Abstract
Despite the benefits associated with radiotherapy and chemotherapy for glioblastoma (GBM) treatment, most patients experience a relapse following initial therapy. Recurrent or progressive GBM usually does not respond anymore to standard therapy, and this is associated with poor patient outcome. GBM stem cells (GSCs) are a subset of cells resistant to radiotherapy and chemotherapy and play a role in tumor recurrence. The targeting of GSCs and the identification of novel markers are crucial issues in the development of innovative strategies for GBM eradication. By differential cell SELEX (systematic evolution of ligands by exponential enrichment), we have recently described two RNA aptamers, that is, the 40L sequence and its truncated form A40s, able to bind the cell surface of human GSCs. Both aptamers were selective for stem-like growing GBM cells and are rapidly internalized into target cells. In this study, we demonstrate that their binding to cells is mediated by direct recognition of the ephrin type-A receptor 2 (EphA2). Functionally, the two aptamers were able to inhibit cell growth, stemness, and migration of GSCs. Furthermore, A40s was able to cross the blood-brain barrier (BBB) and was stable in serum in in vitro experiments. These results suggest that 40L and A40s represent innovative potential therapeutic tools for GBM.
Keywords: glioblastoma, cancer stem cell, aptamer, EphA2, GSCs, RNA, cancer therapy, therapeutic RNAs
Introduction
Among the malignancies of the brain, glioblastoma (GBM) has the worst prognosis, with a median survival of 15 months.1 The disease relapse rate is very high. Very often the disease rapidly evolves and patients succumb in a few months. The current state of care for patients suffering of GBM is surgical resection, when possible, followed by temozolomide (TMZ) chemotherapy combined with radiotherapy. However, in patients undergoing relapse the standard therapy has minimal benefits.1 The improvement in omics approaches to GBM has led to the identification of specific molecular markers able to stratify patient subgroups and to better improve GBM treatment and survival prediction. The genetic loss of chromosomes 1p/19q is predictive of an increase in sensitivity to both radiotherapy and chemotherapy.2 The presence of mutations of IDH1 or IDH2 genes is indicative of an evolution of the GBM from a low-grade glioma, and usually their presence is associated with a more favorable prognosis, whereas the overexpression of epidermal growth factor receptor (EGFR) variant III is usually associated with a worse prognosis. Methylation of the O-6-methylguanine-DNA methyltransferase (MGMT) gene promoter is usually associated with a better response to TMZ treatment and it characterizes a subpopulation with a better prognosis. Identification of new biomarkers is needed for a better sub-stratification of patients and to adopt new therapeutic strategies.3
GBM stem cells (GSCs) are a heterogeneous subpopulation of cells characterized by an increased resistance to conventional GBM therapies and could cause tumor relapse. Therefore, identification and eradication of GSCs are an important challenge to treat GBM.4,5
Within key molecules in GBM development are the Eph (erythropoietin-producing hepatocellular carcinoma) receptors family members. These receptors are mainly expressed in early development and are crucial for embryonic development, regulating processes such as cell migration and adhesion.6 Expression of Eph receptors that is very low in adult and differentiated tissues becomes upregulated in a number of human malignancies such as melanoma and breast, lung, and ovarian cancer, as well as glioma.7,8 In these malignancies, contrary to an Eph receptor, the endogenous ligand ephrin appears downregulated.9 In GBM, Eph receptor A2 (EphA2) has been proposed as a novel molecular marker and therapeutic target since it is strongly overexpressed in GBM cells but not in normal brain.10,11 The overexpression of EphA2 correlates with poor patient outcome, and it is essential in the maintenance of the pool of GSCs, promoting their invasiveness in vivo.12 EphA2 overexpression promotes GSC tumorigenesis in GBM, and its blockage strongly induces a tumor-suppressive phenotype.13 Moreover, EphA2 is co-expressed with other stem cell markers such as CD133 and integrin alpha 7.13,14
Aptamers are short sequences of single-stranded oligonucleotides generated by a SELEX (systematic evolution of ligands by exponential enrichment) approach; they bind to molecular targets in the same fashion as antibodies.15 Due to their intrinsic properties, such as selectivity, easy modification, low immunogenicity, and high affinity for their targets, they appear to be an ideal novel therapeutic agent for GBM treatment to improve the standard of care.
In this study we characterized biological functions of the 2′-fluoropyrimidine-containing RNA aptamer 40L and its truncated form A40s, recently identified by our group.16 These two aptamers (long and short) are able to specifically recognize GSCs and to discriminate them from differentiated cells. Both aptamers appear to be functionally active on GSCs, inhibiting stem properties, growth, and migration. We characterized 40L and A40s as a high-affinity ligand for EphA2, which is overexpressed by a subset of GSCs grown as tumorspheres.
Results
Functional Effects of Aptamers
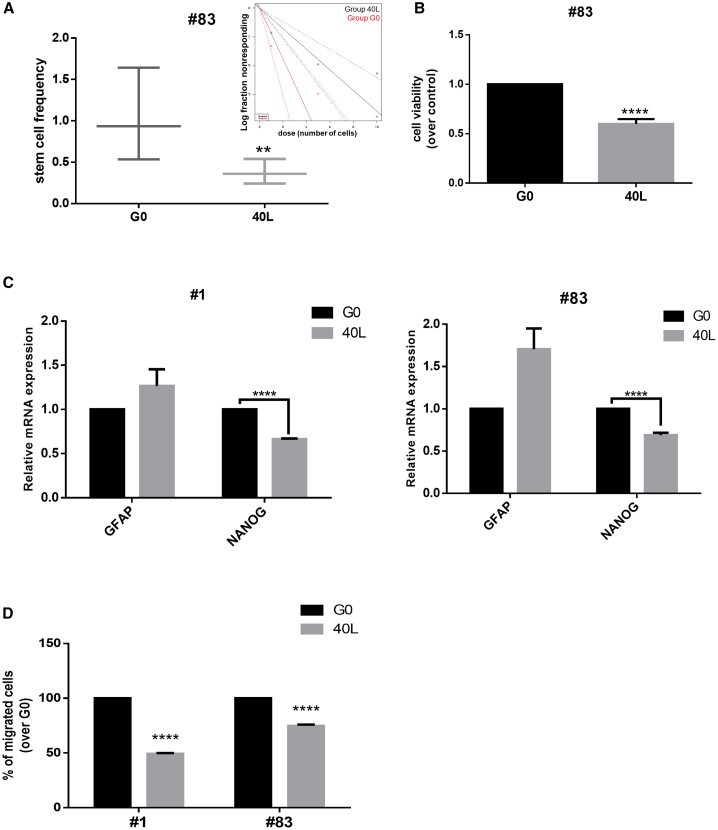
To determine the functional effects of aptamers on GSC growth, we used primary stem cells derived from GBM patients. We first performed a limiting dilution assay (LDA) on GSC#83 upon treatment with the 40L aptamer. Data were analyzed using ELDA (extreme limiting dilution analysis) software.17 Cells treated with 40L for 2 weeks showed an approximately 50% reduction in the estimated stem cell frequency compared to the starting pool of the aptamer selection G0, used as a control (Figure 1A). Furthermore, as determined by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, 40L inhibited cell viability by about 50% after 6 days of treatment (Figure 1B). We also assessed stem cell/differentiation marker expressions on 40L incubation on GSC#83 and GSC#1. We found that 400 nM of 40L induced downregulation of the stem cell-specific transcriptional factor Nanog and upregulation of the astrocyte differentiation marker glial fibrillary acidic protein (GFAP) (Figure 1C). We then compared the effects of 40L on GSC#1 and GSC#83 on cell migration. We found that 40L interfered with the ability of both cell lines to migrate (Figure 1D).
Figure 1.
Functional Activity of 40L
(A) 20 wells per dose of cells were treated with the aptamer, and limiting dilution analysis (LDA) was performed. Confidence intervals for stem cell frequency are shown. Estimates of stem cell frequency are reported in the graph; bars indicate lower and upper confidence intervals for stem cell frequency as calculated by ELDA software. (B) Stem cells were incubated with 400 nM of 40L and cell viability was evaluated by an MTT assay after 6 days of treatment. Results are presented as mean ± SD of three independent experiments. (C) Quantitative real-time PCR was performed to analyze GFAP and Nanog levels in GSCs#1 or GSCs#83 upon 2 weeks of treatment with 40L at 400 nM. Representative experiments are shown and results are expressed relative to the background binding, detected with the starting pool of sequences used for selection. (D) Overnight cell migration was analyzed by a transwell migration assay upon 72 h of treatment with 400 nM of 40L aptamer. A representative experiment is shown. Vertical bars indicate standard deviation values. **p ≤ 0.01, ****p ≤ 0.0001.
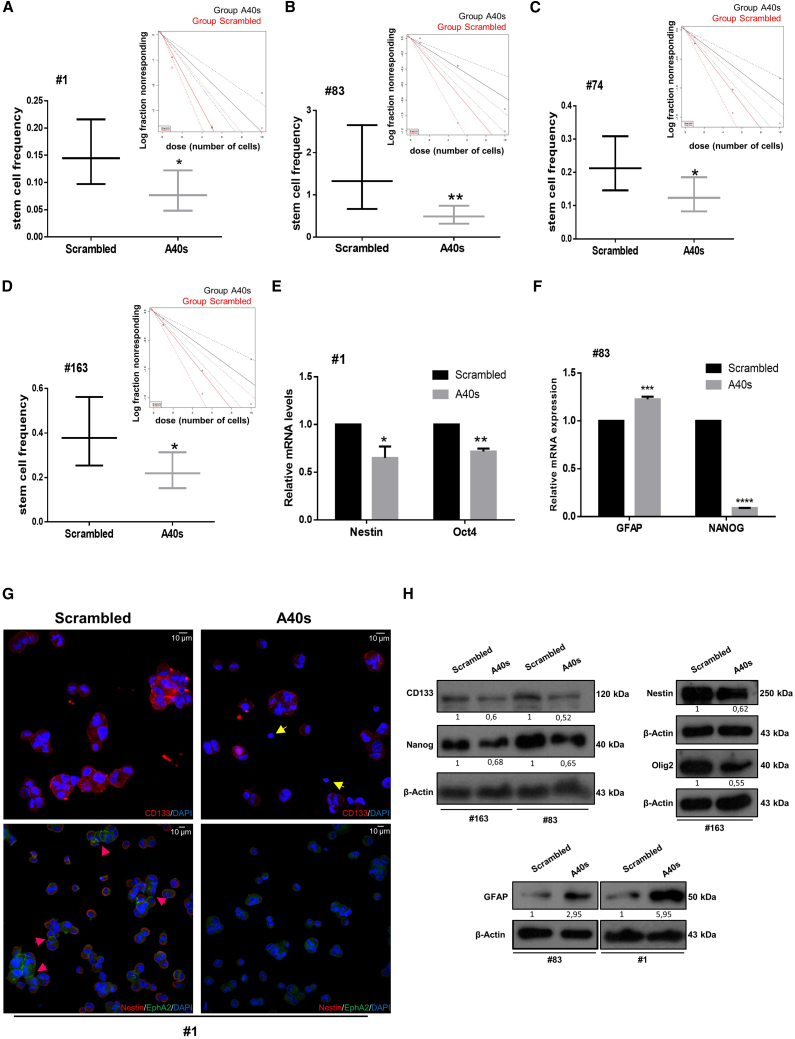
Next, we tested whether the truncated aptamer preserves the functional properties of the long sequence, evaluating the efficacy of A40s to reduce colony formation with a LDA. We found that similar to the long aptamer, A40s reduced stem cell frequency about 40%–50% in GSCs of several patients (#1, #83, #74, and #163) (Figures 2A–2D).
Figure 2.
A40s Effects on GSC Stemness
(A–D) LDA was performed using ELDA (A, #1; B, #83; C, #74; D, #163). Confidence intervals for stem cell frequency are shown. Estimates stem cell frequency are reported in the graph; bars indicate lower and upper confidence intervals for stem cell frequency, as calculated by ELDA software. (E–H) Changes in GSC marker expression are shown after treating the cells for 2 weeks with 400 nM A40s sequences or negative control. (E and F) Quantitative real-time PCR was performed to analyze stemness markers levels in GSCs #1 (E) and #83 (F). Representative experiments are shown, and results are expressed in relationship to the background effect detected by using a scrambled sequence. *p ≤ 0.05, **p ≤ 0.01. (G) Confocal microscopic images display CD133 (red), Nestin (red), and EphA2 (green) staining in GSCs. Yellow arrows show CD133-negative cells, and pink arrows indicate cell surface localization of EphA2. (H) Western blots show downregulation and upregulation of principal GBM stemness and differentiation markers, respectively.
We also assessed stem cell/differentiation marker expression on A40s incubation. A40s is able to affect stemness, inducing downregulation of stem cell markers such as Nestin, Oct4, CD133, Nanog, and Olig2 as detected by qRT-PCR, immunofluorescence, or western blot analysis (Figures 2E–2H). Furthermore, A40s treatment increases the differentiation marker GFAP (Figures 2F and 2H), indicating that the active functional site of 40L is preserved in the shorter A40s aptamer.
Moreover, as for the long aptamer, A40s demonstrated reductions in both GSC viability and migration (Figures S1A and S1B). Conversely, no functional effect was detected on differentiated cells. In fact, as shown in Figures S1C and S1D, no change was observed in viability and migratory ability upon A40s treatment in differentiated cells. Taken together, these results indicate that once bound to GSCs, 40L and A40s elicit an intrinsic biological activity that is expressed by a strong reduction of stemness characteristics, suggesting that these aptamers could be used to target stem cell phenotypes.
EphA2 Is an A40s and 40L Target
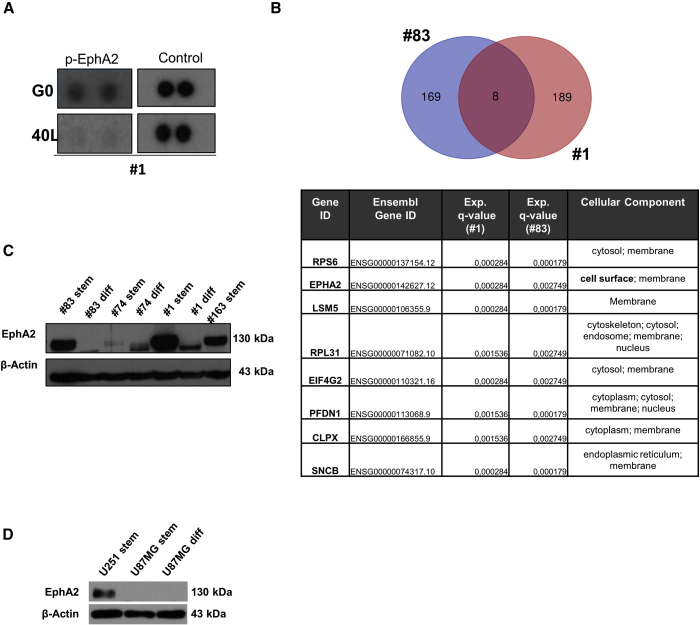
In order to identify a 40L and A40s aptamer target, we first performed a receptor tyrosine kinase (RTK) proteome profile array.18 After incubation of GSC#1 with 40L or G0, the phosphorylation of EphA2 was mainly impaired in 40L-treated cells compared to G0 (Figure 3A). Then, we performed a protein pull-down experiment incubating the protein extract of GSC#1 and GSC#83 with biotinylated A40s and scrambled control. Eluted proteins were analyzed by mass spectrometry (MS). Among the identified proteins, in both groups we restricted the analysis for proteins whose expression was annotated for cellular component as “membrane” and “cell surface” with a false discovery rate (FDR) q-value <0.003. Eight proteins were shared between the two analyzed samples (GSC#1 and GSC#83), and among them EphA2 was the only one with a clear cell component annotation of cell surface, further indicating EphA2 as a candidate target for A40s (Figure 3B). Notably, we found that EphA2 expression levels correlate with the A40s aptamer GSC binding profile reported previously.16 Indeed, EphA2 was overexpressed in GSC#1, GSC#83, GSC#74, and GSC#163 spheres compared to the differentiated counterpart, and it was present at higher levels in the two cell lines used for the aptamer cell SELEX16 (Figure 3C). Moreover, as shown in Figure 3D, EphA2 is overexpressed in U251-derived stem-like cells compared to U87MG-derived stem-like cells in which EphA2 is absent. Indeed, A40s selectively binds U251-derived stem-like cells but not U87MG-derived stem-like cells, as previously demonstrated.16
Figure 3.
EphA2 Is the Target of 40L and A40s
(A) Reduction of EphA2 phosphorylation is shown through a receptor tyrosine kinase (RTK) proteome profile array after treating GSCs with 40L or G0. For control, reference spots of each array are shown. (B) A protein pull-down protocol is optimized for detection of A40s-binding proteins in GSC#1 and GSC#83. Eluted proteins were analyzed by mass spectrometry, and the most significant common membrane proteins are shown in the table. (C) EphA2 levels in several stem cells are compared to a differentiated counterpart. (D) The EphA2 level is shown in U251 derived stem-like cells, U87MG derived stem-like cells, and differentiated cells.
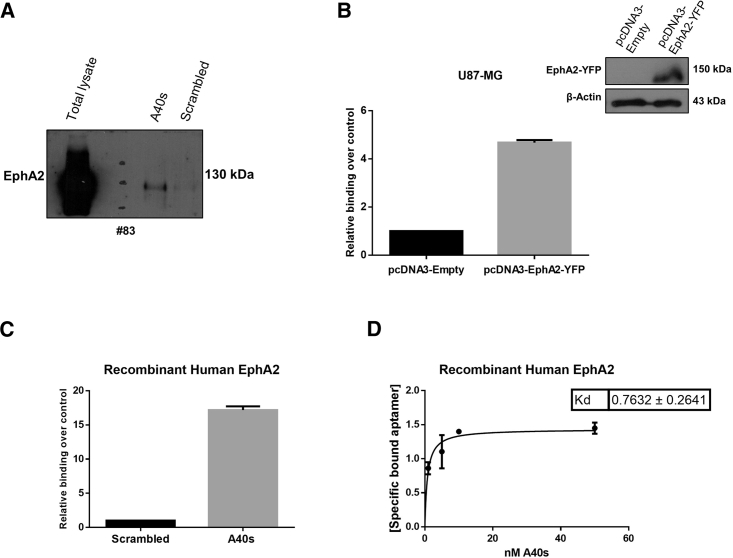
To establish the specific interaction of A40s with EphA2, aptamer pull-down proteins were analyzed by western blot with anti-EphA2 antibodies. Results confirmed A40s binding to EphA2 (Figure 4A). Furthermore, overexpression of EphA2 made the differentiated U87MG cells able to be bound by A40s (Figure 4B). By using enzyme-linked oligonucleotide assays (ELONAs)19 with the recombinant protein, we further proved the direct interaction between the A40s aptamer and EphA2 as a specific target (Figure 4C). Finally, by using ELONAs we also determined the apparent dissociation constant (KD) of A40s for the recombinant EphA2 (KD of 0.76 ± 0.2641 nM) (Figure 4D). Collectively, these data indicate that A40s targets at high affinity and specificity the EphA2 expressed on the cell surface of GSCs.
Figure 4.
A40s Binds EphA2 with High Affinity
(A) Pull-down EphA2 expression is analyzed by SDS-PAGE. (B) EphA2 overexpression in differentiated U87MG cells results in increasing A40s binding ability. (C) ELONA is used to prove A40s binding to recombinant human EphA2 and (D) to calculate KD of the A40s-EphA2 complex. Vertical bars indicate standard deviation values.
Serum Stability and In Vivo Functional Aspects of A40s
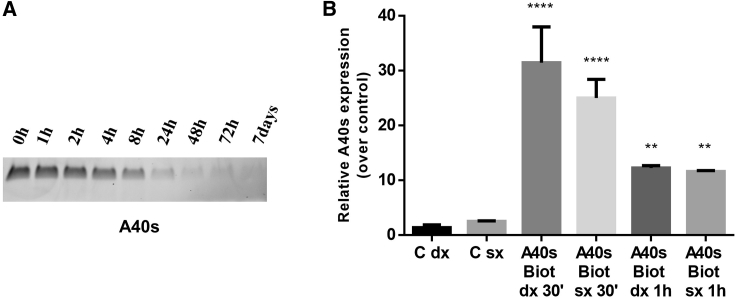
An important feature for clinical translation of new therapeutics is in vivo stability. Therefore, we evaluated the stability of A40s, incubating the aptamer in human serum for up to 1 week. Serum RNA aptamer samples were recovered at different time points and analyzed through 15% polyacrylamide/urea (7 M) denaturing gel (Figure 5A). The aptamer was found to have good stability; it is gradually degraded, but at least the 45% of the sequence remains stable in 90% serum for 8 h.
Figure 5.
A40s In Vivo Distribution
(A) Time course analyzed through denaturing polyacrylamide gel electrophoresis illustrates A40s stability in 90% human serum at 37°C. (B) Biotinylated A40s in mice brain were quantified 30 min and 1 h after aptamer intracardiac injection in left (sx) and right (dx) brain hemispheres. A representative experiment is shown. Vertical bars indicates standard deviation values. ***p<0.001, ****p<0.0001.
Moreover, in order to evaluate the aptamer use in vivo for future applicability of this molecule, we investigated the capability of A40s to cross a healthy blood-brain barrier (BBB) after systemic injection. A40s proved to be able to reach the brain and to be present until 1 h upon its systemic injection (Figure 5B). As a result, our in vivo and in vitro data altogether demonstrate that A40s is able to reach the tumor, overcoming the BBB when systemically injected.
Discussion
The presence of GSCs within GBM represents a major impairment for the treatment of this tumor. It is well established that GSCs are usually more resistant to conventional therapy and give rise to recurrence and more aggressive tumors.4,5 Therefore, their targeting is an important goal for cancer therapy. The use of specific “bullets” targeting the GSCs in combination with conventional therapy for the differentiated population could represent a more effective approach to treat GBM, ameliorate the patient’s condition, and prolong survival by reducing tumor recurrences. Several proteins have been shown to be overexpressed in GBM and in particular in the GSC population. Among them, EphA2 and EphA3 are the most investigated, showing a role in self-renewal of the GSCs compartment and blocking their differentiation.20
In this study, we demonstrated that A40s targets specifically EphA2 both as a recombinant protein and when expressed on the cell surface of the stem-like population of GBM. Indeed, EphA2 is a transmembrane receptor tysosine kinase overexpressed in stem-like cells and is required for self-renewal and GBM tumor propagation.13 We showed that EphA2 expression was restricted to GSCs, indicating that EphA2 may represent a good candidate to discriminate between GSCs and differentiated cells. Other investigation14 has also reported this same observation on EphA2 expression and its inverse correlation with cell differentiation, supporting our idea that EphA2 is a marker for discriminating between GSCs and differentiated cells. Moreover, EphA2 knockdown has been demonstrated to suppress both self-renewal and tumorigenicity, and several intracellular pathways such as AKT, JNK, and mTORC1 have been reported to crosstalk with EphA2 signaling, regulating the GSC proprieties.7 In its mechanisms of action, the A40s aptamer could induce an internalization of the EphA2 receptor, decreasing the amount of the receptor on the cellular surface, or it could impair the activation of the intracellular crosstalk responsible of the maintenance of GSCs.
EphA2 has been reported as a promising therapeutic and molecular target for GBM diagnosis and therapy.21,22 Indeed, we showed that upon binding to EphA2, the A40s aptamer downregulated self-renewal and the expression of the stem-like phenotype, reducing the cell viability of GSCs. The ability of A40s to bind and specifically recognize EphA2 highlights this aptamer as a great candidate for selective inhibition and targeting of GSCs.
Our finding strongly enlarges the possibility of using the A40s as a novel targeting molecule for EphA2. To our knowledge, our aptamer is the first example of a novel molecule able to recognize and inhibit specifically EphA2 and, through this, to impair GSC expansion in GBM tumor. Different strategies are under investigation in order to block its signaling13 or to develop novel immunotherapeutic vaccines.23 For example, GLPG1790 is a small inhibitor molecule shown to inhibit various Eph receptor kinases, resulting in reducing tumor growth and stem cell populations.24 In fact, in a preclinical model it was shown to stop EphA2 receptor signaling, mimicking the ephrin A1-mediated phosphorylation.25 However, GLPG1790 lacks specificity, since it shows an inhibitory activity also on other members of the Eph receptor family such as EphA3 and EphB4.24 Another pan-inhibitor of the Eph receptor family, UniPR1331, was also shown in a xenograft model to inhibit GBM angiogenesis and vasculogenesis through EphA2 blockage.26 A more specific approach targeting indirectly EphA2-overexpressing GSC populations was done by Arnold et al.27 who developed an antibody against CD44, conjugated with an antisense oligonucleotide against EphA2 mRNA, able to be internalized in GSC populations and to reduce EphA2 expression. Highlighting the importance of EphA2 targeting, several attempts have also been done in the reeducation of the immune system with a generation of panel of chimeric antigen receptor (CAR)-T cells for EphA2 as a novel GBM immunotherapy strategy.28 As we have previously reported, A40s is able to serve as a targeting moiety for therapeutic small interfering RNAs (siRNAs)/microRNAs (miRNAs),29 thus combining the intrinsic inhibitory potential of EphA2 intracellular signaling to the selective silencing of GBM tumor targets as STAT3 and miR-10b.30,31
An important limit for the development of therapeutic strategy for GBM is the passage of the molecules through the BBB. Even if only molecules smaller than 400 Da and lipophilic are considered able to cross the BBB,32 recent evidence supports the ability of aptamers to overcome the BBB;31,33 in fact, receptor- and/or channel-mediated endocytosis, fluid-phase pinocytosis, and transcytosis could be implicated in BBB permeability to aptamers.34 In this study, we demonstrated that A40s was able to reach the brain in healthy mice. Moreover, previously we have demonstrated that A40s and CD133 were co-stained in sections from human brain tumor, indicating that A40s localizes in human brain cancer stem cells.
As a result, our in vivo and in vitro data indicate that not only A40s can be stable in human serum, allowing systemic administration, but it is also able to reach the brain, where it can potentially give this anti-tumoral effect by targeting GSCs, halting tumor growth, and reducing tumor relapse. Taken together, these data suggest that A40s is a good candidate to selectively target EphA2 on GSCs and show potential applicability as a therapeutic tool to block a GSC population and thus GBM recurrence.
Materials and Methods
Patient-Derived Tumor Stem Cells and Continuous Cell Lines
GBM tissue-derived stem cells were obtained from the Institute of Neurosurgery,35,36 School of Medicine, Università Cattolica, Rome, Italy, as indicated previously.16 U251MG and U87MG cell lines were from ATCC and were cultured in DMEM (Sigma-Aldrich, Milan, Italy) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotic and antimicotic (Thermo Fisher Scientific, Milan, Italy)
Western Blot Analysis
Western blot analysis was performed as described previously.37 Band intensity quantification was performed using ImageJ software.38 Primary antibodies were as follows: anti-β3-tubulin, anti-Sox2, anti-Nestin, anti-EphA2, anti-GFAP, anti-Nanog, anti-OLIG2 (Santa Cruz Biotechnology, MA, USA), anti-β actin (Sigma-Aldrich), and anti-CD133 (Proteintech, Manchester, UK). Secondary antibodies were goat anti-rabbit and anti-mouse immunoglobulin G (IgG) (from Santa Cruz Biotechnology).
RNA Extraction and Quantitative Real-Time PCR
Total RNAs (miRNAs and mRNAs) were extracted using EuroGold TriFast (EuroClone, Milan, Italy) according to the manufacturer’s protocol. All RNAs were reverse transcribed as described previously.39 To amplify genes of interest we used the following primers: β-actin forward, 5′-TGCGTGACATTAAGGAGAAG-3′, reverse, 5′-GCTCGTAGCTCTTCTCCA-3′; NANOG forward, 5′-CAAAGGCAAACAACCCACTT-3′, reverse, 5′-TCTGGAACCAGGTCTTCACC-3′; GFAP forward, 5′-CTGCGGCTCGATCAACTCA-3′, reverse, 5′-TCCAGCGACTCAATCTTCCTC-3′; OCT4 forward, 5′-CGAAAGAGAAAGCGAACCAG-3′, reverse, 5′-GCCGGTTACAGAACCACACT-3′; Nestin forward, 5′-ACCTCAAGATGTCCCTCAGC-3′, reverse, 5′-ACAGGTGTCTCAAGGGTAGC-3′.
Immunofluorescence Analysis
Mechanically disaggregated stem cells were forced to adhere on polylysine-coated glass coverslips for 15 min, or, alternatively, differentiated cells were directly seeded on coverslips after treating them for 2 weeks with 200 nM A40s or scrambled sequence. Fixed cells with 4% paraformaldehyde were permeabilized with PBS/0.5% Triton X-100 for 15 min at room temperature. Thereafter, they were blocked in PBS/1% BSA for 30 min at room temperature, and, after two washes with PBS, incubated with phycoerythrin (PE) mouse anti-human anti-CD133 (BD Biosciences), mouse anti-Nestin, and rabbit anti-EphA2 (Santa Cruz Biotechnology) diluted in PBS/1% BSA for 1 h at 37°C. Anti-Nestin and anti-EphA2 coverslips were treated with allophycocyanin (APC) goat anti-mouse Ig and fluorescein isothiocyanate (FITC) goat anti-rabbit IgG (BD Biosciences), respectively, for 30 min at 37°C. Coverslips were washed three times with PBS, mounted with Gold antifade reagent with DAPI (Thermo Fisher Scientific), and the cells were visualized by confocal microscopy (LSM 700, Zeiss, Milan, Italy). Images were captured at the same settings, enabling direct comparison of staining patterns.
Proteome Profiler Array
A human phosphorylated (phospho-)receptor tyrosine kinase (RTK) array kit (R&D Systems) was used according to the manufacturer’s protocol. GSC#1 (45 × 104) cells were treated with 400 nM 40 L or with the negative control G0 for 3 h at 37°C and then incubated with 20% FBS for 20 min at 37°C. Cells were harvested and lysed, and 75 μg of proteins was incubated with the human phospho-RTK array (R&D Systems). Finally, EphA2 phosphorylation spots were evaluated by aligning them with the pairs of reference spots.
In Vitro Limiting Dilution Assay
The assay was performed as previously described by Adamo et al.40 1, 5, or 10 cells per well were seeded in stem cell medium onto a 96-well plate. Two weeks after seeding, the number of wells containing spheroids for each cell plating density was counted, and extreme limiting dilution analysis was performed using software available at http://bioinf.wehi.edu.au/software/. For clear and unambiguous understanding, the reciprocal of 95% confidence intervals for 1/(stem cell frequency) generated by ELDA software was calculated and is shown in the corresponding graph. Given the long period of treatment, aptamers were renewed in wells twice a week at a concentration of 100 nM.
Aptamer-Based Pull-Down Assay
Protein identification was performed with an adapted protocol described previously by Berezovski et al.41 To isolate protein targets, 6 million GSCs (#1 and #83) were lysed with sodium deoxycholate (Sigma-Aldrich) at 0.1% (w/v) in 10 mM PBS with Ca2+, Mg2+ (Sigma-Aldrich) and incubated with a biotinylated-scrambled oligonucleotide (TriLinK Biotechnologies, San Diego, CA, USA) at 200 nM for 30 min at room temperature, as a counterselection step. Then, lysates were incubated with streptavidin MagneSphere paramagnetic particles (Promega, Milan, Italy) for 30 min at 4°C. Afterward, unbound proteins were incubated with biotinylated A40s (TriLinK Biotechnologies) at 200 nM for 30 min at room temperature. A40s-protein complexes were incubated with streptavidin MagneSphere paramagnetic particles (Promega) for 30 min at 4°C. Collected beads were washed twice with cold 10 mM PBS with Ca2+, Mg2+ (Sigma-Aldrich). Thus, incubation with 8 M urea for 1 h at 4°C led to protein denaturation and release from the aptamer and magnetic beads. Pull-down proteins were separated by SDS-PAGE (10% polyacrylamide gel), transferred to nitrocellulose membranes (Millipore, Bedford, MA, USA), and immunoblotted for EphA2 antibody (Santa Cruz Biotechnology).
Mass Spectrometry Identification of Aptamer Target
Putative targets of A40s in GSC#1 and GSC#83 were pulled down by affinity chromatography as indicated in “Aptamer-Based Pull-Down Assay” above. Samples were analyzed by an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) coupled to an UltiMate 3000 nanoRLSC (Dionex, Thermo Fisher Scientific). Peptides were separated on an in-house column (Polymicro Technologies), 15 cm × 70 μm inner diameter (ID), packed with Luna C18(2) (3 μm, 100 Å) (Phenomenex) and employing a water/acetonitrile/0.1% formic acid gradient. Samples were loaded onto the column for 105 min at a flow rate of 0.30 μL/min. Peptides were separated using 2% acetonitrile in the first 7 min and then using a linear gradient from 2% to 38% acetonitrile for 70 min, followed by a gradient from 38% to 98% acetonitrile for 9 min, then at 98% of acetonitrile for 10 min, followed by a gradient from 98% to 2% acetonitrile for 3 min and a 10-min wash at 2% acetonitrile. Eluted peptides were directly sprayed into a mass spectrometer using positive electrospray ionization (ESI) at an ion source temperature of 250°C and an ion spray voltage of 2.1 kV. The Orbitrap Fusion Tribrid was run in top speed mode. Full-scan MS spectra (m/z [mass-to-charge ratio] of 350–2,000) were acquired at a resolution of 60,000. Precursor ions were filtered according to monoisotopic precursor selection, charge state (+2 to +7), and dynamic exclusion (30 s with a ±10 ppm window). The automatic gain control settings were 4e5 for full Fourier transform MS (FTMS) scans and 1e4 for MS/MS scans. Fragmentation was performed with collision-induced dissociation (CID) in the linear ion trap. Precursors were isolated using a 2 m/z isolation window and fragmented with a normalized collision energy of 35%. Proteome Discoverer 2.1 (Thermo Fisher Scientific) was used for protein identification. The precursor mass tolerance was set at 10 ppm and 0.6 Da mass tolerance for fragment ions. The search engine SEQUEST-HT implemented in Proteome Discoverer was applied for all MS raw files. Search parameters were set to allow for dynamic modification of methionine oxidation, acetyl on the N terminus, and static modification of cysteine carbamidomethylation. The search database consisted of nonredundant/reviewed human (20,326 proteins) protein sequences in FASTA file format from the UniProt/SwissProt database. The FDR was set to 0.05 for both peptide and protein identifications.
Aptamer-Based ELONA
Assays were performed as previously described.19 Nunc-Immuno Plate MaxiSorp 96-well plates (Thermo Fisher Scientific) were left untreated or were coated with 30 nM purified EphA2 extracellular domain (R&D Systems, Milan, Italy, CF 3035-A2) overnight at 4°C. Wells were blocked for 2 h at room temperature with PBS containing 3% BSA, washed twice with PBS, and then incubated for 2 h at room temperature in PBS with 200 nM biotinylated A40s aptamer (TriLinK Biotechnologies) or an unrelated aptamer (scrambled) as a negative control. Following two washes with PBS, samples were incubated with horseradish peroxidase (HRP)-conjugated streptavidin (Thermo Fisher Scientific) for 1 h at room temperature and washed twice with PBS. Signals were reveled with tetramethylbenzidine (TMB) substrate solution (Thermo Fisher Scientific) and stopped with the stop solution for TMB substrate (Thermo Fisher Scientific). Absorbance at 450 nm was measured with a Multiskan FC microplate photometer (Thermo Fisher Scientific).
Determination of a KD
The KD for the A40s-GSC complex was determined by performing custom TaqMan small RNA assays (Thermo Fisher Scientific) or, alternatively, the A40s-recombinant human EphA2 complex KD was determined by performing an aptamer-based ELONA, as previously described. Fitting curves were designed by using GraphPad Prism 6 software.
Stability in Human Serum
2′-F-RNA A40s was incubated until 7 days in 90% human serum (type AB human serum, EuroClone) at a starting concentration of 4 μM. At each time point, A40s (16 pmol) was recovered and incubated for 1 h at 37°C with 0.5 μL of proteinase K solution (600 mAU/mL) in order to remove serum proteins, which interfere with electrophoretic migration. Then, 9 μL of the denaturing gel loading buffer (1× TBE, 95% formamide, 10 mM EDTA, and bromophenol blue) were added to each sample before storing at −80°C. All time point samples were loaded on 15% polyacrylamide/urea (7 M) denaturing gel. The gel was visualized by UV exposure after ethidium bromide staining.
In Vivo Experiments
BBB crossing was assessed with intracardiac injection of 1,600 pmol of biotinylated A40s or saline solution in healthy, housed athymic CD-1 nude mice (nu/nu). The aptamer amount was determined by performing qRT-PCR.
Statistical Analysis
Continuous variables are given as mean ± 1 standard deviation (SD) or standard error of the mean (SEM). Statistical values were defined using GraphPad Prism 6 (GraphPad, San Diego, CA, USA) software, by Student’s t test (two variables), or one-way ANOVA (more than two variables). A p value ≤0.05 was considered significant for all analyses.
Plasmid Transfections
EphA2-yellow fluorescent protein (YFP) was a gift from Kalina Hristova (Addgene, plasmid #108852).42 EphA2-YFP or control vector were transfected by using Lipofectamine 3000 reagent (Thermo Fisher Scientific).
Ethics Statement
Animal studies were conformed to internationally accepted standards according to D.lgs 26/2014 and approved by “Servizio Veterinario Area C,” ASL Roma E, with authorization no. 259/2017-PR (prot. D9997.35 on February 21, 2017).
Author Contributions
Experiment Design and Performance, C.Q. and A.A. (bindings and target discovery); Technical Support, C.G. and I.S.; Data Interpretations, S.N., A.A., C.Q., C.L.E., and G.R.; In Vivo Assays, L.R.V. and R.P.; Manuscript Preparation, A.A., C.Q., G.C., and V.d.F.; Figure Assembly, A.A. and S.N.; Proteomic Experiments, Z.M. and M.V.B. All authors reviewed the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was partially supported by the Associazione Italiana Ricerca sul Cancro (AIRC) (IG 2013 N.14046 and IG 2016 N. 8473 to G.C.; IG N.15584 2014 AIRC to L.R.V.; N.13345 and 9980 to V.d.F.), the Italian Ministry of Health (GR-2011-02352546 to C.L.E.), CANCER (H2020-MSCA-RISE-2017 Grant no. 777682), MSCA (IF N.838988 to C.Q.), Fondazione Umberto Veronesi individual fellowship (to G.R.), and SATIN grant 2018-2020 (to G.C.). We are thankful to Fortunato Moscato for technical support.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.02.005.
Contributor Information
Cristina Quintavalle, Email: cristina.quintavalle@gmail.com.
Gerolama Condorelli, Email: gecondor@unina.it.
Supplemental Information
References
- 1.Taylor O.G., Brzozowski J.S., Skelding K.A. Glioblastoma multiforme: an overview of emerging therapeutic targets. Front. Oncol. 2019;9:963. doi: 10.3389/fonc.2019.00963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizoguchi M., Yoshimoto K., Ma X., Guan Y., Hata N., Amano T., Nakamizo A., Suzuki S.O., Iwaki T., Sasaki T. Molecular characteristics of glioblastoma with 1p/19q co-deletion. Brain Tumor Pathol. 2012;29:148–153. doi: 10.1007/s10014-012-0107-z. [DOI] [PubMed] [Google Scholar]
- 3.Gao W.Z., Guo L.M., Xu T.Q., Yin Y.H., Jia F. Identification of a multidimensional transcriptome signature for survival prediction of postoperative glioblastoma multiforme patients. J. Transl. Med. 2018;16:368. doi: 10.1186/s12967-018-1744-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bao S., Wu Q., McLendon R.E., Hao Y., Shi Q., Hjelmeland A.B., Dewhirst M.W., Bigner D.D., Rich J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 5.Bovenberg M.S., Degeling M.H., Tannous B.A. Advances in stem cell therapy against gliomas. Trends Mol. Med. 2013;19:281–291. doi: 10.1016/j.molmed.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Liu D.P., Wang Y., Koeffler H.P., Xie D. Ephrin-A1 is a negative regulator in glioma through down-regulation of EphA2 and FAK. Int. J. Oncol. 2007;30:865–871. [PubMed] [Google Scholar]
- 7.Chen J., Song W., Amato K. Eph receptor tyrosine kinases in cancer stem cells. Cytokine Growth Factor Rev. 2015;26:1–6. doi: 10.1016/j.cytogfr.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Day B.W., Stringer B.W., Boyd A.W. Eph receptors as therapeutic targets in glioblastoma. Br. J. Cancer. 2014;111:1255–1261. doi: 10.1038/bjc.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakada M., Hayashi Y., Hamada J. Role of Eph/ephrin tyrosine kinase in malignant glioma. Neuro-oncol. 2011;13:1163–1170. doi: 10.1093/neuonc/nor102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puttick S., Stringer B.W., Day B.W., Bruce Z.C., Ensbey K.S., Mardon K., Cowin G.J., Thurecht K.J., Whittaker A.K., Fay M. EphA2 as a diagnostic imaging target in glioblastoma: a positron emission tomography/magnetic resonance imaging study. Mol. Imaging. 2015;14:385–399. [PubMed] [Google Scholar]
- 11.Wykosky J., Gibo D.M., Stanton C., Debinski W. EphA2 as a novel molecular marker and target in glioblastoma multiforme. Mol. Cancer Res. 2005;3:541–551. doi: 10.1158/1541-7786.MCR-05-0056. [DOI] [PubMed] [Google Scholar]
- 12.Miao H., Gale N.W., Guo H., Qian J., Petty A., Kaspar J., Murphy A.J., Valenzuela D.M., Yancopoulos G., Hambardzumyan D. EphA2 promotes infiltrative invasion of glioma stem cells in vivo through cross-talk with Akt and regulates stem cell properties. Oncogene. 2015;34:558–567. doi: 10.1038/onc.2013.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binda E., Visioli A., Giani F., Lamorte G., Copetti M., Pitter K.L., Huse J.T., Cajola L., Zanetti N., DiMeco F. The EphA2 receptor drives self-renewal and tumorigenicity in stem-like tumor-propagating cells from human glioblastomas. Cancer Cell. 2012;22:765–780. doi: 10.1016/j.ccr.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas T.L., Sciuto M.R., Brunetto L., Valvo C., Signore M., Fiori M.E., di Martino S., Giannetti S., Morgante L., Bo A. Integrin α7 is a functional marker and potential therapeutic target in glioblastoma. Cell Stem Cell. 2017;21:35–50.e9. doi: 10.1016/j.stem.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 15.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 16.Affinito A., Quintavalle C., Esposito C.L., Roscigno G., Vilardo C., Nuzzo S., Ricci-Vitiani L., De Luca G., Pallini R., Kichkailo A.S. The discovery of RNA aptamers that selectively bind glioblastoma stem cells. Mol. Ther. Nucleic Acids. 2019;18:99–109. doi: 10.1016/j.omtn.2019.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y., Smyth G.K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 18.Cerchia L., Esposito C.L., Camorani S., Rienzo A., Stasio L., Insabato L., Affuso A., de Franciscis V. Targeting Axl with an high-affinity inhibitory aptamer. Mol. Ther. 2012;20:2291–2303. doi: 10.1038/mt.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iaboni M., Fontanella R., Rienzo A., Capuozzo M., Nuzzo S., Santamaria G., Catuogno S., Condorelli G., de Franciscis V., Esposito C.L. Targeting insulin receptor with a novel internalizing aptamer. Mol. Ther. Nucleic Acids. 2016;5:e365. doi: 10.1038/mtna.2016.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barquilla A., Pasquale E.B. Eph receptors and ephrins: therapeutic opportunities. Annu. Rev. Pharmacol. Toxicol. 2015;55:465–487. doi: 10.1146/annurev-pharmtox-011112-140226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qazi M.A., Vora P., Venugopal C., Adams J., Singh M., Hu A., Gorelik M., Subapanditha M.K., Savage N., Yang J. Cotargeting ephrin receptor tyrosine kinases A2 and A3 in cancer stem cells reduces growth of recurrent glioblastoma. Cancer Res. 2018;78:5023–5037. doi: 10.1158/0008-5472.CAN-18-0267. [DOI] [PubMed] [Google Scholar]
- 22.Tandon M., Vemula S.V., Mittal S.K. Emerging strategies for EphA2 receptor targeting for cancer therapeutics. Expert Opin. Ther. Targets. 2011;15:31–51. doi: 10.1517/14728222.2011.538682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatano M., Eguchi J., Tatsumi T., Kuwashima N., Dusak J.E., Kinch M.S., Pollack I.F., Hamilton R.L., Storkus W.J., Okada H. EphA2 as a glioma-associated antigen: a novel target for glioma vaccines. Neoplasia. 2005;7:717–722. doi: 10.1593/neo.05277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gravina G.L., Mancini A., Colapietro A., Delle Monache S., Sferra R., Vitale F., Cristiano L., Martellucci S., Marampon F., Mattei V. The small molecule ephrin receptor inhibitor, GLPG1790, reduces renewal capabilities of cancer stem cells, showing anti-tumour efficacy on preclinical glioblastoma models. Cancers (Basel) 2019;11:359. doi: 10.3390/cancers11030359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Megiorni F., Gravina G.L., Camero S., Ceccarelli S., Del Fattore A., Desiderio V., Papaccio F., McDowell H.P., Shukla R., Pizzuti A. Pharmacological targeting of the ephrin receptor kinase signalling by GLPG1790 in vitro and in vivo reverts oncophenotype, induces myogenic differentiation and radiosensitizes embryonal rhabdomyosarcoma cells. J. Hematol. Oncol. 2017;10:161. doi: 10.1186/s13045-017-0530-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Festuccia C., Giorgio C., Gravina G.L., Castelli R., Vacondio F., Rusnati M., Chiodelli P., Barocelli E., Lodola A., Tognolini M. Targeting glioblastoma with UniPR1331, a new and stable bioavailable small molecule inhibiting Eph-ephrin interaction: in vitro and in vivo evidence. Eur. J. Cancer. 2016;69(Suppl 1):S30–S31. [Google Scholar]
- 27.Arnold A.E., Malek-Adamian E., Le P.U., Meng A., Martínez-Montero S., Petrecca K., Damha M.J., Shoichet M.S. Antibody-antisense oligonucleotide conjugate downregulates a key gene in glioblastoma stem cells. Mol. Ther. Nucleic Acids. 2018;11:518–527. doi: 10.1016/j.omtn.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yi Z., Prinzing B.L., Cao F., Gottschalk S., Krenciute G. Optimizing EphA2-CAR T cells for the adoptive immunotherapy of glioma. Mol. Ther. Methods Clin. Dev. 2018;9:70–80. doi: 10.1016/j.omtm.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nuzzo S., Roscigno G., Affinito A., Ingenito F., Quintavalle C., Condorelli G. Potential and challenges of aptamers as specific carriers of therapeutic oligonucleotides for precision medicine in cancer. Cancers (Basel) 2019;11:1521. doi: 10.3390/cancers11101521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esposito C.L., Nuzzo S., Catuogno S., Romano S., de Nigris F., de Franciscis V. STAT3 gene silencing by aptamer-siRNA chimera as selective therapeutic for glioblastoma. Mol. Ther. Nucleic Acids. 2018;10:398–411. doi: 10.1016/j.omtn.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esposito C.L., Nuzzo S., Kumar S.A., Rienzo A., Lawrence C.L., Pallini R., Shaw L., Alder J.E., Ricci-Vitiani L., Catuogno S. A combined microRNA-based targeted therapeutic approach to eradicate glioblastoma stem-like cells. J. Control. Release. 2016;238:43–57. doi: 10.1016/j.jconrel.2016.07.032. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y., Liu L. Modern methods for delivery of drugs across the blood-brain barrier. Adv. Drug Deliv. Rev. 2012;64:640–665. doi: 10.1016/j.addr.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 33.Cheng C., Chen Y.H., Lennox K.A., Behlke M.A., Davidson B.L. In vivo SELEX for identification of brain-penetrating aptamers. Mol. Ther. Nucleic Acids. 2013;2:e67. doi: 10.1038/mtna.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanss B., Leal-Pinto E., Bruggeman L.A., Copeland T.D., Klotman P.E. Identification and characterization of a cell membrane nucleic acid channel. Proc. Natl. Acad. Sci. USA. 1998;95:1921–1926. doi: 10.1073/pnas.95.4.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pallini R., Ricci-Vitiani L., Banna G.L., Signore M., Lombardi D., Todaro M., Stassi G., Martini M., Maira G., Larocca L.M. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin. Cancer Res. 2008;14:8205–8212. doi: 10.1158/1078-0432.CCR-08-0644. [DOI] [PubMed] [Google Scholar]
- 36.Ricci-Vitiani L., Pallini R., Biffoni M., Todaro M., Invernici G., Cenci T., Maira G., Parati E.A., Stassi G., Larocca L.M., De Maria R. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468:824–828. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- 37.Roscigno G., Puoti I., Giordano I., Donnarumma E., Russo V., Affinito A., Adamo A., Quintavalle C., Todaro M., Vivanco M.D., Condorelli G. miR-24 induces chemotherapy resistance and hypoxic advantage in breast cancer. Oncotarget. 2017;8:19507–19521. doi: 10.18632/oncotarget.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quintavalle C., Di Costanzo S., Zanca C., Tasset I., Fraldi A., Incoronato M., Mirabelli P., Monti M., Ballabio A., Pucci P. Phosphorylation-regulated degradation of the tumor-suppressor form of PED by chaperone-mediated autophagy in lung cancer cells. J. Cell. Physiol. 2014;229:1359–1368. doi: 10.1002/jcp.24569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adamo A., Fiore D., De Martino F., Roscigno G., Affinito A., Donnarumma E., Puoti I., Ricci Vitiani L., Pallini R., Quintavalle C., Condorelli G. RYK promotes the stemness of glioblastoma cells via the WNT/ β-catenin pathway. Oncotarget. 2017;8:13476–13487. doi: 10.18632/oncotarget.14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berezovski M.V., Lechmann M., Musheev M.U., Mak T.W., Krylov S.N. Aptamer-facilitated biomarker discovery (AptaBiD) J. Am. Chem. Soc. 2008;130:9137–9143. doi: 10.1021/ja801951p. [DOI] [PubMed] [Google Scholar]
- 42.Singh D.R., Ahmed F., King C., Gupta N., Salotto M., Pasquale E.B., Hristova K. EphA2 receptor unliganded dimers suppress EphA2 pro-tumorigenic signaling. J. Biol. Chem. 2015;290:27271–27279. doi: 10.1074/jbc.M115.676866. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.