Abstract
In this work, we used a whole-genome sequencing (WGS) approach to study the features of KPC-producing Klebsiella pneumoniae (KPC-Kp) spreading in a large Italian long-term acute-care rehabilitation facility (LTACRF), and to track the dynamics of dissemination within this setting. Thirty-eight, non-replicated, KPC-Kp isolates from colonized patients (either already colonized at admission or colonized during admission), collected during 2016, were subjected to antimicrobial-susceptibility testing and WGS. All isolates were resistant to β-lactams, with the exception of ceftazidime/avibactam (97.4 % susceptible). The second most effective agent was fosfomycin, followed by colistin, trimethoprim/sulfamethoxazole, gentamicin and amikacin (92.1, 86.8, 60.5, 44.7 and 50 % of susceptibility, respectively). A large proportion of isolates (n=18/38, 47.4%) belonged to clonal group (CG) 101, and most of them (n=15) to a new sequence type (ST) designated as ST2502. All the CG101 isolates had a capsule locus type KL17. The ST2502 harboured the genes encoding for the yersiniabactin siderophore and the ArmA methylase, conferring high-level resistance to aminoglycosides. The second most represented lineage of isolates (16/38, 42.1%) belonged to ST512 of CG258. Analysing WGS data, we were able to ascertain the common origin of some isolates imported from other hospitals, and to track several clusters of in-LTACRF cross-transmissions. The results revealed that, in peculiar epidemiological settings such as LTACRF, new KPC-Kp clones different from those prevailing in acute-care hospitals and associated with uncommon resistance and virulence determinants can successfully emerge and disseminate.
Keywords: Klebsiella pneumoniae, molecular epidemiology, virulence determinants, KPC-type carbapenemases, sequence type 101
Data Summary
The sequencing project data have been deposited in the International Nucleotide Sequence Database Collaboration databases under the National Center for Biotechnology Information BioProject number PRJNA431724. Sequence read files and genome assemblies have been deposited in SRA and GenBank, respectively. A complete list of BioSample, SRA and GenBank accession numbers is available in Table S1 (available with the online version of this article).
Outcome.
In this work, we describe the clonal expansion of a novel KPC-producing Klebsiella pneumoniae clone [i.e. sequence type (ST)2502] in a large Italian long-term acute-care rehabilitation facility (LTACRF). ST2502 exhibits a peculiar set of resistance and virulence traits infrequent among KPC-Kp clones prevailing in acute-care hospitals. A whole-genome-sequencing based phylogenetic analysis enabled us to trace some dynamics of KPC-Kp dissemination within the LTACRF.
Introduction
Carbapenem-resistant Enterobacterales (CRE) have emerged worldwide among the most challenging resistant pathogens in the clinical setting, due to their difficult-to-treat resistance phenotypes and ability to rapidly disseminate in healthcare facilities [1, 2]. Dissemination of CRE has mostly been caused by intercontinental expansion of some high-risk clones of Klebsiella pneumoniae [e.g. members of clonal groups (CGs) 258, 101, 147 and 307], which have acquired carbapenemase determinants of various types (mostly of the KPC, OXA-48-like, NDM and VIM types) [1–4]. The epidemiological success of these difficult-to-treat K. pneumoniae clones is likely contributed to by a number of features, such as the ability to acquire new genetic material (including antimicrobial-resistance genes and virulence determinants), to efficiently colonize the intestinal tract, and to persist in the hospital environment [5].
In endemic areas, the molecular epidemiology of CRE can be variable, with different combinations of clones and enzymes. In Italy, CRE have been established at a level of high endemicity since 2010, and this has mostly been caused by a countrywide, inter-hospital dissemination of KPC-Kp strains of CG258 [4, 6]. Other clonal lineages have remained less prevalent, but clusters of expansion of sequence type (ST)101, ST307 and ST273 strains have locally been described [7–10]. Long-term acute-care rehabilitation facilities (LTACRFs) were previously shown to be hubs for CRE dissemination in settings of high endemicity. In ths setting, the occurrence of large and difficult to control outbreaks is likely facilitated by the prolonged length of stay of patients (including those colonized by KPC-Kp) and by the sharing of common spaces (gyms, pools, etc.) [11–15].
In a recent study, carried out in a large Italian LTACRF, we observed an overall high burden in terms of KPC-Kp dissemination, with KPC-Kp carriage and infection rates being notably higher among patients with severe brain injuries (SBI) admitted to the SBI ward and requiring a higher intensity of care. This condition resulted from both a higher rate of KPC-Kp carriage at admission and from a high rate of KPC-Kp cross-transmission after admission among these patients [16]. In this work, we investigated the molecular epidemiology of KPC-Kp circulating in the SBI ward of that LTACRF, and found a peculiar profile in the population structure of these pathogens, suggesting that specific clones could evolve to successfully disseminate in similar settings.
Methods
Bacterial strains
Bacterial strains investigated in this work included all the available CRE isolates (n=38) obtained from surveillance cultures (rectal swabs) of colonized patients from the SBI ward of the Don Carlo Gnocchi Foundation LTCRF of Florence (Italy), during 2016. Each isolate was from a different patient. The studied isolates represented approximately half of the total cases of carriage (n=74) observed in the SBI ward during 2016, and had been isolated throughout the whole study period (1–5 per month).
Rectal swabs were collected using the FecalSwab system (Copan). The specimens (in 10 µl medium) were cultured on ChromID CARBA SMART plates (bioMérieux) to screen for CRE, within 48 h of collection. Plates were inspected for growth after 18–24 h incubation at 35±2 °C. Colonies grown on the selective medium were identified using the MALDI-TOF MS Vitek 2 system (bioMérieux). Suspect CRE isolates with a meropenem minimum inhibitory concentration >0.125 mg l−1 were included in the study.
Antimicrobial-susceptibility testing
Antimicrobial-susceptibility testing for amoxicillin/clavulanate, piperacillin/tazobactam, ceftazidime/avibactam, cefepime, ceftazidime, meropenem, ertapenem, ciprofloxacin, levofloxacin, colistin, amikacin, gentamicin, trimethoprim/sulfamethoxazole and tigecycline was carried out by broth microdilution [17], using lyophilized custom plates (Thermo Fisher Scientific). Fosfomycin susceptibility was tested by agar dilution in the presence of 25 mg glucose-6-phosphate l−1 [18]. Results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints (version 9, www.eucast.org).
Whole-genome sequencing (WGS) and bioinformatics
Total DNA was extracted from each strain by processing 400 µl of a 0.5 McFarland suspension in normal saline (0.9% w/v)) using the Complex 400_V3_DSP protocol with the DSP virus/pathogen midikit on a QIAsymphony SP instrument (Qiagen). Sequencing libraries were prepared using the NEBNext Ultra DNA library prep kit (NEB) and Illumina data was generated by an Illumina HiSeq 250 (at Novogene) following a 2×250 bp paired-end approach. Draft genome assemblies were generated using SPAdes 3.11 [19]. A complete list of accession numbers for Illumina reads sets and assemblies is detailed in.
Kaptive software was used to associate isolate sequencing data with known capsule locus type/serotypes [20]. Kleborate software was used for analysis of the O antigen locus, acquired genes encoding resistance mechanisms to antibiotics (aminoglycosides, colistin, fosfomycin, tetracyclines, sulfonamides, trimethoprim and β-lactams), genes encoding virulence determinants associated with hypervirulence (yersiniabactin, colibactin, aerobactin, salmochelin and regulators of the mucoid phenotype), for multi-locus sequence typing (MLST) and yersiniabactin ST (Ybt-ST) determination [21]. For isolates resistant to colistin, the presence of mgrB and pmrAB gene variants previously associated in the literature with colistin resistance was inspected manually using the National Center for Biotechnology Information (NCBI) blast tool (coverage and identity cut-offs: 85 %) [22].
The genetic relatedness among isolates was evaluated through SNP-based phylogenetic trees using Snippy v 4.4.3 with default parameters (minimum read mapping quality to consider, 60; minimum base quality to consider, 13; minimum site depth for calling alleles, 10; minimum proportion for variant evidence, auto; minimum quality in VCF column 6, 100; minimum soft clipping to allow, 10) [23]. Maximum-likelihood (ML) phylogenetic trees were inferred from core SNP alignments by iq-tree v1.6.12 [24], using the K3P model with the ascertainment bias correction (ASC) for SNP data that typically do not contain constant sites. Branch supports were assessed by standard non-parametric bootstrap employing 100 replicate trees. The final ML tree with assigned support was visualized and edited using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/) and Phandango [25]. The global phylogenetic relatedness was evaluated using the genome of the NTUH-K2044 strain (accession number AP006725) as a reference and samples’ raw reads as input. In order to better explore the genetic relatedness of isolates belonging to the same clonal lineage, a fine-scale analysis was performed following the same pipeline and using the oldest isolated strain within each lineage as the reference (Table 1) and raw reads of the remaining isolates as the input.
Table 1.
Statistics of core-genome analysis of locally sequenced isolates
|
Clone (n) |
Reference genome (size) |
Percentage coverage of reference genome* |
Core sites† |
SNP distance from reference‡ |
|---|---|---|---|---|
|
CG258 (16) |
DG5547 (5 493 002 bp) |
97.91–99.97 (99.99) |
233 |
3–57 (38; 44) |
|
ST2502 (15) |
DG5544 (5 631 301 bp) |
98.77–99.94 (99.92) |
25 |
0–7 (4; 4) |
|
ST101 (3) |
DG5546 (5 545 304 bp) |
98.13–99.94 (99.03) |
440 |
0–440 (220; 220) |
*Data are expressed as minimum–maximum (mean) values.
†Number of genomic positions present in all samples showing at least one polymorphism (i.e. excluding invariant sites from the core-genome portion).
‡Data are expressed as minimum–maximum (mean; median) values.
Sequence comparisons with publicly available genomes were performed using blastn of the blast 2.2.26+ package with Kleborate software. K. pneumoniae genomes publicly available at the NCBI-NIH database (accessed on 12 February 2019) were used for comparison analysis, with the following inclusion criteria: (i) organism name ‘ Klebsiella pneumoniae ’, taxid:573; (ii) presence of a RefSeq accession number. A total of 6439 genomes available at the date of analysis (12 February 2019) were selected and analysed (Table S2).
Patients’ data
Patients’ data (admission, discharge and transfer dates, hospital of provenance) were obtained from the software used to store and manage clinical records data at the LTACRF. The screening programme for CRE ongoing at the LTACRF, and the criteria used for the definition of incident and imported colonization, were previously described [16]. Briefly, screening for CRE carriage was performed with all patients at admission and, if the patient was found to be negative, was repeated on a weekly basis until the patient was discharged or became colonized. Colonization cases identified at admission were considered ‘imported’, while cases of colonization occurring during admission in a patient found to be not colonized at admission were defined as ‘incident’.
Ethical committee clearance
All the isolates were obtained during a surveillance programme compliant with a nationwide CRE containment strategy recommended by the Italian Ministry of Health since 2013, following the recommendations of the European Centre for Disease Prevention and Control [26, 27]. An informed written consent for the use of surveillance samples was obtained from all patients.
Results
Features of the KPC-Kp isolates investigated in this study
During 2016, a total of 74 cases of colonization by CRE, either imported (n=41) or incident (n=33), were detected at the SBI ward of the LTCRF by the screening programme [16]. CRE isolates from 38 of these cases were available for further analysis, including 15 from imported cases and 23 from incident cases. The identification of all isolates investigated in the study was confirmed as K. pneumoniae by MALDI-TOF MS and as K. pneumoniae sensu stricto by WGS analysis (see below).
Antimicrobial-susceptibility testing revealed that the studied isolates were resistant to amoxicillin/clavulanate, piperacillin/tazobactam, cefepime, ceftazidime and carbapenems. Most of them retained susceptibility to ceftazidime/avibactam (97.4 %), fosfomycin (92.1 %), colistin (86.8 %) and trimethoprim/sulfamethoxazole (60.5 %), while some of them retained susceptibility to aminoglycosides (gentamicin, 44.7 %, and amikacin, 50.0 %). All except one isolate were resistant to levofloxacin and ciprofloxacin. The European Committee on Antimicrobial Susceptibility Testing (EUCAST) doesn’t provide a breakpoint for K. pneumoniae and tigecycline; however, 13.1 % of isolates retained susceptibility adopting the tigecycline breakpoint proposed for Escherichia coli (MIC90=2; MIC90: Minimum Inhibitory Concentration required to inhibit the growth of 90% of organisms).
Population structure of the KPC-Kp isolates
Analysis of the clonal relationships among the 38 KPC-Kp isolates from the SBI ward was carried out based on WGS analysis, to which all isolates were subjected. A global phylogenetic analysis, including all isolates, revealed the presence of two major lineages and four singletons (Fig. 1). The most populous lineage, including almost half of the isolates (18 of 38, 47.4 %), comprised members of CG101, with 3 belonging to ST101 and 15 to a novel ST, which was a single locus variant of ST101 (by a different rpoB allele), designed ST2502. The novel ST was submitted to the BIGSdb multi-locus sequence typing database (https://bigsdb.pasteur.fr). Comparison analysis with K. pneumoniae genomes, available in the NCBI-NIH database (accessed on 12 February 2019), revealed that no K. pneumoniae genome of ST2502 had been previously deposited .
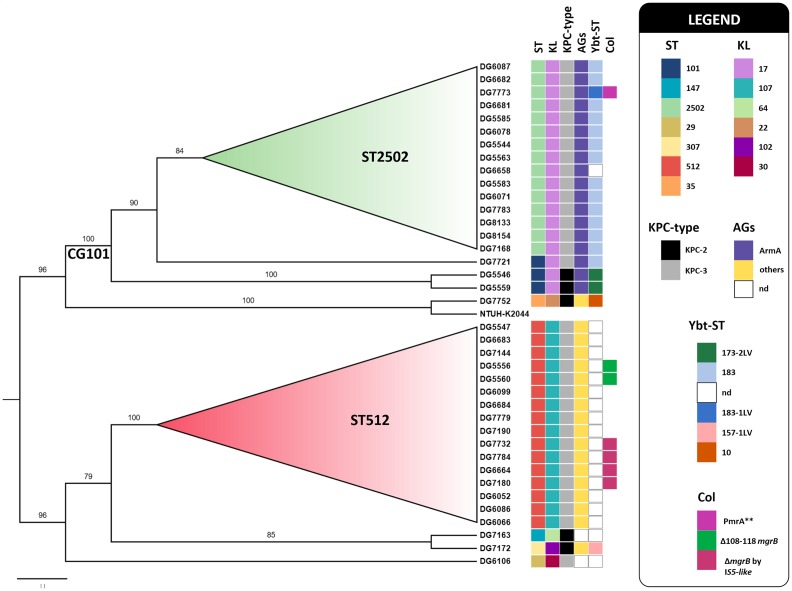
Fig. 1.
Global core-genome SNP phylogeny inferred for the 38 K . pneumoniae isolates analysed in this study. A ML phylogenetic tree (midpoint rooted) was reconstructed using Snippy and iq-tree (see Methods), using K. pneumoniae NTUH-K2044 (accession number AP006725) as a reference genome [36]. Bootstrap-based branch support values, obtained from 100 replicate trees, are indicated as branch labels. Branches showing bootstrap values less than 50 were collapsed. Bar, substitutions per nucleotide position. Coloured squares provide details about ST, capsule locus (KL), and about the presence of selected resistance mechanisms and genetic markers associated with virulence, as detailed in the legend. AGs, Aminoglycosides; Col, colistin.
The second most populous lineage included isolates of ST512, a member of CG258 (16/38, 42.1 %). The remaining four singletons were of ST29, ST35, ST147 and ST307 (Fig. 1).
Features, comparative genomics and epidemiology of the various clonal lineages of KPC-Kp
CG101 isolates
All the CG101 isolates had a KL17 capsule locus type and an O1v1 O locus. All but one (the isolate DG6658) carried the yersiniabactin siderophore gene cluster of the ybt9 lineage, associated with an ICEKp3 element and including different Ybt-ST (Fig. 1, (Table S3). None of them carried other virulence determinants, such as those for colibactin, aerobactin and salmochelin siderophores, or the rmpA and rmpA2 genes associated with the hypermucoviscous phenotype (Table S3).
With respect to the resistome (Fig. 1), in addition to the bla KPC carbapenemase gene, which was either bla KPC-3 or bla KPC-2 (see below), all the CG101 isolates carried the armA gene, which accounted for resistance to aminoglycosides, and were lacking sul and dfrA genes, in agreement with their overall susceptibility to trimethoprim/sulfamethoxazole (Table S3). Three CG101 isolates were colistin resistant. In one isolate of ST2502 (the isolate DG7773), resistance could be attributed to the presence of two mutations in the pmrA gene, which resulted in a mutant PmrA protein (G53V and A217V) that was previously shown to be associated with colistin resistance [28]. In the other two isolates of ST101 (isolates DG5546 and DG5559), the resistance mechanism remained unknown.
A core-genome SNP-based analysis performed with the ST101 isolates revealed that two were identical (differing by 0 SNPs), while the remaining one was more divergent (differing by 440 SNPs vs the others) (Table 1). Inference of phylogenetic relatedness of all CG101 members further confirmed that ST101 isolates clustered in two different sublineages, indicated as A and B (Fig. S1). Interestingly, these two ST101 sublineages also differed by the bla KPC allele (bla KPC-3vs bla KPC-2) and by the subtype of the yersiniabactin siderophore (Ybt-ST 173-2LV vs 183). A comparison with previously sequenced ST101 K. pneumoniae available at the NCBI-NIH database revealed that the ST101 isolates investigated in this work were similar overall to the majority of ST101 strains in terms of carbapenemase gene content and capsule locus type (Table 2).
Table 2.
Summary of the main genetic features of the genomes available in the NCBI-NIH database (accessed on 12 February 2019), belonging to the same STs of those found in this work
|
ST |
Proportion of carbapenemase-positive genomes (no. of isolates/ total no. of sequenced isolates belonging to the same ST) |
Most frequent carbapenemase (no. of isolates) |
Other carbapenemases (no. of isolates) |
Most frequent capsule locus type associated with carbapenemase-positive genomes (no. of isolates) |
Other capsule locus types associated with carbapenemase-positive genomes (no. of isolates) |
Capsule locus types associated with KPC-positive isolates* (no. of isolates) |
Association found in this work* (no. of locally sequenced isolates) |
|---|---|---|---|---|---|---|---|
|
29 |
4.4 % (2/45) |
bla OXA-48-like (2) |
– |
KL54 (2) |
– |
– |
KL30 [bla KPC-3] (1) |
|
35 |
15.5 % (9/58) |
bla OXA-48-like (7) |
bla KPC-2 (1), bla NDM-5 (1) |
KL22 (3) |
KL2 (1), KL16 (3), KL103 (1), KL124 (1) |
KL22 [bla KPC-2] (1) |
KL22 [bla KPC-2] (1) |
|
101 |
68.1 % (130/191) |
bla OXA-48-like (85) |
bla KPC-type (14), bla NDM-type (24), bla VIM-type (5), bla GES-5 (8) |
KL17 (130) |
– |
KL17 [bla KPC-2] (7); KL17 [bla KPC-3] (7) |
KL17 [bla KPC-2] (2); KL17 [bla KPC-3] (1) |
|
147 |
67.4 % (93/138) |
bla OXA-48-like (48) |
bla KPC-type (17), bla NDM-type (28), bla VIM-type (10), bla IMP-1 (1) |
KL64 (73) |
KL10 (17), KL14 (1), KL35 (1), KL107 (1) |
KL64 [bla KPC-2] (11); KL10 [bla KPC-2] (5) |
KL64 [bla KPC-2] (1) |
|
307 |
50.3 % (91/181) |
bla KPC-type (67) |
bla OXA-48-like (18), bla NDM-type (5), bla VIM-type (1) |
KL102 (91) |
– |
KL102 [bla KPC-2] (60); KL102 [bla KPC-3] (6); KL102 [bla KPC-4] (1) |
KL102 [bla KPC-2] (1) |
|
512 |
98.3 % (289/294) |
bla KPC-type (289) |
– |
KL107 (289) |
– |
KL107 [bla KPC-3] (289) |
KL107 [bla KPC-3] (16) |
*[KPC-type].
A core-genome SNP-based analysis performed with ST2502 isolates revealed that these were closely related to each other overall (range 0–7 SNPs distance from reference; mean and median, 4 and 4, respectively) and all carried the bla KPC-3 allele (Table 1, Figs 1 and 2).
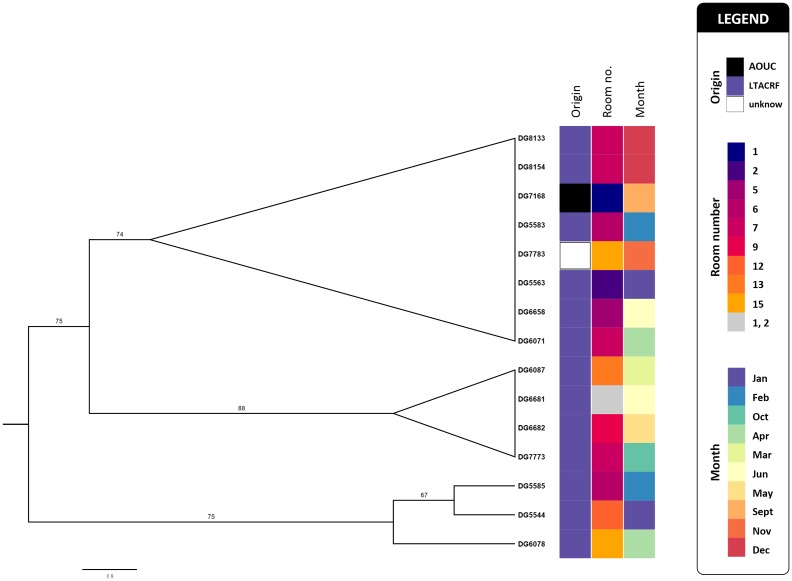
Fig. 2.
A SNP-derived ML phylogenetic tree (midpoint rooted) of K. pneumoniae isolates belonging to ST2502, reconstructed using Snippy and iq-tree (see Methods) and using K. pneumoniae DG5544 as the reference genome (see Table 1 for details). Bootstrap-based branch support values, obtained from 100 replicate trees, are indicated as branch labels. Branches showing bootstrap values less than 50 were collapsed. Bar, substitutions per nucleotide position. Coloured squares give details about the origin of analysed strains, the isolation month during the study period (2016) and about the patients’ room numbers.
With respect to epidemiology, the two ST101 of sublineage A were linked to each other, being from two patients with incident colonization sharing the same room(Figure S1); the ST101 of sublineage B (isolate DG7721) was from an independent, imported case from Florence University Hospital (i.e. the major acute-care hospital referring patients to this LTCRF). The ST2502 isolates were mostly obtained (13 of 15, 86.7 %) from incident colonization cases that occurred during admission to the facility, while only two were imported from other hospitals of the area (Fig. 2).
ST512 isolates
All the ST512 isolates had a KL107 capsule locus type and an O2v2 O antigen locus. None of them carried yersiniabactin, colibactin, aerobactin, salmochelin, rmpA nor rmpA2 genes (Table S3). With respect to the resistome (Fig. 1), all the ST512 isolates carried the bla KPC-3 allele. Most of them also carried genes for resistance to sulfonamides and trimethoprim (sul and/or dfrA types), in overall agreement with their resistance profiles. Aminoglycoside-resistance genes were found in all ST512 isolates. In particular, the aac6-Ib gene, corresponding to the allelic variant b1 in the BIGSdb resistance genes database (https://bigsdb.pasteur.fr/cgi-bin/bigsdb/bigsdb.pl?db=pubmlst_klebsiella_seqdef&page=alleleQuery&locus=aac6p_Ib&submit=1), was always present. However, this did not translate into a homogeneous pattern of aminoglycoside resistance phenotype (Table S3). Six isolates with alterations in the mgrB gene were found, two with an 11 bp deletion (Δ108–118, associated with frameshift and premature termination), and four others with an IS5-like insertion sequence at position 75. However, only two out of these six isolates were phenotypically resistant to colistin.
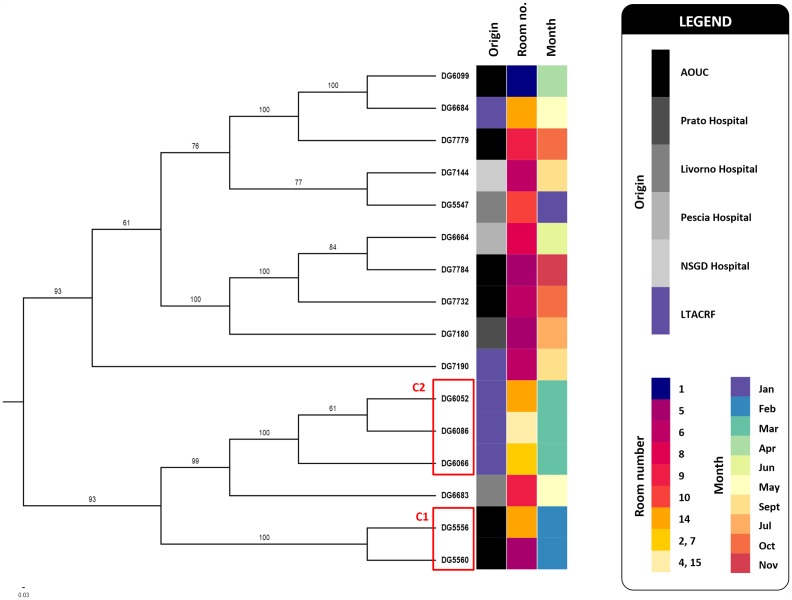
Comparative analysis of the core-genome SNPs revealed some diversity among the ST512 isolates (range 3–57 SNPs distance from the reference; mean and median, 38 and 44, respectively) (Table 1). The ST512 isolates were more frequent among patients already colonized at admission (11/16, 68.8 %), coming from other acute-care hospitals of the area (Fig. 3). One clusters (C1) of closely related ST512, differing by at most 5 SNPs, was apparently imported from the major tertiary-care hospital of the Florence metropolitan area [i.e. AOUC (Florence Careggi University Hospital)]. Some microepidemic events, traceable to cases of incident colonizations by clusters of ST512 isolates, were also detected. An example is one that occurred in March 2016, caused by three isolates belonging a second cluster (C2), which differed from each other by a maximum of 4 SNPs (Fig. 3).
Fig. 3.
A SNP-derived ML phylogenetic tree (midpoint rooted) of K. pneumoniae isolates belonging to ST512, reconstructed using Snippy and iq-tree (see Methods) and using K. pneumoniae DG5547 as the reference genome (see Table 1 for details). Bootstrap-based branch support values, obtained from 100 replicate trees, are indicated as branch labels. Branches showing bootstrap values less than 50 were collapsed. Bar, substitutions per nucleotide position. Coloured squares give details about the origin of analysed strains, the isolation month during the study period (2016) and about the patients’ room numbers. Isolates’ clusters (C), likely reflecting the import sources (C1) or microepidemic events (C2), are indicated by red boxes. NSGD, Nuovo San Giovanni di Dio Hospital.
A comparison with previously sequenced KPC-encoding ST512 isolates, publicly available in the NCBI-NIH database, revealed that they were uniformly very similar to those analysed in this work, being all of KL107 capsule locus type and carrying the bla KPC-3 carbapenemase (Table 2).
Isolates of other lineages
The remaining four isolates were singletons of ST29, ST35, ST147 and ST307. Only the patient with the ST29 (isolate DG6106) was already colonized at admission, while the others acquired the colonization during LTARF admission. The ST29 isolate had a KL30 capsule locus type, as for other previously sequenced ST29 isolates. The isolate was associated with the bla KPC-3 variant (Table 2). At the time of the analysis, there were no other ST29 carrying bla KPC-type genes available in the NCBI-NIH database.
The ST35 isolate had a KL22 capsule locus type, as for the majority of ST35 sequenced isolates. Like the only other previously sequenced KPC-encoding ST35 isolate (from the USA, in 2013), our isolate was associated with the bla KPC-2 variant (Table 2). The ST147 isolate had a KL64 capsule locus type and carried a bla KPC-2 variant, as the majority of the ST147 sequenced isolates (Table 2).
The ST307 isolate had a KL102 capsule locus type and carried a bla KPC-2 variant, as for the majority of the sequenced ST307 isolates. However, by manually inspecting the sequence of this isolate, we found that several genes (wzi, wza, wzb, wzc, wbaP) were missing, apparently due to recombination mediated by an ISKpn14 insertion sequence [29]. Such deletion has never been reported before, and was responsible for a novel deletion variant of KL102. A second capsular gene cluster was found in this isolate, being identical to the one recently described in the ST307 K. pneumoniae strain 48 (KY271410.1) [29]. A similar genetic set of capsular genes, arranged in two capsular loci, was recently found to be highly conserved within the ST307 genetic lineage [29].
Discussion
Altogether, our findings suggest that, in some specific settings, the molecular epidemiology of KPC-Kp can be significantly different from that predicted basing on Italian CRE larger surveillance studies (mostly performed with isolates collected from acute-care hospitals), which demonstrated an overall prevalence of KPC-Kp CG258. [6–10, 30–35]. In particular, our study revealed that new clones of KPC-Kp, infrequent and different from those previously reported in acute-care hospitals [4], can emerge and disseminate in the LTACRF setting. In fact, we identified a new CG101 lineage, named ST2502, which has never been previously reported elsewhere, and that was apparently adapted to this setting where it has apparently established a condition of hyperendemicity. ST2502 could be considered as a new potential high-risk clone, harbouring an extensive set of resistance genes (including the armA methylase, infrequent among KPC isolates circulating in Italy and conferring high-level resistance to all aminoglycosides) and the yersiniabactin virulence determinant.
The study of the fine-tuned clonal relatedness among the isolates allowed by WGS analysis enabled us to trace some pathways of transmission. In particular, we were able to ascertain the common origin of some isolates imported from other hospitals, and to track several clusters of in-LTACRF cross-transmissions, further underscoring the powerful role of WGS analysis for fine-tuned molecular epidemiology of similar resistant strains. An extensive evaluation of the potential reservoirs, other than the gut of colonized patients, of the new clone ST2502 (environment, healthcare workers, etc.) will be performed and will be the subject of further studies.
Data bibliography
1. Raw Illumina sequences have been deposited at the NCBI under the BioProject ID number PRJNA431724 (2018).
2. Full accompanying metadata and phenotypic resistance characterization are available in Table S3 (2019).
3. Accession numbers of publicly available (RefSeq) genomes of K. pneumoniae used for the comparative analysis are reported in Table S2 (2019).
Supplementary Data
Funding information
This work was partially supported by a grant from the Don Carlo Gnocchi Foundation to G.M.R. (project no. 1345).
Acknowledgements
We are grateful to Dr Chiara Marraccini for assistance with bacterial isolate storage.
Author contributions
F.A. (ORCID ID: https://orcid.org/0000-0002-7265-3698): conceptualization, methodology, investigation, data curation, writing – original draft preparation, visualization. V.D.P. (ORCID ID: https://orcid.org/0000-0002-5863-5805): methodology, formal software analysis, investigation, data curation, writing – original draft preparation. F.V.: conceptualization, resources, project administration. L.F.: formal software analysis, data curation. A.A.: investigation. M.C.: investigation. R.P.: supervision. C.M.: supervision. G.M.R.: conceptualization, writing – original draft preparation, writing – review and editing, visualization, supervision, funding.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AOUC, Florence Careggi University Hospital; CG, clonal group; CRE, carbapenem-resistant Enterobacterales; LTACRF, long-term acute-care rehabilitation facility; ML, maximum-likelihood; NCBI, National Center for Biotechnology Information; SBI, severe brain injuries; ST, sequence type; WGS, whole-genome sequencing; Ybt-ST, yersiniabactin sequence type.
All supporting data, code and protocols have been provided within the article or through supplementary data files. One supplementary figure and three supplementary tables are available with the online version of this article.
References
- 1.Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215:S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kadri SS, Adjemian J, Lai YL, Spaulding AB, Ricotta E, et al. Difficult-to-treat resistance in gram-negative bacteremia at 173 US hospitals: retrospective cohort analysis of prevalence, predictors, and outcome of resistance to all first-line agents. Clin Infect Dis. 2018;67:1803–1814. doi: 10.1093/cid/ciy378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gomez-Simmonds A, Uhlemann A-C. Clinical implications of genomic adaptation and evolution of carbapenem-resistant Klebsiella pneumoniae . J Infect Dis. 2017;215:S18–S27. doi: 10.1093/infdis/jiw378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David S, Reuter S, Harris SR, Glasner C, Feltwell T, et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 19 doi: 10.1038/s41564-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broberg CA, Palacios M, Miller VL. Klebsiella: a long way to go towards understanding this enigmatic jet-setter. F1000Prime Rep. 2014;6:64. doi: 10.12703/P6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conte V, Monaco M, Giani T, D'Ancona F, Moro ML, et al. Molecular epidemiology of KPC-producing Klebsiella pneumoniae from invasive infections in Italy: increasing diversity with predominance of the ST512 clade II sublineage. J Antimicrob Chemother. 2016;71:3386–3391. doi: 10.1093/jac/dkw337. [DOI] [PubMed] [Google Scholar]
- 7.Mezzatesta ML, Gona F, Caio C, Adembri C, Dell'utri P, et al. Emergence of an extensively drug-resistant ArmA- and KPC-2-producing ST101 Klebsiella pneumoniae clone in Italy. J Antimicrob Chemother. 2013;68:1932–1934. doi: 10.1093/jac/dkt116. [DOI] [PubMed] [Google Scholar]
- 8.Bonura C, Giuffrè M, Aleo A, Fasciana T, Di Bernardo F, et al. An update of the evolving epidemic of bla KPC carrying Klebsiella pneumoniae in Sicily, Italy, 2014: emergence of multiple non-ST258 clones. PLoS One. 2015;10:e0132936. doi: 10.1371/journal.pone.0132936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Del Franco M, Paone L, Novati R, et al. Molecular epidemiology of carbapenem resistant Enterobacteriaceae in Valle d'Aosta region, Italy, shows the emergence of KPC-2 producing Klebsiella pneumoniae clonal complex 101 (ST101 and ST1789) BMC Microbiol. 2015;15:260. doi: 10.1186/s12866-015-0597-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wyres KL, Hawkey J, Hetland MAK, Fostervold A, Wick RR, et al. Emergence and rapid global dissemination of CTX-M-15-associated Klebsiella pneumoniae strain ST307. J Antimicrob Chemother. 2019;74:577–581. doi: 10.1093/jac/dky492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayden MK, Lin MY, Lolans K, Weiner S, Blom D, et al. Prevention of colonization and infection by Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae in long-term acute-care hospitals. Clin Infect Dis. 2015;60:1153–1161. doi: 10.1093/cid/ciu1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchaim D, Chopra T, Bogan C, Bheemreddy S, Sengstock D, et al. The burden of multidrug-resistant organisms on tertiary hospitals posed by patients with recent stays in long-term acute care facilities. Am J Infect Control. 2012;40:760–765. doi: 10.1016/j.ajic.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Lin MY, Lyles-Banks RD, Lolans K, Hines DW, Spear JB, et al. The importance of long-term acute care hospitals in the regional epidemiology of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae . Clin Infect Dis. 2013;57:1246–1252. doi: 10.1093/cid/cit500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossini A, Di Santo SG, Libori MF, Tiracchia V, Balice MP, et al. Risk factors for carbapenemase-producing Enterobacteriaceae colonization of asymptomatic carriers on admission to an Italian rehabilitation hospital. J Hosp Infect. 2016;92:78–81. doi: 10.1016/j.jhin.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Prabaker K, Lin MY, McNally M, Cherabuddi K, Ahmed S, et al. Transfer from high-acuity long-term care facilities is associated with carriage of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae: a multihospital study. Infect Control Hosp Epidemiol. 2012;33:1193–1199. doi: 10.1086/668435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arena F, Vannetti F, Di Pilato V, Fabbri L, Colavecchio OL, et al. Diversity of the epidemiology of carbapenemase-producing Enterobacteriaceae in long-term acute care rehabilitation settings from an area of hyperendemicity, and evaluation of an intervention bundle. J Hosp Infect. 2018;100:29–34. doi: 10.1016/j.jhin.2018.05.025. [DOI] [PubMed] [Google Scholar]
- 17.CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, Approved Standard M07. 11th edn. Wayne, PA: Clinical and Laboratory Standards Institute; 2018. [Google Scholar]
- 18.Camarlinghi G, Parisio EM, Antonelli A, Nardone M, Coppi M, et al. Discrepancies in fosfomycin susceptibility testing of KPC-producing Klebsiella pneumoniae with various commercial methods. Diagn Microbiol Infect Dis. 2019;93:74–76. doi: 10.1016/j.diagmicrobio.2018.07.014. [DOI] [PubMed] [Google Scholar]
- 19.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom. 2016;2:e000102. doi: 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wick RR, Wyres KL, Gorrie CL, Judd LM, MMC L, et al. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom. 2018;4:mgen.0.000196. doi: 10.1099/mgen.0.000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seemann T. Snippy: fast bacterial variant calling from NGS reads. 2018 https://github.com/tseemann/snippy
- 24.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hadfield J, Croucher NJ, Goater RJ, Abudahab K, Aanensen DM, et al. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics. 2018;34:292–293. doi: 10.1093/bioinformatics/btx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.ECDC Risk Assessment on the Spread of Carbapenemase-Producing Enterobacteriaceae (CPE) Through Patient Transfer Between Healthcare Facilities, with Special Emphasis on Cross-Border Transfer. Stockholm: European Centre for Disease Prevention and Control; 2011. [Google Scholar]
- 27.Ministero della Salute Rome: Ministero della Salute; 2013. Circolare Ministeriale ‘Sorveglianza e Controllo delle Infezioni da Batteri Produttori di Carbapenemasi (CPE)’. [Google Scholar]
- 28.Esposito EP, Cervoni M, Bernardo M, et al. Molecular epidemiology and virulence profiles of colistin-resistant Klebsiella pneumoniae blood isolates from the hospital agency "Ospedale dei Colli," Naples, Italy. Front Microbiol. 2018;16:1463. doi: 10.3389/fmicb.2018.01463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villa L, Feudi C, Fortini D, Brisse S, Passet V, et al. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb Genom. 2017;3:mgen.0.000110. doi: 10.1099/mgen.0.000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mammina C, Palma DM, Bonura C, Anna Plano MR, Monastero R, et al. Outbreak of infection with Klebsiella pneumoniae sequence type 258 producing Klebsiella pneumoniae carbapenemase 3 in an intensive care unit in Italy. J Clin Microbiol. 2010;48:1506–1507. doi: 10.1128/JCM.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richter SN, Frasson I, Franchin E, Bergo C, Lavezzo E, et al. KPC-mediated resistance in Klebsiella pneumoniae in two hospitals in Padua, Italy, June 2009-December 2011: massive spreading of a KPC-3-encoding plasmid and involvement of non-intensive care units. Gut Pathog. 2012;4:4. doi: 10.1186/1757-4749-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giani T, Arena F, Vaggelli G, Conte V, Chiarelli A, et al. Large nosocomial outbreak of colistin-resistant, carbapenemase-producing Klebsiella pneumoniae traced to clonal expansion of an mgrB deletion mutant. J Clin Microbiol. 2015;53:3341–3344. doi: 10.1128/JCM.01017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calia C, Pazzani C, Oliva M, Scrascia M, Lovreglio P, et al. Carbapenemases-producing Klebsiella pneumoniae in hospitals of two regions of Southern Italy. APMIS. 2017;125:491–498. doi: 10.1111/apm.12666. [DOI] [PubMed] [Google Scholar]
- 34.Giani T, Antonelli A, Caltagirone M, Mauri C, Nicchi J, et al. Evolving beta-lactamase epidemiology in Enterobacteriaceae from Italian nationwide surveillance, October 2013: KPC-carbapenemase spreading among outpatients. Euro Surveill. 2017;22:30583. doi: 10.2807/1560-7917.ES.2017.22.31.30583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sotgiu G, Are BM, Pesapane L, Palmieri A, Muresu N, et al. Nosocomial transmission of carbapenem-resistant Klebsiella pneumoniae in an Italian university hospital: a molecular epidemiological study. J Hosp Infect. 2018;99:413–418. doi: 10.1016/j.jhin.2018.03.033. [DOI] [PubMed] [Google Scholar]
- 36.Wu KM, Li LH, Yan JJ, Tsao N, Liao TL, et al. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol. 2009;191:4492–4501. doi: 10.1128/JB.00315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.