Abstract
A prospective single-arm clinical trial was conducted to determine whether 18F-choline PET/mpMRI can improve the specificity of multiparametric MRI (mpMRI) of the prostate for Gleason ≥ 3+4 prostate cancer. Methods: Before targeted and systematic prostate biopsy, mpMRI and 18F-choline PET/CT were performed on 56 evaluable subjects with 90 Likert score 3–5 mpMRI target lesions, using a 18F-choline target-to-background ratio of greater than 1.58 to indicate a positive 18F-choline result. Prostate biopsies were performed after registration of real-time transrectal ultrasound with T2-weighted MRI. A mixed-effects logistic regression was applied to measure the performance of mpMRI (based on prospective Likert and retrospective Prostate Imaging Reporting and Data System, version 2 [PI-RADS], scores) compared with 18F-choline PET/mpMRI to detect Gleason ≥ 3+4 cancer. Results: The per-lesion accuracy of systematic plus targeted biopsy for mpMRI alone was 67.8% (area under receiver-operating-characteristic curve [AUC], 0.73) for Likert 4–5 and 70.0% (AUC, 0.76) for PI-RADS 3–5. Several PET/MRI models incorporating 18F-choline with mpMRI data were investigated. The most promising model selected all high-risk disease on mpMRI (Likert 5 or PI-RADS 5) plus low- and intermediate-risk disease (Likert 4 or PI-RADS 3–4), with an elevated 18F-choline target-to-background ratio greater than 1.58 as positive for significant cancer. Using this approach, the accuracy on a per-lesion basis significantly improved to 88.9% for Likert (AUC, 0.90; P < 0.001) and 91.1% for PI-RADS (AUC, 0.92; P < 0.001). On a per-patient basis, the accuracy improved to 92.9% for Likert (AUC, 0.93; P < 0.001) and to 91.1% for PI-RADS (AUC, 0.91; P = 0.009). Conclusion: 18F-choline PET/mpMRI improved the identification of Gleason ≥ 3+4 prostate cancer compared with mpMRI, with the principal effect being improved risk stratification of intermediate-risk mpMRI lesions.
Keywords: 18F-fluoromethylcholine, PET/MRI, prostate cancer, targeted prostate biopsy, interrater agreement
Multiparametric MRI (mpMRI) has emerged as a useful tool for the detection and risk stratification of primary prostate cancer that can cost-effectively identify a significantly greater fraction of clinically important cancers (Gleason ≥ 3+4) than standard biopsy alone while minimizing detection of low-risk cancer (1,2). These outcomes are important because accurate risk stratification of primary prostate cancer has been fraught with overdiagnosis and overtreatment (3,4). The inadequacies and harms of screening and the indolence of most prostate cancer led the U.S. Preventive Services Task Force to give prostate-specific antigen testing a C rating (i.e., do not use except in individual circumstances) for men aged 55–69 y (5).
mpMRI improves prostate cancer screening by better selecting which patients need biopsy and subsequent treatment, but it is limited by a large false-positive fraction (6,7), only moderate interrater agreement (8), and a steep learning curve (9), each contributing to unnecessary biopsies that drive complication rates and unwanted detection of low-risk disease. Efforts to standardize performance and interpretation of mpMRI have resulted in the Prostate Imaging Reporting and Data System (PI-RADS), which is now in version 2 (10). Though the system effectively stratifies risk of Gleason ≥ 3+4 cancer, the positive predictive values for some commonly biopsied lesions are low (e.g., PI-RADS 3, ∼15%, and PI-RADS 4, ∼45%) (6,11–13). Additionally, it has been reported that the majority (60%–70%) of mpMRI examinations contain at least 1 lesion assigned a PI-RADS score of 3 or higher (7), indicating that most patients undergoing mpMRI also eventually will likely undergo a targeted biopsy.
Hybrid PET/mpMRI is a promising option to improve identification of primary prostate cancer by better stratifying low- and intermediate-risk mpMRI lesions. Such results were suggested in the interim analysis of this trial, which demonstrated that the addition of PET with 18F-choline to mpMRI significantly improved the identification of Gleason ≥ 3+4 prostate cancer over mpMRI alone (14). We now present the final analysis of this trial testing specific PET/mpMRI scenarios to optimize risk stratification before prostate biopsy.
MATERIALS AND METHODS
Patient Population
This prospective single-arm open-label observational diagnostic clinical trial was registered at clinicaltrials.gov (NCT01751737) and approved by the institutional review board. Informed consent was obtained from all subjects. The study population comprised 63 subjects recruited between March 2013 and June 2016 with rising prostate-specific antigen values, suspected or known untreated localized adenocarcinoma of the prostate (Gleason 3+3 or 3+4), and at least 1 low-, intermediate-, or high-risk mpMRI lesion scheduled to undergo prostate biopsy. Five subjects were not evaluable because of failure of radiotracer synthesis (n = 2) or declining prostate biopsy after imaging (n = 3), and 2 were screening failures. The exclusion criteria are listed in a previous publication (14).
The characteristics of 56 evaluable subjects with 90 lesions are summarized in Supplemental Table 1 (supplemental materials are available at http://jnm.snmjournals.org). The interim analysis (36 subjects, 52 lesions) provided the technical details of PET/mpMRI and established the 18F-choline target-to-background ratio (TBR) threshold used in our analysis (14). In this report, we detail the diagnostic accuracy of 18F-choline PET/mpMRI on the complete study population in specific diagnostic scenarios.
Forty-seven subjects underwent at least 1 prior prostate biopsy procedure. Twenty-four had benign findings, 20 showed Gleason 3+3 cancer, and 3 showed Gleason 3+4 cancer. Nine subjects had no prior biopsy. Although the number of prior biopsies ranged from 0 to 6, most subjects underwent 1 (n = 27) or 2 (n = 13) prior procedures (Supplemental Table 1).
Prostate mpMRI and Target Selection for Biopsy
All MRI pelvis imaging was performed on the same 3-T MR unit (Ingenia; Philips Healthcare) without an endorectal coil using a 16-channel phased-array surface coil (14). Briefly, MR sequences included axial 2-dimensional and 3-dimensional (3D) T2-weighted fast spin echo imaging, axial diffusion-weighted imaging, and axial T1-weighted unenhanced and dynamic contrast-enhanced imaging. An apparent diffusion coefficient map was reconstructed.
Images were reviewed and classified prospectively on the basis of a 5-point Likert scale by 1 fellowship-trained expert genitourinary radiologist (reader 1). At the time the study was designed, no widely accepted scoring system for prostate mpMRI interpretation existed. The selected Likert scale was derived from references and clinical experience (1,15). Scores of 1 (negative) and 2 (likely benign) were not targeted for biopsy. Risk scores of 3 (low), 4 (intermediate), and 5 (high) were targeted for biopsy. The reader had full knowledge of the clinical history, including prior imaging and pathology results, but did not know the results from 18F-choline PET/CT. After identification of candidate lesions (Likert 3–5), individual target volumes of interest were segmented on the basis of visual perception of the lesion borders (14).
Because the trial began before release of the PI-RADS technical parameters (2015), high b-value imaging (b ≥ 1,400 s/mm2) and prospective PI-RADS scoring were not used. Of note, Likert scales have been shown to have diagnostic performance similar to the PI-RADS schema (1,15). We added a retrospective analysis of mpMRI data using PI-RADS scoring performed by the same radiologist with knowledge of clinical data before biopsy but masked to 18F-choline results. A second fellowship-trained expert genitourinary radiologist (reader 2) retrospectively assessed the mpMRI examinations using Likert and PI-RADS scales to determine interreader variability and result consistency across readers. Interreader agreement was based on whether a given score would result in a recommendation for targeted biopsy. A biopsy was recommended if a Likert 4–5 or PI-RADS 3–5 lesion was found.
18F-Choline PET/CT
18F-fluoromethylcholine (18F-choline) was synthesized under good-manufacturing-practice conditions (16). mpMRI and 18F-choline PET/CT were acquired on separate days for 27 subjects (range, 1–70 d) and on the same day for 29 subjects. PET/CT was performed on a Biograph TrueV mCT scanner (Siemens). After CT transmission scanning without intravenous contrast medium, a 10-min emission scan of the lower abdomen and pelvis was performed 20 min after injection of 230 ± 31 MBq of 18F-choline (continued table motion speed, 0.4 mm/s). Data were corrected for decay, scatter, and attenuation. Image reconstruction involved an iterative ordered-subset expectation maximization algorithm (14).
Image Registration Tasks
Registration of PET/CT images onto T2-weighted MR images was achieved using commercially available software (MIM Maestro; MIM Software Inc.) by rigidly aligning the pelvic bones on both modalities. In the few cases in which the position of the prostate differed because of variable rectal content or bladder filling, constrained intensity-based, free-form deformable registration was applied (17). As shown in Figure 1, mpMRI-defined biopsy target volumes of interest were mirrored to the contralateral side of the prostate to create a background volume of interest for subsequent data analysis (14).
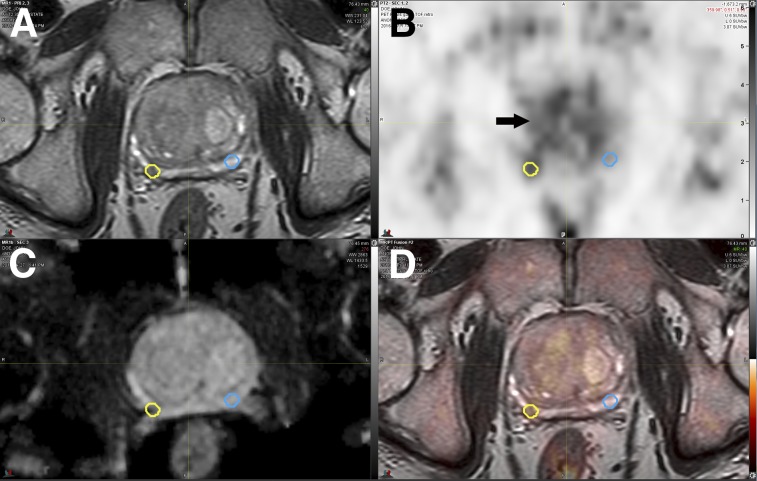
FIGURE 1.
Coregistered axial 3D T2-weighted fast-spin echo MRI (A), 18F-choline PET (SUV range, 0–6) (B), apparent diffusion coefficient map (C), and PET/mpMRI (D). Small 0.16 cm3 right peripheral zone intermediate-risk lesion (yellow circles) was confirmed as Gleason 4+4 cancer. Respective mirrored background volume of interest (blue circles) is shown, resulting in 18F-choline TBR of 1.61. Diffusely increased 18F-choline uptake is noted in benign prostate hyperplasia (arrow).
Transrectal Biopsy Procedure
The biopsy procedure was performed on average 17.8 ± 14.9 d (mean ± SD) after PET or mpMRI using a navigated 3D ultrasound system (Logiq E9; GE Healthcare). After the ultrasound transducer was inserted into the rectum, a rigid registration of transrectal ultrasound and 3D T2-weighted MRI with embedded target information was performed (14). Using an 18-gauge biopsy needle, targeted and standard (systematic 12-core) prostate biopsy samples were obtained.
Pathologic Evaluation
Each tumor focus identified on 3-μm hematoxylin- and eosin-stained sections was assigned a primary and secondary Gleason grade. The final pathologic determination was defined as the highest Gleason score of any cancer detected (per side) from any tissue (prior reviewed biopsy, standard and targeted biopsy, or from the prostatectomy specimen as a reference standard). Clinically significant prostate cancer was defined conservatively as any Gleason ≥ 3+4 cancer (18).
Registration of Histopathologic Findings to In Vivo PET/mpMRI
We used a validated multistep process to register the histopathologic findings to in vivo 3D T2-weighted MR images (19,20). Briefly, the prostate specimen underwent a 3-T ex vivo MRI scan for accurate segmentation of the prostate (Vitrea; Vital Images) to allow subsequent 3D printing of a plastic mold (Dimension Elite 3D; Stratasys). The specimen was placed into the mold for a second high-resolution MRI scan. Then, the prostate was cross-sectioned within the mold at 3-mm intervals. Hematoxylin- and eosin-stained whole-mount histologic sections of all prostate cancer foci were then registered to ex vivo MR images to allow a consistent registration of the two (20).
Data Analysis
From mpMRI-defined target and mirrored background volumes of interest, we calculated the lesion 18F-choline SUVmean and its TBR (Fig. 1). An mpMRI-only base-case (for Likert or PI-RADS) was compared with 8 respective synthetic PET/mpMRI combinations. The 18F-choline TBR (>1.58) indicating a positive 18F-choline result was derived from a retrospective analysis of 36 subjects (14).
Likert Score–Based
mpMRI-only base-case (Likert 4–5): IF high-risk (Likert 5) lesion on mpMRI OR intermediate-risk lesion (Likert 4) on mpMRI THEN biopsy ELSE no biopsy,
Model L1) IF Likert 5 OR (Likert 3–4 with SUVmean TBR > 1.58) THEN biopsy ELSE no biopsy,
Model L2) IF Likert 5 OR (Likert 4 with SUVmean TBR > 1.58) THEN biopsy ELSE no biopsy.
PI-RADS Score–Based
mpMRI-only base-case (PI-RADS scores 3–5): IF PI-RADS score > 2 THEN biopsy ELSE no biopsy,
Model P1) IF high-risk lesion on mpMRI (PI-RADS 5) OR (PI-RADS 3–4 with SUVmean TBR > 1.58) THEN biopsy ELSE no biopsy,
Model P2) IF PI-RADS 5 OR (PI-RADS 4 with SUVmean TBR > 1.58) THEN biopsy ELSE no biopsy.
Additional models were investigated classifying mpMRI-identified lesions based on 18F-choline uptake (SUVmean TBR > 1.58; models L3 and P3) or selecting only intermediate- and high-risk lesions (Likert 4–5 or PI-RADS 4–5) with a 18F-choline SUVmean TBR greater than 1.58 (models L4 and P4).
Statistics
Data are expressed as mean ± SD. For subject-based biopsy results, we compared the continuity-corrected odds ratio of targeted biopsy to final pathology versus the ratio of standard biopsy to final pathology. Detection of Gleason ≥ 3+4 cancer was the primary outcome. A permutation-based test was used to address repeated measures (targeted plus standard biopsies) within each subject. We compared the observed difference in odds ratio between targeted biopsy and standard biopsy with the difference in permuted data, which were generated by shuffling the standard biopsy and targeted biopsy labels repeated 10,000 times to construct an approximate permutation distribution under the null hypothesis of nondifference.
To account for multiple lesions within each subject, a mixed-effects logistic regression model was used to assess the value of PET/mpMRI models. A random intercept was included to account for the individual variability of lesion-level risk. The area under the receiver-operating-characteristic curve (AUC) was estimated by leave-one-out cross-validation to measure the diagnostic performance of PET/mpMRI combinations. Leave-one-out cross-validation was used to minimize the risk of statistically overfitting the data to the TBR threshold. To assess the additional value of 18F-choline PET to mpMRI, we compared the AUC of the regression models obtained from synthetic PET/mpMRI combinations with that of an mpMRI-only model (i.e., base-case model). We conducted contingency table and AUC analyses to assess the performance of synthetic PET/mpMRI combinations. Interreader agreement statistics were assessed by the Krippendorff α for the Likert and PI-RADS scores and by the Cohen κ for the binary variables.
RESULTS
Biopsy Results
In total, 224 targeted and 672 standard biopsy cores were obtained. On average, 2.4 ± 1.6 biopsy cores were obtained per target. Prostate biopsies performed within this trial identified 27 Gleason ≥ 3+4 cancers in 24 of 56 subjects.
Targeted biopsies detected significantly more Gleason ≥ 3+4 prostate cancers (23 of 27; odds ratio, 241.4) than did nontargeted standard biopsies (12 of 27; odds ratio, 47.6; P = 0.0037). Gleason ≥ 3+4 prostate cancer was found at 15 targeted biopsies alone, 4 standard biopsies alone, and 8 with both biopsy methods. The number of low-risk cancers (Gleason 3+3) found at standard biopsy did not significantly differ from that found at targeted biopsy (P = 0.706).
Comparison of Gleason ≥ 3+4 Cancer Detection Rates
In total, 90 lesions were prospectively identified by mpMRI (40 low-risk [Likert 3], 30 intermediate-risk [Likert 4], and 20 high-risk [Likert 5]). Target volumes classified as Likert 5 or PI-RADS 5 were significantly larger than Likert 3–4 or PI-RADS 2–4 lesions; also, lesions positive for Gleason ≥ 3+4 cancer were significantly larger than those negative for Gleason ≥ 3+4 cancer (Supplemental Table 2). However, the target volume of low- and intermediate-risk lesions was similar regardless of whether they were positive (Likert, 0.36 ± 0.21; PI-RADS, 0.42 ± 0.35) or negative (Likert, 0.34 ± 0.35; PI-RADS, 0.34 ± 0.34) for Gleason ≥ 3+4 cancer.
Most Gleason ≥ 3+4 cancer targets were either of small volume at mpMRI (≤0.25 cm3; n = 4; example in Fig. 1) or located in areas of the prostate typically undersampled by standard (12-core) biopsy (e.g., anterior gland [n = 18] or apical [n = 7]). The positive predictive value for Gleason ≥ 3+4 cancer was 10% (4/40) for low-risk lesions, 27% (8/30) for intermediate-risk lesions, and 85% (17/20) for high-risk lesions. Targeting intermediate- and high-risk mpMRI lesions (n = 50) resulted in 25 true-positives, 36 true-negatives, 25 false-positives, and 4 false-negatives (Supplemental Table 3A). Targeting only high-risk mpMRI lesions (n = 20) resulted in 17 true-positives, 58 true-negatives, 3 false-positives, and 12 false-negatives. mpMRI was a strong lesion-level predictor of Gleason ≥ 3+4 cancer in the base-case model for both readers (Supplemental Tables 3A and 3B).
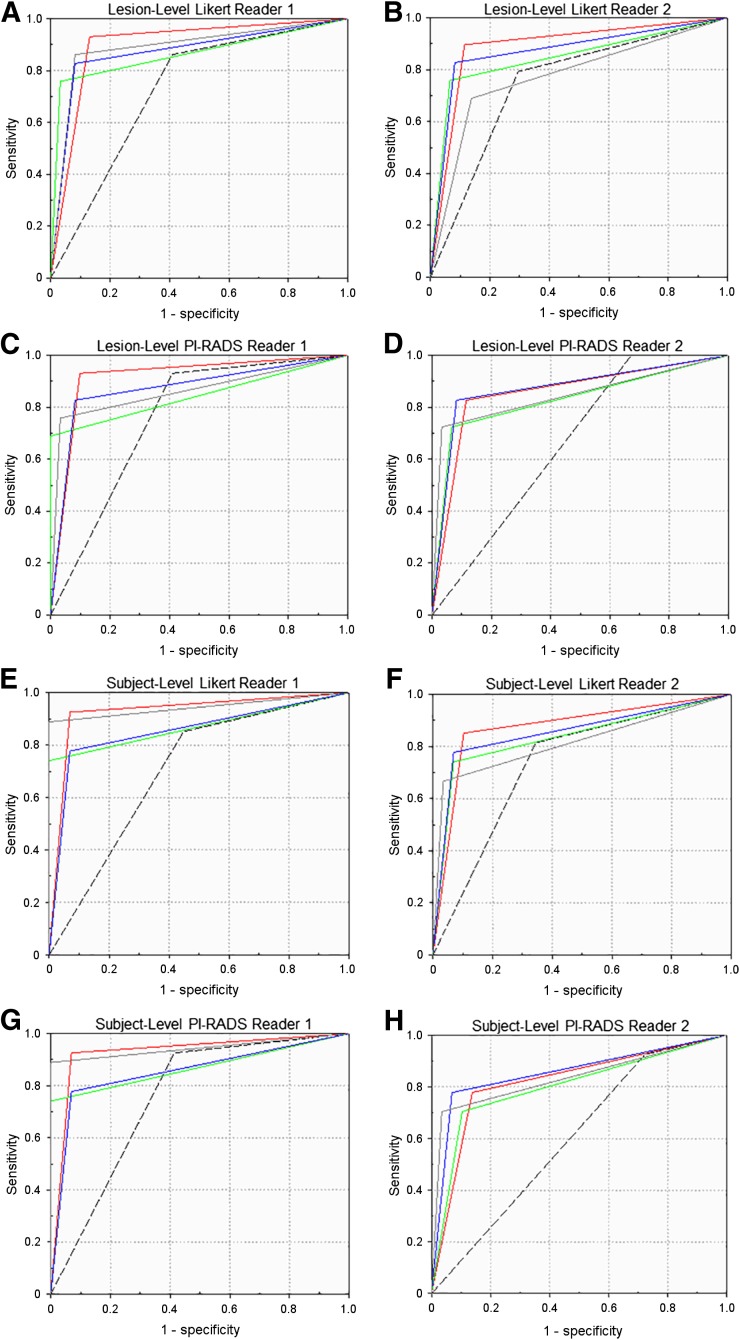
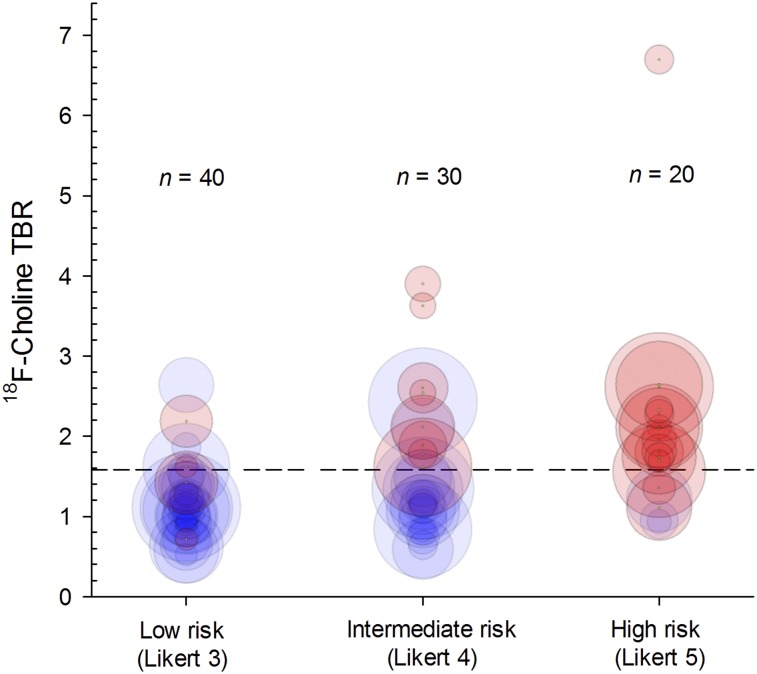
18F-choline SUVmean was not a significant predictor of Gleason ≥ 3+4 cancer (P = 0.81) (Supplemental Tables 2A–2C). Regardless of the scoring system used, the 18F-choline TBR threshold (>1.58; models L3 and P3) significantly improved the diagnostic accuracy over the mpMRI base-case model on a per-lesion and per-patient basis. In fact, for both readers, all evaluated PET/mpMRI models (Likert L1–L4, PI-RADS P1–P4) significantly outperformed their respective mpMRI-only base-case models for the identification of Gleason ≥ 3+4 prostate cancer on a per-lesion basis (Figs. 2A–2D, Supplemental Tables 3A and 3B). On a per-patient basis, the most promising models were L1 (AUC range, 0.87–0.93) and P1 (AUC range, 0.82–0.91), for which all high-risk lesions based on MRI and low- and intermediate risk lesions with increased 18F-choline uptake are selected (Figs. 2E–2H, Supplemental Tables 4A and 4B). The primary effect was an improvement in specificity and positive predictive value. For both readers, the positive predictive value of all PET/mpMRI evaluated models was significantly higher than the respective base-case on a per-lesion and per-patient basis (Supplemental Tables 3 and 4). Figure 3 highlights the relationship between 18F-choline TBR, target volume, and final histologic result stratified by prospective Likert results (reader 1).
FIGURE 2.
Receiver-operating-characteristic curves of models L1/P1 (red line), L2/P2 (green line), L3/P3 (blue line), and L4/P4 (gray line) relative to mpMRI base-case (dashed line) for lesion-based and subject-based data, separate for readers 1 and 2 as well as Likert and PI-RADS scoring systems.
FIGURE 3.
Bubble plot of mpMRI-identified lesions stratified by 18F-choline PET TBR and presence (red circles) or absence (blue circles) of Gleason ≥ 3+4 cancer on per-lesion basis. Circle diameter represents target volume (Supplemental Table 2). Dashed line indicates 18F-choline TBR threshold of 1.58.
Interreader Agreement
Supplemental Tables 5A–5D show the Likert or PI-RADS scores of both readers on a lesion or subject basis relative to their respective final histologic result (positive or negative for Gleason ≥ 3+4). Likert and PI-RADS scores had moderate interrater agreement (α) on a per-lesion (Likert, 0.578; PI-RADS, 0.452) and per-patient (Likert, 0.582, PI-RADS, 0.439) basis. Agreement (κ) for the decision to biopsy was moderate for Likert scores (per-lesion, 0.494; per-patient, 0.521) and fair for PI-RADS (per-lesion, 0.222; per-patient, 0.290). Greater disagreement for PI-RADS was related mainly to a larger number of low-risk lesions rated as PI-RADS 3 (instead of PI-RADS 2) by reader 2.
Postbiopsy Management
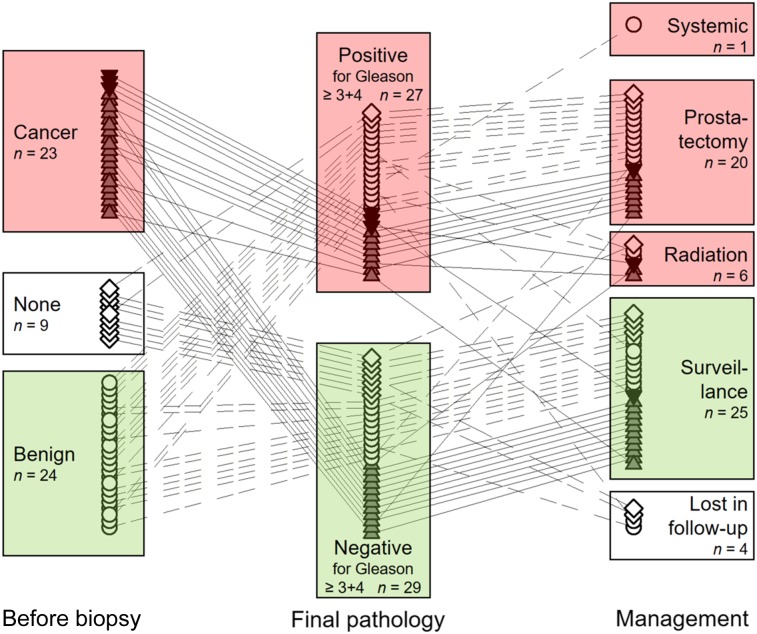
The relationship among diagnosis before enrollment, final pathology, and clinical management after biopsy is shown in Figure 4. Twenty-seven subjects underwent postbiopsy therapy (prostatectomy [n = 20], radiation [n = 6], or systemic treatment [n = 1]). On a per-patient basis, the histologic results obtained from prostatectomy did not change the final outcome assessment (Gleason ≥ 3+4 or not) in all but 1 patient—a patient for whom a known Gleason 3+3 cancer was upgraded to Gleason 3+4.
FIGURE 4.
For 56 evaluable subjects, relationship of diagnosis before study enrollment to final pathology and subsequent clinical management. Connecting lines (plain dashed: benign; solid: cancer; long-short dashed: no prior biopsy) indicate management decisions related to final pathology. ▲ = Gleason 3+3 cancer; ▼ = Gleason 3+4 cancer.
Four outcome changes were seen on a per-lesion basis in subjects undergoing prostatectomy for confirmed Gleason ≥ 3+4 cancer identified on template biopsy. We identified 2 mistargeted biopsies of Gleason ≥ 3+4 lesions in 2 subjects (included in Supplemental Tables 3A and 3B), and 2 unidentified Gleason ≥ 3+4 cancer lesions in 2 additional subjects (not in Supplemental Tables 3A and 3B).
Two subjects underwent prostatectomy for Gleason 3+3 cancer (both of low risk at mpMRI and without elevated 18F-choline TBR); Gleason 3+3 cancer was confirmed in both at prostatectomy. Five subjects with Gleason ≥ 3+4 cancer on biopsy had their final Gleason score modified at prostatectomy (upgrade, n = 3; downgrade, n = 2; both within the Gleason ≥ 3+4 group).
Of the 29 subjects who did not undergo postbiopsy therapy, 25 were followed an average of 28.5 ± 10.8 mo (median, 27.5 mo; range, 12–51 mo) and 4 were lost in follow-up. Eighteen subjects underwent repeat mpMRI, and 14 subjects underwent repeat combined biopsy. None were newly diagnosed with Gleason ≥ 3+4 prostate cancer, and the mpMRI risk scores in the reimaged subjects remained unchanged.
DISCUSSION
mpMRI has transformed the diagnosis of prostate cancer by enabling improved identification of clinically significant disease and avoidance of indolent disease. However, mpMRI suffers from high false-positive rates for low- and intermediate-risk lesions that lead to downstream costs and unnecessary biopsies. We found that when 18F-choline data were used to augment risk stratification of mpMRI-identified lesions, the diagnostic accuracy of imaging for the detection of Gleason ≥ 3+4 prostate cancer significantly improved. The main effect was an improvement in specificity and positive predictive value for mpMRI-designated intermediate-risk lesions.
There are various strategies that can be considered in a hypothetical future state that uses PET/mpMRI for the detection of primary prostate cancer. These include simultaneous PET/mpMRI in all subjects (using a variety of decision rules to indicate positive and negative results; for example, models L1, L3, and L4; lesion-level AUC: Likert 0.83–0.90, PI-RADS 0.83–0.92) and sequential PET/mpMRI, in which PET is performed only to better stratify intermediate-risk lesions (e.g., lesion-level model L2 AUC: Likert 0.85–0.86, PI-RADS 0.83–0.84). On a patient basis, the most promising PET/mpMRI models, L1 and P1, were more accurate for both readers for the prediction of Gleason ≥ 3+4 prostate cancer than their respective Likert or PI-RADS mpMRI-only base-case scenarios. Our MRI base-case performance data (AUC: Likert 0.70–0.75, PI-RADS 0.60–0.76) are in the expected range for mpMRI, particularly for studies that use whole-gland pathology as the reference standard (1,21).
For example, in the PROMIS trial (6), which was a prospective paired validating confirmatory study of mpMRI in 576 men using template mapping biopsy as a reference standard, the sensitivity (93%) and specificity (41%) of mpMRI were similar to the diagnostic accuracy of mpMRI we observed. Because both the PROMIS trial and our trial were initiated before PI-RADS was released, both used Likert scoring. Likewise, high b-value imaging was not yet part of routine mpMRI and was not used. To our knowledge, there is no high-level evidence indicating that high b-value imaging is necessary, but our results might have been different had this been incorporated. When our data were retrospectively evaluated using PI-RADS criteria, the results were similar to the Likert data—a finding that has been shown previously for PI-RADS version 1 (15).
We found moderate interrater agreement for Likert and PI-RADS scoring. The performance for distinguishing low- from intermediate- or high-grade disease was similar between readers for Likert scores but was lower for PI-RADS. These results are within the range of other PI-RADS agreement studies (11,22,23). The difficulties in accurately characterizing low- and intermediate-risk lesions on the basis of PI-RADS criteria underscore known limitations of mpMRI and highlight the benefit we observed from adding 18F-choline for disease stratification in this trial.
The success of newer prostate-specific membrane antigen (PSMA)–based radioligands in biochemically recurrent and metastatic prostate cancer suggests a role for the identification of primary prostate cancer (24,25). Although the overall immunohistochemical PSMA expression of primary prostate cancer increases with aggressiveness (grading), PSMA expression is also present in low-grade prostate cancer (26). Such uptake, as seen with 68Ga-PSMA-11 (27–29), is not desired. Hicks et al. studied 68Ga-PSMA PET/MRI before prostatectomy for mostly high-risk prostate cancer and found that low-grade (Gleason 3+3) cancers might be differentiated from intermediate- or high-grade tumors by absolute 68Ga-PSMA uptake (SUV), but there was overlap between the groups (30). Similar overlap has also been seen in other work with choline radiotracers (14,31).
Our study has limitations. The results could be biased by unequal target volumes across MRI risk groups or pathology. Partial-volume effects limit quantification of the 18F-choline uptake of small lesions. Lesion volumes classified as high-risk by MRI were larger than those of other risk groups. However, and more important for the results of this study, the target volumes of low- and intermediate-risk groups were similar irrespective of the final pathology (positive or negative for Gleason ≥ 3+4). This is relevant because the gain in specificity based on 18F-choline noted in the favored PET/MRI models L1 and P1 resulted exclusively from the reclassification of low- and intermediate-risk lesions. Partial-volume effects are therefore unlikely to have influenced the results of this study.
All subjects enrolled in our trial had an mpMRI-identified low-, intermediate-, or high-risk lesion. This was intentional to test whether 18F-choline PET/mpMRI can improve the specificity of mpMRI for Gleason ≥ 3+4 prostate cancer. In this context, PET was being used to modify the positive predictive value of mpMRI-identified lesions rather than to identify lesions that mpMRI had missed. mpMRI has excellent sensitivity but only moderate specificity for clinically important prostate cancer (6). In addition, 18F-choline PET is known to have common false-positive findings in the prostate such as benign prostatic hyperplasia (20,32). Starting with mpMRI-identified lesions allowed us to mitigate this weakness. Therefore, we are not able to evaluate the negative predictive value of PET/mpMRI or to determine the significance of mpMRI-invisible lesions that might be positive on 18F-choline PET. The TBR threshold we used to indicate a positive result on 18F-choline PET was established in a previous study (14). To minimize the risk of statistical overfitting, we used a leave-one-out cross-validation procedure. Although our data show significant improvements in diagnostic accuracy for multiple 18F-choline PET/mpMRI models compared with an mpMRI-only base-case, our study was underpowered for subgroup analysis. For that, a larger independent study would be needed.
In sum our data indicate that in the future patients with elevated prostate cancer risk might undergo 18F-choline PET/mpMRI rather than mpMRI alone, followed by confirmatory histologic sampling only if PET/mpMRI suggests the presence of significant disease. This diagnostic approach not only improves detection of clinically significant prostate cancer and decreases the number of unnecessary prostate biopsy procedures but also, as will be demonstrated in part 2 of this article, is cost-effective (33).
CONCLUSION
The study results demonstrate that 18F-choline PET/mpMRI improves the identification of Gleason ≥ 3+4 prostate cancer compared with mpMRI alone, mainly due to superior risk stratification of intermediate-risk mpMRI lesions. A multisite trial is needed to confirm these findings.
DISCLOSURE
This work was supported by grants from the U.S. Department of Defense (PC110389) and NIH/NCI (P01CA87634 and P50CA069568). No other potential conflict of interest relevant to this article was reported.
KEY POINTS
QUESTION: What is the value of 18F-choline PET/mpMRI for the identification of significant prostate cancer?
PERTINENT FINDINGS: 18F-choline PET/mpMRI improved the identification of Gleason ≥ 3+4 prostate cancer compared with mpMRI alone, with the principal effect being improved risk stratification of intermediate-risk mpMRI lesions.
IMPLICATIONS FOR PATIENT CARE: In a hypothetical future state, patients with elevated prostate cancer risk might undergo 18F-choline PET/mpMRI rather than mpMRI alone, followed by confirmatory histologic sampling only if PET/mpMRI suggests the presence of significant disease.
Supplementary Material
Acknowledgments
We thank the many dedicated individuals of the radiology staff and Shawn O’Grady from the University of Michigan 3D Lab for their excellent support.
REFERENCES
- 1.Siddiqui MM, Rais-Bahrami S, Turkbey B, et al. Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA. 2015;313:390–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett CL, Davenport MS, Montgomery JS, Wei JT, Montie JE, Denton BT. Cost-effectiveness of magnetic resonance imaging and targeted fusion biopsy for early detection of prostate cancer. BJU Int. 2018;122:50–58. [DOI] [PubMed] [Google Scholar]
- 3.Rosenkrantz AB, Taneja SS. Prostate MRI can reduce overdiagnosis and overtreatment of prostate cancer. Acad Radiol. 2015;22:1000–1006. [DOI] [PubMed] [Google Scholar]
- 4.Aizer AA, Gu X, Chen MH, et al. Cost implications and complications of overtreatment of low-risk prostate cancer in the United States. J Natl Compr Canc Netw. 2015;13:61–68. [DOI] [PubMed] [Google Scholar]
- 5.Prostate cancer: screening. U.S. Preventive Services Task Force website. https://www.uspreventiveservicestaskforce.org/Page/Document/UpdateSummaryFinal/prostate-cancer-screening1. Published May 2018. Accessed October 16, 2019.
- 6.Ahmed HU, El-Shater Bosaily A, Brown LC, et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet. 2017;389:815–822. [DOI] [PubMed] [Google Scholar]
- 7.Abd-Alazeez M, Ahmed HU, Arya M, et al. The accuracy of multiparametric MRI in men with negative biopsy and elevated PSA level: can it rule out clinically significant prostate cancer? Urol Oncol. 2014;32:45.e17–45.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller BG, Shih JH, Sankineni S, et al. Prostate cancer: interobserver agreement and accuracy with the revised Prostate Imaging Reporting and Data System at multiparametric MR imaging. Radiology. 2015;277:741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenkrantz AB, Ayoola A, Hoffman D, et al. The learning curve in prostate MRI interpretation: self-directed learning versus continual reader feedback. AJR. 2017;208:W92–W100. [DOI] [PubMed] [Google Scholar]
- 10.Weinreb JC, Barentsz JO, Choyke PL, et al. PI-RADS Prostate Imaging – Reporting and Data System: 2015, version 2. Eur Urol. 2016;69:16–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shankar PR, Kaza RK, Al-Hawary MM, et al. Impact of clinical history on maximum PI-RADS version 2 score: a six-reader 120-case sham history retrospective evaluation. Radiology. 2018;288:158–163. [DOI] [PubMed] [Google Scholar]
- 12.Curci NE, Lane BR, Shankar PR, et al. Integration and diagnostic accuracy of 3T nonendorectal coil prostate magnetic resonance imaging in the context of active surveillance. Urology. 2018;116:137–143. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Tang M, Chen S, Lei X, Zhang X, Huan Y. A meta-analysis of use of Prostate Imaging Reporting and Data System version 2 (PI-RADS V2) with multiparametric MR imaging for the detection of prostate cancer. Eur Radiol. 2017;27:5204–5214. [DOI] [PubMed] [Google Scholar]
- 14.Piert M, Montgomery J, Kunju LP, et al. 18F-choline PET/MRI: the additional value of PET for MRI-guided transrectal prostate biopsies. J Nucl Med. 2016;57:1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenkrantz AB, Kim S, Lim RP, et al. Prostate cancer localization using multiparametric MR imaging: comparison of Prostate Imaging Reporting and Data System (PI-RADS) and Likert scales. Radiology. 2013;269:482–492. [DOI] [PubMed] [Google Scholar]
- 16.Rodnick ME, Brooks AF, Hockley BG, Henderson BD, Scott PJ. A fully-automated one-pot synthesis of [18F]fluoromethylcholine with reduced dimethylaminoethanol contamination via [18F]fluoromethyl tosylate. Appl Radiat Isot. 2013;78:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirby N, Chuang C, Ueda U, Pouliot J. The need for application-based adaptation of deformable image registration. Med Phys. 2013;40:011702. [DOI] [PubMed] [Google Scholar]
- 18.Van der Kwast TH, Roobol MJ. Defining the threshold for significant versus insignificant prostate cancer. Nat Rev Urol. 2013;10:473–482. [DOI] [PubMed] [Google Scholar]
- 19.Meyer C, Ma B, Kunju LP, Davenport M, Piert M. Challenges in accurate registration of 3-D medical imaging and histopathology in primary prostate cancer. Eur J Nucl Med Mol Imaging. 2013;40(suppl 1):S72–S78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piert M, Shankar PR, Montgomery J, et al. Accuracy of tumor segmentation from multi-parametric prostate MRI and 18F-choline PET/CT for focal prostate cancer therapy applications. EJNMMI Res. 2018;8:23–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chamie K, Sonn GA, Finley DS, et al. The role of magnetic resonance imaging in delineating clinically significant prostate cancer. Urology. 2014;83:369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glazer DI, Mayo-Smith WW, Sainani NI, et al. Interreader agreement of Prostate Imaging Reporting and Data System version 2 using an in-bore MRI-guided prostate biopsy cohort: a single institution’s initial experience. AJR. 2017;209:W145–W151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenkrantz AB, Ginocchio LA, Cornfeld D, et al. Interobserver reproducibility of the PI-RADS version 2 lexicon: a multicenter study of six experienced prostate radiologists. Radiology. 2016;280:793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grubmüller B, Baltzer PA, Hartenbach S, et al. PSMA ligand PET/MRI for primary prostate cancer: staging performance and clinical impact. Clin Cancer Res. 2018;24:6300–6307. [DOI] [PubMed] [Google Scholar]
- 25.Park SY, Zacharias C, Harrison C, et al. Gallium 68 PSMA-11 PET/MR imaging in patients with intermediate- or high-risk prostate cancer. Radiology. 2018;288:495–505. [DOI] [PubMed] [Google Scholar]
- 26.Bravaccini S, Puccetti M, Bocchini M, et al. PSMA expression: a potential ally for the pathologist in prostate cancer diagnosis. Sci Rep. 2018;8:4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eiber M, Weirich G, Holzapfel K, et al. Simultaneous Ga-PSMA HBED-CC PET/MRI improves the localization of primary prostate cancer. Eur Urol. 2016;70:829–836. [DOI] [PubMed] [Google Scholar]
- 28.Zamboglou C, Schiller F, Fechter T, et al. 68Ga-HBED-CC-PSMA PET/CT versus histopathology in primary localized prostate cancer: a voxel-wise comparison. Theranostics. 2016;6:1619–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woythal N, Arsenic R, Kempkensteffen C, et al. Immunohistochemical validation of PSMA expression measured by 68Ga-PSMA PET/CT in primary prostate cancer. J Nucl Med. 2018;59:238–243. [DOI] [PubMed] [Google Scholar]
- 30.Hicks RM, Simko JP, Westphalen AC, et al. Diagnostic accuracy of 68Ga-PSMA-11 PET/MRI compared with multiparametric MRI in the detection of prostate cancer. Radiology. 2018;289:730–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piert M, Park H, Khan A, et al. Detection of aggressive primary prostate cancer with 11C-choline PET/CT using multimodality fusion techniques. J Nucl Med. 2009;50:1585–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piert M, El Naqa I, Davenport MS, Incerti E, Mapelli P, Picchio M. PET/MRI and prostate cancer. Clin Transl Imaging. 2016;4:473–485. [Google Scholar]
- 33.Barnett CL, Davenport MS, Montgomery JS, Kunju LP, Denton BT, Piert M. 18F-choline PET/mpMRI for detection of significant prostate cancer: part 2. Cost-effectiveness analysis. J Nucl Med. July 26, 2019 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.