Candida auris is a newly emerged fungal pathogen of humans. This species was first reported in 2009 when it was identified in an ear infection of a patient in Japan. However, despite intense interest in this organism as an often multidrug-resistant fungus, there is little knowledge about its cellular biology. During infection of human patients, fungi are able to change cell shape from ellipsoidal yeast cells to elongated filaments to adapt to various conditions within the host organism. There are different types of filaments, which are triggered by reactions to different cues. Candida auris fails to form filaments when exposed to triggers that stimulate yeast filament morphogenesis in other fungi. Here, we show that it does form filaments when its DNA is damaged. These conditions might arise when Candida auris cells interact with host immune cells or during growth in certain host tissues (kidney or bladder) or during treatment with antifungal drugs.
KEYWORDS: Candida auris, filamentous growth, S phase checkpoint, Rad51, Rad9, Mrc1
ABSTRACT
The morphogenetic switching between yeast cells and filaments (true hyphae and pseudohyphae) is a key cellular feature required for full virulence in many polymorphic fungal pathogens, such as Candida albicans. In the recently emerged yeast pathogen Candida auris, occasional elongation of cells has been reported. However, environmental conditions and genetic triggers for filament formation have remained elusive. Here, we report that induction of DNA damage and perturbation of replication forks by treatment with genotoxins, such as hydroxyurea, methyl methanesulfonate, and the clinically relevant fungistatic 5-fluorocytosine, cause filamentation in C. auris. The filaments formed were characteristic of pseudohyphae and not parallel-sided true hyphae. Pseudohyphal growth is apparently signaled through the S phase checkpoint and, interestingly, is Tup1 independent in C. auris. Intriguingly, the morphogenetic switching capability is strain specific in C. auris, highlighting the heterogenous nature of the species as a whole.
IMPORTANCE Candida auris is a newly emerged fungal pathogen of humans. This species was first reported in 2009 when it was identified in an ear infection of a patient in Japan. However, despite intense interest in this organism as an often multidrug-resistant fungus, there is little knowledge about its cellular biology. During infection of human patients, fungi are able to change cell shape from ellipsoidal yeast cells to elongated filaments to adapt to various conditions within the host organism. There are different types of filaments, which are triggered by reactions to different cues. Candida auris fails to form filaments when exposed to triggers that stimulate yeast filament morphogenesis in other fungi. Here, we show that it does form filaments when its DNA is damaged. These conditions might arise when Candida auris cells interact with host immune cells or during growth in certain host tissues (kidney or bladder) or during treatment with antifungal drugs.
INTRODUCTION
The emergence of novel multidrug-resistant pathogens poses a recurrent global threat to health care settings. This is the case for the fungus Candida auris discovered as a new human pathogen only 10 years ago (1), albeit a retrospective review of strain collections dated the first case back to 1996 (2). Since its first identification, C. auris has been found across all continents, causing clonal outbreaks in hospital settings (3). It shows mortality rates in systemic disease close to 50% (4), is one of the most drug-resistant yeast pathogens (5), and has been described as a skin colonizer able to undergo nosocomial spread (6), thus becoming a major concern for medical mycology.
Due to the recent emergence of this pathogen, we are largely ignorant about its general biological traits. This lack of fundamental understanding about the origin (7, 8) and the life cycle of C. auris impedes our capacity to explain its sudden emergence, rapid global spread, and unique phenotypic characteristics. An example is the lack of information about its ability to undergo morphogenetic switches as described for other fungi. In fungi, a morphogenetic switch enables cells to change from growing as unicellular yeasts to pseudohyphae or true hyphae and can be triggered by a multitude of environmental factors, such as nutrient limitation, temperature, and pH changes (reviewed in references 9, to ,12). Filamentous growth allows the exploration of new environments and is considered a virulence trait in pathogenic fungi (reviewed in references 13 and 14). However, most cues causing filamentation in the best-studied and only distantly related pathogen Candida albicans do not induce filamentous growth in C. auris (15).
In contrast to growth of true hyphae, pseudohyphal growth has been associated with a delay in cell cycle progression and the subsequent extension of the apical growth period (16). Indeed, it has been demonstrated that drugs causing genotoxic stress, such as hydroxyurea (HU) or methyl methanesulfonate (MMS), trigger S phase arrest via a cell cycle checkpoint (reviewed in reference 17); this results in pseudohyphal growth in C. albicans and Saccharomyces cerevisiae (18, 19). The S phase checkpoint is a surveillance system, which responds to DNA damage or DNA replication fork arrest, and involves the sensor kinases Mec1 and Tel1, the mediator proteins Rad9 and Mrc1, and an effector kinase, Rad53. Rad9 acts as the main mediator for the DNA damage response, whereas Mrc1 functions as a DNA replication arrest responder (17). Once activated, the S phase checkpoint modulates multiple biological processes, including the repression of late-firing replication origins, cell cycle progression, the production of deoxynucleotide triphosphates (dNTPs), the transcription of DNA damage response genes, and inhibition of homologous recombination. Accordingly, activation of Rad53 triggers pseudohyphal growth since in S. cerevisiae or C. albicans mutants deficient in this kinase showed a drastic reduction of filamentation under genotoxic stress conditions (19, 20).
Here, we demonstrate that many, but not all, clinical isolates of C. auris are capable of pseudohyphal growth when treated with genotoxins such as HU, MMS, or the clinically relevant fungistatic 5-fluorocytosine (5-FC). Deletion mutants of genes involved in the S phase checkpoint, RAD9 and MRC1, or homologous recombination, RAD51 and RAD57, allowed us to probe whether a functional S phase checkpoint is required for filamentous growth in C. auris. Our work provides the first insight into how genome stability maintenance supports cell growth and proliferation and what triggers morphogenetic switching in the newly emerged fungal pathogen of humans, C. auris.
RESULTS
C. auris produces pseudohyphae under genotoxic stress.
Because the ability to switch between unicellular and filamentous forms plays a role in pathogenesis in some fungi (reviewed in references 11 and 12), we were interested in whether C. auris has the capability to form filaments as well. Several conditions which induce hyphal growth in C. albicans, such as incubation at 37°C and in Lee’s medium at pH 3.5 or pH 6.5, as well as medium containing serum, isoamyl alcohol, or Bleocin, were tested, but none of these triggered filamentous growth in the South Asian (clade I) C. auris strain UACa11 (data not shown) (15). Likewise, growth at 25°C did not produce filaments as described previously for a C. auris clinical isolate (21). However, using strain UACa11, we observed filamentous growth in the presence of sublethal concentrations of genotoxic drugs affecting DNA replication progression or inducing DNA damage (HU, MMS, and 5-FC) (Fig. 1) (see also Fig. S1 in the supplemental material; all supplemental figures and tables can be found at https://doi.org/10.6084/m9.figshare.11378550). HU inhibits the activity of ribonucleotide reductase (22) and thus induces a depletion of the dNTP pools (23). MMS creates bulky adducts by alkylating DNA that interferes with fork progression (24). 5-FC is converted into fluorouracil in the cell and perturbs RNA and DNA biosynthesis (25). The filaments observed in C. auris show characteristics attributed to pseudohyphae (9, 11). In contrast to true hyphae, filaments in C. auris are wider than the diameter of a yeast cell and do not present parallel sides, septa between neighboring cells show visible indentations, and actin patches are not accumulating at the growing tip (Fig. 1). Moreover, nuclear divisions seem to occur at mother-daughter junctions, but this phenotype is often complicated by nuclear division defects due to the genotoxin treatments (Fig. 1).
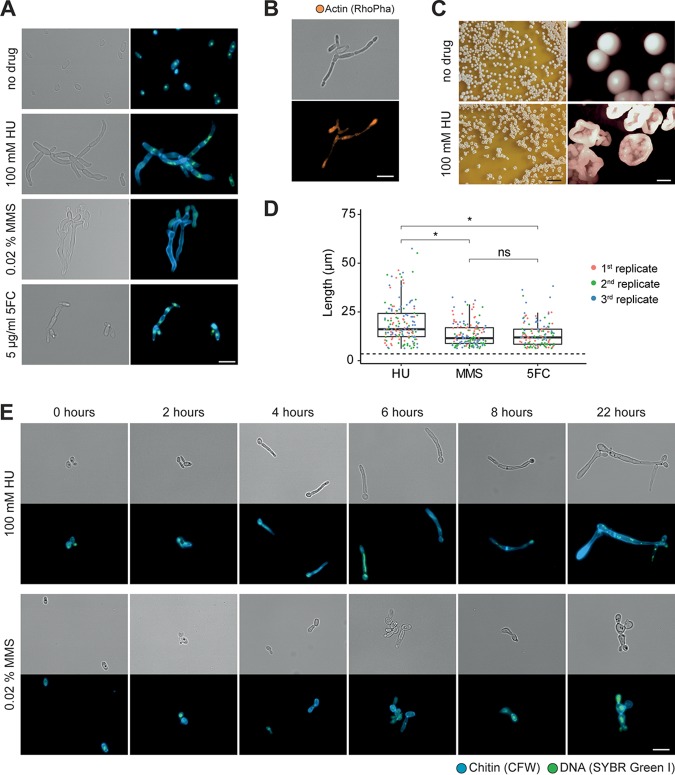
FIG 1.
Filamentation of Candida auris (UACa11) in the presence of genotoxic drugs. (A) Microscopy images of C. auris filamentation after growing strain UACa11 cells on YPD plates with or without the addition of the indicated drug after 3 days at 30°C. A bright-field image is shown on the left, and a merged fluorescent image (chitin stained by calcofluor white [blue] and DNA stained by SYBR green I [green]) is shown on the right. (B) Cells grown on YPD plates containing 100 mM HU for 3 days at 30°C stained for actin using rhodamine-phalloidin (RhoPha) (bottom image). The bright-field image is on top. (C) Details of colonies from strain UACA11 grown on YPD plates in the absence and presence of 100 mM hydroxyurea (HU) for 6 days at 30°C. Scale bars, 10 mm (left) and 1 mm (right). (D) Lengths of filaments of wild-type (UACa11) cells grown in YPD medium containing 100 mM HU, 0.02% MMS, or 5 μg/ml 5-FC for 18 to 20 h at 30°C (n = 50 for each replicate). Only cells longer than 6 μm were considered filaments. The dotted line indicates the average length of 300 yeast cells (wild-type UACa11 grown in YPD medium for 18 to 20 h at 30°C). *, P < 0.05 (by Wilcoxon rank sum test); ns, not significant. (E) Microscopy images of representative filaments of C. auris UACa11 formed in liquid culture. Cells were stained as described for panel A. After arrest in G1, cultures were grown for 165 min in YPD medium before the indicated drugs were added (time point 0 h). Bright-field images are shown in the top rows, and fluorescent images are shown at bottom. Scale bar, 10 μm.
Within 6 to 8 h of growth in yeast extract-peptone-glucose (YPD) medium containing 100 mM HU, daughter cells started to show hyperpolarized growth. This results in almost 100% filamentation after overnight culture (Fig. 1E; see Fig. S1A at the URL mentioned above), similar to C. albicans (18, 20). In this early filamentation phase, generally one or two nuclei (SYBR green-stained DNA signals) were observed within the same cell, and occasionally very large, high-intensity or stretched out DNA signals could be seen (Fig. 1E). After 22 h constrictions at septa became evident throughout the filaments, and new buds emerged from seemingly random locations along the filament; these cultures contained almost no separated (yeast) cells, indicating a defect in cell separation as expected for pseudohyphal growth (Fig. 1E; Fig. S1A). Cells were often multi- or anucleate, suggestive of karyokinesis defects. A similar phenotype was generated by the addition of 0.02% MMS (Fig. 1E) although the formation of filaments occurred more slowly, and the resulting pseudohyphae tended to be shorter and more irregular than those with HU treatment (Fig. 1D). Moreover, after 22 h in MMS, round and elongated yeast cells were still present in the cultures. Importantly, the number and length of filaments decreased over time when cells were grown on plates containing genotoxins (Fig. S2). This means either that long filaments might not be viable and that surviving yeast cells start growing after adapting their checkpoint or that filaments bud off yeast cells, which then form a new population. It should also be kept in mind that drugs could be degraded over time and that after several days the environment on solid medium containing genotoxic drugs could be less challenging. Filaments can also be observed without stress, as previously reported (26), but these are rare and could be explained by sporadic DNA damage (see Fig. S1A at the URL mentioned above). Strikingly, sporadically giant round cells were also observed in the presence of genotoxic drugs (Fig. S1A and S2). In accordance with the cellular phenotype, the colony morphology after 5 to 6 days on solid medium is rougher when HU is present, lacking the typical smooth-colony appearance (Fig. 1C). This was not true in the presence of MMS or 5-FC, either because these filaments were shorter or less abundant or because they were more short-lived than the ones treated with HU (Fig. S2).
C. auris lacks key genes associated with hyphal growth.
Filamentation is a complex mechanism, usually triggered by environmental conditions, involving hundreds of genes in S. cerevisiae and C. albicans (27–29). Among them is a group of well-studied key regulators known as hypha-specific genes (HSGs). Previously, Muñoz and coworkers reported that some HSGs essential for true hyphal growth, such as ECE1 or HWP1, are absent in the C. auris genome (30). We further explored the presence of key genes in filamentation in the C. auris genome (see Table S1 at https://doi.org/10.6084/m9.figshare.11378550). Besides ECE1 and HWP1, FLO11, EED1, and HWP2 were also absent in C. auris. In S. cerevisiae Flo11 is a key factor for pseudohyphal growth in response to nutrient limitation (31, 32). Intriguingly, in C. albicans, which also lacks Flo11, Hwp2 seems to cover this function (33). UME6, which codes for a Zn(II)2Cys6 transcription factor involved in filamentation regulation, is present in Candida species capable of forming true hyphae (C. albicans, C. tropicalis, and C. dubliniensis) and species unable to form true hyphae (C. parapsilosis, C. orthopsilosis, and C. lusitaniae). Importantly, in the latter group of species only the C-terminal Zn(II)2Cys6 domain is well conserved; this is also true for C. auris (see Fig. S3 at the URL mentioned above).
Tup1 is a transcriptional repressor for filamentous growth in C. albicans acting on several HSGs, and tup1Δ strains grow constitutively as true hyphae (34, 35). In a C. auris tup1Δ strain, no filaments were observed when cells were grown without stress (Fig. 2). However, strings of yeast cells were frequently observed, indicating that cells cannot separate properly. Under genotoxic stress, the tup1Δ mutant formed pseudohyphae similar to those of the parental strain. Altogether, this suggests that filamentation in C. auris is likely regulated differently than that in C. albicans.
FIG 2.
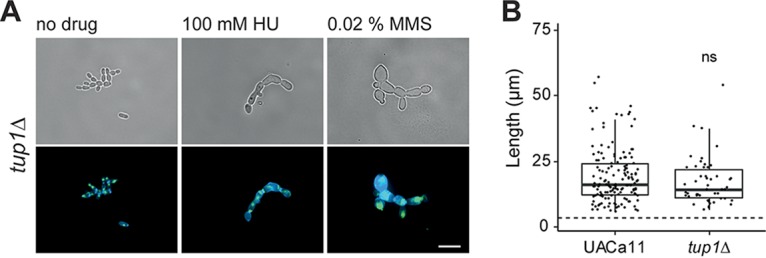
Microscopic analysis of filament formation in Candida auris tup1 mutant (UACa11 background). (A) Representative microscopic images of C. auris filaments after growing tup1Δ cells in YPD broth with or without the addition of the indicated drug for 18 to 20 h at 30°C (top, bright-field images; bottom, merged fluorescent images showing chitin stained by calcofluor white [blue] and DNA stained by SYBR green I [green]). Scale bar, 10 μm. (B) Length of filaments observed after growth of wild-type (UACa11) (n = 150) and tup1Δ (n = 50) cells in YPD broth containing 100 mM HU for 18 to 20 h at 30°C. Only cells longer than 6 μm were considered filaments. The dotted line indicates the average length of 300 yeast cells (wild-type UACa11 grown in YPD for 18 to 20 h at 30°C). ns, not significant (Wilcoxon rank sum test).
C. auris has a functional S phase checkpoint.
Genotoxic drugs induce replication fork perturbations and/or DNA damage, leading to cell cycle arrest by triggering the S phase checkpoint. Genes encoding S phase checkpoint factors, MEC1 (GenBank accession number XP_028890424), RAD9 (XP_028889586), MRC1 (XP_028891779), and RAD53 (XP_028891118), and homologous recombination proteins, RAD51 (GenBank accession number XP_028892133) and RAD57 (KND99929), were found in the C. auris genome (see Fig. S4 and S5 and Table S1 at the URL mentioned above). Null mutants of RAD9, MRC1, RAD51, and RAD57 were obtained by deleting the open reading frames (ORFs) in the C. auris UACa11 strain (Fig. S6). Unfortunately, three attempts to generate mec1Δ or rad53Δ null mutants with >300 transformants each were unsuccessful, possibly because these genes are essential in C. auris. The rad51Δ and rad57Δ strains showed very similar phenotypes (see Fig. S7 at the URL mentioned above); for detailed analysis we focused on the rad51Δ strain.
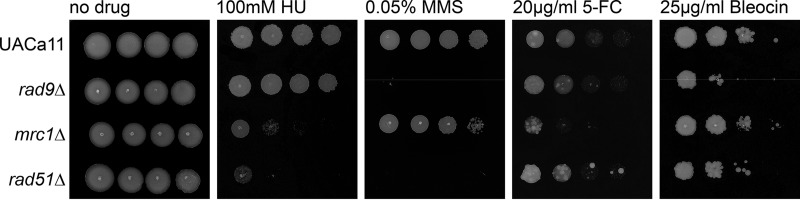
The mutant phenotypes were characterized using sublethal concentrations of various genotoxic drugs (Fig. 3). None of the mutants showed a conspicuous growth defect in the absence of genotoxins (Fig. 3). The rad9Δ strain showed sensitivity only to MMS and Bleocin, indicating a role in responding to double-stranded DNA breaks (Fig. 3). In contrast, the mrc1Δ strain displayed sensitivity to drugs inducing replication-associated damage (HU, MMS, and 5-FC) but grew like the parental control strain on Bleocin (Fig. 3). Growth of rad51Δ was severely affected in the presence of HU and MMS but only moderately so in the presence of Bleocin, and growth was almost indistinguishable from that of the wild type on 5-FC (Fig. 3). These mutant C. auris phenotypes are similar to the ones observed in the corresponding S. cerevisiae and C. albicans mutants (20, 36–39). However, Mrc1 seems to play a lesser role in C. auris since in the other two yeasts mrc1Δ strains were more sensitive to MMS, and in C. albicans this mutant showed a growth defect even when genotoxic challenges were absent (20, 36–39). Taken together, these results suggest that the S phase checkpoint is conserved in C. auris, albeit with a somewhat reduced importance of Mrc1.
FIG 3.
Candida auris rad9, mrc1, and rad51 mutants show sensitivity to genotoxic drugs. Growth analysis by spot assays of the wild type (UACa11) and the indicated deletion mutants in the presence and absence of genotoxic drugs. Ten-fold serial dilutions of C. auris cells were grown on YPD plates containing the indicated drug for 3 days at 30°C.
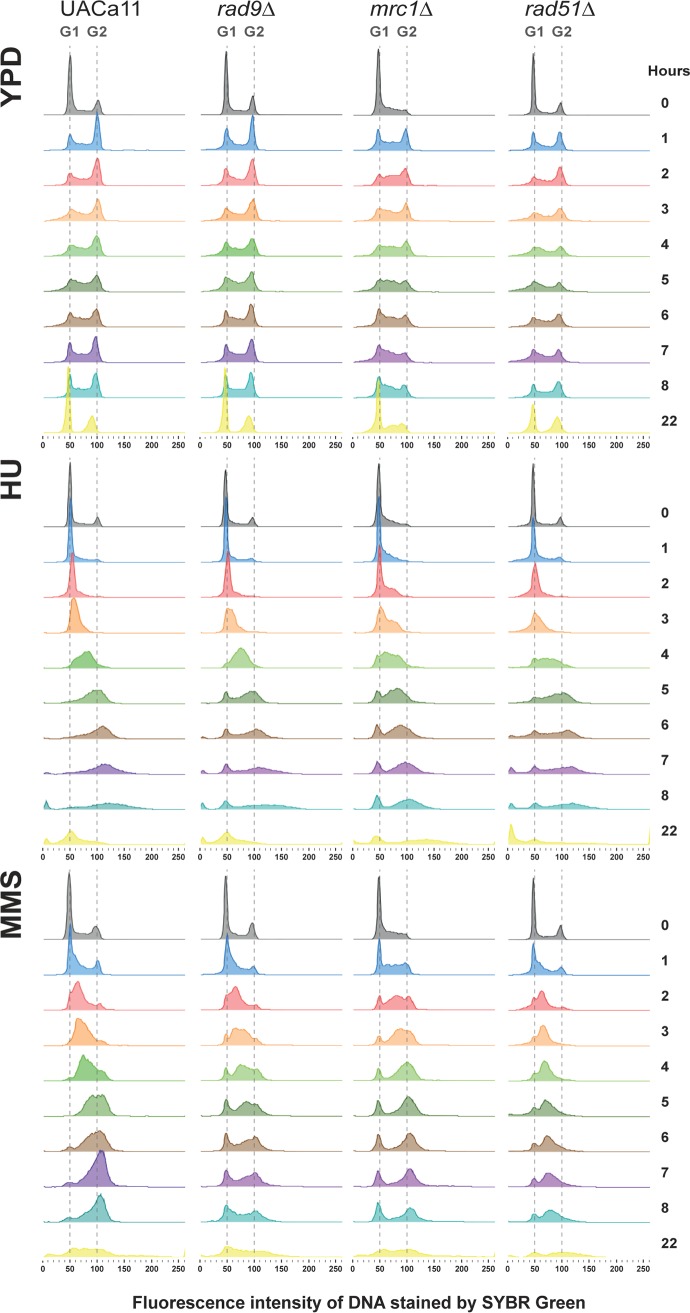
The S phase checkpoint slows down the cell cycle in response to DNA damage or DNA replication inhibition. The role of C. auris Rad51, Rad9, and Mrc1 for cell cycle arrest in response to genotoxic stress was further studied. Wild-type C. auris (UACa11) cultures can be arrested in G1 by nitrogen starvation; almost 100% of cells grown without a nitrogen source were arrested in G1 after 7 h and remained in G1 during prolonged (24 h) starvation (see Fig. S8A at https://doi.org/10.6084/m9.figshare.11378550). Upon return to favorable growth conditions (YPD), wild-type cultures had a lag phase of ∼2.5 to 3 h (Fig. S8B). Therefore, we grew wild-type and mutant yeast cultures for 165 min in YPD medium after G1 arrest before we started the experiments (time point 0 h) by adding 100 mM HU or 0.02% MMS as a genotoxic challenge; no drugs were added for the control. Cells were harvested at different time points, and the replication status of the cells was determined by flow cytometry (Fig. 4). Under unperturbed conditions, wild-type (UACa11), rad9Δ, and rad51Δ cells progressed from G1 to G2 within the first hour after reentering the cell cycle (Fig. 4). This is in line with these mutants showing wild-type growth in solid medium in the absence of genotoxins (Fig. 3). In mrc1Δ cells a larger fraction of the cell population was in S phase, which suggests that S phase might last longer in this mutant, which is similar to results in S. cerevisiae and C. albicans (20, 40, 41). Intriguingly, in C. auris this does not result in a notable growth defect on solid medium (Fig. 3).
FIG 4.
Cell cycle progression of Candida auris mutants under genotoxic stress. Histograms show cell cycle profiles obtained by flow cytometry of the wild type (UACa11) and the indicated mutant derivatives. Cells, previously arrested in G1 by nitrogen starvation, were transferred to fresh YPD broth and grown for 165 min at 30°C to restart the cycle before addition of 100 mM HU, 0.02% MMS, or no drug (time point 0 h). Cells were harvested at the indicated time points and DNA stained using SYBR green I. The amount of DNA is expressed as fluorescence intensity. Approximate positions of G1 and G2 peaks are indicated with dotted lines.
In the parental wild-type strain, the S phase checkpoint is functional, and in the presence of HU and MMS cell cycle progression is slower than that under unperturbed conditions, with a large fraction of cells in S phase between 4 and 6 h (Fig. 4). Eventually, the checkpoint adapts, and cells move to G2 (Fig. 4). This is also largely true for rad9Δ and rad51Δ cells in the presence of HU, indicating a minor role in the HU response (Fig. 4). The mrc1 null mutant behaves differently and shows a slower progression than the parental strain since after 6 h the majority of cells are still in S phase (Fig. 4). In the rad51Δ mutant most of the cells were arrested in S phase under MMS treatment, whereas cell cycle progression in the rad9Δ and mrc1Δ mutants was similar to that of the parental strain under these conditions (Fig. 4). However, in all the mutants, but not the wild type, a small population of G1 cells was always present during genotoxin treatments (Fig. 4). This could mean that some cells never enter the cell cycle after the G1 arrest or that they escape the S phase checkpoint early on and progress through the cell cycle to G1 quickly; these possibilities are not mutually exclusive. Indications that the latter possibility actually occurs are (i) that these G1 populations grow over time in rad9Δ and mrc1Δ mutants under both genotoxic conditions and (ii) that it also occurs in the wild type when cells are challenged with MMS. This indicates that rad9Δ and mrc1Δ cells are indeed defective in the S phase checkpoint. At 22 h the cycle seems to be partially restored (Fig. 4) though the presence of filaments and increased cell death after genotoxic treatment confound the interpretation of this result. Overall, these results support the idea that a functional S phase checkpoint exists in C. auris.
The S phase checkpoint is involved in pseudohyphal growth.
HU and MMS treatments induce pseudohyphal growth in C. albicans and S. cerevisiae which is dependent on activation of the S phase checkpoint because a reduction of filamentation was described for strains carrying mutations in the gene coding for the S phase checkpoint kinase Rad53 (19, 20). The ability of mutant C. auris to produce filaments was tested in liquid medium (Fig. 5A; see Fig. S1 at the URL mentioned above) and on solid medium (Fig. 5B; Fig. S2). As described above, wild-type cells of strain UACa11 form pseudohyphal filaments upon treatment with various genotoxins (Fig. 1).
FIG 5.
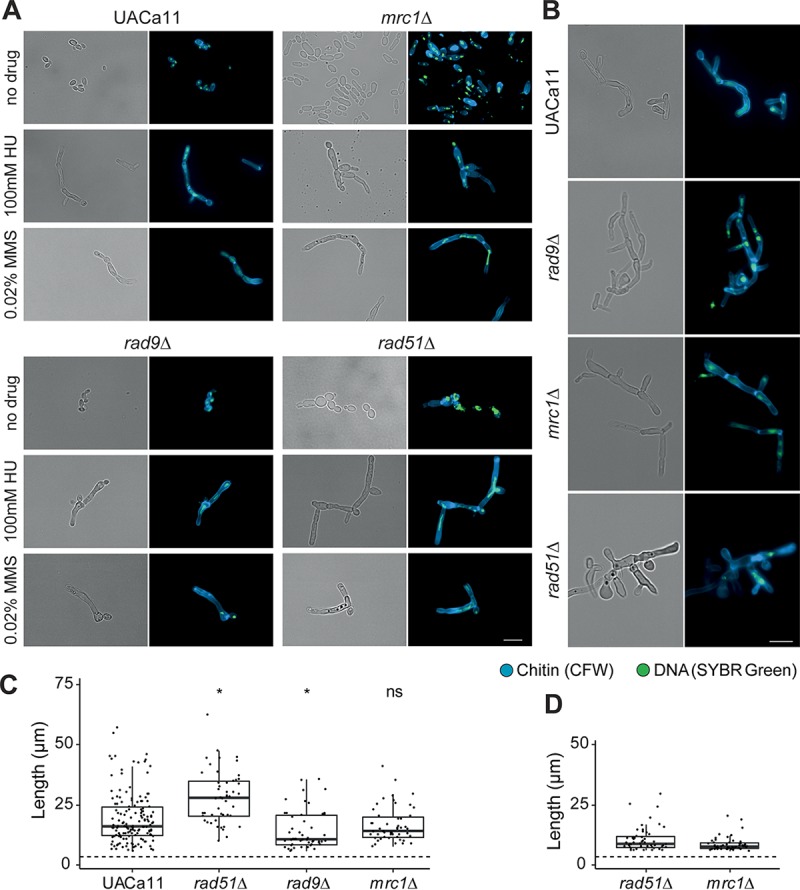
Microscopic analysis of filament formation in Candida auris rad9, mrc1, and rad51 mutants (UACa11 background). (A) Representative microscopy images of C. auris filaments after growth of wild-type (UACa11), rad9Δ, mrc1Δ, and rad51Δ cells in YPD broth with or without the addition of the indicated drug for 18 to 20 h at 30°C. (B) Representative microscopic images of C. auris filaments after growth of wild-type (UACa11), rad9Δ, mrc1Δ, and rad51Δ cells on YPD plates containing 100 mM HU after 3 days at 30°C. In panels A and B, bright-field images are shown in the left columns, and merged fluorescent images (chitin stained by calcofluor white [CFW] and DNA stained by SYBR green I) are shown in the right columns. Scale bar, 10 μm. (C) Length of filaments observed after growth of wild-type (UACa11) (n = 150), rad51Δ (n = 50), rad9Δ (n = 50), and mrc1Δ (n = 50) cells in YPD broth containing 100 mM HU for 18 to 20 h at 30°C. Only cells longer than 6 μm were considered filaments. The dotted line indicates average length of 300 yeast cells (wild-type UACa11 grown in YPD for 18 to 20 h at 30°C). *, P < 0.05, for results relative to those of the wild type (UACa11) (Wilcoxon rank sum test); ns, not significant. (D) Length of filaments observed after growth of rad51Δ (n = 50) and mrc1Δ (n = 50) cells in YPD medium for 18 to 20 h at 30°C without genotoxic stress. Only cells longer than 6 μm were considered filaments. The dotted line indicates average length of 300 yeast cells (wild-type UACa11 grown in YPD for 18 to 20 h at 30°C).
C. auris rad9Δ cells grew as yeasts without genotoxic stress and as pseudohyphae in the presence of HU, similar to growth of the wild type (Fig. 5) and of C. albicans rad9Δ (20). Filaments formed in C. auris rad9Δ are significantly shorter than those of the wild type. Intriguingly, rad9Δ cells produced filaments growing in liquid medium containing MMS (Fig. 5; see Fig. S1 at the URL mentioned above), as observed in the wild-type parent, but failed to form filaments on solid medium with MMS even after 2 days (Fig. S2). That partially contrasts with the situation in C. albicans, where rad9Δ cells were somewhat compromised for filamentous growth in response to MMS compared to those of the wild type but would still form short pseudohyphae (20). After 3 days on solid medium containing 0.02% MMS, a noticeable fraction of rad9Δ cells became very large and round (giant cells); most of these giant cells are gone after 7 days of culture (presumably because giant cells have a low viability) (Fig. S2). These giant cells were also observed in the wild type and other mutants, especially under genotoxic stress, although at much lower frequency (Fig. S1 and S2), indicating that the lack of Rad9 enhances the production of this cell type.
C. auris mrc1Δ cells were elongated but not fully filamented after growth of 18 to 20 h in liquid medium without genotoxins, in contrast to the round or oval shape of wild-type cells (Fig. 5). This differs somewhat from growth of C. albicans where Mrc1-defective cells displayed pseudohyphal growth without stress (20). However, on solid medium without genotoxic stress, pseudohyphae were observed in mrc1Δ cells (see Fig. S2 at the URL mentioned above). In the presence of HU or MMS, the phenotype of the mrc1Δ mutant was almost identical to that of the wild-type parent in both liquid and solid media.
As observed in rad52Δ and rad51Δ cells in C. albicans (37, 42), a sizeable fraction of cells in the C. auris rad51Δ mutant showed constitutive pseudohyphal growth under unstressed conditions (Fig. 5; see Fig. S1 and S2 at the URL mentioned above). The remainder of the cells showed a yeast phenotype or were somewhat elongated and often of aberrant shape (e.g., giant cells or very wide and elongated cells). When treated with HU or MMS, this mutant formed pseudohyphae to a similar extent as the wild type, but the filaments tended to be longer (Fig. 5C).
Overall, this suggests a role of the S phase checkpoint in C. auris filamentous growth although the involvement of this pathway slightly differs from what has been observed in C. albicans.
C. auris filamentation is strain dependent.
Strikingly, filamentation in C. auris was strain dependent. We tested the filamentation of 22 different C. auris clinical isolates (see Table S2 at https://doi.org/10.6084/m9.figshare.11378550) covering the four major clades (43) by treatment with HU (Fig. 6; Fig. S9). Among them, different grades of filamentation were observed: some strains showed longer filaments (UACa11 or UACa24) than others (UACa6 or UACa23); some were straight and thin (UACa11 or UACa24) or wider and shorter, growing as chains of (elongated) cells (bubbles) (UACa7 or UACa22); some formed aberrant cell shapes more frequently (UACa4 or UACa25). The clade III strain UACa10 seems to be unable to produce filaments when treated with genotoxins; it tended to produce bigger cells than in those produced under unperturbed conditions, however. The grade of filamentation was not obviously correlated with a particular clade since isolates from the same clade showed different phenotypes.
FIG 6.
Different degrees of filamentation in Candida auris clinical isolates. Representative microscopy images of selected clinical isolates (see Table S2 at https://doi.org/10.6084/m9.figshare.11378550) after growth in YPD broth containing 100 mM HU for 18 to 20 h at 30°C. Bright-field images are shown in the left columns, and merged fluorescent images (chitin stained by calcofluor white [CFW] and DNA stained by SYBR green I) are shown in the right columns. Scale bar, 10 μm.
To evaluate if these strain-dependent filamentation phenotypes are due to differences in cell cycle progression under genotoxic stress, we selected five strains (UACa10, UACa11, UACa18, UACa22, and UACa25) representing all four clades and showing different grades of filamentation (Fig. 6). As before, these strains were subjected to nitrogen starvation to arrest them in G1, and after pregrowth in liquid medium for 165 min, genotoxins were added. In the presence of HU, the cell cycle in strains UACa10, UACa22, and UACa25 progressed ∼1 h faster than the cell cycle in strain UACa11 (see Fig. S10 at the URL mentioned above). When UACa25 cells were treated with MMS, the cell cycle progression in was ∼2 h faster (Fig. S10). In the slow-growing clade II isolate UACa18, a fraction of the cell population remained in G1 after MMS treatment (Fig. S10), which indicates that the restart of the cell cycle after G1 arrest might be slower in this strain. Although we observed some subtle differences between the C. auris isolates, these do not explain their substantial differences in filamentation.
Therefore, we looked for single nucleotide polymorphisms (SNPs) between two strains from the same clade showing different grades of filamentation, which might explain this differential cellular behavior (isolates from different clades are too diverse, and it would be too difficult to filter out the noise from these data) (43–45). We selected two strains from clade I (South Asia), UACa1 and UACa4, with different capabilities of forming pseudohyphae, which differ by 298 SNPs. Intriguingly, of a total of 99 nonsynonymous SNPs within open reading frames, we found 23 genes with a predicted/demonstrated role in filamentation and one hypothetical protein of unknown function with an SNP causing a premature stop codon in UACa1 (see Table S3 at the URL mentioned above). Unfortunately, this makes it unfeasible to determine a single target gene as the reason for the phenotypic differences between these two strains and suggests that several pathways could be involved in enabling or preventing a particular strain from forming filaments under stress conditions.
DISCUSSION
Due to the recent emergence of C. auris, there is a lack of understanding about its life cycle, which impedes full comprehension of its origin, cellular behavior, and pathogenic traits. Due to the evolutionary distance to the best-studied Candida species, C. albicans (30), inferences from research on C. albicans are not transferable to C. auris. This certainly is the case for the morphological switch between yeast cells and filaments. Most cues causing filamentation in C. albicans do not work in C. auris (15; this study). Accordingly, several genes essential for filamentation in C. albicans and S. cerevisiae, such as EED1, FLO11, HWP1, HWP2, or ECE1 (11, 12) are missing from the C. auris genome (see Table S1; all supplemental figures and tables can be found at https://doi.org/10.6084/m9.figshare.11378550) (30), and some important regulatory determinants of filamentation, such as Ume6, show conspicuous differences between C. auris and C. albicans (Fig. S3). Accordingly, tup1Δ cells do not trigger constitutive filamentation in C. auris (Fig. 2A) as they do in C. albicans (34). However, untreated tup1Δ cells showed defective cell separation. This phenotype might be due to overexpression of HGC1, encoding a G1 cyclin, which controls the expression of septum-degrading enzymes in C. albicans (46), and its expression is kept off by the repressor complex Tup1/Nrg1 (47). Altogether, these results indicate that C. auris is incapable of inducing filamentous growth under the same stimuli as C. albicans.
However, we observed that genotoxic stress triggers filamentation in most C. auris isolates tested (Fig. 1 and 6; see Fig. S9 at the URL mentioned above). These filaments displayed characteristics attributed to pseudohyphae (reviewed in references 9 and 11), as previously described in the presence of HU (48). Indeed, the presence of genotoxic drugs, such as MMS or HU, also triggers pseudohyphal growth in C. albicans and S. cerevisiae (18–20, 49, 50).
In fungi, the switch from yeast to filamentous growth is signaled through the mitogen-activated protein kinase (MAPK) and the fungal cyclic AMP (cAMP)-protein kinase A (PKA) pathways. Additionally, other pathways, such as the sucrose-nonfermentable (SNF), TOR, Hog1, and Rim101 pH-sensing pathways, influence filamentation (reviewed in references 11 to 13). This process involves ∼700 genes for pseudohypha formation in S. cerevisiae (27, 28) and more than 2,000 genes in C. albicans filamentous growth (29). However, mutation of key genes regulating filamentous growth, such as HGC1, UME6, FLO8, TEC1, NRG1, CPH1 (STE12 in S. cerevisiae), or EFG1, does not affect HU-induced pseudohypha formation (19, 49, 50). Therefore, pseudohyphal growth upon genotoxic stress apparently involves, at least partially, different mechanisms.
Genotoxic drugs can induce a variety of DNA damage types and/or perturbation of DNA replication forks, thus triggering the S phase checkpoint. As part of the checkpoint response, the kinase Rad53 is activated, which leads to a temporal cell cycle delay until the issue is resolved and the cell cycle can continue (reviewed in reference 17). A delay of the cell cycle in S phase and a subsequent transient arrest in G2/M were observed in various C. auris strains in the presence of the genotoxic agents HU and MMS (Fig. 4; see Fig. S10 at the URL mentioned above); this suggests that C. auris has a functional S phase checkpoint. Furthermore, mutation of the S phase checkpoint genes MRC1 and RAD9 leads to a defective cell cycle arrest since some cells seem to progress quickly to G1 under genotoxic stress (Fig. 4), similar to growth of the same C. albicans or S. cerevisiae mutants (20, 38, 40, 41, 51). Although activation of Rad53 by DNA damage or perturbed DNA replication forks is sensed differently, both types of DNA lesions possess the molecular signal that triggers the response: an accumulation of single-stranded DNA (ssDNA) which acts as a signal for recruiting and later activating Mec1 (52–54). Homologous recombination is required to repair the DNA lesions generated under genotoxic stress, and, therefore, Rad51 and Rad52 are required to prevent an excess of ssDNA (52, 55, 56). Indeed, in C. auris rad51Δ, cells were arrested in S phase during MMS treatment, which suggests an inability to restore the DNA lesions and, therefore, a constitutive activation of Rad53. Moreover, similar to the rad9Δ or mrc1Δ strain and in contrast to the wild type, a fraction of the genotoxin-stressed rad51Δ cell population seems unable to restart the cell cycle after the G1 arrest or move through the cell cycle without delay (see above) (Fig. 4). Altogether, our results suggest that, similar to other ascomycetes, C. auris has fully functional S phase checkpoint and homologous recombination pathways.
Pseudohyphal growth in response to genotoxins is S phase checkpoint dependent as rad53 mutants in C. albicans or S. cerevisiae and mec1Δ mutants in S. cerevisiae showed a drastic decrease of filamentation under genotoxin treatment (19, 20). Similarly, strains defective for RAD9 or MRC1 also showed alterations of morphology in the presence of genotoxic stress in C. albicans (20). After MMS treatment, a C. albicans RAD9-defective mutant forms filaments which are considerably shorter than those in the wild type. In contrast, filamentation of a C. auris rad9Δ mutant was not different from that of the wild type after 24 h of MMS treatment in liquid culture (see Fig. S1B at the URL mentioned above). However, on plates, fewer pseudohyphae were observed in the C. auris rad9Δ mutant than in the wild type after long-term exposure to MMS (>2 days), albeit a higher proportion of yeast cells became enlarged in the mutant (Fig. S1A and S2). This cell enlargement was obvious in, but not restricted to, rad9Δ cells and could occasionally be observed in the wild type and other mutants, especially in the presence of MMS (Fig. S1A and S2). In other fungi, the presence of enlarged round cells has been described as Titan cells in Cryptococcus neoformans (57, 58) or Goliath cells in C. albicans (59). Further studies will be necessary to elucidate whether these large cells in C. auris resemble Titan or Goliath cells or are something completely different. Taking these observations together, this would suggest that in C. albicans Rad9 might control different or additional HSGs than those controlled by C. auris (see below). The C. auris mrc1Δ mutant induced, at least partially, pseudohyphal growth in the absence of any genotoxic stress; otherwise, its growth was indiscernible from that of the wild type (Fig. 5; Fig. S1 and S2). This result is similar to observations in C. albicans (20, 60). Interestingly, a C. albicans SGS1 mutant strain, which fails to activate Rad53 via Mrc1, forms filaments under unperturbed conditions (61). As a possible explanation, formation of DNA double-strand breaks in mrc1Δ strains and the subsequent activation of Rad53 through Rad9 have been suggested (40, 62). An S phase-independent role, however, has also been described for Mrc1, regulating the replication initiation through interaction with Cdc7, a conserved kinase that triggers firing at each replication origin (63), which regulates the mitotic exit through interaction with Cdc5 (64). This could explain the filamentation observed in the mrc1Δ mutant (see below) and the larger S phase population observed in this mutant under unperturbed conditions (Fig. 4). Mutation of homologous recombination genes such as RAD51, RAD52, MRE11, or RAD50 causes constitutive pseudohypha formation in C. albicans (37, 42, 61). We observed that deletion of RAD51 and RAD57 also triggered constitutive pseudohyphal growth in C. auris (Fig. 5; see also Fig. S1, S2, and S7 at the URL mentioned above).
The S phase checkpoint is a complex process, including several back-up mechanisms which could explain why MRC1 and RAD9 mutants are still able to arrest the cell cycle and produce filaments under genotoxic stress. Indeed, it has been reported that Mrc1 depends on Rad9 to stay activated for a long period and that Rad53 is rapidly but only transiently activated by Mrc1 in rad9Δ cells and is slowly, but continuously, activated by Rad9 in the absence of MRC1 (17, 51). Accordingly, the double mutant mrc1Δ rad9Δ strain is inviable (40, 62). That would explain our observations, under MMS treatment, that in the short term C. auris rad9Δ produces filaments, due to activation of Rad53 by Mrc1; but in the long term, Mrc1 is not able to maintain this activation, and cells lose their filamentation phenotype.
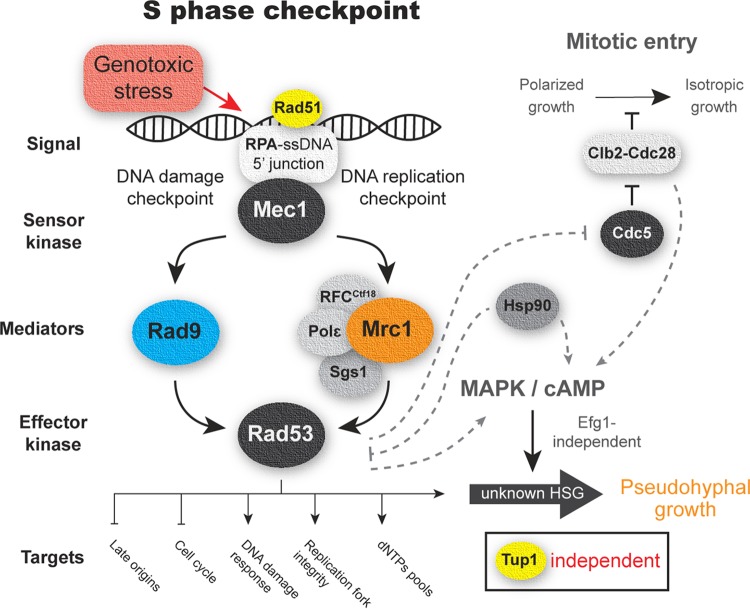
Mechanisms involved in pseudohyphal growth in response to S phase checkpoint activation are not well understood, and further studies will be necessary. However, one mechanism could involve the constitutive activation of the Clb2-Cdc28 complex by Rad53 in response to genotoxic stress through the polo kinase Cdc5 (65–67). The activation of Clb2-Cdc28 prevents the entry into mitosis and the associated switch from polarized to isotropic growth (68, 69); therefore, cells would be stuck in the apical growth phase, thus forming filaments. Hence, mutation of CLB2, CDC28, and CDC5 triggers constitutive pseudohyphal growth in yeasts (49, 70, 71). Furthermore, the cAMP and MAPK pathways have been implicated in pseudohyphal growth in response to genotoxic stress via downstream regulators (18, 42, 50, 72). A plausible model for C. auris is depicted in Fig. 7; our model also takes into account that, in C. auris, constitutive pseudohyphal growth is triggered by downregulation of HSP90, a heat shock family protein, which acts as a chaperone and influences a diverse range of signal transducers (48). Inhibition of HSP90 induces pseudohyphal growth via cAMP-PKA signaling in an Efg1-independent way in C. albicans (73), and, interestingly, a direct inhibition of Rad53 by Hsp90 has been observed in S. cerevisiae (74).
FIG 7.
Schematic representation S phase checkpoint expected for Candida auris. A working model of the S phase checkpoint based on results of this study and related literature in yeast is shown. Proteins tested in this study are in color. Possible S phase checkpoint downstream pathways related to filamentous growth are connected by dotted lines. Tup1 is a major negative regulator of filamentation in C. albicans; filamentation in C. auris is unaffected by deletion of TUP1. HSGs, hyphal-specific genes.
During infection, cells may encounter various conditions that lead to cell cycle arrest, produced either by the host or by other microorganisms cohabiting a given niche. Switching to filamentous growth might be advantageous, allowing cells to escape from such perturbations. However, to our knowledge, filamentous growth in the context of pathogenesis and colonization has only been described once in C. auris (21). This strain recovered from an infected mouse displayed filamentous growth at temperatures below 25°C. However, none of our strains have developed filamentation under these conditions. In any case, there currently are not enough data available for C. auris infection to fully appreciate the potential role of morphogenetic switching, which contributes to full virulence in C. albicans (reviewed in references 11, 13, and 14). The different capacities of different strains to form filaments (21; this study) and the plasticity of the C. auris genome (43, 75) are indications of the flexibility and adaptability of this fungus, suggesting that different clinical isolates could use morphogenetic switching during different phases of pathogenesis.
MATERIALS AND METHODS
Yeast strains and culture conditions.
Candida strains used in this study are listed in Table S2 in the supplemental material (all supplemental figures and tables can be found at https://doi.org/10.6084/m9.figshare.11378550). Yeast cells were grown at 30°C on YPD plates (1% yeast extract, 2% mycological peptone, 2% glucose, 2% agar; Oxoid, Basingstoke, UK) or with shaking at 200 rpm in YPD broth (same constituents as plates, but without agar). Cell concentrations were determined by measuring the optical density of the culture at a wavelength of 600 nm (OD600) on an Ultraspec 2000 (Pharmacia Biotech, Uppsala, Sweden) spectrometer (calibration defined an OD600 of 1 to contain ∼3 × 107 C. auris cells/ml).
Spot assays were carried out on YPD agar adding the indicated drug where necessary (Fig. 3; see also Fig. S7A). Cell concentrations were determined by OD600 after overnight growth in YPD broth, and four 1:10 serial dilutions containing 102 to 105 cells were plated and grown at 30°C for 3 days (unless indicated otherwise).
For filament visualization, ∼1 × 107 cells were grown for 18 to 20 h in YPD broth, unless indicated otherwise, with addition of 100 μM HU, 0.02% MMS, or 5 μg/ml 5-FC. Other potential filamentation conditions tested were the following: Lee’s medium at pH 3.5 and pH 6.5 (76); YPD (see above) or RPMI 1640 medium containing 20% fetal calf serum (Sigma-Aldrich Corp., St. Louis, MO); YPD medium at a temperature of 25, 30, or 37°C; presence of 0.5% isoamyl alcohol in YPD medium; and 25 μg/ml Bleocin in YPD medium. To check filamentation on solid medium, fresh cells were streaked on YPD plates containing 100 μM HU, 0.02% MMS, or 5 μg/ml 5-FC and grown at 30°C for the times indicated (Fig. 1 and 5; see also Fig. S2).
Cell cycle analysis and microscopy.
To synchronize cultures in G1, cells were grown in YPD broth overnight at 30°C and 200 rpm. Then 1 × 107 cells were washed with sterile water and transferred to nitrogen starvation conditions (6.7 g/liter yeast nitrogen base [YNB] without amino acids and without ammonium sulfate [Sigma-Aldrich] and 20 g/liter glucose) and grown at 30°C and 200 rpm. Samples were harvested by centrifugation (1,000 × g, 2 min) and fixed by addition of 70% ethanol (EtOH) at the indicated time points (Fig. 4; see also Fig. S8 and S10). Cells were stored at +4°C before proceeding to flow cytometry (see below).
The cell cycle of cells previously arrested in G1 from different C. auris strains in the presence of genotoxic drugs was determined. For this purpose, 1 × 107 cells, obtained after growth in YPD broth overnight at 30°C and 200 rpm, were harvested, washed with sterile water, and transferred to nitrogen starvation conditions (see above) for 15 to 17 h at 30°C and 200 rpm. Then, 1 × 108 cells were transferred to 10 ml of fresh YPD broth. To restart the cell cycle, cells were grown for 165 min at 30°C and 200 rpm, after which 100 mM HU or 0.02% MMS were added (a third culture without any additions served as a control). Cells were harvested by centrifugation (1,000 × g, 2 min)and fixed by addition of 70% ethanol every hour from 0 to 8 h and at a final time point of 22 h. Cells were stored at +4°C before proceeding to flow cytometry (see below). For flow cytometry, yeast samples were prepared as previously described (75, 77).
For microscopy, cells were fixed by addition of 1 ml 70% EtOH and incubated for at least 2 h at 4°C. Fixed cells were pelleted (2 min at 1,000 × g) and resuspended in sodium citrate buffer (14.7 g/liter sodium citrate, pH 7.5). Cells were then treated with RNase A (250 μg per 1 × 107 cells) and proteinase K (1,000 μg per 1 × 107 cells) for 1 h at 50°C; subsequently, Triton X-100 (Sigma-Aldrich) was added to a final concentration of 0.25%. Cells were stained for DNA (SYBR green I, 1:5000; Sigma-Aldrich) and for chitin (1 μg/ml calcofluor white; Sigma) at +4°C overnight. For time course experiments, the same samples were used for flow cytometry and microscopy (adding 1 μg/ml calcofluor white for microscopy). Rhodamine-phalloidin staining was carried out as previously described (78) with a few modifications. Approximately 1 × 107 cells from an overnight culture were fixed for 1 h in 4% formaldehyde (phosphate-buffered saline [PBS] buffered) at room temperature, harvested by centrifugation (2 min at 1,000 ×g), and resuspended in PBS before addition of 10 μl of rhodamine-phalloidin (6.6 μM in methanol) and 1 μl of 4′,6′-diamidino-2-phenylindole (DAPI; 10 μg/ml). Samples were incubated in the dark for 1 h and washed in PBS (2 min, 1,000 × g) twice. Finally, cells were resuspended in ProLong Diamond antifade (Thermo Fisher Scientific, Waltham, MA).
Fluorescent images were taken on a Zeiss Axio Imager M2 microscope with a Zeiss 503 camera and analyzed using ZEN 2 blue edition software (Carl Zeiss Microscopy, Jena, Germany). Standard bright-field microscopy was performed on a BX50 microscope (Olympus, Tokyo, Japan) equipped with an Infinity 1 camera (Lumenera, Ottawa, Canada) and analyzed using the Infinity capture software, version 6.5.7 (Lumenera, Ottawa, Canada). Colony morphology was imaged on a Zeiss Stemi 2000-c binocular (Carl Zeiss Microscopy, Jena, Germany) equipped with an Infinity 1 camera (Lumenera, Ottawa, Canada).
Phenotype quantifications and filament length measurements were done from bright-field microscopy images using the cell counter and the measuring tools included in ImageJ (version 1.47; National Institutes of Health, Bethesda, MD). Box plots, bar charts, and statistical analysis were done in R (version 3.6.0) (https://www.r-project.org/) using the ggplot2 and the ggpubr libraries in the RStudio environment (version 1.2.1335; RStudio, Inc., Boston, MA). Kruskal-Wallis and Wilcoxon rank sum tests at an alpha level of 0.05 were used to identify statistical differences between data sets.
Nucleic acid manipulations.
For fast genomic DNA (gDNA) extraction, cells from a single colony were suspended in 20 mM NaOH, boiled at 100°C for 10 min, and incubated on ice for 10 min. Samples were then centrifuged (2 min at 1,000 × g), and a small amount of supernatant was used for PCRs.
For high-quality gDNA extractions, cells from overnight cultures were harvested (2 min at 1,000 × g) and washed with double-distilled water (ddH2O). After the supernatant was removed, acid-washed glass beads (Sigma-Aldrich) and DNA extraction buffer (1 M NaCl, 2% Triton x-100, 1 M Tris-HCl, pH 8, 0.5 M EDTA, 1% SDS) were added. Cells were disrupted in a Vibrax VXR (IKA GmbH & Co. KG, Staufen, Germany) multitube mixer for 3 min before addition of 1 volume of phenol-chloroform-isoamyl alcohol (25:24:1) (Sigma-Aldrich). Samples were mixed vigorously and centrifuged (10 min at 12,000 × g). The supernatant was transferred to a fresh reaction tube, and 0.1 volume of 3 M sodium acetate, pH 5.2, and 2 volumes of ice-cold 100% ethanol were added. After precipitation (30 to 60 min at –20°C), samples were centrifuged (5 min at 12,000 × g) and washed with 70% ethanol. Pellets were airdried and resuspended in ddH2O, and 5 μl of RNase (10 mg/ml) was added, followed by an incubation of 30 to 60 min at 37°C. A small amount of a 1:10 dilution was used for subsequent PCRs.
Standard PCRs were carried out using DreamTaq PCR Master Mix (Thermo Fisher Scientific) according to the manufacturer’s instructions. All primers used in this study (see Table S4 at https://doi.org/10.6084/m9.figshare.11378550) were designed using the C. auris reference genome (https://www.ncbi.nlm.nih.gov/genome/?term=txid498019) and synthesized by Sigma-Aldrich.
Gene deletion constructs and transformation.
All gene deletion constructs were amplified using Phusion High-Fidelity PCR Master Mix (Thermo Fisher Scientific) as recommended by the supplier.
Genes were deleted using the nourseothricin resistance marker CaNAT1 flanked by 1.5- to 2-kb regions of homology to up- and downstream sequences of the target gene. CaNAT1, including TEF promoter and terminator sequences, was PCR amplified from pV1025 (79) using oligonucleotides oUA315 and oUA316 (see Table S4 at the URL mentioned above) and cloned into a BamHI-linearized pFA6a backbone by NEBuilder DNA Assembly Master Mix (New England BioLabs, Ipswich, MA). The assembly mix was transformed into NEB 10-beta Escherichia coli cells (New England BioLabs) according to the manufacturer’s instructions. The resulting plasmid, pALo218, was then used to amplify CaNAT1 with the oligonucleotide oUA353 and oUA354 (Table S4). Up- and downstream homologous regions of the target gene were PCR amplified from gDNA of UACa11 (Table S2) using specific primers for each target gene (Table S4). The oligonucleotide primers on the 3′ end of the upstream region and at the 5′ end of the downstream region include short homologies to the CaNAT1 marker to assemble transformation cassettes via fusion PCR (80). The three PCR products (CaNAT1 and up- and downstream homology regions) were combined in roughly equimolar amounts (∼0.15 pmols) together with Phusion High-Fidelity PCR Master Mix (Thermo Fisher Scientific) containing 2.5% dimethyl sulfoxide (DMSO) and subjected to eight PCR cycles (10 s at 98°C, 30 s at 55°C, and 60 s at 72°C). Nested primers (see Table S4 at the URL mentioned above) under standard PCR conditions were used to amplify the final transformation cassettes.
Gene deletion constructs were transformed into C. auris strain UACa11 using a protocol developed for C. albicans (81), with small modifications. Briefly, cells from a culture grown in YPD medium to stationary phase were diluted 1:100 into fresh YPD broth and incubated with shaking at 30°C until they reached mid-exponential phase (OD600 of 0.5 to 0.8). Cells were centrifuged (2 min, 1,000 × g), washed once with ddH2O, and resuspended in 100 mM lithium acetate. Transformation mixtures contained ∼1 μg of DNA of the transformation cassette, 100 μg of carrier DNA (herring sperm DNA solution; Thermo Fisher Scientific), 37% polyethylene glycol (PEG) 3350, 100 mM lithium acetate, and ∼1.5 × 108 C. auris cells. The transformation mixture was incubated overnight (16 to 20 h) at 30°C. After a heat shock of 15 min at 44°C, cells were pelleted by centrifugation (2 min at 1,500 × g), resuspended in YPD medium, and incubated with shaking for 4 h at 30°C. Cells were then plated onto selective YPD agar containing 100 μg/ml nourseothricin (clonNAT; Werner BioAgents GmbH, Jena, Germany) and incubated at 30°C until transformants appeared. Gene deletion and correct integration were confirmed by PCR using primers located within the ORF of the target gene and primers located within and without the transformation cassette (see Fig. S5 and Table S4 at the URL mentioned above).
In silico analysis.
Gene and protein sequences were obtained from the C. auris reference genome (assembly Cand_auris_B11221_V1) in the NCBI database (https://www.ncbi.nlm.nih.gov/genome/?term=txid498019). Interestingly, RAD57 was not present in this reference genome but could be found in the draft genome of strain Ci6684 (82). Protein sequences of Candida spp. were obtained from the Candida Genome Database (http://www.candidagenome.org/) or from Uniprot (https://www.uniprot.org/), and the Saccharomyces Genome Database (https://www.yeastgenome.org/) served as source for S. cerevisiae protein sequences. Homology scores of C. auris proteins against C. albicans and S. cerevisiae proteins (E value, length aligned; identities are shown in Table S1, available at the URL mentioned above) were generated using the BLAST tools of the Candida or the Saccharomyces Genome Databases. Alignments were carried out by the MSAprobs method (default settings) using Jalview, version 2.10.5 (83).
For SNP calling, Illumina sequencing reads from two clade I strains (UACa1 and UACa4) were aligned (Z. K. Ross, N. A. R. Gow, and A. Lorenz, unpublished results) using BWA-MEM, version 0.7.12 (84), and processed with Samtools, version 0.1.19, view, sort, rmdup, and index (85). SNPs were then detected using Pilon, version 1.22 (86), filtering the resulting variant call format (VCF) file for genotype 1/1 only. Low coverage (less than 10% of mean coverage), ambiguous positions, and deletions were removed. The reference genome was annotated with Augustus, version 3.3.1 (87), ab initio gene prediction software, and VCF annotator (http://vcfannotator.sourceforge.net) was used to predict the effect of the SNPs called on the annotated genes.
ACKNOWLEDGMENTS
We are grateful to Arunaloke Chakrabarti, Anuradha Chowdhary, Elizabeth Johnson (PHE), Takashi Kubota, and Shawn Lockhart (CDC) for providing strains. We thank Fei Long for skillful technical assistance. Flow cytometry was performed at the Iain Fraser Cytometry Centre (IFCC), University of Aberdeen (Raif Yuecel). Microscopy was done at the Microscopy & Histology Facility, University of Aberdeen (Kevin S. Mackenzie).
This work was supported by a Wellcome Trust Seed Award to A.L. (grant number 212524/Z/18/Z) and the Medical Research Council (MRC) Centre for Medical Mycology at the University of Exeter (grant numbers MR/P501955/1 and MR/N006364/1).
G.B.R., N.A.R.G., and A.L. conceived and designed the study; G.B.R. and Z.K.R. collected the data; G.B.R., Z.K.R., and A.L. performed the analysis; G.B.R. and A.L. drafted the manuscript; G.B.R., Z.K.R., N.A.R.G. and A.L. revised the manuscript.
We have no conflicts of interest to declare.
REFERENCES
- 1.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 2.Kwon YJ, Shin JH, Byun SA, Choi MJ, Won EJ, Lee D, Lee SY, Chun S, Lee JH, Choi HJ, Kee SJ, Kim SH, Shin MG. 2019. Candida auris clinical isolates from South Korea: identification, antifungal susceptibility, and genotyping. J Clin Microbiol 57:e01624-18. doi: 10.1128/JCM.01624-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhodes J, Fisher MC. 2019. Global epidemiology of emerging Candida auris. Curr Opin Microbiol 52:84–89. doi: 10.1016/j.mib.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Lone SA, Ahmad A. 2019. Candida auris—the growing menace to global health. Mycoses 62:620–637. doi: 10.1111/myc.12904. [DOI] [PubMed] [Google Scholar]
- 5.Kean R, Ramage G. 2019. Combined antifungal resistance and biofilm tolerance: the global threat of Candida auris. mSphere 4:e00458-19. doi: 10.1128/mSphere.00458-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forsberg K, Woodworth K, Walters M, Berkow EL, Jackson B, Chiller T, Vallabhaneni S. 2019. Candida auris: the recent emergence of a multidrug-resistant fungal pathogen. Med Mycol 57:1–12. doi: 10.1093/mmy/myy054. [DOI] [PubMed] [Google Scholar]
- 7.Jackson BR, Chow N, Forsberg K, Litvintseva AP, Lockhart SR, Welsh R, Vallabhaneni S, Chiller T. 2019. On the origins of a species: what might explain the rise of Candida auris? J Fungi (Baseo) 5:E58. doi: 10.3390/jof5030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casadevall A, Kontoyiannis DP, Robert V. 2019. On the emergence of Candida auris: climate change, azoles, swamps, and birds. mBio 10:e01397-19. doi: 10.1128/mBio.01397-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sudbery P, Gow N, Berman J. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol 12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Berman J. 2006. Morphogenesis and cell cycle progression in Candida albicans. Curr Opin Microbiol 9:595–601. doi: 10.1016/j.mib.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sudbery PE. 2011. Growth of Candida albicans hyphae. Nat Rev Microbiol 9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- 12.Cullen PJ, Sprague GF. 2012. The regulation of filamentous growth in yeast. Genetics 190:23–49. doi: 10.1534/genetics.111.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noble SM, Gianetti BA, Witchley JN. 2017. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat Rev Microbiol 15:96–108. doi: 10.1038/nrmicro.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson DS, Carlisle PL, Kadosh D. 2011. Coevolution of morphology and virulence in Candida Species. Eukaryot Cell 10:1173–1182. doi: 10.1128/EC.05085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Bing J, Zheng Q, Zhang F, Liu J, Yue H, Tao L, Du H, Wang Y, Wang H, Huang G. 2018. The first isolate of Candida auris in China: clinical and biological aspects. Emerg Microbes Infect 7:93. doi: 10.1038/s41426-018-0095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kron SJ, Gow NA. 1995. Budding yeast morphogenesis: signalling, cytoskeleton and cell cycle. Curr Opin Cell Biol 7:845–855. doi: 10.1016/0955-0674(95)80069-7. [DOI] [PubMed] [Google Scholar]
- 17.Pardo B, Crabbé L, Pasero P. 2017. Signaling pathways of replication stress in yeast. FEMS Yeast Res 17:fow101. doi: 10.1093/femsyr/fow101. [DOI] [PubMed] [Google Scholar]
- 18.Bachewich C, Nantel A, Whiteway M. 2005. Cell cycle arrest during S or M phase generates polarized growth via distinct signals in Candida albicans. Mol Microbiol 57:942–959. doi: 10.1111/j.1365-2958.2005.04727.x. [DOI] [PubMed] [Google Scholar]
- 19.Jiang YW, Kang CM. 2003. Induction of S. cerevisiae filamentous differentiation by slowed DNA synthesis involves Mec1, Rad53 and Swe1 checkpoint proteins. Mol Biol Cell 14:5116–5124. doi: 10.1091/mbc.e03-06-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi Q-M, Wang Y-M, De Zheng X, Lee RTH, Wang Y. 2007. Critical role of DNA checkpoints in mediating genotoxic-stress–induced filamentous growth in Candida albicans. Mol Biol Cell 18:815–826. doi: 10.1091/mbc.e06-05-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yue H, Bing J, Zheng Q, Zhang Y, Hu T, Du H, Wang H, Huang G. 2018. Filamentation in Candida auris, an emerging fungal pathogen of humans: passage through the mammalian body induces a heritable phenotypic switch. Emerg Microbes Infect 7:188. doi: 10.1038/s41426-018-0187-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nyholm S, Thelander L, Gräslund A. 1993. Reduction and loss of the iron center in the reaction of the small subunit of Mouse ribonucleotide reductase with hydroxyurea. Biochemistry 32:11569–11574. doi: 10.1021/bi00094a013. [DOI] [PubMed] [Google Scholar]
- 23.Poli J, Tsaponina O, Crabbé L, Keszthelyi A, Pantesco V, Chabes A, Lengronne A, Pasero P. 2012. dNTP pools determine fork progression and origin usage under replication stress. EMBO J 31:883–894. doi: 10.1038/emboj.2011.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vázquez MV, Rojas V, Tercero JA. 2008. Multiple pathways cooperate to facilitate DNA replication fork progression through alkylated DNA. DNA Repair (Amst) 7:1693–1704. doi: 10.1016/j.dnarep.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 25.Grem JL. 2000. 5-Fluorouracil: forty-plus and still ticking. A review of its preclinical and clinical development. Invest New Drugs 18:299–313. doi: 10.1023/a:1006416410198. [DOI] [PubMed] [Google Scholar]
- 26.Sherry L, Ramage G, Kean R, Borman A, Johnson EM, Richardson MD, Rautemaa-Richardson R. 2017. Biofilm-forming capability of highly virulent, multidrug-resistant Candida auris. Emerg Infect Dis 23:328–331. doi: 10.3201/eid2302.161320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin R, Dobry CJ, McCown PJ, Kumar A. 2008. Large-scale analysis of yeast filamentous growth by systematic gene disruption and overexpression. Mol Biol Cell 19:284–196. doi: 10.1091/mbc.e07-05-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryan O, Shapiro RS, Kurat CF, Mayhew D, Baryshnikova A, Chin B, Lin ZY, Cox MJ, Vizeacoumar F, Cheung D, Bahr S, Tsui K, Tebbji F, Sellam A, Istel F, Schwarzmüller T, Reynolds TB, Kuchler K, Gifford DK, Whiteway M, Giaever G, Nislow C, Costanzo M, Gingras AC, Mitra RD, Andrews B, Fink GR, Cowen LE, Boone C. 2012. Global gene deletion analysis exploring yeast filamentous growth. Science 337:1352–1356. doi: 10.1126/science.1224339. [DOI] [PubMed] [Google Scholar]
- 29.Azadmanesh J, Gowen AM, Creger PE, Schafer ND, Blankenship JR. 2017. Filamentation involves two overlapping, but distinct, programs of filamentation in the pathogenic fungus Candida albicans. G3 (Bethesda) 7:3797–3808. doi: 10.1534/g3.117.300224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munoz JF, Gade L, Chow NA, Loparev VN, Juieng P, Farrer RA, Litvintseva AP, Cuomo CA. 2018. Genomic basis of multidrug-resistance, mating, and virulence in Candida auris and related emerging species. Nat Commun 9:5346. doi: 10.1038/s41467-018-07779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lo W, Dranginis AM. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol Biol Cell 9:161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vinod PK, Sengupta N, Bhat PJ, Venkatesh KV. 2008. Integration of global signaling pathways, cAMP-PKA, MAPK and TOR in the regulation of FLO11. PLoS One 3:e1663. doi: 10.1371/journal.pone.0001663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Younes SS, Khalaf RA. 2013. The Candida albicans Hwp2p can complement the lack of filamentation of a Saccharomyces cerevisiae flo11 null strain. Microbiology 159:1160–1164. doi: 10.1099/mic.0.067249-0. [DOI] [PubMed] [Google Scholar]
- 34.Braun BR, Johnson AD. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- 35.Braun BR, Head WS, Wang MX, Johnson AD. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Legrand M, Chan CL, Jauert PA, Kirkpatrick DT. 2007. Role of DNA mismatch repair and double-strand break repair in genome stability and antifungal drug resistance in Candida albicans. Eukaryot Cell 6:2194–2205. doi: 10.1128/EC.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.García-Prieto F, Gómez-Raja J, Andaluz E, Calderone R, Larriba G. 2010. Role of the homologous recombination genes RAD51 and RAD59 in the resistance of Candida albicans to UV light, radiomimetic and anti-tumor compounds and oxidizing agents. Fungal Genet Biol 47:433–445. doi: 10.1016/j.fgb.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang G, Tong X, Weng S, Zhou H. 2012. Multiple phosphorylation of Rad9 by CDK is required for DNA damage checkpoint activation. Cell Cycle 11:3792–3800. doi: 10.4161/cc.21987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.González-Prieto R, Muñoz-Cabello AM, Cabello-Lobato MJ, Prado F. 2013. Rad51 replication fork recruitment is required for DNA damage tolerance. EMBO J 32:1307–1321. doi: 10.1038/emboj.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJH, Bousset K, Furuya K, Diffley JFX, Carr AM, Elledge SJ. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol 3:958–965. doi: 10.1038/ncb1101-958. [DOI] [PubMed] [Google Scholar]
- 41.Osborn AJ, Elledge SJ. 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev 17:1755–1767. doi: 10.1101/gad.1098303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andaluz E, Ciudad T, Gómez-Raja J, Calderone R, Larriba G. 2006. Rad52 depletion in Candida albicans triggers both the DNA-damage checkpoint and filamentation accompanied by but independent of expression of hypha-specific genes. Mol Microbiol 59:1452–1472. doi: 10.1111/j.1365-2958.2005.05038.x. [DOI] [PubMed] [Google Scholar]
- 43.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chakrabarti A, Sood P, Rudramurthy SM, Chen S, Kaur H, Capoor M, Chhina D, Rao R, Eshwara VK, Xess I, Kindo AJ, Umabala P, Savio J, Patel A, Ray U, Mohan S, Iyer R, Chander J, Arora A, Sardana R, Roy I, Appalaraju B, Sharma A, Shetty A, Khanna N, Marak R, Biswas S, Das S, Harish BN, Joshi S, Mendiratta D. 2015. Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med 41:285–295. doi: 10.1007/s00134-014-3603-2. [DOI] [PubMed] [Google Scholar]
- 45.Rhodes J, Abdolrasouli A, Farrer RA, Cuomo CA, Aanensen DM, Armstrong-James D, Fisher MC, Schelenz S. 2018. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg Microbes Infect 43:1–12. doi: 10.1038/s41426-018-0045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng X, Wang Y, Wang Y. 2004. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J 23:1845–1856. doi: 10.1038/sj.emboj.7600195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braun BR, Kadosh D, Johnson AD. 2001. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J 20:4753–4761. doi: 10.1093/emboj/20.17.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim SH, Iyer KR, Pardeshi L, Muñoz JF, Robbins N, Cuomo CA, Wong KH, Cowen LE. 2019. Genetic analysis of Candida auris implicates Hsp90 in morphogenesis and azole tolerance and Cdr1 in azole resistance. mBio 10:e02529-18. doi: 10.1128/mBio.02529-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bachewich C, Thomas DY, Whiteway M. 2003. Depletion of a Polo-like kinase in Candida albicans activates Cyclase-dependent hyphal-like growth. Mol Biol Cell 14:2163–2180. doi: 10.1091/mbc.02-05-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C, Zeng G, Wang Y. 2018. G1 and S phase arrest in Candida albicans induces filamentous growth via distinct mechanisms. Mol Microbiol 110:191–203. doi: 10.1111/mmi.14097. [DOI] [PubMed] [Google Scholar]
- 51.Bacal J, Moriel‐Carretero M, Pardo B, Barthe A, Sharma S, Chabes A, Lengronne A, Pasero P. 2018. Mrc1 and Rad9 cooperate to regulate initiation and elongation of DNA replication in response to DNA damage. EMBO J 37:1–18. doi: 10.15252/embj.201899319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopes M, Cotta-Ramusino C, Liberi G, Foiani M. 2003. Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms. Mol Cell 12:1499–1510. doi: 10.1016/s1097-2765(03)00473-8. [DOI] [PubMed] [Google Scholar]
- 53.Zou L, Elledge SJ. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 54.Hashimoto Y, Chaudhuri AR, Lopes M, Costanzo V. 2010. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol 17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. 2010. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prado F. 2018. Homologous recombination: to fork and beyond. Genes (Basel) 9:603. doi: 10.3390/genes9120603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zaragoza O, Nielsen K. 2013. Titan cells in Cryptococcus neoformans: cells with a giant impact. Curr Opin Microbiol 16:409–413. doi: 10.1016/j.mib.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dambuza IM, Drake T, Chapuis A, Zhou X, Correia J, Taylor-Smith L, LeGrave N, Rasmussen T, Fisher MC, Bicanic T, Harrison TS, Jaspars M, May RC, Brown GD, Yuecel R, MacCallum DM, Ballou ER. 2018. The Cryptococcus neoformans Titan cell is an inducible and regulated morphotype underlying pathogenesis. PLoS Pathog 14:e1006978. doi: 10.1371/journal.ppat.1006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malavia D, Lehtovirta-Morley LE, Alamir O, Weiß E, Gow NAR, Hube B, Wilson D. 2017. Zinc limitation induces a hyper-adherent Goliath phenotype in Candida albicans. Front Microbiol 8:2238. doi: 10.3389/fmicb.2017.02238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loll-Krippleber R, Enfert C, Feri A, Diogo D, Perin A, Marcet-Houben M, Bougnoux M, Legrand M. 2014. A study of the DNA damage checkpoint in Candida albicans: uncoupling of the functions of Rad53 in DNA repair, cell cycle regulation and genotoxic stress-induced polarized growth. Mol Microbiol 91:452–471. doi: 10.1111/mmi.12471. [DOI] [PubMed] [Google Scholar]
- 61.Legrand M, Chan CL, Jauert PA, Kirkpatrick DT. 2011. The contribution of the S-phase checkpoint genes MEC1 and SGS1 to genome stability maintenance in Candida albicans. Fungal Genet Biol 48:823–830. doi: 10.1016/j.fgb.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 63.Masai H, Yang CC, Matsumoto S. 2017. Mrc1/Claspin: a new role for regulation of origin firing. Curr Genet 63:813–818. doi: 10.1007/s00294-017-0690-y. [DOI] [PubMed] [Google Scholar]
- 64.Miller CT, Gabrielse C, Chen YC, Weinreich M. 2009. Cdc7p-Dbf4p regulates mitotic exit by inhibiting Polo kinase. PLoS Genet 5:e1000498. doi: 10.1371/journal.pgen.1000498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Asano S, Park JE, Sakchaisri K, Yu LR, Song S, Supavilai P, Veenstra TD, Lee KS. 2005. Concerted mechanism of Swe1/Wee1 regulation by multiple kinases in budding yeast. EMBO J 24:2194–2204. doi: 10.1038/sj.emboj.7600683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang T, Nirantar S, Lim HH, Sinha I, Surana U. 2009. DNA damage checkpoint maintains Cdh1 in an active state to inhibit anaphase progression. Dev Cell 17:541–551. doi: 10.1016/j.devcel.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 67.Simpson-Lavy KJ, Brandeis M. 2010. Clb2 and the APC/CCdh1 regulate Swe1 stability. Cell Cycle 9:3046–3053. doi: 10.4161/cc.9.115.12457. [DOI] [PubMed] [Google Scholar]
- 68.Pruyne D, Bretscher A. 2000. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J Cell Sci 113:365–375. [DOI] [PubMed] [Google Scholar]
- 69.Lew DJ. 2003. The morphogenesis checkpoint: how yeast cells watch their figures. Curr Opin Cell Biol 15:648–653. doi: 10.1016/j.ceb.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 70.Ahn SH, Acurio A, Kron SJ. 1999. Regulation of G2/M progression by the STE mitogen-activated protein kinase pathway in budding yeast filamentous growth. Mol Biol Cell 10:3301–3316. doi: 10.1091/mbc.10.10.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bensen ES, Clemente-Blanco A, Finley KR, Correa-Bordes J, Berman J. 2005. The mitotic cyclins Clb2p and Clb4p affect morphogenesis in Candida albicans. Mol Biol Cell 16:3387–3400. doi: 10.1091/mbc.e04-12-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu X, Jiang YW. 2005. Possible integration of upstream signals at Cdc42 in filamentous differentiation of S. cerevisiae. Yeast 22:1069–1077. doi: 10.1002/yea.1294. [DOI] [PubMed] [Google Scholar]
- 73.Shapiro RS, Cowen LE. 2010. Coupling temperature sensing and development: hsp90 regulates morphogenetic signaling in Candida albicans. Virulence 1:45–48. doi: 10.4161/viru.1.1.10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khurana N, Laskar S, Bhattacharyya MK, Bhattacharyya S. 2016. Hsp90 induces increased genomic instability toward DNA-damaging agents by tuning down RAD53 transcription. Mol Biol Cell 27:2463–2478. doi: 10.1091/mbc.E15-12-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bravo Ruiz G, Ross ZK, Holmes E, Schelenz S, Gow NAR, Lorenz A. 2019. Rapid and extensive karyotype diversification in haploid clinical Candida auris isolates. Curr Genet 65:1217–1228. doi: 10.1007/s00294-019-00976-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee KL, Buckley HR, Campbell CC. 1975. An amino acid liquid synthetic medium for the development of mycellal and yeast forms of Candida albicans. Sabouraudia 13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 77.Fortuna M, Sousa MJ, Corte-Real M, Leao C, Salvador A, Sansonetty F. 2000. Cell cycle analysis of yeasts. Curr Protoc Cytom Chapter 11:Unit 11.13. doi: 10.1002/0471142956.cy1113s13. [DOI] [PubMed] [Google Scholar]
- 78.Burke D, Dawson D, Stearns T. 2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 79.Vyas VK, Barrasa MI, Fink GR. 2015. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci Adv 1:e1500248. doi: 10.1126/sciadv.1500248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shevchuk NA, Bryksin AV, Nusinovich YA, Cabello FC, Sutherland M, Ladisch S. 2004. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res 32:e19. doi: 10.1093/nar/gnh014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Walther A, Wendland J. 2003. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr Genet 42:339–343. doi: 10.1007/s00294-002-0349-0. [DOI] [PubMed] [Google Scholar]
- 82.Chatterjee S, Alampalli SV, Nageshan RK, Chettiar ST, Joshi S, Tatu US. 2015. Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris. BMC Genomics 16:686. doi: 10.1186/s12864-015-1863-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stanke M, Steinkamp R, Waack S, Morgenstern B. 2004. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res 32:W309–W312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]