Abstract
Purpose:
To develop and demonstrate the efficacy of a novel head-and-neck multimodality image registration technique using deep-learning based cross-modality synthesis.
Methods and Materials:
25 head-and-neck patients received MR and CT (CTaligned) scans on the same day with the same immobilization. 5-fold cross validation was used with all of the MR-CT pairs to train a neural network to generate synthetic CTs from MR images. 24 of the 25 patients also had a separate CT without immobilization (CTnon-aligned) and were used for testing. CTnon-aligned’s were deformed to the synthetic CT, and compared to CTnon-aligned registered to MR. The same registrations were performed from MR to CTnon-aligned and from synthetic CT to CTnon-aligned. All registrations used B-splines for modeling the deformation, and mutual information for the objective. Results were evaluated using the 95% Hausdorff distance among spinal cord contours, landmark error, inverse consistency, and Jacobian determinant of the estimated deformation fields.
Results:
When large initial rigid misalignment is present, registering CT to MRI-derived synthetic CT aligns the cord better than a direct registration. The average landmark error decreased from 9.8±3.1mm in MR→CTnon-aligned to 6.0±2.1mm in CTsynth→CTnon-aligned deformable registrations. In the CT to MR direction, the landmark error decreased from 10.0±4.3mm in CTnon-aligned→MR deformable registrations to 6.6±2.0 mm in CTnon-aligned→ CTsynth deformable registrations. The Jacobian determinant had an average value of 0.98. The proposed method also demonstrated improved inverse consistency over the direct method.
Conclusions:
We showed that using a deep learning derived synthetic CT in lieu of an MR for MR→CT and CT→MR deformable registration offers superior results to direct multimodal registration.
Keywords: multi-modal registration, synthetic CT, deep learning
Introduction
Image registration is often used in medicine for diagnostic and therapeutic purposes1. The registration can take place between a single image modality, or different modalities (multimodal registration), which aggregates complementary data from different sources into a spatially unified context2. A common multimodal registration problem is magnetic resonance (MR) and computed tomography (CT) registration2. MR imaging has superior soft tissue contrast while computed tomography (CT) has better bone contrast and spatial integrity3. Specifically, CT is the foundation of modern radiotherapy by providing anatomical information as well as the electron density for treatment planning and dose calculation4. In image guided radiation therapy, CT or cone beam CT is instrumental to guide patient set-up. Because of their complementary strengths, MR-CT registration is often needed for accurate tumor and organ-at-risk (OAR) delineation, targeting and sparing5–9.
Relevant to the current study, head-and-neck radiotherapy benefits from the superior soft tissue contrast provided by the MR images. Studies have demonstrated that MR images in addition to CT improve delineation of head-and-neck target volumes, and reduce interobserver variation10–14. Consensus guidelines recommend that MRI be used for primary tumors of the nasopharynx, oral cavity, and oropharynx to contour head-and-neck normal tissues15. However, these guidelines also acknowledge the challenges associated with MR-CT registration. While MR-CT registration is a common practice in head-and-neck radiotherapy, the process and results are not satisfactory due to the different imaging mechanisms and contrast, as well as the unavoidable patient non-rigid motion between scans, such as neck flexion2. Deformable registration using commercial algorithms can produce difficult-to-validate distortion and is regarded as unreliable in clinical practice. Instead, rigid registration is performed as a trade-off to avoid the uncertain deformation16. Subsequently, delineation based on rigid MR-CT registration is limited to a small volume of interest, without using other information about OARs and lymph nodes from the MR images due to the increasing misalignment with the distance from the volume of interest.
Efforts have been made to address some of the technical issues in multi-modality image registration17,18. For example, instead of directly matching the image voxel values, mutual information is used to determine the image similarity based on joint entropy17. However, mutual information based on an image histogram cannot resolve tissue types with similar image intensities, such as the bones and air cavities in MR and various soft tissues in CT2. The problem is further complicated by the common presence of MR shading and susceptibility artifacts19. Some have endeavored to overcome this difference by translating one image into the other, or a third domain. For example, Heinrich et al. used a new image descriptor describing similarities between adjacent patches as features for registration2. Researchers have previously used an atlas-based synthetic CT to replace the MR in MR-CT registration in the brain20,21; however, the brain experiences considerably less deformation than the head and neck, and thus a domain-translating deformable registration has yet to be proven in this challenging location. Recently, Cao et al. proposed using a patchwise random forest to translate MR and CT into the others’ domains for improved pelvic registration22. We propose to build on these existing domain-translating registration techniques by incorporating recent advances in deep learning imaging synthesis.
Specifically, Generative Adversarial Networks (GAN)23 are capable of converting images of one modality into another24–26. For example Wolterink et al. used the CycleGAN implementation27 to convert brain MR images into synthetic CT images26. In the current study, GANs’ capability to create synthetic images with the geometry of one image modality and the contrast of the other is used to improve multimodality registration in the head-and-neck. A GAN trained to generate head and neck images learns to estimate realistic anatomy in its image synthesis, and can leverage image features to determine low-confidence regions such as bone air interfaces, which are otherwise invisible in standard T1 weighted MR images. The head and neck is a particularly challenging site in this regard, as there are many bone and air regions in close proximity that can move several centimeters with neck flexion. Deep learning patient-specific image synthesis takes the field beyond atlas-based approaches which try to fit a patient to a standard anatomical layout. By extending modality-translating registration techniques with patient specific deep learning image synthesis, we provide a valuable new technique and prove its performance in the challenging head and neck region.
Methods and Materials
Data
In order to train a network capable of generating synthetic CT’s and subsequently test registration accuracy, 25 head-and-neck patients were selected, each with a paired MR and CT volume acquired on the same day with the same immobilization mask and headrest. The original dataset before processing had 126 to 336 512×512 axial slices with voxel sizes of 1×1×3(or 1.5) mm3, and 288 334×300 axial slices with 1.5×1.5×1.5mm3 voxel sizes for CT and MR images, respectively. The MR images were acquired on an MR guided radiotherapy system with a 0.35T B0 using a balanced steady state free-precession sequence. Due to the same rigorous immobilization being used for CT and MR acquisition, deformation between the two image sets was small, providing a unique opportunity for comparison and validation. In this study, we will refer to this MR-aligned CT as CTaligned. A five-fold cross validation technique was employed for machine learning purposes. 20 patients were used for training the synthetic CT generating network, and 5 were left out to test replacing an MR image with a synthetic CT during MR-CT registration. This split was rotated through the data, allowing us to include all patients in our analysis.
In addition to the paired images, each test patient also had a diagnostic CT from another time point ranging from 4 days prior to almost 3 years after. One patient did not have a usable separate CT volume, leaving us with 24 patients total for registration testing. The goal was to have patient positions substantially different from the paired images to challenge the registration. For the remainder of this paper, we will refer to these images as CTnon-aligned.
Data Processing
All CTaligned datasets were first automatically rigidly registered in Elastix28,29 to their corresponding MR image using mutual information as the similarity metric. To better show the posture during network training, volumes were resliced into 2D sagittal slices. All images were resampled to slices of size 256×256, and each voxel was 1.76×1.76×1.5 mm3. The images were quantized to 256 greyscale values. For CT, this was accomplished by renormalizing and quantizing the image into 256 levels between intensity values −600 and 1400, and between 0 and 600 for MR. In all, this gave 8,350 CT and 8,350 MR sagittal slices to be used for training and testing.
Deep Learning Networks
In a conventional GAN, two networks are used; a generator network attempts to generate realistic images, while a discriminator network attempts to distinguish between real images and those created by the generator. When successful training is complete, the generator is able to create an image that appears to come from the domain of the training set. This study used an adversarial network utilizing cycle-consistency (CycleGAN)27 with two GAN’s: one attempted to generate a realistic synthetic CT (CTsynth) slice given a real MR slice, and the other attempted to generate a realistic synthetic MR slice given a real CT slice. The generators were then switched and applied to the synthetic outputs, so that the synthetic MR was translated back into a CT slice, and vice versa. Ideally, the original CT or MR slice should be recovered, and hence this network architecture has cycle consistency. The loss function for CycleGAN therefore has an adversarial loss term for generating realistic CT images, an adversarial loss term for generating realistic MR images, and a cycle consistency loss term to prevent the network from assigning any random realistic-looking image from the other domain.
Overall, the full loss function can be written as:
| (1) |
where λ (set to 10 in this work) is a relative weighting coefficient, and G and D are the generator and discriminator networks with subscripts describing the direction of image translation and discrimination domain, respectively. The adversarial loss is given by:
| (2) |
where the discriminator gives an output between 0 (image determined to be fake) and 1 (image determined to be real). In the minmax optimization problem, the generator attempts to create a realistic image by minimizing the second term towards a large negative value while the discriminator is trained to maximize the objective by correctly differentiating real images from fake. A similar loss is used for . The cycle consistency loss using L1 norm is then given as:
| (3). |
The generator networks follow the Resnet architecture described in Johnson et al30. The discriminator uses a patch-based network described in31. Because it is patch-based, this allows greater flexibility for different sized images, as well as forces the discriminator to focus on smaller-scale details. The network hyperparameters used in our study are the same as those in the pytorch-CycleGAN-and-pix2pix repository (https://github.com/junyanz/pytorch-CycleGAN-and-pix2pix).
Registration
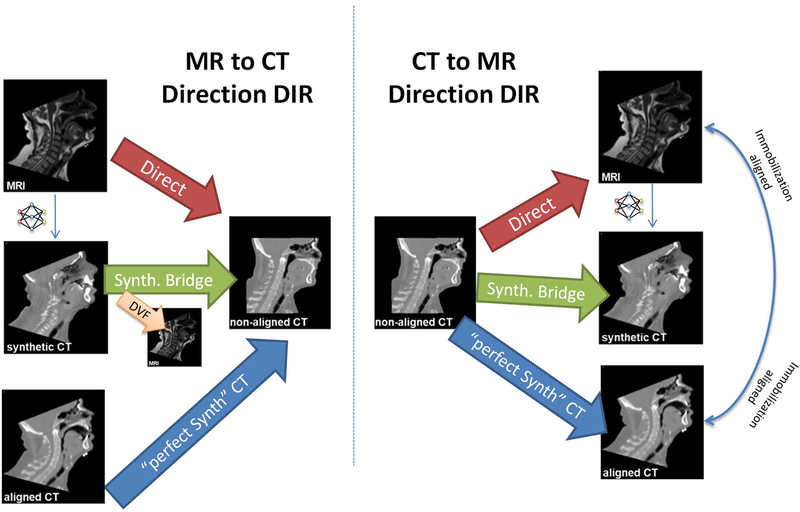
We first used the trained CycleGAN to generate a synthetic CT given a head and neck MR image. We then registered CTnon-aligned to the synthetic MR-derived CTsynth, which reduced the multimodal registration problem to a mono-modal one. Conversely, we registered the MR to CTnon-aligned by first registering the CTsynth to CTnon-aligned, then applying the resulting deformation vector field (DVF) to the original MR. For comparison, direct registrations between MR and CT were also performed. CTnon-aligned was additionally registered to CTaligned to characterize the behavior of a typical mono-modality (CT vs CT) registration. This paper will denote deformable registration with an arrow (→) pointing from source to target. Figure 1 gives an overview of the registrations performed in this study.
Figure 1.
The deformable registrations performed in this study. Registrations were performed in both CT-to-MR and MR-to-CT directions to test for inverse consistency, as well as both directly (multi-modal) and with a synthetic CT bridge (synthetic mono-modal). The DVF from CTsynth→ CTnon-aligned was applied to the MR to generate a deformed MR image. The non-aligned CT was also registered to the aligned CT (and vice versa) to see an approximate best-case synthetic mono-modal registration.
The multi-resolution registration using B-splines and mutual information was performed using Elastix28,29 with six Gaussian blurring levels, repeated. These levels allow for a hierarchical approach to the registration, starting at a coarse resolution with large-scale deformations, and gradually progressing towards a finer resolution for fine-detailed deformations. The B-spline grid spacings for each resolution level were 128, 64, 32, 8, and 4 mm, sequentially. The Gaussian sigmas were 8, 4, 2, 1, 0.5, and 0.5 voxels isotropically. The registration was optimized using gradient descent32. The gradient descent gain factor, ak, was set to: , where k is the iteration number, and a is set for each resolution level to be: 50000, 10000, 2000, 500, 100, 100. Large values of a in the coarse resolutions allow the registration to capture large deformations, which were necessary when registering to CTnon-aligned. The maximum number of iterations at each resolution level was: 500, 500, 500, 500, 100, and 100.
Analysis
To evaluate the registration, the following tests were performed. The spinal cord was manually contoured on the original MR, CTaligned, and CTnon-aligned image volumes for all patients. The cord is an appealing anatomical landmark structure, as it is present throughout the head-and-neck region, reflective of the neck flex, and conspicuous in both modalities. The resulting cord contours from the deformable registration were compared to their respective target volumes’ contours using 95% Hausdorff Distance33, measured in mm.
The Euclidean distance between a set of 11 landmarks (Dens of C2, center of the vertebral bodies of C2–C7, center of left and right eyes, the mental protuberance of the mandible, and the tip of the nose) was evaluated between deformed and target images. We performed a 2-way repeated measures anova with a post hoc Tukey’s multiple comparison to test the null hypothesis that all registrations had the same mean error, and identify the significantly different registrations if the null hypothesis was rejected. The two factors in the anova analysis were registration direction and landmark.
Additionally, the quality of the registration itself was evaluated by calculating the Jacobian determinant from each resulting transformation. This was calculated using the Insight Toolkit’s implementation, and is given as the scalar determinant of the derivative of the deformation vector field at each point . A reasonable registration in the head and neck anatomical region should have most voxels experiencing small shrinking and expansion with the average Jacobian determinant close to 1.
The quality of the registration is also reflected in its inverse consistency, which was evaluated by comparing the composition of transformation pairs in the opposite direction on a standard CT image to reduce input from the background, then calculating the mean square error (MSE) between that image and the initial image. The MSE was calculated using the Insight Toolkit’s implementation, and is the sum of squared differences between intensity values between the images. A lower MSE indicates better inverse consistency. We elected to use this method since we did not expect true inverse consistency in the DVF due to the occasional appearance and disappearance of tissue with different patient positioning (e.g. arms up versus arms down). This is a known challenge in head and neck image registration. However, we can compare directional bias between direct and our proposed registration techniques. Therefore we emphasize that while a 0 MSE would indicate a perfect recovery of the original image, our inverse consistency study was a relative comparison.
Results
Synthetic CT
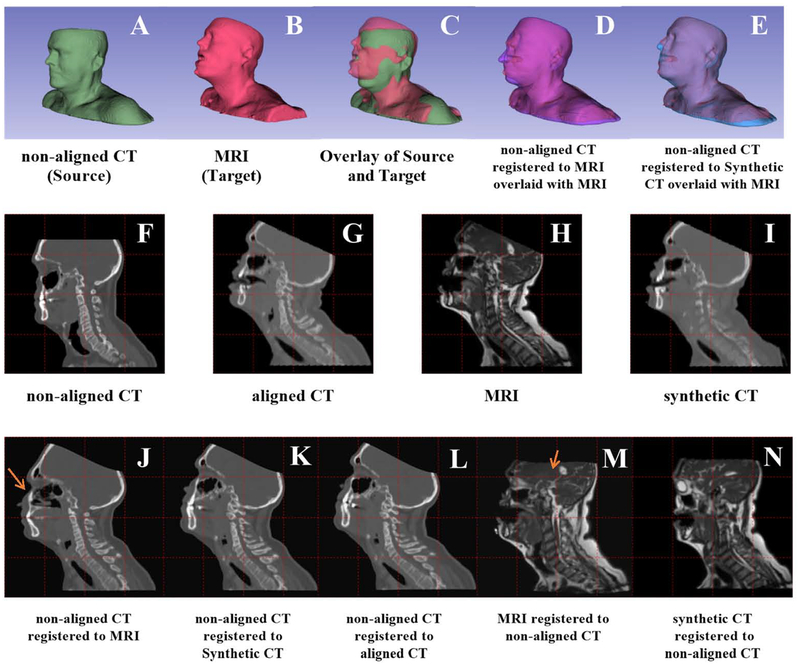
Figure 2I is a typical synthetic CT achieved in this work. The CTsynth image preserves bulk anatomy, distinguishes bones and sinuses, but is missing certain anatomical details of a real CT (e.g. accurate description of individual vertebrae).
Figure 2.
An example patient shows the various registrations studied in this paper. First row: registration of non-aligned CT (green) to an MR image (red). Box D shows the directly registered non-aligned CT (purple); Box E shows registration of the corersponding synthetic CT to the nonaligned CT. Second row: sagittal view. Boxes F-I are the non-deformed volumes. The third row shows the results for various registrations. Note that the images used in the registration were downsampled to match the 256×256 resolution output from the neural network. All slices shown were in the same location. Arrows denote unanatomical deformation in direct multimodel registration.
Registration Accuracy: Qualitative Evaluation
Figure 2 A–E illustrate an example of a CTnon-aligned (green) registered to an MRI (red) using a synthetic CT. The 3D surface rendering in Figure 2C shows a large initial discrepancy in the head pose. Figure 2D shows how the CTnon-aligned pose was deformed (purple) to match the MRI when the CT is directly registered to the MRI (red). Fig 2E shows how closely the registered CTnon-aligned’s pose (blue) matches the target MRI (red) when using a synthetic CT bridge. The head tilt matches better when using a synthetic CT, as can be seen by the improved match in the nose.
Figures 2 J–N show the interior anatomy of registration results in sagittal slices. Looking at the gridlines, Figure 2K shows that the CTnon-aligned matches the MR’s pose when CTsynth is used as the target. CTnon-aligned registered to MR matched the pose but produced slight unrealistic tissue deformation, as in the stretched sinuses indicated by the red arrow in Figure 2J. Comparing Figure 2N and Figure 2M, the CTsynth registered to CTnon-aligned shows even better improvement over direct registration. This is evident in the MR to CTnon-aligned registration’s relatively greater stretching in the skull and brain anatomy indicated by the red arrow in Figure 2M, and also the better positioning of the orbit. While the registrations including CTsynth matched overall pose, there is a residual discrepancy due to different mouth opening with or without a bite block. Also, the registration accuracy is similar for CTsynth registered to CTnon-aligned and CTnon-aligned registered to CTsynth, while there is a noticeable decline in quality for MR registered to CTnon-aligned relative to CTnon-aligned registered to MR, showing improved inverse consistency using a synthetic CT bridge.
Registration Accuracy: Spinal Cord Contour Comparisons
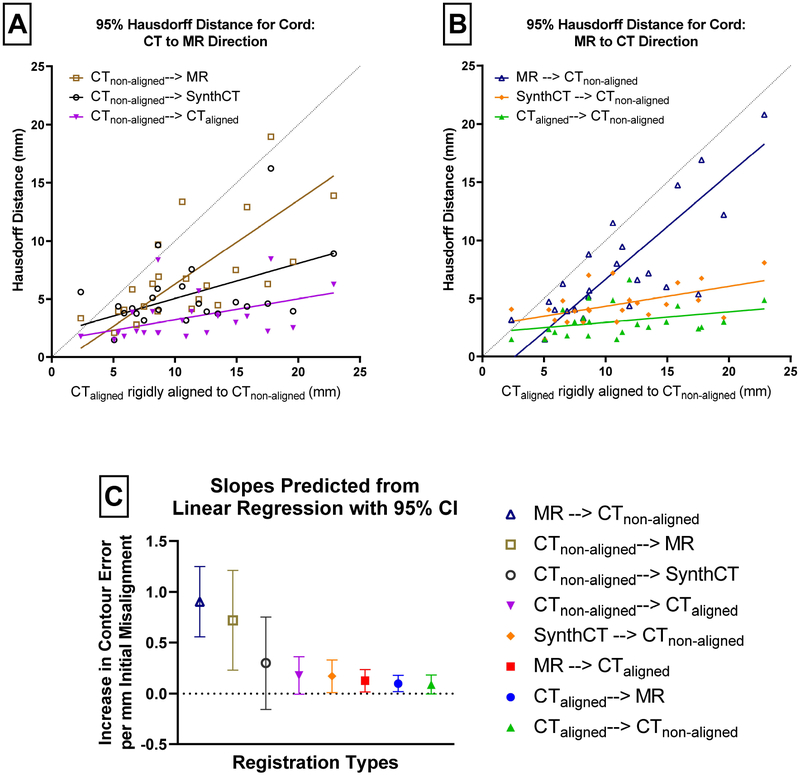
The spinal cord contour comparison results are shown in Figure 3. In Figure 3A–B, the initial 95% Hausdorff distance of the MR and CTnon-aligned rigid alignment is on the horizontal axis, and the vertical axis shows their 95% Hausdorff distance33 after registration, to reflect the quality of registration and its dependency on the original level of rigid misalignment. The fitted lines are a result of Deming linear regression, where a lower slope means the registration is more robust to initial misalignment. The dotted reference line indicates equal rigid and deformable cord error. For small initial misalignment in the bottom left, the deformable results using CTsynth and MR are similar to the rigid results. With larger initial misalignments, there is an increasing divergence in the results with and without CTsynth. In the presence of large initial misalignment, compared with the direct registration, our proposed method results in a larger improvement. Using a Deming linear regression and comparing estimated slopes, we found that the error using our method is lower than that of the direct registration in both the MR to CT direction (p=0.0002) and the CT to MR direction (p=0.08). Unsurprisingly, replacing the MRI with the aligned CT results in the lowest contour misalignment but the difference with our proposed method is not statistically significant (p=0.34 and p=0.65, respectively).
Figure 3.
The spinal cord 95% Hausdorff distance for the deformations is plotted in the top two figures as a function of the initial rigid alignment Hausdorff distance. Thus, the error in cord alignment can be evaluated in terms of how misaligned the images were initially. The diagonal line shows where the deformable image registration’s cord error would equal the initial rigid alignement’s cord error. The top figure is divided in the CT-to-MR direction on the left and the MR-to-CT direction on the right. The bottom figure shows the slopes of the best fit lines with their 95% confidence intervals.
The slopes of the fitted lines and their 95% confidence intervals are plotted in Figure 3C. The direct registrations show a clear increase in sensitivity to initial misalignment. The wide confidence intervals on the non-aligned CT registered to the synthetic CT reveal the larger spread when using our method in the CT to MR direction. In fact, all of the CT to MR direction registrations show increased sensitivity relative to their opposite-direction pairs. The registrations between MR and the aligned CT represent the residual sensitivity to initial misalignment inherent to our registration algorithm, as the MR and aligned CT should already have overlapping cord contours.
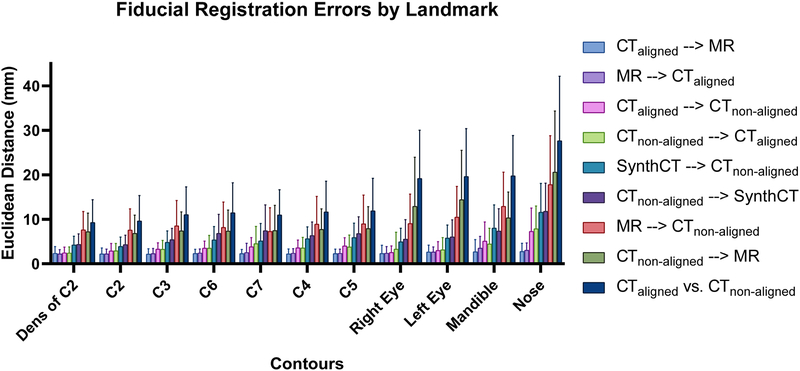
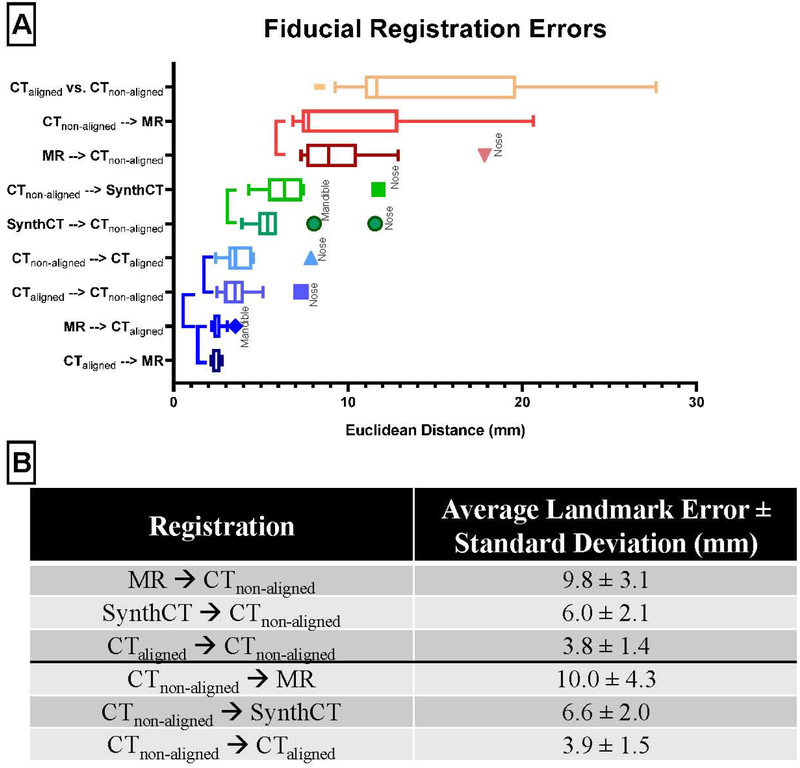
Registration Accuracy: Landmark Analysis
Figure 4 summarizes the landmark analysis. The plot is ordered from the lowest average landmark error to the highest. The vertebrae landmarks tended to be lower than those on the head. This is caused by the distances of landmarks to the head rotating motion axis, as a small head tilt could lead to large distances in the eyes, nose, and mandible. The registrations between the aligned CT and MR are consistent across all landmarks, as these images were already closely aligned. Interestingly, when the MR was replaced with the aligned CT in the non-aligned CT registrations, the landmark error also stays relatively consistent. This aligned CT and MR registration error defines the performance upper bound of the proposed method if we were able to generate perfect synthetic CTs.
Figure 4.
The patient-average Euclidean landmark error for the various registrations investigated in this study. The bars are ordered by the average error across all landmarks. From this figure, we can see that for the large CTnon-aligned registrations our proposed method has an overall lower landmark error than the direct registration method. The rigid alignment between CTaligned and CTnon-aligned is denoted by CTaligned vs. CTnon-aligned.
The direct registration shows the second largest variation in performance after the rigid alignment with respect to landmark type. Figure 5 shows the landmark error by registration type and the statistically significant groupings from the anova post-hoc Tukey test. There are four groups in order of increasing average error,
Group 1: deformable registrations between the CTaligned and MR, and the CTaligned and CTnon-aligned;
Group 2: the registrations of our proposed method between CTnon-aligned and the Synthetic CT;
Group 3: the direct CTnon-aligned and MR registrations;
Group 4: the rigid alignment between CTnon-aligned and CTaligned.
The outliers in the different registration groups were either nose or mandible landmarks. Figure 5B tabulates the average and standard deviation of the landmarks per registration type. There is an average reduction of 3.8mm in average landmark error (from 9.8mm to 6.0mm) by replacing the MR in the MR registered to CTnon-aligned registration with a synthetic CT.
Figure 5.
Combining all landmarks together to better visualize the post hoc results by registration type. The vertical bars to the left of the boxplots indicate registrations which were not significantly different. The rigid alignment between CTaligned and CTnon-aligned is denoted by CTaligned vs. CTnon-aligned. Below is a table to more easily display the decrease in average landmark error with the proposed method. The bottom rows in each section show the average error if CTaligned is used as a surrogate for the MR. This represents a “best-case” scenario.
If the synthetic CT were replaced with a CTaligned, the error decreases further by 2.2mm (from 6.0mm to 3.8mm). The trend is similar in the CT to MR direction. The average landmark error is reduced by 3.4 mm (from 10.0mm to 6.6mm) when replacing MR with a synthetic CT in the CTnon-aligned registered to MR registration. The error is further reduced by 2.7mm (from 6.6mm to 3.9mm) with registration to a CTaligned. The error reduction from direct registration to synthetic CT bridged registration is not only significant (2way ANOVA with Tukey’s multiple comparisons test, p<0.001) but also greater than half the potential improvement with registration to CTaligned, which is typically unavailable.
Registration Accuracy: Jacobian Determinant
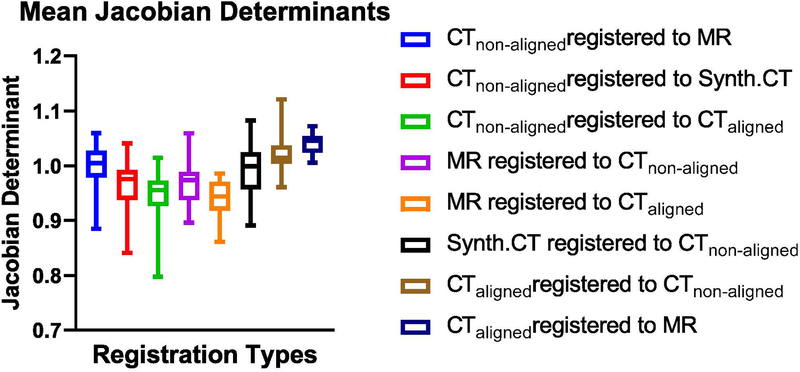
The Jacobian determinant was calculated for each transformation. The mean across each resulting 3D matrix was found. Figure 6 shows the descriptive statistics averaged across all 24 test patients, for each registration investigated. All of the registrations have mean Jacobian determinants around 1.0. This shows that the majority of the deformed images did not experience large expansion or shrinking, consistent with the head-and-neck anatomy.
Figure 6.
Patient-averaged descriptive statistics of Jacobian determinants across different deformable registration types. Error bars show the range.
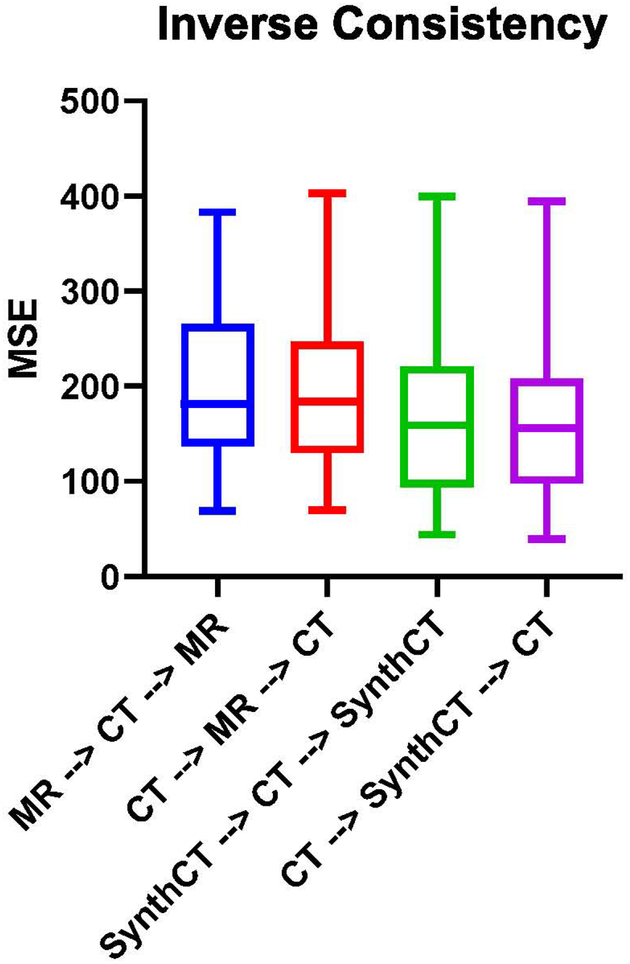
Inverse Consistency
There is an improved inverse consistency when using the synthetic CT bridge in both the CT to MR direction (MSE of 193.9 to 165.1, p=0.04) and the MR to CT direction (MSE of 197 to 168, p=0.04). Statistical comparisons were made using a paired T-test.
Discussion
Multi-modal deformable registration is an important technique yet a challenging problem due to its ill-conditioned nature. It is made more difficult by different material-to-imaging-value mapping and large deformations. Both compounding problems are seen in head-and-neck MR- and CT-scanned cancer patients and have hampered the utility of multimodal imaging for their radiotherapy. In the current study, we showed that the difficulty can be effectively mitigated using deep-learning generated synthetic images, with which the multimodel registration problem is reduced to a monomodal one. For large deformation, this novel registration pipeline is able to significantly improve the deformable registration results versus direct registration. The anatomy is more accurately morphed to the target images as shown in the quantitative results of spinal cord contours and the landmark tests. An additional benefit of this method is that the pipeline can be fully automated.
Current clinical practice often uses rigid registration to align CT and MR images in the head-and-neck, which is clearly suboptimal given the discrepancy in patient posture as shown in Figure 2. We show a method that offers significant improvement over the current standard of care. In addition, we show that our method is more robust than traditional, direct deformable image registration (DIR) methods. A significant challenge in this work was ensuring accurate deformation even in the presence of large head motion. Through careful parameter tuning, a balance was able to be struck which was both accurate and robust. It was noted during this tuning process that the direct multimodal registration results were more sensitive to choice of parameters and initial conditions relative to our CTsynth method. Future work will endeavor to discover better ways to automatically choose these parameters for both head-and-neck, as well as other anatomical sites.
It was also observed that some of the artifacts in the MR would disappear during the process of generating synthetic CT’s. Artifact reduction in MR using deep learning has been previously studied34,35 therefore, while not the focus of this study, we are unsurprised at this result. It is well known that the variability in MRI intensity values can make image registration more challenging, and numerical techniques exist to mitigate these issues36. A possible added benefit of our technique may be an implicit correction in MRI intensity variations during the process of generating a synthetic CT, thus further aiding the registration. Our study was not designed to pursue this question, but would be an interesting future pursuit.
This study’s analysis process closely follows the recommendations for DIR quality assurance put forward by the AAPM Task Group 1321. They recommend evaluating registrations with landmark error, contour error, the Jacobian determinant, and inverse consistency. Our proposed method demonstrates superior landmark error and cord contour conformation, while also showing reasonable Jacobian determinant values and improved inverse consistency. These results make us confident that a CTsynth based deformable registration in the head-and-neck is a valuable tool, even in the setting of large neck flexion. We saw average landmark improvements of 3–4mm for our method, which is more than half of the 6mm improvement seen in registrations between the MR and aligned CT. The aligned CT acts as a surrogate for a more realistic synthetic CT since its anatomy closely matches the MR’s. The improvements are on the same order of margins (~3mm) used in head-and-neck radiotherapy, thus they are considered clinically relevant. Note that the utilized registration algorithm does not explicitly penalize a registration for violating inverse consistency, thus the non-zero MSE. The result indicates that the registration is biased with the choice of the target image, which is also seen in the directionality-dependent differences in registration. In fact, most registration algorithms used are asymmetric17 and this is an ongoing avenue of research.
There are a few limitations in the current study. First, although the data is unique to offer rigidly aligned CT/MR for validation, the patient number is relatively small, which has limited the power of statistical analysis. It may also have limited the quality of generated synthetic images. We performed five-fold cross validation to allow all cases to contribute to the performance analysis. Second, the MR images are from a low field scanner for MR-guided radiation therapy that provides inferior quality to the diagnostic images for head-and-neck registration. It is possible that the quality of the synthetic images can be improved based on diagnostic and multiparametric MR images. Improvements to the CTsynth generation could lead to further accuracy, as seen in the CTaligned registrations. Additionally, it is important to note that the MR and CT images were acquired with different resolutions, although they were resampled to be the same. Changing this additional variable could lead to different registration accuracies.
In our work, we used PET attenuation correction CT’s as the source of our large misalignement CT’s. In this way, the positioning would be very different from the immbolized planning CT. Previous work37 examined the difficulty of registering a PET attenuation correction CT with a treatment planning CT. They found large variability in alignment of the spinal cord (5.3mm) and mandible (5.4mm) post DIR, which were still superior to rigid registration (10.6mm and 5.5mm, for spinal cord and mandible, respectively). Our direct registration from the PET CT (CTnon-aligned) to the planning CT (CTaligned) resulted in an average landmark error of 4.5mm for the mandible and 4.7mm for the spinal cord, while the CTsynth bridge method had a 7.4 mm mandible error and 6.6mm error for the cord. These values are consistent with what was shown in the referenced study. While we only have one landmark and one contour in common with this study, it shows that even in the setting of CT-CT registration, large deformations in the head-and-neck can be difficult to register. In synergy with using a CTsynth bridge, improvements in mono-modality registration would also lead to better multimodal registration in the head-and-neck. Currently, research using neural networks offers some exciting new avenues in this regard, including completely learning-based unsupervised DVF generation38–41. However, the performance of these methods depends on the availability and quality of training sets, which are particularly challenging for multimodel registration. The proposed synthetic image bridge can work well with new deformable registration techniques optimized for single modality registration.
Conclusion
Multi-modality deformable registration is challenging, especially in regions of large deformation. CT and MR are important, complementary modalities in the treatment of head-and-neck cancer. By first transforming the MR into a CTsynth and running a synthetic mono-modal registration, we showed that we were able to produce improved registration results in the form of lower landmark error and more accurate contour warping. Furthermore, we showed that our DIR method improves inverse consistency and has realistic Jacobian determinant values. Continued efforts to improve CTsynth generation could advance this technique further.
Figure 7.
Transformation pairs in the opposite direction (e.g. non-aligned CT→MR and MR → non-aligned CT) were composed together and a single baseline CT was transformed under this composition. Given perfect inverse consistency, the original image should be recovered. The mean square error (MSE) was calculated between the original and transformed CT for the direct and synthetic CT bridged registrations, and in both directions. The boxplot bars show 5–95% range. A paired T-test was performed to evaluate significant difference.
Acknowledgement:
The study is supported in part by NIH R01CA188300 and R44CA183390.
Footnotes
Conflict of Interest: None
References
- 1.Brock KK, Mutic S, McNutt TR, Li H, Kessler ML. Use of image registration and fusion algorithms and techniques in radiotherapy: Report of the AAPM Radiation Therapy Committee Task Group No. 132: Report. Med Phys. 2017;44(7):e43–e76. doi: 10.1002/mp.12256 [DOI] [PubMed] [Google Scholar]
- 2.Heinrich MP, Jenkinson M, Bhushan M, et al. MIND: Modality independent neighbourhood descriptor for multi-modal deformable registration. Med Image Anal. 2012;16(7):1423–1435. doi: 10.1016/j.media.2012.05.008 [DOI] [PubMed] [Google Scholar]
- 3.Edmund JM, Nyholm T. A review of substitute CT generation for MRI-only radiation therapy. Radiat Oncol. 2017;12(1):28. doi: 10.1186/s13014-016-0747-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan FM. The Physics of Radiation Therapy. 4th ed. (Pine J, Murphy J, Panetta A, Larkin J, eds.). Baltimore, MD: Lippincott Williams & Wilkins; 2010. [Google Scholar]
- 5.Ulin K, Urie MM, Cherlow JM. Results of a multi-institutional benchmark test for cranial CT/MR image registration. Int J Radiat Oncol Biol Phys. 2010;77(5):1584–1589. doi: 10.1016/j.ijrobp.2009.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberson PL, McLaughlin PW, Narayana V, Troyer S, Hixson GV., Kessler ML. Use and uncertainties of mutual information for computed tomography/magnetic resonance (CT/MR) registration post permanent implant of the prostate. Med Phys. 2005;32(2):473–482. doi: 10.1118/1.1851920 [DOI] [PubMed] [Google Scholar]
- 7.Dean CJ, Sykes JR, Cooper RA, et al. An evaluation of four CT-MRI co-registration techniques for radiotherapy treatment planning of prone rectal cancer patients. Br J Radiol. 2012;85(1009):61–68. doi: 10.1259/bjr/11855927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daisne JF, Sibomana M, Bol A, Cosnard G, Lonneux M, Grégoire V. Evaluation of a multimodality image (CT, MRI and PET) coregistration procedure on phantom and head and neck cancer patients: Accuracy, reproducibility and consistency. Radiother Oncol. 2003;69(3):237–245. doi: 10.1016/j.radonc.2003.10.009 [DOI] [PubMed] [Google Scholar]
- 9.Nyholm T, Nyberg M, Karlsson MG, Karlsson M. Systematisation of spatial uncertainties for comparison between a MR and a CT-based radiotherapy workflow for prostate treatments. Radiat Oncol. 2009;4(1). doi: 10.1186/1748-717X-4-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weltens C, Menten J, Feron M, et al. Interobserver variations in gross tumor volume delineation of brain tumors on computed tomography and impact of magnetic resonance imaging. Radiother Oncol. 2001;60(1):49–59. doi: 10.1016/S0167-8140(01)00371-1 [DOI] [PubMed] [Google Scholar]
- 11.Rasch C, Keus R, Pameijer FA, et al. The potential impact of CT-MRI matching on tumor volume delineation in advanced head and neck cancer. Int J Radiat Oncol Biol Phys. 1997;39(4):841–848. doi: 10.1016/S0360-3016(97)00465-3 [DOI] [PubMed] [Google Scholar]
- 12.Emami B, Sethi A, Petruzzelli GJ. Influence of MRI on target volume delineation and IMRT planning in nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys. 2003;57(2):481–488. doi: 10.1016/S0360-3016(03)00570-4 [DOI] [PubMed] [Google Scholar]
- 13.Chung NN, Ting LL, Hsu WC, Lui LT, Wang PM. Impact of magnetic resonance imaging versus CT on nasopharyngeal carcinoma: Primary tumor target delineation for radiotherapy. Head Neck. 2004;26(3):241–246. doi: 10.1002/hed.10378 [DOI] [PubMed] [Google Scholar]
- 14.Chuter R, Prestwich R, Bird D, et al. The use of deformable image registration to integrate diagnostic MRI into the radiotherapy planning pathway for head and neck cancer. Radiother Oncol. 2017;122(2):229–235. doi: 10.1016/J.RADONC.2016.07.016 [DOI] [PubMed] [Google Scholar]
- 15.Brouwer CL, Steenbakkers RJHM, Bourhis J, et al. CT-based delineation of organs at risk in the head and neck region: DAHANCA, EORTC, GORTEC, HKNPCSG, NCIC CTG, NCRI, NRG Oncology and TROG consensus guidelines. Radiother Oncol. 2015;117(1):83–90. doi: 10.1016/j.radonc.2015.07.041 [DOI] [PubMed] [Google Scholar]
- 16.Fortunati V, Verhaart RF, Angeloni F, et al. Feasibility of multimodal deformable registration for head and neck tumor treatment planning. Int J Radiat Oncol Biol Phys. 2014;90(1):85–93. doi: 10.1016/j.ijrobp.2014.05.027 [DOI] [PubMed] [Google Scholar]
- 17.Sotiras A, Davatzikos C, Paragios N. Deformable Medical Image Registration: A Survey. IEEE Trans Med Imaging. 2013;32(7):1153–1190. doi: 10.1109/TMI.2013.2265603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnabel JA, Heinrich MP, Papież BW, Brady SJM. Advances and challenges in deformable image registration: From image fusion to complex motion modelling. Med Image Anal. 2016;33:145–148. doi: 10.1016/j.media.2016.06.031 [DOI] [PubMed] [Google Scholar]
- 19.Maes F, Vandermeulen D, Suetens P. Medical image registration using mutual information. Proc IEEE. 2003;91(10):1699–1722. doi: 10.1109/JPROC.2003.817864 [DOI] [PubMed] [Google Scholar]
- 20.Roy S, Carass A, Jog A, Prince JL, Lee J. MR to CT registration of brains using image synthesis. Med Imaging 2014 Image Process. 2014;9034:903419. doi: 10.1117/12.2043954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen M, Carass A, Jog A, Lee J, Roy S, Prince JL. Cross contrast multi-channel image registration using image synthesis for MR brain images. Med Image Anal. 2017;36(3):2–14. doi: 10.1016/j.media.2016.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao X, Yang J, Gao Y, Guo Y, Wu G, Shen D. Dual-core steered non-rigid registration for multi-modal images via bi-directional image synthesis. Med Image Anal. 2017;41:18–31. doi: 10.1016/j.media.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodfellow IJ, Pouget-Abadie J, Mirza M, et al. Generative Adversarial Networks. 2014:1–9. doi: 10.1001/jamainternmed.2016.8245 [DOI] [Google Scholar]
- 24.Hiasa Y, Otake Y, Takao M, et al. Cross-Modality Image Synthesis from Unpaired Data Using CycleGAN In: International Workshop on Simulation and Synthesis in Medical Imaging. Vol 1 Springer; 2018:31–41. doi: 10.1007/978-3-030-00536-8_4 [DOI] [Google Scholar]
- 25.Tanner C, Ozdemir F, Profanter R, Vishnevsky V. Generative Adversarial Networks for MR-CT Deformable Image Registration. arXiv Prepr arXiv180707349. 2018:1–11.
- 26.Wolterink JM, Dinkla AM, Savenije MHF, Seevinck PR, van den Berg CAT, Išgum I. Deep MR to CT synthesis using unpaired data. Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics). 2017;10557 LNCS:14–23. doi: 10.1007/978-3-319-68127-6_2 [DOI] [Google Scholar]
- 27.Zhu JY, Park T, Isola P, Efros AA. Unpaired Image-to-Image Translation Using Cycle-Consistent Adversarial Networks. Proc IEEE Int Conf Comput Vis. 2017;2017-Octob:2242–2251. doi: 10.1109/ICCV.2017.244 [DOI] [Google Scholar]
- 28.Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW. elastix : A Toolbox for Intensity-Based Medical Image Registration. 2010;29(1):196–205. [DOI] [PubMed] [Google Scholar]
- 29.Shamonin D, Bron E, Lelieveldt B, Smits M, Klein S, Staring M. Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer’s disease. Front Neuroinform. 2014;7(January):1–15. doi: 10.3389/fninf.2013.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson J, Alahi A, Fei-Fei L. Perceptual losses for real-time style transfer and super-resolution. Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics). 2016;9906 LNCS:694–711. doi: 10.1007/978-3-319-46475-6_43 [DOI] [Google Scholar]
- 31.Isola P, Zhu JY, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks Proc - 30th IEEE Conf Comput Vis Pattern Recognition, CVPR 2017; 2017;2017-Janua:5967–5976. doi: 10.1109/CVPR.2017.632 [DOI] [Google Scholar]
- 32.Klein S, Staring M, Pluim JPW. Comparison of gradient approximation techniques for optimisation of mutual information in nonrigid registration. 2005;(2):192. doi: 10.1117/12.595277 [DOI] [Google Scholar]
- 33.Dubuisson M-P, Jain AK. A modified Hausdorff distance for object matching. 2002;(1):566–568. doi: 10.1109/icpr.1994.576361 [DOI] [Google Scholar]
- 34.Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys. 2019;29(2):102–127. doi: 10.1016/j.zemedi.2018.11.002 [DOI] [PubMed] [Google Scholar]
- 35.Higaki T, Nakamura Y, Tatsugami F, Nakaura T, Awai K. Improvement of image quality at CT and MRI using deep learning. Jpn J Radiol. 2019;37(1):73–80. doi: 10.1007/s11604-018-0796-2 [DOI] [PubMed] [Google Scholar]
- 36.Baǧci U, Udupa JK, Bai L. The role of intensity standardization in medical image registration. Pattern Recognit Lett. 2010;31(4):315–323. doi: 10.1016/j.patrec.2009.09.010 [DOI] [Google Scholar]
- 37.Hwang AB, Bacharach SL, Yom SS, et al. Can Positron Emission Tomography (PET) or PET/Computed Tomography (CT) Acquired in a Nontreatment Position Be Accurately Registered to a Head-and-Neck Radiotherapy Planning CT? Int J Radiat Oncol Biol Phys. 2009;73(2):578–584. doi: 10.1016/j.ijrobp.2008.09.041 [DOI] [PubMed] [Google Scholar]
- 38.Krebs J, Mansi T, Delingette H, et al. Robust Non-rigid Registration Through Agent-Based Action Learning. Med Image Comput Comput Assist Interv − MICCAI 2017. 2017;10433:344–352. doi: 10.1007/978-3-319-66182-7 [DOI] [Google Scholar]
- 39.Simonovsky M, Gutierrez-Becker B, Mateus D, Navab N, Komodakis N. A Deep Metric for Multimodal Registration. Med Image Comput Comput Interv -- MICCAI 2016. 2016;9902:10–18. doi: 10.1007/978-3-319-46726-9 [DOI] [Google Scholar]
- 40.Dalca AV, Balakrishnan G, Guttag J, Sabuncu MR. Unsupervised learning of probabilistic diffeomorphic registration for images and surfaces. Med Image Anal. 2019;57:226–236. doi: 10.1016/j.media.2019.07.006 [DOI] [PubMed] [Google Scholar]
- 41.Balakrishnan G, Zhao A, Sabuncu MR, Dalca AV, Guttag J. An Unsupervised Learning Model for Deformable Medical Image Registration In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition. IEEE; 2018:9252–9260. doi: 10.1109/CVPR.2018.00964 [DOI] [Google Scholar]