Abstract
Majority of the imported malaria cases in Korea is attributed to Plasmodium falciparum and P. vivax infections, whereas P. malariae and P. ovale infections are very rare. Falciparum and ovale malaria are mostly imported from Africa, while most of the vivax malaria cases are imported from Southeast Asia. Here, we report 6 Korean imported ovale malaria cases (4 males and 2 females) who had visited in Africa during 2013–2016. These subjects were diagnosed with P. ovale based on microscopic findings, Plasmodium species-specific nested-PCR, and phylogenetic clade using 18S rRNA gene sequences. We identified 2 P. ovale subtypes, 1 P. ovale curtisi (classic type) and 5 P. ovale wallikeri (variant type). All patients were treated with chloroquine and primaquine, and no relapse or recrudescence was reported for 1 year after treatment. With increase of travelers to the countries where existing Plasmodium species, the risk of Plasmodium infection is also increasing. Molecular monitoring for imported malaria parasites should be rigorously and continuously performed to enable diagnosis and certification of Plasmodium spp.
Keywords: Plasmodium ovale, malaria, imported case, molecular analysis
Malaria is caused by the infection of human red blood cells by Plasmodium spp. and is transmitted through mosquitoes as vector. Five species of Plasmodium are known to be infectious to humans [1]. According to 2018 World Malaria Report, 2.4 billion people worldwide are exposed to the malaria infection, and more than 219 million are infected annually, with about 435,000 deaths [2]. According to the disease web statistics system of the Korea Centers for Disease Control and Prevention (KCDC), 33,115 malaria cases have been reported between 1993 and 2016, and 3.2% of them were imported. In last 10 years (2005–2016), most of the malaria were influxed from Africa (63.7%) and followed by Asia (33.3%). species of the imported malaria was P. falciparum (50.4%), P. vivax (28.7%), P. ovale (3.0%), and P. malariae (1.1%) [3]. Three cases of ovale malaria were reported between 2005 and 2012, all of which occurred in patients who had visited West Africa (Cote d’Ivoire and Ghana) [4–6]. Malariae malaria has also been reported in 2 patients who had been infected in West Africa (Guinea and Ghana) in 2011 and 2013, respectively [7,8].
According to the World Malaria Report 2018 [2], it is reported that most cases of falciparum malaria occurring in Africa, and recently, the spread of ovale malaria from Africa has also increasing. Occurrence of ovale malaria has been increasingly reported in people returning from Africa in Henan, Shandong, and Jiangsu provinces in China [9–11].
In this study, we report the diagnosis and genetic analysis of P. ovale imported from West and South Africa between 2013 and 2016. Six patients (4 males and 2 females; 25–57 years old) infected with P. ovale were collected for this study (Table 1). The patients had recently visited Equatorial Guinea, Angola, Cameroon, Zambia, or Uganda for missionary activity or business trips. According to the US CDC’s Yellow Book [12], malaria with chloroquine resistance have been reported in all these countries. P. ovale represents 5 to 15% of all Plasmodium spp. in these countries.
Table 1.
Epidemiological factors in 6 cases with Plasmodium ovale infection
| No. | Code | Gender | Age | Diagnosis | Travel country | Date of return from travel | Diagnosis date | Latency period (days) | Accession No. (Genbank) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Methods | Type | |||||||||
| 1 | 13Po-1 | Male | 45 | Microscopy, PCR | Pow | Equatorial Guinea | 20121009 | 20130114 | 97 | MN515106 |
| 2 | 14Po-1 | Male | 47 | PCR | Pow | Angola | 20131017 | 20140407 | 172 | MN515108 |
| 3 | 15Po-1 | Male | 25 | PCR | Poc | Cameroon | 20140820 | 20150116 | 149 | MN515109 |
| 4 | 15Po-2 | Male | 36 | PCR | Pow | Zambia | 20141028 | 20150128 | 92 |
MN518342 MN515138 |
| 5 | 15Po-3 | Female | 57 | PCR | Pow | Zambia | 20150303 | 20150407 | 35 | MN515141 |
| 6 | 16Po-2 | Female | 45 | Microscopy, PCR | Pow | Uganda | 20160814 | 20161129 | 107 | MN515142 |
Po, Plasmodium ovale; Pow, P. ovale wallikeri; Poc, P. ovale curtisi; PCR, Polymerase Chain Reaction.
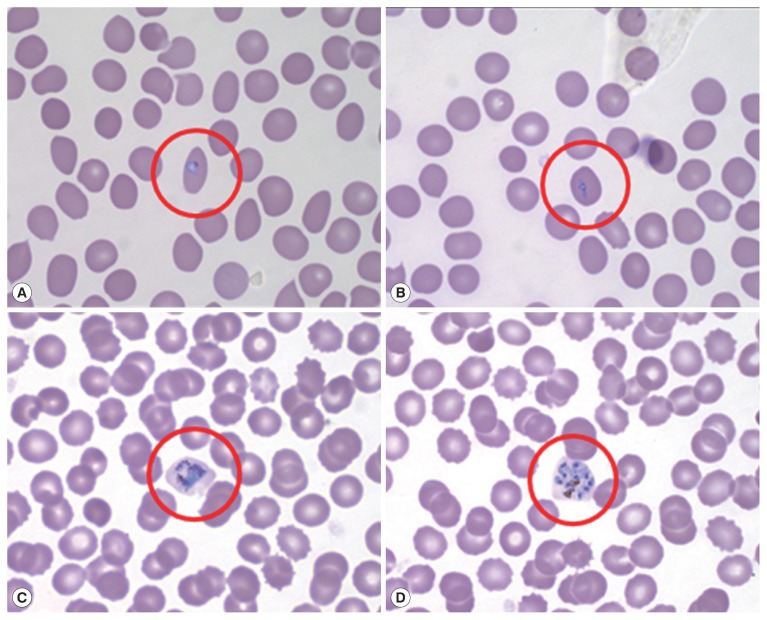
Microscopic examination with Wright-Giemsa staining revealed that the parasites were morphologically similar to P. vivax; the infected RBCs were slightly enlarged and oval in shape with fimbriated edges characteristic of P. ovale (Fig. 1). The results of the rapid diagnostic test (RDT) and SD Bioline Malaria Antigen P.f/Pan rapid kits were negative in all patients. A recent report shows that PCR testing is also necessary due to the low sensitivity of ovale malaria RDT testing [13]. Thus, nested polymerase chain reaction (PCR) targeting 18S rRNA genes was performed for molecular discrimination of Plasmodium spp., as previously described by Snounou et al. [14].
Fig. 1.
Peripheral blood thin smear stained with Wright-Giemsa before treatment. Red circle shows RBC infected by aplaomodial parasite, young trophozoite (ring form; A, B), old trophozoite (C), and schizont (D).
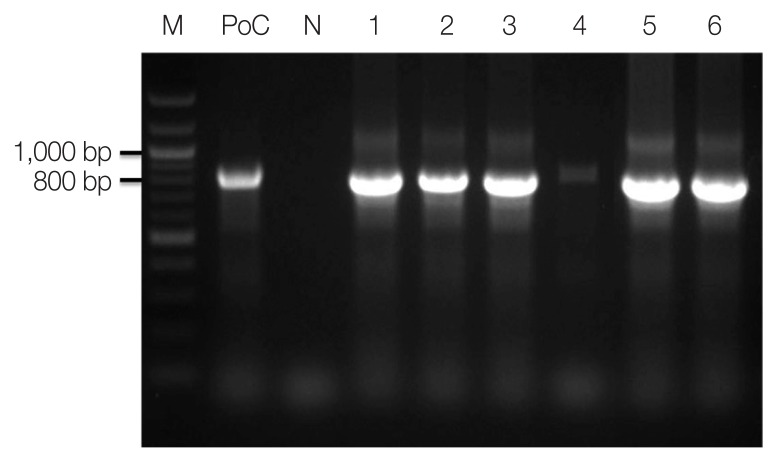
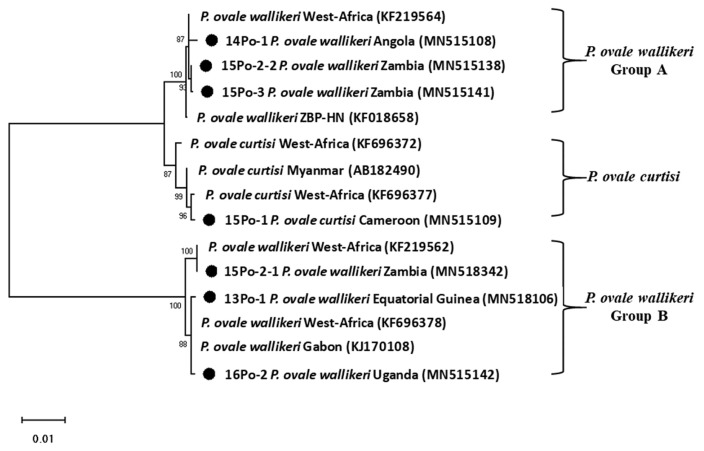
Genomic DNAs of ovale malaria patients were extracted from blood samples using the QIAamp DNA Blood Mini kit (Qiagen, Valencia, California, USA). As described by Snounou et al. [14], namely, the Plasmodium specific primer (rPLU5/rPLU6) was used for the first-round PCR and ovale specific primer (rOVA1/rOVA2) were used for the second-round PCR. Thermal cycling was performed with the following parameters: 1st PCR: pre-denaturation at 95°C for 1 min, denaturation at 95°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1 min (35 cycles); and final extension at 72°C for 10 min; 2nd PCR: pre-denaturation at 95°C for 5 min, denaturation at 95°C for 1 min, annealing at 60°C for 30 sec, and extension at 72°C for 30 sec (35 cycles); and final extension at 72°C for 10 min. The amplified DNA products corresponding to P. ovale (800 bp) were visualized on a 1.5% agarose gel stained ethidium bromide (Fig. 2). The PCR products were sequenced by Macrogen Inc. (Seoul, Korea). The Bioedit Sequence Alignment Editor (version 7.2.5) was used to alignment and edit the sequence data [15]. A phylogenetic analysis was performed based on the neighbor-joining methods with a bootstrap value of 1,000 and analyzed using the MEGA (version 6.0) [16] (Fig. 3). Sequence genbank enrolled MN515106, MN515108, MN515109, MN518342, MN515138, MN515141, and MN515142 (Table 1).
Fig. 2.
Amplicons of nested PCR using primenr pair of Plasmodium ovale. M, 100 bp size ladder; PoC, P. ovale positive control DNA; lane 1, 13Po-1; lane 2, 14Po-1; lane 3, 15Po-1; lane 4, 15Po-2; lane 5, 15Po-3; lane 6, 16Po-2.
Fig. 3.
Phylogenetic tree of Plasmodium ovale constructed using 18S rRNA sequence. Number on node is bootstrap value of 1,000.
DNA sequence analysis revealed that one of the patients was infected with P. ovale curtisi and 5 were infected with P. ovale wallikeri. P. ovale curtisi and P. ovale wallikeri have been reported previously in Angola, Bangladesh, Equatorial Guinea, Republic of Congo, and Uganda [17]. The one patient in our study with P. ovale curtisi infection (15Po-1) had visited middle Africa (Cameroon). Among the 5 patients with P. ovale wallikeri infections, 3 patients with P. ovale wallikeri (14Po-1, 15Po-2-1, 15Po-2-2, and 15Po-3) had visited southern Africa (Angola and Zambia). 15Po-2-1 (1,090 bp) and 15Po-2-2 (1,062 bp) have a base sequence difference of 248 bp, and 28 bp are deleted. Therefore, 15Po-2 patients were estimated to be multi-infection. 14Po-1 and 15Po-3 are showed single infection with group A, hwever, 15Po-2 have showed multi-infection with P. ovale wallikeri groups A and B, while the other 2 (13Po-1 and 16Po-2) had visited western and eastern Africa (Equatorial Guinea and Uganda) and showed single infection with group B. Although specific country names were not listed, they are quite homologous to those infected and isolated in most West Africa. In this study, the isolated clones were found to be more than 99% consistent with those reported in West Africa. In particular, 15Po-2-1 was found to be 100% consistent (Fig. 3). Seven 18S rRNA gene variants (GenBank MN515106, MN 515108, MN515109, MN518342, MN515138, MN515141, and MN515142) of P. ovale curtisi and P. ovale wallikeri were registered in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) (Table 1).
P. ovale was described in 1922 and was named after the characteristic oval morphology with fimbriated edges of erythrocytes infected with it [18]. Its morphological resemblance and to P. vivax and its tertian periodicity make it easy to be mistaken for P. vivax in microscopic analysis [19]. The incubation period of P. ovale has been reported to be 10–17 days [1]. P. ovale is primarily concentrated in sub-Saharan Africa and islands in the western Pacific [20]. Recently, it was subdivided into 2 genetic types: P. ovale curtisi (classic type) and P. ovale wallikeri (variant type). Although the 2 types are not morphologically distinguishable through microscopy [19], their latency periods were reported significantly different; the latency period of P. ovale curtisi (85.7 days) is 2 times longer than that of P. ovale wallikeri (40.6 days) [21]. Our epidemiological investigation revealed that latency periods of P. ovale curtisi and P. ovale wallikeri ranged 149 days and 100.6 days, respectively. Only 1 case (14Po-1) showed 172 days the latency period longer than P. ovale curtisi. The latency period was calculated by subtracting the return date from the travel from the date of diagnosis (Table 1). In terms of clinical manifestations, P. ovale wallikeri has been reported to cause higher levels of parasitemia and more severe symptoms than P. ovale curtisi [17]. As a result of confirming the parasite count of P. ovale wallikeri 2 patients (14Po-1 and 16Po-2), they appeared in 0.62% and 0.67% parasitemia, respectively, and could not be confirmed in the other patients.
All 6 patients reported in this study recovered well. They were treated with chloroquine and primaquine for malaria, and none of them has been diagnoses and treated again because of relapse or drug resistance. To combat the frequent occurrence of imported malaria parasites such as P. ovale, rapid and effective PCR-based molecular diagnosis should be performed regularly. Further, routine surveillance of drug susceptibility and resistance among imported malaria parasites will help efforts to effectively eradicate malaria. In particular, molecular monitoring and differential diagnoses such as that for P. ovale infection should be routinely performed to understand the multigenic diversities, drug susceptibilities, and drug resistance of malarial parasite.
In conclusion, to the best of our knowledge, this is the first study conducted in the Republic of Korea to report the molecular discrimination between P. ovale curtisi and P. ovale wallikeri. The low parasite of P. ovale and the morphological features similar to P. vivax are not easy to differentiate unless a microscopy expert. In addition, Plasmodium ovale has low sensitivity in the RDT and is displayed almost in negative. Therefore, after a trip to Africa, PCR must be performed even if malaria is suspected and RDT is negative. The latency period of P. ovale infection was found to be similar to that of P. vivax infection. P. ovale was latent in all patients for at least 30 days, and in some patients, it remained latent for up to 6 months. Thus, it is necessary to epidemiologically assess the latency periods of imported malaria as confirmed in the 6 patients enrolled in this study. Furthermore, travelers who plan to travel to areas endemic for malaria with long latency periods, such as vivax and ovale malaria, will need to take preventive medicines before and after their travels. In the current scenario, owing to the rise in tourism and the associated increase in cases of imported malaria, continuous genetic monitoring would be essential for controlling malaria in the Republic of Korea.
ACKNOWLEDGMENT
This work was supported by the Endemic Parasitic Diseases Control Program (4800-4847-311) from Centers for Disease Control and Prevention, Republic of Korea.
Footnotes
All authors declare no conflict of interest.
REFERENCES
- 1.Garcia LS. Malaria. Clin Lab Med. 2010;30:93–129. doi: 10.1016/j.cll.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organizaion . World Malaria Report 2018. Geneva, Switzerland: World Health Organizaion; 2018. [Google Scholar]
- 3.Korean Centers for Disease Control and Prevention . A Guideline for Malaria Prevention and Control 2019. Cheongju, Republic of Korea: Korean Centers for Disease Control and Prevention; 2019. [Google Scholar]
- 4.Han TH, Kim BN, Seong HK. A case of imported Plasmodium ovale malaria. J Korean Med Sci. 2006;21:932–935. doi: 10.3346/jkms.2006.21.5.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moon S, Kim BN, Kuak EY, Han TH. A case of Plasmodium ovale malaria imported from West Africa. Lab Med Online. 2012;2:51–54. (Korean). [Google Scholar]
- 6.Kang Y, Yang J. A case of Plasmodium ovale malaria imported from West Africa. Korean J Parasitol. 2013;51:213–218. doi: 10.3347/kjp.2013.51.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong YJ, Yang SY, Lee K, Kim TS, Kim HB, Park KU, Song JH, Kim EC. A case of imported Plasmodium malariae malaria. Ann Lab Med. 2012;32:229–233. doi: 10.3343/alm.2012.32.3.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang YJ, Shim MJ, Kim JY, Ji SY, Lee WJ, Yang J. A Case of Plasmodium malariae Infection Imported from Guinea. Lab Med Online. 2015;5:33–37. (Korean). [Google Scholar]
- 9.Zhou R, Li S, Zhao Y, Yang C, Liu Y, Qian D, Wang H, Lu D, Zhang H. Characterization of Plasmodium ovale spp. imported from Africa to Henan Province, China. Sci Rep. 2019;9:2191. doi: 10.1038/s41598-019-38629-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun H, Li J, Xu C, Xiao T, Wang L, Kong X, Wang Y, Zhang B, Zhao C, Huang B, Wei Q. Increasing number of imported Plasmodium ovale wallikeri malaria in Shandong Province, China, 2015–2017. Acta Trop. 2019;191:248–251. doi: 10.1016/j.actatropica.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 11.Cao YY, Wang WM, Zhou HY, Zhu GD, Xu S, Gu YP, Zhang C, Liu YB, Cao J. Cases diagnosis of imported malaria in Jiangsu province, 2014–2016. Zhonghua liu xing bing xue za zhi. 2018;39:218–221. doi: 10.3760/cma.j.issn.0254-6450.2018.02.016. (in Chinese). [DOI] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention . CDC Yellow Book: Health Information for International Travel. Oxford, UK: Oxford University Press; 2017. [Google Scholar]
- 13.Tang J, Tang F, Zhu H, Lu F, Xu S, Cao Y, Gu Y, He X, Zhou H, Zhu G, Cao J. Assessment of false negative rates of lactate dehydrogenase-based malaria rapid diagnostic tests for Plasmodium ovale detection. PLoS Negl Trop Dis. 2019;13:e0007254. doi: 10.1371/journal.pntd.0007254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, Thaithong S, Brown KN. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- 15.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 16.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaw MT, Lin Z. Two sympatric types of Plasmodium ovale and discrimination by molecular methods. J Microbiol Immunol Infect. 2017;50:559–564. doi: 10.1016/j.jmii.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Stephens JWW. A new malaria parasite of man. Ann Trop Med Parasitol. 1922;16:383–388. [Google Scholar]
- 19.Fuehrer HP, Noedl H. Recent advances in detection of Plasmodium ovale: implications of separation into the two species Plasmodium ovale wallikeri and Plasmodium ovale curtisi . J Clin Microbiol. 2014;52:387–391. doi: 10.1128/JCM.02760-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins WE, Jeffery GM. Plasmodium ovale: parasite and disease. Clin Microbiol Rev. 2005;18:570–581. doi: 10.1128/CMR.18.3.570-581.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nolder D, Oguike MC, Maxwell-Scott H, Niyazi HA, Smith V, Chiodini PL, Sutherland CJ. An observational study of malaria in British travellers: Plasmodium ovale wallikeri and Plasmodium ovale curtisi differ significantly in the duration of latency. BMJ Open. 2013;3:e002711. doi: 10.1136/bmjopen-2013-002711. [DOI] [PMC free article] [PubMed] [Google Scholar]