Abstract
Hematological malignancies are the most common type of pediatric cancers, and acute lymphocytic leukemia (ALL) is the most frequently occurring hematological malignancy during childhood. A major cause of mortality in leukemia is bloodstream infection (BSI). The aim of the current study was to explore the gut microbiota in ALL and its potential functional alterations. High‐throughput sequencing was used to characterize the bacterial and fungal microbiota in feces and their predicted functional characteristics in a xenotransplant pediatric ALL mouse model. Our work shows that gut microbiota significantly changes in leukemia, which may result in functional alterations. This study may provide potential therapeutic or preventive strategies of BSI in ALL.
Keywords: high‐throughput sequencing, leukemia, metagenomics, microbiota
High‐throughput sequencing was used to characterize the bacterial and fungal microbiota in feces and their predicted functional characteristics in a xenotransplant pediatric ALL mouse model. Our work shows that the gut microbiota composition changes significantly in leukemia, and this change is mainly caused by an increase in opportunistic pathogens.

Abbreviations
- ALL
acute lymphocytic leukemia
- BSI
bloodstream infection
- GALT
gut‐associated lymphoid tissue
- T‐ALL
T‐cell acute lymphocytic leukemia
1. BACKGROUND
Leukemia is a group of cancers resulting in nonfunctional white blood cells. Over 400,000 people are diagnosed with leukemia worldwide, and approximately 309,000 people die of this disease annually (Bray et al., 2018). Hematological malignancies are the most common cancers during childhood, and leukemia comprises of 30% of all pediatric cancers. Acute lymphocytic leukemia (ALL) is the most frequently occurring pediatric leukemia, comprising 80% of childhood leukemia, with the highest incidence between 2 and 5 years of age Hutter (2010).
Bloodstream infection (BSI) is a commonly occurring life‐threatening complication and a major cause of mortality in leukemia due to underlying conditions and chemotherapy‐induced neutropenia. Several studies indicate that bacterial translocation (BT) from the gut is the main cause of BSI due to disruptions in the intestinal barrier and the gut‐associated lymphoid tissue (GALT; Belkaid & Hand, 2014; Cornely & Schirmacher, 2001; Gyarmati et al., 2016; Wiest & Rath, 2003). In addition, the gut microbiota also plays an important role in antitumor autoimmunity and affects the efficiency of anticancer treatments (Viaud, Daillere, Boneca, Lepage, & Langella, 2015).
Despite of their importance, alterations of the microbiota in the small intestine and feces are poorly explored in controlled leukemia models. We used a well‐established xenotransplant pediatric T‐ALL mouse model to characterize the changes in diversity and function in the bacterial and fungal microbiota in leukemia. We also assessed differences between feces and intestinal content in the small intestine.
2. METHODS
2.1. Mice
Three‐week‐old female Nod/Scid mice were used in this study (Jackson Laboratories, Bar Harbor, ME, USA). All experimental procedures were approved by the University of Illinois College of Medicine at Peoria Institutional Animal Care and Use Committee review board. Leukemia was induced (n = 8 leukemic and n = 8 controls) as previously described (Nijmeijer et al., 2001) using CCL‐119 cells (lymphoblasts from a pediatric acute lymphocytic leukemia patient, from ATCC) and equal volume of PBS for controls. Mice were housed in a barrier room with a 12‐hr dark–light cycle, in cages (four mice/cage) with UV‐treated shred bedding. Water and regular chow were provided ad libitum. Fecal samples were freshly collected and pooled before inducing leukemia and for 4 weeks after induction twice a week (Figure A1), and DNA was extracted using the QIAmp DNA Stool mini kit (Qiagen). Mice were sacrificed on the 5th week, small intestines were removed, and intestinal contents were washed with PBS. Three leukemic mice died on the 5th week before the end of the experimental period. Euthanasia was performed with cervical dislocation in anesthesia induced by 5% Isothesia in 1.0 L/min oxygen. The following humane endpoints were used for immediate euthanasia: severe weight loss (>15% from starting weight), body condition score 2, ascites, moribund condition, and dehydration. Body weight was measured twice a week. The mouse model was validated on mouse blood samples using TPOX in STR typing (Huang, Schumm, & Budowle, 1995).
2.2. Sequencing
The V4 region of the 16S gene was amplified using the 515F‐806R primer pairs (5′‐GTGYCAGCMGCCGCGGTAA ‐ 5′‐GGACTACNVGGGTWTCTAAT) to characterize bacteria, and the ITS3‐4 regions (5′‐GCATCGATGAAGAACGCAGC ‐ 5′‐TCCTCCGCTTATTGATATGC) were amplified to identify fungi using the Fluidigm assay. The amplicons were sequenced on a MiSeq instrument using 2*250 base pair paired‐end reads. Picrust (Langille et al., 2013) was used to estimate functional profiling of microbial communities. No template control (water) and DNA extraction reagents were used as negative controls, and none resulted in amplification in library preparation.
Sequencing data have been uploaded to the Metagenomics Analysis Server (mg‐rast.org) under submission number MGP88128.
2.3. Data analysis
Mothur v 1.39.5 (Schloss et al., 2009) pipeline (Kozich, Westcott, Baxter, Highlander, & Schloss, 2013) was used to process the sequencing data with the Greengenes 13_5_99 database (DeSantis et al., 2006) for the 16S reads, and the UNITE database (Kõljalg et al., 2003) was used for ITS reads. The parsimony method was used to compare the structure of microbial communities (Schloss et al., 2009). Student's t test was used to estimate statistical significance unless otherwise noted, with significance level set to 0.05.
3. RESULTS
3.1. Bacterial composition changes significantly in ALL in feces, but not in SI
Bacterial compositions were determined on the genus level based on sequencing the V4 region of the 16S rRNA gene, with an average read number of 189,433 per sample. Compositions were compared in feces (Figure 1, Figure A1) and the small intestine (Table 1) between control and leukemic mice.
Figure 1.

Relative abundances (>0.01) of microbiota compositions in control and leukemic feces on the genus level
Table 1.
Parsimony method (Schloss et al., 2009) determines distances in community structures
| Small intestine control | Small intestine leukemic | Feces control | Feces leukemic | |
|---|---|---|---|---|
| Small intestine control | • | 1 | 0.039 | 0.036 |
| Small intestine leukemic | 1 | • | 1 | 1 |
| Feces control | 1 | 1 | • | 0.019 |
| Feces leukemic | 0.036 | 1 | 0.019 | • |
Table shows the differences represented by p‐values based on the 16S analysis of bacterial compositions. SI = small intestine, F = feces. Red = p < .05.
The composition of bacterial microbiota in feces differed significantly (p < .05) between leukemic and control mice (Table 1). However, bacterial compositions showed no changes in the SI between control and leukemic mice. As expected, the microbiota in the small intestine differed from the microbiota in feces in control mice (Gu et al., 2013; Marteau et al., 2001; Onishi et al., 2017). Interestingly, there was no difference between microbiota compositions in SI and feces in leukemic mice.
Alpha diversity, measured by the Shannon diversity index, did not differ significantly (3.47 in leukemic mice, and 3.37 in control mice [p > .05]), although the distribution of the number of OTUs was broader in the leukemic group (Figure A2).
3.2. The change in bacterial communities in feces is mainly caused by opportunistic pathogens
The composition changes of the microbiota in leukemic feces showed a decreased proportion of bacteria compared to the control group, which are commonly considered beneficial for the host (Clostridiaceae_02d06, Lactobacillus, Streptomyces [Seipke, Kaltenpoth, & Hutchings, 2012; Walter, 2008]; Figure 2). Population‐level analysis using metastats (White, Nagarajan, & Pop, 2009) revealed that the main driver of compositional difference was a lower proportion of Clostridiaceae in leukemic feces (p < .001). Leukemic samples had an increased ratio of pathogenic bacteria (Staphylococcus, Streptococcus, Ralstonia, Lactococcus), which are opportunistic pathogens present in the microbiota. These pathogens are commonly detected in bloodstream infections in leukemia and can lead to outbreaks in ICUs and immunocompromised patients (Chen et al., 2017; Gyarmati et al., 2016).
Figure 2.

Ratio of relative abundances between the fecal compositions of control and leukemic mice
Principal component analysis (PCA) was performed to analyze differences in microbiota distributions. PCA graph shows that leukemic and control samples clustered differently (Figure 3). However, it also indicates an even greater difference between batches (Figure A3), implying the necessity to use multiple batches and/or littermates for metagenomics comparisons (Stappenbeck & Virgin, 2016).
Figure 3.

PCA graph shows clustering of control (red dots) and leukemic (blue dots) microbiota
3.3. Bacterial diversity in the small intestine
The Shannon diversity index in the SI was 2.02 for leukemic mice and 2.38 in control mice (p > .05). There was no significant difference in the SI bacterial diversity between control and leukemia mice. A significant difference was shown in control mice between feces and SI in Shannon diversities (p < .05) as observed in physiological conditions (Gu et al., 2013; Marteau et al., 2001; Onishi et al., 2017). However, there was no significant difference in diversities in leukemic mice between feces and SI.
Similarly to the fecal compositions, the abundances of Lactobacillus, Adlercreutzia, and Turicibacter in SI were higher in control mice than that in leukemic (Figure 4). Our results also show increased proportions of Butyrivibrio, Anaeroplasma, and Oscillospira in leukemia (Figure 4). Butyrivibrio is a butyrate‐producing Clostridia and commonly found in the mammalian microbiota (Fabbiano, 2017). Oscillospira is elevated in colorectal cancer (Hibberd et al., 2017) and associated as an early marker for small intestinal damage and metabolic disorder (Hamilton, Boudry, Lemay, & Raybould, 2015). Anaeroplasma is an opportunistic pathogen and has been reported in inflammatory gut (Zeng, Ishaq, Liu, & Bukowski, 2018).
Figure 4.

Relative abundances (a) and the ratio of relative abundances (b) of the bacterial microbiota between the fecal compositions in the small intestine of control and leukemic mice
3.4. Functional prediction of the bacterial microbiota
We used Picrust (Langille et al., 2013) for predictive functional profiling of the microbiota. The analysis indicates an increase in bacterial chemotaxis and flagellar assembly in the SI in leukemia (Figure A4) and was not observed in feces (Figure A5). This may also suggest that the composition change of the gut microbiota in leukemia, associated with its functional change, contributes to bacterial translocation (Wiest & Rath, 2003). All functions shown in Figures A4, A5, A6 were significantly different (p < .05) between control and leukemic mice.
In the small intestine, the proportion of bacteria with the function of the conversion of dietary flavonoids increased in leukemia (Figure A4). Flavonoids may contribute to the pathogenesis of pediatric leukemia as a natural topoisomerase II inhibitor through the cleavage of the MLL gene (Strick, Strissel, Borgers, Smith, & Rowley, 2000). Flavonoids may also have a beneficial effect on host health (Braune & Blaut, 2016), although this effect is poorly explored. In addition, flavonoids may benefit cancer prevention and oncotherapy in leukemias as well (Spagnuolo et al., 2012). In feces, the response to bacterial infection was also increased in leukemia (Figure A5).
3.5. Fungal microbiota did not change significantly in ALL mouse model
ITS analysis indicated no significant difference in fungal fecal or SI compositions between leukemic and control mice. However, in feces, there was a significant difference in fungal compositions between batches as observed in bacterial compositions (p < .05, Figure A3). Shannon diversity index was 2.48 ± 0.67 (mean ± SD) in leukemic and 2.56 ± 0.59 (p > .05) for control mice (Figure A6). Interestingly, there were no fungi detected in the small intestine of leukemic mice, while all the samples from control mice contained fungal reads.
3.6. Weight of leukemic and control mice
Weight was measured for all mice twice a week, firstly to provide humane endpoint, and secondly as weight loss could be a possible confounder for microbiota composition. There was no significant difference in weight between the control and leukemic groups.
4. DISCUSSION
BSI is a severe complication and a major cause of mortality in ALL. SI is the primary site for BSI due to its permeability as absorption of nutrients takes place in the SI (Belkaid & Hand, 2014; Wiest & Rath, 2003). Therefore, most of the detected pathogens in BSI are opportunistic pathogens (Gyarmati et al., 2015), as a result to a loss of immune cells in GALT and damaged intestinal barrier (Song & Gyarmati, 2019). Bacteria are more abundant in the colon; however, changes of the colonic microbiota influences bacterial composition in the SI (Guarner & Malagelada, 2003; Figures 2 and 4) and further are related to BSI. Thus, both the feces and the intestinal content in SI were analyzed in this study.
This project aimed to characterize changes in the leukemic microbiota by using 16S and ITS metagenomics sequencing as the changes in the gut microbiota could reveal mechanistic insights into the pathophysiology of BSI. We identified an increase of opportunistic pathogens in the gut, which may explain the frequent occurrence of certain pathogens detected in BSI in ALL (Hakim et al., 2018; Viaud et al., 2015). Several reports also indicated that the microbiota is a major origin of causative pathogens in BSI in immunocompromised patients (Belkaid & Hand, 2014; Cornely & Schirmacher, 2001; Gyarmati et al., 2016; Wiest & Rath, 2003) and our results suggest a selection of opportunistic pathogens in the microbiota in ALL.
Specifically, our findings support the increase of pathogenic bacteria in the feces of ALL mice (Clostridiaceae and Streptococcaceae), which was also reported in pediatric patients with ALL during chemotherapy (Hakim et al., 2018; Taur et al., 2012). In addition, there was an elevation of Lactobacilli in controls compared to leukemic feces in our study, which was also demonstrated in a different ALL mouse model system using denaturing gradient gel electrophoresis (Bindels et al., 2012).
We have also shown predicted functional changes in gut bacteria, which may promote BT and subsequent BSI (e.g., bacterial chemotaxis and flagellar assembly in the small intestine, Figure A4). Even though leukemic and control mice had a significant difference in microbial composition, there was also a significant difference between batches of mice (Figure A4). Therefore, this study also highlights the importance of batch and littermate controls in rodent metagenomics studies.
Short‐chain fatty acids (SCFAs) are microbial metabolites derived from dietary fiber in colon and play an essential role in body health. Butyrate is one of the most important SCFAs for host gastrointestinal health, by providing nutritional source to epithelial cells and stimulating regulatory T cells (Zhang et al., 2016). In addition, butyrate may also affect cell differentiation in ALL (Miller, Kurschel, Osieka, & Schmidt, 1987). Our results show that both of butyrate‐producing genera, Anaerostipes and Coprococcus, had increased proportion both in feces and SI in leukemic animals (Figures 2 and 4). Anaerostipes is also elevated in colitis (Zhang et al., 2016). As a propionate and acetate producing genus, Akkermansia is elevated in controls in feces and decreased in SI (Figure 2; Koh, De Vadder, Kovatcheva‐Datchary, & Bäckhed, 2016; Louis & Flint, 2017); however, there is an elevated proportion in colon cancer (Weir et al., 2013).
Fungi consist of <0.1% of the microbiota and are often overlooked in microbiota analyses, even though they may perform important physiological functions (Hillman, Lu, Yao, & Nakatsu, 2017). Based on ITS sequencing, our results confirmed that the murine SI contains a variety of fungal microbiota (Scupham et al., 2006) in physiological conditions, but there were no fungi detected in the SI of leukemic mice. Bacterial diversity also showed a decrease in SI in leukemic mice, although the change was not significant. Possibly this change of fungal diversity is due to an increased intestinal permeability (Allert et al., 2018; Song & Gyarmati, 2019) in ALL. Fungal communities in the feces did not show a significant difference between leukemic and control mice, possibly due to the low concentration of fungi in feces in physiological status (Hillman et al., 2017).
There are several limitations of this study. We have used a limited number of mice (n = 8 for control and n = 8 for leukemic group); however, mice are coprophagic and were co‐housed to ensure effective microbiota transfer, and there were no significant differences in microbiota compositions in individual mice. In addition, we have used a mouse model which utilizes immunocompromised mice. As all mouse models, this model has a limitation of modeling the human clinical case (e.g., there are considerable differences between the human and mouse gastrointestinal tracts), but the xenograft model is a well‐established ALL mouse model, allowing to model patient‐specific disease characteristics (Nijmeijer et al., 2001).
5. CONCLUSIONS
Taken together, our results show that (a) the bacterial microbiota changes significantly in feces in ALL mouse model, (b) this change is dominantly due to an increase of opportunistic pathogens in leukemic mice, and (c) and the change in bacterial composition may results in a functional change of the fecal microbiota in pediatric ALL mouse model. These findings may contribute to characterize alterations in the microbiota caused by ALL and can extend our knowledge into the pathogenesis of BSI and the dynamics of BT.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
Yajing Song: Conceptualization‐Equal, Data curation‐Equal, Formal analysis‐Equal, Investigation‐Equal, Methodology‐Equal, Writing‐review & editing‐Equal; Peter Gyarmati: Conceptualization‐Equal, Data curation‐Equal, Formal analysis‐Equal, Funding acquisition‐Equal, Investigation‐Equal, Methodology‐Equal, Project administration‐Equal, Supervision‐Equal, Validation‐Equal, Visualization‐Equal, Writing‐original draft‐Equal.
ETHICS STATEMENT
All experimental procedures were approved by the University of Illinois College of Medicine at Peoria Institutional Animal Care and Use Committee review board.
ACKNOWLEDGMENTS
This study was supported by the University of Illinois College of Medicine start‐up fund to PG. The sponsor had no role in study design, collection, analysis and interpretation of the data, and writing and submission of the manuscript.
Appendix 1.
Figure A1.

Relative abundances in control and leukemic samples over a 5‐week experimental period. (C = control, Wx = week number, B2 = batch 2)
Figure A2.

Number of OTUs in control and leukemic groups
Figure A3.

PCA graph shows clustering of control (red dots) and leukemic (blue dots) microbiota and the differences between batches of mice
Figure A4.
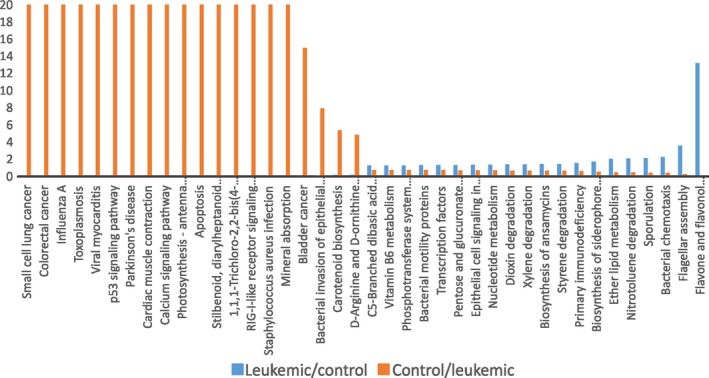
Functional analysis of the fecal material in the small intestine microbiota based on 16S sequencing
Figure A5.

Functional analysis of the fecal microbiota based on 16S sequencing
Figure A6.

Ratio of relative abundances of the fungal microbiota between the fecal compositions of control and leukemic mice
Song Y, Gyarmati P. Microbiota changes in a pediatric acute lymphocytic leukemia mouse model. MicrobiologyOpen. 2020;9:e982 10.1002/mbo3.982
DATA AVAILABILITY STATEMENT
Sequencing data have been uploaded to the Metagenomics Analysis Server (mg‐rast.org) under submission number MGP88128 (https://www.mg-rast.org/linkin.cgi?project=mgp88128).
REFERENCES
- Allert, S. , Förster, T. M. , Svensson, C. M. , Richardson, J. P. , Pawlik, T. , Hebecker, B. , … Hube, B. (2018). Candida albicans‐induced epithelial damage mediates translocation through intestinal barriers. MBio, 9(3), e00915‐e00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid, Y. , & Hand, T. W. (2014). Role of the microbiota in immunity and inflammation. Cell, 157(1), 121–141. 10.1016/j.cell.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindels, L. B. , Beck, R. , Schakman, O. , Martin, J. C. , De Backer, F. , Sohet, F. M. , … Delzenne, N. M. (2012). Restoring specific lactobacilli levels decreases inflammation and muscle atrophy markers in an acute leukemia mouse model. PLoS ONE, 7(6), e37971 10.1371/journal.pone.0037971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braune, A. , & Blaut, M. (2016). Bacterial species involved in the conversion of dietary flavonoids in the human gut. Gut Microbes, 7(3), 216–234. 10.1080/19490976.2016.1158395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, F. , Ferlay, J. , Soerjomataram, I. , Siegel, R. L. , Torre, L. A. , & Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 68(6), 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Chen, Y. Y. , Huang, W. T. , Chen, C. P. , Sun, S. M. , Kuo, F. M. , Chan, Y. J. , … Wang, F. D. (2017). An Outbreak of ralstonia pickettii bloodstream infection associated with an intrinsically contaminated normal saline solution. Infection Control and Hospital Epidemiology, 38(4), 444–448. [DOI] [PubMed] [Google Scholar]
- Cornely, O. A. , & Schirmacher, P. (2001). Clinical picture: Bacterial translocation in neutropenic sepsis. The Lancet, 358, 1842 10.1016/S0140-6736(01)06884-2 [DOI] [PubMed] [Google Scholar]
- DeSantis, T. Z. , Hugenholtz, P. , Larsen, N. , Rojas, M. , Brodie, E. L. , Keller, K. , … Andersen, G. L. (2006). Greengenes, a chimera‐checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environment Microbiology, 72(7), 5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbiano, S. et al (2017). Host‐microbiota mutualism in metabolic diseases. Frontiers in Endocrinology, 8, 267 10.3389/fendo.2017.00267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, S. , Chen, D. , Zhang, J.‐N. , Lv, X. , Wang, K. , Duan, L.‐P. , … Wu, X.‐L. (2013). Bacterial community mapping of the mouse gastrointestinal tract. PLoS ONE, 8(10), e74957 10.1371/journal.pone.0074957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner, F. , & Malagelada, J. R. (2003). Gut flora in health and disease. Lancet, 361(9356), 512–519. 10.1016/S0140-6736(03)12489-0 [DOI] [PubMed] [Google Scholar]
- Gyarmati, P. , Kjellander, C. , Aust, C. , Kalin, M. , Öhrmalm, L. , & Giske, C. G. (2015). Bacterial landscape of bloodstream infections in neutropenic patients via high throughput sequencing. PLoS ONE, 10(8), e0135756 10.1371/journal.pone.0135756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyarmati, P. , Kjellander, C. , Aust, C. , Song, Y. , Öhrmalm, L. , & Giske, C. G. (2016). Metagenomic analysis of bloodstream infections in patients with acute leukemia and therapy‐induced neutropenia. Scientific Reports, 6, 23532 10.1038/srep23532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim, H. , Dallas, R. , Wolf, J. , Tang, L. , Schultz‐Cherry, S. , Darling, V. , … Rosch, J. W. (2018). Gut microbiome composition predicts infection risk during chemotherapy in children with acute lymphoblastic leukemia. Clinical Infectious Diseases, 67(4), 541–548. 10.1093/cid/ciy153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, M. K. , Boudry, G. , Lemay, D. G. , & Raybould, H. E. (2015). Changes in intestinal barrier function and gut microbiota in high‐fat diet‐fed rats are dynamic and region dependent. American Journal of Physiology‐Gastrointestinal and Liver Physiology, 308(10), G840–G851. 10.1152/ajpgi.00029.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd, A. A. , Lyra, A. , Ouwehand, A. C. , Rolny, P. , Lindegren, H. , Cedgård, L. , & Wettergren, Y. (2017). Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterology, 4(1), e000145 10.1136/bmjgast-2017-000145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman, E. T. , Lu, H. , Yao, T. , & Nakatsu, C. H. (2017). Microbial ecology along the gastrointestinal tract. Microbes and Environments, 32(4), 300–313. 10.1264/jsme2.ME17017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, N. E. , Schumm, J. W. , & Budowle, B. (1995). Chinese population data on three tetrameric short tandem repeat loci–HUMTH01, TPOX, and CSF1PO–derived using multiplex PCR and manual typing. Forensic Science International, 71, 131–136. [DOI] [PubMed] [Google Scholar]
- Hutter, J. J. (2010). Childhood Leukemia. Pediatrics in Review, 31, 6. [DOI] [PubMed] [Google Scholar]
- Koh, A. , De Vadder, F. , Kovatcheva‐Datchary, P. , & Bäckhed, F. (2016). From dietary fiber to host physiology: short‐chain fatty acids as key bacterial metabolites. Cell, 165(6), 1332–1345. 10.1016/j.cell.2016.05.041 [DOI] [PubMed] [Google Scholar]
- Kõljalg, U. , Nilsson, R. H. , Abarenkov, K. , Tedersoo, L. , Taylor, A. F. S. , Bahram, M. , … Larsson, K.‐H. (2003). Towards a unified paradigm for sequence‐based identification of fungi. Molecular Ecology, 22(21), 5271–5277. 10.1111/mec.12481 [DOI] [PubMed] [Google Scholar]
- Kozich, J. J. , Westcott, S. L. , Baxter, N. T. , Highlander, S. K. , & Schloss, P. D. (2013). Development of a dual‐index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Applied and Environmental Microbiology, 79(17), 5112–5120. 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille, M. G. I. , Zaneveld, J. , Caporaso, J. G. , McDonald, D. , Knights, D. , Reyes, J. A. , … Huttenhower, C. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology, 1, 10 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis, P. , & Flint, H. J. (2017). Formation of propionate and butyrate by the human colonic microbiota. Environmental Microbiology, 19(1), 29–41. 10.1111/1462-2920.13589 [DOI] [PubMed] [Google Scholar]
- Marteau, P. , Pochart, P. , Doré, J. , Béra‐Maillet, C. , Bernalier, A. , & Corthier, G. (2001). Comparative study of bacterial groups within the human cecal and fecal microbiota. Applied and Environment Microbiology, 67(10), 4939–4942. 10.1128/AEM.67.10.4939-4942.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, A. A. , Kurschel, E. , Osieka, R. , & Schmidt, C. G. (1987). Clinical pharmacology of sodium butyrate in patients with acute leukemia. European Journal of Cancer and Clinical Oncology, 23(9), 1283–1287. 10.1016/0277-5379(87)90109-X [DOI] [PubMed] [Google Scholar]
- Nijmeijer, B. A. , Mollevanger, P. , van Zelderen‐Bhola, S. L. , Kluin‐Nelemans, H. C. , Willemze, R. , & Falkenburg, J. H. (2001). Monitoring of engraftment and progression of acute lymphoblastic leukemia in individual NOD/SCID mice. Experimental Hematology, 29(3), 322–329. 10.1016/S0301-472X(00)00669-X [DOI] [PubMed] [Google Scholar]
- Onishi, J. C. , Campbell, S. , Moreau, M. , Patel, F. , Brooks, A. I. , Zhou, Y. X. , … Storch, J. (2017). Bacterial communities in the small intestine respond differently to those in the caecum and colon in mice fed low‐ and high‐fat diets. Microbiology, 163(8), 1189–1197. 10.1099/mic.0.000496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss, P. D. , Westcott, S. L. , Ryabin, T. , Hall, J. R. , Hartmann, M. , Hollister, E. B. , … Weber, C. F. (2009). Introducing mothur: Open‐source, platform‐independent, community‐supported software for describing and comparing microbial communities. Applied and Environment Microbiology, 75(23), 7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scupham, A. J. , Presley, L. L. , Wei, B. , Bent, E. , Griffith, N. , McPherson, M. , … Borneman, J. (2006). Abundant and diverse fungal microbiota in the murine intestine. Applied and Environment Microbiology, 72(1), 793–801. 10.1128/AEM.72.1.793-801.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipke, R. F. , Kaltenpoth, M. , & Hutchings, M. I. (2012). Streptomyces as symbionts: An emerging and widespread theme? FEMS Microbiology Reviews, 36(4), 862–876. [DOI] [PubMed] [Google Scholar]
- Song, Y. , & Gyarmati, P. (2019). Bacterial translocation in acute lymphocytic leukemia. PLoS ONE, 14(4), e0214526 10.1371/journal.pone.0214526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnuolo, C. , Russo, M. , Bilotto, S. , Tedesco, I. , Laratta, B. , & Russo, G. L. (2012). Dietary polyphenols in cancer prevention: The example of the flavonoid quercetin in leukemia. Annals of the New York Academy of Sciences, 1259, 95–103. 10.1111/j.1749-6632.2012.06599.x [DOI] [PubMed] [Google Scholar]
- Stappenbeck, T. S. , & Virgin, H. W. (2016). Accounting for reciprocal host–microbiome interactions in experimental science. Nature, 534, 191–199. 10.1038/nature18285 [DOI] [PubMed] [Google Scholar]
- Strick, R. , Strissel, P. L. , Borgers, S. , Smith, S. L. , & Rowley, J. D. (2000). Dietary bioflavonoids induce cleavage in the MLL gene and may contribute to infant leukemia. Proceedings of the National Academy of Sciences United States of America, 97(9), 4790–4795. 10.1073/pnas.070061297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taur, Y. , Xavier, J. B. , Lipuma, L. , Ubeda, C. , Goldberg, J. , Gobourne, A. , … Pamer, E. G. (2012). Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clinical Infectious Diseases, 55(7), 905–914. 10.1093/cid/cis580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaud, S. , Daillere, R. , Boneca, I. G. , Lepage, P. , Langella, P. et al (2015). Gut microbiome and anticancer immune response: Really hot Sh*t! Cell Death and Differentiation, 22, 199–214. 10.1038/cdd.2014.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, J. (2008). Ecological role of lactobacilli in the gastrointestinal tract: Implications for fundamental and biomedical research. Applied and Environment Microbiology, 74(16), 4985–4996. 10.1128/AEM.00753-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir, T. L. , Manter, D. K. , Sheflin, A. M. , Barnett, B. A. , Heuberger, A. L. , & Ryan, E. P. (2013). Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS ONE, 8(8), e70803 10.1371/journal.pone.0070803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, J. R. , Nagarajan, N. , & Pop, M. (2009). Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Computational Biology, 5, e1000352 10.1371/journal.pcbi.1000352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiest, R. , & Rath, H. C. (2003). Gastrointestinal disorders of the critically ill. Bacterial translocation in the gut. Best Practice & Research Clinical Gastroenterology, 7(3), 397–425. 10.1016/S1521-6918(03)00024-6 [DOI] [PubMed] [Google Scholar]
- Zeng, H. , Ishaq, S. L. , Liu, Z. , & Bukowski, M. R. (2018). Colonic aberrant crypt formation accompanies an increase of opportunistic pathogenic bacteria in C57BL/6 mice fed a high‐fat diet. Journal of Nutritional Biochemistry, 54, 18–27. 10.1016/j.jnutbio.2017.11.001 [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Wu, Y. , Wang, J. , Wu, G. , Long, W. , Xue, Z. , … Zhang, C. (2016). Accelerated dysbiosis of gut microbiota during aggravation of DSS‐induced colitis by a butyrate‐producing bacterium. Scientific Reports, 6, 27572 10.1038/srep27572 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequencing data have been uploaded to the Metagenomics Analysis Server (mg‐rast.org) under submission number MGP88128 (https://www.mg-rast.org/linkin.cgi?project=mgp88128).


