG132 or prevented protein synthesis with cycloheximide (CHX) (Figures 2A and S2C). Inhibiting protein degradation resulted in a higher fold-change for OKSMS [PL] (2.7-fold) compared with OKLMS [P] (1.9-fold), indicating that N-terminal PL-modified KLF4S is prone to proteasomal degradation. In accordance, blocking new protein synthesis depleted OKSMS [PL] (0.2-fold) but had a minimal effect on OKLMS [P] (0.95-fold), suggesting reduced stability of KLF4S and increased resistance to proteasomal degradation for KLF4L.
Figure 2.
N-Terminal Glutamate Stabilizes the KLF4S Isoform
(A) The top depicts schedule of transient transfection of HEK293T cells with OKSMS [PL] and OKLMS [P]. Cells were treated with 20 μM MG132, with 100 μg/mL CHX or both for 24 h before western blot analysis. The left side shows conditions, polycistronic constructs, and sequences of the 2A peptide-KLF4 junctions. The right side depicts western blot analysis. The KLF4/GAPDH ratio was normalized to DMSO conditions.
(B) The top panel depicts schedule of transient transfection of MEF cells with 5 μM MG132 treatment for 2 h before harvesting. The left visualizes polycistronic constructs and sequences of the 2A peptide-KLF4 junctions. The right shows western blot analysis on reprogramming day 2 (d2). The KLF4/GAPDH ratio was normalized to DMSO conditions. Means ± SEM for three independent experiments. ∗p < 0.05 of two-sided unpaired t tests against OKSMS [PL].
To check the effect of individual preceding non-native amino acids on KLF4S degradation in early reprogramming, transfected MEFs were treated with MG132 (Figures 2B, S2D, and S2E). For all constructs tested, MG132 enriched ubiquitinylation (Figure S2E) and enhanced OCT3/4 expression independent of N-terminal KLF4 modification (Figure S2D). Furthermore, MG132 did not show any detectable enhancement of endogenous KLF4 or OCT3/4 in untreated MEFs (Figure S2D). Proteasome inhibition enhanced KLF4S expression in OKSMS [P] and [PL] demonstrating indeed that N-terminal [P] and [PL] together with the native N-terminal hydrophobic amino acids destabilize KLF4S. In contrast, MG132 treatment had significantly less effect on KLF4 levels for constructs with reduced N-terminal hydrophobicity scores, such as OKLMS [P] and OKSMS [PLE] and [PE] vectors protected with the hydrophilic N-terminal glutamate.
Because hydrophilic and negatively charged glutamate enhances KLF4 stability we also tested the effect of hydrophilic non-charged serine [PS] versus positively charged lysine [PK] on KLF4 expression (Figure S3A). Interestingly, only [PK] significantly enhanced KLF4 expression in HEK293T cells, indicating that a combination of charged and hydrophilic N-terminal amino acids leads to increased KLF4 expression. Finally, the preceding N-terminal amino acids did not appear to impact 2A peptide cleavage efficiency, since similar levels of uncleaved higher-molecular-weight protein were detected from all constructs (Figure S3B).
KLF4 N-Terminal Modifications Correlate with Reprogramming Intermediates and Outcomes
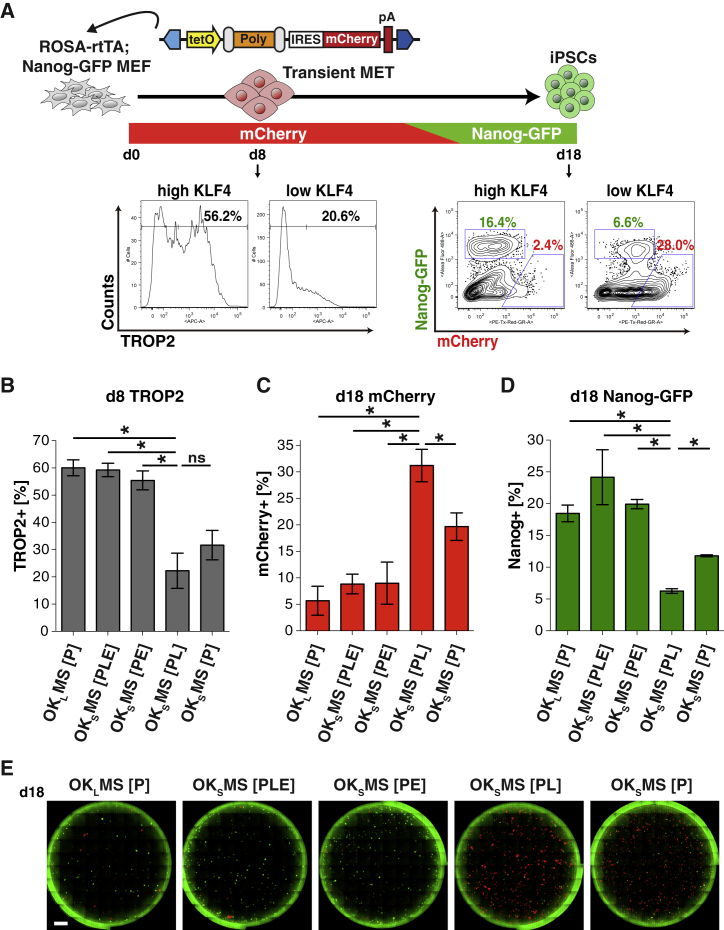
Finally, we assessed how the wide range of KLF4 levels induced by OKMS variants affect reprogramming paths and outcomes (Figure 3A). Reprogramming is largely influenced by the KLF4 stoichiometry (Kim et al., 2015, Carey et al., 2011). Specific to high-KLF4 reprogramming, an early population of cells undergoing a transient mesenchymal-to-epithelial transition (MET) emerges, which is marked by the cell surface marker TROP2 (Kagawa et al., 2018). Moreover, transient MET induces genes that suppress the expansion of partially reprogrammed cells––an alternative cell lineage marked by retained expression of the factor-linked mCherry reporter and failure to activate the Nanog-GFP reporter, which indicates pluripotency––such that high-KLF4 results in a majority of mCherry−, Nanog-GFP+ iPSCs. On the other hand, low-KLF4 reprogramming populations do not activate TROP2 and result in an overwhelming population of mCherry+, Nanog-GFP– partially reprogrammed cells.
Figure 3.
KLF4 Stability Correlates with Reprogramming Intermediates and Outcomes
(A) Schematic depicting the reprogramming process and expression pattern of transient MET marker TROP2 and reprogramming markers mCherry and Nanog-GFP induced by different polycistronic cassettes. Representative flow cytometry analyses for TROP2 (d8) and mCherry versus Nanog-GFP (d18) using OKLMS [P] and OKSMS [PL] (d18).
(B) Proportions of TROP2+ cells by flow cytometry analysis on reprogramming d8 using polycistronic cassettes with N-terminal KLF4 modifications described in Figure 1B. Means ± SEM for three independent experiments. ∗p < 0.05 of two-sided unpaired t tests against OKSMS [PL].
(C) Proportions of mCherry+ cells by flow cytometry analysis on reprogramming d18 using polycistronic cassettes with N-terminal KLF4 modifications described in Figure 1B. Means ± SEM for three independent experiments. ∗p < 0.05 of two-sided unpaired t tests against OKSMS [PL].
(D) Proportions of Nanog-GFP+ cells by flow cytometry analysis on reprogramming d18 using polycistronic cassettes with N-terminal KLF4 modifications described in Figure 1B. Means ± SEM for three independent experiments. ∗p < 0.05 of two-sided unpaired t tests against OKSMS [PL].
(E) Whole-well fluorescence microscopy images of Nanog-GFP and mCherry on reprogramming d18. Scale bar, 4,000 μm.
As predicted in accordance with their KLF4 expression (Figure 1B), OKLMS [P] induced a higher fraction of TROP2+ cells than OKSMS [PL] (60% versus 22%) (Figure 3B). Consistent with KLF4 protein expression levels detected by western blot analysis (Figure 1B), high-KLF4 polycistronic cassettes, including OKSMS [PE] and [PLE] produced significantly greater proportions of TROP2+ cells equally as well as OKLMS [P], while the moderate KLF4 expression levels of OKSMS [P] induced TROP2+ cells at an intermediate frequency (31%) (Figure 3B). Assessing reprogramming outcomes, we found that OKSMS [P] and [PL] caused a robust expansion of mCherry+ partially reprogrammed cells and reduced activation of Nanog-GFP (Figures 3C–3E). OKSMS [PL], the most destabilized form of KLF4S, presented the lowest percentage of TROP2+ (d8) Nanog-GFP+ (d18) cells. Importantly, similar to OKLMS [P], OKSMS [PLE] and [PE] induced expression of Nanog-GFP+ efficiently (>18%) with reduced mCherry+ (<10%), indicating a high purity reprogramming despite employing the short KLF4 isoform (Figures 3C–3E). These data support a tight correlation between KLF4 N-terminal modifications, protein stoichiometry, and the characteristics of reprogramming intermediates and outcomes.
Discussion
In this study, we illustrate how additional non-native amino acids introduced by cloning sites and the 2A peptide-derived proline affects protein stoichiometry in polycistronic constructs exemplified by KLF4S isoforms in OSKM and OKMS reprogramming vectors. Both monocistronic isoforms, which only differ in length by nine N-terminal residues, show similar expression. Until now it remained unclear why the two variants of KLF4 could give rise to different protein expression solely in polycistronic systems (Kagawa et al., 2018, Kim et al., 2015). In the current study, inhibited proteasomal degradation led to enhanced accumulation of KLF4 from OKSMS [P] and [PL] vectors, demonstrating that the preceding non-native [P] and [PL] introduced by cloning or the 2A peptide destabilize KLF4S in a polycistronic system.
The “N-end” rule proposes the effect of N-terminal amino acids on protein stability. In the P-N-end rule, [P] and also penultimate residues are involved in recruitment of the GID ubiquitin ligase subunit GID4, while adjacent [E] residues are prone to escape from the recognition (Chen et al., 2017). The principle of these rules is still not fully understood, but hydrophobicity of N-terminal residues is one of the regulatory factors in the protein stability (Abe et al., 2014). Consistent with these previous reports, we found an enhanced hydrophobicity at the N-terminus of KLF4S suggesting that addition of [P] and [PL] triggers its proteasomal degradation. Unavoidably, other 2A-linked polycistronic reprogramming vectors (STEMCCA, WTSI, EB-C5) also possess N-terminal [P] residues and show low KLF4S expression and predictably altered reprogramming outcomes (Kim et al., 2015). Interestingly, just one additional [E] in OKSMS and OSKSM [PE] or [PLE] can enhance KLF4S expression to a similar extent as the nine hydrophilic N-terminal amino acids of KLF4L. In addition, lysine [PK] significantly increases KLF4S expression but not hydrophilic non-charged serine [PS] suggesting that the combination of charged and hydrophilic N-terminus supports KLF4s stability. Our observations suggest that a stretch of hydrophilic amino acids or even a singular glutamate or lysine could be used to counteract instability of other susceptible proteins with hydrophobic N-termini in polycistronic constructs.
Various reports suggest that gene order in 2A-peptide constructs affects protein stoichiometry (Liu et al., 2017, Wang et al., 2015). Using fluorescent reporter proteins, Liu and colleagues concluded that protein expression decreased if a cDNA was placed closer to the end in a polycistronic construct. However, apart from gene order, N-terminal [PLE] derived from an Xho1 restriction site following T2A correlated with enhanced GFP expression (Liu et al., 2017). Furthermore, N-terminal [PLE] increased Gata4 and Tbx5 expression when linked by T2A to Mef2c (Wang et al., 2015). These results are consistent with our findings that altering N-terminal hydrophobicity or amino acid charge during 2A-peptide linkage can alter the stability of KLF4. In addition, C-terminal alterations (Hasegawa et al., 2007, Lengler et al., 2005, Minskaia and Ryan, 2013), and elimination of the starting methionine (Momose and Morikawa, 2016) have been shown to affect protein expression or 2A cleavage efficiency, and likely influence studies of gene order and desired experimental outcomes. These data emphasize the importance of seemingly minor modifications to protein sequence on protein stability, and indicate the importance of pre-screening polycistronic constructs for optimal protein stoichiometry to achieve a desired effect.
Experimental Procedures
Plasmid Construction
A three-fragment InFusion (Clontech) cloning strategy combining two PCR fragments overlapping the F2A-Klf4 junction with a restriction enzyme-digested polycistronic cDNA construct was used to modify the N-terminal codons of Klf4. Primers used for cloning are listed in Table S1. Each primer pair consists of an External Fwd and Internal Rev for the 5′ fragment or an Internal Fwd and External Rev for the 3′ fragment, where the overlapping InFusion sequences of the Internal primers modify the relevant F2A-Klf4 codons, and External primers overlap the restriction sites in the recipient plasmid. To remove the nine N-terminal codons from Klf4 in OSKM and create PB-TAC-OSK-9M, a BspHI-BstZ17I restriction fragment in pENTR-OSKM was replaced by three-fragment InFusion, followed by Gateway (Invitrogen) cloning to PB-TAC (Kim et al., 2015). For additional modification of Klf4 in OSK-9M, an AatII-AfeI restriction fragment was replaced in PB-TAC-OSK-9M. Modification of Klf4 in OKMS an AflII-AfeI fragment was replaced in PB-TAC-OKMS (Kim et al., 2015) or PB-TAC-OKMS [–L + E] (this study). To create PB-TAC-OKMS [–L], a SalI-BstZ17I fragment of pENTR[kan]-OKMS (Kagawa et al., 2018) was replaced by three-fragment InFusion followed by Gateway cloning to PB-TAC. The resulting plasmids, along with previously reported plasmids used in this study, are listed in Table S2. Complete sequences of plasmids are available upon request.
Cell Transfection and Chemical Treatment
HEK293T cells were cultured in DMEM containing 10% FBS, penicillin-streptomycin, and L-glutamine. Cells at 90% confluency were detached with 0.025% trypsin-EDTA for 4 min at 37°C. Next 3 × 105 cells were transfected with 500 ng of mono- or polycistronic KLF4 constructs and 500 ng of PB-CAG-rtTA (Woltjen et al., 2009) using FuGENE HD (Promega, cat. no. E2312) at a FuGENE/DNA ratio of 4:1 in accordance with the manufacturer's protocol. Cells were plated in a 6-well plate and 1 μg/mL dox was added after 24 h culture. After an additional 24 h, transfected cells were harvested with ice-cold PBS for western blot analysis. For inhibition of the proteasome or protein synthesis, 20 μM MG132 (Wako, cat. no. 135-16,253) and/or 100 μg/mL CHX (Calbiochem, cat. no. 239763) was added to the cells 24 h after dox treatment. After an additional 24 h cells were harvested with ice-cold PBS for western blot analysis.
MEF Isolation and PB Reprogramming
MEFs were isolated from E13.5 mouse embryos resulting from the mating of homozygous Nanog-GFP (Okita et al., 2007) transgenic males and homozygous ROSA26-rtTA (Ohnishi et al., 2014) transgenic females on a C57BL/6 background, or wild-type C57BL/6 mice (without ROSA26-rtTA or Nanog-GFP transgenes), and cultured as described previously (Woltjen et al., 2016). Animal experiments were approved by the CiRA Animal Experiment Committee in accordance with Kyoto University guidelines. MEFs were seeded in DMEM containing 10% FBS, penicillin-streptomycin, and L-glutamine on gelatin-coated 6-well dishes at a density of 1 × 105 cells per well. After 24 h culture, FuGENE HD (Promega, Cat.E2312) was used to transfect cells at a FuGENE/DNA ratio of 4:1. A total of 500 ng of transposons and 1,000 ng of pCyL43 PB transposase plasmid was used. After 24 h the medium was replaced with ESC medium (DMEM containing 15% FBS, penicillin-streptomycin, GlutaMAX, β-mercaptoethanol, sodium-pyruvate, non-essential amino acids, leukemia inhibitory factor, and 1 μg/mL dox). After transfection, cells were fed daily with dox-containing ESC medium. On day 8 (d8), cells were detached by using TrypLE Select (1×) (Thermo Fisher Scientific, cat. no. 12563011) and re-seeded at 3 × 105 cells per well of gelatin-coated 6-well dishes for analysis at d18. For proteasome inhibition cells were treated with 5 μM MG132 on d2 for 2 h before harvesting.
Western Blot Analysis
Reprogrammed cells were collected on d2 using 0.25% trypsin-EDTA (3 min, 37°C), neutralized with 2% FBS-PBS, and washed once using PBS before freezing at −80°C. Total cell lysates were prepared by lysing 1 × 105 cells in 7.5 μL lysis buffer (50 mM HEPES [pH 8], 200 mM NaCl, 0.1 M EDTA [pH 8], 0.5% NP-40, 10% glycerol, protease inhibitors) (Letourneau et al., 2015), ultrasonication for 5 min in ice water, followed by addition of 2.5 μL NuPAGE LDS Sample Buffer (1×) (Thermo Fisher Scientific, cat. no. NP0008) containing 50 mM DTT and denaturation at 70°C for 10 min.
HEK293T cell pellets were solubilized in 100 μL lysis buffer and ultrasonicated for 5 min in ice water. Protein concentrations were determined by Bradford assay (Apro Science, KY-1030). Total protein (100 or 200 ng) was solubilized in NuPAGE LDS Sample Buffer (1×) (Thermo Fisher Scientific, cat. no. NP0008) containing 50 mM DTT and denature at 70°C for 10 min. Lysates were resolved on NuPAGE 10% Bis-Tris gels (Thermo Fisher Scientific, cat. no. NP0316BOX), and blotted on a 0.45-μm PVDF membrane (Millipore, no. IPVH00010) using NuPAGE Transfer Buffer (1×) (Thermo Fisher Scientific no. NP00061) that was probed with antibodies described in Table S3. Signals were raised using ECL Prime Western Blotting Detection Reagent (GE Healthcare, cat. no. RPN2232), detected on an ImageQuant LAS 4000 imaging system (GE Healthcare) and analyzed with ImageQuant TL software (GE Healthcare, version 7.0).
Flow Cytometry
For cell surface marker detection, TrypLE Select (1×) (Thermo Fisher Scientific, cat.no. 12563011) was used for cell dissociation. Cells (3 × 105) were re-suspended in 100 μL of fluorescence-activated cell sorting (FACS) buffer (PBS contained 2% of FBS) and incubated with primary antibodies and appropriate secondary antibodies on ice for 30 min each. Antibodies used in this study are described in Table S3. The samples were washed with 1 mL of FACS buffer two times and analyzed using a BD LSRFortessa Cell Analyzer (BD Biosciences) with BD FACSDiva software (BD Biosciences). Flow cytometry data were analyzed and generated by FlowJo software (Tree Star, v.9.9.6). The boundaries between positive and negative were defined using un-stained samples or non-reprogrammed MEFs as negative controls.
Whole-Well Fluorescence Microscopy Imaging
Mouse fibroblasts were plated on standard tissue culture 6-well plastic plates (Greiner, cat. no. 657160). Images were acquired with a Nikon BioStation CT (Nikon) equipped with GFP and mCherry fluorescence filters and phase contrast using 2× lenses. The single-plane images of each channel were stitched automatically using the automated image analysis software CL-Quant 3.0 (Nikon).
Statistical Analysis
The data are presented as the means ± SEM or mean ± SD from indicated numbers of independent experiments.
Author Contributions
A.R. and K.W. conceived the study. A.R., H.K., and K.W. designed experiments and interpreted the data. A.R. performed western blot analysis. H.K. performed reprogramming experiments. The article was written and approved by all authors.
Acknowledgments
We would like to acknowledge the technical assistance of Michiko Nakamura and Ryoko Hirohata for plasmid construction, and Yanjun Lan for plasmid construction, transgene overexpression, and western blotting. We acknowledge Shin-Il Kim for valuable advice and constructive discussion. This work was funded in part by grants to K.W. from AMED (nos. JP17jm0210039 and JP17bm0104001), and to H.K. who is supported by a Grant-in-Aid for JSPS Research Fellows (18J14431). A.R. is supported by iPS Cell Research Fund.
Published: February 27, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.01.014.
Supplemental Information
References
- Abe T., Kojima M., Akanuma S., Iwashita H., Yamazaki T., Okuyama R., Ichikawa K., Umemura M., Nakano H., Takahashi S. N-Terminal hydrophobic amino acids of activating transcription factor 5 (ATF5) protein confer interleukin 1beta (IL-1beta)-induced stabilization. J. Biol. Chem. 2014;289:3888–3900. doi: 10.1074/jbc.M113.491217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Markoulaki S., Hanna J., Saha K., Gao Q., Mitalipova M., Jaenisch R. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc. Natl. Acad. Sci. USA. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Markoulaki S., Hanna J.H., Faddah D.A., Buganim Y., Kim J., Ganz K., Steine E.J., Cassady J.P., Creyghton M.P. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;9:588–598. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Chen S.J., Wu X., Wadas B., Oh J.H., Varshavsky A. An N-end rule pathway that recognizes proline and destroys gluconeogenic enzymes. Science. 2017;355 doi: 10.1126/science.aal3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felipe P. Skipping the co-expression problem: the new 2A "CHYSEL" technology. Genet. Vaccines Ther. 2004;2:13. doi: 10.1186/1479-0556-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly M.L., Luke G., Mehrotra A., Li X., Hughes L.E., Gani D., Ryan M.D. Analysis of the aphthovirus 2A/2B polyprotein 'cleavage' mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal 'skip'. J. Gen. Virol. 2001;82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- Doronina V.A., Wu C., de Felipe P., Sachs M.S., Ryan M.D., Brown J.D. Site-specific release of nascent chains from ribosomes at a sense codon. Mol. Cell. Biol. 2008;28:4227–4239. doi: 10.1128/MCB.00421-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Sinha L.A., Eberspaecher H., Seldin M.F., de Crombrugghe B. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J. Biol. Chem. 1996;271:31384–31390. doi: 10.1074/jbc.271.49.31384. [DOI] [PubMed] [Google Scholar]
- Hasegawa K., Cowan A.B., Nakatsuji N., Suemori H. Efficient multicistronic expression of a transgene in human embryonic stem cells. Stem Cells. 2007;25:1707–1712. doi: 10.1634/stemcells.2006-0813. [DOI] [PubMed] [Google Scholar]
- Imamura K., Izumi Y., Watanabe A., Tsukita K., Woltjen K., Yamamoto T., Hotta A., Kondo T., Kitaoka S., Ohta A. The Src/c-Abl pathway is a potential therapeutic target in amyotrophic lateral sclerosis. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aaf3962. [DOI] [PubMed] [Google Scholar]
- Kagawa H., Shimamoto R., Kim S.I., Oceguera-Yanez F., Yamamoto T., Schroeder T., Woltjen K. OVOL1 influences the determination and expansion of iPSC reprogramming intermediates. Stem Cell Reports. 2018;12:1–14. doi: 10.1016/j.stemcr.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K., Norrby K., Paca A., Mileikovsky M., Mohseni P., Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.I., Oceguera-Yanez F., Hirohata R., Linker S., Okita K., Yamada Y., Yamamoto T., Yamanaka S., Woltjen K. KLF4 N-terminal variance modulates induced reprogramming to pluripotency. Stem Cell Reports. 2015;4:727–743. doi: 10.1016/j.stemcr.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengler J., Holzmuller H., Salmons B., Gunzburg W.H., Renner M. FMDV-2A sequence and protein arrangement contribute to functionality of CYP2B1-reporter fusion protein. Anal. Biochem. 2005;343:116–124. doi: 10.1016/j.ab.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Letourneau A., Cobellis G., Fort A., Santoni F., Garieri M., Falconnet E., Ribaux P., Vannier A., Guipponi M., Carninci P. HSA21 single-minded 2 (Sim2) binding sites co-localize with super-enhancers and pioneer transcription factors in pluripotent mouse ES cells. PLoS One. 2015;10:e0126475. doi: 10.1371/journal.pone.0126475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Chen O., Wall J.B.J., Zheng M., Zhou Y., Wang L., Ruth Vaseghi H., Qian L., Liu J. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci. Rep. 2017;7:2193. doi: 10.1038/s41598-017-02460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minskaia E., Ryan M.D. Protein coexpression using FMDV 2A: effect of "linker" residues. Biomed. Res. Int. 2013;2013:291730. doi: 10.1155/2013/291730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose F., Morikawa Y. Polycistronic expression of the influenza A virus RNA-dependent RNA polymerase by using the thosea asigna virus 2A-like self-processing sequence. Front. Microbiol. 2016;7:288. doi: 10.3389/fmicb.2016.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K., Semi K., Yamamoto T., Shimizu M., Tanaka A., Mitsunaga K., Okita K., Osafune K., Arioka Y., Maeda T. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell. 2014;156:663–677. doi: 10.1016/j.cell.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Okita K., Nakagawa M., Hyenjong H., Ichisaka T., Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- Okita K., Yamakawa T., Matsumura Y., Sato Y., Amano N., Watanabe A., Goshima N., Yamanaka S. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells. 2013;31:458–466. doi: 10.1002/stem.1293. [DOI] [PubMed] [Google Scholar]
- Shields J.M., Christy R.J., Yang V.W. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J. Biol. Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak-Workman A.L., Vignali K.M., Vignali D.A. Design and construction of 2A peptide-linked multicistronic vectors. Cold Spring Harb. Protoc. 2012;2012:199–204. doi: 10.1101/pdb.ip067876. [DOI] [PubMed] [Google Scholar]
- Wang L., Liu Z., Yin C., Asfour H., Chen O., Li Y., Bursac N., Liu J., Qian L. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ. Res. 2015;116:237–244. doi: 10.1161/CIRCRESAHA.116.305547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltjen K., Kim S.I., Nagy A. The piggyBac transposon as a platform technology for somatic cell reprogramming studies in mouse. Methods Mol. Biol. 2016;1357:1–22. doi: 10.1007/7651_2015_274. [DOI] [PubMed] [Google Scholar]
- Woltjen K., Michael I.P., Mohseni P., Desai R., Mileikovsky M., Hamalainen R., Cowling R., Wang W., Liu P., Gertsenstein M. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Shen W., Zhang J., Yang B., Liu Y.N., Qi H., Yu X., Lu S.Y., Chen Y., Xu Y.Z. CRISPR interference-based specific and efficient gene inactivation in the brain. Nat. Neurosci. 2018;21:447–454. doi: 10.1038/s41593-018-0077-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.