Abstract
Tissue-resident lymphocytes that lack expression of rearranged antigen receptors and are lineage negative for classical T and B cell markers are collectively known as innate lymphoid cells (ILCs). The ILC family is remarkably heterogeneous and exhibits plasticity; however, mature ILCs can be grouped based on their steady state expression of distinct surface receptors and transcription factors as well as production of signature cytokines following activation. The study of ILC subsets in mouse and human tissues has revealed that the elicitation and magnitude of their effector functions is determined by a combination of extrinsic cues specific to the niches in which they reside. In this short review, we will summarize some recent findings related to tissue-specific signals that govern ILC responses and localization.
Keywords: innate lymphoid cell, natural killer cell, lymphoid tissue inducer, Stroma
ILC2 circuits: stromal- and tuft cell-derived cytokines in ILC2 activation
The subgrouping of ILCs tracks with the established paradigm for adaptive T helper cell subsets. ILC2s are the innate counterparts of Th2 cells as they highly express Gata3 and produce IL-5 and IL-13. Secretion of IL-25, IL-33 and TSLP following epithelial damage during injury or parasitic infections acts as a trigger for production of IL-5 and IL-13 by ILC2s. Recent efforts have been made to define the specific cellular sources of these ILC2-activating cytokines and what factors control their production. Reporter mice revealed that mucosal tuft cells, which are rare chemosensory epithelial cells, are the main producers of IL-25 in the intestines [1]. During helminth infection, tuft cell-derived IL-25 triggers IL-13 production by ILC2s that acts in a circuit to promote increased differentiation of tuft and goblet cells [1,2]. Dietary/microbiota-derived succinate was recently identified as an upstream signal that promotes IL-25 production by tuft cells to initiate the tuft cell-ILC2 circuit and provides protective barrier immunity [3,4]. It will be interesting to test whether modulation of intestinal succinate via alterations in diet or microbiota can be harnessed to combat parasitic infections or counteract aberrant type 1/3 inflammation.
Stromal cells have recently gained recognition as an important source of IL-33 that regulates ILC2 expansion and function within numerous compartments. In the mouse lung, ILC2s localize with adventitial stromal cells (ASCs) which are important for tissue remodeling and inflammatory responses [6]. ASC-derived IL-33 acts on ILC2s and ILC2-derived IL-13 acts on ASCs thereby forming a circuit (reminiscent of that formed between tuft cells and ILC2s in the intestines) that enables reciprocal cell expansion following helminth infection. Similar examples of stromal-ILC2 cell interactions have been described in other tissues in mice, including pericryptal fibroblasts and colonic mesenchymal cells in the intestines, cardiac fibroblasts in the heart, and adipose stem and progenitor cells in white adipose tissue [5–9]. What are the stimuli that direct stromal cell production of IL-33? Are they different across tissues? Such information could be used to target ILC2 activation or inhibition within specific compartments. Elucidating those signals could also inform about the circumstances in which ILC2 activation is immunologically required in particular tissues and the conditions in which ILC2s are the culprits of pathologic type 2 inflammation.
In addition to epithelial- and stromal-derived cytokines, recent work has brought to light the importance of neuro-immune crosstalk in lung and intestinal ILC2 programs. ILC2s highly and selectively express the neuropeptide receptor NMUR1 at steady state and co-localize with enteric neurons [10–12]. Furthermore, addition of the NMUR1 ligand neuromedin U (NmU) promotes ILC2 proliferation and amplifies the effects of ILC2-activating cytokines. In addition to neuropeptides, ILC2s can also sense vasoactive intestinal peptide (VIP) which, in combination with IL-7, can increase expression of IL-5 in vitro [13]. Therefore, tissue-specific signals that regulate ILC2 effector functions include not only cytokines but also peptides that are likely dynamically regulated across tissues and immune states.
Although IL-25, IL-33 and TSLP are clearly important for activating ILC2s, ILC2 functionality is not strictly based on these signals. Deletion of IL-25, IL-33 and TLSP receptors in ILC2s does not affect their numbers nor their production of IL-5 in the skin [14]. In this setting, IL-18 is the key factor that activates type 2 inflammation. The importance of the microanatomical niche in ILC2 regulation is perhaps best exemplified in single-cell RNAseq data. Although ILC2s isolated from different tissues share a common program of activation following exposure to IL-25/IL-33/TSLP in vitro, with unique expression programs induced by each cytokine [10], their steady-state transcriptional identity is inextricably linked to the tissues in which they reside even in the absence of a microbiota and deficiency in IL-25/IL-33/TSLP signaling [14]. Presently, the tissue-specific signals that guide the establishment and maintenance of resident ILC2 programs are largely undefined.
Location, location, location: importance of the ILC3 microanatomical niche
Similar to ILC2s, ILC3s also function within several known circuits. ILC3s mirror Th17 cells with their expression of RORγt and production of IL-17 and IL-22. The prototypical fetal lymphoid tissue inducer (LTi) cells are group 3 ILC family members that are critical for lymphoid tissue development. Secondary lymphoid organ (SLO) formation depends on LTi-derived lymphotoxin (LT) α1β2 ligation of LTβ-receptor on stromal cells [15–17]. The activation of LTβ-receptor on the stromal cells in turn induces their maturation into lymphoid tissue organizer cells that feedback to LTi by promoting their continued recruitment within the SLO. Intense focus has been paid to the role of adult LTi-like ILC3 in lymphoid tissue development in the intestines. Cryptopatches (CP) and isolated lymphoid follicles (ILFs) are tertiary lymphoid tissues that contain most of the adult LTi-like ILC3 in the small intestines and colon [18,19]. Clusters of LTi-like ILC3s are surrounded by dendritic cells and stromal cells in CPs as well as B cells in ILFs. Recently is was revealed that recruitment of ILC3s into colonic CPs and ILFs is a function of fibroblastic stromal cells whereby their production of the oxysterol 7α,25-hydroxycholesterol acts as chemoattractant by binding to GPR183 expressed by LTi-like ILC3s [20]. Whether diet, the microbiota, or inflammatory conditions regulate production of 7α,25-hydroxycholesterol in the CP niche and the consequent dynamics on ILC3 localization remain to be elucidated.
In addition to LTi-like ILC3-stromal interactions within CPs, ILC3s also form a regulatory circuit all on their own. Expression of the tumor necrosis factor superfamily member receptor activator of nuclear factor κB ligand (RANKL) and RANK expressed on LTi-like ILC3s functions as a sort of quorum sensing mechanism [21]. Loss of either RANKL or RANK on ILC3s results in an expansion of LTi-like ILC3s as well as enhanced RORγt expression and production of IL-17 and IL-22. Therefore, intestinal LTi-like ILC3 populations are controlled by their self-interaction and is a particularly relevant autoregulatory mechanism for those cells residing within CPs and ILFs where their population density is highest.
Other ILC3 circuits include the maintenance of intestinal stem cells via IL-22-mediated action that limits tissue damage by preserving stem cells [22]. Additionally, IL-22 production by ILC3s following intestinal colonization with segmented filamentous bacteria (SFB) induces serum amyloid A (SAA) production by epithelial cells that drives recruitment of SFB-specific Th17 cells [23]. Overall, ILC3 functionality within the intestines depends on the microanatomical niche of where the cells reside. Whether it be within CPs and ILFs where effector responses to pathogens are mounted, in proximity to intestinal stem cells in crypts where damage responses are prompted, or poised within the intraepithelial space ready to receive signals from DCs to drive the SAA-Th17 axis – the many roles of ILC3s in the intestines are choreographed via strategic recruitment and positioning of these lymphocytes within microanatomical niches. Despite this ILC3 pattern of circuitry in the intestines and the role of neuro-immune crosstalk in ILC2 function, we know very little about ILC3 interaction with enteric neurons and such studies will be an interesting line of future inquiry. Furthermore, the function of ILC3s beyond production of IL-17 and IL-22 in regulating gut homeostasis is an active area of investigation as recent work reported that ILC3-derived IL-2 is essential for maintaining intestinal regulatory T cells [24].
Group 1 ILC identity crisis
The group 1 ILC family includes ILC1s as well as cNK cells. ILC1s mirror Th1 cells as they express the transcription factor T-bet and produce IFN-γ. ILC1s are closely related to conventional NK (cNK) cells but are not dependent on Eomes for their development and are not as cytotoxic. Depending on the tissue, ILC1s have differential dependencies on specific transcription factors. In mice, Nfil3 is required for ILC1 development in all tissues examined except for the salivary gland [25]. Hobit, a transcription factor important for tissue-residency programs of T cells, is required specifically for liver ILC1s but not ILC1s in other tissues or liver cNK cells, which instead depend on Blimp1 [26–28]. Aside from transcription factor dependencies, another feature of ILC1s that distinguishes them from cNK is imprinting by TGF-β family members as a result of their tissue-residency, which has been observed in salivary gland and intestinal intraepithelial ILC1s [29,30].
Recent efforts have been made to better characterize the tissue-specific transcriptional programs of group 1 ILCs in mice and humans [31–35]. It has been challenging to identify a true “core” ILC1 signature versus cNK cells due to the heterogeneity of this family within and across tissues. Although the expression of IL-7R is a hallmark of ILC2 and ILC3 populations, ILC1s exhibit more variability in their expression of this receptor. In mice, the majority of liver and salivary gland ILC1s are IL7R− as well as intraepithelial ILC1s (ielICL1) which aligns them with cNK cells [25,34]. Similarly, IL-7R expression on human ILC1s is variegated, with IL-7R+ and IL-7R− populations identified in tonsils and intestinal intraepithelial populations [30,32,36,37]. Recently, a multi-pronged study integrating single-cell RNAseq and CyTOF analysis found that IL-7R decreases in a gradation as human ILC3 transition into ILC1 [35], although it is unclear why these transitioning cells reduce IL-7R expression. Moreover, the dynamics of ILC1 IL-7R expression across individuals and health states warrants more exploration.
As stated, group 1 ILCs in mice are remarkably heterogenous across tissues. Therefore, in order to more accurately denote cell identities nomenclature for specific subpopulations within tissues will likely be adopted (for instance, the already commissioned “iel-ILC1”). However, in humans the tissue microenvironment appears to be less of a determinant of transcriptional group 1 ILC identity than in mice. Single cell RNA-seq analysis revealed ILC1 and cNK isolated from human lung, jejunum and spleen clustered according to cell ontogeny independent of their tissue origin [32]. There was also extensive transcriptional overlap between human tissue ILC1s and splenic cNK cells, although intestinal ILC1s proved to be the most transcriptionally heterogenous with overlapping and non-overlapping cNK signatures identified similar to what is observed in mice [32,34]. What is the explanation for the discordant degree of diversity of group 1 ILC populations in mice versus humans? Does pathogen exposure, metabolic state, or diet somehow promote a contraction in group 1 ILC diversity as a product of overall life experience that is not reflected in lab mice? Perhaps the diversity in human ILC1s may be managed by a spectrum of cellular plasticity as indicated by a recent study that describes the existence of intermediate ILC3-ILC1 cell identities [35]. Or perhaps, as more human tissues are sampled and sequenced from healthy and diseased donors alike, our appreciation for human group 1 ILC heterogeneity will expand in turn.
Not strictly resident: ILCs on the move
ILC development and subsequent expansion in tissues proceeds following seeding by a progenitor from the bone marrow or fetal liver [38]. As better genetic tools for marking and tracking ILCs become available, it is now becoming clear that ILCs are not strictly tissue-resident populations and that they can traffic from the tissues in which they developed. Accordingly, they can play protective or inflammatory roles at peripheral sites. Some of the first evidence for recirculation of ILCs came from the discovery that CD49a+ liver ILC1s were capable of conferring hapten-specific memory responses in mice [39]. Recent work has delineated the steps of this process, whereby hapten sensitization induces recruitment of IL-7R+ ILC1s into skin-draining lymph nodes (LNs) via CXCR3 where they acquire memory potential [40]. Subsequently, these memory ILC1s egress from LNs via sphingosine-1-phosphate (S1P) receptors, a G protein-coupled receptor family that is also required for T and cNK cell movement across lymphatic endothelial barriers [41,42]. Following egress from the LNs, memory ILC1s are recruited to the liver where they maintain long-term residency until challenge.
The fate and migration of other ILC populations under non-steady-state conditions is an active area of research. During Nippostrongylus brasiliensis infection or following i.p. injection of IL-25, “inflammatory” KLRG1hi ILC2s (iILC2s) appear in the lung, liver, and spleen [43]. iILC2s can proliferate and expand the ILC2 pool or can develop into ILC3-like cells capable of producing IL-17. Recent work has revealed that iILC2s from the small intestine lamina propria (siLP) can migrate to peripheral organs, such as the lung, liver and spleen, transiently expand during acute infection, and subsequently contribute to the steady-state ILC2 pool following pathogen clearance [44]. The accumulation of iILC2s at distant sites was dependent on S1P signaling, similar to the mechanism controlling the circulation of T cells, cNK cells and memory ILC1s [40–42]. Importantly, survival of Rag1−/− mice during early-stage N. brasiliensis lung infection was dependent on the migration of iILC2s indicating that siLP-derived iILC2s provide crucial multiorgan protection to the host.
Future perspectives in ILC biology
The characterization of ILCs in tissues and the identification of the signals, both at steady-state and during inflammation, that guide their trafficking, activation and function has been the focus of this field for the last decade. Emerging areas in ILC biology are now shifting attention from the cellular niche to the intrinsic molecular programs that these lymphocytes utilize. Studies on T and NK cells have revealed the importance of specific metabolic pathways in driving polarization and effector responses as well as in the generation of memory. For example, Th17 cells employ aerobic glycolysis for energy whereas Tregs and memory T cells rely on fatty acid oxidization [45]. The metabolic programs utilized by ILCs and the context-dependent metabolic switches and checkpoints that they confront is an active area of investigation. Recent data on ILC3s has provided evidence that, unlike T cells, these cells simultaneously utilize both glycolysis and mitochondrial ROS to meet the metabolic demands of activation [46]. In addition to metabolism, epigenetics is another emerging area in ILC biology. The functional polarization of ILCs is controlled by the concerted action of enhancers and transcription factors that poise these cells for rapid responses [33,47–49]. However, more work is needed to determine the exact regulatory programs under their control and how they differ from T cell programs. A deeper molecular definition of ILC identity will aid in our understanding of what distinguishes these cells within the immune system and will likely reveal novel aspects of their biology beyond prototypical effector responses.
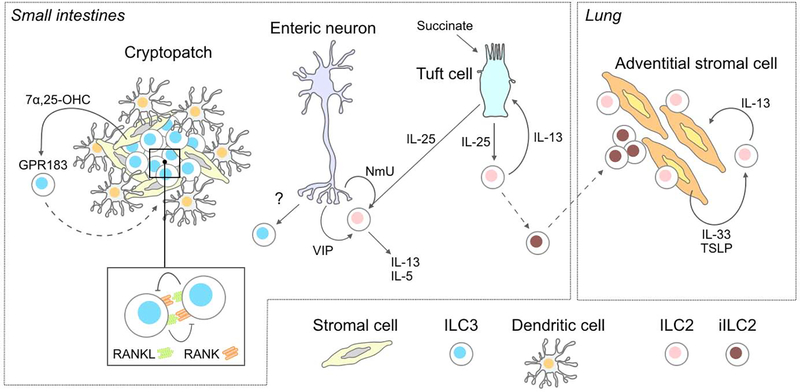
Innate lymphoid cell circuitry.
In the small intestines, stromal cells localized in cryptopatches produce 7α,25-hydroxycholesterol that engages GPR183 expressed by ILC3s. Sensing of this oxysterol promotes chemotaxis of ILC3s into the tertiary lymphoid tissue [20]. Interaction of RANK with RANKL between adjacent ILC3s suppresses effector activity, thereby allowing cell clustering to mediate cell activation [21]. ILC2s are activated by neuropeptides (neuromedin U, NmU) and vasoactive intestinal peptides (VIP) produced by enteric neurons [10–13]. Succinate in the intestinal lumen triggers IL-25 production by chemosensory tuft cells that activates ILC2 expression of IL-13 which signals back onto tuft cells increasing their differentiation [3,4]. During N. brasiliensis infection, IL-25 signaling on ILC2s promotes an inflammatory KLRG1hi ILC2 (iILC2) phenotype [43]. iILC2s in the small intestine lamina propria can migrate to peripheral organs, such as the lung, where they contribute to systemic pathogen control [44]. Lung adventitial stromal cells, which contribute to tissue remodeling and inflammatory responses, form a circuit with ILC2s whereby stromal cell production of IL-33 and TSLP induces activation of ILC2s and expression of IL-13 that promotes ASC expansion [5].
Highlights.
Signaling circuits between ILC2s and stromal cells as well as tuft cells dictates ILC2 activation in different tissue niches.
ILC3 function in the intestines is determined in part by residence in microanatomical niches including tertiary lymphoid tissues and crypts.
Group 1 ILCs are tremendously heterogenous in mice, but this appears to be a discordant phenotype from what is observed in humans.
Under certain conditions, ILC1s and ILC2s can become migratory and contribute to host defense systemically.
Acknowledgments
Declarations of interest: Marco Colonna receives research support from Pfizer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.von Moltke J, Ji M, Liang HE, Locksley RM: Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 2016, 529:221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerbe F, Sidot E, Smyth DJ, Ohmoto M, Matsumoto I, Dardalhon V, Cesses P, Garnier L, Pouzolles M, Brulin B, et al. : Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 2016, 529:226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider C, O’Leary CE, von Moltke J, Liang HE, Ang QY, Turnbaugh PJ, Radhakrishnan S, Pellizzon M, Ma A, Locksley RM: A Metabolite-Triggered Tuft Cell-ILC2 Circuit Drives Small Intestinal Remodeling. Cell 2018, 174:271–284 e214.Although tuft cell-derived IL-25 was reported to trigger IL-13 production by intestinal ILC2s in 2016, this and work by Nadjsombati et al. (Immunity, 2018) identified luminal succinate as the upstream signal that activates tuft cells.
- 4.Nadjsombati MS, McGinty JW, Lyons-Cohen MR, Jaffe JB, DiPeso L, Schneider C, Miller CN, Pollack JL, Nagana Gowda GA, Fontana MF, et al. : Detection of Succinate by Intestinal Tuft Cells Triggers a Type 2 Innate Immune Circuit. Immunity 2018, 49:33–41 e37.Although tuft cell-derived IL-25 was reported to trigger IL-13 production by intestinal ILC2s in 2016, this and work by Schneider et al. (Cell, 2018) identified luminal succinate as the upstream signal that activates tuft cells.
- 5.Mahapatro M, Foersch S, Hefele M, He GW, Giner-Ventura E, McHedlidze T, Kindermann M, Vetrano S, Danese S, Gunther C, et al. : Programming of Intestinal Epithelial Differentiation by IL-33 Derived from Pericryptal Fibroblasts in Response to Systemic Infection. Cell Rep 2016, 15:1743–1756. [DOI] [PubMed] [Google Scholar]
- 6.Dahlgren MW, Jones SW, Cautivo KM, Dubinin A, Ortiz-Carpena JF, Farhat S, Yu KS, Lee K, Wang C, Molofsky AV, et al. : Adventitial Stromal Cells Define Group 2 Innate Lymphoid Cell Tissue Niches. Immunity 2019, 50:707–722 e706.With a focus on physiological tissue niches, this work used quantitative imaging to identify a reciprocal interaction between ILC2s and adventitial stromal cells that supports effector functions and prolfieration, respectively.
- 7.Kinchen J, Chen HH, Parikh K, Antanaviciute A, Jagielowicz M, Fawkner-Corbett D, Ashley N, Cubitt L, Mellado-Gomez E, Attar M, et al. : Structural Remodeling of the Human Colonic Mesenchyme in Inflammatory Bowel Disease. Cell 2018, 175:372–386 e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahlakoiv T, Flamar AL, Johnston LK, Moriyama S, Putzel GG, Bryce PJ, Artis D: Stromal cells maintain immune cell homeostasis in adipose tissue via production of interleukin-33. Sci Immunol 2019, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bracamonte-Baran W, Chen G, Hou X, Talor MV, Choi HS, Davogustto G, Taegtmeyer H, Sung J, Hackam DJ, Nauen D, et al. : Non-cytotoxic Cardiac Innate Lymphoid Cells Are a Resident and Quiescent Type 2-Commited Population. Front Immunol 2019, 10:634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallrapp A, Riesenfeld SJ, Burkett PR, Abdulnour RE, Nyman J, Dionne D, Hofree M, Cuoco MS, Rodman C, Farouq D, et al. : The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature 2017, 549:351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardoso V, Chesne J, Ribeiro H, Garcia-Cassani B, Carvalho T, Bouchery T, Shah K, Barbosa-Morais NL, Harris N, Veiga-Fernandes H: Neuronal regulation of type 2 innate lymphoid cells via neuromedin U. Nature 2017, 549:277–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klose CSN, Mahlakoiv T, Moeller JB, Rankin LC, Flamar AL, Kabata H, Monticelli LA, Moriyama S, Putzel GG, Rakhilin N, et al. : The neuropeptide neuromedin U stimulates innate lymphoid cells and type 2 inflammation. Nature 2017, 549:282–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nussbaum JC, Van Dyken SJ, von Moltke J, Cheng LE, Mohapatra A, Molofsky AB, Thornton EE, Krummel MF, Chawla A, Liang HE, et al. : Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 2013, 502:245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ricardo-Gonzalez RR, Van Dyken SJ, Schneider C, Lee J, Nussbaum JC, Liang HE, Vaka D, Eckalbar WL, Molofsky AB, Erle DJ, et al. : Tissue signals imprint ILC2 identity with anticipatory function. Nat Immunol 2018, 19:1093–1099.Single-cell RNAseq analyses determined that ILC2s group according to their tissue of origin, thereby revealing the importance of tissue-derived signals in guiding the maturation and activities of these ILCs.
- 15.Roozendaal R, Mebius RE: Stromal cell-immune cell interactions. Annu Rev Immunol 2011, 29:23–43. [DOI] [PubMed] [Google Scholar]
- 16.Scandella E, Bolinger B, Lattmann E, Miller S, Favre S, Littman DR, Finke D, Luther SA, Junt T, Ludewig B: Restoration of lymphoid organ integrity through the interaction of lymphoid tissue-inducer cells with stroma of the T cell zone. Nat Immunol 2008, 9:667–675. [DOI] [PubMed] [Google Scholar]
- 17.Benezech C, White A, Mader E, Serre K, Parnell S, Pfeffer K, Ware CF, Anderson G, Caamano JH: Ontogeny of stromal organizer cells during lymph node development. J Immunol 2010, 184:4521–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eberl G, Sawa S: Opening the crypt: current facts and hypotheses on the function of cryptopatches. Trends Immunol 2010, 31:50–55. [DOI] [PubMed] [Google Scholar]
- 19.Tsuji M, Suzuki K, Kitamura H, Maruya M, Kinoshita K, Ivanov, II, Itoh K, Littman DR, Fagarasan S: Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity 2008, 29:261–271. [DOI] [PubMed] [Google Scholar]
- 20.Emgard J, Kammoun H, Garcia-Cassani B, Chesne J, Parigi SM, Jacob JM, Cheng HW, Evren E, Das S, Czarnewski P, et al. : Oxysterol Sensing through the Receptor GPR183 Promotes the Lymphoid-Tissue-Inducing Function of Innate Lymphoid Cells and Colonic Inflammation. Immunity 2018, 48:120–132 e128.This work identified stromal cell-derived 7alpha,25-hydroxycholesterol as a chemotactic factor that recruits LTi-like ILC3s via GPR183 into tertiary lymphoid tissues in the intestines.
- 21.Bando JK, Gilfillan S, Song C, McDonald KG, Huang SC, Newberry RD, Kobayashi Y, Allan DSJ, Carlyle JR, Cella M, et al. : The Tumor Necrosis Factor Superfamily Member RANKL Suppresses Effector Cytokine Production in Group 3 Innate Lymphoid Cells. Immunity 2018, 48:1208–1219 e1204.This work identified stromal cell-derived 7alpha,25-hydroxycholesterol as a chemotactic factor that recruits LTi-like ILC3s via GPR183 into tertiary lymphoid tissues in the intestines.
- 22.Aparicio-Domingo P, Romera-Hernandez M, Karrich JJ, Cornelissen F, Papazian N, Lindenbergh-Kortleve DJ, Butler JA, Boon L, Coles MC, Samsom JN, et al. : Type 3 innate lymphoid cells maintain intestinal epithelial stem cells after tissue damage. J Exp Med 2015, 212:1783–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, Lee JY, Ziel JW, Miraldi ER, Domingos AI, et al. : An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses. Cell 2015, 163:381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou L, Chu C, Teng F, Bessman NJ, Goc J, Santosa EK, Putzel GG, Kabata H, Kelsen JR, Baldassano RN, et al. : Innate lymphoid cells support regulatory T cells in the intestine through interleukin-2. Nature 2019, 568:405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cortez VS, Fuchs A, Cella M, Gilfillan S, Colonna M: Cutting edge: Salivary gland NK cells develop independently of Nfil3 in steady-state. J Immunol 2014, 192:4487–4491. [DOI] [PubMed] [Google Scholar]
- 26.Mackay LK, Minnich M, Kragten NA, Liao Y, Nota B, Seillet C, Zaid A, Man K, Preston S, Freestone D, et al. : Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 2016, 352:459–463. [DOI] [PubMed] [Google Scholar]
- 27.Weizman OE, Adams NM, Schuster IS, Krishna C, Pritykin Y, Lau C, Degli-Esposti MA, Leslie CS, Sun JC, O’Sullivan TE: ILC1 Confer Early Host Protection at Initial Sites of Viral Infection. Cell 2017, 171:795–808 e712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lunemann S, Martrus G, Goebels H, Kautz T, Langeneckert A, Salzberger W, Koch M, M JB, Nashan B, van Gisbergen K, et al. : Hobit expression by a subset of human liverresident CD56(bright) Natural Killer cells. Sci Rep 2017, 7:6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cortez VS, Cervantes-Barragan L, Robinette ML, Bando JK, Wang Y, Geiger TL, Gilfillan S, Fuchs A, Vivier E, Sun JC, et al. : Transforming Growth Factor-beta Signaling Guides the Differentiation of Innate Lymphoid Cells in Salivary Glands. Immunity 2016, 44:1127–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, Cella M, Colonna M: Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity 2013, 38:769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crinier A, Milpied P, Escaliere B, Piperoglou C, Galluso J, Balsamo A, Spinelli L, Cervera-Marzal I, Ebbo M, Girard-Madoux M, et al. : High-Dimensional Single-Cell Analysis Identifies Organ-Specific Signatures and Conserved NK Cell Subsets in Humans and Mice. Immunity 2018, 49:971–986 e975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yudanin NA, Schmitz F, Flamar AL, Thome JJC, Tait Wojno E, Moeller JB, Schirmer M, Latorre IJ, Xavier RJ, Farber DL, et al. : Spatial and Temporal Mapping of Human Innate Lymphoid Cells Reveals Elements of Tissue Specificity. Immunity 2019, 50:505–519 e504.Using flow cytometric and transcirptional profiling, this work catalogs the heterogenetiy of human ILCs from multiple tissues across diseased and aging states.
- 33.Collins PL, Cella M, Porter SI, Li S, Gurewitz GL, Hong HS, Johnson RP, Oltz EM, Colonna M: Gene Regulatory Programs Conferring Phenotypic Identities to Human NK Cells. Cell 2019, 176:348–360 e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, Gilfillan S, Colonna M, Immunological Genome C: Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol 2015, 16:306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cella M, Gamini R, Secca C, Collins PL, Zhao S, Peng V, Robinette ML, Schettini J, Zaitsev K, Gordon W, et al. : Subsets of ILC3-ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat Immunol 2019.This is the first work to show that conversion of human ILC3s into ILC1-like cells occurs in vivo. A specturm of transitional ILC3-ILC1 clusters in human tissues were identified using a combination of single cell RNAseq with velocity analysis, CYTOF and humanized mouse work.
- 36.Bjorklund AK, Forkel M, Picelli S, Konya V, Theorell J, Friberg D, Sandberg R, Mjosberg J: The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol 2016, 17:451–460. [DOI] [PubMed] [Google Scholar]
- 37.Simoni Y, Fehlings M, Kloverpris HN, McGovern N, Koo SL, Loh CY, Lim S, Kurioka A, Fergusson JR, Tang CL, et al. : Human Innate Lymphoid Cell Subsets Possess Tissue-Type Based Heterogeneity in Phenotype and Frequency. Immunity 2017, 46:148–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zook EC, Kee BL: Development of innate lymphoid cells. Nat Immunol 2016, 17:775–782. [DOI] [PubMed] [Google Scholar]
- 39.Peng H, Jiang X, Chen Y, Sojka DK, Wei H, Gao X, Sun R, Yokoyama WM, Tian Z: Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J Clin Invest 2013, 123:1444–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Peng H, Cong J, Wang X, Lian Z, Wei H, Sun R, Tian Z: Memory formation and long-term maintenance of IL-7Ralpha(+) ILC1s via a lymph node-liver axis. Nat Commun 2018, 9:4854.Expanding on their intial 2013 report that liver ILC1s confer hapten-specific memory responses, this study reveals the signals and receptors that guide the establishment of a liver memory IL-7Ralpha+ ILC1 population.
- 41.Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, Cyster JG: Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 2004, 427:355–360. [DOI] [PubMed] [Google Scholar]
- 42.Walzer T, Chiossone L, Chaix J, Calver A, Carozzo C, Garrigue-Antar L, Jacques Y, Baratin M, Tomasello E, Vivier E: Natural killer cell trafficking in vivo requires a dedicated sphingosine 1-phosphate receptor. Nat Immunol 2007, 8:1337–1344. [DOI] [PubMed] [Google Scholar]
- 43.Huang Y, Guo L, Qiu J, Chen X, Hu-Li J, Siebenlist U, Williamson PR, Urban JF Jr., Paul WE: IL-25-responsive, lineage-negative KLRG1(hi) cells are multipotential ‘inflammatory’ type 2 innate lymphoid cells. Nat Immunol 2015, 16:161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Y, Mao K, Chen X, Sun MA, Kawabe T, Li W, Usher N, Zhu J, Urban JF Jr., Paul WE, et al. : S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science 2018, 359:114–119.Although typically considered tissue-resident, this study expands our understanding of ILC biology by demonstrating that inflammaotry ILC2s induced in the intestinal lamina propria migrate systemically to diverse tissues where they contribute to local tissue repair and defense.
- 45.Buck MD, O’Sullivan D, Pearce EL: T cell metabolism drives immunity. J Exp Med 2015, 212:1345–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Luccia B, Gilfillan S, Cella M, Colonna M, Huang SC: ILC3s integrate glycolysis and mitochondrial production of reactive oxygen species to fulfill activation demands. J Exp Med 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mowel WK, McCright SJ, Kotzin JJ, Collet MA, Uyar A, Chen X, DeLaney A, Spencer SP, Virtue AT, Yang E, et al. : Group 1 Innate Lymphoid Cell Lineage Identity Is Determined by a cis-Regulatory Element Marked by a Long Non-coding RNA. Immunity 2017, 47:435–449 e438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cortez VS, Ulland TK, Cervantes-Barragan L, Bando JK, Robinette ML, Wang Q, White AJ, Gilfillan S, Cella M, Colonna M: SMAD4 impedes the conversion of NK cells into ILC1-like cells by curtailing non-canonical TGF-beta signaling. Nat Immunol 2017, 18:9951003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shih HY, Sciume G, Mikami Y, Guo L, Sun HW, Brooks SR, Urban JF Jr., Davis FP, Kanno Y, O’Shea JJ: Developmental Acquisition of Regulomes Underlies Innate Lymphoid Cell Functionality. Cell 2016, 165:1120–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]