Abstract
18F-(2S,4R)-4-fluoroglutamine (18F-FGln) is an investigational PET radiotracer for imaging tumor glutamine flux and metabolism. The aim of this study was to investigate its pharmacokinetic properties in patients with cancer. Methods: Fifty lesions from 41 patients (21 men and 20 women, aged 54 ± 14 y) were analyzed. Thirty-minute dynamic PET scans were performed concurrently with a rapid intravenous bolus injection of 232 ± 82 MBq of 18F-FGln, followed by 2 static PET scans at 97 ± 14 and 190 ± 12 min after injection. Five patients also underwent a second 18F-FGln study 4–13 wk after initiation of therapy with glutaminase, dual TORC1/2, or programmed death-1 inhibitors. Blood samples were collected to determine plasma and metabolite fractions and to scale the image-derived input function. Regions of interest were manually drawn to calculate SUVs. Pharmacokinetic modeling with both reversible and irreversible 1- and 2-tissue-compartment models was performed to calculate the kinetic rate constants K1, k2, k3, and k4. The analysis was repeated with truncated 30-min dynamic datasets. Results: Intratumor 18F-FGln uptake patterns demonstrated substantial heterogeneity in different lesion types. In most lesions, the reversible 2-tissue-compartment model was chosen as the most appropriate according to the Akaike information criterion. K1, a surrogate biomarker for 18F-FGln intracellular transport, was the kinetic rate constant that was most correlated both with SUV at 30 min (Spearman ρ = 0.71) and with SUV at 190 min (ρ = 0.51). Only K1 was reproducible from truncated 30-min datasets (intraclass correlation coefficient, 0.96). k3, a surrogate biomarker for glutaminolysis rate, was relatively low in about 50% of lesions. Treatment with glutaminase inhibitor CB-839 substantially reduced the glutaminolysis rates as measured by k3. Conclusion: 18F-FGln dynamic PET is a sensitive tool for studying glutamine transport and metabolism in human malignancies. Analysis of dynamic data facilitates better understanding of 18F-FGln pharmacokinetics and may be necessary for response assessment to targeted therapies that impact intracellular glutamine pool size and tumor glutaminolysis rates.
Keywords: glutamine, metabolism, glutaminolysis, dynamic PET, kinetic modeling
Glutamine, alongside glucose, is 1 of 2 principal nutrients that support survival, biosynthesis, and cellular homeostasis in mammalian cells, thus playing an essential role in cancer cell metabolism (1–5). The glutaminolytic pathway is highly active in many aggressive cancers (6). 18F-(2S,4R)-4-fluoroglutamine (18F-FGln) is an l-glutamine analog that was developed as an investigational PET radiopharmaceutical for imaging tumor glutamine flux and metabolism (7,8). Noninvasive clinical assays for imaging tumor glutamine metabolic pathways can provide complementary value to 18F-FDG PET in several scenarios (9). In the first scenario, the ability of tumors to preferentially use glutamine suggests that glutaminolysis may be the metabolic pathway present in 18F-FDG–negative tumors (2,8,10). In the second, 18F-FGln may be useful for identifying residual viable tumor in patients on drug regimens that suppress glucose uptake, such as inhibitors of the phosphoinositide-3-kinase–protein kinase B/mammalian target of rapamycin (mTOR) pathway (11,12). Third, visualization of 18F-FDG–avid tumors is difficult if the physiologic 18F-FDG uptake in normal surrounding tissue is comparable to that of the tumor, as is the case in the cerebral cortex (13). Because of low glutamine consumption in normal brain tissue, 18F-FGln provides higher tumor-to-background ratios than 18F-FDG (14). Fourth, imaging of glutamine metabolism might carry prognostic value as a metabolic marker of tumor aggressiveness (15) and, fifth, many different mutations can lead to enhanced glucose uptake. Identifying a subset of tumors with enhanced glutamine metabolism such as those with c-Myc amplification (4,5,16) could allow for the prediction of specific genetic alterations, facilitating patient stratification for targeted therapy trials and personalized treatment monitoring. For example, glutaminase, an amidohydrolase enzyme that generates glutamate from glutamine, is the rate-limiting enzyme in glutaminolysis. It is upregulated by the oncogene c-Myc (9), which can lead to glutamine addiction (2). The exploitation of the glutaminolysis pathway for therapeutic purposes is spurring research into glutaminase inhibitors as potential cancer therapeutic targets (17). Furthermore, cancer cells may use both phosphoinositide-3-kinase–protein kinase B/mTOR and c-Myc pathways to generate energy for growth and survival (8). Adding 18F-FGln PET would provide a more complete picture of tumor metabolism than imaging with 18F-FDG alone (6).
18F-FGln has recently been clinically validated as a promising tumor biomarker in several different cancer types (14,15). The objectives of this study were to investigate the pharmacokinetic properties of 18F-FGln and to evaluate the added benefit of dynamic over simpler static 18F-FGln PET.
MATERIALS AND METHODS
Patient Selection
The data presented in this study were acquired as a part of an open-label, nonrandomized, microdose phase I trial of 18F-FGln. The trial was approved by Memorial Sloan Kettering Cancer Center’s Institutional Review Board and conducted under a Food and Drug Administration–approved Investigational New Drug application (ClinicalTrials.gov identifier NCT01697930). The study was conducted in accordance with the Helsinki Declaration and the Health Insurance Portability and Accountability Act. Patients provided written informed consent before participating in the study. Subject inclusion criteria included an age of 21–90 y; serum renal and hepatic function test values less than 1.5- to 2.5-fold greater than the laboratory-specific upper limit of normal, histologically confirmed cancer; and tumors visualized with standard imaging (CT, MR, or 18F-FDG PET/CT) less than 4 wk before consent. Serum complete blood count and hepatorenal function tests were performed less than 2 wk before study participation. Patients were excluded if they were pregnant, breastfeeding, or had an acute major illness.
PET/CT Imaging
18F-FGln was synthesized by Memorial Sloan Kettering Cancer Center’s Radiochemistry and Molecular Imaging Probe Core Facility as described previously (15). Each 18F-FGln dose met drug product acceptance specifications, including radiochemical purity and identity, residual solvent content, endotoxin content, radionuclidic identity, pH, and appearance.
Dynamic PET scans were performed over a single field of view (15.7 cm axially) on a Discovery STE, 690, or 710 camera (GE Healthcare), concurrent with a rapid intravenous bolus injection of 232 ± 82 MBq of 18F-FGln (range, 29–469 MBq). Images were acquired in list mode and binned into 12 × 10-s, 3 × 60-s, and 5 × 300-s time frames, for a total of 30 min. A CT scan (120 kVp, 70 mA, and 3.8-mm slice thickness) was obtained for attenuation correction, anatomic localization, and coregistration purposes. Dynamic acquisition was followed by 2 static PET scans starting at 97 ± 14 and 190 ± 12 min after injection (15- to 30-min acquisition time). PET emission data were acquired in 3-dimensional mode; corrected for attenuation, scatter, and random events; and iteratively reconstructed into either a 256 × 256 × 47 matrix (voxel dimensions, 1.95 × 1.95 × 3.27 mm) for brain lesions or a 128 × 128 × 47 matrix (voxel dimensions, 2.34 × 2.34 × 3.27 mm) for lesions in the thoracic and abdominal area using the ordered-subset expectation maximization algorithm provided by the manufacturer. For 5 patients, second dynamic 18F-FGln PET scans were also performed on the same scanner, 4–13 wk after starting a new anticancer treatment with either glutaminase, dual raptor-mTOR protein complex (TORC1) and rictor-mTOR protein complex (TORC2), or programmed death-1 (PD-1) inhibitors.
Blood Sample Analysis
Activity in whole blood and plasma specimens was radioassayed with a calibrated well counter (1480 Wallac Wizard 3 automatic γ-counter; Perkin Elmer, Inc.) after separating blood and plasma by centrifuge (4,000 rpm for 10 min at 4°C), as described previously (15). Multiple venous blood samples were obtained between 5 and 180 min after injection. The measured activity concentrations were converted to kBq/cc. Metabolite analysis of activity in plasma was performed by reversed-phase high-performance liquid chromatography with in-line radiation detection on samples obtained up to 65 min after injection.
Image Analysis
All 3 PET segments were spatially coregistered using the rigid-body transformation calculated with General Co-Registration tool (GE Healthcare Advantage Workstation, version 4.7) applied to their corresponding CT scans to form a concatenated 18F-FGln dynamic PET scan. Subsequent processing was performed in PMOD, version 3.604 (RRID:SCR_016547; PMOD Software). Regions of interest were drawn over sites of disease identified by a radiologist with experience in nuclear medicine. Time–activity curves and SUVs corrected by body weight were derived from lesions. For each patient with a brain lesion, analysis was also performed for normal brain tissue by averaging the results from 10 spheric regions of interest, each with a 10-mm radius.
Input function was image-derived by manually defining a region of interest over the internal carotid artery or descending aorta on the early frame with the highest image intensity. For each patient, whole-blood input function time–activity curves were scaled by the whole-blood activity concentration as measured from blood samples and corrected for plasma fraction. Metabolite counts were analyzed in only a subset of patients; therefore, an averaged population-based metabolite correction was applied for all patients.
Pharmacokinetic Modeling
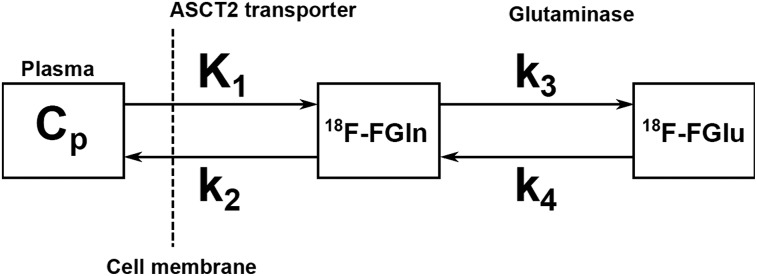
Irreversible and reversible 1-tissue-compartment (1C1K and 1C2K, respectively) and 2-tissue compartment (2C3K and 2C4K, respectively) pharmacokinetic models with a blood fraction component were investigated to calculate the kinetic rate constants K1, k2, k3, and k4. In the 2C4K model (Fig. 1), K1 is assumed to be a surrogate biomarker for perfusion, tumor vascular permeability, and intracellular transport rate mediated by alanine, serine, cysteine transporter 2 (ASCT2). k3, on the other hand, may be a surrogate biomarker for the first and rate-limiting step of glutaminolysis that is catalyzed by glutaminase and yields 18F-fluoroglutamate. Further downstream processes in the glutaminolytic pathway (e.g., metabolization of 18F-fluoroglutamate to 18F-fluoro-α-ketoglutarate) are likely also incorporated into k3. In this framework, k2 represents the efflux back into vasculature, whereas k4 represents the excretion of 18F-fluoroglutamate, efflux of free 18F (a by-product of the metabolization of 18F-fluoroglutamate to α-ketoglutarate by alanine aminotransferase (18)), or conversion of 18F-fluoroglutamate back into 18F-fluoroglutamine by glutamine synthetase (19).
FIGURE 1.
Schematic of 2C4K. Cp is plasma compartment, representing unmetabolized 18F-FGln that is available for transport across vasculature into tissue. First compartment represents nonspecifically bound 18F-fluoroglutamine that has been transported into tumor cells by ASCT2 and other transporters, whereas second compartment represents activity from 18F-fluoroglutamate (18F-FGlu), produced in first and rate-limiting step of glutaminolysis that is catalyzed by glutaminase.
The total concentration of activity measured by the PET scanner as a function of time t after injection, C(t), is given by
| Eq. 1 |
where Cp(t) is the activity concentration of the unmetabolized radiotracer in the plasma, and C1(t) and C2(t) are the activity concentrations associated with the first and second compartments, corresponding to nonspecifically and specifically bound radiotracer in tissue. The rate of change for C1(t) and C2(t) is described by the system of differential equations:
| Eq. 2 |
| Eq. 3 |
Default starting values for the parameters K1, k2, k3, and k4 were 0.1 mL/min/g, 0.1 min−1, 0.1 min−1, and 0.1 min−1, respectively (in all cases, lower and upper bounds were 0 and 8, respectively). Goodness of fit was evaluated with the Akaike information criterion to determine the most appropriate compartmental model. Volume of distribution, VT, was calculated as
| Eq. 4 |
Logan graphical analysis (20), a technique originally developed for calculating the VT of reversible receptor systems, was also performed.
To evaluate the utility of truncated 18F-Gln datasets, tumor time–activity curves derived from the first 30 min of data were refitted with the 2C4K model. All metrics calculated from truncated datasets were compared with those derived from full datasets.
Statistical Analysis
The correlation strength between different indices was analyzed using the Spearman rank correlation coefficient, ρ. Metrics calculated from different patient subgroups were compared using an unpaired 2-tailed t test. Bland–Altman analysis was performed to estimate the mean difference between parameters calculated with full and truncated datasets and 95% limits of agreement. The reproducibility of metrics calculated using different methods was evaluated using a 2-way random single-score intraclass correlation coefficient. A P value of less than 0.05 was assumed to represent statistical significance.
RESULTS
Sixty-five patients were enrolled in the study between January 2013 and October 2018. Of these, 11 patients subsequently withdrew consent, and for 13 patients, dynamic 18F-FGln PET images were not analyzed because there were no lesions within the field of view of the dynamic scan (n = 7) or the dynamic PET data were corrupt or lost (n = 6). Forty-one patients (21 men and 20 women; mean age ± SD, 54 ± 14 y; range, 24–80 y) and 50 lesions in total were included in the analysis (Table 1). For 28 of the 41 patients, the field of view was focused over the brain (subgroup 1, 35 lesions), whereas for the remaining 13 patients, the field of view was focused over the thoracic or abdominal region (subgroup 2, 15 lesions). The lesions measured 4.2 ± 7.5 cm3 (range, 0.2–36.9 cm3). The percentage of activity due to 18F-FGln in plasma was 78% ± 12%, 80% ± 13%, 78% ± 11%, 76% ± 9%, 73% ± 8%, and 69% ± 9% at 2, 6, 16, 30, 65, and 158 min after injection, respectively (44 patients with available blood data). Metabolite analysis at multiple time-points was performed for a subset of patients, from whom a population-based metabolite correction function was derived. Unmetabolized 18F-FGln fraction was 78% ± 10% (n = 9 data points), 75% ± 12% (n = 4), 73% ± 11% (n = 28), and 59% ± 7% (n = 5) at 2, 6, 30, and 65 min after injection.
TABLE 1.
Subject Demographics and Clinical Characteristics (n = 41)
| Characteristic | Data |
| Sex (n) | |
| Male | 21 |
| Female | 20 |
| Age at baseline 18F-FGln PET (y) | |
| <40 | 8 |
| 40–49 | 6 |
| 50–59 | 12 |
| 60–69 | 8 |
| 70–79 | 7 |
| Cancer (n) | |
| Glioblastoma multiforme | 14 |
| Astrocytoma | 6 |
| Lung cancer | 6 |
| Pancreatic cancer | 4 |
| Breast cancer | 3 |
| Oligodendroglioma | 2 |
| Prostate cancer | 2 |
| Colon cancer | 1 |
| Ependymoma | 1 |
| Diffuse large B cell lymphoma | 1 |
| Renal cell carcinoma | 1 |
According to the Akaike information criterion, the 1C2K, 2C3K, and 2C4K models were most appropriate in 9, 15, and 26 lesions, respectively. Across 50 lesions, Akaike information criterion values were 160 ± 30, 146 ± 29, and 141 ± 28, respectively. Lesions for which 1C2K or 2C3K was deemed better than 2C4K exhibited k3 or k4 values close to zero; therefore, adding these fitting parameters did not improve the fit. In most lesions, goodness of fit was perceptibly poorer when the 1C2K or 2C3K model was used. Therefore, only results obtained with the 2C4K model are presented.
Pharmacokinetic modeling of 18F-FGln dynamic PET with a 2C4K model is summarized in Table 2. Also included are the results for all brain lesions (subgroup 1), primary brain lesions (subgroup 1A), brain metastases (subgroup 1B), and all thoracic/abdominal lesions (subgroup 2), as well as for normal brain tissue. 18F-FGln uptake in tumors was rapid and subsequently decreased. Compared with 18F-FGln tumor uptake, 18F-FGln uptake in normal brain tissue was significantly lower at all imaging time-points. Analysis was also repeated for regions of interest encompassing the small area (5 voxels) with the highest 18F-FGln uptake (Supplemental Table 1; supplemental materials are available at http://jnm.snmjournals.org). VT as calculated from Logan graphical analysis and the 1C2K model was strongly correlated (intraclass correlation coefficient, 0.95; VT = 3.7 ± 1.7 and 4.0 ± 2.0 mL/cm3, respectively). Correlation was lower for VT calculated from the 2C4K model (intraclass correlation coefficient, 0.80; VT = 4.5 ± 2.4 mL/cm3). Among kinetic rate constants, K1 was most closely correlated with SUV (Table 3).
TABLE 2.
Mean Intratumor Values for Metrics Derived from Baseline 18F-FGln Dynamic PET, as Calculated with Reversible 2C4K
| Metric | All lesions (n = 50) | All brain lesions (n = 35) | Primary brain lesions (n = 26) | Brain metastases (n = 9) | All thoracic/abdominal lesions (n = 15) | Normal brain tissue (n = 26) |
| SUV1 | 2.5 ± 1.2 (0.6–6.3) | 2.1 ± 0.9 (0.6–4.0) | 2.3 ± 0.9 (0.6–4.0) | 1.7 ± 0.6 (0.6–2.9) | 3.4 ± 1.5 (1.4–6.3) | 0.3 ± 0.1 (0.2–0.4) |
| SUV2 | 2.2 ± 0.9 (0.7–4.8) | 2.1 ± 0.7 (0.7–3.6) | 2.1 ± 0.7 (0.7–3.6) | 2.0 ± 0.6 (1.0–3.0) | 2.5 ± 1.2 (1.0–4.8) | 0.3 ± 0.1 (0.2–0.4) |
| SUV3* | 1.7 ± 0.5 (0.7–2.6) | 1.6 ± 0.5 (0.8–2.6) | 1.6 ± 0.5 (0.8–2.6) | 1.9 ± 0.4 (1.5–2.5) | 1.8 ± 0.5 (0.9–2.3) | 0.4 ± 0.1 (0.3–0.5) |
| vB | 0.07 ± 0.05 (0.01–0.22) | 0.06 ± 0.04 (0.01–0.19) | 0.05 ± 0.04 (0.01–0.19) | 0.07 ± 0.04 (0.02–0.16) | 0.10 ± 0.07 (0.01–0.22) | 0.03 ± 0.01 (0.01–0.07) |
| K1 (mL/min/g) | 0.15 ± 0.10 (0.02–0.42) | 0.10 ± 0.06 (0.02–0.24) | 0.10 ± 0.07 (0.02–0.28) | 0.09 ± 0.05 (0.02–0.15) | 0.27 ± 0.09 (0.18–0.42) | 0.01 ± 0.00 (0.00–0.01) |
| k2 (min−1) | 0.11 ± 0.11 (0.01–0.56) | 0.06 ± 0.06 (0.01–0.24) | 0.06 ± 0.05 (0.01–0.22) | 0.08 ± 0.07 (0.01–0.24) | 0.21 ± 0.14 (0.06–0.56) | 0.05 ± 0.04 (0.02–0.20) |
| k3 (min−1) | 0.06 ± 0.07 (0.00–0.27) | 0.04 ± 0.06 (0.00–0.21) | 0.02 ± 0.04 (0.00–0.15) | 0.08 ± 0.08 (0.01–0.21) | 0.10 ± 0.10 (0.00–0.27) | 0.02 ± 0.01 (0.01–0.04) |
| k4 (min−1) | 0.02 ± 0.03 (0.00–0.10) | 0.02 ± 0.02 (0.00–0.10) | 0.01 ± 0.02 (0.00–0.06) | 0.03 ± 0.03 (0.00–0.10) | 0.03 ± 0.03 (0.00–0.10) | 0.003 ± 0.003 (0.000–0.009) |
| VT (mL/cm3) | 3.7 ± 1.7 (1.8–10.0) | 3.6 ± 1.6 (1.8 ± 10.0) | 3.3 ± 1.6 (1.8–10.0) | 4.3 ± 1.4 (2.8–7.0) | 4.2 ± 2.0 (2.0–9.1) | 0.6 ± 0.2 (0.3–1.3) |
SUV3 was not measured in 10 lesions.
vB = pharmacokinetic model with blood fraction component.
SUV1, SUV2, and SUV3 are SUV, corrected by body weight, as calculated from last 5-min frame of 30-min dynamic PET acquisition, from ∼100-min PET acquisition, and from ∼190-min PET acquisition, respectively. Data are mean ± SD, followed by range in parentheses.
TABLE 3.
Spearman ρ Between Metrics Derived from Dynamic Data (2C4K model) and SUVmax for Small Intratumor Area of Highest 18F-FGln Uptake
| Metric | 30 min (SUV1) | 100 min (SUV2) | 190 min (SUV3) | Δ (SUV3 − SUV1) |
| K1 | 0.71 | 0.63 | 0.51 | −0.65 |
| k2 | 0.38 | 0.36 | 0.48 | −0.28 |
| k3 | 0.13 | 0.25 | 0.38 | 0.13 |
| k4 | 0.14 | 0.26 | 0.26 | −0.10 |
| VT | 0.48 | 0.61 | 0.53 | −0.27 |
SUVmax is corrected by body weight.
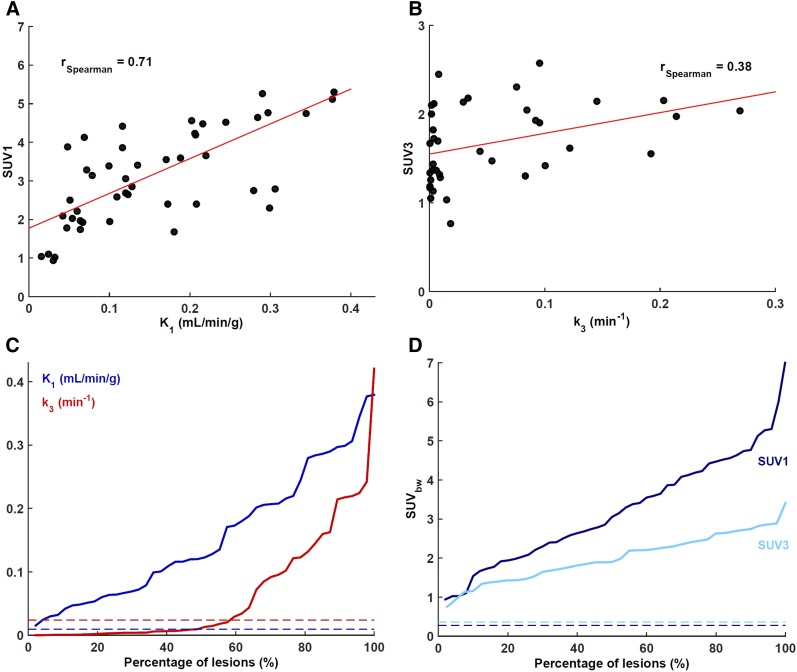
K1-SUV1 and k3-SUV3 scatterplots are presented in Figures 2A and 2B. Waterfall charts for these 4 metrics (Figs. 2C and 2D) revealed a wide range of observed values. k3 was relatively low in about 50% of cases, indicating that glutaminolysis rates are not elevated in all lesions. Two 18F-FGln uptake patterns were observed (Fig. 3 and Supplemental Fig. 1A). In 29 of 42 evaluable lesions (69%; in some patients, 18F-FGln scans were not performed at all imaging time-points), 18F-FGln SUV was highest at around 30 min after injection and decreased afterward (pattern 1). The remaining 13 lesions (31%, all in brain) peaked at around the 100-min imaging time-point, with a subsequent decrease (pattern 2). K1 significantly differed between lesions exhibiting the 2 patterns (K1 = 0.19 ± 0.13 and 0.08 ± 0.05 mL/min/g, respectively; 2-tailed t test, P = 0.01). All 9 lesions in 5 patients with brain metastases exhibited pattern 2 (Supplemental Fig. 1B).
FIGURE 2.
(A) Scatterplot of maximum intratumor K1-SUV1. K1 was the kinetic rate constant most closely correlated with SUV1 (measured on last dynamic 5-min frame, 25–30 min after injection). (B) Scatterplot of maximum intratumor k3-SUV3. (C) Waterfall chart of maximum intratumor K1 and k3. (D) Waterfall chart of maximum intratumor SUV1 and SUV3. In both waterfall charts, averages for normal brain tissue are shown as color-coded dashed lines.
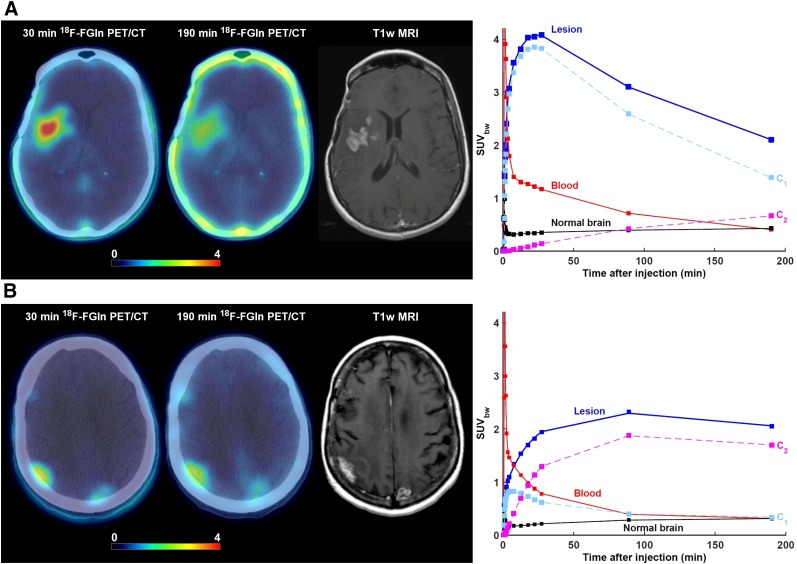
FIGURE 3.
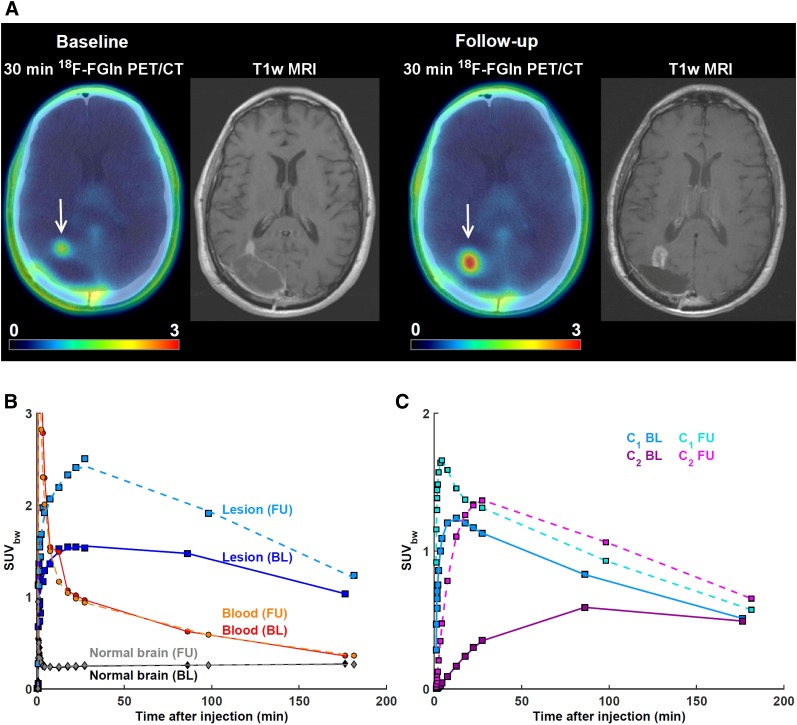
Lesions from 2 patients exhibiting different 18F-FGln pharmacokinetics despite having very similar SUV corrected by body weight (SUVbw) as measured at 190 min after injection. In both cases, axial view of last dynamic PET frame (5-min acquisition time, 25–30 min after injection) and last imaging frame (∼190 min after injection), fused with corresponding CT, is displayed. Gadolinium-enhanced T1-weighted MRI is included for comparison. Three time–activity curves are shown: for whole lesion; image-derived input function scaled by whole-blood activity concentration as measured from blood samples, patient-specific plasma fraction, and population-based metabolite fraction; and normal brain tissue. Contributions to total PET signal for tumor time–activity curves are shown for first compartment (C1) and second compartment (C2). (A) 52-y-old woman with confirmed diagnosis of astrocytoma. 18F-FGln PET/CT shows 2 cm3 lesion in right frontotemporal region. Mean intratumor K1, k2, k3, k4, and VT were 0.28 mL/min/g, 0.08 min−1, 0.002 min−1, 0.001 min−1, and 3.9 mL/cm3, respectively. Corresponding values for normal brain tissue were 0.02 mL/min/g, 0.09 min−1, 0.001 min−1, 0.001 min−1, and 0.5 mL/cm3, respectively. SUVbw at 30, 90, and 190 min was 4.0, 3.1, and 2.1, respectively. At these 3 time points, signal from second compartment contributed 3%, 14%, and 32% of total PET activity concentration. (B) 70-y-old woman with confirmed diagnosis of non–small cell lung cancer that metastasized to brain. PET image shows metastatic lesion in right parietal region. Mean intratumor K1, k2, k3, k4, and VT were 0.13 mL/min/g, 0.13 min−1, 0.09 min−1, 0.02 min−1, and 5.1 mL/cm3, respectively. Corresponding values for normal brain tissue were 0.01 mL/min/g, 0.05 min−1, 0.04 min−1, 0.004 min−1, and 0.7 mL/cm3, respectively. SUVbw at 30, 90, and 190 min was 1.9, 3.1, and 2.1, respectively. At these 3 time points, signal from second compartment contributed 66%, 82%, and 83%, respectively, of total PET activity concentration.
The first compartment (assumed to represent 18F-fluoroglutamine that has been transported from the vasculature into the cell by ASCT2 but has not been converted to 18F-fluoroglutamate) contributed 76% ± 14%, 65% ± 25%, 52% ± 24%, and 46% ± 23% of PET signal at 5, 30, approximately 100, and approximately 190 min after injection, respectively, whereas the contribution from the second compartment (representing 18F-fluoroglutamate but also the incorporation of 18F-fluoroglutamine into proteins) was 10% ± 13%, 29% ± 26%, 44% ± 24%, and 50% ± 23%, respectively (Supplemental Fig. 2).
The reproducibility analysis is summarized in Table 4. The intraclass correlation coefficient was highest for K1, which can be estimated from the initial 30-min segment of time–activity curves; however, none of the other kinetic rate constants were reproducible.
TABLE 4.
Reproducibility of Metrics Derived from Truncated 30-Minute 18F-FGln Dynamic PET Compared with Full 3-Hour Dataset
| Metric | ICC | Mean difference |
| K1 (mL/min/g) | 0.96 | −0.01 (−0.07, 0.05) |
| k2 (min−1) | 0.63 | −0.03 (−0.20, 0.15) |
| k3 (min−1) | −0.01 | −0.06 (−0.42, 0.30) |
| k4 (min−1) | −0.09 | −0.03 (−0.21, 0.15) |
| VT (mL/cm3) | 0.75 | 0.90 (−0.77, 2.57) |
ICC = intraclass correlation coefficient.
Data in parentheses are lower and upper limits of agreement. Kinetic rate constants were derived using 2C4K model.
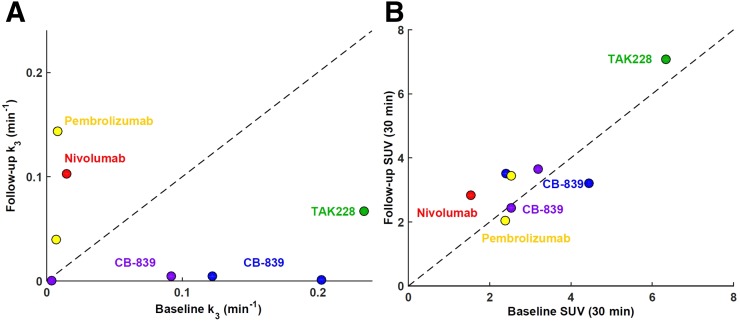
Five patients also underwent a second 18F-FGln dynamic PET scan after therapy with CB-839, a glutaminase inhibitor; TAK-228, a dual TORC1/2 inhibitor; or the PD-1 inhibitors nivolumab or pembrolizumab (Table 5). Eight lesions were analyzed in these 5 patients, with the results summarized in Table 6. The effect of these treatments on glutaminolysis rate as measured by k3 is illustrated in Figure 4A. Therapy with CB-839 resulted in a markedly decreased rate of glutaminolysis (i.e., k3 fell to almost zero). A gradual decrease was also observed for a patient who received therapy with the dual TORC1/2 inhibitor TAK-228. On the other hand, therapy with the PD-1 inhibitors nivolumab and pembrolizumab seems to have increased the rate of glutaminolysis. The corresponding scatterplot for SUV1 (Fig. 4B) indicates greater ambiguity in interpreting the effects of therapies on 18F-FGln uptake. An example of a glioblastoma multiforme patient imaged 13 wk after initiation of treatment with nivolumab and radiotherapy is included in Figure 5. For this patient, elevated 18F-FGln uptake and retention were hypothesized to be due to the increased rate of glutaminolysis (an increased contribution to the signal from the second compartment), because the activity concentration associated with the first compartment remained similar. On the other hand, treatment with CB-839 in a patient with metastatic renal cell carcinoma appears to have reduced the rate of glutaminolysis as assessed by a marked decrease in k3 and decreased signal from the second compartment (Fig. 6).
TABLE 5.
Treatment Information for Patients Who Underwent Second 18F-FGln Dynamic PET Scan
| Patient no. | Cancer | Therapy | Time between first and second scans (d) | Dosing |
| 1 | Renal cell carcinoma | CB-839 | 55 | 400 mg 3 times daily |
| 2 | Non–small cell lung cancer | CB-839 | 27 | 400 mg 3 times daily |
| 3 | Glioblastoma multiforme | Nivolumab + radiotherapy | 92 | 200 mg 1 time daily |
| 4 | Glioblastoma multiforme | Pembrolizumab | 68 | 200 mg 1 time daily |
| 5 | Glioblastoma multiforme | TAK-228 | 49 | 3 mg 1 time daily* |
Patient self-discontinued trial after 1 mo.
FOV = field of view for dynamic PET scan.
TABLE 6.
Mean Intratumor Values for Metrics Derived from Early-Response 18F-FGln Dynamic PET Scans with 2C4K Model
| Metric | All lesions (n = 8) | Corresponding 8 lesions on baseline |
| SUV1 | 3.5 ± 1.5 (2.0–7.1) | 3.2 ± 1.5 (1.8–6.3) |
| SUV2 | 3.0 ± 1.2 (2.0–5.7) | 2.8 ± 1.1 (1.5–4.8) |
| vB | 0.17 ± 0.10 (0.03–0.30) | 0.12 ± 0.06 (0.04–0.19) |
| K1 (mL/min/g) | 0.21 ± 0.16 (0.04–0.46) | 0.23 ± 0.12 (0.07–0.43) |
| k2 (min−1) | 0.09 ± 0.04 (0.03–0.13) | 0.15 ± 0.10 (0.04–0.30) |
| k3 (min−1) | 0.05 ± 0.06 (0.00–0.14) | 0.09 ± 0.09 (0.00–0.23) |
| k4 (min−1) | 0.03 ± 0.03 (0.00–0.10) | 0.02 ± 0.02 (0.00–0.06) |
| VT (mL/cm3) | 3.4 ± 1.5 (1.3–5.6) | 4.7 ± 2.2 (2.2–9.1) |
SUV1 and SUV2 are SUV, corrected by body weight, as calculated from last 5-min frame of 30-min dynamic PET acquisition and from ∼100-min PET acquisition, respectively. SUV3 is not reported because only 2 patients had ∼190-min postinjection acquisitions on both first and second 18F-FGln dynamic PET. Data are mean ± SD, followed by range in parentheses.
vB = pharmacokinetic model with blood fraction component.
FIGURE 4.
(A) Scatterplot of baseline vs. follow-up mean intratumor k3 for 5 patients (8 lesions in total) who underwent 2 18F-FGln dynamic PET scans. Every patient is color-coded and annotated according to therapy received between baseline and follow-up PET. Line of identity is shown as dashed line. (B) Corresponding scatterplot of baseline vs. follow-up mean intratumor SUV as measured at 30 min after injection.
FIGURE 5.
A 68-y-old man with confirmed diagnosis of glioblastoma multiforme. (A) Baseline (BL) 18F-FGln dynamic PET was performed after right parietal occipital craniotomy for resection of heterogeneously enhancing mass centered in right occipital lobe (arrow). Follow-up (FU) 18F-FGln dynamic PET was performed 13 wk after initiation of treatment with nivolumab and radiotherapy. Gadolinium-enhanced T1-weighted MRI scans were performed 3 and 4 d before BL and FU PET, respectively. When compared with BL MRI scan, enhancing nodule extending toward trigone of right lateral ventricle on FU MRI scan is enlarged and exhibits higher uptake at all imaging time points. (B) Time–activity curves from BL and FU PET for tumor, image-derived input function scaled by whole-blood activity concentration as measured from blood samples, patient-specific plasma fraction and population-based metabolite fraction, and normal brain tissue. Mean intratumor K1, k2, k3, k4 and VT as calculated from BL PET was 0.07 mL/min/g, 0.05 min−1, 0.02 min−1, 0.02 min−1, and 2.1 mL/cm3, respectively. Corresponding values as calculated from FU PET were 0.16 mL/min/g, 0.11 min−1, 0.10 min−1, 0.10 min−1, and 2.9 mL/cm3, respectively. (C) Signal from second compartment contributed 23%, 40%, and 47% of total PET activity at 30, 86, and 176 min on BL scan and 50%, 53%, and 53% at 30, 98, and 182 min on FU scan. SUVbw = SUV corrected by body weight.
FIGURE 6.
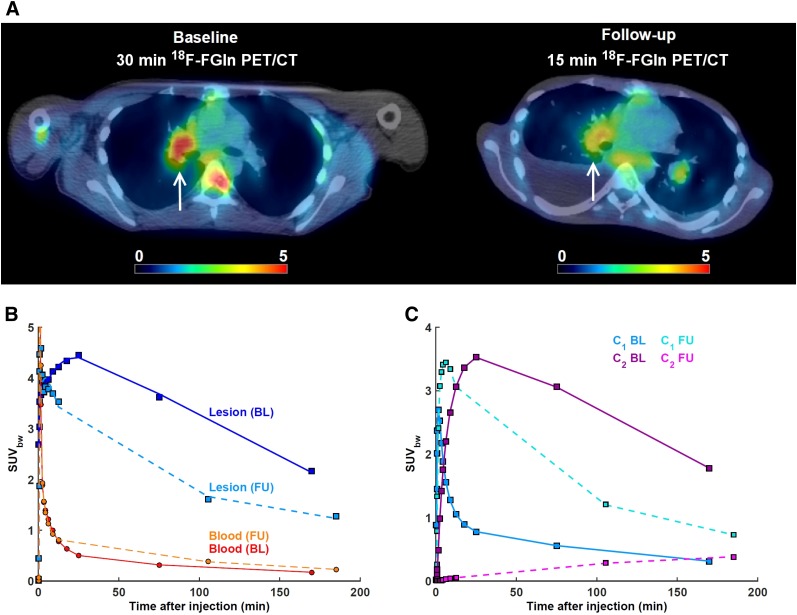
A 23-y-old man with confirmed diagnosis of metastatic renal cell carcinoma. (A) Baseline (BL) and follow-up (FU) 18F-FGln dynamic PET, showing pulmonary metastasis in right lung (arrow). FU scan was performed 4 wk after initiation of therapy with glutaminase inhibitor CB-839. Acquisition was shortened to 15-min because of patient discomfort. (B) Time–activity curves from BL and FU PET for highlighted lesion and image-derived input function scaled by whole-blood activity concentration as measured from blood samples, patient-specific plasma fraction, and population-based metabolite fraction. Mean intratumor K1, k2, k3, k4 and VT as calculated from BL PET were 0.43 mL/min/g, 0.25 min−1, 0.20 min−1, 0.04 min−1, and 9.1 mL/cm3, respectively. Corresponding values as calculated from FU PET were 0.44 mL/min/g, 0.12 min−1, 0.001 min−1, 0.000 min−1, and 4.4 mL/cm3, respectively. (C) Signal from second compartment contributed 80%, 83%, and 83% of total PET activity at 30, 75, and 170 min on BL scan and 2%, 17%, and 31% at 15, 105, and 185 min on FU scan. SUVbw = SUV corrected by body weight.
DISCUSSION
We investigated the pharmacokinetic properties of 18F-FGln across lesions of different etiologies and demonstrated the added benefit of incorporating dynamic 18F-FGln PET acquisitions into analysis. 18F-FGln is readily imported into glutaminolytic tumor cells at rates comparable to 18F-FDG (7). It is transported mainly across the cell membrane by the amino acid transporter ASCT2 (8,21,22). Although 18F-FGln uptake in normal brain was low because of minimal expression of ASCT2 (14), high variability was observed in surrogate metrics of glutamine transport (K1 and SUV1) and retention (k3 and SUV3) in lesions. Additionally, relevant tumor genetic alterations in genes that are key regulators of tumor glutamine flux and metabolism were found in several patients with 18F-FGln–avid tumors (15).
Two 18F-FGln tumor uptake patterns were noted: initially rapid accumulation with a plateau at around 30 min after injection and a subsequent steep decrease; and slower accumulation with a plateau at around 100 min after injection and a more gradual decrease. All brain metastases exhibited the second pattern, in agreement with a recent report (23). Despite similar K1 between primary brain lesions and brain metastases (increased blood–brain barrier permeability does not significantly contribute to 18F-FGln uptake (14)), the latter exhibited 4-fold higher k3, resulting in more sustained retention. Metabolic reprogramming of glutaminolysis was reported to mediate metastatic phenotype in lung cancer (24) and melanoma (25).
18F-FGln uptake patterns might be important in understanding responses to targeted therapies with inhibitors of glutaminase (17) or ASCT2 (26) and cannot be readily elucidated using only static PET. The contributions from different processes to the total PET signal may, however, be uncoupled through analysis of dynamic PET data. A drawback of this approach is a clinically challenging acquisition protocol. Several factors contribute to the intratumor uptake of 18F-FGln, including upregulation of ASCT2 (resulting in higher K1 and SUV1) and increased protein synthesis and glutaminolysis (resulting in higher k3). However, increased glutaminase activity has also been associated with a smaller cellular glutamine pool, which resulted in low tracer retention as 18F-FGln competed with a small pool of native glutamine for efflux (6). Consequently, only a weak correlation was observed between k3 and SUV3. The typical time course of 18F-FGln accumulation in lesions precludes the use of truncated 30-min dynamic acquisitions, as the rate of glutaminolysis cannot be readily estimated from the available temporal information. Of note, k3 was also not found to correlate with the change in SUV (Table 3).
Reversibility of 18F-FGln uptake has been suggested previously (6) and was observed in our study. However, a fraction of the radiotracer might be at least temporarily trapped within cells. Lieberman et al. demonstrated that 18F-FGln showed significant in vivo incorporation into a trichloroacetic acid precipitated fraction, likely associated with intracellular protein or macromolecule synthesis, suggesting that this might be an important mechanism for radiotracer entrapment in tumors (8).
Targeted therapy with CB-839, a glutaminase inhibitor, resulted in a marked decrease in k3, consistent with the hypothesis that k3 is a surrogate biomarker of glutaminolysis. About 50% of all lesions did not demonstrate elevated glutaminolysis levels according to k3, implying that in these cases, targeted therapy with glutaminase inhibitors may not be effective. The patient highlighted in Figure 5 did, however, exhibit high levels of glutaminolysis at baseline (preclinical evidence of addiction of renal cell carcinoma cells to glutamine and glutaminase activity has recently been reviewed (27)). All 4 lesions from the 2 patients who received CB-839 also exhibited a marked decrease in k2 (from 0.19 ± 0.10 min−1 at baseline to 0.08 ± 0.04 min−1 at follow-up) despite no substantial changes in K1 (from 0.29 ± 0.10 to 0.26 ± 0.20 mL/min/g) or SUV at 30 min (from 3.1 ± 0.5 to 3.2 ± 0.5), likely as 18F-FGln competed with a larger pool of native glutamine molecules for efflux after glutaminase inhibition (6). A decrease in k3 was also observed after therapy with a dual TORC1/2 inhibitor, TAK-228, because the inhibition of mTORC1 activity suppresses the conversion of glutamine to α-ketoglutarate (28). On the other hand, treatment with nivolumab and pembrolizumab, human IgG4 anti-PD-1 monoclonal antibodies that work as checkpoint inhibitors and are believed to often provoke tumor inflammation when effective (29), led to an elevated glutaminolysis rate as reflected in higher k3. Albina et al. reported that the intracellular free glutamine concentration is decreased in inflammation and that the cellular components of the inflammatory infiltrate might be capable of active glutaminolysis (30).
Our study has several limitations. First, multiple patients received systemic anticancer treatments during or recently before 18F-FGln PET, potentially reducing tumor 18F-FGln avidity (14). Second, the number of patients undergoing 2 18F-FGln studies was small, lowering the confidence in interpreting the effects of therapies on glutamine flux and metabolism. Third, radiochemical testing before 18F-FGln injection confirmed the presence of less than 20% of stereoisomer, (2R,4R)-4-18F-fluoroglutamine, which, as an analog of d-glutamine, is not avidly accumulated by tumor cells (7). Fourth, since fractions of unmetabolized radiotracer were not determined for all patients, population-derived metabolite correction was implemented instead. Fifth, the 2C4K model assumes that free 18F does not significantly accumulate in tumors. Although metabolite analyses confirmed in vivo production of free 18F metabolite, hindering the analysis of tumors that are close to bone (15), Zhou et al. reported that the contribution of labeled metabolites to the tumor PET signal in mice is small (≤10%) and unlikely to have a significant influence on image-derived metrics (6). When we repeated the analysis using a 3-compartment pharmacokinetic model with 2 input functions that account for nonspecific uptake of radiometabolites, the contribution to the total signal from the third compartment was about 10% (range, 0%–20%), similar to the results reported by Zhou et al. (6). On the other hand, the percentage signal from the third compartment was greater than 85% in bone tissue, as is expected because of accumulation of free 18F. Sixth, the accuracy and precision of kinetic rate constant prediction are susceptible to experimental levels of noise. Exploratory Monte Carlo simulations (Supplemental Methods (31)) indicate that the calculation of K1 and k2 is relatively robust, whereas k3 and k4 exhibit higher variance when their true values are very low. Seventh, the moderate correlation between K1 and k3 (Supplemental Table 2) suggests that ASCT2 and glutaminase activity may both be upregulated in tumors. An alternative interpretation, however, is that the 2C4K model cannot reliably uncouple contributions from different compartments to the total PET signal; that is, the kinetic rate constants are not identifiable.
CONCLUSION
18F-FGln dynamic PET is a sensitive tool for studying glutamine transport and metabolism in human malignancies. Analysis of dynamic data facilitates better understanding of 18F-FGln pharmacokinetics and may be necessary for assessment of response to targeted therapies that have an impact on intracellular glutamine pool size and tumor glutaminolysis rates.
DISCLOSURE
This research was funded in part by the David Mahoney Neuroimaging Program of the Dana Foundation, the Paul Calabresi Career Development Award for Clinical Oncology (K12 CA184746), the National Cancer Institute (P50 CA086438, R01 CA164490, R01 CA172546, R21 CA167803, and R01 CA204093), and Stand Up to Cancer (grant SU2C-AACRDT0509). MSKCC’s core facilities are supported by an NIH/NCI Cancer Center support grant (P30 CA008748). James Harding has received consulting fees unrelated to the current work from Bristol-Myers Squibb, Eiasi, Eli Lilly, and CytomX and research support from Bristol-Myers Squibb. No other potential conflict of interest relevant to this article was reported.
KEY POINTS
QUESTION: Is 18F-FGln suitable as a PET radiotracer for imaging tumor glutamine flux and metabolism?
PERTINENT FINDINGS: Analysis of dynamic 18F-FGln PET data facilitated better understanding of the heterogeneous uptake patterns of 18F-FGln, because of uncoupling of the total PET signal into contributions from, first, perfusion, vascular permeability, and ASCT2-mediated intracellular transport and, second, the rate-limiting step of glutaminolysis that is catalyzed by glutaminase.
IMPLICATIONS FOR PATIENT CARE: Incorporation of pharmacokinetic modeling of dynamic 18F-FGln PET may be necessary for assessment of response to targeted therapies that have an impact on intracellular glutamine pool size and tumor glutaminolysis rates.
Supplementary Material
Acknowledgments
We thank Leah R. Bassity for editorial comments on the manuscript.
REFERENCES
- 1.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wise DR, DeBerardinis RJ, Mancuso A, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA. 2008;105:18782–18787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16:619–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Pavlova NN, Thompson CB. Cancer cell metabolism: the essential role of the nonessential amino acid, glutamine. EMBO J. 2017;36:1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang L, Venneti S, Nagrath D. Glutaminolysis: a hallmark of cancer metabolism. Annu Rev Biomed Eng. 2017;19:163–194. [DOI] [PubMed] [Google Scholar]
- 6.Zhou R, Pantel AR, Li S, et al. [18F](2S,4R)4-fluoroglutamine PET detects glutamine pool size changes in triple-negative breast cancer in response to glutaminase inhibition. Cancer Res. 2017;77:1476–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu W, Zha Z, Ploessl K, et al. Synthesis of optically pure 4-fluoro-glutamines as potential metabolic imaging agents for tumors. J Am Chem Soc. 2011;133:1122–1133. [DOI] [PubMed] [Google Scholar]
- 8.Lieberman BP, Ploessl K, Wang L, et al. PET imaging of glutaminolysis in tumors by 18F-(2S,4R)4-fluoroglutamine. J Nucl Med. 2011;52:1947–1955. [DOI] [PubMed] [Google Scholar]
- 9.Rajagopalan KN, DeBerardinis RJ. Role of glutamine in cancer: therapeutic and imaging implications. J Nucl Med. 2011;52:1005–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao P, Tchernyshyov I, Chang TC, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LoPiccolo J, Blumenthal GM, Bernstein WB, Dennis PA. Targeting the PI3K/Akt/mTOR pathway: effective combinations and clinical considerations. Drug Resist Updat. 2008;11:32–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang C, Sudderth J, Dang T, Bachoo RM, McDonald JG, DeBerardinis RJ. Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 2009;69:7986–7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fink JR, Muzi M, Peck M, Krohn KA. Multimodality brain tumor imaging: MR imaging, PET, and PET/MR imaging. J Nucl Med. 2015;56:1554–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venneti S, Dunphy MP, Zhang H, et al. Glutamine-based PET imaging facilitates enhanced metabolic evaluation of gliomas in vivo. Sci Transl Med. 2015;7:274ra17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunphy MPS, Harding JJ, Venneti S, et al. In vivo PET assay of tumor glutamine flux and metabolism: in-human trial of 18F-(2S,4R)-4-fluoroglutamine. Radiology. 2018;287:667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross MI, Demo SD, Dennison JB, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther. 2014;13:890–901. [DOI] [PubMed] [Google Scholar]
- 18.Cooper AJ, Krasnikov BF, Pinto JT, Kung HF, Ploessl K. Comparative enzymology of (2S,4R)4-fluoroglutamine and (2S,4R)4-fluoroglutamate. Comp Biochem Physiol B Biochem Mol Biol. 2012;163:108–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tardito S, Oudin A, Ahmed SU, et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat Cell Biol. 2015;17:1556–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Logan J, Fowler JS, Ding YS, et al. Strategy for the formation of parametric images under conditions of low injected radioactivity applied to PET studies with the irreversible monoamine oxidase A tracers [11C]clorgyline and deuterium-substituted [11C]clorgyline. J Cereb Blood Flow Metab. 2002;22:1367–1376. [DOI] [PubMed] [Google Scholar]
- 21.Ploessl K, Wang L, Lieberman BP, Qu W, Kung HF. Comparative evaluation of 18F-labeled glutamic acid and glutamine as tumor metabolic imaging agents. J Nucl Med. 2012;53:1616–1624. [DOI] [PubMed] [Google Scholar]
- 22.Hassanein M, Hight MR, Buck JR, et al. Preclinical evaluation of 4-[18F]fluoroglutamine PET to assess ASCT2 expression in lung cancer. Mol Imaging Biol. 2016;18:18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X, Zhu H, Liu F, et al. Imaging brain metastasis patients with 18F-(2S,4R)-4-fluoroglutamine. Clin Nucl Med. 2018;43:e392–e399. [DOI] [PubMed] [Google Scholar]
- 24.Jin L, Chun J, Pan C, et al. The PLAG1-GDH1 axis promotes anoikis resistance and tumor metastasis through CamKK2-AMPK signaling in LKB1-deficient lung cancer. Mol Cell. 2018;69:87–99.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodrigues MF, Obre E, de Melo FH, et al. Enhanced OXPHOS, glutaminolysis and β-oxidation constitute the metastatic phenotype of melanoma cells. Biochem J. 2016;473:703–715. [DOI] [PubMed] [Google Scholar]
- 26.Bröer A, Fairweather S, Bröer S. Disruption of amino acid homeostasis by novel ASCT2 inhibitors involves multiple targets. Front Pharmacol. 2018;9:785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoerner CR, Chen VJ, Fan AC. The ‘Achilles heel’ of metabolism in renal cell carcinoma: glutaminase inhibition as a rational treatment strategy. Kidney Cancer. 2019;3:15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Csibi A, Fendt SM, Li C, et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013;153:840–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vajaitu C, Draghici CC, Solomon I, et al. The central role of inflammation associated with checkpoint inhibitor treatments. J Immunol Res. 2018;2018:4625472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albina JE, Henry W, King PA, et al. Glutamine metabolism in rat skeletal muscle wounded with lambda-carrageenan. Am J Physiol. 1987;252:E49–E56. [DOI] [PubMed] [Google Scholar]
- 31.Grkovski M, Schwartz J, Gonen M, et al. Feasibility of 18F-fluoromisonidazole kinetic modeling in head and neck cancer using shortened acquisition times. J Nucl Med. 2016;57:334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.