Abstract
Gastrointestinal (GI) cancers are known to have a high incidence worldwide and require an early diagnosis to successfully treat them, providing higher survival rates and better quality of life for the patients. MicroRNA-27a is a well-known oncogene that plays a significant role in various GI cancers. It is known to upregulate the expression of numerous oncogenes leading to cancer progression. The miR-27a harbors two polymorphisms rs895819 and rs11671784 which alter the disease susceptibility by interfering with the maturation and expression of miR-27a. In the current study, we aimed to investigate the role played by these polymorphisms in cancers of the GI tract. We conducted a case-control study with 210 GI cancer cases and 210 cancer-free controls to analyze the effect of these polymorphisms. The rs895819 polymorphism was genotyped using PCR-RFLP, and rs11671784 was genotyped on a MassARRAY platform. The association analysis failed to bring out any significant association of the polymorphisms with GI cancer risk. However, genotype-phenotype interaction analysis revealed that the rs895819 was found to increase the risk GI cancers along with the presence of risk factors such as socioeconomic status, diabetes mellitus, hypertension, alcohol consumption, and tobacco chewing.
Keywords: Cancer research, Genetics, Oncology, Gene mutation, Gene regulation, Genomics, Polymerase chain reaction, Biochemistry, Molecular biology, Pathophysiology, Epidemiology, miR-27a, rs895819, rs11671784, Gastrointestinal cancers, MassARRAY, PCR-RFLP
Cancer research; Genetics; Oncology; Gene Mutation; Gene Regulation; Genomics; Polymerase Chain Reaction; Biochemistry; Molecular Biology; Pathophysiology; Epidemiology; miR-27a; rs895819; rs11671784; Gastrointestinal cancers; MassARRAY, PCR-RFLP
1. Introduction
Cancers of the alimentary canal are collectively called as gastrointestinal (GI) cancers and makeup to 23% of deaths caused by cancer worldwide. Although GI cancers are highly curable, they are very challenging due to late diagnosis, incorrect staging, and insensitivity to treatment. The year 2018 has seen a rise in cancer burden to 18.1 million new cases and 9.6 million cancer deaths. Among the common cancers, Colorectal cancer (CRC) was the second most mortal and third most incident cancer and gastric cancer (GC) was the fifth most common cancer diagnosed [1, 2]. The more prevalent cancers of the GI tract such as gastric, colorectal and esophageal cancers have also ranked top positions in terms of incidence and mortality in the Indian sub-continent [3].
MicroRNAs (miRNAs) are tiny non-coding RNA molecules which manage the expression of several thousand genes by binding to the 3’ UTR region of mRNAs [4]. They are well known for their unique roles in regulating cellular processes such as embryonic development, proliferation, differentiation, apoptosis, immunity, and inflammation. Their association with several inflammatory diseases, autoimmune diseases, and cancers are also vastly reported. MicroRNAs are involved in different aspects of cancers, including tumorigenesis, prognosis, metastasis, diagnosis, and therapeutics [5]. Based on the prevailing conditions, MicroRNAs perform dual roles as oncogenes and tumor suppressors. Single nucleotide polymorphisms (SNPs) in these novel molecules have been found to affect cancer risk, serve as markers for susceptibility and predictors of disease outcome and treatment response [6].
MiR-27a belongs to the miR-23a/27a/24-2 cluster and has its genomic location at chromosome 19p13.12 [7]. MiR-27a has been known to control genes involved in cell proliferation, apoptosis, differentiation, and cell cycle regulation. Numerous research findings have acknowledged miR-27a as a critical oncogene in gastric cancer [8, 9], breast cancer [10], cervical cancer [11], ovarian cancer [12] and renal cell carcinoma [13]. Mir-27a also regulates multidrug resistance and resistance to chemotherapy.
The pre microRNA region of miR-27a is known to consist of two polymorphism rs895819 and rs11671784 at positions 40 and 36 relative to the first nucleotide, respectively [14]. These two SNPs are known to have paradoxical effects on the expression of miR-27a. The rs895819 A/G polymorphism was the first to be identified and analyzed. Studies revealed that the mutant genotypes increased the production of mature miR-27a when compared to the ancestral allele and suppressed the expression of its targets significantly [15]. The SNP has been found to adversely alter the susceptibility to several cancers such as breast cancer, gastric cancer, colorectal cancer, esophageal cancer, lung cancer renal cancer, cervical cancer, liver cancer, prostate cancer and nasopharyngeal cancer [16].
The second SNP rs11671784 present just four nucleotides away from rs895819 is a G to A polymorphism. Unlike its neighbor, the SNP does not affect the maturation of miR-27a but is said to lower the expression levels of the mature miR-27a. The SNP has been linked to decreased susceptibility to gastric cancer and prevented lymphatic invasion [17, 18]. All of these findings support that miR-27a is a potential oncogene, and its oncogenicity might be altered by the presence of the two polymorphism, and they might be effective markers for cancer susceptibility. However, the existing studies are limited to Caucasian and Asian (Chinese) populations, which brings the question of consistency of these findings with other ethnicities. In this study, we have attempted to screen the two miR-27a polymorphisms in subjects from the Indian subcontinent and assess its value as an indicator for GI cancer risk using a case-control study.
2. Materials and methods
2.1. Study population
The study was commenced after obtaining ethical clearance from the Institutional Ethics Committee of Sri Ramachandra Institute of Higher Education and Research (Deemed to be University) IEC-NI/13/APR/33/39. A total of 420 subjects; 210 cases and 210 controls were recruited for the study with strict adherence to the declaration of Helsinki and ICMR ethical guidelines for biomedical research.
The subjects were recruited based on convenience sampling from December 2013 to June 2016. The GI cancer patients who were received at the pathology department of Sri Ramachandra Medical Centre for biopsy were identified and recruited after obtaining their informed consent. The GI cancer cases were included after histopathologic diagnosis, and those with secondary tumors were excluded. The controls were recruited from the master health check-up unit of Sri Ramachandra Medical Centre. Subjects with a personal or family history of cancer, autoimmune disorders and other chronic disorders such as epilepsy, asthma were excluded from the study. Such information was obtained by interviewing the patients and confirming the same from the physician's notes.
2.2. Sample and data collection
Around 3 mL of the leftover venous blood, collected for routine clinical investigations was received from the central laboratory of Sri Ramachandra Medical Centre after obtaining proper consent from the subjects.
Clinical data of the subjects were retrieved from the medical records; the subjects were interviewed for information about smoking status, diet, region of residence, past medical history, and family medical history using questionnaires. The identity of all the participants was preserved throughout the study.
2.3. DNA isolation and SNP genotyping
Genomic DNA was isolated from the whole blood by employing the standard Phenol-Chloroform method [19].
2.4. Genotyping of rs895819 using PCR RFLP
The pre miRNA region of miR-27a gene, which contains the rs895819 polymorphism was amplified using primers as described earlier [15]. The 182 bp amplicon was viewed on a 2 % agarose gel to confirm successful amplification. The amplicons were further digested using AdeI restriction enzyme procured from Thermo Fisher Scientific at 37°C overnight. The digested products were resolved on a 2.5 % agarose gel. A single undigested band of size 182 bp represented the GG genotype, while AA genotype was viewed as 155 bp and 27 bp fragments and the AG genotype was observed as 182 bp, 155 bp, and 27 bp fragments. Genotyping was repeated thrice for 10 % of the samples by two different people for consistency and reproducibility.
2.5. Genotyping of rs11671784 using mass ARRAY
The rs11671784 was genotyped using MassARRAY from Agena Bioscience Inc, San Diego, CA, USA which implies the Matrix Assisted Laser Desorption/Ionisation-Time Of Flight methodology (MALDI-TOF) [20]. This technique is based on PCR amplification of the genomic region consisting of the SNP followed by a single base extension reaction to discriminate the alleles using Iplex Gold technique. The end products are finally analyzed using a mass spectrophotometer.
The PCR primers and extension primers were designed using the assay design software provided by Agena Biosciences. The PCR amplification and iPLEX reactions were carried out using Verity 96 PCR thermal cycler following manufacture instructions. The amplified products were spotted onto a SpectroCHIP® bio-array using Nanodispensor -RS1000 and subsequently analyzed with an MA4 mass spectrometer. The alleles are distinguished based on the difference in the mass of the nucleotides. The results were viewed as cluster plots and spectrograms using MassArray Typer 4.0 software.
2.6. Statistical analysis
The concordance of genotype frequencies with Hardy – Weinberg Equilibrium (HWE) was calculated using the chi-square test. The association of the genotypes and alleles with GI cancer was done by calculating the odds ratio with 95% CI through Binary logistic regression. The crude odds ratio was adjusted for covariates using multinomial regression Student's t-test was done to analyze the continuous variables. Univariate Binary Logistic regression was done to check the association of demographic parameters of cases and controls with disease outcome. Association between the tumour characteristics and genotypes was determined by Pearson Chi-square test. The statistical tests were done using SPSS v21.0. Haplotype analysis was done by SNP stats online tool [21]. The interaction between the SNPs and demographic parameters were tested using Multifactor dimensionality reduction (MDR) analysis using MDR software v3.2. MDR analysis can detect gene-gene and gene-environment interactions in data sets which have SNP and environmental data represented as categorical variables. This method is employed to comprehensively detect nonlinear interactions by combining attribute selection, attribute construction, cross-validation and permutation testing [22].
3. Results
There was no significant difference in age between the cases and controls (cases: 56.8 ± 12.38 & controls: 55.01 ± 11.24) and had the same ratio of males and females. The demographic and clinical features about both the groups are listed in Table 1. Diabetes mellitus (P = 0.002), region (P = 0.015) and diet (P = 0.031) was found to be associated with GI cancer. The cases were sub-grouped based on the type of GI cancers. The four subgroups were gastric cancer (GC), colorectal cancer (CRC), esophageal cancer (EC) and others, which contained 82, 84, 24, and 19 cases, respectively. The subgroup ‘others’ consisted of cancers of the periampullary region and small intestine. They were grouped due to their small numbers. The mean age for the subgroups was calculated, and it was found that the GC group had the highest mean age of 60.9 ± 12.41 years followed by CRC (55.94 ± 12.43 years), EC (53.54 ± 12.38 years) and others (52.21 ± 12.26 years).
Table 1.
Demographic and Clinical Parameters of Gastrointestinal Cancer Patients and Cancer free controls.
| Characteristics | Cases, N (%) | Controls, N (%) | OR (CI) | P Value |
|---|---|---|---|---|
| Age† | 56.8 ± 12.38 | 55.01 ± 11.24 | - | 0.122∗ |
|
Gender | ||||
| Male | 137 (65) | 131 (62) | 1.132 (0.76–1.69) | 0.542 |
| Female | 73 (35) | 79 (38) | 1 (reference) | |
|
Region | ||||
| Urban | 124 (59) | 148 (71) | 1.66 (1.11–2.48 | 0.015 |
| Rural | 86 (41) | 62 (30) | 1 (reference) | |
|
Socio-Economic Status | ||||
| Class I | 6 (3) | 18 (9) | 1 (reference) | |
| Class II | 106 (49) | 91 (43) | 0.67 (0.16–2.75) | 0.575 |
| Class III | 93 (44) | 92 (44) | 2.33 (0.77–7.065) | 1.35 |
| Class IV | 5 (2) | 9 (4) | 2.044 (0.67–6.21) | 2.08 |
|
Co morbidities | ||||
| No diabetes | 162 (77) | 138 (66) | 1 (reference) | |
| Diabetes | 42 (20) | 72 (34) | 2.012 (1.29–3.14) | 0.002 |
| missing data | 6 | 0 | ||
| No Hypertension | 165 (79) | 174 (83) | 1 (reference) | |
| Hypertension | 45 (21) | 30 (14) | 1.58 (0.95–2.63) | 0.077 |
| missing data | 0 | 6 | - | |
|
Substance | ||||
| Non Smokers | 158 (75) | 164 (78) | 1 (reference) | |
| Smokers | 38 (18) | 46 (22) | 1.17 (0.72–1.89) | 0.532 |
| missing data | 14 | 0 | ||
| Non Drinkers | 143 (68) | 153 (73) | 1 (reference) | |
| Drinkers | 52 (25) | 57 (27) | 1.03 (0.67–1.59) | 0.914 |
| missing data | 15 | 0 | - | |
|
Diet | ||||
| Mixed | 193 (92) | 180 (72) | 1 (reference) | |
| Vegetarian | 17 (8) | 30 (14) | 2.02 (1.07–3.83) | 0.031 |
Mean ± SD.
P value obtained from t-test, Bold values are significant.
The histologic and cellular differentiation of GI cancers were retrieved from the medical records. It was observed that the majority of the GI cancers were adenocarcinomas (76 %) whereas papillary adenocarcinoma was the least common histologic type consisting of only one case. About 66 % of the tumors were moderately differentiated, whereas well-differentiated tumors were the least common grade, holding 6 % of the cases.
The genotype and allele frequencies were similar among both the groups for the two polymorphisms rs895819 and rs11671784. Both the cases and controls were in concordance with HWE for both rs895819 (Controls P=0.06; Cases P = 0.63) and rs11671784 (Control P = 0.796; Case P = 0.061) polymorphisms. The minor allele frequency of rs11671784 was found to be as low as 0.02 in the controls; the AA genotype was absent in the control group, whereas a single AA genotype was recorded in the case group. The genotype, allele frequencies, HWE P values have been presented in Table 2.
Table 2.
Frequency distribution, minor allele frequency and Hardy Weinberg equilibrium P values of rs895819 and rs11671784.
| Group | rs895819 |
rs11671784 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotypes N (%) |
Alleles N (%) |
MAF | HWE χ2 (P value) | Genotypes N (%) |
Alleles N (%) |
MAF | HWE χ2 (P value) | |||||||
| AA | AG | GG | A | G | GG | GA | AA | G | A | |||||
| Controls | 59 | 114 | 31 | 232 | 176 | 0.43 | 3.48 (0.06) | 183 | 7 | 0 | 373 | 7 | 0.02 | 0.07 (0.796) |
| Cases | ||||||||||||||
| GI cancer cases | 63 | 102 | 36 | 228 | 174 | 0.43 | 0.23 (0.634) | 172 | 10 | 1 | 354 | 12 | 0.03 | 3.51 (0.061) |
| Subgroups | ||||||||||||||
| Gastric cancer | 24 | 44 | 15 | 92 | 74 | 0.45 | 0.44 (0.507) | 68 | 2 | 1 | 138 | 4 | 0.03 | 16.7 (0.000) |
| Colorectal cancer | 27 | 31 | 17 | 85 | 65 | 0.43 | 1.88 (0.170) | 66 | 6 | 0 | 144 | 6 | 0.04 | 0.14 (0.712) |
| Oesophageal cancer | 6 | 14 | 3 | 40 | 20 | 0.44 | 1.31 (0.253) | 22 | 1 | 0 | 45 | 1 | 0.02 | 0.01 (0.915) |
| Others | 5 | 14 | 1 | 24 | 16 | 0.4 | 4.20 (0.040) | 17 | 1 | 0 | 35 | 1 | 0.03 | 0.06 (0.904) |
HWE, Hardy Weinberg Equilibrium, MAF, minor allele frequency.
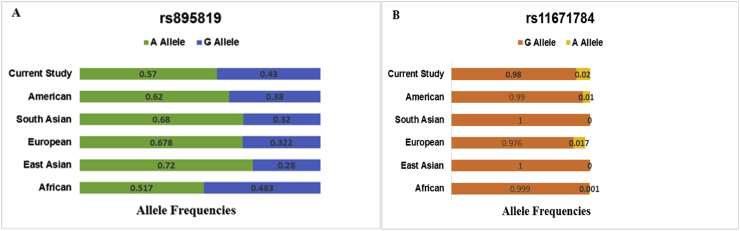
The association results of rs895819 and rs11671784 are given in Tables 3 and 4 respectively. Due to the presence of a zero cell in rs11671784 genotype data, the association tests for specific models were undefined even after zero cell correction. The crude ORs were adjusted for age, sex and associated covariates such as diabetes mellitus, region and diet. There was no remarkable association of both the polymorphisms with GI cancers and the subgroups. Haplotype analysis revealed four combinations, but none of them affected the disease risk significantly. The allele frequencies from control populations of various ethnicities as described in HapMap-dbSNP database were compared to frequencies obtained from our study as in Figure 1.
Table 3.
Association of rs895819 with GI cancer and subgroups.
| Name of Cancer | Genotypes & Alleles | Crude OR (95%CI) | P Value | Adjusted ORa (95%CI) | Pa Value | Adjusted ORb(95% CI) | Pb Value |
|---|---|---|---|---|---|---|---|
| GI cancer | AA | 1 (Reference) | 1 (Reference) | 1 (Reference) | |||
| AG | 0.84 (0.54–1.31) | 0.435 | 1.10 (0.69–1.76) | 0.857 | 1.15 (0.73–1.82) | 0.55 | |
| GG | 1.12 (0.62–2.05) | 0.703 | 0.72 (0.38–1.36) | 0.311 | 0.86 (0.45–1.62) | 0.629 | |
| Dominant | 0.89 (0.58–1.39) | 0.595 | 1 (0.64–1.57) | 0.996 | 1.07 (0.69–1.66) | 0.77 | |
| Recessive | 0.821 (0.49–1.39) | 0.463 | 1.43 (0.85–2.49) | 0.202 | 1.21 (0.71–2.07) | 0.494 | |
| A | 1 (Reference) | 1 (Reference) | 1 (Reference) | ||||
| G | 1.00 (0.76–1.32) | 0.991 | 0.91 (0.68–1.22) | 0.519 | 0.98 (0.73–1.31) | 0.886 | |
| GC | AA | 1 (Reference) | 1 (Reference) | 1 (Reference) | |||
| AG | 0.91 (0.50–1.64) | 0.743 | 1.13 (0.58–2.21) | 0.722 | 0.90 (0.49–1.65) | 0.736 | |
| GG | 1.23 (0.56–2.68) | 0.604 | 2.31 (0.9–5.90) | 0.082 | 1.24 (0.53–2.86) | 0.622 | |
| Dominant | 0.91 (0.55–1.7) | 0.906 | 1.31 (0.69–2.48) | 0.411 | 0.97 (0.55–1.73) | 0.926 | |
| Recessive | 0.79 (0.40–1.55) | 0.492 | 0.51 (0.23–1.07) | 0.075 | 0.784 (0.39–1.57) | 0.491 | |
| A | 1 (Reference) | 1 (Reference) | 1 (Reference) | ||||
| G | 1.05 (0.73–1.52) | 0.794 | 0.74 (0.49–1.12) | 0.154 | 0.948 (0.65–1.38) | 0.78 | |
| CRC | AA | 1 (Reference) | 1 (Reference) | 1 (Reference) | |||
| AG | 0.63 (0.35–1.15) | 0.133 | 1.43 (0.77–2.64) | 0.254 | 1.44 (0.77–2.67) | 0.251 | |
| GG | 1.24 (0.59–2.62) | 0.576 | 0.66 (0.30–1.47) | 0.306 | 0.641 (0.29–1.42) | 0.641 | |
| Dominant | 0.75 (0.43–1.32) | 0.32 | 1.17 (0.66–2.08) | 0.593 | 1.17 (0.66–2.09) | 0.593 | |
| Recessive | 0.63 (0.33–1.22) | 0.174 | 1.83 (0.92–3.65) | 0.84 | 1.70 (0.86–3.33) | 0.126 | |
| A | 1 (Reference) | 1 (Reference) | 1 (Reference) | ||||
| G | 1.01 (0.7–1.47) | 0.955 | 0.897 (0.61–1.32) | 0.582 | 0.91 (0.62–1.34) | 0.649 | |
| EC | AA | 1 (Reference) | 1 (Reference) | 1 (Reference) | |||
| AG | 1.29 (0.37–2.91) | 0.613 | 0.74 (0.27–2.03) | 0.56 | 0.88 (0.32–2.45) | 0.805 | |
| GG | 0.98 (0.23–4.21) | 0.982 | 0.96 (0.21–4.4) | 0.961 | 1.04 (0.23–4.72) | 0.956 | |
| Dominant | 1.22 (0.46–3.23) | 0.688 | 0.77 (0.29–2.05) | 0.595 | 0.93 (0.34–2.5) | 0.882 | |
| Recessive | 0.726 (0.35–4.46) | 0.726 | 0.896 (0.25–3.23) | 0.785 | 0.84 (0.23–3.06) | 0.835 | |
| A | 1 (Reference) | 1 (Reference) | 1 (Reference) | ||||
| G | 1.02 (0.56–1.87) | 0.947 | 0.925 (0.50–1.70) | 0.801 | 1.02 (0.54–1.91) | 0.951 | |
| Others | AA | 1 (Reference) | 1 (Reference) | 1 (Reference) | |||
| AG | 1.04 (0.37–2.91) | 0.948 | 0.981 (0.35–2.78) | 0.972 | 1.07 (0.37–3.09) | 0.904 | |
| GG | 0.33 (0.31–0.33) | 0.312 | 3.39 (0.37–31.6) | 0.282 | 2.91 (0.33–25.9) | 0.339 | |
| Dominant | 0.88 (0.32–2.43) | 0.807 | 1.13 (0.40–3.17) | 0.816 | 1.23 (0.43–3.53) | 0.698 | |
| Recessive | 3.23 (0.42–25.05) | 0.236 | 0.323 (.0.4–2.55) | 0.283 | 0.36 (0.45–2.85) | 0.332 | |
| A | 1 (Reference) | 1 (Reference) | 1 (Reference) | ||||
| G | 0.77 (0.39–1.52) | 0.447 | 1.29 (0.64–2.59) | 0.474 | 1.32 (0.64–2.72) | 0.451 |
ORa, Pa, adjusted for age and sex, ORb, Pb, adjusted for Diabetes mellitus, Diet and Region, GI, Gastrointestinal, GC, gastric cancer, EC, esophageal cancer.
Table 4.
Association of rs11671784 with GI cancer and subgroups.
| Name of Cancer | Genotypes & Alleles | Crude OR (95%CI) | P Value | Adjusted ORa (95%CI) | Pa Value | Adjusted ORb(95% CI) | Pb Value |
|---|---|---|---|---|---|---|---|
| GI cancer | GG | 1 (Reference) | 1 (Reference) | 1 (Reference) | |||
| AG | 1.52 (0.57–4.08) | 0.406 | 0.57 (0.20–1.60) | 0.287 | 0.730 (0.26–2.03) | 0.547 | |
| Dominant | 1.67 (0.63–4.41) | 0.299 | 0.521 (0.19–1.43) | 0.206 | 0.629 (0.23–1.72) | 0.366 | |
| G | 1 (Reference) | 1 (Reference) | 1 (Reference) | ||||
| A | 1.66 (0.64–4.32) | 0.303 | 0.53 (0.21–1.43) | 0.21 | 0.63 (0.24–1.71) | 0.369 | |
| GC | GG | 1 (Reference) | 1 (Reference) | 1 (Reference) | |||
| AG | 0.792 (0.16–3.91) | 0.775 | 1.33 (0.19–9.37) | 0.775 | 1.21 (0.24–6.23) | 0.817 | |
| Dominant | 1.19 (0.31–4.68) | 0.8 | 0.76 (0.15–3.90) | 0.739 | 0.79 (0.18–3.14) | 0.704 | |
| G | 1 (Reference) | 1 (Reference) | 1 (Reference) | ||||
| A | 1.19 (0.30–4.68) | 0.8 | 0.75 (0.15–3.80) | 0.731 | 0.75 (0.18–3.08) | 0.369 | |
| CRC | GG | 1 (Reference) | 1 (Reference) | 1 (Reference) | |||
| AG | 2.34 (0.76–7.21) | 0.139 | 0.36 (0.11–1.14) | 0.82 | 0.44 (0.14–1.40) | 0.165 | |
| Dominant | 2.34 (0.76–7.22) | 0.139 | 0.36 (0.11–1.14) | 0.82 | 0.44 (0.14–1.40) | 0.165 | |
| G | 1 (Reference) | 1 (Reference) | 1 (Reference) | ||||
| A | 2.28 (0.75–6.9) | 0.144 | 0.37 (0.12–1.15) | 0.86 | 0.45 (0.15–1.41) | 0.172 | |
| EC | GG | 1 (Reference) | 1 (Reference) | 1 (Reference) | |||
| AG | 1.29 (0.48–3.51) | 0.613 | 0.74 (0.27–2.03) | 0.56 | 0.88 (0.32–2.45) | 0.805 | |
| Dominant | 1.22 (0.46–3.23) | 0.688 | 0.77 (0.29–2.05) | 0.56 | 0.88 (0.32–2.45) | 0.805 | |
| G | 1 (Reference) | 1 (Reference) | 1 (Reference) | ||||
| A | 1.18 (0.14–9.85) | 0.876 | 0.71 (0.83–1.1) | 0.751 | 0.79 (0.85–7.22) | 0.831 | |
| Others | GG | 1 (Reference) | 1 (Reference) | 1 (Reference) | |||
| AG | 1.54 (0.18–13.25) | 0.695 | 0.69 (0.08–6.1) | 0.742 | 0.60 (0.61–5.94) | 0.665 | |
| Dominant | 1.54 (0.18–13.25) | 0.695 | 0.69 (0.08–6.1) | 0.742 | 0.60 (0.61–5.94) | 0.665 | |
| G | 1 (Reference) | 1 (Reference) | 1 (Reference) | ||||
| A | 1.52 (0.18–12.73) | 0.698 | 0.70 (0.08–5.96) | 0.745 | 0.69 (0.08–5.63) | 0.711 |
ORa, Pa, adjusted for age and sex, ORb, Pb, adjusted for Diabetes mellitus, Diet and Region, GI, Gastrointestinal, GC, gastric cancer, EC, esophageal cancer.
Figure 1.
Comparison of allele frequencies of the polymorphisms A.rs895819 & B.rs11671784 among the various global populations and the current study population.
The frequencies obtained from the current study were concordant with the American population for rs895819 and with European population for rs11671784. The MDR analysis results are given in Table 5. A highly significant model was observed that included rs895819 and risk factors such as age, region, socioeconomic status, diabetes mellitus, hypertension, alcohol consumption, tobacco chewing, suggesting an increased risk for GI cancers. Whereas, the rs11671784 did not display any interaction and was not included in the MDR results output. The chi-square test performed to check the association of histology and cellular differentiation of tumors with the two polymorphism did not reveal any significant outcomes Table 6.
Table 5.
Multifactor dimensionality reduction analysis for the interaction models between SNPs and clinical parameters using cross-validation consistency and prediction error.
| Models | Balanced accuracy CV training | Balanced accuracy CV consistency | CV consistency | P Value |
|---|---|---|---|---|
| Group, DK | 0.6606 | 0.6405 | 10/10 | <0.0001∗ |
| rs895819, Group, S, R, SES, DM, DK | 0.8865 | 0.5395 | 10/10 | <0.0001∗ |
| rs895819, Group, S, R, SES, DM, HT, DK | 0.915 | 0.5624 | 10/10 | <0.0001∗ |
| rs895819, Group, S, R, SES, DM, HT, DK, TC | 0.9275 | 0.5761 | 10/10 | <0.0001∗ |
S, Sex; R, Region; SES, Socioeconomic status; DM, Diabetes mellitus; HT, Hypertension, DK, Drinking, TC, Tobacco chewing, CV, Cross-validation.
Significant P values.
Table 6.
Association between Genotypes and Tumour characteristics.
| Polymorphism | Tumour characteristics | χ2 | P Value |
|---|---|---|---|
| rs895819 | Histology | 8.701 | 0.561 |
| Cell differentiation | 1.581 | 0.812 | |
| rs11671784 | Histology | 6.332 | 0.787 |
| Cell differentiation | 8.338 | 0.08 |
χ2 for histology was calculated with df = 10, χ2 for cell differentiation was calculated with df = 4.
4. Discussion
MiR-27a is widely appreciated for its oncogenic role in several cancers such as gastric cancer, ovarian cancer, cervical cancer, and many more. It belongs to the miR-23ã27ã24-2 cluster and is present sandwiched between miR-23a and miR-24-2 [7]. Apart from being an active oncogene, it also mediates cellular differentiation and promotes cell proliferation. The miR-27a gene is known to house two polymorphisms rs895819 and rs11671784 present four nucleotides apart from each other. These polymorphisms are believed to act antagonistically, wherein rs895819 increases the risk of cancer, and rs11671784 plays a protective role against gastric cancer risk [17].
As per the in silico studies, rs895819 polymorphism is located in the loop region of the miRNA hairpin structure and does not alter the minimum free energy, but blocks the miRNA maturation [23]. Expression of mir-27a was hindered by the presence of mutant genotypes proving that the presence of the SNP negatively affects the expression of miR-27a. This might be because of the SNP being centrally located in the terminal loop decreasing the size of the loop and affecting the binding of DORSHA which reduces the maturation of miR-27a [15].
Despite the mounting evidence for the functionality of the SNP, its association with cancer risk is highly debated due to contrasting results obtained across various diseases and ethnicities. The SNP was found to play a protective role against familial breast cancer in a cohort from Germany; this was probably the first study to bring out the protective role of this SNP [24]. Similarly, the G allele was associated with decreased risk of cervical cancer [25] and breast cancer [26] in different Chinese populations. However, a different scenario was presented with other studies which have stated that rs895819 confers an increased risk of cancer on its carriers. Sun et al. conducted an elaborate study to investigate the role of the polymorphism in gastric cancer and the effect of the SNP on its targets. The study revealed that the SNP elevated the levels of miR-27 which downregulated the expression of Zinc finger and BTB domain containing 10 gene (ZBTB10) which controls genes responsible for gastric cancer angiogenesis and survival like Sp proteins and Sp-dependent genes [15]. The SNP also increased gastric cancer susceptibility in a Romanian population [27]. Xu and his group have reported a similar role of the SNP in CRC patients from a North Han Chinese population. The study revealed that the polymorphism was involved in colorectal carcinogenesis and progression with the G allele conferring an increased risk of CRC [28]. Wang and his group also reported that the G allele increased the risk of CRC and metastasis [29]. Similar results were obtained from two other studies conducted in different Chinese populations [30].
The current study was attempted in order to assess the roles played by the two polymorphisms from an Indian sub population. The analysis of demographic and basic characteristics of the subjects showed that region of stay, diet and diabetes mellitus were associated with GI cancers, but revealed no influence on the association of the polymorphisms with GI cancer. The genotyping results revealed that there were no significant differences in the frequencies of the genotypes and alleles. The association results showed no significant association of the polymorphism with increased risk for GI cancers. The same trend was observed in the subgroup analysis. These results were concordant with various studies in colorectal and gastric cancers which showed the lack of association of the polymorphism with disease risk in Chinese and European studies [31, 32]. All the results mentioned above bring out the three different roles played by rs895819 polymorphism across various cancers and populations. The study reported for the first time, the comparison of the MAFs with other populations observed in the HapMap which revealed that the MAF of rs895819 and rs11671784 from the current study was similar to the American and European population respectively.
There are minimal studies present regarding the rs11671784 polymorphism and its association in human diseases. This might be due to the low minor allele frequencies observed and the difficulty in genotyping it alongside rs896819 using basic techniques [33]. Yang and his group identified that the rs11671784 decreased the risk of gastric cancer by interfering with the maturation of miR-27a and upregulating the expression of Homeobox A10 (HOXA10) gene which is an important transcription factor controlling various cellular pathways [18]. Song et al. also gave similar results showing that the A allele was associated with reduced gastric cancer risk and lymphatic invasion. The rs11671784 also increased susceptibility and chemosensitivity in bladder cancer by upregulating expression of Runt-related transcription factor 1 gene (RUNX 1) an essential regulator of cancer progression pathways [34]. Contrastingly, in a study from a Chinese population, the rs11671784 was not associated with gastric risk and moreover there was only one heterozygous genotype present in the study population [35]. In our study, we observed the same fashion, the minor allele frequency was 0.02, and there was only one AA genotype present, and it belonged to the case group.
We also conducted an MDR analysis to look for genotype-phenotype interactions, which could increase or alter the susceptibility of GI cancers. Parameters such as age, sex, region, socioeconomic status, diabetes mellitus, hypertension, smoking, alcohol consumption, and tobacco chewing were employed for the analysis. MDR is a statistical software which is used to detect the genotype and phenotype combinations which will increase the disease risk [36]. It uses mathematical models to give a various combination of the parameters and gives the best models based on the cross-validation (CV) values. The higher the CV value, the lower the error rate, so generally the CV values of 9/10 and 10/10 are considered effective. From our analysis, we obtained four models with significant CV and p values. The analysis showed that factors such as sex, region, socioeconomic status, diabetes mellitus, hypertension, drinking alcohol, and tobacco chewing, along with the presence of rs895819 increased the risk of GI cancers. Socioeconomic status has been acknowledged as an essential attributing factor for the increased incidence of GI cancers. The socioeconomic status greatly influences the availability of medical care, nutritious food and disease awareness resulting in a higher prevalence of cancers in people belonging to class 2 and class 3 which was also evident in the present study [37]. Alcohol and tobacco chewing are also well-known risk factors for cancer initiation and progression in GI cancers, and our results were also concordant with these findings. Diabetes mellitus has been linked to increased risk of hepatocellular carcinomas and colorectal cancers, and the MDR analysis also revealed similar results in our study [38]. It is important to note that the analysis did not favor the presence of rs11671784 for GI cancer risk. This shows that rs895819 can alter GI cancer risk, and rs11671784 may not have a similar role. This is concordant with the findings of Song and group, which published that both the polymorphisms inversely affect gastric cancer risk [17].
The impact of the environmental factors in genetic studies has been very well observed, thus identifying them as potential risk factors in GI cancers. Although our study supports the combined effects of the risk factors on GI cancer, a detailed investigation of genotype and phenotype interactions are essential to sharpen the focus on risk predicting markers. The study has its own limitations, its’ sample size does not provide significant power to the study, but the fact that this was an exploratory study should be considered and recognised as it has shed light on the polymorphisms, frequency and association with GI cancer in the Indian subpopulation.
MiR-27a is a vital oncogene and has excellent clinical value as a biomarker and therapeutic target. However, the role of its polymorphisms yet remains inconclusive and requires a comprehensive study across various populations to conclude the potential of these SNPs as SNP biomarkers for GI risk prediction. To the best of our knowledge, this is the first attempt to investigate the function of mir-27a polymorphisms in GI cancers from an Indian subpopulation.
5. Conclusion
The study revealed that rs895819 might not directly alter the GI cancer risk, but it will significantly increase the risk of GI cancers along with risk factors like diabetes mellitus, hypertension, region, alcohol drinking, tobacco chewing, and socioeconomic status. The rs11671784 was not associated with GI cancers in our study population.
Declarations
Author contribution statement
TBC C. Walter: Conceived and designed the experiments; Analyzed and interpreted the data.
Z. Shankaran: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
T. Johnson: Conceived and designed the experiments; Wrote the paper.
N. Prakash and K. Ramachandiran: Performed the experiments.
G. Doss: Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We thank all the participants who had consented for the study. We would also like to thank the technical staff of the Pathology department and the Central laboratory for their constant support. Our heartfelt thanks to Dr. Priyanka Venugopal Senior Research Fellow- Genetics, SRIHER, India and Mr. Melvin Joy, Associate Research Officer, Department of Biostatistics, CMC, Vellore, India for their assistance in statistical analysis. We are immensely grateful to the management of Sri Ramachandra Institute of Higher Education and Research (DU) for supporting our study through the Founder Chancellor Research Fellowship for ZS and providing the necessary infrastructure.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.IARC . World Heal. Organ; 2018. Latest Global Cancer Data, 2018; pp. 13–15.http://www.who.int/cancer/PRGlobocanFinal.pdf [Google Scholar]
- 2.WHO | 10 Facts about Cancer. WHO; 2018. https://www.who.int/features/factfiles/cancer/en/ (accessed March 6, 2019) [Google Scholar]
- 3.Cancer statistics - India against cancer. http://cancerindia.org.in/cancer-statistics/ (accessed March 6, 2019)
- 4.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 5.Iorio M.V., Croce C.M. microRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–1133. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayes J., Peruzzi P.P., Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Chhabra R., Dubey R., Saini N. Cooperative and individualistic functions of the microRNAs in the miR-23ã27ã24-2 cluster and its implication in human diseases. Mol. Cancer. 2010;9:232. doi: 10.1186/1476-4598-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou L., Liang X., Zhang L., Yang L., Nagao N., Wu H., Liu C., Lin S., Cai G., Liu J., Zhou L., Liang X., Zhang L., Yang L., Nagao N., Wu H., Liu C., Lin S., Cai G., Liu J. MiR-27a-3p functions as an oncogene in gastric cancer by targeting BTG2. Oncotarget. 2016;7:51943–51954. doi: 10.18632/oncotarget.10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu T., Tang H., Lang Y., Liu M., Li X. MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by targeting prohibitin. Cancer Lett. 2009;273:233–242. doi: 10.1016/j.canlet.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Mertens-Talcott S.U., Chintharlapalli S., Li X., Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G 2 -M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007;67:11001–11011. doi: 10.1158/0008-5472.CAN-07-2416. [DOI] [PubMed] [Google Scholar]
- 11.Sun Y., Yang X., Liu M., Tang H. B4GALT3 up-regulation by miR-27a contributes to the oncogenic activity in human cervical cancer cells. Cancer Lett. 2016;375:284–292. doi: 10.1016/j.canlet.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L.-Y., Zhu X., Wu L.-M., He Y., Jia J., Chen Y. MiR-27a promotes EMT in ovarian cancer through active Wnt/?-catenin signalling by targeting FOXO1. Cancer Biomarkers. 2019;24:1–12. doi: 10.3233/CBM-181229. [DOI] [PubMed] [Google Scholar]
- 13.Peng H., Wang X., Zhang P., Sun T., Ren X., Xia Z. 2015. miR-27a Promotes Cell Proliferation and Metastasis in Renal Cell Carcinoma.www.ijcep.com/ (accessed March 6, 2019) [PMC free article] [PubMed] [Google Scholar]
- 14.Gong J., Liu C., Liu W., Wu Y., Ma Z., Chen H., Guo A.-Y. An update of miRNASNP database for better SNP selection by GWAS data, miRNA expression and online tools. Database. 2015;2015:bav029. doi: 10.1093/database/bav029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Q., Gu H., Zeng Y., Xia Y., Wang Y., Jing Y., Yang L., Wang B. Hsa-mir-27a genetic variant contributes to gastric cancer susceptibility through affecting miR-27a and target gene expression. Cancer Sci. 2010;101:2241–2247. doi: 10.1111/j.1349-7006.2010.01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen M., Fang W., Wu X., Bian S., Chen G., Lu L., Weng Y. Distinct effects of rs895819 on risk of different cancers: an update meta-analysis. Oncotarget. 2017;8:75336–75349. doi: 10.18632/oncotarget.17454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang B. rs11671784 G/A and rs895819 A/G polymorphisms inversely affect gastric cancer susceptibility and miR-27a expression in a Chinese population. Med. Sci. Monit. 2014;20:2318–2326. doi: 10.12659/MSM.892499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Q., Jie Z., Ye S., Li Z., Han Z., Wu J., Yang C., Jiang Y. Genetic variations in miR-27a gene decrease mature miR-27a level and reduce gastric cancer susceptibility. Oncogene. 2014;33:193–202. doi: 10.1038/onc.2012.569. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J., Fritsch E.F., Maniatis T. Cold Spring Harbor Laboratory; 1989. Molecular Cloning: a Laboratory Manual.https://books.google.co.in/books/about/Molecular_Cloning.html?id=G5RqAAAAMAAJ&source=kp_cover&redir_esc=y [Google Scholar]
- 20.Ellis J.A., Ong B. The MassARRAY® system for targeted SNP genotyping. Methods Mol. Biol. 2017:77–94. doi: 10.1007/978-1-4939-6442-0_5. [DOI] [PubMed] [Google Scholar]
- 21.Sole X., Guino E., Valls J., Iniesta R., Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 22.Moore J.H., Andrews P.C. Epistasis analysis using multifactor dimensionality reduction. Methods Mol. Biol. 2015:301–314. doi: 10.1007/978-1-4939-2155-3_16. [DOI] [PubMed] [Google Scholar]
- 23.Ryan B.M., Robles A.I., Harris C.C. Genetic variation in microRNA networks: the implications for cancer research. Nat. Rev. Cancer. 2010;10:389. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang R., Schlehe B., Hemminki K., Sutter C., Bugert P., Wappenschmidt B., Volkmann J., Varon R., Weber B.H.F., Niederacher D., Arnold N., Meindl A., Bartram C.R., Schmutzler R.K., Burwinkel B. A genetic variant in the pre-miR-27a oncogene is associated with a reduced familial breast cancer risk. Breast Cancer Res. Treat. 2010;121:693–702. doi: 10.1007/s10549-009-0633-5. [DOI] [PubMed] [Google Scholar]
- 25.Xiong X.-D., Luo X.-P., Cheng J., Liu X., Li E.-M., Zeng L.-Q. A genetic variant in pre-miR-27a is associated with a reduced cervical cancer risk in southern Chinese women. Gynecol. Oncol. 2014;132:450–454. doi: 10.1016/j.ygyno.2013.12.030. [DOI] [PubMed] [Google Scholar]
- 26.Zhang N., Huo Q., Wang X., Chen X., Long L., Jiang L., Ma T., Yang Q. A genetic variant in pre-miR-27a is associated with a reduced breast cancer risk in younger Chinese population. Gene. 2013;529:125–130. doi: 10.1016/j.gene.2013.07.041. [DOI] [PubMed] [Google Scholar]
- 27.Rogoveanu I., Burada F., Cucu M.G., Vere C.C., Ioana M., Cîmpeanu R.A. Association of microRNA polymorphisms with the risk of gastric cancer in a Romanian population. J. Gastrointestin. Liver Dis. 2017;26:231–238. doi: 10.15403/jgld.2014.1121.263.rog. [DOI] [PubMed] [Google Scholar]
- 28.Xu J., Bian Q., Chen J.-J., Gu J.-P. Association between pre-miR-27a functional polymorphism and risk of colorectal cancer in north Chinese Han population. OncoTargets Ther. 2015;8:3003. doi: 10.2147/OTT.S89754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z., Sun X., Wang Y., Liu X., Xuan Y., Hu S. Association between miR-27a genetic variants and susceptibility to colorectal cancer. Diagn. Pathol. 2014;9:146. doi: 10.1186/1746-1596-9-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alidoust M., Hamzehzadeh L., Rivandi M., Pasdar A. Polymorphisms in non-coding RNAs and risk of colorectal cancer: a systematic review and meta-analysis. Crit. Rev. Oncol. Hematol. 2018;132:100–110. doi: 10.1016/j.critrevonc.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Hezova R., Kovarikova A., Bienertova-Vasku J., Sachlova M., Redova M., Vasku A., Svoboda M., Radova L., Kiss I., Vyzula R., Slaby O. Evaluation of SNPs in miR-196-a2, miR-27a and miR-146a as risk factors of colorectal cancer. World J. Gastroenterol. 2012;18:2827–2831. doi: 10.3748/wjg.v18.i22.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kupcinskas J., Bruzaite I., Juzenas S., Gyvyte U., Jonaitis L., Kiudelis G., Skieceviciene J., Leja M., Pauzas H., Tamelis A., Pavalkis D., Kupcinskas L. Lack of association between miR-27a, miR-146a, miR-196a-2, miR-492 and miR-608 gene polymorphisms and colorectal cancer. Sci. Rep. 2015;4:5993. doi: 10.1038/srep05993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang R., Burwinkel B. A bias in genotyping the miR-27a rs895819 and rs11671784 variants. Breast Cancer Res. Treat. 2012;134:899–901. doi: 10.1007/s10549-012-2140-3. [DOI] [PubMed] [Google Scholar]
- 34.Deng Y., Bai H., Hu H. rs11671784 G/A variation in miR-27a decreases chemo-sensitivity of bladder cancer by decreasing miR-27a and increasing the target RUNX-1 expression. Biochem. Biophys. Res. Commun. 2015;458:321–327. doi: 10.1016/j.bbrc.2015.01.109. [DOI] [PubMed] [Google Scholar]
- 35.Xu Q., Chen T.J., He C.Y., Sun L.P., Liu J.W., Yuan Y. MiR-27a rs895819 is involved in increased atrophic gastritis risk, improved gastric cancer prognosis and negative interaction with Helicobacter pylori. Sci. Rep. 2017;7:1–11. doi: 10.1038/srep41307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hahn L.W., Ritchie M.D., Moore J.H. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19:376–382. doi: 10.1093/bioinformatics/btf869. http://www.ncbi.nlm.nih.gov/pubmed/12584123 [DOI] [PubMed] [Google Scholar]
- 37.Nagel G., Linseisen J., Boshuizen H.C., Pera G., Del Giudice G., Westert G.P., Bueno-de-Mesquita H.B., Allen N.E., Key T.J., Numans M.E., Peeters P.H., Sieri S., Siman H., Berglund G., Hallmans G., Stenling R., Martinez C., Arriola L., Barricarte A., Chirlaque M.D., Quiros J.R., Vineis P., Masala G., Palli D., Panico S., Tumino R., Bingham S., Boeing H., Bergmann M.M., Overvad K., Boutron-Ruault M.-C., Clavel-Chapelon F., Olsen A., Tjonneland A., Trichopoulou A., Bamia C., Soukara S., Sabourin J.-C., Carneiro F., Slimani N., Jenab M., Norat T., Riboli E., Gonzalez C.A. Socioeconomic position and the risk of gastric and esophageal cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC-EURGAST) Int. J. Epidemiol. 2007;36:66–76. doi: 10.1093/ije/dyl275. [DOI] [PubMed] [Google Scholar]
- 38.Wu C.-Y., Lin J.-T. The changing epidemiology of Asian digestive cancers: from etiologies and incidences to preventive strategies. Best Pract. Res. Clin. Gastroenterol. 2015;29:843–853. doi: 10.1016/j.bpg.2015.09.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.