Abstract
Sleep is nearly universal among animals, yet remains poorly understood. Recent work has leveraged simple model organisms, such as Caenorhabditis elegans and Drosophila melanogaster larvae, to investigate the genetic and neural bases of sleep. However, manual methods of recording sleep behavior in these systems are labor intensive and low in throughput. To address these limitations, we developed methods for quantitative imaging of individual animals cultivated in custom microfabricated multi-well substrates and used them to elucidate molecular mechanisms underlying sleep. Here we describe the steps necessary to design, produce, and image these plates, and analyze the resulting behavioral data. We also describe approaches for experimentally manipulating sleep. Following these procedures, after approximately 2 hours of experimental preparation, we are able to simultaneously image 24 C. elegans from the second larval stage to adult stages or 20 Drosophila larvae during the second instar life stage at a spatial resolution of 10 μm or 27 μm, respectively. While this system has been optimized to measure activity and quiescence in Caenorhabditis larvae and adults and in Drosophila larvae, it can also be used to assess other behaviors over short or long periods. Moreover, with minor modifications, it can be adapted for the behavioral monitoring of a wide range of small animals.
Keywords: Drosophila melanogaster, Caenorhabditis elegans, larva, sleep, machine vision, behavior
INTRODUCTION
Evidence from across the animal kingdom indicates that sleep serves vital functions in mature animals as well as during development, and that its functions during development are partly distinct from those during adulthood1–3. Neural circuits modulating sleep are likewise diverse4,5, and it remains to be seen to what degree circuits controlling adult sleep behavior also control developmental sleep. A comprehensive molecular-level understanding of sleep control has remained elusive. For these reasons, studies in animals with simpler nervous systems continue to provide insights into the genetic, molecular, and cellular principles of sleep3,6,7.
The use of Drosophila to study the genetics and neurobiology of sleep began about 20 years ago8,9; the first work on C. elegans sleep followed about 10 years later10. Insights from these models have shaped our understanding of sleep function and control. Worm sleep has been characterized both during development and in adulthood11–13, but studies in Drosophila have focused on adults. While sleep is developmentally regulated in the adult fly (juvenile flies exhibit increased sleep amount and depth compared to mature flies)9,14, it remained unknown whether sleep occurred prior to the adult fly stage. We recently showed that Drosophila larvae also sleep15, and that neurogenetic control of larval sleep is different in part from that of adult fly sleep. C. elegans, with 302 neurons, and Drosophila larvae, with ~10,000 neurons, have smaller and simpler nervous systems than do adult flies (~200,000 neurons) or other model systems like larval zebrafish (~100,000 neurons) or rodents (70–100 million neurons). Indeed, recent work in worms and fly larvae underscore the unique advantages of these systems16–18, which can be leveraged to study sleep. However, an obstacle to studying sleep in these systems is the low-throughput nature of behavioral monitoring. In contrast, sleep monitoring in adult flies has been high in throughput since its infancy, facilitating numerous large genetic screens19–23. Designing systems to allow quantitative and parallel monitoring of quiescence in many animals at once will accelerate the discovery of novel genetic, neurobiological, and developmental perspectives on sleep.
Here we describe protocols for quantitative imaging of C. elegans or D. melanogaster larvae in custom fabricated devices called the WorMotel and LarvaLodge, respectively. The primary advantage of these methods is the ability to measure sleep and activity in many individual animals, as well as other behaviors that occur on a time scale of hours to days. This approach can therefore be utilized to study molecular mechanisms of sleep and other behaviors in worms, flies, and other small animals. Our goal in publishing this protocol is to enable other labs to adopt these technologies. The WorMotel has also been used to study behavior during aging24, and this application is also described. The current protocol is optimized in worms for the L2 stage and later, and in flies for 2nd instar larvae. As our protocol describes how to customize the size and shape of the plate wells, we expect it to be of interest to any worm/fly larva researcher studying behavior, especially those behaviors that unfold over long time periods (from hours to weeks). Recent work has used this slightly modified platform to examine motility in parasitic flatworms (Schistomsoma)25, and in principle, these methods can be used to monitor behavior and perform high-throughput screening of any animal with dimensions from a few hundred microns to a few millimeters (e.g., Trichoplax26–28).
Comparison with other methods
Manual methods of recording sleep behavior in worms require visual tracking of individual animals for the duration of sleep periods (2–12 hours), and are laborious, may lead to sensory disturbance of the animals, and can miss information captured by continuous imaging29,30.
Methods have been reported for monitoring behavior in many worms on a single plate simultaneously31, facilitating high throughput analysis of complex behaviors32. However, most of these methods are optimized for short-term monitoring, in the order of minutes, and thus are not suitable for long-term behavioral analysis, especially when longitudinal behavioral information is needed. A method that utilizes an “artificial dirt” array of posts fabricated from PDMS can monitor up to 10 individual worms for several hours33, but is optimized to a particular animal size and requires growing the worms in enclosed compartments from which the animal cannot be easily recovered. Other platforms allow monitoring of animals throughout multiple developmental stages, but do not lend themselves to easy recovery of the animal, which is desired when undertaking a forward genetic screen. For example, one method consists of agarose hydrogel compartments, which allows parallel monitoring at high optical resolution of up to 25 animals simultaneously34 and throughout larval development35. A second method constrains animals on a polyethylene glycol hydrogel substrate to individual “corrals”, and allows longitudinal imaging of several individuals from embryo to aged adult36. In addition, a microfluidics-based method entails growing the worms in liquid microdroplets, but in this device, longevity is severely compromised and behavior is different from that seen on solid media37.
For fly larvae, numerous methods have been described for monitoring short-term behaviors in a high-throughput manner32,38,39. However, to our knowledge no other method exists to monitor behavior over periods of hours to days.
The WorMotel/LarvaLodge provides a platform to longitudinally monitor behavior of individual animals of various sizes on a time scale that ranges from seconds to days. Moreover, animals can be readily recovered from these devices for further breeding or analysis.
Experimental Design
Behavioral monitoring in the WorMotel/LarvaLodge devices requires four major stages: configuration of imaging system, fabrication of devices, preparation/running of experiments, and data analysis (Figure 1). Each device (described in detail in published work15,24), consists of a molded PDMS array of wells. Each well contains agar media (with slight modifications from standard recipes), food, and a single animal. Animals in the devices are imaged with a digital camera under darkfield illumination, enabling longitudinal imaging and quantitation of sleep behavior in multiple animals simultaneously. Light or mechanical stimuli can be used to induce sleep deprivation and or alternatively conduct arousal threshold measurements. A light-based sleep deprivation system for single animals can also be implemented within a closed-loop system, such that illumination occurs only when an animal is quiescent, for example. All procedures in the protocol are compatible with standard techniques such as RNA interference and optogenetic or thermogenetic manipulations.
Figure 1. Protocol outline.
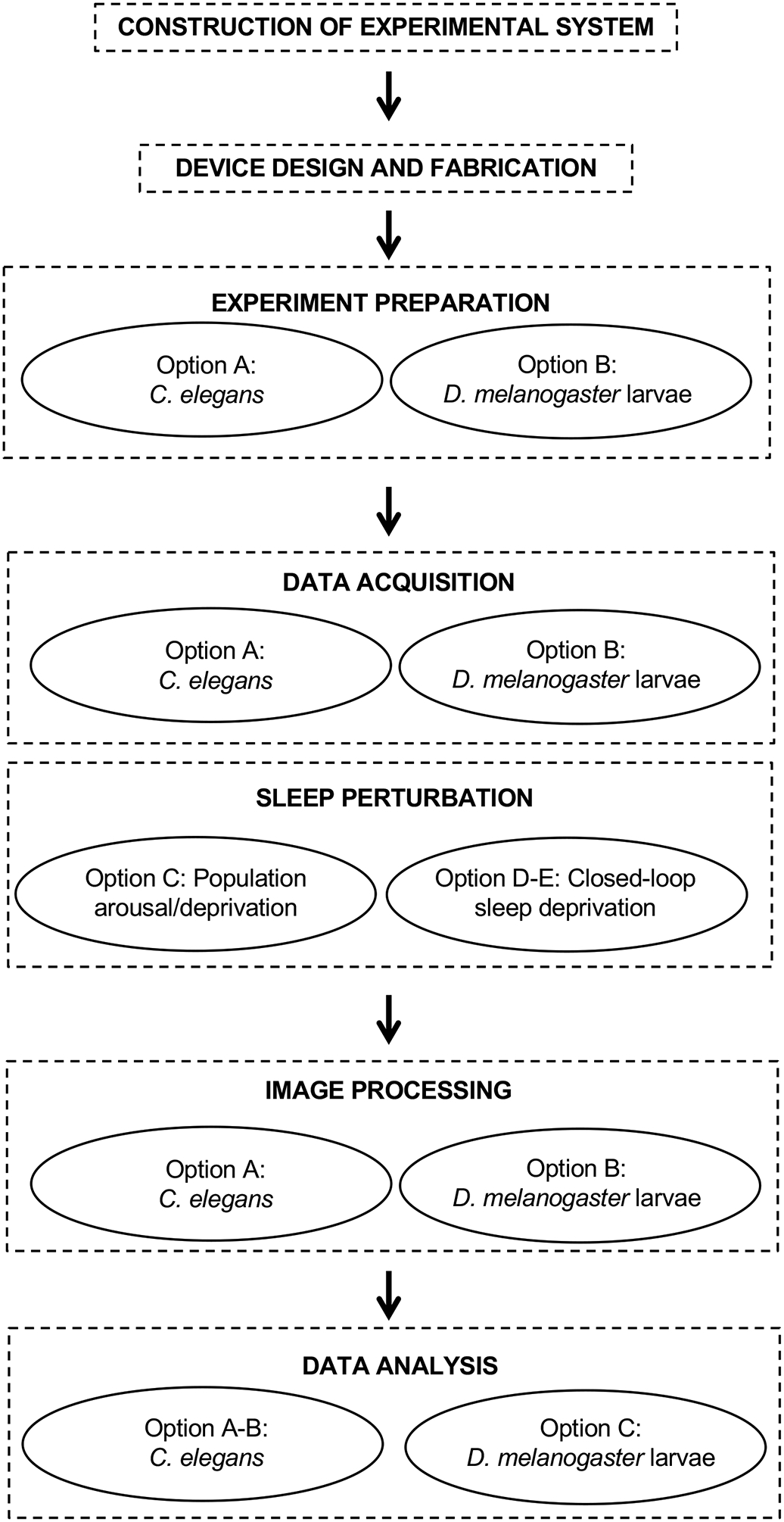
Flowchart indicating the steps outlined in this protocol. Each step may have one or more options depending on the model organism and the specific experimental question under investigation.
Configuration of imaging system.
For all behavioral experiments we use an imaging system that has been previously described40. The primary features of this system are a digital camera, a C-mount lens, a glass imaging platform on which the sample rests, and red or infrared LEDs which provide dark field illumination of the animals. While we describe here a particular dark field imaging system, other imaging systems including those using bright field illumination33 are compatible with the WorMotel and LarvaLodge devices described in this protocol. For sleep deprivation and arousal experiments using a blue light stimulus, it is necessary to separately construct the blue light stimulus delivery apparatus as described below.
Fabrication of devices.
Fabrication of the WorMotel/LarvaLodge requires access to a 3D printer (parts can also be ordered from a commercial 3D printing service), the device design (WorMotel_Mold_48Wells.m or LarvaLodge_Mold_24Wells.m), and materials for casting the device from the 3D-printed mold. The mold STL file (WorMotel.stl or LarvaLodge.stl) can be sent directly to a 3D printer, though the physical dimensions (such as plate size, well size, well dimensions, and well spacing) are easily modifiable via a MATLAB script for specific experimental needs, and should be compatible with the machine vision software for behavioral monitoring. The devices themselves are cast out of polydimethylsiloxane (PDMS), a transparent silicone rubber commonly used in biological applications. The appropriate amount of PDMS is poured into the mold, cured, and the device is removed from the mold (demolded).
Preparation/running of experiments.
After devices are fabricated, each well is loaded individually with agar and food. C. elegans or Drosophila larvae of the desired developmental stage are prepared by standard methods. Animals are manually loaded in individual wells, and moved into the imaging system. Imaging software (IC Capture) and a custom MATLAB Analysis Graphical User Interface (GUI) are then initialized for image acquisition and processing. In general, C. elegans hermaphrodite worms remain in the wells in the presence of food, but adaptations can be made if escape occurs often (see Troubleshooting). In contrast, fly larva invariably escape from the LarvaLodge if a cover is not used. The analysis interface allows for selection of regions of interest (ROIs) and parameter selection, including exposure time and image capture frame rate. If desired, the animals’ sleep can be perturbed using a blue light illuminator (see below).
Data Analysis.
We use custom analysis software to quantify movement of individual worms by identifying pixels for which gray scale value changes between consecutive images by more than a threshold set by the noise in the recording. The total number of pixel changes, termed activity, reflects the amount of movement of the animal. The software identifies sequential frames in which no movement occurs as times when the animal is quiescent, and can be set to consider quiescence periods exceeding a certain minimal duration to be sleep. The data is processed in MATLAB or Excel to calculate total and fractional quiescence, quiescence bout duration and frequency, total activity, and activity bout duration and frequency. No special MATLAB toolboxes are required to run the analysis software.
Extensions and Modifications
Using the same imaging system and image processing pipeline described here, we have successfully used our multi-well devices to study locomotor behavior state41,42 and aging behavior24 in adult worms. For studies in C. elegans, once devices are filled with agar and animals, they are generally stable for weeks to months if they are sealed with Parafilm laboratory film (Bemis Co.) and an appropriate amount of hydrating material is placed inside the dish24. Therefore, experiments need not be confined to a set amount of time. Published work has focused on L2 animals and older, but analysis of L1 stages is also possible (Supplementary Video 1) by moving the camera assembly closer to the WorMotel, which increases the camera pixel resolution and enables visualization of this smaller developmental stage (~150 um body length). Because individual worms can be reared in the WorMotel throughout development and adulthood under either standard or stressful conditions, our method allows the investigation of the relationship between sleep, stress, and aging in individual animals24.
Monitoring of individual fly larvae across multiple developmental periods has not yet been pursued, but we anticipate this can be accomplished with modification to the existing LarvaLodge, with particular focus on a more sustained nutritional source to support larval growth. We have already demonstrated the ability to monitor late 1st instar larva through the molt to 2nd instars15, and short-term experiments with 3rd instars appear possible (Supplementary Figure 1). Prolonged monitoring of 3rd instars will require further modifications, including increased well size and higher percentage agar media to prevent burrowing.
Limitations
Our image processing algorithm tracks activity as the number of pixels that change between successive frames. This metric is attractive for its robustness and its ability to distinguish between sleeping and active animals. More sophisticated machine vision algorithms, which require first identifying the animal using thresholding approaches, can track a large number of features31,43,44 such as centroid and body curvature, and may therefore enable more nuanced behavioral analysis of sleep. Since the primary data acquired by our method are raw images of animals in individual wells, these data may be analyzed using any image processing pipeline if more detailed phenotypic parameters are desired.
Cessation of feeding in addition to body movement is commonly used as a criterion for behavioral quiescence in worms11,45,46. The system described here tracks only body movement and not feeding. However, we have found that the WorMotel chip is a convenient tool for longitudinal monitoring of pharyngeal pumping (feeding) rate by visual observation and/or video recording (Supplementary Video 2). Our system may be compatible with recently described automated methods for tracking pharyngeal pumping in C. elegans47.
Closed loop light stimulation enables well-controlled analysis of the effect of sleep deprivation on individual animals. The closed loop system described here allows for only a single experimental (test) animal at a time due to the lack of spatial control over the light stimulus. Future work will aim to parallelize closed-loop sleep deprivation by employing spatially defined light stimuli.
MATERIALS
Biological materials
Caenorhabditis elegans hermaphrodites.
We have used N2 (wild-type strain, variant Bristol isolate) and ZR1 rbr-2 (tm1231) IV
Drosophila melanogaster.
We have used Iso31 (wild-type strain)48, UAS-TrpA149 (outcrossed >5 generations into Iso31) and the GAL4 line from Rubin Janelia Farm collection50, obtained from Bloomington Drosophila Stock Center
Reagents
WorMotel and LarvaLodge casting
Sylgard 184 Silicone Encapsulant (PDMS) (Dow Corning)
Worm sleep agar and food recipe ingredients
Agar – Agar, granulated; Apex Chemicals & Reagents catalogue #: 20–248 CRITICAL: For reasons that we have not yet identified, heat-shock induction of stress induced sleep is sensitive to the type of agar used. We found that the use of powdered agar resulted in a loss of heat shock stress-induced sleep. We therefore recommend the use of granulated agar from the above manufacturer.
Peptone (powdered, bacteriological grade) - Apex BioResearch catalog #: 20–260
Sodium Chloride (NaCl) – EMD Millipore Corp. catalog #: 567440
Cholesterol - Sigma-Aldrich C3045
Calcium Chloride (CaCl2) – Alfa Aesar catalog #: 33296
Magnesium Sulfate (MgSO4) Sigma Aldrich 230391
Potassium phosphate monobasic (KH2PO4) – Fisher Scientific catalog #: S25499A
Streptomycin – Crystalgen catalog #: 0382–100
Fly larva sleep agar and food recipe ingredients
Agar powder (Fisher Scientific, catalogue #: BP1423–500)
D-Sucrose (Fisher Scientific, catalogue #: BP220–1)
Dry active yeast, granular (LabScientific, catalogue #: FLY-8040–5)
Tween 20 (Fisher Scientific, catalogue #: BP337–100)
Acrylic plate cover (110×60×3 mm)
Embryo collection cage (Genesee Scientific, catalogue #: 59–100)
Equipment
Device fabrication
3D Printing: Objet 30 with VeroBlack material or Projet 6000 with Visijet Clear material (or commercial 3D printing service)
Disposable plastic cups (for mixing PDMS)
Disposable spoon or spatula (for mixing PDMS)
Stainless steel spatula (for demolding) (Fisher #285822)
WorMotel and LarvaLodge preparation
Pipette – P200 Sartorius Biohit mLINE single channel mechanical pipetter Cat. No. 14–559-408
Pipette tips – Bioexpress, Genemate 1250 μL (Cat. No. P1236–1250)
Kimwipes – Kimberly-Clark 11 × 21 cm Cat. No. 34155
Parafilm – Bemis 4 in × 125 ft Cat. No. PM996
Worm pick – Platinum wire (Genesee Scientific, 30 gauge wire 90% platinum 10% iridium Cat No 59–1M30PI) embedded in a glass Pasteur pipette (Fisher Scientific 5 3/4” Cat. No. 13–678-20B)
Plasma cleaner - PDC-32G from Harrick Plasma
Vacuum pump – PDC-VPE from Harrick Plasma (Basic Model)
Imaging system (equipment listed below; please see Churgin and Fang-Yen 201540 for detailed protocol)
Camera DMK 23GP031 (The Imaging Source, USA)
C-mount lens (Fujinon HF12.5SA-1, 1:1.4/12.5 mm, Fujifilm Corp., Japan)
Post-mounting clamps (Thorlabs)
Infrared filter (Hoya 49mm R72)
Infrared LED strips (Ledlightsworld LTD, 850 nm wavelength)
Red LEDs
Aluminum plate (12″ × 12″ × 0.25”)
Acrylic sheet (12″ × 12″ × 0.25”)
Glass plate (8” × 8” × 0.25”)
1.5″ diameter optical posts (Thorlabs P14 and P6)
4 Rubber feet (Thorlabs)
12V power supply
Hex wrench set
Hand drill or drill press with 5/16″ bit
Allen key or hex screwdriver set
Epoxy
Razor blade
Black tape
Wire cutters (optional)
Blue light stimulus (For full information and parts list, see LightBox_Checklist.xlsx, Supplementary Data)
Required Parts
NIDAQ USB-6001 Data Acquisition device
High Power Blue LEDs (72 W)
Power Supply (20A-30V)
Solid state relay (Rated for 20 A Max constant current)
Electrical wire (16 AWG)
Aluminum heat sink
Super-thin electrical tape
Nylon screws
Thermal paste
Thermal fuse
Aluminum bar (1/4″ × 1/2″ × 1′)
Quick connectors (16 AWG)
Required tools
Soldering iron
Wire cutters
Drill press and drill bits
Tap wrench
Taps (4–40)
Computer screwdriver set
Multimeter (optional)
Safety glasses
For constructing the blue light stimulus apparatus
Drill press
Drill bits #43, #30, and #16
Tap wrench
4–40 tap
Soldering Iron
Solder
Equipment for exposing worms to heat or ultraviolet light
Circulating water bath – Lauda Ecoline E200 with E100 controller with capacity of 18 L UV Crosslinker: Spectroline XL-1500 UV, with 254 nm UVC bulb
Mirror box:
Mirrored Acrylic (1/4″ thick): 2 pieces (9″ × 8″), 1 piece (9″ × 4.5″) and 1 piece (6.5″ × 4.5″)
Acrylic cement
Heavy duty tape (for assembly)
To calibrate blue light stimulus:
Optical power meter (optional)
Software
MATLAB 2014 or newer (Mathworks)
IC Capture 2.4 or newer (https://www.theimagingsource.com/support/downloads-forwindows/end-user-software/iccapture/)
Custom Matlab software (Supplementary Data)
Meshlab (http://www.meshlab.net/)
Reagent Setup
For 4 liters of NGM agar:
Combine 68g Agar, 10g Peptone, 12g NaCl and 3900ml DI H2O. Add a stir bar and autoclave, then cool to 55 °C in a water bath before adding the following while stirring: 4ml Cholesterol (5 mg/mL), 4ml 1M CaCl2, 4ml 1M MgSO4, 100ml 1M KH2PO4 and 4ml Streptomycin (200 mg/mL). The NGM is poured into Petri dishes and, after solidification, can be stored for a few weeks at 4°C.
Procedure
CONSTRUCTION OF IMAGING SYSTEM (TIMING 2–4 hours)
CRITICAL A full protocol for constructing the imaging system used herein has been previously published33. Here we briefly recapitulate the main steps required to construct the imaging system (Figure 2).
Figure 2. Imaging system.
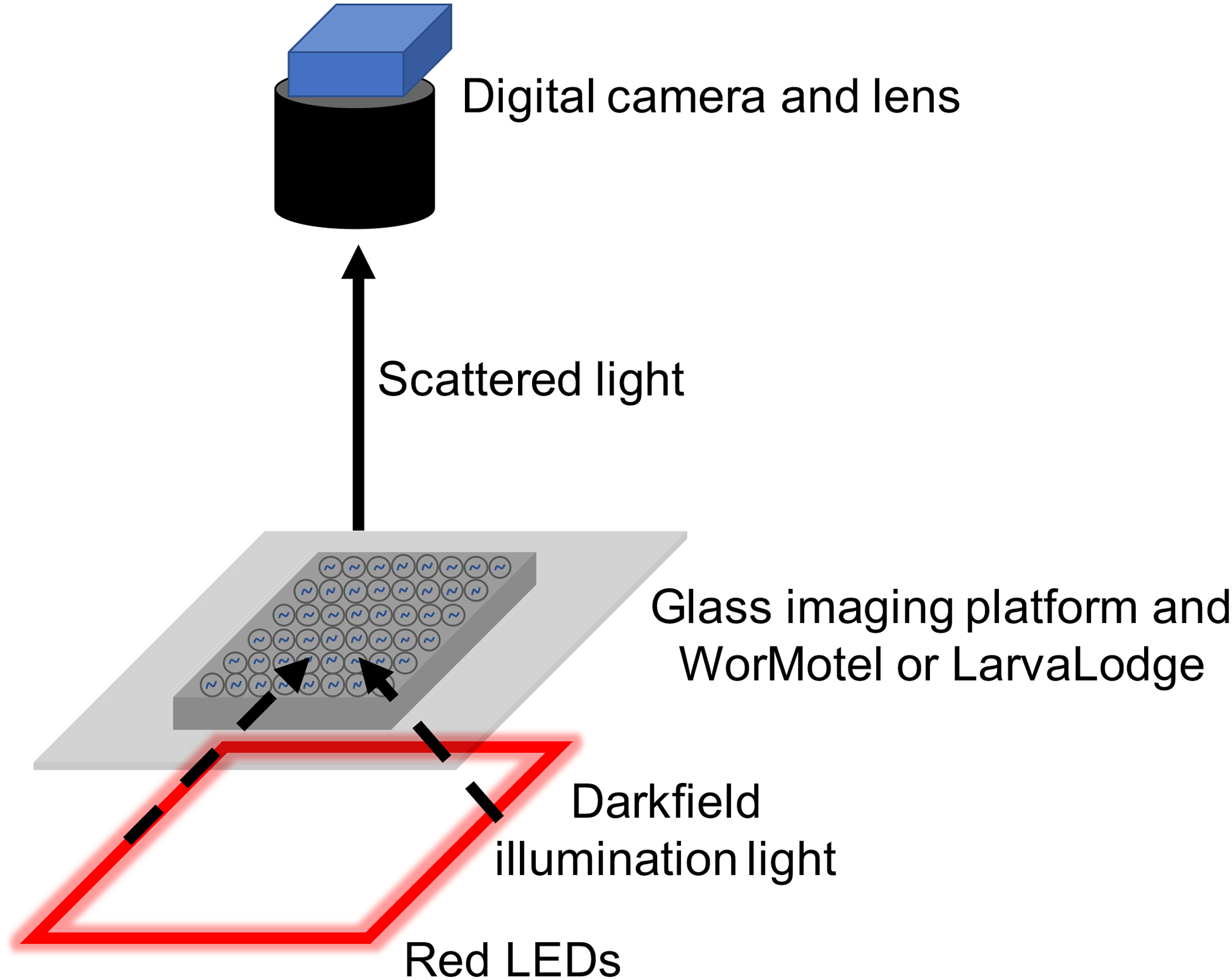
Schematic of imaging system and light path.
-
1
Drill a single 5/16″ hole in the aluminum and acrylic sheets such that the hole is centered along one axis and approximately 1.5″ from the edge. Wrap the aluminum sheet with black fabric. Tape the fabric so that it is secured to the aluminum sheet. Find the hole in the aluminum sheet by feeling through the fabric and use a razor blade to cut the fabric locally and expose the drilled hole.
-
2
Apply four rubber feet to the corners of the fabric-wrapped aluminum sheet. Fasten a 14″ long 1.5″ diameter optical post to the aluminum using a 1/4″−20 screw by placing the screw through the bottom of the aluminum plate and securing it to the post on the other side.
-
3
Use epoxy to secure an L-bracket to the glass plate. Let epoxy cure on a stable surface overnight.
-
4
Fasten the glass plate to a post-mounting clamp using a 1/4″−20 screw and hex nut. Fasten the glass-plate assembly to the 14″ optical post about 1.5″ from the aluminum plate.
-
5
Screw the C-mount lens onto the camera. Secure the camera to a 6″ long 1.5″ diameter optical post using a 1/4″−20 set screw. Mount the camera assembly to a post-mounting clamp and secure to the 14″ long optical post.
-
6
Secure the acrylic plate to the top of the 14″ long optical post. Cut black fabric to create 4 curtains in order to block all light out of the imaging rig. Secure the fabric to the top acrylic plate using tape. Curtains can be fastened to each other with binder clips, which allow for opening and closing of the imaging rig to place and retrieve samples.
-
7
For worms experiments, place four 4.7″ red LED strips below the glass imaging platform. Connect the four LED strips in parallel to a 12-V adjustable power supply. It may be necessary to strip the cladding on the leads of the LEDs using wire cutters in order to connect them in parallel. In order to minimize heat production, set the power supply to the lowest voltage that illuminates the LEDs. For fly experiments, we use the same approach but using infrared LEDs driven by a Tekpower DC variable power supply (2.2–15V), set to approximately 7.5V.
Crucial Step: Setting the voltage as low as possible ensures the LEDs do not generate excessive heat when left continuously on. Use a thermal probe to monitor temperature of the chips during prolonged imaging experiments.
Constructing the Blue Light Stimulus Apparatus for Sleep Perturbation (TIMING 2–4 hours)
-
8
Note that the back of the blue LED chip is electrically connected to one of the terminals. Place two layers of super-thin polyimide electrical tape across the heat sink (Figure 3a–b). The tape should cover an area large enough to prevent the blue LEDs from being in electrical contact with the aluminum.
-
9
Position the blue LEDs on either side of the heat sink and mark where holes should be drilled to secure each LED to the heat sink (Figure 3a). Drill and tap two holes for each of the two blue LEDs. Use a #43 drill bit and a 4–40 tap. Drill another hole towards the top of the heat sink with a #43 drill bit and tap it with a 4–40 tap. Secure the LEDs to the heat sink using nylon 4–40 screws. Nylon screws are used to prevent electrical contact between the LEDs and heat sink.
-
10
Cut the aluminum bar into 3/4″ long pieces. The final dimensions of one piece should be 1/4″ × 1/2″ × 3/4″. Drill two holes on the 1/4″ × 1/2″ face of the aluminum with #16 drill bit. Drill one hole in the 1/2″ × 3/4″ face of the aluminum with a #37 drill bit. Attach the aluminum piece to the primary heat sink using a 4–40 screw (metal or nylon) (Figure 3b).
-
11
Insert a thermal fuse into each hole in the accessory aluminum piece fastened to the primary heat sink (Figure 3b). Solder both thermal fuse ends together alongside a ~4″ length of 16 AWG wire on each side. Be careful not to overheat the thermal fuse during soldering. Attach a quick connector to the ends of each wire by crimping. Slide a quick connector to the negative lead of the right LED. Slide the other quick connector to the positive lead of the left LED.
-
12
Test the LEDs by connecting to the 20A/30V power supply. Using 16 AWG wire, connect the positive lead of the right LED to power and the negative lead of the left LED to ground. Ensure that both LEDs turn on when the power supply is turned on and current is increased to 20 A.
Caution: High power blue LEDs are extremely bright when driven at high current. Avoid viewing LEDs directly when testing.
?Troubleshooting
-
13
To enable software control of the blue LEDs (Figure 3c) connect the NIDAQ and solid-state relay to the LEDs. Connect analog output channel 0 on the NIDAQ to terminal 3 on the solid-state relay. Connect analog ground on the NIDAQ to terminal 4 on the solid-state relay. Connect the positive terminal of the power supply to terminal 2 on the solid-state relay. Connect terminal 1 on the solid-state relay to the positive terminal of the right LED. Connect the negative terminal of the left LED to the power supply ground.
Figure 3. Blue light stimulus assembly.
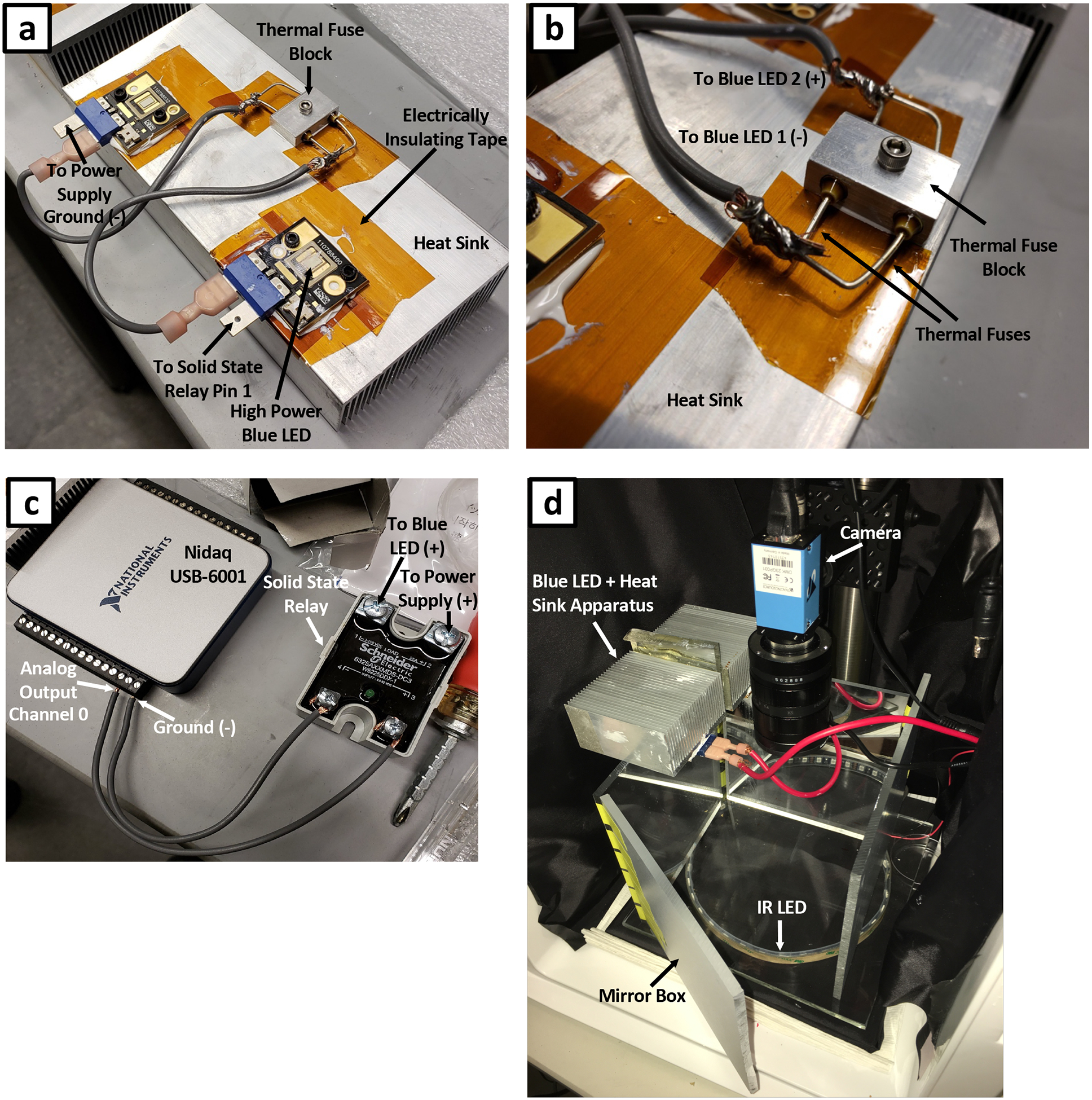
(a) High power blue LEDs attached to an aluminum heat sink. LEDs are wired in series through a set of thermal fuses. LEDs are electrically isolated from the heat sink using electrical insulating tape. (b) Magnified view of thermal fuse block secured to the heat sink. (c) Electrical connections between NIDAQ I/O device and solid state relay. This assembly allows for computer control of blue LEDs. (d) Complete imaging rig outfitted with blue LED stimulation. An acrylic mirror box is used to generate uniform blue light illumination.
Constructing the Mirror Box (TIMING 10 minutes plus 1 day for acrylic cement to set)
CRITICAL: This section explains how to construct the acrylic mirror box, which serves to deliver light from the blue stimulus LEDs onto the WorMotel or LarvaLodge and make the illumination more uniform.
-
14
Remove the protective paper from each panel of mirrored acrylic.
-
15
Stand each panel of acrylic up with the mirrored side facing inward. Arrange the four panels in a rectangular box formation on a stable work surface. One of the panels will be slightly shorter than the others; this panel is for placement and retrieval of experimental plates into and out of the mirror box (Figure 3d) Secure the panels using tape applied to the outside of the box.
-
16
Apply acrylic cement along the inside and outside of each joint between the panels.
-
17
Let the acrylic cement set overnight. The tape may be removed the following day, or left on.
-
18
Rest the blue LED + heat sink apparatus on top of the mirror box with LEDs pointing downward (Figure 3d).
Testing Blue Stimulation LEDs (TIMING 5–15 minutes)
-
19
Install the NIDAQ software (available in Supplementary Data) and MATLAB plugin for NIDAQ.
-
20
Ensure that blue LEDs are wired correctly and the power supply is turned on and operating in constant current mode.
-
21
Ensure MATLAB is operating in cell mode. In a .m file (MATLAB script file), two percent symbols in a row (%%) indicate the beginning of a new cell. Each cell in a script can be run in isolation.
-
22
Execute the first cell of BlueLEDTestWithNidaq.m (Supplementary Data: blue_light_setup). When successfully executed, this cell establishes a connection with Nidaq analog output channel 0.
-
23
Execute the second cell of BlueLEDTestWithNidaq.m. When successfully executed, this cell turns the blue LEDs on for three seconds before shutting them off.
-
24
You may wish to measure and report the irradiance of the blue light at the sample plane. If so, do this. We recommend generating a calibration curve for irradiance as a function of LED current. The light intensity at 20A current should be approximately 0.5–1 mW/mm2.
DESIGNING AND PRINTING DEVICES (TIMING 1–2 hours, plus time to print and cure devices)
CRITICAL: To fabricate a mold for a standard device, download the appropriate STL file (Supplementary Data: Device Fabrication) and proceed to step 28 (“Choice of 3D Printer”.) To design a custom mold, proceed with step 25.
-
25
Download the device fabrication zip file (moldGeneration.zip, Supplementary Data: Device Fabrication), which contains scripts for designing WorMotel (WorMotel_Mold_48Wells.m) and LarvaLodge (LarvaLodge_Mold_24Wells.m) devices in addition to a few additional functions used by these scripts. Ensure all files are in the current MATLAB path.
-
26
Execute the code contained within the .m file for the device you wish to design (either WorMotel_Mold_48Wells.m or LarvaLodge_Mold_24Wells.m). You may adjust the physical dimensions, number of wells, well size, and well spacing by editing the .m file, as specified in the comments. The primary output of each script is an STL (.stl) file for the three-dimensional mold design.
-
27
STL files may be visualized with the free software Meshlab (http://www.meshlab.net/). Once inspected to ensure that the device shape and dimensions are as desired, the .stl file may be sent to a 3D Printer or 3D printing service for fabrication.
-
28
Print the device. CRITICAL STEP: Fabrication of molds requires a 3D printer with adequate resolution. We have successfully used two 3D printers to fabricate our device molds: (1) an Objet30 Photopolymer Printer (0.0011″ layer resolution) with VeroBlack material and (2) a ProJet 6000 Stereolithography Printer (0.002″ layer resolution) with VisiJet Clear material. For the Objet30 printer, specify a glossy finish (no support material). Devices printed on the ProJet printer are slightly sturdier and have a less smooth surface. However, this does not affect device performance. The use of 3D printers with substantially lower resolution than those above, including many fused-filament printers, is not recommended.
Casting Devices (5 minutes, plus curing overnight)
-
29
Prepare the PDMS in a disposable plastic cup or large weighing boat by vigorously mixing the elastomer base and curing agent in a ratio of 10:1 by weight, according to the manufacturer’s instructions. The amount of PDMS you prepare will depend on the size of the device mold as well as the number of devices you plan on casting. The amount of PDMS required to fill each device is 5 g for the 48-well WorMotel and 25 g for the 24-well LarvaLodge. Prepare a generous amount as it is difficult to transfer all of the highly viscous PDMS from the mixing container.
Caution: PDMS can create a mess when not handled with care. Wear gloves when handling uncured PDMS and cover work spaces with a liquid-impermeable liner to aid in clean-up.
-
30
Place the empty mold you wish to cast on a balance and pour the appropriate amount of PDMS into the mold.
-
31
For fabricating devices with a flat bottom, see Box 1,otherwise proceed with this step. If you have a vacuum chamber, you may de-gas (remove bubbles from) the PDMS until no bubbles remain in the device (~5–10 minutes). However, if you cure the device at room temperature (<25° C), there is no need to de-gas in a vacuum chamber, as PDMS cures slowly enough that air bubbles have time to rise to the top and escape. If the poured device has been degassed, it can be cured at a higher temperature (40° to 50° C), and it may be de-molded after 8 hours. If the device is cured at room temperature, it may be de-molded after 24 hours.
Box 1 Fabricating devices with flat bottoms.
The procedures described in this protocol are the simplest and most common method for casting devices. However, these methods result in a curved meniscus of PDMS at the perimeter of the device. Therefore, the bottom of the device is not perfectly flat. This is not generally a problem. However, in certain cases it may be advantageous to fabricate devices with flat bottoms. To do so, use the following fabrication procedure.
Prepare PDMS mixture and de-gas either in a vacuum or by placing in a refrigerator overnight.
Pour PDMS into a device mold almost to the top
Wait ~30 min. for bubbles to rise. Pop bubbles by gently blowing on them.
Top off the molds by slowly pouring additional PDMS such that the surface is just above the sides of the mold.
Let settle ~5 min. until top of PDMS has equilibrated.
Carefully place a clean piece of clear acrylic sheet or overhead transparency film on the top of the mold, tilting slightly as you lower it so that no air becomes trapped. Place a weight on top of the acrylic or transparency film. No release agent is required. The work surface needs to be level and flat for this step to work.
Allow the PDMS to cure (Step 31).
Peel off the acrylic or transparency film and demold the device. Acetone may be used to clean residue off the acrylic or transparency film.
Demolding (1–5 minutes per device)
-
32
Using a thin metal spatula, gently pry the edges of the PDMS away from the walls of the mold, slowly working your way around the perimeter of the device. As you pry the PDMS away from the mold using the metal spatula, gently lift up to separate the PDMS from the mold.
Crucial step: Do not pry too strongly against the mold, as the walls of the mold may crack.
-
33
Once the entire perimeter of the device has been de-molded, gently peel the remainder of the device out of the mold.
Crucial step: Be sure to de-mold slowly, especially when peeling the PDMS off the mold, as PDMS is prone to tearing.
CRITICAL STEP: The first 1–3 devices made from a mold may release poorly. After several demoldings, the devices should release from the molds easily.
PREPARATION OF ANIMALS AND DEVICES FOR SLEEP ASSAYS
-
34If studying C. elegans sleep, follow option A. If studying Drosophila larvae sleep, follow option B. If monitoring aging in C. elegans follow option A but with the modifications discussed in Box 2.
-
Sleep studies in C. elegansTIMING 30–45 min for selection of test animals; 45–60 min to prepare the WorMotel for use; 20–40 min to load worm.
- Selection of test animals for a developmentally timed sleep (DTS) assay. Pick mid third larval stage (L3) animals in order to monitor the worms through two lethargus periods (at the end of the L3 and L4 stages), until the early adult stage. L3 animals are identified by selecting animals approximately 24 hours after feeding first larval stage (L1) diapause animals cultivated at 20°C. L1 diapause animals are made by the alkaline bleach method51. Alternatively, L3 animals can be selected based on size and the appearance of the developing uterus and vulva. For a stress-induced sleep (SIS) assay transfer L4 larvae, which are identified by morphology33, age, and/or size, to a new NGM plate seeded with bacteria 16–24 hours before the assay in order to test 1-day old adults on the day of the assay.
-
Preparing the WorMotel for use. Place the WorMotel chip well-side up in the chamber of the plasma oxygen cleaner (Harrick Plasma Cleaner PDC-32G) shown in Figure 4a. Seal the door (Arrow 1, Figure 4a), turn the power on (Arrow 2, Figure 4a), turn the vacuum pump on (Arrow 3, Figure 4a), and set the RF level to medium (Arrow 4, Figure 4a).CRITICAL STEP: PDMS is hydrophobic, resulting in beading of liquid agar added to the device and therefore undesirable convexity of the agar surface after gelling. Thus the material needs to be rendered more hydrophilic. There are a number of methods available for rendering the material more hydrophilic52–56. We expose the chip to plasma oxygen57, as described here, which introduces polar functional groups to the PDMS surface. This reduces the contact angle between the agar and PDMS and ensures the agar surface is flat once gelled.An alternative to plasma treating is to add 0.1% Tween 20 to the agar solution, which reduces its surface tension and again reduces its contact angle with the PDMS.
- Run the vacuum with the plasma oxygen cleaner radio frequency (RF) set to medium. Once the chamber reaches a pressure of 40–200 mTorr, pressures at which plasma oxygen can form, atmospheric oxygen inside the chamber is converted to plasma oxygen, which acts on the surface of the WorMotel chip. You will see a light blue/purple glow through the chamber window when plasma oxygen forms, typically visible after ~10 seconds (Arrow 1 in Figure 4b).
-
While the WorMotel is undergoing plasma activation, use a microwave oven to heat a ~225 mL volume of water housed in a 400-mL beaker for 2 minutes or until the water is boiling. This water is used to moderate heating of the agar in the in order to minimize boiling over of the agar. The water also helps to keep the agar solution hot while the researcher is filling the wells of the WorMotel chip.Caution! Wear protective gloves and eye protection when handling hot liquids.
- Microwave a 50 mL Erlenmeyer flask containing ~11.5 mL solidified NGM agar (removed with a spatula from a plate used for routine animal cultivation, Figure 4c) together in the beaker of hot water for 30 seconds or until the agar boils. Place a folded paper towel in the mouth of the Erlenmeyer flask to prevent molten agar from bubbling out of the flask and to prevent debris from settling into the molten agar (Figure 4c). Once the agar has melted, set the flask inside the beaker containing the recently boiled water (Figure 4d) and place next to a stereomicroscope.
- Remove the WorMotel from the plasma oxygen cleaner (after a 3–5 minute treatment) and place it in a Petri dish of 100 mm diameter. Add 1–2 wet twisted Kimwipes to the edges of the Petri dish (Arrow 1 in Figure 4e) to provide hydration during the experiment.
- Place the WorMotel on the stereo microscope and fill the wells using a pipette while viewing through the microscope. Using a pipette, dispense 15 μL of molten agar to fill each well to its top (Figure 4e).
-
Loading worms. Place one worm in each well along with a small amount of bacteria using a platinum wire pick; use the platinum wire to spread the bacteria to a thin layer on the agar surface, taking care not to scratch the agar surface.Crucial Step: If this WorMotel chip will be used in a heat shock assay, leave some wells without worms to house control untreated worms, which are added after the heat shock treatment.
- Make note of where each genotype is loaded on the WorMotel chip for analysis later. For DTS assays, there is no further treatment of the worms on the WorMotel. Proceed to step 36, “Setting up image acquisition”.
-
Sleep studies in Drosophila larvaeTIMING 5–10 min per plate to select test animals; 30–45 min to prepare the LarvaLodge for use; 20–40 min to load larvae.
-
Using blunt forceps, move larvae to the agar surface of a fresh petri dish. We use a 3% agar, 2% sucrose, 2.5% apple juice gel with yeast paste on top, but other media can be used as long as larvae are well nourished. During your preparation of the LarvaLodge, these larvae complete the molt to enter the 2nd instar stage.Crucial Step: Be careful not to injure the larvae during the procedure, avoid pinching or puncturing by using only one arm of the forceps to lift the animals up with a scooping motion.
- Preparing the LarvaLodge for use. Prepare the LarvaLodge while the larvae complete their first molt. To do this, first mix 3 g of agar and 2 g of Sucrose with 100 ml of H2O in a 200 ml beaker.
-
Boil solution in the microwave. Place a folded paper towel in the mouth of the Erlenmeyer flask to prevent molten agar from bubbling out of the flask and to minimize debris from settling into the molten agar (see Figure 4c).Caution! Wear protective gloves and glasses when handling hot liquids.
-
Using a pipette, fill each well of the LarvaLodge with 120 μl of molten agar (Supplementary Figure 2a; we typically run 4×5 wells). Try to avoid air bubbles and make the gel as even as possible.Crucial Step: In order to reliably work with the viscous solution, cut back the tip of the pipette approximately 5mm using a razor blade or scissors.
- If desired, pour the remaining gel into empty standard Drosophila vials, about 2.5 cm tall. Allow the medium to solidify, wrap in cellophane, and store the vials at 4°C for future experiments.
- Apply 10% Tween solution using a Kimwipe to the acrylic LarvaLodge covers, then wipe it dry with paper towel. The Tween minimizes fogging due to water condensation on the lid during imaging.
- After the gel cools, distribute a thin layer of yeast paste evenly on the agar surface of each well (Supplementary Figure 2b) using a soft paintbrush (#0, red sable).
-
Loading larvae. Transfer one larva into each well using blunt forceps.Crucial Step: Avoid scratching the surface of the gel when releasing the larvae; otherwise the animals will break up the media and burrow into it.
-
After each larva is loaded, cover the LarvaLodge with the acrylic cover. The side treated with Tween should be in contact with the LarvaLodge.Crucial Step: Make sure the cover of the LarvaLodge is tight to prevent escape by pressing down from the top (pushing up from the bottom of the LarvaLodge can dislodge contents of the wells).
- Note the genotype/condition for each well for later analysis.
-
Box 2 Monitoring aging in C. elegans.
To monitor C. elegans adults during aging, WorMotels can be prepared and imaged as described in the main procedure with the following exceptions. First, the agar should be supplemented with 100 ug/ml FUdR, which prevents growth of self-fertilized progeny, prior to filling the device. Second, 5 uL of densely cultured OP50 (or appropriate bacterially resistant strain such as streptomycin-resistant DA837) should be pipetted into each well. This ensures that enough food will be available to each animal for the duration of its lifespan. Third, single mid- to late-L4 stage larvae should be placed in each well. This can be done either manually with a wire pick or automatically using liquid handling instruments such as a COPAS Biosort. Fourth, plates should be sealed tightly with parafilm to prevent dehydration during prolonged (2–8 weeks) imaging. Finally, imaging is typically carried out at a rate of 0.2 Hz in 30-minute imaging epochs which occur once or twice per day. It is also useful to employ a 10-second bright blue light stimulation during each imaging epoch in order to measure evoked movements, which are important for discerning old, lethargic animals from dead ones. The same blue-light setup used in this protocol for Drosophila larva sleep deprivation may be used to stimulate aging C. elegans. Aging analysis software and further details are available (Churgin et al., elife 2017).
Figure 4. Preparing and filling the WorMotel.
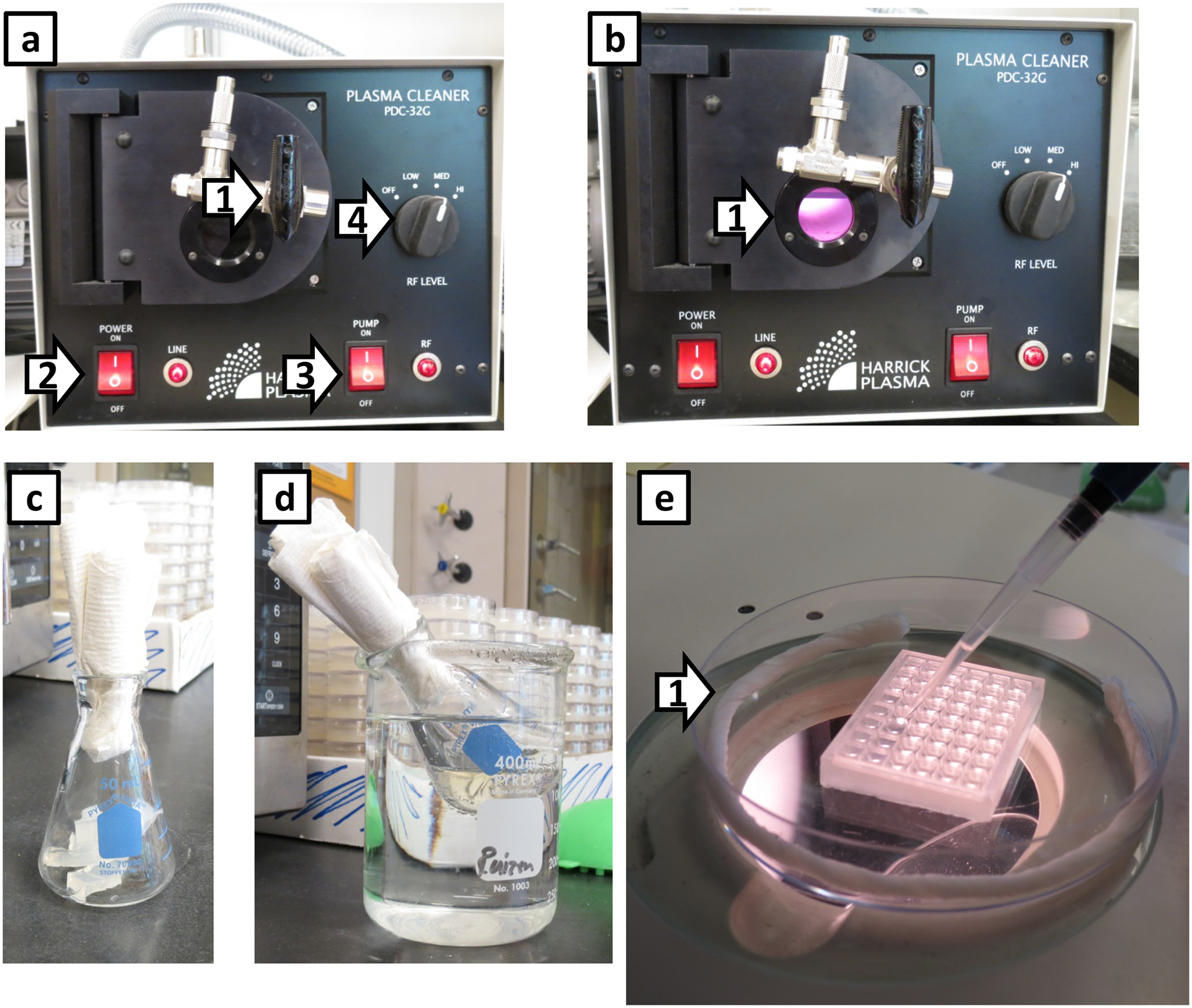
(a) Plasma oxygen cleaner with the vacuum pump turned on before plasma oxygen is formed in the chamber. These are the settings used for plasma oxygen treating the chip. Arrow 1: Valve to control gas into the chamber. Arrow 2: On/Off switch for the plasma cleaner. Arrow 3: On/Off switch for the vacuum pump. The switch on the vacuum pump is always switched to On. Arrow 4: radio frequency (RF) control; we set it on medium. In (b), the reduced pressure produced by the vacuum pump has generated plasma oxygen in the chamber, which has a purple/pink glow (Arrow 1). Running the vacuum pump approximately 10 seconds is sufficient. (c) 50 mL Erlenmeyer flask with ~11.5 mL of solid NGM agar prior to melting. (d) Heated Erlenmeyer flask placed in the beaker of hot water to hold its temperature while filling the WorMotel chip. (e) Filling the WorMotel chip. Arrow 1 indicates a moistened Kimwipe rolled up in the petri dish to prevent the agar from drying out in the WorMotel chip. Depending on the humidity of the lab space, an additional Kimwipe may be required.
INDUCTION OF WORM SLEEP
-
35To treat worms with heat shock, follow option A. To treat worms with UV irradiation, follow option B.
- Treating worms on the WorMotel – heat shock (TIMING 40 minutes)
- Cover the edge of the Petri dish containing the loaded WorMotel with two layers of stretched parafilm and place the Petri dish at the bottom of a water bath set to 35°C. Place a heavy object such as a beaker filled with water (at the same temperature as the water bath) on top of the Petri dish to keep it at the bottom of the water bath.
- After 30 minutes, remove the WorMotel and place it in a fresh 100 mm plate with cool, wet Kimwipes.
- Using a stereo microscope, quickly add untreated control animals to the same WorMotel prior to beginning the image acquisition.
- Treating worms on the WorMotel – UV irradiation (TIMING 6 minutes)
- Cover untreated control animals on the same PDMS device by covering control worm wells with a combination of folded paper and an aluminum foil tent (Supplementary Figure 3a–e) to block UV radiation. CRITICAL STEP: Alternatively the non-treated worms can be added to the WorMotel after the UV irradiation step.
-
Set the UV crosslinker (Spectrolinker XL-1500, Spectroline, equipped with 254 nm UVC bulb) to deliver a 1500 J/m2 dose of UV to the worms.Crucial Step: We find high variability between individual UV crosslinkers in UV power using identical settings, likely due to variable aging of the light source bulb. We therefore recommend performing a dose-response curve of behavioral quiescence as a function of J/m2 dose as reported58, in order determine the optimal setting for each UV crosslinker.
- Place the plate containing the WorMotel in the UV crosslinker with the lid off and press the start button.
SETTING IMAGING SYSTEM PARAMETERS (TIMING 15–60 minutes)
Crucial: It is critical to optimize the settings of the imaging system, such as focus, aperture, and lighting, before acquiring data. Poor image quality will yield noisy and unreliable data.
Crucial: Before imaging, ensure that the camera gain (Device properties → Gain) is minimized in the camera control software. Increasing gain increases the amplification of photocurrent from each pixel, which increases image noise, potentially causing unreliable quiescence and activity data. If image brightness is too low, adjust the position of the LED lights to increase illumination, increase the camera exposure time, or open the lens aperture. Do not increase the camera gain setting.
-
36
Adjust the camera height until the WorMotel or LarvaLodge device containing animals fills the entire camera sensor.
-
37
Open the lens aperture so that it is approximately 75% towards the open position (100% would be fully open).
-
38
Adjust the camera focus until you can clearly see each animal. See Supplementary Figure 4 for examples of optimal and suboptimal exposures.
?Troubleshooting
IMAGE ACQUISITION
(setup requires 5–10 minutes; run time depends on the desired length of the experiment)
-
39Follow option A for C. elegans or option B for Drosophila larvae. To undertake Population arousal and deprivation experiments follow option C. For Closed-loop sleep deprivation follow option D and also undertake option E to carry out the Control experiment for the closed-loop deprivation. Arousal and sleep deprivation systems described for fly larva can also be applied to worms.
- Image acquisition in C. elegans.
- Rub 10% Tween (using a Kimwipe or paper towel) on the inside of the lid of the plate containing the treated WorMotel until you observe many little bubbles form. The Tween minimizes water condensation on the lid of the Petri dish.
-
After the lid dries, place it back on the plate and move the plate under the camera in the imaging system for recording (Figure 5a,b).Crucial Step: The image will be inverted, so remember to turn the plate 180 degrees before placing into the recording chamber in order to keep track of different genotypes in each well.
-
Open a new session of the IC Capture imaging software for each recording session.Crucial Step: The settings for IC capture will depend on the camera. The camera used in this protocol should have the resolution set to “Y800 (2592×1944)” (arrow 1, Figure 5c) and the frames per second (FPS) should be set to 5 when imaging but a higher FPS may be helpful when moving the WorMotel around under the camera (arrow 2, Figure 5c). This FPS is the rate that the image on the screen is updated, not the rate of image acquisition, which is specified elsewhere. When the FPS is set higher, IC capture runs slowly so it is beneficial to lower it to 5 when the WorMotel chip is properly oriented. To optimize the amount of light through the camera for the best image, adjust the aperture of the lens (arrow 2, Figure 5b) or the exposure duration (arrow 3, Figure 5c).
- Set the camera to “Live Mode” and move the chip around until the all 48wells are visible on the computer monitor screen.
- Focus on the worms in the wells by adjusting the lens focus (Figure 5b); aim to get as many worms as possible in focus. Because it is difficult to fill each well to the exact same level and have a perfectly level chip, there may be a few wells that are slightly out of focus; wells in which animals are out of focus can be censored later during analysis. For DTS assays, lower the position of the camera until only 24 wells are visible. Note: smaller lens aperture settings will increase the depth of field, at the expense of image brightness.
- Create a folder on an external hard drive as the destination folder for images acquired during the recording session.
- Set the sequence timer (Figure 5c) to capture one BMP image every 10 seconds for 132 minutes for heat shock SIS experiments, 492 minutes for UV irradiation SIS experiments, or 1452 minutes for DTS experiments and save to the newly created folder (Figure 5d–f). Any of these parameters can be modified depending on the needs of the experiment.
- Create a Notepad text (.txt) file within the picture folder and record the following details pertaining to the assay: the date, the experimental condition (e.g. genotype, drug) of each worm and its location on the WorMotel, how long the recording should be, and the room temperature when the recording started. Also make note of anything atypical that may occur during the recording session (e.g. large fluctuations in room lights, high level of ambient noise, etc).
- Click “start timer” once all of the settings are entered.
- Image acquisition in Drosophila larvae
-
Place the LarvaLodge into the imaging system. We house these systems in temperature-controlled incubators or rooms. Focus on the larvae in the wells by rotating the lower portion of the camera lensCrucial Step: The image will be inverted, so remember to turn the plate 180 degrees before placing into the incubator in order to keep track of different genotypes in each well.
-
Open a new session of the IC Capture imaging software for each recording session.Crucial Step: The settings for IC capture will depend on the camera. We typically set the resolution to “Y800 (2560×1920)” and the frames per second (FPS) should be set to 10. This FPS is the rate that the image on the screen is updated, not the rate of image acquisition. To optimize the amount of light through the camera for the best image, the user can adjust the aperture of the lens or the exposure.
- Set the camera to “Live Mode” by clicking on the camcorder icon and move the chip around until the entire chip is visible on the computer screen.
- Set up imaging using the sequence timer window (Figure 6a). Click on the settings button (Figure 6b) and select the “Automated sequence” tab to set the time between images taken (we typically use 6 seconds) and the length of the experiment (Figure 6c). Set image file type to “BMP” within the “File type” tab (Figure 6d). Next, open the “Filename and Target” tab and set “Image” as “Filename prefix”, “Index” should be 1. Finally, set the “Target directory” to the desired target folder (Figure 6e).
- Click “start timer” on the sequence timer window once all of the settings are entered (Figure 6f).
-
- Population arousal and deprivation experiments (time depends on the length of the experiment)
Figure 5. WorMotel imaging setup.

(a) View of the WorMotel imaging setup under a black cloth cover to block ambient light from the room. (b) Complete view of the imaging system. Arrows 1 and 2 indicate the camera and lens, respectively. Arrow 3 indicates the WorMotel inside a petri dish. Arrow 4 indicates the platform on which the WorMotel is placed, under which are the illuminating red LEDs. Arrow 5 indicates the support post. (c) Screen capture of the IC Capture software for imaging the WorMotel chip. Red box indicates location of Sequence Timer. Arrow 1 indicates resolution setting for camera; we recommend using the highest resolution possible for the camera. Arrow 2 indicates the frames per second (FPS) of the image displayed on the screen; this is the rate at which the image refreshes on the screen and is not the rate of image collection. Arrow 3 is the exposure time of the camera. (d) Screen capture of the Sequence Timer located in the lower right hand corner of the screen. To change the settings of the Sequence Timer, click on “Settings” in the red box. The window in the middle of the screen is used to alter setting of Sequence Timer. It opens on “Automated Sequence” which allows the user to set the frequency of image capture (Interval) and how long the software should collect the images (Automatically End Sequence). (e) Screen capture shows tab of the Sequence Timer “File Type”. The analysis software is designed to handle the BMP (bitmap) image file type. (f) Screen capture shows tab of the Sequence Timer “Filename and Target”. The user sets where images are saved. Set the index to 1 before starting each experiment; otherwise it resumes where the previous experiment left off.
Figure 6. LarvaLodge imaging software setup.
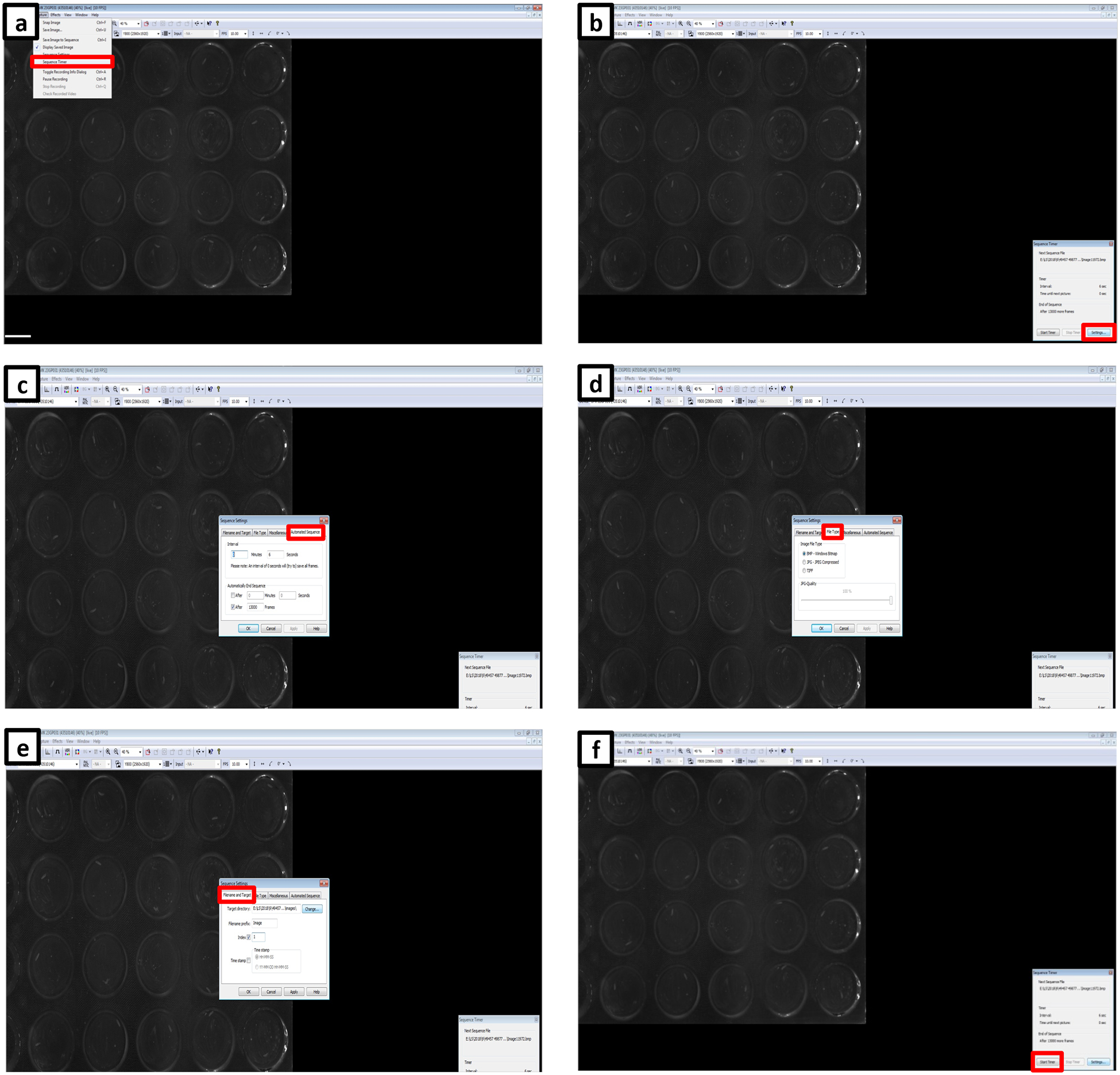
(a) Initiate Sequence Timer from the drop-down menu. (b) Open the Settings menu. (c) Set the imaging sequence interval and the length of the experiment. (d) Select file type and (e) choose file name prefix, the start frame of the imaging, and set the path for the target directory. (f) Initiate the analysis by clicking the Start Timer button.
Critical: While the sleep deprivation experiments described herein using blue light illumination are compatible with both C. elegans and Drosophila larvae, our published sleep deprivation experiments have focused solely on Drosophila larvae. Therefore, we only refer to Drosophila and the LarvaLodge when discussing sleep deprivation here. However, note that these methods are also applicable to C. elegans. We have found experimentally that C. elegans adults are far less sensitive to blue light than Drosophila larvae. Higher LED intensities are therefore required in order to elicit behavioral responses from C. elegans than Drosophila larvae. This observation should be considered when designing experiments.
-
Move the LarvaLodge into an incubator where the blue light LED setup is housed. Place the LarvaLodge under the camera and start imaging.
Crucial step: Remember to set “Filename prefix” to “image” in the sequence timer window.
Initiate the LightArousal.m script (Supplementary Data: drosophila/LightStimulation.zip) in MATLAB by clicking Control+Enter in the upper section of the Editor panel. Set all the variables according to the planned experimental design.
-
Set Arousal and deprivation variables:
line 33: ledontime=x (LED on time in seconds). In our published work15, we used a ledontime of 4 seconds.
line 34: waittime=x (time between successive LED ON times in seconds)
line 35: expruntime=X (the desired experiment run time in hours, the analysis terminates when the set time expires). Most of our experiments are ~24 hours in duration. Caution! Long LED ON time (continuous 2–3 hours) can lead to overheating. Ensure that a thermal fuse (Figure 3a) is installed on your LED heat sink (Figure 3b) to reduce the possibility of fire in the rare case where the LEDs are left on for a prolonged period of time.
Initiate the program by clicking Control+Enter in the second segment of the script in the Editor. Select the path to the folder where the images are being saved. A file named “stimulationFrames will be generated in the target folder.
If required, terminate the experiment manually by clicking Control+c in the MATLAB command window. Otherwise, the experiment runs the duration specified in expruntime variable.
In order to list the exact frames when arousal was employed, open the “stimulationFrames” file in MATLAB, then double-click on the value “stimulationFrames” in the workspace window. The data can be exported from the Editor.
-
Closed-loop sleep deprivation (time depends on the length of the experiment, approximately 3–8 hours)
CRITICAL: In this system, a light pulse is delivered to wake the animal whenever quiescence is detected in real-time.-
Move the LarvaLodge into an incubator where the blue light LED setup is housed (Figure 3d). Place the LarvaLodge under the camera and start imaging.Crucial step: Remember to set “Filename prefix” to blank in the sequence timer window before starting imaging.
- Initiate the ClosedLoopAnalysis.m script (Supplementary Data: drosophila/LightStimulation.zip) in MATLAB by clicking Control+Enter in the upper section of the Editor panel. The program will prompt you to find the first image of the actual experiment. The first row of the ROI Auto-Selection panel will allow the user to select multiple pre-set arrays of experimental wells populated automatically (“type of plate”), type 1 if using previously set ROIs in the next window, otherwise leave it on 0, and finally type 3 if the selection method is the automated web placement (Figure 7a)
- After selecting the web method, follow the instructions to create the ROIs. Click the centers of the following wells: the upper left well, the upper right well, the lower left well and the lower right well (follow arrows, Figure 7b). The GUI then identifies approximately where the center of each well falls on the images.
-
After defining the ROIs, set the path where the analysis folder will be created.Crucial step: Select the same folder where the experimental images are being saved (Figure 7e).
- Enter the test ROI number (Figure 7f; all the other ROIs can be utilized as yoked controls. A new window will open representing the pixel changes between the first two frames. If the thresholding is correct type “1” into the command window, otherwise enter “0” and change the parameters accordingly (Figure 7g).
- After the threshold is set, scroll down in the editor window to the second part of the script and initiate it by clicking Control+Enter. The program will begin the closed-loop analysis at the most recent frame and turn on the LED when the larva in the ROI is quiescent (Figure 7h). Quiescence in these experiments is defined as 6 seconds15.
-
Set the Closed-loop variables (below are the values we typically use):line 52: “spi”=6 (sets how often the images are taken, seconds per images). This variable can be altered as desired.line 53: “AutoSaveFrames”=100 (sets how often the experiment is saved)line 54: “NoiseThres”=40 (the default grayscale threshold in our setups)line 57: “qthresh”=1 (sets the value to label the ROI as quiescent; after thresholding, the presence of any pixel change is considered movement).line 58: “lOT”=4 (LED light ON time in seconds)line 59: “expruntime”=8 (the experiment run time in hours, the analysis stops when the set time expires)
- When the experiment ends, locate and open the “activity” file that has been generated within the “Analysis” folder during the experiment. Open the file in MATLAB. Double-click on the ActVal Value in the Workspace panel and copy the values from the editor panel to obtain the activity data from the experiment.
- Double-click on “lightOnTimes” in the Workspace to export the amount of total LED ON times, as well as the exact frame number when it was initiated.
-
Figure 7. Closed-loop activation setup.
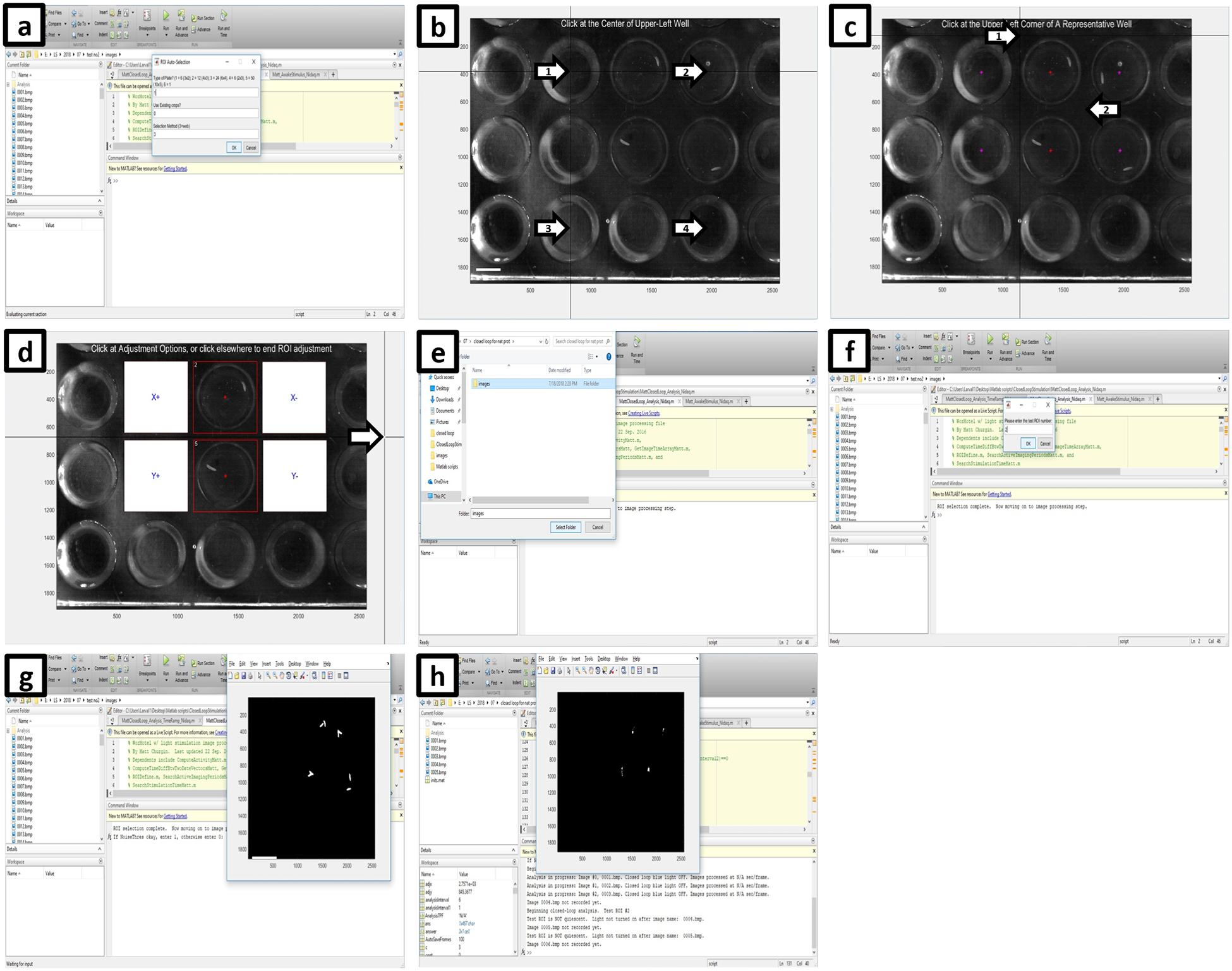
(a) After the first image of the experiment is loaded, use previously set ROIs or decide if new ROIs will be generated using the web method or manually. (b) If using the automatic method, click at the center of the upper left well (arrow number 1), the upper right well (arrow number 2), the bottom left well (arrow number 3) and finally the bottom right well (arrow number 4). (c) If the program successfully designates the center of each well (red stars), click on the upper left, then the bottom right corner of a representative well (arrows). (d) Click outside of the image (arrow) if the ROI placement is satisfactory to complete ROI selection. (e) Next, set the path where the analysis folder will be created, then (f) enter the test ROI number that will be driving the closed-loop system. (g) A new panel labeled Figure1 will appear showing the thresholded view after comparing the first two images to detect movement. Type 1 into the command window of MATLAB if thresholding is satisfactory, or 0 if change is required. (h) After the next part of the script is initiated, the program will begin the closed-loop analysis at the most recent frame and turn on the LED when the larva in the test ROI is quiescent.
-
Control experiment for the closed-loop deprivation (time depends on the length of the experiment, approximately 3–8 hours)
CRITICAL In this system, a light pulse is delivered only to active animals (specifically not during quiescence), allowing for experimental controls that have received the same amount of light pulse exposure without sleep loss.-
Move the LarvaLodge into an incubator where the blue light LED setup is housed. Place the LarvaLodge under the camera and start imaging.Crucial step: Remember to set “Filename prefix” to blank in the sequence timer window.
- Next, initiate the AwakeStimulus.m script in MATLAB by clicking Control+Enter in the upper section of the Editor panel. Follow the instructions as previously described for closed-loop deprivation (steps b–f).
- After the ROI is selected and the threshold is set, initiate the second part of the script by scrolling down in the editor window and clicking Control+Enter. The program will begin the analysis at the most recent frame and initiate the first light pulse when the set time between two arousals will expire AND the larva within the ROI is inactive.
-
Set Awake-stimulus variables (below are the values we typically use):line 47: “spi”=6 (value should be set to the same value as the seconds per frame of image acquisition)line 48: “AutoSaveFrames”=100 (sets how often the experiment is saved)line 49: “NoiseThres”=40 (the default threshold in our setups)line 52: “qthresh”=1 (sets the threshold to label the ROI as quiescent)line 53: “lOT”=4 (light ON time in seconds)line 105: “stimuliPerHour”=30 (number of stimuli per hour)line 106: “timeBetweenStimuli”=3600/stimuliPerHour (time between stimuli)line 107: “continuouslyActive”=3 (the number of frames the test larva must be continuously active before being stimulated)
-
PROCESSING IMAGES (30–45 minutes)
-
40
Process C. elegans data as described in option A and Drosophila larva as described in option B.
Option A) C. elegans.
Start MATLAB and type “guide” in the command window to open all of the GUI options on the computer.
Choose the GUI named “MC_QuiescenceActivity_v1202” Open the GUI and run the WorMotel Analysis GUI by clicking the green arrow head play button (circled in Figure 8a).
In order to open the images within the GUI, click “Preview Images” (Arrow 1 in Figure 8b), then click back to “Process Images” (Arrow 2 in Figure 8b). The GUI gives detailed directions on how to define the ROIs, which step needs to happen next, and when to start the analysis, however, this protocol will also go over the steps to follow.
Click “Load Image” (Red rectangle in Figure 8c) and select the first image collected in the assay.
Click “Select ROI” (Red rectangle in Figure 8c), which offers three options for selecting the regions of interest. Choose “Auto Selection (Web Method)” by typing “1” and clicking “OK.” (Figure 8d)
Enter the number of rows and columns used in the WorMotel; In a typical SIS experiment, the number of rows is six and the number of columns is eight (Figure 8e).
Click the centers of the following wells: the upper left well, the upper right well, the lower left well, and the lower right well (Arrow 1–4, respectively in Figure 8f). The GUI then identifies approximately where the center of each well falls on the images.
Choose a representative well and click the upper left corner and the lower right corner (Arrows 1 and 2, respectively in Figure 9a). Based on this information, the GUI identifies the approximate region each well occupies with a square ROI for each (Figure 9b). If the ROIs are inaccurate, make modifications on the left side of the GUI to the size in pixels of the ROI or the center location of the ROIs. Click “Finish Adjustment” (Red rectangle in Figure 9b) when the ROIs satisfactorily cover each well without extending into adjacent wells.
-
Click “Set Parameters” to specify the settings for the analysis. A window will pop up to enter the parameters for the analysis (Figure 9c). The default “Start Frame” is 1 but this parameter can be altered if desired (for example, if the first few frames were captured while the worms were disturbed). The default “End Frame” is the total number of images collected in the assay, but this parameter too can be adjusted. The “Time/Frame(sec)” is 10 if images were captured every 10 seconds. The “Image File Prefix” is Image; this is the way IC Capture names the collected images. The “BKG Threshold for ActVal Analysis” is 0.2, which is the minimum grayscale fractional change that a pixel must undergo in order to be considered to have changed between two images. The “Spatial Filter Size” is set to 1; the spatial filter size is the standard deviation (in pixels) of a 2-dimensional gaussian kernel used to smooth the difference image prior to thresholding, where a 1 indicates the gaussian filter standard deviation is 1 pixel. The “Frame Skip” is set to some multiple of the frame capture interval (in our case 10) when it is desired to calculate a second difference image to measure activity between images with a longer time window. Note that too short a frame capture interval and subtraction may result in inflated quiescence values whereas too long a frame subtraction interval may result in excessively low quiescence values, since most quiescence bouts are less than 30 seconds10,59. The program always calculates differences between adjacent images, but Frame Skip allows the specification of a second image subtraction interval with the results saved in a separate activity matrix (ActValS). Set “Synchronized Analysis?” to 0 if desired to analyze the images after acquisition is completed and set “Autosave per N frames” to the desired value (we vary this value between 0 and 100), then click “OK.” (Figure 9c)
Crucial Step: The protocol we describe assumes analysis and image acquisition are separate tasks, but if desired, the user can set up the analysis to take place during the experiment. Simply begin the “Processing Images” section as soon as the images are being acquired and set “Synchronized Analysis?” to 1 and set “Autosave per N frames” to an appropriate number of frames after which it will automatically save the analysis. Autosave can be used if it is desired to save the data at regular intervals during acquisition. Note that performing the analysis during image acquisition requires that the computer used for image acquisition to be running MATLAB.
-
Another window will pop up that reads “Click ok to display thresholded image” (Figure 9d). Click “OK” and the thresholded image will appear. The thresholded image shows any pixel whose normalized grayscale value difference between the two frames (first two frames of the experiment) was greater than 0.2 of the average grayscale value at that pixel. Such pixels are caused by worm movement.
Crucial Step: All white spots should correspond to worms in the wells (Figure 9e). If thresholding is set too low (i.e. 0.05), then white spots may appear within the field of view (see Figure 10m for example). In this case, increase the “Threshold for ActVal Analysis” value until all pixel changes can be attributed to the worms. Type “1” to indicate that the parameters are good and click “OK”. Adjust the threshold to best suit your lighting setup, as this significantly influences the background levels. Once you have defined an effective threshold for a particular recording rig, it rarely needs to be adjusted.
Click “Start Analysis” to begin processing the images. The banner above the device image shows the number of the image under analysis as well as the total number of images to be processed (Figure 9f).
When the analysis is finished, click the save button in the GUI, which saves the numbers of pixels changed in each WorMotel well as a function of time in a .mat (MATLAB) file. Save this file in two locations, in the directory location for MATLAB for further analysis and in the file containing the images for the assay. This is done so the file is available for the next step in analysis and also to have a backup to analyze again or differently in the future if necessary.
Figure 8. Screenshots of image processing (WorMotel, part 1).
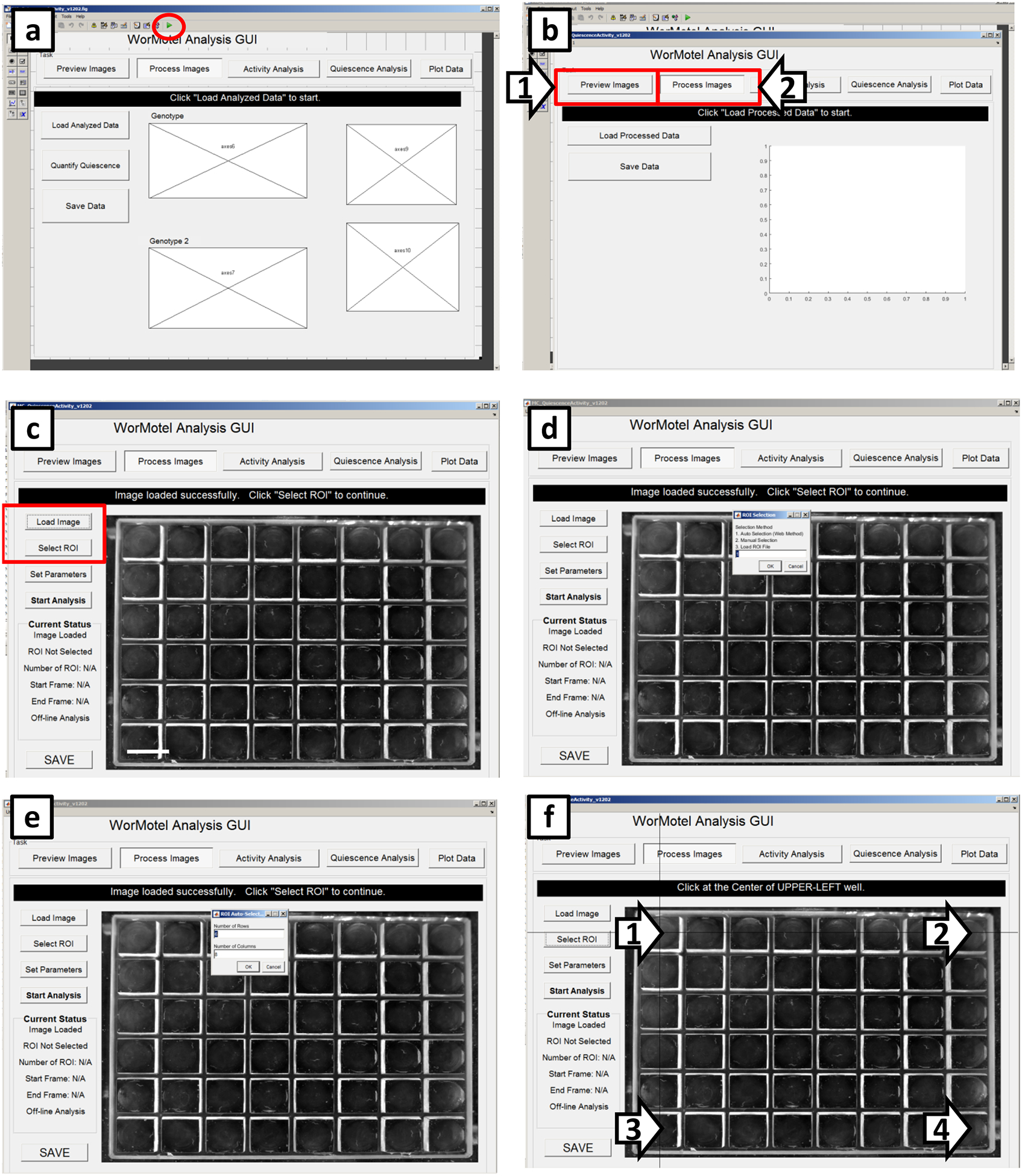
(a) Screen that appears when the GUI has been selected but not initiated. To run the GUI, click the icon within the red circle. (b) To load an image for analysis, first click “Preview Images” (arrow 1) then click back to “Process Images” (arrow 2). (c) To load an image, click “Load Image”. After loading define the regions of interest (ROIs) by clicking on “Select ROI”. (d) To continue ROI selection, type “1” then click “OK”. (e) Define the number of rows and columns to be analyzed. (f) Define the ROIs by clicking on wells in each corner of the imaging range as demarcated by black targets (arrows).
Figure 9. Screenshots of image processing (WorMotel, part 2).
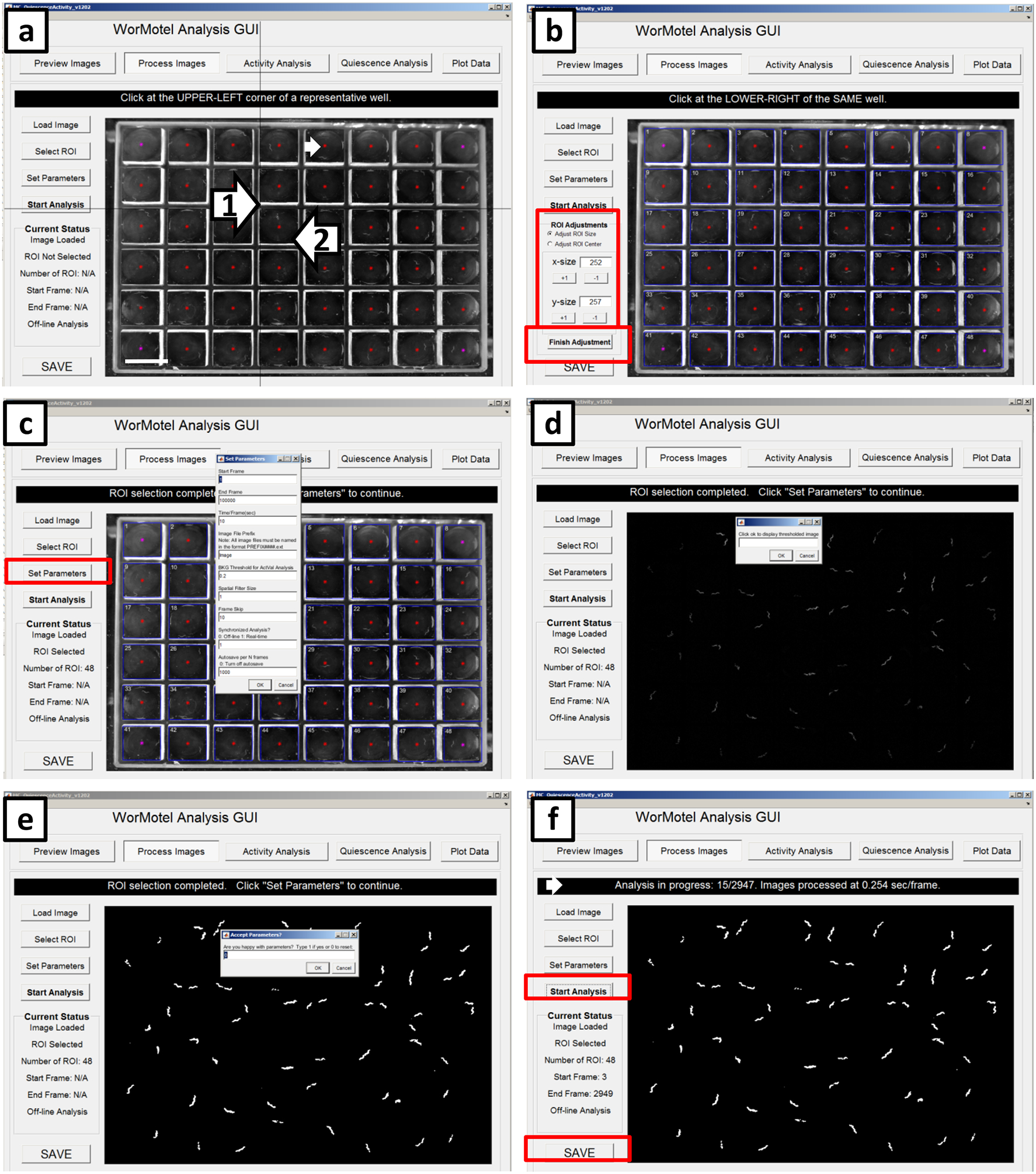
Continuation of defining the ROIs. (a) Following Fig 5f, red stars appear in in the center of each well (small white arrow). Click the upper left corner (arrow 1) of a representative well followed by lower right corner (arrow 2) of the same well. (b) Blue boxes appear around each well; these are the ROIs used for analysis. The center or size of the ROIs can be adjusted if not in the proper location by using the ROI adjustments located in red box. Once satisfied with ROIs, click “Finish Adjustment” in the lower red box. (c) Click “Set Parameters” and enter the appropriate parameters based on experimental details. (d) Click “OK” to show the thresholded image. (e) Screen shot of sample thresholded image that should reflect worm movement. (f) Analysis in progress is displayed after clicking “Start Analysis”. The black banner (small white arrow) shows the image number currently being analyzed and image processing speed. Once the GUI has processed the last image, click “Save” in the bottom left corner then save a .mat file for further analysis.
Figure 10. Screenshots of LarvaLodge image processing.
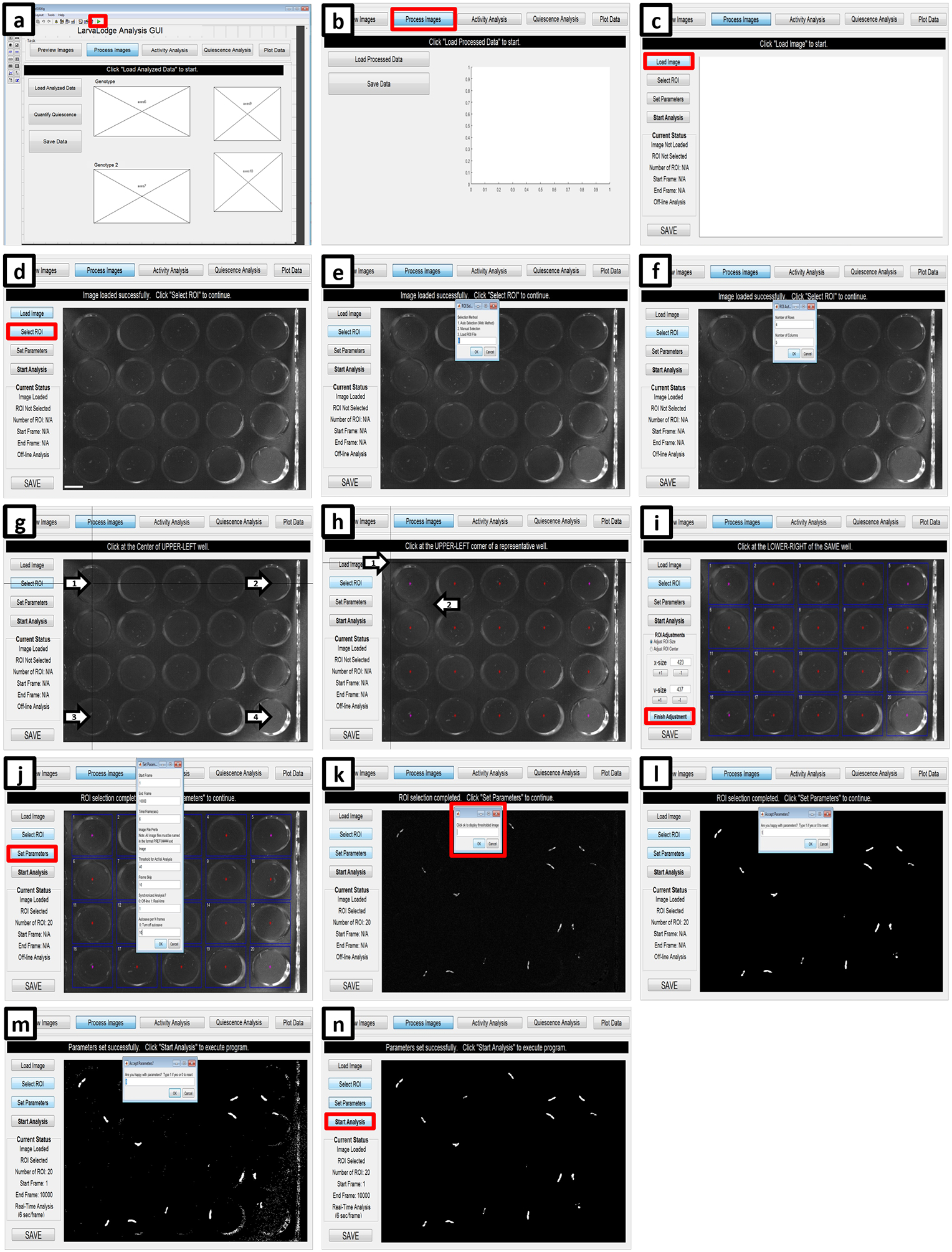
(a) Initiate the LarvaLodge Analysis GUI by clicking on the green play button. (b) On the next window, click process images. (c) Load the first image of the experiment. (d) Click on “Select ROI” and (e) choose ROI detection method. (f) If using the Automated method, fill in the number of rows and columns used in the experiment. (g) Next, click at the center of the upper left well (arrow number 1), the upper right well (arrow number 2), the bottom left well (arrow number 3) and finally the bottom right well (arrow number 4). (h) If the program successfully designates the center of each well (red stars), click on the upper left, then the bottom right corner of a representative well (arrows). (i) On the next panel blue boxes are placed over each ROI. If the placement is not satisfactory, the user can change the dimensions using ROI Adjustments; otherwise click Finish Adjustment. (j) Select Set Parameters (red box) and fill out the required information. (k) Click OK to display the thresholded image (red box). Type 1 if the thresholding is satisfactory (l), or zero if it is too low (m) and change the parameters accordingly. (n) Start the analysis (red box) when all parameters are set successfully.
B) Drosophila larvae (30–45 minutes)
-
Start MATLAB and type “guide” in the command window to open all of the GUI options. Once imaging starts, open up MATLAB and type “guide” in the command window to initiate the LarvaLodge analysis GUI (MC_QuiescenceActivity_Flies_160725).
Caution: Analyzing the images as they are collected requires the computer used for image acquisition to have MATLAB installed with a valid MATLAB license.
Choose the GUI named “MC_QuiescenceActivity_Flies_160725” and run the LarvaLodge Analysis by clicking the play button (Figure 10a).
In order to open the images within the GUI, click “process images” (Figure 10b).
Click “Load Image” (Figure 10c) and select the first image collected in the assay.
Click “Select ROI” (Figure 10d), which offers three options for selecting the regions of interest (ROI). Choose “Auto Selection (Web Method)” by typing “1” and clicking “OK (Figure 10e).”
Enter the number of rows and columns used in the LarvaLodge; in our experiments the typical number of rows is four and the number of columns is five (Figure 10f).
Click the centers of the following wells: the upper left well, the upper right well, the lower left well and the lower right well (Figure 10g). The GUI then identifies approximately where the center of each well falls on the images.
Choose a representative well and click the upper left corner and the lower right corner (Figure 10h). Based on this information, the GUI identifies the approximate region each well occupies with a square ROI for each (Figure 10i). If the ROIs are inaccurate, make modifications on the left-hand side of the GUI to the size or center of the ROIs. Click “Finish Adjustment” when the ROIs satisfactorily cover each well without spilling over into adjacent wells (Figure 10i).
Click “Set Parameters” to specify the settings for the analysis. A window will open to enter the parameters for the analysis (Figure 10j) as follows: “Start Frame” sets where the analysis begins. Set to 1 if it is the first frame. “End Frame” is the last frame analyzed. Set to the total number of images collected in the assay if desired. “Time/Frame(sec)” is 6, if images are to be captured every 6 seconds as in our published work15.”Image File Prefix” is “Image”, which enables IC Capture to name the collected images. “Threshold for ActVal Analysis” sets the number of gray-scale intensity unit a pixel must change by to be considered “changed” between temporally adjacent images. This number is typically set to 40 in our experiments, but this is highly dependent on lighting conditions and camera. See the next step for optimizing this threshold, with the goal of maximizing the signal to noise ratio. “Frame Skip” allows for subtracting images that are not temporally adjacent. For example, if desired, one can measure activity between every 2nd or 3rd frame if analyzing movement and quiescence on a longer time scale than 6 seconds. The program always calculates differences between temporally adjacent images, but frame skip allows the user to specify a second image subtraction interval, and the results are saved in a separate activity matrix (ActValS). Set “Synchronized Analysis?” to 1. This allows the immediate analysis of activity changes between frames while images are acquired. “Autosave per N frames” sets how often the results are saved during the analysis analysis (we usually save after every 100 frames). Finally, click “OK” (Figure 10j).
-
Another window will open that reads “Click ok to display thresholded image” (Figure 10k). Click “OK” and the thresholded image will appear. The thresholded image shows all pixels whose gray scale changed (i.e. greater than the set threshold) between image 1 and image 2, representing movement.
Crucial Step: All white spots should correspond to larvae in the wells (Figure 10l). If thresholding is too low, white spots appear sporadically within the field of view (Figure 10m). In this case, increase the “Threshold for ActVal Analysis” value until all pixel changes can be attributed to the larvae. By contrast, if the threshold for considering a pixel to have “moved” is set too high, little movement will appear in the subtracted image. When using an imaging system for the first time, or after adjustments in the lighting, we recommend calibrating the optimal threshold by starting with thresholding set too low, allowing excess pixels to appear (Figure 10m). Next, increase the grayscale value cutoff with small increments until the thresholded image represents moving animals only. We also leave one well empty to validate thresholding during the analysis. This well should lack any white pixels at this stage. Type “1” to indicate the parameters are sufficient and click “OK”.
Click “Start Analysis” to begin processing the images (Figure 10n). The banner above the image displays the image name in process at that moment as well as the total number of images.
When the analysis is finished, the save button in the GUI saves the pixel changes (the gray scale difference between temporally adjacent images for each pixel in the ROI) for each well in a MATLAB file.
ANALYSIS
-
41
Analyze C. elegans SIS assays as described in option A, C. elegans DTS assays as described in option B, and Drosophila larvae as described in option C.
A) Analyzing C. elegans SIS assays (10–15 minutes)
Start MATLAB and load the saved MATLAB file containing the pixel data to the command window by dragging and dropping.
Run the following command “q=fracQ (ActVal’, 1, 10, 60);”
The first number (1) is the number of pixels required to move to call the frame not quiescent. (Because this data has undergone thresholding, any changed pixel is now counted as activity). The second number (10) is the number of seconds between each frame. The third number (60) is the number of frames for calculating the moving window of quiescence; since we typically capture images every 10 seconds, 60 frames corresponds to 600 seconds or 10 minutes. For UV SIS experiments, we usually use 60-minute bins while for heat shock experiments, we typically use 10-minute bins.
To generate a heatmap of quiescence, run the command “imagesc(q)”. Save the image that appears in the folder containing the raw images from the assay for future reference. To generate a heatmap of activity, run the command “imagesc(ActVal, [0,1000])”; the numbers within brackets represent the minimum and maximum activity values to be shown in the heatmap. Save the image that opens as a JPEG in the same place as the quiescence heat map. These two heat maps help to identify when the worms’ behavior may not be tracked accurately for a variety of reasons (i.e. left the well, died, or some condensation is obstructing the image, Figure 11c,d,g,h, and 12c,d).
Run the command “q=fracQ_SIS_Normalized(ActVal’, .001, 10, 60, ‘Example filename.mat’, 0);”. The “Example file name.mat” needs to be different from the original name of the pixel data .mat file associated with the assay. The first number (0.001) is the minimum normalized activity a worm must exhibit between frames to be considered active. Activity for each worm is normalized to the median activity of unstressed controls. The second number (10) is the number of seconds between frames. The third number (60) is the number of frames for calculating the moving window of quiescence; since we typically capture images every 10 seconds, 60 frames corresponds to 600 seconds or 10 minutes. The final number (0) entered indicates whether the plate was fully filled (enter 0) or not (enter 1). The script will request information on the well locations of each condition on the plate.
The command window will then prompt for the number of conditions in the experiment. Conditions may include different genotypes (e.g. wild-type and mutant), different drug (e.g. histamine or vehicle), or different treatment (animals treated with UV or heat and untreated animals). For example, if testing wildtype and one mutant, enter “2” for number conditions. The software will prompt the user for the well numbers corresponding to each condition. It will also prompt the user to enter the well numbers corresponding to the untreated controls for each condition in the experiment. Well numbering goes left to right and starts at the top left corner of the WorMotel.
Run the following command: “t=totQ_interval_SIS_Normalized(ActVal’, 0.25, 10, 0.001, 48, 10, 60, ‘Example file name.mat’);”. The first number (0.25) is the quiescence threshold, which is the fraction of frames a worm must be quiescent within a 10-minute moving window for those frames to be included in the final quantification of total quiescence. The second number (10) is the number of seconds between the frames. The third number (0.001) is the threshold for considering as “not quiescence” the ratio of pixels moved between two frames of the treated worm divided by the median pixels moved of untreated control animals over the whole experiment. The fourth number (48) indicates the number of wells analyzed. The fifth number (10) is time in minutes relative to the start of the experiment. The sixth number (60) represents the bin size in minutes, and should be the same as the number of minutes in the bins in “q=fracQ…” The “Example file name.mat” should be same as the one used in “q=fracQ…”
vii) The data is stored in the “t” in MATLAB’s workspace. To export this data to a spreadsheet, double click on the variable “t” in the workspace. The raw data will open in a new window. Copy and paste this data to a spread sheet for further analysis and graphing (see Figure 11a–h for example data). Alternatively, enter the command csvwrite (‘quiescence.csv’,t) to export the matrix containing quiescence values into a csv file.
Figure 11. Sample data: UV and heat shock SIS with wildtype and rbr-2.
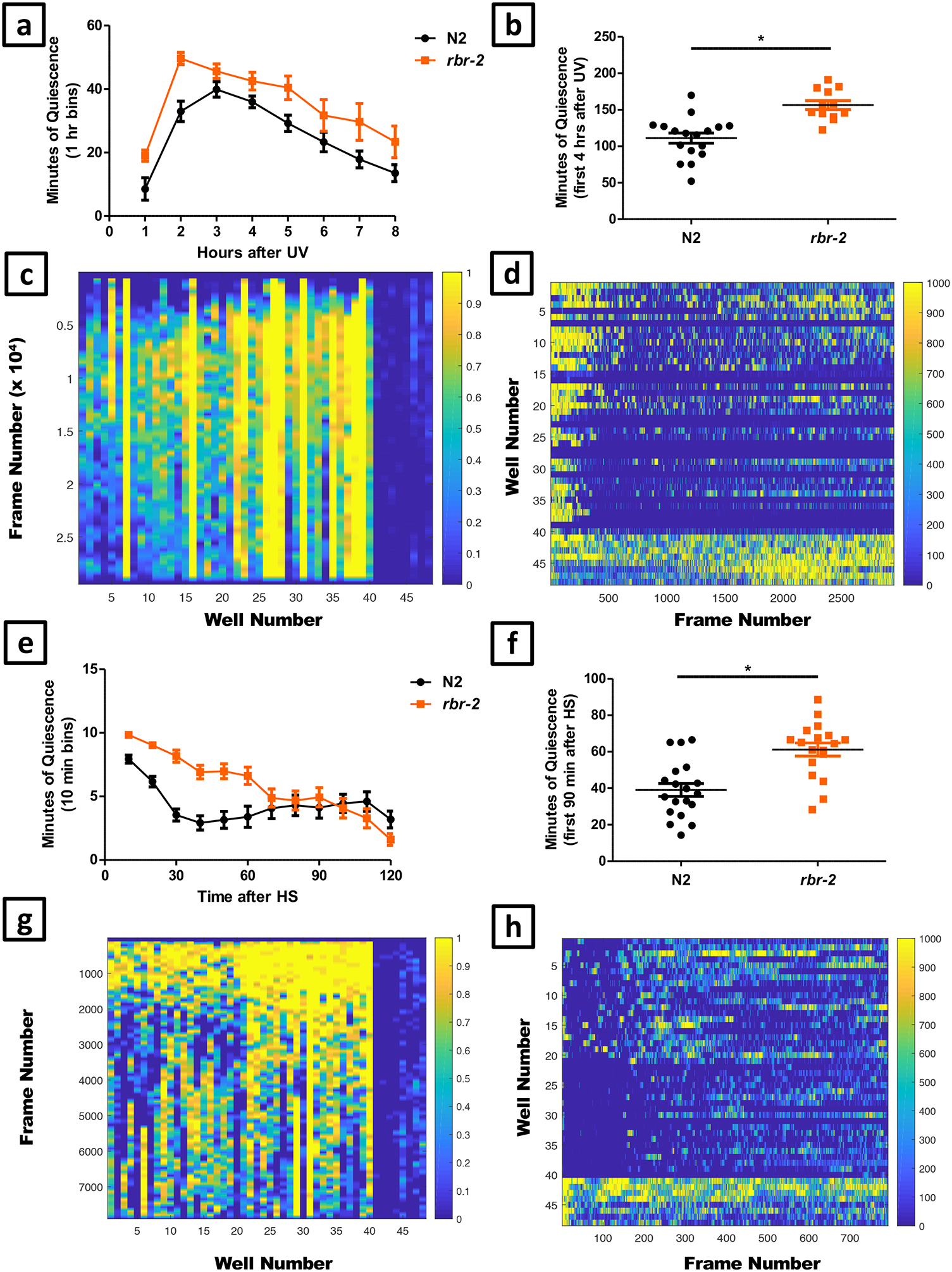
Stress induced sleep (SIS) data obtained from the WorMotel analysis. We compared the behavior of rbr-2 (tm1231) to wildtype (N2) controls. Panels a-d show data from a UV-induced SIS experiment and panels e-h are show data from a heat-shock induced SIS experiment. (a) Average number of minutes of quiescence in each hour following exposure to 1500 J/m2 UV for eight hours. Numbers of animals were N2: n = 17, rbr-2: n = 13. (b) Total minutes of quiescence for each worm in the first four hours after exposure to 1500 J/m2 UV. The rbr-2 mutants were significantly more quiescent than N2 (unpaired t-test, p <0.0001). (c) Quiescence heat map following exposure to 1500 J/m2 UV. 1 (yellow) indicates maximal quiescence while 0 (blue) indicates minimal quiescence. Each column represents the quiescence data of a single animal. Columns that are completely yellow indicate a worm that has died or has escaped from the well, which can be confirmed by reviewing individual images); data from such dead or escaped worms is removed from subsequent analysis. Columns 1–20 are data from treated N2 worms and columns 21–40 are data from treated rbr-2 worms. The right-most eight columns show data from untreated N2 (columns 41–44) and rbr-2 worms (columns 45–48). (d) Activity heat map following exposure to 1500 J/m2 UV. 1000 (yellow) indicates maximum activity, 0 (blue) indicates no activity. Each row represents the data of a single animal. Activity is binned in one hour increments. Rows 1–20 are data from treated N2 worms and rows 21–40 are treated rbr-2 worms. The bottom 8 rows show the activity of untreated N2 worms (rows 41–44) and untreated rbr-2 worms (rows 45–48). rbr-2 mutants appear less active. Data from the following wells were censored from the data presented: 7,16, 23, 26, 27, 28, 31, 35, 38, and 39. (e) Average number of minutes of quiescence in 10 minute increments over a two hour period following a 30 minute heat shock at 35°C. N2 n = 19 rbr-2 n = 18. (f) Total minutes of quiescence for each worm in the first 90 minutes following 30 minutes of heat shock at 35°C. The rbr-2 mutants exhibit more quiescence than N2 (unpaired t-test, p = 0.0001). (g) Quiescence heat map following 30 minutes of heat shock at 35°C (yellow = quiescence). Columns 1–21 include N2 quiescence data, columns 22–40 include rbr-2 quiescence data, and columns 41–48 show data of untreated N2 (41–44) and rbr-2 control animals (45–48). Data from animals that appeared quiescent during the full time course (e.g. columns 14, 29, and 31) were censored. (h) Activity heatmap following 30 minutes of heat shock at 35°C (yellow = activity). The top 20 rows are treated N2 worms and the next set of 20 rows (21–40) are treated rbr-2 worms. The bottom 8 rows show the activity of untreated N2 worms (rows 41–44) and untreated rbr-2 worms (rows 45–48). rbr-2 mutants appear less active. Error bars in this and all other figures indicate standard error of the mean (SEM).
Figure 12. Sample data: DTS with wildtype and rbr-2.
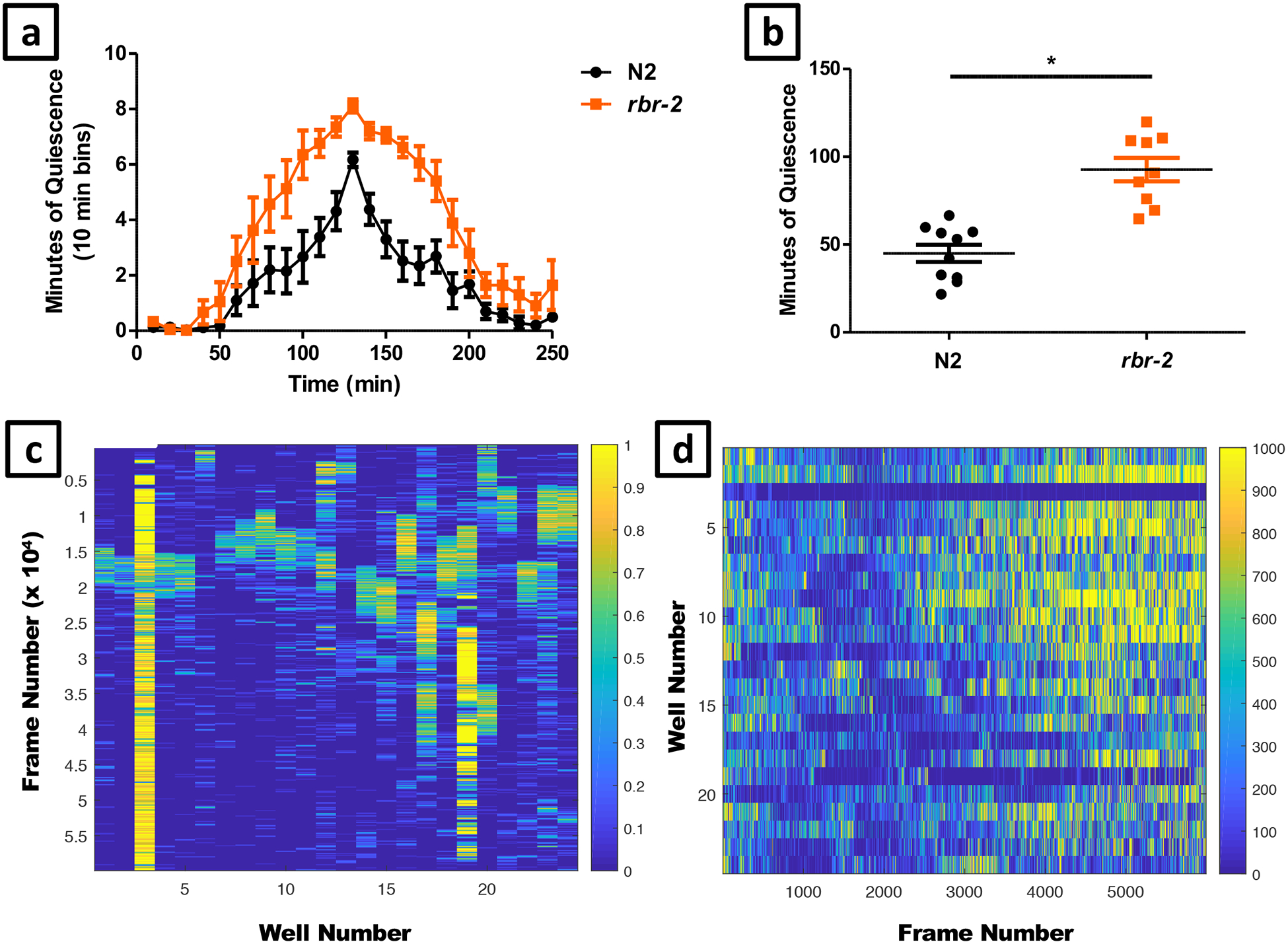
Developmentally timed sleep data obtained from the WorMotel analysis. In this experiment we compared an allele of rbr-2 (tm1231) to wildtype N2 controls. (a) Average number of minutes of quiescence in 10 minute increments during a four-hour period surrounding the peak of lethargus quiescence. N2 n = 10 rbr-2 n = 9. (b) Total number of minutes spent in lethargus for each individual worm. The rbr-2 mutants spent significantly longer in lethargus than N2 (unpaired t-test, p < 0.0001). (c) Quiescence heat map over the course of 16 hours (yellow = quiescence) where time=0 is at the top and time=16 hours is at the bottom of the figure. Columns 1–12 include N2 quiescence data, columns 13–24 include rbr-2 quiescence data. The following wells were censored from the data presented: 3, 6, 13, 19, and 21.The heat map is useful for identifying the time of peak of quiescence (e.g. frames 15000–20000 form worms analyzed in columns 1–2), as well as worms that are injured (e.g. worm analyzed in columns 3 and 19). Quiescence is binned in 10 minute increments. (d) Activity heat map of the same images over the course of 16 hours (yellow = activity), where time=0 is at the left and time=16 hours is at the right of the image. Activity is binned in 10 minute increments.
B) Analyzing C. elegans DTS assays (60 minutes)
Start MATLAB and load the saved .mat file containing the pixel data to the command window by dragging it over.
Run the following command “q=fracQ_DTS_Normalized(ActVal’, .001,10, 4×6×60, 10);”. The first number is the threshold the normalized activity must exceed to be considered active. The second number is the seconds between frame capture. The third parameter (4×6×60) is the moving window of time (in seconds) used to normalize activity; in this case it is 4 hours. i.e. With a 10-second frame capture rate, 6 frames = 1 minute × 60 minutes = 1 hour × 4 hours = 4 hours (a total of 14400 seconds). The last number (10) is the moving window size (in frames) for calculating the fraction of quiescence.
To generate the heatmap of quiescence type the following command “imagesc(q)” and save the image in the pop up as a JPEG for future reference.
Enter and run the following command “t=totQ_interval_DTS_Normalized (ActVal’, 0.25, 10, .001, 24, 10, 10);”. The first number (0.25) is the quiescence threshold as defined above; the activity is normalized so the maximum value is 1. The second number (10) is the number of seconds between frames. The third number (.001) is the normalization threshold for considering movement, which is determined by dividing number of pixels moved by the worm in the frame of interest by the median number of frames in a four-hour moving window. The fourth number (24) is the number of wells analyzed. The fifth number (10) is the start time in minutes for starting the analysis. We often discard the first few minutes of data because the worms’ behavior may have been disturbed during the set-up of acquisition. The final number (10) is the number of minutes in which the data in binned in the output “t”.
The output of step 4 gives the amount of quiescence for each well in “t” in MATLAB’s workspace. Copy and paste these data into Excel or other spreadsheet program to visually identify the peak of lethargus quiescence.
Search through the data for each worm to find the peaks of quiescence based on the amount of sleep in the moving 10-minute bin and compare it with the heatmap. The heatmap should help find the peaks. There should be two peaks, corresponding to L3 and L4 lethargus. The L4 lethargic peak should be larger since worms are more quiescent in L4 lethargus than in L3 lethargus10. Once the peak quiescence is identified, calculate total quiescence in L4 lethargus by summing two hours-worth of data on either side of the peak.
Copy and paste the summarized data as well as the two hours around the lethargic peak into a spread sheet for further analysis and graphing (see Figure 12a–d for example data).
C) Analyzing sleep in Drosophila larvae (10–15 minutes)
Open the excel file template “LarvaLodge Quiescence Analysis.xlsx”. This file processes raw MATLAB data to quantify sleep and activity in hourly bins, as well as over prespecified 6 or 12 hour intervals.
Open the saved file in MATLAB To generate a heatmap of activity, run the command “imagesc(ActVal, [0,1000])”; these numbers represent the minimum and maximum activity values to be show in the heatmap (Figure 13a). Save the image that appears in the folder containing the raw images from the assay for future reference. Next, double-click on ActVal in the workspace, which opens a new window showing raw activity data (each row represents one larva). Copy the raw data and paste into the excel file “raw data” sheet.
Each experimental group is processed on a separate excel sheet. Copy the rows corresponding to the experimental controls and paste→transpose to the corresponding columns on “control” sheet. Do the same for each experimental group, pasting into “experimental-1” and “experimental-2”. These sheets can be copied to accommodate more experimental groups as needed. See Figure 13b–e for example data.
Figure 13. Sample data: Activation of sleep-promoting neurons in fly larvae.
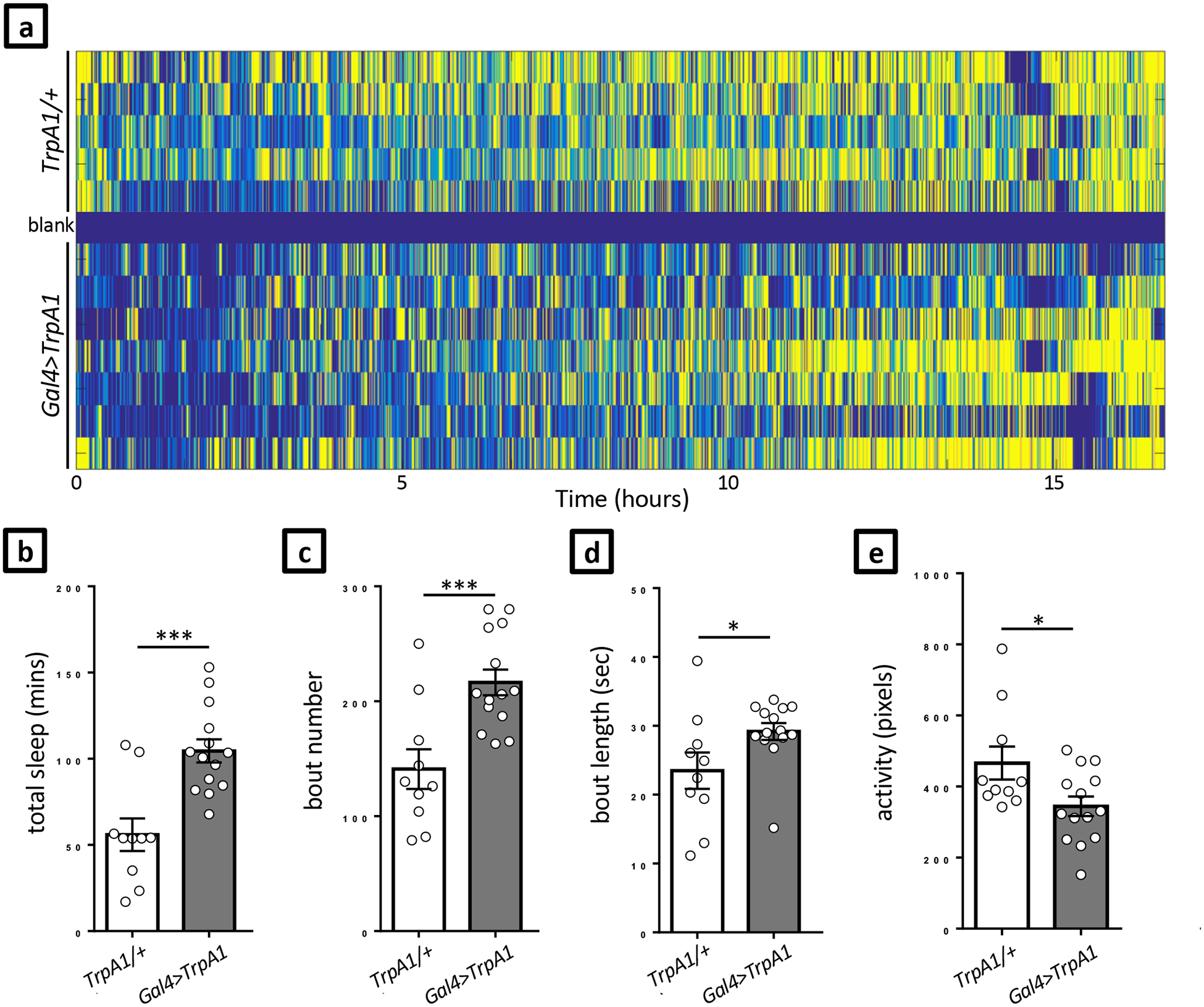
In this example we show thermogenetic activation of a specific population of neurons in second instar Drosophila larvae. We used a restricted GAL4 driver to express the heat-sensitive cation channel TrpA1. (a) Activity heat map showing the activity of control (TrpA1/+) and experimental (Gal4>TrpA1) larvae over >15 hours at 29°C. Yellow indicates maximum activity, blue indicates no activity. Each row represents one larva. One well (blank) was intentionally left empty for thresholding control. Summary of total sleep (b), number of sleep bouts (c), the average length of sleep bouts (d), and the average activity (e) between hours 1–7 of two independent experiments. Unpaired t test, *p<0.05; ***p<0.001; error bars = SEM.
Troubleshooting:
Troubleshooting advice can be found in Table 1
Table 1.
Troubleshooting guidance
| Step | Problem | Possible reason | Possible solution |
|---|---|---|---|
| Preparation/running experiments |
Worms leaving the wells |
|
Fill moats around the wells with copper sulfate |
| Constructing the Blue Light Stimulus for Sleep Perturbation, 12 |
The power supply is turned on but the LEDs do not turn on |
|
|
| Setting Imaging System Parameters, 38 | The image is too dark |
|
|
| Setting Imaging System Parameters, 38 | A white ring is visible at the perimeter of the wells | Scattering from the illumination LEDs | Ensure the illumination LEDs are not pointing upward but instead are directed horizontally, parallel to the sample plane. The angle between the illumination LED direction and the sample should be minimized to reduce scattering of illumination LEDs onto the camera sensor. |
Timing:
Steps 1–24, Construction of experimental system: ~240–480 minutes (plus overnight curing steps)
Steps 25–33, Device design and fabrication: ~60–120 min (plus time to print and overnight curing steps)
Step 34, Experiment preparation:
Option A, C. elegans: ~95–145 min
Option B, D. melanogaster: ~55–95 min
Step 35, Induction of worm sleep:
Option A, Heat shock: ~40 min
Option B, UV irradiation: ~6 min
Steps 36–38, Setting imaging system parameters: ~15–60 min
Step 39, Data/Image Acquisition:
Option A, C. elegans: ~5–15 min (plus run time that depends on the desired length of the experiment)
Option B, D. melanogaster: ~5–15 min (plus run time that depends on the desired length of the experiment)
Option C, Population arousal and deprivation experiments: run time depends on the desired length of the experiment
Option D, Closed-loop sleep deprivation: run time depends on the desired length of the experiment
Option E, Control experiment for the closed-loop deprivation: run time depends on the desired length of the experiment
Step 40, Image processing:
Option A, C. elegans: ~30–45 min
Option B, D. melanogaster: ~30–45 min
Step 41, Data Analysis:
Option A, Analyzing C. elegans SIS assays: ~10–15 min
Option B, Analyzing C. elegans DTS assays: ~60 min
Option C, Analyzing sleep in D. melanogaster larvae: ~10–15 min
Anticipated Results:
Data obtained from the methods described here can be compared to sample data sets provided (Figures 11, 12, and 13). In DTS experiments, quiescence heatmaps of worms in the L3 to adult period should show two peaks of quiescence, corresponding to the L3 and L4 lethargus periods. DTS experiments started in the L4 stage should show 1 peak of quiescence, corresponding to the L4 lethargus period (Figure 12). Wells lacking an animal and wells containing an injured or dead animal should show quiescence at all times (e.g. Lane 3 in Figure 12C). A well that has two worms will typically show a complete lack of quiescence at a time when it is expected (this will occasionally occur due to a worm leaving its well and joining another well that is already occupied). Clicking through the images will confirm the presence of two worms in the same well. For a positive control for the ability to detect strongly reduced quiescence in DTS experiments, use the aptf-1 mutant61. For a positive control to detect increased quiescence in DTS experiments, use the rbr-2 mutant (Figure 12).
In SIS experiments, heat-shocked wild-type animals should show quiescence for about one hour starting at time zero (i.e. immediately after heat shock); UV treated animals should show quiescence for about 6 hours starting at 30–60 minutes after UV exposure, with the peak between 2 to 3 hours after UV exposure. Untreated controls should show no quiescence. A positive control for the ability to detect strongly reduced quiescence in SIS experiments is to use the aptf-1 mutant61 or the ceh-17 mutant11. For a positive control to detect increased quiescence in SIS experiments, use the rbr-2 mutant (Figure 11).
For 2nd instar fly larvae, visual inspection of heat maps should reveal no activity in the blank well, stochastic changes from activity to inactivity in control larva, and evidence of sustained inactivity during the molt to 3rd instar at ~20 hours after loading if experiment is at 25°C (data in Figure 13 is at 29°C, and molt occurs after 15 hours). Unhealthy larva show deviations from these patterns, and are most easily detected based on inspection at conclusion of the experiment (dead or abnormally small/undeveloped) or abnormal timing of the molt (based on heat map data and inspection of collected images). Larva that have burrowed or escaped should be excluded as well. Quantitative sleep and activity measures (Figure 13b–e) are highly dependent on background strain and thresholding settings. Activation of octopaminergic neurons is strongly wake-promoting, so a straightforward approach to verifying the system is accurately detecting changes to sleep is to compare Tdc2-GAL4>UAS-NaChBac flies to genetic controls15.
Supplementary Material
Acknowledgements:
We thank Patrick McClanahan for assistance with the method of fabricating flat WorMotel bases and for recording the video of pharyngeal pumping in the WorMotel. This work was supported by NIH grants K08NS090461 (M.S.K.), R01NS088432 (D.M.R. and C.F-Y.), and R01NS084835 (C. F-Y.), Ellison Medical Foundation (C.F-Y.), European Commission Horizon 2020 (C. F.-Y.), and a Burroughs Wellcome Career Award for Medical Scientists (M.S.K.), March of Dimes Basil O’Connor Scholar Award (M.S.K.), and Sloan Research Fellowship (M.S.K.). Kristen Davis is a trainee on the NIH T32 ES019851 (PI: Trevor Penning, Penn CEET).
Footnotes
EDITORIAL SUMMARY Individual animals are cultivated in custom microfabricated multi-well substrates. This protocol describes how to design, produce, and image the plates, and analyze the resulting behavioral data.
Competing interests:
The authors declare that they have no competing financial and non-financial interests.
Data Availability:
All code is included in Supplementary Data. The sample data presented in this protocol are available from the corresponding author upon reasonable request.
Key references:
Longitudinal imaging of Caenorhabditis elegans in a microfabricated device reveals variation in behavioral decline during aging.
Churgin MA, Jung SK, Yu CC, Chen X, Raizen DM, Fang-Yen C. eLife. 2017 May 31;6. e26652. doi: 10.7554/eLife.26652.
A sleep state in Drosophila larvae required for neural stem cell proliferation.
Szuperak M, Churgin MA, Borja AJ, Raizen DM, Fang-Yen C, Kayser MS. eLife. 2018 Feb 9;7. e33220. doi: 10.7554/eLife.33220.
References
- 1.Joiner WJ Unraveling the Evolutionary Determinants of Sleep. Curr. Biol 26, R1073–R1087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keene AC & Duboue ER The origins and evolution of sleep. J. Exp. Biol 221, jeb159533 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kayser MS & Biron D Sleep and Development in Genetically Tractable Model Organisms. Genetics 203, 21–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber F & Dan Y Circuit-based interrogation of sleep control. Nature 538, 51–59 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Artiushin G & Sehgal A The Drosophila circuitry of sleep–wake regulation. Curr. Opin. Neurobiol 44, 243–250 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubowy C & Sehgal A Circadian Rhythms and Sleep in Drosophila melanogaster. Genetics 205, 1373–1397 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trojanowski NF & Raizen DM Call it Worm Sleep. Trends Neurosci 39, 54–62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendricks JC, et al. Rest in Drosophila is a sleep-like state. Neuron 25, 129–38 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Shaw PJ, Cirelli C, Greenspan RJ & Tononi G Correlates of sleep and waking in Drosophila melanogaster. Science 287, 1834–7 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Raizen DM, et al. Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451, 569–72 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Hill AJ, Mansfield R, Lopez JMNG, Raizen DM & Van Buskirk C Cellular stress induces a protective sleep-like state in C. elegans. Curr. Biol 24, 2399–405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skora S, Mende F & Zimmer M Energy Scarcity Promotes a Brain-wide Sleep State Modulated by Insulin Signaling in C. elegans. Cell Rep 22, 953–966 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y, Masurat F, Preis J & Bringmann H Sleep Counteracts Aging Phenotypes to Survive Starvation-Induced Developmental Arrest in C. elegans. Curr. Biol 28, 3610–3624.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kayser MS, Yue Z & Sehgal A A critical period of sleep for development of courtship circuitry and behavior in Drosophila. Science 344, 269–74 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szuperak M, et al. A sleep state in Drosophila larvae required for neural stem cell proliferation. Elife 7, e33220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nichols ALA, Eichler T, Latham R & Zimmer M A global brain state underlies C. elegans sleep behavior. Science 356, 1247–1256 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Eichler K, et al. The complete connectome of a learning and memory centre in an insect brain. Nature 548, 175–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bringmann H Sleep-Active Neurons: Conserved Motors of Sleep. Genetics 208, 1279–1289 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koh K, et al. Identification of SLEEPLESS, a sleep-promoting factor. Science 321, 372–6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cirelli C, et al. Reduced sleep in Drosophila Shaker mutants. Nature 434, 1087–1092 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Shi M, Yue Z, Kuryatov A, Lindstrom JM & Sehgal A Identification of Redeye, a new sleep-regulating protein whose expression is modulated by sleep amount. Elife 3, e01473 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stavropoulos N & Young MW insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron 72, 964–76 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogulja D & Young MW Control of Sleep by Cyclin A and Its Regulator. Science 335, 1617–1621 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Churgin MA, et al. Longitudinal imaging of Caenorhabditis elegans in a microfabricated device reveals variation in behavioral decline during aging. Elife 6, e26652 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bais S, Churgin MA, Fang-Yen C & Greenberg RM Evidence for Novel Pharmacological Sensitivities of Transient Receptor Potential (TRP) Channels in Schistosoma mansoni. PLoS Negl. Trop. Dis 9, e0004295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senatore A, Reese TS & Smith CL Neuropeptidergic integration of behavior in Trichoplax adhaerens, an animal without synapses. J. Exp. Biol 220, 3381–3390 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith CL, Pivovarova N & Reese TS Coordinated Feeding Behavior in Trichoplax, an Animal without Synapses. PLoS One 10, e0136098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ueda T, Koya S & Maruyama YK Dynamic patterns in the locomotion and feeding behaviors by the placozoan Trichoplax adhaerence. Biosystems. 54, 65–70 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Nagy S, Raizen DM & Biron D Measurements of behavioral quiescence in Caenorhabditis elegans. Methods 68, 500–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi S, Chatzigeorgiou M, Taylor KP, Schafer WR & Kaplan JM Analysis of NPR-1 reveals a circuit mechanism for behavioral quiescence in C. elegans. Neuron 78, 869–80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swierczek NA, Giles AC, Rankin CH & Kerr RA High-throughput behavioral analysis in C. elegans. Nat. Methods 8, 592–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogelstein JT, et al. Discovery of Brainwide Neural-Behavioral Maps via Multiscale Unsupervised Structure Learning. Science 344, 386–392 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Huang H, Singh K & Hart AC Measuring Caenorhabditis elegans Sleep during the Transition to Adulthood Using a Microfluidics-based System. Bio Protocol 7, e2174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bringmann H Agarose hydrogel microcompartments for imaging sleep- and wake-like behavior and nervous system development in Caenorhabditis elegans larvae. J. Neurosci. Methods 201, 78–88 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Turek M, Besseling J & Bringmann H Agarose Microchambers for Long-term Calcium Imaging of Caenorhabditis elegans J. Vis. Exp e52742 (2015). doi: 10.3791/52742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pittman WE, Sinha DB, Zhang WB, Kinser HE & Pincus Z A simple culture system for long-term imaging of individual C. elegans. Lab Chip 17, 3909–3920 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belfer SJ, et al. Caenorhabditis-in-drop array for monitoring C. elegans quiescent behavior. Sleep 36, 689–698G (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomasiunaite U, Widmann A & Thum AS Maggot Instructor: Semi-Automated Analysis of Learning and Memory in Drosophila Larvae. Front. Psychol 9, 1010 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark MQ, McCumsey SJ, Lopez-Darwin S, Heckscher ES & Doe CQ Functional Genetic Screen to Identify Interneurons Governing Behaviorally Distinct Aspects of Drosophila Larval Motor Programs. G3 (Bethesda). 6, 2023–31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Churgin MA & Fang-Yen C An Imaging System for C. elegans Behavior. Methods Mol. Biol 1327, 199–207 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Churgin MA, McCloskey RJ, Peters E & Fang-Yen C Antagonistic Serotonergic and Octopaminergic Neural Circuits Mediate Food-Dependent Locomotory Behavior in Caenorhabditis elegans. J. Neurosci 37, 7811–7823 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCloskey RJ, Fouad AD, Churgin MA & Fang-Yen C Food responsiveness regulates episodic behavioral states in Caenorhabditis elegans. J. Neurophysiol 117, 1911–1934 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagy S, Goessling M, Amit Y & Biron D A Generative Statistical Algorithm for Automatic Detection of Complex Postures. PLoS Comput. Biol 11, e1004517 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Husson SJ, Costa WS, Schmitt C & Gottschalk A Keeping track of worm trackers. WormBook 1–17 (2013). doi: 10.1895/wormbook.1.156.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson MD, et al. FMRFamide-like FLP-13 neuropeptides promote quiescence following heat stress in Caenorhabditis elegans. Curr. Biol 24, 2406–10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.You YJ, Kim J, Raizen DM & Avery L Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metab 7, 249–257 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scholz M, Lynch DJ, Lee KS, Levine E & Biron D A scalable method for automatically measuring pharyngeal pumping in C. elegans. J. Neurosci. Methods 274, 172–178 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Ryder E et al. The DrosDel deletion collection: a Drosophila genomewide chromosomal deficiency resource. Genetics 177, 615–29 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamada FN et al. An internal thermal sensor controlling temperature preference in Drosophila. Nature 454, 217–20 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jennett A, et al. A GAL4-Driver Line Resource for Drosophila Neurobiology. Cell Rep 2, 991–1001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stiernagle T Maintenance of C. elegans. WormBook 1–11 (2006). doi: 10.1895/wormbook.1.101.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dou Y-H, Bao N, Xu J-J & Chen H-Y A dynamically modified microfluidic poly(dimethylsiloxane) chip with electrochemical detection for biological analysis. Electrophoresis 23, 3558–66 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Alcantar NA, Aydil ES & Israelachvili JN Polyethylene glycol-coated biocompatible surfaces. J. Biomed. Mater. Res 51, 343–51 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Ginn BT & Steinbock O Polymer Surface Modification Using Microwave-Oven-Generated Plasma. Langmuir 19, 8117–8118 (2003). [Google Scholar]
- 55.Eddington DT, Puccinelli JP & Beebe DJ Thermal aging and reduced hydrophobic recovery of polydimethylsiloxane. Sensors Actuators B Chem 114, 170–172 (2006). [Google Scholar]
- 56.Xiao D, Zhang H & Wirth M Chemical Modification of the Surface of Poly(dimethylsiloxane) by Atom-Transfer Radical Polymerization of Acrylamide. Langmuir 18, 9971–9976 (2002). [Google Scholar]
- 57.Tan SH, Nguyen N-T, Chua YC & Kang TG Oxygen plasma treatment for reducing hydrophobicity of a sealed polydimethylsiloxane microchannel. Biomicrofluidics 4, 32204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeBardeleben HK, Lopes LE, Nessel MP & Raizen DM Stress-Induced Sleep After Exposure to Ultraviolet Light Is Promoted by p53 in Caenorhabditis elegans. Genetics 207, 571–582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh K, Ju JY, Walsh MB, DiIorio MA & Hart AC Deep conservation of genes required for both Drosphila melanogaster and Caenorhabditis elegans sleep includes a role for dopaminergic signaling. Sleep 37, 1439–51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park Y, Filippov V, Gill SS & Adams ME Deletion of the ecdysis-triggering hormone gene leads to lethal ecdysis deficiency. Development 129, 493–503 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Turek M, Lewandrowski I & Bringmann H An AP2 Transcription Factor Is Required for a Sleep-Active Neuron to Induce Sleep-like Quiescence in C. elegans. Curr. Biol 23, 2215–2223 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


