Abstract
Head and neck squamous cell carcinoma (HNSCC) is the sixth most frequently diagnosed cancer worldwide. However, the clinical outcomes remain unsatisfactory. The aim of this study is to unravel the functional role and regulatory mechanism of HOXA9 in HNSCC. A cohort of 25 HNSCC tumor tissues and normal tissue counterparts was collected. qRT-PCR and western blotting were performed to determine the levels of HOXA9 and epithelial-mesenchymal transition (EMT)-related markers. Cell Counting Kit-8 (CCK-8) and colony formation assays were conducted to monitor cell viability and cytotoxicity. Transwell and wound healing assays were used to determine cell migration and invasion. Annexin V-fluorescein isothiocyanate/propidium iodide (FITC/PI) staining was performed to detect cell apoptosis. Bioinformatic analysis, electrophoretic mobility shift assay and chromatin immunoprecipitation (ChIP) assays were performed to investigate the direct binding between HIF-1α or CCCTC binding factor (CTCF) and HOXA9. Glutathione S-transferase (GST) pull-down and RNA pull-down assays were used to validate the interaction between CTCF and HOTTIP. HOXA9 was upregulated in HNSCC tissues and cells. Knockdown of HOXA9 inhibited cell proliferation, migration, invasion, and chemoresistance but promoted apoptosis in CAL-27 and KB cells. Knockdown of HOXA9 also regulated EMT-related marker via targeting YAP1/β-catenin. Silencing of HOTTIP or CTCF exerted similar tumor-suppressive effects in HNSCC. Mechanistically, HIF-1α or CTCF transcriptionally regulated HOXA9, and HOTTIP/CTCF cooperatively regulated HOXA9 in KB cells. HIF-1α or HOTTIP/CTCF transcriptionally modulates HOXA9 expression to regulate HNSCC progression and drug resistance.
Keywords: head and neck squamous cell carcinoma (HNSCC), HOXA9, HOTTIP, CTCF
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most frequently diagnosed cancer worldwide. The therapeutic outcomes of HNSCC remain poor due to late diagnosis, and the 5-year survival rate for patients with HNSCC is only ∼50%.1 HNSCC, which accounts for ∼90% of head and neck malignancies, can originate from different subsites, including the lip, oral cavity, oropharynx, hypopharynx, and larynx.2 It is well established that tobacco and alcohol abuse, as well as human papilloma virus (HPV) or Epstein-Barr virus (EBV) infections are independent risk factors for HNSCC.3,4 Despite advances in surgery, chemo- and radiotherapies over the past 4 decades, the survival rate of HNSCC has not improved.5 The poor survival rate could be attributed to local recurrence, distant metastases, and chemoresistance. Therefore, a better understanding of the mechanism involved in tumor progression and identification of reliable biomarkers is essential for improving HNSCC survival.
Homeobox (HOX) genes encode homeoproteins that are a family of homeodomain-containing transcription factors. In mammalian cells, the HOX family consists of 39 transcription factors that play important roles in embryonic development.6 In addition, emerging evidence indicates that HOX genes function as either transcriptional activators or repressors in cancers.7 Homeobox A9 (HOXA9) is dysregulated in hematopoietic malignancies and solid tumors, such as acute myeloid leukemia (AML), breast cancer, ovarian cancer, and cervical cancer.8, 9, 10, 11 More important, a recent clinical study has demonstrated that HOXA9 promoter hypermethylation is associated with HNSCC progression and metastasis.12 However, the biological function and the regulatory mechanism of HOXA9 during HNSCC progression remain poorly understood.
Long noncoding RNAs (lncRNAs) are a class of non-coding RNAs that consist of at least 200 nt in length. lncRNAs have been demonstrated to be novel regulators of tumor progression.13 The HOXA transcript at the distal tip (HOTTIP), a lncRNA located at the 5′ tip of the HOXA locus, enhances gene transcription and H3 lysine 4 trimethylation via direct interaction with the WDR5/MLL complex.14 HOTTIP was found to promote HOXA9 expression by binding to WDR5 in pancreatic cancer cells.15 A recent study has reported that HOTTIP physically associates with CCCTC-binding factor (CTCF), a well-known insulator factor, to modulate HOXA gene expression in human foreskin fibroblasts.16 Moreover, a clinical study has revealed that HOTTIP is overexpressed in tongue squamous cell carcinoma (TSCC), and increased HOTTIP expression is positively correlated with distant metastasis and clinical stage,17 indicating the critical role of HOTTIP in HNSCC.
In the present study, we have demonstrated that HOXA9 was significantly upregulated in HNSCC tissues and cells. Silencing of HOXA9 inhibited cell proliferation, migration, invasion, and chemoresistance but promoted apoptosis in CAL-27 and KB cells. Knockdown of HOXA9 regulated EMT-related markers via targeting YAP1/β-catenin. Similarly, HOTTIP or CTCF exerted oncogenic effects in HNSCC. Mechanistically, we found that HIF-1α transcriptionally suppressed HOXA9 expression and that HOTTIP and CTCF cooperatively regulated HOXA9 in KB cells.
Results
HOXA9 Is Upregulated in HNSCC Tissues and Cells
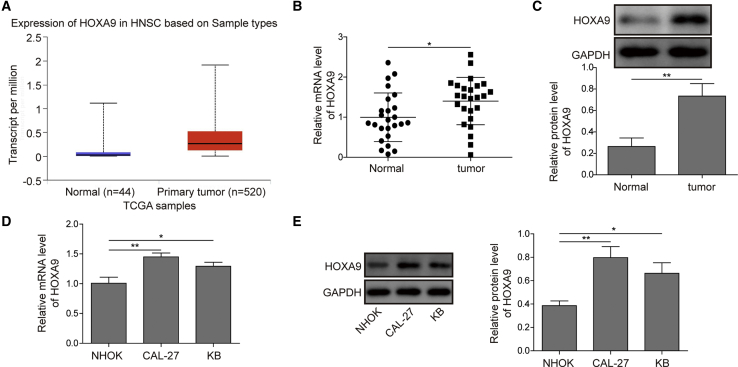
To determine the expression level of HOXA9, we first analyzed the mRNA level of HOXA9 using data from The Cancer Genome Atlas (TCGA) database. As shown in Figure 1A, HOXA9 was significantly elevated in HNSCC tissues. qRT-PCR was further performed with HNSCC tissues and normal counterparts. HOXA9 mRNA was markedly higher in HNSCC tissues compared with that in paired adjacent normal tissues (Figure 1B). A similar expression pattern was also observed at the protein level using western blotting (Figure 1C). In parallel, we examined the level of HOXA9 in normal human oral keratinocyte (NHOK) cells and two HNSCC cell lines. Consistently, the mRNA and protein levels of HOXA9 were significantly upregulated in CAL-27 and KB cells compared to those in NHOK cells (Figures 1D and 1E). Taken together, HOXA9 was significantly upregulated in HNSCC tissues and cells.
Figure 1.
HOXA9 Expression in HNSCC Tissues and Cell Lines
(A) Analysis of HOXA9 expression using TCGA data. (B) HOXA9 mRNA level in HNSCC tissues and normal counterparts were determined by qRT-PCR. GAPDH acted as an internal control. (C) The protein level of HOXA9 in HNSCC tissues and paired adjacent normal tissues were determined by western blotting. GAPDH served as a loading control. (D) HOXA9 mRNA levels in different cell lines were determined by qRT-PCR. GAPDH served as an internal control. (E) The protein level of HOXA9 in different cell lines was determined by western blotting. GAPDH served as a loading control. Data are representative images or expressed as the mean ± SD. ∗p < 0.05; ∗∗p < 0.01.
Knockdown of HOXA9 Inhibits Cell Proliferation, Migration, Invasion, and Chemoresistance but Promotes Apoptosis in CAL-27 and KB Cells
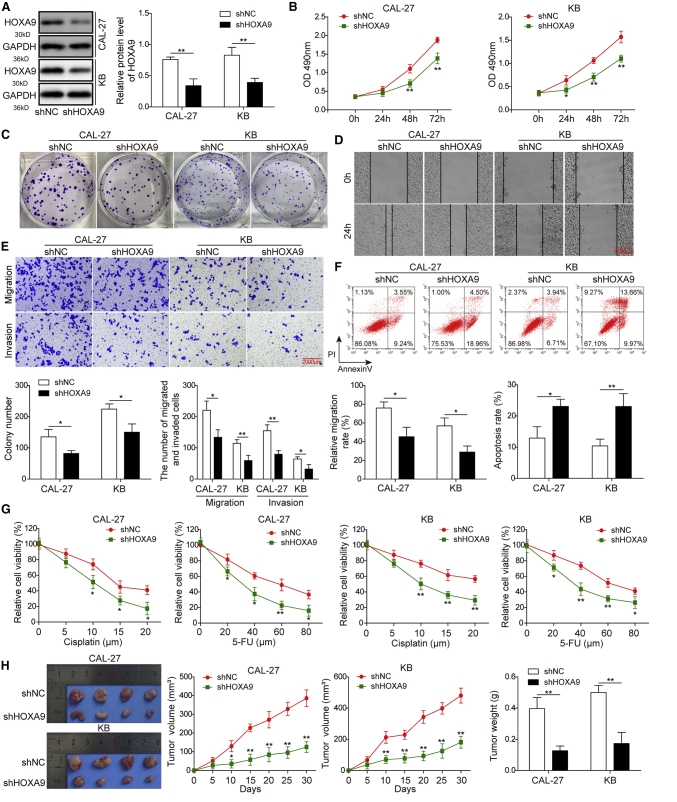
We further explored the potential effects of HOXA9 on cell growth, metastatic properties, chemoresistance, and apoptosis in HNSCC cells. Loss-of-function experiments were performed in CAL-27 and KB cells. As shown in Figure 2A, HOXA9 shRNA (sh-HOXA9 successfully decreased the expression of HOXA9 in both CAL-27 and KB cells. The Cell Counting Kit-8 (CCK-8) and colony formation assays indicated that knockdown of HOXA9 markedly attenuated cell proliferation and impaired clonogenic ability in CAL-27 and KB cells (Figures 2B and 2C). Wound-healing migration and Transwell assays revealed that silencing of HOXA9 significantly decreased migration and invasive capability compared with that in control cells (Figures 2D and 2E). In addition, we next examined the effect of HOXA9 on cell apoptosis and chemoresistance. Annexin V-fluorescein isothiocyanate/propidium iodide (FITC/PI) staining and cytotoxicity assay showed that knockdown of HOXA9 dramatically increased the proportion of apoptotic cells (Figure 2F) and increased sensitivity to cisplatin and 5-fluorouracil (5-FU) in both CAL-17 and KB cells (Figure 2G), respectively. Moreover, an in vivo xenograft study was conducted to validate the function of HOXA9 in cell growth. In accordance with in vitro findings, tumor growth was remarkably slower in the sh-HOXA9 group than in the non-specific sh-negative control (sh-NC) group (Figure 2H). Consistently, tumor weight was significantly lower in the sh-HOXA9 group than in the sh-NC group at 4 weeks after inoculation (Figure 2H). Taken together, these data suggest that knockdown of HOXA9 inhibits cell proliferation, migration, invasion, and chemoresistance but promotes apoptosis in CAL-27 and KB cells.
Figure 2.
HOXA9 Knockdown Inhibits HNSCC Cell Growth, Migration, Invasion, and Chemoresistance but Promote Apoptosis
(A) The protein level of HOXA9 was determined by western blotting. GAPDH served as a loading control. (B) Cell proliferation was monitored by CCK-8 assay. (C) Clonogenic ability was determined by colony formation assay. (D) The migration capacities were detected by wound-healing assay, scale bar: 5000 μm. (E) The migration and invasive capacities were detected by Transwell assays, scale bar: 2000 μm. (F) Cell apoptosis was detected by fluorescence-activated cell sorting (FACS) analysis. Early and late apoptotic cells were defined as PI−/Annexin V+ and PI−/Annexin V+, respectively. (G) CAL-27 or KB cells transfected with sh-NC or sh-HOXA9 were treated with different doses of cisplatin or 5-FU for 48 h. Cell cytotoxicity was monitored by CCK-8 assay. (H) 4 weeks after inoculation of cells transfected with sh-NC or sh-HOXA9, tumors were harvested from nude mice. Representative photographs of tumors at 4 weeks after inoculation. Tumor volumes were measured every week after inoculation. Tumor weights were measured at 4 weeks after inoculation. Error bars represent a mean ± SD of n = 3 experiments. ∗p < 0.05; ∗∗p < 0.01.
Knockdown of HOXA9 Regulates EMT-Related Markers via Targeting YAP1/β-Catenin
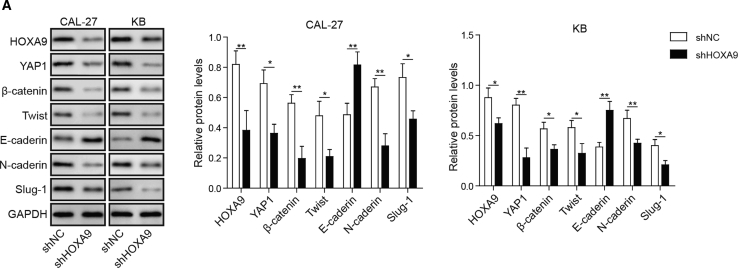
Epithelial-mesenchymal transition (EMT) is a well-characterized process that contributes to the migration and invasion of cancers. In order to further investigate the biological roles of HOXA9 on EMT, several known EMT or mesenchymal-epithelial transition (MET) biomarkers were detected by western blotting, including cell-surface proteins E-cadherin and N-cadherin, cytoskeleton protein β-catenin, and transcription factors Twist and Slug-1. Given the regulatory role of YAP1 on the β-catenin level in laryngeal cancer cells,18 we also examined the effect of YAP1 during EMT in HNSCC cells. The results showed that silencing of HOXA9 led to a significant reduction of YAP1, further inducing downregulation of β-catenin (Figure 3). And we also found that the expression levels of Twist, N-cadherin, and Slug-1 were downregulated, while E-cadherin was upregulated in HOXA9 knockdown in CAL-27 and KB cells (Figure 3). These data indicate that knockdown of HOXA9 regulates EMT-related markers via targeting YAP1/β-catenin.
Figure 3.
Knockdown of HOXA9 Regulates EMT-Related Markers via Targeting YAP1/β-Catenin
CAL-27 or KB cells were transfected with sh-NC or sh-HOXA9. Cells were harvested 48 h post-transfection. The protein levels of HOXA9, YAP1, β-catenin, Twist, E-cadherin, N-cadherin, and Slug-1 were determined by western blotting. GAPDH served as a loading control. Data are representative images or expressed as mean ± SD. ∗p < 0.05; ∗∗p < 0.01.
HIF-1α Transcriptionally Regulates HOXA9
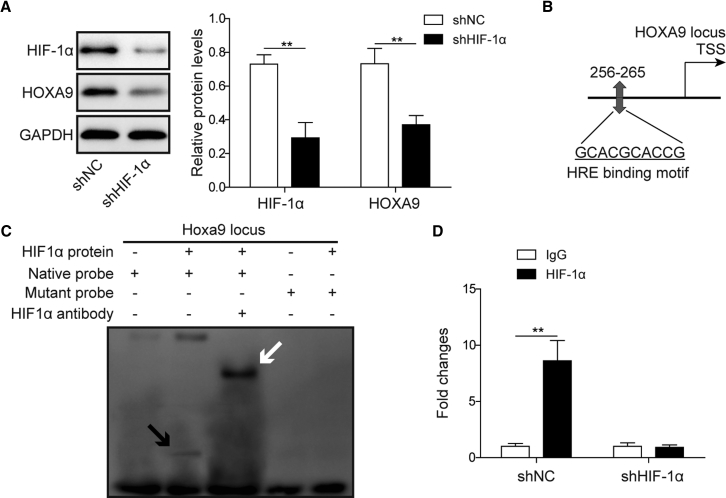
Previous studies have illustrated that HOXA9 regulates HIF-1α on the transcriptional level.19,20 Conversely, bioinformatics analysis predicted hypoxia response elements (HREs) in the HOXA9 promoter region using JASPAR (http://jaspar.genereg.net/). HIF-1α was identified as a putative transcription factor bound to the HOXA9 promoter using the University of California, Santa Cruz (UCSC) genome browser database (http://genome.ucsc.edu/), and the binding site was determined by using the JASPAR database. To further validate the results of bioinformatics analysis, we investigated the effect of sh-HIF-1α on HOXA9 expression. As shown in Figure 4A, HOXA9 expression was significantly decreased by sh-HIF-1α. An electrophoretic mobility shift assay (EMSA) was performed to detect the direct binding between purified HIF-1α protein and the predicted binding motif (Figure 4B). The results of EMSA showed that the DNA-protein complex was formed when the native probe was incubated with purified HIF-1α protein, whereas the mutated probe diminished the binding activity. An antibody supershift assay illustrated that HIF-1α was the transcription factor bound to this motif (Figure 4C). Chromatin immunoprecipitation (ChIP) assay further confirmed that lack of HIF-1α resulted in a significant reduction in the binding enrichment of HIF-1α at the binding site of HOXA9 (Figure 4D). In short, these data suggest that HIF-1α regulates HOXA9 on the transcriptional level.
Figure 4.
HIF-1α Transcriptionally Suppresses HOXA9
(A) KB cells transfected with sh-NC or sh-HIF-1α. The protein levels of HIF-1α and HOXA9 were determined by western blotting. GAPDH served as a loading control. (B) Predicted binding site of HIF-1α in the promoter region of HOXA9 as determined by JASPAR. (C) EMSA of predicted binding motif. The DNA-protein complex is indicated by a black arrow, and the antibody supershift band is indicated by a white arrow. (D) The binding enrichment of HIF-1α on the promoter region of HOXA9 was detected by ChIP-PCR. Data are representative images or expressed as mean ± SD. ∗p < 0.05; ∗∗p < 0.01.
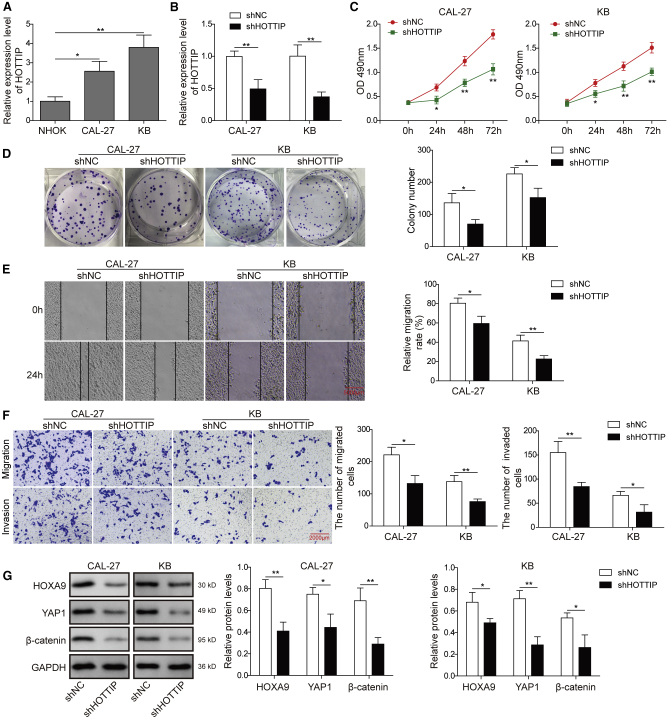
Knockdown of lncRNA HOTTIP Inhibits Cell Proliferation, Migration, and Invasion via Targeting HOXA9 in CAL-27 and KB Cells
Emerging evidence illustrated the critical roles of lncRNAs in a variety of cellular processes during tumor progression, including cell proliferation, migration, and invasion. A recent study has demonstrated that lncRNA HOTTIP promotes HOXA9 in pancreatic cancer stem cells (PCSCs) by binding to WDR5.15 To unravel the regulatory mechanism of HOXA9 in HNSCC cells, functional experiments were conducted to evaluate the biological role of HOTTIP in CAL-27 and KB cells. HOTTIP was significantly elevated in CAL-27 and KB cells compared with normal NHOK cells (Figure 5A). As shown in Figure 5B, sh-HOTTIP-mediated knockdown successfully decreased HOTTIP levels in both CAL-27 and KB cells. Both CCK-8 and colony formation assays showed that knockdown of HOTTIP inhibited cell growth in these two HNSCC cell lines (Figures 5C and 5D). Cell migration capability was monitored by wound-healing assay, and Figure 5E showed significant slower wound closure in sh-HOTTIP knockdown cells at 24 h after creation of the linear wounds. In addition, the Transwell assay revealed that knockdown of HOTTIP also inhibited the invasive and migrated capabilities in CAL-27 and KB cells (Figure 5F). It is apparent that HOTTIP exerted similar effects on cell proliferation, migration, and invasion as HOXA9 in HNSCC cells. To validate whether HOTTIP played oncogenic roles through HOXA9, the expression of HOXA9 and its downstream molecules were examined. As presented in Figure 5G, knockdown of HOTTIP caused a marked reduction of HOXA9. Consistent with previous results, downregulation of HOXA9 reduced YAP1 and EMT biomarker β-catenin in CAL-27 and KB cells (Figure 5G). Taken together, these results suggest that HOTTIP is highly expressed in HNSCC cells and that knockdown of HOTTIP inhibits cell proliferation, migration, and invasion via targeting HOXA9 in CAL-27 and KB cells.
Figure 5.
Knockdown of HOTTIP Inhibits Cell Proliferation, Migration, and Invasion via Targeting HOXA9 in CAL-27 and KB Cells
(A) HOTTIP mRNA level in different cell lines was determined by qRT-PCR. U6 served as an internal control. (B) CAL-27 or KB cells were transfected with sh-NC or sh-HOTTIP. The mRNA level of HOTTIP in different cell lines was determined by qRT-PCR. GAPDH served as an internal control. (C) Cell proliferation was monitored by CCK-8 assay. (D) Clonogenic ability was determined by colony formation assay. (E) Cell migration was monitored by wound-healing assay, scale bar: 5000 μm. (F) Cell-invasive and migrated capacities were detected by Transwell assays, scale bar: 2000 μm. (G) Protein levels of HOXA9, YAP1, and β-catenin were determined by western blotting. GAPDH served as a loading control. Data are representative images or expressed as the mean ± SD. ∗p < 0.05; ∗∗p < 0.01.
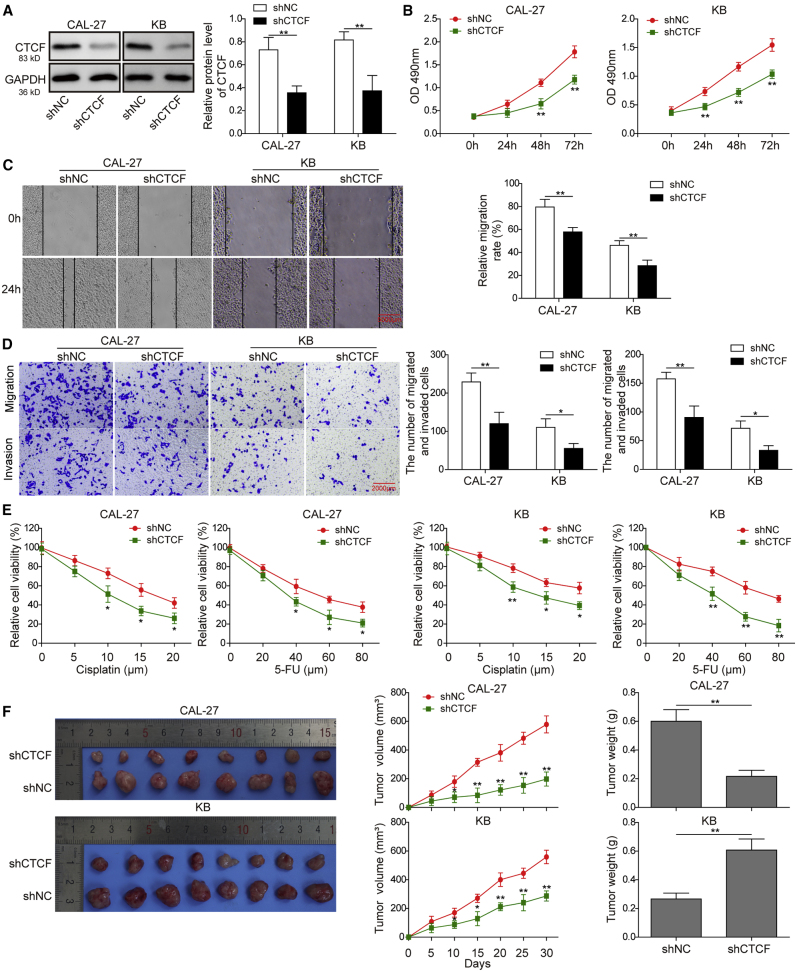
Knockdown of CTCF Inhibits Cell Proliferation, Migration, Invasion, and Chemoresistance in CAL-27 and KB Cells
A previous study has shown that HOTTIP physically associates with CTCF to coordinate HOXA gene expression.16 Hence, it is of interest to investigate the biological role of CTCF in HNSCC cells. As shown in Figure 6A, sh-CTCF dramatically decreased CTCF protein levels in CAL-27 and KB cells. Proliferation curves determined by CCK-8 assays showed that cells transfected with sh-CTCF grew more slowly than control cells (Figure 6B). Wound healing coupled with Transwell assays demonstrated that knockdown of CTCF remarkably inhibited cell migration and invasion in both CAL-27 and KB cells (Figures 6C and 6D). In addition, enhanced sensitivity to cisplatin or 5-FU treatment was found in CTCF-knockdown CAL-27 and KB cells (Figure 6E). Furthermore, we found that in the in vivo experiment tumor growth was suppressed and the tumor volumes and weights were significantly reduced in the sh-CTCF group (Figure 6F). Collectively, knockdown of CTCF inhibits cell proliferation, migration, invasion, and chemoresistance in CAL-27 and KB cells.
Figure 6.
Knockdown of CTCF Inhibits Cell Proliferation, Migration, Invasion, and Chemoresistance in CAL-27 and KB Cells
(A) CAL-27 or KB cells were transfected with sh-NC or sh-CTCF. Protein level of CTCF was determined by western blotting. GAPDH served as a loading control. (B) Cell proliferation was monitored by CCK-8 assay. (C) Cell migration capacity was monitored by wound-healing assay, scale bar: 5000 μm. (D) Cell migration and invasive capacities were detected by Transwell assays, scale bar: 2000 μm. (E) CAL-27 or KB cells transfected with sh-NC or sh-CTCF were treated with different doses of cisplatin or 5-FU for 48 h. Cell cytotoxicity was monitored by CCK-8 assay. (F) 4 weeks after inoculation of CAL-27 and KB cells transfected with sh-NC or sh-CTCF, tumors were harvested from nude mice. Representative photographs of tumors at 4 weeks after inoculation. Tumor volumes were measured every week after inoculation. Tumor weights were measured at 4 weeks after inoculation. Data are representative images or expressed as the mean ± SD. ∗p < 0.05; ∗∗p < 0.01.
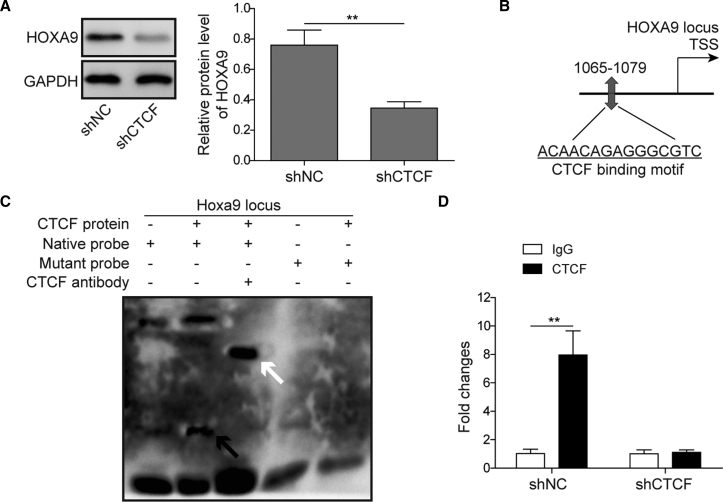
CTCF Transcriptionally Induces HOXA9 Expression
To further test whether CTCF was involved in the transcriptional regulation of HOXA9, a series of experiments were conducted. We found that knockdown of CTCF significantly decreased the HOXA9 protein level in HNSCC cells (Figure 7A). The predicted CTCF binding sites in the HOXA9 promoter region are shown in Figure 7B as determined by JASPAR. A subsequent EMSA showed that CTCF protein and native probe formed a DNA-protein complex, and an antibody supershift assay further confirmed that CTCF directly bound to native probe (Figure 7C). Consistently, a ChIP assay indicated that the binding enrichment of CTCF at the HOXA9 locus was significantly decreased by sh-CTCF (Figure 7D). These findings suggested that CTCF transcriptionally induces HOXA9 expression.
Figure 7.
CTCF Transcriptionally Induces HOXA9 Expression
(A) KB cells were transfected with sh-NC or sh-CTCF. The protein level of HOXA9 was determined by western blotting. GAPDH served as a loading control. (B) Predicted binding site of CTCF in the promoter region of HOXA9 as determined by JASPAR. (C) EMSA of predicted binding motif. The DNA-protein complex is indicated by a black arrow, and the antibody supershift band is indicated by a white arrow. (D) The binding enrichment of CTCF on the promoter region of HOXA9 was detected by ChIP-PCR. Data are representative images or expressed as the mean ± SD. ∗p < 0.05; ∗∗p < 0.01.
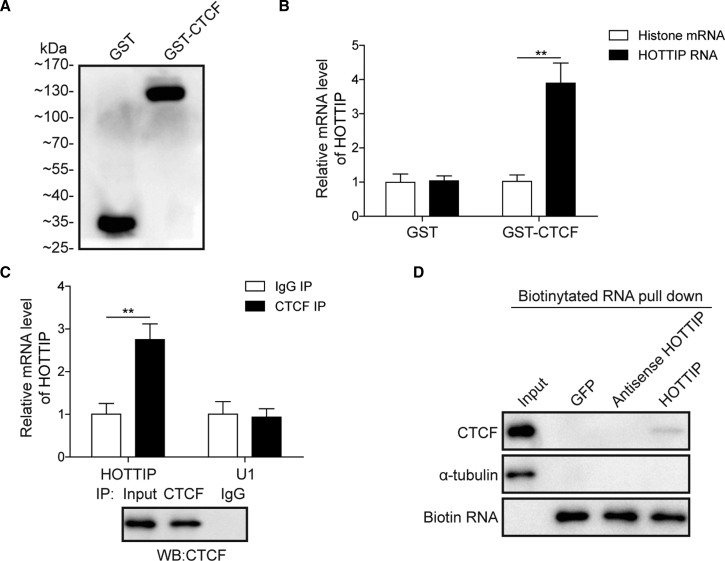
HOTTIP and CTCF Cooperatively Regulate HOXA9 in HNSCC Cells
In order to validate whether HOTTIP physically interacted with CTCF to modulate HOXA9 expression, recombinant glutathione S-transferase (GST)-tagged CTCF (GST-CTCF) was expressed and purified (Figure 8A). Purified GST-CTCF was then used as bait protein to retrieve full-length HOTTIP or control histone RNA. As shown in Figure 8B, HOTTIP RNA specifically bound to GST-CTCF but not to GST. Consistent with the GST pull-down assay, an immunoprecipitation (IP) assay showed that endogenous CTCF successfully retrieved HOTTIP but not negative control U1 spliceosomal RNA (Figure 8C, upper panel). The results of western blotting confirmed the success of IP (Figure 8C, lower panel). An RNA pull-down assay was further performed to verify the direct interaction between HOTTIP and CTCF. Biotinylated HOTTIP specifically retrieved CTCF, whereas GFP or antisense HOTTIP failed to capture CTCF (Figure 8D). These results suggest that HOTTIP physically interacts with CTCF to cooperatively regulate HOXA9 in HNSCC cells.
Figure 8.
HOTTIP and CTCF Cooperatively Regulate HOXA9 in HNSCC Cells
(A) Recombinant GST and GST-tagged CTCF were purified and detected by western blotting. (B) Purified GST and GST-tagged CTCF were used as bait protein to retrieve HOTTIP or control histone mRNA. (C) Interaction between endogenous CTCF and HOTTIP was determined by IP. U1 spliceosomal RNA acted as negative control. Successful IP was confirmed by western blotting. (D) Directed interaction between CTCF and HOTTIP was verified by RNA pull-down assay. α-Tubulin served as negative control. Data are representative images or expressed as the mean ± SD. ∗p < 0.05; ∗∗p < 0.01.
Discussion
HOX genes were dysregulated in most solid tumors, and they acted as either transcriptional activators or repressors to promote tumorigenesis.7 Of the 39 human HOX family members, HOXA9 was reported to be dysregulated in leukemia and in breast and ovarian cancers.9,10,21 Interestingly, our findings showed that HOXA9 was highly expressed in HNSCC compared to their normal tissue counterparts, which was consistent with TCGA data. These results indicate that HOXA9 might play critical roles in HNSCC. In vitro and in vivo experiments showed that HOXA9 promoted cell proliferation, migration, invasion, and chemoresistance in HNSCC cells. In addition, subsequent knockdown experiments revealed that knockdown of HOXA9 decreased YAP1 expression, thereby leading to a reduction of β-catenin. A previous study has demonstrated that YAP1 regulates Hep-2 laryngeal cancer cell growth via mediating EMT and the Wnt/β-catenin pathway.18 These findings suggest that YAP1 plays an important role in regulating EMT in HNSCC cells via modulating β-catenin expression. Future investigations are needed to connect HOXA9 and YAP1 in HNSCC.
Hypoxia has been recognized as a common character of solid tumors due to the increase in tumor volume and poorly formed vasculature. It is well established that HIF-1α plays a crucial role in the adaptive responses of the cancer cells in the harsh microenvironment. For instance, HIF-1α transcriptionally regulates downstream genes that are involved in cell proliferation, migration, invasion, glucose metabolism, and angiogenesis.22 In the present study, bioinformatic analysis coupled with EMSA and ChIP assays showed that HIF-1α positively regulated HOXA9 expression via directly binding to its promoter region. On the contrary, a previous study has illustrated that HOXA9 inhibits glycolysis in cutaneous squamous cell carcinoma by negatively regulating HIF-1α and its downstream effectors.20 Our findings identified a feedback regulation between HOXA9 and HIF-1α.
A number of lncRNAs are dysregulated in HNSCC and correlated with tumor progression, metastasis, clinical stage, and poor prognosis.23 Bioinformatic analysis has illustrated the prognostic value of HOTTIP in HNSCC. HOTTIP is highly expressed in HNSCC tissues and cells and correlated with the clinical stage and histological grade in HNSCC patients,24 indicating that HOTTIP might act as a key candidate biomarker in HNSCC. Consistent with this study, our results showed that HOTTIP was upregulated in CAL-27 and KB cells and that it played an oncogenic role in HNSCC cells. In addition, EMSA, ChIP, GST pull-down, and RNA pull-down assays demonstrated that HOTTIP and CTCF worked in concert to modulate HOXA9 expression in HNSCC cells. These findings were in accordance with a previous study that illustrated the functional cooperation between HOTTIP and CTCF in human foreskin fibroblasts.16
In conclusion, HIF-1α or HOTTIP/CTCF transcriptionally modulates HOXA9 expression to regulate HNSCC progression and drug resistance.
Materials and Methods
Human Tissues and Cells
A cohort of 25 HNSCC tumor tissue and normal tissue counterparts were collected post-operatively from patients with HNSCC in The First Affiliated Hospital of Zhengzhou University. Consents were obtained from all patients.
Human oral squamous cell carcinoma cell line CAL-27 cells, head and neck squamous cell carcinoma cell line KB cells, and NHOK cells were purchased from the Cell Bank of Type Culture Collection, Chinese Academy of Science (Shanghai, China). Cells were grown in DMEM containing 10% fetal bovine serum (FBS) (GIBCO, Thermo Fisher Scientific, Waltham, MA, USA), 10 μg/mL streptomycin and 100 U/mL penicillin. Cultures were maintained at 37°C in humidified atmosphere with 5% CO2 in air.
Cell Transduction and Transfection
sh-NC, sh-HOXA9, sh-HOTTIP, and sh-HIF-1α were designed and synthesized by Hanbio (Shanghai, China). HEK293 cells were used to produce adenovirus using Lipofiter Transfection Reagent (Hanbio) according to the manufacturer’s instructions. Adenoviruses were purified and titered. CAL-27 or KB cells were infected with adenovirus. After 48 h, the infected cells were subjected to subsequent analysis.
qRT-PCR
Total RNA was isolated from tissues or cells using TRIzol Reagent (Invitrogen, Thermo Fisher Scientific) following the manufacturer’s protocols. cDNA was reverse transcripted using SuperScript III reverse transcriptase (Invitrogen), and qPCR was performed using PowerUp SYBR Green Master Mix (Applied Biosystems, Thermo Fisher Scientific) and the following primers targeting the indicated human gene: HOTTIP, forward primer, 5′-CACACTCACATTCGCACACT-3′, and reverse primer 5′-TCCAGAACTAAGCCAGCCATA-3′; HOXA9, forward primer, 5′-TGATTATTTTTGTTTTAGGAGTCGT-3′, and reverse primer, 5′- TAAAAAAATTTATTTCTCACCCGTT-3′; U6 (internal control for lncRNA), forward primer, 5′-CTCGCTTCGGCAGCACA-3′, and reverse primer 5′-AACGCTTCACGAATTTGCGT-3′; and GAPDH (internal control for mRNAs), forward primer, 5′-TGTGGGCATCAATGGATTTGG-3′, and reverse primer, 5′-ACACCATGTATTCCGGGTCAAT-3′. The relative expression of a target gene to that of the internal control was calculated using the 2−ΔΔCt method.
Western Blot Analysis
Protein lysates from tissues or cells were prepared in IP lysis buffer supplemented with protease inhibitor cocktail (Pierce, Thermo Fisher Scientific). Protein estimation was performed using the BCA Protein Assay Kit (Pierce). Equal amounts of protein lysates were resolved by SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membrane for western blotting. Membranes were blocked with 5% non-fat milk, followed by incubation with primary antibody at 4°C overnight: anti-HOXA9 (ab140631, Abcam, Cambridge, UK), anti-β-catenin (71-2700, Invitrogen), anti-Twist (ab50581, Abcam), anti-E-cadherin (3195, Cell Signaling Technologies, Beverly, MA, USA), anti-N-cadherin (33-3900, Invitrogen), anti-Slug-1 (9585, Cell Signaling Technologies), anti-YAP1 (sc-15407, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-HIF-1α (sc-10790, Santa Cruz), anti-CTCF (ab70303, Abcam), and anti-GAPDH (sc-25778, Santa Cruz). Membranes were then incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody (1:5,000, Bioworld Technology, Louis Park, MN, USA) at room temperature for 1 h. Western blots were visualized using Immobilon Western Chemiluminescent HRP Substrate (Merck Millipore, Burlington, MA, USA) followed by film exposure. The relative expression of a target protein was normalized with internal control.
CCK-8 Assay
Cell viability in cell proliferation or cytotoxicity assays was determined by using the CCK-8 assay (Beyotime, Haimen, China) according to the manufacturer’s instructions. In brief, cells (2 × 104 cells per milliliter) were seeded in 96-well plates 24 h prior to the treatment. For cytotoxicity assay, cells were then treated with different doses of cisplatin or 5-FU (Sigma-Aldrich, St. Louis, MO, USA) for 48 h. 10 μL CCK-8 solution was added into each well and incubated for 1 h at 37°C. Absorbance was measured at a wavelength of 490 nm by the use of a microplate reader (Bio-Rad Laboratories, Hercules, CA, USA).
Annexin V-FITC/PI Staining
Cell apoptosis was assessed using the Dead Cell Apoptosis Kit with Annexin V-FITC and PI (Thermo Fisher Scientific). In brief, cells were harvested and resuspended in binding buffer at 24 h post-transfection. Dual staining of Annexin V-FITC and PI was performed according to the manufacturer’s instructions. 100 μL cell suspension (1 × 105 cells) was added into a test tube and stained with 5 μL Annexin V-FITC reagent and 1 μL PI solution for 15 min at room temperature. 400 μL binding buffer was then added into each tube. The stained cells were analyzed by flow cytometry with BD FACSCalibur (BD Biosciences, San Diego, CA, USA).
Colony Formation Assay
CAL-27 or KB cells were seeded on 60-mm culture plates (2 × 102 cells per plate) and transfected with shRNA or corresponding control. After 10 days, the colonies were fixed with 10% formaldehyde for 5 min and stained with 1% crystal violet for 30 s. Viable containing at least 50 cells were counted.
Wound-Healing Migration Assay
CAL-27 and KB cells were seeded in 6-well plates for 100% confluence in 24 h. The cell monolayer was lightly scratched using a sterile 1,000-μL micropipette tip. The cell debris was carefully removed. Wound closure was observed and evaluated in five random fields using an inverted microscope (Carl Zeiss, Jena, Germany) at 24 h. Plates were photographed at 0 and 24 h after scratching at an identical location, respectively. The width, W, of the scratch measured; the wound closure was calculated as (W0h − W24h)/W0h × 100%. All experiments were performed in triplicate.
Transwell Invasion and Migration Assay
For Transwell invasion assay, Transwell chambers (Corning, Lowell, MA, USA) were coated with Matrigel (BD Biosciences). At 24 h post-transfection, cells were deprived of FBS for 6 h. CAL-27 and KB cells were then seeded in the upper chambers. The lower chambers were filled with DMEM supplemented with 10% FBS. After a 48-h incubation, cells remaining in the upper chamber were removed with cotton swabs. Cells that migrated to the lower chambers were fixed with 4% paraformaldehyde (PFA) and stained with 0.2% crystal violet. The number of invading cells was counted under a light microscope. Transwell migration assays were performed with the similar approach without the coating of Matrigel. All experiments were performed in triplicate.
In Vivo Xenograft Assay
Female BALB/c nude mice (6- to 8-week-old; n = 20) were purchased from Shanghai SLAC Laboratory Animal. All mice were housed in a temperature-controlled environment and maintained on a 12-h/12-h light/dark cycle. All experiments were approved by the Animal Care and Use Committee of The First Affiliated Hospital of Zhengzhou University. Stable transfected CAL-27 or KB cells were implanted subcutaneously under the mouse dorsum. The length, L, and width, W, of the tumors were measured weekly; the tumor volume, V, was calculated using the formula: V = L × W2/2. The tumors were harvested and weighted after 4 weeks.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was conducted using the Pierce Agarose ChIP Kit (Pierce) according to the manufacturer’s instructions. In brief, KB cells were transfected with sh-NC or sh-HIF-1α/sh-CTCF. Cells were crosslinked with 1% formaldehyde and lysed to prepare nuclei. Chromatin was sheared using micronuclease digestion. Sheared DNA was then incubated with anti-HIF-1α/anti-CTCF antibody. Normal immunoglobulin G (IgG) was served as a negative control. DNA was purified and analyzed by qRT-PCR.
EMSA
Recombinant HIF-1α and CTCF protein were purchased from Abcam. Native or mutated probe was biotinylated using the Biotin 3′ End DNA Labeling Kit (Pierce) and annealed at room temperature for 1 h. EMSA was performed using the Lightshift Chemiluminescent DNA EMSA Kit (Pierce) according to the manufacturer’s instructions. Briefly, binding reactions were performed with or without anti-HIF-1α/anti-CTCF antibody. DNA-protein complexes were electrophoresed and transferred onto a positive-charged nylon membrane (Pierce), followed by UV light crosslinking. The signal was visualized with chemiluminescent substrate followed by film exposure.
GST Pull-Down Assay
The GST pull-down assay was performed as previously described.16 Briefly, GST-tagged CTCF was expressed in E. coli, purified, and bound to glutathione beads (GE Healthcare, Buckinghamshire, UK) as the bait protein. HOTTIP and histone H2B1 mRNA were transcribed in vitro using T7 polymerase (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The mRNAs were then denatured, refolded, and incubated with the CTCF-bound beads at room temperature for 1 h. The bound RNAs were extracted and determined by qRT-PCR.
RNA Pull-Down Assay
The RNAs were transcribed in vitro using T7 RNA Polymerase (Promega) and biotin labeled using the Pierce RNA 3′ End Desthiobiotinylation Kit (Pierce). RNA pull-down assay was performed using the Pierce Magnetic RNA-Protein Pull-Down Kit (Pierce) according the manufacturer’s instructions. In brief, biotinylated RNA was incubated with streptavidin beads and mixed with cell lysates. The eluted RNA-binding proteins were detected by western blotting.
Statistical Analysis
All experiments were performed at least three times. Data are presented as the means ± SD. Statistical analysis was performed using Student’s t test (two-tailed) between two groups or one-way analysis of variance (ANOVA) followed by Tukey post hoc test for multiple comparison. Statistical analysis was performed using SPSS v.22.0 (SPSS, Chicago, IL, USA). Differences were considered significant if p < 0.05 (∗∗p < 0.01).
Author Contributions
Guarantor of integrity of the entire study: Ming-Lei Sun, Study concepts: Ming-lei Sun, Study design: Qiang Sun, Definition of intellectual content: Qiang Sun, Literature research: Shuai-Yuan Zhang, Clinical studies: Jun-Fang Zhao, Experimental studies: Shuai-Yuan Zhang, Data analysis: Hai-bin Wang, Statistical analysis: Jun-Fang Zhao, Manuscript preparation: Shuai-Yuan Zhang, Manuscript editing: Qiang Sun, Manuscript review: Xin-Guang Han.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the Health Commission of Henan Province Project (2018048), the Basic and Frontier Technology Research Projects by the Science and Technology Department of Henan Province (142300410315 and 172102310262), the Oral and Maxillofacial Surgery Academician Workstation of Zhengzhou (152PYSGZ040), and Key Scientific Research Project for Colleges and Universities by Education Department of Henan Province (13A320452).
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Leemans C.R., Braakhuis B.J., Brakenhoff R.H. The molecular biology of head and neck cancer. Nat. Rev. Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 3.Sturgis E.M., Cinciripini P.M. Trends in head and neck cancer incidence in relation to smoking prevalence: an emerging epidemic of human papillomavirus-associated cancers? Cancer. 2007;110:1429–1435. doi: 10.1002/cncr.22963. [DOI] [PubMed] [Google Scholar]
- 4.Jung A.C., Briolat J., Millon R., de Reyniès A., Rickman D., Thomas E., Abecassis J., Clavel C., Wasylyk B. Biological and clinical relevance of transcriptionally active human papillomavirus (HPV) infection in oropharynx squamous cell carcinoma. Int. J. Cancer. 2010;126:1882–1894. doi: 10.1002/ijc.24911. [DOI] [PubMed] [Google Scholar]
- 5.Gupta S., Kong W., Peng Y., Miao Q., Mackillop W.J. Temporal trends in the incidence and survival of cancers of the upper aerodigestive tract in Ontario and the United States. Int. J. Cancer. 2009;125:2159–2165. doi: 10.1002/ijc.24533. [DOI] [PubMed] [Google Scholar]
- 6.Gehring W.J., Hiromi Y. Homeotic genes and the homeobox. Annu. Rev. Genet. 1986;20:147–173. doi: 10.1146/annurev.ge.20.120186.001051. [DOI] [PubMed] [Google Scholar]
- 7.Bhatlekar S., Fields J.Z., Boman B.M. HOX genes and their role in the development of human cancers. J. Mol. Med. (Berl.) 2014;92:811–823. doi: 10.1007/s00109-014-1181-y. [DOI] [PubMed] [Google Scholar]
- 8.Dorsam S.T., Ferrell C.M., Dorsam G.P., Derynck M.K., Vijapurkar U., Khodabakhsh D., Pau B., Bernstein H., Haqq C.M., Largman C., Lawrence H.J. The transcriptome of the leukemogenic homeoprotein HOXA9 in human hematopoietic cells. Blood. 2004;103:1676–1684. doi: 10.1182/blood-2003-07-2202. [DOI] [PubMed] [Google Scholar]
- 9.Ko S.Y., Barengo N., Ladanyi A., Lee J.S., Marini F., Lengyel E., Naora H. HOXA9 promotes ovarian cancer growth by stimulating cancer-associated fibroblasts. J. Clin. Invest. 2012;122:3603–3617. doi: 10.1172/JCI62229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun M., Song C.X., Huang H., Frankenberger C.A., Sankarasharma D., Gomes S., Chen P., Chen J., Chada K.K., He C., Rosner M.R. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc. Natl. Acad. Sci. USA. 2013;110:9920–9925. doi: 10.1073/pnas.1305172110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alvarado-Ruiz L., Martinez-Silva M.G., Torres-Reyes L.A., Pina-Sanchez P., Ortiz-Lazareno P., Bravo-Cuellar A., Aguilar-Lemarroy A., Jave-Suarez L.F. HOXA9 is underexpressed in cervical cancer cells and its restoration decreases proliferation, migration and expression of epithelial-to-mesenchymal transition genes. Asian Pac. J. Cancer Prev. 2016;17:1037–1047. doi: 10.7314/apjcp.2016.17.3.1037. [DOI] [PubMed] [Google Scholar]
- 12.Zhou C., Li J., Li Q., Liu H., Ye D., Wu Z., Shen Z., Deng H. The clinical significance of HOXA9 promoter hypermethylation in head and neck squamous cell carcinoma. J. Clin. Lab. Anal. 2019;33:e22873. doi: 10.1002/jcla.22873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheetham S.W., Gruhl F., Mattick J.S., Dinger M.E. Long noncoding RNAs and the genetics of cancer. Br. J. Cancer. 2013;108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Z., Chen C., Zhou Q., Wang Y., Zhao Y., Zhao X., Li W., Zheng S., Ye H., Wang L. LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. 2017;410:68–81. doi: 10.1016/j.canlet.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 16.Wang F., Tang Z., Shao H., Guo J., Tan T., Dong Y., Lin L. Long noncoding RNA HOTTIP cooperates with CCCTC-binding factor to coordinate HOXA gene expression. Biochem. Biophys. Res. Commun. 2018;500:852–859. doi: 10.1016/j.bbrc.2018.04.173. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H., Zhao L., Wang Y.X., Xi M., Liu S.L., Luo L.L. Long non-coding RNA HOTTIP is correlated with progression and prognosis in tongue squamous cell carcinoma. Tumour Biol. 2015;36:8805–8809. doi: 10.1007/s13277-015-3645-2. [DOI] [PubMed] [Google Scholar]
- 18.Tang X., Sun Y., Wan G., Sun J., Sun J., Pan C. Knockdown of YAP inhibits growth in Hep-2 laryngeal cancer cells via epithelial-mesenchymal transition and the Wnt/β-catenin pathway. BMC Cancer. 2019;19:654. doi: 10.1186/s12885-019-5832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kocabas F., Xie L., Xie J., Yu Z., DeBerardinis R.J., Kimura W., Thet S., Elshamy A.F., Abouellail H., Muralidhar S. Hypoxic metabolism in human hematopoietic stem cells. Cell Biosci. 2015;5:39. doi: 10.1186/s13578-015-0020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou L., Wang Y., Zhou M., Zhang Y., Wang P., Li X., Yang J., Wang H., Ding Z. HOXA9 inhibits HIF-1α-mediated glycolysis through interacting with CRIP2 to repress cutaneous squamous cell carcinoma development. Nat. Commun. 2018;9:1480. doi: 10.1038/s41467-018-03914-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins C.T., Hess J.L. Role of HOXA9 in leukemia: dysregulation, cofactors and essential targets. Oncogene. 2016;35:1090–1098. doi: 10.1038/onc.2015.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masoud G.N., Li W. HIF-1α pathway: role, regulation and intervention for cancer therapy. Acta Pharm. Sin. B. 2015;5:378–389. doi: 10.1016/j.apsb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guglas K., Bogaczyńska M., Kolenda T., Ryś M., Teresiak A., Bliźniak R., Łasińska I., Mackiewicz J., Lamperska K. lncRNA in HNSCC: challenges and potential. Contemp. Oncol. (Pozn.) 2017;21:259–266. doi: 10.5114/wo.2017.72382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin X., Yang W., Xie J., Wei Z., Tang C., Song C., Wang Y., Cai Y., Xu W., Han W. HOTTIP functions as a key candidate biomarker in head and neck squamous cell carcinoma by integrated bioinformatic analysis. BioMed Res. Int. 2019;2019:5450617. doi: 10.1155/2019/5450617. [DOI] [PMC free article] [PubMed] [Google Scholar]