Abstract
The cycloartane triterpene content in the roots/rhizomes (RR) and aerial parts (PX) of Actaea racemosa (AR), A. podocarpa (AP), and A. cordifolia (AC) have been investigated by quantitative 1H NMR (qHNMR). Thereby, it was demonstrated that qHNMR represents a powerful methodology for the analysis of crude plant extracts as it does not rely on the rarely available identical reference triterpenes. Specifically, the presence of the characteristic C-19 cyclopropane (exo/endo) hydrogen signals made it possible to quantify the less common/not ubiquitously present group of cycloartane triterpenes, directly in extracts. As an example, ARPX and ARRR were shown to contain, 3.8 – 20.8% ± 8.2% and 7.2 – 19.3% ± 4.0% of cycloartane triterpenes, respectively. The cycloartane concentration in ACPX and ACRR was 7.5 – 8.7% ± 0.8% and 13.9 – 28.5% ± 7.3%, respectively, based on the weight of the extract. AP was shown to contain notably lower amounts of the cycloartane triterpenes as compared to AR and AC in the roots/rhizomes. The content for APPX and APRR was only 2.1 – 3.3% ± 0.7% and 1.1 – 4.0% ± 1.5%, respectively. In addition, an example is presented for the identification of specific cycloartanes as marker compounds for AR within crude extracts based on the same qHNMR spectra and 2D NMR methods.
Keywords: Quantitative NMR, qHNMR, Actaea racemosa L., Actaea podocarpa DC., A. cordifolia DC., Cycloartane triterpene
Graphical Abstract
1. Introduction
Black cohosh (Actaea racemosa L., AR) is a popular dietary supplement, widely used for (peri-)menopausal women’s health in the U.S. and Europe. Because plant sourcing continues to rely on natural harvest, adulteration of the plant material by related plant species is of concern. A. podocarpa DC. (AP), A. cordifolia DC. (AC) are related species, for example, that share the same habitat.
With triterpenes being the most abundant as well as the most well-studied class of constituents in Actaea plant materials, common commercial preparations of black cohosh involve powdered dry extracts, which are standardized for their content of triterpene glycosides by high-performance liquid chromatography-evaporative light-scattering detection (HPLC-ELSD); this is typically done with reference to one of the most abundant congeners, (12R)-12-acetoxy-(24R,25R)-24,25-epoxy-3-O-β-D-xylopyranosylacta-(16S,23R)-16,23;23,26-binoxoside (syn. 23-epi-26-deoxyactein, or sometimes 27-deoxyactein) (1) [1] [2]. For example, BNO 1055, a dry extract prepared with 58% ethanol, which is contained in Klimadynon® and Menofem® [3], purportedly contains 2.5% triterpene glycosides using this analytical method. In a clinical study for the management of vasomotor symptoms [4], tested AR extract was standardized to 7.27 mg of triterpenes per daily dose (two capsules) using a similar HPLC method. More precisely, 64 mg of AR 75% EtOH extract was analyzed by HPLC-ELSD and shown to contain 5.6% (3.64 mg) of major cycloartane spiroketal triterpene glycosides, such as 1.42 mg of 1, 0.36 mg of (12R)-12-acetoxy-(24R,25S)-24,25-epoxy-3-O-β-D-xylopyranosyl-acta-(16S,23R)-16,23;23,26-binoxoside (syn. 26-deoxyactein) (2), 1.38 mg of (12R)-12-acetoxy-(24R,25S)-24,25-epoxy-(26S)-26-hydroxy-3-O-β-D-xylopyranosylacta-(16S,23R)-16,23;23,26-binoxoside (syn. 26S-actein) (3), and 0.47 mg (12R)-12-acetoxy-(24R,25S)-24,25-epoxy-(26R)-26-hydroxy-3-O-β-D-xylopyranosylacta-(16S,23R)-16,23;23,26-binoxoside (syn. 26R-actein) (4). AR extracts that were used in a pharmacokinetics study contained 7.0% of the major triterpenoids. More precisely, the extract contained 4.4% of 1, 2.2% of 3, and 0.4% of 4 [5][6]. As seen in the above examples, AR extracts contain 2.5 – 7.0% of the major triterpenes. However, standardization is made to a single or several triterpenes only and typically does not consider other triterpene compounds also present. In contrast, assessment of the total triterpene content in AR extract is not feasible when using classical LC-based standardization methods.
Reports describing the use of qHNMR for analysis of natural product have increased considerably in recent years [7][8]. Together with the growing number of chemical applications, more validation studies on the use of quantitative NMR (qNMR) and quantitative conditions including acquisition and processing parameters have been proposed [9][10]. There are three principal qHNMR methods that are commonly used with regard to their approach to quantitative reference calibration, viz; 1) internal calibration (IC), 2) external calibration (EC), and 3) a hybrid of both (ECIC) [11]. Each method has its own advantages. The major advantage of EC and ECIC methods includes not having to contaminate a precious sample by introduction of a calibrant. The objective of the present study is to investigate the application of qHNMR to the analysis of AR and related species / plant part extracts for the elucidation of the total content of the cycloartane triterpenes.
2. Experimental
2.1. Plant Material
Three North American Actaea species were used for this study: A. racemosa L. (AR), A. podocarpa DC. (AP) and A. cordifolia DC. (AC). Samples were separated into the aerial parts and roots/rhizomes and plant parts coded PX and RR, respectively. A total of 23 samples (ARPX 6, ARRR 6, APPX 3, APRR 3, ACPX 2, ACRR 3) were investigated. The collected samples were originally authenticated as C. racemosa, C. americana, and C. rubifolia, respectively. However, the revised naming system was used here because the genus Cimicifuga has since been reclassified to genus Actaea and many of the species names have changed [12]. These three American species were in part selected due to the high probability of finding them intermixed during the wild collection and because of their shared habitat and morphological similarities.
As shown in Table 1, samples denoted BC624, BC404, BC582, BC626, and BC581 were cultivated and collected from the Dorothy Bradley Atkins Medicinal Plant Garden at UIC. Sample BC629 was cultivated in Chillicothe, Ohio, and the specimen was authenticated and deposited in the Herbarium of the Field Museum of Natural History, Chicago, Illinois. Other American species were acquired through wild collections in Virginia, North Carolina, and Tennessee, and the voucher specimens are housed at the Searle Herbarium, Field Museum of Natural History and the Ramsey-Freer Herbarium, Lynchburg College (Lynchburg, Virginia).
Table 1.
List of plant material used for this study.
| Genus | Species | Plant Parts a | BC number b | Collection date | Collection Site |
|---|---|---|---|---|---|
| Actaea | racemosa | PX | BC026 | 6/29/1999 | Sevier County, TN. Elevation 3500 ft. |
| Actaea | racemosa | PX | BC027 | 6/29/1999 | Sevier County, TN. Elevation 3300 |
| Actaea | racemosa | PX | BC624 | 6/16/2009 | Atkins Garden, Cook County, IL |
| Actaea | racemosa | PX | BC404 | 10/1/2008 | Atkins Garden, Cook County, IL |
| Actaea | racemosa | PX | BC582 | 10/7/2009 | Atkins Garden, Cook County, IL |
| Actaea | racemosa | PX | BC025 | 6/29/1999 | Rockbridge County, VA. Elevation 2481 ft. |
| Actaea | racemosa | RR | BC036 | 10/16/1999 | Elk township, Chester County, PA |
| Actaea | racemosa | RR | BC007 | 6/30/1999 | Sevier County, TN. Elevation 3300 ft. |
| Actaea | racemosa | RR | BC094 | 11/7/2000 | Forbes State Forest, Somerset County. PA |
| Actaea | racemosa | RR | BC629 | 6/3/2010 | Chillicothe, Ross County, OH |
| Actaea | racemosa | RR | BC626 | 6/16/2009 | Atkins Garden, Cook County, IL |
| Actaea | racemosa | RR | BC581 | 10/6/2009 | Atkins Garden, Cook County, IL |
| Actaea | podocarpa | PX | BC029 | 6/30/1999 | Sevier County, TN. Elevation 3300 ft. |
| Actaea | podocarpa | PX | BC020 | 8/26/1999 | Swain County, NC. Elevation 2875 ft. |
| Actaea | podocarpa | PX | BC028 | 6/28/1999 | Rockbridge County, VA. Elevation 3150 ft. |
| Actaea | podocarpa | RR | BC006 | 6/30/1999 | Sevier County, TN. Elevation 3300 ft. |
| Actaea | podocarpa | RR | BC008 | 6/30/1999 | Swain County, NC. Elevation 2875 ft. |
| Actaea | podocarpa | RR | BC011 | 8/26/1999 | Sevier County, TN. Elevation 3300 ft. |
| Actaea | cordifolia | PX | BC022 | 8/27/1999 | Scott County, VA, Elevation 1800 ft. |
| Actaea | cordifolia | PX | BC023 | 6/29/1999 | Scott County, VA. Elevation 1800 ft. |
| Actaea | cordifolia | RR | BC015 | 8/27/1999 | Scott County, VA, Elevation 1800 ft. |
| Actaea | cordifolia | RR | BC003 | 6/29/1999 | Scott County, VA. Elevation 1800 ft. |
| Actaea | cordifolia | RR | BC016 | 8/27/1999 | Scott County, VA, Elevation 1800 ft. |
Plant Parts codes are as follows: PX aerial parts, RR roots/rhizomes.
The BC number is a sole number assigned to each collected plant part at the University of Illinois at Chicago Botanical Center.
2.2. Sample Preparation
The extract samples were prepared by taking a 10 mL aliquot of 70% aqueous MeOH, which was added to 1 g of dried, ground plant material and the mixture sonicated for 30 minutes. The extracts were left overnight at room temperature and then filtered. The residue plant material was similarly extracted and the two filtrates combined. The combined extracts were dried in forced air, to yield the crude extract for metabolomic analysis. A volume of 650 μL of DMSO-d6 (Cambridge Isotopes, lot#8L-052, 99.9% D) was added to between 15 and 27 mg of dried extract in an Eppendorf vial, followed by sonication for 5 min. After sonication, the vial was centrifuged and 600 μL of supernatant were transferred to a 5 mm NMR tube, leaving any insoluble residue in the vial. Additionally, BC036 was also prepared in pyridine-d5 in the same fashion for the purpose of identifying reference compounds. Following sample preparation, the residual extract in the Eppendorf vial was air dried (over a longer period of time, to evaporate DMSO) and then weighed to calculate the sample concentration in the NMR tube. Exact qHNMR sample concentrations were calculated based on using 600.0 μL of the 650 μL solutions.
2.3. qNMR Analysis
One dimensional 1H and homonuclear 2D 1H NMR experiments (gCOSY) were recorded on a Bruker Avance 600 NMR spectrometer equipped with a TXI cryogenic probe with the sample temperature maintained at 25.0 °C (298.0 K). Acquisition parameters were as follows: acquisition time (aq) = 4 s, relaxation delay (d1) = 60 s, pulse width (PI) = 3 us (30° flip angle), time domain data point digitization (TD) = 144k, number of scans (NS) = 64, and dummy scans (DS) = 4.
The NMR data were processed in TopSpin (versions 3.0 and 3.2; Bruker, Karlsruhe, Germany), NUTS (Professional version 20070706; Acorn NMR Inc., San Francisco, CA), and Mnova (version 10.0.2; Mestrelab Research S.L., Santiago de Compostela, Spain). All 1H spectra were individually and identically processed with the following parameters: GM (LB −2.0 Hz, GB 0.1), zero filling to SI=512k and with application of automatic phasing, using Bruker Topspin prior to quantitation. The integral value was obtained following baseline correction with Mnova. Appropriate full auto baseline correction method was chosen within Whittaker smoothing, polynomial, ablative and cubic splines.
Quantitative qHNMR was employed to obtain the quantity of specific types of compounds in the crude extract by using the residual DMSO-d5 resonance as an internal calibrant. To make the integrated intensity of a 1H NMR signal accurately proportional to the number of observed nuclei, acquisition parameters were chosen appropriately. Accordingly, “quantitative experimental conditions” [7], including appropriate parameter selection such as relaxation delay and digitization, were carefully chosen for Actaea extract analysis. The calibration curve was generated as part of our work in the UIC/NIH Botanical Center [10], using dimethylsulfone (DMSO2) as an external calibrant (EC). The residual DMSO-d5 signal was then used as an internal calibrant (IC) for both the DMSO2 EC samples as well as the analyzed extracts. To verify the calibration curve, a mixed sample of DMSO2 and caffeine was used.
Where y is the mM concentration, and x is the ratio of the 1H integral values of the analytes relative to that of the residual DMSO-d5.
2.4. Reference compound identification
Two of the major Actaea triterpenes (12R)-12-acetoxy-(24R,25R)-24,25-epoxy-3-O-β-D-xylopyranosylacta-(16S,23R)-16,23;23,26-binoxoside (syn. 23-epi-26-deoxyactein) (1) and (12R)-12-acetoxy-(24R,25S)-24,25-epoxy-(26R&S)-26-hydroxy-3-O-β-D-xylopyranosylacta-(16S,23R)-16,23;23,26-binoxoside (syn. 26R/S-actein) (3 and 4) were obtained from USP and their spectra in DMSO-d6 were obtained on the same instrument (600 MHz) under the same conditions. Identification was performed by visual comparison of spectra of the crude extract and the reference [13][14].
3. Results and Discussion
3.1. General observations
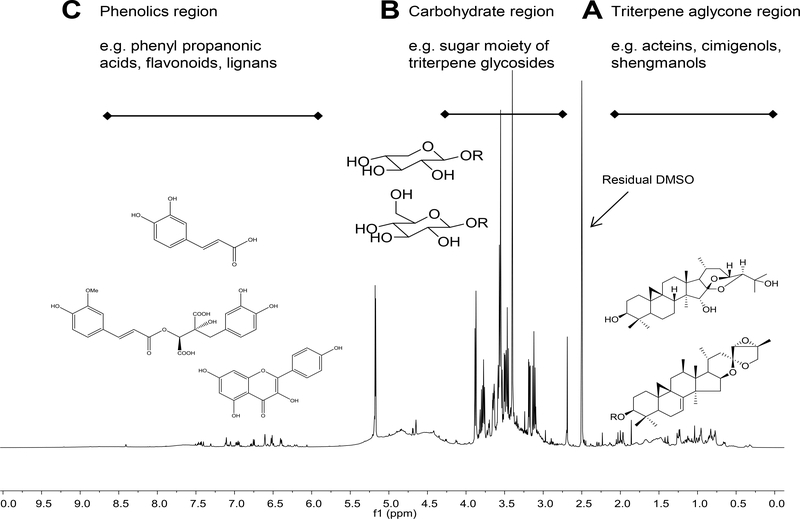
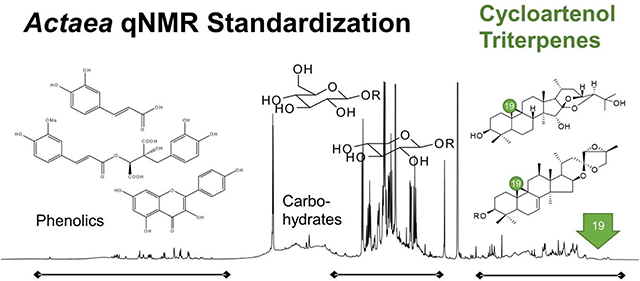
The 1H NMR spectra of Actaea extracts exhibit the presence of numerous constituents, such as cycloartane triterpenes, carbohydrates, phenolic compounds. These three major constituent classes are spectroscopically characterized in the following three regions of a typical 1H NMR spectrum of Actaea extract (BC036) in Figure 1: (A) triterpene aglycone region between 0.0 to 2.2 ppm; (B) carbohydrate region between 3.0 to 4.0 ppm, (C) phenolic region between 6.0 to 8.0 ppm. Other samples exhibited similar profiles. In addition to these three regions, a broad hump in the spectral region between 4.0 and 5.0 ppm was observed in all spectra. This is likely to be associated with overlap of numerous dynamically exchanging hydroxy groups of the highly oxygenated Actaea compounds. Even though the extracts were carefully dried, the H2O/HDO signal was observed as a broadened peak (exchange) overlapped with the carbohydrate signals appearing between 3.3 to 3.6 ppm to different degrees across the samples. The residual DMSO-d5 peak was observed as a 1:2:3:2:1 quintet and the chemical shifts in the spectra were referenced to the center peak at δ = 2.500 ppm. Regardless of the species/plant parts, the most crowded region is B, followed by the A region. The C region tends to be the least crowded.
Fig. 1.
A typical 600 MHz 1H spectrum of a 70% aqueous MeOH extract of ARRR; 12.60 mg of extract (BC036) in 600 μL of DMSO-d6. Chemical shifts were calibrated to 2.500 ppm for the residual DMSO-d5 signal.
3.2. Quantitative Analysis of Cycloartane Triterpenes
The obvious major difficulty with which qHNMR has to contend in the analysis of natural products is the problem of overlapping resonances. In the case of purity assessment, which is one of the most important applications for qHNMR, the target NMR spectrum is relatively clean, and it is not difficult to differentiate signals of the target compounds and those of impurities. On the other hand, the application of qHNMR for the assessment of crude extracts/fractions is more challenging because of their chemically complex nature. However, conditions exist where direct quantitation of whole extracts is possible and include the presence of target signals that are cleanly separated from other signals. For example, Li et al. [15] reported the quantitation of camptothecin and trigonelline in a 1D 1H NMR spectrum. Quantitation in this instance was made possible because the characteristic peaks appeared within a 9.5– 5.5 ppm window, where the spectrum is not crowded. Chauthe et al. [16] demonstrated that the standardization and quantitation of medicinal plant constituents. For example, anthocyanins (delphinidin-3,5-diglucoside, petunidin-3,5-diglucoside and malvidin-3,5-diglucoside) from Eugenia jambolana fruit extract and imperatorin from Aegle marmelos fruit extract were selected as marker constituents. Olefinic or aromatic hydrogen signals of the above-mentioned compounds were used for quantitation. Other food applications have been examined and reviewed [17].
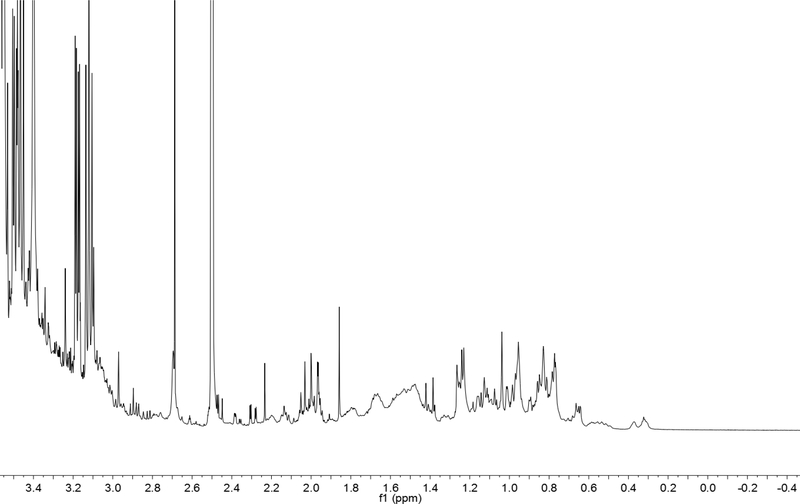
A. racemosa is known to contain a characteristic class of compounds, specifically the cycloartane triterpenes. The quantity of total cycloartane triterpenes can be calculated from the integral value of one of the cyclopropane hydrogen signals using qHNMR. Accumulation of the H-19 exo signals of various cycloartane triterpenes is observed in the spectral region of the samples between 0.20 to 0.45 ppm (Figure2). COSY cross peaks of all of signals between 0.20 to 0.45 ppm are coupled to signals between 0.46 to 0.62 ppm, which confirms that resonances between 0.20 and 0.45 ppm arise only from H-19 exo, and resonances between 0.46 to 0.62 ppm are only from H-19 endo hydrogens. Since no other correlations were observed in the COSY spectrum for signals < 0.62 ppm (note: H-7b resonances can appear at very high fields in pyridine-d5, but not in DMSO-d6) at the current signal-noise ratio, it was reasonable to assume that the total integral of this region represents only the triterpene C-19 hydrogens. The quantity of cycloartane triterpenes was calculated based on the integral value between 0.20 – 0.45 ppm as one hydrogen of a representative cycloartane, for example (12R)-12-acetoxy-(24R,25R)-24,25-epoxy-3-O-β-D-xylopyranosylacta-(16S,23R)-16,23;23,26-binoxoside (1) (C37H56O10 660.83 g/mol), which has been used as a marker compound for AR in HPLC-ELSD method. By using the residual DMSO-d5 signal as an internal calibrant (IC), the quantity of cycloartane triterpene aglycones was calculated as 1. There are two advantages in this method over integrating a broader ppm range of the spectrum: First, as this spectral region (0.20 – 0.45 ppm) contains hydrogen resonances that are very specific to the cycloartanes and are isolated (out in the open), the chance of integrating unrelated signals is low. Secondly, because the 1H resonances are isolated and this region is therefore surrounded by baseline, this makes the baseline correction operation easier and reduces baseline correction errors. Baseline correction was performed using the MNova NMR software using several methods including Whittaker smoother, polynomial, ablative, and cubic splines. Within these four methods, Whittaker smoothing and cubic splines reduced the baseline to zero when focusing on the spectral region that is relevant to the cycloartane C-19 hydrogens. For the DMSO-d5 signal, there was no major difference observed in the four methods. The Whittaker-method was chosen for this study.
Fig. 2.
The structure of a typical cycloartane triterpene and its H-19 exo signal on 1H spectrum of a 70% aqueous MeOH extract of ARRR. Signals between 0.20 – 0.45 ppm arise from the H-19 exo hydrogens as confirmed by 2D COSY.
In addition to ARRR, other representative North American Actaea species/plant parts were quantitated for their cycloartane content in the 70% methanolic extracts (Table 2), including ARPX, APRR, APPX, ACRR, and ACPX.
Table 2.
Calculated triterpene content in 70% methanolic extract of Actaea species.
| Plant code | BC Number | Sample weight [mg] | Integral Value a | Cycloartane Triterpene Weight [mg] | Cycloartane Triterpene Content | Content Range ± St. Dev. |
|---|---|---|---|---|---|---|
| ARPX | BC026 | 17.66 | 3.3% | 1.03 | 5.9% | |
| ARPX | BC027 | 18.36 | 3.3% | 1.03 | 5.6% | |
| ARPX | BC624 | 25.74 | 3.2% | 0.99 | 3.8% | 3.8 – 20.8% |
| ARPX | BC404 | 17.74 | 2.5% | 0.77 | 4.3% | ± 8.2% |
| ARPX | BC582 | 8.48 | 5.6% | 1.75 | 20.7% | |
| ARPX | BC025 | 18.67 | 12.5% | 3.89 | 20.8% | |
| ARRR | BC626 | 18.46 | 6.4% | 1.98 | 10.7% | |
| ARRR | BC036 | 12.60 | 4.3% | 1.34 | 10.7% | |
| ARRR | BC007 | 16.04 | 9.9% | 3.10 | 19.3% | 7.2 – 19.3% |
| ARRR | BC094 | 11.01 | 2.5% | 0.79 | 7.2% | ± 4.0% |
| ARRR | BC629 | 16.31 | 6.9% | 1.99 | 13.2% | |
| ARRR | BC581 | 14.85 | 5.3% | 1.66 | 11.2% | |
| APPX | BC029 | 14.72 | 1.5% | 0.47 | 3.2% | |
| APPX | BC020 | 14.10 | 0.9% | 0.29 | 2.1% | 2.1 – 3.3% |
| APPX | BC028 | 10.21 | 1.1% | 0.32 | 3.3% | ± 0.7% |
| APRR | BC006 | 20.44 | 0.7% | 0.22 | 1.1% | |
| APRR | BC008 | 19.78 | 2.5% | 0.78 | 4.0% | 1.1 – 4.0% |
| APRR | BC011 | 18.97 | 2.1% | 0.65 | 3.4% | ± 1.5% |
| ACPX | BC022 | 12.99 | 3.6% | 1.13 | 8.7% | 7.5 – 8.7% |
| ACPX | BC023 | 15.37 | 3.7% | 1.15 | 7.5% | ± 0.8% |
| ACRR | BC015 | 16.78 | 15.3% | 4.78 | 28.5% | |
| ACRR | BC003 | 19.44 | 8.7% | 2.71 | 13.9% | 13.9 – 28.5% |
| ACRR | BC016 | 18.28 | 15.2% | 4.73 | 26.0% | ± 7.3% |
Integral value was obtained relative to that of DMSO-d5; calculated for 600 μL solvent volume and a cycloartane triterpene molecular weight of 660.8 amu.
As a result, ARPX and ARRR were found to contain 3.8 – 20.8% ± 8.2% and 7.2 – 19.3% ± 4.0%, respectively, of cycloartane triterpenes. Two of the ARPX samples were found to contain over 20% of cycloartane triterpenes. Further experiments might be necessary to substantiate the particularly high values. In contrast, the remainder of the samples were found to contain between 3.8% and 5.9% cycloartane triterpenes. The two samples with the highest content do not have a significant common point, such as collection season or collection site. Sample collections were made in different locations and at different time periods throughout the growing season to specifically analyze for the degree of variation within the botanical. The cycloartane content in ACPX and ACRR was found to be 7.5 – 8.7% ± 0.8% and 13.9 – 28.5% ± 7.3%, respectively. In terms of cycloartane content, the difference between AR and AC was not able to be determined; however, AP showed a consistently lower number.
3.3. Quantitative Results for A. podocarpa
Within three North American species, AP exhibited different features. AP exhibited a lower content of cycloartane triterpenes as compared with AR and AC in the roots/rhizomes. The amount for APPX and APRR was 2.1 – 3.3% ± 0.7% and 1.1 – 4.0% ± 1.5%, respectively. The presence of cycloartane triterpenes in AP has previously been reported [18]. However, 13 representative AR cycloartane triterpenes, such as 1, 3, or 4 were not detected by HPLC–PDA/MS/ELS in a fingerprinting study that examined Actaea species as adulterants of black cohosh samples [19]. Other research [20] compared the phytochemistry of A. racemosa, A. pachypoda, A. podocarpa, and A. rubra. Seven representative AR triterpenes were not found in AP under the conditions of the present study. On the other hand, AP specific triterpenes that are not cycloartanes and lack the characteristic high-field H-19 resonances of the cycloartanes, such as podocarpaside J, podocarpaside H, podocarpaside I, and cimiside A were identified along with 9,10-seco-9,19-cyclolanostanes such as podocarpasides A-G. Several 9,10-seco-9,19-cyclolanostanes have also been reported from AP. The origin of this unique triterpene skeleton was suggested to be biosynthetically derived from a cycloartane triterpene precursor [21][22]. Thus, the present results suggested that AP contains a lower amount of AR specific cycloartane triterpenes and greater amount of the seco-cycloartanes.
AP is also referred to as Cimicifuga americana or yellow cohosh. It shares the same habitat as AR in the eastern United States. Moreover, the dry, dark rhizomes alone are difficult to distinguish outside of the laboratory. The differences found in triterpene content demonstrates that qHNMR can be used for avoiding misidentification of the two species, and hence accidental adulteration of “Black Cohosh”.
3.4. Reference Compound Identification
In addition to the quantitation of a particular compound class as a whole, individual reference compounds specific to A. racemosa could be identified in the hydrogen NMR spectra of the crude extracts. For this identification, the signals of seven skeletal methyl groups in Actaea triterpenes were used. These methyl groups serve as “surveillance units” for the neighboring segments of the molecules. The characteristic patterns (chemical shifts) for methyl group signals in Actaea are very sensitive to their chemical environment so that dereplication without reliance on reference data is possible only using classification binary trees [23].
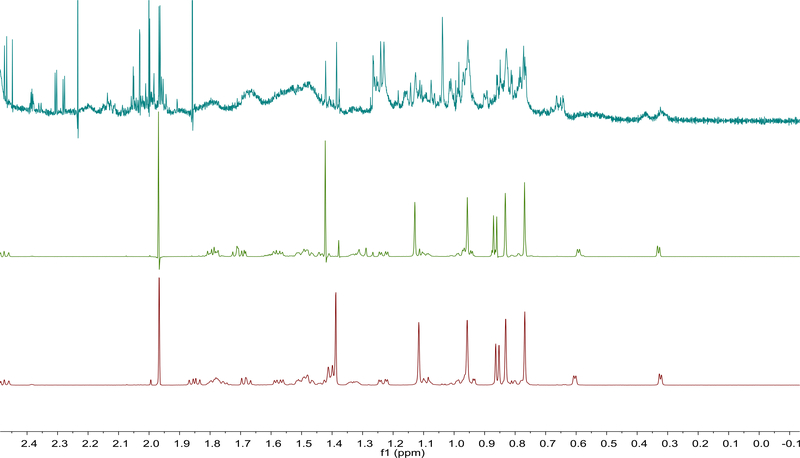
In this study, two of the reference compounds, (12R)-12-acetoxy-(24R,25R)-24,25-epoxy-3-O-β-D-xylopyranosylacta-(16S,23R)-16,23;23,26-binoxoside (1) and (12R)-12-acetoxy-(24R,25S)-24,25-epoxy-(26R/S)-26-hydroxy-3-O-β-D-xylopyranosylacta-(16S,23R)-16,23;23,26-binoxoside (3/4), were identified in the 1H NMR spectrum of the extracts by comparison with the data of reference material. Even though the NMR data of these compounds in DMSO-d6 was not available from the literature, the signal assignments were performed using COSY, HMBC and HSQC correlations as well as the reported data for these compounds in pyridine-d5 as a guide [13] [14]. The NMR data of 1 and 3/4 obtained in DMSO-d6 are summarized in Table 3. When the 1H NMR spectra of the two reference compounds were compared to a resolution enhanced spectrum of the extract (BC036), all of the seven methyl group signals could be identified and matched as shown in Figure 3.
Table 3.
The 1H and 13C NMR assignments of 1 and 3/4 in DMSO-d6.
| Compound 1 |
Compounds 3/4 |
|||||
|---|---|---|---|---|---|---|
| position | δCa | δH | JH-H (Hz); multiplicity b | δCa | δH | JH-H (Hz); multiplicity b |
| 1 | 30.8 | 1.091 | overlapped | 30.8 | 1.105 | overlapped |
| 1.472 | overlapped | 1.493 | overlapped | |||
| 2 | 28.6 | 1.493 | overlapped | 28.6 | 1.508 | overlapped |
| 1.782 | overlapped | 1.796 | overlapped | |||
| 3 | 87.3 | 3.087 | overlapped | 87.3 | 3.086 | overlapped |
| 4 | 40.9 | - | 40.9 | - | ||
| 5 | 46.2 | 1.233 | 4.0, 12.1; dd | 46.2 | 1.232 | 4.0, 12.1; dd |
| 6 | 19.4 | 0.789 | overlapped | 19.4 | 0.780 | overlapped |
| 1.494 | overlapped | 1.508 | overlapped | |||
| 7 | 24.8 | 0.983 | overlapped | 24.8 | 0.988 | overlapped |
| 1.331 | m | 1.332 | m | |||
| 8 | 45.2 | 1.575 | 5.2, 12.1; dd | 45.2 | 1.598 | 5.3, 12.1; dd |
| 9 | 19.5 | - | 19.5 | - | ||
| 10 | 25.9 | - | 25.9 | - | ||
| 11 | 35.6 | 0.946 | overlapped | 35.6 | 0.958 | overlapped |
| 2.481 | overlapped | 2.481 | overlapped | |||
| 12 | 75.3 | 4.725 | 3.4, 8.8; dd, | 76.5 | 4.737 | 3.4, 8.8; dd |
| 13 | 47.7 | - | 47.7 | - | ||
| 14 | 47.2 | - | 47.7 | - | ||
| 15 | 43.0 | 1.407 | overlapped | 43.0 | 1.423 | overlapped |
| 1.851 | 7.5, 13.2; dd | 1.786 | 7.6, 12.8; dd | |||
| 16 | 73.6 | 4.097 | 7.5, 14.7; dd | 71.9 | 4.257 | 7.1, 14.6; q |
| 17 | 55.2 | 1.683 | 8.9; t | 55.2 | 1.711 | overlapped |
| 18 | 12.2 | 1.116 | s | 13,4 | 1.128 | s |
| 19 | 28.8 | 0.323 | 4.2; d | 29.5 | 0.330 | 4.0, d |
| 0.604 | 4.2; d | 0.593 | 4.0, d | |||
| 20 | 21.9 | 1.762 | m | 20.4 | 1.593 | m |
| 21 | 20.1 | 0.858 | 6.5; d | 21.1 | 0.865 | 6.3; d |
| 22 | 36.1 | 1.410 | overlapped | 37.3 | 1.290 | 13.5; t |
| 1.410 | overlapped | 1.697 | overlapped | |||
| 23 | 105.2 | - | 104.7 | - | ||
| 24 | 60.8 | 3.509 | s | 61.8 | 3.556 | s |
| 25 | 61.9 | - | 64.8 | - | ||
| 26 | 67.1 | 3.449 | 10.0; d | 96.5 | 4.996 | 5.7, d |
| 3.710 | 10.0; d | |||||
| 27 | 13.3 | 1.388 | s | 12.9 | 1.423 | s |
| 28 | 18.8 | 0.831 | s | 18.8 | 0.833 | s |
| 29 | 24.5 | 0.957 | s | 24.5 | 0.958 | s |
| 30 | 14.2 | 0.768 | s | 14.2 | 0.770 | s |
| 1’ | 106.7 | 4.125 | 7.8; d | 106.7 | 4.125 | 7.8; d |
| 2’ | 74.0 | 2.946 | m | 74.0 | 2.946 | m |
| 3’ | 77.0 | 3.062 | overlapped | 77.0 | 3.060 | overlapped |
| 4’ | 69.8 | 3.240 | m | 69.8 | 3.239 | m |
| 5’ | 65.7 | 3.005 | 10.6; t | 65.7 | 3.004 | 10.6; t |
| 3.640 | 5.5, 11.5; d, | 3.640 | 5.5, 11.5; dd | |||
| Oac | 21.0/170.4 | 1.967 | s | 21.0/170.4 | 1.969 | s |
The 13C chemical shifts were extracted from the HSQC and HMBC data.
Multiplicities: d doublet; dd double doublet; m multiplet; t triplet.
Fig. 3.
Identification of 1 and 2 by 1H NMR (600 MHz) as Two of the Major constituents of ARRR Extract (BC036). Processing was performed as follows: BC036, GM, LB −3.0 Hz, GB 0.1; compounds 1 and 2, GM, LB −1.0 Hz, GB 0.1.
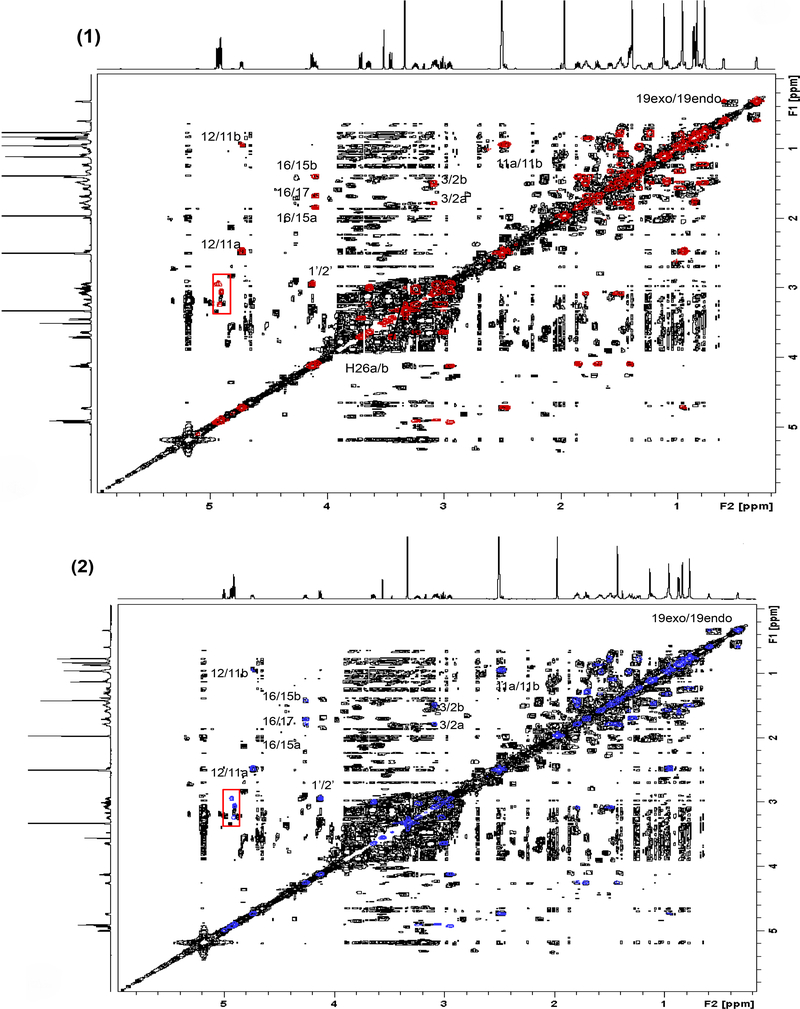
The presence and positions of particular methyl signals is a strong indication of the existence of specific compounds as shown by Qiu et al.[23]. In addition to the 1H spectral evidence, the COSY spectra were examined for compounds 1 and 3/4. The spectra of the reference compounds were superimposed on that of BC036. As a result, all of the key cross peaks of compounds 1 and 3/4 were observed in the BC036 COSY spectrum. Specifically, correlations H-16/H-15a, H-16/H-15b, and H-16/H-17 were clearly observed in both cases. These are characteristic for actein types of side chains. Furthermore, the following COSY correlations were observed: H-12/H-11a COSY and the chemical of C-12 confirmed an OAc moiety at C-12; The H-3/H2a COSY correlation and the chemical shift of the anomeric C-1’ confirmed the sugar position. The H-19 exo/endo COSY correlations confirmed the cycloartane moiety. Altogether, the presence of 1 and 3/4 in the BC036 extract was revealed as each correlation cross peak provides information about part of the structure. COSY cross peaks that did not match with the reference materials were cross peaks between hydroxyl groups and sugar hydrogens that were shown in a solid square in Figure 4. While hydroxyl groups can engage in hydrogen bonding with other components within the mixture and thus may exhibit slightly different behavior than in pure reference compounds, it is quite reasonable to assume that such cross peaks may not match up. Hence, from the 1H, COSY and HMBC evidence, 1 and 3 & 4 were identified in the ARRR extract (BC036).
Fig. 4.
Identification of 1 and 2 in ARRR extracts (BC036). The 600 MHz COSY spectra of compounds 1 and 2 are each superimposed with the ARRR COSY spectrum, respectively. External projections are the 1H spectra of each reference compound. Cross peaks in the solid rectangle are OH-CH couplings.
3.5. Solvent Effects
The extract, BC036, which showed a typical ARRR 1H NMR pattern in DMSO-d6, was dissolved in pyridine-d5 and the 1D 1H and 2D COSY NMR spectra were acquired at 600 MHz (Figure 4). The pyridine-d5 extract sample was prepared in the same manner as the DMSO-d6 sample. As previously noted, the 1H spectrum of the DMSO-d6 sample exhibited the characteristic A, B and C regions which covers the three major classes of structure types: triterpenes, carbohydrates, and phenolics. However, in the pyridine-d5 spectrum of the same sample has triterpene and carbohydrate signals but only a trace of signals associated with the phenolic compounds. Moreover, the residual solvent peaks in pyridine-d5 and their respective 13C satellites appear in the same region as the phenolic peaks and partially overlap with them. A DMSO-d6 solvent signals appears at 2.500 ppm in a less crowded spectral region. Hence the DMSO-d6 is shown to be a more efficient solvent for the Actaea metabolomics studies of species for three reasons: (1) it dissolves the three known major classes of Actaea compounds; (2) the residual solvent peak does not overlap significantly with peaks of interest in the spectrum; and (3) DMSO-d6 is a near-universal solvent and possibly dissolves additional compounds.
Although the diagnostic signals of the triterpene skeleton appear in the same range (0.5 ppm – 2.5 ppm) of both DMSO-d6 and pyridine-d5 spectra, the carbohydrate signals are clearly affected by the solvent. The carbohydrate signals in pyridine-d5 are shifted about 1.2 ppm downfield compared to their position in DMSO-d6. The 1H resonances of the carbohydrate fall in the range of 3.0 – 4.0 ppm in DMSO-d6 vs 4.2 – 5.2 ppm in pyridine-d5.
3.6. Summary
The 1H NMR spectra of crude extracts from a total of 23 Actaea plant samples were obtained and analyzed for their species -and plant part- specific metabolomic characters by qHNMR. This study demonstrates that qHNMR is a powerful methodology for the analysis of crude plant extracts. Specifically, in Actaea species, the presence of the characteristic C-19 cyclopropane signals made it possible to quantify the cycloartane triterpene class as a whole and without the necessity for the identical reference standards. ARPX and ARRR contained 3.8 – 20.8% ± 8.2% and 7.2 – 19.3% ± 4.0%, respectively, of cycloartane triterpenes. The cycloartane content in ACPX and ACRR was 7.5 – 8.7% ± 0.8% and 13.9 – 28.5% ± 7.3%, respectively. AP contains notably lower amounts of cycloartane triterpenes as compared to AR and AC in the roots/rhizomes. The amount for APPX and APRR were 2.1 – 3.3% ± 0.7% and 1.1 – 4.0% ± 1.5%, respectively. In addition to the quantitative ability of 1H NMR, the same spectrum was used for extracting information about the identification of marker compounds. The marker compounds 1 and 3/4 were identified in AR crude extracts by utilizing the seven methyl peaks in the 1D 1H spectrum and 2D COSY.
The use of the residual hydrogen signal from the NMR solvent DMSO-d6 was found to be suitable for the quantitation of cycloartane triterpenes of Actaea extract. While this report was being prepared as part of the dissemination of the 2012 PhD dissertation results of A. Imai [24], Çiçek et al. published a qHNMR study in which the total triterpene content of A. racemosa was assessed using methanol-d4 solution and 1,2,4,5-tetrachloro-3-nitrobenzene as IC [25]. The choice of solvent is a critical parameter in qHNMR as it determines how existing qualitative, structural NMR data can be applied to quantitation. As building of a knowledge base of reliable chemical shift information is laborious, the ability to use historic NMR data is invaluable. Thus, from the perspective of available structural assignments, pyridine-d5 is more suitable than DMSO-d6 for future development of targeted quantitation of individual Actaea triterpenes in crude extracts, because the vast majority of published structural data are based on NMR measurements performed in pyridine-d5. In particular, this knowledge base allows for the mining of Me group chemical shift data for structural barcode patters, as has been shown previously [26]. Future development of targeted qHNMR-based, metabolomic standardization methods will require the translation of the 1H NMR profiles of known triterpenes from pyridine-d5 to DMSO-d6, or their ab initio establishment. Alternatively, external calibration (EC) [27] combined with use of existing pyridine-d5 reference data could be a viable path.
Finally, potentially limiting factors associated with this study should also be mentioned. The plant samples had been collected some time before analysis and stored under ambient conditions. According to our observations working with Actaea botanicals, however, all indications are that the triterpenes are stable, especially in intact plant material. Furthermore, the available number of distinct samples for each species have limited the statistical significance of the species and plant part quantitative triterpene content values. However, the study demonstrated that quantitation of crude black cohosh materials can be achieved by qHNMR without authentic reference materials.
Supplementary Material
Acknowledgements
This work was supported by grant P50 AT000155 from the ODS and NCCIH (formerly NCCAM) of the NIH. We are also indebted to Mike Tortura (UIC) and Katharine Parks (Chillicothe, Ohio) for horticultural support; and Dr. Doel Soejarto (UIC and Herbarium of the Field Museum of Natural History, Chicago, Illinois) for botanical specimen authentication. Furthermore, we wish to acknowledge the field collection efforts in Virginia, North Carolina, and Tennessee by Drs. Daniel Fabricant and Gwynn Ramsey.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Dedication
The authors dedicate this article to the late Dr. Norman Farnsworth and his wife, Priscilla Farnsworth, for their support of botanical research, on the occasion of what would have been Norman’s 90th birthday (March 23, 2020).
Appendix A. Supplementary Data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fitote.xyz.
Furthermore, the original NMR data (FIDs) are made freely available at https://dx.doi.org/10.7910/DVN/TL11QU (Harvard Dataverse).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].He K, Zheng B, Kim CH, Rogers L, Zheng Q. Direct analysis and identification of triterpene glycosides by LC/MS in black cohosh, Cimicifuga racemosa, and in several commercially available black cohosh products. Planta Med 2000;66:635–40. [DOI] [PubMed] [Google Scholar]
- [2].Verbitski SM, Gourdin GT, Ikenouye LM, McChesney JD, Hildreth J. Detection of Actaea racemosa adulteration by thin-layer chromatography and combined thin-layer chromatography-bioluminescence. J AOAC Int 2008;91:268–75. [PMC free article] [PubMed] [Google Scholar]
- [3].Wuttke W, Seidlova-Wuttke D, Gorkow C. The Cimicifuga preparation BNO 1055 vs. conjugated estrogens in a double-blind placebo-controlled study: effects on menopause symptoms and bone markers. Maturitas 2003;44 Suppl 1:S67–77. [DOI] [PubMed] [Google Scholar]
- [4].Geller SE, Shulman LP, van Breemen RB, Banuvar S, Zhou Y, Epstein G, et al. Safety and efficacy of black cohosh and red clover for the management of vasomotor symptoms: a randomized controlled trial. Menopause 2009;16:1156–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].van Breemen RB, Liang W, Banuvar S, Shulman LP, Pang Y, Tao Y, et al. Pharmacokinetics of 23-epi-26-deoxyactein in women after oral administration of a standardized extract of black cohosh. Clin Pharmacol Ther 2010;87:219–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fabricant D Pharmacognostic Investigation of Black Cohosh (Cimicifuga racemosa (L.) Nutt.). Ph.D. Dissertation, University of Illinois at Chicago College of Pharmacy, Department of Medicinal Chemistry, 2005. [Google Scholar]
- [7].Pauli GF, Jaki BU, Lankin DC. Quantitative 1H NMR: development and potential of a method for natural products analysis. J Nat Prod 2005;68:133–49. [DOI] [PubMed] [Google Scholar]
- [8].Pauli GF, Gödecke T, Jaki BU, Lankin DC. Quantitative 1H NMR. Development and Potential of an Analytical Method: An Update. J Nat Prod 2012;75:834–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gödecke T, Napolitano JG, Rodríguez-Brasco MF, Chen SN, Jaki BU, Lankin DC, et al. Validation of a generic quantitative 1H NMR method for natural products analysis. Phytochem Anal 2013;24:581–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gödecke T, Yao P, Napolitano JG, Nikolic D, Dietz BM, Bolton JL, et al. Integrated standardization concept for Angelica botanicals using quantitative NMR. Fitoterapia 2012;83:18–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pauli GF, Chen S-N, Simmler C, Lankin DC, Gödecke T, Jaki BU, et al. Importance of purity evaluation and the potential of quantitative 1H NMR as a purity assay. J Med Chem 2014;57:9220–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Compton JA, Culham A, Jury SL. Reclassification of Actaea to Include Cimicifuga and Souliea (Ranunculaceae): Phylogeny Inferred from Morphology, nrDNA ITS, and cpDNA trnL-F Sequence Variation. Taxon 1998;47:593–634. [Google Scholar]
- [13].Chen SN, Li W, Fabricant DS, Santarsiero BD, Mesecar A, Fitzloff JF, et al. Isolation, structure elucidation, and absolute configuration of 26-deoxyactein from Cimicifuga racemosa and clarification of nomenclature associated with 27-deoxyactein. J Nat Prod 2002;65:601–5. [DOI] [PubMed] [Google Scholar]
- [14].Kusano A, Takahira M, Shibano M, In Y, Ishida T, Miyase T, et al. Studies on the constituents of Cimicifuga Species. XX. Absolute stereostructures of cimicifugoside and actein from Cimicifuga simplex WORMSK. Chem Pharm Bull 1998;46:467–72. [Google Scholar]
- [15].Li C-Y, Lin C-H, Wu T-S. Quantitative analysis of camptothecin derivatives in Nothapodytes foetida using 1H-NMR method. Chem Pharm Bull 2005;53:347–9. [DOI] [PubMed] [Google Scholar]
- [16].Chauthe SK, Sharma RJ, Aqil F, Gupta RC, Singh IP. Quantitative NMR: an applicable method for quantitative analysis of medicinal plant extracts and herbal products. Phytochem Anal 23:689–96. [DOI] [PubMed] [Google Scholar]
- [17].Simmler C, Napolitano JG, McAlpine JB, Chen S-N, Pauli GF. Universal quantitative NMR analysis of complex natural samples. Curr Opin Biotechnol 2014;25:51–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ali Z, Khan SI, Khan IA. New cycloartane-type triterpene arabinosides from the roots of Actaea podocarpa and their biological study. Planta Med 2007;73:699. [DOI] [PubMed] [Google Scholar]
- [19].He K, Pauli GF, Zheng B, Wang H, Bai N, Peng T, et al. Cimicifuga species identification by high performance liquid chromatography-photodiode array/mass spectrometric/evaporative light scattering detection for quality control of black cohosh products. J Chromatogr A 2006;1112:241–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Avula B, Ali Z, Khan IA. Chemical fingerprinting of Actaea racemosa (Black Cohosh) and its comparison study with closely related Actaea species (A. pachypoda, A. podocarpa, A. rubra) by HPLC. Chromatographia 2007;66:757–62. [Google Scholar]
- [21].Ali Z, Khan SI, Fronczek FR, Khan I a. 9,10-seco-9,19-Cyclolanostane arabinosides from the roots of Actaea podocarpa. Phytochemistry 2007;68:373–82. [DOI] [PubMed] [Google Scholar]
- [22].Ali Z, Khan SI, Ferreira D, Khan IA. Podocarpaside, a triterpenoid possessing a new backbone from Actaea podocarpa. Org Lett 2006;8:5529–32. [DOI] [PubMed] [Google Scholar]
- [23].Qiu F, Imai A, McAlpine JB, Lankin DC, Burton I, Karakach T, et al. Dereplication, residual complexity, and rational naming: the case of the Actaea triterpenes. J Nat Prod 2012;75:432–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Imai A Pharmcacognosy of Raw Materials for Black Cohosh Dietary Supplements. Ph.D. Dissertation, University of Illinois at Chicago College of Pharmacy, Department of Medicinal Chemistry, 2012. [Google Scholar]
- [25].Çiçek SS, Girreser U, Zidorn C. Quantification of the total amount of black cohosh cycloartanoids by integration of one specific 1H NMR signal. J Pharm Biomed Anal 2018;155:109–15. [DOI] [PubMed] [Google Scholar]
- [26].Qiu F, McAlpine JB, Lankin DC, Burton I, Karakach T, Chen S-N, et al. 2D NMR Barcoding and Differential Analysis of Complex Mixtures for Chemical Identification: The Actaea Triterpenes. Anal Chem 2014;86:3964–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Phansalkar R, Simmler C, Bisson J, Chen S-N, Lankin D, McAlpine J, Niemitz M, Pauli G. Evolution of Quantitative Measures in NMR: Quantum Mechanical qHNMR Advances Chemical Standardization of a Red Clover (Trifolium pratense) Extract. J Nat Prod 201;80:634–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.