Abstract
The use of advanced data analytics and applications of statistical and machine learning approaches (‘AI’) to materials science is experiencing explosive growth recently. In this prospective, we review recent work focusing on generation and application of libraries from both experiment and theoretical tools, across length scales. The available library data both enables classical correlative machine learning, and also opens the pathway for exploration of underlying causative physical behaviors. We highlight the key advances facilitated by this approach, and illustrate how modeling, macroscopic experiments and atomic-scale imaging can be combined to dramatically accelerate understanding and development of new material systems via a statistical physics framework. These developments point towards a data driven future wherein knowledge can be aggregated and used collectively, accelerating the advancement of materials science.
Introduction
The use of statistical and machine learning algorithms (broadly characterized as ‘Artificial Intelligence’ herein) within the materials science community has experienced a resurgence in recent years.1 However, AI applications to material science have ebbed and flowed through the past few decades.2–7 For instance, Volume 700 of the Materials Research Society’s Symposium Proceedings was entitled “Combinatorial and Artificial Intelligence Methods in Materials Science,” more than 15 years ago,8 and expounds on much of the same topics as those at present, with examples including high-throughput screening, application of neural networks to accelerate particle simulations, and use of genetic algorithms to find ground states. One may ask the question as to what makes this resurgence different, and whether the current trends can be sustainable. In some ways this mirrors the rises and falls of the field of AI, which has had several bursts of intense progress followed by ‘AI winters’.9, 10 The initial interest was sparked in 1956,11 where the term was first coined, and although interest and funding was available, computational power was simply too limited. A rekindling began in the late 1980s, as more algorithms (such as backpropagation for neural networks,12 or the kernel method for classification13) were utilized. The recent spike has been driven in large part by the success of deep learning,14 with the parallel rise in GPU and general computational power.15, 16 The question becomes whether the current, dramatic progress in AI can translate to the materials science community. In fact, the key enabling component of any AI application is the availability of large volumes of structured labeled data – which we term in this prospective “libraries.” The available library data both enables classical correlative machine learning, and also opens a pathway for exploration of underlying causative physical behaviors. We argue in this prospective that, when done in the appropriate manner, AI can be transformative not only in that it can allow for acceleration of scientific discoveries, but also that it can change the way materials science is conducted.
The recent acceleration of adoption of AI/machine learning-based approaches in materials science can be traced back to a few key factors. Perhaps most pertinent is the Materials Genome Initiative, which was launched in 2011 with an objective to transform manufacturing via accelerating materials discovery and deployment.17 This required the advancement of high-throughput approaches to both experiments and calculations, and the formation of online, accessible repositories to facilitate learning. Such databases have by now have become largely mainstream with successful examples of databases including Automatic Flow for Materials Discovery (AFLOWLIB),18 Joint Automated Repository for Various Integrated Simulations (JARVIS-DFT),19 Polymer Genome,20 Citrination,21 Materials Innovation Network,22 etc. that host hundreds of thousands of datapoints from both calculations as well as experiments. The timing of the initiative coincided with a rapid increase in machine learning across commercial spaces, largely driven by the sudden and dramatic improvement in computer vision, courtesy of deep neural networks, and the availability of free packages in R or python (e.g., scikit-learn23) to apply common machine learning methods on acquired datasets. This availability of tools, combined with access to computational resources (e.g., through cloud-based services, or internally at large institutions) was also involved. It can be argued that one of the main driving forces within the materials science community was an acknowledgement that many grand challenges, such as the materials design inverse problem, were not going to be solved with conventional approaches. Moreover, the quantities of data that were being acquired, particularly at user facilities such as synchrotrons or microscopy centers, was accelerating exponentially, rendering traditional analysis methods that relied heavily on human input unworkable. In the face of the data avalanche, it was perhaps inevitable that scientists would turn to the methods provided via data science and machine learning.24–26 Please note commercial software is identified to specify procedures. Such identification does not imply recommendation by the National Institute of Standards and Technology.
Thus, the question becomes, how can these newly found computational capabilities and ‘big’ data be leveraged to gain new insights and predictions for materials? There are already some answers. For example, the torrent of data from first principles simulations has been used for high throughput screening of candidate materials, with notable successes.27–29 Naturally, one asks the question as to what insights can be gained from similar databases based not on theory, but on experimental data, e.g. of atomically resolved structures, along with their functional properties. Of course, microstructures have long been optimized in alloy design.21, 30 Having libraries (equivalently, databases) of these structures, with explicit mentioning of their processing history, can be extremely beneficial not just for alloys but for many other material systems, including soft matter.31 These databases can be used for e.g. utilizing known knowledge of similar systems to accelerate the synthesis optimization process, to train models to automatically classifying structures and defects, and to identify materials with similar behaviors that are exhibited, potentially allowing underlying causal relationships to be established.
In this prospective, we focus on the key areas of library generation of material structures and properties, through both simulations/theory, and imaging. High-throughput approaches enable both simulation and experimental databases to be compiled, with the data used to build models that enable property prediction, determine feature importance, and guide experimental design. In contrast, imaging provides the necessary view of microstates enabling the development of statistical mechanical models that incorporate both simulations and macroscopic characterization to improve predictions and determine underlying driving forces. Combining the available experimental and theoretical libraries in a physics-based framework can accelerate materials discoveries, and lead to lasting transformations of the way materials science research is approached worldwide.
Databases, Libraries and Integration
This prospective will focus on theory-led initiatives for database generation (and subsequent machine learning to predict properties and accelerate material discovery) and contrast them with the equally pressing need for their experimental counterparts. While the theory libraries are well ahead, substantial progress in materials science will rely on experimental validation of theoretical predictions and a tight feedback between data-driven models, first principles and thermodynamic modeling, and experimental outcomes. It is also important to note that theoretical databases and libraries operate with an idealized representation, where all inputs and outputs are known and hence of interest are processes such as data compression, determination of reduced descriptors, and integration into analysis workflows. However, the validity and precision of theoretical models is always evolving. In comparison, experimental data will be characterized by the large number of latent or unknown degrees of freedom that may or may not be relevant to specific phenomena.
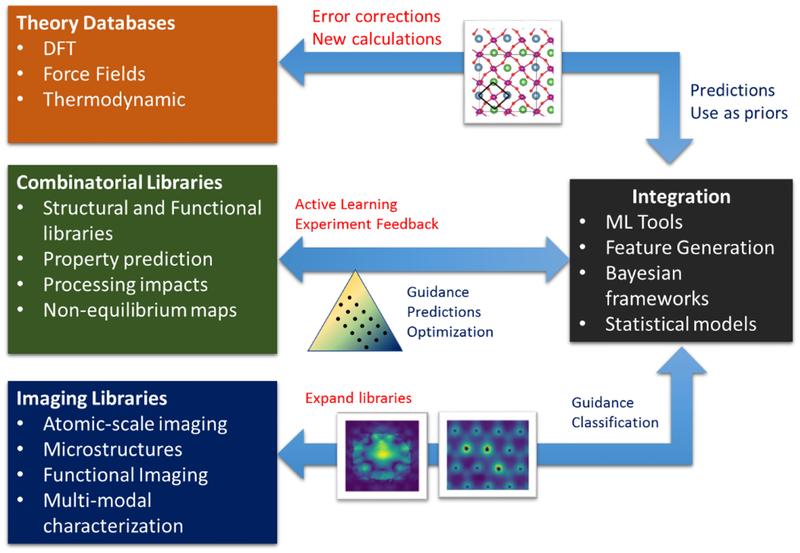
Experimental libraries can be created from combinatorial experiments to rapidly map the composition space and complemented with atomic- and functional imaging to generate libraries that can map local structure to functionality. The broad vision is summarized in Figure 1. The success of any of these individual areas on their own will be limited; experimentally, the search space is much too large to iterate; computationally, prediction of certain properties or the role of defects in e.g. correlated systems remains extremely challenging, and models still need experimental validation. From the imaging standpoint, much work remains to be done in automating the generation of atomic-scale defect libraries, although computer vision and deep-learning based approaches are showing tremendous promise.32, 33 These data, from theory and experiment, across length scales, can then be combined either directly in data-driven models (machine learning), or through more formal methods that consider uncertainty, such as Bayesian methods. This can also be achieved using statistical mechanical models that are refined and fit based on theoretical and experimental data at multiple length scales, allowing understanding of the driving forces for materials behavior, and enabling feedback to experiment and first principles theory.
Figure 1:
Progress in materials science requires understanding driving forces governing phenomena, so that materials can be both discovered and optimized for applications. Fundamentally, accessing the knowledge space to accelerate this cycle requires availability of data from simulations and experiment for materials synthesized under different conditions. Imaging provides a window into local configurations and provides a critical link for understanding the driving forces of observed behavior. Machine learning tools enable the generation of these databases and facilitate rapid prediction of properties from data-driven models. Similarly, the data can be synthesized together in a Bayesian formulation, or using statistical mechanical models, to agglomerate all available sources of information to produce more accurate predictions. Ideally, the knowledge gained will be transferable, enabling more efficient design cycles for similar material systems. These tools all require community efforts for availability of code, data, and workflows, that is critical to realizing this new future.
Our roadmap for this prospective is as follows. We begin with an overview of databases of theoretical calculations, which in many ways catalyzed this field, and which are the most well-established in this area. We then branch from high throughput computations to high throughput experiments, that can be used to generate experimental realizations in rapid time. These are beneficial for exploring macroscopic structure-property relationships. Complementing the macroscopic studies is the need for local imaging libraries, which compare the local atomic or mesoscopic structure with the local functional property. We discuss recent works to address this issue, which has been less well explored, but which are critical for understanding of disordered systems with strong localization. Finally, we explain how these libraries can be utilized in concert and incorporated into a statistical mechanical framework for predictive modeling with quantified uncertainty. We end with a discussion on the challenges at the individual, group and department level, and describe our outlook for material science under this new paradigm.
Theory-based library generation
Whereas for most of humanity materials discovery was largely Edisonian in approach, in the modern era materials design can be facilitated via first principles (and other) simulations that can rapidly explore different candidates in-silico. Computational methods are usually classified in term of length scale, going from quantum atomistic to continuum; however, irrespective of their scale, they all are constrained by the scale of a simulation (length and time), accuracy and transferability. For instance, quantum-based methods, such as density functional theory (DFT), have been phenomenally successful in discovering new materials with important technological applications, such as those used in solid-state batteries,34, 35 dopants for effective strengthening of alloys,36 or 2D materials.37, 38 These methods also aided in explaining physical phenomena such as diffusion mechanisms,39 experimental spectra,40 etc. More recently, DFT41 based high-throughput (HT) approaches have led to the creation of open-source large material property databases such as MaterialsProject,42 AFLOWLIB,18 Open Quantum Materials Database (OQMD),43 Automated Interactive Infrastructure and Database for Computational Science (AiiDA),44 JARVIS-DFT,45 Organic Materials Database (OMDB)46 QM9,47etc. However, DFT is heavily limited by the simulation size to something on the order of a few hundred atoms. Empirical potentials48 help overcome the size issue, as they can simulate millions of atoms. However, they require rigorous potential fitting to simulate reasonable behavior.49, 50 Larger scale methods, such as finite element method and phase field, are limited by depending on critical inputs from experimental data and atomistic simulations.51 Fortunately, machine learning (ML) for materials has evolved to become a promising alternative in solving some of the computational materials science problems mentioned above.52
There are four main components in successfully applying ML to materials: a) acquiring large enough datasets, b) designing feature vectors that can appropriately describe the material, c) implementing a validation strategy for the models, and d) interpreting the machine-learning model where applicable. The first step (a) is facilitated by the generation of the large datasets mentioned above. Step (b) is more complicated: while the databases provide a consistent set of target data, conversion of core material science knowledge to computers require feature vector generation of all those materials in the databases. Chemical descriptors based on elemental properties (for instance, the average of electronegativity and ionization potentials in a compound) have been successfully applied in fields such as alloy-formation53 and have led to for various computational discoveries.53 Nevertheless, this approach is not appropriate when modeling different structure-prototypes with the same composition because ignoring structural information doesn’t allow to differentiate between them. Structural features as descriptors have been recently proposed based on Coulomb matrix,54, 55 partial radial distribution function,56 Voronoi tessellation,57 Fourier-series58 and several others in recent works.59 Features such as classical force-field inspired descriptors (CFID)60 and fragment descriptors61 allow combing structural and chemical descriptors in one unified framework. These are generally a fixed size descriptor of all the samples in the dataset. For example, MagPie53 gives 145 features, while CFID60 gives 1557 descriptors.
A conceptually different way to obtain feature vectors is to generate them automatically using approaches like convolution neural networks,62 SchNet63 and Crystal Graph Convolutional Neural Networks (CGCNN)64 for instance, which extracts the important features by themselves taking advantage of a deep neural network architecture. Most of these methods are applied to specific classes of materials because of presence or absence of periodicity in one or more crystallographic directions, such as crystalline inorganic solids, molecules or proteins, but features such as Coulomb-matrix,54 CFID,60 SchNet,65 MegNet66 and GCNN62, 67 hold a generalized appeal for all classes of materials. Luckily, some of these feature-generators are available in general ML-framework code such as Matminer.68 A comprehensive set of feature vector types, their applications and corresponding resource links are provided in Table 1. The validation strategy consists of reporting accuracy metrics such as the mean absolute error, root mean square error, and R2. Importantly, plots like learning curves and cross-validation plots are standard ways of testing ML models from the data science perspective. Although these are some of the common data-science metrics, physics-inspired validation strategies such as integrating evolutionary approaches with machine learning to map a generalized energy landscape,60, 69 or testing energy-volume curve beyond the training set70 have recently drawn much attention.
Table 1:
Examples of AI based material-property predictions for different types of materials. The types of materials consist of A) 3D inorganic crystalline solids, B) Stable 3D inorganic crystalline solids, C) 2D materials/surfaces, D) Molecules, E) 3D Organic crystals and F) Crystalline polymers.
| Models | Properties trained | Materials (datapoints) | Links |
|---|---|---|---|
| ML based materials screening | |||
| AFLOW-ML61 | Bandgaps, Bulk and shear modulus, Debye temperature, Specific heat, thermal expansion coefficient | A (26,674) | http://aflow.org/aflow-ml/ |
| GBML78 | Bulk and shear modulus | A (1940) | https://github.com/materialsproject/gbml |
| MagPie53 | Volume, band gap energy and formation energy | B (228676) | https://bitbucket.org/wolverton/magpie |
| Matminer68 | Formation energies | A (>3938) | https://hackingmaterials.github.io/matminer, https://github.com/hackingmaterials/matminer |
| JARVIS-ML60 | Formation energies, bandgaps, static refractive indices, magnetic moment, modulus of elasticity and exfoliation energies | A (24549), C (647) | https://www.ctcms.nist.gov/jarvisml, https://github.com/usnistgov/jarvis |
| GCNN62,67 | Zero-point vibrational energy, dipole moment, internal energy, formation energies, bandgaps, elastic properties, etc. | C (20000) | https://github.com/deepchem/deepchem |
| CGCNN64 | Formation and absolute energies, bandgap, Fermi energy, bulk and shear mod, Poisson ratio | A (28046) | https://github.com/txie-93/cgcnn |
| MegNet66 | Zero-point vibrational energy, dipole moment, internal energy, formation energies, bandgaps, elastic properties, etc. | A, C | https://github.com/materialsvirtuallab/megnet |
| Coulomb-matrix54 | Atomization Energies | D (7000) | http://quantum-machine.org/ |
| SchNet65 | Zero-point vibrational energy, dipole moment, internal energy, formation energies, bandgaps, elastic properties, etc. | A, C | https://github.com/atomistic-machine-learning/schnetpack |
| CVAE82 | logP, Quantitative Estimation of Drug-likeness (QED), Highest Occupied Molecular Orbital (HOMO), Lowest Unoccupied Molecular Orbital (LUMO), bandgap | D (>108000) | https://github.com/aspuru-guzik-group/chemical_vae |
| OMDB88 | Bandgap | E (12500) | https://omdb.mathub.io/ |
| KHAZANA89 | Bandgap, dielectric constant (electronic and ionic) | F(284) | https://www.polymergenome.org |
| MolML90 | Atomization energy | C | https://github.com/crcollins/molml |
| QML91 | Atomization energy | C | https://github.com/qmlcode/qml |
| ElemNet | Formation energy | B | https://github.com/dipendra009/ElemNet |
| ML based atomistic potential | |||
| AMP83 | Energy and force | A, C, D | https://amp.readthedocs.io/en/latest/ |
| PINN70 | Energy | A | |
| GAP84 | Energy and force | A | https://github.com/libAtoms/QUIP |
| AGNI85 | Energy and force | A | https://lammps.sandia.gov/doc/pair_agni.html |
| SNAP86 | Energy and force | A, C | https://lammps.sandia.gov/doc/pair_snap.html, https://github.com/materialsvirtuallab/snap |
| PROPhet92 | Energy, force, charge-density | A | https://github.com/biklooost/PROPhet |
| TensorMol93 | Energy and force | C | https://github.com/jparkhill/TensorMol |
| ANI94 | Energy and force | A | https://github.com/isayev/ASE_ANI |
| AENET69 | Energy and force | A | http://ann.atomistic.net/ |
| DeePMD-kit95 | Energy and force | A | https://github.com/deepmodeling/deepmd-kit |
| sGDML96 | Energy and force | A | https://github.com/stefanch/sGDML |
| VAMPnet97 | Energy and force | A | https://github.com/markovmodel/deeptime |
The correlation-based ML models perform well in interpolation but poorly for extrapolation tasks. When combined with the non-differentiability of chemical spaces, it limits the application of classical ML in materials science. An alternative is offered physics-inspired ML, where the extrapolation and interpolation is performed along manifolds corresponding to physically-possible atomic configurations and satisfying basic physical laws and constraints. However, although there has been a lot of work in developing databases and feature vectors, coming up with strategies for physics-based ML models71 still needs much detailed work. Additionally, the interpretability of a model can be vitally important from a scientific understanding perspective. This is motivated by a relatively new area in AI: so-called “Explainable AI”.72 The explainability and the interpretability of the model mainly depend on the type of machine learning algorithm. For example, in refs.60, 61 feature importance plots revealed some of the important machine learning descriptors that guide the model. In models such as graph networks, elemental embeddings can reveal chemical similarity.64, 66
Presently ML models have been primarily used for screening of materials. This is because an ML model for a physical quantity allows to estimate such a quantity much faster than computing it. This allows to probe a much larger space of materials than possible when performing actual calculations, and it is true for any type of computational methodology (DFT, phase field, and continuum modelling, for instance). Once ML has identified the sub-space of materials that likely have the desired property, then those, and only those, are probed using the computational technique of choice, DFT, for instance. In this sense, if traditional methods, such as DFT, have been used as a screening tool for experiments, then ML can act as the screening tool for DFT methods (standard DFT options as well as its hybrid-functional or higher-order corrections). Some of these material screening applications are drug-discovery,73 finding new binary compounds,74 new perovskites,75 full-Heusler compounds,76 ternary oxides,77 hard materials,78 inorganic solid electrolytes,79 high photo-conversion efficiency (PCE) materials,80 2D-materials,60 and superconducting materials.81 Some material science-related ML tools (GBML,78 AFLOW-ML,61 JARVIS-ML60 and OMDB,82 for instance) allow web-based prediction of static properties to further accelerate material screening.
A second, major application of ML techniques to material science is in the realm of developing interatomic potentials, aka force-fields, to simulate the dynamics of a system or to run Monte-Carlo simulations. In this instance, ML is used to determine the parameters used in the phenomenological expression of the energy. Such expressions for the energy are then used to derive all other properties. Finding the right parameters (i.e. fitting the potential) is usually a computationally expensive task because of the very large configurational and parameter multi-dimensional spaces that need to be probed simultaneously while respecting all relevant physical constraints. Some of the atomistic potentials developed using ML are: Atomistic Machine-learning Package (AMP),83 Physically-informed artificial neural networks (PINN),70 Gaussian Approximation Potentials (GAP),84 Agni85 and spectral neighbor analysis potential (SNAP).86 These potentials are shown to surpass conventional interatomic potentials both in accuracy and versatility.87 These models are mainly developed for elemental solids, such as Ta, Mo, W, Al etc., or for few binaries, such as Ni-Mo. Developing force fields for multicomponent systems is still limited due to an exponential increase in the number of ML parameters. However, unlike conventional fitting, these parameters can be optimized in a relatively more systematic way. Importantly, a standard force-field evaluation work-flow, like JARVIS-forcefield (JARVIS-FF)49, 50, still needs to be developed for such ML based force-fields, to understand their generalizability. In fact, verification and validation of these ML-based models is a critical challenge of the field.
Combinatorial libraries and high throughput experimentation
Complementing the theoretical libraries listed above requires experimental libraries that map structures, processing and compositions to functionality. By now numerous outlets exist including Polymer Genome,20 Citrination,21 Dark Reactions,98 Materials Data Facility99 and Materials Innovation Network.22 This again needs to be accomplished at different length scales: microscopic, to better understand the links between microstructure or atomic configurations and macroscopic properties, as well as through macroscopic experiments that explore large regions of the composition space to rapidly map functional phase diagrams. The latter is made possible through high throughput experimentation (HTE).
High throughput experimentation (HTE) and AI tools have been linked since HTE was re-discovered in the early 1990’s. The origins of HTE can be traced back to the early 20th century with the discovery of the Haber-Bosch catalyst100 and the Hanak multi-sample concept.101 In both cases, the investigators realized that the search for new materials with outstanding properties and new mechanisms required a broader search through composition-processing-structure-property space than could be afforded by conventional one-sample-at-a-time techniques. At the time automation and computational resources were limited and so liberal usage of “elbow grease” was required both for performing the experiments and data analysis. It took several decades, the publication of the landmark HTE paper by Xiang et. al.102, and the ready availability of personal computers for this methodology to gain significant traction within the materials community. There have been a number of recent reviews103–106 on the topic and today HTE is largely considered to be a mature field with significant efforts (and discoveries) spanning a large number of fields including catalysis,107 dielectric materials,104 and polymers.108
The creation and deployment of HTE workflows necessarily leads to a bottleneck centered around the need to interpret large (sometimes thousands) of materials data correlated in composition, processing, and microstructure from a single experiment.109, 110 By the early 2000s a single HTE sample containing hundreds of individual samples could be made and measured for a range of characteristics within a week, but the subsequent knowledge extraction of composition, structure, properties of interest, and figure of merit (FOM) often took weeks to months. There were several early international efforts to standardize data formats and create data analysis and interpretation tools for large scale data sets.111 These efforts touched on using AI to enable experimental planning112, 113 and data analysis and visualization.114–117
An unexpectedly difficult exemplar for the field is the mapping of non-equilibrium phase maps through the collection of spectral data as a function of composition and processing, so called “phase mapping.” A great deal of effort has been expended in working with computer scientists to better understand how to effectively correlate diffraction spectra of limited range to phase composition for a given sample. The problem is further exacerbated by peak shift due to alloying, the presence of non-equilibrium phases and distortion of peak intensities due to preferred orientation of crystallites (texturing). The overwhelming majority of this work has focused on using unsupervised techniques such as hierarchical clustering,122 wavelet transformations,123 non-negative matrix factorization124 and constraint programming paired with kernel methods.125 Comparatively little work has been devoted to the use of supervised or semi-supervised techniques.126, 127 A recent review article is available for the interested reader.128 Fully unsupervised techniques face challenges not only from noisy and limited range of experimental data, but also from highly non-linear scaling of the computational resources with number of observations in the dataset. More recent work in the field has sought to impose locality (e.g. that neighboring compositions are likely to include the same phases) into creating the phase map through the use of segmentation techniques129 or by attempting to deconvolve peak shift through the application of convolution nonnegative matrix factorization.130 A common theme for all of these efforts has been the importance of working to translate materials science problems into more general problems that are of interest to computer scientists. These new approaches appear to operate sufficiently rapidly as to permit on-the-fly analysis of diffraction data as it is being taken.131
Once knowledge extraction catches up to HTE synthesis, and characterization, the limit to rate of new materials discovery becomes that of decision making, i.e., what materials to pursue next given the knowledge of materials discovered so far (and processing conditions needed to make them). HTE groups have long worked with theoreticians to identify interesting materials to pursue.132–134 More recently in an effort to decrease the turnover time several HTE groups have turned to the use of AI for hypothesis or lead generation.81, 135, 136 One example of such a AI platform is the Materials-Agnostic Platform for Informatics and Exploration developed at Northwestern, which transforms compositional data into a set of chemical descriptors that can be used to train a ML model that targets a particular property such as the band gap or an alloy’s metallic glass forming capability.53 One additional benefit of HTE experiments is that they produce negative and positive results simultaneously without any additional cost. Thus, the models can use both negative and positive results from HTE experiments to produce less biased models than those based on traditional material discovery campaigns.
A recent example illustrated the power of combining HTE with machine learning models by demonstrating a nearly 1000x acceleration in the rate of the discovery of novel amorphous alloys.136 Amorphous alloys are a particularly apt system to be predicted by ML, as traditional computational approaches like DFT are not particularly effective. From this study several interesting new phenomena were observed. The most notable of which was that the formation of amorphous alloys via physical vapor deposition was more strongly correlated with the presence of complex ordered intermetallic structures than on the traditional presence of deep eutectics. Moreover, the predictions of stability can be coupled with predictions of physical properties (e.g., modulus) and can then be used to guide the discovery of novel high modulus metallic glasses as in Figure 2.
Figure 2:
Illustrating the glass forming ability of a novel Co-V-Zr alloy (left) and its predicted elastic modulus (right).
More recently, the pairing of supervised learning with active learning137–141 – the machine learning implementation of optimal experiment design, has been used to address the dual challenges of hypothesis generation and testing. First a supervised learning method is selected, one that provides uncertainty quantification along with prediction estimates. The output estimate and uncertainty are then exploited by active learning to identify the next experiment to perform that will most rapidy optimize a given objective, e.g., hone in on a material that maximizes or minimizes a functional property. Bayesian optimization, the subset of active learning methods focused on local function optimization, has been used by a number of groups to accelerate the discovery of advanced materials. In these projects, machine learning identifies the material synthesis and fabrication parameter values to investigate next. These values are then used to guide experimentalists in synthesis and characterization and the resulting data is fed back into the machine learning model to select the subsequent experiment. Accelerated materials discovery has been demonstrated for low thermal hysteresis shape memory alloys,71 piezoelectrics with high piezoelectric coupling coefficients,137 and high temperature superconductors.142
Advising systems were the stepping stone to the next level of high throughput experimentation - autonomous systems143, where machine learning is placed in control of the full experiment cycle through direct interfacing with material synthesis and characterization tools. Rather than using a pre-defined grid over which to explore, it would be beneficial to explore the materials space in a more informed manner. Autonomous systems hold great potential, not just in accelerating the experimental cycle by reducing laborious tasks, but also by potentially reducing the amount of prior knowledge and expertise required in synthesis, characterization, and data analysis. Autonomous Research System (ARES) is such a system, capable of optimizing carbon nanotube growth parameters144. ChemOS is another such system, capable of exploring chemical mixtures to achieve a desired optical spectra.145 These systems seek to find the material which optimizes some given properties – a challenge of local optimization. Autonomous systems can also be used for global optimization challenges, e.g. to maximize knowledge gained from a sequence of experiments, as demonstrated by a set of systems capable of autonomous determination of non-equilibrium phase maps across composition and temperature space.146, 147 A similar fusion in chemistry may be the merging chemical robotics systems148 with reaction network models such as CHematica.149
Significant challenges remain before autonomous systems become commonplace. One key challenge is the integration of uncertainty from data collection through machine learning predictions and experiment design. Additionally, many application areas have a wealth of knowledge stored in the literature which can be exploited to accelerate materials exploration and optimization. Extracting this knowledge and making it searchable is another key challenge. Furthermore, researchers are investigating methods for incorporating prior knowledge of materials physics into machine learning frameworks to ensure that predictions are physically realizable. Physical research systems are also susceptible to multiple modes of failure resulting in anomalous data. Anomaly detection and mitigation is thus also required. Integration of physical synthesis and characterization instruments into autonomous platforms is currently restricted by disparate communication protocols and a lack of scriptable interfaces. Accordingly, there is also a need for a data and software platform capable of managing and incorporating diverse data types and communication protocols.
Local structure libraries and functional imaging
The combinatorial libraries above allow rapid scanning of the compositional space. However, for many materials of interest, responses are highly inhomogeneous, for example in materials such as manganites, filamentary superconductors, relaxor ferroelectrics and multiferroic oxides. Due to strong correlations and competing orders, the local atomic and mesoscopic structures, distribution and type of defects and their dynamics are all critically linked to the functionality of these disordered materials. Furthermore, for progress to be made on both understanding the driving forces for their functions, as well as to optimize them for applications, libraries of local atomic-scale structures and ordering are required to complement the macroscopic libraries generated through traditional high throughput experimentation. It should be noted that local imaging studies can provide more evidence as to the structure-property relationships that are of importance. Below, we review some advances in how libraries of atomic-scale defects can be generated using a deep learning approach,24 as well as advances in functional imaging that enable high-throughput local characterization.
Libraries of local structures
Perhaps the most important and least available (at this point) libraries are of atomic-scale structures (configurations) and defects, even in commonly studied materials such as graphene or other 2D materials. This is in comparison to, for instance, libraries of microstructures of alloys, which have been available for years.150 From the statistical physics perspective, access to these microstates should, in principle, enable predictions to be made of the system’s properties as the thermodynamic conditions are varied. Practically, atomic-scale imaging has only become widespread and near routine over the past decade, due in large part due to the proliferation of aberration corrected scanning transmission electron microscopy. Nonetheless, even if atomic scale images are acquired, it is still difficult to manually identify the atomic configurations and classify the types of defects. Indeed, most of the existing “classical” methods of analyzing microscopy data are slow, inefficient and require frequent manual input. Recently, it was demonstrated that deep neural networks151 (aka deep learning) can be trained to perform fast and automated identification of atomic/molecular type and position as well as to spot point-like structural irregularities (atomic defects) in the lattice in static and dynamic scanning transmission electron and scanning tunneling microscopy (STEM and STM) data with varying levels of noise.33, 152, 153 The deep learning approach, and, more generally, machine learning, allows one to generalize from the available labeled images (training set) to make accurate image-level and/or pixel level classification of previously unseen data samples. The training data may come from theoretical simulations, such as a Multislice algorithm154 for electron microscopy or from a (semi-)manual labelling of experimental images by or under a supervision of domain experts.
Fully convolutional neural networks (FCNN),156 which are trained to output a pixel-wise classification maps showing a probability of each pixel in the input image belonging to a certain type of atom and/or atomic defect were shown to be well-suited for the analysis of atomically resolved experimental images. Ziatdinov et al.152 demonstrated that FCNN trained on simulated STEM data of graphene can accurately identify atoms and certain atomic defects in noisy experimental STEM data from a graphene monolayer, including identification of atoms in the regions of the lattice with topological reconstructions that were not a part of the training set. Indeed, these models are eminently transferable. For example, a model based on graphene can perform well on other 2D materials with similar structure, usually without any need for further training. This is particularly important when generating libraries, as continual model training on every system would impede rapid progress.
Furthermore, for the quasi-2D atomic systems, the FCNN output can be mapped onto a graph representing the classical chemical bonding picture, which allows making a transition from classification based on image pixels to classification based on specific chemistry parameters of atomic defects such as bond lengths and bond angles. In such a graph representation, the graph nodes represent the FCNN-predicted atoms of different type, while the graph edges represent bonds between atoms and are constructed using known chemistry constraints, including maximum and minimum allowed bond length between the corresponding pairs of atoms. This FCNN-graphs approach was applied to the analysis of experimental STEM data from a monolayer graphene with Si impurities allowing construction of a library of Si-C atomic defect complexes.155 The FCNNs can also aid studies of solid state reactions on the atomic level observed in dynamic STEM experiments.157 In this case, an FCNN is used in combination with a Gaussian mixture model to extract atomic coordinates and trajectories, and to create a library of the structural descriptors from noisy experimental STEM movies. The associated transition probabilities are then analyzed via a Markov approach to gain insight into the atomistic mechanisms of beam-induced transformations. This was demonstrated for transition probabilities associated with coupling between Mo substitutions and S vacancies in WS2157 and between different Si-C configurations at the edge and in the bulk of graphene.158
While learning the structural properties of atomic defects in materials at the atomic scale is important by itself, it is also critical to understand how the observed structural peculiarities affect electronic and magnetic functionalities at the nanoscale. From the experimental point of view, this requires us to be able to perform both structural (STEM) and functional imaging (STM in the case of electronic properties) on the same sample. Then the goal is to identify the same atomic structures and defects from STEM and STM experiments and to correlate the observed structural properties to measured electronic properties, namely, local density of electronic states at/around the structure of interests. This was recently demonstrated155 via a combined experimental-theoretical approach, where the atomic defects identified via deep learning in STEM structural imaging on graphene with Si dopants were then identified by their density functional theory-calculated electronic fingerprints in the scanning tunneling microscopy measurements of local electronic density of states on the same sample. This work, summarized in Figure 3, shows a realistic path toward the creation of comprehensive libraries of structure-property relationships of atomic defects based on experimental observations from multiple atomically-resolved probes. Such libraries can significantly aid the future theoretical calculations by confining the region of the chemical space that needed to be explored, i.e. by focusing the effort on the experimentally observed atomic defect structures instead of all those that are possible in principle.
Figure 3: Creating local imaging libraries.
(a) Scanning transmission electron microscopy imaging of Si impurities in graphene monolayer. (b) Categorization of defects in (a) based on the number/type of chemical species in their first coordination sphere via deep learning (DL) based approach. (c) The extracted 2d atomic coordinates of these defects are then used as an input into density functional theory (DFT) calculations to obtain a fully-relaxed 3d structure and calculate electronic properties (in this case, the local density of electronic states for the bands below (EV) and above (EC) the Fermi level). (d) The DFT-calculated data can be then used to search for the specific type of defects in the scanning tunneling microscopy (STM) data from the same sample, which measures the local density of states. The search can be performed manually (if the number of STM images is small) or automatically by training a new machine learning (ML) classifier for categorizing the STM data. Image adapted from Ziatdinov et al.155
The current challenges include improvement of infrastructure for cross-platform measurements (sample transfer, automated location of the same nanoscale regions on different platforms) as well as absence of a standard data format for storing and processing these libraries, which is accepted and used by the entire community. There is also the need to collate data across existing platforms, and thus searchability to find the relevant data is another major issue that will need to be addressed.
Functional libraries facilitated with rapid functional imaging
A similar argument can be made for the need for functional property libraries derived from local measurements. Due to the varying local structure in disordered materials, this requires the mesoscopic functionalities to be mapped across the sample, which then facilitates learning the microstructural features that are associated with the observed response. Multiple examples of the imaging techniques that can be applied for these applications are versions of scanning probe microscopy for mapping elastic and electromechanical properties,159 chemical imaging via microRaman160 and time of flight secondary ion mass spectroscopy (ToF-SIMS)161, and nano X ray methods.162, 163 Critical for these applications becomes the issues of physics-based data curation, i.e. the transition from the measured microscope signal to material-specific information. In certain techniques such as Piezoresponse Force Microscopy (PFM), the measured signal is fundamentally quantitative, and with the proper calibrating of the measurement signal can be used as a material-specific descriptor.164, 165 In other techniques such as scanning probe microscopy (SPM)-based elastic measurements or scanning tunneling microscopy the measured signal is a linear or non-linear convolution of the probe and material’s properties, and quantification of materials’ behaviors represents a significantly more complex problem.166 Similarly of interest is the combination of information from multiple sources, realized in multimodal imaging. Here, once the data is converted from microscope-dependent to material dependent, and multiple information sources are spatially aligned, the joint data sets can be mined to extract structure-property relationships.167–169
However, performing experiments is time and labor intensive, and more automated methods of exploring the space and recognizing important areas (such as extended defects, or domain junctions) are necessary. For reducing the labor-intensive portion, ML has been shown to be of substantial utility. For instance, in SPM, an ML-utilizing workflow for a bacterial classification task was originally proposed by Nikiforov et al.4 There, the authors used the measured PFM signal and trained a neural network to enable automatic recognition of bacteria classes, as distinguished by their electromechanical (i.e., PFM) response. Beyond simple classification tasks, the ML methods in SPM have also been useful to extract fitting parameters from noisy hysteresis loop data,170 to enable better functional fits,171 and for phase diagram generation.172, 173 These tools greatly reduce the labor component of acquiring functional imaging, although much work remains.
Still, despite the increasing speed and utility of ML methods in this space, much of the local functional property measurements are inherently time-intensive. For example, traditional spectroscopic methods in SPM, even for seemingly straightforward properties such as the local electrical resistance R(V) where V is the applied voltage to the probe, or the electric-field induced strain S(E) where E is the applied electric field, can take several hours to acquire with conventional atomic force microscopy methods. How can one gain efficiency in this step? One method is to instead collect low-resolution datasets and attempt to reconstruct the high-resolution version with data-fusion methods.174 Recently, large efficiency gains were made via the use of the so-called “General mode” (G-Mode) platform175 in a range of functional imaging by SPM methods. The success of this approach lies largely in the simplicity. The G-mode platform is built on the foundation of complete data acquisition from available sensors, filtering the data via machine learning or statistical methods and subsequent analysis to extract the relevant material parameters. It has since been applied to a raft of SPM modes including current-voltage (I-V) curve acquisition,176 piezoresponse force microscopy175 and spectroscopy,177 and Kelvin Probe force microscopy.178
Consider also the acquisition of local hysteresis loops in ferroelectric materials, typically accomplished via piezoresponse force spectroscopy. Fundamental ferroelectric switching is extremely fast (≈ GHz), and photodetectors can easily operate at ≈ 4 to 10MHz, but heterodyne detection methods average data over time, leading to captures at much lower rates, and typically acquiring one hysteresis loop per second. The reason is that detection and excitation are decoupled, and each excitation is followed by a long (few ms) wave packet for the detection. This problem can be circumvented by using a dynamic mode, where the deflection of the cantilever is continually monitored and stored as a large excitation is applied at a rapid rate (e.g., 10 kHz) to the tip (see Figure 4(a)). If the voltage applied locally exceeds the local coercive voltage of the ferroelectric material, then polarization switching occurs, leading to switching at the excitation frequency. Reconstruction of the signal via signal filtering methods enables generation of the hysteresis loop, as shown in Fig. 4(b). This technique enables acquisition of hysteresis loops while scanning, and ultimately, in a ~1000x increase in throughput, in addition to providing much more statistics on the process.
Figure 4:
(a) General-mode acquisition (G-mode) differs from a standard measurement, in that the raw data is stored without pre-processing such as use of a lock-in amplifier. (b) The raw response of the cantilever deflection signal is Fourier transformed and processed using a band-pass filter and a user-defined noise floor threshold. The cleaned data is then transformed back to the time domain to reconstruct the hysteresis loops. Since many hysteresis loops are captured, the data is better represented as a 2D histogram (bottom right). This enables rapid mapping of relevant material parameters, such as the coercive voltage. This can in principle be stored along with global (macroscopic) characterization to populate libraries of materials behavior. Figure is adapted from Somnath et al.177
As another point, consider the situation for obtaining the resistance R(V) spectra from a single point on a sample in typical scanning tunneling microscopy or atomic force microscopy. Traditionally, the waveform applied to the tip (or sample) is stepped, and a delay time is added after each step to minimize the parasitic capacitance contribution to the measured current. This scheme is shown in Figure 5(a). This is remarkably effective, but also dramatically limits the acquisition speed to ≈ 1 to 2 curves per second for realistic instrument bias waveforms. However, current amplifiers that are used can operate at several kHz without hindrance, suggesting that the fundamental limits lie much higher. Indeed, if one captures an I-V curve by applying a sine wave at several hundred Hz (and measures the raw current from the amplifier at the full bandwidth), it is possible to obtain I-V traces that, although beset with a capacitance contribution, still contain the relevant information. Given that the circuit can be modeled, Bayesian inference can then be used to determine the capacitance contribution and provide the reconstructed resistance curves as a function of voltage, with uncertainty as shown in Fig. 5(b). The reconstructed traces can then be analyzed further, for example to gauge the disorder in the polarization switching within each capacitor (as in Fig. 5(c)), or to analyze the local capacitance contribution. The advantage of this method is not only that it enables functional imaging of electrical properties at hundreds of times the current state of the art; but it also allows to do so with greater spectral and spatial resolution.
Figure 5: G-IV for rapid mapping of local electronic conductance.
(a) Typical I-V measurements on SPM platforms utilize a regimen where after the voltage is stepped to the new value, a delay time is introduced before the current is averaged, as shown in the inset. On the other hand, the G-IV mode utilizes sinusoidal excitation at high frequency (200 Hz in this case), with results shown for a single point on a ferroelectric PbZr0.2Ti0.8O3 (PZT) nanocapacitor in (b). The raw current (Imeas), the reconstructed current (Irec) given the resistance-capacitance (RC) circuit model, and the inferred current without the capacitance contribution (IBayes) are plotted. This method also allows the uncertainty in the inferred resistance traces to be determined, as shown in the respective plots of R(V) with the standard deviation shaded. White space indicates areas where the resistance is too high to be accurately determined. Reconstructing the current after the measurement can facilitate rapid mapping of switching disorder in the nanocapacitors, with the computed parameter for disorder mapped in (c). Figure is adapted from Somnath et al.176
Reiterating, the idea in these experiments is to produce libraries of functionality that can be used synergistically with libraries of the atomic or mesoscopic structures. One can imagine, for example, libraries of defects in 2D materials with corresponding functional property mapping of the opto-electronic properties of the same materials. The challenge is that many of the techniques for functionality mapping with scanning probe are also not necessarily amenable to high-speed and require substantial calibration efforts (e.g., to obtain quantitative maps of the converse piezoelectric coefficient179), but those need either advances in instrumentation180 or automated characterization systems, if large-scale libraries with local functional properties are to be built.
Another major challenge which arises in the formation of these libraries is the choice of format. This is a major topic that is not a portion of this prospective, but which is undoubtedly important and needs mentioning. We envision that the most likely scenario is multiple databases specialized around the specific type of data being housed, e.g. theory calculations, crystallography, mechanical properties, imaging studies, and so forth. Regardless, in all cases we note that it is important to have open, well-documented and standardized data models, to enable better integration.
From libraries to integrated knowledge
Integration of the experiments and simulations across scales is obviously not a simple endeavor, and no universal solution is likely. Numerous efforts have been made in this regard, including for example the very extensive work on microstructural modeling and optimization,182–184 as well as efforts to combine theory and experiment to rationally design new polymers with specific functional properties.185 One can also combine information from multiple sources within a Bayesian framework, to guide experimental design and reduce the time (number of experimental or simulation iterations) to arrive at an optimal result (under uncertainty).140, 186, 187 These methods typically use some objectives based on a desired property of interest. Methods such as transfer learning188 can be useful to combine computational and experimental data, when the data is scarce. Similarly, augmentation can be a useful strategy, as has recently been show for x-ray datasets.189
There is also an alternative view, which is to consider that structure defines all properties, and that imaging and macroscopic experiments can be combined to constrain generative models based on statistical physics. The key to this pathway lies in theory-experiment matching, which should be done in a way that respects the local statistical fluctuations, which contain information about the system’s response to external perturbations. Recently, we have formulated a statistical distance framework that enables this task.190–192 The optimization drives the model to produce data statistically indistinguishable from the experiment, taking into account the inherent sampling uncertainty. The resulting model then allows predicting behavior beyond the measured thermodynamic conditions.
For example, consider a material of a certain composition that has been characterized macroscopically, so that its composition and crystal structure are known. If atomically resolved imaging data is available, then the next step becomes to identify the atomic configurations present, i.e., practically the position and chemical identity of the surface atoms. Chemical identification of atomic elements in scanning tunneling microscopy images can be complicated, but first principles calculations can help guide the classification. Deep convolutional neural networks trained on simulated images from the DFT can then be run on the experimentally observed images to perform the automated atomic identification. From here, local crystallography193 tools can be used to map the local distortions present, and to determine the configurations of nearest and next-nearest neighbors (and higher if need be) of each atom in the image, to produce an atomic configurations histogram. This can then be used to constrain a simple statistical mechanical model (e.g., lattice model) with easily interpretable parameters in the form of effective interaction energies (note this can also be guided by first principles theory). The histograms produced from experiment and theory can be computed and the model can be optimized via minimization of the statistical distance (see Figure 6(a)) between the histograms. As an example, this concept has recently been used to explore phase separation in an FeSe0.45Te0.55 superconductor, for which atomically resolved imaging was available. The image is shown in Fig. 6(b), with red (Te) and blue (Se) atoms determined via a threshold that would preserve the nominal composition of the sample. A simple lattice model that considered the interactions between the Te and Se atoms was setup and optimized based on the statistical distance minimization approach. As can be seen in the histograms of atomic configurations in Fig. 6(d), the model closely matches the observed statistics from experiment. This optimized model can then be used to sample configurations at different temperatures, as shown in Fig. 6(e).
Figure 6: Statistical Distance Framework.
(a) Statistical distance between a model P and a target Q is defined as a distance in probability space of the local configurations. This metric enables estimation of the ability to distinguish samples arising from thermodynamic systems (under equilibrium considerations). (b) Scanning tunneling microscopy image of FeSe0.45Te0.55 system with Se atoms (dark contrast) and Te atoms (bright contrast). The structure of the unit cell is shown in (c). (d) Atomic configurations histogram from both the data and the optimized model in blue and teal colors, as well as from a model that has no interactions (i.e., is random) plotted in red. Once the generative model is optimized, it can be run sampled for different temperatures, as in (e). Note that reduced T units are utilized. Reprinted (adapted) with permission from Vlcek et al.192 Copyright (2017) American Chemical Society.
It is important here to highlight the key points of this approach. The main idea is that by knowing the atomic configurations, we can learn the underpinning physics as these configurations present a detailed probe into the system’s partition function. This can be compared with e.g. time-based spectroscopies, where observations of fluctuations enables mapping the full potential energy surface, as has been done for biomolecules.194 Here, instead of dealing with fluctuations in time, we observe the spatial fluctuations that are quenched within the solid. At the same time, given that the models are physics-based, they are generalizable and should be predictive, thus enabling extrapolation rather than simply interpolation. This may be especially useful for systems where the order parameter is not easily defined, such as relaxors,195 where the goal would be to determine how the statistics of atomic configurations (in particular, the relevant distortions) evolve through phase transitions. The combination of local structure and functional information, macroscopic characterization and first principles theory can therefore be used within this framework to integrate our knowledge and build predictive models that can guide materials discovery and experimental design.
Challenges remain in the areas of uncertainty quantification (how reliable are the predictions as the thermodynamic conditions diverge from those in experiment), as well as how best to choose the appropriate complexity of the model. Moreover, there are challenges associated with non-equilibrium systems that need to be addressed. Practically, there is also much difficulty in actually determining where to retrieve the necessary data, given that it is likely to be strewn across multiple databases. Ideally, these models could be incorporated at the experimental site (e.g., at the microscope) for enabling real-time predictions of sample properties, and guiding the experimenter to maximize information gain, thereby creating efficiencies, whilst automatically adding to the available library. However, this is still a work in progress.
Community Response
Finally, it is worth mentioning that the vision laid out in this prospective requires efforts of individuals, groups and the wider materials community to be successful. Whilst in principle this is no different to the incremental, community-driven progress that has characterized modern science in decades past, there are distinct challenges that deserve attention. One aspect is the sharing of codes and datasets through online repositories, which should be encouraged. Creating curated datasets and well-documented codes takes time, and this should be recognized via appropriate incentives. Sharing codes can be done via use of tools such as Jupyter notebooks run on the cloud. Ensuring that data formats within individual laboratories and organizations are open, documented and standardized requires much work, but pays off in terms of efficiency gains in the long term. Towards this aim, a subset of the authors has created the universal spectral imaging data model (USID181), while the crystallography community is well-versed with the CIF format.196 Logging the correct meta-data with each experiment is critical, and lab notebooks can be digitized to enable searchability and indexing. Perhaps most importantly, teaching and educating the next generation of scientists to be well-versed in data, in addition to machine learning, is essential.
Outlook
The methods outlined in this prospective offer the potential to accelerate materials development via an integrated approach combining high throughput computation and experimentation, imaging libraries and statistical physics-based modeling. In the future, autonomous systems that can utilize this knowledge and perform on the fly optimization (e.g., using reinforcement learning) may become feasible. This would result in ever increasing sizes of the libraries, but also more efficient search and optimization. But perhaps less acknowledged is that given the large libraries that are expected to be built, the chance to learn causal laws197 from this data becomes a reality. Indeed, this is likely to be easier in the case of physics or materials science than in other domains due to the availability of models. In all cases, the availability of such databases and coupling with theoretical and ML methods offers the potential to substantially alter how materials science is approached.
Acknowledgements
The work was supported by the U.S. Department of Energy, Office of Science, Materials Sciences and Engineering Division (R.K.V., S.V.K.). A portion of this research was conducted at and supported (MZ) by the Center for Nanophase Materials Sciences, which is a US DOE Office of Science User Facility.
References
- 1.Agrawal A and Choudhary A: Perspective: Materials informatics and big data: Realization of the “fourth paradigm” of science in materials science. APL Mater. 4, 053208 (2016). [Google Scholar]
- 2.Gakh AA, Gakh EG, Sumpter BG and Noid DW: Neural network-graph theory approach to the prediction of the physical properties of organic compounds. Journal of Chemical Information and Computer Sciences 34, 832 (1994). [Google Scholar]
- 3.Sumpter BG, Getino C and Noid DW: Neural network predictions of energy transfer in macromolecules. The Journal of Physical Chemistry 96, 2761 (1992). [Google Scholar]
- 4.Nikiforov M, Reukov V, Thompson G, Vertegel A, Guo S, Kalinin S and Jesse S: Functional recognition imaging using artificial neural networks: applications to rapid cellular identification via broadband electromechanical response. Nanotechnology 20, 405708 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Currie KR and LeClair SR: Self-improving process control for molecular beam epitaxy. The International Journal of Advanced Manufacturing Technology 8, 244 (1993). [Google Scholar]
- 6.Bensaoula A, Malki HA and Kwari AM: The use of multilayer neural networks in material synthesis. IEEE transactions on semiconductor manufacturing 11, 421 (1998). [Google Scholar]
- 7.Lee KK, Brown T, Dagnall G, Bicknell-Tassius R, Brown A and May GS: Using neural networks to construct models of the molecular beam epitaxy process. IEEE Transactions on Semiconductor Manufacturing 13, 34 (2000). [Google Scholar]
- 8.Takeuchi I, Koinuma H, Amis EJ, Newsam JM, Wille LT and Buelens C: SYMPOSIUM S Combinatorial and Artificial Intelligence Methods in Materials Science. Materials Research Society Symposium Proceedings Series 700 (2002). [Google Scholar]
- 9.Bohannon J: Fears of an AI pioneer. Science 349, 252 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Sejnowski TJ: The deep learning revolution, (MIT Press; 2018). [Google Scholar]
- 11.McCarthy J, Minsky ML, Rochester N and Shannon CE: A proposal for the dartmouth summer research project on artificial intelligence, August 31, 1955. AI magazine 27, 12 (2006). [Google Scholar]
- 12.LeCun Y: A theoretical framework for back-propagation, in Proceedings of the 1988 connectionist models summer school, edited by Touresky D, Hinton G and Sejnowski T (Morgan Kaufmann, CMU, Pittsburgh, Pa, 1988), pp. 21. [Google Scholar]
- 13.Boser BE, Guyon IM and Vapnik VN: A training algorithm for optimal margin classifiers, in Proceedings of the fifth annual workshop on Computational learning theory (ACM, Pittsburgh, Pennsylvania, USA, 1992), pp. 144. [Google Scholar]
- 14.LeCun Y, Bengio Y and Hinton G: Deep learning. Nature 521, 436 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Brodtkorb AR, Hagen TR and Sætra ML: Graphics processing unit (GPU) programming strategies and trends in GPU computing. J. Parallel Distrib. Comput 73, 4 (2013). [Google Scholar]
- 16.Rupp K: 42 Years of Microprocessor Trend Data, (2018). [Google Scholar]
- 17.de Pablo JJ, Jones B, Kovacs CL, Ozolins V and Ramirez AP: The materials genome initiative, the interplay of experiment, theory and computation. Curr. Opin. Solid State Mater. Sci 18, 99 (2014). [Google Scholar]
- 18.Curtarolo S, Setyawan W, Wang S, Xue J, Yang K, Taylor RH, Nelson LJ, Hart GL, Sanvito S and Buongiorno-Nardelli M: AFLOWLIB. ORG: A distributed materials properties repository from high-throughput ab initio calculations. Comput. Mater. Sci 58, 227 (2012). [Google Scholar]
- 19.Choudhary K: Jarvis-DFT, (2014). [Google Scholar]
- 20.Kim C, Chandrasekaran A, Huan TD, Das D and Ramprasad R: Polymer Genome: A Data-Powered Polymer Informatics Platform for Property Predictions. J. Phys. Chem. C 122, 17575 (2018). [Google Scholar]
- 21.C. Informatics: Open Citrination Platform. [Google Scholar]
- 22.Georgia Institute of Technology: Institute for Materials Materials Innovation Network, (2019). [Google Scholar]
- 23.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R and Dubourg V: Scikit-learn: Machine learning in Python. Journal of Machine Learning Research 12, 2825 (2011). [Google Scholar]
- 24.Kalinin SV, Sumpter BG and Archibald RK: Big–deep–smart data in imaging for guiding materials design. Nat. Mater 14, 973 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Kusiak A: Smart manufacturing must embrace big data. Nature News 544, 23 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Bonnet N: Artificial intelligence and pattern recognition techniques in microscope image processing and analysis, in Advances in Imaging and Electron Physics (Elsevier; 2000), pp. 1. [Google Scholar]
- 27.Nyshadham C, Oses C, Hansen JE, Takeuchi I, Curtarolo S and Hart GL: A computational high-throughput search for new ternary superalloys. Acta Mater 122, 438 (2017). [Google Scholar]
- 28.Isayev O, Fourches D, Muratov EN, Oses C, Rasch K, Tropsha A and Curtarolo S: Materials cartography: representing and mining materials space using structural and electronic fingerprints. Chemistry of Materials 27, 735 (2015). [Google Scholar]
- 29.de Pablo JJ, Jackson NE, Webb MA, Chen L-Q, Moore JE, Morgan D, Jacobs R, Pollock T, Schlom DG, Toberer ES, Analytis J, Dabo I, DeLongchamp DM, Fiete GA, Grason GM, Hautier G, Mo Y, Rajan K, Reed EJ, Rodriguez E, Stevanovic V, Suntivich J, Thornton K and Zhao J-C: New frontiers for the materials genome initiative. npj Computational Materials 5, 41 (2019). [Google Scholar]
- 30.Adams BL, Kalidindi S and Fullwood DT: Microstructure sensitive design for performance optimization, (Butterworth-Heinemann; 2012). [Google Scholar]
- 31.Huan TD, Mannodi-Kanakkithodi A, Kim C, Sharma V, Pilania G and Ramprasad R: A polymer dataset for accelerated property prediction and design. Scientific data 3, 160012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ziatdinov M, Jesse S, Vasudevan RK, Sumpter BG, Kalinin SV and Dyck O: Tracking atomic structure evolution during directed electron beam induced Si-atom motion in graphene via deep machine learning. arXiv preprint arXiv:1809.04785 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Madsen J, Liu P, Kling J, Wagner JB, Hansen TW, Winther O and Schiøtz J: A Deep Learning Approach to Identify Local Structures in Atomic-Resolution Transmission Electron Microscopy Images. Advanced Theory and Simulations 1 (2018). [Google Scholar]
- 34.Kang B and Ceder G: Battery materials for ultrafast charging and discharging. Nature 458, 190 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Richards WD, Miara LJ, Wang Y, Kim JC and Ceder G: Interface stability in solid-state batteries. Chem. Mater 28, 266 (2015). [Google Scholar]
- 36.Kirklin S, Saal JE, Hegde VI and Wolverton C: High-throughput computational search for strengthening precipitates in alloys. Acta Mater. 102, 125 (2016). [Google Scholar]
- 37.Mounet N, Gibertini M, Schwaller P, Campi D, Merkys A, Marrazzo A, Sohier T, Castelli IE, Cepellotti A and Pizzi G: Two-dimensional materials from high-throughput computational exfoliation of experimentally known compounds. Nat. Nanotechnol 13, 246 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Choudhary K, Kalish I, Beams R and Tavazza F: High-throughput Identification and Characterization of Two-dimensional Materials using Density functional theory. Sci. Rep 7, 5179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mo Y, Ong SP and Ceder G: Insights into diffusion mechanisms in P2 layered oxide materials by first-principles calculations. Chem. Mater 26, 5208 (2014). [Google Scholar]
- 40.Beams R, Cançado LG, Krylyuk S, Kalish I, Kalanyan B, Singh AK, Choudhary K, Bruma A, Vora PM and Tavazza F.A.n.: Characterization of Few-Layer 1T′ MoTe2 by Polarization-Resolved Second Harmonic Generation and Raman Scattering. ACS nano 10, 9626 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sholl D and Steckel JA: Density functional theory: a practical introduction, (John Wiley & Sons; 2011). [Google Scholar]
- 42.Jain A, Ong SP, Hautier G, Chen W, Richards WD, Dacek S, Cholia S, Gunter D, Skinner D and Ceder G: Commentary: The Materials Project: A materials genome approach to accelerating materials innovation. APL Materials 1, 011002 (2013). [Google Scholar]
- 43.Kirklin S, Saal JE, Meredig B, Thompson A, Doak JW, Aykol M, Rühl S and Wolverton C: The Open Quantum Materials Database (OQMD): assessing the accuracy of DFT formation energies. npj Comp. Mater 1, 15010 (2015). [Google Scholar]
- 44.Pizzi G, Cepellotti A, Sabatini R, Marzari N and Kozinsky B: AiiDA: automated interactive infrastructure and database for computational science. Comput. Mater. Sci 111, 218 (2016). [Google Scholar]
- 45.Choudhary K, Cheon G, Reed E and Tavazza F: Elastic properties of bulk and low-dimensional materials using van der Waals density functional. Phys. Rev. B 98, 014107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geilhufe RM, Olsthoorn B, Ferella A, Koski T, Kahlhoefer F, Conrad J and Balatsky AV: Materials Informatics for Dark Matter Detection. arXiv preprint arXiv:.06040 (2018). [Google Scholar]
- 47.Ramakrishnan R, Dral PO, Rupp M and Von Lilienfeld OA: Quantum chemistry structures and properties of 134 kilo molecules. Sci. Data 1, 140022 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allen MP and Tildesley DJ: Computer simulation of liquids, (Oxford university press; 2017). [Google Scholar]
- 49.Choudhary K, Biacchi AJ, Ghosh S, Hale L, Walker ARH and Tavazza F: High-throughput assessment of vacancy formation and surface energies of materials using classical force-fields. J. Phys. Cond. Mat 30, 395901 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Choudhary K, Congo FYP, Liang T, Becker C, Hennig RG and Tavazza F: Evaluation and comparison of classical interatomic potentials through a user-friendly interactive web-interface. Sci. Data 4, 160125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogata S, Lidorikis E, Shimojo F, Nakano A, Vashishta P and Kalia RK: Hybrid finite-element/molecular-dynamics/electronic-density-functional approach to materials simulations on parallel computers. Comput. Phys. Commun 138, 143 (2001). [Google Scholar]
- 52.Butler KT, Davies DW, Cartwright H, Isayev O and Walsh A: Machine learning for molecular and materials science. Nature 559, 547 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Ward L, Agrawal A, Choudhary A and Wolverton C: A general-purpose machine learning framework for predicting properties of inorganic materials. npj Comp. Mater 2, 16028 (2016). [Google Scholar]
- 54.Rupp M, Tkatchenko A, Müller K-R and Von Lilienfeld OA: Fast and accurate modeling of molecular atomization energies with machine learning. Phys. Rev. Lett 108, 058301 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Faber F, Lindmaa A, O.A.v. Lilienfeld and R. Armiento: Crystal structure representations for machine learning models of formation energies. Int. J. Quantum Chem 115, 1094 (2015). [Google Scholar]
- 56.Schütt K, Glawe H, Brockherde F, Sanna A, Müller K and Gross E: How to represent crystal structures for machine learning: Towards fast prediction of electronic properties. Phys. Rev. B 89, 205118 (2014). [Google Scholar]
- 57.Ward L, Liu R, Krishna A, Hegde VI, Agrawal A, Choudhary A and Wolverton C: Including crystal structure attributes in machine learning models of formation energies via Voronoi tessellations. Phys. Rev. B 96, 024104 (2017). [Google Scholar]
- 58.Bartók AP, Kondor R and Csányi G: On representing chemical environments. Phys. Rev. B 87, 184115 (2013). [Google Scholar]
- 59.Faber FA, Hutchison L, Huang B, Gilmer J, Schoenholz SS, Dahl GE, Vinyals O, Kearnes S, Riley PF and von Lilienfeld OA: Prediction errors of molecular machine learning models lower than hybrid DFT error. J. Chem. Theory Comput 13, 5255 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Choudhary K, DeCost B and Tavazza F: Machine learning with force-field inspired descriptors for materials: fast screening and mapping energy landscape. arXiv preprint arXiv:.07325 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Isayev O, Oses C, Toher C, Gossett E, Curtarolo S and Tropsha A: Universal fragment descriptors for predicting properties of inorganic crystals. Nat. Commun 8, 15679 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kearnes S, McCloskey K, Berndl M, Pande V and Riley P: Molecular graph convolutions: moving beyond fingerprints. J. Comput. Aided Mol. Des 30, 595 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schütt K, Kindermans P-J, Felix HES, Chmiela S, Tkatchenko A and Müller K-R: SchNet: A continuous-filter convolutional neural network for modeling quantum interactions, in Advances in Neural Information Processing Systems (2017), pp. 991. [Google Scholar]
- 64.Xie T and Grossman JC: Crystal Graph Convolutional Neural Networks for an Accurate and Interpretable Prediction of Material Properties. Phys. Rev. Lett 120, 145301 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Schütt KT, Sauceda HE, Kindermans P-J, Tkatchenko A and Müller K-R: SchNet–A deep learning architecture for molecules and materials. J. Chem. Phys 148, 241722 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Chen C, Ye W, Zuo Y, Zheng C and Ong SP: Graph Networks as a Universal Machine Learning Framework for Molecules and Crystals. arXiv preprint arXiv:.05055 (2018). [Google Scholar]
- 67.Gilmer J, Schoenholz SS, Riley PF, Vinyals O and Dahl GE: Neural message passing for quantum chemistry. arXiv preprint arXiv:.01212 (2017). [Google Scholar]
- 68.Ward L, Dunn A, Faghaninia A, Zimmermann NE, Bajaj S, Wang Q, Montoya J, Chen J, Bystrom K and Dylla M: Matminer: An open source toolkit for materials data mining. Comput. Mater. Sci 152, 60 (2018). [Google Scholar]
- 69.Artrith N, Urban A and Ceder G: Efficient and accurate machine-learning interpolation of atomic energies in compositions with many species. Phys. Rev. B 96, 014112 (2017). [Google Scholar]
- 70.Pun G, Batra R, Ramprasad R and Mishin Y: Physically-informed artificial neural networks for atomistic modeling of materials. arXiv preprint arXiv:.01696 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xue D, Balachandran PV, Hogden J, Theiler J, Xue D and Lookman T: Accelerated search for materials with targeted properties by adaptive design. Nat. Commun 7, 11241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gunning D: Explainable artificial intelligence (xai), in Defense Advanced Research Projects Agency (DARPA; 2017). [Google Scholar]
- 73.Mayr A, Klambauer G, Unterthiner T, Steijaert M, Wegner JK, Ceulemans H, Clevert D-A and Hochreiter S: Large-scale comparison of machine learning methods for drug target prediction on ChEMBL. Chem. Sci 9, 5541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Curtarolo S, Morgan D, Persson K, Rodgers J and Ceder G: Predicting crystal structures with data mining of quantum calculations. Phys. Rev. Lett 91, 135503 (2003). [DOI] [PubMed] [Google Scholar]
- 75.Pilania G, Balachandran PV, Kim C and Lookman T: Finding new perovskite halides via machine learning. Front. in Mater 3, 19 (2016). [Google Scholar]
- 76.Oliynyk AO, Antono E, Sparks TD, Ghadbeigi L, Gaultois MW, Meredig B and Mar A: High-throughput machine-learning-driven synthesis of full-Heusler compounds. Chem. Mater 28, 7324 (2016). [Google Scholar]
- 77.Hautier G, Fischer CC, Jain A, Mueller T and Ceder G: Finding nature’s missing ternary oxide compounds using machine learning and density functional theory. Chem. Mater 22, 3762 (2010). [Google Scholar]
- 78.De Jong M, Chen W, Notestine R, Persson K, Ceder G, Jain A, Asta M and Gamst A: A statistical learning framework for materials science: application to elastic moduli of k-nary inorganic polycrystalline compounds. Sci. Rep 6, 34256 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ahmad Z, Xie T, Maheshwari C, Grossman JC and Viswanathan V: Machine Learning Enabled Computational Screening of Inorganic Solid Electrolytes for Suppression of Dendrite Formation in Lithium Metal Anodes. ACS Cent. Sci 4, 996 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pyzer-Knapp EO, Li K and Aspuru-Guzik A: Learning from the harvard clean energy project: The use of neural networks to accelerate materials discovery. Adv. Func. Mater 25, 6495 (2015). [Google Scholar]
- 81.Stanev V, Oses C, Kusne AG, Rodriguez E, Paglione J, Curtarolo S and Takeuchi I: Machine learning modeling of superconducting critical temperature. npj Comp. Mater 4, 29 (2018). [Google Scholar]
- 82.Gómez-Bombarelli R, Wei JN, Duvenaud D, Hernández-Lobato JM, Sánchez-Lengeling B, Sheberla D, Aguilera-Iparraguirre J, Hirzel TD, Adams RP and Aspuru-Guzik A: Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci 4, 268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khorshidi A and Peterson AA: Amp: A modular approach to machine learning in atomistic simulations. Comput. Phys. Commun 207, 310 (2016). [Google Scholar]
- 84.Bartók AP and Csányi G: Gaussian approximation potentials: A brief tutorial introduction. Int. J. Quantum Chem 115, 1051 (2015). [Google Scholar]
- 85.Huan TD, Batra R, Chapman J, Krishnan S, Chen L and Ramprasad R: A universal strategy for the creation of machine learning-based atomistic force fields. npj Comp. Mater 3, 37 (2017). [Google Scholar]
- 86.Thompson AP, Swiler LP, Trott CR, Foiles SM and Tucker GJ: Spectral neighbor analysis method for automated generation of quantum-accurate interatomic potentials. J. Comput. Phys 285, 316 (2015). [Google Scholar]
- 87.Botu V, Batra R, Chapman J and Ramprasad R: Machine learning force fields: construction, validation, and outlook. J. Phys. Chem. C 121, 511 (2016). [Google Scholar]
- 88.Olsthoorn B, Geilhufe RM, Borysov SS and Balatsky AV: Band gap prediction for large organic crystal structures with machine learning. arXiv preprint arXiv:.12814 (2018). [Google Scholar]
- 89.Mannodi-Kanakkithodi A, Pilania G, Huan TD, Lookman T and Ramprasad R: Machine learning strategy for accelerated design of polymer dielectrics. Sci. Rep 6, 20952 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Collins CR, Gordon GJ, von Lilienfeld OA and Yaron DJ: Constant size descriptors for accurate machine learning models of molecular properties. J. Chem. Phys 148, 241718 (2018). [DOI] [PubMed] [Google Scholar]
- 91.Christensen A, Faber F, Huang B, Bratholm L, Tkatchenko A, Müller K and von Lilienfeld O: QML: A Python Toolkit for Quantum Machine Learning, (2017). [Google Scholar]
- 92.Kolb B, Lentz LC and Kolpak AM: Discovering charge density functionals and structure-property relationships with PROPhet: A general framework for coupling machine learning and first-principles methods. Sci. Rep 7, 1192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yao K, Herr JE, Toth DW, Mckintyre R and Parkhill J: The TensorMol-0.1 model chemistry: a neural network augmented with long-range physics. Chem. Sci 9, 2261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smith JS, Isayev O and Roitberg AE: ANI-1: an extensible neural network potential with DFT accuracy at force field computational cost. Chem. Sci 8, 3192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang H, Zhang L, Han J and Weinan E: DeePMD-kit: A deep learning package for many-body potential energy representation and molecular dynamics. Comput. Phys. Commun 228, 178 (2018). [Google Scholar]
- 96.Chmiela S, Tkatchenko A, Sauceda HE, Poltavsky I, Schütt KT and Müller K-R: Machine learning of accurate energy-conserving molecular force fields. Science advances 3, e1603015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mardt A, Pasquali L, Wu H and Noé F: VAMPnets for deep learning of molecular kinetics. Nat. Commun 9, 5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kalinin SV, Rodriguez BJ, Budai JD, Jesse S, Morozovska A, Bokov AA and Ye Z-G: Direct evidence of mesoscopic dynamic heterogeneities at the surfaces of ergodic ferroelectric relaxors. Phys. Rev. B 81, 064107 (2010). [Google Scholar]
- 99.Blaiszik B, Chard K, Pruyne J, Ananthakrishnan R, Tuecke S and Foster I: The Materials Data Facility: Data services to advance materials science research. JOM 68, 2045 (2016). [Google Scholar]
- 100.Sheppard D: Robert Le Rossignol, 1884–1976: engineer of the ‘Haber’process. Notes Rec. 71, 263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hanak JJ: The “multiple-sample concept” in materials research: Synthesis, compositional analysis and testing of entire multicomponent systems. J. Mater. Sci 5, 964 (1970). [Google Scholar]
- 102.Xiang X-D, Sun X, Briceno G, Lou Y, Wang K-A, Chang H, Wallace-Freedman WG, Chen S-W and Schultz PG: A combinatorial approach to materials discovery. Science 268, 1738 (1995). [DOI] [PubMed] [Google Scholar]
- 103.Barber Z and Blamire M: High throughput thin film materials science. Mat. Sci. Tech 24, 757 (2008). [Google Scholar]
- 104.Green ML, Choi C, Hattrick-Simpers J, Joshi A, Takeuchi I, Barron S, Campo E, Chiang T, Empedocles S and Gregoire J: Fulfilling the promise of the materials genome initiative with high-throughput experimental methodologies. Appl. Phys. Rev 4, 011105 (2017). [Google Scholar]
- 105.Maier WF, Stoewe K and Sieg S: Combinatorial and high-throughput materials science. Angew. Chem 46, 6016 (2007). [DOI] [PubMed] [Google Scholar]
- 106.Green ML, Takeuchi I and Hattrick-Simpers JR: Applications of high throughput (combinatorial) methodologies to electronic, magnetic, optical, and energy-related materials. J. Appl. Phys 113, 231101 (2013). [Google Scholar]
- 107.Dubois J-L, Duquenne C, Holderich W and Kervennal J: Process for dehydrating glycerol to acrolein, (Google Patents; 2010). [Google Scholar]
- 108.Arriola DJ, Carnahan EM, Hustad PD, Kuhlman RL and Wenzel TT: Catalytic production of olefin block copolymers via chain shuttling polymerization. Science 312, 714 (2006). [DOI] [PubMed] [Google Scholar]
- 109.Meguro S, Ohnishi T, Lippmaa M and Koinuma H: Elements of informatics for combinatorial solid-state materials science. Meas. Sci. Technol 16, 309 (2004). [Google Scholar]
- 110.Takeuchi I, Lippmaa M and Matsumoto Y: Combinatorial experimentation and materials informatics. MRS bulletin 31, 999 (2006). [Google Scholar]
- 111.Koinuma H: Combinatorial materials research projects in Japan. Appl. Surf. Sci 189, 179 (2002). [Google Scholar]
- 112.Smotkin ES and Diaz-Morales RR: New electrocatalysts by combinatorial methods. Ann. Rev. Mater. Res 33, 557 (2003). [Google Scholar]
- 113.Watanabe Y, Umegaki T, Hashimoto M, Omata K and Yamada M: Optimization of Cu oxide catalysts for methanol synthesis by combinatorial tools using 96 well microplates, artificial neural network and genetic algorithm. Catalysis Today 89, 455 (2004). [Google Scholar]
- 114.Dell’Anna R, Lazzeri P, Canteri R, Long CJ, Hattrick-Simpers J, Takeuchi I and Anderle M: Data Analysis in Combinatorial Experiments: Applying Supervised Principal Component Technique to Investigate the Relationship Between ToF-SIMS Spectra and the Composition Distribution of Ternary Metallic Alloy Thin Films. QSAR Comb. Sci 27, 171 (2008). [Google Scholar]
- 115.Takeuchi I, Long C, Famodu O, Murakami M, Hattrick-Simpers J, Rubloff G, Stukowski M and Rajan K: Data management and visualization of x-ray diffraction spectra from thin film ternary composition spreads. Rev. Sci. Instr 76, 062223 (2005). [Google Scholar]
- 116.Yomada Y and Kobayashi T: Utilization of combinatorial method and high throughput experimentation for development of heterogeneous catalysts. J. Jpn. Petrol Inst 49, 157 (2006). [Google Scholar]
- 117.Rodemerck U, Baerns M, Holena M and Wolf D: Application of a genetic algorithm and a neural network for the discovery and optimization of new solid catalytic materials. Appl. Surf. Sci 223, 168 (2004). [Google Scholar]
- 118.Koinuma H and Takeuchi I: Combinatorial solid-state chemistry of inorganic materials. Nat. Mater 3, 429 (2004). [DOI] [PubMed] [Google Scholar]
- 119.Bassim N, Schenck PK, Otani M and Oguchi H: Model, prediction, and experimental verification of composition and thickness in continuous spread thin film combinatorial libraries grown by pulsed laser deposition. Rev. Sci. Instr 78, 072203 (2007). [DOI] [PubMed] [Google Scholar]
- 120.Bunn JK, Metting C and Hattrick-Simpers J: A semi-empirical model for tilted-gun planar magnetron sputtering accounting for chimney shadowing. JOM 67, 154 (2015). [Google Scholar]
- 121.Gregoire J, Lobovsky M, Heinz M, DiSalvo F and Van Dover R: Resputtering phenomena and determination of composition in codeposited films. Phys. Rev. B 76, 195437 (2007). [Google Scholar]
- 122.Long C, Hattrick-Simpers J, Murakami M, Srivastava R, Takeuchi I, Karen VL and Li X: Rapid structural mapping of ternary metallic alloy systems using the combinatorial approach and cluster analysis. Rev. Sci. Instr 78, 072217 (2007). [DOI] [PubMed] [Google Scholar]
- 123.Gregoire JM, Dale D and Van Dover RB: A wavelet transform algorithm for peak detection and application to powder x-ray diffraction data. Rev. Sci. Instr 82, 015105 (2011). [DOI] [PubMed] [Google Scholar]
- 124.Long C, Bunker D, Li X, Karen V and Takeuchi I: Rapid identification of structural phases in combinatorial thin-film libraries using x-ray diffraction and non-negative matrix factorization. Rev. Sci. Instr 80, 103902 (2009). [DOI] [PubMed] [Google Scholar]
- 125.LeBras R, Damoulas T, Gregoire JM, Sabharwal A, Gomes CP and Van Dover RB: Constraint reasoning and kernel clustering for pattern decomposition with scaling, in International Conference on Principles and Practice of Constraint Programming (Springer; 2011), pp. 508. [Google Scholar]
- 126.Bunn JK, Han S, Zhang Y, Tong Y, Hu J and Hattrick-Simpers JR: Generalized machine learning technique for automatic phase attribution in time variant high-throughput experimental studies. J. Mater. Res 30, 879 (2015). [Google Scholar]
- 127.Bunn JK, Hu J and Hattrick-Simpers JR: Semi-Supervised approach to phase identification from combinatorial sample diffraction patterns. JOM 68, 2116 (2016). [Google Scholar]
- 128.Hattrick-Simpers JR, Gregoire JM and Kusne AG: Perspective: Composition–structure–property mapping in high-throughput experiments: Turning data into knowledge. APL Materials 4, 053211 (2016). [Google Scholar]
- 129.Kusne AG, Keller D, Anderson A, Zaban A and Takeuchi I: High-throughput determination of structural phase diagram and constituent phases using GRENDEL. Nanotechnology 26, 444002 (2015). [DOI] [PubMed] [Google Scholar]
- 130.Suram SK, Xue Y, Bai J, Le Bras R, Rappazzo B, Bernstein R, Bjorck J, Zhou L, van Dover RB and Gomes CP: Automated phase mapping with AgileFD and its application to light absorber discovery in the V–Mn–Nb oxide system. ACS Comb. Sci 19, 37 (2016). [DOI] [PubMed] [Google Scholar]
- 131.Kusne AG, Gao T, Mehta A, Ke L, Nguyen MC, Ho K-M, Antropov V, Wang C-Z, Kramer MJ, Long C and Takeuchi I: On-the-fly machine-learning for high-throughput experiments: search for rare-earth-free permanent magnets. Sci. Rep 4, 6367 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cui J, Chu YS, Famodu OO, Furuya Y, Hattrick-Simpers J, James RD, Ludwig A, Thienhaus S, Wuttig M and Zhang Z: Combinatorial search of thermoelastic shape-memory alloys with extremely small hysteresis width. Nat. Mater 5, 286 (2006). [DOI] [PubMed] [Google Scholar]
- 133.Zakutayev A, Stevanovic V and Lany S: Non-equilibrium Alloying controls optoelectronic properties in Cu2O thin films for photovoltaic absorber applications. Appl. Phys. Lett 106, 123903 (2015). [Google Scholar]
- 134.Yan Q, Yu J, Suram SK, Zhou L, Shinde A, Newhouse PF, Chen W, Li G, Persson KA and Gregoire JM: Solar fuels photoanode materials discovery by integrating high-throughput theory and experiment. Proc. Natl. Acad. Sci. U.S.A 114, 3040 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hattrick-Simpers JR, Choudhary K and Corgnale C: A simple constrained machine learning model for predicting high-pressure-hydrogen-compressor materials. Mol. Sys. Des. Eng (2018). [Google Scholar]
- 136.Ren F, Ward L, Williams T, Laws KJ, Wolverton C, Hattrick-Simpers J and Mehta A: Accelerated discovery of metallic glasses through iteration of machine learning and high-throughput experiments. Sci. Adv 4, eaaq1566 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yuan R, Liu Z, Balachandran PV, Xue D, Zhou Y, Ding X, Sun J, Xue D and Lookman T: Accelerated Discovery of Large Electrostrains in BaTiO3-Based Piezoelectrics Using Active Learning. Adv. Mater 30, 1702884 (2018). [DOI] [PubMed] [Google Scholar]
- 138.Bassman L, Rajak P, Kalia RK, Nakano A, Sha F, Sun J, Singh DJ, Aykol M, Huck P and Persson K: Active learning for accelerated design of layered materials. npj Computational Materials 4, 74 (2018). [Google Scholar]
- 139.Podryabinkin EV, Tikhonov EV, Shapeev AV and Oganov AR: Accelerating crystal structure prediction by machine-learning interatomic potentials with active learning. Phys. Rev. B 99, 064114 (2019). [Google Scholar]
- 140.Talapatra A, Boluki S, Duong T, Qian X, Dougherty E and Arróyave R: Autonomous efficient experiment design for materials discovery with Bayesian model averaging. Physical Review Materials 2, 113803 (2018). [Google Scholar]
- 141.Lookman T, Balachandran PV, Xue D and Yuan R: Active learning in materials science with emphasis on adaptive sampling using uncertainties for targeted design. npj Computational Materials 5, 21 (2019). [Google Scholar]
- 142.Meredig B, Antono E, Church C, Hutchinson M, Ling J, Paradiso S, Blaiszik B, Foster I, Gibbons B and Hattrick-Simpers J: Can machine learning identify the next high-temperature superconductor? Examining extrapolation performance for materials discovery. Mol. Sys. Des. Eng 3, 819 (2018). [Google Scholar]
- 143.King RD, Rowland J, Aubrey W, Liakata M, Markham M, Soldatova LN, Whelan KE, Clare A, Young M and Sparkes A: The robot scientist Adam. Computer 42, 46 (2009). [Google Scholar]
- 144.Nikolaev P, Hooper D, Webber F, Rao R, Decker K, Krein M, Poleski J, Barto R and Maruyama B: Autonomy in materials research: a case study in carbon nanotube growth. npj Comp. Mater 2, 16031 (2016). [Google Scholar]
- 145.Roch LM, Häse F, Kreisbeck C, Tamayo-Mendoza T, Yunker LP, Hein JE and Aspuru-Guzik A: ChemOS: Orchestrating autonomous experimentation. Sci. Robot 3, eaat5559 (2018). [DOI] [PubMed] [Google Scholar]
- 146.DeCost B and Kusne G: Deep Transfer Learning for Active Optimization of Functional Materials Properties in the Data-Limited Regime, (MRS Fall; 2018). [Google Scholar]
- 147.Kusne G, DeCost B, Hattrick-Simpers J and Takeuchi I: Autonomous Materials Research Systems—Phase Mapping, (MRS Fall; 2018). [Google Scholar]
- 148.Caramelli D, Salley D, Henson A, Camarasa GA, Sharabi S, Keenan G and Cronin L: Networking chemical robots for reaction multitasking. Nat. Commun 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Klucznik T, Mikulak-Klucznik B, McCormack MP, Lima H, Szymkuć S, Bhowmick M, Molga K, Zhou Y, Rickershauser L and Gajewska EP: Efficient syntheses of diverse, medicinally relevant targets planned by computer and executed in the laboratory. Chem 4, 522 (2018). [Google Scholar]
- 150.https://www.asminternational.org/materials-resources/online-databases/-https://www.asminternational.org/materials-resources/online-databases/-/journal_content/56/10192/15468789/DATABASE/journal_content/56/10192/15468789/DATABASE, (2019).
- 151.LeCun Y, Bengio Y and Hinton G: Deep learning. Nature 521, 436 (2015). [DOI] [PubMed] [Google Scholar]
- 152.Ziatdinov M, Dyck O, Maksov A, Li X, Sang X, Xiao K, Unocic RR, Vasudevan R, Jesse S and Kalinin SV: Deep Learning of Atomically Resolved Scanning Transmission Electron Microscopy Images: Chemical Identification and Tracking Local Transformations. ACS nano 11, 12742 (2017). [DOI] [PubMed] [Google Scholar]
- 153.Ziatdinov M, Maksov A and Kalinin SV: Learning surface molecular structures via machine vision. npj Computational Materials 3, 31 (2017). [Google Scholar]
- 154.Barthel J: Dr. Probe: A software for high-resolution STEM image simulation. Ultramicroscopy (2018). [DOI] [PubMed] [Google Scholar]
- 155.Ziatdinov M, Dyck O, Sumpter BG, Jesse S, Vasudevan RK and Kalinin SV: Building and exploring libraries of atomic defects in graphene: scanning transmission electron and scanning tunneling microscopy study. arXiv preprint arXiv:1809.04256 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Long J, Shelhamer E and Darrell T: Fully convolutional networks for semantic segmentation, in Proceedings of the IEEE conference on computer vision and pattern recognition (2015), pp. 3431. [DOI] [PubMed] [Google Scholar]
- 157.Maksov A, Dyck O, Wang K, Xiao K, Geohegan DB, Sumpter BG, Vasudevan RK, Jesse S, Kalinin SV and Ziatdinov M: Deep learning analysis of defect and phase evolution during electron beam-induced transformations in WS2. npj Comp. Mater 5, 12 (2019). [Google Scholar]
- 158.Ziatdinov M, Dyck O, Jesse S and Kalinin SV: Atomic mechanisms for the Si atom dynamics in graphene: chemical transformations at the edge and in the bulk. arXiv preprint arXiv:1901.09322 (2019). [Google Scholar]
- 159.Yablon DG, Gannepalli A, Proksch R, Killgore J, Hurley DC, Grabowski J and Tsou AH: Quantitative viscoelastic mapping of polyolefin blends with contact resonance atomic force microscopy. Macromolecules 45, 4363 (2012). [Google Scholar]
- 160.Schlücker S, Schaeberle MD, Huffman SW and Levin IW: Raman microspectroscopy: a comparison of point, line, and wide-field imaging methodologies. Analytical Chemistry 75, 4312 (2003). [DOI] [PubMed] [Google Scholar]
- 161.Ievlev AV, Maksymovych P, Trassin M, Seidel J, Ramesh R, Kalinin SV and Ovchinnikova OS: Chemical state evolution in ferroelectric films during tip-induced polarization and electroresistive switching. ACS applied materials & interfaces 8, 29588 (2016). [DOI] [PubMed] [Google Scholar]
- 162.Hruszkewycz S, Folkman C, Highland M, Holt M, Baek S, Streiffer S, Baldo P, Eom C and Fuoss P: X-ray nanodiffraction of tilted domains in a poled epitaxial BiFeO3 thin film. Appl. Phys. Lett 99, 232903 (2011). [Google Scholar]
- 163.Cai Z, Lai B, Xiao Y and Xu S: An X-ray diffraction microscope at the Advanced Photon Source, in Journal de Physique IV (Proceedings) (EDP sciences2003), pp. 17. [Google Scholar]
- 164.Kalinin SV, Karapetian E and Kachanov M: Nanoelectromechanics of piezoresponse force microscopy. Phys. Rev. B 70, 184101 (2004). [Google Scholar]
- 165.Eliseev EA, Kalinin SV, Jesse S, Bravina SL and Morozovska AN: Electromechanical detection in scanning probe microscopy: Tip models and materials contrast. J. Appl. Phys 102, 014109 (2007). [Google Scholar]
- 166.Monig H, Todorovic M, Baykara MZ, Schwendemann TC, Rodrigo L, Altman EI, Perez R and Schwarz UD: Understanding scanning tunneling microscopy contrast mechanisms on metal oxides: a case study. ACS nano 7, 10233 (2013). [DOI] [PubMed] [Google Scholar]
- 167.Ievlev AV, Susner MA, McGuire MA, Maksymovych P and Kalinin SV: Quantitative analysis of the local phase transitions induced by laser heating. ACS nano 9, 12442 (2015). [DOI] [PubMed] [Google Scholar]
- 168.Dönges SA, Khatib O, O’Callahan BT, Atkin JM, Park JH, Cobden D and Raschke MB: Ultrafast nanoimaging of the photoinduced phase transition dynamics in VO2. Nano Lett. 16, 3029 (2016). [DOI] [PubMed] [Google Scholar]
- 169.Kim Y, Strelcov E, Hwang IR, Choi T, Park BH, Jesse S and Kalinin SV: Correlative multimodal probing of ionically-mediated electromechanical phenomena in simple oxides. Sci. Rep 3, 2924 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ovchinnikov O, Jesse S, Bintacchit P, Trolier-McKinstry S and Kalinin SV: Disorder identification in hysteresis data: Recognition analysis of the random-bond–random-field ising model. Phys. Rev. Lett 103, 157203 (2009). [DOI] [PubMed] [Google Scholar]
- 171.Borodinov N, Neumayer S, Kalinin SV, Ovchinnikova OS, Vasudevan RK and Jesse S: Deep neural networks for understanding noisy data applied to physical property extraction in scanning probe microscopy. npj Computational Materials 5, 25 (2019). [Google Scholar]
- 172.Pradhan DK, Kumari S, Strelcov E, Pradhan DK, Katiyar RS, Kalinin SV, Laanait N and Vasudevan RK: Reconstructing phase diagrams from local measurements via Gaussian processes: mapping the temperature-composition space to confidence. NPJ Computational Materials 4, 1 (2018). [Google Scholar]
- 173.Li L, Yang Y, Zhang D, Ye Z-G, Jesse S, Kalinin SV and Vasudevan RK: Machine learning–enabled identification of material phase transitions based on experimental data: Exploring collective dynamics in ferroelectric relaxors. Science Advances 4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Shah VP, Younan NH and King RL: An efficient pan-sharpening method via a combined adaptive PCA approach and contourlets. IEEE transactions on geoscience and remote sensing 46, 1323 (2008). [Google Scholar]
- 175.Somnath S, Belianinov A, Kalinin SV and Jesse S: Full information acquisition in piezoresponse force microscopy. Appl. Phys. Lett 107 (2015). [Google Scholar]
- 176.Somnath S, Law KJ, Morozovska A, Maksymovych P, Kim Y, Lu X, Alexe M, Archibald R, Kalinin SV and Jesse S: Ultrafast current imaging by Bayesian inversion. Nat. Commun 9, 513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Somnath S, Belianinov A, Kalinin SV and Jesse S: Rapid mapping of polarization switching through complete information acquisition. Nat. Commun 7, 13290 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Collins L, Belianinov A, Somnath S, Balke N, Kalinin SV and Jesse S: Full data acquisition in Kelvin Probe Force Microscopy: Mapping dynamic electric phenomena in real space. Sci. Rep 6, 30557 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Balke N, Jesse S, Yu P, Carmichael B, Kalinin SV and Tselev A: Quantification of surface displacements and electromechanical phenomena via dynamic atomic force microscopy. Nanotechnology 27, 425707 (2016). [DOI] [PubMed] [Google Scholar]
- 180.Labuda A and Proksch R: Quantitative measurements of electromechanical response with a combined optical beam and interferometric atomic force microscope. Appl. Phys. Lett 106, 253103 (2015). [Google Scholar]
- 181.Somnath S, Smith CR, Laanait N, Vasudevan RK, Ievlev A, Belianinov A, Lupini AR, Shankar M, Kalinin SV and Jesse S: USID and Pycroscopy--Open frameworks for storing and analyzing spectroscopic and imaging data. arXiv preprint arXiv:1903.09515 (2019). [Google Scholar]
- 182.Kalidindi SR and De Graef M: Materials data science: current status and future outlook. Ann. Rev. Mater. Res 45, 171 (2015). [Google Scholar]
- 183.Fullwood DT, Niezgoda SR and Kalidindi SR: Microstructure reconstructions from 2-point statistics using phase-recovery algorithms. Acta Mater. 56, 942 (2008). [Google Scholar]
- 184.Kalidindi SR, Niezgoda SR and Salem AA: Microstructure informatics using higher-order statistics and efficient data-mining protocols. Jom 63, 34 (2011). [Google Scholar]
- 185.Sharma V, Wang C, Lorenzini RG, Ma R, Zhu Q, Sinkovits DW, Pilania G, Oganov AR, Kumar S, Sotzing GA, Boggs SA and Ramprasad R: Rational design of all organic polymer dielectrics. Nat. Commun 5, 4845 (2014). [DOI] [PubMed] [Google Scholar]
- 186.Gopakumar AM, Balachandran PV, Xue D, Gubernatis JE and Lookman T: Multi-objective Optimization for Materials Discovery via Adaptive Design. Sci. Rep 8, 3738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bassman L, Rajak P, Kalia RK, Nakano A, Sha F, Sun J, Singh DJ, Aykol M, Huck P, Persson K and Vashishta P: Active learning for accelerated design of layered materials. npj Comp. Mater 4, 74 (2018). [Google Scholar]
- 188.Hutchinson ML, Antono E, Gibbons BM, Paradiso S, Ling J and Meredig B: Overcoming data scarcity with transfer learning. arXiv preprint arXiv:1711.05099 (2017). [Google Scholar]
- 189.Oviedo F, Ren Z, Sun S, Settens C, Liu Z, Hartono NTP, Ramasamy S, DeCost BL, Tian SIP, Romano G, Gilad Kusne A and Buonassisi T: Fast and interpretable classification of small X-ray diffraction datasets using data augmentation and deep neural networks. npj Computational Materials 5, 60 (2019). [Google Scholar]
- 190.Vlcek L, Ziatdinov M, Maksov A, Tselev A, Baddorf AP, Kalinin SV and Vasudevan RK: Learning from Imperfections: Predicting Structure and Thermodynamics from Atomic Imaging of Fluctuations. ACS Nano 13, 718 (2019). [DOI] [PubMed] [Google Scholar]
- 191.Vlcek L, Vasudevan RK, Jesse S and Kalinin SV: Consistent integration of experimental and ab initio data into effective physical models. Journal of chemical theory and computation 13, 5179 (2017). [DOI] [PubMed] [Google Scholar]
- 192.Vlcek L, Maksov A, Pan M, Vasudevan RK and Kalinin SV: Knowledge Extraction from Atomically Resolved Images. ACS nano 11, 10313 (2017). [DOI] [PubMed] [Google Scholar]
- 193.Belianinov A, He Q, Kravchenko M, Jesse S, Borisevich A and Kalinin SV: Identification of phases, symmetries and defects through local crystallography. Nat. Commun 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ross D, Strychalski EA, Jarzynski C and Stavis SM: Equilibrium free energies from non-equilibrium trajectories with relaxation fluctuation spectroscopy. Nat. Phys (2018). [Google Scholar]
- 195.Kutnjak Z, Petzelt J and Blinc R: The giant electromechanical response in ferroelectric relaxors as a critical phenomenon. Nature 441, 956 (2006). [DOI] [PubMed] [Google Scholar]
- 196.Hall SR, Allen FH and Brown ID: The crystallographic information file (CIF): a new standard archive file for crystallography. Acta Crystallographica Section A: Foundations of Crystallography 47, 655 (1991). [Google Scholar]
- 197.Pearl J: Theoretical impediments to machine learning with seven sparks from the causal revolution. arXiv preprint arXiv:1801.04016 (2018). [Google Scholar]