Abstract
Vertebrate DNA crosslink repair excises toxic replication-blocking DNA crosslinks. Numerous factors involved in crosslink repair have been identified, and mutations in their corresponding genes cause Fanconi anemia (FA). A key step in crosslink repair is monoubiquitination of the FANCD2–FANCI heterodimer, which then recruits nucleases to remove the DNA lesion. Here, we use cryoEM to determine the structure of recombinant chicken FANCD2 and FANCI complexes. FANCD2–FANCI adopts a closed conformation when the FANCD2 subunit is monoubiquitinated, creating a channel that encloses double-stranded DNA. Ubiquitin is positioned at the interface of FANCD2 and FANCI, and acts as a covalent molecular pin to trap the complex on DNA. In contrast, isolated FANCD2 is a homodimer unable to bind DNA, suggestive of an autoinhibitory mechanism that prevents premature activation. Together, our work suggests that FANCD2–FANCI is a clamp that is locked onto DNA by ubiquitin, with distinct interfaces that may recruit other DNA repair factors.
Introduction
Vertebrates repair DNA interstrand crosslinks through a complex and essential process1. Genetic inactivation of crosslink repair in humans results in Fanconi anemia (FA), a disorder characterized by abnormal development, loss of blood production, and marked cancer susceptibility2. Remarkably, autosomal recessive mutations in any one of 20 genes (FANCA to FANCQ) result in this genetic illness, and collectively the FANC gene products function in a FA DNA crosslink repair pathway3. An understanding of this pathway is emerging from cell biological and biochemical reconstitution studies4–9. These indicate that a number of the FANC gene products form a large nuclear E3 monoubiquitin ligase complex (the FA core complex). When DNA replication is stalled, the FA core complex is activated to monoubiquitinate the FANCD2–FANCI heterodimer (ubD2–I)6,7,10–12. Activated ubD2–I localizes to sites of DNA damage and is thought to recruit a nuclease incision complex, consisting of FANCP (SLX4) and FANCQ (XPF)–ERCC1, to remove the crosslinked DNA13–20. Genetic studies also indicate that reversal of the monoubiquitination step by the deubiquitinating enzyme USP1, in complex with UAF1, is required to complete DNA crosslink repair21–23.
Most cases of FA are due to mutations in components of the FA core complex, impairing its activity and resulting in a failure to monoubiquitinate FANCD224. Loss of FANCI also abolishes FANCD2 modification11,12. Monoubiquitination is a tightly controlled process, and is regulated by phosphorylation11,12,25–27. Altogether this suggests that heterodimerization of FANCD2 with FANCI is essential for this key step in the pathway.
Monoubiquitination of FANCD2 can be recapitulated in cell free systems in the presence of DNA8,9,28,29. Many different DNA substrates stimulate the reaction to a similar extent despite a modest preference of D2–I for forked or crosslinked DNA structures in DNA binding assays30,31. FANCI is not substantially monoubiquitinated by purified chicken FA core complexes8,29 and is only inefficiently monoubiquitinated by a human complex in vitro9. DNA crosslink repair in cells can be largely rescued by a FANCI variant with a mutated monoubiquitination site, suggesting that FANCI ubiquitination is not essential25. Together, both in vitro and in vivo data suggest that monoubiquitination of FANCD2 (not FANCI) is the critical and predominant function of the FA core complex.
An x-ray crystal structure of the FANCD2–FANCI dimer revealed that the monoubiquitination sites, as well as regulatory FANCI phosphorylation sites, map to the dimerization interface, and that FANCI harbors a binding-site for DNA31. Still, a longstanding mystery is why D2–I monoubiquitination and its subsequent reversal are so fundamental for crosslink repair. Moreover, the mechanistic details of FANCD2 and FANCI interaction with DNA, and how ubD2–I might ultimately enable DNA crosslink removal are not known.
Here, we expressed and purified Gallus gallus FANCD2 and FANCI complexes, and, using a recombinant FA core complex, we obtained monoubiquitinated D2–I. Using cryoEM and biochemical assays, we delineated details of how these factors interact with DNA. We determined the structure of ubD2–I, which we show adopts a tight clamp on DNA. We also find that FANCD2 itself forms a homodimer that is unable to interact with DNA, but this can interchange with FANCI, to create a D2–I heterodimer that interacts with DNA. Our finding that the D2–I dimer transforms into a locked DNA clamp upon monoubiquitination of FANCD2 provides insight into how crosslink repair could be initiated.
Results
CryoEM structure of unmodified D2–I
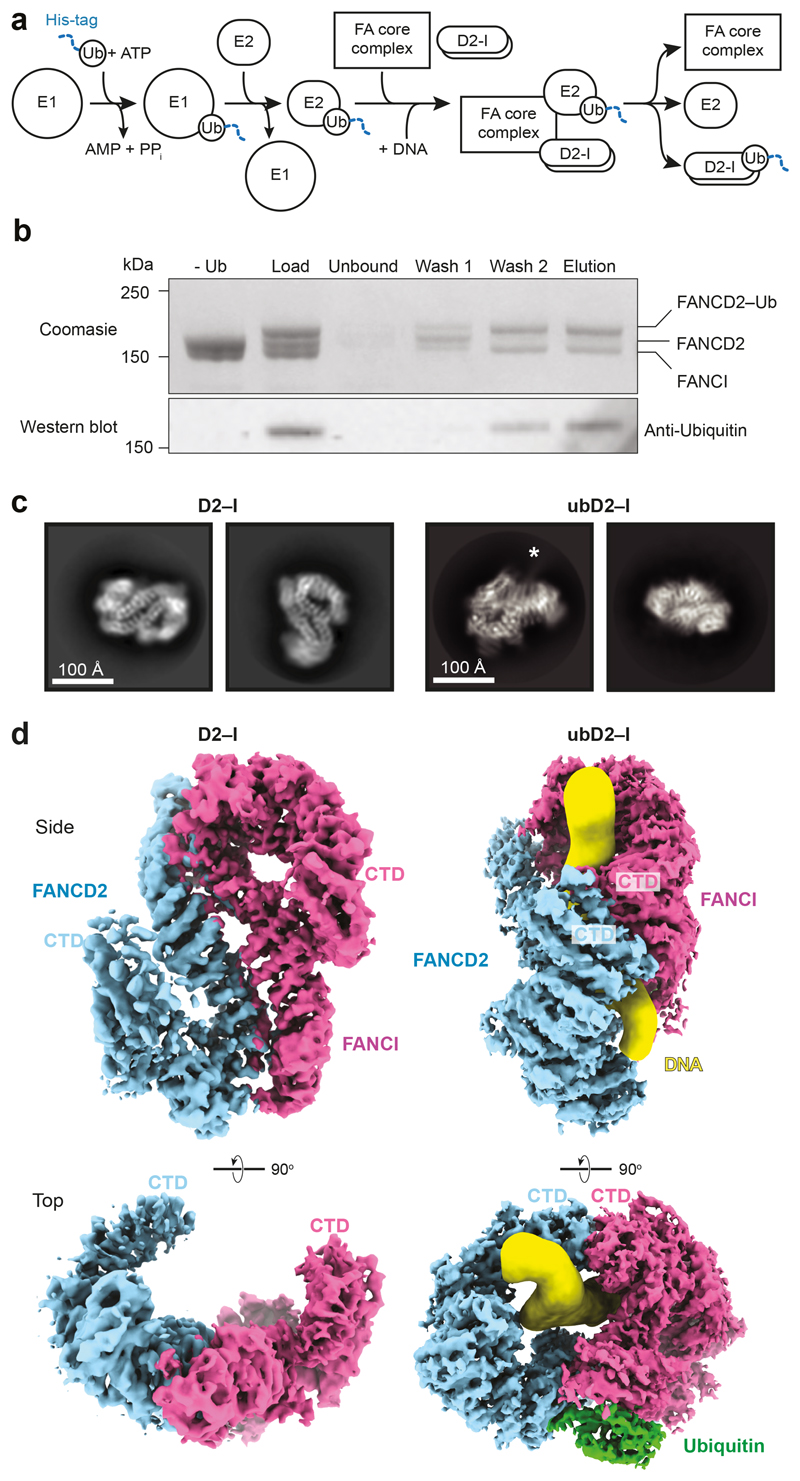
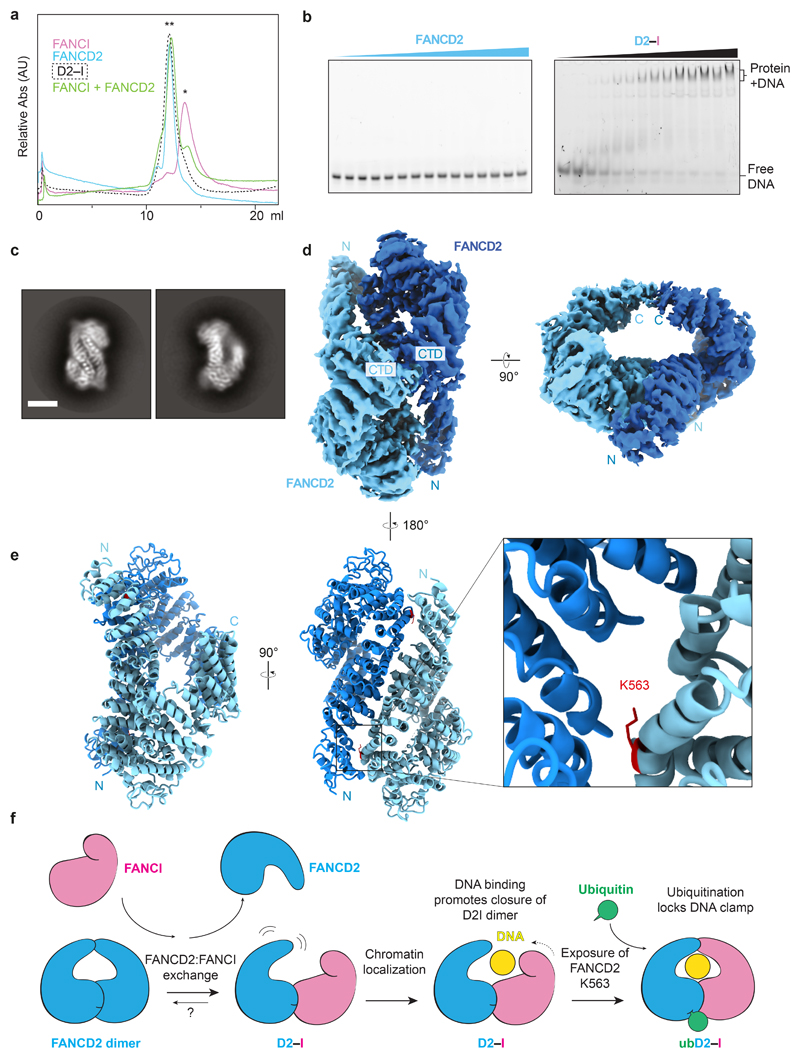
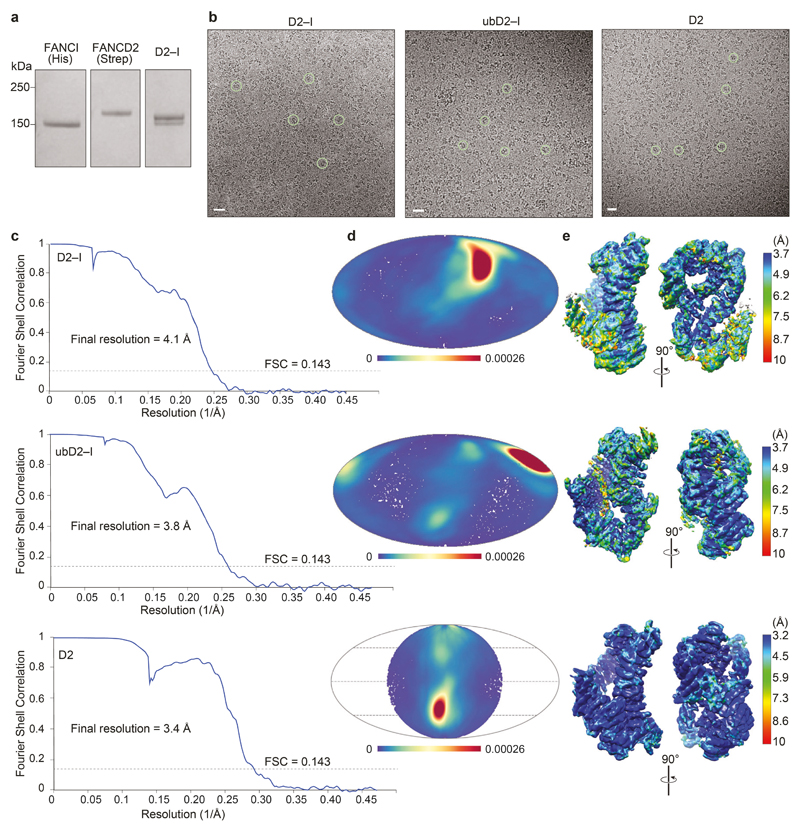
First, we purified a recombinant D2–I complex after co-expression of both proteins in insect cells (Extended Data Fig. 1a). The purified complex functions as a monoubiquitination substrate in vitro (Fig. 1a,b) and interacts with DNA (see below). We used cryoEM to investigate the architecture of unmodified D2–I in the presence of four-fold excess double stalled-fork DNA, and obtained a 3D structure at an overall resolution of ~4.1 Å (Fig. 1c, d; Extended Data Fig. 1b-e; Table 1). We then built homology models of FANCD2 and FANCI, and fitted these into the map (Extended Data Fig. 2a-c). The structure of chicken D2–I is highly similar to the previously-reported crystal structure of the Mus musculus proteins31, in agreement with the high degree of structural and functional conservation of the FA pathway across vertebrates (Extended Data Fig. 2d, Supplementary Table 1; Supplementary Notes 1 and 2). Although the cryoEM sample was prepared in the presence of DNA, DNA was not visible in the maps, consistent with a fast off-rate.
Fig. 1. Purification and structures of D2–I and ubD2–I.
a Scheme for monoubiquitination of D2–I using fully recombinant components. The His-tag on ubiquitin is shown as a blue dashed line. b ubD2–I was enriched from the monoubiquitination mixture by purification of His-tagged ubiquitin on Ni-NTA. The load, unbound fraction, two wash fractions and elution were analyzed by Coomassie-stained SDS-PAGE (top) and Western blotting with an anti-Ubiquitin antibody (bottom). A control reaction lacking ubiquitin is also shown. Uncropped gel and blot are available in Supplementary Fig. 1. These results are representative of experiments performed three times. c Selected 2D reference-free class averages of D2–I (left) and ubD2–I (right). Both samples were prepared with DNA. An asterisk marks density extending from the side of the ubD2–I complex that we assign to DNA. d CryoEM maps of D2–I (left) and ubD2–I (right), segmented into FANCD2 (blue), FANCI (magenta), ubiquitin (green) and DNA (yellow).
Table 1. Cryo-EM data collection, refinement and validation statistics.
| D2–I (EMD-10532, PDB 6TNG) |
ubD2–I (EMD-10531, PDB 6TNF) |
D2 (EMD-10534, PDB 6TNI) |
|
|---|---|---|---|
| Data collection and processing | |||
| Magnification | 81,000 X | 81,000 X | 81,000 X |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e–/Å2) | ~70 | ~70 | ~70 |
| Defocus range (μm) | -1.8 to -3.5 | -1.8 to -3.5 | -1.8 to -3.5 |
| Pixel size (Å) | 1.11 (LMB) | 1.06 (eBIC), 1.11 (LMB) | 1.11 (eBIC) |
| Symmetry imposed | C1 | C1 | C2 |
| Initial particle images (no.)* | ~2,000,000 | ~2,500,000 | ~3,000,000 |
| Final particle images (no.) | 171,936 | 146,245 | 901,085 |
| Map resolution (Å) | 4.1 | 3.8 | 3.4 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 4.1 to > 10 | 3.8 to > 10 | 3.4 to > 10 |
| Refinement | |||
| Initial model used (PDB code) | 3S4W | 3S4W | 3S4W |
| Model resolution (Å) | 4.1 | 3.8 | 3.4 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Model resolution range (Å) | n/a | n/a | n/a |
| Map sharpening B factor (Å2) | -103 | -77 | -108 |
| Model composition | |||
| Non hydrogen atoms | 8,241 | 10,194 | 11,625 |
| Protein residues | 2,060 | 2,206 | 2,134 |
| Ligands | 0 | 0 | 0 |
| B factors (Å2) | |||
| Protein | not estimated | not estimated | not estimated |
| Ligand | |||
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.008 | 0.008 | 0.008 |
| Bond angles (°) | 1.39 | 1.41 | 1.40 |
| Validation | |||
| MolProbity score | 2.25 | 2.19 | 2.81 |
| Clashscore | 10.0 | 8.76 | 4.51 |
| Poor rotamers (%) | 0.00 | 0.00 | 15.63 |
| Ramachandran plot | |||
| Favored (%) | 80.22 | 81.51 | 84.33 |
| Allowed (%) | 18.94 | 17.83 | 15.19 |
| Disallowed (%) | 0.84 | 0.65 | 0.47 |
Initial number of particles after autopicking. This number includes many blank picks.
As previously reported31, the FANCD2 and FANCI proteins share a striking structural similarity containing a series of ɑ-helices arranged into four superhelical ɑ-solenoid structures (S1–S4) and two helical domains (HD1, HD2). They fold into an overall shape akin to two antiparallel saxophones. In this arrangement, the ubiquitination sites (D2K563 and IK525 for the chicken proteins) are buried within the dimerization interface, indicating that remodeling of the complex is very likely required for access by the FA monoubiquitin ligase complex. In the cryoEM structure, the C-terminal domain of FANCD2 was at a lower resolution due to flexibility of this region (Fig. 1d). Flexibility of the C-terminal domains was also observed in a previous negative-stain EM study32.
CryoEM of ubD2–I reveals that it is a DNA clamp
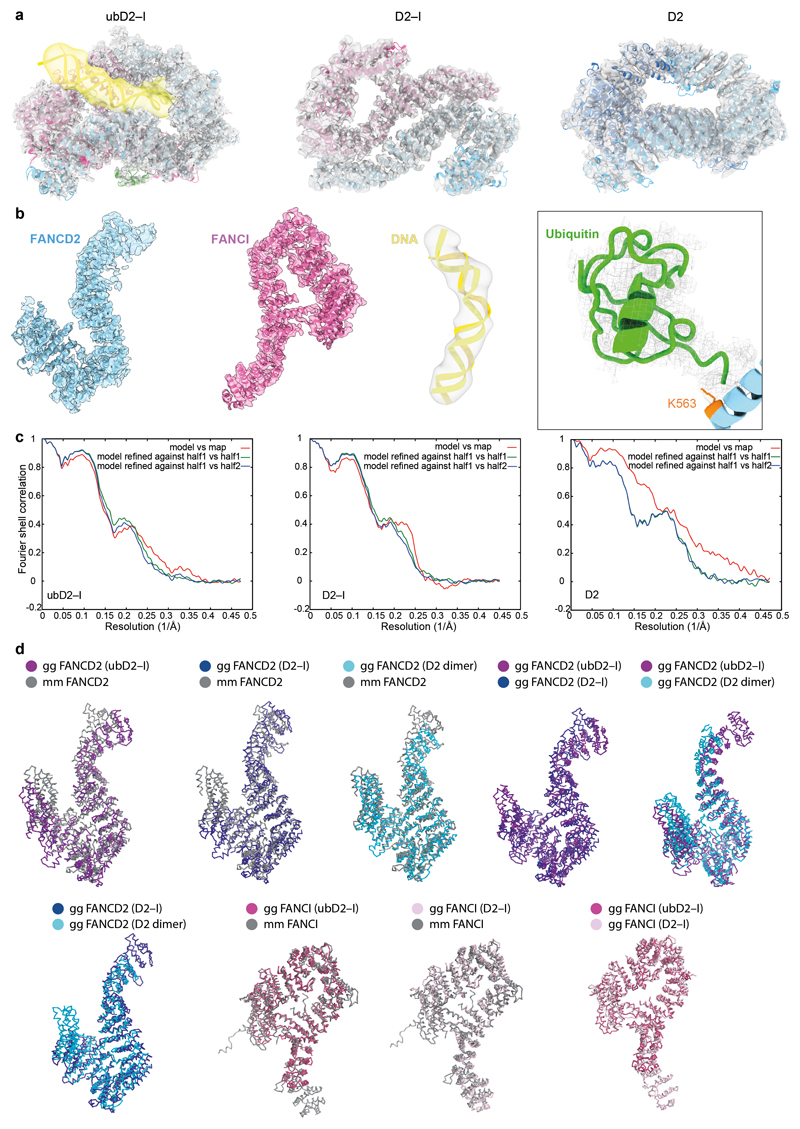
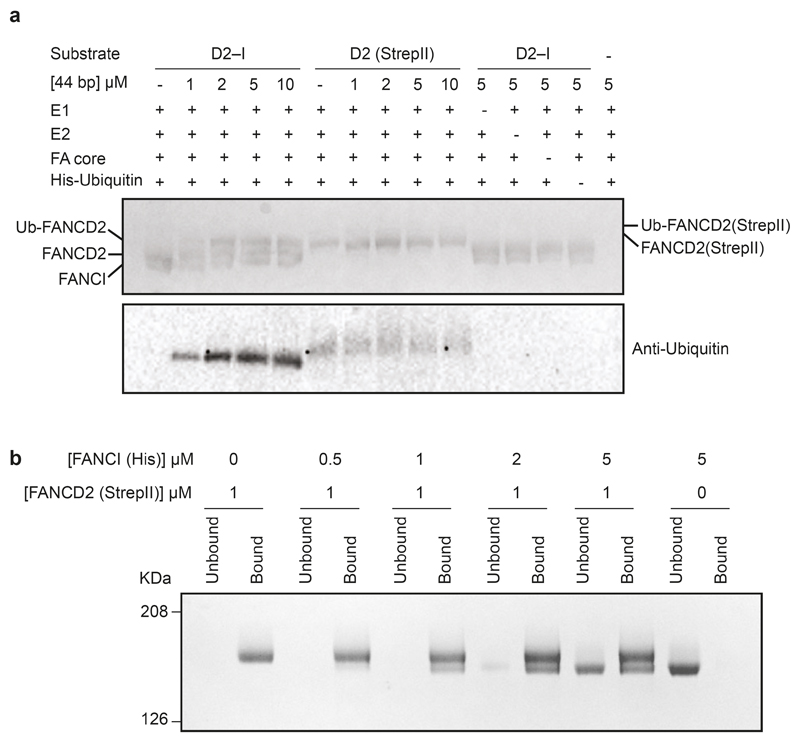
Next, to purify a monoubiquitinated complex, we incubated D2–I with His-tagged ubiquitin, E1, E2 (UBE2T), recombinant FA core complex29 and DNA (Fig. 1a). We used a double stalled-fork DNA, but other DNA structures are also effective in stimulating monoubiquitination8,28. FANCD2 was specifically monoubiquitinated on K563, but FANCI was not ubiquitinated, as previously observed29. We enriched for the ubiquitinated complex by affinity chromatography using the His-tag on ubiquitin (Fig. 1b). Using cryoEM, we obtained a 3D structure of ubD2–I at an overall resolution of ~3.8 Å (Fig. 1c, d; Extended Data Fig. 1b-e; Table 1), allowing us to build models for FANCD2, FANCI and ubiquitin (Fig. 2a; Extended Data Fig. 2).
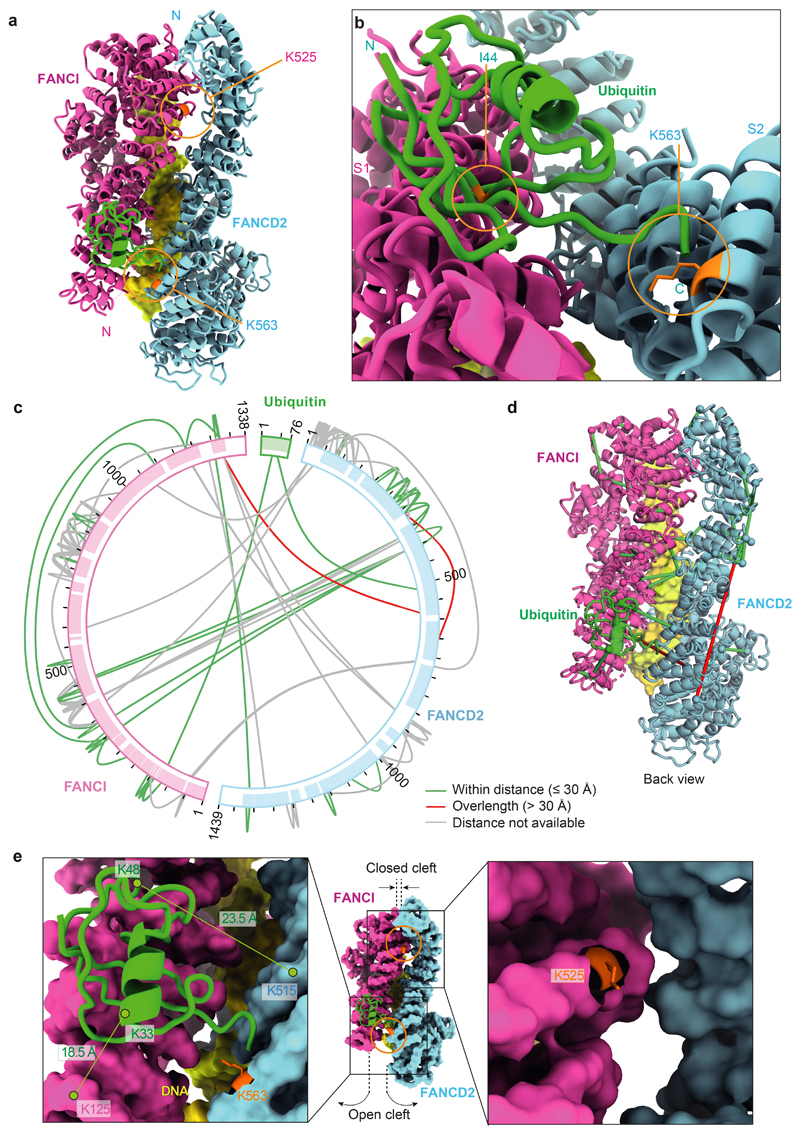
Fig. 2. Ubiquitin is anchored to K563 on FANCD2 but also contacts FANCI.
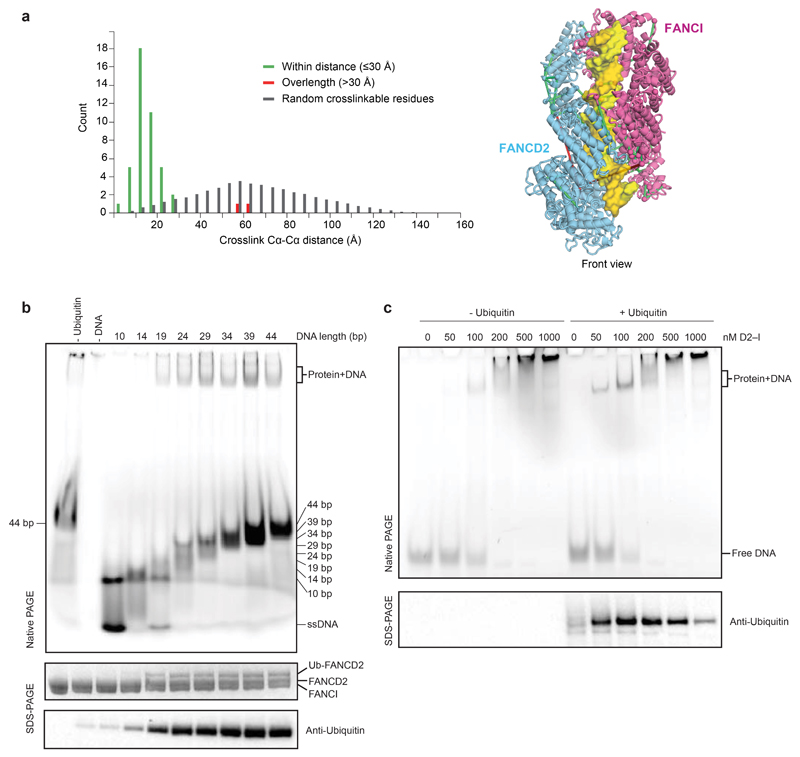
a The ubiquitin moiety attached to FANCD2 makes extensive contacts with FANCI. b Close-up view of the monoubiquitination site. Ile44 (orange) of ubiquitin is shown. c Map of crosslinks identified in the ubD2–I complex. Crosslinking mass spectrometry revealed 122 crosslinks (1% false discovery rate) between residues that are in close proximity. Crosslinks are colored by Cα-Cα distance between linked residue pairs measured in the 3D model of ubD2–I. Two crosslinks are not compatible with the model but are consistent with the flexibility observed within this complex (Supplementary Video 1). Proteins are shown as curved bars and residues that are present in the 3D model are highlighted. d Crosslinks within expected distance (green) and exceeding expected distance (overlength, red) mapped onto the ubD2–I structure. e Details of lysines K563 in FANCD2 (left) and K525 in FANCI (right) within the ubD2–I structure, shown in sticks for lysines (orange), as a surface representation of the model for FANCD2 (blue) and FANCI (magenta), and in cartoon for ubiquitin (green). The residues crosslinked between ubiquitin and FANCD2 and FANCI are labeled.
The structure of ubD2–I is markedly different from the unmodified complex. Symmetry was not obvious in 2D class averages (Fig. 1c), but the 3D reconstruction showed that the overall ubD2–I complex retains some pseudo-symmetric features (Fig. 1d). In the monoubiquitinated form, FANCD2 and FANCI are folded towards each other so the C-terminal domains of each monomer (akin to the bells of the saxophone), wrap around density that we assigned to double-stranded DNA (see below). In agreement with subunit flexibility and remodeling of the complex, multi-body refinement of ubD2–I showed that there is variability in the relative positions of FANCD2 and FANCI (Supplementary Video 1).
DNA binding exposes the FANCD2K563 monoubiquitination site at the dimer interface
Superimposition of the D2–I and ubD2–I models showed that the dimer interface acts as a hinge, allowing the C-termini of FANCD2 and FANCI to swing towards each other, rotating by almost 70° in the monoubiquitinated complex (Supplementary Videos 2-4). This is largely a rigid body movement of both subunits, but the N- and C-terminal regions of FANCD2 are also remodeled (Fig. 1d; Extended Data Fig. 2d). This conformational change opens a cleft to expose the K563 monoubiquitination site in FANCD2 on the backside of the hinge, where we unambiguously identified density for ubiquitin in the cryoEM map (Fig. 1d; Extended Data Fig. 2b). Ubiquitin is covalently anchored onto FANCD2 (Fig. 1b) but it docks onto the surface of FANCI via the hydrophobic Ile44 patch on ubiquitin, and likely stabilizes the complex (Fig. 2a-b). Crosslinking mass spectrometry also showed that ubiquitin is in close proximity to both FANCD2 and FANCI (Fig. 2c-e; Extended Data Fig. 3a; Supplementary Table 2).
In contrast, the FANCI monoubiquitination site (K525) is only partially exposed because the cleft where it is located remains closed (Fig. 2e), explaining why it is inefficiently monoubiquitinated with recombinant or native FA core complex in vitro8,9,29 (Fig. 1b). In cells, FANCI monoubiquitination is stimulated by phosphorylation (which may further open up the dimerization interface31) but it is not essential for DNA crosslink repair12,25.
The C-terminal domains of FANCD2 and FANCI form a channel to embrace DNA
The C-terminal solenoids (S4) of FANCD2 and FANCI, as well as additional unmodeled C-terminal regions, mediate contacts at the new interface (Fig. 3a,b). These interactions likely stabilize the C-terminal region of FANCD2, which is flexible in D2–I but becomes well-ordered in the ubD2–I map (Fig. 1d).
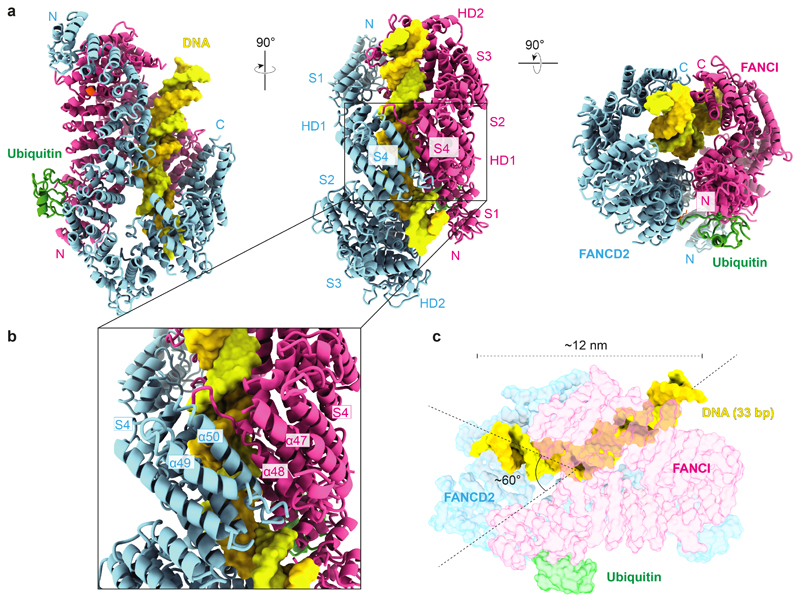
Fig. 3. ubD2–I is a DNA clamp.
a Models for FANCD2 (blue), FANCI (magenta) and ubiquitin (green) built into the ubD2–I cryoEM map. The N- and C-termini, solenoids 1–4 (S1–S4) and helical domains (HD1, HD2) of FANCD2 and FANCI are indicated. The monoubiquitinated lysine in FANCD2 and the lysine that can be monoubiquitinated in FANCI are shown in orange. b Close-up view of the C-terminal domains (S4) of ubD2–I that clamp around DNA. c Model of 33 bp double-stranded DNA (yellow) in the ubD2–I map. DNA is kinked within the ubD2–I complex.
The repositioned C-terminal domains of FANCD2 and FANCI in the mono-ubiquitinated complex form a new channel between the two protomers. In agreement with an important role for the C-terminal domain of FANCD2, deletion of residues 1147–1451 in the human protein results in loss of FANCD2 monoubiquitination in vitro and in cells, loss of in vitro DNA binding, and disruption of crosslink repair33. The channel contained density for ~15–20 bp double-stranded DNA (Fig. 1d). The DNA extends beyond the channel and we could model a total of 33 bp (Fig. 3c). The DNA was at lower resolution than the rest of the map, likely because of heterogeneity in the DNA position, and inherent flexibility. DNA makes contacts with both proteins, but it appears to be shifted towards FANCI, adopting a binding mode similar to that in a previously reported low-resolution FANCI–DNA crystal structure31. Contacts are likely mediated largely through interactions between the negatively-charged phosphate backbone and positively-charged surface residues31. Intriguingly, the bound DNA is kinked next to the C-terminal domains within the ubD2–I complex (Fig. 3c).
Ubiquitin is a covalent molecular pin that locks the ubD2–I clamp on DNA
D2–I interacts with DNA31,33 and DNA is required for monoubiquitination of D2–I in vitro8,28, but the role of DNA in this reaction was unclear. Our data suggested that DNA binding is required to close the D2–I clamp and expose the monoubiquitination site. To further investigate this, we performed monoubiquitination assays of D2–I in the presence of a series of double-stranded DNAs of varying lengths. We used linear double-stranded DNA since this allowed us to specifically analyze the length dependence. D2–I was monoubiquitinated in the presence of a 19 bp DNA, but not with a 14 bp DNA (Extended Data Fig. 3b). We also analyzed protein–DNA interactions within these reactions by loading the monoubiquitination assays performed in the presence of different DNAs directly onto native gels for analysis by electrophoretic mobility shift assays (EMSAs). D2–I did not bind to a 14 bp DNA, but interacted efficiently with DNA substrates that were 19 bp or longer (Extended Data Fig. 3b). Therefore, these biochemical data show that the minimum DNA length required for ubiquitination (15–19 bp) correlates with the minimum length required for D2–I binding. This length also agrees well with the length of DNA encircled by ubD2–I in the cryoEM structure.
Together, our structures and biochemical studies explain why and how DNA stimulates FANCD2 monoubiquitination: D2–I binds double-stranded DNA, promoting closure of the heterodimer to expose the K563 monoubiquitination site in FANCD2 and allow access to the activated E2 enzyme. We also hypothesized that monoubiquitination of FANCD2 on the backside of the hinge acts as a covalent molecular pin, locking the closed heterodimer on DNA and preventing it from rotating back to the open D2–I conformation. In agreement with this, monoubiquitination results in a small, but reproducible, increase in DNA binding affinity, possibly due to a slower off-rate (Extended Data Fig. 3c).
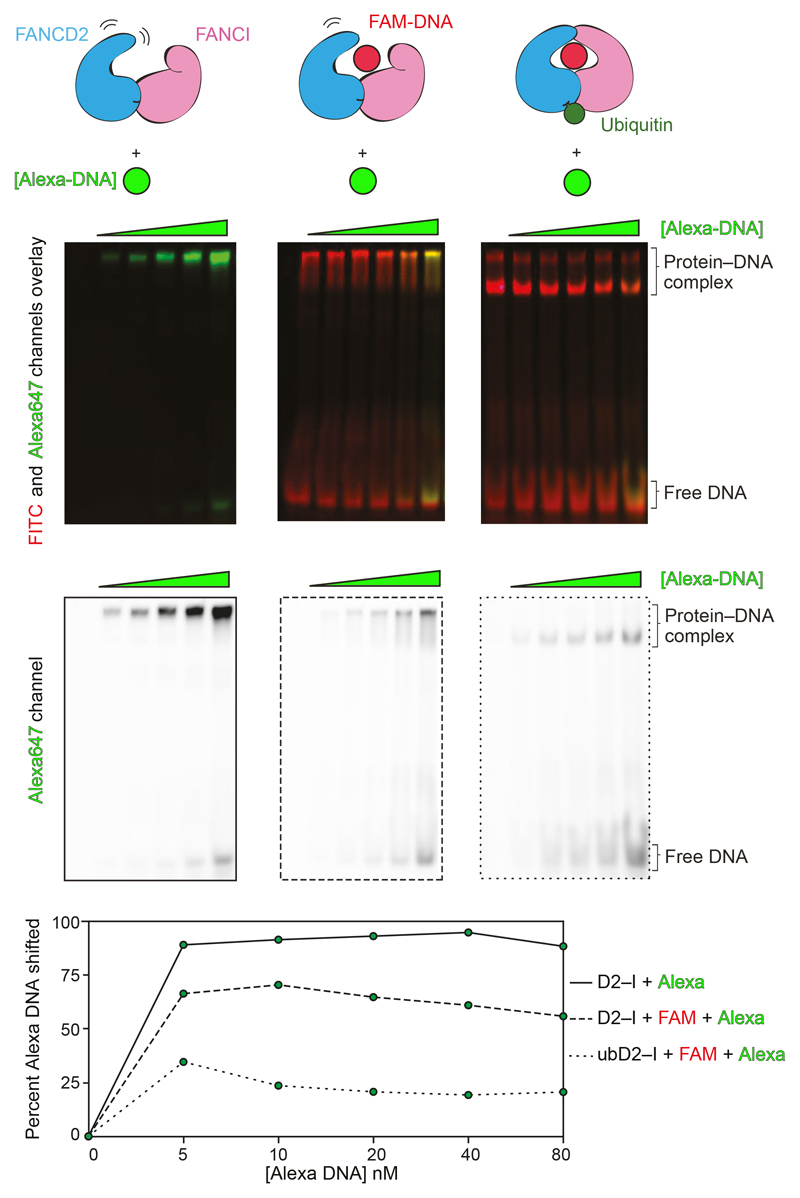
To test whether ubiquitin slows the off-rate and locks the complex on DNA, we performed two-color DNA displacement assays: we incubated D2–I with FAM-labeled DNA and then tested the ability of a second (Alexa-labeled) DNA to be incorporated into D2–I by displacing the FAM-DNA. We found that in the absence of ubiquitination, the second DNA is readily incorporated into D2–I (Fig. 4). In contrast, when FAM-DNA is used to form the ubD2–I complex, a much smaller amount of the competing DNA is incorporated (Fig. 4). Together, these experiments provide evidence that ubiquitin locks the D2–I complex on DNA, effectively slowing the off-rate.
Fig. 4. Monoubiquitination locks the D2–I clamp onto DNA.
A 44-bp FAM-labeled dsDNA (red) was incubated with D2–I (30 nM) or used to form ubD2–I complex (30 nM). Then, an Alexa-labeled DNA (green) of the same length and sequence was added in increasing concentrations (0–80 nM) to these complexes. Overlays of the FITC and Alexa647 channels (top) and the Alexa647 channel alone (middle) are shown.. The intensities of total Alexa-DNA and the fraction incorporated into the complex (shifted) were quantified and the percentage of shifted Alexa-DNA was plotted in the bottom panel. These data are representative of experiments performed twice. Uncropped gels are available in Supplementary Fig. 1. Data for plot is available as source data.
Isolated FANCD2 forms a closed, auto-inhibited homodimer
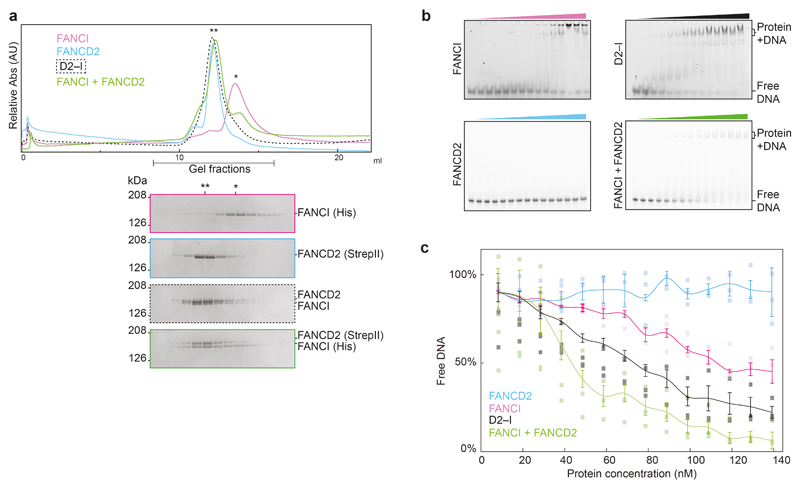
A major remaining question was why FANCD2 was not monoubiquitinated in vivo or in vitro in the absence of FANCI8,16,28. To address this, we purified FANCD2 and FANCI separately. Purified FANCI ran as a monomer on gel filtration chromatography whereas, unexpectedly, FANCD2 ran as a dimer (Fig. 5a; Extended Data Fig. 4a). Interestingly, a recent study showed that purified human FANCD2 also runs as a species of larger size compared to human FANCI, likely corresponding to a dimeric FANCD234.
Fig. 5. FANCI forms a monomer and binds DNA whereas FANCD2 is dimeric and does not bind DNA.
a Size exclusion chromatography analysis of purified FANCI, FANCD2, D2–I, and FANCI mixed with FANCD2. A single asterisk (*) indicates the migration position for monomers. A double asterisk (**) indicates the migration position for dimers. This experiment was performed three times and a representative chromatogram is shown. b DNA binding of FANCD2 and D2–I was analyzed by EMSAs performed with 20 nM 39 bp double-stranded DNA and 0–140 nM protein. Representative gels of experiments independently performed three times are shown. Uncropped gels are available in Supplementary Fig. 1. c Selected 2D reference-free class averages of FANCD2. Scale bar is 100 Å. d CryoEM map of FANCD2 homodimer. The locations of the N- and C-termini are marked. e Model of FANCD2 dimer. The buried K563 residue (red) on FANCD2 is shown in close-up. f Model for regulation of FANCD2 and FANCI in DNA crosslink repair. Isolated FANCD2 purifies as a homodimer that is closed, does not bind DNA, and is not monoubiquitinated. Upon incubation with purified (monomeric) FANCI, this exchanges into a D2–I complex with an open conformation. D2–I binds and encircles DNA, converting the complex into a closed conformation, and thereby acting as a DNA clamp. The ubiquitination site on FANCD2 is exposed in the closed conformation, allowing access to the FA core complex and E2 enzyme. Ubiquitin locks the D2–I clamp in a closed conformation so it is not readily released from DNA.
Next, we assessed the DNA binding capacities of each protein and found that FANCI bound DNA efficiently but FANCD2 did not interact with DNA at any of the concentrations we tested (Fig. 5b; Extended Data Fig. 4b-c). Moreover, isolated FANCD2 was not substantially monoubiquitinated by the FA core complex, and this could not be stimulated by DNA (Extended Data Fig. 5a).
To understand the molecular basis of FANCD2 dimerization and gain insight into why it did not bind DNA, we determined the structure of the FANCD2 homodimer using cryoEM to ~3.4 Å resolution (Fig. 5c-e; Extended Data Fig. 1; Extended Data Fig. 2a, c). The FANCD2 dimer was symmetric, with each protomer containing the same overall domain architecture as in the heterodimeric complex. Surprisingly, the FANCD2 homodimer was in a closed conformation, more similar to ubD2–I than to D2–I. This provides a molecular explanation for the lack of DNA binding by isolated FANCD2: The DNA binding sites were blocked because it was in a closed conformation that presumably does not open to allow entry of a double-stranded helix. In addition, FANCD2 homodimerization occludes K563 on both protomers, likely preventing their ubiquitination (Fig. 5e).
The FANCD2 homodimer was stable over a gel filtration column, indicative of a high affinity interaction. To investigate how the heterodimeric D2–I complex forms, we mixed purified FANCD2 and FANCI and analyzed their oligomerization state. We found that after incubation with FANCD2, the migration position of FANCI on a gel filtration column shifted to the position of a dimer (Fig. 5a; Extended Data Fig 4a). This suggested that the monomeric FANCI displaced one of the FANCD2 protomers in the homodimer. To definitively address this, we immobilized FANCD2 on Streptavidin resin and incubated it with untagged FANCI. After washing, FANCI was bound to the beads in approximately 1:1 stoichiometric ratio with FANCD2 (Extended Data Fig. 5b). This is consistent with FANCI displacing one of the FANCD2 protomers to form the D2–I complex.
Discussion
Until now, the precise role of DNA in monoubiquitination of FANCD2 was unknown. It had also remained unclear how the monoubiquitinated lysine on FANCD2 was accessed by UBE2T and the FA core complex, since it was occluded at the interface of FANCD2 and FANCI in a previous crystal structure31. Here, our structural and biochemical analyses of FANCD2 and FANCI allow us to propose a new model for how DNA crosslink repair is initiated (Fig. 5f): D2–I binds DNA at stalled replication forks, and this causes a hinge-like rotation of FANCD2 and FANCI so they encircle double-stranded DNA (Fig. 5f). Transition into this closed conformation exposes a region on the back of the FANCD2 hinge, allowing the FA core complex and UBE2T to access the monoubiquitination site. Ubiquitin then acts as a covalent molecular pin to lock the ubD2–I DNA clamp into the closed conformation. We are not aware of any other examples where covalently-linked ubiquitin locks a new protein conformation. Thus, our analysis uncovers a new function for ubiquitination.
Unexpectedly, we find that recombinant FANCD2 alone purifies as a homodimer. A recent study showed that purified human FANCD2 migrates as a larger species than human FANCI on a gel filtration column, consistent with human FANCD2 also forming a homodimeric complex34. Here, we show that the FANCD2 homodimer adopts a closed conformation that is unable to engage DNA. It cannot be ubiquitinated because the monoubiquitinated lysines are occluded at the dimer interface. Upon incubation with FANCI, the FANCD2 homodimer readily exchanges into a stable D2–I heterodimer, and adopts an open conformation, permissive for DNA binding. Thus, our results suggest a mechanism of autoinhibition wherein isolated FANCD2 cannot be prematurely recruited to DNA and cannot be monoubiquitinated prior to assembly with FANCI. Since the D2–I heterodimer is favored over the FANCD2 homodimer, it is unlikely that there would be substantial amounts of monomeric FANCI and homodimeric FANCD2 in the cell. However, it remains possible that the dynamics of D2–I association may be modulated by post-translational modifications.
In the physiological context, it is not known if FANCD2 homodimers act in a regulatory capacity to control the amount of D2–I complex in the cell, or if homodimerization is simply a mechanism to sequester any excess FANCD2 if there is an imbalance in the ratio of FANCD2:FANCI proteins produced. Nevertheless, when endogenous FANCD2 is purified from cells, it does not come down with equivalent levels of FANCI12,35. It is therefore possible that the FANCD2–FANCI interaction may be regulated and that a fraction of endogenous FANCD2 is not heterodimerized with FANCI, in agreement with previous data35. FANCI may have additional functions to those shared with FANCD2 in DNA repair, supporting the hypothesis that the proteins might exist as distinct entities36,37. Future experiments will be required to test whether FANCD2 homodimerization regulates the FA pathway.
In monoubiquitination assays, both chicken, human and murine D2–I heterodimers show little selective preference for different forms of double-stranded DNA8,9,28,31,38, although DNA binding assays show a modest preference for certain DNA structures31,33. Thus, it is unclear whether the D2–I dimers are specifically recruited by replication fork structures, crosslinked DNA, the stalled replisome, or other factors. Since the FA pathway plays a more general role in response to stalled replication forks, D2–I is likely to bind a protein or DNA structure common to all pathways. We now show that the ubD2–I dimer locks into a clamp around double-stranded DNA with a footprint of ~20 nt. Repair of a DNA crosslink in a Xenopus cell-free system showed that there is a careful choreography of events that occur after two replication forks converge on a crosslink10,39. The forks stall at a distance of 20–40 nt from the crosslink, and subsequently, one fork advances until it reaches the nucleotide preceding the crosslink. At this point the FANCP–FANCQ nuclease complex cuts around the crosslink (unhooking) on one strand, and this step is abolished when D2–I is depleted10,14,15,40.
We suggest a model whereby D2–I is primed to interact with DNA, possibly when a stalled replication fork is ~20 nt from the crosslink. This could signal to the FA core complex to monoubiquitinate D2–I, locking it around DNA just adjacent to the crosslink site. ubD2–I kinks the double-stranded DNA helix, possibly providing distortion that marks the crosslinked DNA substrate for the nuclease incision complex. Importantly, the ubiquitination of FANCD2 alone is sufficient to lock the D2–I dimer on DNA, consistent with genetic evidence that FANCI monoubiquitination is not required for crosslink repair12. ubD2–I is thought to directly recruit the incision complex10,18, but curiously, the Ile44 patch of ubiquitin that interacts with ubiquitin-binding domains is buried at the interface of ubiquitin and FANCI (Fig. 2b). The covalently-linked ubiquitin could swing out to expose the Ile44 patch, allowing recognition of ubiquitin, or alternatively, the incision complex may recognize an alternate solvent-exposed interface on ubD2–I. Future studies will aim to test these models.
Finally, our work provides an explanation for why it is essential to de-ubiquitinate FANCD2 for the completion of crosslink repair. Genetic evidence in chicken DT40 cells and in mice indicated that knockouts of USP1 lead to the persistence of ubiquitinated FANCD2 tethered to chromatin21,41. This aborts crosslink repair. Since ubiquitin locks the ubD2–I clamp on DNA, ubiquitin must be removed for the clamp to open, releasing the DNA. Failure to carry out this step would lead to the persistence of the clamp on DNA which could promote uncontrolled cutting by the incision complex, prevent repair after incision, or possibly create a protein–DNA obstacle to transcription or replication of DNA. The structures of activated D2–I reported here, as well as the recent structure of the FA core complex29, set the stage for future work to ultimately obtain a complete biochemical and structural understanding of DNA interstrand crosslink repair.
Methods
Cloning, expression and purification
cDNAs encoding full length Gallus gallus FANCI and FANCD2 were synthesized (GeneArt) and cloned into pACEBac1. The individual genes were amplified by PCR and cloned into pBIG1a vector using a modified version of the biGBac system, as previously described42,43. The combined vector was transformed into EMBacY E. coli competent cells for bacmid generation. The purified bacmid was then transfected into Sf9 cells. The virus was passaged twice before a large-scale culture was infected (5 ml of P2 virus into 500 ml of Sf9 cells at 2 million cells/ml). Cell growth and viability were monitored and cells harvested upon growth arrest (typically on day 3 after infection). A similar strategy was used for individual expression of FANCD2 and FANCI except that the FANCD2 gene contained a C-terminal extension with a 3C protease site and double StrepII tag whereas the FANCI gene contained a C-terminal extension with a 3C protease site and 6x His tag.
For D2–I we took advantage of an excellent protein overexpression to devise a tag-free protein purification protocol based on sequential fractionation combining cation exchange and affinity chromatography. Cells were lysed by sonication in lysis buffer (100 mM HEPES pH 7.5, 300 mM NaCl, 1 mM TCEP, 5% glycerol, EDTA-free protease inhibitor, 5 mM benzamidine hydrochloride and 100 U/ml benzonase). Clarified cell lysate was passed through a HiTrap SP HP cation exchange chromatography column (GE Healthcare Life Sciences) to remove impurities. Flow-through containing the unbound D2–I complex was diluted to 150 mM NaCl concentration and loaded onto a HiTrap Heparin HP affinity column (GE Healthcare Life Sciences). Using a shallow NaCl gradient, the D2–I complex eluted around ~ 500 mM NaCl concentration. The complex was run on a Superdex 200 26/60 column (GE Healthcare Life Sciences) in 50 mM HEPES pH 7.5, 150 mM NaCl and 1 mM TCEP. The fractions containing the complex were pooled and concentrated to ~10 mg/ml and flash frozen for storage at -80 °C. Samples from each step were analyzed by SDS-PAGE using 4-12% NuPAGE Bis-Tris gels (ThermoFisher Scientific) to monitor the purification.
His-tagged FANCI was purified by sequential affinity and size exclusion chromatography. Clarified cell lysate produced as for D2–I was loaded onto a HisTrap HP column (GE Healthcare Life Sciences). Using an imidazole gradient, FANCI eluted around ~200 mM imidazole concentration. Collected fractions containing FANCI were diluted to ~100 mM NaCl concentration and loaded onto a HiTrap Heparin HP affinity column (GE Healthcare Life Sciences). Using a shallow NaCl gradient, FANCI eluted around ~500 mM NaCl concentration. FANCI was then run on a Superdex 200 26/60 column (GE Healthcare Life Sciences) in 50 mM HEPES pH 7.5, 150 mM NaCl and 1 mM TCEP. The fractions containing FANCI were pooled and concentrated to ~10 mg/ml and flash frozen for storage at -80 °C.
StrepII-tagged FANCD2 was also purified by sequential affinity and size exclusion chromatography. Clarified cell lysate produced as for D2–I and FANCI was incubated with StrepTactin Sepharose High Performance resin (GE Healthcare Life Sciences) for 60 min. The loaded resin was poured into a glass column and washed twice with lysis buffer before elution with 8 mM D-Desthiobiotin. The elution was then diluted to ~100 mM NaCl concentration and loaded onto a HiTrap Heparin HP affinity column (GE Healthcare Life Sciences). Using a shallow NaCl gradient, FANCD2 eluted around ~500 mM NaCl concentration. FANCD2 was then run on a Superdex 200 26/60 column (GE Healthcare Life Sciences) in 50 mM HEPES pH 7.5, 150 mM NaCl and 1 mM TCEP. The fractions containing FANCD2 were pooled, concentrated to ~10 mg/ml and flash frozen for storage at -80 °C. Throughout purification, we routinely monitored the absorbance at 260 nm and 280 nm. The A260 nm/ A280 nm ratio shows that there is no substantial nucleic acid contamination in the D2–I, FANCD2, or FANCI purifications.
In vitro ubiquitination and purification of ubiquitinated D2–I
Based on previously described ubiquitination assays8,28, we set up a large-scale in vitro reconstitution of the FA core complex-mediated ubiquitination of D2–I. In a total volume of 400 μl, the reaction contained 75 nM E1 ubiquitin activating enzyme (Boston Biochem), 0.8 μM E2 (UBE2T)8, 0.5 μM E3 (FA core complex)29, 1 μM D2–I, 5 μM DNA (double stalled-fork generated by annealing oligonucleotides X1, X2, X3, X4, X5, X6 (Supplementary Table 3)), and 20 μM His-tagged ubiquitin (Enzo Life Sciences) in a reaction buffer of 50 mM HEPES pH 7.5, 64 mM NaCl, 4% glycerol, 5 mM MgCl2, 2 mM ATP and 0.5 mM DTT. The reaction was incubated at 30 °C for 90 min before applying it to 50 μl of Ni-NTA agarose resin (Qiagen) pre-equilibrated in W25 buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP and 25 mM imidazole) in a 1.5 ml centrifuge tube. The resin was incubated under constant rotation at 4 °C for 60 min. The resin was washed twice, first with 400 μl of W25 buffer followed by a wash with 400 μl of W50 buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP and 50 mM imidazole). Each wash was performed for 60 min at 4 °C under rotation. The Ni-NTA bound ubD2–I complex was eluted with W100 buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP and 100 mM imidazole). Samples from each step were analyzed by SDS-PAGE to monitor the purification of ubD2–I complex. Presence of ubiquitin was confirmed by Western blot using anti-Ubiquitin (Millipore; Cat # 07-375). W100 elution fractions were then concentrated using a Vivaspin column (30 kDa MWCO) to a final volume of ~20 μl and ~500 nM concentration.
Electrophoretic mobility shift assays (EMSAs)
Fluorescently-labeled DNA (6FAM- or Alexa647-labeled on 3′ end, purchased from IDT) was prepared by incubating complementary oligonucleotides (Supplementary Table 3) at 95 °C for 5 min and slowly cooling down to room temperature over ~2 h. For EMSAs, a 20 μl reaction containing 20 nM DNA was incubated with the indicated concentration of protein in the presence of 50 mM HEPES pH 8.0, 150 mM NaCl and 1 mM TCEP. The reactions were incubated for 30 min at 22 °C. After incubation, 5 μl were directly loaded on a native polyacrylamide gel (6% DNA Retardation, Thermo Fisher) and run at 4 °C using 0.5 X TBE buffer for 60 min. The gel was then visualized using a Typhoon Imaging System (GE Healthcare). Each binding experiment was repeated two or three times (as indicated in figure legends) and ImageJ44 was used for quantification of percent DNA shifted (Fig. 4) or the mean intensity of free DNA and standard deviation between the measurements (Extended Data Fig. 4c).
Gel filtration assays
A volume of 300 μl of purified D2–I, FANCD2 and FANCI at 1 μM were sequentially run on a Superdex 200 10/300 column (GE Healthcare Life Sciences) equilibrated in 50 mM HEPES pH 7.5, 150 mM NaCl and 1 mM TCEP. To investigate the exchange of FANCD2 and FANCI, we mixed and incubated 0.5 μM FANCD2 with 1 μM FANCI for 30 min at room temperature in a total volume of 300 μl. The mix was then run on a Superdex 200 10/300 column (GE Healthcare Life Sciences) equilibrated in 50 mM HEPES pH 7.5, 150 mM NaCl and 1 mM TCEP. Fractions were analyzed by SDS-PAGE using 4–12% NuPAGE Bis-Tris gels (ThermoFisher Scientific).
Protein binding assays
We used purified StrepII-tagged FANCD2 and His-tagged FANCI to probe their interaction. In a total volume of 50 μl per reaction, FANCD2:FANCI at molar ratios 1:0, 1:0.5, 1:1, 1:2, 1:5 and 0:5 were mixed in 50 mM HEPES pH 7.5, 150 mM NaCl and 1 mM TCEP and incubated for 15 min at room temperature. Each reaction was applied to 20 μl of StrepTactin Sepharose High Performance resin (GE Healthcare Life Sciences) equilibrated in the same buffer. The loaded resin was then incubated for 30 min at 4 °C. The unbound fraction was removed and the resin further washed twice using 250 μl of the same buffer. The bound fraction was analyzed by SDS-PAGE using 4–12% NuPAGE Bis-Tris gels (ThermoFisher Scientific).
Electron microscopy and image processing
Three microliters of ~1 μM ubD2–I, D2–I or D2 complex were blotted on plasma cleaned UltraAufoil R1.2/1.3 grids45 (Quantifoil) for 3–4.5 s and plunged into liquid ethane using a Vitrobot Mark IV. D2–I samples were prepared with four-fold excess double stalled-fork DNA (generated by annealing oligonucleotides X1, X2, X3, X4, X5, X6 (Supplementary Table 3)). The grids were imaged on a Titan Krios using a Gatan K3 detector. All the data collected at eBIC was in super-resolution mode and all the data collected at LMB was in counting mode. Additional data was collected for ubD2–I at a tilt of 23° to overcome preferred orientation.
All image processing was performed using Relion3.0/3.146 unless otherwise stated. The images were drift-corrected using MotionCorr247 and defocus was estimated using CTFFIND448. Particles were initially picked manually and 2D classified. Selected classes from the 2D classification were used to autopick particles from the full datasets. After 2–3 rounds of 2D classification, classes with different orientations were selected for initial model generation in Relion. The initial model was used as reference for 3D classification into 4 classes. The selected classes from 3D classification were subjected to auto-refinement with solvent flattening using a soft mask. The defocus values were further refined using CTF Refinement in Relion followed by Bayesian polishing. Another round of auto-refinement was performed on these polished particles. All maps were post-processed to correct for modulation transfer function of the detector and sharpened by applying a negative B factor as determined automatically by Relion. A soft mask was applied during post processing to generate FSC curves to yield maps of average resolutions of 3.8 Å for ubD2–I, 4.1 Å for D2–I and 3.4 Å for D2.
To further improve the maps, we used focused classification and refinement of the ubD2–I map by dividing them into FANCI, FANCD2 and DNA regions. The density for DNA in ubD2–I was improved by using focused classification without image alignment.
To analyze the conformational heterogeneity in ubD2–I, we applied multi-body refinement using masks around FANCI and FANCD2 protomers. Two major motions were detected using principal component analysis on the optimal orientations of all the bodies for all particle images in the dataset using relion_flex_analyse49 (Supplementary Video 1).
Modelling
The crystal structure of Mus musculus FANDC2-FANCI (PDB: 3S4W)31 was used as a template to generate a homology model of Gallus gallus D2–I in I-TASSER50. The homology model was initially fitted manually by visual inspection of the ubD2–I, D2–I and D2 maps followed by rigid fitting in UCSF Chimera51. The side chains were stubbed except for K563 of FANCD2 in ubD2–I map and the model was iteratively refined in Coot52,53 and Phenix54. Side chains which can be confidently identified in D2 map were added. The crystal structure of ubiquitin (PDB: 1UBQ)55 was rigidly fitted into the unassigned density in the ubD2–I map and refined in Coot. An idealized dsDNA of 33 bp length was placed and refined in the density observed for DNA in ubD2–I using Coot.
Crosslinking mass spectrometry sample preparation
Purified ubD2–I (50 mM HEPES pH 7.5, 150 mM NaCl, 100 mM imidazole and 1 mM TCEP) at a concentration of ~1 μM was crosslinked with 2-fold molar ratio of disulfosuccinimidyl suberate (BS3) for 2 h on ice and the reaction was quenched with 50 mM NH4HCO3 for 30 min at room temperature. Samples were then loaded on 4–12% NuPAGE Bis-Tris (ThermoFisher Scientific). Gel bands of crosslinked ubD2–I complex were excised and digested with trypsin, as previously described56. In brief, proteins were reduced in 10 mM dithiothreitol (Sigma Aldrich, Germany) for 30 min at 37°C and alkylated in 55 mM iodoacetamide (Sigma Aldrich, Germany) for 20 min at ambient temperature in the dark. Proteins were then digested overnight at 37°C with 10 ng μl-1 trypsin (Pierce, Germany) in 45 mM ABC and 10% v/v acetonitrile.
Following digestion, peptides were extracted from the gel and desalted using C18 StageTips as described previously57. Peptides were then eluted from the StageTip using 40 μl 80% v/v acetonitrile (ACN) in 0.1% v/v trifluoroacetic acid (TFA) into a LowBind Eppendorf sample tube. Solvent in the eluate was removed using vacuum centrifugation (Concentrator 5305 plus, Eppendorf, Germany). For LC-MS/MS analysis, peptides were re-suspended with the sample loading buffer (0.1% v/v formic acid (FA), 1.6% v/v ACN) to a concentration of ~ 0.3 μg μl-1, and 3.5 μl was injected for LC-MS/MS acquisition.
LC-MS/MS analysis
LC-MS/MS analysis was performed using an Orbitrap Fusion™ Lumos™ Tribrid™ mass spectrometer (Thermo Fisher Scientific, Germany) coupled on-line to an Ultimate 3000 RSLCnano System (Dionex, Thermo Fisher Scientific). Peptides were separated on a 50-centimetre EASY-Spray C18 LC column (Thermo Scientific) that is operated at 50 ºC column temperature. Mobile phase A consists of water, 0.1% v/v formic acid and mobile phase B consists of 80% v/v acetonitrile and 0.1% v/v formic acid. Peptides were loaded at a flowrate of 0.3 μl/min. With the same flowrate, peptides were eluted using a linear gradient going from 2% mobile phase B to 40% mobile phase B over 110 minutes, followed by a linear increase from 40% to 95% mobile phase B in eleven minutes. Eluted peptides were ionized by an EASY-Spray source (Thermo Scientific).
The MS data is acquired in the data-dependent mode with the top-speed option. For each three-second acquisition cycle, the full scan mass spectrum was recorded in the Orbitrap with a resolution of 120,000. The ions with a charge state from 3+ to 7+ were isolated and fragmented using Higher-energy collisional dissociation (HCD). For each isolated precursor, one of three collision energy settings (26%, 28% or 30%) was selected for fragmentation using data dependent decision tree based on the m/z and charge of the precursor58. The fragmentation spectra were then recorded in the Orbitrap with a resolution of 50,000. Dynamic exclusion was enabled with single repeat count and 60-second exclusion duration.
Identification of crosslinked peptides
The MS2 peak lists were generated from the raw mass spectrometric data files using the MSConvert module in ProteoWizard (version 3.0.11729). The default parameters were applied, except that Top MS/MS Peaks per 100 Da was set to 20 and the de-noising function was enabled. The peak lists were searched against the sequences and the reversed sequences (as decoys) of FANCI, FANCD2, ubiquitin plus 4,514 E.coli proteins using xiSEARCH software (https://www.rappsilberlab.org/software/xisearch)59 for identification of crosslinked peptides. The following parameters were applied for the search: MS accuracy = 5 ppm; MS2 accuracy = 10 ppm; enzyme = trypsin (with full tryptic specificity); allowed number of missed cleavages = two; missing monoisotopic peak=260; crosslinker = BS3 the reaction specificity for BS3 was assumed to be for lysine, serine, threonine, tyrosine and protein N termini); fixed modifications = carbamidomethylation on cysteine; variable modifications = oxidation on methionine, modifications by BS3 that are hydrolyzed or amidated on the end. False discovery rate (FDR) was estimated based on the number of decoy identification in ubID2 sequences. Crosslinking mass spec data was deposited in the ProteomeXchange Consortium via the PRIDE61 partner repository.
FANCD2 and FANCI structure comparisons
The structures of FANCI and FANCD2 determined in this study were compared to each other and to the previously determined Mus musculus FANCI–FANCD2 structure31 using PDBeFOLD62.
Extended Data
Extended Data Fig. 1. Purification of FANCI, FANCD2 and D2–I, and cryoEM of D2–I, ubD2–I and D2 dimer.
a Coomassie gel showing purified His-tagged FANCI, StrepII-tagged FANCD2 and D2–I after gel filtration. b–e CryoEM of D2–I, ubD2–I and D2. b Representative micrographs. Selected individual particles are marked with green circles. Scale bars are 25 nm. c Fourier shell correlation curves for gold-standard refinements. d Angular distribution density plots of particles used in 3D reconstructions calculated using cryoEF63. Every point is a particle orientation and the color scale represents the normalized density of views around this point. The color scale runs from 0 (low, blue) to 0.00026 (high, red). All complexes had a preferred orientation. Note that C2 symmetry was applied for FANCD2. e Local resolution estimates calculated using ResMap64. Uncropped image for panel a is available in Supplementary Fig. 1.
Extended Data Fig. 2. Model fitting.
a Overall fitting of model to map for ubD2–I, D2–I and D2 (FANCD2 is colored blue; FANCI magenta, ubiquitin green, and DNA yellow). b Representative fits of model to map for FANCD2, FANCI, DNA and ubiquitin in the ubD2–I structure. c FSC curves for model versus map. d FANCD2 and FANCI structures from Gallus gallus (gg) were aligned with each other and the with Mus musculus (mm)31 crystal structures using PDBeFOLD62 (http://www.ebi.ac.uk/msd-srv/ssm/) and figures were prepared with Pymol (The PyMOL Molecular Graphics System, Version 2.0, Schrödinger, LLC).
Extended Data Fig. 3. Crosslinking mass spectrometry and analysis of DNA binding by FANCD2 and FANCI.
a Distribution histogram of Cα-Cα distances between linked residue pairs in the 3D model of ubD2–I (left). Crosslinks with Cα-Cα distances below the theoretical crosslinking limit (30 Å) are shown in green. Overlength crosslinks (>30 Å) are shown in red. The distribution of Cα-Cα distances between random crosslinkable residue pairs in the 3D model is shown in grey. Crosslinks mapped onto the front view of the 3D structure are shown on the right. b Monoubiquitination assays were assembled in the presence of 5 μM linear double-stranded DNA of differing lengths (10–44 bp). DNA binding was analyzed by EMSA (top) after loading the reactions onto native gels and imaging of the fluorescently-labeled DNA. Monoubiquitination efficiency was analyzed by Coomassie blue (middle) and Western blotting the His-tagged ubiquitin (bottom). Controls lacking ubiquitin or DNA are indicated. These data are representative of experiments performed three times. c Monoubiquitination assays were assembled without (left) and with (right) ubiquitin, both in the presence of a 39 bp double-stranded DNA at 100 nM and increasing amounts of D2–I (0–1000 nM). Assays were analyzed by EMSA (top, imaging for the fluorescently-labeled DNA) or Western blotting His-tagged ubiquitin (bottom). We cannot exclude that other proteins may interact with DNA in these assays, but the migration positions of the shifted bands are similar to the experiment in Fig. 4 where ubD2–I was purified away from all other proteins. These data are representative of experiments performed three times. Uncropped images for panels b and c are available in Supplementary Fig. 1.
Extended Data Fig. 4. FANCD2 and FANCI oligomerization state and DNA binding activity.
a Size-exclusion chromatogram as shown in Fig. 5a (top). Peak fractions were analyzed by SDS-PAGE (bottom). A single asterisk (*) indicates the migration position for monomers. A double asterisk (**) indicates the migration position for dimers. These results are representative of experiments performed three times. b DNA binding of FANCI, FANCD2, D2–I, and FANCI mixed with FANCD2 was analyzed by EMSAs performed with 20 nM 39-bp double-stranded DNA and 0–140 nM protein. Representative gels of experiments performed independently three times. The FANCD2 and D2–I gels are same as in Fig 5b. c Quantification of mean intensities of free DNA from panel b. Error bars represent the standard deviation. Individual data points (n=3 independent experiments) are shown. The means are connected by lines for clarity. Uncropped images for panels a and b are available in Supplementary Fig. 1. Data for the plot in c are available as source data.
Extended Data Fig. 5. FANCD2 dimers cannot be ubiquitinated and exchange with FANCI to form a D2–I heterodimer.
a Monoubiquitination assays of D2–I and FANCD2 homodimer. The FANCD2 homodimer had a StrepII-tag and was therefore larger than D2 in the D2–I complex. Monoubiquitination efficiency was analyzed by Coomassie blue SDS-PAGE (top) and Western blotting the His-tagged ubiquitin (bottom). b Exchange assay. The FANCD2 homodimer was immobilized on Streptactin resin and incubated with free FANCI. The resin was washed, then bound and unbound fractions were analyzed by SDS-PAGE. These data are representative of experiments performed twice. Uncropped images are available in Supplementary Fig. 1.
Supplementary Material
Acknowledgements
We are grateful to T. Nakane, S.H.W. Scheres, F. O’Reilly, M. Babu, P. Emsley, J. Pruneda, T. Sijacki and members of the Passmore lab for assistance and advice; the LMB EM facility, J. Grimmett and T. Darling (LMB scientific computation), and J.G. Shi (baculovirus) for support. This work was supported by the Medical Research Council, as part of United Kingdom Research and Innovation, MRC file reference numbers MC_U105192715 (L.A.P) and MC_U105178811 (K.J.P.), and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) grant no. 329673113 (J.R.). The Wellcome Centre for Cell Biology is supported by core funding from the Wellcome Trust (grant no. 203149; J.R.). P.A. is supported by an EMBO Long Term Fellowship (ALTF 692-2018). We acknowledge Diamond Light Source for access to eBIC (proposals EM17434 and BI23268) funded by the Wellcome Trust, MRC, and Biotechnology and Biological Sciences Research Council.
Footnotes
Reporting Summary Statement
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
CryoEM maps generated during this study have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMD-10532 (D2–I), EMD-10531 (ubD2–I), and EMD-10534 (D2–D2). Models generated during this study have been deposited in the protein databank (PDB) with accession codes 6TNG (D2–I), 6TNF (ubD2–I) and 6TNI (D2–D2). Mass spec data has been deposited in the PRIDE database with accession code PXD017020. Original gels and blots in Figures 1b, 4 and 5b, and Extended Data Fig. 1b, 3b, 3c, 4a, 4b and 5 are provided in Supplementary Fig. 1. Data for quantifications in Fig. 4 and Extended Data Fig. 4c are available as Source Data with this paper online.
Author contributions: P.A. and S.S. designed protein expression and purification schemes, performed ubiquitination and binding assays, performed cryoEM, 3D reconstruction and modelling; Z.A.C. and J.R. performed and analyzed crosslinking mass spectrometry. L.A.P. and K.J.P. supervised the research; all authors contributed to writing the paper.
Competing Interests: The authors declare no competing interests.
References
- 1.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crossan GP, Patel KJ. The Fanconi anaemia pathway orchestrates incisions at sites of crosslinked DNA. J Pathol. 2012;226:326–337. doi: 10.1002/path.3002. [DOI] [PubMed] [Google Scholar]
- 3.Walden H, Deans AJ. The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annu Rev Biophys. 2014;43:257–278. doi: 10.1146/annurev-biophys-051013-022737. [DOI] [PubMed] [Google Scholar]
- 4.Knipscheer P, Raschle M, Scharer OD, Walter JC. Replication-coupled DNA interstrand cross-link repair in Xenopus egg extracts. Methods Mol Biol. 2012;920:221–243. doi: 10.1007/978-1-61779-998-3_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alpi A, et al. UBE2T, the Fanconi anemia core complex, and FANCD2 are recruited independently to chromatin: a basis for the regulation of FANCD2 monoubiquitination. Mol Cell Biol. 2007;27:8421–8430. doi: 10.1128/MCB.00504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Higuera I, et al. Interaction of the fanconi anemia proteins and BRCA1 in a common pathway. Molecular Cell. 2001;7:249–262. doi: 10.1016/S1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 7.Meetei AR, et al. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol Cell Biol. 2003;23:3417–3426. doi: 10.1128/mcb.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajendra E, et al. The genetic and biochemical basis of FANCD2 monoubiquitination. Mol Cell. 2014;54:858–869. doi: 10.1016/j.molcel.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Twest S, et al. Mechanism of Ubiquitination and Deubiquitination in the Fanconi Anemia Pathway. Mol Cell. 2017;65:247–259. doi: 10.1016/j.molcel.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Knipscheer P, et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sims AE, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 12.Smogorzewska A, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montes de Oca R, et al. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood. 2005;105:1003–1009. doi: 10.1182/blood-2003-11-3997. [DOI] [PubMed] [Google Scholar]
- 14.MacKay C, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 16.Smogorzewska A, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kratz K, et al. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 18.Klein Douwel D, et al. XPF-ERCC1 acts in Unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell. 2014;54:460–471. doi: 10.1016/j.molcel.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodskinson MR, et al. Mouse SLX4 is a tumor suppressor that stimulates the activity of the nuclease XPF-ERCC1 in DNA crosslink repair. Mol Cell. 2014;54:472–484. doi: 10.1016/j.molcel.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto KN, et al. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc Natl Acad Sci U S A. 2011;108:6492–6496. doi: 10.1073/pnas.1018487108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oestergaard VH, et al. Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol Cell. 2007;28:798–809. doi: 10.1016/j.molcel.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JM, et al. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell. 2009;16:314–320. doi: 10.1016/j.devcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nijman SM, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 24.Tan W, Deans AJ. A defined role for multiple Fanconi anemia gene products in DNA-damage-associated ubiquitination. Exp Hematol. 2017;50:27–32. doi: 10.1016/j.exphem.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Ishiai M, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol. 2008;15:1138–1146. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheung RS, et al. Ubiquitination-Linked Phosphorylation of the FANCI S/TQ Cluster Contributes to Activation of the Fanconi Anemia I/D2 Complex. Cell Rep. 2017;19:2432–2440. doi: 10.1016/j.celrep.2017.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez-Martinez D, et al. Phosphorylation of FANCD2 Inhibits the FANCD2/FANCI Complex and Suppresses the Fanconi Anemia Pathway in the Absence of DNA Damage. Cell Rep. 2019;27:2990–3005 e2995. doi: 10.1016/j.celrep.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato K, Toda K, Ishiai M, Takata M, Kurumizaka H. DNA robustly stimulates FANCD2 monoubiquitylation in the complex with FANCI. Nucleic Acids Res. 2012;40:4553–4561. doi: 10.1093/nar/gks053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shakeel S, et al. Structure of the Fanconi anaemia monoubiquitin ligase complex. Nature. 2019;575:234–237. doi: 10.1038/s41586-019-1703-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang CC, Cohn MA. UHRF1 is a sensor for DNA interstrand crosslinks. Oncotarget. 2016;7:3–4. doi: 10.18632/oncotarget.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joo W, et al. Structure of the FANCI-FANCD2 complex: insights into the Fanconi anemia DNA repair pathway. Science. 2011;333:312–316. doi: 10.1126/science.1205805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swuec P, et al. The FA Core Complex Contains a Homo-dimeric Catalytic Module for the Symmetric Mono-ubiquitination of FANCI-FANCD2. Cell Rep. 2017;18:611–623. doi: 10.1016/j.celrep.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang CC, et al. The FANCD2–FANCI complex is recruited to DNA interstrand crosslinks before monoubiquitination of FANCD2. Nat Commun. 2016;7 doi: 10.1038/ncomms12124. 12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaugule VK, Arkinson C, Toth R, Walden H. Enzymatic preparation of monoubiquitinated FANCD2 and FANCI proteins. Methods Enzymol. 2019;618:73–104. doi: 10.1016/bs.mie.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 35.Zhi G, et al. Purification of FANCD2 sub-complexes. Br J Haematol. 2010;150:88–92. doi: 10.1111/j.1365-2141.2010.08217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson EL, et al. FANCI and FANCD2 have common as well as independent functions during the cellular replication stress response. Nucleic Acids Res. 2017;45:11837–11857. doi: 10.1093/nar/gkx847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dubois EL, et al. A Fanci knockout mouse model reveals common and distinct functions for FANCI and FANCD2. Nucleic Acids Res. 2019;47:7532–7547. doi: 10.1093/nar/gkz514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Longerich S, et al. Regulation of FANCD2 and FANCI monoubiquitination by their interaction and by DNA. Nucleic Acids Res. 2014;42:5657–5670. doi: 10.1093/nar/gku198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raschle M, et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crossan GP, et al. Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat Genet. 2011;43:147–152. doi: 10.1038/ng.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parmar K, et al. Hematopoietic stem cell defects in mice with deficiency of Fancd2 or Usp1. Stem Cells. 2010;28:1186–1195. doi: 10.1002/stem.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weissmann F, et al. biGBac enables rapid gene assembly for the expression of large multisubunit protein complexes. Proc Natl Acad Sci U S A. 2016;113:E2564–2569. doi: 10.1073/pnas.1604935113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hill CH, et al. Activation of the Endonuclease that Defines mRNA 3' Ends Requires Incorporation into an 8-Subunit Core Cleavage and Polyadenylation Factor Complex. Mol Cell. 2019 doi: 10.1016/j.molcel.2018.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rueden CT, et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017;18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russo CJ, Passmore LA. Electron microscopy: Ultrastable gold substrates for electron cryomicroscopy. Science. 2014;346:1377–1380. doi: 10.1126/science.1259530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7 doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakane T, Kimanius D, Lindahl E, Scheres SH. Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. Elife. 2018;7 doi: 10.7554/eLife.36861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J, et al. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 54.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 56.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 57.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 58.Kolbowski L, Mendes ML, Rappsilber J. Optimizing the Parameters Governing the Fragmentation of Cross-Linked Peptides in a Tribrid Mass Spectrometer. Anal Chem. 2017;89:5311–5318. doi: 10.1021/acs.analchem.6b04935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mendes ML, et al. An integrated workflow for crosslinking mass spectrometry. Mol Syst Biol. 2019;15:e8994. doi: 10.15252/msb.20198994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lenz S, Giese SH, Fischer L, Rappsilber J. In-Search Assignment of Monoisotopic Peaks Improves the Identification of Cross-Linked Peptides. J Proteome Res. 2018;17:3923–3931. doi: 10.1021/acs.jproteome.8b00600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perez-Riverol Y. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 63.Naydenova K, Russo CJ. Measuring the effects of particle orientation to improve the efficiency of electron cryomicroscopy. Nat Commun. 2017;8 doi: 10.1038/s41467-017-00782-3. 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.