Abstract
Proteins interacting with DNA are fundamental for mediating processes such as gene expression, DNA replication and maintenance of genome integrity. Accumulating evidence suggests that the chromatin of apicomplexan parasites, such as Plasmodium falciparum, is highly organized, and this structure provides an epigenetic mechanism for transcriptional regulation. To investigate how parasite chromatin structure is being regulated, we undertook comparative genomics analysis using 12 distinct eukaryotic genomes. We identified conserved and parasite-specific chromatin-associated domains (CADs) and proteins (CAPs). We then used the chromatin enrichment for proteomics (ChEP) approach to experimentally capture CAPs in P. falciparum. A topological scoring analysis of the proteomics dataset revealed stage-specific enrichments of CADs and CAPs. Finally, we characterized, two candidate CAPs: a conserved homologue of the structural maintenance of chromosome 3 protein and a homologue of the crowded-like nuclei protein, a plant-like protein functionally analogous to animal nuclear lamina proteins. Collectively, our results provide a comprehensive overview of CAPs in apicomplexans, and contribute to our understanding of the complex molecular components regulating chromatin structure and genome architecture in these deadly parasites.
Keywords: chromatin structure, proteome, chromatin-associated proteins, Plasmodium falciparum, topological data analysis
Data Summary
The MS dataset (raw, peak, search and result files) can be obtained from the MassIVE database via ftp://massive.ucsd.edu/MSV000082520/, ftp://massive.ucsd.edu/MSV000082521/ and ftp://massive.ucsd.edu/MSV000084671/ for the chromatin enrichment for proteomics (ChEP), SMC3_IPs and CRWN-L_IPs datasets, respectively; from the ProteomeXChange (http://www.proteomexchange.org/) via PXD010262, PXD010263 and PXD016684; and from the Stowers Original Data Repository (http://www.stowers.org/research/publications/libpb-1239). ChIP-seq datasets generated and analysed during the current study are available from the National Center for Biotechnology Information Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE116219.
Impact Statement.
Malaria still remains one of the deadliest infectious diseases worldwide. Parasite resistance to antimalarial drugs and the absence of an effective vaccine emphasizes the need for novel therapeutic approaches. Targeting mechanisms regulating virulence factors and gene expression throughout the parasite life cycle is a promising strategy towards developing novel antimalarials. Much like in complex metazoans, chromatin structure of Plasmodium falciparum plays important roles in gene expression regulation and parasite development. It is becoming increasingly apparent that parasite nuclear architecture and chromosome dynamics are quite complex and are regulated by many proteins. Therefore, to determine the proteins that may be critical to chromatin organization and function within the nucleus, we developed a new computational and proteomic pipeline. We identify parasite-specific proteins that may regulate chromatin architecture processes in Plasmodium. While these proteins will need to be further validated at the mechanistic level, they represent ideal therapeutic targets that can be used to disrupt parasite development with high specificity.
Introduction
Apicomplexans are obligate protozoan parasites that are responsible for a wide range of diseases in humans and animals. Among apicomplexan parasites that infect humans, Plasmodium spp., the causative agents of malaria, have the largest health and economic impact. While the most prevalent and deadly human malaria parasite, Plasmodium falciparum, is responsible for an estimated 445 000 deaths per year [1], Plasmodium vivax and Plasmodium knowlesi also infect humans. Other apicomplexan parasites relevant to humans include Babesia microti [2], the causative agent of human babesiosis, a malaria-like illness endemic in the USA but with worldwide distribution, and Toxoplasma gondii, the causative agent of toxoplasmosis, an opportunistic parasite that cause infections in immunocompromised individuals [3]. The kinetoplastid parasites, another class of human-relevant protozoan pathogens, also contribute to the global burden of disease and include Trypanosoma brucei (causing African sleeping sickness), Trypanosoma cruzi (causing Chagas disease) and Leishmania spp. (causing leishmaniasis) [4–6].
Given the absence of a protective vaccine and the alarming spread of multidrug-resistant parasites [7–9], there is a desperate need for new therapeutic approaches. One promising strategy towards the development of novel and effective antiparasitic compounds is to inhibit DNA replication and gene expression in these parasites. Since the publication of the first parasite genomes, such as the P. falciparum genome that was published over 15 years ago [10], researchers have attempted to explore the transcriptional machinery of parasites in detail. The distinct developmental stages of the parasite life cycles are characterized by coordinated changes in gene expression [11–17]. However, a surprisingly low number of specific transcription factors have been identified in their genomes [18–20], and only a few stage-specific transcription factors have been characterized and validated in Plasmodium spp. or Toxoplasma gondii [21–27]. Therefore, the coordinated cascade of transcripts observed throughout the parasite life cycles is likely to be regulated by additional components and mechanisms, such as post-transcriptional [28–32], translational and post-translational regulation [28, 33, 34], as well as changes in epigenetics and chromatin structure.
Recently, several groups, including ours, have developed chromosome conformation capture (3C) coupled to next-generation sequencing methods (called Hi-C) as a way of understanding genome organization of the nucleus and its role in regulating biological processes [35–38]. Our work identified distinct chromatin features during parasite life cycles [39]. We also observed a strong association between genome architecture and gene expression in the apicomplexan parasites we analysed [39, 40]. These results suggest that changes in chromatin structure may control, at least partially, gene expression and parasite development. Additionally, Hi-C results demonstrate that the parasite nucleus is highly organized. In particular, telomere ends of the chromosomes cluster together in heterochromatin area(s) in close proximity to the nuclear membrane, while the centromeres cluster at the opposite end of the large heterochromatin cluster, much like the genome organization observed in the similarly sized budding and fission yeasts [41, 42]. However, the parasite genome exhibits a higher degree of organization than the budding yeast genome, as genes involved in immune evasion (e.g. var, rifin and stevor genes) add a striking complexity and act as structural elements that shape global genome architecture [35]. Such observations were confirmed by chromatin structure analysis in different Plasmodium species, demonstrating that spatial genome organization in apicomplexan parasites is often constrained by the colocalization of virulence genes that have a unique effect on chromosome folding. We also identified a potential link between genome organization and gene expression in more virulent pathogens [40]. Based on these observations, we hypothesize that architectural proteins that interact with chromatin and have a strong influence on genome organization may represent novel targets for antiparasitic interventions.
Architectural proteins involved in the maintenance of chromatin structure have been studied in organisms ranging from yeast to human [43]. Among these proteins are RNA polymerase associated factors, cohesin, condensin and CCCTC-binding factor (CTCF) [43–46]. CTCF is an insulator protein conserved in vertebrates that is enriched at chromosome domain boundaries and interacts with the nuclear lamina [47]. Some of these components have homologues in the parasite genomes, but only a few have been characterized at the functional level. Furthermore, many conserved chromatin architectural proteins or chromatin-associated proteins (CAPs) involved in chromatin organization and maintenance (e.g. lamina proteins, CTCF) are missing in parasite genomes [48]. As an example, lamina proteins in metazoans are essential for many nuclear functions including nuclear shape maintenance and architecture, chromatin organization, DNA replication, transcription and cell cycle progression [47, 49].
Although most of our understanding of proteins involved in chromatin structure and their functions comes from studies on model organisms, their importance in the development and virulence of apicomplexan parasites including Plasmodium has recently been appreciated for a small number of candidates [50–53]. Yet a large number of these proteins still need to be identified and functionally characterized. Given the potential roles of CAPs in almost all aspects of parasite biology, we performed a comprehensive computational and comparative genomics approach to generate an extended atlas of chromatin-associated domains (CADs) in 12 eukaryotic organisms. We identified conserved, as well as apicomplexan parasite-specific, euglenid parasite-specific, unicellular yeast-specific and several multicellular organism-specific CADs. We provide functional annotations based on homology, domain organization, domain clustering and expression pattern analysis for 1190 well-defined and putative CAPs in the P. falciparum genome, of which 162 proteins (13.6 %) have been previously described as having chromatin-related functions [54].
To validate some of our candidate CAPs, we employed an unbiased chromatin proteomics approach termed chromatin enrichment for proteomics (ChEP). ChEP has been successfully used to identify chromatin-bound molecules and predict their function and regulation in a number of organisms [55–57]. We further characterized, using standard cellular and molecular approaches, two of the candidate proteins detected by proteomics analysis. These two candidates were the conserved structural maintenance of chromosomes protein 3 (SMC3) and an atypical crowded-like nuclei (CRWN) protein. CRWN proteins are present in plant nuclei and have a function similar to lamina in animal and fungi. These proteins participate in chromatin organization, and are essential for DNA replication and cell division. Altogether, our results validate that while mechanisms regulating chromatin structure in apicomplexan parasites are most likely complex, some of our candidates have plant-like features that could be specifically targeted by new antimalarial strategies. A better understanding of these CAPs not only will provide a comprehensive view of the complex molecular components that control chromatin organization and genome architecture in these deadly parasites, but also will assist the identification of novel targets for therapeutic strategies.
Methods
CAD search
Protein sequences were obtained from the following sources: PlasmoDB version 29.0 (P. falciparum strain 3D7 and P. vivax strain Sal I), ToxoDB version 24.0 (Toxoplasma gondii strain ME49), TriTrypDB version 24.0 (Trypanosoma brucei strain TREU927, Trypanosoma cruzi strain CL Brener Esmeraldo-like and Leishmania major strain Friedlin), Saccharomyces Genome Database (Saccharomyces cerevisiae strain S288C genome assembly R64-2-1), PomBase (Schizosaccharomyces pombe, downloaded on 25 June 2015), Araport (Arabidopsis thaliana 11, downloaded on 10 January 2017), Ensembl release 80 (Homo sapiens genome assembly GRCh38.p2, Caenorhabditis elegans genome assembly WBcel235 and Drosophila melanogaster genome assembly BDGP6).
Protein sequences were searched for the presence of all possible CADs using Hmmscan (hmmer, version 3.16, February 2015) against Pfam hmm profiles (v 30.0) as described elsewhere [30], and using National Center for Biotechnology Information (NCBI) Reversed Position Specific blast (rps-blast version 2.6.0) against NCBI Conserved Domain Database (CDD) (pre-calculated position-specific scoring matrices (PSSMs) originating from CDD from various alignment collections version 3.16). For rps-blast domain searches, if multiple rps-blast hits were reported for the same conserved domain, only the one with the highest per cent identity was maintained for each protein. An E value of 0.001 was used for both approaches, and if a protein had multiple isoforms, only the first isoform was kept. Candidate CAPs were filtered using 3870 pre-filtered CADs. The list of CADs (Table S1a, available with the online version of this article) was generated based on domain annotation. Domains with annotations containing keywords that are related to nucleus or chromatin regulation (e.g. nucleoporin, nuclear pore complex, chromatin remodelling, histone modification, etc) were systematically selected from the NCBI CDD (cddid_all.tbl) and Pfam database (Table S1a). Using manual curation, domains without clear definition of chromatin or nuclear-related functions were excluded from the final list resulting in a total of 3870 CADs regardless of organism source.
Once proteins with CADs were identified from hmm and rps-blast searches, results were merged and Pfam domain identifiers were converted into NCBI PSSMS identifiers, giving rise to the final list of candidate CAPs for each of the proteomes mentioned above. To obtain the final list candidate CAPs in P. falciparum, both manual curation and a list of exported proteins were used to rule out potential false-positive proteins. Exported proteins were defined as proteins with an Export Prediction (ExportPred) score above 5, as well as proteins with a PEXEL or HT motif for export to the red blood cell (RBC) membranes (downloaded from PlasmoDB). The export proteins are likely to be exported into the RBC rather than to be transported into the nucleus; thus, were excluded from the final list of candidate CAPs in P. falciparum (Table S2a).
Barplot comparison of CAPs
For each organism, both Hmmscan and rps-blast approaches were used and merged as previously described. Since not all genomes have been annotated at the same level, manual curation was avoided to eliminate bias and to ensure a fair comparison between organisms; therefore, we systematically calculated the number of proteins containing any of the filtered CADs (n=3870) irrespective of protein annotation. For each organism, the calculated value was then corrected by the proteome size and expressed as the percentage of CAPs in the full proteome of that organism (Table S1b).
Domain heatmap
For each CAD present in any of the 12 organisms, we first calculated its frequency of occurrence in all organisms. Next, the abundance value was corrected by genome size and expressed as the number per 10 000 genes. The relative abundance values were scaled to the domain frequency in the organism with the highest relative abundance of that domain (Table S1c). Finally, all CADs (n=2867) obtained in at least one of the organisms were clustered using the k-mean clustering algorithm with a maximum of 1000 iterations (R v3.31). The number of clusters was selected based on percentage of variance captured, in which a minimum of 60 % variance was required and an increase in number of clusters did not capture an additional 2 % of the variance. Domains associated gene ontology (GO) enrichment analysis was performed with dcGO (http://supfam.org/SUPERFAMILY/dcGO/index.html) with default parameters and Pfam domain IDs (Table S1d).
Protein classification
Candidate proteins were classified based on their general function using existing annotations and known functions of homologues in other species from various sources including PlasmoDB, UniProt and the NCBI gene database. Proteins with no annotation details were classified based on their domain functionality.
Gene expression analysis for Plasmodium CAPs
The gene expression profiles and boxplots were generated using steady-state mRNA expression profiles downloaded from https://plasmodb.org/. The expression profiles were pre-processed using standardized pipelines and are reads per kilobase of transcript per million mapped reads (RPKM)-transformed. For boxplots, gene groups were generated based on gene annotation. Transcription factors were removed from the list of P. falciparum CAPs (PfCAPs) prior to gene expression analysis. The list of RNA binding proteins was obtained from published work [30].
Parasite cultures
The P. falciparum strain 3D7 was cultured in human O+ erythrocytes at 5 % haematocrit, as described elsewhere [58]. Cultures were synchronized at the ring stage with 5 % (w/v) d-sorbitol treatments [59]. Parasite cultures (8 % parasitaemia in 5 % haematocrit) were harvested 48 h after the first sorbitol treatment (ring stage), and 18 h (trophozoite stage) and 36 h thereafter (schizont stage).
ChEP
CAPs were isolated at different stages of the parasite erythrocytic cycle (early ring, early trophozoite and late schizont stages) using a protocol adapted from published work [56]. Briefly, synchronized parasites were cross-linked with 1 % formaldehyde for 15 min at 37 °C. Cross-linking was quenched by adding 0.125 M glycine for 5 min at room temperature. The parasites were then washed with 1x PBS, incubated in nuclear extraction buffer [10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 0.5 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF), protease inhibitor cocktail (Roche) and 0.25 % Igepal CA-630 (Sigma-Aldrich)] for 30 min and needle sheared using a 25-gauge needle. Extracted nuclei were spun at 1300 relative centrifugal force (rcf) for 20 min at 4 °C. The nuclear pellet was incubated in nuclear extraction buffer containing 200 μg RNase A ml−1 for 15 mins at 37 °C, followed by two washes with 1× PBS. Nuclei were further washed with highly denaturing extraction buffers containing 4 % SDS and 8M urea to wash away non-cross-linked proteins. Chromatin was solubilized and genomic DNA was sheared by sonication (Covaris ultrasonicator; 5 % duty cycle, 140 intensity peak incident power, 200 cycles per burst).
As a negative control, protein from the cytoplasmic fractions of early ring, early trophozoite and late schizont stage parasites were also extracted. For the isolation of cytoplasmic fractions, synchronized parasite cultures were collected and subsequently lysed by incubating in 0.15 % saponin for 10 min on ice. Parasites were centrifuged at 1500 rcf for 10 min at 4 °C and washed three times with 1x PBS. For each wash, parasites were resuspended in cold 1x PBS and centrifuged for 10 min at 1500 rcf at 4 °C. After the last wash, parasites were resuspended in 1x PBS, transferred to a microcentrifuge tube and centrifuged for 5 min at 2100 rcf at 4 °C. Subsequently, the parasite pellet was resuspended in a 1.5× volume of cytoplasmic lysis buffer [0.65 % Igepal CA-630 (Sigma-Aldrich), 10 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 2 mM AEBSF and EDTA-free protease inhibitor cocktail (Roche)], and lysed by passing through a 26G ½ inch needle 15 times. Parasite nuclei were centrifuged at 14 500 rcf for 15 min at 4 °C and the supernatant containing the cytoplasmic extract was collected.
Western blotting
Mixed-stage 3D7 P. falciparum parasite cultures were collected and lysed using 0.15 % saponin for 10 min on ice. After subsequent washes, the parasite pellet was resuspended in a 1.5× volume of cytoplasmic lysis buffer [0.65 % Igepal CA-360 (Sigma-Aldrich), 10 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 2 mM AEBSF and EDTA-free protease inhibitor cocktail (Roche)] and lysed by passing through a 26G ½ inch needle 15 times. Parasite nuclei were centrifuged at 14 500 rcf for 15 min at 4 °C and the supernatant containing the cytoplasmic extract was collected. To extract proteins from the parasite nucleus, the nuclear pellet was resuspended in 1 ml shearing buffer (0.1 % SDS, 1 mM EDTA, 10 mM Tris pH 7.5, protease inhibitors, phosphatase inhibitors), lysed by passing through a 26 G ½ inch needle seven times and sonicated seven times 10 s on/30 s off using a probe sonicator. Extracted nuclear protein lysates were incubated for 10 mins at room temperature with DNase I to remove DNA and centrifuged for 10 mins at 14 500 rcf to remove cell debris.
Twenty micrograms parasite cytoplasmic and nuclear protein lysates were diluted 1 : 1 in 2× Laemmli buffer and heated at 95 °C for 10 mins. The protein lysates there then loaded on an Any-KD SDS-PAGE gel (Bio-Rad) and run for 1 h at 125 V. Proteins were transferred to a PVDF membrane for 1 h at 18 V, stained using commercial antibodies anti-histone H3 (Abcam ab1791; 1 : 3000) and anti-Plasmodium aldolase (Abcam ab207494; 1 : 1000) or custom antibodies generated against PF3D7_1325400 (Thermo Fisher; 1 : 100) and PF3D7_0414000 (Thermo Fisher; 1 : 100), followed by an incubation with secondary antibody, goat anti-rabbit IgG horseradish peroxidase conjugate (Bio-Rad; 1 : 10 000). The membranes were visualized using a Bio-Rad Chemidoc MP gel imager.
Custom antibody generation
Custom peptide antibodies were designed to target the C-terminal domain of two proteins: PF3D7_1325400 and PF3D7_0414000 (Thermo Fisher Scientific). For PF3D7_1325400, a 17 amino acid peptide (sequence: KEANKNIKLLQKYNKKM), and for PF3D7_0414000, an 18 amino acid peptide (sequence: KNEAYEIISIEEKHALEN) was used to immunize two rabbits. Antisera from day 72 post-immunization were collected and affinity-purified to obtain antibodies specifically targeting the proteins of interest [30, 56].
Immunofluorescence microscopy
P. falciparum asexual stage parasites were fixed onto slides using 4 % paraformaldehyde for 30 min at room temperature. Slides were washed three times using 1× PBS. Parasites were permeabilized with 0.2 % Triton X for 30 min at room temperature, followed by a wash step with 1× PBS. Samples were blocked overnight at 4 °C in immunofluorescence assay buffer (2 % BSA, 0.05 % Tween-20, 100 mM glycine, 3 mM EDTA, 150 mM NaCl and 1× PBS). Cells were incubated with custom anti-SMC3 (Thermo Fisher; 1 : 500) or anti-CRWN (Thermo Fisher; 1 : 500) antibodies for 1 h at room temperature, followed by anti-rabbit Alexa Fluor 488 (Life Technologies A11008; 1 : 500) secondary antibody for 1 h at room temperature. Slides were mounted in Vectashield mounting medium with DAPI. Images were acquired using the Olympus BX40 epifluorescence microscope.
For the sequential double-staining immunofluorescence methodology, P. falciparum asexual stage parasites were fixed, permeabilized and blocked as described above. Parasites were incubated with custom anti-SMC3 or anti-CRWN antibodies for 1 h at room temperature, followed by anti-rabbit DyLight 550 (Abcam ab98489; 1 : 500) secondary antibody for 1 h at room temperature. Slides were washed seven times using 1× PBS/0.01 % Tween-20 to remove any unbound secondary antibody and incubated with anti-H3K9me3 antibody, Alexa Fluor 488 conjugate (Millipore 07–442-AF488; 1 : 100) for 1 h at room temperature. Slides were mounted in Vectashield mounting medium with DAPI. Images were acquired using an Olympus BX40 epifluorescence microscope.
Immunoprecipitation (IP) of protein complexes
Mixed-stage 3D7 P. falciparum parasite cultures were collected and lysed using 0.15 % saponin for 10 min on ice. After subsequent washes, the parasite pellet was resuspended in a 2.5× volume of IP buffer [0.65 % Igepal CA-360 (Sigma-Aldrich), 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM EDTA, 1 % Triton X, 1 mM AEBSF, 5 µM E-64 and EDTA-free protease inhibitor cocktail (Roche)], lysed by passing through a 26G ½ inch needle ten times and sonicated seven times 10 s on/30 s off using a probe sonicator. Extracted nuclear protein lysates were incubated for 10 mins at room temperature with DNase I to remove DNA and centrifuged for 10 mins at 14 500 rcf to remove cell debris.
Washed protein A magnetic beads (Pure Proteome) were added to the protein sample and incubated for 1 h at 4 °C to preclear the lysate. Precleared lysate was transferred to a new microcentrifuge tube and split equally for the antibody and no antibody control. The anti-SMC3 or anti-CRWN custom antibodies were added at a 1 : 50 ratio and incubated overnight at 4 °C. The negative control with no antibody was also incubated overnight. Antibody–protein complexes were recovered using protein A magnetic beads (Pure Proteome), followed by extensive washes with wash buffer A (1 % Triton X, 1 mM EDTA in 1× PBS), wash buffer B (wash buffer A, 0.5 M NaCl) and wash buffer C (1 mM EDTA, 1× PBS). Proteins were eluted using 0.1 M glycine, pH 2.8, and the eluent was neutralized using 2 M Tris-HCl, pH 8.0.
Chromatin immunoprecipitation (ChIP)
Synchronized parasite cultures were collected at the early trophozoite stage and subsequently lysed by incubating in 0.15 % saponin for 10 min on ice. Parasites were centrifuged at 1500 rcf for 10 min at 4 °C and washed three times with 1x PBS. For each wash, parasites were resuspended in cold 1x PBS and centrifuged for 10 min at 1500 rcf at 4 °C. Subsequently, parasites were cross-linked for 10 min with 1 % formaldehyde in 1x PBS at 37 °C. Glycine was added to a final concentration of 0.125 M to quench the cross-linking reaction, and incubated for 5 min at 37 °C. Parasites were centrifuged for 5 min at 2100 rcf at 4 °C, washed twice with cold 1x PBS and stored at −80 °C.
Parasites were incubated on ice in nuclear extraction buffer [10 mM HEPES, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 0.5 mM AEBSF, EDTA-free protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Roche)]. After 30 min, Igepal CA-360 (Sigma-Aldrich) was added to a final concentration of 0.25 % and the parasites were lysed by passing the suspension through a 26G ½ inch needle seven times. Parasite nuclei were centrifuged at 4 °C for 20 min at 2100 rcf. Parasite nuclei were resuspended in shearing buffer (0.1 % SDS, 1 mM EDTA, 10 mM Tris HCl pH 7.5, EDTA-free protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Roche)]. Chromatin was fragmented using the Covaris ultra-sonicator (S220) for 8 min with the following settings: 5 % duty cycle, 140 intensity peak incident power, 200 cycles per burst. To remove insoluble material, samples were centrifuged for 10 min at 16 800 rcf at 4 °C.
Fragmented chromatin was diluted 1 : 1 in ChIP dilution buffer [30 mM Tris-HCl pH 8, 3 mM EDTA, 0.1 % SDS, 300 mM NaCl, 1.8 % Triton X-100, EDTA-free protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Roche)]. Samples were precleared with protein A agarose beads to reduce non-specific background and incubated overnight at 4 °C with 2 µg custom anti-SMC3 and anti-CRWN antibodies (Thermo Fisher Scientific). A sample with no antibody was also incubated overnight at 4 °C to be used as the negative control. Antibody–protein complexes were recovered using protein A agarose beads, followed by extensive washes with low-salt immune complex wash buffer [1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8, 150 mM NaCl], high-salt immune complex wash buffer [1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8, 500 mM NaCl], LiCl immune complex wash buffer [0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl pH 8] and TE buffer [10 mM Tris-HCl pH 8, 1 mM EDTA]. Chromatin was eluted from the beads by incubating twice with freshly prepared elution buffer (1 % SDS, 0.1 M NaHCO3) for 15 min at room temperature. Samples were reverse cross-linked overnight at 45 °C by adding NaCl to a final concentration of 0.5 M. RNase A (final concentration 0.6 µg ml−1, Life Technologies) was added to the samples and incubated for 30 min at 37 °C, followed by a 2 h incubation at 45 °C with the addition of EDTA (final concentration 8 mM), Tris-HCl pH 7 (final concentration 33 mM) and proteinase K (final concentration 66 µg ml−1; New England Biolabs). DNA was extracted by phenol:chloroform:isoamylalcohol and ethanol precipitation. Extracted DNA was purified using Agencourt AMPure XP beads (Beckman Coulter).
Libraries from the ChIP samples were prepared using the KAPA library preparation kit (KAPA Biosystems). Libraries were amplified for a total of 12 PCR cycles (12 cycles of 15 s at 98 °C, 30 s at 55 °C, 30 s at 62 °C) using the KAPA HiFi HotStart ready mix (KAPA Biosystems). Libraries were sequenced with a NextSeq500 DNA sequencer (Illumina). Raw read quality was first analysed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), the last base was removed using Trimmomatic. Any base with a quality score below 25 was trimmed using Sickle (https://github.com/najoshi/sickle). Trimmed reads were then mapped to the P. falciparum genome using Bowtie2 (v2.3.4.1) [60]. Uniquely mapped reads were further filtered, resulting in a total of 25×106 reads for SMC3 ChIP-seq replicate 1, 17.9×106 reads for SMC3 ChIP-seq replicate 2, 25×106 reads for CRWN ChIP-seq replicate 1, 42×106 reads for CRWN ChIP-seq replicate 2, 16.8×106 reads for the no-antibody negative-control replicate 1 and 25×106 reads for the no-antibody negative-control replicate 2. Read coverage per nucleotide was determined using BEDTools. Both positive and negative libraries were then normalized by dividing through the numbers of million mapped reads. As the anti-CRWN antibody was too weak to immunoprecipitate any specific proteins including the CRWN protein and the ChIP-seq data were similar to the ‘no-antibody’ control, we treated these datasets as additional negative-control libraries using a rabbit IgG antibody. For each nucleotide, the signal from the negative-control library was then subtracted from the SMC3 ChIP-seq libraries and any negative value was replaced with a zero. Genome browser tracks were generated and viewed using the Integrative Genomic Viewer (IGV) from the Broad Institute. Centromere locations were obtained from a published paper [61] for all chromosomes except chromosome 10. The centromere location for chromosome 10 was obtained from [62], as this information was missing from [61].
Proteomics analysis by multidimensional protein identification technology (MudPIT)
Proteins were precipitated with 20 % trichloroacetic acid (TCA) and the resulting pellet was washed once with 10 % TCA and twice with 100% cold acetone. TCA-precipitated protein pellet was solubilized using Tris-HCl pH 8.5 and 8 M urea, followed by addition of TCEP [Tris(2-carboxyethyl)phosphine hydrochloride; Pierce] and chloroacetamide (Sigma) were added to a final concentration of 5 mM and 10 mM, respectively. The protein samples were digested using endoproteinase Lys-C at 1 : 100 (w/w) (Roche) at 37 °C overnight. The samples were brought to a final concentration of 2 M urea and 2 mM CaCl2, and a second digestion was performed overnight at 37 °C using trypsin (Promega) at 1 : 100 (w/w). The reactions were stopped using formic acid (5 % final). The samples were loaded on a split-triple-phase fused-silica micro-capillary column and placed in-line with a linear ion trap mass spectrometer (LTQ; Thermo Scientific), coupled with a Quaternary Agilent 1100 series HPLC system. A fully automated 10-step chromatography run (for a total of 20 h) was carried out, as described elsewhere [63]. Each full MS scan (400–1600 m/z) was followed by five data-dependent MS/MS scans. The number of the micro scans was set to 1 both for MS and MS/MS. The dynamic exclusion settings used were as follows: repeat count 2; repeat duration 30 s; exclusion list size 500; and exclusion duration 120 s; while the minimum signal threshold was set to 100.
The MS/MS data set was processed as described in Fig. S1(a). Briefly, RawDistller [64] extracted peak files were searched using ProLuCID (v. 1.3.3) [65] against a database consisting of 5538 P. falciparum non-redundant proteins (PlasmoDB 9.1), 34 521 H. sapiens non-redundant proteins (downloaded from NCBI 27/08/12), 177 usual contaminants (such as human keratins, IgGs and proteolytic enzymes), and, to estimate false-discovery rates (FDRs), 36 179 randomized amino acid sequences derived from each non-redundant protein entry. To account for alkylation by chloroacetamide, 57 Da was added statically to the cysteine residues. To account for the oxidation of methionine residues to methionine sulfoxide (which can occur as an artefact during sample processing), 16 Da was added as a differential modification to the methionine residue. Peptide/spectrum matches were sorted and selected using DTASelect/Contrast [66]. Proteins had to be detected by one peptide with two independent spectra, leading to mean FDRs of 0.53±0.22 and 0.12±0.05 at the protein and spectral levels, respectively. To estimate relative protein levels and to account for peptides shared between proteins, distributed normalized spectral abundance factors (dNSAFs) were calculated for each detected protein, as described elsewhere [67].
Statistical analysis of proteomics data
Two biological replicates with two technical replicates each were prepared by ChEP at the ring, trophozoite and schizont stages along with their corresponding cytoplasmic control samples, and analysed by MudPIT (Table S3a). To assess the enrichment of proteins in nuclear ChEP fractions, we applied qspec statistics on the distributed spectral counts (dS) measured for the P. falciparum proteins detected in the nuclear fractions [68, 69], with the cytoplasmic fractions used as negative controls. Proteins were considered significantly enriched in a nuclear ChEP fraction compared to the corresponding cytoplasmic fraction of a particular stage if they had a Z-statistics value ≥ 2 and/or an FDR ≤0.05 in at least one stage (Table S3b, Fig. S1a).
To determine whether there was a non-random association between a protein being predicted or known to contain a CAD and its specific detection in a nuclear ChEP fraction, enrichment of CAPs or CADs in the proteins deemed specific to the nuclear ChEP fractions was assessed using the one-tailed Fisher's exact test by defining contingency tables for each categorical variable and calculating the probability hypergeometric distribution in Excel (Tables S3bc and S4).
To determine stage-specificity of protein expression, proteins passing the nuclear ChEP enrichment criteria described above were next subjected to topological scoring (TopS) analysis [70]. To validate TopS results, qspec was also used recursively to compare the dS (distributed spectral count) values measured in one stage against the other two time-points as background. Proteins with both TopS and qspec Z statistics values ≥2 in at least one of the three blood stages were considered specific (Table S3c).
To separate proteins based on their TopS scores observed in the three erythrocytic stages (Table S3c), we used the Ayasdi software platform (Ayasdi) [70–72] to perform topological data analysis (TDA) on TopS values. Filters with the Norm correlation metric were used with Neighbourhood lens 1 and lens 2 (resolution 30, gain 5.0). Proteins were coloured based on the principal component analysis (PCA) 1 metric [68, 69].
Two biological replicate co-immunoprecipitations using the SMC3 antibody, along with two control IPs, were analysed by MudPIT (Table S4a). To determine the enrichment of proteins in the SMC3 IPs compared to control IPs, we applied qprot statistics on the measured dNSAF values [68, 69]. Proteins were deemed specific to the SMC3 IP if they had a log2(fold change) ≥2 and a Z score ≥1.645 (significant at the 0.05 level) or a Z score ≥1.45 to include the bait protein, SMC3.
Results
Comparative in silico analysis of CADs in apicomplexan parasites and other eukaryotes
To obtain a list of all possible domains associated with chromatin-bound proteins, referred to as CADs, we first filtered the NCBI CDD and Pfam database for domains with chromatin-related cellular functions, including heterochromatin regulation, chromosome organization, nucleic acid binding and histone modifications. A total of 3870 CADs was found regardless of their organism sources (Table S1a). We next performed a genomic comparative analysis of proteins containing such domains (CAPs) among a variety of organisms, including three apicomplexan parasites (P. falciparum, P. vivax and Toxoplasma gondii), three euglenid parasites (Trypanosoma brucei, Trypanosoma cruzi and L. major), two unicellular organisms (Saccharomyces cerevisiae and Schizosaccharomyces pombe) and four multicellular organisms (H. sapiens, C. elegans, D. melanogaster and A. thaliana). Generally, we observed relatively similar numbers of CAPs in apicomplexan parasites (~22 % of the full proteome), and a higher number of CAPs in Saccharomyces cerevisiae (29 %) and higher eukaryotes (25 % in human) (Fig. S2a, Table S1b). In higher eukaryotes, increasing amounts of hierarchical chromatin elements such as compartments, topologically associating domains (TADs) and insulated domains have been described [73]. It is, therefore, not surprising that these more complex eukaryotes require more CAPs to regulate their chromatin structure.
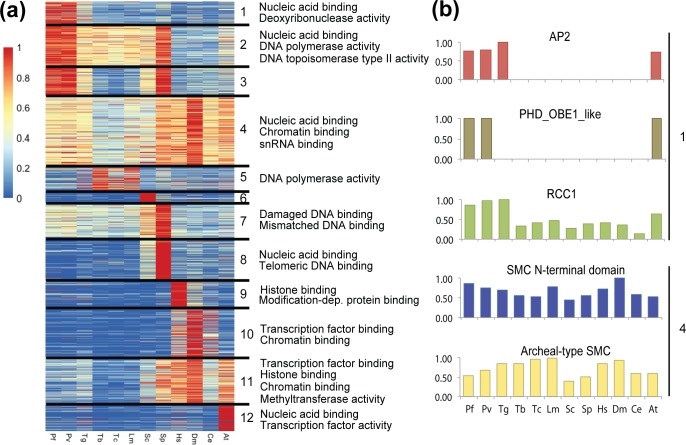
To identify functional differences in chromatin-associated processes, the candidate CADs were clustered based on their relative abundance in all investigated species. Twelve distinct clusters were obtained (Fig. 1a, Table S1c). Each cluster was then analysed for GO enrichment (Fig. S2b, Table S1d). Clusters 1–3 contained CADs that are relatively abundant in apicomplexans, of which the domains in cluster 1 were almost exclusively enriched in apicomplexan parasites (Fig. 1b). These domains showed enrichment for GO terms associated with nucleic acid binding and specifically AP2 domain-containing transcription factors (Fig. 1b). While highly abundant in apicomplexans, AP2 domain containing proteins were also abundant in plant species. AP2 family transcription factors play an essential role in floral development in A. thaliana [74]. In P. falciparum, proteins containing AP2 binding domains (ApiAp2) have been identified as sequence-specific transcription factors [75] and are believed to be master regulators of transcription during parasite development [21, 26]. The enrichment of AP2 domains in apicomplexan parasites and A. thaliana in our cluster analysis further validates our classification methods.
Fig. 1.
In silico comparative analysis of CADs across 12 organisms. (a) k-means clustering of the relative abundance of CADs (Table S1a) among 12 organisms (Fig. S2a, Table S2b). CAD abundance was first normalized for each organism by proteome size and then scaled to the CAD frequency with the highest relative abundance of that CAD, which was given an arbitrary abundance value of 1 (Table S1c). For each cluster, a subset of the GO enriched terms associated with the Pfam domains (FDR <0.01) are shown on the right (Table S1d). Cluster 1 (n=145 CADs); cluster 2 (n=274); cluster 3 (n=183); cluster 4 (n=452); cluster 5 (n=168); cluster 6 (n=64); cluster 7 (n=223); cluster 8 (n=282); cluster 9 (n=168); cluster 10 (n=331); cluster 11 (n=298); cluster 12 (n=166). (b) Relative abundance among 12 organisms of CADs selected from k-means apicomplexan-specific cluster 1 and conserved cluster 4 (Table S1c). Pf, Plasmodium falciparum; Pv, Plasmodium vivax; Tg, Toxoplasma gondii; Tb, Trypanosoma brucei; Tc, Trypanosoma cruzi; Lm, Leishmania major; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe; Hs, Homo sapiens; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; At, Arabidopsis thaliana.
Cluster 1 also includes the PHD_OBE1_like domain that is present in A. thaliana PHD finger proteins (Fig. 1b). This domain, enriched in plant species, is also conserved in Plasmodium species, which highlights the presence of plant-like protein domains in apicomplexan parasites. In addition, cluster 1 harboured the RCC1 domain (Fig. 1b), which is found in chromosome condensation regulating proteins. While these proteins are conserved among unicellular and multicellular organisms, a highly divergent ortholog of the regulator of chromosome condensation 1 (RCC1) that is critical for parasite pathogenesis has been identified in apicomplexan parasites [76].
CADs in cluster 4 were abundant in all 12 organisms. One of the major domains enriched in this cluster was the SMC (structural maintenance of chromosomes) N-terminal domain (Fig. 1b). SMC domain-containing proteins are a large family of ATPases that play a role in many aspects of chromosome organization, including chromosome assembly and segregation [77–79]. Enrichment of this domain across many eukaryotes suggests that proteins containing SMC domains are highly conserved, and are important for maintaining and regulating chromatin structure in a variety of organisms.
In silico identification and classification of CAPs in P. falciparum
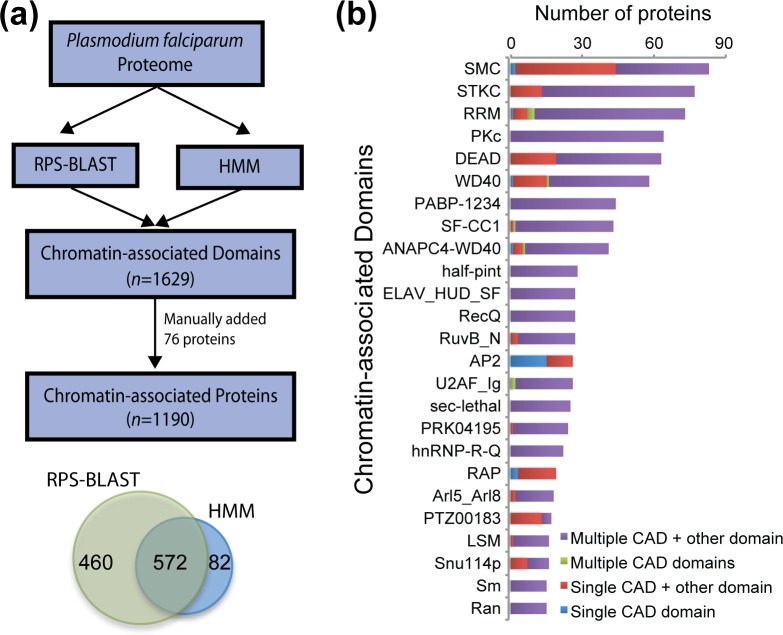
In P. falciparum, we identified a total of 1629 unique CADs (42 % of total CADs) present in 1114 candidate CAPs (20.1 % of P. falciparum proteome, n=5548). Out of these 1114 candidate CAPs, 460 proteins were identified using rps-blast, 82 proteins were identified using Hmmscan and 572 proteins were identified using both approaches (Fig. 2a, Table S2a). Additionally, 76 Plasmodium proteins that lacked any of the CADs, but had chromatin-associated functions based on their protein annotation, were manually added to the final CAP candidates list (Fig. 2a). Importantly, RNA-interacting proteins were not filtered out to ensure that proteins interacting with chromatin in an RNA-dependent manner were not excluded from the list. Among the final list of 1190 candidate CAPs, 162 proteins (13.6 %) have been previously described as having chromatin-related functions in the parasite [54], 877 (73.7 %) have non-chromatin related annotation and 151 proteins (12.7 %) are unknown proteins for which functions have yet to be discovered.
Fig. 2.
Overview of CAPs in P. falciparum. (a) In silico methodology. A total of 1190 proteins (20.1 % of proteome) in P. falciparum were predicted to be CAPs, covering 1629 unique CADs (Table S2a). (b) Structural organization of the domains that were found in eight or more candidate CAPs (Table S2b).
To better define Plasmodium CAPs, we further characterized the CADs that they carried (Fig. 2b, Table S2b). We performed a hierarchical clustering using Ward's minimum variance method with Euclidean distance as dissimilarity metric and multiple-fragment heuristic (MF) as seriation rule on a binary matrix consisting of 1629 CAD IDs × 1114 PfCAPs (Fig. S3a, Table S2b). The RNA recognition motif (RRM) and serine/threonine kinase catalytic (STKC) families were represented by a large number of domain IDs grouping together in two main large clusters consisting of 45 and 65 PfCAPs, respectively (Fig. S3a, bottom right). The 45 RRM PfCAPs also contained a distinct group of RRM IDs in a more sparsely populated cluster (Fig. S3a, top right). Other domain families created more discrete clusters, such as the ones shown in an enlargement of the top left corner of the hierarchical cluster (Fig. S3b). However, the large majority of the 1629 CADs did not cluster with other domains (Fig. S3a) and 74 % were detected in five PfCAPs or less (Table S2b). Going forward, we focused on the 218 CADs (13 % of total) that were detected in at least 15 PfCAPs.
The most abundant CADs, with 83 members (Fig. 2b, Table S2b), were SMC domains and domains from the STKC family associated with cell cycle progression, chromatin remodelling, DNA binding, transcription regulation or other nuclear activities (77 members). Transcription or mRNA processing-associated RNA-binding domains (RRM; 73 members), the catalytic domain of the dual-specificity protein kinases (PKc; 64 members), DEAD box helicase domains (63 members), WD40 domains (58 members), polyadenylate binding domains (PABP; 44 members) and splicing factor, CC1-like domains (SFD-CC1; 43 members) were also found to be abundant in the parasite's genome, along with other domains such as the anaphase-promoting complex unit (ANAPC4), AP2 transcription factor domains, small nuclear ribonucleoprotein domains (Sm/LikeSm) and GTP-binding nuclear protein Ran domains (Fig. 2b). When investigating the structural features of the proteins with CADs present in 15 or more candidate proteins, we observed that many of these CAD-containing proteins consisted of either a single CAD in combination with non-chromatin-related domains or multiple CADs in combination with non-chromatin-related domains (Fig. 2b). In other words, CADs were rarely observed to be the uniquely defining domain(s) of a protein. This finding suggests that PfCAPs may likely have multiple functional roles in the biology of the parasite.
To explore the potential function of these candidate PfCAPs, we further categorized these proteins based on their functional annotations obtained from PlasmoDB (Fig. S3c, Table S2c). We found that a large number of the proteins are likely to be nucleic acid binding proteins (n=172, 14.5 %) or proteins involved in transcriptional regulation (n=151, 12.8 %). Among these protein candidates are high mobility group B1–B4 proteins, proteins that form the transcription initiation factor TFIID subunit, and known transcriptional regulators such as Sir2A/B proteins and transcriptional coactivator ADA2. Another large group of the proteins are found to be structurally or functionally related to chromatin and chromosome structure (n=146, 12.3 %). These proteins include histones, histone modification proteins, nucleosome assembly proteins, chromatin remodelling proteins and chromosomal structural proteins. Lastly, we investigated the overall gene expression of the identified candidate PfCAPs. We observed that the pattern of mRNA expression of the candidate CAPs was similar to that of transcription factors suggesting that the majority of these candidate CAPs are likely to be important for facilitating transcription in the parasite (Fig. S3d).
Isolation and identification of CAPs in P. falciparum
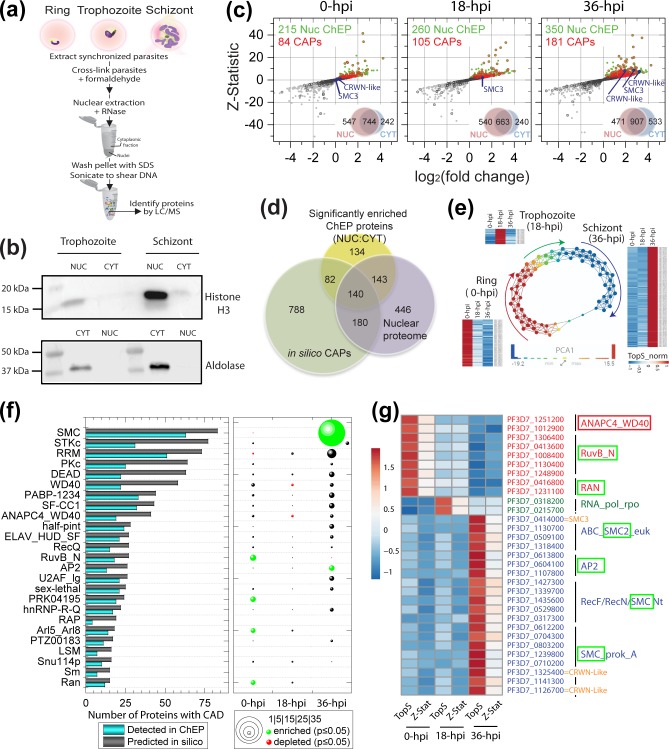
To validate in silico identified candidate PfCAPs and potentially discover additional CAPs without any known CADs, we next used ChEP (Fig. 3a) to isolate, in an unbiased manner, all proteins associated with chromatin (adapted from elsewhere [56]). Briefly, parasites were extracted from infected RBCs at the ring, trophozoite or schizont stages and cross-linked with formaldehyde to preserve protein–nucleic acid interactions. Parasite nuclei were then extracted in the presence of RNase A to avoid enrichment of proteins associated with nascent RNA. Non-cross-linked proteins were washed away using a highly denaturing buffer. As a negative control, we isolated proteins from the cytoplasmic fractions of each of the three stages. By Western blotting, we observed a clear enrichment of the nuclear and cytoplasmic markers in their respective fractions (Fig. 3b).
Fig. 3.
Stage-specific enrichment of CAPs and CADs. (a) Outline of the ChEP procedure. (b) Validation of protein enrichment in the nuclear fraction from ChEP by Western blotting. Nuclear (NUC) and cytoplasmic (CYT) fractions isolated from trophozoite and schizont were probed with anti-histone H3 antibody (top gel) and with anti-aldolase antibody (bottom gel). (c) Significance plots for all proteins detected in each of the three Plasmodium stages analysed. qspec-derived log2(fold change) and Z-statistic values between NUC:CYT (Table S3c) are shown as scatter plots with grey dots. Green dots highlight proteins with a Z score ≥2 and/or P value ≤0.05. Dots circled in black and red denote putative PfCAPs (Table S2a) that were not-significantly or significantly enriched in the nuclear ChEP fractions, respectively. Protein overlaps between the nuclear and cytoplasmic fractions at ring, trophozoite and schizont stages are shown as a Venn diagram (insets). The proteomics dataset was processed and analysed as described in Fig. S1(a). (d) Plasmodium proteins that were significantly enriched in the nuclear ChEP fractions (Table S3c) were compared to all putative PfCAPs (Table S2a) and to proteins detected in a previous analysis of the P. falciparum nuclear proteome [83] (Table S2d). Stage-specific overlaps between these proteins are shown in Fig. S1(a). (e) TDA was applied to the TopS scores calculated for 466 PfChEP proteins that passed two statistical criteria (i.e. TopS score and Z-statistic from qspec; Table S3c). TDA generated three main groups of protein nodes corresponding to ring, trophozoite and schizont stages. Protein nodes are coloured such that red nodes denote high PCA1 values and blue nodes denote low PCA1 values (see inset histogram scale). Node size is proportional with the number of proteins within the node. Heat maps for three groups of proteins are provided on the outside of the TDA map to illustrate high TopS values in one stage and not the other two (TopS >10 in one stage and <3 in the other two). To plot these heat maps, the TopS values were normalized (‘TopS_norm’) to the highest and lowest values (see scale from −1 to 1). (f) Stage-specific enrichment of domains. In the bar graph on the left, for each of the main CAD families (defined in Figs 2b and S3b), the number of CADs predicted in silico are compared to the numbers detected when merging all ChEP fractions analysed by MudPIT (Table S3a). CADs present in proteins that were deemed significantly enriched in one or two stages by TopS and qspec analyses were assessed for enrichment using Fisher’s exact test (Table S3c). On the right, bubble plots for each stage report on the number of significant proteins bearing these domains (bubble size) and on their specific enrichment or depletion (green and red bubbles, respectively). (g) A subset of the heat maps in (a) is shown for PfCAPs with their associated domains. CADs statistically enriched or depleted in each stage are marked with green or red boxes, respectively (Table S3c). Hpi, hours post infection.
Proteins isolated from the parasite nucleus following the ChEP methodology, as well as their corresponding cytoplasmic fractions, were analysed by MudPIT. Two biological replicates, with two technical replicates each, were analysed for each of the three intra-erythrocytic stages. After processing of the proteomics dataset (Fig. S1a), we identified 1291, 1203 and 1378 proteins in the nuclear ChEP fraction at the ring, trophozoite and schizont stages, respectively (Table S3a). A total of 633 of the predicted PfCAPs (Table S2a) were captured, 94 % of which were detected in the nuclear fraction, demonstrating that ChEP nuclear fractions were enriched with CAPs (Fig. S1a, Table S3a).
CAPs detected in the nuclear ChEP fractions showed strong enrichment for GO terms associated with typical chromatin-associated processes such as histone and histone modifying, DNA binding, transcription, RNA processing and splicing (Fig. S1b). Proteins functioning in translation-related processes were also enriched in the nuclear ChEP samples, which points to the existence of nuclear translation in the parasite [80, 81]. Additionally, rRNA processing proteins were enriched in the ChEP samples. Ribosome biogenesis takes place in the nucleus [82], and considering the biology of the parasite, the majority of ribosomes will need to be assembled in preparation for the higher levels of translation that takes place at the later trophozoite and schizont stages. Finally, we have also identified many proteins of unknown function as likely interacting with chromatin.
To identify proteins enriched in the nuclear samples, we next applied qspec statistical analysis [68] on the spectral counts measured for each protein in the replicate analyses and identified 215, 260 and 350 proteins that were detected with Z-statistic ≥2 in the nuclear fraction at the ring, trophozoite and schizont stages, respectively (Fig. 3c), adding up to 499 proteins from all stages (Table S3b). These significantly enriched candidate CAPs were compared to the computationally detected CAPs. A total of 222 candidate CAPs were captured by the ChEP–MudPIT analysis (Fig. 3d), which validated 18.6 % of the CAP candidates identified computationally. Of the 499 proteins significantly enriched by ChEP, 283 had also been previously detected in an analysis of the nuclear proteome [83] (Table S2d). Finally, 134 proteins that did not contain any discernible CADs were uniquely detected in our ChEP–MudPIT analysis (Fig. 3d). Such ChEP-enriched proteins either could contain as yet to be defined CADs or could more simply have been captured during the ChEP cross-linking because they interact with PfCAPs.
Stage-specific enrichment of Plasmodium CADs and CAPs identified by ChEP
Qualitatively, the nuclear schizont samples contributed the largest number of ChEP proteins significantly enriched in the nuclear fractions (Fig. S1a, Table S3b), which led us to further investigate the stage-specific expression of these proteins. To assess preference in stage expression, we applied a novel algorithm [70] that calculates TopS values from protein spectral counts. TopS is based on likelihood ratios and is computed across the aggregate information contained in the entire dataset (Table S3c). In other words, instead of comparing dNSAF levels calculated for each of the stages, TopS values were calculated considering quantitative information from all stages. To emulate the TopS calculations [70], qspec statistics [68] were also computed for each stage using the other two stages as background (Table S3c). Of the 499 proteins on which we applied TopS, 456 were considered significantly enriched in one or two stages (Table S3c). Of the 211 PfCAPs assigned significant TopS values, only 12 % were deemed enriched in two stages. Topological scoring allowed us to readily classify ChEP proteins with strong stage-specific expression.
To further cluster nuclear ChEP proteins, we used their calculated TopS values as input to perform TDA. TDA uses binning a point cloud of data based on several filter functions such as lenses and generates a high-dimensional topological network that can be further inspected. Interestingly, we observed that TDA depicted our dataset as a very characteristic circular network (Fig. 3e) with protein nodes separated by PCA1 values in three different regions corresponding to the three stages. To illustrate this observation, we plotted the topological scores for a subset of proteins as heat maps adjacent to each region of the TDA network (Figs 3 and S1c). For example, proteins in the schizont stage (in blue) had high TopS values in this stage, and lower scores in the ring and trophozoite stages. The combination of TopS analysis with TDA captured that our proteomics dataset was acquired from three cycling time-points of the parasite intra-erythrocytic life cycle, with the trophozoite stage following ring, schizont following trophozoite, and ring following schizont. This emphasizes that different groups of CAPs are expressed in different morphological forms of the parasite.
To assess whether some CAD families also showed stage-specificity, we estimated enrichment of the most abundant domains (Fig. 2b) using a one-tailed Fisher’s exact test by calculating probability hypergeometric distribution (Table S3c). While, on average, 62 % of the proteins bearing the most frequent CADs were detected in the merged dataset (Fig. 3f, left bar plot), the topological scoring and Fisher’s statistical testing of our dataset revealed some clear stage-specific enrichment and depletion. No CADs were specifically enriched in the trophozoite stage, although WD40 (NCBI CDD ID no. 238121) and ANAPC4_WD40 (NCBI CDD ID no. 315554) were particularly depleted in this stage (Fig. 3f, right bubble plots). Six of the main CAD families displayed statistically supported enrichment (Table S3c). With 7 out of 13, the AP2 proteins detected in our dataset were significantly expressed in the schizont. Out of the 12 proteins containing the Ran GTP-binding domain detected in our analysis, half were enriched in the ring stage. Conversely, none of these GTP-binding CAPs were enriched in the schizont. The inverse correlation between domain enrichment in the ring and schizont stages was also observed for the RuvB (NCBI CDD ID no. 310239), PRK04195 (NCBI CDD ID no. 235250) and Arl5_Arl8 (NCBI CDD ID no. 133353) domains.
Finally, the SMC domain was depleted in the ring stage, but statistically enriched in the schizont. The SMC family is diverse and contains several sub-classes, of which three were detected in the schizont samples (Fig. 3g). The conserved SMC3 protein (PF3D7_0414000), which belongs to the ABC cassette SMC class, while detected in all three stages, displayed strong enrichment in the schizont (Figs 3g and S1d). Other proteins contributing to SMC enrichment in the schizont samples (Fig. S1d) are two conserved Plasmodium proteins of unknown function (PF3D7_1325400 and PF3D7_1126700). Additionally, among AP2-domain-containing proteins displaying stage-specific enrichment (Fig. 3g) is PF3D7_0604100, a previously characterized chromatin remodeller involved in heterochromatin formation and genome integrity [84]. Overall, our in-depth statistically supported analysis of the ChEP–MudPIT dataset allowed us to establish stage specificity for a subset of CADs and candidate CAPs that warranted further investigation.
Functional validation of candidate CAPs in P. falciparum
Candidate CAPs with domain homology for chromatin components were selected for further molecular and cellular characterization. Candidate CAPs identified using the ChEP methodology and showing stage-specific enrichment (Table S3c) were searched for even distantly related homologues using psi-blast HHPred [85]. To this end, proteins in the ChEP enriched fractions and annotated as Plasmodium proteins of unknown function were blast searched against protein domains known to be involved in nuclear function in metazoans, eukaryotic pathogens or plants. These included domains found in nuclear lamina or lamina-like proteins, cohesin, condensin, CTCF insulator and insulator-like proteins. Our analysis identified two putative homologues (PF3D7_1325400 and PF3D7_1126700) of the coiled-coil proteins that are among the nuclear matrix constituent proteins found in plants. In A. thaliana, these proteins are encoded by CRWN genes [86]. PF3D7_1126700 was more abundant in the ChEP samples at the schizont stage (dNSAF=0.000692) compared to PF3D7_1325400 (dNSAF=0.000176). However, PF3D7_1325400 was identified with higher confidence in the blast search (E value=0.01) and, therefore, was selected for further analysis. Hereafter, PF3D7_1325400 will be referred to as ‘CRWN-like’ protein. A second protein, PF3D7_0414000, annotated as structural maintenance of chromosome 3 protein (SMC3), was also selected for further validation. SMC3, a subunit of the cohesin complex, although annotated as such, had not yet been characterized in P. falciparum.
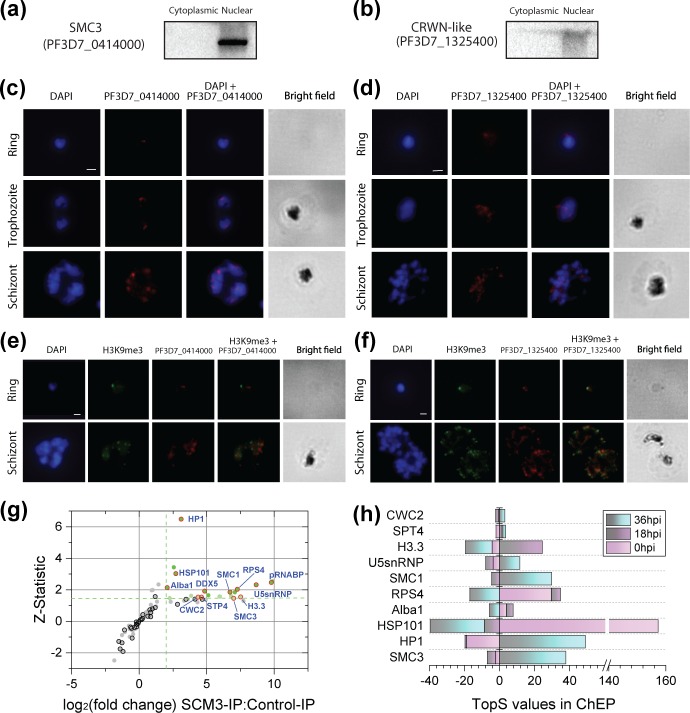
Custom antibodies were generated for each protein by designing peptide antigens targeting the C-terminal end of each protein. To validate these antibodies, we first performed Western blots using nuclear and cytoplasmic protein lysates from mixed blood stage Plasmodium parasites. We observed a clear enrichment of SMC3 (~140 kDa) and CRWN-like (~419 kDa) proteins in the nuclear fraction (Fig. 4a,b). Our results validate the use of these custom antibodies to detect the P. falciparum CRWN-like and SMC3 proteins.
Fig. 4.
Nuclear localization of candidate CAPs. Western blots show enrichment of SMC3 (a) and CRWN-like (b) proteins in the nuclear fraction [lane 1, marker; lane 2, cytoplasmic protein lysate (CYT); lane 3, nuclear protein lysate (NUC)]. (c) Subcellular localization of the SMC3 protein (PF3D7_0414000) during the asexual life stages of the parasite. (d) Subcellular localization of the CRWN-like protein (PF3D7_1325400) during the asexual life stages of the parasite. (e) Immunofluorescence analysis showing the localization of SMC3 at the periphery of the parasite nucleus away from the heterochromatin regions marked by H3K9me3 (Millipore; 07–442-AF488). (f) Colocalization of CRWN-like proteins to the heterochromatin regions of the parasite nucleus marked by H3K9me3. Bars indicate 2 µm in (c–f). (g) Significance plot of Plasmodium proteins interacting with SMC3. For all proteins co-immunoprecipitated with SMC3, qprot-derived log2(fold change) and Z-statistic values between SMC3-IP and control-IP (Table S4) are shown as scatter plots with grey dots. Coloured dots highlight proteins with a log2(fold change) ≥2 and Z score ≥1.645 (bright green) or Z score ≥1.45 (light green). Dots circled in black and red denote putative CAPs that were not-significantly or significantly enriched in the SMC3-IPs, respectively (Table S4). (h) Stage-specific enrichment of SCM3 interaction partners. TopS values calculated from the nuclear ChEP dataset (Table S4) are plotted for candidate PfCAPs potentially interacting with SMC3. Hpi, hours post infection.
We further investigated the subcellular localization of the CRWN-like and SMC3 proteins in intraerythrocytic parasites using immunofluorescence assays. A single focus per nucleus was observed for the SMC3 protein at ring, trophozoite and schizont stages (Fig. 4c). At all three asexual stages, the CRWN-like protein localized to the nuclear compartment (Fig. 4d). In particular, we observed a single focus per nucleus at the ring and schizont stages. At the trophozoite stage, the number of foci varied, in line with the increased level of DNA replication and nuclear expansion that takes place during this stage. In A. thaliana, CRWN proteins localize to the nuclear periphery and play a role in regulating heterochromatin environments in the nucleus [86]. It is possible that the CRWN-like protein in Plasmodium is similarly localizing to the heterochromatin regions of the nucleus.
To localize the SMC3 and CRWN-like proteins more precisely within the parasite nucleus, we next assessed their location in relation to histone H3K9 trimethylation, a marker of heterochromatin. We performed double immunofluorescence staining using a commercially available anti-H3K9me3 antibody. As all the antibodies were raised in the same host species, we performed sequential immunofluorescence staining to avoid cross-reactivity between antibodies. Briefly, fixed and permeabilized parasites were first incubated with the custom anti-SMC3 or anti-CRWN antibodies, followed by the corresponding fluorescent secondary antibody. After extensive washing, the parasites were incubated with the fluorescently conjugated anti-H3K9me3 antibody. At the ring and schizont stages, the SMC3 proteins localized to the nuclear periphery away from the heterochromatin regions marked by H3K9me3 (Fig. 4e). However, co-localization of H3K9me3 and CRWN-like proteins was observed (>90 % of ring-stage parasites), although the localization of CRWN-like proteins appeared to extend to regions adjacent to heterochromatin regions of the nucleus as well (Fig. 4f). Further experiments will be needed to validate the exact function of this CRWN-like protein in heterochromatin maintenance during parasite development.
Co-immunoprecipitation of candidate CAPs
To investigate the molecular components interacting with the SMC3 and CRWN-like proteins, we performed co-immunoprecipitation experiments using the custom generated antibodies. Briefly, protein lysates isolated from mixed erythrocytic stage parasite were incubated with anti-SMC3 or anti-CRWN-like custom antibodies conjugated to protein A magnetic beads and the resulting antibody–protein complexes were collected. Two biological replicate preparations of the proteins interacting with SMC3 or CRWN-like proteins, as well as two negative controls, were analysed by MudPIT (Table S4). While 16 proteins, including 5 putative CAPs, were reproducibly detected in both CRWN-L replicate IPs and significantly enriched over controls, we were unable to recover the bait protein itself from the eluted proteins (Table S4b). This might indicate that the antibody–protein interaction was too weak to immunoprecipitate the large CRWN-like protein (~419 kDa) or too strong to release it from the antibody-conjugated beads. These inconclusive results will not be discussed any further. Alternative epitope-tagging strategies will be needed to identify the interacting partners of the CRWN-like protein in the parasite.
However, a total of 102 proteins were detected combining two biological replicate analyses of SMC3 IPs (Table S4a), 96 % of which had also been detected in the PfChEP nuclear fractions (Table S3a). Negative-control IPs were used as the background for a qprot statistical analysis [68, 69], from which 20 proteins, in addition to the SMC3 bait, were deemed significantly enriched in the SMC3 IPs (Fig. 4g). As assessed by a Fisher’s exact test, these proteins were significantly enriched for PfCAPs (12 out of 21 proteins), with functional classifications falling in the ‘chromatin/chromosome-associated’ (SMC3, HP1, SMC1 and H3.3) ‘nucleic acid binding’ (Alba1, DDX5 and a putative RNA-binding protein) and ‘mRNA processing’ (U5 snRNP, CWC2 and PF3D7_1415400) categories. PF3D7_1415400 was annotated as ‘conserved Plasmodium protein, unknown function’, yet a search of PlasmoDB recovered a match to the Btz_2 InterPro domain (Table S4a). This domain is found in CASC3/Barentsz, a component of the exon junction complex (EJC). EJC is involved in mRNA post-transcriptional regulation and contains four proteins [eIF4AIII, Barentsz (Btz), Mago and Y14]. PF3D7_1003800, a protein annotated as a U5 snRNP, but which also contains several elongation factor domains, was also specifically identified. Importantly, we successfully recovered SMC1 (PF3D7_1130700), which forms a dimer with SMC3 in the cohesin complex; hence, validating our methodology.
To assess the expression profiles of the proteins significantly detected in the SMC3 IP, we used the TopS analysis performed on the ChEP fractions from three blood stages (Table S3c). TopS and Z-statistic values were available for 10 of the 12 PfCAPs significantly enriched in the SMC3 IPs (Table S4a). Plotting the stage-specific TopS values for each protein highlighted that the known interactors, SMC3 and SMC1, indeed shared a very similar expression profile, with highly positive TopS scores in the schizont stage and negative scores in the other two stages (Fig. 4h). Four additional proteins showed the same expression pattern (HP1, U5 snRNP, SPT4 and CWC2), which provides an additional layer of validation for these proteins to potentially interact with SMC3 and SMC1.
Genomic distribution of SMC3
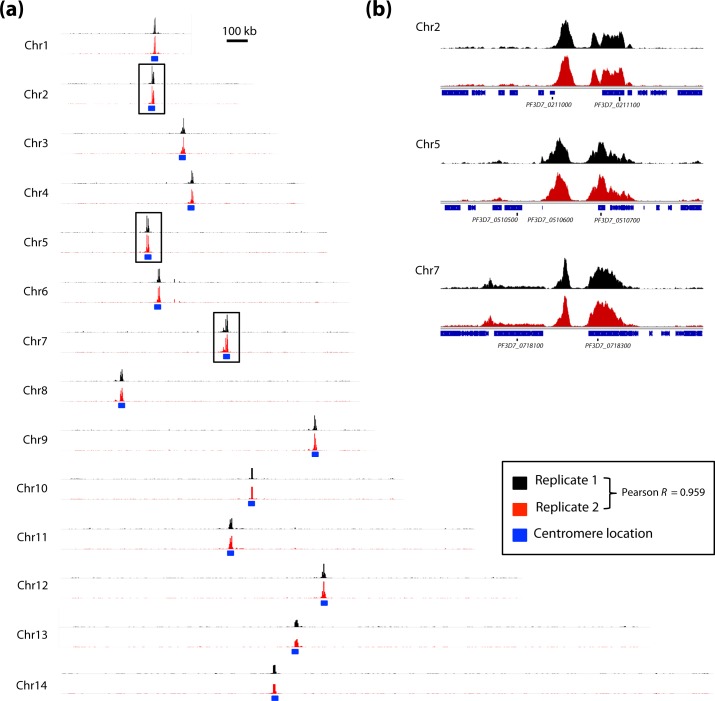
In order to determine the genome-wide distribution of SMC3, we next performed a ChIP-seq analysis. In duplicate experiments, trophozoite stage parasites were cross-linked with formaldehyde. Sonicated chromatin was incubated with the anti-SMC3 antibody and the resulting DNA–protein–antibody complexes were recovered using agarose beads. Purified DNA fragments were sequenced using next-generation sequencing technology. A rabbit IgG antibody and a no-antibody sample were used as negative controls (Fig. S4). In trophozoites, SMC3 binding was restricted to the centromere region on all 14 chromosomes (Fig. 5a). High Pearson correlation between biological replicates (R=0.959) confirmed the reproducibility of our ChIP-seq experiments. Additionally, the SMC3 peaks appear as doublets (Fig. 5b), likely a result of the extremely high AT content and repetitive nature of the region in-between the peaks, which can be a challenge to sequence and capture computationally.
Fig. 5.
ChIP-seq analysis showing genome-wide distribution of SMC3 in trophozoites. (a) SMC3 distribution across all 14 chromosomes. The blue box indicates the location of the centromere on each chromosome. The regions depicted in (b) are indicated with black boxes. (b) Zoomed in regions on chromosomes 2, 5 and 7 depicting SMC3 distribution.
Cohesin consists of four protein subunits (SMC1, SMC3, SCC1 and SCC3) and the enrichment of this complex in genomic locations is observed in all eukaryotes. In mammalian cells, cohesin sites are found near transcription start sites and co-localize with the CCCTC-binding factor (CTCF), where they play multiple roles in chromatin organization [87, 88]. In yeast, cohesin localizes to centromeres and extends to nearby pericentromeric regions [89, 90]. Preferential loading of cohesin at centromeres is a kinetochore-dependent process [91]. The parasite SMC3 distribution during the trophozoite stage, hence, resembles the yeast cohesin occupancy. At the trophozoite stage, the parasite prepares for mitosis and our results suggest that cohesin has a possible role in sister chromatid separation and cell cycle regulation at this developmental time-point. However, in comparison with the yeast cohesin distribution, the parasite SMC3 occupancy does not extend to nearby pericentromeric regions, which suggests that the SMC3 subunit in particular might be important for sister chromatid cohesion.
Discussion
Increasing evidence points towards genome architecture and chromatin structure regulation playing an important role in gene expression throughout the life cycle of apicomplexan parasites [40, 51, 92–95]. To better understand how the 3D structure of the genome is being maintained, it is vital to identify proteins and protein complexes that associate with chromatin throughout parasite development. Comparative genomics analyses have been performed previously to discover proteins involved in transcriptional control in apicomplexan parasites. In 2004, Coulson and Hall identified 156 P. falciparum transcription-associated proteins using homology and hidden Markov model profile searches [19]. Balaji et al., in 2005, conducted a systematic analysis of putative apicomplexan nuclear proteins, which allowed the discovery of a novel family of apicomplexan DNA-binding transcription factors [18]. Similarly in 2008, a comparative analysis by Iyer et al. surveyed transcription factors and chromatin proteins in several protists, and in particular identified 86 chromatin proteins in P. falciparum [96]. In 2010, a bioinformatic study by Volz et al. explored potential nuclear factors important for P. falciparum gene expression, which resulted in the identification of 24 putative nuclear proteins [97]. Specifically, this approach was used to identify putative proteins containing domains involved in gene expression. Collectively, these studies contributed greatly to our knowledge of the chromatin proteome in apicomplexan parasites. However, more than a decade later, some classes of chromatin maintenance proteins (i.e. lamina-like proteins) have yet to be identified in P. falciparum. Therefore, with the use of more updated protein domain databases, we performed comprehensive computational and comparative genomics analyses to generate an extended list of CADs and CAPs in apicomplexan parasites with the goal of identifying otherwise previously unrecognized chromatin-associating elements. In an unbiased comparison with other eukaryotic organisms, we observed that apicomplexan parasites encode a relatively large number of CAPs. Some of these candidate CAPs contain domains that are found almost exclusively in apicomplexan parasites (Figs 1 and S2, Table S1a–d). While the exact function of these proteins will have to be validated at the molecular level, this finding likely reflects the importance of CAPs for parasite biology.
By searching the P. falciparum proteome against a large collection of Pfam (hmm) and NCBI (rps-blast) CADs, we have attempted to identify all Plasmodium CAPs. Since the P. falciparum genome is relatively distant from more traditional model organisms, we used less-stringent parameters for the hmm search to be able to identify PfCAPs (Figs 2 and S3, Table S2a–d). In addition, we have tried to account for false-positive hits by using information from the current genome annotation to filter our initial broad search to proteins that specifically interact with chromatin. Among the final list of 1190 candidate PfCAPs, only 162 proteins (13.6%) have been previously described as having chromatin-related functions in the parasite [54]. While our in silico approach has generated an extensive list of potential CAPs, this less stringent methodology was needed to uncover previously unidentified chromatin-bound elements. Therefore, we combined our in silico results with a more targeted experimental approach to generate a smaller but more confident list of putative chromatin-bound proteins in P. falciparum.
Out of the 1190 in silico-identified P. falciparum proteins that contain a CAD, 633 proteins (53 %) were experimentally confirmed via the ChEP approach (Figs 3 and S1, Table S3a–c). Proteins that were not identified in the proteomics analysis of ChEP fractions isolated from ring, trophozoite and schizont life stages may only be transiently expressed, may have low expression levels making them difficult to detect by MS or may not be expressed during these three time-points of the parasite life cycle. It is also important to note that the presence of a protein in a ChEP sample is not sufficient to conclude that it has a function in chromatin structure, since a number of proteins with no expected chromatin function can be found in these fractions. However, the preservation of in vivo chromatin characteristics through cross-linking is vital for studying chromatin-associated processes by proteomics. Thus, by implementing the ChEP methodology, we have attempted to enrich for chromatin-bound factors by minimizing the loss of transiently bound factors and reducing the risk of purification artefacts that can be introduced following cell lysis. Previous studies have explored the nuclear proteome of P. falciparum during the asexual developmental cycle [83, 98]. It is important to note that these methodologies were used to identify all proteins localizing to the parasite nucleus. While the ChEP-identified dataset is complementary to and overlaps with the existing P. falciparum nuclear proteome datasets, we aimed to specifically identify proteins interacting with chromatin. For example, many proteins with non-chromatin-related functions, such as transporter activity, were enriched in the nuclear proteome dataset [83] and were not identified using our ChEP methodology. Furthermore, we applied a new scoring algorithm (TopS) [70] combined with TDA [70–72] to the ChEP dataset to highlight proteins and domains with stage-specific expression (Table S3c). Many DNA- and RNA-binding proteins such as high mobility group protein B2, putative structural maintenance of chromosome proteins and the putative CRWN-like protein that might be an integral part of the parasite nucleus were identified as enriched in the schizont stage (Fig. S1d), and were significantly enriched in our ChEP sample as compared to the nuclear proteome dataset. Taken together, these results demonstrate the power of this methodology and data analysis pipeline to identify chromatin-bound components in the parasite in an unbiased manner.
Cohesin and condensin protein complexes, composed of SMC subunits, were enriched in the ChEP samples and showed statistically supported enrichment in the schizont stage (Fig. S1d). SMC proteins are an integral part of the eukaryotic nucleus. While SMC proteins are annotated in P. falciparum, further characterization of these proteins is lacking. Here, we have explored the expression, localization, interaction network and genome-wide distribution of the SMC3 protein in the parasite. Using immunofluorescence, we observed a single SMC3 focus at the ring, trophozoite and schizont stages (Fig. 4c). This result was validated by our ChIP-seq analysis showing the distribution of SMC3 at the trophozoite stage to be confined to the centromeric regions on all chromosomes (Fig. 5). According to previously published P. falciparum nuclear architecture data [35], the centromeres of all chromosomes cluster together near the periphery of the parasite nucleus; therefore, proteins such as SMC3 localizing to the centromeric regions of chromosomes would appear as a single focus in immunofluorescence experiments. Double immunofluorescence staining further confirmed that SMC3 proteins localize to the periphery of the parasite nucleus away from the heterochromatin regions of the nucleus (Fig. 4e). Co-immunoprecipitation with the antibody we designed against SMC3 specifically pulled down its known interactor, SMC1, as well as additional PfCAPs (annotated as HP1, H3.3, SPT4 and CWC2) whose expression patterns in the ChEP samples matched the cohesin subunits (Fig. 4g, h). While HP1 has been shown to be absent from centromeric regions in P. falciparum [99], this protein has a significant role in centromere cohesion in mammals [100, 101]. If we cannot exclude potential contamination, it is possible that HP1 may interact weakly with the centromeres in the parasite. Similarly, histone H3.3 has been detected in centromeric and pericentromeric regions of chromosomes in mice leading to mitotic defects [102]. Additional mechanistic insight into how the cohesin complex functions in the parasite is lacking and warrants further investigation.
The crowded nuclei (CRWN) proteins, which are plant-related SMC-domain-containing proteins, are not as widely conserved in other eukaryotes. In A. thaliana, CRWN proteins are among the coiled-coil proteins belonging to the nuclear matrix constituent protein (NMCP) family of proteins and were originally identified as residing at the nuclear periphery in carrots [103]. Previous studies have demonstrated the importance of CRWN proteins in plant viability, as evidenced by the inability to recover mutants with disruptions in CRWN genes [86]. Additionally, mutants deficient in CRWN proteins exhibit altered nuclear organization, including reduced nuclear size, abnormal nuclear shape and heterochromatin organization. The coiled-coil domain and nuclear periphery localization suggest that these NMCP-related proteins might be functionally analogous to components of the animal nuclear lamina [104]. Despite the critical role in providing structure to the metazoan nucleus, lamina proteins have not been identified in plants or unicellular eukaryotes. While lamina-like protein (NUP-1) has been detected in kinetoplastids [105], lamina-like proteins have not been detected in Plasmodium species [48]. Here, we identified two distinct CRWN-like isoforms enriched in the schizont ChEP samples (Fig. 3g). An antibody we generated against one of these isoforms (PF3D7_1325400) allowed us to localize, for what is believed to be the first time in P. falciparum, a potential CRWN-like protein that might be an integral part of the parasite nucleus. This CRWN-like protein localizes to a single focus inside the nucleus at ring and schizont stages (Fig. 4d). Additionally, using double immunofluorescence staining, we observed that the CRWN-like protein co-localizes to heterochromatin and nearby regions of the parasite nucleus marked by the repressive histone mark H3K9me3 (Fig. 4f). It is possible that this protein regulates heterochromatin regions of the parasite nucleus, much like what has been observed in plant species [86].
A recent high-throughput transposon insertional mutagenesis study was performed to distinguish essential and dispensable genes in the P. falciparum genome [106]. According to this study, both genes identified in our analysis as putative homologues of the plant coiled-coil proteins (PF3D7_1325400 and PF3D7_1126700) were classified as non-essential genes. However, both putative CRWN protein homologues may be an integral part of the parasite nucleus and one CRWN-like protein compensates for the loss of the other. A previous study exploring P. falciparum invasion pathways made a similar discovery where the loss of one family of invasion genes resulted in the activation of a separate invasion gene family [107]. Furthermore, in A. thaliana, which harbours four CRWN genes, quadruple CRWN mutants are non-viable, indicating that CRWN proteins participate in essential processes, but single, double and even some triple mutants are viable, indicating a degree of complementation between different CRWN genes [86]. Therefore, while it might be possible to make viable single CRWN-like mutants in the parasite, attempting to disrupt multiple CRWN-like genes will likely provide more information about the essentiality of these nuclear proteins. Further characterization of CRWN-like proteins in Plasmodium could improve our understanding of how telomere and antigenic variation genes cluster at the nuclear periphery. More importantly, such novel plant-related proteins that play an important role in parasite nuclear organization can serve as ideal drug targets that can disrupt the parasite 3D nuclear structure with high specificity and low toxicity to the host.
Conclusions
This study presents a comprehensive overview of potential CAPs in apicomplexan parasites. We have computationally identified candidate chromatin-binding proteins based on the presence of chromatin-binding domains, and further classified these candidate proteins into functional categories. We have also provided experimental evidence for CAPs during P. falciparum development using a new methodology termed ChEP. We have further validated cellular localization and expression for two candidate chromatin-bound proteins. The function of many CAPs is still unknown and further characterization of CAPs will be needed to increase our understanding of parasite biology. It is likely that our results will not only boost the understanding of chromatin structure and chromatin-based processes, but also help to identify key players in pathogenesis and gene regulation in parasites.
Data bibliography
1. Batugedara G et al. MassIVE database (MS datasets), accession numbers MSV000082520, MSV000082521 and MSV000084671 (2019).
2. Batugedara G et al. ProteomeXChange (MS datasets), accession numbers PXD010262, PXD010263 and PXD016684 (2019).
3. Batugedara G et al. Stowers Original Data Repository (MS datasets), accession number LIBPB-1239 (2019).
4. Batugedara G et al. NCBI Gene Expression Omnibus (ChIP-seq datasets), accession number GSE116219 (2019).
Supplementary Data
Funding information
This work was supported by the National Institutes of Allergy and Infectious Diseases (1 R01 AI06775-01 and 1 R01 AI136511 to K.G.L.R.) and the University of California, Riverside (NIFA-Hatch-225935). This work was supported by the Stowers Institute for Medical Research and the National Institute of General Medical Sciences of the National Institutes of Health (RO1GM112639 to M.P.W.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Acknowledgements
The following reagent was obtained through the MR4 as part of the BEI Resources Repository, NIAID, NIH, USA: P. falciparum strain 3D7 (MRA-102) deposited by D.J. Carucci.
Author contributions
G.B., E.M.B. and K.G.L.R. designed the project; G.B., X.M.L., A.S. and A.C. performed experiments; G.B., M.L., A.S., S.A., M.E.S. and L.F. performed data analysis; M.E.S., L.F. and M.P.W. contributed analytical tools; J.P. maintained P. falciparum cultures and assisted in experimental procedures; G.B., X.M.L., L.F. and K.G.L.R. wrote the manuscript. All authors edited and approved the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AEBSF, 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride; CAD, chromatin-associated domain; CAP, chromatin-associated protein; CDD, Conserved Domain Database; ChEP, chromatin enrichment for proteomics; ChIP, chromatin immunoprecipitation; dNSAF, distributed normalized spectral abundance factor; FDR, false-discovery rate; GO, gene ontology; IP, immunoprecipitation; MudPIT, multidimensional protein identification technology; NCBI, National Center for Biotechnology Information; PCA, principal component analysis; PSSM, position-specific scoring matrix; RBC, red blood cell; RCF, relative centrifugal force; RPKM, reads per kilobase of transcript per million mapped reads; RRM, RNA recognition motif; TCA, trichloroacetic acid; TDA, topological data analysis.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Four supplementary figures and four supplementary tables are available with the online version of this article.
References
- 1.WHO The World Malaria Report 2017 (www.who.int/malaria/publications/worldmalaria-report-2017/en/) Geneva: World Health Organization; 2017. [Google Scholar]
- 2.Beugnet F, Moreau Y. Babesiosis. Rev Sci Tech. 2015;34:627–639. doi: 10.20506/rst.34.2.2385. [DOI] [PubMed] [Google Scholar]
- 3.Flegr J, Prandota J, Sovičková M, Israili ZH. Toxoplasmosis – a global threat. Correlation of latent toxoplasmosis with specific disease burden in a set of 88 countries. PLoS One. 2014;9:e90203. doi: 10.1371/journal.pone.0090203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, et al. Leishmaniasis worldwide and global estimates of its incidence. PLoS One. 2012;7:e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bern C. Chagas' disease. N Engl J Med. 2015;373:456–466. doi: 10.1056/NEJMra1410150. [DOI] [PubMed] [Google Scholar]
- 6.Kennedy PGE. Clinical features, diagnosis, and treatment of human African trypanosomiasis (sleeping sickness) Lancet Neurol. 2013;12:186–194. doi: 10.1016/S1474-4422(12)70296-X. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton WL, Amato R, van der Pluijm RW, Jacob CG, Quang HH, et al. Evolution and expansion of multidrug-resistant malaria in southeast Asia: a genomic epidemiology study. Lancet Infect Dis. 2019;19:943–951. doi: 10.1016/S1473-3099(19)30392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sibley CH. Understanding drug resistance in malaria parasites: basic science for public health. Mol Biochem Parasitol. 2014;195:107–114. doi: 10.1016/j.molbiopara.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Takala-Harrison S, Jacob CG, Arze C, Cummings MP, Silva JC, et al. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. J Infect Dis. 2015;211:670–679. doi: 10.1093/infdis/jiu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardner MJ, Hall N, Fung E, White O, Berriman M, et al. Genome sequence of the human malaria parasite Plasmodium falciparum . Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bozdech Z, Llinás M, Pulliam BL, Wong ED, Zhu J, et al. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum . PLoS Biol. 2003;1:e5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bunnik EM, Chung D-W, Hamilton M, Ponts N, Saraf A, et al. Polysome profiling reveals translational control of gene expression in the human malaria parasite Plasmodium falciparum . Genome Biol. 2013;14:R128. doi: 10.1186/gb-2013-14-11-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lapp SA, Mok S, Zhu L, Wu H, Preiser PR, et al. Plasmodium knowlesi gene expression differs in ex vivo compared to in vitro blood-stage cultures. Malar J. 2015;14:110. doi: 10.1186/s12936-015-0612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 15.Otto TD, Böhme U, Jackson AP, Hunt M, Franke-Fayard B, et al. A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biol. 2014;12:86. doi: 10.1186/s12915-014-0086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radke JR, Behnke MS, Mackey AJ, Radke JB, Roos DS, et al. The transcriptome of Toxoplasma gondii . BMC Biol. 2005;3:26. doi: 10.1186/1741-7007-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sacci JB, Ribeiro JMC, Huang F, Alam U, Russell JA, et al. Transcriptional analysis of in vivo Plasmodium yoelii liver stage gene expression. Mol Biochem Parasitol. 2005;142:177–183. doi: 10.1016/j.molbiopara.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 18.Balaji S, Babu MM, Iyer LM. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 2005;33:3994–4006. doi: 10.1093/nar/gki709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coulson RMR, Hall N. Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum . Genome Res. 2004;14:1548–1554. doi: 10.1101/gr.2218604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Silva EK, Gehrke AR, Olszewski K, Leon I, Chahal JS, et al. Specific DNA-binding by apicomplexan AP2 transcription factors. Proc Natl Acad Sci USA. 2008;105:8393–8398. doi: 10.1073/pnas.0801993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwanaga S, Kaneko I, Kato T, Yuda M. Identification of an AP2-family protein that is critical for malaria liver stage development. PLoS One. 2012;7:e47557. doi: 10.1371/journal.pone.0047557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kafsack BFC, Rovira-Graells N, Clark TG, Bancells C, Crowley VM, et al. A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature. 2014;507:248–252. doi: 10.1038/nature12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lesage KM, Huot L, Mouveaux T, Courjol F, Saliou J-M, et al. Cooperative binding of ApiAP2 transcription factors is crucial for the expression of virulence genes in Toxoplasma gondii . Nucleic Acids Res. 2018;46:6057–6068. doi: 10.1093/nar/gky373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radke JB, Worth D, Hong D, Huang S, Sullivan WJ, et al. Transcriptional repression by ApiAP2 factors is central to chronic toxoplasmosis. PLoS Pathog. 2018;14:e1007035. doi: 10.1371/journal.ppat.1007035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinha A, Hughes KR, Modrzynska KK, Otto TD, Pfander C, et al. A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium . Nature. 2014;507:253–257. doi: 10.1038/nature12970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuda M, Iwanaga S, Shigenobu S, Kato T, Kaneko I. Transcription factor AP2-Sp and its target genes in malarial sporozoites. Mol Microbiol. 2010;75:854–863. doi: 10.1111/j.1365-2958.2009.07005.x. [DOI] [PubMed] [Google Scholar]
- 27.Yuda M, Iwanaga S, Shigenobu S, Mair GR, Janse CJ, et al. Identification of a transcription factor in the mosquito-invasive stage of malaria parasites. Mol Microbiol. 2009;71:1402–1414. doi: 10.1111/j.1365-2958.2009.06609.x. [DOI] [PubMed] [Google Scholar]
- 28.Kirchner S, Power BJ, Waters AP. Recent advances in malaria genomics and epigenomics. Genome Med. 2016;8:92. doi: 10.1186/s13073-016-0343-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balu B, Maher SP, Pance A, Chauhan C, Naumov AV, et al. CCR4-associated factor 1 coordinates the expression of Plasmodium falciparum egress and invasion proteins. Eukaryot Cell. 2011;10:1257–1263. doi: 10.1128/EC.05099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bunnik EM, Batugedara G, Saraf A, Prudhomme J, Florens L, et al. The mRNA-bound proteome of the human malaria parasite Plasmodium falciparum . Genome Biol. 2016;17:147. doi: 10.1186/s13059-016-1014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vembar SS, Macpherson CR, Sismeiro O, Coppée J-Y, Scherf A. The PfAlba1 RNA-binding protein is an important regulator of translational timing in Plasmodium falciparum blood stages. Genome Biol. 2015;16:212. doi: 10.1186/s13059-015-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eshar S, Altenhofen L, Rabner A, Ross P, Fastman Y, et al. PfSR1 controls alternative splicing and steady-state RNA levels in Plasmodium falciparum through preferential recognition of specific RNA motifs. Mol Microbiol. 2015;96:1283–1297. doi: 10.1111/mmi.13007. [DOI] [PubMed] [Google Scholar]
- 33.Caro F, Ahyong V, Betegon M, DeRisi JL. Genome-wide regulatory dynamics of translation in the Plasmodium falciparum asexual blood stages. eLife. 2014;3:e04106. doi: 10.7554/eLife.04106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foth BJ, Zhang N, Mok S, Preiser PR, Bozdech Z. Quantitative protein expression profiling reveals extensive post-transcriptional regulation and post-translational modifications in schizont-stage malaria parasites. Genome Biol. 2008;9:R177. doi: 10.1186/gb-2008-9-12-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ay F, Bunnik EM, Varoquaux N, Bol SM, Prudhomme J, et al. Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression. Genome Res. 2014;24:974–988. doi: 10.1101/gr.169417.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dekker J. Gene regulation in the third dimension. Science. 2008;319:1793–1794. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 38.Fudenberg G, Getz G, Meyerson M, Mirny LA. High order chromatin architecture shapes the landscape of chromosomal alterations in cancer. Nat Biotechnol. 2011;29:1109–1113. doi: 10.1038/nbt.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bunnik EM, Cook KB, Varoquaux N, Batugedara G, Prudhomme J, et al. Changes in genome organization of parasite-specific gene families during the Plasmodium transmission stages. Nat Commun. 2018;9:1910. doi: 10.1038/s41467-018-04295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bunnik EM, Venkat A, Shao J, McGovern KE, Batugedara G, et al. Comparative 3D genome organization in apicomplexan parasites. Proc Natl Acad Sci USA. 2019;116:3183–3192. doi: 10.1073/pnas.1810815116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanizawa H, Iwasaki O, Tanaka A, Capizzi JR, Wickramasinghe P, et al. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic Acids Res. 2010;38:8164–8177. doi: 10.1093/nar/gkq955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ong C-T, Corces VG. Ctcf: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hiraga S, Botsios S, Donze D, Donaldson AD. TFIIIC localizes budding yeast ETC sites to the nuclear periphery. Mol Biol Cell. 2012;23:2741–2754. doi: 10.1091/mbc.E11-04-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, et al. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17:635–640. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, et al. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 48.McCulloch R, Navarro M. The protozoan nucleus. Mol Biochem Parasitol. 2016;209:76–87. doi: 10.1016/j.molbiopara.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 49.Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SWM, Solovei I, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brancucci NMB, Bertschi NL, Zhu L, Niederwieser I, Chin WH, et al. Heterochromatin protein 1 secures survival and transmission of malaria parasites. Cell Host Microbe. 2014;16:165–176. doi: 10.1016/j.chom.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 51.Volz JC, Bártfai R, Petter M, Langer C, Josling GA, et al. PfSET10, a Plasmodium falciparum methyltransferase, maintains the active var gene in a poised state during parasite division. Cell Host Microbe. 2012;11:7–18. doi: 10.1016/j.chom.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 52.Malmquist NA, Moss TA, Mecheri S, Scherf A, Fuchter MJ. Small-molecule histone methyltransferase inhibitors display rapid antimalarial activity against all blood stage forms in Plasmodium falciparum . Proc Natl Acad Sci USA. 2012;109:16708–16713. doi: 10.1073/pnas.1205414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malmquist NA, Sundriyal S, Caron J, Chen P, Witkowski B, et al. Histone methyltransferase inhibitors are orally bioavailable, fast-acting molecules with activity against different species causing malaria in humans. Antimicrob Agents Chemother. 2015;59:950–959. doi: 10.1128/AAC.04419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bischoff E, Vaquero C. In silico and biological survey of transcription-associated proteins implicated in the transcriptional machinery during the erythrocytic development of Plasmodium falciparum . BMC Genomics. 2010;11:34. doi: 10.1186/1471-2164-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujita T, Fujii H. Direct identification of insulator components by insertional chromatin immunoprecipitation. PLoS One. 2011;6:e26109. doi: 10.1371/journal.pone.0026109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kustatscher G, Hegarat N, Wills KLH, Furlan C, Bukowski-Wills J-C, et al. Proteomics of a fuzzy organelle: interphase chromatin. EMBO J. 2014;33:648–664. doi: 10.1002/embj.201387614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Franklin S, Chen H, Mitchell-Jordan S, Ren S, Wang Y, et al. Quantitative analysis of the chromatin proteome in disease reveals remodeling principles and identifies high mobility group protein B2 as a regulator of hypertrophic growth. Mol Cell Proteomics. 2012;11:M111.014258. doi: 10.1074/mcp.M111.014258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trager W, Jensen JB. Human malaria parasites in continuous culture. 1976. J Parasitol. 2005;91:484–486. doi: 10.1645/0022-3395(2005)091[0484:HMPICC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 59.Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65:418–420. doi: 10.2307/3280287. [DOI] [PubMed] [Google Scholar]
- 60.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Böhme U, Otto TD, Sanders M, Newbold CI, Berriman M. Progression of the canonical reference malaria parasite genome from 2002–2019. Wellcome Open Res. 2019;4:58. doi: 10.12688/wellcomeopenres.15194.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoeijmakers WAM, Flueck C, Françoijs K-J, Smits AH, Wetzel J, et al. Plasmodium falciparum centromeres display a unique epigenetic makeup and cluster prior to and during schizogony. Cell Microbiol. 2012;14:1391–1401. doi: 10.1111/j.1462-5822.2012.01803.x. [DOI] [PubMed] [Google Scholar]
- 63.Florens L, Washburn MP. Proteomic analysis by multidimensional protein identification technology. Methods Mol Biol. 2006;328:159–175. doi: 10.1385/1-59745-026-X:159. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y, Wen Z, Washburn MP, Florens L. Improving proteomics mass accuracy by dynamic offline lock mass. Anal Chem. 2011;83:9344–9351. doi: 10.1021/ac201867h. [DOI] [PubMed] [Google Scholar]
- 65.Xu T, Park SK, Venable JD, Wohlschlegel JA, Diedrich JK, et al. ProLuCID: an improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J Proteomics. 2015;129:16–24. doi: 10.1016/j.jprot.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tabb DL, McDonald WH, Yates JR. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal Chem. 2010;82:2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- 68.Choi H, Fermin D, Nesvizhskii AI. Significance analysis of spectral count data in label-free shotgun proteomics. Mol Cell Proteomics. 2008;7:2373–2385. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi H, Larsen B, Lin Z-Y, Breitkreutz A, Mellacheruvu D, et al. SAINT: probabilistic scoring of affinity purification–mass spectrometry data. Nat Methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sardiu ME, Gilmore JM, Groppe BD, Dutta A, Florens L, et al. Topological scoring of protein interaction networks. Nat Commun. 2019;10:1118. doi: 10.1038/s41467-019-09123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lum PY, Singh G, Lehman A, Ishkanov T, Vejdemo-Johansson M, et al. Extracting insights from the shape of complex data using topology. Sci Rep. 2013;3:1236. doi: 10.1038/srep01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sardiu ME, Gilmore JM, Groppe B, Florens L, Washburn MP. Identification of topological network modules in perturbed protein interaction networks. Sci Rep. 2017;7:43845. doi: 10.1038/srep43845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jeninga M, Quinn J, Petter M. ApiAP2 transcription factors in apicomplexan parasites. Pathogens. 2019;8:47. doi: 10.3390/pathogens8020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frankel MB, Mordue DG, Knoll LJ. Discovery of parasite virulence genes reveals a unique regulator of chromosome condensation 1 ortholog critical for efficient nuclear trafficking. Proc Natl Acad Sci USA. 2007;104:10181–10186. doi: 10.1073/pnas.0701893104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Strunnikov AV, Jessberger R. Structural maintenance of chromosomes (SMC) proteins: conserved molecular properties for multiple biological functions. Eur J Biochem. 1999;263:6–13. doi: 10.1046/j.1432-1327.1999.00509.x. [DOI] [PubMed] [Google Scholar]
- 78.Freeman L, Aragon-Alcaide L, Strunnikov A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol. 2000;149:811–824. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haering CH, Löwe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. doi: 10.1016/S1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 80.Iborra FJ, Jackson DA. Coupled transcription and translation within nuclei of mammalian cells. Science. 2001;293:1139–1142. doi: 10.1126/science.1061216. [DOI] [PubMed] [Google Scholar]
- 81.Iborra FJ, Jackson DA. The case for nuclear translation. J Cell Sci. 2004;117:5713–5720. doi: 10.1242/jcs.01538. [DOI] [PubMed] [Google Scholar]
- 82.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/S0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 83.Oehring SC, Woodcroft BJ, Moes S, Wetzel J, Dietz O, et al. Organellar proteomics reveals hundreds of novel nuclear proteins in the malaria parasite Plasmodium falciparum . Genome Biol. 2012;13:R108. doi: 10.1186/gb-2012-13-11-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Flueck C, Bartfai R, Niederwieser I, Witmer K, Alako BTF, et al. A major role for the Plasmodium falciparum ApiAP2 protein PfSIP2 in chromosome end biology. PLoS Pathog. 2010;6:e1000784. doi: 10.1371/journal.ppat.1000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang H, Dittmer TA, Richards EJ. Arabidopsis crowded nuclei (CRWN) proteins are required for nuclear size control and heterochromatin organization. BMC Plant Biol. 2013;13:200. doi: 10.1186/1471-2229-13-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 88.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, et al. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Glynn EF, Megee PC, Yu H-G, Mistrot C, Unal E, et al. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae . PLoS Biol. 2004;2:e259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weber SA, Gerton JL, Polancic JE, DeRisi JL, Koshland D, et al. The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2004;2:e260. doi: 10.1371/journal.pbio.0020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, et al. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum . Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 93.Dzikowski R, Li F, Amulic B, Eisberg A, Frank M, et al. Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep. 2007;8:959–965. doi: 10.1038/sj.embor.7401063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ponts N, Harris EY, Prudhomme J, Wick I, Eckhardt-Ludka C, et al. Nucleosome landscape and control of transcription in the human malaria parasite. Genome Res. 2010;20:228–238. doi: 10.1101/gr.101063.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tonkin CJ, Carret CK, Duraisingh MT, Voss TS, Ralph SA, et al. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum . PLoS Biol. 2009;7:e84. doi: 10.1371/journal.pbio.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Iyer LM, Anantharaman V, Wolf MY, Aravind L. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int J Parasitol. 2008;38:1–31. doi: 10.1016/j.ijpara.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 97.Volz J, Carvalho TG, Ralph SA, Gilson P, Thompson J, et al. Potential epigenetic regulatory proteins localise to distinct nuclear sub-compartments in Plasmodium falciparum . Int J Parasitol. 2010;40:109–121. doi: 10.1016/j.ijpara.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 98.Briquet S, Ourimi A, Pionneau C, Bernardes J, Carbone A, et al. Identification of Plasmodium falciparum nuclear proteins by mass spectrometry and proposed protein annotation. PLoS One. 2018;13:e0205596. doi: 10.1371/journal.pone.0205596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Flueck C, Bartfai R, Volz J, Niederwieser I, Salcedo-Amaya AM, et al. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog. 2009;5:e1000569. doi: 10.1371/journal.ppat.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 101.Yi Q, Chen Q, Liang C, Yan H, Zhang Z, et al. HP1 links centromeric heterochromatin to centromere cohesion in mammals. EMBO Rep. 2018;19:e45484. doi: 10.15252/embr.201745484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jang C-W, Shibata Y, Starmer J, Yee D, Magnuson T. Histone H3.3 maintains genome integrity during mammalian development. Genes Dev. 2015;29:1377–1392. doi: 10.1101/gad.264150.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mochizuki R, Tsugama D, Yamazaki M, Fujino K, Masuda K. Identification of candidates for interacting partners of the tail domain of DcNMCP1, a major component of the Daucus carota nuclear lamina-like structure. Nucleus. 2017;8:312–322. doi: 10.1080/19491034.2017.1280210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ciska M, Masuda K, Moreno Díaz de la Espina S. Lamin-like analogues in plants: the characterization of NMCP1 in Allium cepa . J Exp Bot. 2013;64:1553–1564. doi: 10.1093/jxb/ert020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.DuBois KN, Alsford S, Holden JM, Buisson J, Swiderski M, et al. NUP-1 Is a large coiled-coil nucleoskeletal protein in trypanosomes with lamin-like functions. PLoS Biol. 2012;10:e1001287. doi: 10.1371/journal.pbio.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang M, Wang C, Otto TD, Oberstaller J, Liao X, et al. Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science. 2018;360:eaap7847. doi: 10.1126/science.aap7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stubbs J, Simpson KM, Triglia T, Plouffe D, Tonkin CJ. Molecular mechanism for switching of P. falciparum invasion pathways into human erythrocytes. Science. 2005;309:1384–1387. doi: 10.1126/science.1115257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.