Abstract
Ischemic reperfusion injury after a stroke is a leading cause of mortality and disability due to neuronal loss and tissue damage. Mitochondrial dysfunction plays a major role in the reperfusion-injury sequelae, and offers an attractive drug target. Mitochondrial derived reactive oxygen species (ROS) and resultant apoptotic cascade are among the primary mechanisms of neuronal death following ischemia and reperfusion injury. Here we optimized a nanoparticle formulation for the mitoNEET ligand NL-1, to target mitochondrial dysfunction post ischemic reperfusion (IR) injury. NL-1, a hydrophobic drug, was formulated using PLGA polymers with a particle size and entrapment efficiency of 123.9 ± 17.1 nm and 59.7 ± 10.1 %, respectively. The formulation was characterized for physical state of NL-1, in vitro release, uptake and nanoparticle localization. A near complete uptake of nanoparticles was found to occur by three hours, with the process being energy-dependent and occurring via caveolar mediated endocytosis. The fluorescent nanoparticles were found to localize in the cytoplasm of the endothelial cells. An in vitro oxygen glucose deprivation (OGD) model to mimic IR was employed for in vitro efficacy testing in murine brain vascular endothelium cells (bEND.3 cells). Efficacy studies showed that both NL-1 and the nanoparticles loaded with NL-1 had a protective activity against peroxide generation, and displayed improved cellular viability, as seen via reduction in cellular apoptosis. Finally, PLGA nanoparticles were found to have a non-toxic profile in vitro, and were found to be safe for intravenous administration. This study lays the preliminary work for potential use of mitoNEET as a target and NL-1 as a therapeutic for the treatment of cerebral ischemia and reperfusion injury.
Keywords: Stroke; cisd; ferroptosis; blood-brain barrier, BBB
Graphical Abstract
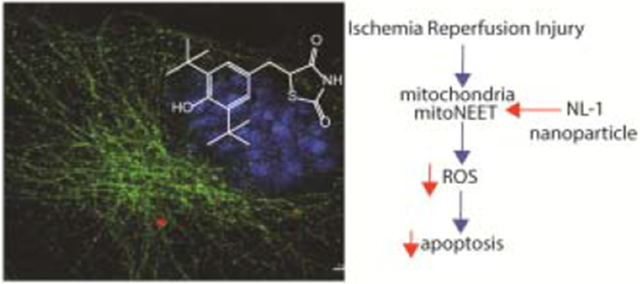
1. Introduction:
Stroke is a leading cause of mortality and morbidity worldwide, with no proven therapies to slow down the neuronal cell death after a stroke.1 About 80% of stroke cases consist of ischemic stroke, wherein the blood supply to a brain region is impeded. As a result, the brain tissue is starved of vital nutrients such as oxygen and glucose. Unless the occlusion is removed and blood supply is restored, prolonged deprivation of nutrients leads to formation of an infarct in the brain tissue which is a result of cell damage to neurons, microglia, astrocytes and endothelial cells. Multiple mechanisms of cell damage are activated following cerebral ischemia and reperfusion which can be a function of location in the brain as well as the time elapsed since the onset of ischemia. Although it might seem counterintuitive, restoration of blood supply, known as reperfusion, can cause further damage. Effects of reperfusion, also known as reperfusion injury, are the causative factors for cerebral edema and blood brain barrier (BBB) disruption2.
Energy failure is the immediate consequence of this ischemic event, as the brain cells are unable to generate ATP molecules due to lack of glucose and oxygen. A decline in ATP generation results in many downstream events such as the dysfunction of ATP pumps, loss of membrane potential, prevention of neurotransmitter reuptake leading to excitotoxicity of neurotransmitters such as glutamate. A direct effect of lack of impeded glucose and oxygen delivery is the alternation of mitochondrial function. The mitochondria are involved in two important physiological functions that include the generation of ATP and regulation of cellular apoptotic pathways. Oxidative stress is a principle mechanism of cell damage following cerebral ischemia and reperfusion, mediated by reactive oxygen species (ROS) such as superoxide radical, hydrogen peroxide, and the product of nitric oxide and superoxide interaction: peroxynitrite. Ischemic events are known to activate enzymes that produce ROS, which also see a surge in generation following reperfusion. These ROS can interact with matrix metalloproteases leading to BBB disruption, and also recruit neutrophils and other leukocytes, adding an inflammatory dimension to the cellular damage3. Mitochondria are the upstream regulators of the intrinsic pathway of neuronal cellular apoptosis, with changes in the mitochondrial membrane integrity following ischemia and reperfusion injury lead to the initiation of cellular apoptosis, resulting in eventual cell death.
MitoNEET is a protein present in the outer mitochondrial membrane, which was discovered as an off-target binding site for the thiazolidinedione category of anti-diabetic drug pioglitazone4,5. It is a 14 kD protein that exists as a homodimer. MitoNEET contains two iron-sulfur clusters (2Fe-2S) that are responsible for its physiological actions6,7. The iron-sulfur cluster is labile, and its stability within the protein is dependent on its oxidation state8. It participates in redox processes thereby regulating the bioenergetics of the cell, and can act as a sensor for ROS9. MitoNEET interacts with endogenous molecules that are involved in cellular respiration10,11. MitoNEET has been discovered to play a role in metabolic conditions, cancer as well as in neurodegenerative diseases12,13. Loss of MitoNEET has been implicated to cause neurodegeneration in Parkinson’s Disease and in spinal cord injury14,15. Pioglitazone, a mitoNEET ligand has shown to have neuroprotective effects after spinal cord injury16. However, keeping in mind about the anti-diabetic actions of pioglitazone, a mitoNEET specific ligand, NL-1, has been previously developed in our lab17. NL-1 has been shown to reduce rotenone induced cell death, and caused uncoupling of mitochondrial respiration. When tested with cardiac stem cells, it has been found to improve their survival under an environment of oxidative stress18. With the current background about mitoNEET and its ligand NL-1, we investigated the potential of NL-1 in treatment of cerebral ischemia reperfusion injury, by targeting the mitochondrial pathways of cell damage such as oxidative stress and apoptosis. The work presented involves development and characterization of polymeric nanoparticles for NL-1 delivery, and the in vitro efficacy testing. Although this study focuses only on the in vitro efficacy of NL-1 nanoparticles, the long term aim of this project is delivery of NL-1 to the brain for treatment of cerebral ischemia. This is the first report of the formulation of NL-1, a first in class mitoNEET ligand into a nanoparticle drug delivery system.
2. Materials and Methods
2.1. Materials
Acid end-capped PLGA Resomer RG 530H (50:50), amiloride hydrochloride hydrate, chlorpromazine hydrochloride, genistein, and thiazolyl blue tetrazolium bromide (MTT) were purchased from Sigma-Aldrich (St. Louis, MO). Polyvinyl alcohol was obtained from MP Biochemicals (Solon, OH). Kolliphor P188 was a generous gift from BASF Chemicals Company (Ludwigshafen, Germany). Rhodamine B was purchased from Fisher Chemicals (Fair Lawn, NJ). Solvents such as dichloromethane, ethyl acetate, methanol and LC-MS grade acetonitrile were also obtained from Fisher Chemicals. Triton-X 100 detergent was purchased from Acros Organics. B-tubulin primary antibody for immunostaining was purchased from Abcam (ab15568). AlexaFluor 488-conjugated Affinipure donkey anti-rabbit secondary antibody was purchased from Jackson Immunoresearch Laboratories (West Grove, PA). Amplex Red Hydrogen Peroxide Kit was purchased from Invitrogen (A22188) (Carlsbad, CA). FITC Annexin V Apoptosis Detection Kit was obtained from BD Biosciences (catalog number: 556547) (Franklin Lane, NJ). The bEnd.3 cells and Dulbecco’s Modified Eagle’s Medium (DMEM) (catalog number 30-2002) were purchased from American Type Cell Culture (ATCC, Manasas, VA). TrypLE Express and Fetal Bovine Serum (FBS) were purchased from Gibco.
2.2. Methods
2.2.1. Preparation of nanoparticles:
PLGA nanoparticles loaded with NL-1 were prepared using emulsification and solvent evaporation technique. Acid-end capped PLGA was dissolved (2% w/v) in organic phase consisting of dichloromethane and ethyl acetate in a 1:1 ratio. This was emulsified with the aqueous phase consisting of 1% w/v PVA and 0.5% w/v Kolliphor P188 as emulsifying agents. The primary emulsion was achieved using ultrasonication process (Heat Systems Ultrasonics Inc., Model W-225). The nanoparticles were obtained after evaporation of the solvent by overnight stirring. The nanoparticles were separated from the unentrapped drug using a 10 cm Sephadex G-50 column. Finally, for long term storage, the nanoparticles were freeze dried using 5% mannitol as a cryoprotectant. The blank nanoparticles were prepared using the same procedure without adding NL-1 to the organic phase.
2.2.2. Characterization PLGA nanoparticles:
Both blank and NL-1 loaded nanoparticles were characterized for their particle size. The particle size and polydispersity index analyses were carried out using the Malvern Zetasizer. The nanoparticle samples were diluted 1:100, and each formulation was measured in triplicate. The zeta potential of the nanoparticles was also measured using the Malvern Zetasizer. The particle size of freeze-dried nanoparticles was measured by resuspending the nanoparticle powder in pure water at a concentration of 1 mg/mL. The stability of resuspended nanoparticles was determined by measuring the particle size of the suspension every week, for 60 days. Upon optimization of the formulations, Scanning Electron Microscopy (SEM) was used to determine the particle morphology as well as the size. The nanoparticle suspension was coated on a silica wafer and allowed to air dry. The dry film of nanoparticles was sputter coated with gold-palladium thin film using Denton Desk V sputter coater (Denton Vacuun, Moorestown, NJ). Subsequently, the particles were visualized using the Hitachi S4700 scanning electron microscope, at an operating voltage of 5 KV.
The physical state of nanoparticles was assessed using X-ray diffraction (XRD) technique. Four separate samples were analyzed that included the NL-1 drug in its pure form, blank PLGA nanoparticles, NL-1 loaded PLGA nanoparticles and a physical mixture of NL-1 drug and NL-1 loaded PLGA nanoparticles. Powder X-ray diffraction measurement was performed at room temperature using a PANalytical X’Pert Pro XRD equipped with Copper k-alpha 8047.2 eV X-ray source.
A modified LC-MS/MS method for NL-1 quantification was developed based on a previously described method19. The system comprised of ExionLC (SCIEX, Farmingham, MA) and an ABSciEx QTrap 5500 mass spectrometer. A Phenomenex Luna Omega UPLC column (100 x 2.1 mm, 1.6 μM) was used to elute NL-1 with an isocratic flow of acetonitrile and water (85:15) at 0.3 mL/minute. NL-1 fragment at 263.1 Daltons was used for quantification.
The entrapment efficiency of NL-1 loaded PLGA nanoparticles was calculated by extracting the drug from about 10 mg lyophilized nanoparticles. One milliliter acetonitrile was added to the nanoparticles and subjected to ultrasonication for two minutes. The solution was then centrifuged at 12,000 rpm for ten minutes and the supernatant was used to quantify NL-1 content. The percent entrapment efficiency of NL-1 was calculated using the following formula:
2.2.3. In vitro release of NL-1
The in vitro release of NL-1 from the lyophilized nanoparticles was studied using Slide-A-Lyzer MINI Dialysis Units (Thermo Scientific, Waltham, MA), with a molecular weight cut-off of 3,500 Daltons. The release medium consisted of 20% v/v methanol in 0.1 M ammonium acetate buffer pH 7.4. About 10 mg nanoparticles were weighed and dispersed in 600 μL of release medium. Five hundred microliters of this suspension was packed in the dialysis units (donor compartment), and the units were set afloat in 10 mL of release medium in a 50 mL tube (recipient compartment). The tubes were incubated at 37 °C. For pure NL-1, an equivalent amount of NL-1 was prepared as a dilution in the release medium, using a methanol stock solution (5 mg/mL) of the drug. Five hundred microliters of the NL-1 dilution was added to the dialysis units, using the same set-up used for NL-1 nanoparticles. At fixed time points, 500 μL of liquid was sampled from the recipient compartments of both, pure NL-1 and NL-1 nanoparticles. An equivalent volume of release medium was replaced to keep the volume constant. Sink conditions were maintained throughout the release study. The sampled release medium was analyzed for NL-1 content using the LC-MS/MS method. The amount of NL-1 was plotted as percent cumulative released from the nanoparticles versus time.
2.2.4. Nanoparticle uptake studies
The nanoparticle uptake study was performed in mouse endothelial bEnd.3 cells (ATCC, Manassas, VA). Rhodamine labelled fluorescent nanoparticles were prepared using the aforementioned procedure, with the addition of rhodamine B to aqueous phase prior to ultrasonication. The nanoparticles were gel filtered to remove excess rhodamine, and were further cleaned by centrifugation. The pellet obtained after second wash was used for uptake studies on the cells. This optimization to reduce background fluorescence was performed using Nanosight NS300 (Malvern Panalytical) with a 532 nm laser, and 565 nm filter. For the uptake study, cells were plated in 6-well plates at a seeding density of 150,000 cells per plate. Cells were incubated with rhodamine nanoparticles dispersed in cell culture medium, for different time periods of 1, 3, 6, and 24 hours. Once the incubation time was complete, the cells were washed thrice with PBS, trypsinized and collected in flow cytometry tubes. The cells were further washed and pelleted thrice with PBS within the tubes, and fixed with 0.4% paraformaldehyde. The samples were analyzed for rhodamine fluorescence by flow cytometry using the BD LSRFortessa 4 laser analyzer (Becton, Dickinson and Company, Franklin Lanes, NJ). The data were analyzed using the BD FACS Diva 8.0 software. To determine the effect of energy dependence of nanoparticle uptake, one plate of cells was pre-incubated at 4°C for 1 hour, after which the cells were treated with rhodamine nanoparticles at 4°C for a single time point of 6 hours. The rhodamine nanoparticle uptake was analyzed using flow cytometry, following the same procedure. The mechanism of nanoparticle uptake was studied using specific uptake mechanism inhibitors. These included amiloride hydrochloride hydrate (100 μM) for phagocytosis, chlorpromazine hydrochloride (3 μM) for clathrin mediated endocytosis, and genistein (400 μM) for lipid raft/ caveolar mediated endocytosis. The cells were pre-treated with the inhibitors separately for 1 hour, after which they were treated with rhodamine nanoparticles for a single time period of 6 hours, and analyzed for cellular uptake using flow cytometry.
2.2.5. Nanoparticle localization studies
Rhodamine nanoparticle localization was studied qualitatively by with immunohistochemistry. bEnd.3 cells were plated on coverslips and incubated with rhodamine labelled nanoparticles for a period of six hours. Upon completion of the incubation period, the cells were washed three times with PBS, fixed with 4% paraformaldehyde for 15 minutes, and permeabilized with 0.1% Triton-X100 for 10 minutes. Upon permeabilization, the cells were blocked with 10% v/v goat serum in 5% w/v bovine serum albumin (BSA). Subsequently, the cells were incubated in β-tubulin primary antibody (Abcam ab15568) overnight at 4°C. Alexa Fluor 488-conjugated AffiniPure donkey anti-rabbit IgG (Jackson Immuno Research Laboratories, West Grove, PA) was used as the secondary antibody in 5% BSA for two hours at room temperature. Finally, the cell nuclei were stained with DAPI (Thermo Scientific) for ten minutes, prior to mounting the coverslips on to slides. Cells were washed thrice with PBS between each step. The cells were imaged using Nikon A1R/SIM confocal and super resolution imaging microscope (Nikon, Tokyo, Japan).
2.2.6. In vitro ischemia model
The efficacy of NL-1 loaded nanoparticles to achieve neuroprotection following stroke was determined as a function of their ability to reduce generation of hydrogen peroxide and occurrence of apoptosis. An in vitro cerebral ischemia and reperfusion model was employed by exposing the cells to Oxygen and Glucose Deprivation (OGD). The process involved removal of regular cell culture medium and replacing the medium with the glucose deficient Hank’s Balanced Salt Solution (HBSS), containing 10% FBS. The cell plates were then transferred to the BioSpherix X3 Xvivo System hypoxia chamber, with 0.1-0.2% oxygen, 5% carbon dioxide with nitrogen making up the rest of gaseous flow. The temperature of hypoxia chamber was maintained at 37°C. The cells were incubated in hypoxia chamber for a specific time period of either 3 or 6 hours. After the desired period of hypoxia, the glucose-free medium was replaced with normal cell growth medium and cells were incubated in normal growth environment for 24 hours, mimicking the reperfusion following cerebral ischemia.
2.2.7. Effect of NL-1 nanoparticles on hydrogen peroxide generation following OGD
The in vitro efficacy of NL-1 nanoparticles in reducing generation of ROS was studied using Amplex Red assay (Thermo Scientific). bEnd.3 cells were plated in a black 96-well plate at a density of 10,000 cells per well. After 24 hours, cells were subjected to OGD. The cells were treated with NL-1 drug and an equivalent dose of NL-1 nanoparticles during the period of recovery. After the 24-hour recovery, 50 μL of Amplex Red working reagent was added to each well and allowed to react in dark at room temperature for 30 minutes. The amount of hydrogen peroxide in each well was measured by fluorimetry using the BioTek Synergy H1 plate reader (BioTek Instruments, Winooski, VT), at an excitation and emission wavelength of 530 nm and 590 nm, respectively. The data of each plate was normalized for the control wells of that plate, and then compared with other plate replicates.
2.2.8. Effect of NL-1 nanoparticles on cellular apoptosis following OGD
The ability of NL-1 nanoparticles to reduce apoptosis and enhance cell survival was determined in bEnd.3 cells using the Annexin V assay. Cells were plated in 6-well plates at a seeding density of 150,000 cells per well. After approximately 24 hours, cells were subjected to OGD, and allowed to recover with concurrent drug and nanoparticle treatments. 1 μM and 10 μM were the two concentrations of NL-1 and an equivalent dose of nanoparticles tested. After about 24 hours, cells were stained with Annexin V-FITC and propidium iodide, following which, they were sorted into live, early apoptotic, late apoptotic and necrotic populations using BD LSRFortessa analyzer, and analyzed using the BD FACS Diva 8.0 software. The cytoprotective ability of NL-1 nanoparticles was determined as a function of percentage of live cells present.
2.2.9. Toxicity studies on PLGA nanoparticles
The cytotoxicity of blank and NL-1 loaded PLGA nanoparticles was tested on hepatic HepG2 cells, using the MTT assay. Cells were plated in clear 96-well plates at a seeding density of 10,000 cells per well. The cells were treated with blank nanoparticle suspensions over a range of concentrations. A highest concentration of 10 mg/mL freeze dried nanoparticles was used for treatment. The cells were allowed to grow for 72 hours after treatments. Upon completion of the treatment incubation period, 50 μL of 1 mg/mL MTT solution was added to each well and the plates were incubated for two hours. After two hours, the MTT and media mix was aspirated, and the formazan crystals were dissolved in DMSO at 37°C, for one hour on a shaker at 150 rpm (Benchmark Incu-shaker mini). The analysis was carried out by measuring the absorbance at 570 nm using the BioTek Synergy H1 plate reader (BioTek Instruments, Winooski, VT). All treatments were performed in triplicates.
Hemolysis studies on PLGA nanoparticles were performed based on a previously used method20, using mouse blood. The blood was centrifuged to remove plasma and serum. The blood was subsequently washed thrice with 0.9% normal saline, and the volume was made up to original volume after the third wash. The blood solution was further diluted 1:10 in normal saline for the assay. An approximate dose of 5 mg/kg for a mouse was calculated, and the required amount of freeze dried nanoparticles were weighed and resuspended in 0.9% normal saline. Following treatments were used: blank nanoparticles, NL-1 loaded nanoparticles, 10 μM NL-1 drug, negative control (0.9% normal saline), and positive control (1% Triton X-100). One hundred microliters of the treatment solutions were diluted with 700 μL 0.9% normal saline, to which 200 μL of blood cells were added. The tubes were incubated at 37°C for 1 hour. After incubation, the tubes were centrifuged, and the supernatant was used for analysis. The sample absorbance was measured at 405 nm and the percent hemolysis was calculated using the following formula:
2.2.10. Statistical analysis:
Statistical analysis of data obtained was carried out by Graphpad Prism 7.04 (Graphpad Software, CA) using one-way ANOVA followed by Dunnett’s post hoc test for comparison of multiple groups. The graphs were prepared using GraphPad Prism 7.04. Data are presented as mean with standard deviations. A statistically significant difference between groups was considered if the p value was <0.05.
3. Results and Discussion
The goal of the current study was to formulate the mitoNEET ligand, NL-1, into a nanoparticle dosage form, for use in ischemia reperfusion injury. Targeting mitochondrial dysfunction poststroke is an attractive drug target, as both neuronal cells and blood—brain barrier endothelium cells are affected with ischemia reperfusion injury. Here we were able to develop formulation strategy for NL-1, with an optimized activity profile in contrast to the drug alone.
3.1. Preparation and characterization of NL-1 loaded PLGA nanoparticles
The blank and NL-1 loaded PLGA nanoparticles were prepared successfully using emulsification and solvent evaporation technique. The formulation employed acid end capped PLGA, a biodegradable polymer, which hydrolyzes into the inert lactic acid and glycolic acid residues21. Emulsification and solvent evaporation is a commonly used technique for the preparation of PLGA nanoparticles. The polymer is dissolved in organic solvent that is present as globules in the continuous aqueous phase, stabilized on the interface by the surfactants (PVA and Kolliphor P188). During solvent evaporation, the solvent diffuses into the continuous external phase, causing the formation of nanoparticles upon aggregation and desolvation of the globules22. Formulation parameters such as solvent, surfactants and their concentrations, polymer concentrations were studied to arrive at the final formulation. The choice of solvent used to dissolve polymer can be critical, as the nanoparticle size is a function of solvent diffusion out of the droplets during evaporation23. Dichloromethane, ethyl acetate and acetone were tried as solvents alone and in combination. Surfactants align at the organic-aqueous interface and stabilize the system by reducing the interfacial tension24. Thus, the use of an ideal surfactant would dictate the stability of the formulation. Non-ionic surfactants such as PVA, Kolliphor P188, and Kolliphor P407 were tried at concentrations of 0.5% w/v and 1% w/v, as well as in combinations. The surfactant combination yielding the most stable formulation, without affecting the particle size range was found to be 1% w/v PVA and 0.5% w/v Kolliphor P188. Finally, the concentration of PLGA to be used (2% w/v) was decided based upon the gradual increase in particle size observed with increasing PLGA concentration. Higher aggregation observed at PLGA concentrations above 3% w/v made it imperative to limit the PLGA concentrations at 2%. Size exclusion chromatography is a frequently used technique for the separation of the unentrapped drug from the obtained nanoparticles25–27. The free NL-1 was separated from the PLGA nanoparticles using the Sephadex G-50 column for size exclusion. PLGA nanoparticles in aqueous solution could be vulnerable to hydrolytic cleavage, resulting in instability of the nanoparticles. Hence, the formulation was freeze dried for long-term storage. The use of a cryoprotectant such as mannitol during freeze drying was warranted in order to prevent physical damage to the nanoparticles due to mechanical stress of freezing and dehydration28,29. The use of cryoprotectant also allows for rapid redispersion of the nanoparticles back into an aqueous solution28.
3.1.1. Particle size and zeta potential
The nanosuspension and the freeze-dried nanoparticles were characterized for their particle size and zeta potential. The particle size of blank and NL-1 loaded nanoparticles was found to be 121.9 ± 20.6 nm (n=5) and 123.9 ± 17.1 nm (n=12), respectively. The polydispersity index (PDI) of blank nanoparticles was found to be 0.17 ± 0.09, whereas the NL-1 loaded nanoparticles have a greater PDI of 0.27 ± 0.08. The particle size of freeze-dried nanoparticles upon resuspension was found to have increased to 175.6 ± 6.8 nm (n=12), while the PDI was found to be 0.10 ± 0.02. The zeta potential of blank and NL-1 loaded nanoparticles was found to be −16.8 ± 5.8 m V (n=3) and −26.2 ± 1.3 mV (n=3), respectively. The results for particle size and zeta potential results are summarized in Table 1. The freeze-dried nanoparticles in suspension were found to maintain their particle size in the range of 168-180 nm over a period of 60 days (data not shown). SEM imaging showed a spherical morphology for blank and NL-1 loaded nanoparticles. Figure 1 shows the SEM images for blank and NL-1 loaded nanoparticles. SEM confirmed the particle sizing results, as the nanoparticles were found to be in the general size range of 100 nm. The NL-1 nanoparticles are intended for delivery to treat ischemic stroke in the long term, which would require them to traverse the blood brain barrier (BBB). A nanoparticle size of under 200 nm has been found to be desirable for effective treatment of diseases of the CNS30–32. The particle size of NL-1 loaded PLGA nanoparticles in suspension as well as upon redispersion was found to be under 200 nm. The zeta potential of the nanoparticles was found to be in the moderately negative range (anionic). This was expected due to the use of acid-end capped PLGA during nanoparticle synthesis. For the purposes of formulation stability, a zeta potential further away from the neutral range is preferred, as repulsion between similarly charged particles is likely to prevent their aggregation. Nanoparticle surface charge has been found to have an impact on the BBB and their brain uptake as well. A low concentration anionic nanoparticles were found to be non-toxic to the BBB, and also showed a higher brain uptake33. This formulation thus has a potential for efficient delivery of NL-1 to the ischemic region in the brain, and at the same time not have a global BBB disruptive effect.
Table 1:
Particle size, polydispersity index and zeta potential of blank, NL-1 loaded and freeze dried nanoparticles
| Particle Size (nm) | Polydispersity Index | Zeta Potential (mV) | |
|---|---|---|---|
| Blank PLGA NP | 121.9 ± 20.6 (n=5) | 0.17 ± 0.09 | −16.8 ± 5.8 |
| NL-1 loaded PLGA NP | 123.9 ± 17.1 (n=12) | 0.27 ± 0.08 | −26.2 ± 1.3 |
| Freeze dried nanoparticles upon re-dispersion | 175.6 ± 6.8 (n=12) | 0.10 ± 0.02 | −26.2 ± 1.3 |
Figure 1:
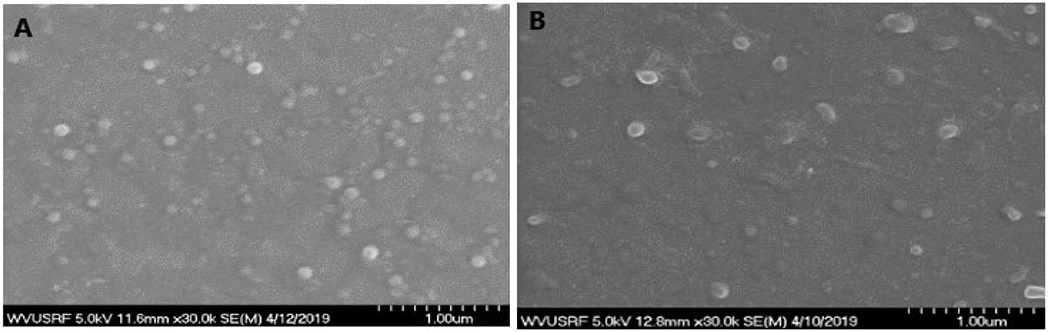
SEM images of blank PLGA nanoparticles (A) and NL-1 loaded PLGA nanoparticles (B). Each unit on the scale represents 100 nm.
3.1.2. Analytical method and entrapment efficiency
The LC-MS/MS method for analysis of NL-1 was successfully modified and used for studying the entrapment efficiency of NL-1 and the in vitro release. The entrapment efficiency of NL-1 in the current PLGA formulation was found to be 59.7 ± 10.1 %.
3.1.3. X-ray diffraction analysis
To determine the physical state of NL-1 in the nanoparticles, X-ray diffraction was performed on NL-1 drug, blank nanoparticles, NL-1 loaded nanoparticles, and a physical mixture of NL-1 drug and blank nanoparticles, as shown in figure 2. NL-1 drug was found to show a very specific X-ray diffraction pattern, with multiple sharp peaks. The patterns of blank and NL-1 loaded nanoparticles were found to almost coincide, showing that these have similar physical character. The pattern obtained for physical mixture of NL-1 drug and blank nanoparticles showed an intermediate pattern. The NL-1 peaks were clearly seen, along with the peaks seen with the nanoparticle only samples, albeit of lesser intensity. These results indicate that NL-1 is present in a different physical state in the nanoparticles, in contrast to its native crystalline form, and there is a definite reduction in crystallinity of the drug. The physical state of drug can impact the release of the drug from the nanoparticles, with crystalline form offering impedance34. The presence of NL-1 in a non-crystalline form (amorphous or dissolved) could lead to an efficient drug release.
Figure2:
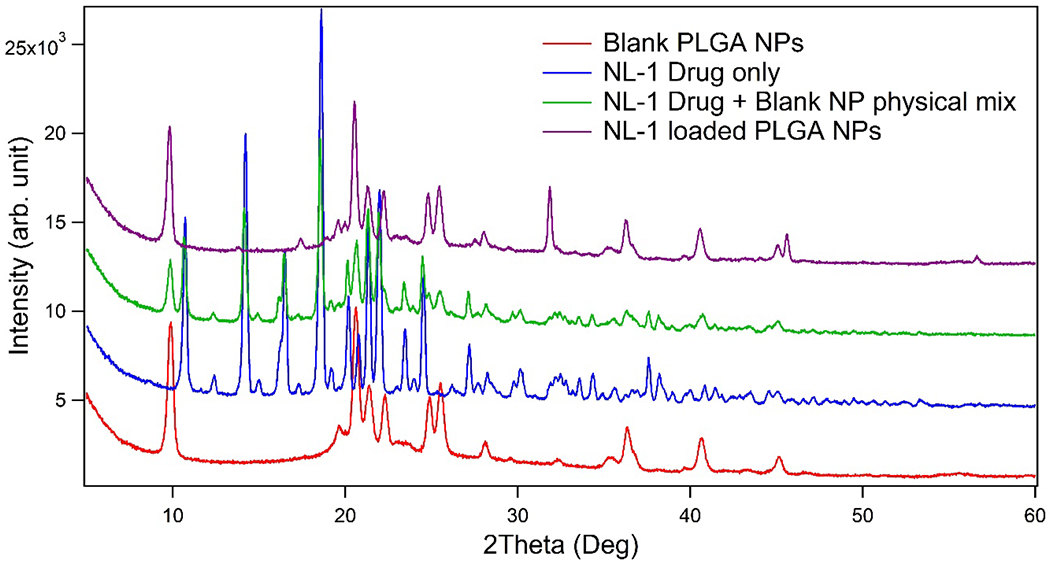
Powder X-ray diffraction pattern for NL-1 drug, NL-1 drug and blank nanoparticles physical mixture, blank PLGA nanoparticles, and NL-1 loaded PLGA nanoparticles.
3.2. In vitro release of NL-1 from nanoparticles
The in vitro release of NL-1 and NL-1 from the PLGA nanoparticles was plotted as cumulative percent release as a function of time, shown in figure 3. It was seen that NL-1 drug and NL-1 nanoparticles had a 78.7 ± 21.6% and 79.1 ± 7.2% drug release after a time period of 24 hours, respectively. NL-1 drug was found to have a faster diffusion as compared to the release of NL-1 from formulation up to a 12 hour period. NL-1 and NL-1 nanoparticles have a 50% release at about 4 and 14 hours, respectively. The in vitro release studies demonstrate that the PLGA nanoparticles have a slower release of NL-1 as compared to the pure NL-1 drug. The release is slower during the phase up to 24 hours. The nanoparticles do not sustain the release of NL-1, as the eventual plateau phase of release is reached concurrently for both, the pure drug and the nanoparticles, at 48 hours. The non-sustained release of NL-1 is suited for the acute injury in this type of neurodegeneration following stroke as it is a time dependent phenomenon. A successful rescue of greater percentage of cells would require a quicker drug treatment.
Figure 3:
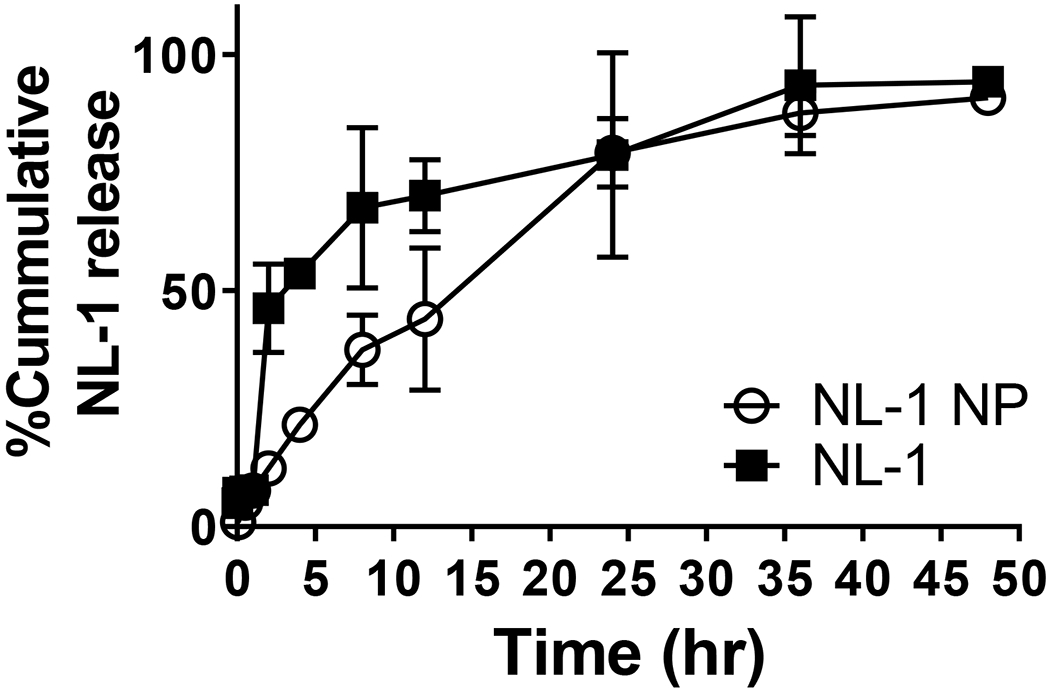
In vitro drug release profile for NL-1 drug and NL-1 from PLGA nanoparticles. Data are plotted as total percent cumulative NL-1 released versus time.
The release models that showed the best fit include Korsmeyer-Peppas (R2: 0.9933), and first order release model (R2: 0.9812). The R2 values obtained for the other models are summarized in table 2. The exponent for Korsmeyer-Peppas fit was found to be 0.768. The Korsmeyer-Peppas model of drug release is used for the first 60% of the release data, and can be helpful in determining the mechanism of drug release35,36. The exponent of 0.768 in the Korsmeyer-Peppas fit of the release kinetics points toward non-Fickian or anomalous release being the primary release mechanism of NL-1. The release of NL-1 from PLGA nanoparticles is a possible combination of NL-1 diffusion and PLGA erosion. A drawback of Korsmeyer-Peppas release model is that fact that it is used only for first 60% of the drug released. When the entire release data was fit to a model, it was found to be close to first-order model. First order kinetics imply that the release of drug from the nanoparticles is dependent on the concentration of the drug present in the system.
Table 2:
Summary of plots used for release modelling and the correlation coefficients obtained for the corresponding plots
| Model name | Graph plotted | R2 value |
|---|---|---|
| Zero order | % cumulative released vs time | 0.9049 |
| First order | Log(cumulative % drug remaining) vs time | 0.9812 |
| Hixson-Crowell cube root model | Cube root of % drug remaining vs time | 0.9674 |
| Higuchi model | % cumulative released vs square root of time | 0.9798 |
| Korsmeyer-Peppas model (first 60%) | Log(% cumulative released) vs log(time) | 0.9933 |
3.3. Cellular uptake of nanoparticles
The first step toward effective treatment of cells by nanoparticles would require uptake of the nanoparticles by the cells. Rhodamine nanoparticles were prepared and characterized for particle size under a fluorescent filter on the Nanosight (data not shown). This was useful in optimizing the wash-outs needed for the nanoparticles to eliminate background fluorescence, and to confirm the presence of fluorescent nanoparticles. The studies were performed in bEnd.3 endothelial cells, a commonly used model for in vitro stroke studies37–39. The time-dependent uptake of rhodamine nanoparticles in bEnd.3 cells was analyzed using flow cytometry analysis. Figure 4 shows an overlay for the representative flow cytometry histograms for each time point, along with the uptake quantification. Two negative controls, untreated cells and cells treated with blank nanoparticles showed low fluorescence intensities as seen in the histograms. The positive control of rhodamine nanoparticles had high fluorescence intensity, indicated with a right shift in the histogram (data not shown). The rhodamine nanoparticle treated cells showed a marginal shift beyond the threshold value. Upon quantification, it was found that 1 hour incubation showed a significantly higher rhodamine fluorescence as compared to the negative controls. The uptake was found to be significantly higher at 3 hours as compared to 1 hour. However, after 3 hours the uptake showed a plateauing as there was no statistically significant difference in fluorescence intensities at 3, 6 and 24 hours. The significantly higher uptake after 1 hour could possibly be beneficial for cell rescue, which can be a time sensitive phenomenon.
Figure 4:
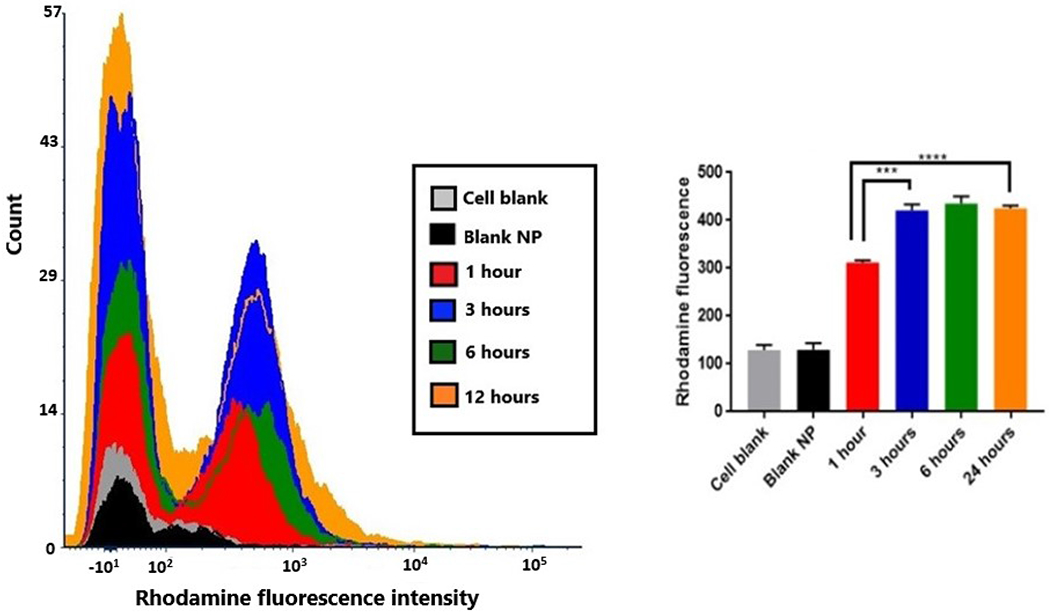
Cellular uptake of rhodamine nanoparticles in bEnd.3 cells. The flow cytometry histograms (left) are representatives for cell control, blank nanoparticles, and each time point for rhodamine nanoparticles. The graph (right) shows the quantification of rhodamine nanoparticle uptake (***p<0.001, ****p<0.0001).
We tested the energy dependence of rhodamine nanoparticle uptake by comparing uptake at 37°C to that of 4°C (figure 5). Compared to the nanoparticle uptake at 37°C, the uptake was significantly lowered during treatment at 4°C, as seen from the left shift in the fluorescence histogram. The fluorescence intensity at 4°C was similar to the negative control with no significant difference between the two. This could suggest that the functioning of proteins and enzymes involved in endocytosis is inhibited at lower temperatures and is an energy-dependent active process.
Figure 5:
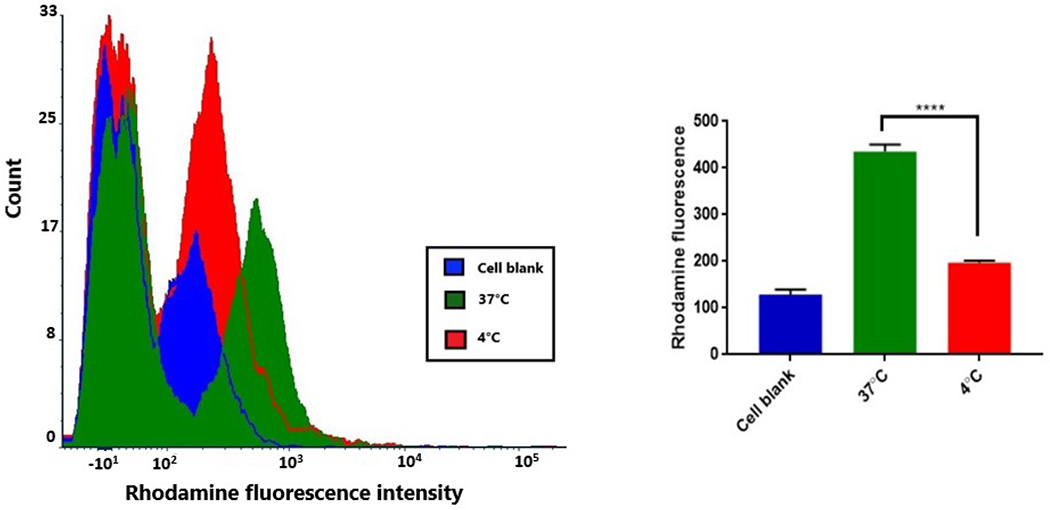
Energy dependence of nanoparticle uptake in bEnd.3 cells. Comparative flow cytometry histograms (left) for cell control, and rhodamine nanoparticle uptake at 37°C and at 4°C. Graphical representation (right) of fluorescence intensities shows a significantly decreased uptake at 4°C (****p<0.0001).
We further tried to delineate the mechanism of nanoparticle uptake by treating cells with specific uptake inhibitors, such as amiloride hydrochloride hydrate for phagocytosis, chlorpromazine hydrochloride for receptor mediated endocytosis, and genistein for caveolar mediated endocytosis. The uptake was quantified by measuring the rhodamine fluorescence by flow cytometry (figure 6). The most prominent mechanisms of nanoparticle uptake into cells include phagocytosis, clathrin mediated endocytosis, and caveolar/lipid raft endocytosis. Phagocytosis is an actin-dependent uptake mechanism, which is generally mediated by specialized cells such as macrophages, suitable to particles around 500 nm in size40. Clathrin mediated endocytosis involves binding of cargo (nanoparticles) to specific receptors on the cell surface, leading to downstream recruitment of proteins that help internalize the nanoparticles into a ‘coated pits’41. Caveolar endocytosis is mediated by components of lipid rafts and occurs in cholesterol rich regions of the plasma membrane. These are vesicles formed by cell membrane invaginations42. Each of the above mechanism was individually inhibited by using specific inhibitors. A single concentration of inhibitors was used for each mechanism, based on literature41–43.The caveolar endocytosis inhibitor genistein was found to significantly reduce the uptake of rhodamine nanoparticles into bEnd.3 cells to 61.8% as compared to the uptake into untreated cells. Amiloride HCl and Chlorpromazine HCl which inhibit phagocytosis and clathrin mediated endocytosis, respectively, were found to have no significant effect on rhodamine nanoparticle uptake. Lipid raft or caveolar mediated endocytosis was thus found to be the prominent mode of nanoparticle uptake. Genistein is an inhibitor of receptor associated tyrosine kinases, involved in the caveolar uptake mechanism44. The caveolar invaginations can be in the size range of 50-200 nm, which would explain the endocytosis of these nanoparticles through this mechanism. Moreover, nanoparticles with an anionic surface charge are known to interact with the cationic lipid components of the lipid rafts in the cell membrane which further explains the mechanism used by the nanoparticles for their uptake45.
Figure 6:
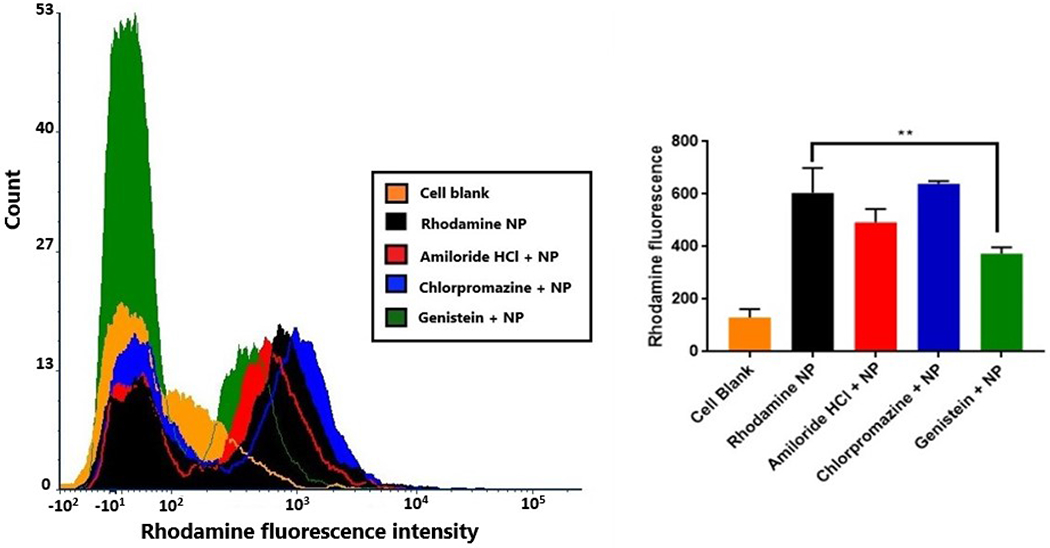
Mechanism of rhodamine nanoparticle uptake in bEnd.3 cells. Flow cytometry histograms (left) are representatives of cell blank, rhodamine nanoparticles alone and with uptake inhibitor treatments. The plot (right) indicates the quantification of fluorescence intensity with respective uptake inhibitors, with genistein treatment showing significant reduction in uptake (**p<0.01).
3.4. Cellular localization of nanoparticles
Upon their uptake, it is important that the nanoparticles localize around the site of action of the drug they intend to deliver. The cellular localization studies were performed using fluorescent rhodamine nanoparticles as seen in figure 7. Confocal microscopy was used to image the nanoparticles in bEnd.3 cells that were immunostained for β-tubulin (green) and nucleus was stained with DAPI (blue). The advantage of using confocal microscopy was the ability to image z-stacks, which provides a better understanding of the position of the nanoparticles within the cell, as compared to a two-dimensional image. The images are shown as individual channels of DAPI for nucleus, FITC for β-tubulin and red for rhodamine. The merged image shows localization of nanoparticles in the cytoplasmic region of the cells, within the tubulin cytoskeleton. The second panel (figure 7, E–H) shows the z-stack images of cells, which confirm the presence of rhodamine nanoparticles in the cytoplasm of the cells. Figure 7I shows a super resolution image, with nanoparticle localization in the cytoplasm. Cytoplasmic localization of nanoparticles is necessary for the activity of NL-1 as a mitoNEET ligand. MitoNEET is present in the outer mitochondrial membrane, and is oriented toward the cytoplasm46.
Figure 7:

Confocal microscopy z-stack images for cellular localization of rhodamine nanoparticles in bEnd.3 cells showing nanoparticle localization in the cytoplasm. Cell nuclei stained blue with DAPI (A and E), tubulin stained green (B and F), red rhodamine nanoparticles (C and G). Images D and H show a merge of individual channels. Image I shows a super resolution image of rhodamine nanoparticles being localized in the cytoplasmic tubulin cytoskeleton.
3.5. In vitro ischemia model for efficacy studies
The efficacy studies for NL-1 loaded PLGA nanoparticles were performed using the in vitro OGD model to mimic stroke. The studies were performed in bEnd.3 endothelial cells, a commonly used model for in vitro stroke studies37–39. HBSS supplemented with 10% FBS was used as the glucose-free medium for cell incubation in the hypoxia chamber47,48. Cellular toxicity following ischemic stroke is multimodal, mediated via multiple mechanisms. Cell damage due to generation of ROS and induction of apoptosis cascade are two of the primary causes of cytotoxicity. Both of these mechanisms are mitochondrial in origin and relevant sites of intervention for NL-1, since it binds the mitochondrial protein mitoNEET. We investigated the effect of NL-1 nanoparticle treatment on both of these physiologic events.
3.6. NL-1 nanoparticles reduce hydrogen peroxide generation
An important ROS mediator generated in mitochondria is the superoxide radical. However, measurement of superoxide produced in the mitochondria can be challenging due to the inability of probes to permeate the outer mitochondrial membrane. Hydrogen peroxide is formed as a dismutation product of superoxide radical by mitochondrial manganese superoxide dismutase enzyme49. It diffuses across the outer mitochondrial membrane into the extra mitochondrial space, wherein it can be quantified50. We determined the effect of NL-1 nanoparticles on peroxide generation using the sensitive fluorimetric Amplex Red assay51. Amplex Red is a colorless substance that reacts with hydrogen peroxide in presence of horseradish peroxidase in a 1:1 stoichiometry to yield a stable fluorescent compound called resorufin52,53. The primary challenge with this assay involves prevention of light exposure of Amplex red reagent for longer periods, and artificial generation of hydrogen peroxide by light exposure of resorufin54,55.
The efficacy of NL-1 nanoparticles in reducing ROS generation was studied by subjecting bEnd.3 cells to OGD conditions for a period of 3 or 6 hours, and a reperfusion period of 24 hours (figure 8). The hydrogen peroxide produced was measured using the Amplex Red assay, and each treatment was compared to control. It was found that the NL-1 nanoparticles had a significant effect in reducing the peroxide produced at a NL-1 dose of 2.5 μM, for a stroke period of 3 hours. However, the efficacy of NL-1 nanoparticles was seen to decrease for a stroke period of 6 hours, as significant reduction in peroxide levels was only seen at 10 μM and 20 μM NL-1 dose. Figures 8C and 8D compare the effect of nanoparticles against that of pure NL-1 drug. For 3 hour OGDR, NL-1 loaded nanoparticles were found to be more efficacious as compared to NL-1 drug, while such an effect was not seen at 6 hour ischemia period. The results indicate that NL-1 nanoparticles are more effective at the shorter ischemic period of 3 hours as seen from the dose response of NL-1 nanoparticles in reducing the peroxide generation. A possible explanation for this could be that other mechanisms of cell damage might predominate for a longer duration of ischemia. ROS generation could likely be the predominant mode of cell toxicity at the shorter ischemic period. Cell damage due to ROS has been implicated in the earlier time period of ischemia56. We also compared the NL-1 nanoparticles to pure NL-1 drug in their ability to reduce the peroxide generation. NL-1 nanoparticles had a greater efficacy as compared to NL-1 drug during the shorter ischemia period. However, this effect was not seen for 6 hour ischemia period.
Figure 8:
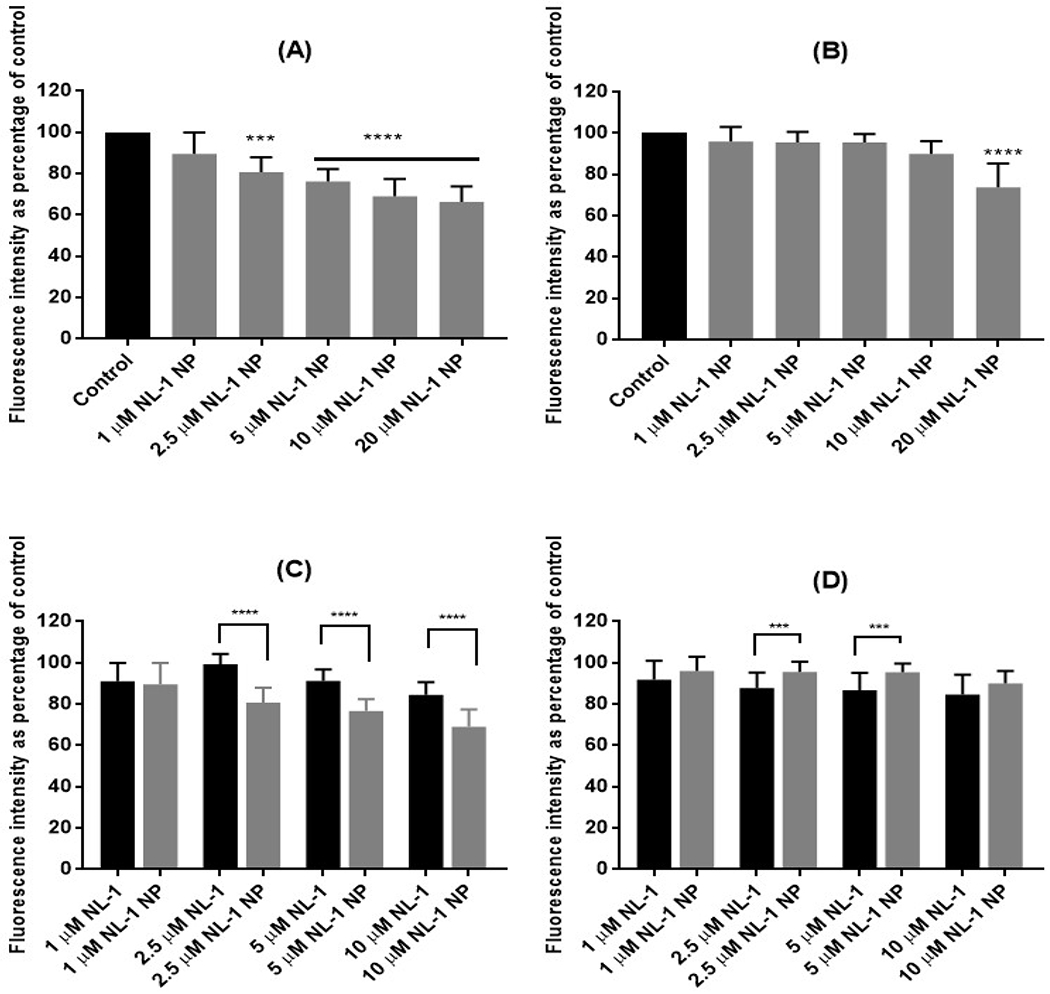
Effect of NL-1 nanoparticle treatment on generation of hydrogen peroxide in bEnd.3 cells after 3 hours (A) and 6 hours (B) of OGD. The control was considered as 100% in order to normalize values for multiple plates. The bottom panel shows a comparison in activity of NL-1 drug and NL-1 nanoparticles after 3 hours (C) and 6 hours (D) of OGD.
3.7. NL-1 nanoparticles improve cell survival and reduce apoptosis
The second mechanism of cell death that has a mitochondrial dimension is apoptosis. The ability of NL-1 nanoparticles in checking apoptosis and in turn improving the cell survival was determined with the Annexin V apoptosis assay (figure 9), which is commonly employed to detect apoptotic and necrotic cell populations57,58. The cells were exposed to either 3 or 6 hours of ischemia, with subsequent treatment with 1 μM and 10 μM of NL-1 and equivalent dose of NL-1 nanoparticles. The cells were differentiated into four categories based on their staining. The live cells are unstained, the cells in early apoptosis pick up a stronger signal of Annexin V-FITC, while the cells in late apoptosis and necrotic phase have a strong PI signal. The results are expressed as percent survival of cells and percent of cells in late apoptotic phase. Figure 9 A–D are representative images showing flow cytometry data, and quantified in Figure 9E–H. The cells undergoing apoptosis were predominantly found to be in the late apoptotic stage. The cell survival for control for 3 hour ischemia was found to be 44.4 ± 2.8%. NL-1 drug improved cells survival to 58.3 ± 2.3% at a concentration of 10 μM, however no significant improvement in cell survival was seen at 1 μM concentration. In contrast, the equivalent dose of nanoparticles significantly improved cell survival at both 1 μM (57.5 ± 3.0%) and 10 μM dose (65.7 ± 0.2 %). Correspondingly, there was a decrease in the percent of cells in late apoptosis for 10 μM NL-1, 1 μM NL-1 nanoparticles and 10 μM NL-1 nanoparticles. The NL-1 nanoparticles showed a significantly higher efficacy as compared to the corresponding concentration of NL-1 drug. For an ischemic period of 6 hours, the control showed a survival of 40.4 ± 1.1%. NL-1 drug showed improved cell survival at both 1 μM (56.2 ± 1.4%) and 10 μM (71.0 ± 1.4%). Enhanced cell survival was also seen for NL-1 nanoparticles at 1 μM (57.3 ± 2.3%) and 10 μM (69.8 ± 4.3%) concentrations. A corresponding significant decrease was seen in the percentage of cells in late apoptosis for all treatments. However, there was no significant difference in the efficacy of NL-1 nanoparticles and the NL-1 drug for the 6 hour ischemic period.
Figure 9:
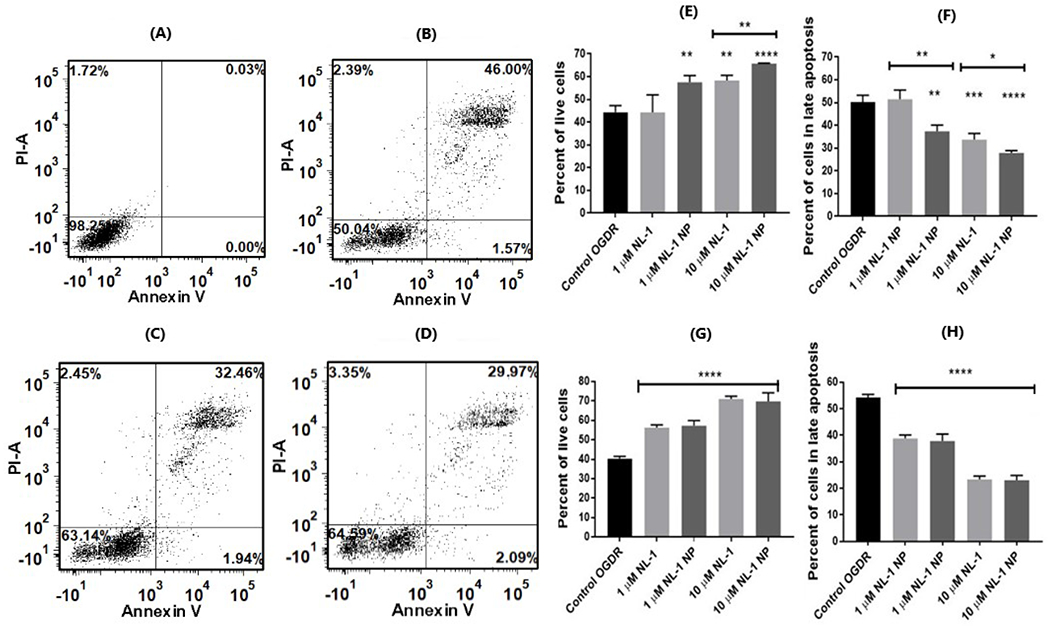
Effect of NL-1 and NL-1 nanoparticles on apoptosis in bEnd.3 cells following ischemia and reperfusion. Figures A-D are representative flow cytometry data for cell blank (A), 3 hour OGD (B), and 3 hour OGD with NL-1 (C) and 3 hour OGD with NL-1 nanoparticles (D). Each figure shows four quadrants: live (bottom left), early apoptotic phase (bottom right), late apoptotic phase (top right), and necrotic (top left) cell populations. Results are indicated in terms of improvement in cell survival at 3 hours (E) and 6 hours (F) ischemic period, as well as the reduction in late stage apoptosis after 3 hours (G) and 6 hours (H) of ischemia (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
We found higher protective activity of the NL-1 nanoparticles as compared to NL-1 drug-alone at the 3 hour ischemia period, however these results are not seen at the longer ischemia duration of 6 hours. Although the NL-1 nanoparticles showed a greater cell survival at both concentrations, the drug by itself was not effective at 1 μM for 3 hour ischemia period. Apoptosis in cerebral ischemia has previously been described to be a biphasic event, in which the apoptosis mediated by mitochondria occurs at a later stage59. It can be speculated that improved activity of NL-1 for longer ischemia period is a possible result of increased mitochondrial involvement in the apoptotic mechanisms. Further studies are needed to confirm this hypothesis. Overall, the system helped reduce apoptotic populations of bEnd.3 cells.
3.8. NL-1 nanoparticle formulation was non-toxic
The inherent toxicity of blank and NL-1 loaded PLGA nanoparticles was studied using MTT cytotoxicity assay on hepatic HepG2 cells (figure 10A). Since liver is a primary metabolic organ, we chose to perform this test on liver cells. A wide concentration range of blank nanoparticles was used for the assay, with the highest concentration of 10 mg/mL. HepG2 cell survival was found to be about 95.9 ± 5.6% at a nanoparticle concentration of 1 mg/mL and was found to reduce to 68.2 ± 2.5% at the highest treatment concentration of 10 mg/mL, in blank PLGA nanoparticles. For NL-1 loaded nanoparticles, the cell survival was found to be 80.4 ± 11.4%, and 58.1 ± 7.8% at 1 mg/mL and 10 mg/mL, respectively. The survival curve shows that even the highest concentration of both, blank and NL-1-loaded PLGA nanoparticles did not induce a 50% cell death, and an IC50 value could not be established. The concentration of 10 mg/mL was at least five times higher than the highest concentration of nanoparticles used for efficacy studies. This is indicative that the results obtained were not affected by any inherent nanoparticle toxicity.
Figure 10:
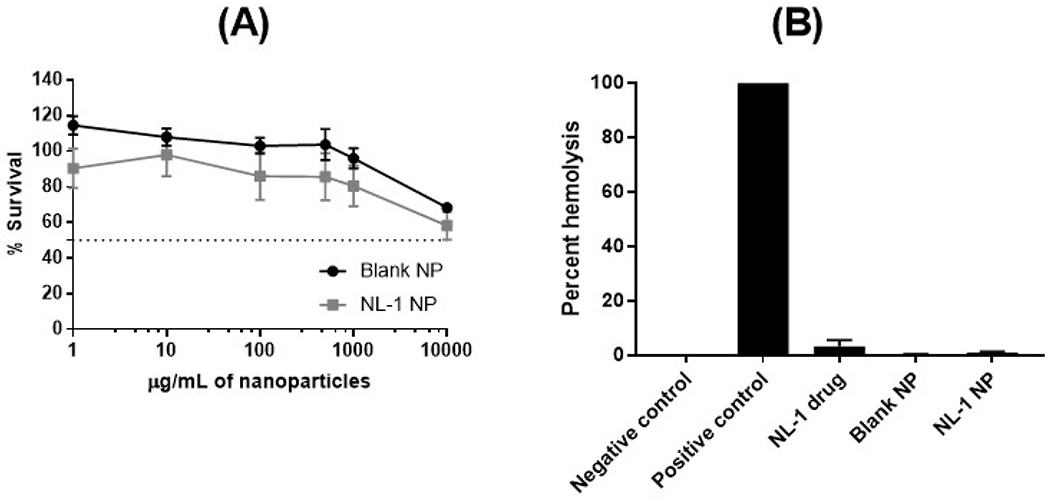
Toxicity profile of PLGA nanoparticles. MTT cytotoxicity assay results indicate a relatively safe profile for blank and NL-1 loaded PLGA nanoparticles at the highest concentration of 10 mg/mL (A). NL-1, blank and NL-1 loaded nanoparticles showed a negligible hemolytic activity (B).
The second toxicity parameter that was investigated was the potential of PLGA nanoparticles to cause hemolysis of erythrocytes, since the nanoparticles are intended for intravenous administration (figure 10B). This assay is indicative of effect of the nanoparticles on cell membrane integrity60. The blood cells were incubated with NL-1 drug, blank nanoparticles and NL-1 loaded nanoparticles, and the absorbance was measured as a function of hemolysis. The positive control of 1% v/v Triton X-100 NL-1 showed 100% hemolysis. The NL-1 drug was found to have a slightly higher hemolysis as compared to blank and NL-1 loaded nanoparticles, which showed an almost zero percent hemolysis. The results obtained show that neither the blank nor the NL-1 loaded nanoparticles showed any hemolytic activity. PLGA nanoparticles have been found to have non-existent hemolysis60, which was confirmed with the current assay. Thus, the formulation was deemed safe for intravenous administration.
Conclusion:
In this study, we developed nanoparticles drug delivery vehicle for the delivery of NL-1 to the mitochondrial target mitoNEET, in treatment of cerebral ischemia and reperfusion injury. NL-1 loaded PLGA nanoparticles were prepared using emulsification and solvent evaporation technique, and subsequently characterized for size, zeta potential, physical state, and entrapment. In vitro analysis of nanoparticles was performed to understand the release kinetics of NL-1. Nanoparticle uptake studies were performed in bEnd.3 cells, with emphasis on mechanism of uptake and its energy dependence. The NPs were effective in reducing hydrogen peroxide generation after ischemia in cells. Also, NL-1 nanoparticles were also successful in enhancing the cell survival by reducing cell apoptosis, following OGD conditions. This study helps to establish the basis for the use of NL-1 as a stroke therapeutic agent for future in vivo stroke studies. However, further investigation is needed in the aspect of mechanism of protection by NL-1 to the differences seen in the cell death mechanisms based on time of ischemic exposure in vivo.
Supplementary Material
Acknowledgements
The authors would like to acknowledge the following West Virginia University core facility managers and the respective funding sources: (1) Dr. Amanda Ammer, WVU imaging facilities core, grants U54GM104942 and P20GM103434; (2) Dr. Kathleen Brundage, WVU flow core, WVUCTS grant GM104942 and WV-INBRE grant GM103434; (3) Dr. Terence McManus, WVU metabolomics core and (4) Dr. Marcela Redigolo and Dr. Qiang Wang, WVU shared research facilities core. Funding in part from the NIH grants P30GM103488, S10OD016165, R42AR074812, R44CA221554, R41NS110070 and P20 GM109098.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Stroke Facts | cdc.gov. https://www.cdc.gov/stroke/facts.htm Accessed September 6, 2019.
- 2.Jung JE, Kim GS, Chen H, et al. Reperfusion and Neurovascular Dysfunction in Stroke: from Basic Mechanisms to Potential Strategies for Neuroprotection. Mol Neurobiol. 2010;41(2-3):172–179. doi: 10.1007/s12035-010-8102-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doyle KP, Simon RP, Stenzel-Poore MP. Mechanisms of ischemic brain damage. Neuropharmacology. 2008;55(3):310–318. doi: 10.1016/J.NEUROPHARM.2008.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colca JR, McDonald WG, Waldon DJ, et al. Identification of a novel mitochondrial protein (“mitoNEET”) cross-linked specifically by a thiazolidinedione photoprobe. Am J Physiol - Endocrinol Metab. 2003;286(2). http://ajpendo.physiology.org/content/286/2/E252.long Accessed June 10, 2017. [DOI] [PubMed] [Google Scholar]
- 5.Lin J, Zhou T, Ye K, Wang J. Crystal structure of human mitoNEET reveals distinct groups of iron-sulfur proteins. Proc Natl Acad Sci U S A. 2007;104(37):14640–14645. doi: 10.1073/pnas.0702426104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiley Sandra E., Paddock Mark L. ECA, Gross Larry, van der Geer Peter RN, Murphy Anne N., Jennings Patricia A. and JED. The Outer Mitochondrial Membrane Protein mitoNEET Contains a Novel Redox-active 2Fe-2S Cluster. J Biol Chem. 282(33):23745–23749. doi: 10.1074/jbc.C700107200 [DOI] [PubMed] [Google Scholar]
- 7.Tamir S, Paddock ML, Darash-Yahana-Baram M, et al. Structure-function analysis of NEET proteins uncovers their role as key regulators of iron and ROS homeostasis in health and disease. Biochim Biophys Acta - Mol Cell Res. 2015;1853(6): 1294–1315. doi: 10.1016/j.bbamcr.2014.10.014 [DOI] [PubMed] [Google Scholar]
- 8.Golinelli-Cohen MP, Lescop E, Mons C, et al. Redox control of the human iron-sulfur repair protein MitoNEET activity via its iron-sulfur cluster. J Biol Chem. 2016;291(14):7583–7593. doi: 10.1074/jbc.M115.711218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landry AP, Ding H. Redox control of human mitochondrial outer membrane protein MitoNEET [2Fe-2S] clusters by biological thiols and hydrogen peroxide. J Biol Chem. 2014;289(7):4307–4315. doi: 10.1074/jbc.M113.542050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou T, Lin J, Feng YWJ. Binding of Reduced Nicotinamide Adenine Dinucleotide Phosphate Destabilizes the Iron-Sulfur Clusters of Human MitoNEET. Biochemistry. 49(44):9604–9612. doi: 10.1021/bi101168c [DOI] [PubMed] [Google Scholar]
- 11.Landry AP, Wang Y, Cheng Z, Crochet RB, Lee YHDH. Flavin nucleotides act as electron shuttles mediating reduction of the [2Fe- 2S] clusters in mitochondrial outer membrane protein mitoNEET. Free Radic Biol Med. 102:240–247. doi: 10.1016/j.freeradbiomed.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mittler R, Darash-Yahana M, Sohn YS, et al. NEET Proteins: A new link between iron metabolism, reactive oxygen species, and cancer. Antioxidants Redox Signal. 2019;30(8):1083–1095. doi: 10.1089/ars.2018.7502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipper CH, Paddock ML, Onuchic JN, Mittler R, Nechushtai R, Jennings PA. Cancer-Related NEET Proteins Transfer 2Fe-2S Clusters to Anamorsin, a Protein Required for Cytosolic Iron-Sulfur Cluster Biogenesis. Levy YK, ed. PLoS One. 2015;10(10):e0139699. doi: 10.1371/journal.pone.0139699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Q-Q, Xiong L-L, Liu F, et al. MicroRNA-127 targeting of mitoNEET inhibits neurite outgrowth, induces cell apoptosis and contributes to physiological dysfunction after spinal cord transection. Sci Rep. 2016;6:35205. doi: 10.1038/srep35205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geldenhuys WJ, Benkovic SA, Lin L, et al. MitoNEET (CISD1) Knockout Mice Show Signs of Striatal Mitochondrial Dysfunction and a Parkinson’s Disease Phenotype. ACS Chem Neurosci. 2017;8(12):2759–2765. doi: 10.1021/acschemneuro.7b00287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel SP, Cox DH, Gollihue JL, et al. Pioglitazone treatment following spinal cord injury maintains acute mitochondrial integrity and increases chronic tissue sparing and functional recovery. Exp Neurol. 2017;293:74–82. doi: 10.1016/j.expneurol.2017.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geldenhuys WJ, Funk MO, Barnes KF, Carroll RT. Structure-based design of a thiazolidinedione which targets the mitochondrial protein mitoNEET. Bioorganic Med Chem Lett. 2010;20(3):819–823. doi: 10.1016/j.bmcl.2009.12.088 [DOI] [PubMed] [Google Scholar]
- 18.Logan SJ, Yin L, Geldenhuys WJ, et al. Novel thiazolidinedione mitoNEET ligand-1 acutely improves cardiac stem cell survival under oxidative stress. Basic Res Cardiol. 2015;110(2):19. doi: 10.1007/s00395-015-0471-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pedada KK, Zhou X, Jogiraju H, et al. A quantitative LC?MS/MS method for determination of thiazolidinedione mitoNEET ligand NL-1 in mouse serum suitable for pharmacokinetic studies. J Chromatogr B. 2014;945-946:141–146. doi: 10.1016/j.jchromb.2013.11.048 [DOI] [PubMed] [Google Scholar]
- 20.Saralkar P, Dash AK. Alginate Nanoparticles Containing Curcumin and Resveratrol: Preparation, Characterization, and In Vitro Evaluation Against DU145 Prostate Cancer Cell Line. AAPS PharmSciTech. 2017;18(7). doi: 10.1208/sl2249-017-0772-7 [DOI] [PubMed] [Google Scholar]
- 21.Lemoine D, Francois C, Kedzierewicz F, Preat V, Hoffman M, Maincent P. Stability study of nanoparticles of poly(ε-caprolactone), poly(D,L-lactide) and poly(D,L-lactide-coglycolide). Biomaterials. 1996;17(22):2191–2197. doi: 10.1016/0142-9612(96)00049-X [DOI] [PubMed] [Google Scholar]
- 22.Kwon H-Y, Lee J-Y, Choi S-W, Jang Y, Kim J-H. Preparation of PLGA nanoparticles containing estrogen by emulsification-diffusion method. Colloids Surfaces A Physicochem Eng Asp. 2001; 182(1-3): 123–130. doi: 10.1016/S0927-7757(00)00825-6 [DOI] [Google Scholar]
- 23.Cooper DL, Harirforoosh S. Design and Optimization of PLGA-Based Diclofenac Loaded Nanoparticles. Sem DS, ed. PLoS One. 2014;9(1):e87326. doi: 10.1371/journal.pone.0087326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma N, Madan P, Lin S. Effect of process and formulation variables on the preparation of parenteral paclitaxel-loaded biodegradable polymeric nanoparticles: A co-surfactant study. Asian J Pharm Sci. 2016; 11(3):404–416. doi: 10.1016/J.AJPS.2015.09.004 [DOI] [Google Scholar]
- 25.Dangi RS, Shakya S. Preparation, optimization and characterization of PLGA nanoparticle. Int J Pharm Life Sci. 2013;4(7):2810–2818. [Google Scholar]
- 26.Guo J, Gao X, Su L, et al. Aptamer-functionalized PEG–PLGA nanoparticles for enhanced anti-glioma drug delivery. Biomaterials. 2011;32(31):8010–8020. doi: 10.1016/J.BIOMATERIALS.2011.07.004 [DOI] [PubMed] [Google Scholar]
- 27.Wen Z, Yan Z, Hu K, et al. Odorranalectin-conjugated nanoparticles: Preparation, brain delivery and pharmacodynamic study on Parkinson’s disease following intranasal administration. J Control Release. 2011;151(2):131–138. doi: 10.1016/J.JCONREL.2011.02.022 [DOI] [PubMed] [Google Scholar]
- 28.Fonte P, Soares S, Costa A, et al. Effect of cryoprotectants on the porosity and stability of insulin-loaded PLGA nanoparticles after freeze-drying. Biomatter. 2012;2(4):329–339. doi: 10.4161/biom.23246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abdelwahed W, Degobert G, Stainmesse S, Fessi H. Freeze-drying of nanoparticles: Formulation, process and storage considerations. Adv Drug Deliv Rev. 2006;58(15):1688–1713. doi: 10.1016/J.ADDR.2006.09.017 [DOI] [PubMed] [Google Scholar]
- 30.Cruz LJ, Stammes MA, Que I, et al. Effect of PLGA NP size on efficiency to target traumatic brain injury. J Control Release. 2016;223:31–41. doi: 10.1016/J.JCONREL.2015.12.029 [DOI] [PubMed] [Google Scholar]
- 31.Hu K, Shi Y, Jiang W, Han J, Huang S, Jiang X. Lactoferrin conjugated PEG-PLGA nanoparticles for brain delivery: Preparation, characterization and efficacy in Parkinson’s disease. Int J Pharm. 2011;415(1-2):273–283. doi: 10.1016/J.IJPHARM.2011.05.062 [DOI] [PubMed] [Google Scholar]
- 32.Saraiva C, Praça C, Ferreira R, Santos T, Ferreira L, Bernardino L. Nanoparticle-mediated brain drug delivery: Overcoming blood-brain barrier to treat neurodegenerative diseases. J Control Release. 2016;235:34–47. doi: 10.1016/J.JCONREL.2016.05.044 [DOI] [PubMed] [Google Scholar]
- 33.Lockman PR, Koziara JM, Mumper RJ, Allen DD. Nanoparticle Surface Charges Alter Blood-Brain Barrier Integrity and Permeability. J Drug Target. 2004;12(9-10):635–641. doi: 10.1080/10611860400015936 [DOI] [PubMed] [Google Scholar]
- 34.Mohanty C, Sahoo SK. The in vitro stability and in vivo pharmacokinetics of curcumin prepared as an aqueous nanoparticulate formulation. Biomaterials. 2010;31(25):6597–6611. doi: 10.1016/j.biomaterials.2010.04.062 [DOI] [PubMed] [Google Scholar]
- 35.Gautam Singhvi MS. In-vitro drug release characterization models. Int J Pharm Stud Res. 2011;2(1):77–84. [Google Scholar]
- 36.Jose S, Fangueiro JF, Smitha J, et al. Predictive modeling of insulin release profile from cross-linked chitosan microspheres. Eur J Med Chem. 2013;60:249–253. doi: 10.1016/J.EJMECH.2012.12.011 [DOI] [PubMed] [Google Scholar]
- 37.Comajoan P, Gubern C, Huguet G, Serena J, Kádár E, Castellanos M. Evaluation of common housekeeping proteins under ischemic conditions and/or rt-PA treatment in bEnd.3 cells. J Proteomics. 2018;184:10–15. doi: 10.1016/J.JPROT.2018.06.011 [DOI] [PubMed] [Google Scholar]
- 38.Wang L, Niu Y, He G, Wang J. Down-regulation of lncRNA GAS5 attenuates neuronal cell injury through regulating miR-9/FOXO3 axis in cerebral ischemic stroke. RSC Adv. 2019;9(28):16158–16166. doi: 10.1039/C9RA01544B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao M, Wang J, Xi X, Tan N, Zhang L. SNHG12 Promotes Angiogenesis Following Ischemic Stroke via Regulating miR-150/VEGF Pathway. Neuroscience. 2018;390:231–240. doi: 10.1016/J.NEUROSCIENCE.2018.08.029 [DOI] [PubMed] [Google Scholar]
- 40.Oh N, Park J-H. Endocytosis and exocytosis of nanoparticles in mammalian cells. Int J Nanomedicine. 2014;9 Suppl 1:51–63. doi: 10.2147/IJN.S26592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.dos Santos T, Varela J, Lynch I, Salvati A, Dawson KA. Effects of Transport Inhibitors on the Cellular Uptake of Carboxylated Polystyrene Nanoparticles in Different Cell Lines. Schnur JM, ed. PLoS One. 2011;6(9):e24438. doi: 10.1371/journal.pone.0024438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ha S-W, Weitzmann MN, Beck GR. Bioactive Silica Nanoparticles Promote Osteoblast Differentiation through Stimulation of Autophagy and Direct Association with LC3 and p62. ACS Nano. 2014;8(6):5898–5910. doi: 10.1021/nn5009879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Díaz-Moscoso A, Vercauteren D, Rejman J, et al. Insights in cellular uptake mechanisms of pDNA-polycationic amphiphilic cyclodextrin nanoparticles (CDplexes). J Control Release. 2010;143(3):318–325. doi: 10.1016/J.JCONREL.2010.01.016 [DOI] [PubMed] [Google Scholar]
- 44.Parton RG, Richards AA. Lipid Rafts and Caveolae as Portals for Endocytosis: New Insights and Common Mechanisms. Traffic. 2003;4(11):724–738. doi: 10.1034/j.1600-0854.2003.00128.x [DOI] [PubMed] [Google Scholar]
- 45.Adjei IM, Sharma B, Labhasetwar V. Nanoparticles: Cellular Uptake and Cytotoxicity. In: Springer, Dordrecht; 2014:73–91. doi: 10.1007/978-94-017-8739-0_5 [DOI] [PubMed] [Google Scholar]
- 46.Conlan AR, Paddock ML, Axelrod HL, et al. The novel 2Fe-2S outer mitochondrial protein mitoNEET displays conformational flexibility in its N-terminal cytoplasmic tethering domain. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65(7):554–559. doi: 10.1107/S1744309109019605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farajdokht F, Mohaddes G, Karimi-Sales E, et al. Inhibition of PTEN protects PC12 cells against oxygen-glucose deprivation induced cell death through mitoprotection. Brain Res. 2018;1692:100–109. doi: 10.1016/J.BRAINRES.2018.05.026 [DOI] [PubMed] [Google Scholar]
- 48.Minaei Beyrami S, Khadem Ansari MH, Rasemi Y, Shakib N, Karimi P. Complete inhibition of phosphatase and tensin homolog promotes the normal and oxygen-glucose deprivation/reperfusion-injured PC12 cells to cell death. J Cardiovasc Thorac Res. 2018;10(2):83–89. doi: 10.15171/jcvtr.2018.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan SHH, Tai M-H, Li C-Y, Chan JYH. Reduction in molecular synthesis or enzyme activity of superoxide dismutases and catalase contributes to oxidative stress and neurogenic hypertension in spontaneously hypertensive rats. Free Radic Biol Med. 2006;40(11):2028–2039. doi: 10.1016/J.FREERADBIOMED.2006.01.032 [DOI] [PubMed] [Google Scholar]
- 50.Staniek K, Nohl H. Are mitochondria a permanent source of reactive oxygen species? Biochim Biophys Acta - Bioenerg. 2000;1460(2-3):268–275. doi: 10.1016/S0005-2728(00)00152-3 [DOI] [PubMed] [Google Scholar]
- 51.Zhou M, Diwu Z, Panchuk-Voloshina N, Haugland RP. A Stable Nonfluorescent Derivative of Resorufin for the Fluorometric Determination of Trace Hydrogen Peroxide: Applications in Detecting the Activity of Phagocyte NADPH Oxidase and Other Oxidases. Anal Biochem. 1997;253(2):162–168. doi: 10.1006/ABIO.1997.2391 [DOI] [PubMed] [Google Scholar]
- 52.Liu Y, Ai K, Ji X, et al. Comprehensive Insights into the Multi-Antioxidative Mechanisms of Melanin Nanoparticles and Their Application To Protect Brain from Injury in Ischemic Stroke. J Am Chem Soc. 2017;139(2):856–862. doi: 10.1021/jacs.6b11013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodrigues JV, Gomes CM. Enhanced superoxide and hydrogen peroxide detection in biological assays. Free Radic Biol Med. 2010;49(1):61–66. doi: 10.1016/J.FREERADBIOMED.2010.03.014 [DOI] [PubMed] [Google Scholar]
- 54.Kalyanaraman B, Darley-Usmar V, Davies KJA, et al. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic Biol Med. 2012;52(1):1–6. doi: 10.1016/J.FREERADBIOMED.2011.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao B, Summers FA, Mason RP. Photooxidation of Amplex red to resorufin: Implications of exposing the Amplex red assay to light. Free Radic Biol Med. 2012;53(5):1080–1087. doi: 10.1016/J.FREERADBIOMED.2012.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dirnagl U, Simon RP, Hallenbeck JM. Ischemic tolerance and endogenous neuroprotection. Trends Neurosci. 2003;26(5):248–254. doi: 10.1016/S0166-2236(03)00071-7 [DOI] [PubMed] [Google Scholar]
- 57.Ma Q, Zhao H, Tao Z, et al. MicroRNA-181c Exacerbates Brain Injury in Acute Ischemic Stroke. Aging Dis. 2016;7(6):705–714. doi: 10.14336/AD.2016.0320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Niu F, Zhang X, Hu X, et al. Targeted mutation of Fas ligand gene attenuates brain inflammation in experimental stroke. Brain Behav Immun. 2012;26(1):61–71. doi: 10.1016/J.BBI.2011.07.235 [DOI] [PubMed] [Google Scholar]
- 59.Benchoua A, Guégan C, Couriaud C, et al. Specific caspase pathways are activated in the two stages of cerebral infarction. J Neurosci. 2001;21(18):7127–7134. doi: 10.1523/JNEUROSCI.21-18-07127.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fornaguera C, Calderó G, Mitjans M, Vinardell MP, Solans C, Vauthier C. Interactions of PLGA nanoparticles with blood components: protein adsorption, coagulation, activation of the complement system and hemolysis studies. Nanoscale. 2015;7(14):6045–6058. doi: 10.1039/c5nr00733j [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


