Abstract
Tumor progression, including metastasis, is significantly influenced by factors in the tumor microenvironment (TME) such as mechanical force, shear stress, chemotaxis, and hypoxia. At present, most cancer studies investigate tumor metastasis by conventional cell culture methods and animal models, which are limited in data interpretation. Although patient tissue analysis, such as human patient-derived xenografts (PDX), can provide important clinical relevant information, they may not be feasible for functional studies as they are costly and time-consuming. Thus, in vitro three-dimensional (3D) models are rapidly being developed that mimic TME and allow functional investigations of metastatic mechanisms and drug responses. One of those new 3D models is tumor-on-a-chip technology that provides a powerful in vitro platform for cancer research, with the ability to mimic the complex physiological architecture and precise spatiotemporal control. Tumor-on-a-chip technology can provide integrated features including 3D scaffolding, multicellular culture, and a vasculature system to simulate dynamic flow in vivo. Here, we review a select set of recent achievements in tumor-on-a-chip approaches and present potential directions for tumor-on-a-chip systems in the future for areas including mechanical and chemical mimetic systems. We also discuss challenges and perspectives in both biological factors and engineering methods for tumor-on-a-chip progress. These approaches will allow in the future for the tumor-on-a-chip systems to test therapeutic approaches for individuals through using their cancerous cells gathered through approaches like biopsies, which then will contribute toward personalized medicine treatments for improving their outcomes.
1. Introduction
Cancer remains a worldwide health issue and is the second leading cause of death in the United States with approximately 1,735,350 new cancer cases and 609,640 cancer deaths in 2018.1 The efforts in cancer research have resulted in a tremendous number of successes on a diversity of fronts, with researchers from varying backgrounds including biologists, surgeons, biomedical engineers and even mechanical engineers contributing to new directions.
One major objective in cancer research is to understand tumor biology related to metastasis mechanisms and TME, which could support the development of anti-cancer drugs and treatment approaches. This is important as it is appreciated that factors such as mechanical force, shear stress, chemotaxis, and hypoxia influence tumor progression, invasion, metastasis, and lastly drug metabolism. 2–4 Through a 3D approach, important features of metastasis mechanisms will be uncovered and hopefully decrease patient death due to metastasis.
Metastasis is still the leading cause of mortality for most cancer patients and is increasingly the most studied topic in cancer research.5 Cancer models, including in vitro cell culture and animal models, 6–8 have contributed tremendously to developing diagnostics and treatments for cancer over the past several decades. In vitro models mostly utilize 2D platforms which are easy to use, but do have limitations due to a lack of mimicking the physiological in vivo environment.9 There have been advances toward 3D systems in cancer research as scientists strive to make the systems more physiologically relevant. For example, cancer cells were cultured and investigated in a 3D hydrogel scaffold and were able to begin to mimic structure in vivo.10–12 One typical phenomenon is regaining epithelial cells’ apicobasal polarity and lumen formation of glandular cells and in 3D culture.13 Thus, 3D cell culture could regenerate certain cell differentiation and reorganizations of multiple cell clusters that were not found in 2D. Although this approach lacked vasculature, which is found in tumor tissue and thus can be limited in recreating multi-cell systems with complex spatial distributions.14–16 This issue can be overcome by utilizing animal models; however drawbacks include cost, lack of tumor heterogeneity (if cell lines transplantation is used) and time.17, 18 Animal models also lack immune system functionality and its interaction with tumor, which could be an important research goal as well.19 Species-specific drug responses also can create a gap between animal models and clinical trials.19, 20 Therefore, animal models such as mouse models may perform differently to drug response when compared to humans as has been shown previously.21 Besides, animal models can be harder to control with experimental manipulation during tumor progression, and the variability with tumor intravasation in animal models may increase the ability to conduct high fidelity real time imaging.20 These limitations lead to the need for a more flexible, controllable and high throughput model for tumor study.
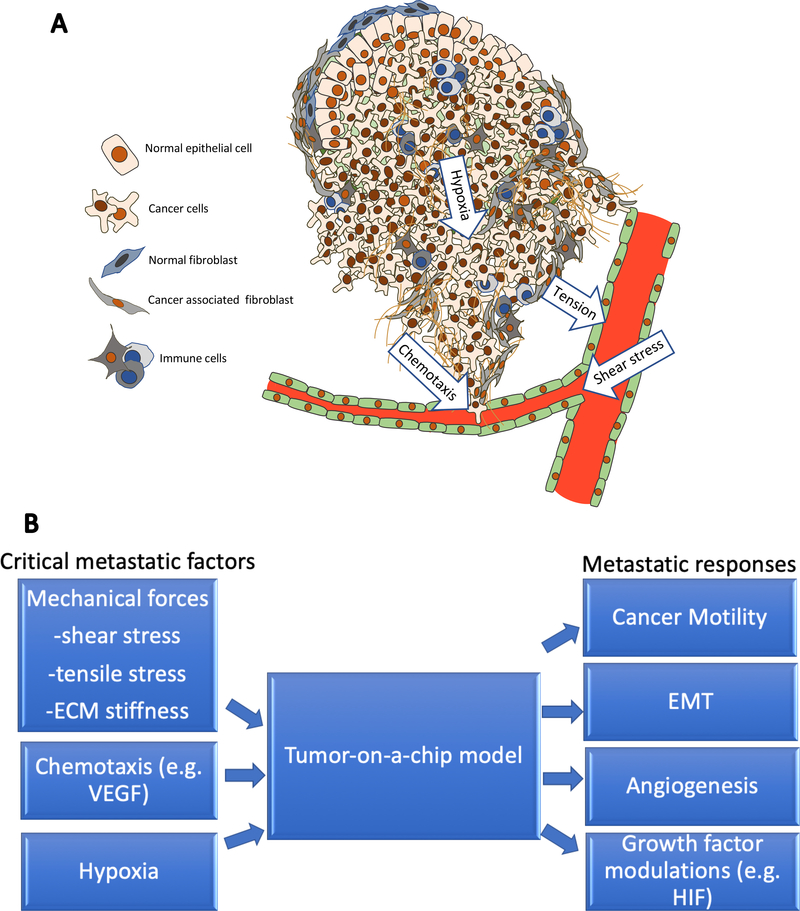
To develop a more reliable tumor model that could mimic TME in vivo, one approach is to consider developing mimetic systems from an engineering perspective. The motivation behind this includes the knowledge that tumors are complex systems with different functional units such as different cell types, extracellular matrix (ECM), vasculature system, and multiple chemical factors (Figure 1A). An engineered system offers many advantages for meaningful analyses of cancer progression as it can mimic physiological relevant tumor-promoting mechanical forces such as shear stress from the dynamic flow in the vasculature, tension from the solid tumor, and stiffness variation of ECM22. Besides, it can also provide chemical factors such as chemotaxis due to nutrient diffusion and growth factor transduction23, as well as hypoxia gradient due to oxygen diffusion limit24. Moreover, cell interaction between tumor cells and cancer-associated fibroblast, endothelial cells, etc. are also considered critical factors in metastasis, and thus having a system that can incorporate multiple cell types interacting with each other could be quite advantageous25(Figures 1A and 1B). All these factors are potentially able to be recreated and examined in tumor-on-a-chip approaches through integrated designs and approaches.2
Figure.1.
A. Schematic of a tumor system with a complex organization of cancer cells, fibroblasts, extracellular matrix, vasculature system, and multiple chemical factors. Tumor progression is affected by microenvironment factors including mechanical and chemical such as shear stress, hypoxia, chemotaxis, cell-cell interactions, etc. These factors could be incorporated into tumor-on-a-chip models. B. To generate a tumor-on-a-chip model for mimicking metastasis, multiple critical metastatic factors could be recreated in vitro for different metastatic responses to be found.
Engineering techniques have been developed that enable better mimicking of physiological environments on-chip systems. These approaches include the development of soft lithography26, which has allowed microfluidic channels and dynamic flow systems to be fabricated and mimics physiological systems such as blood flow. Besides, 3D bioprinting27 has been able to print multiple cell types into ECM systems with high spatially precision. Also, computational models have contributed to these advances including studies in diffusion theory28 and fluid dynamics29, which have contributed to the development of tumor-on-a-chip systems that allow for high flexibility and multiple functionalities due to their rapid computational simulation with a diversity of parameters, at least to provide a rough prediction for multiple experimental results. Through these different techniques, a system can be designed that combines important factors of tumor progression. Even further, tumor-on-a-chip systems can be analyzed in conventional petri dish assays, which would then allow fluorescent staining and real-time measurements to observe cell motility, epithelial-to-mesenchymal transition (EMT), angiogenesis and growth factor modulations (Figure 1B).
In this review, we will discuss the importance of tumor-on-a-chip systems and a selection of recent achievements in designing and engineering of factors related to TME mimicking. We also discuss future directions, including multifunctional tumor-on-a-chip system development.
2. Chemical factors for tumor-on-a-chip systems
The chemical microenvironment in cancer is critical to understand as it tremendously affects the tumor response. During cancer progression, chemotaxis is known to guide many critical processes.30 For example, endothelial cell sprouting (angiogenesis) provides nutrients and oxygen for cancer cell proliferation as well as creating a pathway for metastasis, which is primarily regulated by crosstalk between cancer and endothelial cells through growth factors such as the vascular endothelial growth factor (VEGF).31 Therefore, it is essential to incorporate the chemical factor control that mimics the in vivo environment to study how varying chemicals affect cancer cells and other associated cells. In this section, we will review recent achievements for controlling the chemical TME with a focus on chemotaxis in a physiologically relevant condition, realized by tumor-on-a-chip technologies.
2.1. Chemical gradient generation in a tumor model
Cancer cells and associated cells in the ECM interact primarily through the transmission of growth factors and chemokines. In cancer development, multiple growth factors and chemokines are known to be involved, such as epidermal growth factor (EGF), vascular endothelial growth factor (VEGF), and transforming growth factor beta (TGFβ.23 Although the functions of these factors were investigated and characterized in vitro and in vivo, they are interconnected through complex signaling pathways that are difficult to decipher in a physiologically relevant manner in vitro and in vivo. The interactions in the crosstalk are difficult to quantify in specific cellular responses. Conventional 2D in vitro experiments have added to a better understanding of the TME crosstalk. However, the lack of a 3D environment with precise quantification of cellular responses under the spatiotemporal control of chemokines and growth factors is lacking. All in all, a better understanding of cancer cell response and how these affect therapeutic approaches is needed.
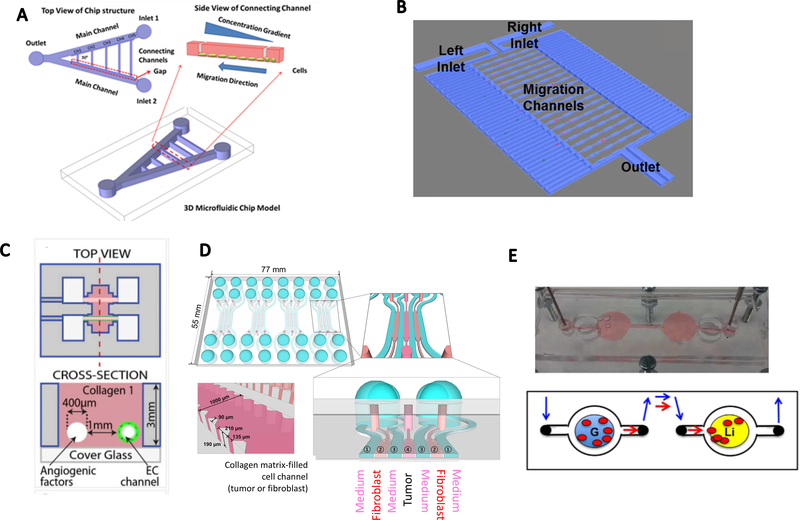
One approach toward the tumor-on-a-chip model that allows control of the chemical environment is in microfluidics. Chemical gradient generation in microfluidic devices can be controlled by two channels with inlets, where one provides a target chemical channel and the other presents buffer in the channel. At the interface, a mid-zone is building where gradients are formed and stabilized. In this midzone, cancer cells are then seeded. For example, Zou et al32 designed a V-shaped microfluidic device to track the migration of lung cancer stem cells (LCSC) and differentiated LCSC triggered by a serum gradient (Figure 2A). This approach provided precise control of multiple gradient profiles as the cells showed a consistent increase in migration distance. This device provided several key features enabled by microfluidics such as high precision of spatiotemporal control of the chemical environment and direct real-time imaging of cells.
Figure.2.
Tumor-on-a-chip models to induce chemotaxis and study cell-cell crosstalk. A. A V-shape double-channel microfluidic device for chemotaxis. Inlet 1 was designed for media flow and inlet 2 for chemokine perfusion. (reproduced with permission from ref. 32) B. Microfluidic channel design for single-cell chemotaxis. Chemical gradients were generated from the right channel to the left channel. (reproduced with permission from ref. 36) C. Parallel-channels in a 3D collagen scaffold to induce a response of endothelial cells in an angiogenic factors gradient. (reproduced with permission from ref. 39) D. Tumor spheroids and CAFs co-cultured in a 3D hydrogel-based microfluidic chip, separated with medium channels. (reproduced with permission from ref. 46) E. A metastasis-on-a-chip model with a gut organoid and a liver organoid located in two connected PDMS chambers. (reproduced with permission from ref. 50)
Besides, microfluidic devices can be used that contain microchannels with dimensions smaller than 100 μm, in which cells can be cultured and continuously infused. Microfluidics can enable accurate control of the TME by providing an incessant supply of nutrients and growth factors.33 A recent study also showed paper-based cell culture serving as a diffusion substrate.34 A confined hydrogel environment for slow-moving cell chemotaxis detection was also developed with similar diffusion scheme.35 An obvious advantage of microfluidic chip applications stems from the possibility to easily adapt the device to the research question asked by adjusting number, directionality, and diameter of the microfluidics together with varying amounts and types of chemicals and cell types tested.
Another powerful readout obtained by a tumor-on-a-chip system is the ability to analyze single-cell migration. Chen et al36 developed a single-cell migration chip that could track the migration of selected single cells (Figure 2B). Here, microfluidic channels were fabricated with dimensions comparable to the cells investigated. Continuous medium flow with and without chemokines (e.g., hepatocyte growth factor) perfused two channels on either side of the migration zone to generate a serum gradient to observe the migration of single cells over time. This single-cell migration chip allowed the discovery of heterogeneity in cell motility and the cells could be sorted for further investigation, such as for expression of mRNA of migration and metastasis-associated genes. Compared with the generation of massive cell chemotaxis, single-cell experiments require high chip resolution, which can be implemented using soft lithography in 2D. Challenges remain though for the fabrication of 3D channels (e.g. circular channels, etc.) with methods such as 3D bioprinting for these types of experiments in three-dimensional controlled environments.
An important factor in examining ECM conditions ex vivo is the choice of the embedding medium. Instead of using conventional soft lithography technique and Polydimethylsiloxane (PDMS) as a platform, Wan et al37 used collagen type I that also could create a 3D environment where the chemical microenvironment could be controlled. They fabricated microfluidic channels directly in ECM scaffolding through micromilling and molding methods. Creating the microchannels in ECM to mimic vessel or lymphatic systems offers a more physiologically relevant environment for cancer cells to grow and respond. Furthermore, this approach is promising for understanding cell response around the channels since both the geometry and biological conditions of the channel simulates vessels more realistically than conventional microfluidic platforms constructed with PDMS. Another promising approach using ECM was developed by Kolesky et al38, who successfully created perfusable channels within ECM made of gelatin and fibrinogen through 3D bioprinting. Although this approach was size limited to larger dimensions, it provided another method to control geometry and complexity of the vasculature in artificial ECM. Building off the 3D ECM work, using tumor-on-a-chip systems for studying angiogenesis is promising as well. Nguyen et al39 fabricated parallel channels in collagen type I and coated one channel with endothelial cells while perfusing the other end with angiogenic factors (Figure 2C). Endothelial cell sprouting was observed and characterized relative to cell morphology and length. Although this method failed to produce more complex vascular structure, the approach indicates the significance of in vitro vasculature in angiogenesis simulation. In recent years, the use of hydrogels has emerged in cancer biology to recreate a 3D environment for cancer cells. For example, photo-reactive hydrogels with tunable stiffness were used in microwell assays to cultures cancer cell lines as well as primary tissues.40 Collagen scaffolds with well-defined physiological components also served as platforms to cause the transition of primary human breast epithelial cells into mature breast tissues.41 However, several issues are limiting factors. For example, given the precisely determined composition of hydrogels, batch to batch differences exist that influence phenotypes studied. Besides, hydrogels do not necessarily provide mechanical properties that will mimic cancer progression as they can degrade in some cases as well as causing immunogenic reactions.42 3D environment is of great interest for probing cell behavior, and microfluidic devices can be created using fabrication techniques for designing complex structures, especially in developing mimetic vessel systems that could overcome the size limitations naturally occurring due to diffusion limitations in 3D biological systems.
2.2. Controlling biochemical interactions for cell crosstalk
Tumors are heterogenic tissues that are composed of multiple different cell types such as cancer cells, stromal cells, endothelial cells, and immune cells. By adding cell types of interest, tumor-on-a-chip mimetics allow the analysis of their specific interactions. This type of approach, when coupled with the proper 3D cell culture system together with real-time imaging, allows the examination of the complex biology causing metastasis. That way, cell-cell interactions that are closely tied to chemotaxis can be examined.
For example, Bruce et al43 developed a tri-culture model, including bone marrow stromal cells, osteoblasts, and leukemic cells to study cell interactions in acute lymphoblastic leukemia. This integration of a 3D environment and co-culture system showed that a complex biological environment could be created on-chip with precise control over the microenvironment and cell interactions.
The tumor-on-a-chip approach also allows for spatial control of both cells and the chemical environment. Spatial distributions of chemical factors are necessary for organism development and functioning. Similarly, extravasation (exiting of cancer cells out of the bloodstream) of cancer cells is stimulated by chemotactic mechanisms generated by the TME involving endothelial cells. To mimic this in 3D tumor-on-a-chip devices, endothelial cells would need to form a monolayer on the surface while cancer cells are located in the ECM. This design is more complex than simply mixing cells and thus generally requires multi-step fabrication. To address this, Bersini et al44 designed a tri-culture system to study cancer cell metastasis in bone with step-by-step cell seeding. Spatial control of multiple cell types could be used to reconstruct in vivo conditions and provided the ability to integrate and examine cell responses in a more physiologically relevant environment compared to conventional methods. Eventually, these approaches will enable direct and more biologically relevant studies of tumor metastasis in vitro. For example, the generation of a multi-cell system in microfluidic devices has allowed researchers to directly study how chemokines affect the metastatic process. Jeon et al45 generated a bone-mimicking microenvironment to study the extravasation of breast cancer cells. They compared cell extravasations in bone, muscle, and acellular mimicking conditions by adding corresponding cell types into the system. Flexible approaches using these with multi-cellular system create new directions for biologists to design and implement stimulation and response experiments to understand how each component including cell response over time. These studies presented the ability of tumor-on-a-chip models to investigate different aspects of cancer behavior.
Highly interesting topics that were previously unclear could be tested using these on-chip designs, with the purpose-oriented organization of multi-cell system. More specifically, cell-cell interactions that are known to contribute to cell progression and cancer drug resistance, such as cancer-cancer associated fibroblast (CAF) interactions, could be examined in microfluidic channels. Co-culturing of tumor spheroids and CAFs in a 3D hydrogel-based microfluidic chip separated by 100μm channels (Figure 2D) could reveal significant interactions between CAF and tumor cells.46 These features were tested and observed in microfluidic channels and compared to animal models through immunostaining and microscope imaging.
2.3. Tumor-on-a-chip with different tissue models
Besides cell-based study, microfluidic systems could be extended for multiple tissue forms. One approach that has been gaining significant attention is organoid-on-a-chip. The difference compared to a cell-based on-chip system is its natural self-organization ability to potentially have a better 3D structure. To approach a more in vivo mimicking system, organoid-on-a-chip is a recently developed technique that can merge with tumor-on-a-chip systems for a variety of directions, including stem cell-derived tissue culture.47 In contrast to cell line grown 3D structures, organoids are a miniaturized and simplified version of an organ obtained by culturing pluripotent stem cells that are able to differentiate and self-organize three-dimensionally with realistic micro-anatomy.47, 48 Although these organoids may have imperfect structures compared with adult human organs, they do capture certain key features that are important for drug screening or disease modeling purposes.49 For example, Skardal et al50 recently developed a metastasis-on-a-chip model to track metastatic tumor cell migration in a microfluidic system (Figure 2E). They inserted a gut organoid and a liver organoid in two independent chambers and connected them with circulating microfluidic channels, and showed influences of ECM geometry and drug treatments on the metastatic cell migration. This model revealed promising potential for generating more mimetic human metastasis responses. Another important area includes spheroids and organoids to constructed 3D models. Organoids often respond in more biologically relevant manners when compared to constructed 3D models. Since they can be derived from stem cells, organoids are often self-organized and generally have self-renewing abilities as well as more functionality than constructed 3D models. 3D constructed models, such as those that are created in hydrogels, are often easier to design and control especially when implementing chip-based approaches, yet they may be a less effective model for recapitulating in vivo features.
Another common 3D model is a spheroid, which can include 3D cell aggregations but with less complex architectures when compared to organoid models. These also provide 3D features that are cell relevant but may not present as physiologically relevant cell organization, and maybe scaffold free.51 In general, all of these model systems are compatible with microfluidic systems and thus can have intersections with tissue-on-a-chip approaches, which may lead to better emulation of model complexity toward higher physiological relevance, and the choice of model mainly depends on research purpose and question to address.
Besides, tumor slices may also be integrated into microfluidic devices. As an example, Chang et al52 developed a PDMS based drug screening device using a xenograft mouse brain tissue slices, which were placed in 96-well plates with controlled drug dosing. More specifically, they fabricated PDMS channels underneath 96-well plates and used a porous membrane to culture tissue slices and apply drugs in a controlled system to the tissue. This well-controlled system allowed for high throughput sample testing. A similar setup was applied to a biopsy tissue by Hattersley et al53.
The adaptation of organoids, spheroids, and primary tissues into on-chip systems require control of the environment beyond just cell lines environmental control. These chambers affect how tissues respond and how substrate responds with respect to tissue attachment. In addition, the delivery of chemicals including media, stimulants, etc. would affect the tissue response.
In summary, the ability to control the chemical microenvironment in tumor-on-a-chip systems will provide exciting new physiologically relevant approaches. These approaches have enabled the generation of well-controlled chemical gradients across 3D ECMs and multi-cell systems in ECMs with spatiotemporal distributions successfully mimicking in vivo conditions. These achievements allow for further development of more sophisticated tumor-on-a-chip systems.
2.4. Hypoxia characteristics for tumor-on-a-chip systems
As chemical forces influence the TME and directly affect cancer growth, one critical factor in this scenario is oxygen. Oxygen deficiency mainly occurs when the oxygen demand at a tumor exceeds the supply being provided from the adjacent vessel system, known as hypoxia. Most healthy organs reside in 3–6% oxygen while conditions lower than 3% oxygen are described as hypoxia.54 Hypoxia occurs widely in malignant tumors and is known to cause tumor progression through multiple mechanisms.5 The response typically includes abnormal growth of the vascular system in angiogenesis and EMT of cancer cells, which eventually leads to metastasis.55 Multiple studies have investigated the complex signaling pathways activated by hypoxia, including the regulation of hypoxia-inducible factors (HIF), phosphatidylinositide 3-kinases (PI3K), VEGF, etc. However, the direct responses of cancer cells to hypoxia remain unclear. Tumor-on-a-chip approaches may provide an efficient approach to examine hypoxia in vitro.
2.4.1. Cellular adaptation to hypoxia: the biological background
Multiple studies have revealed the complex signaling pathways activated by hypoxia, including the regulation of hypoxia-inducible factors (HIF1s). Under hypoxia, HIFs are activated and induce tumor-promoting gene circuits within the TME regulating vessel formation, growth and survival, glucose metabolism, invasion, and metastasis.56 While the effects of hypoxia on the TME are known, the direct responses of individual cell types in the TME to hypoxia remain unclear and require further investigation. Notably, several studies exist that implicate the upregulation of HIFs to be higher in intermittent hypoxia than when compared to acute hypoxia.57 Not surprisingly then, intermittent hypoxia has been closely linked to increased tumor invasion and metastasis.58, 59
Besides the activation of HIF-mediated gene expression, generation of oxygen-derived free radicals, commonly referred to as reactive oxygen species (ROS), is another consequence of hypoxia. As excessive ROS damage cellular components such as proteins, lipids, and DNA and thereby promote many diseases, including cancer, ROS adds to the HIF-mediated invasion and metastasis properties of cancer cells. A direct source of ROS stems from processes of reoxygenation after periods of hypoxia, a process which is comparable to reperfusion injury which follows restored blood flow after periods of ischemia.60 Compared to normal cells, cancer cells tend to contain higher levels of ROS. Therefore, antioxidant proteins are expressed at higher levels. This includes the transcription factor NRF-2 that induces cytoprotective genes and consequently enhances proliferation via metabolic reprogramming and apoptosis.61 The direct influence of hypoxia in cancer cells and signaling regulation can be investigated with tumor-on-a-chip systems, through the integration of multi-cell systems, in vivo mimicking microenvironments, and hypoxia generation schemes.
2.4.2. Methods of hypoxia generation on-chip
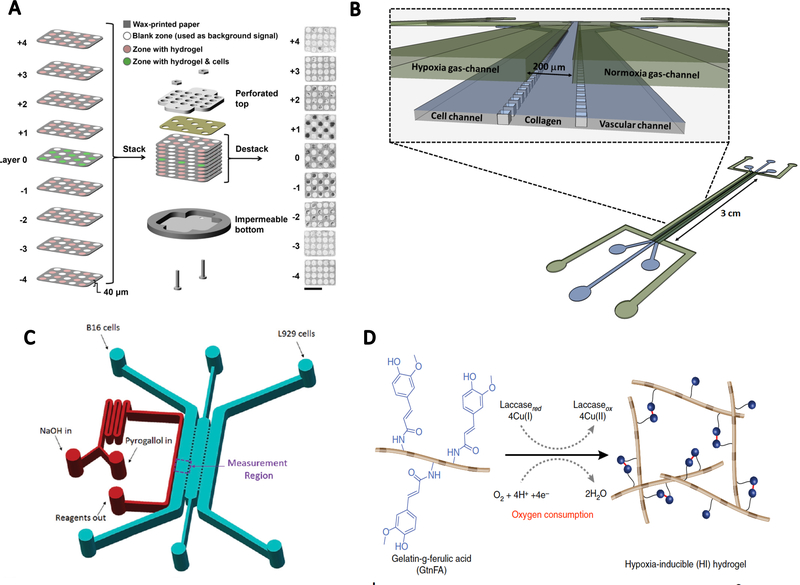
The ability to develop model systems with precise control of the oxygen level is critical to understand the effects of oxygen for cancer models. One approach to generating hypoxia through an oxygen gradient is to utilize the oxygen permeability of certain materials such as ECM or other fibers. Mosadegh et al62 developed a paper-based device to create an oxygen and nutrient gradient for assessing chemotaxis response. Through using multiple layers of hydrogel coated paper, the oxygen and nutrient concentrations could be controlled through diffusion from the top and bottom layers (Figure 3A). However, determining the local oxygen gradient for more quantitative analysis is challenging, and current methods use various fluorescent oxygen probes63, 64, which can be applied to determine oxygen levels in the ECM for calculating the oxygen permeability of scaffold materials such as collagen type I.
Figure.3.
Tumor-on-a-chip approach to generate hypoxia. A. A paper-based hypoxia generation system. Multiple layers of hydrogel coated paper blocked oxygen supply from the air. (reproduced with permission from ref. 62) B. A typical hypoxia control design for cell migration. The top layer was designed for hypoxia gradient generation, and the bottom layer for cell migration. (reproduced with permission from ref. 65). C. Hypoxia-on-a-chip design with a side-channel for chemical oxygen depletion by NaOH and pyrogallol reaction (reproduced with permission from ref. 69). D. Hypoxia-inducible hydrogel produced through laccase-mediated dimerization of ferulic acid molecules with oxygen consumption (reproduced with permission from ref. 72).
Another approach to control oxygen is through a parallel channel system. An oxygen gradient is like a chemical gradient but requires control of a gas flow system along with the ability to seal the device from outside gasses. Acosta et al65 created a PDMS based parallel-channel gradient generator system, with two gas channels above a cell-culture layer (Figure 3B). Oxygen driven migration was observed across the collagen. This design realized both spatial and temporal control of the hypoxic environment, allowing for the introduction of chronic and intermittent hypoxia environments as well. Long-term cancer cell culture was also achieved in the device, thus allowing for metastasis and intravasation study of the cancer cells.
Although PDMS is widely applied in microfluidic applications, it has potential limitations in certain applications. For example, its high permeability to oxygen makes hypoxia control more challenging. Thus another approach in fabricating oxygen gradient systems is to control the fabrication material using material differences in the oxygen permeability.20 Ayuso et al66 created a polystyrene-based microdevice instead of PDMS where the polystyrene oxygen permeability is its relatively low to isolate oxygen from the system. Instead of gas flow, the chemicals and oxygen were introduced across the hydrogel through medium flowing into the system to mimic blood flow, which could create a more physiological approach. This model provided an approach for microenvironment construction to study hypoxia and metastasis with more physiologically mimetic systems. Besides high permeability to oxygen, PDMS could have limitations such as adsorption of hydrophobic molecules and traditional rectangular channel fabrication causing non-physiological geometry. Thus a substitute material such as poly(methyl methacrylate) (PMMA) with lower permeability to oxygen and small molecules might be applied as well.67
Beyond using mechanical gas control with pumps or using approaches based on differences of oxygen permeability, another chemical method that has been used is to apply oxygen-scavenging chemicals such as pyrogallol combined with sodium hydroxide (NaOH).68 As an example, Sun et al69 fabricated a typical PDMS based microfluidic system with co-culture cells, but also added a side channel to inject NaOH and pyrogallol. This approach allowed mixing and reacting next to the main channel and inhibited local oxygen in the main channel (Figure 3C). They quantified the oxygen gradient and examined this with a relationship to cell death and migration. Shih et al70 applied the same method to their system and integrated it into a drug treatment assay. A similar design with an additional chemical channel component was implemented by Wang et al71, with a difference of the integration of a bottom layer instead.
Another physiologically relevant approach is to generate hypoxia directly in cell-cultured hydrogels. Park et al72 reported on a hypoxia-inducible (HI) hydrogel that was constructed with gelatin and ferulic acid to consume oxygen during gelation through a laccase-mediated reaction (Figure 3D). This approach could create a low oxygen environment for up to 50 h. Hypoxia generated by chemical reactions required the ability to refresh the chemicals to maintain the hypoxia for long times during the experiments, but it provided active and precise control over oxygen in the device.
In summary, hypoxia generation on-chip could be realized either through physical methodology (through oxygen pumping and diffusion) or chemical methodology (oxygen absorption by reaction). The former provides more flexible and long-term control over hypoxia, but generally requires additional equipment such as a pumping system. The latter may control hypoxia more accurately but lacks the ability for flexible hypoxia generation (e.g., multiple cycles of hypoxia and reoxygenation). The choice of method mainly depends on the desired hypoxia pattern and precision. Since oxygen is generally less controllable than chemicals with its fast 3D diffusion, a cost-effective and flexible chip system still needs to be designed in the future.
2.4.3. Understanding signaling regulation in hypoxic conditions
The ability to probe and examine signaling in hypoxia could be enhanced by tumor-on-a-chip systems, especially in the area of understanding signaling responses. A range of important signaling pathways is directly related to hypoxia including apoptosis, ROS production, and epithelial-to-mesenchymal transition. 73 For example, to examine in vitro cancer response to hypoxia, Zhang et al74 fabricated a cell migration microfluidic device and tested mesenchymal-mode SUM-159 cell migration under 1% pO2 and 21% pO2 environments. Also, Xu et al75 developed a cell migration chip to test U87 cell migration along a 3D collagen-based surface, under both hypoxia and normal conditions. This type of approach may provide a more physiologically relevant environment as well as allowing many standard biomolecular tracking and signaling pathway assays. A difference in the microenvironment between in vivo and in vitro conditions can be a major challenge that could cause unexpected cell responses, and a tumor-on-a-chip system may provide an approach to overcome this gap.
As we previously discussed, ROS generation is an important cause of hypoxia, and its influence on tumor cells could be examined through chips that can generate ROS gradients. For example, Chittiboyina et al76 developed a gradient-on-a-chip with H2O2 mixed with media, which caused oxidative DNA damage and protective (AOP2) response, along with influencing nuclei morphology. While this is technically a chemical gradient generator, this approach also was meaningful in studying hypoxia.
Another possibility to extend hypoxia studies is to investigate the mechanical effects. As an example, a recent study with co-culture of MDA-MB-231 cells and HUVECs in collagen type I gels under oxygen tension showed that gel contraction-induced mainly by HUVECs was attenuated in hypoxia condition, possibly through control of MMP-7 expression.77 This work shows great potential for integrating cancer or tumor models into chips for having multi-input environment control and measuring this intricate response.
Due to the difficulty in the design and fabrication of hypoxia-on-a-chip systems, studying hypoxia-related signal regulation remains challenging with tumor-on-a-chip approaches. Since the direct interaction between the level of oxygen and tumor response might be critical to cancer progression, we anticipate more collaborations between bioengineers and cancer biologists to investigate this challenge in vitro. Developing flexibility for hypoxia generation and integrating multi-cell systems may be powerful approaches to provide additional information versus just traditional methods. This also might be a key to developing oxygen-related treatments as a supplement to chemotherapy.
3. Mechanical factors and tumor-on-a-chip systems
Mechanics play a diversity of roles in cancer ranging from the changes in mechanical properties in tumors, such as stiffness, to the delivery of the nutrient supply through blood flow. Mechanical force is an important factor to consider since it can induce tumor metastasis by triggering mechanoreceptors of cells, change cytoskeleton structure and actomyosin-mediated contractility78, which leads to mechanotransduction, to changing multiple protein regulations and eventually alter cell behavior.79 The effect in mechanotransduction and specific signaling pathways involved in this process continues to be heavily investigated and is presently unclear.80 Mechanical forces are also known to create physical effects in the microenvironment through compression stemming from rapid tumor growth that reduces perfusion rates of nutrients and promotes hypoxia.22 Mechanical forces can be easily mimicked in tumor-on-a-chip approaches as there are many micro- and nano-fabrication approaches, which enable these factors to be implemented in chip-based systems.80 Mechanical inputs, such as shear stress, compression, tension, and ECM stiffness, can be implemented and tested through the use of stretchable substrates (e.g., PDMS), dynamic flows, and the applying of mechanical loads directly. Here, we discuss recent work involving mechanical stimulation and effects for tumor-on-a-chip approaches.
3.1. Shear stress affects cell signaling regulation
Shear stress is defined as the frictional force between moving layers in laminar flow, which mainly comes from fluid viscosity and shear rate.81 In a tumor microenvironment, shear stresses are affected mainly by blood flow conditions in the vasculature. Solid tumor growth requires nutrient and oxygen supply. Thus the importance of vascularization cannot be overstated. Vascularization comes from a variety of biological responses, including vascular vessel sprouting from adjacent vessel systems and control through proangiogenic factors. Since most tumor-on-a chip mimetics lack the control of vascular organization, most induced vasculature networks would likely be highly disordered, involving multiple junctions, self-loops, and diverse vessel dimensions.82, 83 This could cause an irregular vessel network near the solid tumor which could experience compression from the tumor during its growth84, resulting in complex flow conditions and shear stress distribution in the vessels that would inhibit the mimetics and analysis of the results.85, 86
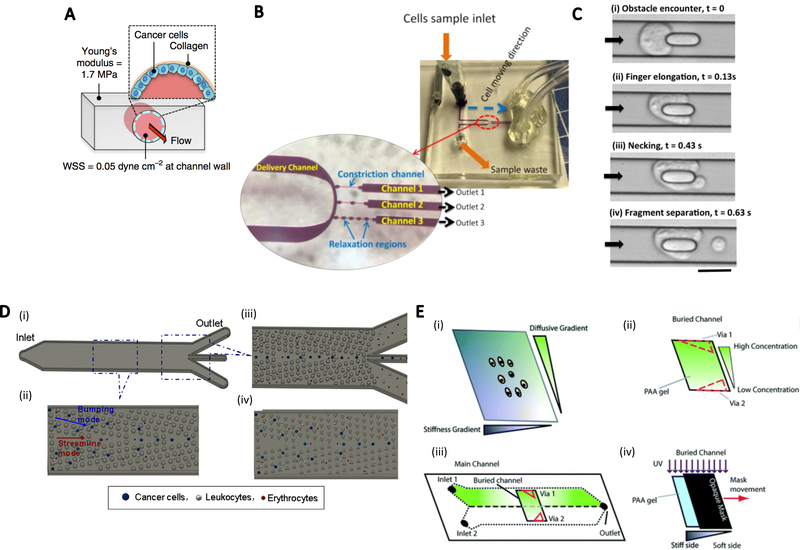
Since the vessel system and tumor morphology are very complex to analyze, a simplified model with calculable shear stress, flow rate, and other parameters is advantageous for examining the general approach. For example, it has been shown that YAP1 upregulation of cancer cells is driven by fluid flow through lymphatics87. To study this using a tumor-on-a chip mimetic, a circular PDMS channel coated with collagen and a monolayer of PC3 prostate cells was fabricated so that the shear stress induced by media flow was directly exerted on the cell surface (Figure 4A). This setup was applied to show regulation of cancer cell behavior with vascular flow and thus underlined the influence of mechanics on cancer cell behavior. Shear stress also affects tumor shedding. For example, Trianafillu et al88 found that increased cancer stem cell features were present on MCF7 and MDA-MB-231 cells after exposure to shear stress in vessels with fluid flow. Syringes and tubing were used to apply shear stress to cells.
Figure.4.
Tumor-on-a-chip models to generate mechanical factors. A. A PDMS cylindrical channel coated with collagen to grow a monolayer of cancer cells. Media flow generated wall shear stress and induced cell filopodia extension. (reproduced with permission from ref. 87) B. Microfluidic design for multiple constriction-relaxation regions along the channels with comparable size to cells for cell deformation testing. (reproduced with permission from ref. 94) C. Channels with micropillars in the center blocked cell motion with flow. (reproduced with permission from ref. 96) D. Micropost array for CTC separation from a blood sample. (reproduced with permission from ref. 99) E. 2D chemical-ECM stiffness gradient setup. A chemical gradient was generated through double laminar flows in a Y-shape channel with different chemical concentrations. ECM stiffness gradients were created by photo-polymerization of ECM with a moving opaque mask. (reproduced with permission from ref. 114)
In addition to shear flow, computational approaches can allow for rapid examination of a diversity of interrelated factors.89, 90 Shirinifard et al91 developed a 3D multi-cell computational simulation model for tumor growth and angiogenesis. Through control of critical parameters such as partial pressure of oxygen, proangiogenic factors, and chemoattractants, the growth rate of tumor and vessel sprouting was projected and allowed the recreation and comparison of multiple features in angiogenesis. This way, minimalistic modeling can provide great insight through a controlled examination of fewer parameters, which helps to understand complex biological phenomena during metastasis and provides a promising tool for using fluid mechanics to study metastasis.
3.2. Deformability as an indicator of cancer cells
Along with external mechanical stimulation, as described above, the mechanical properties of cancer cells and the relationship with their biological response is of great interest. Compared to non-cancer cells, cancer cells possess differences in mechanical features such as cell stiffness and deformability. Stiffness of cells refers to the ability of cells to resist deformation when subjected to stress, while cell deformability means the degree to which applied stress changes the cell shape.92 For example, metastatic cancer cells generally have lower stiffness (thus generally higher deformability) and softer membranes, while non-cancerous cells often present higher stiffness related to the cytoskeleton.93, 94 Therefore, one approach for the characterization of cancer cell deformability has been to detect the migration ability of cells through a microfluidic “squeezer” channel where cells must deform to migrate through the channel.95, 96 This technique led to the development of cell sorters based on the migration ability of cancer cells in a constriction channel.97, 98 Ren et al94 reported a single-cell sorter using multi-constriction channels to detect velocity profiles of cancer cells. They designed up to five constriction-relaxation regions along a microfluidic channel for cells to deform and relax while migrating (Figure 4B).
Another cell mechanical constriction channel was developed by Kamyabi et al96, who applied a similar channel design, but tested the fragmentation of cells with an additional micropillar in the channel (Figure 4C). When the cancer cells that passed through the channel were blocked at the micropillar, they would deform and eventually either squeeze through the obstacle or break into fragments. Fragmentation time and fragment excess area were important evaluation references for different cell types.
Deformability of cancer cells could also contribute to circulating tumor cell (CTC) sorting with deterministic lateral displacement arrays. Liu et al99 fabricated such an array for CTC separation from blood samples (Figure 4D). They specifically simulated cancer cell deformation when hitting the pillars in the array and proved that cell deformation in the array could reduce effective cell radius, which needed to be higher than critical particle size of the array for sorting to occur. Thus, an increase in flow rate caused an increase in shear stress and cell deformation, resulting in a reduction of sorting efficiency.
A similar method to examine the unique deformability of cancer cells is to combine them with inertial effects in microfluidic systems. In an inertial microfluidic cell sorting device100, cancer cells flowing in microfluidic channels experience an inertial lift force that induces lateral migration. In balance with the viscoelasticity induced force in the opposite direction, cancer cells are examined at a lateral position, which is defined mainly by the deformability and size. More deformable cancer cells stay closer to the centreline of the channel compared with benign cells and blood cells for examining specific cells.
Many recent studies on tumor deformability have focused on single-cell analysis, which tends to be in the category of cancer-on-a-chip rather than tumor-on-a-chip systems. Tumor-on-a-chip systems usually refer to applications of 3D cancer models. The effort to study the deformability of a tumor or 3D organoids very likely involves not only the deformation of cells, but also are related to cell adhesion molecules such as cadherin, and the single-cell studies are linked to larger-scale responses. Multiscale systems are intricately related to each other and strongly affect how they respond. The study of single cells provides meaningful tools and foundations for understanding more complex bulk tumor response. For example, in the mechanical arena related to the deformation of the bulk tumor, mathematical models have been developed to estimate solid tumor stress by treating the tumor as a heterogeneous elastic body, yet critical parameters such as bulk modulus are highly dependent on single-cell properties.101, 102 There is still a lack of effective experimental models that will need to be developed to link single-cell responses to larger-scale 3D responses.103
3.3. Examine cell-ECM interaction in metastasis related to mechanics
Another critical component in cancer is the relationship between the cells and the ECM, which is also related to mechanics. The ECM provides tumors with both structural/mechanical support, thereby fostering biochemical interactions.104 Cancer cell interactions with the ECM have been widely studied in 2D and 3D in vitro culture, and evidence has shown active participation of ECM in regulating cell differentiation, invasion, and migration.22, 105–108 For example, human breast cancer transformation is generally correlated with collagen deposition and linearization, which also triggers stromal cells activity and macrophage infiltration.109
The transfer from 2D to 3D cell culture could significantly change cell behavior as it may better mimic physiological conditions. For example, in a virus-infected tumor study, the growth of KSHV-infected BJAB cells in a 3D microwell assay enabled a higher genome copy number and higher lytic reactivation rate.110 EMT could also be induced in 3D platforms instead of just 2D environments. Kuo et al111 developed a hydrated layer of flexible copolymer-based chains (nano-cilia) to reduce the 2D adhesion between putative cancer stem cell spheroids and the polystyrene dish surface. They successfully detected the dynamic regulation of multiple EMT markers. Also, recently gastric cancer cells encapsulated in collagen beads showed the upregulation of EMT and metastatic genes, and downregulation of E-cadherin.112
More complex effects such as ECM geometry, including its fibril alignment direction and convergence, could also affect cell behavior since it might regulate local adhesion force distribution and cell alignment orientation. Microfluidic approaches have the potential to be very useful in studying these effects. Pathak et al113 developed a microfluidic platform to study the influence of matrix confinement in addition to ECM stiffness. By changing the width of the microfluidic channel where the ECM and tumor cells were located, they discovered that a decreased channel width correlated with increased cell migration. They suggested this was due to the narrower channel which induced a polarized cells-ECM traction force.
ECM stiffness was further examined by Garcia et al114 who designed a two-dimensional platform to simultaneously test cell respond to ECM stiffness gradient and chemical gradient perpendicular to each other (Figure 4E). The chemical gradient was generated by diffusion of hepatocyte growth factor (HGF) through a passive approach. The stiffness gradient was created using an acrylamide/bis-acrylamide mix through a photo-polymerization method. By placing an opaque mask on top of the gel mix while irradiating it with UV light, the covered region will be less stiff than the exposed region due to a difference of irradiation time. This device not only provided detection of cell migration and morphology change but also traction force measurements, which enabled a greater understanding of matrix stiffness and cancer cell response.
Cell motility is a prerequisite for cancer cell invasion and metastasis and therefore has been extensively studied. 115, 116 Anguiano et al117 applied a PDMS microfluidic device to examine lung cell migration in mixed collagen-hydrogel (Matrigel) scaffold. The Matrigel was found to facilitate cell migration in relatively low concentrations by providing growth factors retaining the scaffold. This system though slowed down migration at high concentrations due to increased attachments and thus force interactions. Cells also presented a mesenchymal phenotype in the collagen-only scaffold and transitioned to lobopodial in the collagen-matrigel mix matrix.
These examples show the ability of tumor-on-a-chip systems to simulate various conditions and change different parameters when adjusting to specific experimental goals. One major strength of the microfluidic system is quantitative control over microenvironments, substrate properties, and other critical parameters involved in cell stimulation and response. Mechanical stimulations, which naturally occur in many in vivo environments, could be introduced and analyzed on-chip with fluid mechanics models, and in addition programmable systems in the future. Mechanical properties are critical in understanding cancer and are great places where micro- and nano-technologies can be integrated to test these questions for tumor-on-a-chip systems.
4. Tumor-on-a-chip analysis techniques
The ability to analyze cell responses using tumor-on-a-chip technology has become increasingly important, given the negative effects of cancer heterogeneity on drug response. A clear advantage of the tumor-on-a-chip approach is its availability for us to capture the cells after experiments. Since cancer cells are cultured in 3D hydrogel scaffolds, immunofluorescent staining is not as efficient as in 2D cell cultures because of a) less diffusion of the antibody and b) microscopical limitations in the depth of focus. However, the construction out of ECM of this type of chip has advantages. A 3D collagen scaffold can be digested with collagenase after the experiments for extracting the cells. These cells can then be stained and assayed by conventional analysis techniques. Furthermore, the tumor-on-a-chip system can be snap-frozen in liquid nitrogen, embedded, sliced with a microtome and further processed for immunofluorescent staining and imaging.
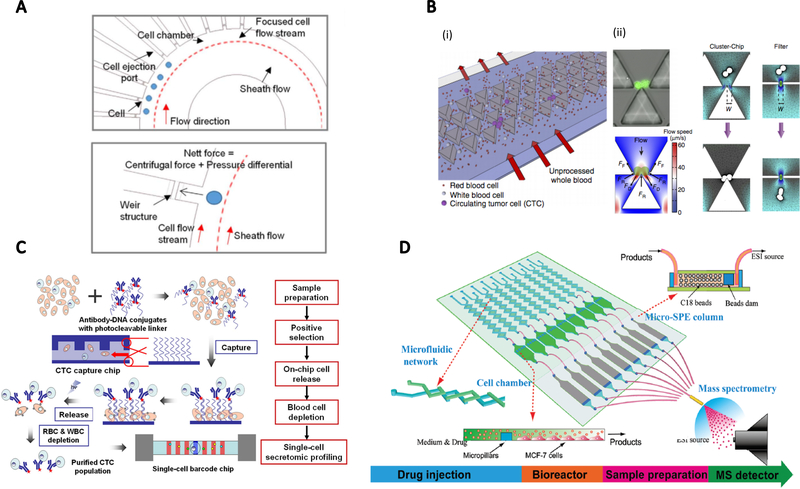
Single-cell analysis is also very important to accurately address tumor heterogeneity.118 For example, there is a relatively low population of CTCs within a large population of non-cancerous cells. The low frequency of CTCs could cause inaccurate results.119 To sort CTCs out from blood samples, Yeo et al120 designed a microfluidic sorter to capture single cells from flow into multiple side chambers (Figure 5A). The initial cell flow channel was designed with a curvature so centrifugal force could be generated. With an inherent differential pressure, cells were trapped into each chamber with a capture size on the scale of a single cell which was then later released using positive pressure applied from the other side of the chamber. Another design to filter CTC clusters from a blood sample was proposed by Sarioglu et al121 using a size exclusion concept. The cell filter was fabricated in triangular blocker arrays, with a gap that could release blood cells and single CTCs but could also capture CTC clusters when finding a matching cell size (Figure 5B). The geometry of the traps was designed so that double-cell cluster could be captured with a balance of forces. Deng et al122 proposed a different approach to purify CTCs from whole blood, with photocleavable ssDNA-encoded antibody conjugates attached to the channel substrate to capture low-density CTCs in blood flow. These cells could be detached later for purification and single cell secretomic profiling (Figure 5C). This approach made use of high chemical reaction efficiency in microfluidic channels through high surface-to-volume ratio.
Figure.5.
Tumor-on-a-chip filters and analyzers. A. A circulating tumor cell (CTC) sorter to capture single cells from flows. The channel curvature generated centrifugal force to push cells into side chambers. (reproduced with permission from ref. 120) B. A filter to capture CTC clusters. Single cells pass through and only cell clusters are captured with the balance of force based on triangular blocker tips. (reproduced with permission from ref. 121) C. The strategy of CTCs isolation on photocleavable ssDNA-encoded antibody conjugates attached to a channel substrate. CTCs could then be released by photoirradiation for single-cell secretomic profiling. (reproduced with permission from ref. 122) D. Tumor cell metabolism inspector. This system had a microfluidic network for media and drug injection, a cell culture chamber, a micro-solid-phase extraction column for sample pretreatment, and an electrospray ionization source for mass spectrometry detection. (reproduced with permission from ref. 123)
Tumor-on-a-chip systems can also be used for metabolomic analysis of tumor cells through the integration of labeling and detection techniques. Chen et al123 developed a tumor cell culture chip integrated with electrospray ionization mass spectrometry to detect drug-induced cell apoptosis and measure metabolism simultaneously (Figure 5D). Kalfe et al124 embedded a U-shape microfluidic tube containing tumor spheroids directly into a miniaturized micro slot NMR detector so they could monitor up to 23 metabolites.
Gene chip analysis of tumor biology provides a tremendous amount of information for understanding genetic changes, which are especially helpful in addressing cancer heterogeneity. The gene chip analysis approach has been integrated with microfabricated systems in the past, including molecular analysis and genomic processing platforms. Spurgeon et al125 developed a dynamic microfluidic array that functioned as a gene expression platform and allowed for 45 genes in 18 tissues to be tested on a single chip. Cheng et al126 developed a protein digestion system for rapid proteolysis that showed much higher efficiency compared to conventional overnight protein digestion.
The techniques discussed above could be used as detection tools in conventional tumor studies, but also could be implemented as a post-treatment approach with other tumor-on-a-chip experiments by direct assembly or integration at the output end of regular tumor-on-a-chip devices. In the future, they could be integrated directly into tumor-on-a-chip systems to allow for instant genomic and proteomic profiling, etc. With the flexibility of chip design, various analysis tools can be integrated into the systems for targeted experiments for tumor-on-a-chip system.
5. Future perspectives on tumor-on- a-chip systems
Tumor-on-a-chip approaches have the potential to bridge the gap between more traditional in vitro cell culture and in vivo experiments. These systems are focused on mimicking the physiologically relevant environments through control of the materials, dimensionality, and microenvironmental variables, including chemical and mechanical factors. In the future, these approaches will provide high reproducibility, precise parameter control, and cost reduction. One of the ultimate goals of tumor-on-a-chip is to create an artificial tumor model that captures the key features of a real tumor so that it can be used for diagnostics and therapeutics while minimizing the need for animal and human testing. Current studies are focusing on developing functionalities of the features of the tumor including chemotaxis, shear stress, and other factors. These systems are using the advantages from micro- and nano-fabrication toward these ends. However, questions remain on how these tissue-on-a-chip models will provide a complete landscape for tumor understanding.
Another important ability of tumor-on-a-chip systems is to integrate TME, multiple cell types, ECM, and vasculature with complex spatiotemporal distribution. Integrating these different features will help mimic more realistic physiological conditions. The addition of more variables though and their integration will increase the number of controllable parameters and variations in the system, which are challenges with any systems integration whether it is biological or non-biological. Therefore, some questions for tumor-on-a-chip design are i) how to screen for the necessary key features that truly affect tumor development; ii) how to integrate these features into one model while simplifying the construction as much as possible. Answers to these questions could be investigated through understanding and mimicking human clinical cancer studies, which would then help designing new engineering techniques.
Besides, a more direct application to tumor-on-a-chip is to provide high throughput, in vivo mimicking models for drug screening. 3D hydrogel environment, spatiotemporal drug distribution, and cell-cell interactions are all crucial factors to be considered. Integration of enough complexity to the system and adapting patients’ tumor tissue for personalized medicine are interesting topics as well.127
While there are many great advances in tissue-on-a-chip systems, there are challenges with these approaches too. These concerns include materials selection as the most widely applied microfabrication approaches are constructed with PDMS, which serves well due to its flexibility and high gas permeability. However, PDMS is not a particularly physiologically relevant material compared to materials mimicking human structures such as the ECM. A more physiologically relevant approach may be to construct microfluidic channels embedded in hydrogels through approaches such as for sacrificial template molding37 and 3D bioprinting128.
Besides, the choice of biocompatible ECM materials such as collagen, Matrigel129, and other hydrogels depends on factors including ECM stiffness, cell adhesion, chemical compositions, etc. For example, collagen type I is widely used due to its abundance in many tissues, thus providing better in vivo mimicking. Matrigel, as a more complex animal product130, has been applied in studies such as cell differentiation, angiogenesis, and tumor growth.129 However Matrigel is not well defined and may be variable from experiment to experiment as well as having a great cost than other ECM related materials.130
Overall, the idea of tumor-on-a-chip is an important step toward future therapeutics and diagnostics that aims to bridge between 2D petri dish experiments and animal and human models. The integration of important components of mechanical, chemical, and oxygen factors will move these 3D systems towards more physiological implementations. This new field will have impacts in different areas ranging from cancer research to microfabrication to mechanical engineering.
Footnotes
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Siegel RL, Miller KD and Jemal A, CA: A Cancer Journal for Clinicians, 2018, 68, 7–30. [DOI] [PubMed] [Google Scholar]
- 2.Tsai H-F, Trubelja A, Shen AQ and Bao G, Journal of the Royal Society Interface, 2017, 14, 20170137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn J, Sei YJ, Jeon NL and Kim Y, Bioengineering, 2017, 4, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Portillo-Lara R and Annabi N, Lab on a chip, 2016, 16, 4063–4081. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D and Weinberg RA, Cell, 2011, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- 6.Reddy BS, Lipids, 1992, 27, 807–813. [DOI] [PubMed] [Google Scholar]
- 7.Lee GY, Kenny PA, Lee EH and Bissell MJ, Nature Methods, 2007, 4, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kashaninejad N, Nikmaneshi M, Moghadas H, Kiyoumarsi Oskouei A, Rismanian M, Barisam M, Saidi M and Firoozabadi B, Micromachines, 2016, 7, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tibbitt MW and Anseth KS, Biotechnology and Bioengineering, 2009, 103, 655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huh D, Hamilton GA and Ingber DE, Trends in Cell Biology, 2011, 21, 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pampaloni F, Reynaud EG and Stelzer EH, Nature reviews Molecular cell biology, 2007, 8, 839. [DOI] [PubMed] [Google Scholar]
- 12.Abbott AJ and Vita GD, Scottish Journal of Political Economy, 2003, 50, 69–89. [Google Scholar]
- 13.Yamada KM and Cukierman E, Cell, 2007, 130, 601–610. [DOI] [PubMed] [Google Scholar]
- 14.Ma H, Xu H and Qin J, Biomicrofluidics, 2013, 7, 011501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loessner D, Stok KS, Lutolf MP, Hutmacher DW, Clements JA and Rizzi SC, Biomaterials, 2010, 31, 8494–8506. [DOI] [PubMed] [Google Scholar]
- 16.Szot CS, Buchanan CF, Freeman JW and Rylander MN, Biomaterials, 2011, 32, 7905–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esch EW, Bahinski A and Huh D, Nature Reviews Drug Discovery, 2015, 14, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huh D, Torisawa Y.-s., Hamilton GA, Kim HJ and Ingber DE, Lab on a Chip, 2012, 12, 2156–2164. [DOI] [PubMed] [Google Scholar]
- 19.Asghar W, El Assal R, Shafiee H, Pitteri S, Paulmurugan R and Demirci U, Materials Today, 2015, 18, 539–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hachey SJ and Hughes CC, Lab on a Chip, 2018, 18, 2893–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Begley CG and Ellis LM, Nature, 2012, 483, 531. [DOI] [PubMed] [Google Scholar]
- 22.Jain RK, Martin JD and Stylianopoulos T, Annual Review of Biomedical Engineering, 2014, 16, 321–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roussos ET, Condeelis JS and Patsialou A, Nature Reviews Cancer, 2011, 11, 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan DA and Giaccia AJ, Cancer and Metastasis Reviews, 2007, 26, 333–339. [DOI] [PubMed] [Google Scholar]
- 25.Kalluri R and Zeisberg M, Nature Reviews Cancer, 2006, 6, 392. [DOI] [PubMed] [Google Scholar]
- 26.Whitesides GM, Ostuni E, Takayama S, Jiang X and Ingber DE, Annual Review of Biomedical Engineering, 2001, 3, 335–373. [DOI] [PubMed] [Google Scholar]
- 27.Murphy SV and Atala A, Nature Biotechnology, 2014, 32, 773. [DOI] [PubMed] [Google Scholar]
- 28.Preziosi L, Cancer modelling and simulation, CRC Press, 2003. [Google Scholar]
- 29.Bagchi P, Johnson PC and Popel AS, Journal of Biomechanical Engineering, 2005, 127, 1070–1080. [DOI] [PubMed] [Google Scholar]
- 30.Hughes-Alford SK and Lauffenburger DA, Current Opinion in Cell Biology, 2012, 24, 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Folkman J, Nature Medicine, 1995, 1, 27. [DOI] [PubMed] [Google Scholar]
- 32.Zou H, Yue W, Yu W-K, Liu D, Fong C-C, Zhao J and Yang M, Analytical Chemistry, 2015, 87, 7098–7108. [DOI] [PubMed] [Google Scholar]
- 33.Bhatia SN and Ingber DE, Nature Biotechnology, 2014, 32, 760. [DOI] [PubMed] [Google Scholar]
- 34.Lei KF, Goh A and Huang C-H, Talanta, 2019, 205, 120124. [DOI] [PubMed] [Google Scholar]
- 35.Tomasova L, Guttenberg Z, Hoffmann B and Merkel R, PloS One, 2019, 14, e0219708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y-C, Allen SG, Ingram PN, Buckanovich R, Merajver SD and Yoon E, Scientific Reports, 2015, 5, 9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan L, Skoko J, Yu J, LeDuc P and Neumann C, Scientific Reports, 2017, 7, 16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolesky DB, Homan KA, Skylar-Scott MA and Lewis JA, Proceedings of the National Academy of Sciences, 2016, 113, 3179–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen D-HT, Stapleton SC, Yang MT, Cha SS, Choi CK, Galie PA and Chen CS, Proceedings of the National Academy of Sciences, 2013, 110, 6712–6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casey J, Yue X, Nguyen TD, Acun A, Zellmer VR, Zhang S and Zorlutuna P, Biomedical Materials, 2017, 12, 025009. [DOI] [PubMed] [Google Scholar]
- 41.Sokol ES, Miller DH, Breggia A, Spencer KC, Arendt LM and Gupta PB, Breast Cancer Research, 2016, 18, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoarau-Véchot J, Rafii A, Touboul C and Pasquier J, International Journal of Molecular Sciences, 2018, 19, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bruce A, Evans R, Mezan R, Shi L, Moses BS, Martin KH, Gibson LF and Yang Y, PloS One, 2015, 10, e0140506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bersini S, Jeon JS, Dubini G, Arrigoni C, Chung S, Charest JL, Moretti M and Kamm RD, Biomaterials, 2014, 35, 2454–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeon JS, Bersini S, Gilardi M, Dubini G, Charest JL, Moretti M and Kamm RD, Proceedings of the National Academy of Sciences, 2015, 112, 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeong S-Y, Lee J-H, Shin Y, Chung S and Kuh H-J, PloS One, 2016, 11, e0159013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takebe T, Zhang B and Radisic M, Cell Stem Cell, 2017, 21, 297–300. [DOI] [PubMed] [Google Scholar]
- 48.Skardal A, Shupe T and Atala A, Drug Discovery Today, 2016, 21, 1399–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang R-R, Ueno Y, Zheng Y-W and Koike N, Nature, 2013, 499, 481. [DOI] [PubMed] [Google Scholar]
- 50.Skardal A, Devarasetty M, Forsythe S, Atala A and Soker S, Biotechnology and Bioengineering, 2016, 113, 2020–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y, Sun W, Kang L, Wang Y, Zhang M, Zhang H and Hu P, Analyst, 2019. [DOI] [PubMed] [Google Scholar]
- 52.Chang TC, Mikheev AM, Huynh W, Monnat RJ, Rostomily RC and Folch A, Lab on a Chip, 2014, 14, 4540–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hattersley SM, Dyer CE, Greenman J and Haswell SJ, Lab on a Chip, 2008, 8, 1842–1846. [DOI] [PubMed] [Google Scholar]
- 54.Cater DB and Silver IA, Acta Radiologica, 1960, os-53, 233–256. [DOI] [PubMed] [Google Scholar]
- 55.Muz B, de la Puente P, Azab F and Azab AK, Hypoxia, 2015, 3, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petrova V, Annicchiarico-Petruzzelli M, Melino G and Amelio I, Oncogenesis, 2018, 7, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Semenza GL and Prabhakar NR, Antioxidants & Redox Signaling, 2007, 9, 1391–1396. [DOI] [PubMed] [Google Scholar]
- 58.Martinive P, Defresne F, Bouzin C, Saliez J, Lair F, Grégoire V, Michiels C, Dessy C and Feron O, Cancer Research, 2006, 66, 11736–11744. [DOI] [PubMed] [Google Scholar]
- 59.Yao K, Gietema J, Shida S, Selvakumaran M, Fonrose X, Haas N, Testa J and O’Dwyer P, British Journal of Cancer, 2005, 93, 1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Granger DN and Kvietys PR, Redox Biology, 2015, 6, 524–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leinonen HM, Kansanen E, Pölönen P, Heinäniemi M and Levonen A-L, in Advances in Cancer Research, Elsevier, 2014, vol. 122, pp. 281–320. [DOI] [PubMed] [Google Scholar]
- 62.Mosadegh B, Lockett MR, Minn KT, Simon KA, Gilbert K, Hillier S, Newsome D, Li H, Hall AB and Boucher DM, Biomaterials, 2015, 52, 262–271. [DOI] [PubMed] [Google Scholar]
- 63.Cheema U, Brown R, Alp B and MacRobert A, Cellular and Molecular Life Sciences, 2008, 65, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kellner K, Liebsch G, Klimant I, Wolfbeis OS, Blunk T, Schulz MB and Göpferich A, Biotechnology and Bioengineering, 2002, 80, 73–83. [DOI] [PubMed] [Google Scholar]
- 65.Acosta MA, Jiang X, Huang P-K, Cutler KB, Grant CS, Walker GM and Gamcsik MP, Biomicrofluidics, 2014, 8, 054117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ayuso JM, Virumbrales-Muñoz M, Lacueva A, Lanuza PM, Checa-Chavarria E, Botella P, Fernández E, Doblare M, Allison SJ and Phillips RM, Scientific Reports, 2016, 6, 36086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vidi P-A, Maleki T, Ochoa M, Wang L, Clark SM, Leary JF and Lelièvre SA, Lab on a Chip, 2014, 14, 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sleeboom JJ, Amirabadi HE, Nair P, Sahlgren CM and Den Toonder JM, Disease Models & Mechanisms, 2018, 11, dmm033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun W, Chen Y, Wang Y, Luo P, Zhang M, Zhang H and Hu P, Analyst, 2018, 143, 5431–5437. [DOI] [PubMed] [Google Scholar]
- 70.Shih H-C, Lee T-A, Wu H-M, Ko P-L, Liao W-H and Tung Y-C, Scientific Reports, 2019, 9, 8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang W, Li L, Ding M, Luo G and Liang Q, BioChip Journal, 2018, 12, 93–101. [Google Scholar]
- 72.Park KM and Gerecht S, Nature Communications, 2014, 5, 4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang J, Tang Y.-l. and Liang X.-h., Cancer Biology & Therapy, 2011, 11, 714–723. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y, Wen J, Zhou L and Qin L, Integrative Biology, 2015, 7, 672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu H, Rahimpour S, Nesvick CL, Zhang X, Ma J, Zhang M, Zhang G, Wang L, Yang C and Hong CS, Oncotarget, 2015, 6, 11882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chittiboyina S, Rahimi R, Atrian F, Ochoa M, Ziaie B and Lelièvre SA, ACS Biomaterials Science & Engineering, 2017, 4, 432–445. [DOI] [PubMed] [Google Scholar]
- 77.Yoshino D and Funamoto K, AIP Advances, 2019, 9, 045215. [Google Scholar]
- 78.Murrell M, Oakes PW, Lenz M and Gardel ML, Nature Reviews Molecular Cell Biology, 2015, 16, 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chin L, Xia Y, Discher DE and Janmey PA, Current Opinion in Chemical Engineering, 2016, 11, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Przybyla L, Muncie JM and Weaver VM, Annual Review of Cell and Developmental Biology, 2016, 32, 527–554. [DOI] [PubMed] [Google Scholar]
- 81.Huang Q, Hu X, He W, Zhao Y, Hao S, Wu Q, Li S, Zhang S and Shi M, American Journal of Cancer Research, 2018, 8, 763. [PMC free article] [PubMed] [Google Scholar]
- 82.Vakoc BJ, Lanning RM, Tyrrell JA, Padera TP, Bartlett LA, Stylianopoulos T, Munn LL, Tearney GJ, Fukumura D and Jain RK, Nature Medicine, 2009, 15, 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Less JR, Skalak TC, Sevick EM and Jain RK, Cancer Research, 1991, 51, 265–273. [PubMed] [Google Scholar]
- 84.Stylianopoulos T, Martin JD, Chauhan VP, Jain SR, Diop-Frimpong B, Bardeesy N, Smith BL, Ferrone CR, Hornicek FJ and Boucher Y, Proceedings of the National Academy of Sciences, 2012, 109, 15101–15108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Steward RL, Cheng C-M, Wang DL and LeDuc PR, Cell Biochemistry and Biophysics, 2010, 56, 115–124. [DOI] [PubMed] [Google Scholar]
- 86.Steward RL, Cheng C-M, Jonathan DY, Bellin RM and LeDuc PR, Scientific Reports, 2011, 1, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee HJ, Diaz MF, Price KM, Ozuna JA, Zhang S, Sevick-Muraca EM, Hagan JP and Wenzel PL, Nature Communications, 2017, 8, 14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Triantafillu UL, Park S, Klaassen NL, Raddatz AD and Kim Y, International Journal of Oncology, 2017, 50, 993–1001. [DOI] [PubMed] [Google Scholar]
- 89.Zeng Y, Lai T, Koh CG, LeDuc PR and Chiam K-H, Biophysical Journal, 2011, 101, 2122–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kang J, Steward RL, Kim Y, Schwartz RS, LeDuc PR and Puskar KM, Journal of Theoretical Biology, 2011, 274, 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shirinifard A, Gens JS, Zaitlen BL, Popławski NJ, Swat M and Glazier JA, PloS One, 2009, 4, e7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chien S, Annual Review of Physiology, 1987, 49, 177–192. [DOI] [PubMed] [Google Scholar]
- 93.Hou HW, Li Q, Lee G, Kumar A, Ong C and Lim CT, Biomedical Microdevices, 2009, 11, 557–564. [DOI] [PubMed] [Google Scholar]
- 94.Ren X, Ghassemi P, Babahosseini H, Strobl JS and Agah M, ACS Sensors, 2017, 2, 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Khan Z and Vanapalli S, Biomicrofluidics, 2013, 7, 011806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kamyabi N and Vanapalli SA, Biomicrofluidics, 2016, 10, 021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Karimi A, Yazdi S and Ardekani A, Biomicrofluidics, 2013, 7, 021501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vanapalli SA, Duits MH and Mugele F, Biomicrofluidics, 2009, 3, 012006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu Z, Huang F, Du J, Shu W, Feng H, Xu X and Chen Y, Biomicrofluidics, 2013, 7, 011801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hur SC, Henderson-MacLennan NK, McCabe ER and Di Carlo D, Lab on a Chip, 2011, 11, 912–920. [DOI] [PubMed] [Google Scholar]
- 101.Roose T, Netti PA, Munn LL, Boucher Y and Jain RK, Microvascular Research, 2003, 66, 204–212. [DOI] [PubMed] [Google Scholar]
- 102.Ciarletta P, Physical Review Letters, 2013, 110, 158102. [DOI] [PubMed] [Google Scholar]
- 103.Ahn J, Sei Y, Jeon N and Kim Y, Bioengineering, 2017, 4, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Malik R, Lelkes PI and Cukierman E, Trends in Biotechnology, 2015, 33, 230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lu P, Weaver VM and Werb Z, J Cell Biol, 2012, 196, 395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Swaminathan V, Mythreye K, O’Brien ET, Berchuck A, Blobe GC and Superfine R, Cancer Research, 2011, 71, 5075–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kubicek JD, Brelsford S, Ahluwalia P and LeDuc PR, Langmuir, 2004, 20, 11552–11556. [DOI] [PubMed] [Google Scholar]
- 108.Leight JL, Drain AP and Weaver VM, Annual Review of Cancer Biology, 2017, 1, 313–334. [Google Scholar]
- 109.Acerbi I, Cassereau L, Dean I, Shi Q, Au A, Park C, Chen Y, Liphardt J, Hwang E and Weaver V, Integrative Biology, 2015, 7, 1120–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.El Assal R, Gurkan UA, Chen P, Juillard F, Tocchio A, Chinnasamy T, Beauchemin C, Unluisler S, Canikyan S and Holman A, Scientific Reports, 2016, 6, 39144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kuo C-T, Chiang C-L, Chang C-H, Liu H-K, Huang G-S, Huang RY-J, Lee H, Huang C-S and Wo AM, Biomaterials, 2014, 35, 1562–1571. [DOI] [PubMed] [Google Scholar]
- 112.Jang M, Koh I, Lee SJ, Cheong J-H and Kim P, Scientific Reports, 2017, 7, 41541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pathak A and Kumar S, Proceedings of the National Academy of Sciences, 2012, 109, 10334–10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Garcia S, Sunyer R, Olivares A, Noailly J, Atencia J and Trepat X, Lab on a Chip, 2015, 15, 2606–2614. [DOI] [PubMed] [Google Scholar]
- 115.Sahai E, Current Opinion in Genetics & Development, 2005, 15, 87–96. [DOI] [PubMed] [Google Scholar]
- 116.Tzvetkova-Chevolleau T, Stéphanou A, Fuard D, Ohayon J, Schiavone P and Tracqui P, Biomaterials, 2008, 29, 1541–1551. [DOI] [PubMed] [Google Scholar]
- 117.Anguiano M, Castilla C, Maška M, Ederra C, Peláez R, Morales X, Muñoz-Arrieta G, Mujika M, Kozubek M and Muñoz-Barrutia A, PloS One, 2017, 12, e0171417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Marusyk A and Polyak K, Biochimica et Biophysica Acta (BBA)-Reviews on Cancer, 2010, 1805, 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, Telli ML, Advani RH, Carlson RW and Mollick JA, PloS One, 2012, 7, e33788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yeo T, Tan SJ, Lim CL, Lau DPX, Chua YW, Krisna SS, Iyer G, San Tan G, Lim TKH and Tan DS, Scientific Reports, 2016, 6, 22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sarioglu AF, Aceto N, Kojic N, Donaldson MC, Zeinali M, Hamza B, Engstrom A, Zhu H, Sundaresan TK and Miyamoto DT, Nature Methods, 2015, 12, 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Deng Y, Zhang Y, Sun S, Wang Z, Wang M, Yu B, Czajkowsky DM, Liu B, Li Y and Wei W, Scientific Reports, 2014, 4, 7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen Q, Wu J, Zhang Y and Lin J-M, Analytical Chemistry, 2012, 84, 1695–1701. [DOI] [PubMed] [Google Scholar]
- 124.Kalfe A, Telfah A, J. r. Lambert and R. Hergenröder, Analytical Chemistry, 2015, 87, 7402–7410. [DOI] [PubMed] [Google Scholar]
- 125.Spurgeon SL, Jones RC and Ramakrishnan R, PloS One, 2008, 3, e1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cheng G, Hao S-J, Yu X and Zheng S-Y, Lab on a Chip, 2015, 15, 650–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Caballero D, Blackburn SM, de Pablo M, Samitier J and Albertazzi L, Lab on a Chip, 2017, 17, 3760–3771. [DOI] [PubMed] [Google Scholar]
- 128.Kolesky DB, Truby RL, Gladman A, Busbee TA, Homan KA and Lewis JA, Advanced Materials, 2014, 26, 3124–3130. [DOI] [PubMed] [Google Scholar]
- 129.Kleinman HK and Martin GR, Seminars in Cancer Biology, 2005, 15, 378–386. [DOI] [PubMed] [Google Scholar]
- 130.Hughes CS, Postovit LM and Lajoie GA, Proteomics, 2010, 10, 1886–1890. [DOI] [PubMed] [Google Scholar]