The causative agent of melioidosis, Burkholderia pseudomallei, a tier 1 select agent, is endemic in Southeast Asia and northern Australia, with increased incidence associated with high levels of rainfall. Increasing reports of this condition have occurred worldwide, with estimates of up to 165,000 cases and 89,000 deaths per year. The ecological niche of the organism has yet to be clearly defined, although the organism is associated with soil and water. The culture of appropriate clinical material remains the mainstay of laboratory diagnosis.
KEYWORDS: Burkholderia pseudomallei, melioidosis
SUMMARY
The causative agent of melioidosis, Burkholderia pseudomallei, a tier 1 select agent, is endemic in Southeast Asia and northern Australia, with increased incidence associated with high levels of rainfall. Increasing reports of this condition have occurred worldwide, with estimates of up to 165,000 cases and 89,000 deaths per year. The ecological niche of the organism has yet to be clearly defined, although the organism is associated with soil and water. The culture of appropriate clinical material remains the mainstay of laboratory diagnosis. Identification is best done by phenotypic methods, although mass spectrometric methods have been described. Serology has a limited diagnostic role. Direct molecular and antigen detection methods have limited availability and sensitivity. Clinical presentations of melioidosis range from acute bacteremic pneumonia to disseminated visceral abscesses and localized infections. Transmission is by direct inoculation, inhalation, or ingestion. Risk factors for melioidosis include male sex, diabetes mellitus, alcohol abuse, and immunosuppression. The organism is well adapted to intracellular survival, with numerous virulence mechanisms. Immunity likely requires innate and adaptive responses. The principles of management of this condition are drainage and debridement of infected material and appropriate antimicrobial therapy. Global mortality rates vary between 9% and 70%. Research into vaccine development is ongoing.
INTRODUCTION
The genus Burkholderia contains over 80 formally named species (1). Only Burkholderia pseudomallei, B. mallei, B. cepacia complex, and B. gladioli are generally recognized as human pathogens (2). These organisms are aerobic, non-spore-forming, nonfermenting Gram-negative bacilli. All are environmental organisms, with the exception of the host-adapted pathogen B. mallei (2).
B. pseudomallei causes melioidosis in both humans and animals and is designated a tier 1 select agent by the U.S. Centers for Disease Control and Prevention (CDC) (3). It is a saprophytic environmental organism found predominantly in the rhizophere, moist soil, and both surface water and groundwater (4–6). This infection was first recognized in Rangoon by Whitmore and Krishnaswami in 1911 (7). Infection with B. pseudomallei is most commonly associated with an inoculating injury, ingestion, or inhalation of aerosolized bacteria and occurs more frequently in the wet season or following extreme weather events such as tropical storms (8–11). Community-acquired pneumonia is the most frequent clinical presentation and bacteremia the most common microbiological diagnosis (9, 12). B. mallei is the etiological agent of glanders, a disease predominantly affecting solipeds and livestock but also occasionally humans (2, 13, 14). In contrast to B. pseudomallei, B. mallei is a host-adapted pathogen predominantly of solipeds and has little environmental persistence. The organism is considered eradicated from North America, Australia, and most of Europe, with only one reported case in the United States since 1945 (2, 14, 15).
Melioidosis is predominantly a disease of subtropical and tropical regions. It is endemic in northern Australia and parts of Southeast Asia and the Indian subcontinent (16). The incidence of melioidosis appears to be increasing, although mortality appears to be improving in Australia, with an average mortality rate of 14% overall in a prospective Australian study (8). Mortality rates from patients admitted to a hospital in northeast Thailand for melioidosis remained high over the period of 1997 until 2006, with an average annual rate of 42.6% (17). Further estimates, as of 2018, indicate overall melioidosis case fatality rates of 30 to 35% in admitted patients to public hospitals in Thailand (18).
EPIDEMIOLOGY
B. pseudomallei appears to have originated in Australia, and dispersal into Southeast Asia is likely to have occurred during a recent glacial period across what is now the Malay Archipelago (19). Emerging evidence supports the anthropogenic dispersal hypothesis, which proposes that the distribution of the organism was influenced by human migration (20). Statistical analysis of multilocus sequence typing (MLST) of isolates from individual islands in the Torres Strait demonstrates nonrandomlocalization of sequence types (STs). This information suggests specific localization of B. pseudomallei STs by biogeographical niches and not random dispersal (20).
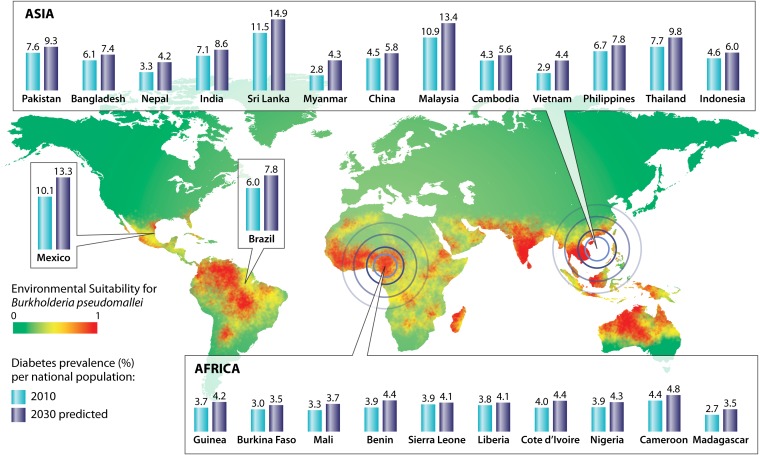
Melioidosis is endemic in approximately 46 countries and is potentially endemic in a further 33 countries yet to report autochthonous cases (21, 22). Figure 1 demonstrates areas of both known and predicted melioidosis endemicity based on environmental suitability, as well as the predicted change in prevalence of the major risk factor for infection, diabetes mellitus, by 2030 (21, 23). Although Thailand and Australia have the highest reported rates of melioidosis worldwide, the true worldwide incidence is unknown, as the majority of infection is likely to occur in rural tropical locations without resources to adequately diagnose cases (9, 18, 24, 25). Current estimates based upon a modelling study suggest that 165,000 cases of melioidosis result in 89,000 deaths worldwide per year (21). A recent publication entitled “Global Burden and Challenges of Melioidosis” encompasses a comprehensive series of region-specific articles on melioidosis (26). Outside of human infection, melioidosis has been found to affect a variety of animals from livestock to domestic pets (9). This may represent a potential for epizootics to result in human infection, but reports describing this are rare, with uncertainty over whether transmission occurred from a zoonotic origin (25, 27).
FIG 1.
Twenty-five countries with the highest predicted melioidosis incidence and predicted increase in prevalence of the major risk factor, diabetes mellitus.
The Environment
B. pseudomallei has been recognized as an environmental saprophyte for a long time, with ongoing investigations defining our understanding of the interactions between the environment and both human and animal hosts (28). Early investigators believed that a rodent host served as a zoonotic reservoir (29). Subsequently, several observations were made leading to our current understanding of B. pseudomallei being widely distributed in the environment, with exposure to soil and surface water being risks for subsequent invasive disease (28). Several factors complicate the study of B. pseudomallei in the environment, including different techniques for environmental surveys. Early studies used animal inoculation techniques, which only detected virulent B. pseudomallei. Later studies used artificial media, detecting both virulent B. pseudomallei and potentially B. pseudomallei-like organisms (28). Sensitivities of various culture techniques for environmental isolation have been noted to vary widely (30). Other challenges include consistency of sampling between studies, including depth of soil sampling, as well as a limited ability to accurately quantify B. pseudomallei in soil samples (28).
B. pseudomallei was cultured from surface water samples in French Indochina in 1955 (31). Subsequently, multiple studies have demonstrated environmental exposure as a risk factor for infection (32, 33). The majority of reported cases occur in regions with environmental factors favorable for survival of B. pseudomallei, most frequently between the tropical latitudes 20°N and 20°S, although multiple cases have been found outside these parameters, including regions of Australia and Taiwan (9, 21, 34, 35). Research regarding the reason for the current geographic distribution of B. pseudomallei has demonstrated that both the occurrence of clinical cases and the presence of the organism in the environment are related to factors such as ambient temperature, soil moisture content, water drainage, soil type, pH, salinity, iron content, and geomorphic position (36–41). The ideal temperature for organism survival ranges between 24 and 32°C, and a soil moisture content of ≥40% results in organism survival within soil for longer than 2 years, compared with only 30 days in soil with 0% water content (38). Acrisol and luvisol soils, which consist of a clay-rich subsoil layer and are associated with tropical climates, appear to have a positive association with B. pseudomallei isolation, whereas ferralsols are negatively associated with organism isolation (42). In the context of flooding, the low water permeability of acrisol and luvisol results in altered physiochemical conditions, including reduced pH, which may favor mobilization of iron and therefore provide a survival advantage (37, 42–46). However, the association between soil iron content and isolation of B. pseudomallei is conflicting, with evidence demonstrating both positive and negative associations (47–50). High saline content in soil appears to induce various virulence genes and may therefore directly correlate with the organism’s pathogenicity in these environments (39). In water samples, recovery of B. pseudomallei is correlated with turbidity, which is a marker of bacterial particulate attachment (51). The organism can be transported via waterways and is increased in the setting of eroded soil during periods of heavy rainfall (42, 52). The annual incidence of infection is also affected by humidity, rainfall, and severe weather events such as monsoons and tropical cyclones (4, 12, 53, 54). Rainfall is thought to increase bacterial concentration in topsoil via the rise in the water table, and severe weather events and wind are associated with bacterially contaminated aerosols resulting in acquisition via inhalation (11, 54–56). A study in Laos and Cambodia reported a specific association between high humidity and a 3-fold increased incidence in children compared with that in adults (54). It remains unclear as to why this association exists, with current theories including increased environmental exposure via swimming in contaminated water, or a shorter incubation period and subsequent presentation in children compared to those in adults (54, 57).
The potential for environmental interaction with Acanthamoeba was first described by Inglis et al., who postulated an association with B. pseudomallei survival (58). The presence of Acanthamoeba astronyxis enhanced survival of B. pseudomallei when subjected to disinfection by chlorine, monochloramine, and UV light (59). However, these results may be organism specific, as multiple taxa of free-living amoebae have demonstrated the ability to internalize but subsequently digest B. pseudomallei bacilli. This suggests that free-living amoebae are unlikely to be a significant environmental reservoir (60).
Oceania
Melioidosis in Australia was described first for sheep in 1949 and subsequently for humans in 1950 (61, 62). Australian epidemiological studies have reported an average annual incidence of 19.6 cases per 100,000 population, ranging from 5.4 to 41.7 during severe weather events in the Northern Territory (NT) (9). An above-average rainfall year in the NT during 2009 to 2010 resulted in the highest annual documented incidences in the world, 50.2 cases per 100,000 population and 102.4 in the indigenous population (63). The annual incidences in the Torres Strait Islands, Cape York, and Cairns have been reported to be 42.7, 12.1, and 1.7 cases per 100,000 population, respectively (12, 64). In western Papua New Guinea, the annual incidence is approximately 20.0 per 100,000 population, and 2.6% of environmental soil samples from other regions were positive (65, 66). This percentage of positive environmental samples is substantially lower than that found in northern Australia, where soil samples taken from around the roots of native grasses have B. pseudomallei direct molecular detection rates using real-time PCR of 32% in the wet season and 20% in the dry season (6). Notably, exotic grasses had constant high prevalences, 71% in the wet and 62% in the dry season, and B. pseudomallei was also found to colonize the rhizosphere and aerial parts of some grasses, which may suggest a mechanism of dispersal by grazing animals via either the oral-fecal route or roaming (6). Furthermore, this environmental interaction may aid in further understanding the incidence in certain geographical settings. Additionally, B. pseudomallei infection has been associated with outbreaks related to contaminated water supplies (67). Four outbreaks have been reported, two of which involved animals, namely, pigs and parrots, and two involving human cases which were associated with unchlorinated water in Western Australia and the Northern Territory (67–70).
Southeast Asia
In Thailand, the true incidence is difficult to assess due to lack of diagnostic resources, potentially high seroprevalence due to B. thailandensis, and incomplete epidemiological data (71). Melioidosis is a notifiable condition by law in Thailand; however, current official reports appear to substantially underreport melioidosis-associated deaths (18). A study performed in northeast Thailand between 1987 and 1991 suggested an incidence of 4.4 cases per 100,000 population per year (72). More recent observations demonstrate a peak incidence of 21.3 per 100,000 population in 2006 and an average of 12.7 per year for the period from 1997 to 2006 (17). Furthermore, a serological study from northeast Thailand suggested an increasing seroprevalence with age, with more than 80% of the population over the age of 4 testing seropositive (73). This result may be due to a number of factors, including repeated exposure in early childhood and the use of the indirect hemagglutination assay (IHA) for diagnosis. The IHA for B. pseudomallei has been reported to have cross-reactivity to the less virulent and rarely pathogenic Burkholderia thailandensis (74, 75). Notably, despite the fact IHA antigens are not standardized, the rate of cross-reactivity was demonstrated to be very low (76). Therefore, exposure to B. thailandensis, specifically to strains expressing a B. pseudomallei-like capsular polysaccharide, is unlikely to account for a significant proportion of seropositive patients (75–77). Outside of the high-endemicity northeast region of Thailand, recent studies provide evidence of high incidence in east and south Thailand (78, 79). A bacteremia study in the eastern province of Sa Kaeo reported an annual melioidosis incidence of 4.9 cases per 100,000 population (78). Extrapolating these data to include all cases of melioidosis suggests that the annual incidence is likely to be 10 cases per 100,000 population (78). The prevalence of melioidosis among patients admitted during a 10-year retrospective study in a southern Thailand hospital was 36.8 per 100,000 inpatients (79). B. pseudomallei is likely endemic in every region of Thailand and is currently underreported (18).
Recent Vietnamese reports suggest that all geographical regions of the country have either positive environmental or clinical isolates (80–83). The seroprevalence in Hanoi from a 1993 study was 6.4 to 31.8%, and more recent environmental surveys indicate that over 80% of soil samples in Southern Vietnam are positive for B. pseudomallei (84, 85). A recent prospective study in 5 central Vietnam hospitals reported a B. pseudomallei detection rate in blood cultures of 3.4 to 10.2% of all bacteremias during the 7-month study period (83).
The first documented cases of melioidosis in Malaysia occurred in a 1913 outbreak among laboratory guinea pigs and rabbits, with the first human cases recognized in subsequent years (29, 86). Currently, Malaysia has the second highest reported incidence of melioidosis in the region. In the northeastern state of Kelantan, 158 proven cases were documented from 2001 to 2015 from a single tertiary center (87). Current reports suggest a range of 6.1 to 16.4 cases per 100,000 population per year (87, 88). Malaysian pediatric melioidosis infections have been reported at 0.6 to 4.1 cases per 100,000 children annually, with central Sarawak having the highest rate (89, 90). Serosurveillance data on multiple population groups from 1969 reported a seroprevalence of 1.9 to 15.8% (91). A 1992 survey of army personnel in Sabah and Sarawak reported a prevalence of up to 65.7% (92). Notably, these studies appear to have included predominantly men and used different serological assays (91, 92). Melioidosis is thought to account for approximately 2,000 deaths annually in Malaysia, which surpasses the mortality rates of both dengue and tuberculosis infections (93).
In Singapore, the first case of melioidosis was reported in 1920, and it has been a notifiable condition since 1989 (94). A study from 2003 to 2014 demonstrated an overall annual melioidosis incidence of 1.1 cases per 100,000 population, with incidence decreasing by 10% annually during this time frame (95). Furthermore, studies of severe community-acquired pneumonia have demonstrated a decreased proportion of the total of microbiologically confirmed bacterial causes from 24% between 1989 and 1993 to 13% in 2003 to 2005 (96, 97). One reason for this decrease is purported to be the improved infrastructure regarding water sanitation, rainwater drainage, and flood reduction (95). A seroprevalence of 0.2% in Singapore has been determined using an IHA with a positive titer defined as ≥1:16 (98). While this study did not appear to stratify for sex or comorbidities, the overall seroprevalence in Singapore and specifically among local construction workers (1.6%) was significantly lower than in samples from foreign construction workers (28.3%) from Thailand, Malaysia, and the Indian subcontinent as determined by the same method to (94). One theory regarding the reportedly lower seroprevalence in Singapore is that it may be due to a combination of Singapore’s highly urbanized environment and the comparatively lower rates of B. pseudomallei in soil and water samples, with the organism able to be recovered from 5.9% of surface water samples and 1.8% of 395 soil samples (94, 99). However, it should be noted that these studies were performed prior to a published international consensus method for environmental sampling and recovery of B. pseudomallei (100). A notable difference in Singapore isolates is a lack of genetic diversity, with only 3 STs from 13 environmental samples, compared with 9 STs from a single sampling point in Thailand, 33 from Cambodia, 32 from Malaysia, and 13 from Laos (101–105).
In Cambodia, microbiologically proven human melioidosis was first diagnosed by local institutions in 2008 (106). Prior to this, a case report described a Cambodian refugee in Canada who presented with pulmonary melioidosis in 1983 (107). Notably, regular identification of B. pseudomallei in Cambodian hospitals commenced only following the establishment of a microbiology laboratory at Angkor Hospital for Children in 2005, and while the laboratory was becoming established, it is possible that cases of melioidosis were misidentified for some time, prior to the correct identification of B. pseudomallei (105, 106, 108). A prospective adult sepsis study of 139 patients in Takeo Province reported that 5% were culture positive for B. pseudomallei over a duration of 1 year (109). However, this is likely to be an underestimate of the true burden of melioidosis, as the study primarily included blood culture specimens only from adults with sepsis (109). A recent pediatric study estimated the annual incidence of melioidosis at 28 to 35 cases per 100,000 children per year (110). This may be an underestimate due to several limitations of the study, including collection of microbiological samples at the discretion of attending clinicians from only one of two pediatric referral centers in Siem Reap and a limited ability to account for children who may have died prior to hospitalization (108, 110). A seroprevalence survey of children demonstrated a 16% seropositivity rate, and furthermore, 30% of soil samples from rice fields were culture positive (111). In a comparison of the bacterial soil burdens, Cambodian samples had a median of 90 CFU/g of soil, whereas Thai samples had a reported count of 230 CFU/g (111, 112). This may account to some extent for the lower seroprevalence, but the fact that only one geographical region in Cambodia was evaluated may have resulted in an inaccurate representation of the burden of B. pseudomallei in the environments of other regions.
In Laos, 36% of environmental soil samples were positive for B. pseudomallei in a 1998 survey, with a mean quantitative value of 39 CFU/g of soil. More recent information from a nationwide survey of 23 rivers reports 9% culture-positive samples in the dry season and 57% positive in the wet season (52, 113). Interestingly, there appears to be a clear north-south divide with regard to environmental isolation of B. pseudomallei, with the two most northern sample sites being negative using both conventional culture and nucleic acid detection (52). Current theories for this contrast include differing climates, soil types, and land uses (42, 52, 114). Clinical cases of melioidosis were first recognized in Laos in 1999 (115, 116). The timing of the first clinical case in Laos is likely directly related to the introduction of diagnostic services specifically for the identification of melioidosis in 1999 through the Lao-Oxford-Mahosot Hospital-Wellcome Trust Research Unit (117). Initial analysis revealed a prevalence of 3% in positive blood cultures (115). However, since 2004, the yearly total of culture-confirmed cases has more than quadrupled, and hence overall prevalence is likely to be much higher (117). Melioidosis has been rarely reported from Indonesia. A recent review of Indonesian cases has summarized 146 culture-confirmed cases ever reported from this region. The authors acknowledge that limited diagnostic capability would be the most likely cause of this potential underreporting (118).
China and Taiwan
The People’s Republic of China reported its first case of melioidosis in 1990 (119). Subsequently, it was shown to be endemic in multiple tropical southern provinces, including Hainan, Guangdong, and Guangxi (119). As with many parts of the world, the number of reported cases has increased dramatically, with 170 confirmed cases reported between 2002 and 2013 in Hainan (120). In Taiwan, cases of melioidosis increased following a 2005 typhoon (121). This was further corroborated by seroprevalence data revealing seropositivity as high as 36.6% in certain regions within the Erren River Basin. Notably, this survey also demonstrated localization of environmental B. pseudomallei distribution, which surprisingly did not correlate with seroprevalence or case incidence (35).
South Asia
Bangladesh has only reported 14 cases of melioidosis from 1988 to 2016 (122). Serological survey results demonstrate a seroprevalence of 9.8% in regions with no reported clinical cases of melioidosis and a seroprevalence of 22.6 to 30.8% in regions with proven cases. Additionally, as may be expected, there is an association between the highest seroprevalence areas and number of cases (123, 124). Environmental sampling isolated B. pseudomallei from only 1% of soil samples; however, the authors of this study recognize that their methods differed from the current international consensus guidelines (124, 125). To date, cases have been described only from north and east Bangladesh, and there is no formal notification policy (123). In Sri Lanka, the first culture-confirmed case of melioidosis occurred in a European resident in 1927 (126). Cases of melioidosis in Sri Lanka have predominated in western provinces, with a notable exception of no cases being reported from areas higher than 500 m above sea level (127, 128). Seroprevalence results from 32 blood banks across Sri Lanka showed a 7.4% seropositivity using an IHA titer of ≥1:40, with a preponderance for the North Western Province (127). These results also revealed a substantially greater number of seropositive females (12.2%) than of seropositive males (6.2%), which is unexpected considering that over 70% of culture-confirmed cases were in males (127). In India, most states have reported confirmed cases of melioidosis (129). From 1991 to 2018, 583 cases were reported, with the southern coastal region of Karnataka and Tamil Nadu representing almost 80% of these cases (122, 130, 131). These regions may not truly reflect the areas of greatest incidence, as current diagnosis and reporting may be influenced by superior resources in these areas (129). Currently, the Udupi district in southwest India has the highest reported annual incidence, estimated to be 1.0 per 100,000 population, with a seroprevalence of approximately 29% using an IHA titer of ≥1:20 (129, 132). Similar to the case with other regions, there has been an increase in case detection following improvement in laboratory diagnostic capabilities coupled with local awareness campaigns for medical practitioners (129). At present, there is limited environmental data regarding the geographical distribution and prevalence of B. pseudomallei infection in India (133, 134). MLST data suggest that Indian isolates are distinct from international isolates, specifically Australasian and Southeast Asian strains. However, some appear closely related to Sri Lankan isolates, representing single-locus variants as determined by BURST analysis ST phylogenetic software (129, 135). While there is no comprehensive South Asia prevalence reporting to date, the continuing rise in case reporting suggests that melioidosis is underreported (136).
The Americas
Five South American countries have reported cases of proven melioidosis: Brazil, Colombia, Venezuela, Ecuador, and Peru (137). Currently, Brazil accounts for two-thirds of melioidosis cases in South America (138). Northeastern Brazil reported the first proven cases of melioidosis in two outbreaks occurring in 2003 and 2004, with sporadic cases shortly thereafter (139, 140). The estimated annual incidence in South America is 1,200 cases and 500 deaths (21). However, the true incidence is uncertain, as Ceará, a state in Brazil, is the only region with compulsory notification of cases, and few laboratories in this region can identify B. pseudomallei (137).
There are potentially two cases of melioidosis without travel to a country of endemicity reported in the United States, but in both cases the organism’s origin was not identified (141–143). To date there is no evidence of environmental B. pseudomallei in the United States (144). The U.S. territory of Puerto Rico is a melioidosis region of endemicity, with multiple cases of autochthonous infection (145–147). Limited population and environmental analyses demonstrated a seropositivity of 6 to 25% and isolation of B. pseudomallei from a soil sample (145). Furthermore, a study of Puerto Rican wildlife described a B. pseudomallei-seropositive terrestrial monkey (148). In Central America, cases have been reported from every country except for Nicaragua and Belize (138). Although no formal incidence is available, predicted annual incidences are 550 cases in Mexico, 114 cases in El Salvador, and 24 cases in Haiti (21). Within the region of Latin America and the Caribbean, the population at risk is 246 million people, with an estimate of 2,000 cases of melioidosis and up to 1,000 deaths annually (21, 149). This discrepancy in incidence compared to that in the rest of the world may be a combination of predictive variables, including environment, climate, and patient risk factors, as well as worldwide underreporting (21, 137). Genetic analysis has demonstrated a diversity of molecular types suggesting potential endemicity (139). In Brazil, theories regarding the organism’s origin include importation through Caribbean livestock or other agricultural products such as rice (139). Whole-genome sequencing (WGS) analysis had provided evidence of anthropogenic B. pseudomallei introduction into South and Central America between 1650 and 1850 CE. Interestingly, the authors of this study implicate the slave trade via transatlantic routes as a potential source (150).
Africa
B. pseudomallei has been isolated from environmental, animal, and human samples from multiple locations in Africa (151). Current modelling predicts approximately 24,000 cases with 15,000 deaths annually in sub-Saharan Africa (21). There are only case reports of proven infection acquired in Africa. However, these demonstrate that infection appears to occur in multiple geographical locations across the continent (151–153). There is only one documented case from North Africa, occurring in a horse from Egypt (154). Additionally, in 2013 a prospective analysis of bloodstream infections in Gabon detected the first case of B. pseudomallei (155). Although Nigeria is speculated to have the greatest environmental suitability and hence burden of infection, only 1 case has been reported to date (156). Four Indian Ocean islands, Madagascar, Mauritius, Reunion Island, and Seychelles, have had confirmed human cases since 2004 (157). Genetic analysis of three African isolates revealed both genetic diversity and an ancestral relationship to an Asian clade, furthermore supporting the evidence of both anthropogenic dissemination and endemicity in Africa (158). The African Melioidosis Network (AMENET) was established in 2014, with the aim of serological and environmental surveillance as well as diagnostic laboratory development for identification of B. pseudomallei (159). With increased awareness and active surveillance on the continent with a more robust data set, more accurate prevalence predictions may be feasible in the coming years.
BACTERIOLOGY AND LABORATORY IDENTIFICATION
Originally termed Whitmore’s bacillus or Bacillus pseudomallei, the organism’s taxonomy was changed to Bacillus whitmori, Malleomyces pseudomallei, Loefflerella whitmori, and Pfeifferella whitmori until 1992, when Pseudomonas pseudomallei was reclassified into the genus Burkholderia (13, 160, 161). B. pseudomallei is an environmental opportunistic saprophyte capable of utilizing at least 80 different compounds tested as a nutritional carbon source (162). It is thus able to persist in a nutritionally depleted environment for substantial periods, with a reported 16 years in distilled water (163).
Specimen Collection
The culture of B. pseudomallei from any specimen in a patient with suspected melioidosis remains the diagnostic “gold standard.” Specimens include blood, respiratory secretions, urine, and, when available, cerebrospinal fluid (CSF), pus, and swabs from wounds or lesions. B. pseudomallei grows well on most routine laboratory media. Improved isolation of the organism from nonsterile sites can be achieved by the use of selective media such as Ashdown’s media and selective enrichment broth (164).
Bacteremia has been found to occur in 38 to 73% of cases (87, 88, 122, 130, 131, 165–168). In one study isolating B. pseudomallei from blood using the BacT/Alert (bioMérieux, Marcy l’Etoile, France) automated blood culture system, 62.5% of isolates were detected in 24 h and 93% were detected within 48 h of incubation. The time for the system to signal positive (mean ± standard error) was 23.9 ± 14.9 h (169). When comparing the BacT/Alert system with conventional culture, which utilized in-house brain heart infusion media and visual detection, the automated system was found to have a sensitivity of 73.5%, compared to 90.3% for conventional culture (170). The major benefit of the automated system was a shorter time to positivity, approximately 1 day (170). Additionally, a study evaluating the sensitivity of the BacT/Alert FA aerobic bottle and BacT/Alert MB bottle with those of Middlebrook 7H9 broth, glycerol, and sodium polyanethol sulfonate demonstrated improved organism recovery with the MB bottle for patients with prior antimicrobial exposure (171). The Bactec (Becton Dickinson, Sparks, MD) automated blood culture system has also been used in laboratories for the isolation of B. pseudomallei from blood and sterile fluid. There are, however, limited data regarding the comparative sensitivity and time to positivity (172, 173). The urine culture of patients with melioidosis is estimated to be positive for 28% of cases in Thailand (174). Centrifugation of the sample and culture of the pellet improve sensitivity. Additionally, a quantitative urine culture revealed a comparatively higher mortality rate with increasing counts, with in-hospital mortality of 39% in culture-negative patients, 58% with a quantification of <103 CFU/ml, and up to 71% with a quantification of >105 CFU/ml (174). Throat swabs are an effective method of organism recovery, and routine screening of suspected melioidosis patients is performed in certain centers (175, 176).
Culture
B. pseudomallei organisms are small Gram-negative bacilli with bipolar staining giving them a safety pin appearance (Fig. 2) (2). This feature, which is not specific to B. pseudomallei, is due to central accumulation of polyhydroxybutyrate (PHB) granules, which do not retain the staining reagents (40). The Gram stain appearance alone is not sufficient to make a presumptive diagnosis. The organism grows well on MacConkey, blood, and chocolate agars. Improved isolation of B. pseudomallei from sites with normal flora can be achieved using the selective Ashdown’s medium which contains Trypticase soy agar with 4% glycerol, 50 mg/liter of neutral red indicator, 5 mg/liter of crystal violet, and 4 mg/liter of gentamicin as selective agents (2, 164, 177, 178). Additionally, the use of a selective enrichment broth for throat, wound, and rectal swabs is likely to increase organism isolation (164). Although Ashdown’s agar is an effective selective agar, it may inhibit persistently mucoid strains and the glycerol in the agar may inhibit smooth strains (179, 180). In addition, rare gentamicin-susceptible strains from Sarawak have been described (181). Due to these limitations, new media have been developed. Burkholderia pseudomallei selective agar (BPSA), which includes maltose as a carbon source, excludes crystal violet, utilizes Nile blue as an indicator, and has a lower concentration of glycerol than Ashdown’s agar, produced large wrinkled colonies faster, allowing for earlier differentiation (180). Furthermore, Francis medium was developed to improve both detection and differentiation between B. pseudomallei and B. cepacia. In one in vitro study, Francis medium had a sensitivity of 78.4% and a specificity of 92.2% (182). Due to the advent of multiple medium options, a trial was performed on clinical isolates, including urine, respiratory samples, pus, and throat and wound swabs, to assess comparative performances (183). This study found no difference in organism isolation but found BPSA to be significantly less selective (183). Subsequently, a modified Ashdown’s agar including norfloxacin, ampicillin, and polymyxin B (NAP-A) was evaluated (184). This agar demonstrated increased specificity compared to that of Ashdown’s agar in a mouse model of gastrointestinal samples. With human clinical isolates this medium had improved selectivity but equal recovery of B. pseudomallei (184). As there have been limited comparative evaluations of selective media, Ashdown’s medium remains the standard selective medium in regions where melioidosis is endemic. Even in regions of low prevalence, the use of selective media has demonstrated cost-effectiveness (185).
FIG 2.
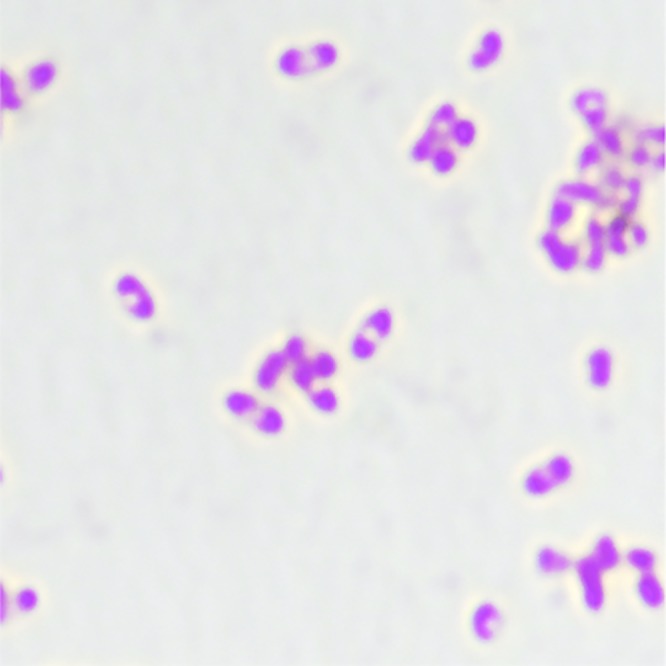
Gram stain demonstrating “safety pin” appearance. Magnification, ×100.
A study evaluating the utility of throat swabs in the diagnosis of melioidosis including 4,535 patients (1,011 proven melioidosis patients and 3,524 controls) demonstrated a sensitivity of 36% and specificity of 100% using Ashdown’s medium (186). Additional analysis of selective and nonselective enrichment broth demonstrated improved sensitivity, 24.2%, with modified Ashdown’s broth containing colistin (50,000 U/liter, equivalent to 50 mg/liter) and crystal violet, as opposed to 10% with tryptic soy broth (175, 187). Modified Ashdown’s broth is therefore considered the standard for B. pseudomallei isolation from throat swabs (175). Overall, the sensitivity of culture in the setting of melioidosis has been reported at 60.2%. Therefore, culture can be said to have low sensitivity and low negative predictive value (NPV) (188). Additionally, there is a lack of quantitative organism correlation between blood and other specimen types such as urine, sputum, or pus. This suggests, for example, that organism isolation in urine is consistent with renal parenchymal infection and not passive filtration into the urine (189).
Presumptive Bench Identification
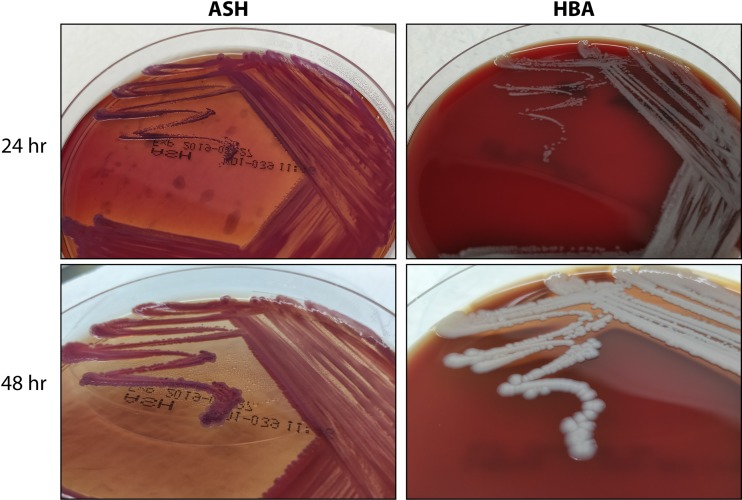
Colony morphology in the first 24 to 48 h of growth on blood agar reveals smooth, creamy colonies (2, 190). On Ashdown’s agar, the colonies are pinpoint, flat, dry, and purple and may be wrinkled (Fig. 3) (2, 177). Phenotypic differentiation of B. pseudomallei includes the ability to grow at 42°C, motility, oxidase activity, and nitrate reduction (2). Additionally, the organism is indole negative, methyl red negative, Voges-Proskauer negative, and H2S negative. The characteristic triple sugar iron reaction is acid/alkaline with gas production (191). B. pseudomallei is a phenotypically heterogeneous organism with various morphologies, particularly after prolonged incubation (192). Despite this, experienced laboratory personnel can often readily identify colonies that may represent B. pseudomallei in areas of endemicity. Simple bench testing using the Gram stain, a metallic sheen on blood agar, oxidase positivity, and a resistance pattern demonstrating gentamicin and colistin resistance and amoxicillin-clavulanate sensitivity can presumptively identify B. pseudomallei (80, 193). It should be noted that, while a rare occurrence worldwide, gentamicin-susceptible B. pseudomallei accounts for 86% of isolates located in Sarawak, Malaysia (181).
FIG 3.
Colonial morphology of B. pseudomallei. Shown are B. pseudomallei cultures on ASH (left) and HBA (right) at 24 h (top) and 48 h (bottom).
Rapid Antigen Detection
Many latex agglutination assays have been developed for the rapid identification of B. pseudomallei. Polyclonal and monoclonal antibodies targeting lipopolysaccharide (LPS), 30-kDa antigen, and exopolysaccharide have been trialed (173, 194–198). Similar to the case with biochemical profiles, the sensitivity and specificity of agglutination assays differ between regions. One assay studied in an Australian laboratory and performed on bacterial colony suspensions showed a sensitivity of 94%, a specificity of 83%, and false-positive cross-reaction with multiple Burkholderia species, including B. thailandensis (197). Monoclonal antibodies recognizing exopolysaccharide have a reported sensitivity of 98.7% for B. pseudomallei and a specificity of 97.2% on direct colony testing (199). A prospective study of direct detection from positive blood cultures using a monoclonal antibody 4B11 immunofluorescence assay (IFA) targeting the exopolysaccharide reported a sensitivity of 97.4% and a specificity of 100% (173). This assay was subsequently prospectively evaluated at another facility using 545 positive blood cultures identified as containing Gram-negative bacilli. The results indicated a sensitivity of 100%, a specificity of 99.6%, and a negative predictive value of 100% (200). Unfortunately, with nonblood clinical samples the same IFA showed sensitivities ranging from 32.7% on respiratory samples to 50% on pus (201). Identification of B. pseudomallei directly from blood culture samples is feasible with latex agglutination assays. Two different monoclonal antibodies have demonstrated a sensitivity of 100% and a specificity of 85 to 100% (195, 196). With this diagnostic method, organism identification may occur up to 2 days earlier than by using traditional phenotypic and biochemical methods (196). This latex agglutination assay is currently not commercially available and is best utilized on culture amplified blood cultures that have signalled positive. This would unfortunately negate the advantage of early, preamplification detection.
The Active Melioidosis Detect (AMD; InBios International, USA) lateral flow assay (LFA) detecting B. pseudomallei capsular polysaccharide (CPS) via a monoclonal antibody was recently developed. An initial laboratory study using cultured organism demonstrated a sensitivity of 98.7% and a specificity of 97.2% (202). Notably, a false-negative result occurred for an isolate with a frameshift mutation in the wcbR gene, which is known to decrease production of CPS (203). The low limit of detection (approximately 2 ng/ml) was felt to be a notable feature of the assay (202–204). An additional study using stored whole unamplified blood from culture-positive patients reported a sensitivity of 40% (205). Subsequently, a prospective clinical trial was undertaken and demonstrated 99% sensitivity and 100% specificity on culture amplified turbid blood culture bottles and a positive predictive value (PPV) of 94% on urine samples (206). The LFA was easy to perform, provided a result in 15 min, and cost approximately $2 (U.S. dollars) per test (206).
With continued improvement in sensitivity and specificity of rapid antigen detection testing, coupled with ease of use and low cost per test, it is conceivable that direct antigen detection from clinical isolates will become a mainstay for diagnosis in resource-limited regions where melioidosis is endemic.
Serology
The serodiagnosis of melioidosis is difficult, with a lack of international standardization and high seropositivity rates in healthy individuals from regions of endemicity (207, 208). Additionally, a number of different antigens have been evaluated, with a wide range of reported sensitivities and specificities among multiple assays (207, 209, 210). It can also be challenging to determine if a seropositive patient has acute, chronic, or past infection or exposure without infection.
The serum indirect hemagglutination assay (IHA) has previously been considered the clinical standard serological test for melioidosis, although 19% and 26% of culture-confirmed cases never seroconverted in two studies (211, 212). The IHA is performed by using poorly defined antigens from strains of B. pseudomallei adsorbed to sheep red blood cells (2). In previous Thai studies, an IHA cutoff titer of less than 1:80 was deemed unlikely to indicate a true positive, as 21% of healthy blood donors were found to have a titer of ≥1:40, titers of 1:80 to 1:320 were suggestive of infection, and a titer of >1:320 was very likely to indicate infection with a specificity of 97% (213, 214). A recent repeat study of blood donors in northeast Thailand reported 38% seropositivity with titers of ≥1:80, further demonstrating the limitation of this test in regions of endemicity (215). In Australia, the cutoff used for positive results is 1:40, which was determined due to a lower seroprevalence, ranging from approximately 2.5 to 8.7%, compared to 35 to 38% in Thailand (215–217).
In 1989 Ashdown et al. reported on a B. pseudomallei inactivated cell suspension IgG enzyme-linked immunosorbent assay (ELISA) developed in Australia with a sensitivity of 90% and a specificity of 99% (218). This study also demonstrated a greater sensitivity than that of the IHA (74%) and a similar sensitivity to that of the IgG IFA (91%) in the acute phase (218). A rapid immunochromatography test (ICT) strip assay for both B. pseudomallei IgM and IgG was evaluated in 1999 and demonstrated sensitivities of 93% and 100%, respectively. The sensitivity for both assays was 95% (219). A similar assay from the same manufacturer was subsequently created in the form of a cassette kit. This assay demonstrated lower IgM and IgG sensitivities, reported as 88% and 77%, respectively. The specificity for IgM was 69%, and that for IgG was 90%. The calculated PPVs of this cassette assay in the Northern Territory, Australia, were 18% for IgM and 32% for IgG, although another study performed in northern Queensland, Australia, had a PPV of 90.5% (220, 221). Due to these conflicting results, an additional study was performed, and it reiterated the low sensitivities for for IgG (50.6%) and for IgM (72%) (220). This test is no longer commercially available. Because of the ease of use, the utility of this type of cassette kit would be high in low-resource regions where melioidosis is endemic, despite relatively low sensitivity (222).
Novel antigen targets to improve diagnostic performance have been described and assessed. Recently, hemolysin-coregulated protein (Hcp1) was determined to be a virulence factor associated with the type VI secretion system, highly expressed in the infected host and therefore a potential diagnostic target (223–225). Additionally, O polysaccharide (OPS) has also been considered a potential serodiagnostic target, as the antigen is specific for B. pseudomallei and conserved across strains (209). However, an ELISA comparison between these two antigens demonstrated a significantly greater diagnostic sensitivity for Hcp1 (226). Furthermore, a retrospective serum analysis from a melioidosis-infected Malaysian cohort using a recombinant Hcp1 ELISA reported a sensitivity of 93.7% and a specificity of 100% (225). Subsequently, this antigen was used as a target in an ICT, which demonstrated a sensitivity of 88.3% and a specificities of 86.1% in Thai samples and 100% in healthy donors from the United States (226, 227). Interestingly, this study reported no significant difference in sensitivity between bacteremic and nonbacteremic patients (226). While this result is in contrast to an older immunoaffinity-purified IgG ELISA demonstrating a higher sensitivity than those of both IgM ELISA and IHA for bacteremic patients, both assays appear to have a greater ability to identify acute infection than that of the IHA (226, 228). Not only are Hcp1 titers significantly elevated in early infection, but also these may be used to monitor disease progress, with an expected decrease over time (226). Two more novel antigen candidates, including heat shock protein (a chaperone in GroEL protein), and outer membrane protein A (OmpA), have been evaluated. Recombinant GroEL protein had a sensitivity of 92.1%, a specificity of 88.3%, and a less cross-reactive antibody response in healthy individuals than did OmpA and may be a potential serodiagnostic antigen in regions of endemicity (210). Another potential method of improving serology diagnostic performance is combining available assays. The combination of IHA and IgM ELISA in the diagnosis of acute melioidosis in an area of endemicity demonstrated a sensitivity of 100% and a specificity of 95.4% (229). Table 1 summarizes the serological methods described in this section.
TABLE 1.
Serological diagnosis of melioidosis
| Serologic test | % sensitivity (reference) | % specificity (reference) | Country(ies) |
|---|---|---|---|
| Serum indirect hemagglutination assay (IHA)a | 51 (212)–95 (213) | 74 (213)–97 (213) | Australia and Thailand |
| Inactivated cell suspension IgG ELISA | 90 (218) | 99 (218) | Australia |
| IgM ICT test (PanBio) | 72 (220)–93 (219) | 69 (221)–95 (219) | Australia |
| IgG ICT test (PanBio) | 51 (220)–100 (219) | 90 (221)–97 (220) | Australia |
| Recombinant Hcp1 ELISA | 93.7 (225) | 100 (225) | Malaysia |
| Recombinant Hcp1 ICT | 88.3 (227) | 86.1 (227) | Thailand |
| Recombinant GroEL ELISA | 92.1 (210) | 88.3 (210) | Thailand |
| Combination of IHA and IgM ELISA | 100 (229) | 95.4 (229) | Thailand |
Cutoff values of ≥1:40 to ≥1:320 were used in studies, with results depending on cutoff value used.
The serodiagnosis of melioidosis remains a challenge but still has a role to play in the diagnosis of chronic melioidosis and where culture may not always be possible, such as in neuromelioidosis or with deep-seated abscesses. Perseverance in research and development may yield a fast, easy-to-use, and cost-efficient method specifically beneficial to resource-limited settings.
Identification by Semiautomated and Automated Phenotypic Methods
Further verification of the identification of an isolate presumptively identified as B. pseudomallei is recommended (177, 192).
Identification of B. pseudomallei has been an ongoing challenge since its initial discovery. The API 20NE (bioMérieux, Marcy l’Etoile, France) has probably had the widest use and in some settings of endemicity performs very reliably, correctly identifying up to 98% of isolates (192). Other studies have not found it so consistent (193). Chromobacterium violaceum was the most common misidentification, which was thought to relate to errant interpretation of the biochemical tests with an opacity endpoint (230). However, it is notable that all isolates identified as C. violaceum had identical repetitive extragenic palindromic sequences patterns, suggestive of a possible local strain (230, 231). Following these discrepant results, the largest identification study was performed on 800 isolates from environmental, animal, and human samples collected from 8 countries. This study reported the sensitivity of the API 20NE to be 99% (95% confidence interval [CI], 98.0 to 99.6%) (232). Automated colorimetry-based identification, such as Vitek 2 (bioMérieux), has an improved capacity for correct organism identification; however, it, too, has a wide margin of error, with only 63 to 81% of isolates accurately identified (233, 234). Using this method, the most common misidentifications were a variety of nonfermenting gram-negative bacilli, including Acinetobacter and Pseudomonas species, followed by B. cepacia complex (231, 235). Performance of the Vitek 2 system appears to be geographically variable. Comparison between Malaysian and Australian isolates revealed a greater number of B. pseudomallei isolates misidentified as B. cepacia in the Malaysian samples (236). These misidentified isolates appeared to cluster with biochemical profiles distinct from that of the correctly identified isolates. The enzyme β-N-acetylglucosaminidase was found in 88% of correctly identified isolates, compared to only 13% in the misidentified isolates (236). With phenotypically similar species, a notable difference between the potentially avirulent B. thailandensis and B. pseudomallei is the assimilation of arabinose (237). The BD Phoenix (Becton Dickinson, Sparks, MD) automated identification system does not have B. pseudomallei in the database and consequently will most commonly misidentify the organism as B. cepacia with 95 to 99% confidence (238, 239). Although not used in the laboratory diagnosis of melioidosis, gas chromatography has also demonstrated a reliable ability to differentiate these species via analysis of fatty acid derivatives, including 2-hydroxymyristic acid (240).
Identification by Mass Spectrometry (MALDI-TOF MS)
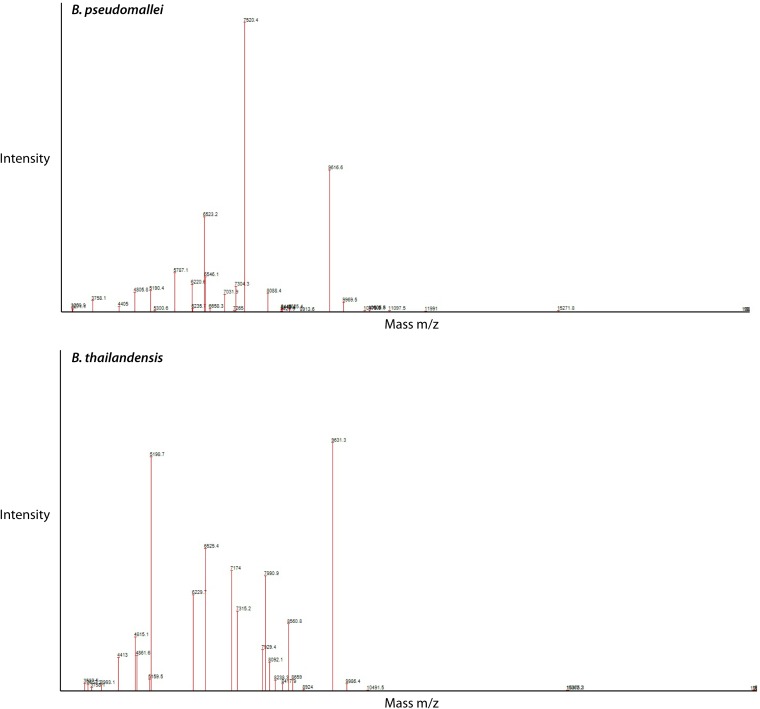
A novel diagnostic method is matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). Compared with other techniques, the advantages include reduced analysis time, high sensitivity and specificity, minimal technical requirements, and relatively simple staff training for implementation (241–244). Two commercially available MALDI-TOF MS instruments, Bruker Microflex Biotyper (Bruker Daltonik GmbH, Bremen, Germany) and biomérieux Vitek MS (biomérieux, Marcy l’Etoile, France), are both certified for identification of clinical isolates. Currently, neither instrument’s routine diagnostic database includes the reference spectra required for identification of B. pseudomallei (245, 246). The current Vitek MS in vitro diagnostic (IVD) database is able to accurately match the acquired organism mass spectrum to the genus but not species level (247). The Vitek MS Research Use Only (RUO) database does include B. pseudomallei spectra, but it is not yet FDA approved, nor has it been assessed in a clinical context (248). Using the conventional Bruker database the organism identification may be valid only to the genus level and identify the isolate as B. thailandensis (248). In this scenario, the security-relevant library, which includes potential agents of bioterrorism, can identify B. pseudomallei (248–250). It appears that there are five conserved biomarkers which are species specific for B. pseudomallei (251). The biomarker for Burkholderia spp. is a mass/charge ratio (m/z) of 4,410. For the B. pseudomallei complex, including B. mallei, B. pseudomallei, and B. thailandensis, m/z 9,713 is required. The mass peak at m/z 6,551 differentiates B. thailandensis from the first two species. The peak mass intensity of m/z 5,794 and 7,553 can be used to differentiate B. pseudomallei from B. mallei, respectively (245, 251). The addition of phage-based diagnostics to MALDI-TOF MS, specifically φX216 (which is found in both B. pseudomallei and B. mallei) and addition of testing for φ1026b (which is B. mallei specific), can aid in rapid identification and indirect susceptibility testing for ceftazidime resistance (252). Using the Bruker MS, in-house reference libraries have been constructed and determined to be accurate in the identification of B. pseudomallei both from primary isolates and directly from positive blood culture broth (253, 254). Similarly, a study using the Vitek MS RUO to create an in-house B. pseudomallei spectrum reported 100% organism identification from culture with a specificity of 99.8% (Fig. 4) (244). These studies demonstrate a viable laboratory alternative that would decrease time to identification by up to 24 h.
FIG 4.
Comparison of B. pseudomallei and B. thailandensis spectra using the Vitek MS.
A potential obstacle to the routine use of MALDI-TOF MS is the requirement for organism inactivation due to the potential risk of laboratory exposure (248). Various inactivation techniques have been trialed, including 70% ethanol, formic acid, trifluoroacetic acid, gamma irradiation, centrifugation, and filtration, with various success (255, 256). With regard to formic acid, one study demonstrated 100% reduction in viable organism when on-plate 70% formic acid was applied (257). The VITEK method currently uses 25% formic acid, compared to 70% in the Bruker method. Further experimental results for comparison of 70% formic acid with a tube extraction method consisting of ethanol-formic acid-acetonitrile and centrifugal filtration demonstrated superior inactivation (255). An additional barrier to the use of mass spectrometry for organism identification is that melioidosis is predominantly endemic in resource-limited settings and is therefore unlikely to be a practical alternative (21).
Molecular Confirmation and Direct Molecular Detection
PCR testing of B. pseudomallei clinical isolates is an option for confirmatory identification. However, due to the genetic variability, recombination, and lack of validation across large data sets, its use has been limited (258, 259). More recent research has increased current knowledge regarding specific genetic targets. At present, there are several real-time PCR assays available for species-specific identification of B. pseudomallei (259–262). The type III secretion system gene cluster, specifically cluster 1 (T3SS-1), orf2, and orf11, appear to be useful in discriminating B. pseudomallei from other Burkholderia species (263, 264). The dual-probe TaqMan single nucleotide polymorphism (SNP) assay BurkDiff has been rigorously tested on known environmental and clinical isolates and appears to have 100% specificity (259). Given the nature of the infection and high mortality, rapid diagnosis is imperative, and to that end, direct identification of the organism from clinical specimens would aid in early directed therapy (8, 12). The T3SS-1 real-time assay has demonstrated 100% sensitivity and specificity on urine, sputum, wound swabs, and drained pus (265). However, overall sensitivity and specificity on all clinical isolates were 73.2% and 89.2%, respectively. Performance of the assay on blood samples was less impressive, with 74% of septic bacteremic patients positive by PCR and only 17% of patients PCR positive in the nonseptic bacteremic cohort (265, 266). Notably, the sensitivity of the assay in septic patients is in keeping with previous evidence that sepsis in melioidosis is associated with a higher blood bacterial burden (267). Differentiating burden of bacteremia by number of CFU counted (≤1 CFU/ml, 1 to 50 CFU/ml, and >50 CFU/ml) is associated with prognosis, with one study demonstrating 50% in-hospital mortality with 1 to 50 CFU/ml, compared to 79% with >50 CFU/ml (268, 269). A study by Wuthiekanun et al. suggests the median concentration of bacteria in blood to be 1.1 CFU/ml (270). Therefore, the limit of detection (LOD) of the aforementioned T3SS-1 assay may play a role in its altered sensitivity in blood compared to other clinical samples (265). Further research on spiked blood using a more targeted T3SS-1 orf2 region primer demonstrated a 95% probability of detection at an organism concentration of 8.4 × 103 CFU/ml (263). However, this probability decreased to just 12.5% for a concentration of 500 CFU/ml. The authors of this research suggest that by increasing the sample volume used 5-fold, which extrapolates a result of approximately 500 CFU/ml, and performing the assay in triplicate, the probability of detection would be approximately 100% (263). This may not be a feasible testing methodology, and a recently described single-tube multiplex PCR may be the alternative. Using the flagellar structural protein gene fliC to identify the entire B. pseudomallei complex (B. mallei, B. pseudomallei, and B. thailandensis), in combination with orf11 specifically for B. pseudomallei, this multiplex assay demonstrated a sensitivity and specificity of 100% (260). Previous PCR assays as described above are based on phylogenetic or virulence genes. Alternative identification using species-specific β-lactamase genes in a multiplex assay has shown promise, although notably not on clinical isolates (271).
Although there remain concerns regarding species detection and differentiation due to potential genetic mutations or deletion, this has been rare, and the most concerning reports of false-negative results are associated with nonseptic bacteremic patients (262, 265). Given the current body of research, there are several potential genetic markers for genus and species identification (262, 264, 272). To overcome issues of genetic variation and specificity, a multiplex approach may be better (262). Several of the mentioned targets in combination have excellent specificity, but the limit of detection in blood as a specimen is a significant limitation (263, 265). Newer molecular markers are available including bucl16, for which an assay was able to detect 50 CFU/ml in a mouse model (273). The method of DNA extraction from clinical isolates may well be the key to improved LOD. Comparison of 7 DNA extraction kits revealed an LOD of 5.5 × 103 CFU/ml. The High Pure kit revealed the best sensitivity and technically the lowest LOD, with 1 positive result from 9 at 4.9 × 102 CFU/ml and a cycle threshold of 37.3 (274). With regard to blood, plasma appears to have a higher rate of nucleic acid recovery than other blood fractions (275). Furthermore, centrifugation of whole blood and then DNA extraction may increase diagnostic yield (276). In a true clinical scenario blood cultures would likely be performed, and detection of an organism directly from blood culture broth (timing of aspiration and analysis to be determined) may furthermore improve detection (277). Improved detection of genetic material from clinical urine samples may also be feasible. A filter-capture DNA isolation method has demonstrated the ability to detect 102 CFU/ml from 0.45 ml of synthetic urine. This method appears to be faster and more sensitive than the QIAamp protocol comparator (278).
While there are multiple methods for the molecular detection of B. pseudomallei, currently none are used in routine diagnostics. With the highest mortality rates occurring in septic and bacteremic patients, the ideal platform for molecular detection must produce a result within hours and require minimal handling. Automation, cost-effectiveness, and a reproducible limit of detection of 1 CFU/ml directly from blood will be essential to achieve this (270).
Antimicrobial Susceptibility Testing and Antimicrobial Resistance
Ceftazidime and meropenem are the preferred antibiotics for the initial parenteral phase of treatment, while co-trimoxazole (TMP-SMX), doxycycline, and amoxicillin-clavulanic acid are used for long-term oral eradication therapy and postexposure prophylaxis (279). No international interpretive guidelines exist for disk diffusion testing of B. pseudomallei. Currently, the only available guidelines are those of the Clinical and Laboratory Standards Institute (CLSI) (280). This is by a broth dilution method and is calibrated for imipenem, ceftazidime, TMP-SMX, tetracycline, and amoxicillin-clavulanic acid. While there is limited evidence for disk diffusion methodology and interpretation currently, it is convenient, easy to perform, and maybe an alternative in cases where laboratories are unequipped to perform CLSI-recommended methods (281, 282). Caution needs to be exercised in interpreting zone diameters for TMP-SMX, as an indistinct endpoint can lead to the incorrect reporting of resistance (Fig. 5) (282).
FIG 5.
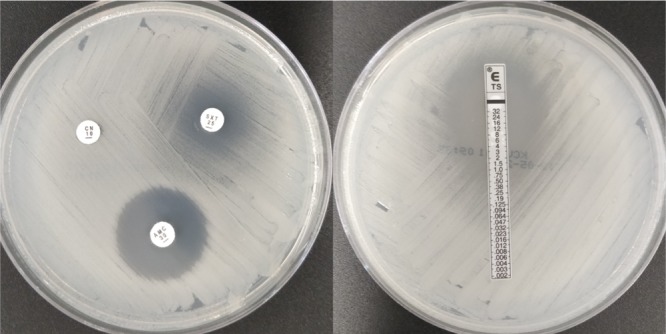
Susceptibility characteristics of B. pseudomallei. AMC, amoxicillin-clavulanate; CN, gentamicin; SXT and TS, trimethoprim-sulfamethoxazole. Double zone of susceptibility were seen with SXT.
Unlike other Gram-negative bacteria, B. pseudomallei is intrinsically resistant to most antimicrobial agents, including penicillin, ampicillin, first- and second-generation cephalosporins, the aminoglycosides gentamicin, tobramycin, and streptomycin, and polymyxin (191, 283–285). There are multiple factors associated with intrinsic resistance, including antimicrobial cell entry, expulsion, and enzymatic degradation (286–288). In B. pseudomallei, efflux pumps of the resistance nodulation cell division (RND) family are the most clinically relevant (286). B. pseudomallei genomes encode up to 10 RND efflux pumps, 7 on chromosome 1 and 3 on chromosome 2. Currently, only 3 RND efflux pumps have been characterized, namely, AmrAB-OprA, BpeAB-OprB, and BpeEF-OprC (286). Interestingly, omeprazole and phenothiazines appear to have synergistic antimicrobial effects against efflux pumps, reducing the MIC of erythromycin and providing a protective benefit to lung epithelium and macrophages via mitigated cytotoxicity (289). Another intrinsic mechanism of resistance is that of reduced outer membrane permeability to antimicrobial agents (288). Polymyxin is poorly bound, and the self-promoted uptake pathway (in which a cationic antimicrobial displaces lipopolysaccharide cations and aids in uptake) of this antimicrobial is blocked (288). Polymyxin resistance specifically is multifactorial and includes the isoprenoid synthesis enzyme IspH, metalloproteases ZmpA and ZmpB, periplasmic protein MucD, efflux pump NorM, and many others (290).
Resistance to first-line antimicrobial therapy is uncommon (284, 291–293). In the majority of primary β-lactam-resistant isolates, the etiology is a class A β-lactamase encoded by the gene penA, located on chromosome 2 (287, 294). Multiple reported penA mutations are associated with β-lactam resistance (287, 294, 295). Amino acid substitutions of Cys69Tyr, Pro167Ser, and Asp240Gly lead to ceftazidime resistance, Ser72Phe leads to clavulanic acid resistance, and Thr147Ala results in resistance to both amoxicillin-clavulanic acid and imipenem (287, 294, 295). Mutations, deletion, duplication, and overexpression of penA result in increased resistance (294, 296). Further β-lactam resistance has been associated with potential selective pressure and genetic rearrangement of chromosome 2 with resultant alteration of penicillin-binding protein 3 (PBP3) in a clinical isolate. This isolate furthermore did not grow on standard media and required Ashdown’s agar supplemented with 4% glycerol (297). Such isolates might easily be missed using standard laboratory methods.
It is important to note that a study of over 4,000 isolates in Thailand and over 600 isolates in Singapore reported ceftazidime resistance at 0.5%, while multiple smaller studies demonstrated 100% susceptibility (284, 291, 293, 298–300). Detailed molecular analysis has demonstrated the appearance of novel single nucleotide polymorphisms selected during ceftazidime therapy. Additionally, these novel mutations appear to create a fitness cost to the organism and may improve susceptibility to other agents. Notably, screening of 2,400 isolates from patients with nonrecurrent melioidosis was negative for these SNPs. Investigators have postulated that as ceftazidime is a synthetic antibiotic, there would be limited environmental selection pressure and therefore these mutations should be rare (301–303). Factors governing penA gene expression are still poorly understood, and further research is required (287).
TMP-SMX resistance was previously thought to be more common and has been reported at 0 to 13% (282, 284, 304–306). However, a study performed on two separate isolate collections in Thailand totaling 3,293 isolates, including repeated testing of the original collection with 13% resistance, revealed a total TMP-SMX susceptibility of 99.7%, which is similar to the rates of 99.1% in Australia, 99.2% in Laos, 99.4% in Malaysia, and 100% in Cambodia, Bangladesh, Brazil, and Taiwan (282, 291, 298, 299, 307, 308). The authors suggest that the incorrect results from a prior study were likely due to inaccurate reading of the 80% inhibition zone (304, 305). Meropenem has a reported susceptibility rate of 100% (284, 293, 298, 299, 307). It is notable that there is evidence of decreased meropenem susceptibility development while on treatment, not only in patients treated with meropenem, although these isolates did not appear to have cross-resistance to imipenem (309).
Fluoroquinolone resistance in B. pseudomallei is commonly associated with alteration of the site of activity, DNA gyrase. As with other Gram-negative organisms, this is via a Thr83Ile mutation. This alters the gene gyrA, which is responsible for DNA gyrase, an enzyme that catalyzes the supercoiling of DNA (310). With regard to aminoglycoside and macrolide resistance, AmrAB-OprA and BpeAB-OprB efflux pumps play a major role (311, 312). Rare gentamicin-susceptible isolates have been reported among clinical isolates and have demonstrated AmrAB-OprA operon deletion or nonsynonymous single nucleotide polymorphism within the amrB gene (181, 313).
In contrast to planktonic organisms, B. pseudomallei biofilms are associated with high-level resistance to multiple antimicrobials, including ceftazidime, imipenem, and TMP-SMX (314, 315). The mechanism of resistance is associated with decreased penetration and therefore reduced effect of ceftazidime and imipenem (315). Notably, differing strains of B. pseudomallei may have altered biofilm formation capacity (316). Capsule and O-side chain LPS-defective strains are more likely to form a biofilm, whereas flagellin-defective mutants produce a lower quantity of biofilm than do wild-type (WT) strains. This suggests that flagellin may have a more substantive role in biofilm formation (314). Another altered growth condition is that of the anaerobic environment. In this environment, such as an abscess, B. pseudomallei can undergo adaptation with altered gene expression, enabling both anaerobic and acidic environmental survival (317). Under these conditions, the total population is tolerant of traditional melioidosis therapy. Interestingly, although resistant to standard therapy, these organisms become susceptible to nitroimidazole antimicrobials. Furthermore, a small subpopulation (0.1%) in this environment is resistant to all antimicrobial therapy (317).
ENVIRONMENTAL SAMPLING
For environmental soil sampling, culture-based methods may be influenced by factors including soil sampling depth, bacterial soil attachment, soil sample volume, incubation environment, and selective media used (84, 100, 318, 319). Multiple sampling methods have been trialed, with various degrees of success (100, 113, 116). Based on a review of 69 articles, a consensus guideline has been proposed (125). A few specific aspects of soil sampling strategy include a soil sampling depth of 30 cm, a 10-g soil sample volume, and transport of sample at room temperature away from direct sunlight. For organism isolation, an extraction broth is suggested, such as Ashdown’s broth containing colistin or l-threonine-buffered salt solution, with vortexing of the solution, incubation at 40°C for 48 h, and subculture of 10 μl of supernatant onto Ashdown’s agar (125). Soil samples taken at a depth of 35 to 45 cm have previously demonstrated the greatest environmental persistence, and more recent evidence suggests that a soil sampling depth of 60 cm is likely to yield greater recovery of B. pseudomallei (114, 320). Timing of environment sampling appears to have contrasting results with regard to organism isolation; however, a number of studies demonstrated greater isolation in the wet season (50, 321, 322). Interestingly, one study demonstrated greater isolation from residential properties in the dry season (323). The authors theorized that this was due to increased use of bore water for garden irrigation, of which 33% of water samples tested were culture positive for B. pseudomallei (323, 324). There are several limitations of culture-based techniques for environmental isolation of B. pseudomallei. Under certain stress conditions, including low pH or high osmolarity, the organism may persist in a viable but nonculturable state in the environment and therefore produce a false-negative culture result (40). Culture is further limited by overgrowth of other environmental flora, decreasing both isolation and true quantification of B. pseudomallei (114). A further challenge to environmental sampling is the lack of a consensus guideline for isolation of B. pseudomallei from water (113, 125, 324). The identification of the organism from water samples previously included intraperitoneal inoculation of guinea pigs or hamsters, followed by plating of the dying animal’s heart blood onto selective agar and subsequent identification of resultant colonies (99, 325). Subsequently, multiple methods for bacterial concentration from water samples have been trialed, including centrifugation, chemical precipitation, and filtration (113, 125, 277, 326). The use of Moore’s swabs to detect B. pseudomallei in flowing water has proven successful in one setting, and although it is an inexpensive and simple method, it does not provide the ability for quantification, and as with soil culture-based methods, the environmental burden may be underestimated (113).
Although no current standard exists, real-time PCR following an enrichment culture has demonstrated a sensitivity nearly double that of culture alone, as well as 100% specificity (277, 327, 328). This technique is also both quicker and less labor-intensive than culture. The current major limitation for molecular detection is cost (277). A second limitation is the inability to perform phylogenetic analysis without cultured organisms, therefore limiting the ability to compare strains between samples and locations (277). The increasing research and use of molecular techniques on environmental samples are likely to improve and expand the current epidemiological data with regard to regions of previously unidentified endemicity, and accurate quantification.
TYPING
In the setting of epidemiological investigation and typing, multiple techniques have been trialed, including ribotyping, multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), randomly amplified polymorphic DNA (RAPD), multiple-locus variable number tandem repeat analysis (MLVA), repetitive element PCR (rep-PCR), variable amplicon typing (VAT), and central intermediary metabolism (CIM) (329, 330). The original ribotyping method was developed in Australia by Lew and Desmarchelier, comparing patterns of restriction fragment length polymorphisms in rRNA genes hybridized to Escherichia coli 16S and 23S rRNA (331). Using this method, Currie et al. were able to demonstrate persistence of an identical ribotype in Western Australia over 25 years (332), therefore confirming the utility of ribotyping as an epidemiological tool (332). Shortly thereafter, ribotyping was used to effectively differentiate B. pseudomallei and the yet-to-be-named B. thailandensis (237, 333). However, these ribotyping studies reported only a few ribotypes, which were subsequently determined to be heterogeneous when assessed by RAPD analysis (334). In relation to the amount of genome visualized by these techniques, PFGE allows approximately 50% visualization, as opposed to 0.1% with ribotyping (335). Therefore, PFGE was introduced to improve discrimination between strains and was subsequently used in outbreak investigations (67, 336, 337). Due to a long turnaround time and expertise requirements, PFGE was not widely adopted. One attempt to improve the efficiency of typing was via an automated ribotyping method (338). Compared to PFGE, automated ribotyping was double the cost, but it produced similar discrimination with a faster turnaround time (338). In an outbreak setting, automated ribotyping may be preferable to PFGE; however, it is important to recognize that PFGE has greater discriminatory power than most typing modalities (330, 339). More recently, LPS strain typing has been developed using a monoclonal antibody immunoassay (340). This method is based on the premise that there are three LPS types which may confer differing severities of disease and may also have distinct epidemiologies (341–343). The use of LPS typing in an Australian setting revealed a distinct geographical relationship and correlated with MLST (344). Further international analysis is required to better understand the future utility of LPS typing both from epidemiological and clinical perspectives.
Compared with the aforementioned methods, molecular typing has greater portability and a robust ability for interlaboratory comparison (339, 345). MLST using seven housekeeping genes, ace (acetoacetyl coenzyme A reductase), gltB (glutamate synthase), gmhD (ADP-l-glycero-d-manno-heptose 6-epimerase), lepA (GTP-binding elongation factor), lipA (lipoic acid synthetase), narK (nitrite extrusion protein), and ndh (NADH dehydrogenase), is able to discriminate B. pseudomallei from B. mallei and B. thailandensis (339, 346). The MLST narK locus appears to be specific for the B. pseudomallei complex, as it is absent in all other Burkholderia species with the exception of Burkholderia ubonensis, which has a unique sequence (346). A significant advantage of MLST is the ability to compare all strains in a single online database, resulting in the capability for rapid comparison and international epidemiology analysis (345). A drawback to MLST is the limited number of genes analyzed and the potential to not identify gene rearrangement outside these regions. The eBURST algorithm is unreliable in inferring geographic origin of STs (347, 348). A supplement to MLST in the setting of isolates of unknown origin may be internal transcribed spacer (ITS) sequencing (349). Analysis of ITS length polymorphisms of Burkholderia spp. indicated 10 types, of which types C, CE, and E predominate in Australia and Southeast Asia, while type G is associated with isolates from the Western Hemisphere (158, 349, 350). Additionally, in vitro and murine virulence data suggest no clear difference between types (351). Therefore, this typing method may assist in determining isolate origins but is likely to be superseded by whole-genome sequencing (WGS) (350).
Similar to MLST, CIM is regulated by conserved housekeeping genes that encode metabolic function. One study selected 12 CIM genes, as they represented 100% coverage across the 48 strains tested. This study demonstrated a slightly greater discriminatory index for CIM than for MLST, although this was not statistically significant and may be associated with greater sequence lengths of CIM genes than of MLST genes (330). It appears that CIM analyses produce greater discriminatory capacity across different geographical regions, specifically, the ability to differentiate Australian and Asian strains with greater resolution (330).
The whole-genome sequence of B. pseudomallei strain K96243 was reported in 2004. It is composed of two chromosomes consisting of 4.07 and 3.17 Mbp, which places it into the largest 5% of sequenced microbial genomes (352, 353). Chromosome 1, the larger of the two, contains a greater proportion of coding sequences involved in core cell function, such as metabolism, biosynthesis, and motility. Chromosome 2 contains coding sequences primarily associated with organism environmental adaptation, including siderophore activity. However, it also contains an rRNA gene cluster involved in amino acid biosynthesis (353). In comparison to the equivalent B. mallei chromosomes, B. pseudomallei has genetic differences of 16% in chromosome 1 and 31% in chromosome 2. The whole-genome size is 1.31 Mb larger in B. pseudomallei (353).
B. pseudomallei is considered to be a highly recombinogenic organism, with an open genome expected to result in new gene discovery (19, 354). The genome demonstrates substantial diversity among strains, with 14% of the accessory genome of the K96243 strain variably absent from 94 strains tested (355). Another study analyzing 37 strain genomes revealed that 74% of genes appear to be associated with the core (354). Furthermore, this reference genome contains 16 large variable chromosome segments called genomic islands (GIs) (356). Sequencing of five reference strains has identified 71 distinct GIs. These GIs contain specific G+C content compared to the rest of the genome and usually contain mobile genetic elements (357). Most GIs are located adjacent to tRNA genes, and these insertions may in fact be mediated by tRNA. This allows site-specific integration and recombination (SSR) and has been termed “tRNA-mediated site specific recombination of tRNA-SSR” (357). Acquisition of GIs appears to be a major source of genetic diversity among bacterial strains. While GIs may play a role in virulence, the current literature is inconclusive. An analysis of severe neurological melioidosis in Australian strains demonstrated a specific absence of two GIs (358). Additionally, human melioidosis cases clustered based on accessory gene content and specific GIs compared to those in animal and environmental isolates (355). Further characterization of GIs and associated clinical manifestations may aid in diagnosis and management in the future.
In the future, WGS is likely to supersede the aforementioned methods of typing, and it has recently demonstrated the ability to resolve the origin of two isolates with identical STs from different continents (347). Within recent years, accessibility to WGS has increased with the arrival of high-throughput next-generation sequencing and the associated rapid decrease in instrument and sequence cost. This technology is becoming a routine part of public health epidemiological and outbreak analysis. With our current understanding of the ability for B. pseudomallei to either mutate rapidly under external pressure or persist unchanged in specific environmental niches for years, the discriminatory power of WGS is likely to substantially improve our understanding of this organism in clinical and epidemiological contexts (359, 360).
PATHOGENESIS AND VIRULENCE
B. pseudomallei is an environmental organism, with many factors facilitating its persistence and survival in often harsh environmental conditions (40). These factors may contribute to facilitating contact with susceptible hosts. Survival has been demonstrated in a large range of pH differences, salt concentrations, a range of temperatures, and in the presence of detergent (361, 362). Once B. pseudomallei leaves the external environment and enters a host, the pathogenesis of disease often follows a defined sequence of events. In vivo animal experiments have been extensively utilized, as they reflect the natural disease process and parallel events that occur in human hosts. Numerous animal models of infection with B. pseudomallei have been developed. These include the nematode Caenorhabditis elegans, small mammals such as rats, hamsters, and commonly used mice (including BALB/c for acute and C57BL/6 for chronic infection modelling), and large mammals such as goats, pigs, and nonhuman primates (363–366). The murine model has been found to be the most applicable, with the type of mouse, route of infection, and infecting dose all adaptable to help mimic various disease states encountered in humans (367, 368). Additionally, genetic and immunological techniques can be employed within these experiments to further our understanding of the disease process (369).
Host Cell Attachment
Intracellular invasion with subsequent survival is a crucial component of the pathogenesis of B. pseudomallei. Multiple virulence factors enhance the ability of B. pseudomallei to evade host defenses and replicate in host cells. To successfully invade human hosts, B. pseudomallei in its environmental reservoir must attach to and invade epithelial cells and macrophages (370). Initial adhesion, demonstrated in free-living protozoan species Acanthamoeba astronyxis, is facilitated by polar flagella, with viable bacteria being observed in both vacuoles and the cytoplasm after engulfment (58, 371). It is postulated that a similar process occurs in the human host. Mutations in the flagellar structural gene fliC stop endocytosis into amoebae in the experimental setting (371). Type 4 pili likely also play a role in B. pseudomallei adherence and virulence. A strain with a mutated gene encoding a pilus structural protein, PilA, showed reduced adherence and virulence compared to those of the wild-type pilA strains (372). Attachment to human pharyngeal epithelial cells is mediated via a thin polysaccharide layer around the bacteria, which binds to the asialoganglioside aGM1-aGM2 receptor complex (373, 374). This attachment is purported as one of the initial steps in the pathogenesis of colonization of pharyngeal epithelial cells and subsequent respiratory tract infection (373).
Intracellular Invasion
B. pseudomallei can be internalized by both phagocytes and nonphagocytes (375). In nonphagocytic cells, PilA and the adhesins BoaA and BoaB are essential components for uptake (376). A type III secretion system cluster 3 (T3SS-3) and type VI secretion system cluster 1 (T6SS-1) are essential for intracellular invasion, survival, and subsequent growth of B. pseudomallei and therefore are highly conserved (224, 377–379). The T3SS-3 and T6SS-1 genes are regulated by a TetR-type regulator, BspR (380). Further, BsaN functions as a transcriptional regulator of BspR, activating a subset of T3SS-3 and T6SS-1 loci (377). Genes regulated by BsaN are essential for transcriptional activation (377). Additionally, BicA acts as a chaperone to control the expression of the T3SS-3 translocon and effector, as well as associated regulatory genes. The BsaN/BicA complex, by altering gene expression, likely contributes significantly to the adaptation and intracellular survival of B. pseudomallei within host cells (377).
The function of the T3SS is to insert a multitude of effector proteins into the target eukaryotic cell which can undermine host cell function and therefore immunity (381, 382). The structure of the T3SS traverses the inner and outer bacterial cell membranes, forming an external needle-like projection which enables the export of effector proteins from the bacterial cytoplasm into host cells via a pore created in the host cell membrane by translocator proteins (381, 382). Three translocator proteins have been described, Burkholderia invasion proteins (Bip) BipB, -C, and -D (369). Specifically, bipB and bipD mutations may result in impaired transfer of effector proteins, reduced intracellular replication, reduced formation of multinucleated giant cells (MNGC), and induction of infected macrophage apoptosis (383, 384). BipC also plays a substantial role in B. pseudomallei virulence not only as a translocator but also as an effector (385, 386). BipC appears to affect adhesion, invasion, actin formation associated with motility and therefore both inter- and intracellular spread, endosomal membrane lysis and thus endosomal escape, and, finally, direct macrophage cytotoxicity (385, 386). Another key effector protein, BopE, has in vitro activity as a guanine nucleotide exchange factor, allowing alteration of the host cytoskeleton, with bopE mutants showing decreased epithelial invasion (387).
Once in the intracellular environment, B. pseudomallei is able to survive within the endosome by production of a protease inhibitor, ecotin (388). The organism then escapes the primary endosome via T3SS, replicates in the cytosol, and localizes to the nuclei of infected cells, suggesting a location of potential intracellular persistence (389–391).
Survival within Macrophages
B. pseudomallei multiplies in phagocytes often without activating a bactericidal response (375). When lysosome fusion does occur, proliferation of surviving bacteria overwhelms the phagocyte (392). For replication within the cytosol, B. pseudomallei upregulates the purine, histidine, fatty acid, and amino acid biosynthesis pathways (393). Reactive oxygen intermediates play an important role in controlling intracellular replication. B. pseudomallei suppresses inducible nitric oxide synthase (iNOS) expression by activating expression of two negative regulators, a suppressor of cytokine signaling 3 (SOCS3) and cytokine-inducible Src homology 2-containing protein (CIS) (394). The importance of iNOS intracellular pathogenesis is highlighted by the observation that gamma interferon (IFN-γ) induction of iNOS in activated macrophages is crucial for optimal clearance of the pathogen (395). Additionally, it has been hypothesized that B. pseudomallei delays polymorphonucleocyte apoptosis to facilitate ongoing intracellular survival and propagation of infection (396).
Intercellular Spread
French et al. propose that the primary means of intercellular spread is via cell fusion, with T3SS playing a fundamental role in escape from the primary endosome (390). Within the host cell cytoplasm, B. pseudomallei induces host actin polymerization, and the organism spreads to neighboring cells by either actin- or flagellum (fla2)-mediated motility (390, 397, 398). Induction of actin polymerization occurs through mechanisms that differ from those observed in Listeria, Shigella, and Rickettsia species (399). This is largely facilitated by recruitment of host actin-associated proteins Arp3, p21 (Arp2/3 complex), and alpha-actinin, resulting in an actin tail (399). In the absence of actin motility, the flagellar system is able to compensate and advance intercellular spread (390). Unique to B. pseudomallei is the employment of other virulence genes, such as bimA, in facilitating intercellular spread (400). BimA is a protein involved in actin polymerization, with bimA mutants unable to form actin tails (400). In an Australian study of virulence factors, Sarovich et al. established that 12% of B. pseudomallei isolates possessed a bimA variant with 95% homology with the Burkholderia mallei gene (bimABm) (401). This study reported a greater proportion of pneumonia in patients with bimABp and a 14-fold increased risk of neurological disease associated with the bimABm variant (401). As aforementioned, BipC appears to have an essential role in pathogenicity and virulence. Kang et al. demonstrated bipC mutants having decreased invasion, adherence, and intracellular survival in vitro (386). The bipC mutant also showed delayed endosomal escape and actin-based motility, a key component of intercellular spread (386).
Six T6SS gene clusters are encoded in the B. pseudomallei genome (402). T6SS-5 has repeatedly demonstrated a significant role in both intercellular spread and virulence (223, 379, 402). There are two genes which encode vital components of the T6SS, hcp and vgrG (403). Hcp creates tubules which facilitate protein translocation across the membranes of host cells. Valine-glycine repeat protein (VgrG) is required for cell fusion and therefore intercellular organism spread (403, 404). Impairment of T6SS-5 in B. thailandensis results in marked attenuation of virulence in wild-type strains but not in mice lacking the Toll-like receptor (TLR)-dependent central innate immune adapter protein MyD88. This finding suggests that T6SS-5 is utilized by the bacteria to surmount the innate immune response (405).
Formation of Multinucleated Giant Cells
A feature of B. pseudomallei and related species is the ability to stimulate host cell fusion, thought to be partly related to their actin polymerization phenotype (384). A result of cell fusion is the formation of MNGC (397). The theoretical rationale for this feature is to promote localized dissemination and immune system escape (406). These giant cells have been demonstrated in infected tissues (407). Inactivation of the T6SS prevents MNGC formation and results in impaired virulence and intercellular spread (379, 406). Several other virulence factors appear to be essential for successful formation of MNGC, including expression of the gene lfpA and functional T3SS-3, with T3SS-3 mutants exhibiting delayed MNGC formation (408, 409). Further studies have implicated the T3SS-3 effector protein BipB and the sigma factor RpoS in formation of MNGC (384, 410).
Secondary Spread
Secondary spread refers to the dissemination of an infecting organism from its primary site of infection. While not confined to B. pseudomallei, nevertheless, this organism commonly displays this feature. Respiratory melioidosis is a common initial infection presentation, often preceding bacteremic spread to a variety of sites, such as the prostate, liver, and occasionally the central nervous system (CNS) (8). A study utilizing intubation-mediated intratracheal inoculation in mice has identified key virulence factors for respiratory melioidosis. Transposon sequencing with mutagenesis was used to determine key virulence genes required for in vivo fitness, by determining phenotypic outcomes after disabling selected genes. T3SS-3, T6SS-5, and capsular polysaccharide were identified as essential virulence factors in respiratory melioidosis (411).
B. pseudomallei produces a capsular polysaccharide (CPS) (412). The genes involved in the CPS production demonstrate significant sequence homology to those genes that produce a capsule in Haemophilus influenzae and Neisseria meningitidis (412). B. pseudomallei CPS impairs opsonization, reduces complement efficacy, and therefore is antiphagocytic and enables organism persistence in blood, resulting in increased ability to infect end organs (412–414).
Virulence
Multiple virulence factors accounting for the pathogenicity of B. pseudomallei have been described, with many factors thought to play only a minor individual role in virulence (370). The production of a capsule, as previously described, and biofilm formation have significant phenotypic diversity, with the capsule contributing to the initial biofilm deposition (415, 416). Multiple other described virulence factors include LPS, flagella, pili, quorum sensing (QS), T3SS, T6SS, and morphotype switching (370, 405).
LPS is an immune stimulating antigen, and LPS of B. pseudomallei is comprised of a core, lipid A, and O-polysaccharide (OPS) components (417). B. pseudomallei has been characterized as having 3 district lipopolysaccharide antigenic types, with smooth serotypes A and B and a rarer rough serotype (418). No immunological cross-reactivity occurs between serotypes, and they share similar macrocyte activation and endotoxic potency, with serotype A accounting for 97% of clinical isolates (418). LPS from a high-virulence strain significantly activated the innate immune response, suggesting that the immunopathogenesis of these strains is distinct from that of infections with less clinically virulent strains (419). Further assessment of the lipid A portions of the LPS using MALDI-TOF MS were performed and demonstrated substantial structural differences that may account for various host responses to LPS (419). This study was limited by the number of isolates used, and subsequent reports suggest that the structure of lipid A is highly conserved in a multitude of clinical and environmental isolates and that the presence of OPS may regulate LPS-associated innate immune responses in melioidosis (417).
Various in vitro and animal studies have shown mixed results for the contribution of flagella and pili to virulence in human infections, and this needs to be further defined (370). QS, a population density-mediated cell-to-cell communication mechanism, is mediated by signaling molecules, such as N-acyl-homoserine lactones. B. pseudomallei strains with inactivated genes encoding QS pathways have been shown to have reduced virulence in animal models (420, 421). T3SS-3 is thought to have various roles in virulence in human infection, as described previously. Mutants have been observed to have reduced pathogenicity (370). However, our understanding is likely incomplete, with a study characterizing the vacuolar escape defect in the bsaZ T3SS-3 mutant as having delayed rather than complete abrogation of virulence (409). The K96243 B. pseudomallei genome encodes six T6SS gene clusters, which is greater than for other bacteria (402, 422). A murine model with BPSS1504 deletion, encoded in T6SS-1, resulted in decreasing intracellular replication and formation of MNGC (423).
B. pseudomallei secretes various exoproducts, such as proteases, lipases, and phospholipases, via the type 2 general secretory pathway (gsp). There is no clear correlation of reduced virulence looking at gsp mutants lacking secreting ability, therefore suggesting that exoproduct secretion has a minor role in virulence (424, 425). The rpoS gene is involved in the response to nutrient restriction in the stationary phase of bacterial cell growth (426, 427). rpoS is associated with regulation of proteins involved in maintaining the integrity of the cell envelope, and rpoS mutants have reduced ability to repair cell wall damage, leading to decreased intracellular survival in macrophages and attenuation in animal models (428, 429).
Morphotype switching or variation in colonial morphology is associated with different potential virulence factor expression. Mechanisms allowing the bacteria to survive under adverse conditions may have key implications for host-pathogen interactions and intracellular persistence (430). Similar to other bacterial species, B. pseudomallei exhibits small-colony variants (SCVs) with unique phenotypic and pathological features (431, 432). SCVs often have reduced susceptibility to antibiotics and may have an enhanced ability to cause latent or recurrent infection, which may in part be due to greater biofilm producing ability and intracellular persistence (432, 433). Certain colony variants, such as yellow colony variant B, are capable of survival in the stomach environment (434). Approximately 8% of clinical samples of B. pseudomallei show colonial variation on Ashdown’s media (435). In vitro studies have shown increased cellular adherence with SCVs compared to wild-type (WT) isolates, although cellular invasion and damage were lower in SCVs. Further, using electrophoresis analysis, protein expression was significantly different between SCVs and WT variants (436). Different B. pseudomallei morphotypes are likely to have differential epithelial adherence and environmental and intracellular survival abilities. Differential expression of virulence-associated proteins during the mid-logarithmic growth phase plays a key component of morphotype switching and variation (436). Further studies will be required to increase our understanding of how this relates to pathogen-host interaction and clinical presentation.
Genomics of Virulence and Pathogenesis
B. pseudomallei has a complex genome with a high rate of horizontal gene transfer (19). Substantial variation among genomes likely contributes to differential virulence, which is often geographically defined (19). B. pseudomallei genomic analysis shows variable numbers of genomic islands, with the strain K96243 having 16 (353). Furthermore, Thai strains, including K96243, contain the horizontally acquired Yersinia-like fimbrial (ylf) gene cluster. In comparison, Australian strains contain the B. thailandensis-like flagellum and chemotaxis gene cluster (437). There are several possible metabolic, virulence, and regulatory genes present in a more virulent Australian strain of B. pseudomallei (MSHR668) compared to two Thai strains (K96243 and 1106a) in a mouse model (438). Determination of virulence gene expression using DNA microarrays has been performed using a murine model for both chronic infection phenotype (C57BL/6 Th1 phenotype with moderate cytokine elevation, relatively resistant to B. pseudomallei infection) and acute infection phenotype (BALB/c Th2 phenotype with high cytokine elevation, relatively susceptible to B. pseudomallei infection). These murine models are thought to broadly correlate to disease patterns of melioidosis seen in human disease. Upregulation of bprD, a transcriptional regulator in the T3SS-3 operon, occurred in C57BL/6 mice. Further, BALB/c mice infected with a bprD mutant strain of B. pseudomallei had decreased survival time. This information sheds light on the complex interactions of host and bacterial factors, which lead to different disease outcomes (439). Ongoing studies are required to further define the genetic basis of virulence of different strains.
Host Response
Several host factors have been observed to increase the risk of contracting melioidosis, including diabetes mellitus, renal failure, and excess alcohol consumption. Disease outcomes—whether asymptomatic, acute, chronic, or latent disease—are thought to be determined largely by host response (440). An exaggerated immune response with hyperproduction of proinflammatory cytokines can result in tissue destruction and organ failure (441, 442). While extensive work has occurred in this area, it is important to realize the contrasting data between human and animal models.
Neutrophils play a critical role in host response by killing up to 90% of intracellular B. pseudomallei organisms (443–445). Intracellular neutrophil function occurs in a T3SS-dependent manner, as B. pseudomallei mutants lacking the T3SS do not induce macroautophagy or bacterial endosome escape into the cytosol (446). Furthermore, neutrophils are able to eliminate extracellular bacteria via neutrophil extracellular traps and promote indirect generation of the host cytokine response (443, 447). Importantly, neutrophil phagocytic function in older patients or those with poor glycemic control is likely to be reduced (444).
The alternative complement pathway is activated by B. pseudomallei, although opsonization with complement does not appear to be essential for uptake into phagocytes or subsequent bacterial killing (397). Antibody-enhanced complement activation is adequate for neutrophil clearance of B. pseudomallei, unlike the case with macrophages, which are ineffective at clearing serum-opsonized B. pseudomallei unless preactivated with IFN-γ (448). Neutrophils are able to effectively destroy B. pseudomallei and B. thailandensis organisms that attain a critical threshold of complement deposition. Phenotypic virulence demonstrated in vivo may be attributed to the contrasting ability to resist surface opsonization (449). Reduced levels of lysosomal fusion are seen in macrophages of melioidosis patients, with resultant high bacterial burden. Depletion of both neutrophils and macrophages has been shown to enhance infection and mortality rates in animal models (447, 450).
TLRs recognize conserved pathogen-associated molecular patterns (PAMPs) and mediate the inflammatory response via various signaling pathways. Myd88 knockout mice have increased susceptibility to B. pseudomallei infection as a result of reduced neutrophil activity (451). It is likely that TLRs in humans play a role in pathogenesis, with increased expression of TLR1, TLR2, and TLR4 noted in patients with melioidosis and subsequent decreased expression observed on recovery from acute illness (452).
The role of antibodies in protection against infection remains uncertain. There is no clear correlation between disease severity and survival with regard to level of antibodies measured against LPS (453). Recurrent disease has been seen in the setting of high antibody levels in patients residing in areas of endemicity (454). In contrast, robust cell-mediated immunity appears to be essential for halting progression of disease. In animal models, depletion of CD4 cells but not CD8 cells leads to increased susceptibility to infection (455). This, however, does not translate to humans in the setting of HIV infection with depleted CD4 cell counts, where there is no known increased incidence (456).
Cytokines play a significant role in the pathogenesis of melioidosis, with levels of IFN-γ, interleukin 6 (IL-6), and IL-18 associated with increased severity of disease and mortality in patients with melioidosis (457, 458). Additionally, severe bacteremic melioidosis is associated with high levels of proinflammatory cytokines and is correlated with poor clinical outcomes (459). It is also thought that higher cytokine levels correspond to inoculation burden, rather than to certain virulence factors of the infecting strain (460). IFN-γ, often derived from natural killer (NK) cells, plays a key role in the immune response against melioidosis, with IFN-γ knockout mice demonstrating higher susceptibility to the organism (447). The CXC chemokines IFN-γ-inducible protein 10 (IP-10) and monokine induced by IFN-γ (Mig) bind to the CXCR3 receptor and specifically target activated T lymphocytes and NK cells (461). Although not unique to melioidosis, patients infected with B. pseudomallei had persistently elevated IP-10 and Mig levels which correlated with elevated IFN-gamma levels and mortality (461). IP-10 and Mig may aid in coordination of Th1-mediated host defense during infection by attracting CXCR3-positive Th1 cells to the site of inflammation (461). These results contrast animal models showing infection with a moderate cytokine response leading to a more chronic disease course and potential development of adaptive immunity, as well as a protective effect of IFN-γ in murine models (441, 442, 462).
The current literature indicates that acute melioidosis results from ineffective innate cellular immune response and also suggests that both antibodies and Th1-adaptive responses are necessary for the successful prevention/eradication of melioidosis (448, 463). Although challenging, given the limitations of correlating animal model data, additional human research is required to truly understand the complex host-organism immunopathological relationship. Figure 6 is a simplified overview of the main stages in the pathogenesis of melioidosis.
FIG 6.
A simplified schematic representation of the B. pseudomallei intra- and intercellular life cycles. Initial nonphagocyte host cell attachment occurs via flagella, type 4 pili, and adhesins BoaA and BoaB. Cellular invasion is facilitated by the T3SS, which injects effector proteins, including BopA, BopE, BipB, BipC, and BipD. During internalization the bacterium is enveloped by the host cell in an endocytic vesicle or endosome. Survival within the endosome occurs via multiple processes, including production of a protease inhibitor, ecotin. Escape from the endosome is mediated by the T3SS and subsequent upregulation of biosynthesis pathways, including purine, histidine, fatty acid, and amino acid, aid in replication within the cytosol. Bacilli may localize to nuclei or form a BimA-dependent actin tail used for motility and intercellular spread. The T6SS forms a bridge between host cells and transfers effector proteins which aid the formation of MNGC.
CLINICAL PRESENTATION
The clinical presentation of melioidosis depends not only on the route of infection, including inoculation, inhalation, and (rarely) ingestion, but also on bacterial load, strain virulence, and host risk factors (279). Most commonly, exposure to B. pseudomallei does not result in an infection, and only 1 in 4,600 seroconversion-associated exposures results in clinical disease (464). Approximately 4% of cases have been attributed to latency as opposed to acute infection, although this is well described (279, 465, 466). The majority, 85%, of patients present with an acute infection, defined as less than 2 months of symptoms (8). The general incubation period of B. pseudomallei has a mean of 9 days (range, 1 to 21) (467). Notably, there are reported cases of late reactivation, with one occurring 29 years postexposure (468). Pneumonia is the most commonly reported presentation, ranging from 51 to 61%. Bacteremia is commonly associated with pneumonia and is found in 55 to 74% of patients with pneumonia (Table 2) (8, 12). These patients presented with dyspnea and a productive cough (279). Septic shock was diagnosed in 21 to 34% of patients (8, 12). Previously reported data demonstrate rainfall 2 weeks prior to presentation as an independent risk factor for pneumonia, septic shock, and death (4, 53). Although severe weather events have been linked with outbreaks, only 2 of 13 events were associated with case clusters in one Australian study, and no association was found in another (55, 469). Pneumonia after an inoculating event is recognized and thought to occur via hematogenous seeding (279). Bacteremic patients often present profoundly unwell, with high fevers and often limited localizing symptoms. Chest X-ray features may rapidly progress from limited infiltrates to diffuse consolidation, abscess formation, and cavitation (470).
TABLE 2.
Comparison of clinical presentation by region
| Clinical presentation | No./total (%) |
||||
|---|---|---|---|---|---|
| Australia (8, 12, 622) | Malaysia (87, 88, 166–168) | Singapore (94, 95) | India (122, 130, 131) | Thailand (17, 33, 72, 79, 473, 623) | |
| Fever | —a | 119/128 (93) | 293/372 (79) | 21/32 (66) | 95/134 (71) |
| Pulmonary infection | 412/763 (54) | 225/537 (42) | 203/614 (33) | 53/226 (23) | 121/247 (49) |
| Skin/soft tissue infection | 123/761 (16) | 99/402 (25) | 65/372 (17) | 23/180 (13) | 35/247 (14) |
| Bone and joint infection | 22/597 (4) | 35/402 (9) | 2/372 (1) | 36/226 (16) | 35/247 (14) |
| Genitourinary infectionb | — | 10/360 (3) | — | 5/180 (3) | 8/134 (6) |
| Neurological infection | 22/751 (3) | 21/370 (6) | 2/372 (1) | 19/180 (11) | 2/30 (7) |
| Liver abscess | 17/597 (3) | 46/537 (9) | — | 20/180 (11) | 93/247 (38) |
| Splenic abscess | 30/597 (5) | 45/537 (8) | — | 20/226 (9) | 68/247 (28) |
| Prostate abscess | 83/408 (20) | 5/225 (2) | — | 8/226 (4) | 13/155 (8) |
| Parotid abscess | 2/794 (0) | — | 77/226 (3) | 5/134 (4) | |
| Mycotic pseudoaneurysm | 3/597 (1) | 4/67 (6) | — | — | — |
| Pericardial effusion/pericarditis | 4/540 (1) | 1/67 (1) | 1/372 (0) | 3/180 (2) | — |
| No clinical focus | 66/597 (11) | 69/392 (18) | — | 9/95 (9) | — |
| Septic shock | 174/715 (24) | 93/225 (41) | — | 32/180 (18) | 26/134 (19) |
| Bacteremia | 477/794 (60) | 391/537 (73) | 592/986 (60) | 87/226 (38) | 491/874 (56) |
| Mortality | 118/794 (15) | 212/527 (40) | 260/614 (42) | 32/180 (18) | 1,205/2,913 (41) |
—, no data reported.
Excluding prostate.
Other common clinical manifestations include abscess formation (most commonly prostate, spleen, liver, and kidney) and genitourinary, skin and soft tissue, musculoskeletal, and neurological involvement (8). In Australian male patients, prostatic abscesses are present in 15 to 21% of cases. This is a higher rate than the 1 to 13% observed in Southeast Asia. It is possible that decreased clinical and radiographic detection in Southeast Asian patients contributes to this difference (8, 79, 87, 166, 167, 471–473). Of note, an absence of urinary symptoms has a reported negative predictive value (NPV) of 96%, and a urinary leukocyte count of <50 × 106 cells/liter has an NPV of 100% (471). Furthermore, digital rectal examination was negative in 55% of proven prostatic abscesses.
A recent literature review revealed just 43 reported cases of primary cutaneous melioidosis (474). The majority of cases (67%) occurred in travelers, with Thailand (48%) being the most common destination (474). Interestingly, in Australian children, primary cutaneous melioidosis is a common presentation (49%), with only 20% presenting with pneumonia and 16% bacteremic (475). In Cambodia and Thailand, children most frequently present with skin and soft tissue infection, suppurative parotitis, or cervical lymphadenopathy (110). Cervical lymphadenopathy accounts for up to 40% of cases of localized melioidosis, which is thought to be associated with contaminated water sources (105, 110, 476, 477). In contrast, parotitis accounted for only 1% of cases in a Malaysian cohort. Similar to adult presentations, disseminated disease and pneumonia were most common in this cohort (89). An Australian report details the largest nosocomial melioidosis outbreak to date, secondary to contaminated saline and associated with soft tissue infections. All 6 cases developed superficial infection that resolved with parenteral and oral antimicrobial therapy (478). Cutaneous manifestations of melioidosis appear to be common in relation to contaminated medical or cleaning products (362, 478, 479).
In Australia, central nervous system involvement has been reported in 3 to 4% of cases in adults and 7 to 33% in children (8, 12, 475, 480). There appear to be fewer reported cases in Asia, with studies from Thailand suggesting an incidence of 1.5 to 3% and a Malaysian study reporting 7.5% (167, 481, 482). The most common presenting symptoms of neuromelioidosis include fever (82%), headache (54%), and cranial nerve palsy (52%) (483). Additional symptoms may include weakness, ataxia, seizures, decreased level of consciousness, and flaccid paralysis (475, 480, 484). The two most common disease processes involving the CNS include encephalomyelitis and brain abscess (483). Diagnosis of neuromelioidosis can be challenging. In one series, only 29% of cerebrospinal fluid (CSF) samples were culture positive (480). CSF protein was elevated in 71 to 93% and monocytosis predominated in 64 to 67% of samples evaluated (480, 483).
B. pseudomallei septic arthritis and osteomyelitis are relatively uncommon in Australia, with a reported incidence of 8% (485). In this cohort, 25.4% had primary septic arthritis and 31.7% had primary osteomyelitis. Septic arthritis of the lower limb is associated with a 27.5% risk of associated osteomyelitis of contiguous bone (485). Results from Thailand reported that 48% of all cases of bacterial-culture-proven septic arthritis were due to B. pseudomallei, and 8.4% of patients had bone or joint involvement (486, 487). The most common joint affected was the knee (47 to 53%), while osteomyelitis most commonly affected the tibia (44%) (485, 486).
Imaging
Various imaging modalities play an important role in the diagnosis and management of melioidosis. In acute bacteremic melioidosis, the most common chest X-ray features are multifocal nodular lesions which are indicative of hematogenous seeding (470). Chest radiographs may also demonstrate rapidly enlarging coalescent nodules and subsequent cavitation in this patient group (470). In acute nonbacteremic melioidosis, the most common chest X-ray findings are that of upper lobe consolidation and possible cavity or, less commonly, abscess formation (470, 488). Subacute and chronic pulmonary infections most often present with slowly progressive upper lobe consolidation which may mimic tuberculosis (470, 488). Notably, in chronic pulmonary infection, resolution is less likely to result in scarring and calcification (470, 489). Additionally, the finding of hilar lymphadenopathy is rare in melioidosis and is more likely to represent tuberculosis in regions of endemicity (470, 488, 490, 491). Computer tomographic (CT) scans of the chest may aid in delineating abscess formation and early cavitation but may otherwise be of limited additional diagnostic utility in pulmonary infection (470).
Extrapulmonary infections, specifically visceral abscesses, are common, and multiple modalities can aid in their diagnosis (488, 490, 492). On ultrasound, visceral abscesses appear as hypoechoic lesions, and multiple studies have demonstrated the role of ultrasound in diagnosis of melioidosis (492, 493). Liver and spleen abscesses are most common, and small “target-like” lesions may be suggestive of melioidosis (490, 492). For prostatic abscesses, ultrasound has a sensitivity of approximately 85%, compared with 99% by CT (472). With regard to liver abscesses, the CT scan “honeycomb sign,” described as an abscess with multiple similar-size loculations separated by thin septa, has a sensitivity of 85%, a tion of melioidosis visceral abscesses appears to favor CT scan (472, 494). However, in resource-limited settings, ultrasound evaluation of all bacteremic melioidosis cases may be warranted.
For patients with neurological melioidosis, a CT scan was negative in 50 to 73% of cases (480, 495). Magnetic resonance imaging (MRI) demonstrated either ring-enhancing lesions or leptomeningeal enhancement in all patients (480, 495). A retrospective analysis of the MRI of 10 patients with CNS culture-confirmed melioidosis showed that there was a predilection for involvement of the trigeminal nerves with contiguous spread to brain stem trigeminal nuclei (496). MRI is comparatively more sensitive for diagnosis of CNS infection, including detection of cerebral edema and microabscesses, and in the setting of clinical features would be the appropriate imaging modality (490).
RISK FACTORS FOR DISEASE
The age of patients affected is wide, ranging from 2 days to 92 years, with a median of 50 (8, 12, 166). Worldwide there is a male predominance of melioidosis cases, ranging from 58.5% in Thailand to 84% in Singapore (Table 3) (17, 95). In Australia, up to 59% of infections occur in aboriginal and Torres Strait Islanders (ATSI), which is thought in part to be associated with an increased prevalence of risk factors in this population (8, 12). Additionally, ATSI females account for 38.8% of those affected, in contrast to only 16.6% of females in the non-ATSI population (4). Precise data on the mode of acquisition of disease are limited. A case-controlled study by Limmathurotsakul et al. has clarified this to a considerable extent (497). The main modes of acquisition in a cohort of 414 patients were determined by interviewing subjects. Ingestion, inhalation, and inoculation were all implicated, with exposure to rainwater being an independent risk factor (497).
TABLE 3.
Comparison of melioidosis risk factors by region
| Risk factor | No./total (%) |
||||
|---|---|---|---|---|---|
| Australia (8, 12, 622) | Malaysia (87, 88, 166–168) | Singapore (94, 95) | India (122, 130, 131) | Thailand (17, 33, 72, 79, 473, 623) |
|
| Sex (male) | 555/794 (70) | 425/549 (77) | 821/986 (83) | 152/226 (67) | 1722/2887 (60) |
| ATSI | 414/794 (52) | —a | — | — | — |
| Diabetes | 339/779 (44) | 360/537 (67) | 562/986 (57) | 131/226 (58) | 1275/2464 (52) |
| Renal disease | 99/777 (13) | 48/547 (9) | 115/986 (12) | 37/180 (21) | 83/874 (9) |
| Lung disease | 181/766 (24) | 8/280 (3) | 61/986 (6) | — | 1/30 (3) |
| Chronic heart disease | 39/540 (7) | — | 9/372 (2) | — | |
| Cancer | 49/765 (6) | 8/225 (4) | 62/986 (6) | — | 23/368 (6) |
| Immunosuppression | 55/765 (7) | 24/370 (6) | — | 12/95 (13) | 25/791 (3) |
| HIV | — | 8/312 (3) | — | — | 2/83 (2) |
| Alcohol excess | 317/754 (42) | 3/202 (1) | — | 22/226 (10) | 25/204 (12) |
| Thalassemia | — | 1/67 (1) | — | — | 117/874 (13) |
| Occupational exposure | 96/540 (18) | 73/317 (23) | — | — | 262/368 (71) |
| Kava | 27/540 (5) | — | — | — | — |
| Smoking | — | — | 245/372 (66) | — | 92/204 (45) |
—, no data reported.
Patients with diabetes mellitus have a significantly greater risk of infection than nondiabetics, with a relative risk for an infection-related hospitalization of 2.2 (99% CI, 2.10 to 2.23) (498, 499). Additionally, the relative risks (RR) of cellulitis (1.8) and sepsis (2.5) attributable to any pathogen are substantially higher than those for nondiabetics (498). Diabetes is an important risk factor for melioidosis and has been reported at 37 to 56% in Australian cases, 17 to 47% in Thai cases, and up to 75% in Malaysian cases (4, 12, 72, 79, 87, 88). Because diabetes is such a pervasive risk factor, it is critical to understand that the prevalence of diabetes is expected to increase by 20% in developed nations and 69% in developing nations by 2030 (Fig. 1) (23). The adjusted odds ratio (OR) in case-controlled studies of diabetes in melioidosis cases compared with controls has been reported at between 5.9 (95% CI, 4.0 to 8.9) and 12.9 (95% CI, 5.1 to 37.2) (32, 33). In a population analysis, the relative risk was reported at 13.1 (95% CI, 9.4 to 18.1) (4). India is the country with the highest predicted number of annual melioidosis cases, and it has a predicted rise in diabetes prevalence of the total adult population from 7.1% in 2010 to 8.6% in 2030, an increase from approximately 51 million to 87 million diabetics (Fig. 1) (21, 23). Diabetes specifically alters immune function through decreased chemotaxis, phagocytosis, cytokine response, and bacterial killing and thus increases risk of infection (500, 501). Polymorphonuclear leukocytes (PMN) are a vital component of innate immunity and in prevention of B. pseudomallei infection directly and via neutrophil-derived chemokine signaling of macrophages (445, 502–504). Both the PMN response to and release of the neutrophil signaling chemokine IL-8 are delayed in diabetic patients, and given that even in healthy individuals B. pseudomallei is a poor activator of IL-8 from lung epithelial cells, diabetics may therefore be at greater risk of infection via inhalation (445). PMN phagocytosis of B. pseudomallei is reduced, and the ability to delay apoptosis is lost in diabetic patients. While not statistically significant in one study, oxidative burst also appeared to be reduced in diabetic patients with melioidosis (445). Aside from PMN activity, diabetic patients also exhibit impaired T cell immunity to specific antigens during acute melioidosis and also reduced activity of IL-23 and IL-10, both of which are likely to be important cytokines in the innate immune response to B. pseudomallei (505, 506). It is interesting that the diabetic medication glibenclamide impairs cytokine production and migration of polymorphonuclear cells, via intracellular depletion of glutathione and glutathione peroxidase, after exposure to B. pseudomallei (507). More than 50% of Thai diabetics are prescribed glibenclamide, and this may contribute to the burden of melioidosis in that country (507, 508). Additionally, while glibenclamide may increase the susceptibility of diabetic patients to infection, it may also have a mortality benefit associated with a decreased inflammatory response (509). Further research into this mechanism of cellular alteration is required both for prognostication and potential therapy (507).
Alcohol intake appears to be a significant risk factor in Australia, but less so in other countries (136, 486). This may in part be due to reporting bias and has also been attributed to religious beliefs, as alcohol is prohibited in Islam (17, 88, 510). Australian studies report that 37 to 52% of melioidosis cases are affected by hazardous alcohol intake at time of diagnosis, with an adjusted RR of 2.1 (95% CI, 1.6 to 2.6) (4, 12). Similarly, chronic lung disease appears to have the greatest representation in Australian data, with studies reporting a prevalence of 15 to 26% and RR of 6.7 (95% CI, 4.7 to 9.6) (4, 12). One Malaysian study reported the prevalence of chronic lung disease to be 3% (88).
The prevalence of chronic renal disease in melioidosis cases has been reported at 6.0 to 11.4% in Malaysia, 6 to 18% in Thailand, and 9 to 16% in Australia, with a reported RR of 3.2 (95% CI, 2.2 to 4.8) (4, 12, 79, 87, 88, 486). Other less common risk factor include malignancy (4 to 8%), rheumatic heart disease or cardiac failure (2.5 to 7%), immunosuppression (10%), and kava (Piper methysticum) use (8, 12, 79, 87). It is interesting that there does not appear to be a direct correlation between human immunodeficiency virus (HIV) infection and risk for melioidosis (456, 511). Independent risk factors for presentation with pneumonia include recent rainfall, chronic lung disease, hazardous alcohol consumption, and rheumatic heart disease and/or congestive cardiac failure (8, 53). Thalassemia is another significant risk factor for melioidosis. In a Thai study, patients with beta-thalassemia had an 11-fold risk of disease (33). A Malaysian cohort of pediatric beta-thalassemia major patients had an annual incidence of 140 cases per 100,000 population, compared with 0.33 case per 100,000 population in patients without thalassemia (512). Notably, patients presenting with chronic melioidosis, representing approximately 11% of all cases, are less likely to have diabetes (8). Reactivation of melioidosis is associated with chronic lung disease and rheumatic heart disease and/or congestive cardiac failure (8).
In a region where B. pseudomallei is endemic, risk factors may include simple activities of daily living. A matched case-control study in Thailand identified several factors associated with increased risk of infection. With regard to skin inoculation, working in rice fields, an open wound, walking barefoot more than once per week, and bathing with pond water all increased risk of infection (33, 497). Acquisition of infection via ingestion was most commonly associated with eating food contaminated by soil or dust and drinking untreated water. Inhalation events were associated with outdoor exposure to rain or dust and current history of smoking (497).
Information regarding genetic risk factors has increased in recent years. Nucleotide binding oligomerization domain 2 (NOD2) is a receptor able to recognize pathogens and aid in immune function. NOD2 genetic variation has been implicated in inflammatory and infective conditions such as pulmonary tuberculosis (513, 514). With respect to B. pseudomallei infection, NOD2-deficient mice are more susceptible to pulmonary infection and dissemination (515). Additionally, in a review of 1,562 Thai patients, a specific polymorphism in the NOD2 region, rs7194886, appeared to have an association with B. pseudomallei infection, which was surprisingly greater in females, with an odds ratio of 12.56 (515). Furthermore, polymorphisms in TLRs may have beneficial or detrimental effects. A case-control study of melioidosis patients demonstrated both protective benefit and increased risk of disease with different TLR4 genetic polymorphisms. Interestingly, this study also revealed an association between TLR6-1-10 region genetic variants in diabetic patients with melioidosis, where homozygous diabetic patients had an 8-fold-lower rate of B. pseudomallei infection specifically (516).
Recurrence
Recurrent melioidosis is defined as the new onset of signs and symptoms of infection with culture-proven B. pseudomallei after response to therapy (517). Recurrence may be further stratified into relapse, where primary and repeat B. pseudomallei culture results are genetically indistinguishable, and reinfection, where isolates do not share an identical genetic composition (517, 518). In Australia, the recurrence rate is reported at 5.7% with a median time to relapse of 9.4 months (range, 3.6 to 28.0). Relapse is commonly associated with poor antimicrobial compliance (8, 519). However, reevaluation suggests that an overall improvement in relapse rates may be due to an increased length of intravenous therapy (519). In a Darwin-based study, notably, only 1 (0.4%) episode of relapse was identified from 2010 to 2012 in a cohort of 223 melioidosis survivors, suggesting an improvement in eradication therapy (519). In Northern Thailand, the most recent study reported a recurrence rate of 6%, which is substantially lower than in previous studies, which reported that the rate of recurrence was 13 to 17% (517, 518, 520). Of the available samples genotyped, 75% of these cases were defined as relapse; therefore, 25% were reinfection with a different strain (510). In this study, the key determinants of relapse were antimicrobial agent prescribed and duration of therapy. Risk of recurrence was decreased by 90% when therapy was prescribed for greater than 12 weeks, compared with 8 weeks or less (510). Furthermore, a predictive scoring system was developed to differentiate relapse from reinfection. Taking into account duration of oral therapy, interval between primary infection and recurrence, season, and renal function, a quantifiable score was created using the area under the receiver operator curve. A score of less than 5 was able to correctly identify 85% of patients with relapse and therefore potentially impact treatment (517). While most members of the at-risk population do not develop melioidosis in regions of endemicity, there is a subset of patients that are susceptible to recurrence and may therefore represent a yet-unknown host-related association with recurrent infection.
Latency in melioidosis is also described in the literature, and periods of latency between 18 and 29 years are reported (466, 468, 521). One case suggested a latency of 62 years; however, genetic analysis of this isolate demonstrated a likely Western Hemisphere clade that was distinct from the patient’s Southeast Asia exposure history (465, 522). Additionally, it was not determined if the patient had symptomatic disease with the initial infection, and therefore, it is unclear if the presentation represented reactivation or prolonged incubation (465). Current evidence suggests that clinicians should be aware of the risk of reactivation in immunosuppressed patients, irrespective of time frame, from B. pseudomallei regions of endemicity (523, 524).
MANAGEMENT
Up until the late 1980s, conventional therapy for melioidosis included chloramphenicol, doxycycline, trimethoprim-sulfamethoxazole (TMP-SMX), and kanamycin (525). The overall in-hospital mortality rate for bacteremic patients with multiple foci was up to 87% (526). At that time, trials of third-generation cephalosporins such as ceftazidime, broad-spectrum penicillins such as piperacillin, and carbapenems, including imipenem, were underway and were showing promising in vitro results (525).
Since then, many therapeutic options have been trialed for melioidosis. Ceftazidime (120 mg/kg of body weight/day) has been shown to provide a mortality benefit compared to “conventional therapy” which included chloramphenicol at 100 mg/kg/day, doxycycline at 4 mg/kg/day, and TMP-SMX at 10/50 mg/kg/day. In the initial study by White et al., in-hospital mortality was halved from 74 to 37% (527). Further evidence of efficacy followed in a trial of severe melioidosis treatment with combination ceftazidime (100 mg/kg/day) and TMP-SMX (8/40 mg/kg/day), which revealed a decrease in cumulative day 7 mortality from 47.0% to 18.5% compared with that with conventional therapy overall (528). Notably, there is currently no evidence to support the use of combination ceftazidime and TMP-SMX, as subsequent trials have demonstrated no significant difference in short- or longer-term outcomes (529, 530). B. pseudomallei is susceptible to amoxicillin-clavulanic acid, and both intravenous and oral preparations may be used as a second-line agent in patients intolerant to TMP-SMX or with sulfonamide allergy, or where other agents may be contraindicated due to pregnancy or young age (531, 532). Notably, clavulanic acid appears to be integral in the efficacy of this therapy, and pharmacokinetic and pharmacodynamics assessment suggests a need for increased frequency of dosing to maintain therapeutic levels (intravenous, 20/5 mg/kg every 4 hours; oral, 20/5 mg/kg every 8 hours) (532, 533). Caution is required when using this therapy given an increased risk of relapse and potential decreased efficacy (531, 532, 534). Retrospective reviews of other third-generation cephalosporins, including ceftriaxone and cefotaxime, have been performed, but these agents are less active in vitro and are associated with higher mortality (285, 535, 536). Carbapenems, including meropenem and imipenem, appear to have the greatest in vitro activity against B. pseudomallei (537). Furthermore, they provide a strain-dependent postantibiotic effect that may last up to 3.66 h (538).
Current therapy guidelines recommend an initial intensive phase followed by an eradication phase (Table 4) (539, 540). The 2014 Revised Royal Darwin Hospital guideline recommends an intravenous intensive phase including therapy with either ceftazidime at 50 mg/kg of body weight (up to 2 g) every 6 to 8 h if the patient is on the ward or meropenem at 25 mg/kg (up to 1 g) every 8 h if in the intensive care unit (ICU) (540). The duration of this phase is 10 to 14 days for uncomplicated infection or 4 to 6 weeks for persistent septic shock, deep-seated or organ abscesses, extensive lung disease, septic arthritis, osteomyelitis, or neurological melioidosis. The oral eradication phase includes therapy with TMP-SMX, dose depending on weight and age, for a period of 3 to 6 months. For a child of <40 kg, a dosage of 8/40 mg every 12 h is recommended. Dosage recommendations for adults include the following: <40 kg, 160/800 mg every 12 hours; 40 to 60 kg, 240/1,200 mg every 12 hours; and >60 kg, 320/1,600 mg every 12 hours (283, 539, 540).
TABLE 4.
Melioidosis treatment and prophylaxisa
| Phase or adjustment | Drug and dose | Clinical manifestation(s) (duration of treatment) |
|---|---|---|
| Intensive | Ceftazidime, 2 g i.v. (child, 50 mg/kg up to 2 g), every 6 hours, or meropenem, 1 g i.v. (child, 25 mg/kg up to 1 g), every 8 hours (2 g for CNS infection) | Pneumonia (2–4 wks); bacteremia, no focus (2 wks); skin and soft tissue (2 wks); abscess, deep tissue (4 wks); septic arthritis, single joint (4 wks); osteomyelitis (6 wks); neurological (8 wks); mycotic aneurysm (8 wks) |
| Renal dose adjustment | ||
| Ceftazidime | ||
| CLCR of (ml/min): | ||
| 31–50 | ≤60 kg, 1 g q8h; >60 kg, 2 g q8h | |
| 15–30 | ≤60 kg, 1 g q12h; >60 kg, 2 g q12h | |
| <15 | ≤60 kg, 1 g q24h | |
| Dialysis | ||
| HD | As for CLCR of <15, dose post-HD | |
| CAPD | As for CLCR of <15, dose post-HD, may administer intraperitoneally with dwell time of >6 h and 25% extra dose for convenience | |
| CRRT | 2 g q12h | |
| Meropenem | ||
| CLCR of (ml/min): | ||
| 31–50 | 1 g q12h | |
| 15–30 | 1 g q12h | |
| <15 | 1 g q24h | |
| Dialysis | ||
| HD | As for CLCR of <15, dose post-HD | |
| CAPD | As for CLCR of <15 | |
| CRRT | 1 g q12h | |
| Trimethoprim-sulfamethoxazole | ||
| CLCR of (ml/min): | ||
| 31–50 | ≤60 kg, 240/1,200 mg every 12 hours; >60 kg, 320/1,600 mg every 12 hours | |
| 15–30 | ≤60 kg, 240/1,200 mg every 24 hours; >60 kg, 320/1,600 mg every 24 hours | |
| <15 | ≤60 kg, 240/1,200 mg every 24 hours; >60 kg, 320/1,600 mg every 24 hours | |
| Dialysis | ||
| HD | As for CLCR of <15, dose post-HD | |
| CAPD | As for CLCR of <15 | |
| CRRT | As for CLCR of 15–30 | |
| Eradication | Trimethoprim-sulfamethoxazole (child, 6/30 mg/kg up to 240/1,200 mg; adult 40–60 kg, 240/1,200 mg; >60 kg, 320/1,600 mg orally, every 12 hours) and folic acid, 5 mg (child, 0.1 mg/kg up to 5 mg) orally, daily | Pneumonia (3 mo); bacteremia, no focus (3 mo); skin and soft tissue (3 mo); abscess, deep tissue (3 mo); septic arthritis, single joint (3 mo); osteomyelitis (6 mo); neurological (6 mo); endovascular (6 mo) |
| Prophylaxis | Trimethoprim-sulfamethoxazole (<40kg, 160/800 mg; 40–60 kg, 240/1,200 mg; >60 kg, 320/1,600 mg orally, every 12 hours) and folic acid, 5 mg (child, 0.1 mg/kg up to 5 mg) orally, daily, or doxycycline (2.5 mg/kg/dose up to 100 mg orally, every 12 hours) or amoxicillin-clavulanate (≤60 kg, 1,000/250 mg; >60 kg, 1,500/375 mg, every 8 hours) | High probability postexposure (3 wks) |
| Trimethoprim-sulfamethoxazole (160/800 mg orally, daily) and folic acid (5 mg orally, daily) | Hemodialysis patients, wet season, high-incidence region (26 wks) |
Adapted from the work of Peacock et al., Currie, Lipsitz et al., Majoni et al., Jabbar et al., and Inglis (3, 279, 539, 540, 560, 624, 625). i.v., intravenous; q8h, every 8 h; CLCR, creatiine clearance; CNS, central nervous system; HD, hemodialysis; CAPD, continuous ambulatory peritoneal dialysis; CCRT, continuous renal replacement therapy.
Although TMP-SMX is thought to be a bacteriostatic antimicrobial with time-dependent action, a time-kill study demonstrated that achievable in vivo drug concentrations appear to have a concentration-dependent bactericidal effect (541). Second-line oral agents include doxycycline and amoxicillin-clavulanic acid. The recommended amoxicillin-clavulanic acid dosage is 20/5 mg per kilogram of body weight three times per day (9, 283, 532). Previous Thai recommendations for oral therapy included a four-drug regimen consisting of TMP-SMX, chloramphenicol, and doxycycline (542). However, one open-label randomized trial demonstrated no difference in efficacy of this regimen and poorer tolerability than with the combination of TMP-SMX and doxycycline. Furthermore, this trial revealed that duration of therapy was critical, with patients receiving less than 12 weeks of therapy incurring a 5.7-fold-increased risk of relapse or death (543). More recent evidence has shown noninferiority and improved tolerability when comparing TMP-SMX alone and TMP-SMX with doxycycline (520). Thus, the current Thai recommendation for duration of oral therapy is 12 to 20 weeks, compared to the Australian recommendation of 3 to 6 months (9, 510).
Research in the Northern Territory of Australia has revealed decreased rates of relapse or recrudescence depending on duration of intensive-phase therapy (544). With a median intensive phase of 26 days, the relapse or recrudescence rate decreased from 5.2% to 0.5% irrespective of compliance to eradication/oral phase (544). Current Darwin guidelines recommend a minimum intensive phase of 2 weeks for skin abscess, bacteremia without focus, and pneumonia without lymphadenopathy or ICU admission (540). Four weeks is required for pneumonia with lymphadenopathy or ICU admission or deep-seated collection (abscess anywhere other than skin), 6 weeks for osteomyelitis, and 8 weeks for CNS or arterial infection (544). This study alters traditional thinking that choice and duration of eradication therapy are the most important predictors of relapse (544). Further analysis of septic arthritis and osteomyelitis management suggests that 5 weeks of intravenous therapy or 4 weeks for an isolated single joint without osteomyelitis will suffice. Three months of oral eradication-phase therapy appears to demonstrate a similar rate of relapse or complications. However, there is insufficient evidence to recommend a shorter course of therapy for osteomyelitis (485). A relapse rate of approximately 9.7% between 1986 and 2004 was reported from Thailand (510). This has improved to approximately 6% with an unspecified intravenous phase of at least 10 days (510, 520). There is a role for 3 months of oral-only therapy for uncomplicated soft tissue infection, provided that investigations for deep-seated collections are negative (475, 545).
Adjunctive therapy may include the addition of granulocyte colony-stimulating factor (G-CSF), which has been studied in multiple locations with various results (546–548). Initial observational data suggested that decreased in-hospital mortality from 95 to 10% was attributable to the use of G-CSF (548). However, it was subsequently argued that concomitant improvement in other management factors confounded these results (279). A randomized controlled trial of G-CSF use in severe melioidosis sepsis demonstrated a longer duration of survival when measured in hours, but no overall mortality benefit (547). The current Darwin guidelines recommend 300 μg of intravenous G-CSF daily for patients with septic shock, initiated as soon as a probable microbiological diagnosis of melioidosis is made and continuing for either 10 days or the duration of intensive care unit stay contingent on clinical progress. Contraindications to commencement or continuation include an acute coronary event or total blood white cell count of >50,000 ×106/liter (540).
Source control is an important feature in overall management (526). The majority of visceral abscesses, other than prostatic, responded to antimicrobial therapy alone in one study; prostatic abscesses greater than 1 cm should be considered for drainage (472, 549). Fever clearance may be slow, with a median of 9 days in one study, and therefore, this may not necessarily stand as an indication for surgical intervention (550). While ideal, source control may not be achieved in every circumstance, particularly in resource-limited settings. It is important to note that treatment success has been achieved with prolonged therapy in the setting of undrained abscesses (8, 472).
Novel therapeutic agents for the treatment of melioidosis are emerging. A novel cephalosporin/β-lactamase inhibitor, ceftolozane-tazobactam, was developed with the intention of treating Pseudomonas and specifically AmpC β-lactamase-producing strains, and it also has activity against a number of Enterobacteriaceae (551, 552). Results from an Australian in vitro study report the MIC against B. pseudomallei to be 0.75 to 4 μg/ml, with an MIC90 of 2 μg/ml (553). This study lends support to further research of this agent as a therapeutic option. While fluoroquinolone therapy has not been recommended based on data demonstrating a 29% failure rate, finafloxacin is a C-8-cyanofluoroquinolone containing a unique chiral C-7 substituent which enhances its activity in low pH environments (554). Compared with those of ciprofloxacin, finafloxacin demonstrates greater bactericidal activity under acidic conditions and superior time-kill assays. Furthermore, in an inhalational mouse model, finafloxacin was comparable to TMP-SMX (555). Lipid A biosynthesis inhibitors prevent formation of lipopolysaccharide. Inhibitors of the enzyme UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase or LpxC have potent bactericidal effects (556). LpxC-4 is an inhibitor of the LPS biosynthesis pathway and has demonstrated in vitro activity against B. pseudomallei, with a growth inhibition concentration of 2 μg/ml persisting to 24 h (557).
Currently there is limited evidence to demonstrate effective prevention of infection in humans (3, 526). Amoxicillin-clavulanic acid, doxycycline, and TMP-SMX have been assessed in a 10-day postexposure prophylaxis mouse model. TMP-SMX was the only agent to achieve 100% survival at 21 days in this study; however, additional murine research demonstrated 44% and 83% survival following a 14- or 21-day course (558, 559). Current international consensus guidelines recommend a 21-day course of TMP-SMX. Alternative prophylactic therapy includes doxycycline or amoxicillin-clavulanic acid (3, 539). Prophylaxis for specific at-risk populations may be considered. One study reported that TMP-SMX prophylaxis for hemodialysis patients during the wet season in a region of endemicity is likely to be effective in reducing incidence of infection and that an oral dose of 160/800 mg daily appears to be well tolerated and safe in this patient cohort (560). Notably, compared to that in a neighboring region, the study appears to have included a higher proportion of indigenous hemodialysis patients in rural dialysis centers. This region had a lower incidence of melioidosis among their hemodialysis cohort, and therefore, rates of adverse effects, and cost-effectiveness modelling, would not support universal prophylaxis of their population (561).
PREVENTION
Prevention of infection is a critical component of a holistic treatment strategy. Intervention targeting risk factors, both behavioral and medical, is crucial. Further, accurate epidemiological data are valuable in a prevention strategy. Australia has a standard Public Health Laboratory Network case definition and melioidosis is a reportable condition, therefore enabling public health to monitor disease burden and identify outbreaks (562). Additionally, the Northern Territory Centre for Disease Control regularly embarks on awareness campaigns to increase community understanding of melioidosis (563). The key features of the 2015-2016 campaign were to increase the use of shoes in the wet season and use of gloves when working outdoors and to encourage people to remain indoors during storms, decrease alcohol intake, and wear a mask when working with a high-pressure water hose (563). The unit provided additional education to all patients diagnosed with melioidosis, with the aim of reducing the likelihood of reinfection (563). In Darwin, Australia, surveys of bore water demonstrated 33% B. pseudomallei contamination, and a subsequent study revealed the efficacy of UV irradiation as a method to disinfect untreated bore water supplies (324, 564). Furthermore, chlorination is an effective method of disinfection of potable water and was effectively used in control of an outbreak (59, 67, 565). Repurposing of current biological agents is also being considered. Chitosan, the polysaccharide derived from chitin, has wide-ranging antimicrobial activity (566). Experiments using chitosan on environmental B. pseudomallei isolates reveal a bactericidal effect via disruption of the cell membrane and release of intracellular content (567), demonstrating potential for control in soil as well as suggesting the need for further research in clinical models. An in vitro study demonstrated the significant and prolonged bactericidal effect of calcium oxide in decreasing the risk of infection from contaminated rice field soil (568). Additional information regarding soil treatment with calcium oxide in a Thai zoo further underlines the potential benefit for environmental control (569). This strategy has yet to be implemented due to the unknown effects on the crop, potential ecological impact, and the large volume required (568).
In northeast Thailand, a region where melioidosis accounts for more deaths than the combined effects of malaria, diarrheal disease, and measles, the awareness of melioidosis is lacking (17, 497, 570). Reporting of culture-confirmed cases of melioidosis has been mandated only since June 2016 (18). A study of over 4,000 Thais revealed that 74% of respondents have never heard of melioidosis (570). This is likely to be associated with a lack of education in schools and limited media reporting (18, 570). Further investigation into barriers to prevention utilizing focus group discussion revealed that 97% of a rural, diabetic, predominantly rice-farming population had no knowledge of melioidosis. This study found that providing information alone would be unlikely to lead to recommendation adherence. Barriers to change included time constraints in relation to boiling water and not wearing protective footwear due to discomfort (571). Therefore, a stepwise multifaceted approach at both the community and government levels will be required to improve long-term and lasting prevention of infection.
Although the risk of laboratory-acquired melioidosis is low, there have been two documented cases (3, 572, 573). Consequently, a guideline was published in order to prevent infection in diagnostic and research laboratories (3). Laboratory staff working with B. pseudomallei should undergo training regarding handling of this organism in a biosafety level 3 (BSL3) facility within a biosafety cabinet (3). Additionally, staff are required to wear appropriate personal protective equipment, which includes a gown, gloves, and a respiratory mask if handling infected animal material or during sample centrifugation (3). It is also important to note that country-specific guidelines may differ, and the United States mandates registration and clearance from federal agencies prior to handling or work with any select agents (3).
OUTCOMES
Overall survival is affected by multiple factors, including premorbid host determinants. Diabetes, ATSI status, chronic renal disease, and older age negatively impact mortality (8, 12, 166, 574). In southern Thailand, septic shock on admission has a reported in-hospital mortality odds ratio of 29.14 to 68.20 (79). It has been shown that 73.7% of patients with a positive blood culture occurring within 24 h of incubation died during their admission. This compared with a 40.9% in-hospital mortality rate for patients whose blood cultures signaled positive more than 24 h from incubation (169). A bacteremia quantification of ≤1 CFU/ml has been associated with a 42% mortality rate, while a positive blood culture with >100 CFU/ml resulted in a fatal outcome in 96% of patients (267).
Although mortality remains high in developing nations such as Thailand, two centers in Australia have demonstrated a remarkable decrease in overall mortality over the preceding two decades (8, 12, 17). The initial peak mortality of 30% in one facility has decreased to 9% in all patients monitored throughout treatment (8). When comparing presentation with septic shock and death over time, the improvement has been found to be dramatic, improving from 100% mortality to 27% (8). Patients over 50 years of age have an increased mortality, with an odds ratio of 2.0 (1.2 to 3.3), compared with the rate for those under 50. The presence of any risk factor has a reported mortality OR of 9.4 (2.3 to 39.0) (8). With regard to bacteremia, repeated positive blood culture after the first or second week of therapy is a strong predictor of mortality, with an OR of 4.2 after adjustment for age, sex, diabetes, blood pressure, pneumonia, and duration of intravenous antibiotics. Importantly, repeated culture of B. pseudomallei from nonsterile sites did not confer increased risk of death (575). The mortality of chronic melioidosis is low and is estimated at 2% in Australia (8).
In children, the mortality rates range from 7% overall in a prospective Australian study to 16.8% in-hospital mortality from a retrospective Cambodian study (110, 475). This Cambodian study revealed a 71.8% in-hospital mortality rate in bacteremic children, and another study revealed a similarly high in-hospital mortality rate (73%) in a Thai neonatal population who presented with either neonatal meningitis or bacteremia (110, 576). Additional prospective data from Cambodia reveal an overall mortality rate of 62% (577). Specifically in Cambodia, potential reasons for a high mortality rate may be a combination of inadequate microbiology diagnostic services coupled with empirical therapy without adequate B. pseudomallei activity (578). Combined Malaysian data report an overall in-hospital mortality rate of 33 to 54% (87, 88, 93, 166–168, 579). Bacteremic patients have a mortality rate of of 48 to 65%, and nonbacteremic patients had a substantially lower case-fatality rate, 19%. Additionally, the mortality rate was 100% among those patients, while patients without septic shock had a mortality rate of 30% (87, 167).
Genetic risk factors for melioidosis-associated mortality have recently been described (505). In a study of melioidosis patients in northeast Thailand, HLA-B*46 and HLA-C*01 were associated with an increased risk of death (OR, 2.8 and 3.1, respectively) (505). A recent evaluation of the TLR1 variants common in East Asian populations demonstrated an association with a severe bacteremic phenotype and potentially worse outcome (580). Conversely, genetic polymorphisms for two flagellin-sensing receptors, TLR5 and NLRC4, have demonstrated survival benefit in patients with melioidosis (581–584). A review of 600 Thai patients with melioidosis infection reported a protective effect with regard to in-hospital death and organ failure in patients with a TLR5 genetic variant resulting in decreased function (581). A specific NLRC4 region polymorphism was also associated with survival in patients with pulmonary involvement. Furthermore, coinheritance of both TLR5 and NLRC4 polymorphisms has a cumulative effect on survival (583). It is interesting that while these receptors are required for functional immunity, in the setting of B. pseudomallei infection they appear to have a potentially detrimental immunomodulatory effect resulting in inflammation-associated organ failure (581).
VACCINE DEVELOPMENT
Due to the current burden of disease and potential bioterrorism threat, effective vaccines for melioidosis are imperative (585). Based on numerous reports it is likely that both cellular and humoral immunity will be required to induce complete protection against B. pseudomallei (586). As inhalation is a common route of infection, it is important to note that antibodies are sufficient to protect against lethal aerosol infection with B. pseudomallei and B. mallei (587). However, an additional consideration is that vaccination from a public health perspective would aim to protect against natural infection and therefore target the population at greatest risk, such as diabetics (585, 588). This population is at greater risk of infection via inoculation than are healthy individuals from the likely bioterrorism threat of inhalational acquisition (585).
Multiple vaccine candidates, including killed whole-cell (KWC), live attenuated, glycoconjugate, subunit, outer membrane vesicle (OMV), plasmid DNA, and dendritic cell, have been pursued (588). Unfortunately, due to lack of standardization of organism strains and dose tested, animal models, route of inoculation, and duration of follow-up, comparison of vaccine efficacy is challenging (585). Yet no vaccine candidate has been trialed in a human model (585, 588, 589).
Numerous live attenuated vaccines have been developed, including (but not limited to) mutations of genes for biosynthetic pathways, such as purN, purM, aroA, aroC, and serC, or virulence factors, including bipD, tonB, and hcp1 (383, 455, 590–593). These vaccines have demonstrated protection against both inhalation and inoculation; however, efficacy appears to be dependent on the same route of challenge as vaccine administration (588). In a live attenuated vaccine animal model, Scott et al. demonstrated that nonpathogenic B. thailandensis E555 used as a vaccine showed complete bacterial clearance from the lungs, liver, and spleen on day three after mice were challenged with B. pseudomallei K96243 (594). B. thailandensis E555 produces B. pseudomallei-like manno-heptose capsule, which is thought to be a significant factor in developing immunity, with higher B. pseudomallei-specific IgG levels and survival times noted in mice vaccinated with the E555 strain compared to a vaccination with a nonencapsulated control strain (594). A further study utilized a subcutaneous vaccine of a highly attenuated purM mutant of B. pseudomallei strain, 1026b (Bp82), with immunized mice showing high survival rates after a wild-type (WT) challenge (592). Higher levels of IgM and IgG were noted in the immunized group than in controls (592). Recent evidence corroborates the necessity of a robust humoral immune response for vaccine-induced immunity (593). Using a ΔtonB Δhcp1 mutant in a mouse model, Khakhum et al. demonstrated almost complete sterilizing immunity predominantly via humoral immunity (593). However, due to conflicting results, the role of cellular immunity remains unclear (455, 593). The major concern for live attenuated vaccination is the potential for reversion to a virulent WT strain capable of causing infection.
The potential advantages of KWC vaccines are the low production cost and, similar to the case with live attenuated vaccines, the potential to induce immunity via multiple antigens (589). However, as with live vaccines, KWC vaccines are potentially reactogenic, and most KWC vaccines require multiple doses (589, 595). Results regarding induction of protective immunity with killed vaccines are conflicting. Variable protection associated with heat-killed vaccines may be attributable to differences in immunization organism dose and route of infection challenge (596–598). One study demonstrated significantly superior protection with paraformaldehyde-killed B. pseudomallei vaccine given intramuscularly compared to that with heat-killed vaccines (597). However, a considerable disadvantage to killed vaccines is the potential for altered production of protective antigens in vitro and therefore limited protective immunity in vivo (589).
Subunit vaccines contain only protective antigens, may be less reactogenic, and may be more readily reproducible (589, 599–601). Many of the antigens examined directly relate to organism virulence, such as CPS, LPS, or constituents of the secretion systems such as BimA and BopA (383, 400, 589). These have included CPS covalently linked to a CRM197 diphtheria toxin mutant, producing CPS-CRM197, and highly purified recombinant B. pseudomallei proteins Hcp1 and TssM (599). Inoculation of C57BL/6 mice with CPS-CRM197 resulted in high IgG titers as well as an opsonizing antibody response against the CPS constituent of the glycoconjugate. Inoculation with Hcp1 and TssM similarly achieved a high IgG antibody titer and substantial IFN-γ-secreting T cell responses against these antigens (599). In a murine model of combined CPS-CRM197 and Hcp1 vaccination, mice challenged with a lethal inhalational dose of B. pseudomallei exhibited 100% survival. Additionally, this vaccine composition demonstrated the ability to generate a sterilizing immune response with no culturable bacteria in the lungs, livers, or spleens in 70% of the survivors (599).
Several studies utilizing OMVs (noninfectious particles containing LPS and other immunogenic proteins) have shown increased survival in mice but may not provide complete protection, as splenic persistence of bacteria was demonstrated (602, 603). Safety and immunogenicity have been demonstrated in nonhuman primates, resulting in OMVs being a leading candidate for further vaccine development (604).
Antigenic proteins used alone as vaccine components have demonstrated incomplete protection against B. pseudomallei infection in murine models (601). However, conjugate vaccines have subsequently demonstrated a more robust immune response (605, 606). Unfortunately, although safe and immunogenic, these vaccines failed to provide complete protection (606). As there are only limited known vaccine antigen candidates, an alternative approach using in silico-predicted reverse vaccinology was performed (607). Candidates were chosen according to predicted antigenicity, physiochemical and adhesive properties, and affinity for major histocompatibility complex (MHC) class I and class II (607). Two proteins which demonstrated seroreactivity with convalescent-phase human sera, together with Hcp1, were linked to LPS and incorporated with the surface of a gold nanoparticle (AuNP) (608). A murine model of AuNP glycoconjugate vaccines demonstrated high protein and polysaccharide-specific antibody titers, and a combination of the novel flagellar protein FlgL, hemagglutinin, and HCP1 yielded 100% survival and reduced lung colonization following a lethal intranasal challenge with B. pseudomallei (607).
Cost-effectiveness analysis has determined that vaccination could be a practical intervention in Thailand, especially in at-risk populations, such as diabetics (588). In a region with an annual melioidosis incidence of 25 per 100,000 population, a vaccine with a protective duration of only 3 years and efficacy of 50% costing $2 (U.S. dollars) would be cost-effective. Additionally, a vaccine with the same efficacy and a 10-year protective duration would remain cost-effective for use in all diabetics at over $25 per course (588). While further development is required, vaccination could play an important part in reducing the global burden of melioidosis (585). However, the development of a safe and effective vaccine against B. pseudomallei remains currently unresolved. The major barriers are the limited efficacy of candidate vaccines in animal models, the method of inoculation in the animal models that most reflects acquisition in humans, and the logistic and financial issues relating to the establishment of phase 3 trials in an area of endemicity. The efficacy of any candidate vaccine will have to be assessed in the relevant risk groups (589).
BIOTERRORISM
Burkholderia mallei, the causative agent of glanders, has already been used as a bioweapon against both animals and humans (609). It has also been implicated in major outbreaks resulting in the deaths of thousands of horses (610). Both the former Soviet Union and the United States have considered the potential for use of B. pseudomallei as a bioweapon, but there appears to have been less research into this potential than with B. mallei (609). B. pseudomallei is considered a biothreat due to the high mortality rate with melioidosis, difficulty in diagnosis and treatment in regions where it is not endemic, and ability to survive outside its natural environment (611, 612). Additional features relating to the suitability of this organism as a bioterrorism agent are its availability and intrinsic antibiotic resistance (609). It is classified as a select agent by the U.S. Centers for Disease Control and Prevention (CDC) and should be handled within a biosafety level 3 facility or equivalent (3, 611). Studies have demonstrated the ability of B. pseudomallei to survive not only in distilled water for 16 years but also on paper, stainless steel, and polyethylene coupons for hours to days (163, 613, 614). It is proposed that with a dense inoculum, inhalation could have an incubation period of less than 7 days (609). Therefore, there is concern with regard to weaponization in the form of aerosolization (615).
THE ECONOMIC IMPLICATIONS OF DISEASE
Due to its prevalence, morbidity, and mortality, melioidosis creates a substantial financial burden in high-endemicity regions. Hospitals and health districts require the resources to diagnose and treat melioidosis. The direct cost of melioidosis cases includes specific medical expenses such as medication, health professional time, hospital admission, laboratory services, and patient transportation. Indirect costs relate to productivity. This can be a loss of productivity of both the patient and the potential caregivers (616–618). Some regions that are likely to have underdiagnosed and underreported cases of melioidosis are not equipped to adequately manage the true burden of infection. At a provincial referral center in Cambodia, only 17% of surveyed physicians had experience in treating melioidosis, and the hospital’s annual supply of ceftazidime would be sufficient to treat up to 14 patients (109, 578). Additional Cambodian data reveal a mean direct cost to the patient of $565 and that two-thirds of households would have incurred debt (577). The mean cost almost equaled the gross domestic product (GDP) per capita of Cambodia in 2008. To add further context to the direct cost on a household, $565 equates to more than a 1-year supply of food (619). In a Thai study of two provinces, the estimated annual total cost of bacteremic melioidosis hospitalizations was between $152,159 and $465,303. The majority, 75 to 85%, of the financial burden was because of premature mortality. The average cost per fatal case of melioidosis was 2.7 to 2.8 times greater than Thailand’s GDP per capita (616). Empirical therapy with a carbapenem, such as meropenem, is estimated to cost $140 per day in Thailand. Ceftazidime has an estimated cost of $5 per day (620). There is limited evidence to suggest that empirical carbapenem therapy may be cost-effective in the setting of presumed severe melioidosis, as the mortality benefit is likely to be modest (550, 620, 621).
With the worldwide increase in prevalence of diabetes mellitus, a major risk factor for melioidosis, the financial burden is likely to be compounded (23, 616, 617).
CONCLUSION
B. pseudomallei is increasingly being recognized as a significant human pathogen worldwide. It presents challenges to both the diagnostic laboratory and the clinician, and early diagnosis is fundamental to appropriate management and survival. Direct molecular detection from blood remains challenging, and the mainstay of laboratory diagnosis is still culture. The organism is well adapted for intracellular survival, particularly in hosts with defined risk factors such as diabetes and excessive alcohol intake. The clinical presentation of melioidosis can be varied, although pneumonia with or without bacteremia is most common. Management and outcomes depend upon an early institution of appropriate, directed intravenous antibiotic therapy followed by a lengthy maintenance course of antibiotics. Vaccine prevention currently remains elusive. In this era of globalization and widespread tourism, an awareness of this condition will be important.
ACKNOWLEDGMENTS
We declare no financial or other relationships that may lead to conflict of interest.
The manuscript has been read and approved by all authors.
We gratefully acknowledge the review of the manuscript by Patrick Harris.
Biographies

I. Gassiep completed medical school through Monash University Australia & Malaysia. His early career aspirations in infectious diseases were shaped by his upbringing in South Africa, and his interest in tropical infectious diseases was sparked by the time he spent in the Malaysian health care system. He is currently an Infectious Diseases Physician and Clinical Microbiologist working in The Mater Hospital, Brisbane, Queensland, Australia.

M. Armstrong received his medical degree from the University of Otago, New Zealand, and is a registrar training in Infectious Diseases and Clinical Microbiology. Currently he is based in Townsville, Australia, to complete training in Clinical Microbiology. Before Townsville he worked in diverse settings, including Timor-Leste. He has an interest in tropical medicine, having just completed his master of public health and tropical medicine through James Cook University.

R. Norton trained as a clinical microbiologist at The Institute of Medical and Veterinary Science, Adelaide, Australia. He has worked in a variety of clinical positions, including 5 years in Australian indigenous communities. He is currently Director of Pathology and Microbiology at Townsville Hospital, Queensland, Australia. In his current capacity he has collaborated with researchers locally and internationally on projects relating to melioidosis. This is a disease which is endemic in this region, and he has had an interest in this condition for the last 20 years. He was a coauthor in the CDC-sponsored guideline (2010) on Diagnostics for Melioidosis.
REFERENCES
- 1.Depoorter E, Bull MJ, Peeters C, Coenye T, Vandamme P, Mahenthiralingam E. 2016. Burkholderia: an update on taxonomy and biotechnological potential as antibiotic producers. Appl Microbiol Biotechnol 100:5215–5229. doi: 10.1007/s00253-016-7520-x. [DOI] [PubMed] [Google Scholar]
- 2.LiPuma JJ, Currie BJ, Peacock SJ, Vandamme PWE. 2015. Burkholderia, Stenotrophomonas, Ralstonia, Cupriavidus, Pandoraea, Brevundimonas, Comamonas, Delftia, and Acidovorax, p 791–812. In Jorgensen JH, Pfaller MA, Carroll KC, Funke G, Landry ML, Richter SS, Warnock DW (ed), Manual of clinical microbiology, 11th ed ASM Press, Washington, DC. [Google Scholar]
- 3.Peacock SJ, Schweizer HP, Dance DAB, Smith TL, Gee JE, Wuthiekanun V, DeShazer D, Steinmetz I, Tan P, Currie BJ. 2008. Management of accidental laboratory exposure to Burkholderia pseudomallei and B. mallei. Emerg Infect Dis 14:e2. doi: 10.3201/eid1407.071501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Currie BJ, Jacups SP, Cheng AC, Fisher DA, Anstey NM, Huffam SE, Krause VL. 2004. Melioidosis epidemiology and risk factors from a prospective whole-population study in northern Australia. Trop Med Int Health 9:1167–1174. doi: 10.1111/j.1365-3156.2004.01328.x. [DOI] [PubMed] [Google Scholar]
- 5.Vuddhakul V, Tharavichitkul P, Na-Ngam N, Jitsurong S, Kunthawa B, Noimay P, Noimay P, Binla A, Thamlikitkul V. 1999. Epidemiology of Burkholderia pseudomallei in Thailand. Am J Trop Med Hyg 60:458–461. doi: 10.4269/ajtmh.1999.60.458. [DOI] [PubMed] [Google Scholar]
- 6.Kaestli M, Schmid M, Mayo M, Rothballer M, Harrington G, Richardson L, Hill A, Hill J, Tuanyok A, Keim P, Hartmann A, Currie BJ. 2012. Out of the ground: aerial and exotic habitats of the melioidosis bacterium Burkholderia pseudomallei in grasses in Australia. Environ Microbiol 14:2058–2070. doi: 10.1111/j.1462-2920.2011.02671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitmore A, Krishnaswami CS. 1912. A hitherto undescribed infective disease in Rangoon. Ind Med Gaz 47:262–267. [PMC free article] [PubMed] [Google Scholar]
- 8.Currie BJ, Ward L, Cheng AC. 2010. The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study. PLoS Negl Trop Dis 4:e900. doi: 10.1371/journal.pntd.0000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng AC, Currie BJ. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev 18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Currie BJ, Price EP, Mayo M, Kaestli M, Theobald V, Harrington I, Harrington G, Sarovich DS. 2015. Use of whole-genome sequencing to link Burkholderia pseudomallei from air sampling to mediastinal melioidosis, Australia. Emerg Infect Dis 21:2052–2054. doi: 10.3201/eid2111.141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen P-S, Chen Y-S, Lin H-H, Liu P-J, Ni W-F, Hsueh P-T, Liang S-H, Chen C, Chen Y-L. 2015. Airborne transmission of melioidosis to humans from environmental aerosols contaminated with B. pseudomallei. PLoS Negl Trop Dis 9:e0003834. doi: 10.1371/journal.pntd.0003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart JD, Smith S, Binotto E, McBride WJ, Currie BJ, Hanson J. 2017. The epidemiology and clinical features of melioidosis in Far North Queensland: implications for patient management. PLoS Negl Trop Dis 11:e0005411. doi: 10.1371/journal.pntd.0005411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White NJ. 2003. Meliodosis. Lancet 361:1715–1722. doi: 10.1016/s0140-6736(03)13374-0. [DOI] [PubMed] [Google Scholar]
- 14.Van Zandt KE, Greer MT, Gelhaus HC. 2013. Glanders: an overview of infection in humans. Orphanet J Rare Dis 8:131. doi: 10.1186/1750-1172-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. 2000. Laboratory-acquired human glanders—Maryland. MMWR Morb Mortal Wkly Rep 49:532–535. [PubMed] [Google Scholar]
- 16.Currie BJ, Dance DAB, Cheng AC. 2008. The global distribution of Burkholderia pseudomallei and melioidosis: an update. Trans R Soc Trop Med Hyg 102:S1–S4. doi: 10.1016/S0035-9203(08)70002-6. [DOI] [PubMed] [Google Scholar]
- 17.Limmathurotsakul D, Wongratanacheewin S, Teerawattanasook N, Wongsuvan G, Chaisuksant S, Chetchotisakd P, Chaowagul W, Day NPJ, Peacock SJ. 2010. Increasing incidence of human melioidosis in northeast Thailand. Am J Trop Med Hyg 82:1113–1117. doi: 10.4269/ajtmh.2010.10-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinjoy S, Hantrakun V, Kongyu S, Kaewrakmuk J, Wangrangsimakul T, Jitsuronk S, Saengchun W, Bhengsri S, Akarachotpong T, Thamthitiwat S, Sangwichian O, Anunnatsiri S, Sermswan R, Lertmemongkolchai G, Sitthidet Tharinjaroen C, Preechasuth K, Udpaun R, Chuensombut P, Waranyasirikul N, Anudit C, Narenpitak S, Jutrakul Y, Teparrukkul P, Teerawattanasook N, Thanvisej K, Suphan A, Sukbut P, Ploddi K, Sirichotirat P, Chiewchanyon B, Rukseree K, Hongsuwan M, Wongsuwan G, Sunthornsut P, Wuthiekanun V, Sachaphimukh S, Wannapinij P, Chierakul W, Chewapreecha C, Thaipadungpanit J, Chantratita N, Korbsrisate S, Taunyok A, Dunachie S, Palittapongarnpim P, Sirisinha S, Kitphati R, Iamsirithaworn S, Chaowagul W, Chetchotisak P, Whistler T, Wongratanacheewin S, Limmathurotsakul D. 2018. Melioidosis in Thailand: present and future. Trop Med Infect Dis 3:38. doi: 10.3390/tropicalmed3020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearson T, Giffard P, Beckstrom-Sternberg S, Auerbach R, Hornstra H, Tuanyok A, Price EP, Glass MB, Leadem B, Beckstrom-Sternberg JS, Allan GJ, Foster JT, Wagner DM, Okinaka RT, Sim SH, Pearson O, Wu Z, Chang J, Kaul R, Hoffmaster AR, Brettin TS, Robison RA, Mayo M, Gee JE, Tan P, Currie BJ, Keim P. 2009. Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biol 7:78. doi: 10.1186/1741-7007-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker A, Mayo M, Owens L, Burgess G, Norton R, McBride WJH, Currie BJ, Warner J. 2013. Biogeography of Burkholderia pseudomallei in the Torres Strait Islands of Northern Australia. J Clin Microbiol 51:2520–2525. doi: 10.1128/JCM.00418-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Limmathurotsakul D, Golding N, Dance DA, Messina JP, Pigott DM, Moyes CL, Rolim DB, Bertherat E, Day NP, Peacock SJ, Hay SI. 2016. Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat Microbiol 1:15008. doi: 10.1038/nmicrobiol.2015.8. [DOI] [PubMed] [Google Scholar]
- 22.Almog Y, Yagel Y, Geffen Y, Yagupsky P. 2016. A Burkholderia pseudomallei infection imported from Eritrea to Israel. Am J Trop Med Hyg 95:997–998. doi: 10.4269/ajtmh.16-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaw JE, Sicree RA, Zimmet PZ. 2010. Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes Res Clin Pract 87:4–14. doi: 10.1016/j.diabres.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 24.Smith S, Hanson J, Currie BJ. 2018. Melioidosis: an Australian perspective. Trop Med Infect Dis 3:27. doi: 10.3390/tropicalmed3010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dance DA. 1991. Melioidosis: the tip of the iceberg? Clin Microbiol Rev 4:52–60. doi: 10.1128/cmr.4.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dance DAB, Limmathurotsakul D. 2018. Global burden and challenges of melioidosis. Trop Med Infect Dis 3:13. doi: 10.3390/tropicalmed3010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mollaret HH. 1988. “L’affaire du Jardin des plantes” ou comment la melioidose fit son apparition en France. Med Mal Infect 18:643–654. doi: 10.1016/S0399-077X(88)80175-6. [DOI] [Google Scholar]
- 28.Dance DAB. 2000. Ecology of Burkholderia pseudomallei and the interactions between environmental Burkholderia spp. and human-animal hosts. Acta Trop 74:159–168. doi: 10.1016/S0001-706X(99)00066-2. [DOI] [PubMed] [Google Scholar]
- 29.Stanton AT, Fletcher W. 1925. Melioidosis: a disease of rodents communicable to man. Lancet 205:10–13. doi: 10.1016/S0140-6736(01)04724-9. [DOI] [Google Scholar]
- 30.Ashdown LR, Clarke SG. 1992. Evaluation of culture techniques for isolation of Pseudomonas pseudomallei from soil. Appl Environ Microbiol 58:4011–4015. doi: 10.1128/AEM.58.12.4011-4015.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chambon L. 1955. Isolement du bacille de Whitmore a partir du milieu exterieur. Ann Inst Pasteur 89:229–235. [PubMed] [Google Scholar]
- 32.Merianos A, Patel M, Lane JM, Noonan CN, Sharrock D, Mock PA, Currie B. 1993. The 1990-1991 outbreak of melioidosis in the Northern Territory of Australia: epidemiology and environmental studies. Southeast Asian J Trop Med Public Health 24:425–435. [PubMed] [Google Scholar]
- 33.Suputtamongkol Y, Chaowagul W, Chetchotisakd P, Lertpatanasuwun N, Intaranongpai S, Ruchutrakool T, Budhsarawong D, Mootsikapun P, Wuthiekanun V, Teerawatasook N, Lulitanond A. 1999. Risk factors for melioidosis and bacteremic melioidosis. Clin Infect Dis 29:408–413. doi: 10.1086/520223. [DOI] [PubMed] [Google Scholar]
- 34.Nasner-Posso KM, Cruz-Calderon S, Montufar-Andrade FE, Dance DA, Rodriguez-Morales AJ. 2015. Human melioidosis reported by ProMED. Int J Infect Dis 35:103–106. doi: 10.1016/j.ijid.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su HP, Yang HW, Chen YL, Ferng TL, Chou YL, Chung TC, Chen CH, Chiang CS, Kuan MM, Lin HH, Chen YS. 2007. Prevalence of melioidosis in the Er-Ren River Basin, Taiwan: implications for transmission. J Clin Microbiol 45:2599–2603. doi: 10.1128/JCM.00228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corkeron ML, Norton R, Nelson PN. 2010. Spatial analysis of melioidosis distribution in a suburban area. Epidemiol Infect 138:1346–1352. doi: 10.1017/S0950268809991634. [DOI] [PubMed] [Google Scholar]
- 37.Palasatien S, Lertsirivorakul R, Royros P, Wongratanacheewin S, Sermswan RW. 2008. Soil physicochemical properties related to the presence of Burkholderia pseudomallei. Trans R Soc Trop Med Hyg 102(Suppl 1):S5–S9. doi: 10.1016/S0035-9203(08)70003-8. [DOI] [PubMed] [Google Scholar]
- 38.Tong S, Yang S, Lu Z, He W. 1996. Laboratory investigation of ecological factors influencing the environmental presence of Burkholderia pseudomallei. Microbiol Immunol 40:451–453. doi: 10.1111/j.1348-0421.1996.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 39.Pumirat P, Cuccui J, Stabler RA, Stevens JM, Muangsombut V, Singsuksawat E, Stevens MP, Wren BW, Korbsrisate S. 2010. Global transcriptional profiling of Burkholderia pseudomallei under salt stress reveals differential effects on the Bsa type III secretion system. BMC Microbiol 10:171. doi: 10.1186/1471-2180-10-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inglis TJ, Sagripanti J-L. 2006. Environmental factors that affect the survival and persistence of Burkholderia pseudomallei. Appl Environ Microbiol 72:6865–6875. doi: 10.1128/AEM.01036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Musa HI, Hassan L, Shamsuddin ZH, Panchadcharam C, Zakaria Z, Abdul Aziz S. 2016. Physicochemical properties influencing presence of Burkholderia pseudomallei in soil from small ruminant farms in peninsular Malaysia. PLoS One 11:e0162348. doi: 10.1371/journal.pone.0162348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ribolzi O, Rochelle-Newall E, Dittrich S, Auda Y, Newton PN, Rattanavong S, Knappik M, Soulileuth B, Sengtaheuanghoung O, Dance DA, Pierret A. 2016. Land use and soil type determine the presence of the pathogen Burkholderia pseudomallei in tropical rivers. Environ Sci Pollut Res Int 23:7828–7839. doi: 10.1007/s11356-015-5943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang HM, Chaowagul W, Sokol PA. 1991. Siderophore production by Pseudomonas pseudomallei. Infect Immun 59:776–780. doi: 10.1128/IAI.59.3.776-780.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kvitko BH, Goodyear A, Propst KL, Dow SW, Schweizer HP. 2012. Burkholderia pseudomallei known siderophores and hemin uptake are dispensable for lethal murine melioidosis. PLoS Negl Trop Dis 6:e1715. doi: 10.1371/journal.pntd.0001715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker AL, Ezzahir J, Gardiner C, Shipton W, Warner JM. 2015. Environmental attributes influencing the distribution of Burkholderia pseudomallei in northern Australia. PLoS One 10:e0138953. doi: 10.1371/journal.pone.0138953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stopnisek N, Bodenhausen N, Frey B, Fierer N, Eberl L, Weisskopf L. 2014. Genus-wide acid tolerance accounts for the biogeographical distribution of soil Burkholderia populations. Environ Microbiol 16:1503–1512. doi: 10.1111/1462-2920.12211. [DOI] [PubMed] [Google Scholar]
- 47.Wang-Ngarm S, Chareonsudjai S, Chareonsudjai P. 2014. Physicochemical factors affecting the growth of Burkholderia pseudomallei in soil microcosm. Am J Trop Med Hyg 90:480–485. doi: 10.4269/ajtmh.13-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hantrakun V, Rongkard P, Oyuchua M, Amornchai P, Lim C, Wuthiekanun V, Day NPJ, Peacock SJ, Limmathurotsakul D. 2016. Soil nutrient depletion is associated with the presence of Burkholderia pseudomallei. Appl Environ Microbiol 82:7086–7092. doi: 10.1128/AEM.02538-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ngamsang R, Potisap C, Boonmee A, Lawongsa P, Chaianunporn T, Wongratanacheewin S, Rodrigues JL, Sermswan RW. 2015. The contribution of soil physicochemical properties to the presence and genetic diversity of Burkholderia pseudomallei. Southeast Asian J Trop Med Public Health 46:38–50. [PubMed] [Google Scholar]
- 50.Suebrasri T, Wang-ngarm S, Chareonsudjai P, Sermswan R, Chareonsudjai S. 2013. Seasonal variation of soil environmental characteristics affect the presence of Burkholderia pseudomallei in Khon Kaen, Thailand. Afr J Microbiol Res 7:1940–1945. [Google Scholar]
- 51.Draper AD, Mayo M, Harrington G, Karp D, Yinfoo D, Ward L, Haslem A, Currie BJ, Kaestli M. 2010. Association of the melioidosis agent Burkholderia pseudomallei with water parameters in rural water supplies in Northern Australia. Appl Environ Microbiol 76:5305–5307. doi: 10.1128/AEM.00287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zimmermann RE, Ribolzi O, Pierret A, Rattanavong S, Robinson MT, Newton PN, Davong V, Auda Y, Zopfi J, Dance DAB. 2018. Rivers as carriers and potential sentinels for Burkholderia pseudomallei in Laos. Sci Rep 8:8674. doi: 10.1038/s41598-018-26684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Currie BJ, Jacups SP. 2003. Intensity of rainfall and severity of melioidosis, Australia. Emerg Infect Dis 9:1538–1542. doi: 10.3201/eid0912.020750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bulterys PL, Bulterys MA, Phommasone K, Luangraj M, Mayxay M, Kloprogge S, Miliya T, Vongsouvath M, Newton PN, Phetsouvanh R, French CT, Miller JF, Turner P, Dance DAB. 2018. Climatic drivers of melioidosis in Laos and Cambodia: a 16-year case series analysis. Lancet Planet Health 2:e334–e343. doi: 10.1016/S2542-5196(18)30172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng AC, Jacups SP, Gal D, Mayo M, Currie BJ. 2006. Extreme weather events and environmental contamination are associated with case-clusters of melioidosis in the Northern Territory of Australia. Int J Epidemiol 35:323–329. doi: 10.1093/ije/dyi271. [DOI] [PubMed] [Google Scholar]
- 56.Ko WC, Cheung BM, Tang HJ, Shih HI, Lau YJ, Wang LR, Chuang YC. 2007. Melioidosis outbreak after typhoon, southern Taiwan. Emerg Infect Dis 13:896–898. doi: 10.3201/eid1306.060646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanderson C, Currie BJ. 2014. Melioidosis: a pediatric disease. Pediatr Infect Dis J 33:770–771. doi: 10.1097/INF.0000000000000358. [DOI] [PubMed] [Google Scholar]
- 58.Inglis TJ, Rigby P, Robertson TA, Dutton NS, Henderson M, Chang BJ. 2000. Interaction between Burkholderia pseudomallei and Acanthamoeba species results in coiling phagocytosis, endamebic bacterial survival, and escape. Infect Immun 68:1681–1686. doi: 10.1128/iai.68.3.1681-1686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Howard K, Inglis TJ. 2005. Disinfection of Burkholderia pseudomallei in potable water. Water Res 39:1085–1092. doi: 10.1016/j.watres.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 60.Noinarin P, Chareonsudjai P, Wangsomnuk P, Wongratanacheewin S, Chareonsudjai S. 2016. Environmental free-living amoebae isolated from soil in Khon Kaen, Thailand, antagonize Burkholderia pseudomallei. PLoS One 11:e0167355. doi: 10.1371/journal.pone.0167355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cottew GS. 1950. Melioidosis in sheep in Queensland. A description of the causal organism. Aust J Exp Biol Med Sci 28:677–683. doi: 10.1038/icb.1950.70. [DOI] [PubMed] [Google Scholar]
- 62.Rimington RA. 1962. Melioidosis in north Queensland. Med J Aust 49:50–53. [DOI] [PubMed] [Google Scholar]
- 63.Parameswaran U, Baird RW, Ward LM, Currie BJ. 2012. Melioidosis at Royal Darwin Hospital in the big 2009-2010 wet season: comparison with the preceding 20 years. Med J Aust 196:345–348. doi: 10.5694/mja11.11170. [DOI] [PubMed] [Google Scholar]
- 64.Faa AG, Holt PJ. 2002. Melioidosis in the Torres Strait Islands of Far North Queensland. Commun Dis Intell 26:279–283. [DOI] [PubMed] [Google Scholar]
- 65.Warner JM, Pelowa DB, Currie BJ, Hirst RG. 2007. Melioidosis in a rural community of Western Province, Papua New Guinea. Trans R Soc Trop Med Hyg 101:809–813. doi: 10.1016/j.trstmh.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 66.Warner JM, Pelowa DB, Gal D, Rai G, Mayo M, Currie BJ, Govan B, Skerratt LF, Hirst RG. 2008. The epidemiology of melioidosis in the Balimo region of Papua New Guinea. Epidemiol Infect 136:965–971. doi: 10.1017/S0950268807009429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Inglis TJ, Garrow SC, Henderson M, Clair A, Sampson J, O’Reilly L, Cameron B. 2000. Burkholderia pseudomallei traced to water treatment plant in Australia. Emerg Infect Dis 6:56–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ketterer PJ, Webster WR, Shield J, Arthur RJ, Blackall PJ, Thomas AD. 1986. Melioidosis in intensive piggeries in south eastern Queensland. Aust Vet J 63:146–149. doi: 10.1111/j.1751-0813.1986.tb02953.x. [DOI] [PubMed] [Google Scholar]
- 69.Currie BJ, Mayo M, Anstey NM, Donohoe P, Haase A, Kemp DJ. 2001. A cluster of melioidosis cases from an endemic region is clonal and is linked to the water supply using molecular typing of Burkholderia pseudomallei isolates. Am J Trop Med Hyg 65:177–179. doi: 10.4269/ajtmh.2001.65.177. [DOI] [PubMed] [Google Scholar]
- 70.Hampton V, Kaestli M, Mayo M, Choy JL, Harrington G, Richardson L, Benedict S, Noske R, Garnett ST, Godoy D, Spratt BG, Currie BJ. 2011. Melioidosis in birds and Burkholderia pseudomallei dispersal, Australia. Emerg Infect Dis 17:1310–1312. doi: 10.3201/eid1707.100707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trakulsomboon S, Vuddhakul V, Tharavichitkul P, Na-Gnam N, Suputtamongkol Y, Thamlikitkul V. 1999. Epidemiology of arabinose assimilation in burkholderia pseudomallei isolated from patients and soil in Thailand. Southeast Asian J Trop Med Public Health 30:756–759. [PubMed] [Google Scholar]
- 72.Suputtamongkol Y, Hall AJ, Dance DA, Chaowagul W, Rajchanuvong A, Smith MD, White NJ. 1994. The epidemiology of melioidosis in Ubon Ratchatani, northeast Thailand. Int J Epidemiol 23:1082–1090. doi: 10.1093/ije/23.5.1082. [DOI] [PubMed] [Google Scholar]
- 73.Kanaphun P, Thirawattanasuk N, Suputtamongkol Y, Naigowit P, Dance DA, Smith MD, White NJ. 1993. Serology and carriage of Pseudomonas pseudomallei: a prospective study in 1000 hospitalized children in northeast Thailand. J Infect Dis 167:230–233. doi: 10.1093/infdis/167.1.230. [DOI] [PubMed] [Google Scholar]
- 74.Smith MD, Angus BJ, Wuthiekanun V, White NJ. 1997. Arabinose assimilation defines a nonvirulent biotype of Burkholderia pseudomallei. Infect Immun 65:4319–4321. doi: 10.1128/IAI.65.10.4319-4321.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gilmore G, Barnes J, Ketheesan N, Norton R. 2007. Use of antigens derived from Burkholderia pseudomallei, B. thailandensis, and B. cepacia in the indirect hemagglutination assay for melioidosis. Clin Vaccine Immunol 14:1529–1531. doi: 10.1128/CVI.00197-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tiyawisutsri R, Peacock SJ, Langa S, Limmathurotsakul D, Cheng AC, Chierakul W, Chaowagul W, Day NPJ, Wuthiekanun V. 2005. Antibodies from patients with melioidosis recognize Burkholderia mallei but not Burkholderia thailandensis antigens in the indirect hemagglutination assay. J Clin Microbiol 43:4872–4874. doi: 10.1128/JCM.43.9.4872-4874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hantrakun V, Thaipadungpanit J, Rongkard P, Srilohasin P, Amornchai P, Langla S, Mukaka M, Chantratita N, Wuthiekanun V, Dance DAB, Day NPJ, Peacock SJ, Limmathurotsakul D. 2018. Presence of B. thailandensis and B. thailandensis expressing B. pseudomallei-like capsular polysaccharide in Thailand, and their associations with serological response to B. pseudomallei. PLoS Negl Trop Dis 12:e0006193. doi: 10.1371/journal.pntd.0006193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bhengsri S, Baggett HC, Jorakate P, Kaewpan A, Prapasiri P, Naorat S, Thamthitiwat S, Tanwisaid K, Chantra S, Salika P, Dejsirilert S, Peruski LF, Maloney SA. 2011. Incidence of bacteremic melioidosis in eastern and northeastern Thailand. Am J Trop Med Hyg 85:117–120. doi: 10.4269/ajtmh.2011.11-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Churuangsuk C, Chusri S, Hortiwakul T, Charernmak B, Silpapojakul K. 2016. Characteristics, clinical outcomes and factors influencing mortality of patients with melioidosis in southern Thailand: a 10-year retrospective study. Asian Pac J Trop Med 9:256–260. doi: 10.1016/j.apjtm.2016.01.034. [DOI] [PubMed] [Google Scholar]
- 80.Trinh TT, Hoang TS, Tran DA, Trinh VT, Göhler A, Nguyen TT, Hoang SN, Krumkamp R, Nguyen LTN, May J, Doan PM, Do CD, Que TA, Steinmetz I. 2018. A simple laboratory algorithm for diagnosis of melioidosis in resource-constrained areas: a study from north-central Vietnam. Clin Microbiol Infect 24:84.e1–84.e4. doi: 10.1016/j.cmi.2017.07.029. [DOI] [PubMed] [Google Scholar]
- 81.Phuong DM, Trung TT, Breitbach K, Tuan NQ, Nubel U, Flunker G, Khang DD, Quang NX, Steinmetz I. 2008. Clinical and microbiological features of melioidosis in northern Vietnam. Trans R Soc Trop Med Hyg 102(Suppl 1):S30–S36. doi: 10.1016/S0035-9203(08)70009-9. [DOI] [PubMed] [Google Scholar]
- 82.Parry CM, Wuthiekanun V, Hoa NT, Diep TS, Thao LT, Loc PV, Wills BA, Wain J, Hien TT, White NJ, Farrar JJ. 1999. Melioidosis in southern Vietnam: clinical surveillance and environmental sampling. Clin Infect Dis 29:1323–1326. doi: 10.1086/313479. [DOI] [PubMed] [Google Scholar]
- 83.Trinh TT, Nguyen DL, Nguyen VT, Tran XC, Le VA, Nguyen VH, Assig K, Lichtenegger S, Wagner EG, Do DC, Steinmetz I. 2018. Melioidosis in Vietnam: recently improved recognition but still an uncertain disease burden after almost a century of reporting. Trop Med Infect Dis 3:39. doi: 10.3390/tropicalmed3020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Göhler A, Trung TT, Hopf V, Kohler C, Hartleib J, Wuthiekanun V, Peacock SJ, Limmathurotsakul D, Tuanyok A, Steinmetz I. 2017. Multitarget quantitative PCR improves detection and predicts cultivability of the pathogen Burkholderia pseudomallei. Appl Environ Microbiol 83:e03212-16. doi: 10.1128/AEM.03212-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Van Phung L, Quynh HT, Yabuuchi E, Dance DAB. 1993. Pilot study of exposure to Pseudomonas pseudomallei in northern Vietnam. Trans R Soc Trop Med Hyg 87:416. doi: 10.1016/0035-9203(93)90017-K. [DOI] [PubMed] [Google Scholar]
- 86.Stanton AT, Flectcher W, Kanagarayer K. 1924. Two cases of melioidosis. J Hyg (Lond) 23:268–276. doi: 10.1017/s0022172400034197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zueter AR, Yean CY, Abumarzouq M, Rahman ZA, Deris ZZ, Harun A. 2016. The epidemiology and clinical spectrum of melioidosis in a teaching hospital in a North-Eastern state of Malaysia: a fifteen-year review. BMC Infect Dis 16:333. doi: 10.1186/s12879-016-1583-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.How SH, Ng KH, Jamalludin AR, Shah A, Rathor Y. 2005. Melioidosis in Pahang, Malaysia. Med J Malaysia 60:606–613. [PubMed] [Google Scholar]
- 89.Mohan A, Podin Y, Tai N, Chieng CH, Rigas V, Machunter B, Mayo M, Wong D, Chien SL, Tan LS, Goh C, Bantin R, Mijen A, Chua WY, Hii KC, Wong SC, Ngian HU, Wong JS, Hashim J, Currie BJ, Ooi MH. 2017. Pediatric melioidosis in Sarawak, Malaysia: epidemiological, clinical and microbiological characteristics. PLoS Negl Trop Dis 11:e0005650. doi: 10.1371/journal.pntd.0005650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.How HS, Ng KH, Yeo HB, Tee HP, Shah A. 2005. Pediatric melioidosis in Pahang, Malaysia. J Microbiol Immunol Infect 38:314–319. [PubMed] [Google Scholar]
- 91.Strauss JM, Alexander AD, Rapmund G, Gan E, Dorsey AE. 1969. Melioidosis in Malaysia. Am J Trop Med Hyg 18:703–707. doi: 10.4269/ajtmh.1969.18.703. [DOI] [PubMed] [Google Scholar]
- 92.Embi N, Suhaimi A, Mohamed R, Ismail G. 1992. Prevalence of antibodies to Pseudomonas pseudomallei exotoxin and whole cell antigens in military personnel in Sabah and Sarawak, Malaysia. Microbiol Immunol 36:899–904. doi: 10.1111/j.1348-0421.1992.tb02092.x. [DOI] [PubMed] [Google Scholar]
- 93.Nathan S, Chieng S, Kingsley P, Mohan A, Podin Y, Ooi M-H, Mariappan V, Vellasamy K, Vadivelu J, Daim S, How S-H. 2018. Melioidosis in Malaysia: incidence, clinical challenges, and advances in understanding pathogenesis. Trop Med Infect Dis 3:25. doi: 10.3390/tropicalmed3010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heng BH, Goh KT, Yap EH, Loh H, Yeo M. 1998. Epidemiological surveillance of melioidosis in Singapore. Ann Acad Med Singapore 27:478–484. [PubMed] [Google Scholar]
- 95.Pang L, Harris PNA, Seiler RL, Ooi PL, Cutter J, Goh KT, Cook AR, Fisher D, Chai LYA. 2018. Melioidosis, Singapore, 2003–2014. Emerg Infect Dis 24:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tan YK, Khoo KL, Chin SP, Ong YY. 1998. Aetiology and outcome of severe community-acquired pneumonia in Singapore. Eur Respir J 12:113–115. doi: 10.1183/09031936.98.12010113. [DOI] [PubMed] [Google Scholar]
- 97.Poulose V. 2008. Severe community-acquired pneumonia requiring intensive care: a study of 80 cases from Singapore. Singapore Med J 49:458–461. [PubMed] [Google Scholar]
- 98.Yap EH, Chan YC, Ti TY, Thong TW, Tan AL, Yeo M, Ho LC, Singh M. 1991. Serodiagnosis of melioidosis in Singapore by the indirect haemagglutination test. Singapore Med J 32:211–213. [PubMed] [Google Scholar]
- 99.Thin RNT, Groves M, Rapmund G, Mariappan M. 1971. Pseudomonas pseudomallei in the surface water of Singapore. Singap Med J 12:181–182. [PubMed] [Google Scholar]
- 100.Limmathurotsakul D, Wuthiekanun V, Amornchai P, Wongsuwan G, Day NP, Peacock SJ. 2012. Effectiveness of a simplified method for isolation of Burkholderia pseudomallei from soil. Appl Environ Microbiol 78:876–877. doi: 10.1128/AEM.07039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nandi T, Holden MTG, Didelot X, Mehershahi K, Boddey JA, Beacham I, Peak I, Harting J, Baybayan P, Guo Y, Wang S, How LC, Sim B, Essex-Lopresti A, Sarkar-Tyson M, Nelson M, Smither S, Ong C, Aw LT, Hoon CH, Michell S, Studholme DJ, Titball R, Chen SL, Parkhill J, Tan P. 2015. Burkholderia pseudomallei sequencing identifies genomic clades with distinct recombination, accessory, and epigenetic profiles. Genome Res 25:129–141. doi: 10.1101/gr.177543.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chantratita N, Wuthiekanun V, Limmathurotsakul D, Vesaratchavest M, Thanwisai A, Amornchai P, Tumapa S, Feil EJ, Day NP, Peacock SJ. 2008. Genetic diversity and microevolution of Burkholderia pseudomallei in the environment. PLoS Negl Trop Dis 2:e182. doi: 10.1371/journal.pntd.0000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zueter AR, Rahman ZA, Abumarzouq M, Harun A. 2018. Multilocus sequence types of clinical Burkholderia pseudomallei isolates from peninsular Malaysia and their associations with disease outcomes. BMC Infect Dis 18:5. doi: 10.1186/s12879-017-2912-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rachlin A, Dittrich S, Phommasone K, Douangnouvong A, Phetsouvanh R, Newton PN, Dance DAB. 2016. Investigation of recurrent melioidosis in Lao People’s Democratic Republic by multilocus sequence typing. Am J Trop Med Hyg 94:1208–1211. doi: 10.4269/ajtmh.15-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pagnarith Y, Kumar V, Thaipadungpanit J, Wuthiekanun V, Amornchai P, Sin L, Day NP, Peacock SJ. 2010. Emergence of pediatric melioidosis in Siem Reap, Cambodia. Am J Trop Med Hyg 82:1106–1112. doi: 10.4269/ajtmh.2010.10-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Overtoom R, Khieu V, Hem S, Cavailler P, Te V, Chan S, Lau P, Guillard B, Vong S. 2008. A first report of pulmonary melioidosis in Cambodia. Trans R Soc Trop Med Hyg 102(Suppl 1):S21–S25. doi: 10.1016/S0035-9203(08)70007-5. [DOI] [PubMed] [Google Scholar]
- 107.Chan CK, Hyland RH, Leers WD, Hutcheon MA, Chang D. 1984. Pleuropulmonary melioidosis in a Cambodian refugee. Can Med Assoc J 131:1365–1367. [PMC free article] [PubMed] [Google Scholar]
- 108.Bory S, Daily F, Khim G, Letchford J, Sok S, Kol H, Seang Lak M, Tuseo L, Vibol C, Oeng S, Turner P. 2018. A report from the Cambodia Training Event for Awareness of Melioidosis (C-TEAM), October 2017. Trop Med Infect Dis 3:23. doi: 10.3390/tropicalmed3010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schully KL, Berjohn CM, Prouty AM, Fitkariwala A, Som T, Sieng D, Gregory MJ, Vaughn A, Kheng S, Te V, Duplessis CA, Lawler JV, Clark DV. 2017. Melioidosis in lower provincial Cambodia: a case series from a prospective study of sepsis in Takeo Province. PLoS Negl Trop Dis 11:e0005923. doi: 10.1371/journal.pntd.0005923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Turner P, Kloprogge S, Miliya T, Soeng S, Tan P, Sar P, Yos P, Moore CE, Wuthiekanun V, Limmathurotsakul D, Turner C, Day NPJ, Dance DAB. 2016. A retrospective analysis of melioidosis in Cambodian children, 2009–2013. BMC Infect Dis 16:688. doi: 10.1186/s12879-016-2034-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wuthiekanun V, Pheaktra N, Putchhat H, Sin L, Sen B, Kumar V, Langla S, Peacock SJ, Day NP. 2008. Burkholderia pseudomallei antibodies in children, Cambodia. Emerg Infect Dis 14:301–303. doi: 10.3201/eid1402.070811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smith MD, Wuthiekanun V, Walsh AL, White NJ. 1995. Quantitative recovery of Burkholderia pseudomallei from soil in Thailand. Trans R Soc Trop Med Hyg 89:488–490. doi: 10.1016/0035-9203(95)90078-0. [DOI] [PubMed] [Google Scholar]
- 113.Vongphayloth K, Rattanavong S, Moore CE, Phetsouvanh R, Wuthiekanun V, Sengdouangphachanh A, Phouminh P, Newton PN, Buisson Y. 2012. Burkholderia pseudomallei detection in surface water in southern Laos using Moore’s swabs. Am J Trop Med Hyg 86:872–877. doi: 10.4269/ajtmh.2012.11-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Manivanh L, Pierret A, Rattanavong S, Kounnavongsa O, Buisson Y, Elliott I, Maeght J, Xayyathip K, Silisouk J, Vongsouvath M, Phetsouvanh R, Newton PN, Lacombe G, Ribolzi O, Rochelle-Newall E, Dance DAB. 2017. Burkholderia pseudomallei in a lowland rice paddy: seasonal changes and influence of soil depth and physico-chemical properties. Sci Rep 7:3031. doi: 10.1038/s41598-017-02946-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Phetsouvanh R, Phongmany S, Soukaloun D, Rasachak B, Soukhaseum V, Soukhaseum S, Frichithavong K, Khounnorath S, Pengdee B, Phiasakha K, Chu V, Luangxay K, Rattanavong S, Sisouk K, Keolouangkot V, Mayxay M, Ramsay A, Blacksell SD, Campbell J, Martinez-Aussel B, Heuanvongsy M, Bounxouei B, Thammavong C, Syhavong B, Strobel M, Peacock SJ, White NJ, Newton PN. 2006. Causes of community-acquired bacteremia and patterns of antimicrobial resistance in Vientiane, Laos. Am J Trop Med Hyg 75:978–985. doi: 10.4269/ajtmh.2006.75.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wuthiekanun V, Mayxay M, Chierakul W, Phetsouvanh R, Cheng AC, White NJ, Day NP, Peacock SJ. 2005. Detection of Burkholderia pseudomallei in soil within the Lao People’s Democratic Republic. J Clin Microbiol 43:923–924. doi: 10.1128/JCM.43.2.923-924.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dance DAB, Luangraj M, Rattanavong S, Sithivong N, Vongnalaysane O, Vongsouvath M, Newton PN. 2018. Melioidosis in the Lao People’s Democratic Republic. Trop Med Infect Dis 3:21. doi: 10.3390/tropicalmed3010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tauran PM, Wahyunie S, Saad F, Dahesihdewi A, Graciella M, Muhammad M, Lestari DC, Aryati A, Parwati I, Loho T, Pratiwi DIN, Mutiawati VK, Loesnihari R, Anggraini D, Rahayu SI, Wulan WN, Antonjaya U, Dance DAB, Currie BJ, Limmathuthurotsakul D, Arif M, Aman AT, Budayanti NNS, Iskandriati D. 2018. Emergence of melioidosis in Indonesia and today’s challenges. Trop Med Infect Dis 3:32. doi: 10.3390/tropicalmed3010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang S. 2000. Melioidosis research in China. Acta Trop 77:157–165. doi: 10.1016/s0001-706x(00)00139-x. [DOI] [PubMed] [Google Scholar]
- 120.Chen H, Lianxu X, Xiong Z, Wei L, Xiaoli D, Duorong W, Rong H, Xiaona S, Ying L, Hong C, Xiao Z. 2015. Burkholderia pseudomallei sequence type 562 in China and Australia. Emerg Infect Dis 21:166. doi: 10.3201/eid2101.140156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen Y-L, Yen Y-C, Yang C-Y, Lee MS, Ho C-K, Mena KD, Wang P-Y, Chen P-S. 2014. The concentrations of ambient Burkholderia pseudomallei during typhoon season in endemic area of melioidosis in Taiwan. PLoS Negl Trop Dis 8:e2877. doi: 10.1371/journal.pntd.0002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tipre M, Kingsley P, Smith T, Leader M, Sathiakumar N. 2018. Melioidosis in India and Bangladesh: a review of case reports. Asian Pac J Trop Med 11:320–329. doi: 10.4103/1995-7645.233179. [DOI] [Google Scholar]
- 123.Chowdhury RF, Jilani AMS, Barai L, Rahman T, Saha RM, Amin MR, Fatema K, Islam MK, Faiz AM, Dunachie JS, Dance AD. 2018. Melioidosis in Bangladesh: a clinical and epidemiological analysis of culture-confirmed cases. Trop Med Infect Dis 3:40. doi: 10.3390/tropicalmed3020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jilani MSA, Robayet JAM, Mohiuddin M, Hasan MR, Ahsan CR, Haq JA. 2016. Burkholderia pseudomallei: its detection in soil and seroprevalence in Bangladesh. PLoS Negl Trop Dis 10:e0004301. doi: 10.1371/journal.pntd.0004301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Limmathurotsakul D, Dance DA, Wuthiekanun V, Kaestli M, Mayo M, Warner J, Wagner DM, Tuanyok A, Wertheim H, Yoke Cheng T, Mukhopadhyay C, Puthucheary S, Day NP, Steinmetz I, Currie BJ, Peacock SJ. 2013. Systematic review and consensus guidelines for environmental sampling of Burkholderia pseudomallei. PLoS Negl Trop Dis 7:e2105. doi: 10.1371/journal.pntd.0002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Denny CR, Nicholls L. 1927. Melioidosis in a European. Ceylon J Sci 2:37–40. [Google Scholar]
- 127.Corea EM, de Silva AD, Thevanesam V. 2018. Melioidosis in Sri Lanka. Trop Med Infect Dis 3:22. doi: 10.3390/tropicalmed3010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Corea EM, Merritt AJ, Ler Y-H, Thevanesam V, Inglis TJJ. 2016. Sri Lankan national melioidosis surveillance program uncovers a nationwide distribution of invasive melioidosis. Am J Trop Med Hyg 94:292–298. doi: 10.4269/ajtmh.15-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mukhopadhyay C, Shaw T, Varghese G, Dance D. 2018. Melioidosis in South Asia (India, Nepal, Pakistan, Bhutan and Afghanistan). Trop Med Infect Dis 3:51. doi: 10.3390/tropicalmed3020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gopalakrishnan R, Sureshkumar D, Thirunarayan MA, Ramasubramanian V. 2013. Melioidosis: an emerging infection in India. J Assoc Physicians India 61:612–614. [PubMed] [Google Scholar]
- 131.Vidyalakshmi K, Lipika S, Vishal S, Damodar S, Chakrapani M. 2012. Emerging clinico-epidemiological trends in melioidosis: analysis of 95 cases from western coastal India. Int J Infect Dis 16:e491–e497. doi: 10.1016/j.ijid.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 132.Vandana KE, Mukhopadhyay C, Tellapragada C, Kamath A, Tipre M, Bhat V, Sathiakumar N. 2016. Seroprevalence of Burkholderia pseudomallei among adults in coastal areas in southwestern India. PLoS Negl Trop Dis 10:e0004610. doi: 10.1371/journal.pntd.0004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Peddayelachagiri BV, Paul S, Nagaraj S, Gogoi M, Sripathy MH, Batra HV. 2016. Prevalence and identification of Burkholderia pseudomallei and near-neighbor species in the Malabar coastal region of India. PLoS Negl Trop Dis 10:e0004956. doi: 10.1371/journal.pntd.0004956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Prakash A, Thavaselvam D, Kumar A, Kumar A, Arora S, Tiwari S, Barua A, Sathyaseelan K. 2014. Isolation, identification and characterization of Burkholderia pseudomallei from soil of coastal region of India. SpringerPlus 3:438. doi: 10.1186/2193-1801-3-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tellapragada C, Kamthan A, Shaw T, Ke V, Kumar S, Bhat V, Mukhopadhyay C. 2016. Unravelling the molecular epidemiology and genetic diversity among Burkholderia pseudomallei isolates from South India using multi-locus sequence typing. PLoS One 11:e0168331. doi: 10.1371/journal.pone.0168331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sathkumara HD, Merritt AJ, Corea EM, Krishnananthasivam S, Natesan M, Inglis TJJ, De Silva AD. 2018. Clinical, bacteriologic, and geographic stratification of melioidosis emerges from the Sri Lankan national surveillance program. Am J Trop Med Hyg 98:607–615. doi: 10.4269/ajtmh.17-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rolim D, Lima R, Ribeiro A, Colares R, Lima L, Rodríguez-Morales A, Montúfar F, Dance D. 2018. Melioidosis in South America. Trop Med Infect Dis 3:60. doi: 10.3390/tropicalmed3020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Benoit TJ, Blaney DD, Doker TJ, Gee JE, Elrod MG, Rolim DB, Inglis TJ, Hoffmaster AR, Bower WA, Walke HT. 2015. A review of melioidosis cases in the Americas. Am J Trop Med Hyg 93:1134–1139. doi: 10.4269/ajtmh.15-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rolim DB, Vilar DCFL, Sousa AQ, Miralles IS, Almeida de Oliveira DC, Harnett G, O’Reilly L, Howard K, Sampson I, Inglis TJJ. 2005. Melioidosis, northeastern Brazil. Emerg Infect Dis 11:1458–1460. doi: 10.3201/eid1109.050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Aardema H, Luijnenburg EM, Salm EF, Bijlmer HA, Visser CE, Van’T Wout JW. 2005. Changing epidemiology of melioidosis? A case of acute pulmonary melioidosis with fatal outcome imported from Brazil. Epidemiol Infect 133:871–875. doi: 10.1017/S0950268805004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Inglis TJ, Rolim DB, Sousa Ade Q. 2006. Melioidosis in the Americas. Am J Trop Med Hyg 75:947–954. doi: 10.4269/ajtmh.2006.75.947. [DOI] [PubMed] [Google Scholar]
- 142.Nussbaum JJ, Hull DS, Carter M. 1980. Pseudomonas pseudomallel [sic] in an anophthalmic orbit. Arch Ophthalmol 98:1224–1225. doi: 10.1001/archopht.1980.01020040076008. [DOI] [PubMed] [Google Scholar]
- 143.Stewart T, Engelthaler DM, Blaney DD, Tuanyok A, Wangsness E, Smith TL, Pearson T, Komatsu KK, Keim P, Currie BJ, Levy C, Sunenshine R. 2011. Epidemiology and investigation of melioidosis, southern Arizona. Emerg Infect Dis 17:1286–1288. doi: 10.3201/eid1707.100661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hall CM, Busch JD, Shippy K, Allender CJ, Kaestli M, Mayo M, Sahl JW, Schupp JM, Colman RE, Keim P, Currie BJ, Wagner DM. 2015. Diverse Burkholderia species isolated from soils in the southern United States with no evidence of B. pseudomallei. PLoS One 10:e0143254. doi: 10.1371/journal.pone.0143254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Doker TJ, Ellis EM, Hoffmaster AR, Beesley CA, Blaney DD, Walke HT, Gee JE, Elrod MG, Galloway RL, Traxler RM, Shadomy SV, Benoit TJ, Bower WA, Perez-Padilla J, Sharp TM, Rivera-Garcia B, Ryff KR, Shieh W-J, Haberling DL, Waller LA. 2014. Contact investigation of melioidosis cases reveals regional endemicity in Puerto Rico. Clin Infect Dis 60:243–250. doi: 10.1093/cid/ciu764. [DOI] [PubMed] [Google Scholar]
- 146.Dorman SE, Vee JG, Gallin JI, Holland SM. 1998. Burkholderia pseudomallei infection in a Puerto Rican patient with chronic granulomatous disease: case report and review of occurrences in the Americas. Clin Infect Dis 26:889–894. doi: 10.1086/513928. [DOI] [PubMed] [Google Scholar]
- 147.Christenson B, Fuxench Z, Morales JA, Suarez-Villamil RA, Souchet LM. 2003. Severe community-acquired pneumonia and sepsis caused by Burkholderia pseudomallei associated with flooding in Puerto Rico. Bol Asoc Med P R 95:17–20. [PubMed] [Google Scholar]
- 148.Hemme RR, Lopez-Ortiz R, Garcia BR, Sharp TM, Galloway RL, Elrod MG, Hunsperger EA. 2016. Serological evidence of infection with endemic human pathogens among free-ranging Old World monkeys in Puerto Rico. Am J Trop Med Hyg 94:1095–1099. doi: 10.4269/ajtmh.15-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sanchez-Villamil JI, Torres AG. 2018. Melioidosis in Mexico, Central America, and the Caribbean. Trop Med Infect Dis 3:24. doi: 10.3390/tropicalmed3010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chewapreecha C, Holden MTG, Vehkala M, Välimäki N, Yang Z, Harris SR, Mather AE, Tuanyok A, De Smet B, Le Hello S, Bizet C, Mayo M, Wuthiekanun V, Limmathurotsakul D, Phetsouvanh R, Spratt BG, Corander J, Keim P, Dougan G, Dance DAB, Currie BJ, Parkhill J, Peacock SJ. 2017. Global and regional dissemination and evolution of Burkholderia pseudomallei. Nat Microbiol 2:16263. doi: 10.1038/nmicrobiol.2016.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Birnie E, Wiersinga WJ, Limmathurotsakul D, Grobusch MP. 2015. Melioidosis in Africa: should we be looking more closely? Future Microbiol 10:273–281. doi: 10.2217/fmb.14.113. [DOI] [PubMed] [Google Scholar]
- 152.Morelli F, Smeets L, Hobijn M, Boom H. 2015. Melioidosis and renal failure in a Dutch man after a trip to Gambia. Neth J Med 73:296–298. [PubMed] [Google Scholar]
- 153.Morosini MI, Quereda C, Gil H, Anda P, Núñez-Murga M, Cantón R, López-Vélez R. 2013. Melioidosis in traveler from Africa to Spain. Emerg Infect Dis 19:1656–1659. doi: 10.3201/eid1910.121785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.MacLennan IS. 1953. Melioidosis in the horse. J R Army Vet Corps 24:130–134. [Google Scholar]
- 155.Wiersinga WJ, Birnie E, Weehuizen TA, Alabi AS, Huson MA, Huis in ‘t Veld RA, Mabala HK, Adzoda GK, Raczynski-Henk Y, Esen M, Lell B, Kremsner PG, Visser CE, Wuthiekanun V, Peacock SJ, van der Ende A, Limmathurotsakul D, Grobusch MP. 2015. Clinical, environmental, and serologic surveillance studies of melioidosis in Gabon, 2012–2013. Emerg Infect Dis 21:40. doi: 10.3201/eid2101.140762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Salam AP, Khan N, Malnick H, Kenna DTD, Dance DAB, Klein JL. 2011. Melioidosis acquired by traveler to Nigeria. Emerg Infect Dis 17:1296–1298. doi: 10.3201/eid1707.100502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rakotondrasoa A, Issack IM, Garin B, Biot F, Valade E, Wattiau P, Allou N, Belmonte O, Bibi J, Price PE, Collard J-M. 2018. Melioidosis in the western Indian Ocean and the importance of improving diagnosis, surveillance, and molecular typing. Trop Med Infect Dis 3:30. doi: 10.3390/tropicalmed3010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sarovich DS, Garin B, De Smet B, Kaestli M, Mayo M, Vandamme P, Jacobs J, Lompo P, Tahita MC, Tinto H, Djaomalaza I, Currie BJ, Price EP. 2016. Phylogenomic analysis reveals an Asian origin for African Burkholderia pseudomallei and further supports melioidosis endemicity in Africa. mSphere 1:e00089-15. doi: 10.1128/mSphere.00089-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Steinmetz I, Wagner EG, Kanyala E, Sawadogo M, Soumeya H, Teferi M, Andargie E, Yeshitela B, Yaba Atsé-Achi L, Sanogo M, Bonfoh B, Rakotozandrindrainy R, Pongombo Shongo C, Shongoya Pongombo M, Kasamba Ilunga E, Lichtenegger S, Assig K, May J, Bertherat E, Owusu M, Owusu-Dabo E, Adu-Sarkodie Y. 2018. Melioidosis in Africa: time to uncover the true disease load. Trop Med Infect Dis 3:62. doi: 10.3390/tropicalmed3020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Govan B. 2012. Molecular characterisation and classification of Burkholderia pseudomallei, p 60–67. In Ketheesan N. (ed), Melioidosis: a century of observation and research. Elsevier, Amsterdam, the Netherlands. [Google Scholar]
- 161.Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa M. 1992. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol Immunol 36:1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x. [DOI] [PubMed] [Google Scholar]
- 162.Redfearn MS, Palleroni NJ, Stanier RY. 1966. A comparative study of Pseudomonas pseudomallei and Bacillus mallei. J Gen Microbiol 43:293–313. doi: 10.1099/00221287-43-2-293. [DOI] [PubMed] [Google Scholar]
- 163.Pumpuang A, Chantratita N, Wikraiphat C, Saiprom N, Day NPJ, Peacock SJ, Wuthiekanun V. 2011. Survival of Burkholderia pseudomallei in distilled water for 16 years. Trans R Soc Trop Med Hyg 105:598–600. doi: 10.1016/j.trstmh.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wuthiekanun V, Dance DAB, Wattanagoon Y, Supputtamongkol Y, Chaowagul W, White NJ. 1990. The use of selective media for the isolation of Pseudomonas pseudomallei in clinical practice. J Med Microbiol 33:121–126. doi: 10.1099/00222615-33-2-121. [DOI] [PubMed] [Google Scholar]
- 165.White NJ, Dance DAB. 1988. Clinical and laboratory studies of malaria and melioidosis. Trans R Soc Trop Med Hyg 82:15–20. doi: 10.1016/0035-9203(88)90249-0. [DOI] [PubMed] [Google Scholar]
- 166.Hassan MRA, Pani SP, Peng NP, Voralu K, Vijayalakshmi N, Mehanderkar R, Aziz NA, Michael E. 2010. Incidence, risk factors and clinical epidemiology of melioidosis: a complex socio-ecological emerging infectious disease in the Alor Setar region of Kedah, Malaysia. BMC Infect Dis 10:302. doi: 10.1186/1471-2334-10-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kingsley PV, Leader M, Nagodawithana NS, Tipre M, Sathiakumar N. 2016. Melioidosis in Malaysia: a review of case reports. PLoS Negl Trop Dis 10:e0005182. doi: 10.1371/journal.pntd.0005182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pagalavan L. 2005. Melioidosis: the Johor Bahru experience. Med J Malaysia 60:599–605. [PubMed] [Google Scholar]
- 169.Tiangpitayakorn C, Songsivilai S, Piyasangthong N, Dharakul T. 1997. Speed of detection of Burkholderia pseudomallei in blood cultures and its correlation with the clinical outcome. Am J Trop Med Hyg 57:96–99. doi: 10.4269/ajtmh.1997.57.96. [DOI] [PubMed] [Google Scholar]
- 170.Teerawattanasook N, Limmathurotsakul D, Day NPJ, Wuthiekanun V. 2014. Failure of Burkholderia pseudomallei to grow in an automated blood culture system. Am J Trop Med Hyg 91:1173–1175. doi: 10.4269/ajtmh.14-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jorakate P, Higdon M, Kaewpan A, Makprasert S, Yuenprakhon S, Tawisaid K, Dejsirilert S, Whistler T, Baggett HC. 2015. Contribution of the BacT/Alert MB Mycobacterium bottle to bloodstream infection surveillance in Thailand: added yield for Burkholderia pseudomallei. J Clin Microbiol 53:910–914. doi: 10.1128/JCM.02008-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Udayan U, Dias M. 2014. Evaluation of BACTEC™ blood culture system for culture of normally sterile body fluids. Indian J Crit Care Med 18:829–830. doi: 10.4103/0972-5229.146331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chantratita N, Tandhavanant S, Wongsuvan G, Wuthiekanun V, Teerawattanasook N, Day NPJ, Limmathurotsakul D, Peacock SJ. 2013. Rapid detection of Burkholderia pseudomallei in blood cultures using a monoclonal antibody-based immunofluorescent assay. Am J Trop Med Hyg 89:971–972. doi: 10.4269/ajtmh.13-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Limmathurotsakul D, Wuthiekanun V, Chierakul W, Cheng AC, Maharjan B, Chaowagul W, White NJ, Day NP, Peacock SJ. 2005. Role and significance of quantitative urine cultures in diagnosis of melioidosis. J Clin Microbiol 43:2274–2276. doi: 10.1128/JCM.43.5.2274-2276.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cheng AC, Wuthiekanun V, Limmathurosakul D, Wongsuvan G, Day NP, Peacock SJ. 2006. Role of selective and nonselective media for isolation of Burkholderia pseudomallei from throat swabs of patients with melioidosis. J Clin Microbiol 44:2316. doi: 10.1128/JCM.00231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hoffmaster AR, AuCoin D, Baccam P, Baggett HC, Baird R, Bhengsri S, Blaney DD, Brett PJ, Brooks TJ, Brown KA, Chantratita N, Cheng AC, Dance DA, Decuypere S, Defenbaugh D, Gee JE, Houghton R, Jorakate P, Lertmemongkolchai G, Limmathurotsakul D, Merlin TL, Mukhopadhyay C, Norton R, Peacock SJ, Rolim DB, Simpson AJ, Steinmetz I, Stoddard RA, Stokes MM, Sue D, Tuanyok A, Whistler T, Wuthiekanun V, Walke HT. 2015. Melioidosis diagnostic workshop, 2013. Emerg Infect Dis doi: 10.3201/eid2102.141045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gilad J, Schwartz D, Amsalem Y. 2007. Clinical features and laboratory diagnosis of infection with the potential bioterrorism agents Burkholderia mallei and Burkholderia pseudomallei. Int J Biomed Sci 3:144–152. [PMC free article] [PubMed] [Google Scholar]
- 178.Ashdown LR. 1979. An improved screening technique for isolation of Pseudomonas pseudomallei from clinical specimens. Pathology 11:293–297. doi: 10.3109/00313027909061954. [DOI] [PubMed] [Google Scholar]
- 179.Rogul M, Carr SR. 1972. Variable ammonia production among smooth and rough strains of Pseudomonas pseudomallei: resemblance to bacteriocin production. J Bacteriol 112:372–380. doi: 10.1128/JB.112.1.372-380.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Howard K, Inglis TJJ. 2003. Novel selective medium for isolation of Burkholderia pseudomallei. J Clin Microbiol 41:3312–3316. doi: 10.1128/jcm.41.7.3312-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Podin Y, Sarovich DS, Price EP, Kaestli M, Mayo M, Hii K, Ngian H, Wong S, Wong I, Wong J, Mohan A, Ooi M, Fam T, Wong J, Tuanyok A, Keim P, Giffard PM, Currie BJ. 2014. Burkholderia pseudomallei isolates from Sarawak, Malaysian Borneo, are predominantly susceptible to aminoglycosides and macrolides. Antimicrob Agents Chemother 58:162–166. doi: 10.1128/AAC.01842-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Francis A, Aiyar S, Yean CY, Naing L, Ravichandran M. 2006. An improved selective and differential medium for the isolation of Burkholderia pseudomallei from clinical specimens. Diagn Microbiol Infect Dis 55:95–99. doi: 10.1016/j.diagmicrobio.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 183.Peacock SJ, Chieng G, Cheng AC, Dance DAB, Amornchai P, Wongsuvan G, Teerawattanasook N, Chierakul W, Day NPJ, Wuthiekanun V. 2005. Comparison of Ashdown’s medium, Burkholderia cepacia medium, and Burkholderia pseudomallei selective agar for clinical isolation of Burkholderia pseudomallei. J Clin Microbiol 43:5359–5361. doi: 10.1128/JCM.43.10.5359-5361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Goodyear A, Strange L, Rholl DA, Silisouk J, Dance DAB, Schweizer HP, Dow S. 2013. An improved selective culture medium enhances the isolation of Burkholderia pseudomallei from contaminated specimens. Am J Trop Med Hyg 89:973–982. doi: 10.4269/ajtmh.13-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Roesnita B, Tay ST, Puthucheary SD, Sam IC. 2012. Diagnostic use of Burkholderia pseudomallei selective media in a low prevalence setting. Trans R Soc Trop Med Hyg 106:131–133. doi: 10.1016/j.trstmh.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 186.Wuthiekanun V, Suputtamongkol Y, Simpson AJH, Kanaphun P, White NJ. 2001. Value of throat swab in diagnosis of melioidosis. J Clin Microbiol 39:3801–3802. doi: 10.1128/JCM.39.10.3801-3802.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Walsh AL, Wuthiekanun V, Smith MD, Suputtamongkol Y, White NJ. 1995. Selective broths for the isolation of Pseudomonas pseudomallei from clinical samples. Trans R Soc Trop Med Hyg 89:124. doi: 10.1016/0035-9203(95)90685-1. [DOI] [PubMed] [Google Scholar]
- 188.Limmathurotsakul D, Jamsen K, Arayawichanont A, Simpson JA, White LJ, Lee SJ, Wuthiekanun V, Chantratita N, Cheng A, Day NPJ, Verzilli C, Peacock SJ. 2010. Defining the true sensitivity of culture for the diagnosis of melioidosis using Bayesian latent class models. PLoS One 5:e12485. doi: 10.1371/journal.pone.0012485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wongsuvan G, Limmathurotsakul D, Wannapasni S, Chierakul W, Teerawattanasook N, Wuthiekanun V. 2009. Lack of correlation of Burkholderia pseudomallei quantities in blood, urine, sputum and pus. Southeast Asian J Trop Med Public Health 40:781–784. [PubMed] [Google Scholar]
- 190.Hemarajata P, Baghdadi JD, Hoffman R, Humphries RM. 2016. Burkholderia pseudomallei: challenges for the clinical microbiology laboratory. J Clin Microbiol 54:2866–2873. doi: 10.1128/JCM.01636-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Eickhoff TC, Bennett JV, Hayes PS, Feeley J. 1970. Pseudomonas pseudomallei: susceptibility to chemotherapeutic agents. J Infect Dis 121:95–102. doi: 10.1093/infdis/121.2.95. [DOI] [PubMed] [Google Scholar]
- 192.Dance DA, Wuthiekanun V, Naigowit P, White NJ. 1989. Identification of Pseudomonas pseudomallei in clinical practice: use of simple screening tests and API 20NE. J Clin Pathol 42:645–648. doi: 10.1136/jcp.42.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hodgson K, Engler C, Govan B, Ketheesan N, Norton R. 2009. Comparison of routine bench and molecular diagnostic methods in identification of Burkholderia pseudomallei. J Clin Microbiol 47:1578–1580. doi: 10.1128/JCM.02507-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Anuntagool N, Naigowit P, Petkanchanapong V, Aramsri P, Panichakul T, Sirisinha S. 2000. Monoclonal antibody-based rapid identification of burkholderia pseudomallei in blood culture fluid from patients with community-acquired septicaemia. J Med Microbiol 49:1075–1078. doi: 10.1099/0022-1317-49-12-1075. [DOI] [PubMed] [Google Scholar]
- 195.Dharakul T, Songsivilai S, Smithikarn S, Thepthai C, Leelaporn A. 1999. Rapid identification of Burkholderia pseudomallei in blood cultures by latex agglutination using lipopolysaccharide-specific monoclonal antibody. Am J Trop Med Hyg 61:658–662. doi: 10.4269/ajtmh.1999.61.658. [DOI] [PubMed] [Google Scholar]
- 196.Pongsunk S, Thirawattanasuk N, Piyasangthong N, Ekpo P. 1999. Rapid identification of Burkholderia pseudomallei in blood cultures by a monoclonal antibody assay. J Clin Microbiol 37:3662–3667. doi: 10.1128/JCM.37.11.3662-3667.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Inglis TJ, Merritt A, Chidlow G, Aravena-Roman M, Harnett G. 2005. Comparison of diagnostic laboratory methods for identification of Burkholderia pseudomallei. J Clin Microbiol 43:2201–2206. doi: 10.1128/JCM.43.5.2201-2206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wuthiekanun V, Anuntagool N, White NJ, Sirisinha S. 2002. Short report: a rapid method for the differentiation of Burkholderia pseudomallei and Burkholderia thailandensis. Am J Trop Med Hyg 66:759–761. doi: 10.4269/ajtmh.2002.66.759. [DOI] [PubMed] [Google Scholar]
- 199.Duval BD, Elrod MG, Gee JE, Chantratita N, Tandhavanant S, Limmathurotsakul D, Hoffmaster AR. 2014. Evaluation of a latex agglutination assay for the identification of Burkholderia pseudomallei and Burkholderia mallei. Am J Trop Med Hyg 90:1043–1046. doi: 10.4269/ajtmh.14-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Dulsuk A, Paksanont S, Sangchankoom A, Ekchariyawat P, Phunpang R, Jutrakul Y, Chantratita N, West TE. 2017. Validation of a monoclonal antibody-based immunofluorescent assay to detect Burkholderia pseudomallei in blood cultures. Trans R Soc Trop Med Hyg 110:670–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tandhavanant S, Wongsuvan G, Wuthiekanun V, Teerawattanasook N, Day NPJ, Limmathurotsakul D, Peacock SJ, Chantratita N. 2013. Monoclonal antibody-based immunofluorescence microscopy for the rapid identification of Burkholderia pseudomallei in clinical specimens. Am J Trop Med Hyg 89:165–168. doi: 10.4269/ajtmh.13-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Houghton RL, Reed DE, Hubbard MA, Dillon MJ, Chen H, Currie BJ, Mayo M, Sarovich DS, Theobald V, Limmathurotsakul D, Wongsuvan G, Chantratita N, Peacock SJ, Hoffmaster AR, Duval B, Brett PJ, Burtnick MN, AuCoin DP. 2014. Development of a prototype lateral flow immunoassay (LFI) for the rapid diagnosis of melioidosis. PLoS Negl Trop Dis 8:e2727. doi: 10.1371/journal.pntd.0002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Price EP, Sarovich DS, Mayo M, Tuanyok A, Drees KP, Kaestli M, Beckstrom-Sternberg SM, Babic-Sternberg JS, Kidd TJ, Bell SC, Keim P, Pearson T, Currie BJ. 2013. Within-host evolution of Burkholderia pseudomallei over a twelve-year chronic carriage infection. mBio 4:e00388-13. doi: 10.1128/mBio.00388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Cuccui J, Milne TS, Harmer N, George AJ, Harding SV, Dean RE, Scott AE, Sarkar-Tyson M, Wren BW, Titball RW, Prior JL. 2012. Characterization of the Burkholderia pseudomallei K96243 capsular polysaccharide I coding region. Infect Immun 80:1209–1221. doi: 10.1128/IAI.05805-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Robertson G, Sorenson A, Govan B, Ketheesan N, Houghton R, Chen H, AuCoin D, Dillon M, Norton R. 2015. Rapid diagnostics for melioidosis: a comparative study of a novel lateral flow antigen detection assay. J Med Microbiol 64:845–848. doi: 10.1099/jmm.0.000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Woods KL, Boutthasavong L, NicFhogartaigh C, Lee SJ, Davong V, AuCoin DP, Dance DAB. 2018. Evaluation of a rapid diagnostic test for detection of Burkholderia pseudomallei in the Lao People’s Democratic Republic. J Clin Microbiol 56:e02002-17. doi: 10.1128/JCM.02002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Suttisunhakul V, Wuthiekanun V, Brett PJ, Khusmith S, Day NP, Burtnick MN, Limmathurotsakul D, Chantratita N. 2016. Development of rapid enzyme-linked immunosorbent assays for detection of antibodies to Burkholderia pseudomallei. J Clin Microbiol 54:1259–1268. doi: 10.1128/JCM.02856-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wuthiekanun V, Chierakul W, Langa S, Chaowagul W, Panpitpat C, Saipan P, Thoujaikong T, Day NP, Peacock SJ. 2006. Development of antibodies to Burkholderia pseudomallei during childhood in melioidosis-endemic northeast Thailand. Am J Trop Med Hyg 74:1074–1075. doi: 10.4269/ajtmh.2006.74.1074. [DOI] [PubMed] [Google Scholar]
- 209.Suttisunhakul V, Chantratita N, Wikraiphat C, Wuthiekanun V, Douglas Z, Day NPJ, Limmathurotsakul D, Brett PJ, Burtnick MN. 2015. Evaluation of polysaccharide-based latex agglutination assays for the rapid detection of antibodies to Burkholderia pseudomallei. Am J Trop Med Hyg 93:542–546. doi: 10.4269/ajtmh.15-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kritsiriwuthinan K, Wajanarogana S, Choosang K, Homsian J, Rerkthanom S. 2018. Production and evaluation of recombinant Burkholderia pseudomallei GroEL and OmpA proteins for serodiagnosis of melioidosis. Acta Trop 178:333–339. doi: 10.1016/j.actatropica.2017.10.019. [DOI] [PubMed] [Google Scholar]
- 211.Alexander AD, Huxsoll DL, Warner AR Jr, Shepler V, Dorsey A. 1970. Serological diagnosis of human melioidosis with indirect hemagglutination and complement fixation tests. Appl Microbiol 20:825–833. doi: 10.1128/AEM.20.5.825-833.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Harris PN, Ketheesan N, Owens L, Norton RE. 2009. Clinical features that affect indirect-hemagglutination-assay responses to Burkholderia pseudomallei. Clin Vaccine Immunol 16:924–930. doi: 10.1128/CVI.00026-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Leelarasamee A. 1985. Diagnostic value of indirect hemagglutination method for melioidosis in Thailand. J Infect Dis Antimicrob Agents 2:213–214. [Google Scholar]
- 214.Appassakij H, Silpapojakul KR, Wansit R, Pornpatkul M. 1990. Diagnostic value of the indirect hemagglutination test for melioidosis in an endemic area. Am J Trop Med Hyg 42:248–253. doi: 10.4269/ajtmh.1990.42.248. [DOI] [PubMed] [Google Scholar]
- 215.Chaichana P, Jenjaroen K, Amornchai P, Chumseng S, Langla S, Rongkard P, Sumonwiriya M, Jeeyapant A, Chantratita N, Teparrukkul P, Limmathurotsakul D, Day NPJ, Wuthiekanun V, Dunachie SJ. 2018. Antibodies in melioidosis: the role of the indirect hemagglutination assay in evaluating patients and exposed populations. Am J Trop Med Hyg 99:1378–1385. doi: 10.4269/ajtmh.17-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Cheng AC, O’Brien M, Freeman K, Lum G, Currie BJ. 2006. Indirect hemagglutination assay in patients with melioidosis in northern Australia. Am J Trop Med Hyg 74:330–334. doi: 10.4269/ajtmh.2006.74.330. [DOI] [PubMed] [Google Scholar]
- 217.Lazzaroni SM, Barnes JL, Williams NL, Govan BL, Norton RE, LaBrooy JT, Ketheesan N. 2008. Seropositivity to Burkholderia pseudomallei does not reflect the development of cell-mediated immunity. Trans R Soc Trop Med Hyg 102(Suppl 1):S66–S70. doi: 10.1016/S0035-9203(08)70018-X. [DOI] [PubMed] [Google Scholar]
- 218.Ashdown LR, Johnson RW, Koehler JM, Cooney CA. 1989. Enzyme-linked immunosorbent assay for the diagnosis of clinical and subclinical melioidosis. J Infect Dis 160:253–260. doi: 10.1093/infdis/160.2.253. [DOI] [PubMed] [Google Scholar]
- 219.Cuzzubbo AJ, Chenthamarakshan V, Vadivelu J, Puthucheary SD, Rowland D, Devine PL. 2000. Evaluation of a new commercially available immunoglobulin M and immunoglobulin G immunochromatographic test for diagnosis of melioidosis infection. J Clin Microbiol 38:1670–1671. doi: 10.1128/JCM.38.4.1670-1671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Chuah SC, Gilmore G, Norton RE. 2005. Rapid serological diagnosis of melioidosis: an evaluation of a prototype immunochromatographic test. Pathology 37:169–171. doi: 10.1080/00313020500058516. [DOI] [PubMed] [Google Scholar]
- 221.O’Brien M, Freeman K, Lum G, Cheng AC, Jacups SP, Currie BJ. 2004. Further evaluation of a rapid diagnostic test for melioidosis in an area of endemicity. J Clin Microbiol 42:2239–2240. doi: 10.1128/jcm.42.5.2239-2240.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Chantratita N, Lertmemongkolchai G, Wuthiekanun V, Norton R. 2012. The serological diagnosis of melioidosis, p 160–167. In Ketheesan N. (ed), Melioidosis: a century of observation and research. Elsevier, Amsterdam, the Netherlands. [Google Scholar]
- 223.Burtnick MN, Brett PJ, Harding SV, Ngugi SA, Ribot WJ, Chantratita N, Scorpio A, Milne TS, Dean RE, Fritz DL, Peacock SJ, Prior JL, Atkins TP, Deshazer D. 2011. The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect Immun 79:1512–1525. doi: 10.1128/IAI.01218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Chen Y, Wong J, Sun GW, Liu Y, Tan G-YG, Gan Y-H. 2011. Regulation of type VI secretion system during Burkholderia pseudomallei Infection. Infect Immun 79:3064–3073. doi: 10.1128/IAI.05148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Chieng S, Mohamed R, Nathan S. 2015. Transcriptome analysis of Burkholderia pseudomallei T6SS identifies Hcp1 as a potential serodiagnostic marker. Microb Pathog 79:47–56. doi: 10.1016/j.micpath.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 226.Pumpuang A, Dunachie SJ, Phokrai P, Jenjaroen K, Sintiprungrat K, Boonsilp S, Brett PJ, Burtnick MN, Chantratita N. 2017. Comparison of O-polysaccharide and hemolysin co-regulated protein as target antigens for serodiagnosis of melioidosis. PLoS Negl Trop Dis 11:e0005499. doi: 10.1371/journal.pntd.0005499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Phokrai P, Karoonboonyanan W, Thanapattarapairoj N, Promkong C, Dulsuk A, Koosakulnirand S, Canovali S, Indrawattana N, Jutrakul Y, Wuthiekanun V, Limmathurotsakul D, Brett PJ, Burtnick MN, Lertmemongkolchai G, Chantratita N. 2018. A rapid immunochromatography test based on Hcp1 is a potential point-of-care test for serological diagnosis of melioidosis. J Clin Microbiol 56:e00346-18. doi: 10.1128/JCM.00346-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Dharakul T, Songsivilai S, Anuntagool N, Chaowagul W, Wongbunnate S, Intachote P, Sirisinha S. 1997. Diagnostic value of an antibody enzyme-linked immunosorbent assay using affinity-purified antigen in an area endemic for melioidosis. Am J Trop Med Hyg 56:418–423. doi: 10.4269/ajtmh.1997.56.418. [DOI] [PubMed] [Google Scholar]
- 229.Kunakorn M, Boonma P, Khupulsup K, Petchclai B. 1990. Enzyme-linked immunosorbent assay for immunoglobulin M specific antibody for the diagnosis of melioidosis. J Clin Microbiol 28:1249–1253. doi: 10.1128/JCM.28.6.1249-1253.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Inglis TJ, Chiang D, Lee GS, Chor-Kiang L. 1998. Potential misidentification of Burkholderia pseudomallei by API 20NE. Pathology 30:62–64. doi: 10.1080/00313029800169685. [DOI] [PubMed] [Google Scholar]
- 231.Lowe P, Engler C, Norton R. 2002. Comparison of automated and nonautomated systems for identification of Burkholderia pseudomallei. J Clin Microbiol 40:4625–4627. doi: 10.1128/jcm.40.12.4625-4627.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Amornchai P, Chierakul W, Wuthiekanun V, Mahakhunkijcharoen Y, Phetsouvanh R, Currie BJ, Newton PN, van Vinh Chau N, Wongratanacheewin S, Day NP, Peacock SJ. 2007. Accuracy of Burkholderia pseudomallei identification using the API 20NE system and a latex agglutination test. J Clin Microbiol 45:3774–3776. doi: 10.1128/JCM.00935-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zong Z, Wang X, Deng Y, Zhou T. 2012. Misidentification of Burkholderia pseudomallei as Burkholderia cepacia by the VITEK 2 system. J Med Microbiol 61:1483–1484. doi: 10.1099/jmm.0.041525-0. [DOI] [PubMed] [Google Scholar]
- 234.Zakharova I, Lopasteyskaya Y, Toporkov A, Viktorov D. 2018. Influence of biochemical features of Burkholderia pseudomallei strains on identification reliability by Vitek 2 system. J Glob Infect Dis 10:7–10. doi: 10.4103/jgid.jgid_39_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Lowe P, Haswell H, Lewis K. 2006. Use of various common isolation media to evaluate the new VITEK 2 colorimetric GN card for identification of Burkholderia pseudomallei. J Clin Microbiol 44:854–856. doi: 10.1128/JCM.44.3.854-856.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Podin Y, Kaestli M, McMahon N, Hennessy J, Ngian HU, Wong JS, Mohana A, Wong SC, William T, Mayo M, Baird RW, Currie BJ. 2013. Reliability of automated biochemical identification of Burkholderia pseudomallei is regionally dependent. J Clin Microbiol 51:3076–3078. doi: 10.1128/JCM.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Brett PJ, DeShazer D, Woods DE. 1998. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int J Syst Bacteriol 4:317–320. doi: 10.1099/00207713-48-1-317. [DOI] [PubMed] [Google Scholar]
- 238.Koh TH, Yong Ng LS, Foon Ho JL, Sng L-H, Wang GCY, Valentine Tzer Pin Lin R. 2003. Automated identification systems and Burkholderia pseudomallei. J Clin Microbiol 41:1809. doi: 10.1128/jcm.41.4.1809.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Weissert C, Dollenmaier G, Rafeiner P, Riehm J, Schultze D. 2009. Burkholderia pseudomallei misidentified by automated system. Emerg Infect Dis 15:1799. doi: 10.3201/eid1511.081719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Inglis TJ, Aravena-Roman M, Ching S, Croft K, Wuthiekanun V, Mee BJ. 2003. Cellular fatty acid profile distinguishes Burkholderia pseudomallei from avirulent Burkholderia thailandensis. J Clin Microbiol 41:4812–4814. doi: 10.1128/jcm.41.10.4812-4814.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Welker M, Moore ER. 2011. Applications of whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry in systematic microbiology. Syst Appl Microbiol 34:2–11. doi: 10.1016/j.syapm.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 242.Krásný L, Hynek R, Hochel I. 2013. Identification of bacteria using mass spectrometry techniques. Int J Mass Spectrom 353:67–79. doi: 10.1016/j.ijms.2013.04.016. [DOI] [Google Scholar]
- 243.Holland RD, Wilkes JG, Rafii F, Sutherland JB, Persons CC, Voorhees KJ, Lay JO Jr.. 1996. Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 10:1227–1232. doi:. [DOI] [PubMed] [Google Scholar]
- 244.Gassiep I, Armstrong M, Norton RE. 2019. Identification of Burkholderia pseudomallei by use of the Vitek mass spectrometer. J Clin Microbiol 57:e00081-19. doi: 10.1128/JCM.00081-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Karger A, Stock R, Ziller M, Elschner MC, Bettin B, Melzer F, Maier T, Kostrzewa M, Scholz HC, Neubauer H, Tomaso H. 2012. Rapid identification of Burkholderia mallei and Burkholderia pseudomallei by intact cell matrix-assisted laser desorption/ionisation mass spectrometric typing. BMC Microbiol 12:229. doi: 10.1186/1471-2180-12-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Wang H, Chen YL, Teng SH, Xu ZP, Xu YC, Hsueh PR. 2016. Evaluation of the Bruker Biotyper matrix-assisted laser desorption/ionization time-of-flight mass spectrometry system for identification of clinical and environmental isolates of Burkholderia pseudomallei. Front Microbiol 7:415. doi: 10.3389/fmicb.2016.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Luo Y, Siu GK, Yeung AS, Chen JH, Ho PL, Leung KW, Tsang JL, Cheng VC, Guo L, Yang J, Ye L, Yam WC. 2015. Performance of the VITEK MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for rapid bacterial identification in two diagnostic centres in China. J Med Microbiol 64:18–24. doi: 10.1099/jmm.0.080317-0. [DOI] [PubMed] [Google Scholar]
- 248.Dingle TC, Butler-Wu SM, Abbott AN. 2014. Accidental exposure to Burkholderia pseudomallei in the laboratory in the era of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 52:3490–3491. doi: 10.1128/JCM.01238-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Walewski V, Mechai F, Billard-Pomares T, Juguet W, Jaureguy F, Picard B, Tandjaoui-Lambiotte Y, Carbonnelle E, Bouchaud O. 2016. MALDI-TOF MS contribution to diagnosis of melioidosis in a nonendemic country in three French travellers. New Microbes New Infect 12:31–34. doi: 10.1016/j.nmni.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Jang HR, Lee CW, Ok SJ, Kim MJ, Bae MJ, Song S, Yi J, Kim KH. 2015. Melioidosis presenting as a mycotic aneurysm in a Korean patient, diagnosed by 16S rRNA sequencing and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Int J Infect Dis 38:62–64. doi: 10.1016/j.ijid.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 251.Niyompanich S, Jaresitthikunchai J, Srisanga K, Roytrakul S, Tungpradabkul S. 2014. Source-identifying biomarker ions between environmental and clinical Burkholderia pseudomallei using whole-cell matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). PLoS One 9:e99160. doi: 10.1371/journal.pone.0099160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Cox CR, Saichek NR, Schweizer HP, Voorhees KJ. 2014. Rapid Burkholderia pseudomallei identification and antibiotic resistance determination by bacteriophage amplification and MALDI-TOF MS. Bacteriophage 4:e29011. doi: 10.4161/bact.29011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Inglis TJ, Healy PE, Fremlin LJ, Golledge CL. 2012. Use of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis for rapid confirmation of Burkholderia pseudomallei in septicemic melioidosis. Am J Trop Med Hyg 86:1039–1042. doi: 10.4269/ajtmh.2012.11-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Suttisunhakul V, Pumpuang A, Ekchariyawat P, Wuthiekanun V, Elrod MG, Turner P, Currie BJ, Phetsouvanh R, Dance DA, Limmathurotsakul D, Peacock SJ, Chantratita N. 2017. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for the identification of Burkholderia pseudomallei from Asia and Australia and differentiation between Burkholderia species. PLoS One 12:e0175294. doi: 10.1371/journal.pone.0175294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Rudrik JT, Soehnlen MK, Perry MJ, Sullivan MM, Reiter-Kintz W, Lee PA, Pettit D, Tran A, Swaney E. 2017. Safety and accuracy of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of highly pathogenic organisms. J Clin Microbiol 55:3513–3529. doi: 10.1128/JCM.01023-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Cunningham SA, Patel R. 2013. Importance of using Bruker’s security-relevant library for Biotyper identification of Burkholderia pseudomallei, Brucella species, and Francisella tularensis. J Clin Microbiol 51:1639–1640. doi: 10.1128/JCM.00267-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Cunningham SA, Patel R. 2015. Standard matrix-assisted laser desorption ionization-time of flight mass spectrometry reagents may inactivate potentially hazardous bacteria. J Clin Microbiol 53:2788–2789. doi: 10.1128/JCM.00957-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Merritt A, Inglis TJJ, Chidlow G, Harnett G. 2006. PCR-based identification of Burkholderia pseudomallei. Rev Inst Med Trop Sao Paulo 48:239–244. doi: 10.1590/s0036-46652006000500001. [DOI] [PubMed] [Google Scholar]
- 259.Price EP, Dale JL, Cook JM, Sarovich DS, Seymour ML, Ginther JL, Kaufman EL, Beckstrom-Sternberg SM, Mayo M, Kaestli M, Glass MB, Gee JE, Wuthiekanun V, Warner JM, Baker A, Foster JT, Tan P, Tuanyok A, Limmathurotsakul D, Peacock SJ, Currie BJ, Wagner DM, Keim P, Pearson T. 2012. Development and validation of Burkholderia pseudomallei-specific real-time PCR assays for clinical, environmental or forensic detection applications. PLoS One 7:e37723. doi: 10.1371/journal.pone.0037723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Lowe CW, Satterfield BA, Nelson DB, Thiriot JD, Heder MJ, March JK, Drake DS, Lew CS, Bunnell AJ, Moore ES, O’Neill KL, Robison RA. 2016. A quadruplex real-time PCR assay for the rapid detection and differentiation of the most relevant members of the B. pseudomallei complex: B. mallei, B. pseudomallei, and B. thailandensis. PLoS One 11:e0164006. doi: 10.1371/journal.pone.0164006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.U’Ren JM, Van Ert MN, Schupp JM, Easterday WR, Simonson TS, Okinaka RT, Pearson T, Keim P. 2005. Use of a real-time PCR TaqMan assay for rapid identification and differentiation of Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol 43:5771–5774. doi: 10.1128/JCM.43.11.5771-5774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Bowers JR, Engelthaler DM, Ginther JL, Pearson T, Peacock SJ, Tuanyok A, Wagner DM, Currie BJ, Keim PS. 2010. BurkDiff: a real-time PCR allelic discrimination assay for Burkholderia pseudomallei and B. mallei. PLoS One 5:e15413. doi: 10.1371/journal.pone.0015413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Novak RT, Glass MB, Gee JE, Gal D, Mayo MJ, Currie BJ, Wilkins PP. 2006. Development and evaluation of a real-time PCR assay targeting the type III secretion system of Burkholderia pseudomallei. J Clin Microbiol 44:85–90. doi: 10.1128/JCM.44.1.85-90.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Thibault FM, Valade E, Vidal DR. 2004. Identification and discrimination of Burkholderia pseudomallei, B. mallei, and B. thailandensis by real-time PCR targeting type III secretion system genes. J Clin Microbiol 42:5871–5874. doi: 10.1128/JCM.42.12.5871-5874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Meumann EM, Novak RT, Gal D, Kaestli ME, Mayo M, Hanson JP, Spencer E, Glass MB, Gee JE, Wilkins PP, Currie BJ. 2006. Clinical evaluation of a type III secretion system real-time PCR assay for diagnosing melioidosis. J Clin Microbiol 44:3028–3030. doi: 10.1128/JCM.00913-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Gal D, Mayo M, Spencer E, Cheng AC, Currie BJ. 2005. Short report: application of a polymerase chain reaction to detect Burkholderia pseudomallei in clinical specimens from patients with suspected melioidosis. Am J Trop Med Hyg 73:1162–1164. doi: 10.4269/ajtmh.2005.73.1162. [DOI] [PubMed] [Google Scholar]
- 267.Walsh AL, Smith MD, Wuthiekanun V, Suputtamongkol Y, Chaowagul W, Dance DA, Angus B, White NJ. 1995. Prognostic significance of quantitative bacteremia in septicemic melioidosis. Clin Infect Dis 21:1498–1500. doi: 10.1093/clinids/21.6.1498. [DOI] [PubMed] [Google Scholar]
- 268.Kluge RM, Du Pont HL. 1973. Factors affecting mortality of patients with bacteremia. Crit Care Med 1:291. doi: 10.1097/00003246-197309000-00022. [DOI] [PubMed] [Google Scholar]
- 269.DuPont HL, Spink WW. 1969. Infections due to gram-negative organisms: an analysis of 860 patients with bacteremia at the University of Minnesota Medical Center, 1958–1966. Medicine 48:307–332. doi: 10.1097/00005792-196907000-00003. [DOI] [PubMed] [Google Scholar]
- 270.Wuthiekanun V, Limmathurotsakul D, Wongsuvan G, Chierakul W, Teerawattanasook N, Teparrukkul P, Day NP, Peacock SJ. 2007. Quantitation of B. pseudomallei in clinical samples. Am J Trop Med Hyg 77:812–813. doi: 10.4269/ajtmh.2007.77.812. [DOI] [PubMed] [Google Scholar]
- 271.Zakharova I, Teteryatnikova N, Toporkov A, Viktorov D. 2017. Development of a multiplex PCR assay for the detection and differentiation of Burkholderia pseudomallei, Burkholderia mallei, Burkholderia thailandensis, and Burkholderia cepacia complex. Acta Trop 174:1–8. doi: 10.1016/j.actatropica.2017.06.016. [DOI] [PubMed] [Google Scholar]
- 272.Lee MA, Wang D, Yap EH. 2005. Detection and differentiation of Burkholderia pseudomallei, Burkholderia mallei and Burkholderia thailandensis by multiplex PCR. FEMS Immunol Med Microbiol 43:413–417. doi: 10.1016/j.femsim.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 273.Bachert BA, Choi SJ, Snyder AK, Rio RV, Durney BC, Holland LA, Amemiya K, Welkos SL, Bozue JA, Cote CK, Berisio R, Lukomski S. 2015. A unique set of the Burkholderia collagen-like proteins provides insight into pathogenesis, genome evolution and niche adaptation, and infection detection. PLoS One 10:e0137578. doi: 10.1371/journal.pone.0137578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Podnecky NL, Elrod MG, Newton BR, Dauphin LA, Shi J, Chawalchitiporn S, Baggett HC, Hoffmaster AR, Gee JE. 2013. Comparison of DNA extraction kits for detection of Burkholderia pseudomallei in spiked human whole blood using real-time PCR. PLoS One 8:e58032. doi: 10.1371/journal.pone.0058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Richardson LJ, Kaestli M, Mayo M, Bowers JR, Tuanyok A, Schupp J, Engelthaler D, Wagner DM, Keim PS, Currie BJ. 2012. Towards a rapid molecular diagnostic for melioidosis: comparison of DNA extraction methods from clinical specimens. J Microbiol Methods 88:179–181. doi: 10.1016/j.mimet.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Merk S, Meyer H, Greiser-Wilke I, Sprague LD, Neubauer H. 2006. Detection of Burkholderia cepacia DNA from artificially infected EDTA-blood and lung tissue comparing different DNA isolation methods. J Vet Med B Infect Dis Vet Public Health 53:281–285. doi: 10.1111/j.1439-0450.2006.00956.x. [DOI] [PubMed] [Google Scholar]
- 277.Knappik M, Dance DA, Rattanavong S, Pierret A, Ribolzi O, Davong V, Silisouk J, Vongsouvath M, Newton PN, Dittrich S. 2015. Evaluation of molecular methods to improve the detection of Burkholderia pseudomallei in soil and water samples from Laos. Appl Environ Microbiol 81:3722–3727. doi: 10.1128/AEM.04204-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Michel PA, Lascols C, Gee JE, Weigel LM, Sue D. 2017. Rapid filter-based detection and culture of Burkholderia pseudomallei from small volumes of urine. J Clin Microbiol 55:2698–2707. doi: 10.1128/JCM.00764-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Currie BJ. 2015. Melioidosis: evolving concepts in epidemiology, pathogenesis, and treatment. Semin Respir Crit Care Med 36:111–125. doi: 10.1055/s-0034-1398389. [DOI] [PubMed] [Google Scholar]
- 280.CLSI. 2010. Performance standards for antimicrobial susceptibility testing; twentieth informational supplement M100-S20. CLSI, Wayne, PA. [Google Scholar]
- 281.Inglis TJ, Rodrigues F, Rigby P, Norton R, Currie BJ. 2004. Comparison of the susceptibilities of Burkholderia pseudomallei to meropenem and ceftazidime by conventional and intracellular methods. Antimicrob Agents Chemother 48:2999–3005. doi: 10.1128/AAC.48.8.2999-3005.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Dance DA, Davong V, Soeng S, Phetsouvanh R, Newton PN, Turner P. 2014. Trimethoprim/sulfamethoxazole resistance in Burkholderia pseudomallei. Int J Antimicrob Agents 44:368–369. doi: 10.1016/j.ijantimicag.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Wiersinga WJ, Currie BJ, Peacock SJ. 2012. Melioidosis. N Engl J Med 367:1035–1044. doi: 10.1056/NEJMra1204699. [DOI] [PubMed] [Google Scholar]
- 284.Jenney AWJ, Lum G, Fisher DA, Currie BJ. 2001. Antibiotic susceptibility of Burkholderia pseudomallei from tropical northern Australia and implications for therapy of melioidosis. Int J Antimicrob Agents 17:109–113. doi: 10.1016/S0924-8579(00)00334-4. [DOI] [PubMed] [Google Scholar]
- 285.Dance DA, Wuthiekanun V, Chaowagul W, White NJ. 1989. The antimicrobial susceptibility of Pseudomonas pseudomallei. Emergence of resistance in vitro and during treatment. J Antimicrob Chemother 24:295–309. doi: 10.1093/jac/24.3.295. [DOI] [PubMed] [Google Scholar]
- 286.Podnecky NL, Rhodes KA, Schweizer HP. 2015. Efflux pump-mediated drug resistance in Burkholderia. Front Microbiol 6:305. doi: 10.3389/fmicb.2015.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Rhodes KA, Schweizer HP. 2016. Antibiotic resistance in Burkholderia species. Drug Resist Updat 28:82–90. doi: 10.1016/j.drup.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Burtnick MN, Woods DE. 1999. Isolation of polymyxin B-susceptible mutants of Burkholderia pseudomallei and molecular characterization of genetic loci involved in polymyxin B resistance. Antimicrob Agents Chemother 43:2648–2656. doi: 10.1128/AAC.43.11.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Chan YY, Ong YM, Chua KL. 2007. Synergistic interaction between phenothiazines and antimicrobial agents against Burkholderia pseudomallei. Antimicrob Agents Chemother 51:623–630. doi: 10.1128/AAC.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Loutet SA, Valvano MA. 2011. Extreme antimicrobial peptide and polymyxin B resistance in the genus Burkholderia. Front Microbiol 2:159. doi: 10.3389/fmicb.2011.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Dutta S, Haq S, Hasan MR, Haq JA. 2017. Antimicrobial susceptibility pattern of clinical isolates of Burkholderia pseudomallei in Bangladesh. BMC Res Notes 10:299. doi: 10.1186/s13104-017-2626-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Khosravi Y, Vellasamy KM, Mariappan V, Ng S-L, Vadivelu J. 2014. Antimicrobial susceptibility and genetic characterisation of Burkholderia pseudomallei isolated from Malaysian patients. Sci World J 2014:9. doi: 10.1155/2014/132971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Wuthiekanun V, Amornchai P, Saiprom N, Chantratita N, Chierakul W, Koh GC, Chaowagul W, Day NP, Limmathurotsakul D, Peacock SJ. 2011. Survey of antimicrobial resistance in clinical Burkholderia pseudomallei isolates over two decades in Northeast Thailand. Antimicrob Agents Chemother 55:5388–5391. doi: 10.1128/AAC.05517-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Rholl DA, Papp-Wallace KM, Tomaras AP, Vasil ML, Bonomo RA, Schweizer HP. 2011. Molecular investigations of PenA-mediated beta-lactam resistance in Burkholderia pseudomallei. Front Microbiol 2:139. doi: 10.3389/fmicb.2011.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Bugrysheva JV, Sue D, Gee JE, Elrod MG, Hoffmaster AR, Randall LB, Chirakul S, Tuanyok A, Schweizer HP, Weigel LM. 2017. Antibiotic resistance markers in Burkholderia pseudomallei strain Bp1651 identified by genome sequence analysis. Antimicrob Agents Chemother 61:e00010-17. doi: 10.1128/AAC.00010-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Chirakul S, Somprasong N, Norris MH, Wuthiekanun V, Chantratita N, Tuanyok A, Schweizer HP. 2019. Burkholderia pseudomallei acquired ceftazidime resistance due to gene duplication and amplification. Int J Antimicrob Agents 53:582–588. doi: 10.1016/j.ijantimicag.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 297.Chantratita N, Rholl DA, Sim B, Wuthiekanun V, Limmathurotsakul D, Amornchai P, Thanwisai A, Chua HH, Ooi WF, Holden MT, Day NP, Tan P, Schweizer HP, Peacock SJ. 2011. Antimicrobial resistance to ceftazidime involving loss of penicillin-binding protein 3 in Burkholderia pseudomallei. Proc Natl Acad Sci U S A 108:17165–17170. doi: 10.1073/pnas.1111020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Crowe A, McMahon N, Currie BJ, Baird RW. 2014. Current antimicrobial susceptibility of first-episode melioidosis Burkholderia pseudomallei isolates from the Northern Territory, Australia. Int J Antimicrob Agents 44:160–162. doi: 10.1016/j.ijantimicag.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 299.Shih HI, Chuang YC, Cheung BMH, Yan JJ, Chang CM, Chang K, Lee NY, Lee HC, Wu CJ, Chen PL, Lee CC, Wang LR, Ko NY, Ko WC. 2008. Sporadic and outbreak cases of melioidosis in southern Taiwan: clinical features and antimicrobial susceptibility. Infection 37:9. doi: 10.1007/s15010-008-7324-8. [DOI] [PubMed] [Google Scholar]
- 300.Tan AL, Tan M-L. 2008. Melioidosis: antibiogram of cases in Singapore 1987–2007. Trans R Soc Trop Med Hyg 102:S101–S102. doi: 10.1016/S0035-9203(08)70024-5. [DOI] [PubMed] [Google Scholar]
- 301.Sarovich DS, Price EP, Von Schulze AT, Cook JM, Mayo M, Watson LM, Richardson L, Seymour ML, Tuanyok A, Engelthaler DM, Pearson T, Peacock SJ, Currie BJ, Keim P, Wagner DM. 2012. Characterization of ceftazidime resistance mechanisms in clinical isolates of Burkholderia pseudomallei from Australia. PLoS One 7:e30789. doi: 10.1371/journal.pone.0030789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.O’Callaghan CH, Acred P, Harper PB, Ryan DM, Kirby SM, Harding SM. 1980. GR 20263, a new broad-spectrum cephalosporin with anti-pseudomonal activity. Antimicrob Agents Chemother 17:876. doi: 10.1128/AAC.17.5.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Sarovich DS, Price EP, Limmathurotsakul D, Cook JM, Von Schulze AT, Wolken SR, Keim P, Peacock SJ, Pearson T. 2012. Development of ceftazidime resistance in an acute Burkholderia pseudomallei infection. Infect Drug Resist 5:129–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Saiprom N, Amornchai P, Wuthiekanun V, Day NPJ, Limmathurotsakul D, Peacock SJ, Chantratita N. 2015. Trimethoprim/sulfamethoxazole resistance in clinical isolates of Burkholderia pseudomallei from Thailand. Int J Antimicrob Agents 45:557–559. doi: 10.1016/j.ijantimicag.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Wuthiekanun V, Cheng AC, Chierakul W, Amornchai P, Limmathurotsakul D, Chaowagul W, Simpson AJ, Short JM, Wongsuvan G, Maharjan B, White NJ, Peacock SJ. 2005. Trimethoprim/sulfamethoxazole resistance in clinical isolates of Burkholderia pseudomallei. J Antimicrob Chemother 55:1029–1031. doi: 10.1093/jac/dki151. [DOI] [PubMed] [Google Scholar]
- 306.Piliouras P, Ulett GC, Ashhurst-Smith C, Hirst RG, Norton RE. 2002. A comparison of antibiotic susceptibility testing methods for cotrimoxazole with Burkholderia pseudomallei. Int J Antimicrob Agents 19:427–429. doi: 10.1016/S0924-8579(02)00016-X. [DOI] [PubMed] [Google Scholar]
- 307.Ahmad N, Hashim R, Mohd Noor A. 2013. The in vitro antibiotic susceptibility of Malaysian isolates of Burkholderia pseudomallei. Int J Microbiol 2013:121845. doi: 10.1155/2013/121845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Bandeira TDJPG, Brilhante RSN, Rocha MFG, Moreira CA, Cordeiro RdA, Ribeiro JF, Castelo-Branco DDSCM, Sidrim JJC. 2013. In vitro antimicrobial susceptibility of clinical and environmental strains of Burkholderia pseudomallei from Brazil. Int J Antimicrob Agents 42:375–377. doi: 10.1016/j.ijantimicag.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 309.Sarovich DS, Webb JR, Pitman MC, Viberg LT, Mayo M, Baird RW, Robson JM, Currie BJ, Price EP. 2018. Raising the stakes: loss of efflux pump regulation decreases meropenem susceptibility in Burkholderia pseudomallei. Clin Infect Dis 67:243–250. doi: 10.1093/cid/ciy069. [DOI] [PubMed] [Google Scholar]
- 310.Viktorov DV, Zakharova IB, Podshivalova MV, Kalinkina EV, Merinova OA, Ageeva NP, Antonov VA, Merinova LK, Alekseev VV. 2008. High-level resistance to fluoroquinolones and cephalosporins in Burkholderia pseudomallei and closely related species. Trans R Soc Trop Med Hyg 102(Suppl 1):S103–S110. doi: 10.1016/S0035-9203(08)70025-7. [DOI] [PubMed] [Google Scholar]
- 311.Chan YY, Tan TM, Ong YM, Chua KL. 2004. BpeAB-OprB, a multidrug efflux pump in Burkholderia pseudomallei. Antimicrob Agents Chemother 48:1128–1135. doi: 10.1128/aac.48.4.1128-1135.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Moore RA, DeShazer D, Reckseidler S, Weissman A, Woods DE. 1999. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother 43:465–470. doi: 10.1128/AAC.43.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Trunck LA, Propst KL, Wuthiekanun V, Tuanyok A, Beckstrom-Sternberg SM, Beckstrom-Sternberg JS, Peacock SJ, Keim P, Dow SW, Schweizer HP. 2009. Molecular basis of rare aminoglycoside susceptibility and pathogenesis of Burkholderia pseudomallei clinical isolates from Thailand. PLoS Negl Trop Dis 3:e519. doi: 10.1371/journal.pntd.0000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Sawasdidoln C, Taweechaisupapong S, Sermswan RW, Tattawasart U, Tungpradabkul S, Wongratanacheewin S. 2010. Growing Burkholderia pseudomallei in biofilm stimulating conditions significantly induces antimicrobial resistance. PLoS One 5:e9196. doi: 10.1371/journal.pone.0009196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Pibalpakdee P, Wongratanacheewin S, Taweechaisupapong S, Niumsup PR. 2012. Diffusion and activity of antibiotics against Burkholderia pseudomallei biofilms. Int J Antimicrob Agents 39:356–359. doi: 10.1016/j.ijantimicag.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 316.Anuntagool N, Wuthiekanun V, White NJ, Currie BJ, Sermswan RW, Wongratanacheewin S, Taweechaisupapong S, Chaiyaroj SC, Sirisinha S. 2006. Lipopolysaccharide heterogeneity among Burkholderia pseudomallei from different geographic and clinical origins. Am J Trop Med Hyg 74:348–352. doi: 10.4269/ajtmh.2006.74.348. [DOI] [PubMed] [Google Scholar]
- 317.Hamad MA, Austin CR, Stewart AL, Higgins M, Vázquez-Torres A, Voskuil MI. 2011. Adaptation and antibiotic tolerance of anaerobic Burkholderia pseudomallei. Antimicrob Agents Chemother 55:3313–3323. doi: 10.1128/AAC.00953-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Thomas AD, Forbes-Faulkner J, Parker M. 1979. Isolation of Pseudomonas pseudomallei from clay layers at defined depths. Am J Epidemiol 110:515–521. doi: 10.1093/oxfordjournals.aje.a112832. [DOI] [PubMed] [Google Scholar]
- 319.Trung TT, Hetzer A, Topfstedt E, Gohler A, Limmathurotsakul D, Wuthiekanun V, Peacock SJ, Steinmetz I. 2011. Improved culture-based detection and quantification of Burkholderia pseudomallei from soil. Trans R Soc Trop Med Hyg 105:346–351. doi: 10.1016/j.trstmh.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 320.Thomas AD, Forbes-Faulkner JC. 1981. Persistence of Pseudomonas pseudomallei in soil. Aust Vet J 57:535–536. doi: 10.1111/j.1751-0813.1981.tb05804.x. [DOI] [PubMed] [Google Scholar]
- 321.Wuthiekanun V, Smith MD, Dance DA, White NJ. 1995. Isolation of Pseudomonas pseudomallei from soil in north-eastern Thailand. Trans R Soc Trop Med Hyg 89:41–43. doi: 10.1016/0035-9203(95)90651-7. [DOI] [PubMed] [Google Scholar]
- 322.Kaestli M, Harrington G, Mayo M, Chatfield MD, Harrington I, Hill A, Munksgaard N, Gibb K, Currie BJ. 2015. What drives the occurrence of the melioidosis bacterium Burkholderia pseudomallei in domestic gardens? PLoS Negl Trop Dis 9:e0003635. doi: 10.1371/journal.pntd.0003635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Kaestli M, Mayo M, Harrington G, Ward L, Watt F, Hill JV, Cheng AC, Currie BJ. 2009. Landscape changes influence the occurrence of the melioidosis bacterium Burkholderia pseudomallei in soil in northern Australia. PLoS Negl Trop Dis 3:e364. doi: 10.1371/journal.pntd.0000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Mayo M, Kaesti M, Harrington G, Cheng AC, Ward L, Karp D, Jolly P, Godoy D, Spratt BG, Currie BJ. 2011. Burkholderia pseudomallei in unchlorinated domestic bore water, tropical northern Australia. Emerg Infect Dis 17:1283–1285. doi: 10.3201/eid1707.100614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Ellison DW, Baker HJ, Mariappan M. 1969. Melioidosis in Malaysia. I. A method for isolation of Pseudomonas pseudomallei from soil and surface water. Am J Trop Med Hyg 18:694–697. doi: 10.4269/ajtmh.1969.18.694. [DOI] [PubMed] [Google Scholar]
- 326.Zanetti F, De Luca G, Stampi S. 2000. Recovery of Burkholderia pseudomallei and B. cepacia from drinking water. Int J Food Microbiol 59:67–72. doi: 10.1016/s0168-1605(00)00255-5. [DOI] [PubMed] [Google Scholar]
- 327.Trung TT, Hetzer A, Göhler A, Topfstedt E, Wuthiekanun V, Limmathurotsakul D, Peacock SJ, Steinmetz I. 2011. Highly sensitive direct detection and quantification of Burkholderia pseudomallei bacteria in environmental soil samples by using real-time PCR. Appl Environ Microbiol 77:6486–6494. doi: 10.1128/AEM.00735-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Kaestli M, Mayo M, Harrington G, Watt F, Hill J, Gal D, Currie BJ. 2007. Sensitive and specific molecular detection of Burkholderia pseudomallei, the causative agent of melioidosis, in the soil of tropical northern Australia. Appl Environ Microbiol 73:6891–6897. doi: 10.1128/AEM.01038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Antonov VA, Tkachenko GA, Altukhova VV, Savchenko SS, Zinchenko OV, Viktorov DV, Zamaraev VS, Ilyukhin VI, Alekseev VV. 2008. Molecular identification and typing of Burkholderia pseudomallei and Burkholderia mallei: when is enough enough? Trans R Soc Trop Med Hyg 102:S134–S139. doi: 10.1016/S0035-9203(08)70030-0. [DOI] [PubMed] [Google Scholar]
- 330.Zulkefli NJ, Mariappan V, Vellasamy KM, Chong CW, Thong KL, Ponnampalavanar S, Vadivelu J, Teh CSJ. 2016. Molecular evidence of Burkholderia pseudomallei genotypes based on geographical distribution. PeerJ 4:e1802. doi: 10.7717/peerj.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Lew AE, Desmarchelier PM. 1993. Molecular typing of Pseudomonas pseudomallei: restriction fragment length polymorphisms of rRNA genes. J Clin Microbiol 31:533–539. doi: 10.1128/JCM.31.3.533-539.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Currie B, Smith-Vaughan H, Golledge C, Buller N, Sriprakash KS, Kemp DJ. 1994. Pseudomonas pseudomallei isolates collected over 25 years from a non-tropical endemic focus show clonality on the basis of ribotyping. Epidemiol Infect 113:307–312. doi: 10.1017/s0950268800051736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Trakulsomboon S, Dance DAB, Smith MD, White NJ, Pitt TL. 1997. Ribotype differences between clinical and environmental isolates of Burkholderia pseudomallei. J Med Microbiol 46:565–570. doi: 10.1099/00222615-46-7-565. [DOI] [PubMed] [Google Scholar]
- 334.Haase A, Smith-Vaughan H, Melder A, Wood Y, Janmaat A, Gilfedder J, Kemp D, Currie B. 1995. Subdivision of Burkholderia pseudomallei ribotypes into multiple types by random amplified polymorphic DNA analysis provides new insights into epidemiology. J Clin Microbiol 33:1687–1690. doi: 10.1128/JCM.33.7.1687-1690.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Pitt TL, Trakulsomboon S, Dance DAB. 2000. Molecular phylogeny of Burkholderia pseudomallei. Acta Trop 74:181–185. doi: 10.1016/s0001-706x(99)00068-6. [DOI] [PubMed] [Google Scholar]
- 336.Vadivelu J, Puthucheary SD, Mifsud A, Drasar BS, Dance DAB, Pitt TL. 1997. Ribotyping and DNA macrorestriction analysis of isolates of Burkholderia pseudomallei from cases of melioidosis in Malaysia. Trans R Soc Trop Med Hyg 91:358–360. doi: 10.1016/S0035-9203(97)90107-3. [DOI] [PubMed] [Google Scholar]
- 337.Inglis TJ, Garrow SC, Adams C, Henderson M, Mayo M, Currie BJ. 1999. Acute melioidosis outbreak in Western Australia. Epidemiol Infect 123:437–443. doi: 10.1017/s0950268899002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Inglis TJ, O’Reilly L, Foster N, Clair A, Sampson J. 2002. Comparison of rapid, automated ribotyping and DNA macrorestriction analysis of Burkholderia pseudomallei. J Clin Microbiol 40:3198–3203. doi: 10.1128/jcm.40.9.3198-3203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Godoy D, Randle G, Simpson AJ, Aanensen DM, Pitt TL, Kinoshita R, Spratt BG. 2003. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol 41:2068–2079. doi: 10.1128/jcm.41.5.2068-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Nualnoi T, Norris MH, Tuanyok A, Brett PJ, Burtnick MN, Keim PS, Settles EW, Allender CJ, AuCoin DP. 2017. Development of immunoassays for Burkholderia pseudomallei typical and atypical lipopolysaccharide strain typing. Am J Trop Med Hyg 96:358–367. doi: 10.4269/ajtmh.16-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Tuanyok A, Stone JK, Mayo M, Kaestli M, Gruendike J, Georgia S, Warrington S, Mullins T, Allender CJ, Wagner DM, Chantratita N, Peacock SJ, Currie BJ, Keim P. 2012. The genetic and molecular basis of O-antigenic diversity in Burkholderia pseudomallei lipopolysaccharide. PLoS Negl Trop Dis 6:e1453. doi: 10.1371/journal.pntd.0001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Stone JK, Mayo M, Grasso SA, Ginther JL, Warrington SD, Allender CJ, Doyle A, Georgia S, Kaestli M, Broomall SM, Karavis MA, Insalaco JM, Hubbard KS, McNew LA, Gibbons HS, Currie BJ, Keim P, Tuanyok A. 2012. Detection of Burkholderia pseudomallei O-antigen serotypes in near-neighbor species. BMC Microbiol 12:250. doi: 10.1186/1471-2180-12-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Webb JR, Sarovich DS, Price EP, Ward LM, Mayo M, Currie BJ. 2019. Burkholderia pseudomallei lipopolysaccharide genotype does not correlate with severity or outcome in melioidosis: host risk factors remain the critical determinant. Open Forum Infect Dis 6:ofz091. doi: 10.1093/ofid/ofz091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Webb JR, Rachlin A, Rigas V, Sarovich DS, Price EP, Kaestli M, Ward LM, Mayo M, Currie BJ. 2019. Tracing the environmental footprint of the Burkholderia pseudomallei lipopolysaccharide genotypes in the tropical “Top End” of the Northern Territory, Australia. bioRxiv doi: 10.1101/603886. [DOI] [PMC free article] [PubMed]
- 345.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Price EP, MacHunter B, Spratt BG, Wagner DM, Currie BJ, Sarovich DS. 2016. Improved multilocus sequence typing of Burkholderia pseudomallei and closely related species. J Med Microbiol 65:992–997. doi: 10.1099/jmm.0.000312. [DOI] [PubMed] [Google Scholar]
- 347.De Smet B, Sarovich DS, Price EP, Mayo M, Theobald V, Kham C, Heng S, Thong P, Holden MTG, Parkhill J, Peacock SJ, Spratt BG, Jacobs JA, Vandamme P, Currie BJ. 2015. Whole-genome sequencing confirms that Burkholderia pseudomallei multilocus sequence types common to both Cambodia and Australia are due to homoplasy. J Clin Microbiol 53:323–326. doi: 10.1128/JCM.02574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Turner KME, Hanage WP, Fraser C, Connor TR, Spratt BG. 2007. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol 7:30. doi: 10.1186/1471-2180-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Liguori AP, Warrington SD, Ginther JL, Pearson T, Bowers J, Glass MB, Mayo M, Wuthiekanun V, Engelthaler D, Peacock SJ, Currie BJ, Wagner DM, Keim P, Tuanyok A. 2011. Diversity of 16S-23S rDNA internal transcribed spacer (ITS) reveals phylogenetic relationships in Burkholderia pseudomallei and its near-neighbors. PLoS One 6:e29323. doi: 10.1371/journal.pone.0029323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Gee JE, Allender CJ, Tuanyok A, Elrod MG, Hoffmaster AR. 2014. Burkholderia pseudomallei type G in Western Hemisphere. Emerg Infect Dis 20:682–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Lewis ERG, Kilgore PB, Mott TM, Pradenas GA, Torres AG. 2017. Comparing in vitro and in vivo virulence phenotypes of Burkholderia pseudomallei type G strains. PLoS One 12:e0175983. doi: 10.1371/journal.pone.0175983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Nandi T, Tan P. 2012. The Burkholderia pseudomallei genome—an emerging model for microbial complexity and pathogen virulence, p 68–81. In Ketheesan N. (ed), Melioidosis: a century of observation and research. Elsevier, Amsterdam, the Netherlands. [Google Scholar]
- 353.Holden MT, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, Sebaihia M, Thomson NR, Bason N, Beacham IR, Brooks K, Brown KA, Brown NF, Challis GL, Cherevach I, Chillingworth T, Cronin A, Crossett B, Davis P, DeShazer D, Feltwell T, Fraser A, Hance Z, Hauser H, Holroyd S, Jagels K, Keith KE, Maddison M, Moule S, Price C, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Simmonds M, Songsivilai S, Stevens K, Tumapa S, Vesaratchavest M, Whitehead S, Yeats C, Barrell BG, Oyston PC, Parkhill J. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci U S A 101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Spring-Pearson SM, Stone JK, Doyle A, Allender CJ, Okinaka RT, Mayo M, Broomall SM, Hill JM, Karavis MA, Hubbard KS, Insalaco JM, McNew LA, Rosenzweig CN, Gibbons HS, Currie BJ, Wagner DM, Keim P, Tuanyok A. 2015. Pangenome analysis of Burkholderia pseudomallei: genome evolution preserves gene order despite high recombination rates. PLoS One 10:e0140274. doi: 10.1371/journal.pone.0140274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Sim SH, Yu Y, Lin CH, Karuturi RKM, Wuthiekanun V, Tuanyok A, Chua HH, Ong C, Paramalingam SS, Tan G, Tang L, Lau G, Ooi EE, Woods D, Feil E, Peacock SJ, Tan P. 2008. The core and accessory genomes of Burkholderia pseudomallei: implications for human melioidosis. PLoS Pathog 4:e1000178. doi: 10.1371/journal.ppat.1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Tuanyok A. 2012. Genomic islands in Burkholderia pseudomallei, p 82–86. In Ketheesan N. (ed), Melioidosis: a century of observation and research. Elsevier, Amsterdam, the Netherlands. [Google Scholar]
- 357.Tuanyok A, Leadem BR, Auerbach RK, Beckstrom-Sternberg SM, Beckstrom-Sternberg JS, Mayo M, Wuthiekanun V, Brettin TS, Nierman WC, Peacock SJ, Currie BJ, Wagner DM, Keim P. 2008. Genomic islands from five strains of Burkholderia pseudomallei. BMC Genom 9:566. doi: 10.1186/1471-2164-9-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Duangsonk K, Gal D, Mayo M, Hart CA, Currie BJ, Winstanley C. 2006. Use of a variable amplicon typing scheme reveals considerable variation in the accessory genomes of isolates of Burkholderia pseudomallei. J Clin Microbiol 44:1323–1334. doi: 10.1128/JCM.44.4.1323-1334.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Chapple SNJ, Sarovich DS, Holden MTG, Peacock SJ, Buller N, Golledge C, Mayo M, Currie BJ, Price EP. 2016. Whole-genome sequencing of a quarter-century melioidosis outbreak in temperate Australia uncovers a region of low-prevalence endemicity. Microb Genom 2:e000067. doi: 10.1099/mgen.0.000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Limmathurotsakul D, Holden MTG, Coupland P, Price EP, Chantratita N, Wuthiekanun V, Amornchai P, Parkhill J, Peacock SJ. 2014. Microevolution of Burkholderia pseudomallei during an acute infection. J Clin Microbiol 52:3418–3421. doi: 10.1128/JCM.01219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Robertson J, Levy A, Sagripanti J-L, Inglis TJJ. 2010. The survival of Burkholderia pseudomallei in liquid media. Am J Trop Med Hyg 82:88–94. doi: 10.4269/ajtmh.2010.09-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Gal D, Mayo M, Smith-Vaughan H, Dasari P, McKinnon M, Jacups SP, Urquhart AI, Hassell M, Currie BJ. 2004. Contamination of hand wash detergent linked to occupationally acquired melioidosis. Am J Trop Med Hyg 71:360–362. doi: 10.4269/ajtmh.2004.71.360. [DOI] [PubMed] [Google Scholar]
- 363.Ooi S-K, Lim T-Y, Lee S-H, Nathan S. 2012. Burkholderia pseudomallei kills Caenorhabditis elegans through virulence mechanisms distinct from intestinal lumen colonization. Virulence 3:485–496. doi: 10.4161/viru.21808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Soffler C, Bosco-Lauth AM, Aboellail TA, Marolf AJ, Bowen RA. 2014. Pathogenesis of percutaneous infection of goats with Burkholderia pseudomallei: clinical, pathologic, and immunological responses in chronic melioidosis. Int J Exp Pathol 95:101–119. doi: 10.1111/iep.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Warawa JM. 2010. Evaluation of surrogate animal models of melioidosis. Front Microbiol 1:141. doi: 10.3389/fmicb.2010.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Leakey AK, Ulett GC, Hirst RG. 1998. BALB/c and C57Bl/6 mice infected with virulent Burkholderia pseudomallei provide contrasting animal models for the acute and chronic forms of human melioidosis. Microb Pathog 24:269–275. doi: 10.1006/mpat.1997.0179. [DOI] [PubMed] [Google Scholar]
- 367.Liu B, Koo GC, Yap EH, Chua KL, Gan YH. 2002. Model of differential susceptibility to mucosal Burkholderia pseudomallei infection. Infect Immun 70:504–511. doi: 10.1128/iai.70.2.504-511.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Tan GY, Liu Y, Sivalingam SP, Sim SH, Wang D, Paucod JC, Gauthier Y, Ooi EE. 2008. Burkholderia pseudomallei aerosol infection results in differential inflammatory responses in BALB/c and C57Bl/6 mice. J Med Microbiol 57:508–515. doi: 10.1099/jmm.0.47596-0. [DOI] [PubMed] [Google Scholar]
- 369.Galyov EE, Brett PJ, DeShazer D. 2010. Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu Rev Microbiol 64:495. doi: 10.1146/annurev.micro.112408.134030. [DOI] [PubMed] [Google Scholar]
- 370.Lazar Adler NR, Govan B, Cullinane M, Harper M, Adler B, Boyce JD. 2009. The molecular and cellular basis of pathogenesis in melioidosis: how does Burkholderia pseudomallei cause disease? FEMS Microbiol Rev 33:1079. doi: 10.1111/j.1574-6976.2009.00189.x. [DOI] [PubMed] [Google Scholar]
- 371.Inglis TJ, Robertson T, Woods DE, Dutton N, Chang BJ. 2003. Flagellum-mediated adhesion by Burkholderia pseudomallei precedes invasion of Acanthamoeba astronyxis. Infect Immun 71:2280–2282. doi: 10.1128/iai.71.4.2280-2282.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Essex-Lopresti AE, Boddey JA, Thomas R, Smith MP, Hartley MG, Atkins T, Brown NF, Tsang CH, Ian RAP, Hill J, Beacham IR, Titball RW. 2005. A type IV pilin, PilA, contributes to adherence of Burkholderia pseudomallei and virulence in vivo. Infect Immun 73:1260–1264. doi: 10.1128/IAI.73.2.1260-1264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Ahmed K, Enciso HD, Masaki H, Tao M, Omori A, Tharavichikul P, Nagatake T. 1999. Attachment of Burkholderia pseudomallei to pharyngeal epithelial cells: a highly pathogenic bacteria with low attachment ability. Am J Trop Med Hyg 60:90–93. doi: 10.4269/ajtmh.1999.60.90. [DOI] [PubMed] [Google Scholar]
- 374.Gori AH, Ahmed K, Martinez G, Masaki H, Watanabe K, Nagatake T. 1999. Mediation of attachment of Burkholderia pseudomallei to human pharyngeal epithelial cells by the asialoganglioside GM1-GM2 receptor complex. Am J Trop Med Hyg 61:473–475. doi: 10.4269/ajtmh.1999.61.473. [DOI] [PubMed] [Google Scholar]
- 375.Jones AL, Beveridge TJ, Woods DE. 1996. Intracellular survival of Burkholderia pseudomallei. Infect Immun 64:782–790. doi: 10.1128/IAI.64.3.782-790.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Allwood EM, Devenish RJ, Prescott M, Adler B, Boyce JD. 2011. Strategies for intracellular survival of Burkholderia pseudomallei. Front Microbiol 2:170. doi: 10.3389/fmicb.2011.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Chen Y, Schröder I, French CT, Jaroszewicz A, Yee XJ, Teh B-E, Toesca IJ, Miller JF, Gan Y-H. 2014. Characterization and analysis of the Burkholderia pseudomallei BsaN virulence regulon. BMC Microbiol 14:206. doi: 10.1186/s12866-014-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Warawa J, Woods DE. 2005. Type III secretion system cluster 3 is required for maximal virulence of Burkholderia pseudomallei in a hamster infection model. FEMS Microbiol Lett 242:101–108. doi: 10.1016/j.femsle.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 379.Schwarz S, West TE, Boyer F, Chiang W-C, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. 2010. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog 6:e1001068. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Sun GW, Chen Y, Liu Y, Tan GY, Ong C, Tan P, Gan YH. 2010. Identification of a regulatory cascade controlling type III secretion system 3 gene expression in Burkholderia pseudomallei. Mol Microbiol 76:677–689. doi: 10.1111/j.1365-2958.2010.07124.x. [DOI] [PubMed] [Google Scholar]
- 381.Vander Broek CW, Stevens JM. 2017. Type III secretion in the melioidosis pathogen Burkholderia pseudomallei. Front Cell Infect Microbiol 7:255. doi: 10.3389/fcimb.2017.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Galán JE, Lara-Tejero M, Marlovits TC, Wagner S. 2014. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol 68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Stevens MP, Haque A, Atkins T, Hill J, Wood MW, Easton A, Nelson M, Underwood-Fowler C, Titball RW, Bancroft GJ, Galyov EE. 2004. Attenuated virulence and protective efficacy of a Burkholderia pseudomallei bsa type III secretion mutant in murine models of melioidosis. Microbiology 150:2669–2676. doi: 10.1099/mic.0.27146-0. [DOI] [PubMed] [Google Scholar]
- 384.Suparak S, Kespichayawattana W, Haque A, Easton A, Damnin S, Lertmemongkolchai G, Bancroft GJ, Korbsrisate S. 2005. Multinucleated giant cell formation and apoptosis in infected host cells is mediated by Burkholderia pseudomallei type III secretion protein BipB. J Bacteriol 187:6556–6560. doi: 10.1128/JB.187.18.6556-6560.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Kang WT, Vellasamy KM, Rajamani L, Beuerman RW, Vadivelu J. 2016. Burkholderia pseudomallei type III secreted protein BipC: role in actin modulation and translocation activities required for the bacterial intracellular lifecycle. PeerJ 4:e2532. doi: 10.7717/peerj.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Kang WT, Vellasamy KM, Chua E-G, Vadivelu J. 2015. Functional characterizations of effector protein BipC, a type III secretion system protein, in Burkholderia pseudomallei pathogenesis. J Infect Dis 211:827–834. doi: 10.1093/infdis/jiu492. [DOI] [PubMed] [Google Scholar]
- 387.Stevens MP, Friebel A, Taylor LA, Wood MW, Brown PJ, Hardt W-D, Galyov EE. 2003. A Burkholderia pseudomallei type III secreted protein, BopE, facilitates bacterial invasion of epithelial cells and exhibits guanine nucleotide exchange factor activity. J Bacteriol 185:4992–4996. doi: 10.1128/jb.185.16.4992-4996.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Ireland PM, Marshall L, Norville I, Sarkar-Tyson M. 2014. The serine protease inhibitor Ecotin is required for full virulence of Burkholderia pseudomallei. Microb Pathog 67-68:55–58. doi: 10.1016/j.micpath.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 389.Stevens MP, Wood MW, Taylor LA, Monaghan P, Hawes P, Jones PW, Wallis TS, Galyov EE. 2002. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol Microbiol 46:649–659. doi: 10.1046/j.1365-2958.2002.03190.x. [DOI] [PubMed] [Google Scholar]
- 390.French CT, Toesca IJ, Wu T-H, Teslaa T, Beaty SM, Wong W, Liu M, Schröder I, Chiou P-Y, Teitell MA, Miller JF. 2011. Dissection of the Burkholderia intracellular life cycle using a photothermal nanoblade. Proc Natl Acad Sci U S A 108:12095–12100. doi: 10.1073/pnas.1107183108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Vadivelu J, Vellasamy KM, Thimma J, Mariappan V, Kang W-T, Choh L-C, Shankar EM, Wong KT. 2017. Survival and intra-nuclear trafficking of Burkholderia pseudomallei: strategies of evasion from immune surveillance? PLoS Negl Trop Dis 11:e0005241. doi: 10.1371/journal.pntd.0005241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Nathan SA, Puthucheary SD. 2005. An electronmicroscopic study of the interaction of Burkholderia pseudomallei and human macrophages. Malays J Pathol 27:3. [PubMed] [Google Scholar]
- 393.Willcocks SJ, Denman CC, Atkins HS, Wren BW. 2015. Intracellular replication of the well-armed pathogen Burkholderia pseudomallei. Curr Opin Microbiol 29:94–103. doi: 10.1016/j.mib.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 394.Ekchariyawat P, Pudla S, Limposuwan K, Arjcharoen S, Sirisinha S, Utaisincharoen P. 2005. Burkholderia pseudomallei-induced expression of suppressor of cytokine signaling 3 and cytokine-inducible Src homology 2-containing protein in mouse macrophages: a possible mechanism for suppression of the response to gamma interferon stimulation. Infect Immun 73:7332–7339. doi: 10.1128/IAI.73.11.7332-7339.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Miyagi K, Kawakami K, Saito A. 1997. Role of reactive nitrogen and oxygen intermediates in gamma interferon-stimulated murine macrophage bactericidal activity against Burkholderia pseudomallei. Infect Immun 65:4108–4113. doi: 10.1128/IAI.65.10.4108-4113.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Krakauer T. 2018. Living dangerously: Burkholderia pseudomallei modulates phagocyte cell death to survive. Med Hypotheses 121:64–69. doi: 10.1016/j.mehy.2018.09.028. [DOI] [PubMed] [Google Scholar]
- 397.Kespichayawattana W, Rattanachetkul S, Wanun T, Utaisincharoen P, Sirisinha S. 2000. Burkholderia pseudomallei induces cell fusion and actin-associated membrane protrusion: a possible mechanism for cell-to-cell spreading. Infect Immun 68:5377–5384. doi: 10.1128/iai.68.9.5377-5384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Stevens JM, Galyov EE, Stevens MP. 2006. Actin-dependent movement of bacterial pathogens. Nat Rev Microbiol 4:91–101. doi: 10.1038/nrmicro1320. [DOI] [PubMed] [Google Scholar]
- 399.Breitbach K, Rottner K, Klocke S, Rohde M, Jenzora A, Wehland J, Steinmetz I. 2003. Actin-based motility of Burkholderia pseudomallei involves the Arp 2/3 complex, but not N-WASP and Ena/VASP proteins. Cell Microbiol 5:385–393. doi: 10.1046/j.1462-5822.2003.00277.x. [DOI] [PubMed] [Google Scholar]
- 400.Stevens MP, Stevens JM, Jeng RL, Taylor LA, Wood MW, Hawes P, Monaghan P, Welch MD, Galyov EE. 2005. Identification of a bacterial factor required for actin-based motility of Burkholderia pseudomallei. Mol Microbiol 56:40–53. doi: 10.1111/j.1365-2958.2004.04528.x. [DOI] [PubMed] [Google Scholar]
- 401.Sarovich DS, Price EP, Webb JR, Ward LM, Voutsinos MY, Tuanyok A, Mayo M, Kaestli M, Currie BJ. 2014. Variable virulence factors in Burkholderia pseudomallei (melioidosis) associated with human disease. PLoS One 9:e91682. doi: 10.1371/journal.pone.0091682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Shalom G, Shaw JG, Thomas MS. 2007. In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153:2689–2699. doi: 10.1099/mic.0.2007/006585-0. [DOI] [PubMed] [Google Scholar]
- 403.Toesca IJ, French CT, Miller JF. 2014. The type VI secretion system spike protein VgrG5 mediates membrane fusion during intercellular spread by pseudomallei group Burkholderia species. Infect Immun 82:1436–1444. doi: 10.1128/IAI.01367-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Schwarz S, Singh P, Robertson JD, LeRoux M, Skerrett SJ, Goodlett DR, West TE, Mougous JD. 2014. VgrG-5 is a Burkholderia type VI secretion system-exported protein required for multinucleated giant cell formation and virulence. Infect Immun 82:1445–1452. doi: 10.1128/IAI.01368-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Lennings J, West TE, Schwarz S. 2019. The Burkholderia type VI secretion system 5: composition, regulation and role in virulence. Front Microbiol 9:3339. doi: 10.3389/fmicb.2018.03339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Whiteley L, Meffert T, Haug M, Weidenmaier C, Hopf V, Bitschar K, Schittek B, Kohler C, Steinmetz I, West TE, Schwarz S. 2017. Entry, intracellular survival, and multinucleated-giant-cell-forming activity of Burkholderia pseudomallei in human primary phagocytic and nonphagocytic cells. Infect Immun 85:e00468-17. doi: 10.1128/IAI.00468-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Wong KT, Puthucheary SD, Vadivelu J. 1995. The histopathology of human melioidosis. Histopathology 26:51–55. doi: 10.1111/j.1365-2559.1995.tb00620.x. [DOI] [PubMed] [Google Scholar]
- 408.Boddey JA, Day CJ, Flegg CP, Ulrich RL, Stephens SR, Beacham IR, Morrison NA, Peak IRA. 2007. The bacterial gene lfpA influences the potent induction of calcitonin receptor and osteoclast-related genes in Burkholderia pseudomallei-induced TRAP-positive multinucleated giant cells. Cell Microbiol 9:514–531. doi: 10.1111/j.1462-5822.2006.00807.x. [DOI] [PubMed] [Google Scholar]
- 409.Burtnick MN, Brett PJ, Nair V, Warawa JM, Woods DE, Gherardini FC. 2008. Burkholderia pseudomallei type III secretion system mutants exhibit delayed vacuolar escape phenotypes in RAW 264.7 murine macrophages. Infect Immun 76:2991–3000. doi: 10.1128/IAI.00263-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Utaisincharoen P, Arjcharoen S, Limposuwan K, Tungpradabkul S, Sirisinha S. 2006. Burkholderia pseudomallei RpoS regulates multinucleated giant cell formation and inducible nitric oxide synthase expression in mouse macrophage cell line (RAW 264.7). Microb Pathog 40:184–189. doi: 10.1016/j.micpath.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 411.Gutierrez MG, Yoder-Himes DR, Warawa JM. 2015. Comprehensive identification of virulence factors required for respiratory melioidosis using Tn-seq mutagenesis. Front Cell Infect Microbiol 5:78. doi: 10.3389/fcimb.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Reckseidler SL, DeShazer D, Sokol PA, Woods DE. 2001. Detection of bacterial virulence genes by subtractive hybridization: identification of capsular polysaccharide of Burkholderia pseudomallei as a major virulence determinant. Infect Immun 69:34–44. doi: 10.1128/IAI.69.1.34-44.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Reckseidler-Zenteno SL, DeVinney R, Woods DE. 2005. The capsular polysaccharide of Burkholderia pseudomallei contributes to survival in serum by reducing complement factor C3b deposition. Infect Immun 73:1106–1115. doi: 10.1128/IAI.73.2.1106-1115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Atkins T, Prior R, Mack K, Russell P, Nelson M, Prior J, Ellis J, Oyston PC, Dougan G, Titball RW. 2002. Characterisation of an acapsular mutant of Burkholderia pseudomallei identified by signature tagged mutagenesis. J Med Microbiol 51:539–547. doi: 10.1099/0022-1317-51-7-539. [DOI] [PubMed] [Google Scholar]
- 415.Puthucheary SD, Vadivelu J, Ce-Cile C, Kum-Thong W, Ismail G. 1996. Short report: electron microscopic demonstration of extracellular structure of Burkholderia pseudomallei. Am J Trop Med Hyg 54:313. doi: 10.4269/ajtmh.1996.54.313. [DOI] [PubMed] [Google Scholar]
- 416.Vorachit M, Lam K, Jayanetra P, Costerton JW. 1995. Electron microscopy study of the mode of growth of Pseudomonas pseudomallei in vitro and in vivo. Am J Trop Med Hyg 98:379. [PubMed] [Google Scholar]
- 417.Sengyee S, Yoon SH, Paksanont S, Yimthin T, Wuthiekanun V, Limmathurotsakul D, West TE, Ernst RK, Chantratita N. 2018. Comprehensive analysis of clinical Burkholderia pseudomallei isolates demonstrates conservation of unique lipid A structure and TLR4-dependent innate immune activation. PLoS Negl Trop Dis 12:e0006287. doi: 10.1371/journal.pntd.0006287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Anuntagool N, Aramsri P, Panichakul T, Wuthiekanun VR, Kinoshita R, White NJ, Sirisinha S. 2000. Antigenic heterogeneity of lipopolysaccharide among Burkholderia pseudomallei clinical isolates Southeast Asian J Trop Med Public Health 31(Suppl 1):146. [PubMed] [Google Scholar]
- 419.Norris MH, Schweizer HP, Tuanyok A. 2017. Structural diversity of Burkholderia pseudomallei lipopolysaccharides affects innate immune signaling. PLoS Negl Trop Dis 11:e0005571. doi: 10.1371/journal.pntd.0005571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Ulrich RL, DeShazer D, Brueggemann EE, Hines HB, Oyston PC, Jeddeloh JA. 2004. Role of quorum sensing in the pathogenicity of Burkholderia pseudomallei. J Med Microbiol 53:1053–1064. doi: 10.1099/jmm.0.45661-0. [DOI] [PubMed] [Google Scholar]
- 421.Valade E, Thibault FM, Gauthier YP, Palencia M, Popoff MY, Vidal DR. 2004. The PmlI-PmlR quorum-sensing system in Burkholderia pseudomallei plays a key role in virulence and modulates production of the MprA protease. J Bacteriol 186:2288–2294. doi: 10.1128/jb.186.8.2288-2294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. 2009. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. doi: 10.1186/1471-2164-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Hopf V, Göhler A, Eske-Pogodda K, Bast A, Steinmetz I, Breitbach K. 2014. BPSS1504, a cluster 1 type VI secretion gene, is involved in intracellular survival and virulence of Burkholderia pseudomallei. Infect Immun 82:2006–2015. doi: 10.1128/IAI.01544-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.DeShazer D, Brett PJ, Burtnick MN, Woods DE. 1999. Molecular characterization of genetic loci required for secretion of exoproducts in Burkholderia pseudomallei. J Bacteriol 181:4661–4664. doi: 10.1128/JB.181.15.4661-4664.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Gauthier YP, Thibault FM, Paucod JC, Vidal DR. 2000. Protease production by Burkholderia pseudomallei and virulence in mice. Acta Trop 74:215–220. doi: 10.1016/s0001-706x(99)00073-x. [DOI] [PubMed] [Google Scholar]
- 426.Duangurai T, Indrawattana N, Pumirat P. 2018. Burkholderia pseudomallei adaptation for survival in stressful conditions. BioMed Res Int 2018:11. doi: 10.1155/2018/3039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Subsin B, Thomas MS, Katzenmeier G, Shaw JG, Tungpradabkul S, Kunakorn M. 2003. Role of the stationary growth phase sigma factor RpoS of Burkholderia pseudomallei in response to physiological stress conditions. J Bacteriol 185:7008–7014. doi: 10.1128/jb.185.23.7008-7014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Korbsrisate S, Vanaporn M, Kerdsuk P, Kespichayawattana W, Vattanaviboon P, Kiatpapan P, Lertmemongkolchai G. 2005. The Burkholderia pseudomallei RpoE (AlgU) operon is involved in environmental stress tolerance and biofilm formation. FEMS Microbiol Lett 252:243–249. doi: 10.1016/j.femsle.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 429.Thongboonkerd V, Vanaporn M, Songtawee N, Kanlaya R, Sinchaikul S, Chen S-T, Easton A, Chu K, Bancroft GJ, Korbsrisate S. 2007. Altered proteome in Burkholderia pseudomallei rpoE operon knockout mutant: insights into mechanisms of rpoE operon in stress tolerance, survival, and virulence. J Proteome Res 6:1334. doi: 10.1021/pr060457t. [DOI] [PubMed] [Google Scholar]
- 430.Chantratita N, Wuthiekanun V, Boonbumrung K, Tiyawisutsri R, Vesaratchavest M, Limmathurotsakul D, Chierakul W, Wongratanacheewin S, Pukritiyakamee S, White NJ, Nicholas PJD, Peacock SJ. 2007. Biological relevance of colony morphology and phenotypic switching by Burkholderia pseudomallei. J Bacteriol 189:807–817. doi: 10.1128/JB.01258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol 4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 432.Häußler S, Rohde M, Steinmetz I. 1999. Highly resistant Burkholderia pseudomallei small colony variants isolated in vitro and in experimental melioidosis. Med Microbiol Immunol 188:91–97. doi: 10.1007/s004300050110. [DOI] [PubMed] [Google Scholar]
- 433.Nur Siti KR, Guan CE, Nathan S, Vadivelu J. 2012. The effect of environmental conditions on biofilm formation of Burkholderia pseudomallei clinical isolates. PLoS One 7:e44104. doi: 10.1371/journal.pone.0044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Austin CR, Goodyear AW, Bartek IL, Stewart A, Sutherland MD, Silva EB, Zweifel A, Vitko NP, Tuanyok A, Highnam G, Mittelman D, Keim P, Schweizer HP, Vazquez-Torres A, Dow SW, Voskuil MI. 2015. A Burkholderia pseudomallei colony variant necessary for gastric colonization. mBio 6:e0462-14. doi: 10.1128/mBio.02462-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Tandhavanant S, Thanwisai A, Limmathurotsakul D, Korbsrisate S, Day NP, Peacock SJ, Chantratita N. 2010. Effect of colony morphology variation of Burkholderia pseudomallei on intracellular survival and resistance to antimicrobial environments in human macrophages in vitro. BMC Microbiol 10:303. doi: 10.1186/1471-2180-10-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Al-Maleki AR, Mariappan V, Vellasamy KM, Shankar EM, Tay ST, Vadivelu J. 2014. Enhanced intracellular survival and epithelial cell adherence abilities of Burkholderia pseudomallei morphotypes are dependent on differential expression of virulence-associated proteins during mid-logarithmic growth phase. J Proteom 106:205–220. doi: 10.1016/j.jprot.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 437.Tuanyok A, Auerbach RK, Brettin TS, Bruce DC, Munk AC, Detter JC, Pearson T, Hornstra H, Sermswan RW, Wuthiekanun V, Peacock SJ, Currie BJ, Keim P, Wagner DM. 2007. A horizontal gene transfer event defines two distinct groups within Burkholderia pseudomallei that have dissimilar geographic distributions. J Bacteriol 189:9044–9049. doi: 10.1128/JB.01264-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Challacombe JF, Stubben CJ, Klimko CP, Welkos SL, Kern SJ, Bozue JA, Worsham PL, Cote CK, Wolfe DN. 2014. Interrogation of the Burkholderia pseudomallei genome to address differential virulence among isolates. PLoS One 9:e115951. doi: 10.1371/journal.pone.0115951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Chirakul S, Bartpho T, Wongsurawat T, Taweechaisupapong S, Karoonutaisiri N, Talaat AM, Wongratanacheewin S, Ernst RK, Sermswan RW. 2014. Characterization of BPSS1521 (bprD), a regulator of Burkholderia pseudomallei virulence gene expression in the mouse model. PLoS One 9:e104313. doi: 10.1371/journal.pone.0104313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Gan Y-H. 2005. Interaction between Burkholderia pseudomallei and the host immune response: sleeping with the enemy? J Infect Dis 192:1845–1850. doi: 10.1086/497382. [DOI] [PubMed] [Google Scholar]
- 441.Ulett GC, Ketheesan N, Hirst RG. 2000. Cytokine gene expression in innately susceptible BALB/c mice and relatively resistant C57BL/6 mice during infection with virulent Burkholderia pseudomallei. Infect Immun 68:2034–2042. doi: 10.1128/iai.68.4.2034-2042.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Ulett GC, Ketheesan N, Hirst RG. 2000. Proinflammatory cytokine mRNA responses in experimental Burkholderia pseudomallei infection in mice. Acta Trop 74:229–234. doi: 10.1016/s0001-706x(99)00075-3. [DOI] [PubMed] [Google Scholar]
- 443.Riyapa D, Buddhisa S, Korbsrisate S, Cuccui J, Wren BW, Stevens MP, Ato M, Lertmemongkolchai G. 2012. Neutrophil extracellular traps exhibit antibacterial activity against Burkholderia pseudomallei and are influenced by bacterial and host factors. Infect Immun 80:3921–3929. doi: 10.1128/IAI.00806-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Saengmuang P, Kewcharoenwong C, Tippayawat P, Nithichanon A, Buddhisa S, Lertmemongkolchai G. 2014. Effect of host factors on neutrophil functions in response to Burkholderia pseudomallei in healthy Thai subjects. Jpn J Infect Dis 67:436–440. doi: 10.7883/yoken.67.436. [DOI] [PubMed] [Google Scholar]
- 445.Chanchamroen S, Kewcharoenwong C, Susaengrat W, Ato M, Lertmemongkolchai G. 2009. Human polymorphonuclear neutrophil responses to Burkholderia pseudomallei in healthy and diabetic subjects. Infect Immun 77:456–463. doi: 10.1128/IAI.00503-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Rinchai D, Riyapa D, Buddhisa S, Utispan K, Titball RW, Stevens MP, Stevens JM, Ogawa M, Tanida I, Koike M, Uchiyama Y, Ato M, Lertmemongkolchai G. 2015. Macroautophagy is essential for killing of intracellular Burkholderia pseudomallei in human neutrophils. Autophagy 11:748–755. doi: 10.1080/15548627.2015.1040969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Easton A, Haque A, Chu K, Lukaszewski R, Bancroft GJ. 2007. A critical role for neutrophils in resistance to experimental infection with Burkholderia pseudomallei. J Infect Dis 195:99–107. doi: 10.1086/509810. [DOI] [PubMed] [Google Scholar]
- 448.Mulye M, Bechill MP, Grose W, Ferreira VP, Lafontaine ER, Wooten RM. 2014. Delineating the importance of serum opsonins and the bacterial capsule in affecting the uptake and killing of Burkholderia pseudomallei by murine neutrophils and macrophages. PLoS Negl Trop Dis 8:e2988. doi: 10.1371/journal.pntd.0002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Woodman ME, Worth RG, Wooten RM. 2012. Capsule influences the deposition of critical complement C3 levels required for the killing of Burkholderia pseudomallei via NADPH-oxidase induction by human neutrophils. PLoS One 7:e52276. doi: 10.1371/journal.pone.0052276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Barnes JL, Williams NL, Ketheesan N. 2008. Susceptibility to Burkholderia pseudomallei is associated with host immune responses involving tumor necrosis factor receptor-1 (TNFR1) and TNF receptor-2 (TNFR2). FEMS Immunol Med Microbiol 52:379–388. doi: 10.1111/j.1574-695X.2008.00389.x. [DOI] [PubMed] [Google Scholar]
- 451.Wiersinga WJ, Wieland CW, Joris JTHR, Tom van der P. 2008. MyD88 dependent signaling contributes to protective host defense against Burkholderia pseudomallei. PLoS One 3:e3494. doi: 10.1371/journal.pone.0003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Wiersinga WJ, Wieland CW, Dessing MC, Chantratita N, Cheng AC, Limmathurotsakul D, Chierakul W, Leendertse M, Florquin S, de Vos AF, White N, Dondorp AM, Day NP, Peacock SJ, van der Poll T. 2007. Toll-like receptor 2 impairs host defense in gram-negative sepsis caused by Burkholderia pseudomallei (melioidosis). PLoS Med 4:e248. doi: 10.1371/journal.pmed.0040248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Ho M, Schollaardt T, Smith MD, Perry MB, Brett PJ, Chaowagul W, Bryan LE. 1997. Specificity and functional activity of anti-Burkholderia pseudomallei polysaccharide antibodies. Infect Immun 65:3648–3653. doi: 10.1128/IAI.65.9.3648-3653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Vasu C, Vadivelu J, Puthucheary SD. 2003. The humoral immune response in melioidosis patients during therapy. Infection 31:24–30. doi: 10.1007/s15010-002-3020-2. [DOI] [PubMed] [Google Scholar]
- 455.Haque A, Chu K, Easton A, Stevens MP, Galyov EE, Atkins T, Titball R, Bancroft GJ. 2006. A live experimental vaccine against Burkholderia pseudomallei elicits CD4+ T cell-mediated immunity, priming T cells specific for 2 type III secretion system proteins. J Infect Dis 194:1241–1248. doi: 10.1086/508217. [DOI] [PubMed] [Google Scholar]
- 456.Chierakul W, Rajanuwong A, Wuthiekanun V, Teerawattanasook N, Gasiprong M, Simpson A, Chaowagul W, White NJ. 2004. The changing pattern of bloodstream infections associated with the rise in HIV prevalence in northeastern Thailand. Trans R Soc Trop Med Hyg 98:678–686. doi: 10.1016/j.trstmh.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 457.Brown AE, Dance DA, Suputtamongkol Y, Chaowagul W, Kongchareon S, Webster HK, White NJ. 1991. Immune cell activation in melioidosis: increased serum levels of interferon-gamma and soluble interleukin-2 receptors without change in soluble CD8 protein. J Infect Dis 163:1145–1148. doi: 10.1093/infdis/163.5.1145. [DOI] [PubMed] [Google Scholar]
- 458.Lauw FN, Simpson AJ, Prins JM, Smith MD, Kurimoto M, van Deventer SJ, Speelman P, Chaowagul W, White NJ, van der Poll T. 1999. Elevated plasma concentrations of interferon (IFN)-gamma and the IFN-gamma-inducing cytokines interleukin (IL)-18, IL-12, and IL-15 in severe melioidosis. J Infect Dis 180:1878–1885. doi: 10.1086/315155. [DOI] [PubMed] [Google Scholar]
- 459.Kessler B, Rinchai D, Kewcharoenwong C, Nithichanon A, Biggart R, Hawrylowicz CM, Bancroft GJ, Lertmemongkolchai G. 2017. Interleukin 10 inhibits pro-inflammatory cytokine responses and killing of Burkholderia pseudomallei. Sci Rep 7:42791. doi: 10.1038/srep42791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Ulett GC, Ketheesan N, Clair TW, McElnea CL, Barnes JL, Hirst RG. 2002. Analogous cytokine responses to Burkholderia pseudomallei strains contrasting in virulence correlate with partial cross-protection in immunized mice. Infect Immun 70:3953–3958. doi: 10.1128/iai.70.7.3953-3958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Lauw FN, Simpson AJ, Prins JM, van Deventer SJ, Chaowagul W, White NJ, van der Poll T. 2000. The CXC chemokines gamma interferon (IFN-gamma)-inducible protein 10 and monokine induced by IFN-gamma are released during severe melioidosis. Infect Immun 68:3888–3893. doi: 10.1128/iai.68.7.3888-3893.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Santanirand P, Harley VS, Dance DAB, Drasar BS, Bancroft GJ. 1999. Obligatory role of Gamma interferon for host survival in a murine model of infection with Burkholderia pseudomallei. Infect Immun 67:3593–3600. doi: 10.1128/IAI.67.7.3593-3600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Puthucheary SD, Nathan SA. 2006. Comparison by electron microscopy of intracellular events and survival of Burkholderia pseudomallei in monocytes from normal subjects and patients with melioidosis. Singapore Med J 47:697. [PubMed] [Google Scholar]
- 464.Cheng AC, Wuthiekanun V, Limmathurotsakul D, Chierakul W, Peacock SJ. 2008. Intensity of exposure and incidence of melioidosis in Thai children. Trans R Soc Trop Med Hyg 102(Suppl 1):S37–S39. doi: 10.1016/S0035-9203(08)70010-5. [DOI] [PubMed] [Google Scholar]
- 465.Ngauy V, Lemeshev Y, Sadkowski L, Crawford G. 2005. Cutaneous melioidosis in a man who was taken as a prisoner of war by the Japanese during World War II. J Clin Microbiol 43:970–972. doi: 10.1128/JCM.43.2.970-972.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Koponen MA, Zlock D, Palmer DL, Merlin TL. 1991. Melioidosis. Forgotten, but not gone! Arch Intern Med 151:605–608. doi: 10.1001/archinte.151.3.605. [DOI] [PubMed] [Google Scholar]
- 467.Currie BJ, Fisher DA, Anstey NM, Jacups SP. 2000. Melioidosis: acute and chronic disease, relapse and re-activation. Trans R Soc Trop Med Hyg 94:301–304. doi: 10.1016/s0035-9203(00)90333-x. [DOI] [PubMed] [Google Scholar]
- 468.Chodimella U, Hoppes WL, Whalen S, Ognibene AJ, Rutecki GW. 1997. Septicemia and suppuration in a Vietnam veteran. Hosp Pract 32:219–221. doi: 10.1080/21548331.1997.11443493. [DOI] [PubMed] [Google Scholar]
- 469.Stewart JD, Smith S, Hanson J. 2017. Melioidosis in Far North Queensland is not correlated with severe weather events. Med J Aust 207:394. doi: 10.5694/mja16.01332. [DOI] [PubMed] [Google Scholar]
- 470.Burivong W, Wu X, Saenkote W, Stern EJ. 2012. Thoracic radiologic manifestations of melioidosis. Curr Probl Diagn Radiol 41:199–209. doi: 10.1067/j.cpradiol.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 471.Kozlowska J, Smith S, Roberts J, Pridgeon S, Hanson J. 2018. Prostatic abscess due to Burkholderia pseudomallei: facilitating diagnosis to optimize management. Am J Trop Med Hyg 98:227–230. doi: 10.4269/ajtmh.17-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Morse LP, Morse LP, Moller C-CB, Harvey E, Ward L. 2009. Prostatic abscess due to Burkholderia pseudomallei: 81 cases from a 19-year prospective melioidosis study. J Urol 182:542–547. doi: 10.1016/j.juro.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 473.Waiwarawooth J, Jutiworakul K, Joraka W. 2008. Epidemiology and clinical outcome of melioidosis at Chonburi Hospital, Thailand. J Infect Dis Antimicrob Agents 25:1–11. [Google Scholar]
- 474.Fertitta L, Monsel G, Torresi J, Caumes E. 2019. Cutaneous melioidosis: a review of the literature. Int J Dermatol 58:221–227. doi: 10.1111/ijd.14167. [DOI] [PubMed] [Google Scholar]
- 475.McLeod C, Morris PS, Bauert PA, Kilburn CJ, Ward LM, Baird RW, Currie BJ. 2015. Clinical presentation and medical management of melioidosis in children: a 24-year prospective study in the Northern Territory of Australia and review of the literature. Clin Infect Dis 60:21–26. doi: 10.1093/cid/ciu733. [DOI] [PubMed] [Google Scholar]
- 476.Dance DA, Davis TM, Wattanagoon Y, Chaowagul W, Saiphan P, Looareesuwan S, Wuthiekanun V, White NJ. 1989. Acute suppurative parotitis caused by Pseudomonas pseudomallei in children. J Infect Dis 159:654–660. doi: 10.1093/infdis/159.4.654. [DOI] [PubMed] [Google Scholar]
- 477.Lumbiganon P, Viengnondha S. 1995. Clinical manifestations of melioidosis in children. Pediatr Infect Dis J 14:136–139. doi: 10.1097/00006454-199502000-00010. [DOI] [PubMed] [Google Scholar]
- 478.Clark B, Merritt A, Inglis T, Manning L. 2018. Clinical features and outcome of patients with cutaneous melioidosis during a nosocomial outbreak in a temperate region of Australia. Intern Med J 48:461–465. doi: 10.1111/imj.13752. [DOI] [PubMed] [Google Scholar]
- 479.Sookpranee M, Lumbiganon P, Puapermpoonsiri S, Tattawasatra A, Nopwinyoovongs J. 1989. Contamination of Savlon solution with Pseudomonas pseudomallei at Srinagarind Hospital, p 211–213. In Punyagupta S, Sirisanthana T, Stapatayavong B (ed), Melioidosis. Bangkok Medical Publisher, Bangkok, Thailand. [Google Scholar]
- 480.Deuble M, Aquilina C, Norton R. 2013. Neurologic melioidosis. Am J Trop Med Hyg 89:535–539. doi: 10.4269/ajtmh.12-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Limmathurotsakul D, Chaowagul W, Wongsrikaew P, Narmwong A, Day NP, Peacock SJ. 2007. Variable presentation of neurological melioidosis in Northeast Thailand. Am J Trop Med Hyg 77:118–120. doi: 10.4269/ajtmh.2007.77.118. [DOI] [PubMed] [Google Scholar]
- 482.Punyagupta S. 1989. Review of 686 cases and presentation of a new clinical classification, p 217–229. In Punyagupta S, Sirisanthana T, Stapatayavong B (ed), Melioidosis. Bangkok Medical Publisher, Bangkok, Thailand. [Google Scholar]
- 483.Wongwandee M, Linasmita P. 2019. Central nervous system melioidosis: a systematic review of individual participant data of case reports and case series. PLoS Negl Trop Dis 13:e0007320. doi: 10.1371/journal.pntd.0007320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Chadwick DR, Chadwick DR, Ang B, Sitoh YY, Lee CC. 2002. Cerebral melioidosis in Singapore: a review of five cases. Trans R Soc Trop Med Hyg 96:72–76. doi: 10.1016/s0035-9203(02)90248-8. [DOI] [PubMed] [Google Scholar]
- 485.Shetty RP, Mathew M, Smith J, Morse LP, Mehta JA, Currie BJ. 2015. Management of melioidosis osteomyelitis and septic arthritis. Bone Joint J 97-b:277–282. doi: 10.1302/0301-620X.97B2.34799. [DOI] [PubMed] [Google Scholar]
- 486.Teparrukkul P, Nilsakul J, Dunachie S, Limmathurotsakul D. 2017. Clinical epidemiology of septic arthritis caused by Burkholderia pseudomallei and other bacterial pathogens in northeast Thailand. Am J Trop Med Hyg 97:1695–1701. doi: 10.4269/ajtmh.17-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Teparrakkul P, Tsai JJ, Chierakul W, Gerstenmaier JF, Wacharaprechasgu T, Piyaphanee W, Limmathurotsakul D, Chaowagul W, Day NP, Peacock SJ. 2008. Rheumatological manifestations in patients with melioidosis. Southeast Asian J Trop Med Public Health 39:649–655. [PubMed] [Google Scholar]
- 488.Muttarak M, Peh WC, Euathrongchit J, Lin SE, Tan AG, Lerttumnongtum P, Sivasomboon C. 2009. Spectrum of imaging findings in melioidosis. Br J Radiol 82:514–21. doi: 10.1259/bjr/15785231. [DOI] [PubMed] [Google Scholar]
- 489.Reechaipichitkul W. 2004. Clinical manifestation of pulmonary melioidosis in adults. Southeast Asian J Trop Med Public Health 35:664–669. [PubMed] [Google Scholar]
- 490.Lim KS, Chong VH. 2010. Radiological manifestations of melioidosis. Clin Radiol 65:66–72. doi: 10.1016/j.crad.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 491.Dhiensiri T, Puapairoj S, Susaengrat W. 1988. Pulmonary melioidosis: clinical-radiologic correlation in 183 cases in northeastern Thailand. Radiology 166:711–715. doi: 10.1148/radiology.166.3.3340766. [DOI] [PubMed] [Google Scholar]
- 492.Wibulpolprasert B, Dhiensiri T. 1999. Visceral organ abscesses in melioidosis: sonographic findings. J Clin Ultrasound 27:29–34. doi:. [DOI] [PubMed] [Google Scholar]
- 493.Vatcharapreechasakul T, Suputtamongkol Y, Dance DA, Chaowagul W, White NJ. 1992. Pseudomonas pseudomallei liver abscesses: a clinical, laboratory, and ultrasonographic study. Clin Infect Dis 14:412–417. doi: 10.1093/clinids/14.2.412. [DOI] [PubMed] [Google Scholar]
- 494.Khiangte HL, Vimala LR, Eapen A, Veeraraghavan B, Karuppusami R, Gibikote S. 2018. A retrospective case-control study to evaluate the diagnostic accuracy of honeycomb sign in melioid liver abscess. Am J Trop Med Hyg 99:852–857. doi: 10.4269/ajtmh.18-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Currie BJ, Fisher DA, Howard DM, Burrow JN. 2000. Neurological melioidosis. Acta Trop 74:145–151. doi: 10.1016/s0001-706x(99)00064-9. [DOI] [PubMed] [Google Scholar]
- 496.Hsu CC-T, Singh D, Kwan G, Deuble M, Aquilina C, Korah I, Norton R. 2016. Neuromelioidosis: craniospinal MRI findings in Burkholderia pseudomallei infection. J Neuroimaging 26:75–82. doi: 10.1111/jon.12282. [DOI] [PubMed] [Google Scholar]
- 497.Limmathurotsakul D, Kanoksil M, Wuthiekanun V, Kitphati R, deStavola B, Day NP, Peacock SJ. 2013. Activities of daily living associated with acquisition of melioidosis in northeast Thailand: a matched case-control study. PLoS Negl Trop Dis 7:e2072. doi: 10.1371/journal.pntd.0002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Carey IM, Critchley JA, DeWilde S, Harris T, Hosking FJ, Cook DG. 2018. Risk of infection in type 1 and type 2 diabetes compared with the general population: a matched cohort study. Diabetes Care 41:513. doi: 10.2337/dc17-2131. [DOI] [PubMed] [Google Scholar]
- 499.Shah BR, Hux JE. 2003. Quantifying the risk of infectious diseases for people with diabetes. Diabetes Care 26:510. doi: 10.2337/diacare.26.2.510. [DOI] [PubMed] [Google Scholar]
- 500.Geerlings SE, Hoepelman AI. 1999. Immune dysfunction in patients with diabetes mellitus (DM). FEMS Immunol Med Microbiol 26:259–6265. doi: 10.1111/j.1574-695X.1999.tb01397.x. [DOI] [PubMed] [Google Scholar]
- 501.Graves DT, Kayal RA. 2008. Diabetic complications and dysregulated innate immunity. Front Biosci 13:1227–1239. doi: 10.2741/2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Mayer-Scholl A, Averhoff P, Zychlinsky A. 2004. How do neutrophils and pathogens interact? Curr Opin Microbiol 7:62–66. doi: 10.1016/j.mib.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 503.Hodgson K, Morris J, Bridson T, Govan B, Rush C, Ketheesan N. 2015. Immunological mechanisms contributing to the double burden of diabetes and intracellular bacterial infections. Immunology 144:171–185. doi: 10.1111/imm.12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Hodgson KA, Govan BL, Walduck AK, Ketheesan N, Morris JL. 2013. Impaired early cytokine responses at the site of infection in a murine model of type 2 diabetes and melioidosis comorbidity. Infect Immun 81:470–477. doi: 10.1128/IAI.00930-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Dunachie SJ, Jenjaroen K, Reynolds CJ, Quigley KJ, Sergeant R, Sumonwiriya M, Chaichana P, Chumseng S, Ariyaprasert P, Lassaux P, Gourlay L, Promwong C, Teparrukkul P, Limmathurotsakul D, Day NPJ, Altmann DM, Boyton RJ. 2017. Infection with Burkholderia pseudomallei—immune correlates of survival in acute melioidosis. Sci Rep 7:12143. doi: 10.1038/s41598-017-12331-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Kulsantiwong P, Pudla M, Boondit J, Wikraiphat C, Dunachie SJ, Chantratita N, Utaisincharoen P. 2016. Burkholderia pseudomallei induces IL-23 production in primary human monocytes. Med Microbiol Immunol 205:255–260. doi: 10.1007/s00430-015-0440-z. [DOI] [PubMed] [Google Scholar]
- 507.Kewcharoenwong C, Rinchai D, Nithichanon A, Bancroft GJ, Ato M, Lertmemongkolchai G. 2016. Glibenclamide impairs responses of neutrophils against Burkholderia pseudomallei by reduction of intracellular glutathione. Sci Rep 6:34794. doi: 10.1038/srep34794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Kewcharoenwong C, Rinchai D, Utispan K, Suwannasaen D, Bancroft GJ, Ato M, Lertmemongkolchai G. 2013. Glibenclamide reduces pro-inflammatory cytokine production by neutrophils of diabetes patients in response to bacterial infection. Sci Rep 3:3363. doi: 10.1038/srep03363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Koh GCKW, Maude RR, Schreiber MF, Limmathurotsakul D, Wiersinga WJ, Wuthiekanun V, Lee SJ, Mahavanakul W, Chaowagul W, Chierakul W, White NJ, van der Poll T, Day NPJ, Dougan G, Peacock SJ. 2011. Glyburide is anti-inflammatory and associated with reduced mortality in melioidosis. Clin Infect Dis 52:717–725. doi: 10.1093/cid/ciq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Limmathurotsakul D, Chaowagul W, Chierakul W, Stepniewska K, Maharjan B, Wuthiekanun V, White NJ, Day NP, Peacock SJ. 2006. Risk factors for recurrent melioidosis in northeast Thailand. Clin Infect Dis 43:979–986. doi: 10.1086/507632. [DOI] [PubMed] [Google Scholar]
- 511.Chierakul W, Wuthiekanun V, Chaowagul W, Amornchai P, Cheng AC, White NJ, Day NP, Peacock SJ. 2005. Short report: disease severity and outcome of melioidosis in HIV coinfected individuals. Am J Trop Med Hyg 73:1165–1166. doi: 10.4269/ajtmh.2005.73.1165. [DOI] [PubMed] [Google Scholar]
- 512.Fong SM, Wong KJ, Fukushima M, Yeo TW. 2015. Thalassemia major is a major risk factor for pediatric melioidosis in Kota Kinabalu, Sabah, Malaysia. Clin Infect Dis 60:1802–1807. doi: 10.1093/cid/civ189. [DOI] [PubMed] [Google Scholar]
- 513.Abraham C, Cho JH. 2006. Functional consequences of NOD2 (CARD15) mutations. Inflamm Bowel Dis 12:641–650. doi: 10.1097/01.MIB.0000225332.83861.5f. [DOI] [PubMed] [Google Scholar]
- 514.Pan H, Dai Y, Tang S, Wang J. 2012. Polymorphisms of NOD2 and the risk of tuberculosis: a validation study in the Chinese population. Int J Immunogenet 39:233–240. doi: 10.1111/j.1744-313X.2011.01079.x. [DOI] [PubMed] [Google Scholar]
- 515.Myers ND, Chantratita N, Berrington WR, Chierakul W, Limmathurotsakul D, Wuthiekanun V, Robertson JD, Liggitt HD, Peacock SJ, Skerrett SJ, West TE. 2014. The role of NOD2 in murine and human melioidosis. J Immunol 192:300–307. doi: 10.4049/jimmunol.1301436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.West TE, Chierakul W, Chantratita N, Limmathurotsakul D, Wuthiekanun V, Emond MJ, Hawn TR, Peacock SJ, Skerrett SJ. 2012. Toll-like receptor 4 region genetic variants are associated with susceptibility to melioidosis. Genes Immun 13:38–46. doi: 10.1038/gene.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Limmathurotsakul D, Chaowagul W, Chantratita N, Wuthiekanun V, Biaklang M, Tumapa S, White NJ, Day NPJ, Peacock SJ. 2008. A simple scoring system to differentiate between relapse and re-infection in patients with recurrent melioidosis. PLoS Negl Trop Dis 2:e327. doi: 10.1371/journal.pntd.0000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Chaowagul W, Suputtamongkol Y, Dance DA, Rajchanuvong A, Pattara J, White NJ. 1993. Relapse in melioidosis: incidence and risk factors. J Infect Dis 168:1181–1185. [PubMed] [Google Scholar]
- 519.Sarovich DS, Ward L, Price EP, Mayo M, Pitman MC, Baird RW, Currie BJ. 2014. Recurrent melioidosis in the Darwin Prospective Melioidosis Study: improving therapies mean that relapse cases are now rare. J Clin Microbiol 52:650–653. doi: 10.1128/JCM.02239-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Chetchotisakd P, Chierakul W, Chaowagul W, Anunnatsiri S, Phimda K, Mootsikapun P, Chaisuksant S, Pilaikul J, Thinkhamrop B, Phiphitaporn S, Susaengrat W, Toondee C, Wongrattanacheewin S, Wuthiekanun V, Chantratita N, Thaipadungpanit J, Day NP, Limmathurotsakul D, Peacock SJ. 2014. Trimethoprim-sulfamethoxazole versus trimethoprim-sulfamethoxazole plus doxycycline as oral eradicative treatment for melioidosis (MERTH): a multicentre, double-blind, non-inferiority, randomised controlled trial. Lancet 383:807–814. doi: 10.1016/S0140-6736(13)61951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Newland RC. 1969. Chronic melioidosis: a case in Sydney. Pathology 1:149–152. doi: 10.3109/00313026909061049. [DOI] [PubMed] [Google Scholar]
- 522.Gee JE, Gulvik CA, Elrod MG, Batra D, Rowe LA, Sheth M, Hoffmaster AR. 2017. Phylogeography of Burkholderia pseudomallei isolates, Western Hemisphere. Emerg Infect Dis 23:1133–1138. doi: 10.3201/eid2307.161978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Johnson AB, Ali N. 1990. Reactivation of latent melioidosis. Postgrad Med J 66:732–733. doi: 10.1136/pgmj.66.779.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Shaaban H, Hallit R, Slim J, Sree A, Sensakovic JW. 2014. Reactivation of latent melioidosis presenting with acute pyelonephritis and bacteremia. Avicenna J Med 4:20–21. doi: 10.4103/2231-0770.127418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Leelarasamee A, Bovornkitti S. 1989. Melioidosis: review and update. Rev Infect Dis 11:413–425. doi: 10.1093/clinids/11.3.413. [DOI] [PubMed] [Google Scholar]
- 526.Dance D. 2014. Treatment and prophylaxis of melioidosis. Int J Antimicrob Agents 43:310–318. doi: 10.1016/j.ijantimicag.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.White NJ, Dance DA, Chaowagul W, Wattanagoon Y, Wuthiekanun V, Pitakwatchara N. 1989. Halving of mortality of severe melioidosis by ceftazidime. Lancet ii:697–701. doi: 10.1016/s0140-6736(89)90768-x. [DOI] [PubMed] [Google Scholar]
- 528.Sookpranee M, Boonma P, Susaengrat W, Bhuripanyo K, Punyagupta S. 1992. Multicenter prospective randomized trial comparing ceftazidime plus co-trimoxazole with chloramphenicol plus doxycycline and co-trimoxazole for treatment of severe melioidosis. Antimicrob Agents Chemother 36:158–162. doi: 10.1128/aac.36.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Chierakul W, Anunnatsiri S, Short JM, Maharjan B, Mootsikapun P, Simpson AJ, Limmathurotsakul D, Cheng AC, Stepniewska K, Newton PN, Chaowagul W, White NJ, Peacock SJ, Day NP, Chetchotisakd P. 2005. Two randomized controlled trials of ceftazidime alone versus ceftazidime in combination with trimethoprim-sulfamethoxazole for the treatment of severe melioidosis. Clin Infect Dis 41:1105–1113. doi: 10.1086/444456. [DOI] [PubMed] [Google Scholar]
- 530.Chierakul W, Anunnatsiri S, Chaowagul W, Peacock SJ, Chetchotisakd P, Day NP. 2007. Addition of trimethoprim-sulfamethoxazole to ceftazidime during parenteral treatment of melioidosis is not associated with a long-term outcome benefit. Clin Infect Dis 45:521–523. doi: 10.1086/520010. [DOI] [PubMed] [Google Scholar]
- 531.Suputtamongkol Y, Rajchanuwong A, Chaowagul W, Dance DA, Smith MD, Wuthiekanun V, Walsh AL, Pukrittayakamee S, White NJ. 1994. Ceftazidime vs. amoxicillin/clavulanate in the treatment of severe melioidosis. Clin Infect Dis 19:846–853. doi: 10.1093/clinids/19.5.846. [DOI] [PubMed] [Google Scholar]
- 532.Cheng AC, Chierakul W, Chaowagul W, Chetchotisakd P, Limmathurotsakul D, Dance DA, Peacock SJ, Currie BJ. 2008. Consensus guidelines for dosing of amoxicillin-clavulanate in melioidosis. Am J Trop Med Hyg 78:208–209. doi: 10.4269/ajtmh.2008.78.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Chierakul W, Wangboonskul J, Singtoroj T, Pongtavornpinyo W, Short JM, Maharjan B, Wuthiekanun V, Dance DAB, Teparrukkul P, Lindegardh N, Peacock SJ, Day NP, Chaowagul W, White NJ. 2006. Pharmacokinetic and pharmacodynamic assessment of co-amoxiclav in the treatment of melioidosis. J Antimicrob Chemother 58:1215–1220. doi: 10.1093/jac/dkl389. [DOI] [PubMed] [Google Scholar]
- 534.Rajchanuvong A, Chaowagul W, Suputtamongkol Y, Smith MD, Dance DA, White NJ. 1995. A prospective comparison of co-amoxiclav and the combination of chloramphenicol, doxycycline, and co-trimoxazole for the oral maintenance treatment of melioidosis. Trans R Soc Trop Med Hyg 89:546–549. doi: 10.1016/0035-9203(95)90104-3. [DOI] [PubMed] [Google Scholar]
- 535.Chaowagul W, Simpson AJ, Suputtamongkol Y, White NJ. 1999. Empirical cephalosporin treatment of melioidosis. Clin Infect Dis 28:1328. doi: 10.1086/517787. [DOI] [PubMed] [Google Scholar]
- 536.Ashdown LR. 1988. In vitro activities of the newer beta-lactam and quinolone antimicrobial agents against Pseudomonas pseudomallei. Antimicrob Agents Chemother 32:1435–1436. doi: 10.1128/aac.32.9.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Smith MD, Wuthiekanun V, Walsh AL, White NJ. 1996. In-vitro activity of carbapenem antibiotics against beta-lactam susceptible and resistant strains of Burkholderia pseudomallei. J Antimicrob Chemother 37:611–615. doi: 10.1093/jac/37.3.611. [DOI] [PubMed] [Google Scholar]
- 538.Walsh AL, Smith MD, Wuthiekanun V, White NJ. 1995. Postantibiotic effects and Burkholderia (Pseudomonas) pseudomallei: evaluation of current treatment. Antimicrob Agents Chemother 39:2356–2358. doi: 10.1128/aac.39.10.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Lipsitz R, Garges S, Aurigemma R, Baccam P, Blaney DD, Cheng AC, Currie BJ, Dance D, Gee JE, Larsen J, Limmathurotsakul D, Morrow MG, Norton R, O’Mara E, Peacock SJ, Pesik N, Rogers LP, Schweizer HP, Steinmetz I, Tan G, Tan P, Wiersinga WJ, Wuthiekanun V, Smith TL. 2012. Workshop on treatment of and postexposure prophylaxis for Burkholderia pseudomallei and B. mallei infection, 2010. Emerg Infect Dis 18:e2. doi: 10.3201/eid1812.120638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Currie BJ. 2014. Melioidosis: the 2014 revised RDH guideline. Northern Territ Dis Control Bull 21:4–8. [Google Scholar]
- 541.Cheng AC, McBryde ES, Wuthiekanun V, Chierakul W, Amornchai P, Day NP, White NJ, Peacock SJ. 2009. Dosing regimens of cotrimoxazole (trimethoprim-sulfamethoxazole) for melioidosis. Antimicrob Agents Chemother 53:4193–4199. doi: 10.1128/AAC.01301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Chaowagul W, Simpson AJ, Suputtamongkol Y, Smith MD, Angus BJ, White NJ. 1999. A comparison of chloramphenicol, trimethoprim-sulfamethoxazole, and doxycycline with doxycycline alone as maintenance therapy for melioidosis. Clin Infect Dis 29:375–380. doi: 10.1086/520218. [DOI] [PubMed] [Google Scholar]
- 543.Chaowagul W, Chierakul W, Simpson AJ, Short JM, Stepniewska K, Maharjan B, Rajchanuvong A, Busarawong D, Limmathurotsakul D, Cheng AC, Wuthiekanun V, Newton PN, White NJ, Day NP, Peacock SJ. 2005. Open-label randomized trial of oral trimethoprim-sulfamethoxazole, doxycycline, and chloramphenicol compared with trimethoprim-sulfamethoxazole and doxycycline for maintenance therapy of melioidosis. Antimicrob Agents Chemother 49:4020–4025. doi: 10.1128/AAC.49.10.4020-4025.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Pitman MC, Luck T, Marshall CS, Anstey NM, Ward L, Currie BJ. 2015. Intravenous therapy duration and outcomes in melioidosis: a new treatment paradigm. PLoS Negl Trop Dis 9:e0003586. doi: 10.1371/journal.pntd.0003586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Gibney KB, Cheng AC, Currie BJ. 2008. Cutaneous melioidosis in the tropical top end of Australia: a prospective study and review of the literature. Clin Infect Dis 47:603–609. doi: 10.1086/590931. [DOI] [PubMed] [Google Scholar]
- 546.Cheng AC, Dasari P, Currie BJ. 2004. Granulocyte colony-stimulating factor and an in vitro whole blood model of melioidosis. Eur J Clin Microbiol Infect Dis 23:205–207. doi: 10.1007/s10096-003-1088-y. [DOI] [PubMed] [Google Scholar]
- 547.Cheng AC, Limmathurotsakul D, Chierakul W, Getchalarat N, Wuthiekanun V, Stephens DP, Day NP, White NJ, Chaowagul W, Currie BJ, Peacock SJ. 2007. A randomized controlled trial of granulocyte colony-stimulating factor for the treatment of severe sepsis due to melioidosis in Thailand. Clin Infect Dis 45:308–314. doi: 10.1086/519261. [DOI] [PubMed] [Google Scholar]
- 548.Cheng AC, Stephens DP, Anstey NM, Currie BJ. 2004. Adjunctive granulocyte colony-stimulating factor for treatment of septic shock due to melioidosis. Clin Infect Dis 38:32–37. doi: 10.1086/380456. [DOI] [PubMed] [Google Scholar]
- 549.Currie BJ, Fisher DA, Howard DM, Burrow JN, Lo D, Selva-Nayagam S, Anstey NM, Huffam SE, Snelling PL, Marks PJ, Stephens DP, Lum GD, Jacups SP, Krause VL. 2000. Endemic melioidosis in tropical northern Australia: a 10-year prospective study and review of the literature. Clin Infect Dis 31:981–986. doi: 10.1086/318116. [DOI] [PubMed] [Google Scholar]
- 550.Simpson AJ, Suputtamongkol Y, Smith MD, Angus BJ, Rajanuwong A, Wuthiekanun V, Howe PA, Walsh AL, Chaowagul W, White NJ. 1999. Comparison of imipenem and ceftazidime as therapy for severe melioidosis. Clin Infect Dis 29:381–387. doi: 10.1086/520219. [DOI] [PubMed] [Google Scholar]
- 551.Toda A, Ohki H, Yamanaka T, Murano K, Okuda S, Kawabata K, Hatano K, Matsuda K, Misumi K, Itoh K, Satoh K, Inoue S. 2008. Synthesis and SAR of novel parenteral anti-pseudomonal cephalosporins: discovery of FR264205. Bioorg Med Chem Lett 18:4849–4852. doi: 10.1016/j.bmcl.2008.07.085. [DOI] [PubMed] [Google Scholar]
- 552.Hong MC, Hsu DI, Bounthavong M. 2013. Ceftolozane/tazobactam: a novel antipseudomonal cephalosporin and beta-lactamase-inhibitor combination. Infect Drug Resist 6:215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Slack A, Parsonson F, Cronin K, Engler C, Norton R. 2018. Activity of ceftolozane-tazobactam against Burkholderia pseudomallei. Am J Trop Med Hyg 99:281–282. doi: 10.4269/ajtmh.17-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Chaowagul W, Suputtamongkul Y, Smith MD, White NJ. 1997. Oral fluoroquinolones for maintenance treatment of melioidosis. Trans R Soc Trop Med Hyg 91:599–601. doi: 10.1016/s0035-9203(97)90044-4. [DOI] [PubMed] [Google Scholar]
- 555.Barnes KB, Hamblin KA, Richards MI, Laws TR, Vente A, Atkins HS, Harding SV. 2017. Demonstrating the protective efficacy of the novel fluoroquinolone finafloxacin against an inhalational exposure to Burkholderia pseudomallei. Antimicrob Agents Chemother 61:e00082-17. doi: 10.1128/AAC.00082-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Tomaras AP, McPherson CJ, Kuhn M, Carifa A, Mullins L, George D, Desbonnet C, Eidem TM, Montgomery JI, Brown MF, Reilly U, Miller AA, O’Donnell JP. 2014. LpxC inhibitors as new antibacterial agents and tools for studying regulation of lipid A biosynthesis in Gram-negative pathogens. mBio 5:e01551-14. doi: 10.1128/mBio.01551-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Sengyee S, Saiprom N, Paksanont S, Limmathurotsakul D, Wuthiekanun V, Chantratita N. 2017. Susceptibility of clinical isolates of Burkholderia pseudomallei to a lipid A biosynthesis inhibitor. Am J Trop Med Hyg 97:62–67. doi: 10.4269/ajtmh.16-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Sivalingam SP, Sim SH, Jasper LCW, Wang D, Liu Y, Ooi EE. 2008. Pre- and post-exposure prophylaxis of experimental Burkholderia pseudomallei infection with doxycycline, amoxicillin/clavulanic acid and co-trimoxazole. J Antimicrob Chemother 61:674–678. doi: 10.1093/jac/dkm527. [DOI] [PubMed] [Google Scholar]
- 559.Barnes KB, Steward J, Thwaite JE, Lever MS, Davies CH, Armstrong SJ, Laws TR, Roughley N, Harding SV, Atkins TP, Simpson AJH, Atkins HS. 2013. Trimethoprim/sulfamethoxazole (co-trimoxazole) prophylaxis is effective against acute murine inhalational melioidosis and glanders. Int J Antimicrob Agents 41:552–557. doi: 10.1016/j.ijantimicag.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 560.Majoni SW, Hughes JT, Heron B, Currie BJ. 2018. Trimethoprim+sulfamethoxazole reduces rates of melioidosis in high-risk hemodialysis patients. Kidney Int Rep 3:160–167. doi: 10.1016/j.ekir.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561.Chau KWT, Smith S, Kang K, Dheda S, Hanson J. 2018. Antibiotic prophylaxis for melioidosis in patients receiving hemodialysis in the tropics? One size does not fit all. Am J Trop Med Hyg 99:597–600. doi: 10.4269/ajtmh.18-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Inglis TJ, Sousa AQ. 2009. The public health implications of melioidosis. Braz J Infect Dis 13:59–66. doi: 10.1590/s1413-86702009000100013. [DOI] [PubMed] [Google Scholar]
- 563.Boyd R, McGuinness S, Draper A, Neilson M, Krause V. 2016. melioidosis awareness campaign …….don’t get melioidosis…. Northern Territ Dis Control Bull 23:1–4. [Google Scholar]
- 564.McRobb E, Kaestli M, Mayo M, Price EP, Sarovich DS, Godoy D, Spratt BG, Currie BJ. 2013. Melioidosis from contaminated bore water and successful UV sterilization. Am J Trop Med Hyg 89:367–368. doi: 10.4269/ajtmh.13-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Rose LJ, Rice EW, Jensen B, Murga R, Peterson A, Donlan RM, Arduino MJ. 2005. Chlorine inactivation of bacterial bioterrorism agents. Appl Environ Microbiol 71:566–568. doi: 10.1128/AEM.71.1.566-568.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.Cheung RCF, Ng TB, Wong JH, Chan WY. 2015. Chitosan: an update on potential biomedical and pharmaceutical applications. Mar Drugs 13:5156–5186. doi: 10.3390/md13085156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Kamjumphol W, Chareonsudjai P, Chareonsudjai S. 2018. Antibacterial activity of chitosan against Burkholderia pseudomallei. Microbiologyopen 7:e00534. doi: 10.1002/mbo3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Na-ngam N, Angkititakul S, Noimay P, Thamlikitkul V. 2004. The effect of quicklime (calcium oxide) as an inhibitor of Burkholderia pseudomallei. Trans R Soc Trop Med Hyg 98:337–341. doi: 10.1016/j.trstmh.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 569.Sommanustweechai A, Kasantikul T, Somsa W, Wongratanacheewin S, Sermswan RW, Kongmakee P, Thomas W, Kamolnorranath S, Siriaroonrat B, Bush M, Banlunara W. 2013. Environmental management procedures following fatal melioidosis in a captive chimpanzee (Pan troglodytes). J Zoo Wildl Med 44:475–479. doi: 10.1638/2012-0025R5.1. [DOI] [PubMed] [Google Scholar]
- 570.Chansrichavala P, Wongsuwan N, Suddee S, Malasit M, Hongsuwan M, Wannapinij P, Kitphati R, Day NPJ, Michie S, Peacock SJ, Limmathurotsakul D. 2015. Public awareness of melioidosis in Thailand and potential use of video clips as educational tools. PLoS One 10:e0121311. doi: 10.1371/journal.pone.0121311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Suntornsut P, Wongsuwan N, Malasit M, Kitphati R, Michie S, Peacock SJ, Limmathurotsakul D. 2016. Barriers and recommended interventions to prevent melioidosis in northeast Thailand: a focus group study using the behaviour change wheel. PLoS Negl Trop Dis 10:e0004823. doi: 10.1371/journal.pntd.0004823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Green RN, Tuffnell PG. 1968. Laboratory acquired melioidosis. Am J Med 44:599–605. doi: 10.1016/0002-9343(68)90060-0. [DOI] [PubMed] [Google Scholar]
- 573.Schlech WF, Turchik JB, Westlake RE, Klein GC, Band JD, Weaver RE. 1981. Laboratory-acquired infection with Pseudomonas pseudomallei (melioidosis). N Engl J Med 305:1133–1135. doi: 10.1056/NEJM198111053051907. [DOI] [PubMed] [Google Scholar]
- 574.Vlieghe E, Kruy L, Smet B, Kham C, Veng CH, Phe T, Koole O, Thai S, Lynen L, Jacobs J. 2011. Melioidosis, Phnom Penh, Cambodia. Emerg Infect Dis 17:1289–1292. doi: 10.3201/eid1707.101069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Limmathurotsakul D, Wuthiekanun V, Wongsuvan G, Pangmee S, Amornchai P, Teparrakkul P, Teerawattanasook N, Day NPJ, Peacock SJ. 2011. Repeat blood culture positive for B. pseudomallei indicates an increased risk of death from melioidosis. Am J Trop Med Hyg 84:858–861. doi: 10.4269/ajtmh.2011.10-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Thatrimontrichai A, Maneenil G. 2012. Neonatal melioidosis: systematic review of the literature. Pediatr Infect Dis J 31:1195–1197. doi: 10.1097/INF.0b013e318265ac62. [DOI] [PubMed] [Google Scholar]
- 577.Rammaert B, Beaute J, Borand L, Hem S, Buchy P, Goyet S, Overtoom R, Angebault C, Te V, Try PL, Mayaud C, Vong S, Guillard B. 2011. Pulmonary melioidosis in Cambodia: a prospective study. BMC Infect Dis 11:126. doi: 10.1186/1471-2334-11-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.Om C, Daily F, Vlieghe E, McLaughlin JC, McLaws M-L. 2016. “If it’s a broad spectrum, it can shoot better”: inappropriate antibiotic prescribing in Cambodia. Antimicrob Resist Infect Control 5:58. doi: 10.1186/s13756-016-0159-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579.Puthucheary SD, Parasakthi N, Lee MK. 1992. Septicaemic melioidosis: a review of 50 cases from Malaysia. Trans R Soc Trop Med Hyg 86:683–685. doi: 10.1016/0035-9203(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 580.Wright SW, Emond MJ, Lovelace-Macon L, Ducken D, Kashima J, Hantrakun V, Chierakul W, Teparrukkul P, Chantratita N, Limmathurotsakul D, West TE. 2019. Exonic sequencing identifies TLR1 genetic variation associated with mortality in Thais with melioidosis. Emerg Microbes Infect 8:282–290. doi: 10.1080/22221751.2019.1575172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 581.West TE, Chantratita N, Chierakul W, Limmathurotsakul D, Wuthiekanun V, Myers ND, Emond MJ, Wurfel MM, Hawn TR, Peacock SJ, Skerrett SJ. 2013. Impaired TLR5 functionality is associated with survival in melioidosis. J Immunol 190:3373–3379. doi: 10.4049/jimmunol.1202974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582.Chaichana P, Chantratita N, Brod F, Koosakulnirand S, Jenjaroen K, Chumseng S, Sumonwiriya M, Burtnick MN, Brett PJ, Teparrukkul P, Limmathurotsakul D, Day NPJ, Dunachie SJ, West TE. 2017. A nonsense mutation in TLR5 is associated with survival and reduced IL-10 and TNF-α levels in human melioidosis. PLoS Negl Trop Dis 11:e0005587. doi: 10.1371/journal.pntd.0005587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583.West TE, Myers ND, Chantratita N, Chierakul W, Limmathurotsakul D, Wuthiekanun V, Miao EA, Hajjar AM, Peacock SJ, Liggitt HD, Skerrett SJ. 2014. NLRC4 and TLR5 each contribute to host defense in respiratory melioidosis. PLoS Negl Trop Dis 8:e3178. doi: 10.1371/journal.pntd.0003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Chantratita N, Tandhavanant S, Myers ND, Chierakul W, Robertson JD, Mahavanakul W, Singhasivanon P, Emond MJ, Peacock SJ, West TE. 2014. Screen of whole blood responses to flagellin identifies TLR5 variation associated with outcome in melioidosis. Genes Immun 15:63–71. doi: 10.1038/gene.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.Limmathurotsakul D, Funnell SG, Torres AG, Morici LA, Brett PJ, Dunachie S, Atkins T, Altmann DM, Bancroft G, Peacock SJ, Steering Group on Melioidosis Vaccine Development . 2015. Consensus on the development of vaccines against naturally acquired melioidosis. Emerg Infect Dis doi: 10.3201/eid2106.141480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Hatcher CL, Muruato LA, Torres AG. 2015. Recent advances in Burkholderia mallei and B. pseudomallei research. Curr Trop Med Rep 2:62–69. doi: 10.1007/s40475-015-0042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 587.Hogan RJ, Lafontaine ER. 2019. Antibodies are major drivers of protection against lethal aerosol infection with highly pathogenic Burkholderia spp. mSphere 4:e00674-18. doi: 10.1128/mSphere.00674-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 588.Peacock SJ, Limmathurotsakul D, Lubell Y, Koh GC, White LJ, Day NP, Titball RW. 2012. Melioidosis vaccines: a systematic review and appraisal of the potential to exploit biodefense vaccines for public health purposes. PLoS Negl Trop Dis 6:e1488. doi: 10.1371/journal.pntd.0001488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589.Titball RW, Burtnick MN, Bancroft GJ, Brett P. 2017. Burkholderia pseudomallei and Burkholderia mallei vaccines: are we close to clinical trials? Vaccine 35:5981–5989. doi: 10.1016/j.vaccine.2017.03.022. [DOI] [PubMed] [Google Scholar]
- 590.Cuccui J, Easton A, Chu KK, Bancroft GJ, Oyston PCF, Titball RW, Wren BW. 2007. Development of signature-tagged mutagenesis in Burkholderia pseudomallei to identify genes important in survival and pathogenesis. Infect Immun 75:1186–1195. doi: 10.1128/IAI.01240-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Breitbach K, Kohler J, Steinmetz I. 2008. Induction of protective immunity against Burkholderia pseudomallei using attenuated mutants with defects in the intracellular life cycle. Trans R Soc Trop Med Hyg 102(Suppl 1):S89–S94. doi: 10.1016/S0035-9203(08)70022-1. [DOI] [PubMed] [Google Scholar]
- 592.Silva EB, Goodyear A, Sutherland MD, Podnecky NL, Gonzalez-Juarrero M, Schweizer HP, Dow SW. 2013. Correlates of immune protection following cutaneous immunization with an attenuated Burkholderia pseudomallei vaccine. Infect Immun 81:4626–4634. doi: 10.1128/IAI.00915-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593.Khakhum N, Bharaj P, Myers JN, Tapia D, Kilgore PB, Ross BN, Walker DH, Endsley JJ, Torres AG. 2019. Burkholderia pseudomallei ΔtonB Δhcp1 live attenuated vaccine strain elicits full protective immunity against aerosolized melioidosis infection. mSphere 4:e00570-18. doi: 10.1128/mSphere.00570-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Scott AE, Laws TR, D’Elia RV, Margaret GMS, Nandi T, Williamson ED, Tan P, Prior JL, Atkins TP. 2013. Protection against experimental melioidosis following immunization with live Burkholderia thailandensis expressing a manno-heptose capsule. Clin Vaccine Immunol 20:1041–1047. doi: 10.1128/CVI.00113-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595.Patel N, Conejero L, De Reynal M, Easton A, Bancroft GJ, Titball RW. 2011. Development of vaccines against burkholderia pseudomallei. Front Microbiol 2:198. doi: 10.3389/fmicb.2011.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596.Barnes JL, Ketheesan N. 2007. Development of protective immunity in a murine model of melioidosis is influenced by the source of Burkholderia pseudomallei antigens. Immunol Cell Biol 85:551–557. doi: 10.1038/sj.icb.7100084. [DOI] [PubMed] [Google Scholar]
- 597.Puangpetch A, Anderson R, Huang YY, Saengsot R, Sermswan RW, Wongratanacheewin S. 2014. Comparison of the protective effects of killed Burkholderia pseudomallei and CpG oligodeoxynucleotide against live challenge. Vaccine 32:5983–5988. doi: 10.1016/j.vaccine.2014.08.035. [DOI] [PubMed] [Google Scholar]
- 598.Sarkar-Tyson M, Smither SJ, Harding SV, Atkins TP, Titball RW. 2009. Protective efficacy of heat-inactivated B. thailandensis, B. mallei or B. pseudomallei against experimental melioidosis and glanders. Vaccine 27:4447–4451. doi: 10.1016/j.vaccine.2009.05.040. [DOI] [PubMed] [Google Scholar]
- 599.Burtnick MN, Shaffer TL, Ross BN, Muruato LA, Sbrana E, DeShazer D, Torres AG, Brett PJ. 2018. Development of subunit vaccines that provide high-level protection and sterilizing immunity against acute inhalational melioidosis. Infect Immun 86:e00724-17. doi: 10.1128/IAI.00724-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600.Nelson M, Prior JL, Lever MS, Jones HE, Atkins TP, Titball RW. 2004. Evaluation of lipopolysaccharide and capsular polysaccharide as subunit vaccines against experimental melioidosis. J Med Microbiol 53:1177–1182. doi: 10.1099/jmm.0.45766-0. [DOI] [PubMed] [Google Scholar]
- 601.Whitlock GC, Deeraksa A, Qazi O, Judy BM, Taylor K, Propst KL, Duffy AJ, Johnson K, Kitto GB, Brown KA, Dow SW, Torres AG, Estes DM. 2010. Protective response to subunit vaccination against intranasal Burkholderia mallei and B. pseudomallei challenge. Procedia Vaccinol 2:73–77. doi: 10.1016/j.provac.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.Nieves W, Asakrah S, Qazi O, Brown KA, Kurtz J, AuCoin DP, McLachlan JB, Roy CJ, Morici LA. 2011. A naturally derived outer-membrane vesicle vaccine protects against lethal pulmonary Burkholderia pseudomallei infection. Vaccine 29:8381–8389. doi: 10.1016/j.vaccine.2011.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 603.Nieves W, Petersen H, Judy BM, Blumentritt CA, Russell-Lodrigue K, Roy CJ, Torres AG, Morici LA. 2014. A Burkholderia pseudomallei outer membrane vesicle vaccine provides protection against lethal sepsis. Clin Vaccine Immunol 21:747–754. doi: 10.1128/CVI.00119-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 604.Petersen H, Nieves W, Russell-Lodrigue K, Roy CJ, Morici LA. 2014. Evaluation of a Burkholderia pseudomallei outer membrane vesicle vaccine in nonhuman primates. Procedia Vaccinol 8:38–42. doi: 10.1016/j.provac.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 605.Scott AE, Ngugi SA, Laws TR, Corser D, Lonsdale CL, D’Elia RV, Titball RW, Williamson ED, Atkins TP, Prior JL. 2014. Protection against experimental melioidosis following immunisation with a lipopolysaccharide-protein conjugate. J Immunol Res 2014:392170. doi: 10.1155/2014/392170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606.Scott AE, Burtnick MN, Stokes MGM, Whelan AO, Williamson ED, Atkins TP, Prior JL, Brett PJ. 2014. Burkholderia pseudomallei capsular polysaccharide conjugates provide protection against acute melioidosis. Infect Immun 82:3206–3213. doi: 10.1128/IAI.01847-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Muruato LA, Tapia D, Hatcher CL, Kalita M, Brett PJ, Gregory AE, Samuel JE, Titball RW, Torres AG. 2017. Use of reverse vaccinology in the design and construction of nanoglycoconjugate vaccines against Burkholderia pseudomallei. Clin Vaccine Immunol 24:e00206-17. doi: 10.1128/CVI.00206-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608.Gregory AE, Judy BM, Qazi O, Blumentritt CA, Brown KA, Shaw AM, Torres AG, Titball RW. 2015. A gold nanoparticle-linked glycoconjugate vaccine against Burkholderia mallei. Nanomedicine 11:447–456. doi: 10.1016/j.nano.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609.Dance DAB. 2005. Melioidosis and glanders as possible biological weapons, p 99–145. In Fong IW, Alibek K (ed), Bioterrorism and infectious agents: a new dilemma for the 21st century. Springer US, Boston, MA. [Google Scholar]
- 610.Dvorak GD, Spickler AR. 2008. Glanders. J Am Vet Med Assoc 233:570–577. doi: 10.2460/javma.233.4.570. [DOI] [PubMed] [Google Scholar]
- 611.Rotz LD, Khan AS, Lillibridge SR, Ostroff SM, Hughes JM. 2002. Public health assessment of potential biological terrorism agents. Emerg Infect Dis 8:225–230. doi: 10.3201/eid0802.010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612.Wiersinga WJ, van der Poll T, White NJ, Day NP, Peacock SJ. 2006. Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat Rev Microbiol 4:272. doi: 10.1038/nrmicro1385. [DOI] [PubMed] [Google Scholar]
- 613.Shams AM, Rose LJ, Hodges L, Arduino MJ. 2007. Survival of Burkholderia pseudomallei on environmental surfaces. Appl Environ Microbiol 73:8001–8004. doi: 10.1128/AEM.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 614.Wuthiekanun V, Smith MD, White NJ. 1995. Survival of Burkholderia pseudomallei in the absence of nutrients. Trans R Soc Trop Med Hyg 89:491. doi: 10.1016/0035-9203(95)90080-2. [DOI] [PubMed] [Google Scholar]
- 615.Estes DM, Dow SW, Schweizer HP, Torres AG. 2010. Present and future therapeutic strategies for melioidosis and glanders. Expert Rev Anti-infect Ther 8:325–338. doi: 10.1586/eri.10.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 616.Bhengsri S, Lertiendumrong J, Baggett HC, Thamthitiwat S, Chierakul W, Tisayaticom K, Tanwisaid K, Chantra S, Kaewkungwal J. 2013. Economic burden of bacteremic melioidosis in eastern and northeastern, [sic] Thailand. Am J Trop Med Hyg 89:369–373. doi: 10.4269/ajtmh.13-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 617.Chatterjee S, Riewpaiboon A, Piyauthakit P, Riewpaiboon W, Boupaijit K, Panpuwong N, Archavanuntagul V. 2011. Cost of diabetes and its complications in Thailand: a complete picture of economic burden. Health Soc Care Community 19:289–298. doi: 10.1111/j.1365-2524.2010.00981.x. [DOI] [PubMed] [Google Scholar]
- 618.Liljas B. 1998. How to calculate indirect costs in economic evaluations. Pharmacoeconomics 13:1–7. doi: 10.2165/00019053-199813010-00001. [DOI] [PubMed] [Google Scholar]
- 619.Huy R, Wichmann O, Beatty M, Ngan C, Duong S, Margolis HS, Vong S. 2009. Cost of dengue and other febrile illnesses to households in rural Cambodia: a prospective community-based case-control study. BMC Public Health 9:155. doi: 10.1186/1471-2458-9-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620.Hantrakun V, Chierakul W, Chetchotisakd P, Anunnatsiri S, Currie BJ, Peacock SJ, Day NP, Cheah PY, Limmathurotsakul D, Lubell Y. 2015. Cost-effectiveness analysis of parenteral antimicrobials for acute melioidosis in Thailand. Trans R Soc Trop Med Hyg 109:803. doi: 10.1093/trstmh/trv093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 621.Cheng AC, Fisher DA, Anstey NM, Stephens DP, Jacups SP, Currie BJ. 2004. Outcomes of patients with melioidosis treated with meropenem. Antimicrob Agents Chemother 48:1763–1765. doi: 10.1128/aac.48.5.1763-1765.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 622.Malczewski AB, Oman KM, Norton RE, Ketheesan N. 2005. Clinical presentation of melioidosis in Queensland, Australia. Trans R Soc Trop Med Hyg 99:856–860. doi: 10.1016/j.trstmh.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 623.Chaiwarith R, Patiwetwitoon P, Supparatpinyo K, Sirisanthana T. 2005. Melioidosis at Maharaj Nakorn Chiang Mai Hospital, Thailand. J Infect Dis Antimicrob Agents 22:45–51. [Google Scholar]
- 624.Jabbar Z, Currie BJ. 2013. Melioidosis and the kidney. Nephrology (Carlton) 18:169–175. doi: 10.1111/nep.12024. [DOI] [PubMed] [Google Scholar]
- 625.Inglis TJ. 2010. The treatment of melioidosis. Pharmaceuticals (Basel) 3:1296–1303. doi: 10.3390/ph3051296. [DOI] [PMC free article] [PubMed] [Google Scholar]