Abstract
Previous Mendelian randomization (MR) studies have yielded a conflicting causal relationship between sarcopenia and coronary artery disease (CAD), and lack the association of CAD with sarcopenia. We performed a bi-directional MR approach to clarify the causality and causal direction between sarcopenia-related traits and CAD. In stage 1 analysis, estimates of inverse variance weighting (IVW) and several sensitivity analyses were obtained by applying genetic variants that predict sarcopenia-related traits to CAD. Conversely, we also applied genetic variants that predict CAD to sarcopenia-related traits in stage 2 analyses. IVW analysis showed that higher handgrip strength reduces risk for CAD: A 1-kilogram (kg) increase in genetically determined left handgrip strength reduced odds of CAD by 36% [odds ratio (OR) = 0.64, 95% confidence interval (CI) 0.498 - 0.821, p = 4.56E-04], and right handgrip strength reduced odds of CAD by 41.1% (OR = 0.599, 95% CI 0.476 - 0.753, p = 1.10E-05). However, genetically predicted CAD did not show any causal association with handgrip strength, and no significant causal relationship was detected between genetically instrumented body lean mass and CAD. Our results suggest that decreased muscle strength but not decreased muscle mass leads to the increased risk of CAD in sarcopenia.
Keywords: Mendelian randomization, sarcopenia, coronary artery disease
INTRODUCTION
Sarcopenia is a common clinical condition characterized by reduced muscle mass and muscle strength with aging [1]. Morbidity and mortality from cardiovascular disease (CVD) are increasing worldwide due to aging [2]. Coronary artery disease (CAD) is the most common form of CVD and a major global health problem that imposes a great economic burden [2]. A series of observational studies have examined the association between sarcopenia and CVD; however, no consensus has been reached [3–6]. A recent meta-analysis study illustrated that handgrip strength was an independent predictor of CVD in community-dwelling populations [3], but this association disappeared after adjusting for baseline CVD risk factors in another prospective cohort study [4]. Furthermore, reverse association between muscle mass and CAD has also been observed in traditional studies [5, 6]. A possible mechanism underlying these contradictory results might be the association between both CAD and sarcopenia with high body fat mass [7, 8] and other unmeasured potential confounders. Less muscle mass reduces total energy expenditure, which might lead to higher fat mass and a subsequent increase in risk of CAD. On the other hand, accumulated body fat mass induces chronic inflammation, which further contributes to the development and progression of sarcopenia and CAD [7–9].
Mendelian randomization (MR) analysis has been widely performed to investigate the potential causality between genetically predicted environmental factors and diseases independent of the factors that might confound observational studies [10–12]. MR studies have also yielded a conflicting causal relationship between sarcopenia-related traits (handgrip strength) and CVD [13–16]. A two-sample MR (TSMR) study reported that increased handgrip strength was causally related to lower risk of CAD [13], and similar results were also obtained between handgrip strength and CVD in UK individuals with one-sample MR analysis [15, 16]. However, another TSMR analysis did not find causal evidence between handgrip strength and CAD [14]. To our knowledge, no MR analysis has been conducted between muscle mass and CAD, nor was the MR study focused on the association of CAD on sarcopenia. Therefore, we performed a bi-directional MR approach to clarify the causality and causal direction between sarcopenia-related traits (muscle mass and muscle strength) and CAD.
RESULTS
Stage 1: Influence of genetically predicted sarcopenia-related traits on CAD
In total, we obtained 399, 81, and 95 linkage disequilibrium (LD)-independent (r2 < 0.001) instrumental variables (IVs) that achieved genome-wide significance level (p < 5×10−8) from body lean mass, left handgrip strength, and right handgrip strength, respectively (Supplementary Table 1). F statistics are presented to demonstrate the strength of selected IVs. The F statistics for selected IVs and the variance explained by them for body lean mass, left handgrip strength and right handgrip strength were 81.02 and 8.9%, 50.34 and 1.1%, and 46.55 and 1.3%, respectively. F statistics were larger than 10, indicating that selected IVs were powerful enough to mitigate any potential bias of the causal IVs estimate. The negative control analysis results showed that body lean mass, left handgrip strength and right handgrip strength were not related to myopia, indicating the selected IVs of exposures were appropriate (Supplementary Tables 4–6).
As the heterogeneity test showed significant heterogeneity among selected IVs (p < 0.05, Table 1), inverse variance weight (IVW) in a random-effects model was used. IVW analysis showed that higher handgrip strength reduces risk for CAD: A 1-kg increase in genetically determined left handgrip strength reduced odds of CAD by 36% [odds ratio (OR) = 0.640, 95% confidence interval (CI) 0.498- 0.821, p = 4.56E-04], and a 1-kg increase in genetically determined right handgrip reduced odds of CAD by 41.1% (OR = 0.599, 95% CI 0.476- 0.753, p = 1.10E-05) (Table 1). The MR-Egger analysis did not detect any pleiotropy for the selected IVs (left handgrip strength, intercept = 0.008, p = 0.307; right handgrip strength, intercept = 0.004, p = 0.524) (Table 1). Consistent with the IVW results, all sensitivity analyses identified significant causal effect of handgrip strength on CAD (Tables 2 and 3).
Table 1. Association between sarcopenia-related traits and CAD using MR Egger and IVW analysis.
| Exposures | Outcomes | IVs selection a | No. of IVs | Heterogeneity test | MR Egger | IVW (random-effect model) | |
| Cochran's Q (p) | Intercept (p) | OR (95% CI) | p | ||||
| Body lean mass | CAD | All | 399 | 842.005 (<0.001) | -0.001 (0.65) | 0.929 (0.834, 1.033) | 0.170 |
| Removed 1 | 398 | 830.824 (<0.001) | -0.001 (0.548) | 0.924 (0.831, 1.027) | 0.144 | ||
| Removed 2 | 387 | 805.502 (<0.001) | 0 (0.957) | 0.901 (0.808, 1.005) | 0.061 | ||
| Left handgrip strength | CAD | All | 81 | 156.954 (<0.001) | 0.008 (0.307) | 0.640 (0.498, 0.821) | 4.56E-04* |
| Removed 1 | 79 | 149.928 (<0.001) | 0.007 (0.330) | 0.645 (0.502, 0.829) | 0.001* | ||
| Removed 2 | 76 | 148.771 (<0.001) | 0.009 (0.284) | 0.647 (0.498, 0.841) | 0.001* | ||
| Right handgrip strength | CAD | All | 95 | 180.1231 (<0.001) | 0.004 (0.524) | 0.599 (0.476, 0.753) | 1.10E-05* |
| Removed 1 | 93 | 173.892 (<0.001) | 0.004 (0.513) | 0.606 (0.482, 0.762) | 0* | ||
| Removed 2 | 91 | 172.782 (<0.001) | 0.006 (0.408) | 0.604 (0.477, 0.765) | 0* | ||
| CAD | Body lean mass | All | 39 | 898.685 (<0.001) | 0.003 (0.366) | 0.992 (0.963, 1.023) | 0.618 |
| Removed 1 | 36 | 893.680 (<0.001) | 0.003 (0.379) | 0.992 (0.976, 1.007) | 0.616 | ||
| CAD | Left handgrip strength | All | 39 | 110.459 (<0.001) | 0 (0.913) | 0.997 (0.985, 1.009) | 0.642 |
| Removed 1 | 36 | 109.932 (<0.001) | 0 (0.886) | 0.997 (0.99, 1.003) | 0.62 | ||
| CAD | Right handgrip strength | All | 39 | 102.492 (<0.001) | -0.001 (0.695) | 0.996 (0.984, 1.008) | 0.531 |
| Removed 1 | 36 | 101.680 (<0.001) | 0 (0.667) | 0.996 (0.99, 1.002) | 0.505 | ||
a‘All’ represents analyses with all selected IVs; ‘Removed 1’ represents analyses after removing proxy IVs; ‘Removed 2’ represents analyses after removing potential pleiotropic SNPs (related to body fat mass and height).
Bonferroni corrected significance level (0.05/3 = 0.016) was used to correct for multiple comparisons.
*p<0.016.
CAD: coronary artery disease; MR: mendelian randomization; IVW: inverse variance weighted; IVs: instrumental variables; CI: confidence interval; OR: odds ratio.
Table 2. Association between sarcopenia-related traits and CAD using weighted median and RAPS analysis.
| Exposures | Outcomes | IVs selection a | No. of IVs | Weighted median | RAPS | ||
| OR (95% CI) | p | OR (95% CI) | p | ||||
| Body lean mass | CAD | All | 399 | 0.961 (0.84, 1.097) | 0.554 | 0.919 (0.827, 1.019) | 0.110 |
| Removed 1 | 398 | 0.959 (0.844, 1.091) | 0.524 | 0.916 (0.825, 1.015) | 0.095 | ||
| Removed 2 | 387 | 0.941 (0.827, 1.07) | 0.356 | 0.89 (0.8, 0.991) | 0.033 | ||
| Left handgrip strength | CAD | All | 81 | 0.631 (0.47, 0.849) | 0.002* | 0.642 (0.493, 0.836) | 0.001* |
| Removed 1 | 79 | 0.614 (0.461, 0.817) | 0.001* | 0.649 (0.498, 0.846) | 0.001* | ||
| Removed 2 | 76 | 0.638 (0.478, 0.85) | 0.002* | 0.645 (0.489, 0.851) | 0.002* | ||
| Right handgrip strength | CAD | All | 95 | 0.61 (0.466, 0.799) | 3.32E-04* | 0.593 (0.466, 0.757) | 2.55E-05* |
| Removed 1 | 93 | 0.613 (0.470, 0.800) | 0* | 0.6 (0.471, 0.764) | 0* | ||
| Removed 2 | 91 | 0.673 (0.515, 0.880) | 0.004* | 0.592 (0.46, 0.76) | 0* | ||
| CAD | Body lean mass | All | 39 | 0.998 (0.987, 1.008) | 0.685 | 0.988 (0.968, 1.008) | 0.234 |
| Removed 1 | 36 | 0.997 (0.992, 1.002) | 0.563 | 0.986 (0.976, 0.996) | 0.199 | ||
| CAD | Left handgrip strength | All | 39 | 0.995 (0.983, 1.007) | 0.390 | 0.994 (0.983, 1.005) | 0.261 |
| Removed 1 | 36 | 0.995 (0.989, 1) | 0.382 | 0.993 (0.987, 0.999) | 0.232 | ||
| CAD | Right handgrip strength | All | 39 | 1.002 (0.99, 1.014) | 0.751 | 0.993 (0.982, 1.004) | 0.218 |
| Removed 1 | 36 | 1.001 (0.995, 1.007) | 0.877 | 0.992 (0.987, 0.998) | 0.188 | ||
a‘All’ represents analyses with all selected IVs; ‘Removed 1’ represents analyses after removing proxy IVs; ‘Removed 2’ represents analyses after removing potential pleiotropic SNPs (related to body fat mass and height).
Bonferroni corrected significance level (0.05/3 = 0.016) was used to correct for multiple comparisons.
*p<0.016.
CAD: coronary artery disease; RAPS: robust adjusted profile score; IVs: instrumental variables; CI: confidence interval; OR: odds ratio.
Table 3. Association between sarcopenia-related traits and CAD using MR-PRESSO analysis.
| Exposures | Outcomes | IVs selection a | No. of IVs | MR-PRESSO | ||
| OR (95% CI) | p | IVs outliers | ||||
| Body lean mass | CAD | All | 399 | 0.919 (0.834, 1.012) | 0.0866 | rs11065979, rs216193, rs2289976, rs2678204, rs28391281, rs3843751, rs66922415, rs7097872, rs7781964 |
| Removed 1 | 398 | 0.996 (0.986, 1.007) | 0.494 | rs11065979, rs216193, rs2289976, rs2678204, rs28391281, rs3843751, rs66922415, rs7097872, rs7781964 | ||
| Removed 2 | 387 | 0.894 (0.808, 0.989) | 0.030 | rs11065979, rs216193, rs2678204, rs28391281, rs3843751, rs66922415, rs7097872 | ||
| Left handgrip strength | CAD | All | 81 | 0.663 (0.521, 0.843) | 0.001* | rs11130333 |
| Removed 1 | 79 | 0.669 (0.526, 0.852) | 0.002* | rs11130333 | ||
| Removed 2 | 76 | 0.672 (0.523, 0.865) | 0.003* | rs11130333 | ||
| Right handgrip strength | CAD | All | 95 | 0.599 (0.476, 0.753) | 2.9E-05* | No significant outliers |
| Removed 1 | 93 | 0.606 (0.482, 0.762) | 4.3E-05* | No significant outliers | ||
| Removed 2 | 91 | 0.604 (0.477, 0.765) | 6.81E-05* | No significant outliers | ||
| CAD | Body lean mass | All | 39 | 0.997 (0.986, 1.008) | 0.575 | rs10840293, rs11065979, rs11191416, rs17087335, rs2681472, rs3918226, rs56289821, rs663129, rs7528419 |
| Removed 1 | 36 | 0.995 (0.99, 1) | 0.381 | rs10840293, rs11065979, rs11191416, rs17087335, rs2681472, rs3918226, rs56289821, rs663129, rs7528419 | ||
| CAD | Left handgrip strength | All | 39 | 0.992 (0.983, 1) | 0.052 | rs2519093, rs663129, rs9349379 |
| Removed 1 | 36 | 0.991 (0.987, 0.995) | 0.049 | rs2519093, rs663129, rs9349379 | ||
| CAD | Right handgrip strength | All | 39 | 0.992 (0.983, 1) | 0.064 | rs2519093, rs663129, rs9349379 |
| Removed 1 | 36 | 0.991 (0.987, 0.995) | 0.058 | rs2519093, rs663129, rs9349379 | ||
MR-PRESSO analysis was only carried out for associations with a significant global test p value (p < 0.05).
a‘All’ represents analyses with all selected IVs; ‘Removed 1’ represents analyses after removing proxy IVs; ‘Removed 2’ represents analyses after removing potential pleiotropic SNPs (related to body fat mass and height).
Bonferroni corrected significance level (0.05/3 = 0.016) was used to correct for multiple comparisons.
*p<0.016.
CAD: coronary artery disease; MR-PRESSO: Mendelian Randomization Pleiotropy RESidual Sum and Outlier; IVs: instrumental variables; CI: confidence interval; OR: odds ratio.
After removing the proxy IVs and the potential pleiotropic IVs, both left and right handgrip strength were still negatively associated with CAD (Tables 1–3, Supplementary Table 1). However, our standard IVW and sensitivity analysis did not detect a significant association between genetically determined body lean mass and CAD (Tables 1–3).
Stage 2: Influence of genetically predicted CAD on sarcopenia-related traits
In total, we obtained 39 LD-independent (r2 < 0.001) IVs that achieved genome-wide significance level (p < 5×10−8) from CAD (Supplementary Table 2). F statistics are presented to demonstrate the strength of the relationship between IVs and sarcopenia-related traits. The F statistics for selected IVs of CAD was larger than 10 (F = 64.17), and variance explained by these IVs is shown in Supplementary Table 3. Further, the negative control analysis results showed that CAD was not related to myopia, indicating the selected IVs of exposure were appropriate (Supplementary Tables 4–6).
As the heterogeneity test showed significant heterogeneity among the selected IVs (p < 0.05, Table 1), IVW in random-effects model was used. We did not detect a relationship between the genetically instrumented CAD and sarcopenia-related traits (body lean mass, left handgrip strength and right handgrip strength) in either IVW analysis or sensitivity analysis, even after removing the proxy IVs (Tables 1–3).
DISCUSSION
In the current study, by applying bi-directional MR analysis, we successfully confirmed that increased handgrip strength was associated with decreased risk of CAD. However, no significant causal effect of CAD on handgrip strength was observed. Additionally, no significant causal relationship was detected between genetically determined body lean mass and CAD. To our knowledge, this is the first bi-directional MR study conducted on the topic of sarcopenia and CAD, simultaneously considering both muscle mass and muscle strength and the potential causality and reverse causality between sarcopenia and CAD.
The causal relationship of muscle strength with CAD has been discussed in previous TSMR studies [13, 14]. Xu and Hao’s TSMR analysis applied two genetic variants that predict handgrip strength in a European population to CAD in a mixed population of various ethnicities, indicating that increased handgrip reduced the risk of CAD [13]. However, another MR analysis did not find evidence for causality in the associations between handgrip strength (European population) and CAD (mixed population). A total of 20 genetic variants were selected to predict handgrip strength in this study [14]. These inconsistent results may be caused by different ancestry of exposure (handgrip strength) and outcome (CAD), the number of genetic variants selected as IVs, different methods applied, and even potential pleiotropic confounding IVs (body fat mass-related genetics variables). Compared to these two previous TSMR analyses [13, 14], more selected IVs were utilized to increase the interpretation of exposures. Several sensitivity analyses were performed to reduce the potential pleiotropic IVs, and results indicated a decreased causal effect of handgrip strength on risk of CAD. Epidemiological evidence has also suggested that muscle strength is negatively associated with CVD events and mortality [15], and similar results were also obtained between handgrip strength and CVD in UK individuals with one-sample MR analysis [15, 16].
Furthermore, a serial of traditional studies have frequently examined the association between muscle mass and CVD; however, they reached conflicting conclusions [5, 6, 17]. Previous studies indicated that the conflicting association of muscle mass with CVD might be confounded by body fat mass [7, 8] and subtypes of CVD (CAD, stroke, heart failure and atrial fibrillation). Other studies found evidence for low muscle mass as an independent risk factor of CAD [5], while inverse results were observed in obese postmenopausal women [6]. A possible mechanism underlying these contrasting association results between muscle mass and CVD might be a confounding common risk factor of high body fat mass [7, 8]. Accumulated body fat mass induces chronic inflammation, which further contributes to the development and progression of sarcopenia and CAD [7–9]. Unfortunately, few previous studies have been conducted to determine the causation between muscle mass and CAD, especially in the context of robust MR analysis for causal inference. In this study, we used a bi-directional MR study to make up for the lack of causal relationship between muscle mass and CAD, suggesting there is no significant causal relationship between genetically determined muscle mass and CAD even after removing genetic variants associated with potential confounders (body fat mass, body fat percentage and body fat distribution).
The main strength of this study is the application of bidirectional MR design, which is more robust to confounding and reverse causation and could clarify the causal direction between sarcopenia-related traits (muscle mass and muscle strength) and CAD. Secondly, we included absolute handgrip strength, which might be more appropriate to assess muscle strength than relative handgrip strength (handgrip strength/body weight), as relative grip strength may not only represent a change in muscle strength but also a change in body fat mass and bone mass. Additionally, several sensitivity analyses were performed to ensure the valid estimation of true MR causal effect size. Furthermore, as sarcopenia and CAD are closely related to body fat mass, we also detected and removed the potential pleiotropic effects introduced by those SNPs associated with body fat mass-related traits (body fat mass, body fat distribution and body fat percentage) by obtaining their association effect size from the GWAS Catalog (https://www.ebi.ac.uk/gwas) [18]. Finally, both sarcopenia and CAD unrelated trait (myopia) was used as negative control to demonstrate the validity of selected IVs, further validating our analyses and results.
This study has some potential limitations. Firstly, we used body lean mass rather than appendicular lean mass to measure muscle mass, which might be not exactly appropriate, as the measurement could be biased by other components of non-fat soft tissue, such as lungs, liver, and other viscera. Moreover, compared with absolute muscle mass, relative muscle mass (appendicular lean mass/height2) may be more appropriate to measure muscle mass [1]. Therefore, the conclusion of no significant association between muscle mass and CAD might not be easily generalized. Secondly, it is important to acknowledge that MR analysis has several other potential limitations [19], and while using multiple genetic instruments improves the power of MR, there is always some risk of pleiotropy despite extensive sensitivity analyses. Therefore, Mendelian Randomization Pleiotropy RESidual Sum and Outlier (MR-PRESSO) was performed to identify and remove the possible pleiotropic genetic variables and provide outlier-adjusted estimates, which should have minimized this possibility. Thirdly, we believe that sex- and age-specific analysis would be helpful, given the difference in incidence of sarcopenia and CAD by age and sex. As we only used summary statistics and had no access to the original individual measures, sex- and age-specific analysis may be difficult to perform. Fortunately, age and sex were all adjusted in genetics association of CARDIoGRAMplusCD4 and UK Biobank consortium [20–22], which should have minimized this possibility. Finally, as we only used summary statistics and had no access to the original individual measures, different standards of quality control and selection in individual-level GWAS may affect our results. Therefore, the results may not be easily generalized.
In summary, we found that increased genetically predicted muscle strength was causally associated with decreased risk of CAD, yet we did not identify a significant causal effect of CAD on muscle strength. Moreover, no significant causal association between muscle mass and CAD was observed in bi-directional MR analysis. Our results suggest that decreased muscle strength, but not decreased muscle mass, leads to the increased risk of CAD in sarcopenia.
MATERIALS AND METHODS
Study design
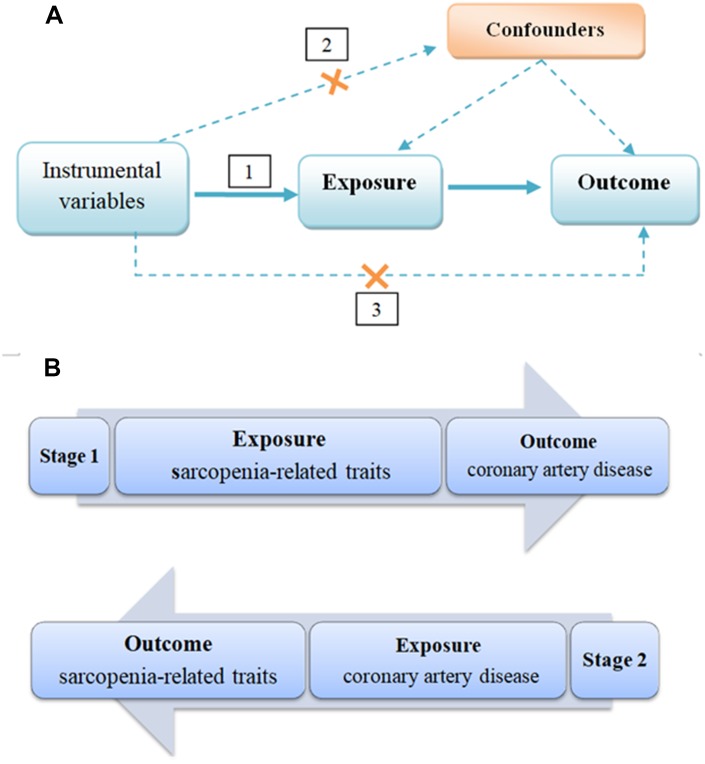
The genetic variants used as IVs in TSMR analysis must satisfy three assumptions as follows (Figure 1A): 1) IVs are strongly associated with exposure. 2) The IVs are independent of any known confounders. 3) The selected IVs are conditionally independent of outcome, given exposure and potential confounders. The second and third assumptions are known as independence from pleiotropy [23]. In the current study, we performed a bi-directional MR study design to clarify the causal association between sarcopenia-related traits (muscle mass and muscle strength) and CAD with independent GWAS summary-level dataset (Figure 1B). In the first stage, we assessed whether sarcopenia-related traits were causally related to CAD. In the second stage, we also examined whether genetically driven CAD was causally associated with sarcopenia-related traits.
Figure 1.
Schematic Representation of Bi-directional MR Analysis. (A) illustrates three assumptions of MR analysis as follows: 1) Instrumental variables must be associated with exposure, 2) instrumental variables must not be associated with confounders, and 3) instrumental variables must influence outcome only through exposure. (B) illustrates bi-directional MR analysis. In stage 1 analysis, influence of genetically predicted sarcopenia-related traits (body lean mass, left handgrip strength and right handgrip strength) on risk of CAD was estimated. In stage 2 analysis, influence of genetically predicted CAD on risk of sarcopenia-related traits (body lean mass, left handgrip strength and right handgrip strength) was estimated. TSMR: two-sample mendelian randomization; CAD: coronary artery disease.
Sarcopenia-related traits
Bioelectrical impedance analysis (BIA) was used to measure body lean mass (kilogram, kg) as a valid alternative for the assessment of body composition [24]. Body lean mass represents lipid-free soft tissue including muscle mass, body water, protein, glycerol and soft tissue mineral mass [25], which is considered to be a valid measure of muscle mass [26].
Handgrip strength (kg) was measured by a calibrated hydraulic hand dynamometer adjusted for hand size [20]. We used absolute rather than relative handgrip strength (absolute handgrip strength/weight) as a proxy for muscle strength, because absolute handgrip strength might have a higher correlation with muscle strength than relative grip strength [27].
Data sources
GWAS summary statistics for body lean mass (n = 331, 291) and handgrip strength (left hand, n = 335, 821; right hand, n = 335, 842) were obtained from the UK biobank study [20], and CAD summary statistics (n = 184, 305) were obtained from the CARDIoGRAMplusC4D consortium [21]. Sarcopenia-related traits’ GWAS summary statistics of UK Biobank were conducted on European populations from population-based cohorts, and age, age2, sex, sex × age and sex × age2 were adjusted for in genetic association analysis [20]. CARDIoGRAMplusC4D Genomes-based GWAS summary statistics is a meta-analysis of 48 GWAS studies involving 60,801 CAD cases and 123,504 controls from European (~ 74%) and Asian (~ 26%) subjects, adjusted for sex and age in analyses of individual GWASs [21, 22].
Selection and validation of IVs
To satisfy the three assumptions of MR (Figure 1A), we firstly selected independent (linkage disequilibrium r2 < 0.001) [28] SNPs that were strongly (p < 5 × 10−8) associated with exposure. Then, we obtained the corresponding effect estimates of these SNPs in outcome GWAS summary statistics. For the SNPs that were not available in the outcome, we used proxy SNPs that were highly correlated (r2 > 0.8) with the requested SNPs.
Additionally, MR-Egger regression was applied to assess the horizontal pleiotropy of selected IVs [23], and the intercept that deviates from the origin may provide evidence for potential pleiotropic effects across the IVs. We also performed MR-PRESSO to identify and remove pleiotropic IVs [29]. To further validate the strength of the selected IVs, we computed the F statistic of selected IVs using an online tool (https://sb452.shinyapps.io/overlap) [30]. F statistics greater than 10 are often considered as powerful enough to mitigate any potential bias of the causal IV estimate. Finally, MR-Steiger test was conducted to calculate the pooled variance explained in exposure and outcome by the selected IVs (r2) [31], and to further assess whether the variance explained in the exposure is larger than that in the outcome.
MR analysis
IVW was conducted to estimate the causal effect between exposure and outcome, which was calculated as the SNP-outcome association effect size divided by the SNP-exposure association effect size [32]. The causal effect β was estimated as wi (αi/γi), where i refers to the ith IV, αi defines as the association effect of IVs on exposure, γi represents the association effect of IVs on outcome, and wi means the weights of the causal effect of exposure on outcome [32]. The IVW method was considered the most reliable indicator if there was no evidence of directional pleiotropy (p for MR-Egger intercept > 0.05) among the selected IVs [33, 34]. Given the multiple testing situation (body lean mass, left handgrip strength and right handgrip strength were included), we used a conservative approach and applied a Bonferroni corrected significance level of 0.016 (0.05/3).
Sensitivity analysis
To further ensure the valid estimation of the true MR causal effect, we conducted several sensitivity analyses. Firstly, weighted median estimate was performed because it tends to provide valid estimates when at least 50% of information is derived from valid IVs [35]. Secondly, the MR might fail if the selected IVs are weak instruments. Therefore, we carried out a recently proposed method called Robust Adjusted Profile Score (RAPS), which considers the measurement error in SNP-exposure effects and is unbiased even when there are many (e.g. hundreds of) weak instruments [36]. Also, RAPS is robust to systematic pleiotropy. Thirdly, MR-PRESSO was performed to identify and remove the possible pleiotropic IVs, assuming that at least 50% of the IVs were valid IVs [29]. MR-PRESSO can also identify outlier IVs and provides outlier-adjusted estimates. MR-PRESSO detects pleiotropy by assessing outliers among the selected IVs contributing to the MR estimate and provides adjusted estimates. MR-PRESSO analysis was only carried out for associations with a significant global test p value (p < 0.05). Next, in order to guarantee that the MR estimates were not influenced by the inclusion of proxy SNPs, we repeated the analysis after excluding proxy SNPs. Finally, as muscle mass and CAD were closely related to body fat mass, we intend to exclude the potential pleiotropic effects introduced by those SNPs associated with body fat-related traits (body fat distribution and body fat percentage) by obtaining their association effect size from GWAS Catalog (https://www.ebi.ac.uk/gwas) [18].
Negative control
To further demonstrate the validity of the selected IVs, we included myopia as negative control in our analysis, as there is little evidence presented that sarcopenia and CAD are associated with myopia. The summary statistics of myopia were derived from UK Biobank imputed genotype data, including 335,700 individuals of European descent [20].
All the analyses were performed using R statistical software (Version 3.4.2) with the R packages “TwosampleMR”, “MendelianRandomization” and “MRPRESSO”.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank MR-base dataset for providing relevant publicly available summary statistics. Thanks to all the consortiums that provide public data, which have been mentioned in the data sources section in this study.
Footnotes
AUTHOR CONTRIBUTIONS: Hui-Min Liu as the first author performed data analysis and wrote the manuscript. Qiang Zhang, Wen-Di Shen, Bo-Yang Li, Wan-Qian Lv and Hong-Mei Xiao gave constructive suggestions during the process. Hong-Mei Xiao provided critical revisions. Hui-Min Liu and Hong-Wen Deng conceived and initiated this project, revised and finalized the manuscript. All authors read and approved the final manuscript.
CONFLICTS OF INTEREST: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflicts of interest.
FUNDING: This study was partially supported by the Fundamental Research Funds for the Central Universities of Central South University and was partially supported by National Key R&D Program of China (2017YFC1001100 and 2016YFC1201805), National Natural Science Foundation of China (81471453 and 81501248), Natural Science Foundation of Hunan Province of China (2015JJ2166 and 2017JJ3425), and construction project of the center of reproductive health, Central South University (164990007). Hong-Wen Deng was partially supported by grants from the NIH (R01-AR069055, U19-AG055373, R01-MH104680, R01-AR059781, and P20-GM109036) and Edward G. Schlieder Endowment fund at Tulane University.
REFERENCES
- 1.Cruz-Jentoft AJ, Bahat G, Bauer J, Boirie Y, Bruyère O, Cederholm T, Cooper C, Landi F, Rolland Y, Sayer AA, Schneider SM, Sieber CC, Topinkova E, et al. , and Writing Group for the European Working Group on Sarcopenia in Older People 2 (EWGSOP2), and the Extended Group for EWGSOP2. Sarcopenia: revised European consensus on definition and diagnosis. Age Ageing. 2019; 48:16–31. 10.1093/ageing/afy169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roth GA, Forouzanfar MH, Moran AE, Barber R, Nguyen G, Feigin VL, Naghavi M, Mensah GA, Murray CJ. Demographic and epidemiologic drivers of global cardiovascular mortality. N Engl J Med. 2015; 372:1333–41. 10.1056/NEJMoa1406656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu Y, Wang W, Liu T, Zhang D. Association of Grip Strength With Risk of All-Cause Mortality, Cardiovascular Diseases, and Cancer in Community-Dwelling Populations: A Meta-analysis of Prospective Cohort Studies. J Am Med Dir Assoc. 2017; 18:551.e17–551.e35. 10.1016/j.jamda.2017.03.011 [DOI] [PubMed] [Google Scholar]
- 4.Gubelmann C, Vollenweider P, Marques-Vidal P. No association between grip strength and cardiovascular risk: the CoLaus population-based study. Int J Cardiol. 2017; 236:478–82. 10.1016/j.ijcard.2017.01.110 [DOI] [PubMed] [Google Scholar]
- 5.Ko BJ, Chang Y, Jung HS, Yun KE, Kim CW, Park HS, Chung EC, Shin H, Ryu S. Relationship Between Low Relative Muscle Mass and Coronary Artery Calcification in Healthy Adults. Arterioscler Thromb Vasc Biol. 2016; 36:1016–21. 10.1161/ATVBAHA.116.307156 [DOI] [PubMed] [Google Scholar]
- 6.Aubertin-Leheudre M, Lord C, Goulet ED, Khalil A, Dionne IJ. Effect of sarcopenia on cardiovascular disease risk factors in obese postmenopausal women. Obesity (Silver Spring). 2006; 14:2277–83. 10.1038/oby.2006.267 [DOI] [PubMed] [Google Scholar]
- 7.Lee K. Muscle Mass and Body Fat in Relation to Cardiovascular Risk Estimation and Lipid-Lowering Eligibility. J Clin Densitom. 2017; 20:247–55. 10.1016/j.jocd.2016.07.009 [DOI] [PubMed] [Google Scholar]
- 8.Kim TN, Choi KM. The implications of sarcopenia and sarcopenic obesity on cardiometabolic disease. J Cell Biochem. 2015; 116:1171–78. 10.1002/jcb.25077 [DOI] [PubMed] [Google Scholar]
- 9.Pearson TA, Mensah GA, Alexander RW, Anderson JL, Cannon RO 3rd, Criqui M, Fadl YY, Fortmann SP, Hong Y, Myers GL, Rifai N, Smith SC Jr, Taubert K, et al. , and Centers for Disease Control and Prevention, and American Heart Association. Markers of inflammation and cardiovascular disease: application to clinical and public health practice: A statement for healthcare professionals from the Centers for Disease Control and Prevention and the American Heart Association. Circulation. 2003; 107:499–511. 10.1161/01.CIR.0000052939.59093.45 [DOI] [PubMed] [Google Scholar]
- 10.Katan MB. Apolipoprotein E isoforms, serum cholesterol, and cancer. 1986. Int J Epidemiol. 2004; 33:9. 10.1093/ije/dyh312 [DOI] [PubMed] [Google Scholar]
- 11.Boef AG, Dekkers OM, le Cessie S. Mendelian randomization studies: a review of the approaches used and the quality of reporting. Int J Epidemiol. 2015; 44:496–511. 10.1093/ije/dyv071 [DOI] [PubMed] [Google Scholar]
- 12.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008; 27:1133–63. 10.1002/sim.3034 [DOI] [PubMed] [Google Scholar]
- 13.Xu L, Hao YT. Effect of handgrip on coronary artery disease and myocardial infarction: a Mendelian randomization study. Sci Rep. 2017; 7:954. 10.1038/s41598-017-01073-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willems SM, Wright DJ, Day FR, Trajanoska K, Joshi PK, Morris JA, Matteini AM, Garton FC, Grarup N, Oskolkov N, Thalamuthu A, Mangino M, Liu J, et al. , and GEFOS Any-Type of Fracture Consortium. Large-scale GWAS identifies multiple loci for hand grip strength providing biological insights into muscular fitness. Nat Commun. 2017; 8:16015. 10.1038/ncomms16015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tikkanen E, Gustafsson S, Ingelsson E. Associations of Fitness, Physical Activity, Strength, and Genetic Risk With Cardiovascular Disease: Longitudinal Analyses in the UK Biobank Study. Circulation. 2018; 137:2583–91. 10.1161/CIRCULATIONAHA.117.032432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farmer RE, Mathur R, Schmidt AF, Bhaskaran K, Fatemifar G, Eastwood SV, Finan C, Denaxas S, Smeeth L, Chaturvedi N. Associations Between Measures of Sarcopenic Obesity and Risk of Cardiovascular Disease and Mortality: A Cohort Study and Mendelian Randomization Analysis Using the UK Biobank. J Am Heart Assoc. 2019; 8:e011638. 10.1161/JAHA.118.011638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanada K, Miyachi M, Tanimoto M, Yamamoto K, Murakami H, Okumura S, Gando Y, Suzuki K, Tabata I, Higuchi M. A cross-sectional study of sarcopenia in Japanese men and women: reference values and association with cardiovascular risk factors. Eur J Appl Physiol. 2010; 110:57–65. 10.1007/s00421-010-1473-z [DOI] [PubMed] [Google Scholar]
- 18.MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, Junkins H, McMahon A, Milano A, Morales J, Pendlington ZM, Welter D, Burdett T, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017; 45:D896–901. 10.1093/nar/gkw1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haycock PC, Burgess S, Wade KH, Bowden J, Relton C, Davey Smith G. Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies. Am J Clin Nutr. 2016; 103:965–78. 10.3945/ajcn.115.118216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015; 12:e1001779. 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, Webb TR, Zeng L, Dehghan A, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015; 47:1121–30. 10.1038/ng.3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, Goldstein BA, Stirrups K, König IR, Cazier JB, et al. , and CARDIoGRAMplusC4D Consortium, and DIAGRAM Consortium, and CARDIOGENICS Consortium, and MuTHER Consortium, and Wellcome Trust Case Control Consortium. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013; 45:25–33. 10.1038/ng.2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015; 44:512–25. 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sergi G, De Rui M, Stubbs B, Veronese N, Manzato E. Measurement of lean body mass using bioelectrical impedance analysis: a consideration of the pros and cons. Aging Clin Exp Res. 2017; 29:591–97. 10.1007/s40520-016-0622-6 [DOI] [PubMed] [Google Scholar]
- 25.Zillikens MC, Demissie S, Hsu YH, Yerges-Armstrong LM, Chou WC, Stolk L, Livshits G, Broer L, Johnson T, Koller DL, Kutalik Z, Luan J, Malkin I, et al. Large meta-analysis of genome-wide association studies identifies five loci for lean body mass. Nat Commun. 2017; 8:80. 10.1038/s41467-017-00031-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Visser M, Fuerst T, Lang T, Salamone L, Harris TB. Validity of fan-beam dual-energy X-ray absorptiometry for measuring fat-free mass and leg muscle mass. Health, Aging, and Body Composition Study--Dual-Energy X-ray Absorptiometry and Body Composition Working Group. J Appl Physiol (1985). 1999; 87:1513–20. 10.1152/jappl.1999.87.4.1513 [DOI] [PubMed] [Google Scholar]
- 27.Wind AE, Takken T, Helders PJ, Engelbert RH. Is grip strength a predictor for total muscle strength in healthy children, adolescents, and young adults? Eur J Pediatr. 2010; 169:281–87. 10.1007/s00431-009-1010-4 [DOI] [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81:559–75. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018; 50:693–98. 10.1038/s41588-018-0099-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmer TM, Lawlor DA, Harbord RM, Sheehan NA, Tobias JH, Timpson NJ, Davey Smith G, Sterne JA. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res. 2012; 21:223–42. 10.1177/0962280210394459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017; 13:e1007081. 10.1371/journal.pgen.1007081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013; 37:658–65. 10.1002/gepi.21758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holmes MV, Ala-Korpela M, Smith GD. Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat Rev Cardiol. 2017; 14:577–90. 10.1038/nrcardio.2017.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White J, Swerdlow DI, Preiss D, Fairhurst-Hunter Z, Keating BJ, Asselbergs FW, Sattar N, Humphries SE, Hingorani AD, Holmes MV. Association of Lipid Fractions With Risks for Coronary Artery Disease and Diabetes. JAMA Cardiol. 2016; 1:692–99. 10.1001/jamacardio.2016.1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016; 40:304–14. 10.1002/gepi.21965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Q, Wang J, Hemani G, Bowden J, Small DS. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. arXiv: Stat. 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.