Abstract
Hepatocellular carcinoma (HCC) is one of the most common types of cancer worldwide with a high morbidity and mortality rate. An increasing number of studies have demonstrated that microRNAs (miRNAs) serve an important role in HCC. The present study investigated the role of miR-939-3p in HCC. It was demonstrated that miR-939-3p was upregulated in HCC cell lines and HCC tissues compared with normal liver cell lines and paired normal tissues, respectively. It was also found that upregulation of miR-939-3p expression levels in HCC tissues was associated with a less favorable prognosis. Moreover, the overexpression of miR-939-3p in LM3 cells enhanced the metastatic capacity of these cells and promoted epithelial-mesenchymal transition (EMT). In contrast, miR-939-3p inhibition decreased the invasive capacity of HCC cells and EMT. Potential binding target of miR-939-3p to estrogen receptor 1 (ESR1) were predicted using TargetScan. The expression levels of miR-939-3p were negatively associated with ESR1 in HCC tissues based on data from The Cancer Genome Atlas. A luciferase reporter assay was used to confirm ESR1 as a direct downstream target of miR-393-3p. The miR-939-3p/ESR1 axis may be a potential novel target for the treatment of HCC.
Keywords: hepatocellular carcinoma, microRNA-939-3p, estrogen receptor 1
Introduction
Hepatocellular carcinoma (HCC) is a type of primary liver cancer and is one of the most common malignant types of cancer worldwide, with high morbidity and cancer associated mortality rates (1). The incidence rate of HCC in China is the highest in the world due to an increased rate of hepatitis B virus infection (2). Furthermore, the overall survival rate has remained unsatisfactory for the last decade at 22–35% (3). Although numerous studies have been performed, the carcinogenesis and progression of HCC remains unclear (4–6). Therefore, identifying and clarifying the molecular mechanisms involved in development and progression of HCC may improve prognostic outcomes.
It has been reported that microRNAs (miRNAs/miRs), which are highly conserved, small non-coding RNAs, 19–25 nucleotides in length and abundantly expressed in animals (7,8), may bind to the 3′-untranslated region (UTR) of target genes and inhibit the expression of these genes through post-transcriptional regulation of mRNAs (9). A number of studies have demonstrated that miRs, including miR-21, miR-197-3p and miR-497-5p, serve an important role in apoptosis, cell proliferation, differentiation and metastasis (10–13). A previous study reported that inhibition of miR-939-3p may suppress the development of human non-small cell lung cancer (NSCLC) via the upregulation of metalloproteinase 2 (14). However, to the best of our knowledge, the function of miR-939-3p in HCC remains unknown.
Estrogen receptor 1 (ESR1), a ligand-activated transcription factor, may directly bind to the transcription factor (TF) complex and lead to altered functions of proteins in the cytoplasm, for examples activation of eNOS or regulation of gene expression through phosphorylation (15). Numerous studies have demonstrated that ESR1 acts as a tumor suppressor in various cancer types. Yang et al (16) reported that ESR1 directly regulates the hypoxia-inducible factor 1 or the pathway associated with the anti-estrogen response in breast cancer. An ESR α inhibitor activated the unfolded protein response, blocked protein synthesis and induced tumor regression in HCC (17). Hishida et al (18) predicted that ESR1 is a tumor suppressor gene in HCC by triple-combination array analysis. Additionally, Tu et al (19) demonstrated that ESR1 overexpression mediated apoptosis in Hep3B cells by binding with SP1 proteins. However, to the best of our knowledge, the effect of ESR1 on the metastasis of HCC cells has not been studied. Therefore, the aim of the present study was to determine the potential gene binding of miR-939-3p and the function of miR-939-3p in HCC.
Materials and methods
Tissue samples
The present study was approved by The Institutional Ethics Committee of Zhejiang Provincial People's Hospital (Hangzhou, China). The clinical data were obtained from The Cancer Genome Atlas (TCGA, portal.gdc.cancer.gov/).
Cell culture
The HCC cell line (HCCLM3) was obtained from the American Type Culture Collection. Cells were cultured at 37°C with 5% CO2 in Minimum Essential Medium (MEM; Thermo Fisher Scientific, Inc.) containing 10% FBS (Thermo Fisher Scientific, Inc.). This cell line was authenticated by short tandem repeats profiling.
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). RNA was then reverse transcribed to cDNA using PrimeScript™ RT Master mix (cat. no. RR036A; Takara Bio, Inc.), according to the manufacturer's protocol. qPCR was performed using an ABI 7500 (Thermo Fisher Scientific, Inc.). The primer sequences were as follows: miR-939 forward, 5′-TGGGGAGCTGAGGCTCTG-3′ and reverse, 3′-AGTGCAGGGTCCGAGGTATT-5′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 3′-AACGCTTCACGAATTTGCGT-5′; and ESR1 forward, 5′-CCGGCTCCGTAAATGCTACG-3′ and reverse, 3′-TCCAGCAGACCCCACTTCAC-5′. U6 was used as the internal control.
Transfection
miR-939-3p mimic, miR-939-3p inhibitor and ESR1 small interfering (si) RNA were obtained from Shanghai GenePharma Co., Ltd. Cells were seeded in 6-well plates (3×105/well) and cultured for 24 h before transfection. Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used for transfection, according to the manufacturer's protocol. The sequences of the miRNAs were as follows: hsa-miR-939 mimic sense, UGGGGAGCUGAGGCUCUGGGGGUG and antisense, CCCCCAGAGCCUCAGCUCCCCAUU; mimics negative control (NC) sense, UUCUCCGAACGUGUCACGUTT and antisense, ACGUGACACGUUCGGAGAATT; and hsa-miR-939 inhibitor, CACCCCCAGAGCCUCAGCUCCCCA; and inhibitor NC, CAGUACUUUUGUGUAGUACAA. The sequences of the ESR1 siRNA were as follows: Sense, GCAAGUUGAUCUUAGUUAAGU and antisense, UUAACUAAGAUCAACUUGCUG; siRNA NC (cat. no. siN05815122147; Guangzhou RiboBio Co., Ltd.,) was used as the siRNA negative control, but the sequence was not provided by the supplier.
Western blot
Tissues or cells were lysed with RIPA lysis buffer (Thermo Fisher Scientific, Inc.) containing 1% protease inhibitor cocktail. The concentration of the extracted protein was analyzed using BCA kit (Beyotime Institute of Biotechnology). Microplate reader and Gen5 software version 2 (BioTek Instruments, Inc) were used to detect the quantification of protein expression.12% SDS-PAGE was used to resolve the proteins, which were then transferred to PVDF membranes. PVDF membranes were blocked with 5% fat-free milk at room temperature for 2 h, followed by incubation with primary antibodies (all 1:1,000) against ESR1 (cat. no. MA5-14501l; Invitrogen; Thermo Fisher Scientific, Inc.), matrix metalloproteinase (MMP) 2 (cat. no. MA5-14186), MMP9 (cat. no. MA5-15886), vimentin (cat. no. MA5-11883) and GAPDH (cat. no. AM4300; all from Invitrogen; Thermo Fisher Scientific, Inc.) at 4°C overnight. Subsequently, the PVDF membranes were incubated with the secondary antibody (1:2,000; cat. no. ab6721; Abcam) for 1 h at room temperature. Signals were visualized using ECL substrate (Pierce; Thermo Fisher Scientific, Inc.).
Dual-luciferase reporter assay
PmirGLO plasmids containing the wild-type (Wt) or mutant (Mut) 3′UTR of ESR1 were purchased from Shanghai GenePharma Co., Ltd. PmirGLO plasmids were transfected into LM3 cells with miR-939-3p mimic or inhibitor with Lipofectamine® 2000 reagent (Thermo Fisher Scientific, Inc.). Cells were cultured for 48 h prior to measurement of luciferase intensity. At 48 h post-transfection, the cells were lysed using radioimmunoprecipitation assay buffer (cat. no. P0013C; Beyotime Institute of Biotechnology) according to the manufacturer's protocol. A F-4500 fluorescence spectrophotometer (Hitachi, Ltd.) was used to measure the luciferase intensity according to the manufacturer's protocol and normalized to that of Renilla luciferase.
Migration and invasion assays
Cell migration and invasion ability was evaluated using a Transwell assay. LM3 cells (5×104) were seeded in the upper chamber with FBS-free MEM and the lower chamber contained MEM supplemented with 10% FBS. For invasion assays, membranes were coated with 50 µl growth factor-reduced Matrigel (BD Biosciences). Cell migration and invasion were measured after incubation for 24 h. Cells were stained with crystal violet dye solution for 5 min at room temperature, and the number of cells were counted in five randomly selected fields with a light inverted microscope at ×200 magnification. Each experiment was repeated three times.
Bioinformation analysis
We predict the target gene of miRNA with TargetScan (version 5.0; genes.mit.edu/targetscan). The level of miR-939-3p in the adjacent normal tissues and HCC tissues and the Kaplan-Meier survival curve analysis in HCC patients in The Cancer Genome Atlas (TCGA) were analyzed with miRpower (kmplot.com/analysis/) (20).
Statistical analysis
Data are presented as the mean ± standard deviation unless otherwise shown. Statistical analysis was performed using SPSS 19.0 (IBM, Corp.). Significance between groups was analyzed using an unpaired Student's t-test. The correlation between miR-939-3p and ESR1 expression levels was examined using a Pearson's correlation coefficient. The log rank test was used for survival analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-939-3p is upregulated in HCC tissues and is associated with prognosis
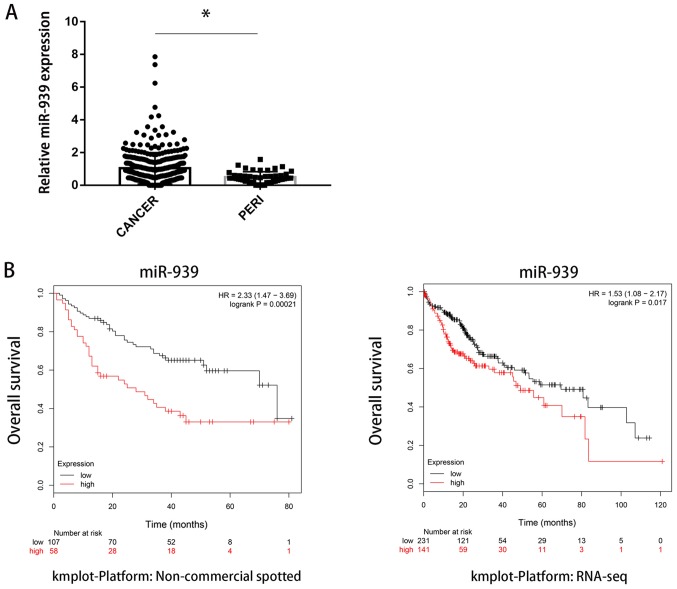
miR-939-3p was upregulated in HCC tissues compared with paired normal tissues based on data obtained from The Cancer Genome Atlas (TCGA; P<0.05; Fig. 1A). Furthermore, patients with upregulated expression levels of miR-939-3p exhibited significantly improved overall survival compared with patients with low expression levels of miR-939-3p in two different datasets (kmplot.com/analysis/index.php?p=background) (Fig. 1B).
Figure 1.
miR-939-3p expression is upregulated in HCC tissues. (A) miR-939-3p expression levels were significantly higher in HCC tissues compared with paired normal tissues. (B) Low miR-939-3p expression levels were associated with improved overall survival in two different datasets obtained from The Cancer Genome Atlas. *P<0.05 vs. normal tissues. HCC, hepatocellular carcinoma; miR, microRNA; PERI, peri-tumoral tissue; HR, Hazard ratio.
miR-939-3p regulates migration, invasion and EMT of LM3 cells
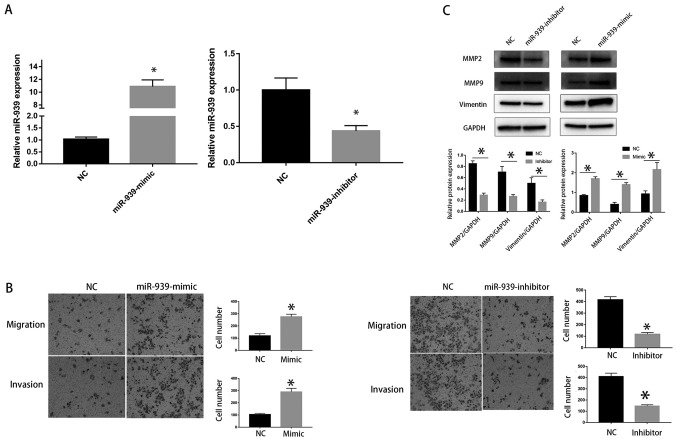
To investigate the function of miR-939-3p, miR-939-3p mimics and inhibitors were used to increase or decrease the expression levels of miR-939-3p, respectively (Fig. 2A). LM3 cells transfected with miR-939-3p mimic exhibited an increased migratory and invasive capacity (P<0.05; Fig. 2B). In addition, western blot analysis demonstrated that overexpression of miR-939-3p significantly upregulated the protein expression levels of MMP2, MMP9 and vimentin (Fig. 2C). Knockdown of miR-939-3p resulted in a reduction of invasion and expression of EMT-associated proteins in LM3 cells (Fig. 2C).
Figure 2.
miR-939-3p promotes invasion and EMT in a HCC cell line. (A) Transfection efficiency of miR-939-3p mimic and inhibitor was detected by reverse transcription quantitative PCR. *P<0.05 vs. NC. (B) Transwell assay were performed to assess the migration of LM3 cells treated with miR-939-mimic or miR-939-inhibitor. *P<0.05 vs. NC. (C) Western blotting was used to examine the effect of miR-939-3p on EMT in LM3 cells. miR, microRNA; HCC, hepatocellular carcinoma; EMT, epithelial-mesenchymal transition; NC, negative control.
ESR1 is downregulated in HCC tissues and cell lines, and is associated with a less favorable prognosis
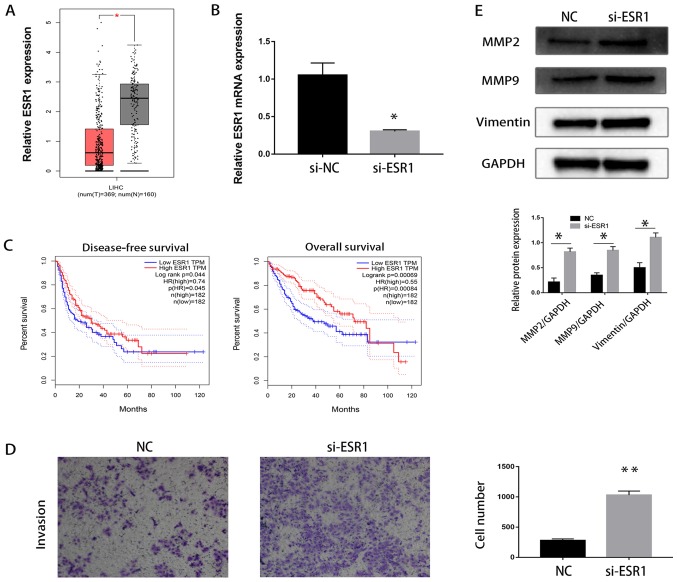
ESR1 expression was downregulated in HCC tissues compared with paired normal tissues based on data obtained from TCGA (P<0.05; Fig. 3A). Furthermore, patients with lower expression levels of ESR1 exhibited improved overall survival and disease-free survival compared with patients who exhibited increased ESR1 expression levels in TCGA dataset (P<0.05; Fig. 3C).
Figure 3.
ESR1 is downregulated in HCC tissues. (A) ESR1 mRNA expression levels in HCC tissues and paired normal tissues. *P<0.05 vs. paired normal tissues. (B) mRNA expression levels of ESR1 in LM3 cells transfected with siRNA-ESR1 and LM3 cells transfected with siRNA-NC. *P<0.05 vs. siRNA-NC. (C) Increased ESR1 expression levels were correlated with improved disease-free survival. (D) ESR1-knockdown enhanced the migration of LM3 cells. (E) Western blotting was used to evaluate the effect of ESR1 on EMT in LM3 cells. **P<0.01 vs. NC. ESR1, Estrogen receptor 1; HCC, hepatocellular carcinoma; NC, negative control; si, small interfering; T, Tumor tissue; N, Normal tissue; LIHC, Liver hepatocellular carcinoma; HR, Hazard ratio; MMP, matrix metalloproteinase.
ESR1 regulates the invasion and EMT of LM3 cells
The mRNA expression levels of ESR1 were lower in LM3 cells transfected with siRNA-ESR1 compared with LM3 cells transfected with siRNA-NC (P<0.05; Fig. 3B). It was demonstrated that ESR1-knockdown significantly increased the invasion of HCCLM3 cells (P<0.01; Fig. 3D). Additionally, western blotting revealed that ESR1-knockdown increased the protein expression levels of MMP2, MMP9 and vimentin in HCCLM3 cells (P<0.05; Fig. 3E).
miR-939-3p directly targets ESR1 and regulates ESR1 expression levels in HCC cells
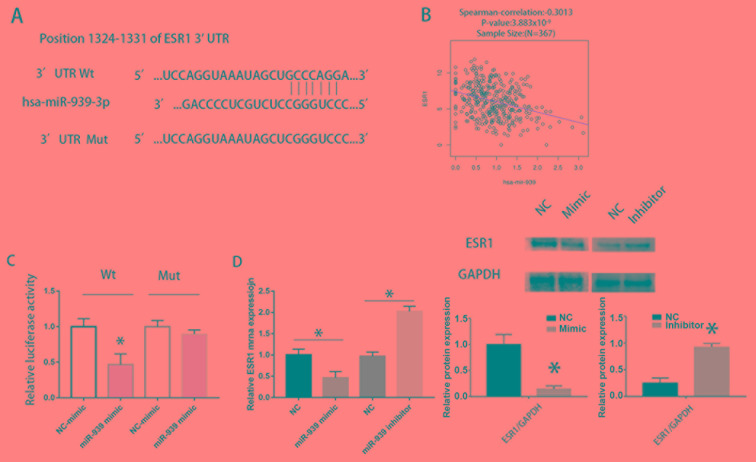
Binding of the ESR1 3′UTR with miR-939-3p was predicted using TargetScan. The Wt and Mut sequences were constructed and inserted into a PmirGLO vector (Fig. 4A). A significantly negative correlation between miR-939-3p and ESR1 was observed in the TCGA dataset (Fig. 4B). A dual-luciferase report assay was performed to detect the effect of miR-939-3p on ESR1 promoter activity. Decreased luciferase activity was observed in the Wt group, whereas no changes were detected in the Mut group compared with the NC group (P<0.05; Fig. 4C). Western blotting demonstrated that the protein expression levels of ESR1 were downregulated following transfection with miR-939-3p mimic in HCCLM3 cells (Fig. 4D), whereas, the reverse was observed following the transfection of miR-939-3p inhibitor in HCCLM3 cells (Fig. 4D).
Figure 4.
miR-939-3p directly binds to the 3′UTR of ESR1. (A) Wt and Mut sequences of the ESR1 3′UTR and the binding sequences of miR-939-3p. (B) Correlation of ESR1 and miR-939-3p in The Cancer Genome Atlas. (C) miR-939-3p suppressed the luciferase activity of the Wt ESR1 3′UTR, whereas the Mut miR-939-3p sequence did not in LM3 cells. *P<0.05 vs. NC-mimic (D) mRNA and protein expression levels of ESR1 were reduced following transfection with miR-939-3p mimic, whereas miR-939-3p inhibitor increased ESR1 expression in LM3 cells. *P<0.05 vs. NC. ESR1, Estrogen receptor 1; Wt, wild type; Mut, mutant; NC, negative control; UTR, untranslated region; miR, microRNA.
Discussion
HCC is one of the most common tumor types worldwide, with high morbidity and mortality rates (1). Although numerous oncogenes and tumor suppressors have been reported in HCC (21–24), the underlying mechanisms of development and recurrence of HCC remain unclear. Over the past decade, the overall survival rate of HCC has remained unsatisfactory and is only 22–35%. A number of genes such as CAV1, SPOCK1 and PRMT1 (25–27) may contribute to the metastasis of HCC cells, aberrant expression of which results in a worse prognosis. Therefore, there is a need to determine the molecular mechanisms underlying metastasis of HCC.
miRNAs may bind to the 3′UTR of target genes and inhibit expression via post-transcriptional regulation (7). A number of studies have demonstrated that miRNAs participate in the occurrence, progression and metastasis of tumors, including gastric cancer and colorectal cancer (28,29). miRNAs are also involved in the pathogenesis and progression of HCC. Hu et al (24) reported that miR-665 promotes HCC cell migration, invasion and proliferation by decreasing Hippo signaling by targeting protein tyrosine phosphatase receptor type B. Wang et al (30) demonstrated that downregulation of circDYNC1H1 is associated with inhibitory effects on cell proliferation and migration in HCC via miR-140-5p. Yu et al (31) demonstrated that miR-501 acts as an independent prognostic factor which promoted EMT via targeting Jun dimerization protein 2 in HCC. However, the functions of miR-939-3p in tumors have not been extensively studied in HCC to the best of our knowledge. The present study demonstrated that the expression levels of miR-939-3p were increased in HCC tissues and HCC cell lines compared with paired normal tissues and normal cell lines, respectively. Furthermore, it was demonstrated that low expression levels of miR-939-3p were correlated with a more favorable prognosis. Inhibition of miR-939-3p decreased the metastatic ability of HCCLM3 cells and western blotting revealed that miR-939-3p may promote EMT via upregulation of MMP2, MMP9 and vimentin.
An increasing number of studies have demonstrated that ESR1 may act as a tumor suppressor in various cancer types (16,19). The present study demonstrated that the expression levels of ESR1 are downregulated in HCC cell lines compared with normal liver cells. It was demonstrated that inhibition of ESR1 decreased the metastatic ability of HCCLM3 cells, therefore ESR1 inhibition was associated with a more favorable prognosis. Finally, through a dual-luciferase report assay and western blotting, a direct binding association was identified between miR-939-3p and ESR1. miR-939-3p may influence EMT via ESR1, although the present study did not directly show this. Limitations of the present study include the fact that rescue experiments were not performed therefore in further studies these experiments should be performed.
In conclusion, the present study demonstrated that miR-939-3p serves a role in HCC cell invasion and metastasis. miR-939-3p negatively regulated ESR1 by binding to its 3′UTR, therefore the miR-939-3p/ESR1 axis may present a potential target for treatment of HCC in the future.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Medicine and Health Research Foundation of Zhejiang Province (grant no. 2017KY018) and the Zhejiang Provincial Natural Science Foundation of China (grant no. LY18H160043).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
JX designed the study. FC, XYN, LXC and XYW performed the experiments and analyzed the data. FC and XYN wrote the manuscript. XYN and XYW revised the manuscript.
Ethics approval and consent to participate
The present study was approved by The Institutional Review Board of the Zhejiang Provincial Peoples' Hospital (Taizhou, China). All patients gave written informed consent to participate in the study and the data were anonymized.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Tan A, Yeh SH, Liu CJ, Cheung C, Chen PJ. Viral hepatocarcinogenesis: From infection to cancer. Liver Int. 2008;28:175–188. doi: 10.1111/j.1478-3231.2007.01652.x. [DOI] [PubMed] [Google Scholar]
- 3.Ren FH, Yang H, He RQ, Lu JN, Lin XG, Liang HW, Dang YW, Feng ZB, Chen G, Luo DZ. Analysis of microarrays of miR-34a and its identification of prospective target gene signature in hepatocellular carcinoma. BMC Cancer. 2018;18:12. doi: 10.1186/s12885-017-3941-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brondfield MN, Dodge JL, Hirose R, Heimbach J, Yao FY, Mehta N. Hepatocellular carcinoma (HCC) patients listed in short wait regions remain advantaged for liver transplant (LT) following 2015 HCC policy change. Liver Transpl. 2019 Dec 13; doi: 10.1002/lt.25701. doi: 10.1002/lt.25701 (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakheet AMH, Zhao C, Chen JN, Zhang JY, Huang JT, Du Y, Gong LP, Bi YH, Shao CK. Improving pathological early diagnosis and differential biomarker value for hepatocellular carcinoma via RNAscope technology. Hepatol Int. 2019 Dec 12; doi: 10.1007/s12072-019-10006-z. doi: 10.1007/s12072-019-10006-z (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 6.Hu J, Wang E, Liu L, Wang Q, Xia D, Bai W, Tie J, Li X, Yuan J, Yang S, et al. Sorafenib may enhance antitumour efficacy in hepatocellular carcinoma patients by modulating the proportions and functions of natural killer cells. Invest New Drugs. 2019 Dec 13; doi: 10.1007/s10637-019-00885-2. doi: 10.1007/s10637-019-00885-2 (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 7.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 8.Rana TM. Illuminating the silence: Understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 9.Loosen SH, Schueller F, Trautwein C, Roy S, Roderburg C. Role of circulating microRNAs in liver diseases. World J Hepatol. 2017;9:586–594. doi: 10.4254/wjh.v9.i12.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang X, Wang L, Liu W, Li F. MicroRNA-497-5p inhibits proliferation and invasion of non-small cell lung cancer by regulating FGF2. Oncol Lett. 2019;17:3425–3431. doi: 10.3892/ol.2019.9954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ni JS, Zheng H, Huang ZP, Hong YG, Ou YL, Tao YP, Wang MC, Wang ZG, Yang Y, Zhou WP. MicroRNA-197-3p acts as a prognostic marker and inhibits cell invasion in hepatocellular carcinoma. Oncol Lett. 2019;17:2317–2327. doi: 10.3892/ol.2018.9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Z, Fang Y, Du R. MicroRNA-107 induces cell cycle arrests by directly targeting cyclin E1 in ovarian cancer. Biochem Biophys Res Commun. 2019;512:331–337. doi: 10.1016/j.bbrc.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Chu Y, Xu M, Zhang X, Zhou Y, Xu M. miR-21 promotes cell migration and invasion of hepatocellular carcinoma by targeting KLF5. Oncol Lett. 2019;17:2221–2227. doi: 10.3892/ol.2018.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen A, Liu S, Lu X, Wei L, Chen Y. Inhibition of microRNA939 suppresses the development of human nonsmall cell lung cancer via the upregulation of tissue inhibitor of metalloproteinases 2. Mol Med Rep. 2018;18:4831–4838. doi: 10.3892/mmr.2018.9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bjornstrom L, Sjoberg M. Mechanisms of estrogen receptor signaling: Convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19:833–842. doi: 10.1210/me.2004-0486. [DOI] [PubMed] [Google Scholar]
- 16.Yang J, AlTahan A, Jones DT, Buffa FM, Bridges E, Interiano RB, Qu C, Vogt N, Li JL, Baban D, et al. Estrogen receptor-alpha directly regulates the hypoxia-inducible factor 1 pathway associated with antiestrogen response in breast cancer. Proc Natl Acad Sci USA. 2015;112:15172–15177. doi: 10.1073/pnas.1422015112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andruska ND, Zheng X, Yang X, Mao C, Cherian MM, Mahapatra L, Helferich WG, Shapiro DJ. Estrogen receptor α inhibitor activates the unfolded protein response, blocks protein synthesis, and induces tumor regression. Proc Natl Acad Sci USA. 2015;112:4737–4742. doi: 10.1073/pnas.1403685112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hishida M, Nomoto S, Inokawa Y, Hayashi M, Kanda M, Okamura Y, Nishikawa Y, Tanaka C, Kobayashi D, Yamada S, et al. Estrogen receptor 1 gene as a tumor suppressor gene in hepatocellular carcinoma detected by triple-combination array analysis. Int J Oncol. 2013;43:88–94. doi: 10.3892/ijo.2013.1951. [DOI] [PubMed] [Google Scholar]
- 19.Tu CC, Kumar VB, Day CH, Kuo WW, Yeh SP, Chen RJ, Liao CR, Chen HY, Tsai FJ, Wu WJ, Huang CY. Estrogen receptor alpha (ESR1) over-expression mediated apoptosis in Hep3B cells by binding with SP1 proteins. J Mol Endocrinol. 2013;51:203–212. doi: 10.1530/JME-13-0085. [DOI] [PubMed] [Google Scholar]
- 20.Nagy Á, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227. doi: 10.1038/s41598-018-29514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan J, Liu H, Yang L, Ma L, Liu J, Ming L. JMJD6 promotes hepatocellular carcinoma carcinogenesis by targeting CDK4. Int J Cancer. 2019;144:2489–2500. doi: 10.1002/ijc.31816. [DOI] [PubMed] [Google Scholar]
- 22.Hu D, Hu Y, Xu W, Yu H, Yang N, Ni S, Fu R. miR203 inhibits the expression of collagenrelated genes and the proliferation of hepatic stellate cells through a SMAD3dependent mechanism. Mol Med Rep. 2017;16:1248–1254. doi: 10.3892/mmr.2017.6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan X, Cheng C, Shao Q, Lin Z, Lu S, Chen Y. CD24 promotes HCC progression via triggering Notch-related EMT and modulation of tumor microenvironment. Tumour Biol. 2016;37:6073–6084. doi: 10.1007/s13277-015-4442-7. [DOI] [PubMed] [Google Scholar]
- 24.Hu Y, Yang C, Yang S, Cheng F, Rao J, Wang X. miR-665 promotes hepatocellular carcinoma cell migration, invasion, and proliferation by decreasing Hippo signaling through targeting PTPRB. Cell Death Dis. 2018;9:954. doi: 10.1038/s41419-018-0978-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Zhang XP, Jiang YB, Zhong CQ, Ma N, Zhang EB, Zhang F, Li JJ, Deng YZ, Wang K, Xie D, Cheng SQ. PRMT1 promoted HCC growth and metastasis in vitro and in vivo via activating the STAT3 signalling pathway. Cell Physiol Biochem. 2018;47:1643–1654. doi: 10.1159/000490983. [DOI] [PubMed] [Google Scholar]
- 26.Yu H, Shen H, Zhang Y, Zhong F, Liu Y, Qin L, Yang P. CAV1 promotes HCC cell progression and metastasis through Wnt/β-catenin pathway. PLoS One. 2014;9:e106451. doi: 10.1371/journal.pone.0106451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Chen L, Chan TH, Liu M, Kong KL, Qiu JL, Li Y, Yuan YF, Guan XY. SPOCK1 is regulated by CHD1L and blocks apoptosis and promotes HCC cell invasiveness and metastasis in mice. Gastroenterology. 2013;144:179–191 e174. doi: 10.1053/j.gastro.2012.09.042. [DOI] [PubMed] [Google Scholar]
- 28.Feng L, Jing L, Han J, Wang G, Liu Y, Zhang X, Wang Y, Wang F, Ma H, Liu Y. MicroRNA 486–3p directly targets BIK and regulates apoptosis and invasion in colorectal cancer cells. Onco Targets Ther. 2018;11:8791–8801. doi: 10.2147/OTT.S180354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang M, Shi B, Liu J, He L, Yi G, Zhou L, Yu G, Zhou X. Downregulation of miR203 induces overexpression of PIK3CA and predicts poor prognosis of gastric cancer patients. Drug Des Devel Ther. 2015;9:3607–3616. doi: 10.2147/DDDT.S85525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang ZY, Zhu Z, Wang HF, Qin B, Liu J, Yao XH, Li WC, Chen KS. Downregulation of circDYNC1H1 exhibits inhibitor effect on cell proliferation and migration in hepatocellular carcinoma through miR-140-5p. J Cell Physiol. 2019;234:17775–17785. doi: 10.1002/jcp.28403. [DOI] [PubMed] [Google Scholar]
- 31.Yu W, Deng W, Zhao Q, Zhuang H, Zhang C, Jian Z. miR-501 acts as an independent prognostic factor that promotes the epithelial-mesenchymal transition through targeting JDP2 in hepatocellular carcinoma. Hum Cell. 2019;32:343–351. doi: 10.1007/s13577-019-00243-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.