Abstract
Background
Joubert syndrome is a recessive neurodevelopmental disorder characterized by clinical and genetic heterogeneity. Clinical hallmarks include hypotonia, ataxia, facial dysmorphism, abnormal eye movement, irregular breathing pattern cognitive impairment and, the molar tooth sign is the pathognomonic midbrain-hindbrain malformation on magnetic resonance imaging. The disorder is predominantly caused by biallelic mutations in more than 30 genes encoding proteins with a pivotal role in morphology and function of the primary cilium. Oligogenic inheritance or occurrence of genetic modifiers has been suggested to contribute to the variability of the clinical phenotype. We report on a family with peculiar clinical spectrum Joubert syndrome molecularly and clinically dissecting a complex phenotype, in which hypogonadism, pituitary malformation and growth hormone deficiency occur as major features.
Case presentation
A 7 year-old male was enrolled in a dedicated “Undiagnosed Patients Program” for a peculiar form of Joubert syndrome complicated by iris and retinochoroidal coloboma, hypogonadism pituitary malformation, and growth hormone deficiency. The molecular basis of the complex phenotype was investigated by whole exome sequencing. The concomitant occurrence of homozygosity for mutations in KIF7 and KIAA0556 was identified, and the assessment of major clinical features associated with mutations in these two genes provided evidence that these two independent events represent the cause underlying the complexity of the present clinical phenotype.
Conclusion
Beside the clinical variability of Joubert syndrome, co-occurrence of mutations in ciliopathy-associated genes may contribute to increase the clinical complexity of the trait.
Keywords: Joubert syndrome, Growth hormone deficiency, Pituitary gland malformation, Oligogenic inheritance
Background
Joubert syndrome (JBTS, MIM PS213300) constitutes a clinically heterogeneous group of developmental delay/multiple congenital anomalies disorders [1]. Major clinical features include hypotonia, ataxia, developmental delay (DD), intellectual disability (ID), oculomotor apraxia, recurrent hyperventilation, and the recognizable molar tooth sign (MTS), which is the obligatory radiological hallmark of the condition. Additional findings include retinal dystrophy, nephronophthisis, hepatic fibrosis and polydactyly, with both inter- and intra-familial variability [2]. JBTS belongs to the ciliopathy spectrum that recognizes defects in genes codifying proteins of the primary cilium and/or basal body and centrosome, where they play a role in formation, structure and function of these organelles. Defective primary cilium is known to impair the cellular chemo- and mechano-sensation as well as signaling, with WNT, SHH and PDGF representing the major involved signaling pathways [3]. These disorders are generally single gene traits, predominantly following recessive inheritance involving more of 30 genes [4]. Biallelic mutations in KIF7 (MIM# 611254) have unfrequently been reported in a clinically variable group of ciliopathies (hydrolethalus syndromes, HLS, MIM# 614120; acrocallosal syndrome, ACLS, and Joubert-12, JBTS12, MIM# 200990; Al-Gazali-Bakalinova syndrome, MIM# 607131). Homozygosity for truncating mutations in KIAA0556 (MIM# 616650), another cilium-associated gene, has recently been identified in a few families with JBTS-related phenotype (Joubert syndrome 26, JBTS26, MIM#616784).
The classification from digenic diseases database (DIDA) distinguishes the ‘true digenic’ event (i.e., variants in two loci that are required for expression of the disease, with none of the variants alone displaying a phenotype), from the “genetic modifier” (GM) effect (i.e., two variants segregating independently and resulting in a combination of two distinctive phenotypes) [5]. Oligogenic inheritance or occurrence of genetic modifiers involving variation in a subset of ciliopathy-related genes (e.g. AHI1, CEP41, CC2D2A, TMEM67, KIAA0556 and KIF7) has been suggested to contribute to phenotype worsening [6–8]. The extensive use of second generation sequencing techniques makes today possible to assess of the relevance of co-occurrence of variants in ciliopathy-related genes underlying JBTS.
Here, we report the concomitant occurrence of biallelic mutations in KIF7 and KIAA0556, two genes encoding proteins individually implicated in ciliopathies, in a subject with a distinctive JBTS phenotype complicated by ocular coloboma, hypogonadism, pituitary dysgenesis, and growth hormone deficiency (GHD).
Case presentation
Clinical features
The patient (OPBG_13–16) was the first child of healthy parents without family history of relevant genetic disorders, who declined consanguinity but stemmed from the same small village in Italy. The mother had a previous spontaneous abortion in first trimester of pregnancy. The delivery occurred spontaneously at 40 + 3 weeks of gestation after an unremarkable pregnancy except for a prenatal ultrasound of cleft lip and palate. Birth weight was 3550 g (0.22 SDS), birth length 51 cm (0.2 SDS), head circumference (OFC) 34 cm (− 0.66 SDS) according to Bertino’s charts [9]. Apgar scores were 5–9 at 1–5 min. At birth, physical evaluation disclosed cleft lip and palate, left microphthalmia, micropenis, cryptorchidism, hand and feet brachydactyly, and positional talipes. He also showed transient hypoglycemia and jaundice. Head ultrasound and magnetic resonance imaging (MRI) revealed dysgenesis of the corpus callosum, ectopic posterior pituitary, hypoplasia of cerebellum, and thin left optic nerve. In the first month, neurological examination revealed truncal hypotonia, and ID/DD with difficulties in mastication and deglutition. Ophthalmological inspection disclosed left microphthalmia and coloboma and right oculomotor nerve palsy; visually evoked potential was absent in the left eye. The auditory brainstem response was bilaterally normal, and heart, abdominal and renal ultrasounds were unremarkable. Blood basal hormone levels performed at 9 days of life resulted in the normal ranges (TSH, fT3, fT4, testosterone, LH, ACTH, prolactin). Serologic tests for CMV and toxoplasma were negative. Evaluation at 2 months showed the weight 4820 g (− 0.2 SDS), length 54 cm (− 0.22 SDS), head circumference 39 cm (− 0.15 SDS). Endocrine assessment documented low IGF1 and IGF-BP3 levels (respectively < 25 ng/ml and < 0.5 mcg/ml). During hypoglycemia (47 mg/dl), growth hormone (GH) was 0.37 ng/ml with a normal cortisol rise (20.28 mcg/dl), while ACTH, TSH, fT4 and prolactin resulted in the normal ranges. Treatment with human GH (hGH) was started (0.035 mg/kg/day). At 6 months, weight and length measurements were 6400 g (− 2.5 SD) and 64 cm (− 2.7 DS), respectively, whereas head circumference fitted adequacy the 50th centile (43 cm). ACTH stimulation test (250 mcg) showed normal adrenal function, while LHRH stimulation test induced a blunted gonadotropin response for age (minipuberty period) (basal LH 0.8, peak LH 2.51 mU/ml; basal FSH 1.6, peak FSH 2.09 mU/ml). Topical application of 2.5% dihydrotestosterone gel (0.2 mg daily for 4 weeks) was prescribed with good response (more the 3.5 cm in length). Endocrine re-evaluation at 2 years, documented normal levels of fT4, TSH, ACTH, and cortisol; IGF1 levels were 92 ng/ml (under GH treatment). At 3 years of age, GH retesting was performed after 1.5 months of washout, indicating GHD; GH peak after clonidine stimulation was 0.17 ng/ml (n.v. > 8) while GH peak after GHRH plus arginine was 1.08 ng/ml (n.v. > 20). IGF1 and IGF-BP3 were low (respectively < 25 ng/ml and < 0.5 mcg/ml), while basal blood levels of fT4, TSH, cortisol, ACTH resulted normal. At 6.5 years, length was 116 cm (− 0.7 SDS), weight was 20.6 Kg (− 0.79 SDS), and the height velocity was 2.4 cm/year (− 3.87 SDS) with 0.030 mg/kg/day GH treatment. For this poor response, GH was discontinued.
At last evaluation (7 years), his height was 116.2 cm (− 1.41 SDS), weight 24.1 Kg (− 0.24 SDS), and his height velocity 0.3 cm/year (− 6.4 SDS) (all SDS were calculated on the Italian Growth Charts) [10]. The stagnation of growth during discontinuation of therapy confirmed GHD. The therapy was hence restarted (0.031 mg/kg/day), and low height velocity was observed. Of note, height velocity could not be precisely calculated due to his spine deformity. Craniofacial dysmorphism (sparse hair, hypertelorism, thick eyebrow, bilateral ptosis, short columella, low-set ears, and signs of cleft lip and palate correction by surgery), flat-overpronated feet with bilateral calcaneovalgus deformity, and severe global ID/DD were confirmed (Fig. 1, panels b and c). He also presented microphthalmia, strabismus, right low vision and left blindness, dysphagia with oral motor dysfunction, and was unable to walk independently. Small retractile gonads and micropenis were also documented. Repeated cerebral MRI confirmed ectopic posterior pituitary and dysgenesis of the corpus callosum, and a small anterior pituitary (maximum height 2 mm), with a thin and partially recognizable stalk. Lateral ventricles were enlarged, particularly in their posterior portions. Both optic nerves and chiasm, and supratentorial deep white matter appeared thinned. Cerebellar vermis hypoplasia was also noticed with high tentorial insertion and large posterior cranial fossa, resulting the classical neuroimaging feature of JBTS, the MTS (Fig. 1, panels d and e). Metabolic analyses for glycosaminoglycan storage and amino acid metabolism disorders ruled out inborn errors of metabolism. Chromosome and high-resolution CGH-array (60 K) analyses excluded the presence of clinically relevant CNVs. Based on its complex phenotype, the proband was enrolled in the Undiagnosed Patients Program at the Bambino Gesù Children’s Hospital for whole exome sequencing (WES) analysis. Collection, use and storage of clinical data, pictures, biological material of the patient and his parents were attained after written informed consent was secured. Clinical and instrumental data of the present subject were consistent with JBTS spectrum; however, the complex phenotype and the occurrence of rare features, including peculiar craniofacial dysmorphism, iris and retinochoroidal coloboma, pituitary malformation, hypogonadism, and GHD, indicated the possibility that the condition might differ from the classical forms of Joubert syndrome (Fig. 1, panels b-e).
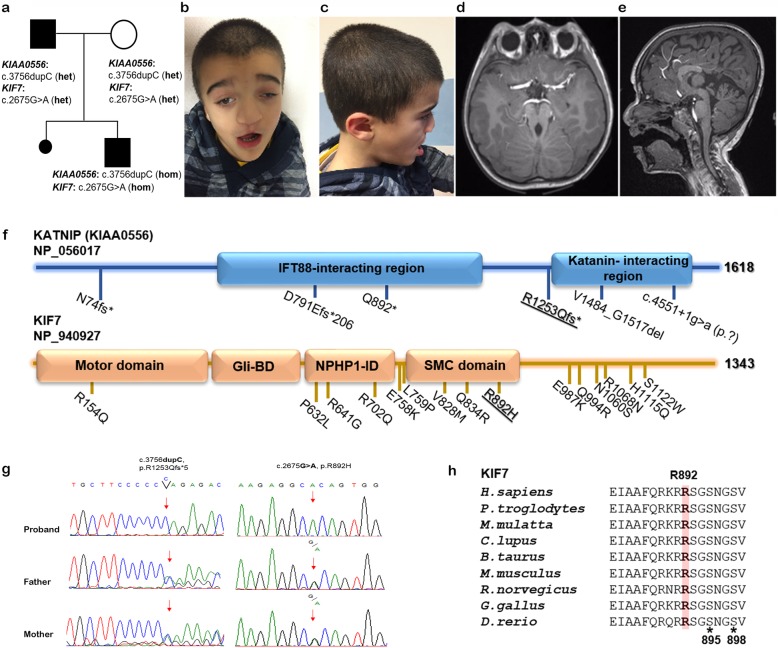
Fig. 1.
a Family tree of the subject with co-occurring mutations in KIAA0556 and KIF7; b, c Proband’s photos at 7 years; d, e MRI shows hypoplasia of cerebellar vermis, molar tooth sign (assial, view), and dysgenesis of the corpus callosum (sagittal view); f Domain structure of human KIAA0556 (KATNIP) (1618 amino acid residues) and KIF7 (1343 amino acid residues) proteins. The location of the truncating disease-causing variant in KATNIP, and missense change in KIF7 identified is displayed (bold and underlined). Functional domains of KATNIP and KIF7 are also shown. (Abbreviations: Gli-BD, Gli-binding domain; NPHP1-ID, Nephoronophthisis-1-interacting domain; SMC, Structural Maintenance of Chromosomes; g Sanger sequencing confirmation of mutations (KIAA0556, left panel; KIF7, right panel) in proband and his parents. Red asterisk (*) indicates the nucleotide changes; h Partial amino acid sequence alignment of the KIF7 SMC domain (NP_940927.2; XP_003314912.2; XP_001094468.1; XP_545852.3; XP_002696621.1 NP_034756.2; 218,828.5; XP_004943897.1; NP_001014816.1). Arginine 892 is highly conserved among vertebrates. Black asterisk (*) indicates phosphorylatable sites, Ser895 and Ser898
Molecular analyses
WES was performed using genomic DNA extracted from leukocytes of the affected subject and his parents, using the Nextera Rapid Capture Exome kit (Illumina) for library preparation, and a NextSeq500 platform (Illumina) for sequencing. WES data analysis was performed using an in-house implemented pipeline [11–13]. WES details and statistics are described in Supplementary Data 1 and Table S1 (Additional files). Validation of variants and their segregation analyses were assessed by Sanger sequencing.
WES data filtering and prioritization allowed to identify a homozygous missense variant in exon 13 of KIF7 (c.2675G>A, p.(Arg892His), NM_198525.2) and a homozygous frameshift in exon 19 of KIAA0556 (c.3756dupC, p.(Arg1253Glnfs*5), NM_015202.2) as the two co-segregating molecular events likely underlying the patient’s condition (Table 1). Heterozygosity in the healthy parents for both variants were confirmed (Fig. 1, panel g). The former variant had previously been reported in public databases (rs143866575; Maximum Allele Frequency, 0.0002213) with uncertain significance based on ACMG classification, but annotated as damaging according to the different in silico predictors (DANN, GERP, MutationAssessor, CADD and MetaDome) (Supplementary Data 1). The latter had not been reported in public and in-house databases. Its functional effect was also predicted to be damaging (CADD score 35), causing premature termination of the protein after five codons.
Table 1.
Molecular and clinical characteristics of the subject with the occurrence of mutations in KIF7 and KIAA0556
| Subject | OPBG_13–16 | |
| Ethnicity | Caucasian | |
| Sex | M | |
| Age (at last evaluation) | 7 years | |
| Auxological parameter (at birth) | ||
| Normal growth | Normal | |
| Length (cm) | 51 | |
| Weight (gr) | 3550 | |
| OFC (cm) | 34 | |
| Auxological parameter (at last evaluation) | ||
| Growth delay | Reduced height velocity 0.3 cm/year | |
| Height (cm) | 116.2 | |
| Weight (gr) | 24,100 | |
| OFC (cm) | 52.5 | |
| Genetic investigation | ||
| Karyotype | 46,XY | |
| CGH array | Normal | |
| Whole Exome Sequencing analysis | See Supplementary data | |
| Gene/es | KIF7 | KIAA0556 |
| Variant in proband (state) | c.2675G>A p.(Arg892His) (homozygous) | c.3756_3757insC, p.(Arg1253Glnfs*5) (homozygous) |
| Genome region (hg38) | Chr15:89633184C>T | Chr16:27772854dupC |
| Reference coding isoform | NM_198525.2 | NM_015202.2 |
| Reference protein isoform | NP_940927 | NP_056017 |
| rsID (dbSNP) | rs143866575 | none |
| Max AF (gnomAD) | T = 0.00022 | none |
| CADD score (PHRED) | 29.4 | 35 |
| MIM#ID | 209900 | 616784 |
| Condition(s) | Joubert syndrome 12, JBTS12 / Acrocallosal syndrome, ACLS | Joubert syndrome 26, JBTS26 |
| Protein | Kinesin-like protein, KIF7 | Katanin-interacting protein, KATNIP |
| Basic function | Cilium-associated protein | Ciliary-base protein |
| Murine model | Kif7 knockout mouse model recapitulates major ACLS features | Kiaa0556 knockout mouse model possess a Joubert syndrome-associated brain-restricted phenotype |
| (References) | [14] | [15] |
Discussion and conclusion
Joubert syndrome is caused by a large repertoire of biallelic variants affecting genes encoding proteins of the primary cilium. Based on the wide inter- and intra-familial phenotypic variability observed in this disorder, the existence of an oligogenic contribution and/or the occurrence of a genetic modifier (GM) effects have been hypothesed [4–8], which has been subsequently confirmed by a recent clinical report [16]. Here, we identify homozygous variants in KIF7 and KIAA0556 as the molecular events contributing to a variegated JBTS phenotype associated with clinically relevant endocrine abnormalities.
Systematic revision of the clinical features in previously reported subjects with mutations of KIF7 and KIAA0556 confirmed that the complexity of the present phenotype is explained by a model of digenic contribution (Table 2). The co-occurrence of variants in these genes has not been reported thus far. Mutations in KIF7 have been associated with a broad spectrum of ciliopathies with variable features ranging from lethal hydrocephalus, with or without polydactyly (HLS), to craniofacial dysmorphism, ID, and brain abnormalities with or without MTS (ACLS or JBTS12) [14, 17]. Of note, KIF7 encodes a member of a family of 14 kinesin motor proteins known to localize from the base to the tip of the cilium governing its structure and length, its architecture and transport [3]. KIF7 protein regulates Gli-Sufu activity thus acting as regulator of hedgehog signaling [18]. The phenotypic variability has been suggested to be the results of a differential impact of individual KIF7 mutations on protein function [14]. While biallelic truncating mutations causing loss of function of KIF7 have been associated to a more severe phenotype (macrocephaly, ID, facial dysmorphism, polydactyly of the hands/feet, and MTS) [14], biallelic missense variants in KIF7 have been identified in a few families with variable JBTS spectrum, from classical ACLS phenotype to a clinical presentation (lacking of MTS and having pachygyria), with an overall phenotype different from what was generally associated with inactivating KIF7 mutations [17, 20]. Notably, endocrine abnormalities including those observed in the present case have never been reported in KIF7-associated phenotypes (Table 2).
Table 2.
Clinical features of the present subject compared with the clinical features individually reported for mutations in KIF7 and KIAA0556
| Clinical features | HPO | KIF7 | KIAA0556 |
|---|---|---|---|
| Growth delay | HP:0001510 | ■ | ■ |
| Craniofacial | |||
| Head | |||
| Macrocephaly | HP:0000256 | ■ | ■ |
| Wide anterior fontanel | HP:0000260 | □ | NR |
| Prominent occiput | HP:0000269 | □ | NR |
| Frontal bossing | HP:0002007 | ■ | ■ |
| Hypertelorism | HP:0000316 | ■ | ■ |
| Nystagmus | HP:0000639 | o | ■ |
| Ptosis | HP:0000508 | o | ■ |
| Face | |||
| Prominent forehead | HP:0011220 | □ | NR |
| Hypertelorism | HP:0000316 | o | o |
| Short philtrum | HP:0000322 | ■ | o |
| Midface retrusion | HP:0011800 | ■ | o |
| Ears | |||
| Malformed ears | HP:0000377 | ■ | o |
| Preauricular skin tag | HP:0000384 | □ | NR |
| Posteriorly rotated ears | HP:0000358 | ■ | o |
| Low-set, posteriorly rotated ears | HP:0000368 | ■ | o |
| Eyes | |||
| Microphthalmia | HP:0000568 | o | o |
| Strabismus | HP:0000486 | ■ | o |
| Thick eyebrow | HP:0000574 | o | o |
| Epicanthal folds | HP:0000286 | □ | NR |
| Ptosis | HP:0000508 | ■ | ■ |
| Downslanting palpebral fissures | HP:0000494 | ■ | o |
| Optic atrophy | HP:0000648 | ■ | o |
| Retinal dystrophy | HP:0000556 | □ | NR |
| Nystagmus | HP:0000639 | ■ | ■ |
| Coloboma | HP:0000589 | ■ | ■ |
| Oculomotor apraxia | HP:0000657 | NR | ■ |
| Nose | |||
| Hypoplastic nose | HP:0003196 | ■ | ■ |
| Anteverted nares | HP:0000463 | ■ | ■ |
| Short columella | HP:0002000 | o | o |
| Mouth | |||
| Cleft lip and palate | HP:0008501 | ■ | ■ |
| Cardiovascular | |||
| Septal defects | HP:0001671 | □ | NR |
| Pulmonary valve defects | HP:0005148 | □ | NR |
| Abdomen | |||
| Abnormality of the kidney | HP:0000077 | NR | NR |
| Abnormality of the liver | HP:0001392 | NR | NR |
| Imperforate anus | HP:0002023 | □ | NR |
| Umbilical hernia | HP:0001537 | □ | NR |
| Inguinal hernia | HP:0000023 | □ | □ |
| Genitourinary | |||
| Hypospadias | HP:0000047 | □ | □ |
| Micropenis | HP:0000054 | ■ | ■ |
| Cryptorchidism | HP:0000028 | ■ | ■ |
| Skeletal | |||
| Abnormality of the hand | HP:0001155 | □ | NR |
| Abnormality of the foot | HP:0001760 | □ | NR |
| Neurology | |||
| Global developmental delay | HP:0001263 | ■ | ■ |
| Intellectual disability | HP:0001249 | ■ | ■ |
| Hypotonia | HP:0001252 | ■ | ■ |
| Seizures | HP:0001250 | □ | □ |
| Cerebellar Ataxia | HP:0001251 | □ | NR |
| Neuromuscular | □ | □ | |
| MRI abnormalities | |||
| Molar tooth sign | HP:0002419 | ■ | ■ |
| Hypoplastic or absent corpus callosum | HP:0007370 | ■ | ■ |
| Ectopic posterior pituitary | HP:0011747 | o | ■ |
| Enlarged ventricles | HP:0002119 | o | ■ |
| Cerebellar hypoplasia | HP:0001321 | NR | □ |
| Optic nerve hypoplasia | HP:0000609 | ■ | o |
| Endocrine features | |||
| Panhypopituitarism | HP:0000871 | NR | □ |
| Growth hormone deficiency | HP:0000824 | o | ■ |
| Hypothyroidism | HP:0000821 | □ | ■ |
| Immunology | |||
| Recurrent infections | HP:0002719 | NR | □ |
| (References) | [14, 17–20] | [15, 16, 21, 22] | |
(■), feature previously reported and present in the proband; (□), feature previously reported but absent in the proband; (o), feature not previously reported but present in the proband; (NR), feature not previously reported and absent in the proband
Biallelic mutations in KIAA0556 have rarely been reported thus far. To our knowledge, only 4 families with biallelic truncating variants in this gene have been identified and the associated clinical phenotype resembled a form of JBTS spectrum with variable brain malformations including pituitary malformations (Joubert syndrome type 26, JBTS26) [15, 16, 21, 22] (Tables 1 and 2). The first family with three affected subjects of Saudi Arabian origin was described by Sanders et al. [15]. Affected members displayed a homozygous inactivating variant and a mild form of JBTS phenotype including global DD with variable hypotonia, transient tachypnea, variable cerebellar hypoplasia, and MTS. Two out of three affected subjects had panhypopituitarism with hypoplasia/aplasia of the anterior pituitary and an ectopic posterior pituitary. The second reported family included an affected subject having a mild form of JBTS phenotype characterized by hypotonia, DD, ataxia, oculomotor apraxia, nystagmus and bilateral ptosis. Brain abnormalities including thin corpus callosum and MTS had been also documented by MRI scan [21]. The third family had recently been reported by Cauley et al. [16]; remarkably, the affected subjects from consanguineous parents showed the co-inheritance of a frameshift KIAA0556 mutation and a nonsense variant in a polymicrogyria-associated gene, ADGRG1. They display central nervous system malformations including diffuse polymicrogyria, lissencephaly and cerebellar vermis, pons, and brain stem hypoplasia, and MTS. This clinical complexity was explained by the additive effect of the two truncating mutations [16]. More recently, a fourth case with two compound heterozygous variants in KIAA0556 (c.[2373del]/c.[4551 + 1G-A]) resulting in an abnormal transcript with an in-frame deletion by skipping of exon 25 (102 bp) has been reported [22]. The affected subject showed hypotonia, DD, hypoplastic pituitary, agenesis of corpus callosum, oculomotor apraxia but not coloboma, nystagmus, microphthalmia and craniofacial dysmorphism, as noted in our patient. Interestingly, hypoplasia/aplasia of corpus callosum, cerebellar defects, enlargement of the ventricles, pituitary malformation, and MTS frequently occurred in affected subjects, indicating that defective KIAA0556 likely impacts specifically in brain development and morphology. KIAA0556 (katanin-interacting protein, KATNIP) is evolutionarily conserved, and highly expressed in brain. It is localized to the ciliary base and axoneme where it aids to regulate microtubule dynamics and ciliary integrity. In C. elegans, its orthologue interacts with an ARL13B orthologue to control the cilium integrity. Homozygous Kiaa0556-null mice show a variable hydrocephalus phenotype with enlargement of the ventricles resulting from a block of cerebrospinal fluid flow in the cerebral aqueduct, confirming a JBTS-associated brain-restricted phenotype [15].
In the present case, homozygosity for KIAA0556 variant is assumed to have a truncating impact on the mature protein. On the other hand, the change p.(Arg892His) in KIF7 affects the highly conserved Arg892 residue within structural maintenance of chromosomes (SMC) domain [23]. Interestingly, Arg892 is predicted to be intolerant to changes, being localized very close to the phosphorylatable sites Ser895 and Ser898 within a conserved consensus motif, which is thought to be implicated in the regulatory activity of the protein [24] (Fig. 1, panel h). Few missense changes affecting conserved residues into the SMC domain of KIF7 have previously been reported, and established to impair cilia stability and function, hence causing of ACLS [14, 23].
The clinical phenotype in the present case is distinctive and likely caused by the co-occurrence of two ciliopathies (ACLS/JBTS12 and JBTS26) in the same individual. Detailed clinical features are listed in Table 2. Basing on the minimal diagnostic criteria suggested by Courtens et al. [25], features in the present subject are consistent with the diagnosis of ACLS/JBTS12 more than JBTS26, satisfying three out of the four following criteria: total or partial absence of the corpus callosum, craniofacial anomalies, moderate-severe ID and polydactyly. Accordingly with ACL phenotype, craniofacial dysmorphism, including macrocephaly with frontal bossing, hypertelorism, downslanting palpebral fissures, ptosis and strabismus, depressed nasal tip and cleft lip and palate, were also noted (Table 2). Same consideration applies for the central nervous system features, such as profound ID, severe psychomotor delay, and brain malformations. Of note, polydactyly and skeletal defects, considered major features of ACLS, were absent in our patient. Divergent from ACLS/JBTS12, the present subject showed eye abnormalities, including oculomotor apraxia, endocrine disorders such as GHD and specific MRI features (enlargement of the ventricles, high T2 signal in the white matter, reduction of cortical thickness, and pituitary malformation), which have been reported in subjects with mutations in KIAA0556. Additional features including microphthalmia, iris and retinochoroidal coloboma, nystagmus, which are rarely seen in both ACLS/JBTS12 and JBTS26, were here present.
In conclusion, our findings document that the complex phenotype occurring in a subset of patient with JBTS can be the result of concomitant mutations in cilium-associated genes. In this specific case, the co-occurrence of two rare/private variants of KIF7 and KIAA0556 likely contributes to additional defects of the primary cilium causing congenital organ malformations that are not individually observed in JBTS12 nor in JBTS26. Notably, GM effect of KIF7 with other cilia-associated loci (i.e. TMEM67, CEP41, and Bardet-Biedl syndrome genes) does not seem to be so infrequent in ciliopathies [14, 19, 26]. Similarity, mutations in KIAA0056 have previously been documented to co-associate with ADGRG1 variants contributing to a complex trait resulting in two distinct phenotypes (JBTS26 and BFPP bilateral frontoparietal polymicrogyria, MIM#606854) [16]. This study corroborates previous reports suggesting that the complexity of the JBTS phenotype can be exacerbated by GM occurrences, more commonly than formerly appreciated.
Supplementary information
Additional file 1: Supplementary Data 1. Whole exome sequencing (WES) data analysis was carried out by using an in-house implemented pipeline. Detailed methods and relative references are provided. Supplementary Table S1. Whole exome sequencing metrics, statistics and output.
Acknowledgements
The authors are grateful for the family participating to this study and the further analyses. The authors acknowledge Alessia Palma and Anna Saveria Rao (OPBG, Rome, Italy) for skillful technical assistance.
Abbreviations
- ACLS
Acrocallosal syndrome
- ACMG
American College of Medical Genetics
- ACTH
Adrenocorticotropic hormone
- AHI1
Abelson Helper Integration Site 1
- CADD
Combined Annotation Dependent Depletion
- CC2D2A
Coiled-Coil and C2 Domains-Containing Protein 2A
- CEP41
Centrosomal Protein, 41-KD
- CMV
Cytomegalovirus
- CNVs
Copy number variations
- DANN
Deep Learning Approach for Annotating of genetic variants
- DD
Developmental delay
- DIDA
Digenic diseases database
- fT3
Free triiodothyronine
- fT4
Free thyroxine
- GERP
Genomic Evolutionary Rate Profiling
- GHD
Growth hormone deficiency
- GM
Genetic modifier
- hGH
Human growth hormone
- HLS
Hydrolethalus syndromes
- ID
Intellectual disability
- IGF1
Insulin-like growth factor 1
- IGF-BP3
Insulin like growth factor binding protein 3
- JBTS
Joubert syndrome
- KATNIP
Katanin-interacting protein
- KIF7
Kinesin Family Member 7
- LH
Luteinizing hormone
- MRI
Magnetic resonance imaging
- MTS
Molar tooth sign
- OFC
Head circumference
- PDGF
Platelet-derived growth factor
- SDS
Standard deviation scores
- SHH
Sonic Hedgehog Homolog
- TMEM67
Transmembrane Protein 67
- TSH
Thyroid-stimulating hormone
- WES
Whole exome sequencing
- WNT
Wingless-type
Authors’ contributions
MLD, MCD, RM, MN, EB, BD, and MC contributed to clinical and instrumental data; SB, AC, FRL, MN, and AN contributed to the laboratory analyses; MN, BD, and MT analyzed the data, and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Fondazione Bambino Gesù (“Vite coraggiose” to M.T.), Rome, Italy and funded by the Italian ministry of health (“Ricerca Corrente 2019” to M.N. and A.C.). Funders were not involved in data analysis or interpretation.
Availability of data and materials
Not applicable. We are submitting the data together with this manuscript.
Ethics approval and consent to participate
The study was approved by the institutional ethical committee (No prot. 1702_OPBG_2018) and have therefore been performed in accordance with the ethical standards of Declaration of Helsinki.
Consent for publication
Written informed consent for publication and any accompanying images was obtained from the patients’ legal guardians for publication of this case report. It is available if requested.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12887-020-2019-0.
References
- 1.Bachmann-Gagescu R, Dempsey JC, Phelps IG, O'Roak BJ, et al. Joubert syndrome: a model for untangling recessive disorders with extreme genetic heterogeneity. J Med Genet. 2015;52:514–522. doi: 10.1136/jmedgenet-2015-103087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valente EM, Brancati F, Dallapiccola B. Genotypes and phenotypes of Joubert syndrome and related disorders. Eur J Med Genet. 2008;51:1–23. doi: 10.1016/j.ejmg.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Lancaster MA, Gleeson JG. The primary cilium as a cellular signaling center: lessons from disease. Curr Opin Genet Dev. 2009;19:220–229. doi: 10.1016/j.gde.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valente EM, Dallapiccola B, Bertini E. Joubert syndrome and related disorders. Handb Clin Neurol. 2013;113:1879–1888. doi: 10.1016/B978-0-444-59565-2.00058-7. [DOI] [PubMed] [Google Scholar]
- 5.Gazzo A, Raimondi D, Daneels D, Moreau Y, Smits G, Van Dooren S, et al. Understanding mutational effects in digenic diseases. Nucleic Acids Res. 2017;45:e140. doi: 10.1093/nar/gkx557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schäffer AA. Digenic inheritance in medical genetics. J Med Genet. 2013;50:641–652. doi: 10.1136/jmedgenet-2013-101713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaki MS, Sattar S, Massoudi RA, Gleeson JG. Co-occurrence of distinct ciliopathy diseases in single families suggests genetic modifiers. Am J Med Genet A. 2011;155A:3042–3049. doi: 10.1002/ajmg.a.34173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deltas C. Digenic inheritance and genetic modifiers. Clin Genet. 2018;93:429–438. doi: 10.1111/cge.13150. [DOI] [PubMed] [Google Scholar]
- 9.Bertino E, Spada E, Occhi L, et al. Neonatal anthropometric charts: the Italian neonatal study compared with other europeanstudies. J Pediatr Gastroenterol Nutr. 2010;51:353–361. doi: 10.1097/MPG.0b013e3181da213e. [DOI] [PubMed] [Google Scholar]
- 10.Cacciari E, Milani S, Balsamo A, et al. Italian cross-sctional growth charts for height, weight and BMI (2 to 20 years) J Endocrinol Investig. 2006;29:581–593. doi: 10.1007/BF03344156. [DOI] [PubMed] [Google Scholar]
- 11.Bauer CK, Calligari P, et al. Mutations in KCNK4 that affect gating cause a recognizable neurodevelopmental syndrome. Am J Hum Genet. 2018;103:621–630. doi: 10.1016/j.ajhg.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flex E, Niceta M, et al. Biallelic mutations in TBCD, encoding the tubulin folding cofactor d, perturb microtubule dynamics and cause early-onset encephalopathy. Am J Hum Genet. 2016;99:962–973. doi: 10.1016/j.ajhg.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niceta M, Stellacci E, et al. Mutations impairing GSK3-mediated MAF phosphorylation cause cataract, deafness, intellectual disability, seizures, and a down syndrome-like facies. Am J Hum Genet. 2015;96:816–825. doi: 10.1016/j.ajhg.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Putoux A, Thomas S, Coene KL, Davis EE, Alanay Y, Ogur G, Uz E, Buzas D, Gomes C, Patrier S, Bennett CL, Elkhartoufi N, et al. KIF7 mutations cause fetal hydrolethalus and acrocallosal syndromes. Nature Genet. 2011;43:601–606. doi: 10.1038/ng.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanders AA, de Vrieze E, Alazami AM, Alzahrani F, Malarkey EB, et al. KIAA0556 is a novel ciliary basal body component mutated in Joubert syndrome. Genome Biol. 2015;16:293. doi: 10.1186/s13059-015-0858-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cauley ES, Hamed A, Mohamed IN, Elseed M, Martinez S, Yahia A, Abozar F, Abubakr R, Koko M, Elsayed L, Piao X, Salih MA, Manzini MC. Overlap of polymicrogyria, hydrocephalus, and Joubert syndrome in a family with novel truncating mutations in ADGRG1/GPR56 and KIAA0556. Neurogenetics. 2019;20:91–98. doi: 10.1007/s10048-019-00577-2. [DOI] [PubMed] [Google Scholar]
- 17.Barakeh D, Faqeih E, Anazi S, Al-Dosari MS, Softah A, Albadr F, Hassan H, Alazami AM, Alkuraya FS. The many faces of KIF7. Hum Genome Var. 2015;26:15006. doi: 10.1038/hgv.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedersen LB, Akhmanova A. Kif7 keeps cilia tips in shape. Nat Cell Biol. 2014;16:623–625. doi: 10.1038/ncb2997. [DOI] [PubMed] [Google Scholar]
- 19.Dafinger C, Liebau MC, Elsayed SM, Hellenbroich Y, Boltshauser E, Korenke GC, et al. Mutations in KIF7 link Joubert syndrome with Sonic Hedgehog signaling and microtubule dynamics. J Clin Invest. 2011;121:2662–2667. doi: 10.1172/JCI43639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tunovic S, Barañano KW, Barkovich JA, Strober JB, Jamal L, Slavotinek AM. Novel KIF7 missense substitutions in two patients presenting with multiple malformations and features of acrocallosal syndrome. Am J Med Genet A. 2015;167A:2767–2776. doi: 10.1002/ajmg.a.37249. [DOI] [PubMed] [Google Scholar]
- 21.Roosing S, Rosti RO, Rosti B, de Vrieze E, Silhavy JL, van Wijk E, Wakeling E, Gleeson JG. Identification of a homozygous nonsense mutation in KIAA0556 in a consanguineous family displaying Joubert syndrome. Hum Genet. 2016;135:919–921. doi: 10.1007/s00439-016-1689-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujita A, Higashijima T, Shirozu H, Masuda H, Sonoda M, Tohyama J, Kato M, Nakashima M, Tsurusaki Y, Mitsuhashi S, Mizuguchi T, Takata A, Miyatake S, Miyake N, Fukuda M, Kameyama S, Saitsu H, Matsumoto N. Pathogenic variants of DYNC2H1, KIAA0556, and PTPN11 associated with hypothalamic hamartoma. Neurology. 2019;93:e237–e251. doi: 10.1212/WNL.0000000000007774. [DOI] [PubMed] [Google Scholar]
- 23.Ali BR, Silhavy JL, Akawi NA, Gleeson JG, Al-Gazali L. A mutation in KIF7 is responsible for the autosomal recessive syndrome of macrocephaly, multiple epiphyseal dysplasia and distinctive facial appearance. Orphanet J Rare Dis. 2012;7:27. doi: 10.1186/1750-1172-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu YC, Couzens AL, Deshwar AR, McBroom-Cerajewski LDB, Zhang X, Puviindran V, Scott IC, Gingras AC, Hui CC, Angers S. The PPFIA1-PP2A protein complex promotes trafficking of Kif7 to the ciliary tip and Hedgehog signaling. Sci Signal. 2014;7:RA117. doi: 10.1126/scisignal.2005608. [DOI] [PubMed] [Google Scholar]
- 25.Courtens W, Vamos E, Christophe C, Schinzel A. Acrocallosal syndrome in an Algerian boy born to consanguineous parents: review of the literature and further delineation of the syndrome. Am J Med Genet. 1997;69:17–22. doi: 10.1002/(SICI)1096-8628(19970303)69:1<17::AID-AJMG4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 26.Lee JE, Silhavy JL, Zaki, et al. CEP41 is mutated in Joubert syndrome and is required for tubulin glutamylation at the cilium. Nature Genet. 2012;44:193–199. doi: 10.1038/ng.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Data 1. Whole exome sequencing (WES) data analysis was carried out by using an in-house implemented pipeline. Detailed methods and relative references are provided. Supplementary Table S1. Whole exome sequencing metrics, statistics and output.
Data Availability Statement
Not applicable. We are submitting the data together with this manuscript.