Abstract
Synucleinopathies are neurodegenerative diseases that are associated with the misfolding and aggregation of α-synuclein, including Parkinson’s disease, dementia with Lewy bodies and multiple system atrophy1. Clinically, it is challenging to differentiate Parkinson’s disease and multiple system atrophy, especially at the early stages of disease2. Aggregates of α-synuclein in distinct synucleinopathies have been proposed to represent different conformational strains of α-synuclein that can self-propagate and spread from cell to cell3–6. Protein misfolding cyclic amplification (PMCA) is a technique that has previously been used to detect α-synuclein aggregates in samples of cerebrospinal fluid with high sensitivity and specificity7,8. Here we show that the α-synuclein-PMCA assay can discriminate between samples of cerebrospinal fluid from patients diagnosed with Parkinson’s disease and samples from patients with multiple system atrophy, with an overall sensitivity of 95.4%. We used a combination of biochemical, biophysical and biological methods to analyse the product of α-synuclein-PMCA, and found that the characteristics of the α-synuclein aggregates in the cerebrospinal fluid could be used to readily distinguish between Parkinson’s disease and multiple system atrophy. We also found that the properties of aggregates that were amplified from the cerebrospinal fluid were similar to those of aggregates that were amplified from the brain. These findings suggest that α-synuclein aggregates that are associated with Parkinson’s disease and multiple system atrophy correspond to different conformational strains of α-synuclein, which can be amplified and detected by α-synuclein-PMCA. Our results may help to improve our understanding of the mechanism of α-synuclein misfolding and the structures of the aggregates that are implicated in different synucleinopathies, and may also enable the development of a biochemical assay to discriminate between Parkinson’s disease and multiple system atrophy.
The misfolding and aggregation of α-synuclein (α-syn) involves a mechanism of seeding and nucleation, in which initial seeds of α-syn recruit other soluble monomers that assemble to form aggregates9,10. Aggregates of α-syn circulate in biological fluids such as the cerebrospinal fluid (CSF) and blood11,12. The process of protein misfolding and aggregation appears to begin years or decades before the onset of clinical signs, and thus detection of α-syn aggregates in easily accessible biological fluids may enable the biochemical diagnosis of synucleinopathies. In previous studies, the PMCA technology has been adapted to enable highly sensitive and specific detection of α-syn aggregates that are produced in vitro6,13,14 or derived from the biological fluids of patients with synucleinopathies7,8. The α-syn-PMCA assay (also referred to as α-syn-RT-QuIC15,16) uses the seeding–nucleation mechanism to cyclically amplify the process of protein misfolding, enabling the efficient amplification of small quantities of α-syn oligomers and thereby facilitating their detection.
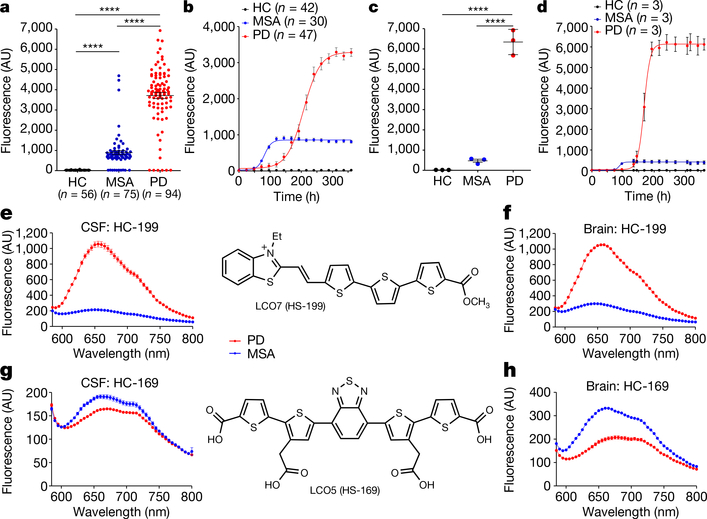
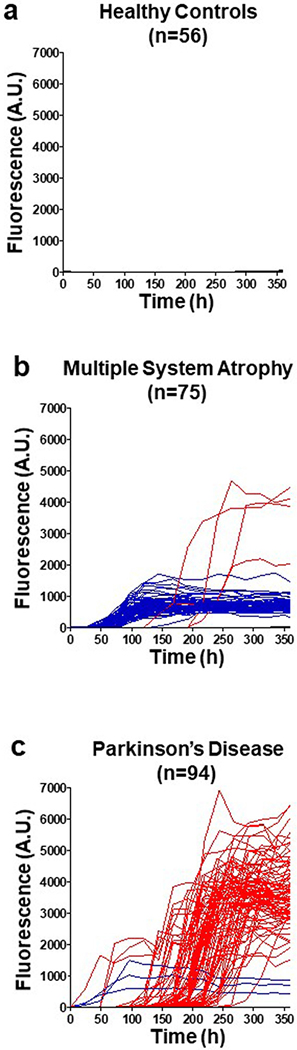
In the α-syn-PMCA assay, the kinetics of aggregation of α-syn are monitored by the fluorescence signal of thioflavin T (ThT)—a dye that is specific to amyloid fibrils17. Previous studies have noted that the maximum fluorescence signal of the α-syn-PMCA product from reactions that were initiated with CSF from patients with multiple system atrophy (MSA) was smaller than the corresponding fluorescence signal for CSF from patients with Parkinson’s disease (PD) or dementia with Lewy bodies7. To further investigate the possibility that PD and MSA can be differentiated by α-syn-PMCA, we performed a study using 94 samples of CSF from patients with PD, 75 from patients with MSA and 56 from control individuals with other neurological diseases (Methods; see Extended Data Table 1 for patient demographics). The maximum ThT fluorescence after α-syn-PMCA was significantly greater in samples from patients with PD than in samples from patients with MSA (Fig. 1a). Products of α-syn-PMCA that were derived from samples from patients with MSA had a maximum fluorescence of less than 1,800 units, whereas for PD this value ranged between 2,000 and 8,000 units. Control samples did not show any fluorescence over the background levels (Fig. 1a). The kinetics of aggregation for all samples in this study are shown in Extended Data Fig. 1. Of the 75 samples from patients with MSA, 4 had an aggregation profile that was compatible with the PD strain and, conversely, 3 of the 94 samples from patients with PD had a profile typical of MSA. From this cohort of samples the overall sensitivity for diagnosis of PD and MSA, as compared to controls calculated by receiving operating curves, was 93.6% and 84.6%, respectively. In both cases, specificity was 100%. Comparing differential diagnosis of PD and MSA, we estimated that of the 88 samples from patients with clinically diagnosed PD that showed α-syn seeds by α-syn-PMCA, 85 were correctly identified as PD in our assay (that is, a sensitivity of 96.6%). Of the 65 samples from patients with MSA that were shown by α-syn-PMCA to contain α-syn aggregates, 61 had the typical signature of MSA (maximum fluorescence of less than 1,800), indicating a sensitivity of 93.8%. Combining all samples, we correctly distinguished PD from MSA in 146 of the 153 samples analysed–an overall sensitivity of 95.4%.
Fig. 1 |. Differential interaction of amyloid-binding dyes with α-syn aggregates derived from patients with PD or patients with MSA.
a, b, Samples of CSF (40 μl) from patients with PD (PD), patients with MSA or healthy control individuals (HC) were subjected to α-syn-PMCA and the extent of aggregation was monitored by ThT fluorescence. a, Maximum fluorescence values (measured at plateau of aggregation) for PD (n = 94; red), MSA (n = 75; blue) and healthy controls (n = 56; black). Each dot represents an individual biological sample measured in duplicate and data are mean ± s.e.m. b, Representative aggregation curves of α-syn in the presence of CSF from patients with PD (n = 47), patients with MSA (n = 30) and healthy controls (n = 42). Data are mean ± s.e.m. of all patients analysed in each group. c, d, Frozen brain samples from patients with pathologically confirmed PD or MSA, or from healthy controls, were homogenized at 10% w/v. A 0.001% dilution of brain homogenate was used for the α-syn-PMCA reaction. c, Maximum fluorescence values for PD (n = 3), MSA (n = 3) and healthy controls (n = 3). Each dot represents an individual biological sample measured in duplicate and data are mean ± s.e.m. of three patients in each group. ****P < 0.0001 by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test (a, c). d, Aggregation profiles of α-syn in the presence of samples from the brain of patients with PD (n = 3), patients with MSA (n = 3) and healthy controls (n = 3). Data are mean ± s.e.m. of three patients in each group. e–h, Differential binding of two amyloid-conformation-specific dyes (HS-199 and HS-169) to α-syn aggregates obtained after two rounds of α-syn-PMCA in samples from the CSF (e, g; n = 43) or the brain (f, h; n = 3) of different patients with PD or MSA. Excitation was at 540 nm and the emission spectrum was recorded between 580 and 800 nm. The chemical structures of HS-199 and HS-169 are also shown. Each experiment was performed in duplicate and data are mean ± s.e.m. (for many points the error bars are smaller than the symbols).
The above data were obtained from different cohorts of patients and across several separate experiments. To illustrate the typical profile of α-syn-PMCA aggregation for samples of PD and MSA, we took the largest individual cohort of samples analysed in Fig. 1a and plotted data from samples that were identified as PD (n = 47) and MSA (n = 30) (Fig. 1b). The maximum fluorescence and the kinetics of aggregation were consistently different for PD and MSA, with samples from patients with MSA aggregating faster but reaching a lower fluorescence plateau than those from patients with PD (Fig. 1b). To determine whether the aggregates present in the CSF are representative of those found in the brain, we also amplified brain samples from three different patients with PD or MSA. To reduce the chance of other brain components interfering in the reaction, we started the PMCA assay with a 10−4 dilution of brain homogenate. Under these conditions, we found that amplified brain-derived α-syn aggregates showed the typical signature of PD or MSA, both in terms of the maximum ThT fluorescence (Fig. 1c) and the kinetics of aggregation (Fig. 1d). These results suggest that the aggregates present in the CSF of patients reflect the aggregates present in the brain.
Notably, the qualitative differences in ThT fluorescence were maintained when the α-syn aggregates that were amplified from samples of CSF from patients with PD or patients with MSA were replicated serially at the expense of monomeric α-syn (Extended Data Fig. 2). For these studies, an aliquot of the final product of the first α-syn-PMCA reaction (starting from CSF samples) was diluted 100-fold into fresh α-syn monomers, and a new α-syn-PMCA assay was performed. This was repeated several times, and the product maintained the high-fluorescence signal for PD and low-fluorescence signal for MSA (Extended Data Fig. 2). To further study the properties of the aggregates that were amplified from patients with PD or with MSA, we selected samples from 43 patients with PD and 43 patients with MSA (see Extended Data Table 2 for the demographic characteristics of these patients). The selection of the 43 samples for each disease was done by eliminating samples that did not aggregate (false negatives) and including those that had the typical signatures of PD or MSA, as indicated above (Fig. 1b, Extended Data Fig. 1). The majority of the characterization studies were done with samples from the second cycle of amplification; this was necessary to generate sufficient material and also to reduce any interference from the CSF, which is important for some of the techniques used (for example, circular dichroism and Fourier-transform infrared (FTIR) spectroscopy).
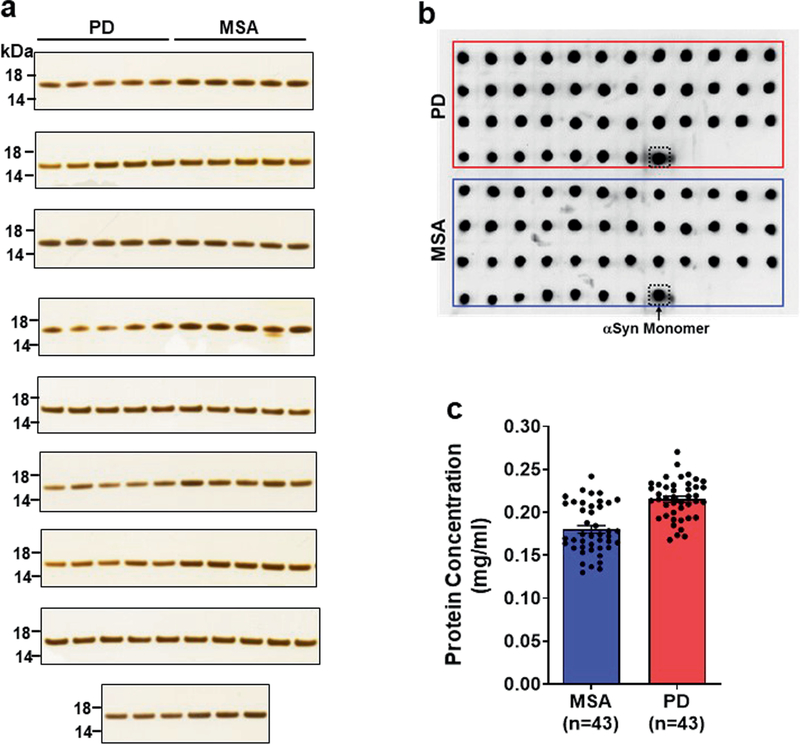
First, we wanted to verify that the differences in ThT fluorescence did not simply reflect different amounts of aggregates at the end of the reaction. To investigate this further, we performed sedimentation assays to separate the pools of soluble and aggregated α-syn. We measured the amount of protein pelleting after centrifugation at 20,000g for 30 minutes, using silver staining after SDS–PAGE (Extended Data Fig. 3a) and dot blot analysis (Extended Data Fig. 3b). We also measured the amount of protein remaining in the supernatant, using the bicinchoninic acid assay (Extended Data Fig. 3c). The results clearly showed that the amount of aggregates produced at the end of the α-syn-PMCA assay was the same in both the PD and the MSA samples. Our interpretation of these results is that either the accessibility or the mode of interaction of ThT with aggregates differs between aggregates derived from patients with PD and those derived from patients with MSA, and that this probably reflects structural differences in the aggregates.
To study the differences between aggregates associated with PD and aggregates associated with MSA in more detail, we first used a panel of thiophene-based ligands that have previously been shown to interact with amyloid aggregates and produce a different spectrum depending on the structural characteristics of the aggregates18,19. The conjugated thiophene backbone is flexible and thus the binding and fluorescence emission of the molecules depends on the conformational properties of the aggregates, providing a specific spectral fingerprint of different aggregates18,19. These compounds have previously been shown to discriminate between different conformational strains of prions, amyloid β and tau proteins20,21. We analysed a set of seven different thiophene-based ligands and found that some of them showed substantially different capacities to interact with α-syn aggregates derived from PD samples compared to those derived from MSA samples (Fig. 1e–h). HS-199 showed a very specific binding affinity and high emission of fluorescence for PD aggregates, whereas the fluorescence of this dye in the presence of MSA aggregates was very low (Fig. 1e). Similar results were obtained when analysing samples derived from brain extracts (Fig. 1f), further supporting the conclusion that aggregates amplified from the CSF and the brain are equivalent. Conversely, the HS-169 dye appeared to bind preferentially to MSA aggregates over PD aggregates, again in samples amplified from both the CSF (Fig. 1g) and the brain (Fig. 1h).
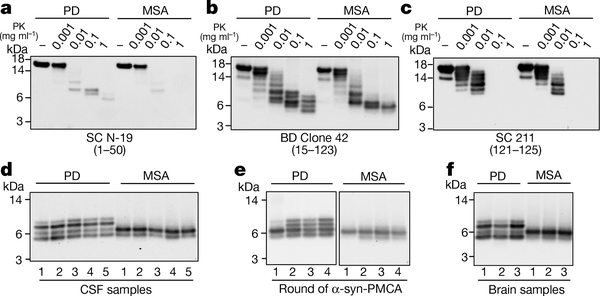
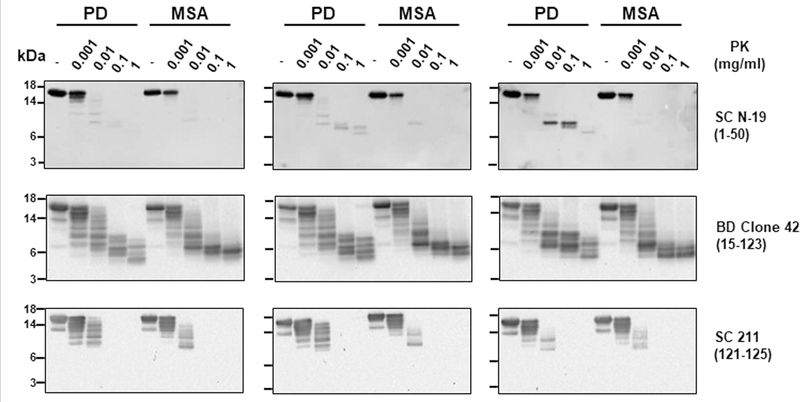
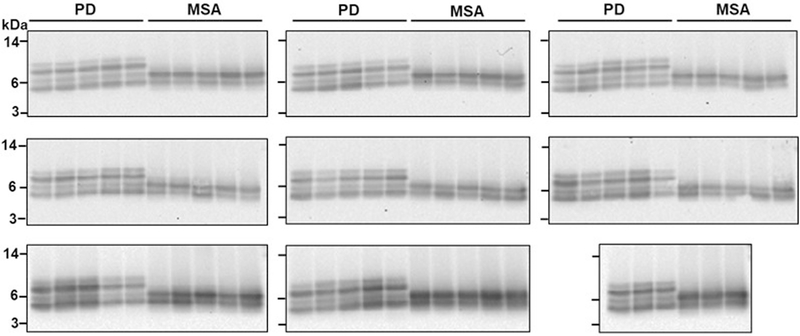
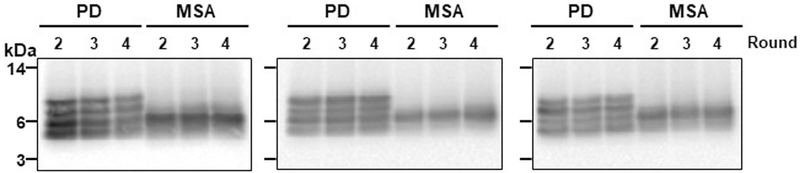
To analyse the biochemical differences between α-syn aggregates derived from patients with PD and from patients with MSA, we examined their resistance to proteolytic degradation and performed epitope-mapping experiments. Limited protease digestion is commonly used to distinguish prion strains22. Aggregates of α-syn derived by seeding and amplification from the CSF of patients with PD or patients with MSA differed in their extent of protease resistance and in the size of the core fragment that was resistant to degradation, as analysed by a panel of different antibodies (Fig. 2a–c; see Extended Data Fig. 4 for the study done with a larger number of samples). Aggregates of α-syn that were amplified from the CSF of patients with PD or patients with MSA were very resistant to degradation, even after treatment with a high concentration of proteinase K (1 mg ml−1) for 1 hour. Under these conditions, protease-resistant fragments mostly mapped to the N-terminal (Fig. 2a) and middle (Fig. 2b) regions of the protein. Conversely, the C-terminal region of α-syn appeared to be fully degraded after incubation with more than 0.01 mg ml−1 of proteinase K (Fig. 2c), which suggests that this part of the protein may not be implicated in the formation of the aggregates (consistent with previous structural studies of α-syn fibrils23–25). Notably, the size and number of protease-resistant bands that were detectable by antibodies directed to the middle region of α-syn (residues 15–123) differed substantially between PD and MSA. Four bands with molecular weights ranging from 4 to10 kDa were detected for samples from patients with PD, whereas only two bands (4 and 6 kDa) were detected for samples from patients with MSA (Fig. 2b, d). This signature was observed across all of the 43 PD and 43 MSA samples that were analysed (Fig. 2d shows 5 representative samples per disease; Extended Data Fig. 5 shows all 86 samples). The signature was maintained after serial replication in vitro by α-syn-PMCA (Fig. 2e, Extended Data Fig. 6), albeit with some small variability in the relative proportions of different bands between rounds of amplification. This result provides further evidence that α-syn-PMCA maintains the biochemical and structural properties of α-syn aggregates. We also analysed the pattern of proteinase K resistance of α-syn aggregates that were amplified from the brain of patients with PD or patients with MSA. The profiles of protease-resistant fragments from brain exhibited the typical signature of PD or MSA (Fig. 4d), again suggesting that the aggregates present in the CSF are equivalent to those that accumulate in the brain.
Fig. 2 |. Protease resistance and epitope mapping of α-syn aggregates derived from the CSF or the brain of patients with PD or patients with MSA.
a–c, α-Syn-PMCA products starting from samples of CSF from patients with MSA or patients with PD were incubated without (−) or in the presence of increasing concentrations of proteinase K (PK; 0.001, 0.01, 0.1 and 1 mg ml−1) at 37 °C for 1 h. Samples were subjected to western blotting using three different antibodies against α-syn: N-19 (Santa Cruz), which recognizes the N-terminal region (residues 1–50) of α-syn (a); anti-α-syn clone 42 (BD Biosciences), which is raised against the middle region of α-syn (residues 15–123) (b); and 211 (Santa Cruz), which is reactive against the C-terminal region of α-syn (residues 121–125) (c). Similar results were obtained for three other patients analysed per disease (Extended Data Fig. 4). d, Profiles of digested fragments from five patients in each group, developed with the BD clone 42 anti-α-syn antibody. The results for all of the PD (n = 43) and MSA (n = 43) samples analysed are shown in Extended Data Fig. 5. For the experiments in a–d, we used the aggregates from the second round of amplification. e, Profile of proteinase- K-resistant fragments after serial rounds of α-syn-PMCA. The first round corresponds to direct amplification from the CSF. For the second round of amplification, aggregates produced in the first round were diluted 100-fold into fresh α-syn monomer substrate and a new round of α-syn-PMCA was performed. The assay was then repeated for the third and fourth rounds using amplified α-syn aggregates (1%) from the previous round. As before, amplified aggregates were treated with proteinase K (1 mg ml−1) and blots were developed with the BD clone 42 anti-α-syn antibody. f, Proteinase K resistance profiles of aggregates amplified from the brain of patients with neuropathologically confirmed PD (n = 3) or MSA (n = 3). Molecular weight markers (kDa) are indicated on the left of each blot.
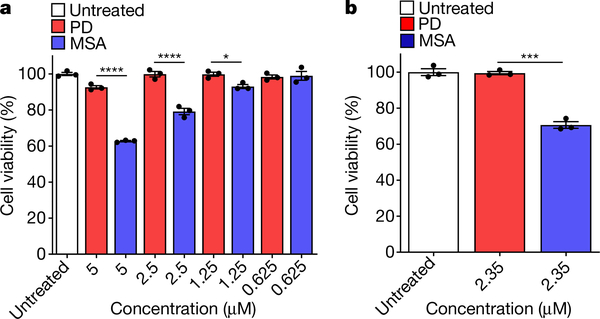
Fig. 4 |. Cytotoxicity of amplified α-syn aggregates from the CSF of patients with PD or patients with MSA.
a, b, RK13 cells (a) (10,000 cells), or neuronal precursor cells derived from human induced pluripotent stem cells generated as previously described28 (b) (5,000 cells), were plated in a 96-well plate. After 24 h, cells were treated for 24 h for RK13 cells and 48 h for neuronal precursor cells with different concentrations of amplified α-syn fibrils from samples of CSF from patients with MSA or patients with PD. Cell viability was determined by MTT assay. Experiments were carried out in triplicate, each dot represents an individual replicate and data are mean ± s.e.m. *P < 0.05, ***P < 0.001, ****P < 0.0001 by one-way ANOVA followed by Tukey’s multiple comparison test.
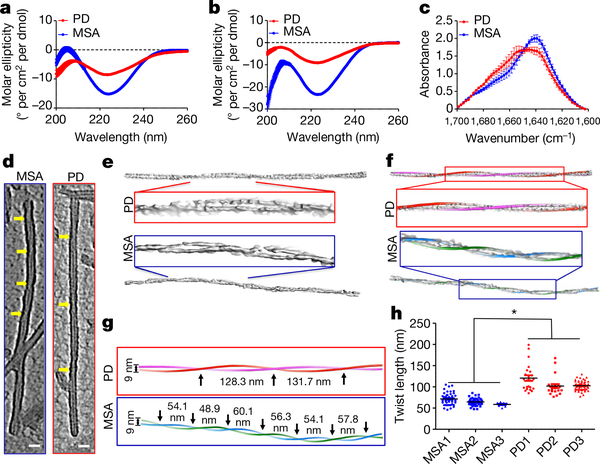
Circular dichroism spectroscopy showed that the secondary structure of α-syn aggregates in both PD and MSA predominantly comprises β-sheets (as illustrated by a negative peak at around 220 nm) (Fig. 3a). Analysis of the spectra indicates that MSA aggregates have a higher proportion of β-sheet structure than PD aggregates. Analogous results were obtained in the three samples from patients with PD and three samples from patients with MSA that were amplified from the brain rather than the CSF (Fig. 3b). To confirm these results using a different methodology, we used FTIR spectroscopy to estimate the secondary structures of α-syn aggregates in samples from a group of randomly selected patients with PD (n = 10) and patients with MSA (n = 10) (Fig. 3c). The MSA-derived aggregates showed a spectrum dominated by parallel β-sheet structure (peak at 1,640 cm−1), whereas for PD-derived aggregates there was also another clear peak at around 1,652 cm−1, which could be assigned to either α-helix- or random-coil-type structures (Fig. 3c).
Fig. 3 |. Structural differences between α-syn aggregates derived from patients with PD or patients with MSA.
a, Circular dichroism spectra of α-syn aggregates from the CSF of patients with PD (red) or patients with MSA (blue), amplified by two rounds of α-syn-PMCA. Spectra were recorded from 35 μM suspensions of α-syn aggregates, as described in Methods. Measurements were taken for all of the PD (n = 43) and MSA (n = 43) samples analysed and data (molar ellipticity) are mean ± s.e.m. b, A similar experiment was performed for α-syn aggregates that were amplified from the brain of patients with PD (n = 3) or patients with MSA (n = 3). c, FTIR spectra of α-syn aggregates that were obtained after two rounds of seeding and amplification of samples of CSF from patients with PD (n = 10) or patients with MSA (n = 10). The solution of aggregated proteins (5 μl; 5 mg ml−1) was analysed with an FTIR-4100 spectrometer (JASCO). d, Cryo-ET was performed to evaluate structural differences between fibrils from patients with PD and fibrils from patients with MSA. Central slices of representative subtomograms of PD-associated fibrils and MSA-associated fibrils are shown. The negative-stained fibrils were imaged with a 300-kV electron microscope (Methods). Yellow arrows indicate twists in the filaments. Scale bar, 20 nm. e, Three-dimensional density maps segmented from the original tomograms. Boxed densities are magnified views. f, Three-dimensional helical models were built that overlapped with the corresponding densities of PD- and MSA-associated fibrils, including a magnification of the central region. g, Helical models showing the periodicity of twisting of PD- or MSA-associated fibrils. Black arrows indicate the twist in the 3D model of the filament. h, Quantification of the periodic spacing (in nm) in many different fibrils derived from samples from patients with PD (n = 3) or patients with MSA (n = 3) samples. Each dot corresponds to a different fibril and data are mean ± s.e.m. *P < 0.05 by one-way ANOVA followed by Tukey’s multiple comparison test.
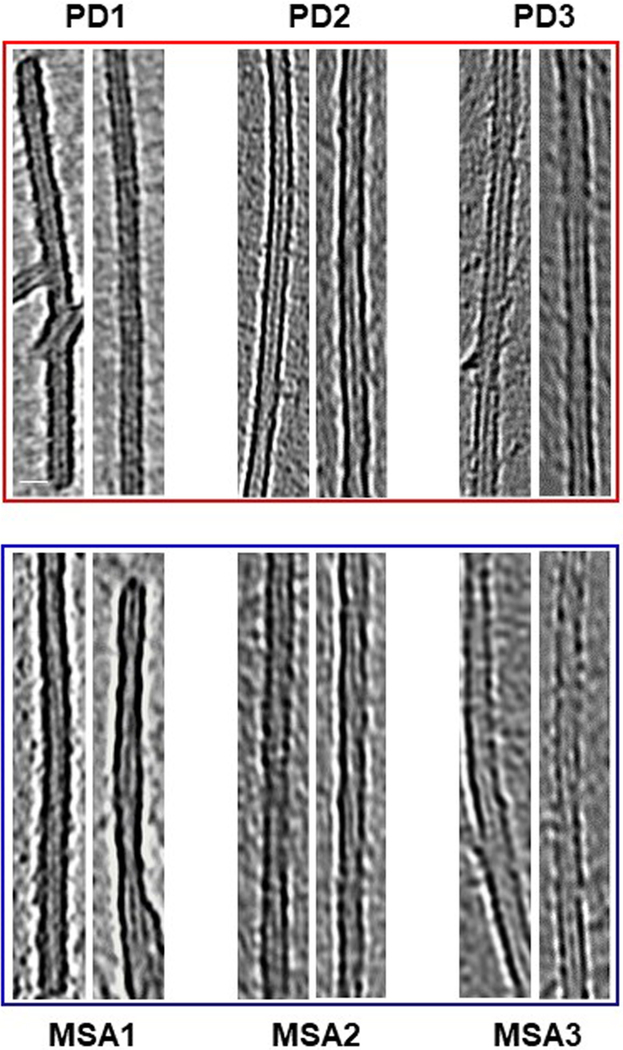
To gain further insight into the structures of both species of α-syn, we performed cryo-electron tomography (cryo-ET) studies. Single-particle cryo-electron microscopy (cryo-EM) has previously been used to determine the high-resolution structure of α-syn aggregates that were generated in vitro24,25. Instead of taking single shots of two-dimensional (2D) images for a given area of a sample grid (as in single-particle cryo-EM), cryo-ET takes multiple shots in the same area by tilting the sample in a series of angles. A three-dimensional (3D) tomogram can be directly reconstructed from the series of tilts. To increase the contrast of the tomographic images, we negatively stained the fibrils amplified from the CSF of patients with PD or patients with MSA. We took 17- and 22-tilt series for PD and MSA samples, respectively (see ‘Cryo-ET analysis and 3D reconstructions’ in Methods for details). The tomograms (Fig. 3d, e) had enough contrast for us to determine that both fibrils were composed of two protofilaments that intertwine in a left-handed helix with a diameter of around 9 nm (see Extended Data Fig. 7 for more images of representative fibrils from three different patients). This is consistent with the high-resolution structure obtained by cryo-EM for full-length α-syn aggregates that were prepared in vitro24. However, the lengths of fibril twists clearly varied between PD and MSA. On the basis of individual measurements of helical diameter and twist lengths, we were able to manually build helical models (Fig. 3f, g) guided by the segmented fibril densities (Fig. 3e). PMCA-derived α-syn aggregates from patients with PD were composed of long stretches of straight filaments with helical twists that generally ranged from 76.6 to 199 nm in length (Fig. 3g). By contrast, α-syn filaments from patients with MSA had shorter twists that mostly ranged from 46 to 105 nm in length (Fig. 3g). In accordance with this, measurements of periodic spacing indicated that the average twisting distance was significantly different between fibrils associated with PD and fibrils associated with MSA (65.2 ± 3.8 nm (mean ± s.e.m.) in MSA fibrils, n = 104 from 3 different patients; 108.5 ± 6.1 nm in PD fibrils, n = 104 from 3 different patients) (Fig. 3h). These data indicate that the structures of α-syn aggregates derived from patients with PD and from patients with MSA are clearly different on the basis of their average periodicities of helical twists. Notably, previous studies using immuno-electron microscopy showed that non-amplified brain-derived α-syn filaments from patients with MSA are predominantly twisted26, whereas those from patients with PD are mostly straight27.
To explore whether aggregates derived from the CSF of patients with PD and patients with MSA have biological differences, we studied their toxicity in cell culture. For these experiments, we used a cell line that is often used in the prion field to study prion replication and toxicity (RK13) (Fig. 4a), together with human neuronal precursor cells derived from induced pluripotent stem cells (Fig. 4b). Induced pluripotent stem cells and neuronal precursors were generated and characterized from fibroblasts obtained from a healthy individual, as previously described28. We tested cytotoxicity by incubating cells with different concentrations of α-syn aggregates derived from the CSF of patients with PD or patients with MSA. MSA-derived aggregates showed highly significant toxicity in RK13 cells, even at concentrations of 1.25 μM; by contrast, PD-derived aggregates began to show significant toxicity only at 5 μM (Fig. 4a), indicating that MSA aggregates are more toxic than PD aggregates. A similar conclusion was obtained in the neuronal precursor cells that were derived from human induced pluripotent stem cells (Fig. 4b).
The prion-like behaviour of α-syn aggregates is a recently recognized principle that may have a central role in the pathological progression of various synucleinopathies29,30. Indeed, the ability of α-syn aggregates to propagate their misfolded abnormalities enables the progressive spreading of damage from cell to cell3–5. One of the tenets of the prion principle is that the misfolded protein can exist in different self-perpetuating conformational strains, which have the ability to faithfully template the misfolding of the normal monomeric protein in the abnormal-strain-specific conformation29. Here we have shown that the prion principle can be used as an effective strategy to cyclically amplify the process of protein misfolding and thereby enable the detection of small amounts of α-syn aggregates in the CSF. Notably, we were able to distinguish—with high sensitivity and specificity—between samples from patients with two clinically similar synucleinopathies (PD and MSA). Moreover, we have shown that the α-syn aggregates present in the CSF of patients are representative of those that accumulate in the brain, indicating that the α-syn-PMCA assay can measure—non-invasively—the pathological species that are associated with different synucleinopathies. Our results demonstrate that α-syn aggregates exist as distinct conformational strains with different biochemical and structural properties, which will help to improve our understanding of the pathogenesis of these diseases. Furthermore, our study shows that patients with distinct synucleinopathies can be distinguished on the basis of the α-syn strain that is present in their CSF. These data may enable the development of a biochemical test for the specific diagnosis of different disorders that involve the misfolding of α-syn, with potential future applications in clinical trials and personalized medicine.
Methods
Data reporting
No statistical methods were used to predetermine sample size, and the experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Patient samples
CSF samples were obtained from 94 patients who were clinically diagnosed with PD, 75 patients who were diagnosed with MSA and 56 control individuals (people with other neurological diseases: epilepsy, cervical spondylosis, polyneuropathy, muscular dystrophy, viral myositis, myelopathy and hydrocephalus). Extended Data Table 1 displays a summary of the demographic characteristics of these patients. Most samples were collected at the Mayo Clinic, as indicated below. The clinical diagnoses of probable PD and MSA were made according to internationally standardized criteria, including the UK Brain Bank guidelines31. CSF samples were collected in the morning using polypropylene tubes following lumbar puncture at the L4/L5 or L3/L4 interspace with atraumatic needles after overnight fasting. The samples were centrifuged at 3,000g for 10 min at room temperature, aliquoted and stored at −80 °C until analysis. Blood cell (red and white) counts and glucose, protein and haemoglobin concentrations were determined as previously described7. The methods of CSF collection were approved by the institutional review boards at the study centres (Mayo Clinic and the University of Texas Health Science Center at Houston), and all study participants provided written informed consent.
Brain tissue from patients with PD and patients with MSA was obtained from the Banner Sun Health Research Institute. Control brain tissue was supplied by NDRI (National Human Tissue Resource Center). Frozen samples of frontal cortex were homogenized using a tissue grinder in 10% w/v ice-cold PBS (HyClone, SH30256.01) with complete protease inhibitor cocktail (Roche). The experiments with human tissue were performed following the universal precautions for working with human specimens and as directed by the Institutional Review Board of The University of Texas Health Science Center at Houston (HSC-MS-14–0608).
Expression and preparation of monomeric α-syn
The purification and characterization of monomeric α-syn was done as previously described7. In brief, the pET-21b plasmid carrying the coding DNA sequence for human α-syn containing a His-tag at the C terminus32 was overexpressed in BL21(DE3) pLysS (Invitrogen) Escherichia coli cells at 25 °C using 0.1 mM IPTG (isopropyl β-d-thiogalactoside) for 6 h. The bacterial pellets were lysed in 50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 10 mM imidazole, 1 mM PMSF, 0.1 mM tris-(2-carboxyethyl) phosphine (TCEP) and 1 mg ml−1 lysozyme, followed by sonication on ice. The lysate was then centrifuged at 12,000g for 15 min at 4 °C, followed by ultracentrifugation at 100,000g for 30 min at 4 °C. The supernatant was filtered through a 0.45-μm filter and loaded onto a nickel-affinity column (Nickel Sepharose Fast flow, GE Healthcare). Proteins were eluted using 250 mM imidazole and α-syn-containing fractions were dialysed overnight at 4 °C against PBS, pH 7.4. To remove any preformed seeds or aggregates, the protein solution was filtered through a 100-kDa cut-off filter (Amicon Ultra, Millipore), separated into small aliquots and stored at −80 °C until use. Protein concentration was determined by bicinchoninic acid (BCA) assay (Pierce). The purity of the protein was evaluated by silver staining.
α-syn-PMCA
The α-syn-PMCA (also known as α-syn-RT-QuIC) assay was performed as previously described7. In brief, samples of seed-free, monomeric α-syn at a concentration of 1 mg ml−1 in 100 mM PIPES, pH 6.5 and 500 mM NaCl were placed in opaque 96-well plates (Costar, REF 3916) in the presence of 5 μM ThT at a final volume of 200 μl. For each test, we added 40 μl of CSF from patients and controls or 40 μl of brain homogenate (at a final concentration of 0.001%). Positive controls consisted of a well-documented and previously screened healthy CSF sample spiked with preformed α-syn oligomeric seeds. Samples were subjected to cyclic agitation (1 min at 500 rpm followed by 29 min without shaking) at 37 °C. The increase in ThT fluorescence was monitored at an excitation of 435 nm and emission of 485 nm, periodically, using a microplate spectrofluorometer Gemini-EM (Molecular Devices).
For serial rounds of amplification, an aliquot from the amplified material was diluted 100-fold into fresh α-syn monomer substrate and a new α-syn-PMCA assay was performed. This was repeated three consecutive times to obtain aggregates corresponding to the second, third and fourth rounds of amplification. The first round of amplification corresponds to the one initiated with the biological samples (CSF or brain homogenate).
Measurement of protein concentration in the aggregated product after amplification
Samples at the end of the PMCA reaction were centrifuged at 20,000g for 30 min at 4 °C. The resultant supernatants were carefully separated from the pellets. The amount of aggregated product was measured in all samples by three different procedures: (1) protein quantity in pellets was measured by silver staining after SDS–PAGE; (2) dot blot analysis of sedimented materials; and (3) BCA measurements of total protein content in the supernatant fraction. For SDS–PAGE, pellets were resuspended in PBS and separated on a 12% Bis-Tris gel and protein bands were visualized by silver staining as per the manufacturer’s protocol. For dot blot analysis, 2 μl of resuspended pellets was spotted onto nitrocellulose membranes (Amersham Biosciences) and air-dried for 30 min at room temperature. Blots were blocked with 5% w/v non-fat dry milk in Tris-buffered saline–Tween 20 (TBS-T) (20 mM Tris, pH 7.2, 150 mM NaCl and 0.05% (v/v) Tween 20) at room temperature for 2 h. After blocking, the membranes were probed with anti-α-syn antibody (BD Bioscience; 1:2,000) and anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibodies (1:5,000). The blots were visualized using enhanced chemiluminescence and a western blotting detection kit (Amersham Biosciences). Finally, the protein concentration in supernatants was determined using a BCA assay kit as per the manufacturer’s recommendations.
Interaction of α-syn aggregates with thiophene-based ligands
A set of seven thiophene-based ligands (p-FTAA, h-FTAA, HS-68, HS-167, HS-169, HS-194 and HS-199) that have previously been shown to discriminate between different conformational strains composed of various proteins19,21,33 was used in this study. These compounds were synthetized and characterized as previously described19,33–36, or as outlined below and in Extended Data Fig. 8 for compound HS-199. The stock solution for each compound was prepared in deionized water or DMSO at 1.5 mM. For our experiments, we diluted these stocks to reach a final concentration of 150 μM. The excitation and emission wavelength range was different depending on the molecule, as previously described18,19.
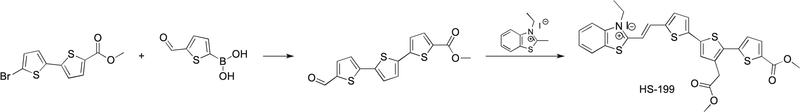
Synthesis and characterization of HS-199
A mixture of methyl 5′-bromo-[2,2′-bithiophene]-5-carboxylate (140 mg, 0.462 mM), (5-formylthiophen-2-yl)boronic acid (80 mg, 0.508 mM) (Extended Data Fig. 8), K2CO3 (192 mg, 1.39 mmol) in 1,4-dioxane/methanol (8: 2, 8 ml, degassed) and PEPPS-IPr (2 mol%) was heated to 80 °C for 30 min. After cooling to room temperature, the pH was adjusted to 4 by addition of 1 M HCl and the residue was extracted with DCM (3 × 20 ml) and washed with water (3 × 20 ml) and brine (30 ml). The combined organic phase was dried over MgSO4 and the solvent was evaporated. The residue was subjected to column chromatography using CH2Cl2 followed by crystallization from DMF to give a trimer (Extended Data Fig. 8) as a yellow solid (115 mg, 74%).
A few drops of pyridine were added to a cold solution of this trimer (0.05 g, 0.150 mM) and the corresponding 2-methyl-3-alkylbenzothiazolium salt (Extended Data Fig. 8) (46 mg, 150 mM) in an anhydrous mixture of MeOH and THF (8:2). The mixture was refluxed until completion of the reaction (monitored by TLC, eluent: DCM/MeOH 1%). The solvent was evaporated in vacuo to provide a dark red solid, which was crystallized from MeOH. The red crystals were collected by filtration, washed with cold MeOH and dried in vacuum to afford HS-199 as a dark red solid (53 mg, 57%). Extended Data Figure 8 provides a summary of this reaction scheme.
The compound was characterized by infrared (IR) spectroscopy, nuclear magnetic resonance and mass spectrometry. IR (neat) 1,697, 7,594, 1,582, 1,525, 1,446, 1,421, 1,304, 1,245, 1,210, 1,165, 1,098, 1,054, 1,035, 926, 806, 786, 758 and 744 cm−1. 1H NMR (300 MHz, DMSO-d6) δ δ 8.48–841 (m, 2H), 8.27 (d, J = 8.6 Hz, 1H), 7.97 (d, J = 3.9 Hz, 1H), 7.91–7.58 (m, 7H), 7.50 (d, J = 3.9 Hz, 1H), 4.92 (q, J = 7.0 Hz, 2H), 3.85 (s, 3H), 1.47 (t, J = 7.0 Hz, 3H).13C NMR (75 MHz, DMSO-d6) δ 170.53, 161.43, 143.01, 142.17, 140.89, 140.83, 138.37, 137.10, 136.14, 136.01, 134.89, 131.30, 129.48, 28.26, 128.16, 127.79, 127.69, 126.66, 125.58, 124.38, 116.44, 111.21, 52.41, 44.29, 14.14. Matrix-assisted laser resorption ionization–time of flight (MALDI–TOF): m/z calculated for C25H20NO2S4 (M+H)+: 495.0. Found: 495.0.
Protease digestion and epitope mapping
Samples containing α-syn aggregates amplified by PMCA were treated with different concentrations of proteinase K at 37 °C for 1 h. The reaction was stopped by heating the sample in NuPAGE LDS buffer at 95 °C for 10 min. The digested products were resolved by 12% Bis-Tris gels (Invitrogen). Proteins were electrophoretically transferred to nitrocellulose membranes (Amersham Biosciences). Membranes were blocked with 5% w/v non-fat dry milk in PBS–Tween 20 (PBS (Hyclone SH.30258.02, pH 7.2, 0.1% (v/v) Tween 20) at room temperature for 1 h. After blocking, the membranes were probed with the following antibodies against α-syn: N-19 (Santa Cruz), which recognizes the N terminus (residues 1–50) of α-syn; anti-α-syn clone 42 (BD Biosciences), which is raised against the middle region (residues 15–123) of the protein; and 211(Santa Cruz),which is reactive against the C-terminal region (residues 121–125) of α-syn. The blots were developed using ECL prime detection western blotting reagents (Amersham Biosciences).
Circular dichroism
Solutions containing around 35 μM of α-syn aggregates amplified by α-syn-PMCA were used for these studies. Circular dichroism spectra were recorded at room temperature using a JASCO J815 spectropolarimeter, with 1-mm path-length cuvette. Circular dichroism data were collected at 0.1-nm resolution and at a scan speed of 200 nm min−1. The portion of the circular dichroism spectrum between 250 and 350 nm was fitted with a quadratic function and the baseline of the whole spectrum was calculated using the function. Then the calculated baseline was subtracted from the circular dichroism spectrum to obtain the baseline-corrected circular dichroism spectrum.
FTIR spectroscopy
FTIR experiments were conducted using an FT/IR-4100 spectrometer from JASCO. The product of α-syn-PMCA (5 μl) was placed on the top of a diamond PRO450-S attenuated total reflectance unit (JASCO) adapted to the FT/IR-4100 system. The system parameters included a resolution of 4.0 cm−1 and an accumulation of 80 scans per sample. The data were processed using cosine apodization and Mertz phase correction. The data were also corrected for attenuated total reflectance and carbon dioxide vapour absorption.
Cryo-ET analysis and 3D reconstructions
The product of α-syn-PMCA (after 2 rounds of amplification from CSF samples from patients with PD or patients with MSA) was sedimented at 20,000g for 30 min at 4 °C, resuspended in 100 mM PIPES, pH 6.5 and 500 mM NaCl, diluted 10-fold in deionized water and loaded onto Formvar/Carbon Copper grids. Samples were negatively stained with 2% uranyl acetate and rapidly frozen in liquid ethane, using a gravity-driven plunger apparatus. Materials were imaged at −170 °C using a Polara G2 electron microscope (FEI) equipped with a field-emission gun and a direct-detection device (Gatan K2 Summit). The microscope was operated at 300 kV with a magnification of ×15,500. We used SerialEM37 to collect tomographic tilt series at a defocus of around 6 μm, with cumulative doses of around 200 e− per Å2. For each dataset, 35 image stacks were collected in a range from −51° to +51°, using increments of 3°. Each stack contained about 10 images, which were first aligned using MotionCor238. The tomograms were reconstructed using IMOD software39 and were further processed by EMAN software40.
The helical models were manually built based on individual fibril density. The twist lengths of fibrils vary from one to another. We were not able to perform subtomogram averaging owing to the heterogeneous nature of the fibrils. The single fibril density used for modelling has very limited resolution. Its density in the z-direction is elongated because of a missing wedge issue (no high-angle tilt images). We rely on the helical parameters (diameter and the twist lengths) to build a helical model, which can be directly measured from the centre slice of the tomogram in the x–y plane. Instead of creating a mathematical helical model on the basis of the two parameters, we manually built the helical model by tracing the filament density using Chimera software41. Model dots were placed along the densities followed by manual adjustment of the dot positions to make the model shape helix-like under the restriction of cryo-ET density. Although density restriction was applied, the constructed pseudo-helical model does not completely fit into the noisy and distorted fibril density.
Cytotoxicity assays
RK13 cells (rabbit kidney cell line, ATCC CCL-37) were grown in DMEM medium supplemented with 10% FBS 1× GLUTAmax, 1× MEM and 1 mM sodium pyruvate. For toxicity, 10,000 cells were plated without antibiotic in a 96-well plate and incubated at 37 °C for 24 h. Neuronal precursors derived from human induced pluripotent stem cells were generated and characterized as previously described28. These cells were maintained in neural precursor expansion medium (NPEM) as previously described. Approximately 5,000 cells were plated per well in a 96-well plate, pre-coated with Geltrex LDEV-free reduced growth factor basement membrane matrix-treated dishes (1:100, Invitrogen) and incubated at 37 °C for 24 h. After 24 h, cells were treated for either 24 h (RK13 cells) or 48 h (neuronal precursors) with different concentration(s) of amplified α-syn fibrils originating from CSF samples from patients with MSA and patients with PD. Cell viability was determined by the MTT assay, following the manufacturer’s protocol.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
All data generated and/or analysed during this study are included in the Article, Supplementary Fig. 1 (uncropped blots) and the Source Data files for Figs. 1, 3, 4 and Extended Data Figs. 1–3. Any additional information required are available from the corresponding author on reasonable request.
Extended Data
Extended Data Fig. 1 |. Kinetics of α-syn aggregation in the presence of CSF from patients with PD, patients with MSA or healthy control individuals.
a–c, Individual α-syn aggregation curves are shown in the presence of CSF samples (40 μl) from all study participants, including healthy controls (a; n = 56), patients with MSA (b; n = 75) and patients with PD (c; n = 94). The α-syn-PMCA assay was started by adding α-syn monomers (1 mg ml−1) and ThT (5 μM) to 100 mM PIPES, pH 6.5 containing 500 mM NaCl. The plate was incubated at 37 °C with intermittent shaking for 1 min every 30 min at 500 rpm. The extent of aggregation was monitored using a fluorometer to measure ThT fluorescence, with an excitation of 435 nm and emission of 485 nm. The colours represent the expected aggregation curves for patients with PD (red), patients with MSA (blue) and healthy controls (black), regardless of clinical diagnosis.
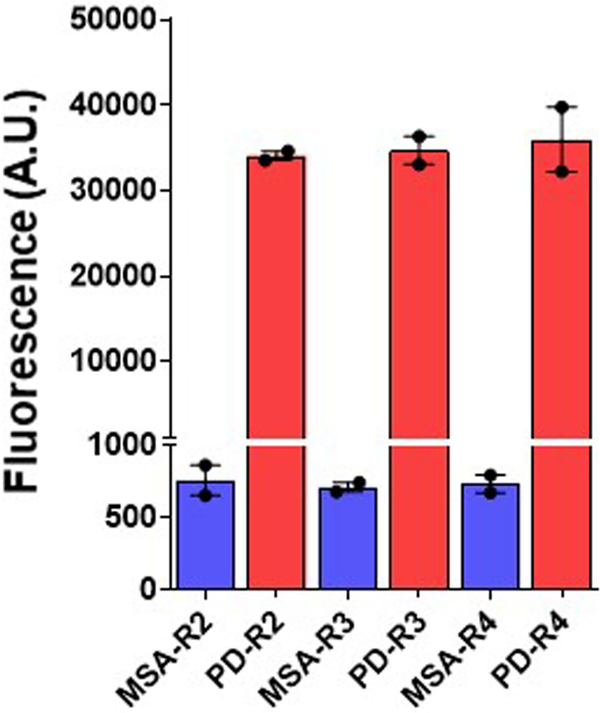
Extended Data Fig. 2 |. Serial propagation of α-syn aggregates derived from patients with MSA and patients with PD.
For serial propagation of α-syn aggregates, an aliquot of the final product of the first α-syn-PMCA reaction (starting from CSF samples) was diluted 100-fold into a solution containing fresh α-syn monomers (1 mg ml−1). A second round of amplification was done in the same buffer (100 mM PIPES, pH 6.5 containing 500 mM NaCl) at 37 °C with intermittent shaking for 1 min every 30 min at 500 rpm. The extent of aggregation was monitored by the increase in ThT fluorescence. The maximum fluorescence value at the plateau of aggregation was recorded and plotted in the graph as the second round of amplification (R2). Similarly, the third and fourth rounds of amplification (R3 and R4) were performed by diluting the product 100-fold on amplification each time into fresh α-syn monomer substrate and repeating the α-syn-PMCA assay. The results shown are from one patient with PD and one patient with MSA. The experiment was carried out in duplicate, each dot represents an individual technical replicate and data are mean ± s.e.m.
Extended Data Fig. 3 |. Analyses of the quantity of α-syn aggregates after amplification from patients with MSA and patients with PD by sedimentation assay.
Aggregates of α-syn that were obtained after two rounds of α-syn-PMCA amplification (starting from CSF samples from patients with MSA (n = 43) and patients with PD (n = 43)) were centrifuged at 20,000g for 30 min. a, The resultant pellets were separated on a 12% Bis-Tris gel, and protein bands were visualized by silver staining as per the manufacturer’s protocol. Molecular weight markers (kDa) are indicated on the left of the gel. b, Resuspended pellets (2 μl) were spotted onto nitrocellulose membranes and air-dried for 30 min at room temperature. After blocking with 5% w/v non-fat dry milk at room temperature for 2 h, membranes were probed with an anti-α-syn antibody (BD Bioscience, 1:2,000) and anti-rabbit HRP-conjugated secondary antibodies (1:5,000). The blots were visualized using enhanced chemiluminescence and a western blotting detection kit. The dot blot shows each of the 86 samples (n = 43, PD; n = 43, MSA) and a positive control using non-aggregated α-syn monomer (dotted box). The results are representative of two independent experiments with similar results. c, Protein concentration in the supernatants was determined by a BCA assay kit as per the manufacturer’s instructions. Each dot represents an individual sample (n = 43, PD; n = 43, MSA) in each disease group and data are mean ± s.e.m.
Extended Data Fig. 4 |. Proteinase K digestion profiles of α-syn aggregates derived from samples of CSF from patients with PD and patients with MSA.
This is the same experiment as Fig. 2a–c, showing proteinase K digestion profiles of other representative samples from patients with PD (n = 3) and patients with MSA (n = 3). The amplified product from the second round of α-syn-PMCA in samples of CSF from patients with MSA or patients with PD was incubated either without (−) or in the presence of increasing concentrations of proteinase K (0.001, 0.01, 0.1 and 1 mg ml−1) at 37 °C for 1 h. Proteins were separated on a 12% Bis-Tris gel and immunoblotted with the same antibodies as in Fig. 2 (SC N-19 (top), BD anti-α-syn clone 42 (middle) and SC 211 (bottom)). Each blot represents an individual sample. Molecular weight markers (kDa) are indicated on the left of the blot.
Extended Data Fig. 5 |. Proteinase K digestion profiles of α-syn aggregates derived from samples of CSF from all 43 patients with PD and 43 patients with MSA.
This is the same experiment as Fig. 2d, showing proteinase K digestion profiles of all 86 (n = 43, PD; n = 43, MSA) biologically independent samples analysed. Aliquots of the product of the second round of the α-syn-PMCA assay were treated with proteinase K (1 mg ml−1) at 37 °C for 1 h. Proteins were separated on a 12% Bis-Tris gel and immunoblotted with the BD anti-α-syn clone 42 antibody. Molecular weight markers (kDa) are indicated on the left of the blot. The third blot on the top row is the same as that shown in Fig. 2d.
Extended Data Fig. 6 |. Proteinase K digestion profiles of α-syn aggregates after several rounds of α-syn-PMCA.
This is the same experiment as Fig. 2e, showing the results obtained with samples from different patients with PD (n = 3) and patients with MSA (n = 3). The first round corresponds to direct amplification from the CSF of the patients. For the second round of amplification, aggregates produced in the first round were diluted 100-fold into fresh α-syn monomer substrate and a new round of α-syn-PMCA was performed. The assay was then repeated for the third and fourth rounds using amplified α-syn aggregates (1%) from the previous round. Amplified aggregates were treated with proteinase K (1 mg ml−1) for 1 h and proteins were separated on a 12% Bis-Tris gel and immunoblotted with the BD anti-α-syn clone 42 antibody. Molecular weight markers (kDa) are indicated on the left of the blot.
Extended Data Fig. 7 |. Electron microscopy images of PD-associated fibrils and MSA-associated fibrils.
Representative images of fibrils produced after two rounds of α-syn-PMCA in samples from different patients with PD (n = 3) and patients with MSA (n = 3). The negative-stained fibrils were imaged with a 300 kV electron microscope. Scale bar, 10 nm (applies to all of the images).
Extended Data Fig. 8 |. Reaction scheme for the chemical synthesis of HS-199.
HS-199 was synthesized by mixing 0.462 mM methyl 5′-bromo-[2,2′- bithiophene]-5-carboxylate with 0.508 mM (5-formylthiophen-2-yl)boronic acid, K2CO3 (1.39 mmol) in 1,4-dioxane/methanol (8: 2, 8 mL/mM, degassed) and PEPPS-IPr (2 mol %).
Extended Data Table 1 |.
Number of samples and basic demographic information for all study participants
| Healthy Controls (HC) | Multiple System Atrophy (MSA) | Parkinson’s Disease (PD) | ||||
|---|---|---|---|---|---|---|
| Number of samples | 56 | 75 | 94 | |||
| Age (Mean ± SD) | 60.14 ± 10.18 | 59.67 ± 6.9 | 66.78 ± 7.91 | |||
| Sex (M,F) | M = 28 | F = 28 | M = 56 | F= 19 | M = 59 | F = 36 |
| Disease Duration (Mean ± SD) | N/A | 3.847 ± 2.6 | 8.13 ± 4.7 | |||
Age and disease duration are given in years.
Extended Data Table 2 |.
Demographic information for the 43 patients with PD and 43 patients with MSA whose CSF samples were used to characterize amplified α-syn aggregates in detail
| Parkinson’s Disease (PD) | Multiple System Atrophy (MSA) | ||||||
|---|---|---|---|---|---|---|---|
| S/No | Clinical Diagnosis | Age/Sex | Disease Duration | S/No | Clinical Diagnosis | Age/Sex | Disease Duration |
| 1 | PD | 71/F | 11 | 1 | MSA-P | 61/M | 2 |
| 2 | PD | 61/M | 8 | 2 | MSA-C | 60/M | 6 |
| 3 | PD | 59/F | 12 | 3 | MSA-C | 61/M | 5 |
| 4 | PD | 63/F | 7 | 4 | MSA-C | 66/F | 3 |
| 5 | PD | 66/F | 8 | 5 | MSA-C | 58/M | 7 |
| 6 | PD | 62/M | 8 | 6 | MSA-C | 53/M | 2 |
| 7 | PD | 72/M | 17 | 7 | MSA-C | 65/F | 2 |
| 8 | PD | 69/M | 12 | 8 | MSA-P | 55/M | 3 |
| 9 | PD | 67/M | 15 | 9 | MSA-P | 64/M | 1 |
| 10 | PD | 58/F | 8 | 10 | MSA-C | 60/M | 6 |
| 11 | PD | 66/M | 6 | 11 | MSA-C | 63/M | 1 |
| 12 | PD | 74/F | 6 | 12 | MSA-C | 51/M | 2.5 |
| 13 | PD | 73/F | 5 | 13 | MSA-P | 59/F | 2 |
| 14 | PD | 61/F | 9 | 14 | MSA-C | 54/M | 6 |
| 15 | PD | 69/M | 5 | 15 | MSA-P | 49/F | 3 |
| 16 | PD | 65/M | 7 | 16 | MSA-C | 55/M | 3 |
| 17 | PD | 69/F | 11 | 17 | MSA-C | 68/M | 5 |
| 18 | PD | 65/M | 10 | 18 | MSA-C | 56/F | 1.5 |
| 19 | PD | 68/M | N/A | 19 | MSA-C | 57/F | 2.5 |
| 20 | PD | 57/F | N/A | 20 | MSA-C | 59/M | 1 |
| 21 | PD | 71/F | N/A | 21 | MSA-C | 49/M | 1 |
| 22 | PD | 68/M | N/A | 22 | MSA-C | 50/M | 2 |
| 23 | PD | 79/M | N/A | 23 | MSA-P | 54/M | 5 |
| 24 | PD | 68/M | 21 | 24 | MSA-P | 64/M | 3 |
| 25 | PD | 70/M | 1 | 25 | MSA-C | 68/F | 6 |
| 26 | PD | 62/M | 3 | 26 | MSA-C | 60/M | 2 |
| 27 | PD | 63/M | 10 | 27 | MSA-C | 61/M | 1 |
| 28 | PD | 74/M | 17 | 28 | MSA-C | 62/M | 2 |
| 29 | PD | 69/M | 6 | 29 | MSA-P | 53/M | 4 |
| 30 | PD | 66/M | 5 | 30 | MSA-P | 54/M | 5 |
| 31 | PD | 73/F | 15 | 31 | MSA-C | 61/M | 8 |
| 32 | PD | 54/M | 6 | 32 | MSA-C | 66/F | 3 |
| 33 | PD | 71/M | 5 | 33 | MSA-C | 54/M | 4 |
| 34 | PD | 72/M | 5 | 34 | MSA-C | 62/M | 9 |
| 35 | PD | 74/M | 5 | 35 | MSA-C | 48/F | 5 |
| 36 | PD | 59/M | 15 | 36 | MSA-P | 54/F | 2 |
| 37 | PD | 53/F | 4 | 37 | MSA-P | 60/F | 2 |
| 38 | PD | 73/M | N/A | 38 | MSA-P | 73/F | 10 |
| 39 | PD | 69/F | N/A | 39 | MSA-P | 57/F | 2.5 |
| 40 | PD | 47/M | N/A | 40 | MSA-C | 68/F | 3.5 |
| 41 | PD | 59/M | N/A | 41 | MSA-C | 52/M | 12 |
| 42 | PD | 61/F | 24 | 42 | MSA-C | 58/M | 5 |
| 43 | PD | 68/M | 12 | 43 | MSA-C | 54/M | 3 |
Age and disease duration are given in years. MSA-C, MSA with cerebellar ataxia; MSA-P, MSA with Parkinsonism.
Supplementary Material
Antibodies.
Polyclonal goat anti-α/β-Synuclein (N-19), Cat. No. sc-7012, Santa Cruz, Dilution 1:2000, Lot. No. K1213.
Monoclonal mouse anti-α-Synuclein (211), Cat. No. sc-12767, Santa Cruz, Dilution 1:4000, Lot. No. B1518.
Monoclonal mouse anti-α-Synuclein (clone 42), Cat. No. 610787, BD Biosciences, Dot Blot Dilution 1:2000, Western Blot Dilution 1:5000, Lot. No. 7243572
Sheep anti-mouse IgG conjugated to horseradish peroxidase (HRP), Cat. No. A5906, Sigma Aldrich, Dot Blot Dilution 1:5000, Western Blot Dilution 1:10000, Lot. No. SLBT9505
Donkey anti-goat IgG conjugated to horseradish peroxidase (HRP), Cat. No. A15999, Invitrogen, Dilution 1:10000
Validation.
All antibodies were validated according to the manufacturer’s instruction and as described in the literature
Commercial antibody, validation data available on manufacturer’s website. Ref: Sharma N, et al. 2001 Acta Neuropathologica 102(4):329–34.
Commercial antibody, validation data available on manufacturer’s website. Ref: Zunke F, Moise AC, et al. 2018 Neuron 97(1):92–107.
Commercial antibody, validation data available on manufacturer’s website. Ref: Liu Y, Fallon L, et al. 2002 Cell 111(2):209–18.
Commercial antibody, validation data available on manufacturer’s website. Ref: Trowitzsch S, Viola C, et al. 2015 Nature Communications 6: 6011.
Validated by Manufacturer, see Certificate of Analysis on website
Bacteria.
BL21 Star™ (DE3)pLysS One Shot™ Chemically Competent E. coli
Cat. No. 44–0054
Lot. No. 1437577
Invitrogen
Eukaryotic cell lines.
Cell line source
RK13 (ATCC® CCL37™) cell lines were purchased from ATCC.
Authentication
ATCC provided certificate of analysis for RK13 (ATCC® CCL37™) cell line.
Cell line source
Neural precursor cells were generated from human iPSC cells reprogrammed from skin fibroblasts obtained from a 66 year old female (cell number: AG08517) purchased from (Coriell, Candem, NJ, USA). The procedure for reprogramming, differentiation and growing of cells is described in detail in our previous publication (Armijo, E. et al. Neurosci. Lett 639, 74–81, 2017).
Authentication
Cells were fully characterized by various procedures as described in detail in our previous publication (Armijo, E. et al. Neurosci. Lett 639, 74–81, 2017).
Mycoplasma contamination
All cell lines were tested routinely negative for Mycoplasma contamination by PCR.
Commonly misidentified lines (See ICLAC register)
No commonly misidentified lines were used.
Acknowledgements
This study was funded in part by grants from the Michael J. Fox Foundation for Parkinson’s disease (to C.S. and S.P.); NIH (R01AG055053, R01AG061069) and Department of Defense (to C.S.); NIH (P01NS44233, U54NS065736, K23NS075141, R01 FD004789, R01 NS092625), Department of Defense and Mayo Funds (to P.A.L.); RO1 NS094535 (to A.-L.T.); and the Swedish Research Council (2016-00748 to H.S. and K.P.R.N.). We are grateful to the Banner Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona for the provision of brain tissue. We also thank N. P. Rocha for providing CSF samples, I. Moreno-Gonzalez for helping with the preparation and characterization of brain homogenate and T. Eckland for editing the manuscript.
Footnotes
Competing interests C.S. and M.S. are inventors on patent applications (US20160077111, WO2016040905, EP3191599A1, US20160077112 and WO2016040907) for the use of PMCA technology for high-sensitive detection of α-syn aggregates in patients affected by synucleinopathies. These applications were filed by the University of Texas Health Science Center at Houston and Amprion Inc. C.S. is an inventor on several patents related to PMCA technology and is a Founder, Chief Scientific Officer and Member of the Board of Directors of Amprion Inc, a biotechnology company that focuses on the commercial use of PMCA (RT-QuIC) for high-sensitivity detection of misfolded protein aggregates that are implicated in a variety of neurodegenerative diseases. The University of Texas Health Science Center at Houston owns some patent applications related to the PMCA (RT-QuIC) technology that have been licensed to Amprion Inc.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-020-1984-7.
Supplementary information is available for this paper at https://doi.org/10.1038/s41586-020-1984-7.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Goedert M, Jakes R & Spillantini MG The synucleinopathies: twenty years on. J. Parkinsons Dis 7, S51–S69 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wenning GK et al. What clinical features are most useful to distinguish definite multiple system atrophy from Parkinson’s disease? J. Neurol. Neurosurg. Psychiatry 68, 434–440 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melki R Role of different alpha-synuclein strains in synucleinopathies, similarities with other neurodegenerative diseases. J. Parkinsons Dis 5, 217–227 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prusiner SB et al. Evidence for α-synuclein prions causing multiple system atrophy in humans with parkinsonism. Proc. Natl Acad. Sci. USA 112, E5308–E5317 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng C et al. Cellular milieu imparts distinct pathological α-synuclein strains in α-synucleinopathies. Nature 557, 558–563 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarutani A, Arai T, Murayama S, Hisanaga SI & Hasegawa M Potent prion-like behaviors of pathogenic α-synuclein and evaluation of inactivation methods. Acta Neuropathol. Commun 6, 29 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shahnawaz M et al. Development of a biochemical diagnosis of Parkinson disease by detection of α-synuclein misfolded aggregates in cerebrospinal fluid. JAMA Neurol. 74, 163–172 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Kang UJ et al. Comparative study of cerebrospinal fluid α-synuclein seeding aggregation assays for diagnosis of Parkinson’s disease. Mov. Disord 34, 536–544 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wood SJ et al. α-synuclein fibrillogenesis is nucleation-dependent. Implications for the pathogenesis of Parkinson’s disease. J. Biol. Chem 274, 19509–19512 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Volles MJ & Lansbury PT Jr. Zeroing in on the pathogenic form of α-synuclein and its mechanism of neurotoxicity in Parkinson’s disease. Biochemistry 42, 7871–7878 (2003). [DOI] [PubMed] [Google Scholar]
- 11.El-Agnaf OM et al. Detection of oligomeric forms of α-synuclein protein in human plasma as a potential biomarker for Parkinson’s disease. FASEB J. 20, 419–425 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Tokuda T et al. Detection of elevated levels of α-synuclein oligomers in CSF from patients with Parkinson disease. Neurology 75, 1766–1770 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Herva ME et al. Anti-amyloid compounds inhibit α-synuclein aggregation induced by protein misfolding cyclic amplification (PMCA). J. Biol. Chem 289, 11897–11905 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung BC et al. Amplification of distinct α-synuclein fibril conformers through protein misfolding cyclic amplification. Exp. Mol. Med 49, e314 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groveman BR et al. Rapid and ultra-sensitive quantitation of disease-associated α-synuclein seeds in brain and cerebrospinal fluid by αSyn RT-QuIC. Acta Neuropathol. Commun 6, 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fairfoul G et al. Alpha-synuclein RT-QuIC in the CSF of patients with alpha-synucleinopathies. Ann. Clin. Transl. Neurol 3, 812–818 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naiki H, Higuchi K, Hosokawa M & Takeda T Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavin T1. Anal. Biochem 177, 244–249 (1989). [DOI] [PubMed] [Google Scholar]
- 18.Sjöqvist J et al. Toward a molecular understanding of the detection of amyloid proteins with flexible conjugated oligothiophenes. J. Phys. Chem. A 118, 9820–9827 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Klingstedt T & Nilsson KP Luminescent conjugated poly- and oligo-thiophenes: optical ligands for spectral assignment of a plethora of protein aggregates. Biochem. Soc. Trans 40, 704–710 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Rasmussen J et al. Amyloid polymorphisms constitute distinct clouds of conformational variants in different etiological subtypes of Alzheimer’s disease. Proc. Natl Acad. Sci. USA 114, 13018–13023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sigurdson CJ et al. Prion strain discrimination using luminescent conjugated polymers. Nat. Methods 4, 1023–1030 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Bessen RA & Marsh RF Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J. Virol 66, 2096–2101 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuttle MD et al. Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat. Struct. Mol. Biol 23, 409–415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y et al. Amyloid fibril structure of α-synuclein determined by cryo-electron microscopy. Cell Res. 28, 897–903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerrero-Ferreira R et al. Cryo-EM structure of alpha-synuclein fibrils. eLife 7, e36402 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grazia Spillantini M et al. Filamentous α-synuclein inclusions link multiple system atrophy with Parkinson’s disease and dementia with Lewy bodies. Neurosci. Lett 251, 205–208 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Crowther RA, Daniel SE & Goedert M Characterisation of isolated α-synuclein filaments from substantia nigra of Parkinson’s disease brain. Neurosci. Lett 292, 128–130 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Armijo E et al. Increased susceptibility to Aβ toxicity in neuronal cultures derived from familial Alzheimer’s disease (PSEN1-A246E) induced pluripotent stem cells. Neurosci. Lett 639, 74–81 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Soto C & Pritzkow S Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci 21, 1332–1340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olanow CW & Prusiner SB Is Parkinson’s disease a prion disorder? Proc. Natl Acad. Sci. USA 106, 12571–12572 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tolosa E, Wenning G & Poewe W The diagnosis of Parkinson’s disease. Lancet Neurol. 5, 75–86 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Roostaee A, Beaudoin S, Staskevicius A & Roucou X Aggregation and neurotoxicity of recombinant α-synuclein aggregates initiated by dimerization. Mol. Neurodegener 8, 5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Åslund A et al. Novel pentameric thiophene derivatives for in vitro and in vivo optical imaging of a plethora of protein aggregates in cerebral amyloidoses. ACS Chem. Biol 4, 673–684 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klingstedt T et al. Distinct spacing between anionic groups: an essential chemical determinant for achieving thiophene-based ligands to distinguish β-amyloid or tau polymorphic aggregates. Chemistry 21, 9072–9082 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shirani H et al. Synthesis of thiophene-based optical ligands that selectively detect tau pathology in Alzheimer’s disease. Chemistry 23, 17127–17135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirani H et al. A palette of fluorescent thiophene-based ligands for the identification of protein aggregates. Chemistry 21, 15133–15137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mastronarde DN & Held SR Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol 197, 102–113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ludtke SJ, Baldwin PR & Chiu W EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol 128, 82–97 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Pettersen EF et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and/or analysed during this study are included in the Article, Supplementary Fig. 1 (uncropped blots) and the Source Data files for Figs. 1, 3, 4 and Extended Data Figs. 1–3. Any additional information required are available from the corresponding author on reasonable request.