Abstract
In addition to their role in regulating transport across the nuclear envelope, increasing evidence suggests nuclear pore complexes (NPCs) function in regulating gene expression. For example, the induction of certain genes (e.g., yeast INO1) is accompanied by their movement from the nuclear interior to NPCs. As sumoylation has been linked to the regulation of chromatin spatial organization and transcriptional activity, we investigated the role of sumoylation in the expression and NPC recruitment of the INO1 gene. We observed that induction of INO1 is accompanied by both increased and decreased sumoylation of proteins associated with specific regions along the INO1 locus. Furthermore, we show that the E3 ligase Siz2/Nfi1 is required for targeting the INO1 locus to the NPC where it interacts with the SUMO isopeptidase Ulp1. Our data suggest that this interaction is required for both the association of INO1 with the NPC and for its normal expression. These results imply that sumoylation is a key regulator of INO1 targeting to the NPC, and a cycle of sumoylation and NPC-associated desumoylation events contribute to the regulation of INO1 expression.
Keywords: nuclear pore complex, gene positioning, gene expression, sumoylation, INO1, Ulp1, Siz2
Introduction
Small ubiquitin-related modifier (SUMO) is an ubiquitin-like peptide that is covalently attached to certain lysines in proteins. Sumoylation of proteins can result in changes to protein stability, subcellular localization, or interactions with other proteins. The vast majority of proteins modified by sumoylation are nuclear proteins and sumoylation and desumoylation events have been linked to the regulation of diverse group of nuclear processes including DNA replication, transcriptional control, and the spatial organization of the genome (Jentsch and Psakhye, 2013; Texari and Stutz, 2015; Zhao, 2018; Rosonina, 2019).
In Saccharomyces cerevisiae, the SUMO polypeptide is encoded by a single gene, SMT3. SUMO is translated with a C-terminal extension and later cleaved at a di-glycine motif by a SUMO peptidase to yield mature SUMO (Li and Hochstrasser, 1999). Like ubiquitination, SUMO conjugation is accomplished by a series of enzymes that first activate the mature peptide via ATP-dependent formation of a thioester bond (E1 enzyme, Aos1/Uba2 heterodimer in S. cerevisiae). Activated SUMO is subsequently handed-off to an active site cysteine in the E2 conjugating enzyme Ubc9. Ubc9 functions to directly transfer SUMO to a lysine in the target protein. In most cases, an E3 ligase aids SUMO-substrate specificity by mediating or stabilizing target interactions with the E2 (Geiss-Friedlander and Melchior, 2007). There are four known SUMO ligases in yeast, Zip3, Mms21, Siz1, and Siz2, the former functioning during meiosis while the latter three function in actively growing cells (Johnson, 2004; Jentsch and Psakhye, 2013). There is a significant overlap and redundancy in E3 ligase targets; however, E3 ligases have also been shown to have specific and independent functions (Makhnevych et al., 2007; Ferreira et al., 2011; Hannan et al., 2015). The SUMO conjugating system components are primarily present in the nucleoplasm (Srikumar et al., 2013) as are the vast majority of proteins modified by sumoylation (Panse et al., 2004; Wohlschlegel et al., 2004; Zhao et al., 2004; Zhou et al., 2004; Hannich et al., 2005; Wykoff and O’Shea, 2005).
Sumoylation is a reversible process. In budding yeast, there are two functionally distinct isopeptidases, Ulp1 and Ulp2. Both Ulp1 and Ulp2 deconjugate SUMO from target proteins; however, Ulp2 also suppresses poly-SUMO chain accumulation (Bylebyl et al., 2003), while Ulp1 carries out the essential role of processing pre-SUMO to mature SUMO (Li and Hochstrasser, 1999; Mossessova and Lima, 2000). ulp1-ts and ulp2Δ mutants exhibit very different overall sumoylation patterns (Li and Hochstrasser, 2000), and the desumoylation of specific sumoylated targets have been shown to be dependent on specific isopeptidases (Makhnevych et al., 2007; Felberbaum et al., 2012), indicating that each enzyme has distinct substrates. Ulp1 and Ulp2 also have distinct locations in the cell; Ulp2 is distributed throughout the nucleoplasm, while Ulp1 is associated with the nucleoplasmic face of nuclear pore complexes (NPCs) (Li and Hochstrasser, 2000; Panse et al., 2003).
The Ulp1 catalytic domain resides in its C-terminus, whereas several N-terminal domains of Ulp1 contribute to its association with NPCs (Li and Hochstrasser, 2003; Panse et al., 2003; Makhnevych et al., 2007). The N-terminal regions also bind to nuclear transport factors; residues 1–150 bind the import karyopherin Kap121, residues 150–340 bind the import karyopherin heterodimer Kap95/Kap60, and residues 340–403 contain a nuclear export signal that appears to interact with the export factor Xpo1 (Panse et al., 2003). Binding of Ulp1 to the NPC appears dependent on structures positioned on the nucleoplasmic face of the NPC, including proteins such as Mlp1, Mlp2, Nup60, and Nup2 (Zhao et al., 2004; Palancade et al., 2007; Srikumar et al., 2013). However, the molecular basis for these interactions has not been established.
The association of Ulp1 with NPCs is particularly intriguing and has led to the analysis of its role in various processes performed by NPCs, including the regulation of nuclear transport (Stade et al., 2002; Panse et al., 2006; Lewis et al., 2007), certain DNA repair pathways (Zhao et al., 2004; Palancade et al., 2007; Freudenreich and Su, 2016), and the regulation of gene expression (Texari et al., 2013; Bonnet et al., 2015; Abraham and Mishra, 2018). Among the various mechanisms by which NPCs can influence gene expression is their direct interactions with chromatin. NPCs interact with both transcriptionally repressed and active genes through interactions that appear to be mediated by transcription factors (TFs) of various types that bind specific chromatin sites and interact with different sets of Nups (Brickner et al., 2007; Ahmed et al., 2010; Light et al., 2010; Ruben et al., 2011; Van de Vosse et al., 2013; Brickner et al., 2019). These interactions influence both the spatial organization of associated genes and contribute to transcriptional state.
In yeast, numerous studies have examined the relocalization to NPCs of inducible genes following activation. Several genes have been shown to reside in the nuclear interior when repressed, but move to NPCs when induced. Well studied among these is the INO1 locus (Brickner and Walter, 2004; Cabal et al., 2006; Brickner et al., 2007; Ahmed et al., 2010; Light et al., 2010). In the presence of inositol (repressive conditions), INO1 is bound by the repressors Opi1 and Ume6, and the Rpd3(L) histone deacetylase complex, which repress INO1 expression and association with the NPC. Following induction (inositol starvation), Opi1, presumably with Ume6 and Rpd3(L), dissociates from INO1. This is thought to be followed by the binding of TFs (Put3 and Cbf1) to cis-acting DNA elements (termed gene GRS1 and GRS2), which exhibit redundant functions in targeting INO1 to an NPC (Loewen et al., 2003; Brickner and Walter, 2004; Shetty and Lopes, 2010; Brickner and Brickner, 2012; Randise-Hinchliff et al., 2016; Brickner et al., 2019). The SAGA complex (involved in transcription initiation) also appears to contribute to the association of active INO1 with the NPC (Lo et al., 2001, 2005). Here, specific NPC components contribute to the efficient binding to INO1 (Light et al., 2010). These interactions have been proposed to promote optimal transcription and mRNA export. However, our current lack of knowledge on the molecular basis for the interactions of INO1, and other active genes, with the NPC has limited our understanding of its significance.
Several Nups that play a role in the NPC-association of INO1 also functionally interact with Ulp1 suggesting it is positioned at or near the site of gene association. Furthermore, alterations in sumoylation events have been shown to impact INO1 transcription (Felberbaum et al., 2012). Consistent with this idea, Ulp1 has been previously shown to contribute to the activation and NPC-binding of the GAL1 gene (Cabal et al., 2006; Texari et al., 2013). These observations and others implicating sumoylation in chromatin association with the NE and the regulation of gene expression has led us to investigate the role of sumoylation and desumoylation in the localization and expression of INO1. Our analysis of the roles of the SUMO ligase Siz2 and Ulp1 have revealed functions for sumoylation and desumoylation events in the NPC targeting and expression of activated INO1. We show that induction of INO1 is accompanied by Siz2-dependent sumoylation of proteins associated with the INO1 locus and propose that these modifications are required for targeting the gene to the NPC. Once at an NPC, Ulp1 interacts with sumoylated proteins associated with the induced INO1 gene, primarily within its ORF. We propose that subsequent Ulp1-mediated desumoylation promotes expression and NPC association of activated INO1.
Materials and Methods
Media, Yeast Strains, and Plasmids
Saccharomyces cerevisiae strains used in this study were derived from YEF473A (Bi and Pringle, 1996) and are listed in Supplementary Table S1. Strains were grown in either fully supplemented synthetic media (SC media) (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% glucose) or in synthetic media lacking inositol (INO– media) or were grown in YPD (1% yeast extract, 2% bactopeptone, and 2% glucose). Plasmid bearing strains were grown in the appropriate synthetic dropout media (0.8% dropout powder, 0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% glucose) that either contained or lacked inositol.
Strain construction employed genome modifications performed using a one-step genomic integration method (Longtine et al., 1998), in which a DNA cassette was transformed into an appropriate strain using the lithium acetate/polyethylene glycol method (Gietz and Woods, 2002). DNA cassettes used to produce C-terminal Protein A, GFP, RFP, and mCherry gene fusions were made using a plasmid/PCR-based method (Longtine et al., 1998). DNA cassettes encoding the various ulp1Δ mutants were made by ligating together PCR generated DNA segments modified with specific restriction enzyme sites. For ulp1Δ1–150 and ulp1Δ1–340, these DNA cassettes (bracketed) and restriction sites include: (ULP1 5′UTR)-EcoRI-(ULP1 nucleotides from 451 within the ORF to 26 after the stop codon for ulp1Δ1–150 and nucleotides from 1021 within the ORF to 26 after the stop codon for ulp1Δ1–340)-BamHI-(marker gene: NATMX for ulp1Δ1–150 and KANMX for ulp1Δ1–340)-SpeI-(ULP1 3′UTR). The EcoRI site introduces Glu-Phe codons after the Met start codon. The ulp1Δ150–340 cassette consisted of (ULP1 5′UTR to ULP1 nucleotide 450 within the ORF)-BssHII-(ULP1 nucleotides 1021–1861 within the ORF)-SpeI-(ULP1 nucleotides 2–26 after the stop codon)-BamHI-(marker gene NATMX)-SpeI-(ULP1 3′UTR). The BssHII restriction site introduces Ala-Arg codons, while the SpeI restriction site at the 3′ end of ulp1Δ150–340 overlaps with the stop codon and introduces a Lys to Asn codon substitution at the 3′ end of the ORF. PCR-based generation of DNA cassettes also introduced a point mutation in, ulp1Δ1–340 resulting in an F610S amino acid residue substitution, and in ulp1Δ150–340 resulting in an E409G residue substitution, that have no apparent effect on the relative function of these deletion derivatives. The NUP53-ulp340–621 DNA cassette consisted of (ULP1 5′UTR)-NdeI-(NUP53)-SalI-(ULP1 nucleotides from 1018 within the ORF to 26 after the stop codon)-SpeI-(ULP1 nucleotides 2–26 after the stop codon)-BamHI-(marker gene NATMX)-SpeI-(ULP1 3′UTR). The SalI restriction site adds Val-Asp codons at the fusion point between NUP53 and ulp1340–621. The SpeI restriction site at the 3′ end of ulp340–621 overlaps with the stop codon and introduces a Lys to Asn codon substitution at the 3′ end of the ORF. PCR-based generation of NUP53-ulp340–621 introduced point mutations resulting in an E409G, and V584A amino acid substitutions in ulp340–621 that have no apparent effect on the relative function of this fusion.
To visualize INO1 gene localization, a previously described genomic tagging system was employed (Straight et al., 1996). GFP-lacI-HIS was integrated at the his-Δ200 locus using the pAFS78 plasmid (Robinett et al., 1996). To tag the INO1 locus, the plasmid pAFS52.INO1 was made by cloning PCR amplified INO1, containing XhoI sites at its 5′ and 3′ ends, into pAFS52 (Straight et al., 1996). BglII digested pAFS52.INO1 was then transformed into yeast to integrate the lacO256-TRP array. This resulted in a duplication of the INO1 locus with lacO256-TRP found between the duplicates.
Plasmids used here are derivatives of pRS315 (pEMPTY) (Sikorski and Hieter, 1989) and include pRS315.Ulp1-GFP and pRS315.ulp1CSDN-GFP (Elmore et al., 2011), as well as pRS315.Ulp1 and pRS315.ulp1CSDN (this work).
INO1 Gene Induction
To induce INO1 gene expression, cell cultures were grown overnight at room temperature in SC media, diluted into fresh SC media to an OD600 = 0.2, and then grown at 30°C until the cultures reached mid-log phase (OD600 = ∼0.8). A sample of these cultures was then taken as the uninduced control, and processed as required. A second sample of cells from these cultures were collected by centrifugation, washed once with water, and then resuspended in INO– media to an OD600 = 0.5 to induce INO1 expression. These cultures were then grown at 30°C. Cells were collected at the stated time points and processed as indicated.
qRT-PCR for INO1 Gene Expression
INO1 induction was carried out as described above and, at each time point, an OD600 = 10 equivalent of cells was pelleted and processed. RNA preparation from these cells and subsequent real-time qRT-PCRs were performed as previously described (Wan et al., 2009). cDNA was amplified using 2 μg of DNAse-treated RNA that was reverse transcribed using 200 units of Superscript II reverse transcriptase (Invitrogen) at 42°C for 50 min and the resulting cDNAs were diluted 100-fold. Reactions were assembled using SYBR green super mix (Quanta), as per the manufacturer’s protocol, and included sense (S) and antisense (AS) primers against ACT1 (S-GGATTCCGGTGATG GTGTTA, AS-TCAAATCTCTACCGGCCAAA) and INO1 (S-CACCAT GGAAAACCTCTTGC, AS-GGGGACACCTTCCAAGATAGA) as previously described (Brickner et al., 2007). Reactions were carried out on an Mx3000P QPCR System (Agilent Technologies). INO1 mRNA levels were normalized relative to ACT1 mRNA levels from three independent qRT-PCR analyses.
ChIP for Protein Localization at INO1
INO1 induction was carried out as described above and, at each time point, an OD600 = 50 equivalent of cells was pelleted and processed. Chromatin immunoprecipitation experiments were performed as previously described (Wan et al., 2009). For immunoprecipitation, 4 μl of rabbit polyclonal anti-PrA (Sigma) antibody or 4 μl of rabbit polyclonal anti-Smt3 (SUMO) antibody (Wozniak lab) was prebound to 100 μl of Protein G Dynabeads (Invitrogen). Immunoprecipitated DNA was recovered and analyzed by qRT-PCR as described above. Sense (S) and antisense (AS) primers used for qRT-PCR included: Chromosome V intergenic region (S-ACATTCTTGGAAA CCCATCG, AS-TCGTATCATGATTTAGCGTCGT); INO1 regions: GRS1 (S-TC GTTCCTTTTGTTC TTCACG, AS-GCCTCCGCATATTTCA CATT), A (S-AAATGCGGCATGTGAAAAGT, AS-AGAG GTG CGCTTTCTCTGC), B (S-AGAGAAAGCGCACCTCTGC, AS- (AGGAACCCGACAACAGAACA), C (S-CGACAAGTGCACG TACAAGG, AS-CAGTGGGCGTTACATCGAA), D (S-CTTC GGCTCC ATGACTCAAT, AS-GCTAACCATGGGCAACAG AG), E (S-GGACTCAAAAGTGGCAATGG, AS-TCAAGGGC GTAGCCAGTAAA), F (S-CGTCTTAAAAGGGGCGTTTT, AS-TTTACTGAGG TGGCCCTTGA). To quantify the ChIP experiments, we first expressed the amount of INO1 or Chromosome V intergenic region sequence immunoprecipitated as a percentage of total input (% of input). Using these values, we calculated ratios comparing the% of input from each region of the INO1 gene to the% of input for the Chromosome V intergenic region for both uninduced (cells grown in the presence of inositol) and induced (cells grown in the absence of inositol for 1 and 3 h) samples. Relative fold change for the induced samples was then calculated by dividing the induced ratio determined for a given region of the INO1 gene by the uninduced ratio for that same region.
Fluorescence Microscopy
To image the GFP-lacI/lacO256 tagged INO1 locus, cell cultures were treated as described above for INO1 gene induction. Cells from 1 ml of culture were pelleted by centrifugation, washed once with the appropriate synthetic media, and then resuspended in the same media; 1.5 μl of cells was then spotted onto a microscope slide for live-cell image acquisition. Epifluorescence images were acquired on a DeltaVision Elite imaging system (GE Healthcare Life Sciences) at 60x magnification using a 1.42 NA oil, Plan Apo N objective (Olympus). Images were collected and saved as 15 × 0.2 μm z-stacks using SoftWoRx software (version 6.5.2, GE Healthcare Life Sciences), then rendered and analyzed using Image J (NIH). INO1 localization (GFP-lacI signal) was assessed relative the nuclear periphery (Nup49-mRFP signal) and was considered to colocalize with NPCs when the GFP-lacI focus fully or partially overlapped with Nup49-mRFP, similar to the previously described method (Brickner and Walter, 2004; Brickner and Brickner, 2010).
To assess the localization of the various Ulp1-GFP and Ulp1-mCherrry derivatives, strains producing these derivatives were grown in YPD media at 30oC to mid-log phase; 1 ml of cells from each culture was then pelleted by centrifugation, washed once with 1 ml of SC media, and resuspended in 20 μl of SC media; 1.5 μl was then spotted onto a microscope slide for epifluorescence imaging. Images were acquired using an Axio Observer.Z1 microscope (Carl Zeiss, Inc.), equipped with an UPlanS-Apochromat 100x/1.40 NA oil objective lens (Carl Zeiss, Inc.) and an AxioCam MRm digital camera with a charge-coupled device (Carl Zeiss, Inc.). Images were acquired in a single focal plane through the center of nuclei. Images were saved using AxioVision (Carl Zeiss, Inc.) software and rendered for display using Image J (NIH) software.
Western Blot
Ulp1-GFP and various ulp1Δ-GFP derivatives were detected using western blot. Proteins from cells lysates were separated by SDS-PAGE and then transferred to nitrocellulose membranes. Membranes were incubated in blocking buffer (TBS containing 0.1% Tween-20 and 5% milk powder) for at least 1 h at room temperature. Blocking buffer was then removed and replaced with fresh blocking buffer supplemented with rabbit polyclonal antibodies directed against GFP, GSP1, or SUMO (Makhnevych et al., 2007) then incubated overnight at 4°C. Membranes were then washed three times using 0.1% Tween-20 in TBS, followed by incubation in blocking buffer supplemented with goat anti-rabbit HRP conjugated antibody (BioRad) at a 1:10,000 dilution for at least 1 h at room temperature. Membranes were then washed three times using 0.1% Tween-20 in TBS. Bound anti-rabbit HRP conjugated antibody was detected by chemiluminescence (Amersham) using an ImageQuant LAS 4000 (GE) imaging system.
Results
Activation of the INO1 Gene Is Accompanied by Sumoylation of Associated Proteins
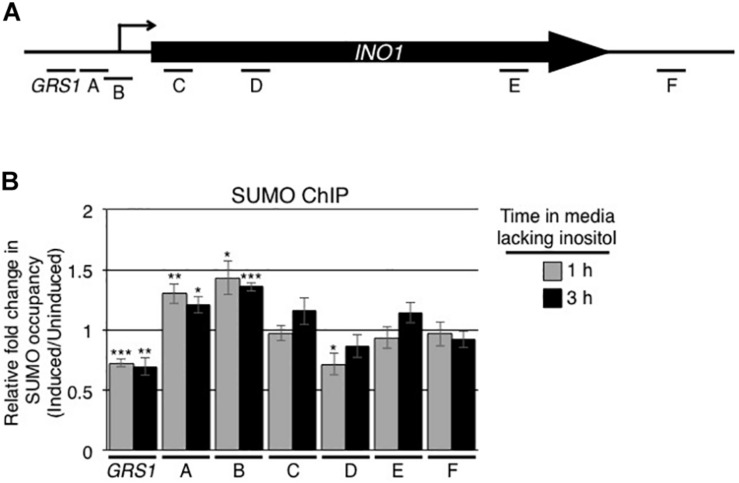
Following their activation, numerous yeast genes are repositioned from the nuclear interior to the nuclear envelope (Brickner and Walter, 2004; Texari et al., 2013; Brickner et al., 2019). A well-studied example is the INO1 gene. When cells are switched from medium containing inositol to medium lacking this carbon source, the INO1 gene is induced and the gene locus is targeted to NPCs (Brickner and Walter, 2004; Cabal et al., 2006; Brickner et al., 2007; Ahmed et al., 2010; Light et al., 2010). Since changes in the expression of genes are often accompanied by changes in the levels of sumoylation of associated TFs and other chromatin-associated proteins, we examined whether the induction of INO1 alters the sumoylation state of proteins associated with the INO1 locus. To test this, antibodies directed against SUMO were used in ChIP analysis targeting the INO1 gene prior to and following induction. For these experiments, various sets of oligonucleotides were used to detect interacting regions along the INO1 gene (Figure 1A). Prior to induction, the sumoylation state of chromatin associated proteins within the GRS1 and ORF regions of the INO1 gene were higher than that detected in a control intergenic region (Supplementary Figure S1B). Following induction, we observed significant changes in the levels of sumoylated proteins associated with specific regions of the gene (Figure 1B). In a 5′ region containing the previously identified INO1 gene recruitment sequence 1 (GRS I), we observed a decrease in sumoylation of associated proteins while adjacent regions containing the transcriptional start site showed increases. Downstream regions within the ORF and the 3′ regions showed little or no change in the levels of associated sumoylated proteins.
FIGURE 1.
Induction of INO1 leads to changes in the sumoylation of INO1-associated proteins. (A) Diagram of the INO1 locus showing the relative positions of the transcriptional start site (arrow), the GRS1 sequence, the open reading frame, and regions targeted for the ChIP analysis shown in B. (B) WT cells were grown in medium containing inositol (repressing/uninduced conditions) to an OD600 of ∼0.8, washed, and transferred to medium lacking inositol (inducing conditions). Cells were then subjected to ChIP analysis using antibodies directed against the SUMO polypeptide (anti-SUMO antibody) prior to and at 1 and 3 h post induction of INO1. qRT-PCR was used to quantify levels of DNA corresponding to the various regions of INO1 (see A) bound to sumoylated proteins. Shown is the relative fold change in the occupancy of sumoylated proteins associated with the various regions of INO1 at the indicated times after induction relative to the uninduced samples (see section “Materials and Methods”). Results are the means ± SEM of five biological replicates. Asterisks indicate a significant difference relative to uninduced as determined by a Student’s paired t-test. *p < 0.05, **p < 0.01, ***p < 0.001.
The SUMO Ligase Siz2 Is Required for Recruitment of the INO1 Locus to the NE
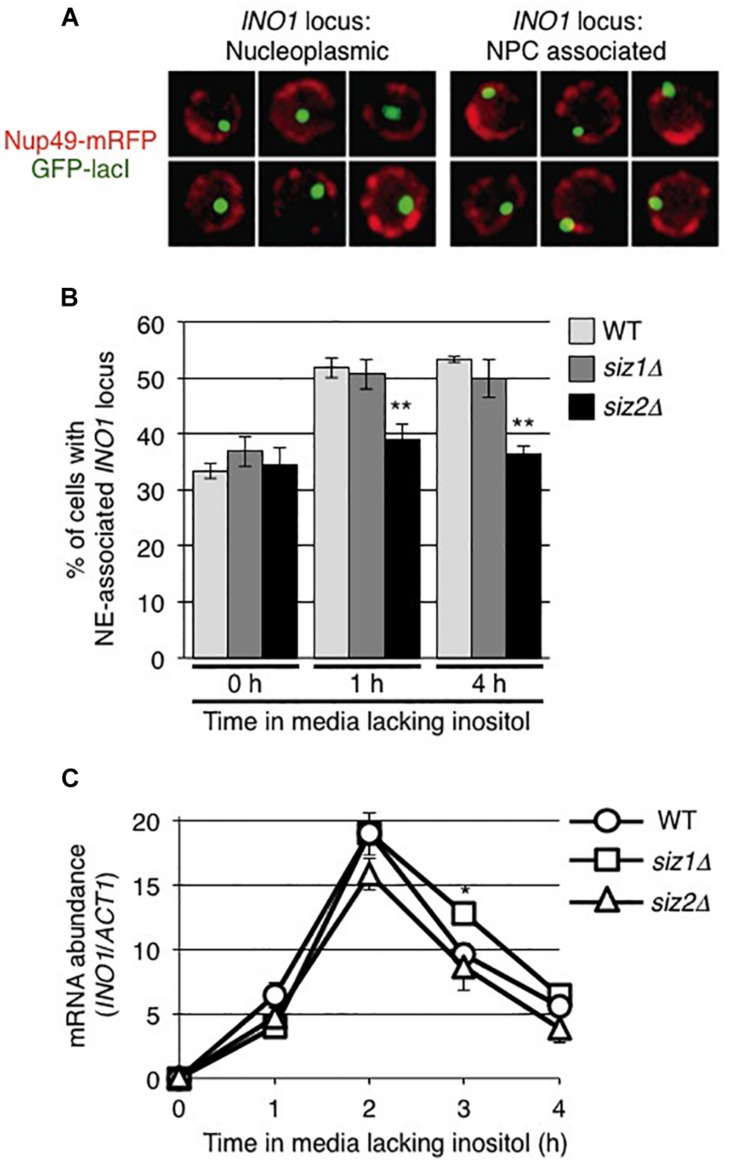
Our observations that the levels of sumoylated proteins bound to the INO1 locus, in particular those associated with 5’ regions containing the GRS1 sequence, changed upon induction led us to investigate the role of sumoylation in the NPC association of INO1. As the SUMO ligase Siz2 had been previously shown to play a role in the nuclear envelope association of telomeres (Ferreira et al., 2011; Churikov et al., 2016; Lapetina et al., 2017), we examined the role of Siz2, and the related SUMO ligase Siz1, in INO1 localization following its induction. The position of INO1 was monitored by tagging with an adjacent lacO256 cassette in cells producing the GFP-lacI protein (Brickner and Walter, 2004; Ahmed et al., 2010; see Figure 2A). Induction of INO1 led to a rapid (within 1 h) accumulation of INO1:lacO256/GFP-lacI foci at the nuclear periphery in WT cells and those lacking Siz1 (siz1Δ) (Figure 2B). By contrast, we observed that in cells lacking Siz2 (siz2Δ), INO1 recruitment to the NE was not observed following induction. We also measured INO1 mRNA levels following induction in these various strains, and observed no differences in the induction profiles (Figure 2C). These results suggest that Siz2 is required for INO1 binding to the nuclear periphery upon induction, but its loss has no significant effect on INO1 expression.
FIGURE 2.
The SUMO ligase Siz2 is required for NE-association, but not transcription, of induced INO1. Localization of the INO1 locus prior to and following INO1 induction was examined in cells containing the INO1-lacO256 construct and producing GFP-lacI (green). The position of the nuclear envelope (NE) in these cells relative to the INO1-lacO locus was determined using Nup49-RFP (red). (A) Examples of cell nuclei in which the INO1 locus (green) was (left panels) or was not (right panels) associated with the nuclear periphery (red) are shown. INO1 was scored to be NE-associated if the GFP-lacI signal fully or partially overlapped with the Nup49-mRFP signal. (B) The percentage of cells showing NE-association of INO1-lacO prior to (0 h) or at 1 and 4 h after induction was determined in the indicated strains. Results are the means ± SD of three biological replicates. At least 100 cells were counted for each sample. (C) Levels of INO1 mRNA were examined in the indicated strains. Cell cultures were grown in medium containing inositol (repressing conditions) to an OD600 of ∼0.8 and then washed and placed in medium lacking inositol (inducing conditions) for the indicated times. Levels of mRNA encoded by the INO1 gene were evaluated by qRT-PCR and their abundance relative to ACT1 mRNA was determined. Results are the means ± SEM of three biological replicates. Asterisks indicate a significant difference from WT as indicated by Student’s unpaired t-test, *p < 0.05, **p < 0.01.
The requirement of Siz2 for the binding of induced INO1 to the NE led us to examine whether Siz2 contributed to sumoylation at the INO1 locus. To test this idea, we examined levels of sumoylated proteins at the INO1 locus in the siz2Δ mutant. In contrast to WT cells, the siz2Δ mutant cells showed reduced levels of sumoylation within the ORF of the INO1 locus in uninduced cells. Moreover, induction of INO1 did not significantly alter sumoylated proteins levels along the INO1 locus (Supplementary Figure S1C). These results are consistent with Siz2 functioning in the sumoylation of proteins associated with regions of the INO1 locus prior to and following INO1 activation.
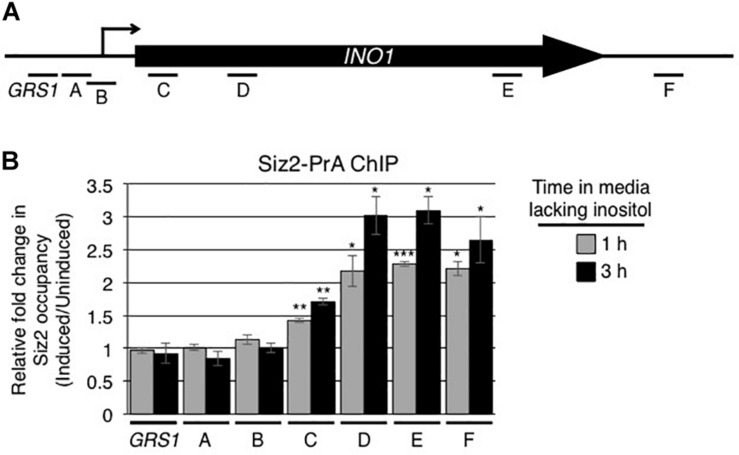
We also examined whether Siz2 physically interacted with the INO1 locus upon activation of the INO1 gene. Using protein A tagged Siz2 (Siz2-PrA) and ChIP analysis, we examined the binding of Siz2-PrA along the INO1 locus prior to and following 1 and 3 h induction (Supplementary Figure S2 and Figure 3B). In uninduced cells, we detected significantly higher levels of Siz2-PrA bound to the GRS1-containing region relative to the intergenic control, while other regions show no enrichment (Supplementary Figure S2). Upon induction of INO1, we observed no significant change in Siz2-PrA binding within 5′ regions of INO1 locus (from site GRS I to site B, see Figure 3A for map). However, Siz2-PrA occupancy within the INO1 ORF (region C to F) increased markedly upon induction (Figure 3B). These results are consistent with Siz2 functioning in the sumoylation of proteins associated with various regions of the INO1 locus both prior to (upstream of the ORF) or in response to INO1 activation (within the ORF).
FIGURE 3.
Induction of INO1 is accompanied by recruitment of Siz2 to the INO1 locus. (A) Diagram of the INO1 locus and regions detected by ChIP analysis (see Figure 1A) in B. (B) Cells expressing protein A tagged Siz2 (Siz2-PrA) were grown as described in the Figure 1 legend, and harvested at 0, 1, and 3 h after INO1 induction. Changes in Siz2-PrA occupancy along the INO1 locus following induction were examined by ChIP and qRT-PCR using primer pairs that amplify regions of INO1 indicated in A. Relative fold change in Siz2-PrA binding was determined as described for SUMO binding in Figure 1. Results are the means ± SEM of at least three biological replicates. Asterisks indicate a significant difference relative to uninduced as determined by a Student’s paired t-test. *p < 0.05, **p < 0.01, ***p < 0.001.
INO1 Interacts With Ulp1 Following Induction
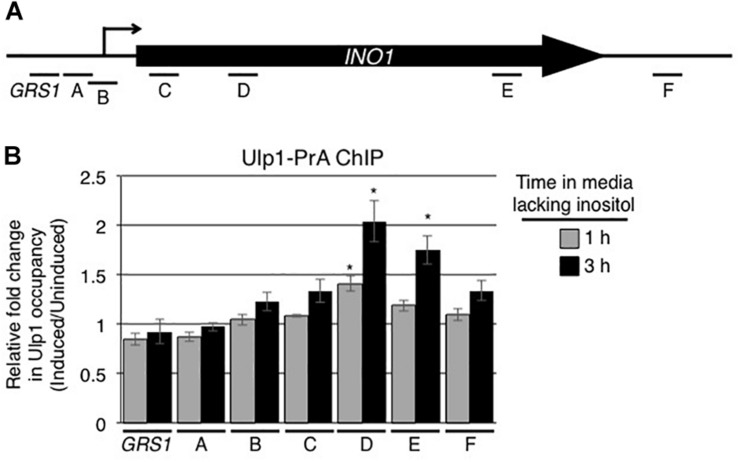
The association of activated genes with NPCs is thought to occur through interactions between the transcriptional machinery and proteins located on the nuclear face of the NPC. Among these, Nup60 and the related proteins Mlp1/Mlp2 are required for INO1 association with NPCs (Brickner et al., 2007; Ahmed et al., 2010; Light et al., 2010; Brickner et al., 2019). These NPC proteins are also required for the association of the desumoylase Ulp1 with NPCs (Zhao et al., 2004; Palancade et al., 2007; Srikumar et al., 2013). As Ulp1 is an important regulator of protein sumoylation, we investigated its role in the expression and NPC targeting of the induced INO1 gene. We tested whether the INO1 gene physically interacts with Ulp1. Using ChIP, no significant enrichment of Ulp1-pA was detected along the INO1 locus prior to induction (Supplementary Figure S2C). However, following activation, we observed that Ulp1 occupancy significantly increased specifically within the INO1 ORF (Figure 4). By contrast, no detectable change was observed in regions upstream of the ORF. These results suggest that induction of INO1 is followed by the association of Ulp1 with specific regions of the INO1 gene.
FIGURE 4.
Ulp1 interacts with the induced INO1 gene. (A) Diagram of the INO1 locus and regions detected by ChIP analysis (see Figure 1A). (B) Cells producing protein A tagged Ulp1 (Ulp1-PrA) were grown as described in the Figure 1 legend, and harvested at 0, 1, and 3 h after INO1 induction. Changes in Ulp1-PrA occupancy throughout the INO1 locus upon induction were examined by ChIP and qRT-PCR using primer pairs that amplify regions of INO1 indicated in A. Relative fold change in Ulp1-PrA binding was determined as described for SUMO binding in Figure 1. Results are the means ± SEM of three biological replicates. Asterisks indicate a significant difference relative to uninduced as determined by a Student’s paired t-test. *p < 0.05.
NPC Recruitment and Expression of INO1 Require NPC-Associated Ulp1
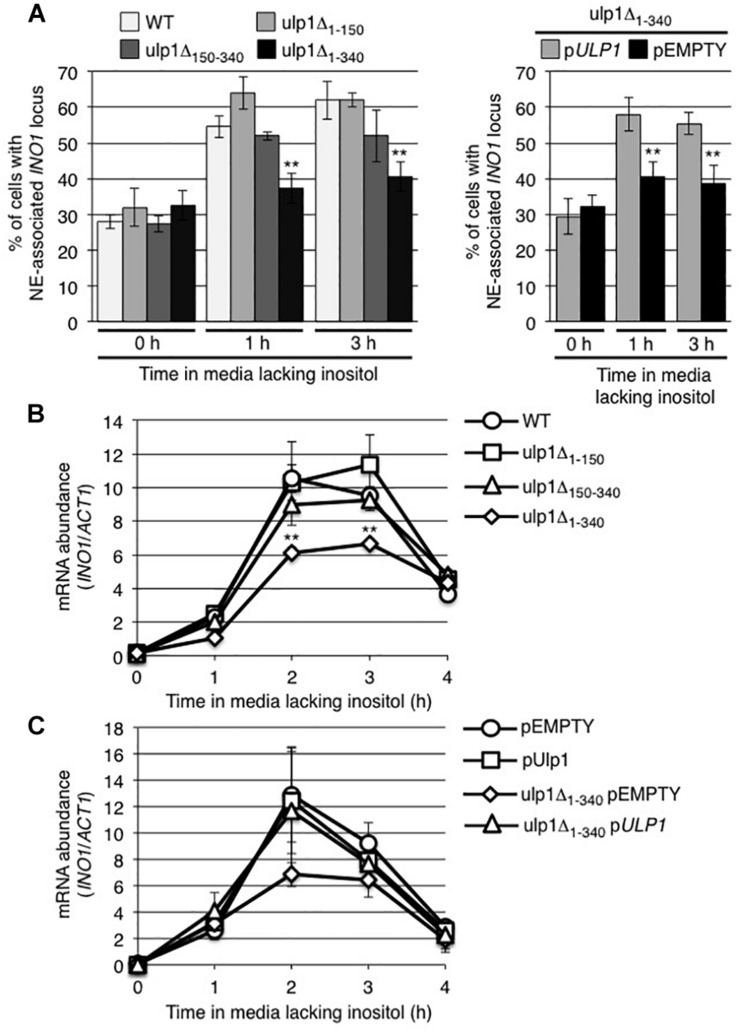
To evaluate the role of Ulp1 in the recruitment of activated INO1 to NPCs, we examined whether ulp1 mutants that lacked domains required for Ulp1 association with NPCs altered the localization of the INO1 gene. Our strategy was to uncouple Ulp1 from NPCs without altering its catalytic domain (contained within amino-acid residues 403-621) and its essential function in SUMO maturation (Li and Hochstrasser, 2003). Mutants lacking either of the two of previously described NPC binding domains of Ulp1, residues 1–150 (ulp1Δ1–150) or 150–340 (ulp1Δ150–340), were previously shown to still bind to NPCs (Li and Hochstrasser, 2003; Panse et al., 2003). However, a mutant lacking both domains, i.e., residues 1–340 (ulp1Δ1–340), showed greatly reduced levels of NPC association (Panse et al., 2003). Each of these truncation mutations was integrated within the context of the endogenous ULP1 locus by replacing the endogenous ORF and thus retaining the endogenous promoter and single copy number of the gene. This approach reduces the potential for artifacts arising from elevated levels of Ulp1 derived from plasmid-encoded genes. An examination of the integrated GFP-tagged versions of these mutants revealed similar protein levels to WT (Supplementary Figure S3B) and a localization pattern consistent with previous reports, with both the ulp1Δ1–150-GFP and the ulp1Δ150–340-GFP showing NE levels similar to WT Ulp1, while ulp1Δ1–340-GFP showed low levels of NE-association with a concomitant increase in cytoplasmic and nucleoplasmic localization relative to WT Ulp1-GFP (Supplementary Figure S3A). Similarly, sumoylation patterns in the ulp1Δ1–150 and the ulp1Δ150–340 mutants appeared largely similar to WT cells. However, levels of sumoylated proteins are slightly reduced in the ulp1Δ1–340 mutant (Supplementary Figure S3C).
We examined the effects of ulp1 truncation mutants on the inducible recruitment of INO1 to NPCs (Figure 5A). Induction of INO1 led to a rapid (within 1 h) accumulation of INO1 foci at the nuclear periphery in cells producing the ulp1Δ1–150 or ulp1Δ150–340 truncation, similar to that observed in WT cells. By contrast, cells producing the ulp1Δ1–340 mutant showed no localization of INO1 to the NE following induction. Furthermore, in monitoring mRNA levels at various times post induction, we found that INO1 transcript levels were reduced in the ulp1Δ1–340 mutant and failed to reach levels detected in the ulp1Δ1–150, ulp1Δ150–340, or WT strains (Figure 5B). Importantly, both the recruitment of INO1 to the NE and WT levels of INO1 mRNA levels could be restored in the ulp1Δ1–340 mutant by the introduction of WT ULP1 (Figures 5A,C), suggesting the phenotype detected in ulp1Δ1–340 mutant arises from the loss of Ulp1 at NPCs, and not the presence of the ulp1Δ1–340 mutant protein outside of the NPC.
FIGURE 5.
Ulp1 is required for INO1 expression and NE-association following induction. (A) The percentage of cells showing NE-association of INO1-lacO256 was determined as described in Figure 2 prior to (0 h) and at 1 and 3 h post induction in WT and the indicated ulp1 mutant strain backgrounds. Shown on the right are the results of similar experiments performed on a ulp1Δ1–340 strain transformed with either an empty plasmid (pEMPTY) or a plasmid containing a version of WT ULP1 (pULP1). Results are the means ± SD of three or more biological replicates. At least 50 cells were counted for each sample. (B,C) Levels of mRNA encoded by the INO1 gene were evaluated by qRT-PCR as described in Figure 2 following induction for the specified times in the indicated strains. Results are the means ± SEM of three biological replicates. Note, C shows data from the indicated strains transformed with either an empty plasmid (pEMPTY) or a plasmid containing a version of WT ULP1 (pULP1). Asterisks indicate a significant difference from WT as indicated by a Student’s unpaired t-test, **p < 0.01.
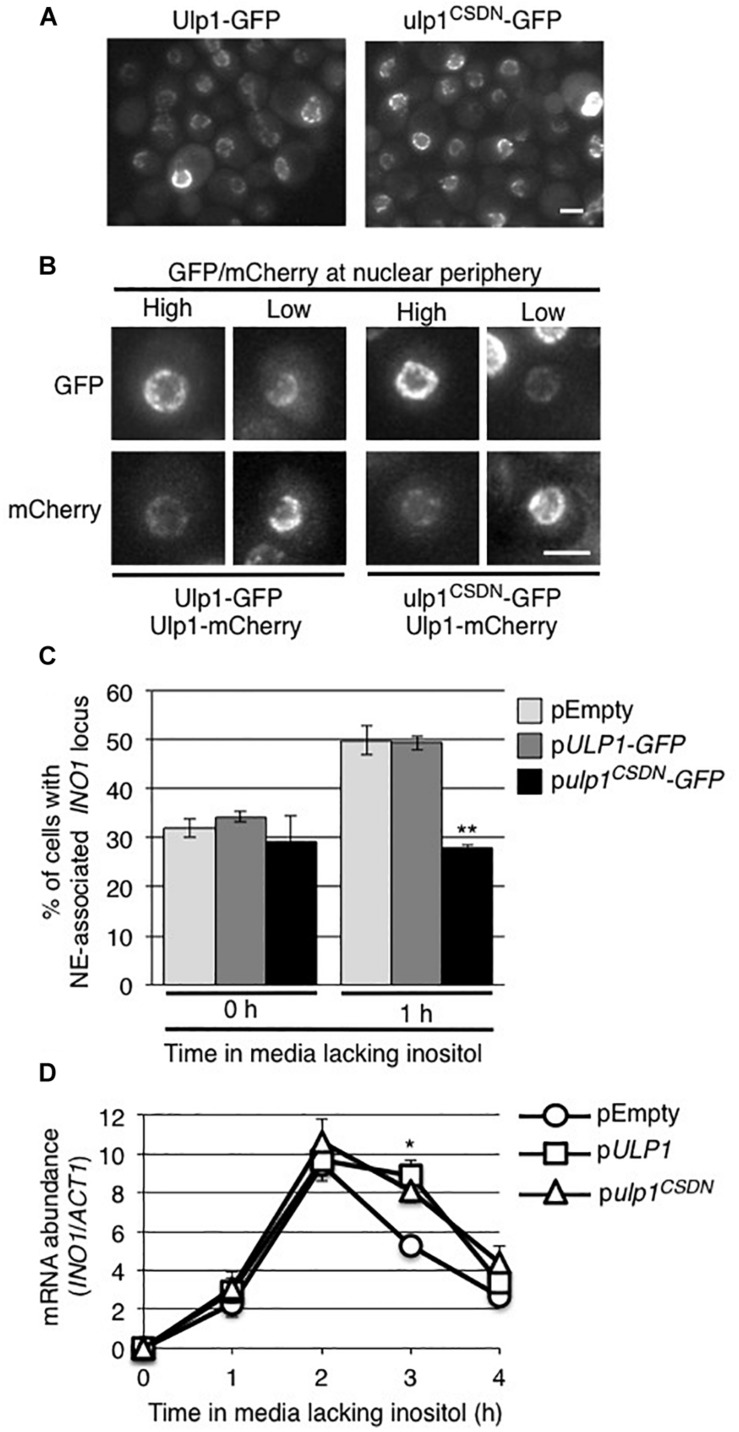
The effects of the ulp1Δ1–340 mutant on the INO1 localization and expression could occur as a consequence of the loss of functions linked to its N-terminal domain (residues 1–340) or the loss of Ulp1 isopeptidase activity at the NPC. To investigate these possibilities, we examined the effects of expressing a ulp1 double point mutant (ulp1CSDN) that abrogates SUMO binding and isopeptidase activity (Mossessova and Lima, 2000; Elmore et al., 2011) but does not alter its N-terminal domain or targeting to NPCs. This catalytically dead ulp1CSDN does not support cell viability in the absence of WT Ulp1 (Elmore et al., 2011), thus we expressed the ulp1CSDN mutant in WT cells and assessed whether the mutant exhibited dominant negative phenotypes. As shown in Figure 6A, ulp1CSDN-GFP localizes to NPCs, consistent with the known functionality of its N-terminus. Levels of the mutant protein at the nuclear periphery varied between cells, likely due to cell-to-cell variations in the amount of ulp1CSDN-GFP (arising from cell-to-cell variation in the plasmid encoded gene). Inspection of these cells revealed that the amount of ulp1CSDN-GFP at the NE appeared inversely proportional to the amount of endogenous WT Ulp1p at the same locale, suggesting the mutant protein was capable of competing with the WT protein for NPC binding sites (Figure 6B).
FIGURE 6.
The ulp1-CSDN mutant inhibits INO1 targeting to the NE. (A) Shown are images of WT cells producing exogenously expressed, plasmid-encoded Ulp1-GFP or Ulp1CSDN-GFP. (B) The localization of plasmid-encoded Ulp1-GFP or Ulp1CSDN-GFP in cells producing mCherry-tagged endogenous wild-type Ulp1 (Ulp1-mCherry) was examined. Shown are cells containing relatively high or low levels of the GFP fusion, likely stemming from cell to cell variability in plasmid copy number. Competition of NE-associated binding between the plasmid-encoded Ulp1-GFP or Ulp1CSDN-GFP and endogenous Ulp1-mCherry proteins is indicated by the relative GFP and mCherry signal intensities at the NE. Scale bars = 2 μm. (C) Recruitment of INO1 to the NPCs was measured as in Figure 2 in a WT strain containing INO1-lacO256/GFP-lacI and the indicated plasmid. Results are the means ± SD of three or more biological replicates. At least 50 cells were counted for each sample. (D) Levels of mRNA encoded by the INO1 gene were evaluated by qRT-PCR as described in Figure 2 following induction for the specified times in the indicated strains. Asterisks indicate a significant difference from WT (pEMPTY) as indicated by a Student’s unpaired t-test, *p < 0.05, **p < 0.01.
The expression of the ulp1CSDN mutant had little effect on cell growth and no striking changes were seen in global protein sumoylation patterns (Supplementary Figures S4A,B). However, when we examined the localization of INO1 following induction, the presence of the ulp1CSDN mutant protein inhibited the INO1 locus from relocating to the NE (Figure 6C). By contrast, plasmid-borne ULP1 did not alter induction-dependent INO1 association with the NE. These results led us to conclude that the association of Ulp1 catalytic activity at the NPC is required for recruitment of the INO1 locus. However, cells producing the ulp1CSDN-GFP mutant did not exhibit altered INO1 expression following induction (Figure 6D), suggesting that the mutant may not exhibit a dominant negative phenotype with respect to INO1 expression.
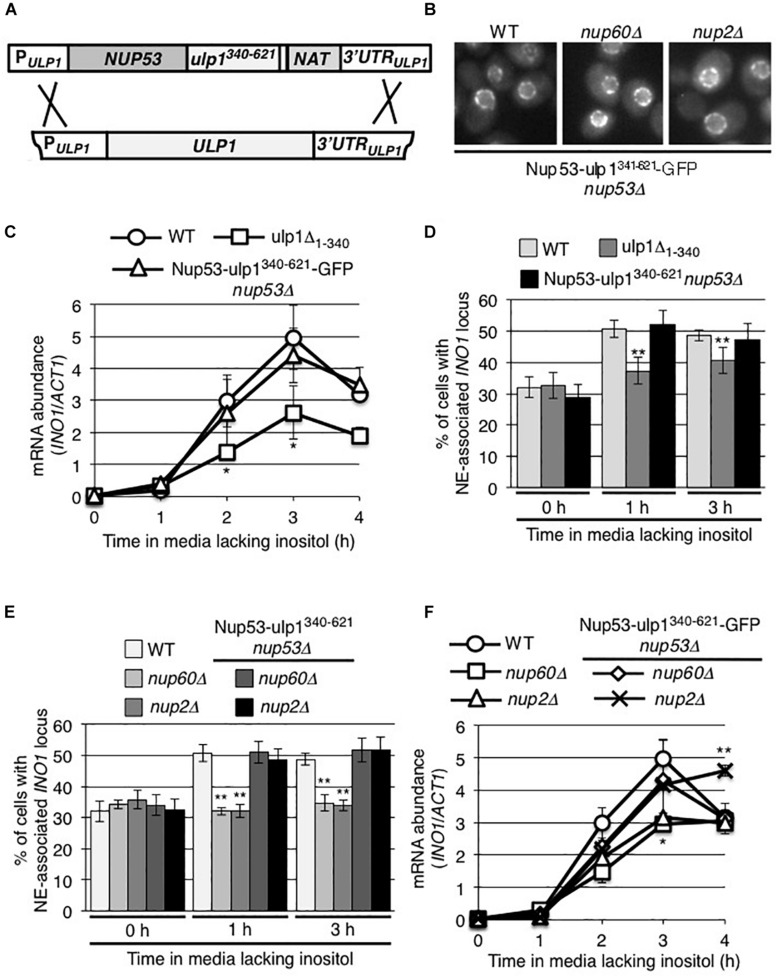
Our analysis of the ulp1CSDN mutant suggests a role for the Ulp1 catalytic activity in the association of induced INO1 with NPCs; however, these data provided no insight into its function in INO1 expression. Therefore, we asked whether positioning of the Ulp1 catalytic domain at NPCs would be sufficient for INO1 expression. To do this, we constructed a chimeric gene encoding the Ulp1 catalytic domain (residues 340–621) fused to the C-terminus of the nucleoporin Nup53 (Figure 7A). The Nup53-ulp1340–621 fusion protein showed a similar localization pattern to that observed for nucleoporins, consistent with its association with NPCs (Figure 7B). Moreover, cells producing the Nup53-ulp1340–621 protein, and lacking endogenous Ulp1, grew similar to WT cells (Supplementary Figure S4C), suggesting that the Ulp1 catalytic domain of the fusion protein could replace the essential function of WT Ulp1. In these Nup53-ulp1340–621 producing cells, we observed levels of INO1 mRNA production following induction similar to that detected in WT cells, and analysis of INO1 localization showed its recruitment to the NE was comparable to that seen in WT cells (Figures 7C,D). Thus, positioning of the Ulp1 catalytic domain at NPCs was sufficient to support INO1 expression and NPC association.
FIGURE 7.
The NPC association of the C-terminal domain of Ulp1 is sufficient to support recruitment of induced INO1. (A) Endogenous ULP1 was replaced by a NUP53-ulp1340– 621 chimera under the control of the ULP1 promoter in a haploid yeast strain. A schematic representation of the construction of this chimeric gene is shown. (B) Localization of the Nup53-ulp1340– 621 fusion protein C-terminally tagged with GFP was examined in an otherwise WT background or in strains lacking NUP60 or NUP2. (C,F) Levels of mRNA encoded by the INO1 gene were evaluated by qRT-PCR following induction for the indicated times; (C) in WT, NUP53-ulp1340– 621, and ulp1Δ1–340 strains and (F) in WT, nup60Δ, nup2Δ, NUP53-ulp1340– 621 nup60Δ, and NUP53-ulp1340– 621 nup2Δ strains as described in Figure 2. Results are the means ± SEM of three biological replicates. (D,E) Localization of the INO1-lacO256 locus was examined prior to (0 h) or at 1 and 3 h after induction in the indicated strain backgrounds. Note that the data shown here for ulp1Δ1–340 in D are the same as that in Figure 5A and are shown here for comparison. Localization of INO1-lacO256 locus was assessed as described in Figure 2. Results are the means ± SD of at least three biological replicates. For D and E, at least 100 cells were counted for each replicate. Asterisks indicate a significant difference from WT samples at the corresponding time points as indicated by Student’s unpaired t-test. *p < 0.05, **p < 0.01.
Several nup mutants have been previously shown to inhibit post-induction INO1 association with the NE, including a strain lacking Nup60 (nup60Δ) or Nup2 (nup2Δ) (Ahmed et al., 2010; Light et al., 2010; also see Figure 7E). Both Nup60 and Nup2 are functionally linked to Ulp1; the loss of Nup60 results in decreased cellular levels of Ulp1 (Palancade et al., 2007) and Nup2 has been reported as a SUMO target and a Ulp1 interacting partner (Hannich et al., 2005; Srikumar et al., 2013; Folz et al., 2019). Therefore, we tested whether the Nup53-ulp1340–621 protein could rescue the INO1 targeting defects in the nup60Δ and nup2Δ mutants. In these mutants, the Nup53-ulp1340–621-GFP fusion was visible at the NE in a characteristic NPC pattern (Figure 7B). Importantly, we observed that production of the Nup53-ulp1340–621 protein in the nup60Δ and nup2Δ mutants rescued inducible INO1 recruitment to the NPC (Figure 7E). Moreover, an examination of the expression of the INO1 gene in these strains revealed that they produced WT levels of INO1 mRNA (Figure 7F). On the basis of these data, we conclude that the defects previously detected in the nup60Δ and nup2Δ mutants are functionally linked to Ulp1.
NPC-Associated Ulp1 Regulates Sumoylation Levels of Proteins Associated With the INO1 ORF
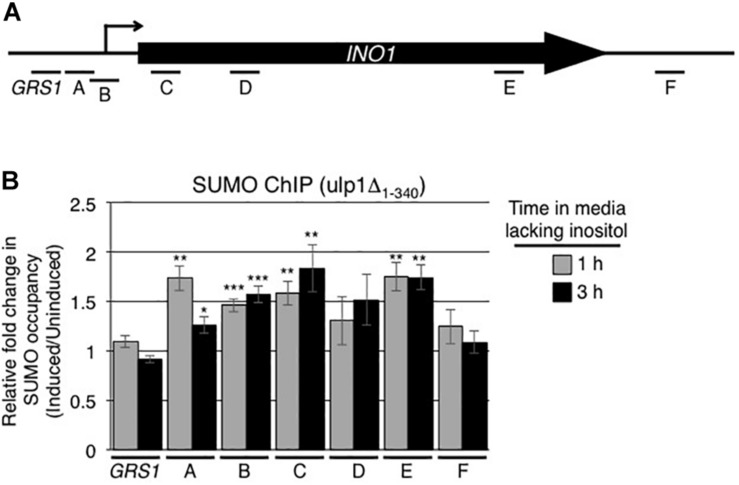
Since the C-terminal domain of Ulp1 (residues 340–621) possesses both SUMO binding and desumoylase activity, we examined the effects of removing this domain of Ulp1 from the NPC on the sumoylation state of INO1 bound proteins. To test this, we examined SUMO occupancy along the induced INO1 locus in the ulp1Δ1–340 mutant. In this mutant, we observed an increase in sumoylation of proteins in regions A and B of the INO1 gene (adjacent to and containing the transcriptional start site) after induction (Figure 8B) similar to that seen in WT cells (Figure 1B). However unlike WT cells, the ulp1Δ1–340 mutant showed no decrease in sumoylation in the GRS1 region and generally higher levels of protein sumoylation within INO1 ORF (Figure 1B) where we detected Ulp1 binding in WT cells (Figure 4B). These data led us to conclude that Ulp1, within the context of the NPCs, functions to bind and desumoylate proteins associated with the induced INO1 GRS1 and ORF.
FIGURE 8.
Ulp1 regulates sumoylation levels of proteins associated with the INO1 ORF. (A) Diagram of the INO1 locus and regions detected by ChIP analysis (see Figure 1A). (B) ulpΔ1–340 cells were subjected to ChIP analysis using antibodies directed against the SUMO polypeptide (anti-SUMO antibody) prior to and at 1 and 3 h post induction of INO1. qRT-PCR was used to quantify levels of DNA corresponding to the various regions of INO1 (see panel A) bound to sumoylated proteins. Shown are the relative fold changes in the occupancy of sumoylated proteins associated with the various regions of INO1 at the indicated times after induction relative to the uninduced samples. Results are the means ± SEM of five biological replicates. Asterisks indicate a significant difference relative to uninduced as determined by a Student’s paired t-test. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
Numerous observations have established that the spatial organization of the yeast genome is dynamic, and the positioning of numerous yeast genes within the nucleoplasm has been shown to be altered by their expression status. For example, transcriptional activation of the INO1 gene, induced by a lack of inositol, is accompanied by its relocalization from the nuclear interior to an NPC. Here we report that these events are dependent on specific regulators of sumoylation, suggesting a role for sumoylation in the expression of the INO1 gene and its targeting to the NPC. Conditions that induce expression of INO1 lead to changes in the sumoylation of proteins associated with the INO1 gene. These sumoylation events are largely mediated by the SUMO ligase Siz2, and, importantly, Siz2 is essential for the relocalization of activated INO1 from the nucleoplasm to the NE. Concomitant with relocalization, INO1 also interacts with NPC-associated Ulp1. Our data suggest that this interaction desumoylates INO1-associated proteins and is required for both targeting of the INO1 locus to an NPC and normal induction of INO1 expression. These results imply that a cycle of sumoylation and NPC-associated desumoylation contribute to INO1 targeting to the NPC and its expression.
The observation that a gene locus relocalizes from the nucleoplasm to an NPC following activation was first described in yeast for the INO1 gene, and it represents one of the most well studied of a growing list of genes exhibiting this behavior (Egecioglu and Brickner, 2011; Randise-Hinchliff and Brickner, 2016). Several factors required for INO1 targeting to the NPC have been identified. Two cis-acting DNA elements 5′ to the INO1 ORF are involved in INO1 recruitment to NPCs following long-term repression (Ahmed et al., 2010; Light et al., 2010; Randise-Hinchliff et al., 2016). Both sites bind TFs, GRS-I binds Put3 and GRS-II binds Cbf1, and each TF is required for directing GRS-containing DNA elements to NPCs (Shetty and Lopes, 2010; Brickner and Brickner, 2012; Randise-Hinchliff et al., 2016; Brickner et al., 2019). Notably, while TFs may be required for NPC association of activated genes (Randise-Hinchliff et al., 2016; Brickner et al., 2019), RNA polymerase II-mediated transcription is not necessarily required, as shown for both GAL1 and INO1 (Schmid et al., 2006; Brickner et al., 2007, 2016).
The functions of Put3 and Cbf1 are not unique as various other TFs, including those functioning in inducible and constitutive expression, or acting as transcriptional regulators (including repressors), also have the potential to target genes to NPCs (Randise-Hinchliff et al., 2016; Brickner et al., 2019). Whether these various factors interact directly or indirectly with Nups is not clear, but it does appear that specific subsets of Nups are required for genes to interact with NPCs. For example, the binding of multiple TFs to NPCs requires Nup2 and Nup100 (Dilworth et al., 2005; Brickner et al., 2019). These and additional Nups positioned on the nucleoplasmic face of NPCs, including Nup60 and Nup1, have been linked to the NPC association of induced INO1 (Brickner et al., 2007; Ahmed et al., 2010; Light et al., 2010). Of note, this function of Nup1 appears to require its phosphorylation by Cdc28 (Brickner et al., 2007; Brickner and Brickner, 2010). This and related observations support the idea that NPC association of activated INO1 is cell cycle regulated, being lost during S-phase and reestablished during G2/M-phase where it primarily resides until the following S-phase (Brickner and Brickner, 2010).
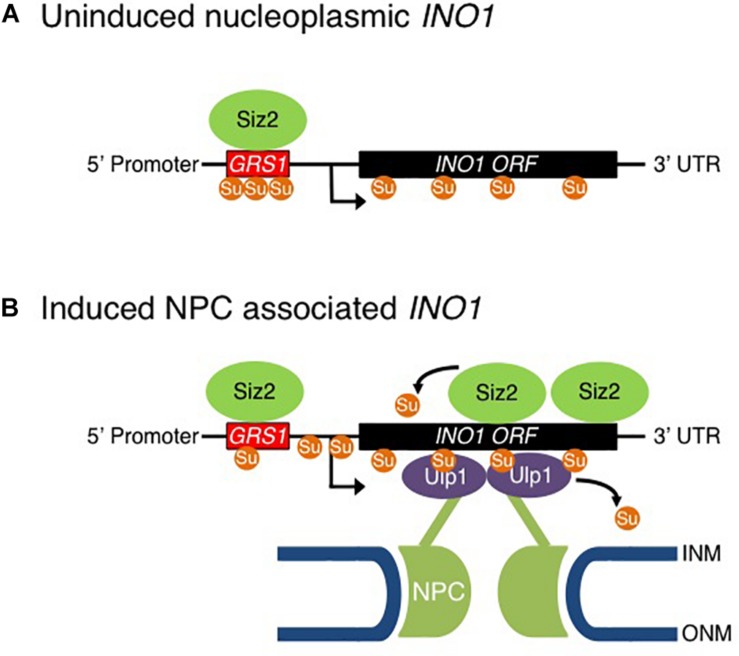
Built upon the various requirements previously established for the expression and NPC-targeting of INO1, our data have led us to conclude that a cycle of sumoylation and desumoylation is essential for the expression and NPC targeting of activated INO1. We envisage a model for these processes that includes multiple steps that we assume initiate in the nucleoplasm (Figure 9). Prior to induction, Siz2 is bound to the GRS1-containing region of INO1 (Supplementary Figure S2B) and Siz2-dependent sumoylation of proteins associated with the ORF is detected (Supplementary Figure S1C). Following induction, Siz2 binding increases within the INO1 ORF (Figure 3B), as does sumoylation of targets associated with the 5′ region of the locus, including the INO1 transcriptional start site (Figure 1, regions A and B). These increases in sumoylation are Siz2 dependent (Supplementary Figure S1C). While we have yet to identify the exact targets of Siz2 sumoylation prior to or following activation of INO1, considering their location within the INO1 gene, it seems likely that these would include TFs that function in INO1 targeting to the NPC, including Put3, which contains consensus sumoylation sites (Zhao et al., 2014), and Cbf1, which has previously been shown to be sumoylated (Wohlschlegel et al., 2004; Denison et al., 2005).
FIGURE 9.
Model for Sumo-mediated recruitment of INO1 to the NPC. Shown is model for the proposed role of sumoylation in the NPC targeting and transcriptional activation of the INO1 gene. (A) In a repressed state, INO1 is positioned in the nucleoplasm. (B) Reducing levels of inositol in the medium leads to recruitment of Siz2 to the INO1 locus and increased Siz2-mediated sumoylation of INO1-associated proteins. These events are required for the targeting of the INO1 locus to the NPC where it interacts with NPC-associated Ulp1. Ulp1 supports the association of INO1 with the NPC and functions to desumoylation of INO1 bound proteins in regions containing the GRS1 and the ORF. Desumoylation of INO1-associated proteins is proposed to promote INO1 transcription.
Siz2-mediated sumoylation events at the INO1 locus may perform several functions. In some contexts, sumoylation of TFs and histones has been shown to contribute to transcriptional repression; whereas, in others, sumoylation has been linked to activation of gene transcription (Rosonina et al., 2010; Chymkowitch et al., 2015). Interestingly, previous reports have also implicated sumoylation in both repression and activation of certain genes, suggesting that the effects of sumoylation at a given gene are dependent on the target proteins bound to the locus. For example, analysis of the expression of the inducible GAL1 gene suggests that it is maintained in a repressed state by sumoylation of two corepressors, Tup1 and Ssn6, that are desumoylated upon activation, reportedly by Ulp1 (Smith and Johnson, 2000; Zhang and Reese, 2004; Texari et al., 2013). Paradoxically, activation of the GAL1 gene is also accompanied by the recruitment of the SUMO conjugating enzyme Ubc9 to the gene locus and increased levels of SUMO-modified proteins within the promoter region (Rosonina et al., 2010).
Similarly, activation of INO1 is accompanied by both increased and decreased levels of bound SUMO-modified proteins at distinct regions of the gene (Figure 1B). In addition to increased sumoylation in regions A and B containing the transcriptional start site, INO1 induction is accompanied by decreased sumoylation levels in the Put3-binding GRS1 region (Figure 1). These events raise the possibility that desumoylation of proteins associated with the GRS1 region contribute to increased INO1 expression. In this regard, increased levels of Siz2-dependent sumoylation within the INO1 gene observed in the ulp1Δ1–340 mutant (Figure 8B) are coincident with reduced levels of INO1 mRNA accumulation following induction (Figure 5B). Whether Siz2 sumoylation may play a repressive role in INO1 expression will require further analysis.
Siz2 is required for targeting the activated INO1 locus to the NE (Figure 2B). This could involve Siz2 directly mediating the binding of INO1 to Nups. However, several observations have led us to conclude that Siz2-mediated sumoylation events direct INO1 relocation to the NPC, including, for example, sumoylation of proteins bound to regions of INO1 such as those near the transcriptional start site (Figure 1B). We speculate that the SUMO polypeptide may function as the NPC targeting signal. This idea is consistent with our observation that Ulp1, a SUMO binding protein, is required for the NE localization of INO1 (Figure 5A). Specifically, we showed that the C-terminal domain of Ulp1, which contains SUMO binding sites within the catalytic pocket and a SIM domain, when alone anchored to the NPC, is sufficient for the accumulation of the activated INO1 gene at the NPC (Figure 7D). Moreover, Ulp1 binds to the INO1 ORF and is required for the desumoylation of Siz2-mediated sumoylation sites positioned within the ORF (Figures 4B, 8B). Finally, we observed that a ulp1CSDN mutant protein, which exhibits reduced SUMO binding and no isopeptidase activity (Elmore et al., 2011) but binds to the NPC and competes with endogenous Ulp1 for NPC-binding (Figure 6B), also inhibits NE localization of INO1 (Figure 6C). Each of these observations is consistent with a role for SUMO, and its association with Ulp1, in the targeting of INO1 to the NPC (see Figure 9).
The requirement for NPC-bound Ulp1 in the targeting of activated INO1 to the NPC provides further insight into previously described defects associated with certain nup mutations, including several encoding Nups positioned on the nucleoplasmic face of the NPC (Ahmed et al., 2010; Light et al., 2010). These include Nup60 and Nup2, both of which physically and functionally interact with Ulp1 (Zhao et al., 2004; Srikumar et al., 2013) and have been shown to play a role in the association of Ulp1 with the NPC. Here we have shown that the INO1 localization defects associated with nup60 and nup2 null mutants can be rescued by positioning the Ulp1 C-terminal catalytic domain at the NPC as part of a fusion protein with Nup53 (Nup53-ulp1340–621; Figure 7) suggesting the role Nup2 and Nup60 play in this process is to position Ulp1 at the NPC.
The positioning of Ulp1 at the NPC is also essential for normal expression of INO1. In cells where NPC association of Ulp1 is inhibited, such as in the ulp1Δ1–340 mutant, levels of INO1 mRNA are reduced (Figures 5A, 7D). Importantly, placing the Ulp1 C-terminal catalytic domain at the NPC using Nup53-ulp1340–621 fusion rescued INO1 expression defects in an otherwise WT background, as well as in the nup60 and nup2 null mutants (Figures 7D,F).
Cumulatively, our observations support a model in which induction of INO1 is followed by increased binding of Siz2 to regions within the INO1 ORF and its 3′ end. We propose that the sumoylation events that arise from the Siz2 binding facilitate binding of the INO1 locus to NPC-bound Ulp1 and desumoylation of ORF-associated targets. The continuous presence of both Siz2 and Ulp1 bound to the INO1 ORF (during the 3 h period of induction examined) may support a cycle of sumoylation and desumoylation of as yet unidentified target proteins that retains these proteins and the associated INO1 gene at the NPC. Furthermore, Ulp1 binding and desumoylation of proteins associated with the GRS1 region are also predicted to facilitate INO1 binding to the NPC and potentially facilitate INO1 transcription. Interestingly, Ulp1 bound near the 3′-end of the INO1 gene might facilitate desumoylation of GRS1 associated proteins as a consequence of INO1 gene looping, which has been previously shown to occur following induction (Kaderi et al., 2009). Such a mechanism could also support Siz2 sumoylation of proteins within the 5′ region of INO1. We envisage that these steps in the NPC targeting and expression of INO1 are built upon other key requirements previously reported for these processes, including specific DNA sequences, TFs, nuclear transport factors, and Nups (Chen et al., 2007; Light and Brickner, 2013; Randise-Hinchliff and Brickner, 2016).
The concepts described here for SUMO-mediated regulation of INO1 localization and expression are likely to apply to other inducible genes. Of note, previous observations made in the analysis of the GAL1 gene revealed sumoylation processes that occur during its activation that parallel events we have observed for INO1. For example, induction of the GAL1 gene is accompanied by sumoylation of associated proteins (Rosonina et al., 2010), and NPC association of activated GAL1 was inhibited when Ulp1 association with NPCs was reduced (Texari et al., 2013). It will be of interest to further test the broader impact of sumoylation and NPC-associated desumoylation.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
NP, NS, and CP performed the experiments. All authors contributed to the conception and design of the study, manuscript revision, and read and approved the submitted version. Figures were assembled by NS, NP, and CP. RW wrote the first draft of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Ben Montpetit (University of California, Davis), Michael Hendzel (University of Alberta), and members of the Wozniak lab for helpful discussions.
Footnotes
Funding. Funding for this work was supported by the Canadian Institutes of Health Research (MOP 106502 and 36519) to RW and the National Institutes of Health, United States (NCDIR: 2P41GM109824-06 and R01: 2R01GM112108-05) to JA. The authors declare no competing financial interests.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00174/full#supplementary-material
Sumoylation of proteins at the uninduced INO1 locus. (A) Diagram of the INO1 locus and regions detected by ChIP analysis (see Figure 1A). (B,C) The graphs show the relative fold enrichment of SUMO modified proteins at each position along the INO1 gene relative to the level of SUMO modified proteins at an intergenic region within chromosome V as assessed by ChIP. B shows ChIP results for uninduced WT samples while C shows results comparing uninduced WT samples (WT 0 h) to samples derived from siz2Δ cells at the indicated times after INO1 induction. Results are the means ± SEM of at least three biological replicates.
Siz2 associates with the GRS1 region of the uninduced INO1 locus. (A) Diagram of the INO1 locus and regions detected by ChIP analysis (see Figure 1A). The graph shows the relative fold enrichment of Siz2-PrA (B) or Ulp1-PrA (C) at each position along the INO1 gene relative to the level at an intergenic region within chromosome V as assessed by ChIP. Results are the means ± SEM of at least three biological replicates.
Characterization of chromosomally encoded ulp1 truncation mutants. (A) Shown are cells producing Ulp1-GFP or the indicated truncation mutants (ulp1Δ1–150-GFP, ulp1Δ150–340-GFP, and ulp1Δ1–340-GFP) encoded by integrated and ULP1 promoter-controlled mutant genes. Camera exposure times are equivalent for each strain. Scale bars = 2 μm. (B) Truncation mutants showed similar expression levels with that of full length Ulp1-GFP. Whole cell lysates of the indicated strains were tested by Western blotting with an anti-GFP antibody. For the loading control, the levels of Gsp1 were tested with an anti-Gsp1p antibody. (C) Whole cell lysates, derived from cultures of the indicated strains, were examined by Western blotting using an anti-Smt3 (SUMO) or an anti-Gsp1 (loading control) antibody. The positions of molecular mass markers are indicated in kilodaltons.
Growth of strains producing ulp1CSDN and Nup53-ulp1341–621. (A) WT cells transformed with an empty plasmid (pEMPTY), a plasmid encoding WT Ulp1 (pULP1), or a plasmid encoding the ulp1CSDN mutant (pulp1CSDN) were grown to mid-log phase in synthetic drop out liquid culture. Tenfold serial dilutions of each culture were made and cells from each dilution spotted onto synthetic drop out plates. Total cells plated ranged between 105 and 102 cells per spot. Plates were then incubated at 30°C for 2 days prior to imaging. (B) Cell lysates, derived from the same cultures described in A, were examined by Western blotting using an anti-Smt3 (SUMO) or an anti-Gsp1 (loading control) antibody. The positions of molecular mass markers are indicated in kilodaltons. (C) WT and NUP53-ulp1341–621 cells were grown in YPD liquid culture to mid-log phase and cells from each culture analyzed as described in A.
Yeast strains used in this study.
References
- Abraham N. M., Mishra K. (2018). Elevated dosage of Ulp1 disrupts telomeric silencing in Saccharomyces cerevisiae. Mol. Biol. Rep. 45 2481–2489. 10.1007/s11033-018-4415-1 [DOI] [PubMed] [Google Scholar]
- Ahmed S., Brickner D. G., Light W. H., Cajigas I., McDonough M., Froyshteter A. B., et al. (2010). DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat. Cell Biol. 12 111–118. 10.1038/ncb2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Pringle J. R. (1996). ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol. 16 5264–5275. 10.1128/mcb.16.10.5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet A., Bretes H., Palancade B. (2015). Nuclear pore components affect distinct stages of intron-containing gene expression. Nucleic Acids Res. 43 4249–4261. 10.1093/nar/gkv280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D. G., Brickner J. H. (2010). Cdk phosphorylation of a nucleoporin controls localization of active genes through the cell cycle. Mol. Biol. Cell 21 3421–3432. 10.1091/mbc.e10-01-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D. G., Brickner J. H. (2012). Interchromosomal clustering of active genes at the nuclear pore complex. Nucleus 3 487–492. 10.4161/nucl.22663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D. G., Cajigas I., Fondufe-Mittendorf Y., Ahmed S., Lee P. C., Widom J., et al. (2007). H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5:e81. 10.1371/journal.pbio.0050081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D. G., Randise-Hinchliff C., Lebrun Corbin M., Liang J. M., Kim S., Sump B., et al. (2019). The role of transcription factors and nuclear pore proteins in controlling the spatial organization of the yeast genome. Dev. Cell 49 936–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D. G., Sood V., Tutucci E., Coukos R., Viets K., Singer R. H., et al. (2016). Subnuclear positioning and interchromosomal clustering of the GAL1-10 locus are controlled by separable, interdependent mechanisms. Mol. Biol. Cell 27 2980–2993. 10.1091/mbc.e16-03-0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner J. H., Walter P. (2004). Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2:e342. 10.1371/journal.pbio.0020342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylebyl G. R., Belichenko I., Johnson E. S. (2003). The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 278 44113–44120. 10.1074/jbc.m308357200 [DOI] [PubMed] [Google Scholar]
- Cabal G. G., Genovesio A., Rodriguez-Navarro S., Zimmer C., Gadal O., Lesne A., et al. (2006). SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441 770–773. 10.1038/nature04752 [DOI] [PubMed] [Google Scholar]
- Chen M., Hancock L. C., Lopes J. M. (2007). Transcriptional regulation of yeast phospholipid biosynthetic genes. Biochim. Biophys. Acta 1771 310–321. 10.1016/j.bbalip.2006.05.017 [DOI] [PubMed] [Google Scholar]
- Churikov D., Charifi F., Eckert-Boulet N., Silva S., Simon M.-N., Lisby M., et al. (2016). SUMO-Dependent relocalization of eroded telomeres to nuclear pore complexes controls telomere recombination. Cell Rep. 15 1242–1253. 10.1016/j.celrep.2016.04.008 [DOI] [PubMed] [Google Scholar]
- Chymkowitch P., Nguea P. A., Enserink J. M. (2015). SUMO-regulated transcription: challenging the dogma. Bioessays 37 1095–1105. 10.1002/bies.201500065 [DOI] [PubMed] [Google Scholar]
- Denison C., Rudner A. D., Gerber S. A., Bakalarski C. E., Moazed D., Gygi S. P. (2005). A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol. Cell. Proteom. 4 246–254. 10.1074/mcp.m400154-mcp200 [DOI] [PubMed] [Google Scholar]
- Dilworth D. J., Tackett A. J., Rogers R. S., Yi E. C., Christmas R. H., Smith J. J., et al. (2005). The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J. Cell. Biol. 171 955–965. 10.1083/jcb.200509061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu D., Brickner J. H. (2011). Gene positioning and expression. Curr. Opin. Cell Biol. 23 338–345. 10.1016/j.ceb.2011.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore Z. C., Donaher M., Matson B. C., Murphy H., Westerbeck J. W., Kerscher O. (2011). Sumo-dependent substrate targeting of the SUMO protease Ulp1. BMC Biol. 9:74. 10.1186/1741-7007-9-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felberbaum R., Wilson N. R., Cheng D., Peng J., Hochstrasser M. (2012). Desumoylation of the endoplasmic reticulum membrane VAP family protein Scs2 by Ulp1 and SUMO regulation of the inositol synthesis pathway. Mol. Cell. Biol. 32 64–75. 10.1128/mcb.05878-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira H. C., Luke B., Schober H., Kalck V., Lingner J., Gasser S. M. (2011). The PIAS homologue Siz2 regulates perinuclear telomere position and telomerase activity in budding yeast. Nat. Cell Biol. 13 867–874. 10.1038/ncb2263 [DOI] [PubMed] [Google Scholar]
- Folz H., Nino C. A., Taranum S., Caesar S., Lorenz L., Waharte F., et al. (2019). SUMOylation of the nuclear pore complex basket is involved in sensing cellular stresses. J. Cell Sci. 132:jcs224279. 10.1242/jcs.224279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenreich C. H., Su X. A. (2016). Relocalization of DNA lesions to the nuclear pore complex. FEMS Yeast Res. 16:fow095. 10.1093/femsyr/fow095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R., Melchior F. (2007). Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8 947–956. 10.1038/nrm2293 [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Woods R. A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350 87–96. 10.1016/s0076-6879(02)50957-5 [DOI] [PubMed] [Google Scholar]
- Hannan A., Abraham N. M., Goyal S., Jamir I., Priyakumar U. D., Mishra K. (2015). Sumoylation of Sir2 differentially regulates transcriptional silencing in yeast. Nucleic Acids Res. 43 10213–10226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannich J. T., Lewis A., Kroetz M. B., Li S. J., Heide H., Emili A., et al. (2005). Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 280 4102–4110. 10.1074/jbc.m413209200 [DOI] [PubMed] [Google Scholar]
- Jentsch S., Psakhye I. (2013). Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu. Rev. Genet. 47 167–186. 10.1146/annurev-genet-111212-133453 [DOI] [PubMed] [Google Scholar]
- Johnson E. S. (2004). Protein modification by SUMO. Annu. Rev. Bioch. 73 355–382. 10.1146/annurev.biochem.73.011303.074118 [DOI] [PubMed] [Google Scholar]
- Kaderi E. B., Medler S., Raghunayakula S., Ansari A. (2009). Gene looping is conferred by activator-dependent interaction of transcription initiation and termination machineries. J. Biol. Chem. 284 25015–25025. 10.1074/jbc.m109.007948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapetina D. L., Ptak C., Roesner U. K., Wozniak R. W. (2017). Yeast silencing factor Sir4 and a subset of nucleoporins form a complex distinct from nuclear pore complexes. J. Cell. Biol. 16 3145–3159. 10.1083/jcb.201609049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A., Felberbaum R., Hochstrasser M. (2007). A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J. Cell. Biol. 178 813–827. 10.1083/jcb.200702154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. J., Hochstrasser M. (1999). A new protease required for cell-cycle progression in yeast. Nature 398 246–251. 10.1038/18457 [DOI] [PubMed] [Google Scholar]
- Li S. J., Hochstrasser M. (2000). The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell Biol. 20 2367–2377. 10.1128/mcb.20.7.2367-2377.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. J., Hochstrasser M. (2003). The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J. Cell. Biol. 160 1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light W. H., Brickner D. G., Brand V. R., Brickner J. H. (2010). Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol. Cell 40 112–125. 10.1016/j.molcel.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light W. H., Brickner J. H. (2013). Nuclear pore proteins regulate chromatin structure and transcriptional memory by a conserved mechanism. Nucleus 4 357–360. 10.4161/nucl.26209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W. S., Duggan L., Emre N. C., Belotserkovskya R., Lane W. S., Shiekhattar R. (2001). Snf1 – a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293 1142–1146. 10.1126/science.1062322 [DOI] [PubMed] [Google Scholar]
- Lo W. S., Gamache E. R., Henry K. W., Yang D., Pillus L., Berger S. L. (2005). Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. EMBO J. 24 997–1008. 10.1038/sj.emboj.7600577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen C. J., Roy A., Levine T. P. (2003). A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22 2025–2035. 10.1093/emboj/cdg201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., Demarini D. J., Shah N. G., Wach A., Brachat A., et al. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953–961. [DOI] [PubMed] [Google Scholar]
- Makhnevych T., Ptak C., Lusk C. P., Aitchison J. D., Wozniak R. W. (2007). The role of karyopherins in the regulated sumoylation of septins. J. Cell Biol. 177 39–49. 10.1083/jcb.200608066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E., Lima C. D. (2000). Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5 865–876. 10.1016/s1097-2765(00)80326-3 [DOI] [PubMed] [Google Scholar]
- Palancade B., Liu X., Garcia-Rubio M., Aguilera A., Zhao X., Doye V. (2007). Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol. Biol. Cell 18 2912–2923. 10.1091/mbc.e07-02-0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse V. G., Hardeland U., Werner T., Kuster B., Hurt E. (2004). A proteome-wide approach identifies sumoylated substrate proteins in yeast. J. Biol. Chem. 279 41346–41351. 10.1074/jbc.m407950200 [DOI] [PubMed] [Google Scholar]
- Panse V. G., Kressler D., Pauli A., Petfalski E., Gnädig M., Tollervey D., et al. (2006). Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic 7 1311–1321. 10.1111/j.1600-0854.2006.00471.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse V. G., Kuster B., Gerstberger T., Hurt E. (2003). Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat. Cell Biol. 5 21–27. 10.1038/ncb893 [DOI] [PubMed] [Google Scholar]
- Randise-Hinchliff C., Brickner J. H. (2016). Transcription factors dynamically control the spatial organization of the yeast genome. Nucleus 7 369–374. 10.1080/19491034.2016.1212797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randise-Hinchliff C., Coukos R., Sood V., Sumner M. C., Zdraljevic S., Meldi Sholl L., et al. (2016). Strategies to regulate transcription factor–mediated gene positioning and interchromosomal clustering at the nuclear periphery. J. Cell Biol. 212 633–646. 10.1083/jcb.201508068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett C. C., Straight A. F., Li G., Willhelm C., Sudlow G., Murray A. W., et al. (1996). In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135 1685–1700. 10.1083/jcb.135.6.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E. (2019). A conserved role for transcription factor sumoylation in binding-site selection. Curr. Genet. 65 1307–1312. 10.1007/s00294-019-00992-w [DOI] [PubMed] [Google Scholar]
- Rosonina E., Duncan S. M., Manley J. L. (2010). SUMO functions in constitutive transcription and during activation of inducible genes in yeast. Genes Dev. 24 1242–1252. 10.1101/gad.1917910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben G. J., Kirkland J. G., MacDonough T., Chen M., Dubey R. N., Gartenberg M. R., et al. (2011). Nucleoporin mediated nuclear positioning and silencing of HMR. PLoS ONE 6:e21923. 10.1371/journal.pone.0021923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Arib G., Laemmli C., Nishikawa J., Durussel T., Laemmli U. K. (2006). Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol. Cell. 21 379–391. 10.1016/j.molcel.2005.12.012 [DOI] [PubMed] [Google Scholar]
- Shetty A., Lopes J. M. (2010). Derepression of INO1 transcription requires cooperation between the Ino2p-Ino4p heterodimer and Cbf1p and recruitment of the ISW2 chromatin-remodeling complex. Eukaryot. Cell 9 1845–1855. 10.1128/ec.00144-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. L., Johnson A. D. (2000). Turning genes off by Ssn-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25 325–330. 10.1016/s0968-0004(00)01592-9 [DOI] [PubMed] [Google Scholar]
- Srikumar T., Lewicki M. C., Raught B. (2013). A global S. cerevisiae small ubiquitin-related modifier (SUMO) system interactome. Mol. Syst. Biol. 9:668. 10.1038/msb.2013.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K., Vogel F., Schwienhorst I., Meusser B., Volkwein C., Nentwig B., et al. (2002). A lack of SUMO conjugation affects cNLS-dependent nuclear protein import in yeast. J. Biol. Chem. 277 49554–49561. 10.1074/jbc.m207991200 [DOI] [PubMed] [Google Scholar]
- Straight A. F., Belmont A. S., Robinett C. C., Murray A. W. (1996). GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6 1599–1608. 10.1016/s0960-9822(02)70783-5 [DOI] [PubMed] [Google Scholar]
- Texari L., Dieppois G., Vinciguerra P., Contreras M. P., Groner A., Letourneau A., et al. (2013). The nuclear pore regulates GAL1 gene transcription by controlling the localization of the SUMO protease Ulp1. Mol. Cell 51 807–818. 10.1016/j.molcel.2013.08.047 [DOI] [PubMed] [Google Scholar]
- Texari L., Stutz F. (2015). Sumoylation and transcription regulation at nuclear pores. Chromosoma 124 45–56. 10.1007/s00412-014-0481-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Vosse D. W., Wan Y., Lapetina D. L., Chen W. M., Chiang J. H., Aitchison J. D., et al. (2013). A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell 152 969–983. 10.1016/j.cell.2013.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Saleem R. A., Ratushny A. V., Roda O., Smith J. J., Lin C. H., et al. (2009). Role of the histone variant H2A.Z/Htz1p in TBP recruitment, chromatin dynamics, and regulated expression of oleate-responsive genes. Mol. Cell Biol. 29 2346–2358. 10.1128/mcb.01233-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel J. A., Johnson E. S., Reed S. I., Yates J. R., III (2004). Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 279 45662–45668. 10.1074/jbc.m409203200 [DOI] [PubMed] [Google Scholar]
- Wykoff D. D., O’Shea E. K. (2005). Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol. Cell. Proteomics 4 73–83. 10.1074/mcp.m400166-mcp200 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Reese J. C. (2004). Redundant mechansims are used by Ssn6-Tup1 in repressing chromosomal gene transcription in Saccharomyces cerevisiae. J. Biol. Chem. 279 39240–39250. 10.1074/jbc.m407159200 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Xie Y., Zheng Y., Jiang S., Liu W., Mu W., et al. (2014). GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 42 W325–W330. 10.1093/nar/gku383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. (2018). SUMO-mediated regulation of nuclear functions and signaling processes. Mol. Cell 71 409–418. 10.1016/j.molcel.2018.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Wu C. Y., Blobel G. (2004). Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 167 605–611. 10.1083/jcb.200405168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Ryan J. J., Zhou H. (2004). Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J. Biol. Chem. 279 32262–32268. 10.1074/jbc.m404173200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sumoylation of proteins at the uninduced INO1 locus. (A) Diagram of the INO1 locus and regions detected by ChIP analysis (see Figure 1A). (B,C) The graphs show the relative fold enrichment of SUMO modified proteins at each position along the INO1 gene relative to the level of SUMO modified proteins at an intergenic region within chromosome V as assessed by ChIP. B shows ChIP results for uninduced WT samples while C shows results comparing uninduced WT samples (WT 0 h) to samples derived from siz2Δ cells at the indicated times after INO1 induction. Results are the means ± SEM of at least three biological replicates.
Siz2 associates with the GRS1 region of the uninduced INO1 locus. (A) Diagram of the INO1 locus and regions detected by ChIP analysis (see Figure 1A). The graph shows the relative fold enrichment of Siz2-PrA (B) or Ulp1-PrA (C) at each position along the INO1 gene relative to the level at an intergenic region within chromosome V as assessed by ChIP. Results are the means ± SEM of at least three biological replicates.
Characterization of chromosomally encoded ulp1 truncation mutants. (A) Shown are cells producing Ulp1-GFP or the indicated truncation mutants (ulp1Δ1–150-GFP, ulp1Δ150–340-GFP, and ulp1Δ1–340-GFP) encoded by integrated and ULP1 promoter-controlled mutant genes. Camera exposure times are equivalent for each strain. Scale bars = 2 μm. (B) Truncation mutants showed similar expression levels with that of full length Ulp1-GFP. Whole cell lysates of the indicated strains were tested by Western blotting with an anti-GFP antibody. For the loading control, the levels of Gsp1 were tested with an anti-Gsp1p antibody. (C) Whole cell lysates, derived from cultures of the indicated strains, were examined by Western blotting using an anti-Smt3 (SUMO) or an anti-Gsp1 (loading control) antibody. The positions of molecular mass markers are indicated in kilodaltons.
Growth of strains producing ulp1CSDN and Nup53-ulp1341–621. (A) WT cells transformed with an empty plasmid (pEMPTY), a plasmid encoding WT Ulp1 (pULP1), or a plasmid encoding the ulp1CSDN mutant (pulp1CSDN) were grown to mid-log phase in synthetic drop out liquid culture. Tenfold serial dilutions of each culture were made and cells from each dilution spotted onto synthetic drop out plates. Total cells plated ranged between 105 and 102 cells per spot. Plates were then incubated at 30°C for 2 days prior to imaging. (B) Cell lysates, derived from the same cultures described in A, were examined by Western blotting using an anti-Smt3 (SUMO) or an anti-Gsp1 (loading control) antibody. The positions of molecular mass markers are indicated in kilodaltons. (C) WT and NUP53-ulp1341–621 cells were grown in YPD liquid culture to mid-log phase and cells from each culture analyzed as described in A.
Yeast strains used in this study.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.