Abstract
Purpose
Early clinical recognition of sepsis can be challenging. With the advancement of machine learning, promising real-time models to predict sepsis have emerged. We assessed their performance by carrying out a systematic review and meta-analysis.
Methods
A systematic search was performed in PubMed, Embase.com and Scopus. Studies targeting sepsis, severe sepsis or septic shock in any hospital setting were eligible for inclusion. The index test was any supervised machine learning model for real-time prediction of these conditions. Quality of evidence was assessed using the Grading of Recommendations Assessment, Development and Evaluation (GRADE) methodology, with a tailored Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) checklist to evaluate risk of bias. Models with a reported area under the curve of the receiver operating characteristic (AUROC) metric were meta-analyzed to identify strongest contributors to model performance.
Results
After screening, a total of 28 papers were eligible for synthesis, from which 130 models were extracted. The majority of papers were developed in the intensive care unit (ICU, n = 15; 54%), followed by hospital wards (n = 7; 25%), the emergency department (ED, n = 4; 14%) and all of these settings (n = 2; 7%). For the prediction of sepsis, diagnostic test accuracy assessed by the AUROC ranged from 0.68–0.99 in the ICU, to 0.96–0.98 in-hospital and 0.87 to 0.97 in the ED. Varying sepsis definitions limit pooling of the performance across studies. Only three papers clinically implemented models with mixed results. In the multivariate analysis, temperature, lab values, and model type contributed most to model performance.
Conclusion
This systematic review and meta-analysis show that on retrospective data, individual machine learning models can accurately predict sepsis onset ahead of time. Although they present alternatives to traditional scoring systems, between-study heterogeneity limits the assessment of pooled results. Systematic reporting and clinical implementation studies are needed to bridge the gap between bytes and bedside.
Electronic supplementary material
The online version of this article (10.1007/s00134-019-05872-y) contains supplementary material, which is available to authorized users.
Keywords: Machine learning, Sepsis, Septic shock, Prediction, Systematic review, Meta-analysis
Take-home message
| Retrospective studies demonstrate that machine learning models can accurately predict sepsis and septic shock onset. Prospective clinical studies at the bedside are needed to assess their effect on patient-relevant outcomes. |
Introduction
Sepsis is one of the leading causes of death worldwide [1], with incidence and mortality rates failing to decrease substantially over the last few decades [2, 3]. While the Surviving Sepsis international consensus guidelines recommend starting antimicrobial treatment within 1 h from sepsis onset given the association between treatment delay and mortality [4–8], early recognition can be difficult due to disease complexity in clinical context [9, 10] and heterogeneity of the septic population [11].
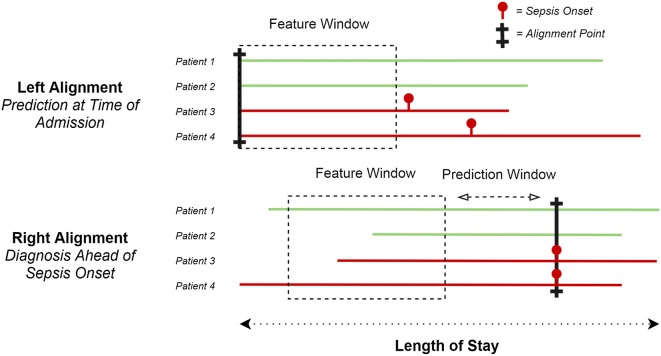
In recent years, medicine has witnessed the emergence of machine learning as a novel tool to analyze large amounts of data [12, 13]. Machine learning models to diagnose sepsis ahead of time are typically left or right aligned (Fig. 1) [14]. Left-aligned models predict the onset of sepsis following a fixed point in time, with varying time points such as on admission [15] or preoperatively [16, 17]. Right-aligned models continuously predict whether sepsis will occur after a distinct period of time and are also known as real-time or continuous prediction models. From a clinical perspective, they are particularly useful as they could trigger direct clinical action such as administration of antibiotics. Given their potential of prospective implementation and the large variety of left-aligned models, we focus on right-aligned models in this paper.
Fig. 1.
Left versus right alignment. Left alignment (top) versus right alignment (bottom). Cases are aligned at the alignment point, in the feature window data are collected, the prediction window is the time of the prediction ahead of sepsis onset. Red sepsis cases, green non-septic cases
Interpretation of machine learning studies predicting sepsis can be confusing, as some predict sepsis at its onset, which may seem counterintuitive and of little practical use. Their goal, however, is to identify whether a patient fulfills a predefined definition of sepsis, including proxies for infection such as antibiotic use or culture sampling. During development, these proxies are available to the model, while in a test set or new clinical patient, these are unknown. A model has therefore trained to predict whether sepsis is present in a new patient based on all other variables. In clinical practice, recognition of sepsis may be delayed and timely detection could expedite diagnosis and treatment. While we prefer the terms identification or detection in this context, we will use the term prediction throughout this work for brevity.
Considering the potential of machine learning in sepsis prediction, we set out to perform a systematic review of published, real-time (i.e. right aligned) machine learning models that predict sepsis including aggravate forms such as septic shock in any hospital setting. We hypothesized that these models show excellent performance retrospectively, but that few prospective studies have been carried out. In addition, we aimed to identify the most important factors that determine predictive performance in a meta-analysis.
Methods
This systematic review was conducted in accordance with the Preferred Reporting Items for a Systematic Review and Meta-analysis of Diagnostic Test Accuracy Studies (PRISMA-DTA) statement [18]. The study protocol was registered and approved on the international prospective register of systematic reviews PROSPERO before the start of the study (reference number CRD42019118716).
Search strategy
A comprehensive search was performed in the bibliographic databases PubMed, Embase.com, and Scopus up until September 13th, 2019, in collaboration with a medical librarian (LS). Search terms included controlled terms (MesH in PubMed and Emtree in Embase), as well as free-text terms. The following terms were used (including synonyms and closely related words) as index terms or free-text words: ‘sepsis’ and ‘machine learning’ and ‘prediction’. A search filter was used to limit the results to humans and adults. Only peer-reviewed articles were included. Conference abstracts were included to identify models that were published in full text elsewhere, but were excluded from the review. The full search strategies for all databases can be found in Online Resource 1.
Two review authors (LF and CZ) independently performed the title-abstract and full text screening. Disagreement was resolved by an independent intensivist (PE) and data scientist (MH). For the full text article screen, reasons for exclusion per article were recorded. References of the identified articles were checked for additional papers. Data were extracted by LF and confirmed by CZ. Discrepancies were revisited by both the authors to guarantee database accuracy.
Eligibility criteria and study selection
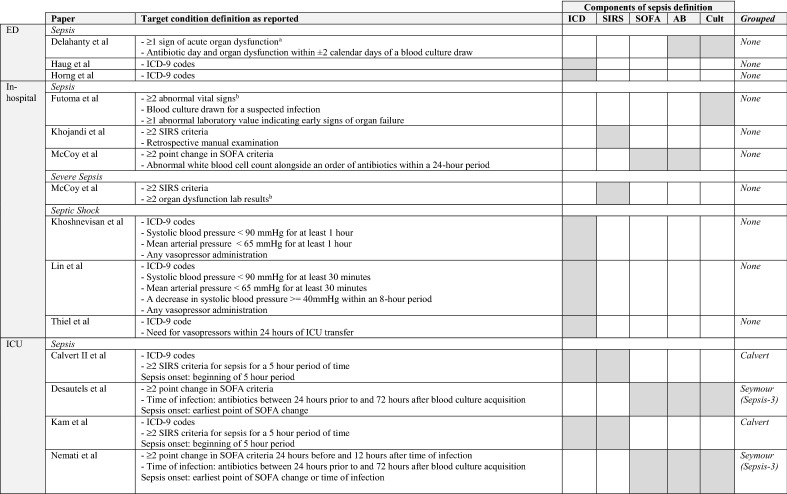
Studies were eligible if they aimed to predict the onset of sepsis in real time, i.e., right alignment, in adult patients in any hospital setting. Both prospective and retrospective studies were eligible for inclusion. The target condition was the onset of sepsis, severe sepsis, or septic shock. Although the 2016 consensus statement abandoned the term severe sepsis [19], papers prior to the consensus statement targeting severe sepsis were included. The target condition (gold standard) is defined per paper and serves to establish model performance (i.e., how well the model predicts sepsis versus non-sepsis cases). We collected these definitions per paper, as well as the components of these definitions: use of international classification of diseases (ICD) codes, SIRS/SOFA criteria, initiation of antibiotics, or sampling of blood cultures.
Supervised machine learning models were the index test of interest, defined as any machine learning classifying technique to predict the onset of the target condition, through some type of learning from presented data in a training dataset. Scikit Learn is one of the most used packages to code machine learning models in the popular programming language Python. Pragmatically, all supervised learning models found in this package were considered machine learning models [20]. A statement that the paper belongs to the machine learning domain, or any of its synonyms, was required for inclusion. An extensive list of commonly used machine learning model names was added to the search to cover any papers that failed to mention machine learning in their title or abstract.
Other items that were collected from the papers included the year of publication, study design, privacy statements, the origin of the model development and test dataset, use of an online database, description of the study population, the country of origin, the dataset split, the inclusion and exclusion criteria used, data granularity, methods for dealing with missing values, size of the database, number of patients with the outcome, the number of hours the model predicted ahead of time, the features used in the model, whether cross-validation was performed and its number of folds and the length of the sliding window, i.e. hours of data that were continuously fed to the model and the type of machine learning model.
Quality of evidence and risk of bias
As of yet, there exists no widely accepted checklist for assessing the quality of diagnostic machine learning papers in a medical setting. This paper used the Grading of Recommendations Assessment, Development and Evaluation (GRADE) methodology to assess the quality of evidence per hospital setting for all studies reporting the area under the curve of the receiver operating characteristic (AUROC) as their performance metric [21]. In line with the GRADE guidelines for diagnostic test accuracy, we included the domains risk of bias (limitations), comparison of patients, setting, and outcome across studies (indirectness of comparisons), and imprecision of the results. As we do not compute point estimates for multiple studies combined, judgment of inconsistency was omitted. One level of evidence was deducted for each domain with serious concerns or high risk of bias, no factors increased the level of evidence (see Online Resource 2). Overall level of evidence is expressed in four categories (high, moderate, low, very low).
To evaluate risk of bias, the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) criteria [22] were combined with an adapted version of the Joanna Briggs Institute Critical Appraisal checklist for analytical cross-sectional studies [23]. The latter has been used in previous work to assess machine learning papers [24]. Domains included patient selection, index test, reference standard, flow and timing, and data management. In line with the recommendations from the QUADAS-2 guidelines, questions per domain were tailored for this paper and can be found in Online Resource 3. Two review authors (LF and CZ) independently piloted the questions to ascertain between-reviewer agreement. If one of the questions was scored at risk of bias, the domain was scored as high risk of bias. At least one domain at high risk of bias resulted in an overall score of high risk of bias, only one domain scored as unclear risk of bias resulted in an overall score of unclear risk of bias for that paper.
Performance metric and meta-analysis
Substantial heterogeneity was observed between studies regarding the setting, index test, and outcome. We therefore refrained from computing a point estimate for overall model performance. However, the large number of studies and models did allow for analysis of study characteristics’ and model parameters’ contribution to model performance. Multiple models were reported per paper, introducing collinearity in their performance. A linear random effect model was built with a paper-specific random effect to account for correlations between models published in the same paper. For clarity, we refer to all study characteristics that served as input to this analysis as covariates, while variables to develop the presented models are referred to as features.
The machine learning field distinguishes numerous metrics to gauge model performance, none of which gives a complete picture. The AUROC, a summary measure of sensitivity and specificity, has been customary to the field of diagnostic test accuracy. Since 24 out of 28 papers (86%) reported the AUROC, this was pragmatically selected as the main performance metric. Other metrics were collected, but unsystematically reported. As AUROCs are constrained to the interval 0.5 to 1.0, they were transformed and linearized to a continuous scale by taking the logit transformation of the result of the formula . Because only 43 models (33%) reported confidence intervals, within-study variability was omitted from the analysis. For studies that did report confidence intervals, one-sided AUROC confidence intervals did not exceed 0.02.
All items collected from the presented studies were added as covariates to the random effects model, including components of the target condition. Missing values in the continuous covariates were imputed with the column median. To account for the high ratio of covariates to number of models, some of the features identified in the models were grouped (lab values, blood gas values, co-morbidities, department information), only covariates with 10% variance in their values were included and models that aimed to predict combined outcomes were removed as they were too scarce in the database. One outlier reference model was excluded [25].
All covariates were first tested in a univariate model for a significant contribution to the transformed AUROC using a likelihood ratio test against an empty model containing only the intercept and the variance components. All significant covariates (p < 0.05) were then considered for a multivariate model. Through backward Akaike information criterion (AIC) selection, a parsimonious model was selected. Covariate coefficients, standard error, and p values are reported. All analyses were carried out in R [26].
Results
Study selection
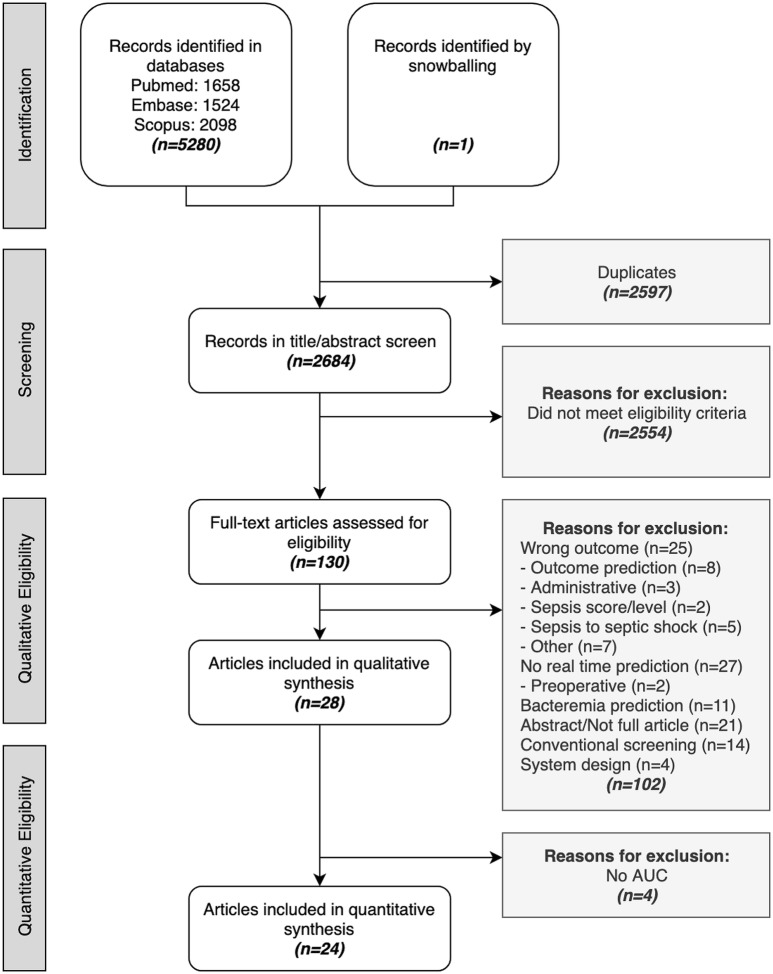
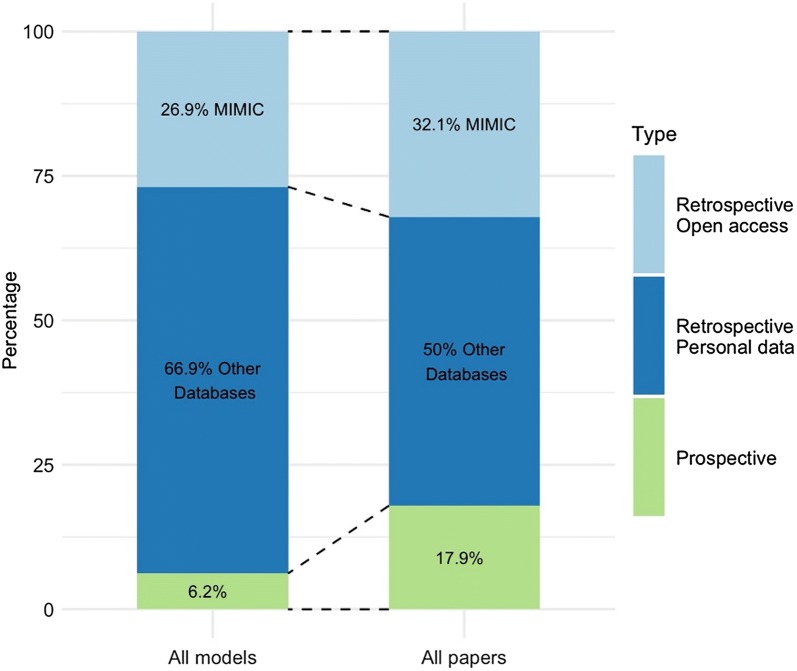
After removing duplicates and reference checking for extra papers, a total of 2.684 papers were screened. Among these, 130 papers were read full text resulting in 28 papers that met the inclusion criteria for synthesis. Reasons for exclusion at this stage were recorded and can be found in the flow diagram in Fig. 2. From these papers, 130 models were retrieved (range 1–16 models per paper). All studies reported retrospective diagnostic test accuracy. In addition, models were prospectively validated in two papers (7%) and clinically implemented in three papers (11%), as depicted in Fig. 3. Out of all papers, 24 reported AUROC as their performance metric.
Fig. 2.
Flow diagram. Papers identified in databases, title/abstract screened, read full text, and included in the synthesis. Reasons for exclusion are listed
Fig. 3.
Prospective versus retrospective models. Percentages specified per paper and for all models
Study characteristics
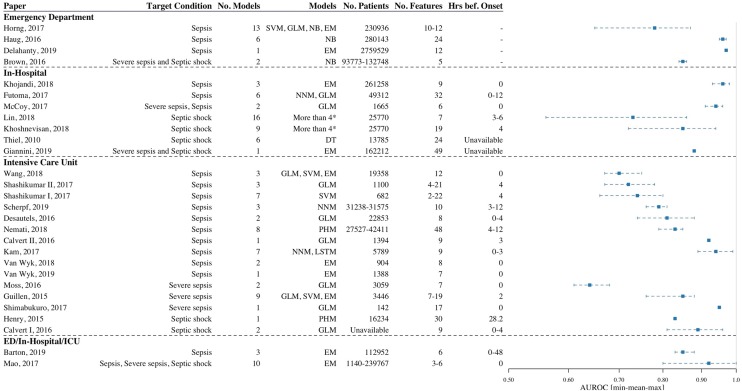
Most of the studies were carried out in the ICU (n = 15; 54%), followed by hospital wards (n = 7; 25%) and the emergency department (ED, n = 4; 14%). Two studies by Barton et al., and Mao et al., examined all of these settings [25, 27]. In the intensive care, most of the studies modeled sepsis as their target condition (n = 10; 67%), compared to severe sepsis (n = 3; 20%) or septic shock (n = 2; 13%). This contrasts the in-hospital studies, where almost half of the papers aimed to predict septic shock (n = 3; 43%). Figure 4 gives an overview of key characteristics per study.
Fig. 4.
Overview of retrospective diagnostic test accuracy studies. Papers are binned per hospital setting, data are sorted in ascending order of AUROC values. AUROC ranges are displayed per paper. AUROC area under the curve of the receiver operating characteristic, SVM support vector machines, GLM generalized linear model, NB Naive Bayes, EM ensemble methods, NNM neural network model, DT decision trees, PHM proportional hazards model, LSTM long short term memory, Hrs bef. onset hours before onset * DT, EM, GLM, LSTM, NB, NNM, SVM
Retrospective diagnostic test accuracy varied per setting and target condition. For the studies that reported AUROCs, best predictions of sepsis ranged from 0.87 to 0.97 in the emergency department, to 0.96–0.98 in-hospital and 0.68–0.99 in the intensive care unit. Septic shock predictions in an in-hospital setting ranged between 0.86–0.94 and 0.83–0.96 in the ICU at best. Other outcome measures such as positive predictive value (n = 11; 39%), accuracy (n = 10; 36%), and negative predictive value (n = 6; 21%) were unsystematically reported. The minimum, mean, and maximum AUROC values with relevant study characteristics are visualized per paper in Fig. 4.
Prospective studies included two clinical validation studies (ED and in-hospital) and three interventional studies (in-hospital and ICU). One clinical validation study in the ED showed the machine learning model outperformed manual scoring by nurses and the SIRS criteria when identifying severe sepsis and septic shock [28], the other study made no comparison [29]. The interventional studies included two pre-post implementation studies (in-hospital) [30, 31] and one ICU randomized controlled trial [32]. All looked at mortality and hospital length of stay, but results are mixed as shown in Table 1.
Table 1.
Prospective models
| Paper | Design | Target condition | Patient encounters | Machine learning model | Comparators | |
|---|---|---|---|---|---|---|
| Validation | ||||||
| ED | Brown et al. | Prospective validation | Severe sepsis and Septic shock | 93,773 (15 months) |
Cut05 Primary outcome Sensitivity: 0.764 False positive rate: 0.47 Secondary outcome AUC: 0.859 |
Nurse triage Primary outcome Sensitivity: 0.543 False positive rate: 0.31 Secondary outcome AUC: 0.756 |
|
SIRS Primary outcome Sensitivity: 0.216 False positive rate: 0.004 Secondary outcome AUC: 0.606 | ||||||
| In-hospital | Thiel et al. | Prospective validation | Septic shock | 27,674 (24 months) |
RPARTa 2006 Primary outcome Misclassification rate: 8.4% |
None |
|
RPARTa 2007 Primary outcome Misclassification rate: 8.8% | ||||||
| Paper | Design | Target condition | Patient encounters | Machine learning group | Control group | |
|---|---|---|---|---|---|---|
| Interventional | ||||||
| In-hospital | Giannini et al. | Pre-post implementation | Severe sepsis and septic shock | 54,464 (6 pre-months, 1 post-month) |
EWS 2.0 Primary/secondary outcome Hospital LOS: 9 days Time to ICU transfer after alert: 8 he In-hospital mortality: 10.3% |
Unclear Primary/secondary outcome Hospital LOS: 9 days Time to ICU transfer after alert: 16 he In-hospital mortality: 10.6% |
| McCoy et al. | Pre-post implementationb | Severe sepsis | 611 (3 pre-months, 2 post-months) |
Linear model (Insight) Primary outcome In-hospital mortality: 2.94% Secondary outcome Hospital LOS: 2.92 days Readmission rate: 7.84% |
Manual nurse scoringc Primary outcome In-hospital mortality: 7.37% Secondary outcome Hospital LOS: 3.35 days Readmission rate: 46.19% |
|
| ICU | Shimabukuro et al. | RCT | Severe sepsis | 142 (3 months) |
Elastic net reg.d (Insight) Primary outcome Hospital LOS: 10.3 dayse Secondary outcome ICU LOS: 6.3 dayse In-hospital mortality: 8.96%e |
SIRS detector Primary outcome Hospital LOS: 13.0 dayse Secondary outcome ICU LOS: 8.4 dayse In-hospital mortality: 21.3%e |
aRecursive partitioning and regression tree (RPART) analysis
bOnly baseline and steady state are reported
cNurses scored patient twice daily to see if they met the SIRS criteria
dElastic net regularization (generalized linear model)
eSignificant results
For the target condition, different definitions of sepsis, severe sepsis, and septic shock were used. Definitions and their components are reported in Table 2. Definitions that had been used before are named according to the first paper they appeared in. Calvert et al. [33] was one of the first to study machine learning to identify sepsis in an ICU population and Seymour et al. [34] assessed the sepsis-3 criteria. Nine studies (32%) employed a definition for sepsis that had been previously used.
Table 2.
Target condition definitions per paper per setting
Group listed when more than one paper used definitions. Combined outcomes are not shown, sorted alphabetically
aOrgan dysfunction: initiation of vasopressors or mechanical ventilation, elevated lactate level, or significant changes in baseline creatinine level, bilirubin level, or platelet count
bUndefine
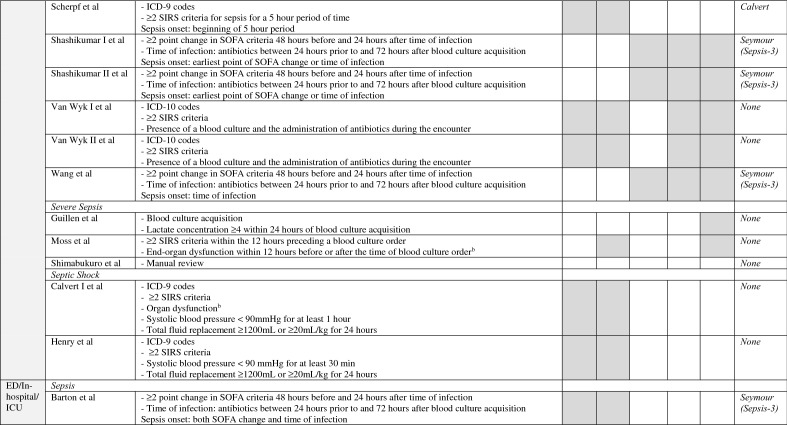
A breakdown of the paper and model characteristics per setting can be found in Table 3. The number of features used in the models ranges from 2 to 49 and the most common features are shown in Fig. 5. Thirty six percent of papers used MIMIC data; others used non-freely available hospital datasets. Three papers using their own hospital data reported inquiries for data sharing were possible [28, 32, 35], two papers reported data would not be shared [25, 31]. None of the studies mentioned their code was released and only one paper reported adhering to a reporting standard [36].
Table 3.
Description of the data per paper and per model
| EDa 4 papers 22 models |
In-hospitala 7 papers 43 models |
ICUa 15 papers 52 models |
||||
|---|---|---|---|---|---|---|
| Absolute | Proportion | Absolute | Proportion | Absolute | Proportion | |
| Per paper | ||||||
| Prospective design | 1 | 0.25 | 2 | 0.29 | 1 | 0.07 |
| Privacy statement | 0 | 0.00 | 3 | 0.43 | 5 | 0.33 |
| MIMICb | – | – | – | – | 9 | 0.60 |
| Description of patients | 4 | 1.00 | 2 | 0.29 | 5 | 0.33 |
| Inclusion criteria | 3 | 0.75 | 4 | 0.57 | 12 | 0.80 |
| Country—USA | 4 | 1.00 | 7 | 1.00 | 15 | 1.00 |
| Per model | ||||||
| Target condition | ||||||
| Sepsis | 20 | 0.91 | 10 | 0.23 | 37 | 0.71 |
| Severe sepsis | 0 | 0.00 | 1 | 0.02 | 12 | 0.23 |
| Severe sepsis & septic shock | 2 | 0.09 | 1 | 0.02 | 0 | 0.00 |
| Septic shock | 0 | 0.00 | 31 | 0.72 | 3 | 0.06 |
| Components of target condition definition | ||||||
| ICD | 20 | 0.91 | 32 | 0.74 | 17 | 0.33 |
| SIRS | 0 | 0.00 | 4 | 0.09 | 19 | 0.37 |
| SOFA | 0 | 0.00 | 1 | 0.02 | 21 | 0.40 |
| Data split design | ||||||
| Train-(validate)-test | 20 | 0.91 | 15 | 0.35 | 21 | 0.40 |
| Cross-validation | 0 | 0.00 | 25 | 0.58 | 28 | 0.54 |
| Data granularity | ||||||
| 1-hourly values | – | – | 6 | 0.14 | 30 | 0.58 |
| > 1/hourly values | – | – | 0 | 0.00 | 18 | 0.35 |
| Not described | – | – | 31 | 0.72 | 4 | 0.08 |
| Missing values strategies | ||||||
| Feedforward | 0 | 0.00 | 8 | 0.19 | 14 | 0.27 |
| Mean imputation | 0 | 0.00 | 9 | 0.21 | 12 | 0.23 |
| Zero imputation | 0 | 0.00 | 16 | 0.37 | 0 | 0.00 |
| Nearest neighbor | 0 | 0.00 | 0 | 0.00 | 16 | 0.31 |
| Physiological imputation | 13 | 0.59 | 0 | 0.00 | 0 | 0.00 |
| Otherc | 7 | 0.32 | 3 | 0.07 | 2 | 0.04 |
| Not described | 2 | 0.09 | 7 | 0.16 | 8 | 0.15 |
| Model | ||||||
| Generalized linear model | 3 | 0.14 | 6 | 0.14 | 15 | 0.29 |
| Naïve Bayes | 11 | 0.50 | 3 | 0.07 | 0 | 0.00 |
| Ensemble methods | 4 | 0.18 | 9 | 0.21 | 7 | 0.13 |
| Proportional hazard | 0 | 0.00 | 0 | 0.00 | 9 | 0.17 |
| Decision tree | 0 | 0.00 | 9 | 0.21 | 0 | 0.00 |
| Support vector machines | 4 | 0.18 | 3 | 0.07 | 11 | 0.21 |
| Neural network | 0 | 0.00 | 8 | 0.19 | 6 | 0.12 |
| Long short-term memory (LSTM) | 0 | 0.00 | 5 | 0.12 | 4 | 0.08 |
ICD International Statistical Classification of Diseases and Related Health Problems, SIRS systemic inflammatory response syndrome, SOFA sequential organ failure assessment
aStudy by Mao et al. (2017) with an ED, In-hospital, ICU setting has been omitted for brevity
bStudies that included MIMIC in at least one of their reported models
cOthers: proxy variable, removal of variable, and predictive mean matching
Fig. 5.
Features used in the papers. Features are grouped by type. ESR erythrocyte sedimentation rate, HR heart rate, MAP mean arterial pressure
Quality of evidence and risk of bias
In accordance with the publication guidelines of the QUADAS-2 criteria, results for the risk of bias for retrospective diagnostic test accuracy studies are shown in Table 4. Nine out of 28 (32%) papers were scored as unclear risk of bias; all other papers were scored as high risk of bias. Papers scored a high risk of bias for failing to describe their study population (patient selection), not reporting their data split or cross-validation strategies (index test), or failing to specify ethical approval (data management). As there exists no gold standard in diagnosing sepsis, the variety in definitions may increase the risk of bias of the models. All papers therefore have an unclear risk of bias concerning the reference standard.
Table 4.
QUADAS-2 risk of bias assessment per setting
| Paper | Setting | Risk of bias | |||||
|---|---|---|---|---|---|---|---|
| Patient selection | Index test | Reference standard | Flow and timing | Data management | |||
| ED | Horng et al. [35] | Sepsis | ? | ||||
| Haug et al. [62] | Sepsis | ? | |||||
| Delahanty et al. [63] | Sepsis | ? | |||||
| Brown et al. [28] | Severe sepsis and septic shock | ? | |||||
| In-hospital | Khojandi et al. [64] | Sepsis | ? | ||||
| Futoma et al. [65] | Sepsis | ? | |||||
| McCoy et al. [31] | Severe sepsis, Sepsis | ? | |||||
| Lin et al. [66] | Septic shock | ? | |||||
| Khoshnevisan et al. [14] | Septic shock | ? | |||||
| Thiel et al. [29] | Septic shock | ? | |||||
| Giannini et al. [30] | Severe sepsis and septic shock | ? | |||||
| ICU | Wang et al. [57] | Sepsis | ? | ||||
| Shashikumar II et al. [67] | Sepsis | ? | |||||
| Shashikumar I et al. [68] | Sepsis | ? | |||||
| Scherpf et al. [59] | Sepsis | ? | |||||
| Desautels et al. [54] | Sepsis | ? | |||||
| Nemati et al. [55] | Sepsis | ? | |||||
| Calvert II et al. [52] | Sepsis | ? | |||||
| Kam et al. [53] | Sepsis | ? | |||||
| Van Wyk I et al. [69] | Sepsis | ? | |||||
| Van Wyk II et al. [70] | Sepsis | ? | |||||
| Moss et al. [36] | Severe sepsis | ? | |||||
| Guillén et al. [58] | Severe sepsis | ? | |||||
| Shimabukuro et al. [32] | Severe sepsis | ? | |||||
| Henry et al. [56] | Septic shock | ? | |||||
| Calvert I et al. [33] | Septic shock | ? | |||||
| ED/In-hospital/ICU | Barton et al. [27] | Sepsis | ? | ||||
| Mao et al. [71] | Sepsis, severe sepsis, septic shock | ? | |||||
 low risk,
low risk,
 high risk, ? unclear risk
high risk, ? unclear risk
The GRADE evidence profile can be found in Table 5. Results are shown when at least two studies reported the same target condition. All study aggregates were considered to be at high risk of bias, only five studies were considered at unclear risk of bias (included in brackets in Table 5). One level of evidence was deducted for high risk of bias and one level was deducted for indirectness of the outcome. Consequently, the quality of evidence for each of the settings was scored as low. Additionally, the outcome column distinguishes AUROC values for high and unclear risk of bias studies. Consistently, high risk of bias studies reported the highest AUROC values, although ranges are wide and relatively few unclear risk of bias studies were identified.
Table 5.
GRADE evidence profile for area under the receiving operating characteristic curve (AUROC)
| Study Characteristics | Quality Assessment | Outcome | |||||||
|---|---|---|---|---|---|---|---|---|---|
| No of studies | Design | Limitations (Unclear risk of bias studies/total) | Indirectness of patients, settingb | Indirectness of outcome | Inconsistencyc | Imprecision | AUROC high risk of bias/unclear risk of bias | Quality of evidence | |
| ED | Sepsis | ||||||||
| 3 studies (3.270.608 patients) | Cohort studies | High risk of bias (2/3) | None | Serious indirectness—differences in outcome definition | Not available | None | 0.95–0.97/0.65–0.97 | ⊕ ⊕ ⊙ ⊙ Low | |
| In-hospital | Septic shock | ||||||||
| 2 studies (51,540 patients) | Cohort studies | High risk of bias (0/2) | None | Serious indirectness—differences in outcome definition | Not available | None | 0.86–0.94 | ⊕ ⊕ ⊙ ⊙ Low | |
| ICU | Sepsis | ||||||||
| 8 studies (125.162 patients) | Cohort studies | High risk of bias (2/8) | None | Serious indirectness—differences in outcome definition | Not available | None | 0.70–0.99/0.81–0.88 | ⊕ ⊕ ⊙ ⊙ Low | |
| Severe sepsis | |||||||||
| 3 studies (6.647 patients) | Cohort studies | High risk of bias (0/3) | None | Serious indirectness—differences in outcome definition | Not available | None | 0.68–0.95 | ⊕ ⊕ ⊙ ⊙ Low | |
| Septic shock | |||||||||
| 2 studies (16.234 patientsa) | Cohort studies | High risk of bias (1/2) | None | Serious indirectness—differences in outcome definition | Not available | None | 0.89–0.96/0.83–0.83 | ⊕ ⊕ ⊙ ⊙ Low | |
Only settings with at least two studies are reported
aCalvert et al. (2016) had no information on total number of patients studied
bEvidence profile is binned per setting
cConfidence intervals were inconsistently reported, and therefore no heterogeneity assessment was performed
Meta-analysis
A total of 111 models were included in the meta-analysis after removal of an outlier (n = 1; 1%), combined outcomes (n = 3; 2%), and models without an AUROC outcome measure (n = 15; 12%). Initially, 103 covariates were included in the model. To reduce the ratio of covariates to the number of models, features used in the models were grouped (n = 41; 40%) and covariates with low variance (n = 24; 23%) and perfectly colinear covariates (n = 1; 1%) removed. This amounted to a total of 39 covariates in the meta-analysis random effect model.
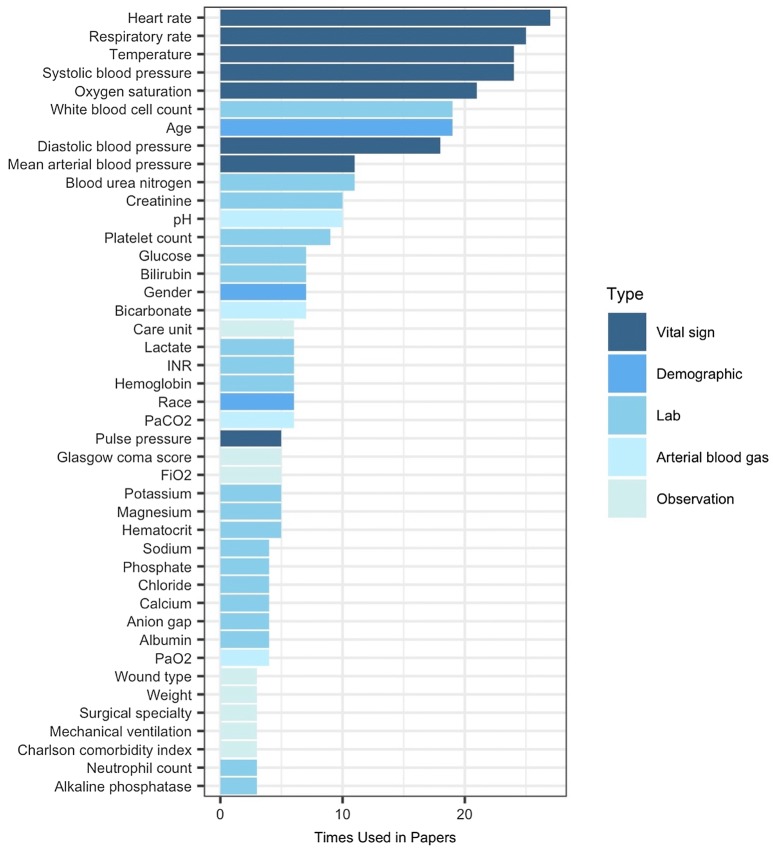
Univariate and multivariate random effect model results are shown in Table 6. Coefficients are logit transformed AUROC values and represent the expected mean change in AUROC, when the sepsis prediction model exhibited the respective characteristic (e.g. used lab values). Univariate analysis of the 39 covariates shows heart rate, respiratory rate, temperature, lab and arterial blood gas values, and neural networks (relative to ensemble methods) positively contributed to the AUROC (range 0.344–0.835). Only temperature, lab values, and model type remained in the multivariate model. On the contrary, defining sepsis using the definition coined by Seymour et al., using SOFA scores in the target condition definition, or any other model but ensemble methods or neural networks negatively impacts AUROC in the univariate analysis (range 0.168–1.039). Since the AUROC was logit transformed, it was back-transformed to the AUROC scale by taking the anti-logit. The relationship between AUROC and the hours before onset of the prediction is visualized for three models in Fig. 6.
Table 6.
Univariate and multivariate outcomes
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| Coeff | SE | p value | Coeff | SE | p value | |
| Temperature as feature | 0.788 | 0.239 | 0.002 | 0.812 | 0.218 | 0.000 |
| Lab values as feature | 0.835 | 0.311 | 0.008 | 0.842 | 0.291 | 0.003 |
| Type of model (ref. = EM) | 0.018 | 0.020 | ||||
| Generalized linear model | − 0.211 | 0.251 | − 0.211 | 0.231 | ||
| Naïve Bayes | − 0.651 | 0.312 | − 0.682 | 0.291 | ||
| Neural network | 0.344 | 0.300 | 0.172 | 0.278 | ||
| Proportional hazard | − 0.464 | 0.851 | − 0.506 | 0.673 | ||
| Support vector machines | − 0.168 | 0.256 | − 0.161 | 0.241 | ||
| Decision trees | − 1.013 | 0.419 | − 1.088 | 0.399 | ||
| Target condition defined as Seymour (Sepsis-3) | − 1.039 | 0.459 | 0.025 | |||
| Target condition definition contains SOFA | − 0.935 | 0.438 | 0.033 | |||
| Respiratory rate as feature | 0.672 | 0.250 | 0.008 | |||
| Heart rate as feature | 0.680 | 0.327 | 0.037 | |||
| Arterial blood gas as feature | 0.802 | 0.313 | 0.011 | |||
Coeff coefficient, SE standard error, ref. reference model, EM ensemble methods, SOFA sequential organ failure assessment
Fig. 6.
Relative effect of hours before sepsis onset on AUROC for different models. Expected change in AUROC for three models at different prediction windows (hours before sepsis onset)
Discussion
This is the first study to systematically review the use of machine learning to predict sepsis in the intensive care unit, hospital wards, and emergency department. Twenty eight papers reporting 130 machine learning models were included, each showing excellent performance on retrospective data. The most predictive covariates in these models are clinically recognized for their importance in sepsis detection. Assessment of overall pooled performance, however, is hampered by varying sepsis definitions across papers. Clinical implementation studies that demonstrate improvement in patient outcomes using machine learning are scarce.
Performance and clinical relevance of individual models
Clinically, accurate identification of sepsis and prediction of patients at risk of developing sepsis is essential to improve treatment [37]. Current approaches to identify septic patients have centered around biomarkers and (automated) clinical decision rules such as the SIRS and (q)SOFA criteria [38, 39]. However, concerns have been raised regarding the poor sensitivity of the qSOFA possibly leading to delays in sepsis identification [40]. The high sensitivity of the SIRS criteria, on the other hand, could lead to over diagnosis of sepsis resulting in inappropriate antibiotics use [41]. Additionally, most of the investigated biomarkers failed to show discriminative power or clinical relevance [42, 43]. The presented machine learning models provide a novel approach to continuously identify sepsis ahead of time with excellent individual performance. These models present an alternative to the widely used SIRS and SOFA criteria and clinicians may be faced with these models in the near future. Therefore, it is important that they understand the strengths and limitations of these models.
Heterogeneity and pooled performance
Ideally AUROC values across all presented models would be pooled to estimate overall machine learning performance. However, considerable heterogeneity in the sepsis definitions between studies hampers such computation. The lack of a gold standard for sepsis allows for a variety of definitions to be adopted. Many studies use ICD coding, which may be an unreliable instrument to identify septic patients [44, 45]. Arguably, all papers should use the most recent consensus definition [19]. Only a minority of papers used the latest sepsis-3 criteria and within these studies, we found differences in the way the sepsis onset time was defined. Due to these varying definitions, we refrained from computing overall performance of machine learning models and we consequently judged the quality of evidence as low for each of the hospital settings. Nonetheless, each of the definitions is a clinically relevant entity that might justify early antibiotic and supportive treatment.
Additionally, heterogeneity is observed in machine learning models, preprocessing of the data, and hospital setting. While this further limits pooling of the overall performance, it does allow for meta-analysis of the models to identify the most important factors that contribute to model performance. Most predictive covariates from our meta-analysis such as heart rate and temperature are recognized for their clinical importance in sepsis detection. Variables that are part of the SIRS and SOFA criteria were expected to correlate with model performance, since they are frequently part of the sepsis definitions. Interestingly, some other factors that are not part of these criteria, such as arterial blood gas variables, were also strong predictors univariately. Lab values are often not considered in early warning scores [46], but our results imply that these scores may miss predictive information.
Clinical model performance
It is important to investigate, whether improved sepsis predictions lead to better clinical outcomes for patients. We distinguish prospective clinical validation studies that assess model performance in a clinical setting and interventional studies, where the effect of exposing healthcare professionals to model predictions on patient outcomes is investigated. Only one study clinically validated their model and showed that these models outperformed nurse triaging and SIRS criteria in the emergency room [47].
Interventional studies using traditional SIRS and SOFA alarm systems have not shown significant changes in clinical outcomes [48–50]. Only three interventional studies have been identified in this review, which were carried out in different clinical settings and show mixed results [31, 32, 51]. None of the studies, however, investigated a direct clinical action associated with the sepsis prediction, but left treatment decisions at the discretion of the clinician. Prior to sepsis onset, however, clinically overt signs of sepsis may be subtle or absent and false positive alerts in these studies may create alarm fatigue. Nonetheless, as of yet, there is no compelling evidence that machine learning predictions lead to better patient outcomes in sepsis.
Future directions and academic contribution
An important message in this paper is that systematic reporting is essential for reliable interpretation and aggregation of results. Almost none of the papers mentioned using a reporting standard and very few papers reported they accept data inquiries [32, 35, 47]. In addition, high bias studies showed highest AUROC values overall. We encourage the authors to strive for the sharing of code and data in compliance with relevant regulations. This would allow for easy data aggregation, model retraining, and comparison as our insight into sepsis definitions evolves.
It should be noted that many models were developed on similar populations. Specifically, numerous models were tested on the freely accessible MIMIC database [27, 33, 52–59] and all models were developed in the United States. The current trend holds risks for promoting inequality in healthcare as no models were developed or validates in middle or low income countries. We encourage developing models on data of different centers and countries to ensure generalizability.
Finally, future research is needed to determine effective integration strategies of these models into the clinical workflow and assess the effect on relevant clinical outcomes. Interestingly, most models only use a small subset of the wealth of available data to clinicians, which may present an opportunity for future models to further increase predictive performance. Lastly, baseline characteristics may lead to clinically relevant heterogeneity in sepsis trials [11]. To administer treatment to more homogenous patient groups, the accurate identification of pre-specified populations by machine learning models could be investigated.
Strengths and limitations
Several strengths can be identified in this study. First of all, this is the first study to systematically list all research in this area. It combines both clinical and more technical work and assesses performance in a clinical light, while studies are scrutinized through a technical and clinical lens. Additionally, a large number of models resulted from the search, which permitted comparison and meta-analysis of the contribution of model components to performance.
This study also has limitations. First, the AUROC was pragmatically chosen as a summary measure, while it may underperform in the setting of imbalanced datasets [60]. Nonetheless, it was the summary measure most frequently reported; other measures would have eroded the possibility to compare performance across studies. Similarly, no contingency tables were feasible for the majority of papers as the necessary data were too infrequently reported and very few papers reported measures of uncertainty such as confidence intervals or standard deviations. In line with a previous machine learning review on imaging [61], we believe reporting of these studies has to be improved to guarantee reliable interpretation and we encourage guideline development in the areas of intensive care and emergency medicine.
Conclusion
This systematic review and meta-analysis show that machine learning models can accurately predict sepsis onset with good discrimination in retrospective cohorts. Important factors associated with model performance include the use of variables that are well recognized for their clinical importance in sepsis. Even though individual models tend to outperform traditional scoring tools, assessment of their pooled performance is limited by heterogeneity of studies. This calls for the development of reporting guidelines for machine learning for intensive care medicine. Clinical implementation of models is currently scarce and is therefore urgently needed across diverse patient populations to determine clinical impact, ensure generalizability, and to bridge the gap between bytes and bedside.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Compliance with ethical standards
Conflicts of interest
The author(s) declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fleischmann C, Scherag A, Adhikari NKJ, et al. Assessment of global incidence and mortality of hospital-treated sepsis. current estimates and limitations. Am J Respir Crit Care Med. 2016;193:259–272. doi: 10.1164/rccm.201504-0781OC. [DOI] [PubMed] [Google Scholar]
- 2.Rhee C, Dantes R, Epstein L, et al. Incidence and trends of sepsis in US hospitals using clinical vs claims data, 2009–2014. JAMA. 2017;318:1241. doi: 10.1001/jama.2017.13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Álvaro-Meca A, Jiménez-Sousa MA, Micheloud D, et al. Epidemiological trends of sepsis in the twenty-first century (2000–2013): an analysis of incidence, mortality, and associated costs in Spain. Popul Health Metr. 2018;16:4. doi: 10.1186/s12963-018-0160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seymour CW, Gesten F, Prescott HC, et al. Time to treatment and mortality during mandated emergency care for sepsis. N Engl J Med. 2017;376:2235–2244. doi: 10.1056/NEJMoa1703058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu VX, Fielding-Singh V, Greene JD, et al. The timing of early antibiotics and hospital mortality in sepsis. Am J Respir Crit Care Med. 2017;196:856–863. doi: 10.1164/rccm.201609-1848OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhodes A, Evans LE, Alhazzani W, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Intensive Care Med. 2017;43:304–377. doi: 10.1007/s00134-017-4683-6. [DOI] [PubMed] [Google Scholar]
- 7.Ferrer R, Martin-Loeches I, Phillips G, et al. Empiric antibiotic treatment reduces mortality in severe sepsis and septic shock from the first hour. Crit Care Med. 2014;42:1749–1755. doi: 10.1097/CCM.0000000000000330. [DOI] [PubMed] [Google Scholar]
- 8.Kumar A, Roberts D, Wood KE, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34:1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- 9.Vincent J-L. The clinical challenge of sepsis identification and monitoring. PLoS Med. 2016;13:e1002022. doi: 10.1371/journal.pmed.1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Talisa VB, Yende S, Seymour CW, Angus DC. Arguing for adaptive clinical trials in sepsis. Front Immunol. 2018;9:1502. doi: 10.3389/fimmu.2018.01502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Grooth H-J, Postema J, Loer SA, et al. Unexplained mortality differences between septic shock trials: a systematic analysis of population characteristics and control-group mortality rates. Intensive Care Med. 2018;44:311–322. doi: 10.1007/s00134-018-5134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beam AL, Kohane IS. Big data and machine learning in health care. JAMA. 2018;319:1317. doi: 10.1001/jama.2017.18391. [DOI] [PubMed] [Google Scholar]
- 13.Yu K-H, Beam AL, Kohane IS. Artificial intelligence in healthcare. Nat Biomed Eng. 2018;2:719–731. doi: 10.1038/s41551-018-0305-z. [DOI] [PubMed] [Google Scholar]
- 14.Khoshnevisan F, Ivy J, Capan M, et al (2018) Recent temporal pattern mining for septic shock early prediction. In: 2018 IEEE international conference on healthcare informatics (ICHI). IEEE, pp 229–240
- 15.Nachimuthu SK, Haug PJ. Early detection of sepsis in the emergency department using dynamic Bayesian networks. AMIA Annu Symp Proc AMIA Symp. 2012;2012:653–662. [PMC free article] [PubMed] [Google Scholar]
- 16.Thottakkara P, Ozrazgat-Baslanti T, Hupf BB, et al. Application of machine learning techniques to high-dimensional clinical data to forecast postoperative complications. PLoS One. 2016;11:e0155705. doi: 10.1371/journal.pone.0155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bihorac A, Ozrazgat-Baslanti T, Ebadi A, et al. My surgery risk. Ann Surg. 2019;269:652–662. doi: 10.1097/SLA.0000000000002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McInnes MDF, Moher D, Thombs BD, et al. Preferred reporting items for a systematic review and meta-analysis of diagnostic test accuracy studies the PRISMA-DTA statement. JAMA J Am Med Assoc. 2018;319:388–396. doi: 10.1001/jama.2017.19163. [DOI] [PubMed] [Google Scholar]
- 19.Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (sepsis-3) JAMA. 2016;315:801. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Supervised learning—scikit-learn 0.21.2 documentation (2019) https://scikit-learn.org/stable/supervised_learning.html. Accessed 8 Jul 2019
- 21.Schünemann H, Brożek J, Guyatt G OA (2013) GRADE handbook for grading quality of evidence and strength of recommendations
- 22.Whiting PF, Rutjes AWS, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 23.Critical Appraisal Tools|Joanna Briggs Institute (2019) https://joannabriggs.org/critical_appraisal_tools. Accessed 8 Jul 2019
- 24.Kwong MT, Colopy GW, Weber AM, et al. The efficacy and effectiveness of machine learning for weaning in mechanically ventilated patients at the intensive care unit: a systematic review. Bio-Design Manuf. 2019;2:31–40. doi: 10.1007/s42242-018-0030-1. [DOI] [Google Scholar]
- 25.Mao Q, Jay M, Hoffman JL, et al. Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and ICU. BMJ Open. 2018;8:1–11. doi: 10.1136/bmjopen-2017-017833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Core Team R. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2019. [Google Scholar]
- 27.Barton C, Chettipally U, Zhou Y, et al. Evaluation of a machine learning algorithm for up to 48-hour advance prediction of sepsis using six vital signs. Comput Biol Med. 2019;109:79–84. doi: 10.1016/j.compbiomed.2019.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown SM, Jones J, Kuttler KG, et al. Prospective evaluation of an automated method to identify patients with severe sepsis or septic shock in the emergency department. BMC Emerg Med. 2016;16:1–7. doi: 10.1186/s12873-016-0095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thiel SW, Rosini JM, Shannon W, et al. Early prediction of septic shock in hospitalized patients. J Hosp Med. 2010;5:19–25. doi: 10.1002/jhm.530. [DOI] [PubMed] [Google Scholar]
- 30.Giannini HM, Ginestra JC, Chivers C, et al. A machine learning algorithm to predict severe sepsis and septic shock. Crit Care Med. 2019 doi: 10.1097/ccm.0000000000003891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy A, Das R. Reducing patient mortality, length of stay and readmissions through machine learning-based sepsis prediction in the emergency department, intensive care unit and hospital floor units. BMJ open Qual. 2017;6:e000158. doi: 10.1136/bmjoq-2017-000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimabukuro DW, Barton CW, Feldman MD, et al. Effect of a machine learning-based severe sepsis prediction algorithm on patient survival and hospital length of stay: a randomised clinical trial. BMJ Open Respir Res. 2017;4:e000234. doi: 10.1136/bmjresp-2017-000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calvert J, Desautels T, Chettipally U, et al. High-performance detection and early prediction of septic shock for alcohol-use disorder patients. Ann Med Surg. 2016;8:50–55. doi: 10.1016/j.amsu.2016.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seymour CW, Liu VX, Iwashyna TJ, et al. Assessment of clinical criteria for sepsis. JAMA. 2016;315:762. doi: 10.1001/jama.2016.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horng S, Sontag DA, Halpern Y, et al. Creating an automated trigger for sepsis clinical decision support at emergency department triage using machine learning. PLoS One. 2017;12:e0174708. doi: 10.1371/journal.pone.0174708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moss TJ, Lake DE, Calland JF, et al. Signatures of subacute potentially catastrophic illness in the ICU: model development and validation. Crit Care Med. 2016;44:1639–1648. doi: 10.1097/CCM.0000000000001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Møller MH, Alhazzani W, Shankar-Hari M. Focus on sepsis. Intensive Care Med. 2019;45:1459–1461. doi: 10.1007/s00134-019-05680-4. [DOI] [PubMed] [Google Scholar]
- 38.Makam AN, Nguyen OK, Auerbach AD. Diagnostic accuracy and effectiveness of automated electronic sepsis alert systems: a systematic review. J Hosp Med. 2015;10:396–402. doi: 10.1002/jhm.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alsolamy S, Al Salamah M, Al Thagafi M, et al. Diagnostic accuracy of a screening electronic alert tool for severe sepsis and septic shock in the emergency department. BMC Med Inform Decis Mak. 2014;14:105. doi: 10.1186/s12911-014-0105-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serafim R, Gomes JA, Salluh J, Póvoa P. A comparison of the quick-SOFA and systemic inflammatory response syndrome criteria for the diagnosis of sepsis and prediction of mortality: a systematic review and meta-analysis. Chest. 2018;153:646–655. doi: 10.1016/J.CHEST.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 41.Hiensch R, Poeran J, Saunders-Hao P, et al. Impact of an electronic sepsis initiative on antibiotic use and health care facility–onset clostridium difficile infection rates. Am J Infect Control. 2017;45:1091–1100. doi: 10.1016/j.ajic.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Parlato M, Philippart F, Rouquette A, et al. Circulating biomarkers may be unable to detect infection at the early phase of sepsis in ICU patients: the CAPTAIN prospective multicenter cohort study. Intensive Care Med. 2018;44:1061–1070. doi: 10.1007/s00134-018-5228-3. [DOI] [PubMed] [Google Scholar]
- 43.Shankar-Hari Manu, Datta Deepankar, Wilson Julie, Assi Valentina, Stephen Jacqueline, Weir Christopher J., Rennie Jillian, Antonelli Jean, Bateman Anthony, Felton Jennifer M., Warner Noel, Judge Kevin, Keenan Jim, Wang Alice, Burpee Tony, Brown Alun K., Lewis Sion M., Mare Tracey, Roy Alistair I., Wright John, Hulme Gillian, Dimmick Ian, Gray Alasdair, Rossi Adriano G., Simpson A. John, Conway Morris Andrew, Walsh Timothy S. Early PREdiction of sepsis using leukocyte surface biomarkers: the ExPRES-sepsis cohort study. Intensive Care Medicine. 2018;44(11):1836–1848. doi: 10.1007/s00134-018-5389-0. [DOI] [PubMed] [Google Scholar]
- 44.Fleischmann-Struzek C, Thomas-Rüddel DO, Schettler A, et al. Comparing the validity of different ICD coding abstraction strategies for sepsis case identification in German claims data. PLoS One. 2018;13:e0198847. doi: 10.1371/journal.pone.0198847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bouza C, Lopez-Cuadrado T, Amate-Blanco JM. Use of explicit ICD9-CM codes to identify adult severe sepsis: impacts on epidemiological estimates. Crit Care. 2016;20:313. doi: 10.1186/s13054-016-1497-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones M. NEWSDIG: the national early warning score development and implementation group. Clin Med. 2012;12:501–503. doi: 10.7861/clinmedicine.12-6-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown SM, Jones J, Kuttler KG, et al. Prospective evaluation of an automated method to identify patients with severe sepsis or septic shock in the emergency department. BMC Emerg Med. 2016;16:31. doi: 10.1186/s12873-016-0095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen SQ, Mwakalindile E, Booth JS, et al. Automated electronic medical record sepsis detection in the emergency department. PeerJ. 2014;2:e343. doi: 10.7717/peerj.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson JL, Smith BL, Jared JD, Younger JG. Prospective trial of real-time electronic surveillance to expedite early care of severe sepsis. Ann Emerg Med. 2011;57:500–504. doi: 10.1016/j.annemergmed.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 50.Hooper MH, Weavind L, Wheeler AP, et al. Randomized trial of automated, electronic monitoring to facilitate early detection of sepsis in the intensive care unit. Crit Care Med. 2012;40:2096–2101. doi: 10.1097/CCM.0b013e318250a887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giannini HM, Ginestra JC, Chivers C, et al. A machine learning algorithm to predict severe sepsis and septic shock: development, implementation, and impact on clinical practice. Crit Care Med. 2019 doi: 10.1097/CCM.0000000000003891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calvert JS, Price DA, Chettipally UK, et al. A computational approach to early sepsis detection. Comput Biol Med. 2016;74:69–73. doi: 10.1016/j.compbiomed.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 53.Kam HJ, Kim HY. Learning representations for the early detection of sepsis with deep neural networks. Comput Biol Med. 2017;89:248–255. doi: 10.1016/j.compbiomed.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 54.Desautels T, Calvert J, Hoffman J, et al. Prediction of sepsis in the intensive care unit with minimal electronic health record data: a machine learning approach. JMIR Med Inf. 2016;4:e28. doi: 10.2196/medinform.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nemati S, Holder A, Razmi F, et al. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit Care Med. 2018;46:547–553. doi: 10.1097/CCM.0000000000002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henry Katharine E., Hager David N., Pronovost Peter J., Saria Suchi. A targeted real-time early warning score (TREWScore) for septic shock. Science Translational Medicine. 2015;7(299):299ra122–299ra122. doi: 10.1126/scitranslmed.aab3719. [DOI] [PubMed] [Google Scholar]
- 57.Wang RZ, Sun CH, Schroeder PH et al (2018) Predictive models of sepsis in adult ICU patients. In: 2018 IEEE international conference on healthcare informatics (ICHI), IEEE, pp 390–391. 10.1109/ICHI.2018.00068
- 58.Guillén J, Liu J, Furr M et al (2015) Predictive models for severe sepsis in adult ICU patients. In: 2015 systems and information engineering design symposium, IEEE, pp 182–187. 10.1109/SIEDS.2015.7116970
- 59.Scherpf M, Gräßer F, Malberg H, Zaunseder S. Predicting sepsis with a recurrent neural network using the MIMIC III database. Comput Biol Med. 2019;113:103395. doi: 10.1016/j.compbiomed.2019.103395. [DOI] [PubMed] [Google Scholar]
- 60.He Haibo, Garcia EA. Learning from imbalanced data. IEEE Trans Knowl Data Eng. 2009;21:1263–1284. doi: 10.1109/TKDE.2008.239. [DOI] [Google Scholar]
- 61.Liu X, Faes L, Kale AU, et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit Heal. 2019;1:e271–e297. doi: 10.1016/s2589-7500(19)30123-2. [DOI] [PubMed] [Google Scholar]
- 62.Haug P, Ferraro J, et al. Using a semi-automated modeling environment to construct a Bayesian, sepsis diagnostic system. BCB '16. 2016 doi: 10.1145/2975167.2985841. [DOI] [Google Scholar]
- 63.Delahanty RJ, Alvarez J, Flynn LM, et al. Development and evaluation of a machine learning model for the early identification of patients at risk for sepsis. Ann Emerg Med. 2018 doi: 10.1016/j.annemergmed.2018.11.036. [DOI] [PubMed] [Google Scholar]
- 64.Khojandi A, Tansakul V, Li X, et al. Prediction of sepsis and in-hospital mortality using electronic health records. Methods Inf Med. 2018;57:185–193. doi: 10.3414/ME18-01-0014. [DOI] [PubMed] [Google Scholar]
- 65.Futoma J, Hariharan S, Heller K (2017) Learning to detect sepsis with a multitask Gaussian process RNN classifier. In: Proceedings of the 34th international conference on machine learning
- 66.Lin C, Zhang Y, Ivy J et al (2018) Early diagnosis and prediction of sepsis shock by combining static and dynamic information using convolutional-LSTM. In: 2018 IEEE international conference on healthcare informatics (ICHI), IEEE, pp 219–228. 10.1109/ICHI.2018.00032
- 67.Shashikumar SP, Stanley MD, Sadiq I, et al. Early sepsis detection in critical care patients using multiscale blood pressure and heart rate dynamics. J Electrocardiol. 2017;50:739–743. doi: 10.1016/j.jelectrocard.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shashikumar SP, Li Q, Clifford GD, Nemati S. Multiscale network representation of physiological time series for early prediction of sepsis. Physiol Meas. 2017;38:2235–2248. doi: 10.1088/1361-6579/aa9772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Wyk F, Khojandi A, Mohammed A, et al. A minimal set of physiomarkers in continuous high frequency data streams predict adult sepsis onset earlier. Int J Med Inform. 2019;122:55–62. doi: 10.1016/j.ijmedinf.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 70.Van Wyk F, Khojandi A, Kamaleswaran R. Improving prediction performance using hierarchical analysis of real-time data : a sepsis case study. IEEE J Biomed Health Inf. 2018;2018:1–9. doi: 10.1109/JBHI.2019.2894570. [DOI] [PubMed] [Google Scholar]
- 71.Mao Q, Jay M, Hoffman JL, et al. Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and ICU. BMJ Open. 2018;8:e017833. doi: 10.1136/bmjopen-2017-017833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.