Abstract
Molecular mechanisms underlying the pathogenesis and progression of malignant thyroid cancers, such as follicular thyroid carcinomas (FTCs), and how these differ from benign thyroid lesions, are poorly understood. In this study, we employed network-based integrative analyses of FTC and benign follicular thyroid adenoma (FTA) lesion transcriptomes to identify key genes and pathways that differ between them. We first analysed a microarray gene expression dataset (Gene Expression Omnibus GSE82208, n = 52) obtained from FTC and FTA tissues to identify differentially expressed genes (DEGs). Pathway analyses of these DEGs were then performed using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) resources to identify potentially important pathways, and protein-protein interactions (PPIs) were examined to identify pathway hub genes. Our data analysis identified 598 DEGs, 133 genes with higher and 465 genes with lower expression in FTCs. We identified four significant pathways (one carbon pool by folate, p53 signalling, progesterone-mediated oocyte maturation signalling, and cell cycle pathways) connected to DEGs with high FTC expression; eight pathways were connected to DEGs with lower relative FTC expression. Ten GO groups were significantly connected with FTC-high expression DEGs and 80 with low-FTC expression DEGs. PPI analysis then identified 12 potential hub genes based on degree and betweenness centrality; namely, TOP2A, JUN, EGFR, CDK1, FOS, CDKN3, EZH2, TYMS, PBK, CDH1, UBE2C, and CCNB2. Moreover, transcription factors (TFs) were identified that may underlie gene expression differences observed between FTC and FTA, including FOXC1, GATA2, YY1, FOXL1, E2F1, NFIC, SRF, TFAP2A, HINFP, and CREB1. We also identified microRNA (miRNAs) that may also affect transcript levels of DEGs; these included hsa-mir-335-5p, -26b-5p, -124-3p, -16-5p, -192-5p, -1-3p, -17-5p, -92a-3p, -215-5p, and -20a-5p. Thus, our study identified DEGs, molecular pathways, TFs, and miRNAs that reflect molecular mechanisms that differ between FTC and benign FTA. Given the general similarities of these lesions and common tissue origin, some of these differences may reflect malignant progression potential, and include useful candidate biomarkers for FTC and identifying factors important for FTC pathogenesis.
Keywords: thyroid cancer, carcinoma, protein-protein interaction, reporter transcription factors, reporter microRNAs, molecular pathways
1. Introduction
Thyroid cancers are the most common type of endocrine malignancy, although they have a relatively low mortality rate compared to most other common metastatic diseases. The United States had 56,460 new diagnoses of thyroid cancer and 1780 related deaths reported in 2012 [1]. The incidence of thyroid cancers is also rising globally at about 5% per year, although some of this increase may be due to improved detection, and it notably affects those in the 20 to 34 year age range [2]. Thyroid cancers include several major types, such as papillary thyroid carcinomas, medullary thyroid carcinoma, anaplastic thyroid carcinoma, and follicular thyroid carcinomas (FTCs) [3]; FTC is one of the more aggressive types, although it accounts for a minority (14%) of total thyroid cancers [4,5,6].
The causes and cellular processes underlying FTC and controlling these tumours’ behaviours are poorly understood; accordingly, these cancers have few effective treatment options [7]. There is therefore a great need to understand the mechanisms that drive development and progression in FTC to identify new approaches to detection, estimate the risk of progression, and find new therapies. In addition, differential diagnosis of FTC is problematic as it can be difficult to distinguish from follicular thyroid adenoma (FTA), a benign and non-invasive lesion. Accordingly, there is more focus on molecular markers that distinguish FTC and FTA (and other types of thyroid lesions). Thus, Wojtas et al. conducted a gene expression comparison of FTC and FTA lesions which identified potential markers that can distinguish FTC from FTA with a sensitivity and specificity of 78% and 80%, respectively [8]. We aim to also use this dataset to identify pathways with different levels of activity in benign and aggressive thyroid tumours (here, FTA and FTC) that may reflect important molecular mechanisms that underlie their behaviour.
For such pathway studies, our starting point is the differential expression of genes (DEGs) between these lesions. We can then use analytic tools to study these DEGs and discover pathways, and pathway hub genes that may affect cell functions and thereby underlie tumour morbidity, growth, and invasion [9,10,11,12]. The repertoire of candidate pathway factors can be extended using approaches such as gene ontology analysis and protein-protein interaction (PPI) studies.
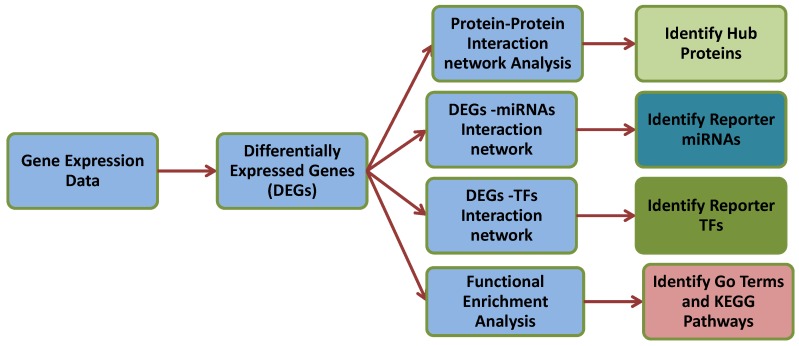
To identify such DEGs, microarray gene expression profiling is widely used [13,14], and several such studies have been performed on thyroid cancer subtypes [15]. For example, Huang et al. [16] studied gene expression profiles of thyroid cancers, although not functional interactions between gene products. To better understand the underlying molecular mechanisms and identify functionally critical biomolecules, integrative analysis within a gene network context is needed [17,18,19,20]. That may lead to candidate FTC biomarkers (the focus of the work by Wojtas et al.), but our main interest here is to obtain key or hub genes distinguishing malignant FTC from benign FTA, as these may be potential therapeutic targets. Such a systems biology approach integrating statistical network and topological analyses of experimental datasets can thus clarify disease mechanisms that are difficult to identify by other approaches [21]. Thus, we used a systems approach (Figure 1) to identify FTC molecular signatures at the miRNA and mRNA levels (and derived from these, the protein-protein interactions) that distinguish FTC tissue from FTA. These analyses should be more meaningful than a comparison between FTC with normal thyroid tissue, as the the two tumour types are similar, while markedly differing in their potential for invasive metastasis.
Figure 1.
The multi-stage analysis methodologies employed in this study. Gene expression datasets related to follicular thyroid carcinoma (FTC) and follicular thyroid adenoma (FTA) tissues were collected from the NCBI Gene Expression Omnibus (NCBI-GEO) database and statistically analysed using GEO2R to identify differential expression of genes (DEGs). Four types of functional enrichment analyses of DEGs were then performed to identify significantly enriched pathways. Thus, we constructed protein-protein interaction networks around DEGs topological analyses to identify putative pathways hub proteins, identified possible micro-RNA (miRNA) and transcription factor (TF) interactors, and used Gene Ontology annotation terms to provide pathways enrichment. TF and miRNA studies employed JASPAR and miRTarbase databases, respectively. DEGs were integrated with those networks, and higher degree and the betweenness centrality were used to designate TFs and miRNAs as the reporter transcriptional regulatory elements. The target DEGs of reporter miRNAs and TFs were subjected to pathway enrichment analyses.
2. Materials and Methods
In this study, the multi-step analysis method we developed and applied is shown in Figure 1. We statistically analysed gene expression datasets to identify the DEGs and their regulatory patterns. We employed these DEGs to identify enriched pathways, biological processes, and annotation terms (i.e., Gene Ontology terms) by using functional enrichment methods. Then, to identify reporter biomolecules, we integrated the intermediate analysis results with biomolecular networks.
2.1. Dataset Employed and Statistical Methods Used
We obtained the gene expression data of FTC and FTA (GSE82208) for our study from the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) [8,22]. This dataset contains analyses of RNA from frozen tumour tissue specimens from 27 FTC and 25 FTA lesions using Affymetrix human genome U133 (Plus 2.0) arrays. The lesions were diagnosed histologically, many with a second diagnosis to confirm, when paraffin embedded material was available [8]. Gene expression analysis using microarrays is a widely used method to develop and refine the molecular determinants of disorders affecting humans. Using these methods, we analysed the gene expression profiles of FTC and FTA. To identify the DEGs between FTC samples and FTA, we ran t-tests using the Limma package [23]. To identify the up-regulated genes, we used the conditions of p-value and (FC, fold change), and for identifying the down-regulated genes, p-value and were used. All identified up-regulated genes and down-regulation genes were considered DEGs. We applied the topological and neighbourhood-based benchmark methods to find gene-gene associations. A gene-gene network was constructed by using the gene-gene associations, where the nodes in the network represent gene [18,24]. This network can also be characterised as a bipartite graph. These topological and neighbourhood-based benchmark methods were adopted from our previous studies [12].
The common neighbours are based on the Jaccard Coefficient method, where the edge prediction score for the node pair is as [25]:
| (1) |
where G is the set of nodes and E is the set of all edges. We used R software packages “comoR” [9] and “POGO” [12] to cross-check the genes-diseases associations.
2.1.1. Functional Enrichment of Gene Sets
We performed gene ontology and pathway analysis on identified up-regulation genes and down-regulation genes using DAVID bioinformatics resources (https://david-d.ncifcrf.gov/) (version v6.8) [26] to obtain further insight into the molecular pathways that differ between FTC and FTA. In these analyses, GO and KEGG pathway databases were used as annotation sources. Enrichment results showing adjusted p-values were considered significant.
2.1.2. Construction and Analysis of Protein-Protein Interaction (PPI) Subnetworks
The PPI network was first constructed with the DEGs and analysed using STRING [27], a web-based visualisation software resource. The constructed PPI network was represented as an undirected graph, where nodes represent the proteins and the edges represent the interactions between the proteins. To construct the PPI network from the STRING database (http://string-db.org) [27], we used database data, data mined from PubMed abstract text. Co-expression, gene fusion, and neighbourhood were active interaction sources and a combined score that was greater than 0.4 was set as the level of significance. The PPI network was then visualised and analysed using Cytoscape (v3.5.1) [28,29]. A topological analysis was applied to identify highly connected proteins (i.e., hub proteins) by using the Cyto-Hubba plugin [30] where betweenness centrality and higher degree were employed. The top three modules (i.e., the three most highly interconnected protein clusters) in the PPI subnetwork were identified using the MCODE plug-in [30]. These modules were further analysed and characterised using enrichment analyses by NetworkAnalyst [31]. The KEGG pathway enrichment analysis of the PPI networks involving DEGs were performed by NetworkAnalyst [31].
2.1.3. Identifying TFs and miRNAs that Influence the Expression of Candidate Genes
To identify TFs and miRNAs that affect transcript levels around which significant changes occur at the transcriptional level, we obtained experimentally verified TF-target genes from the JASPAR database [32] and miRNA-target gene interactions from TarBase [33] and miRTarBase [34] by using NetworkAnalyst tools [31] where betweenness centrality and higher degree filters were used. At the time, there were many techniques to measure the topological properties. We used the degree centrality (DC) and betweenness centrality (BC) to find out network’s topological properties. We can define the DC of a node v in a network as the total number of nodes which are directly connected to node v in that network. The definition can also be written as follows [35]:
whereas n represents total number of nodes in the network, and represents that the node v and the node j are directly connected. In the case of betweenness centrality (BC), the total number of times of node v appearing in the shortest path between other nodes is quantified. It is also defined as follows [35]:
where = total number of shortest paths from node i to node j, and = total number of paths through node v.
3. Results
3.1. Results of DEG Analyses
Transcriptomic Signatures: Differentially Expressed Genes
FTA and FTC tissue gene expression patterns were analysed using oligonucleotide microarrays from the NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE82208) [8]. Gene expression profiling was performed in 27 malignant FTCs and 25 FTAs: 598 genes were differentially expressed ( log2 fold change) relative to FTAs, of which 465 genes were significantly lower expression and 133 genes were higher expression levels in FTC lesions.
3.2. Pathway and Functional Correlation Analysis
By combining large scale and state of the art transcriptome and proteome analysis, we performed a regulatory analysis to gain further insight into the molecular pathways associated with the FTC and predicted links to pathways that differ relative to the benign FTAs. DEGs and pathways were analysed using KEGG pathway database (http://www.genome.jp/kegg/pathway.html) and functional annotation tool DAVID version 6.8 (http://niaid.abcc.ncifcrf.gov) to identify overrepresented pathway groups, and it was observed that four pathways were associated with DEGs with higher expression in FTC compared to FTA; namely, the “one carbon pool by folate” pathway, the p53 signalling pathway, the cell cycle, and the progesterone-mediated oocyte maturation signalling pathway. Fold enrichment, adjusted p-values, and genes associated with these pathways are presented in Table 1 (a).
Table 1.
(a) Part A shows pathways associated with the genes showing higher expression in FTC than FTA. This set showed an enrichment of four KEGG pathways, annotated using DAVID. Part B shows pathways associated with genes showing lower expression levels in FTC. This set showed in an enrichment of eight KEGG pathways using DAVID. Fold enrichment, adjusted p-values and genes associated with these pathways are indicated. (b) Summary of reporter biomolecules (including putative hub genes, TFs, and miRNAs) in that differ between FTC and FTA.
(a)
| KEGG ID | Pathway | Genes in the Pathway | Fold Enrichment | Adjusted p-Value |
|---|---|---|---|---|
| A. | ||||
| hsa00670 | One carbon pool by folate | MTHFD2, TYMS, SHMT2, ALDH1L1 | 24 | 5.21E-04 |
| hsa04115 | p53 signaling pathway | CDK1, CCNB2, RRM2, PMAIP1, IGFBP3 | 9 | 2.03E-03 |
| hsa04110 | Cell cycle | CDK1, MAD2L1, CCNB2, BUB1, BUB1B | 5 | 1.77E-02 |
| hsa04914 | Progesterone-mediated oocyte maturation | CDK1, MAD2L1, CCNB2, BUB1 | 6 | 3.31E-02 |
| B. | ||||
| hsa04978 | Mineral absorption | MT1M, MT2A, CYBRD1, MT1E, MT1H, MT1X, MT1G, MT1F | 6 | 3.44E-04 |
| hsa04918 | Thyroid hormone synthesis | SLC26A4, TG, HSP90B1, PAX8, TPO, HSPA5, LRP2, TSHR, IYD | 4 | 9.55E-04 |
| hsa05200 | Pathways in cancer | FGFR2, BCR, EPAS1, PGF, RXRB, GNAI1, RUNX1T1, FZD1, CDH1, CDK6, KIT, CTNNA1, MMP2, CXCL12, TCF7L1, AGTR1, FOS, HSP90B1, JUN, PAX8, SOS2, LAMB1 | 2 | 5.48E-03 |
| hsa04915 | Estrogen signaling pathway | HSPA1L, FOS, HSP90B1, GNAI1, JUN, SOS2, HSPA1A, HSPA1B, MMP2 | 3 | 8.41E-03 |
| hsa00600 | Sphingolipid metabolism | SPTLC1, SGMS2, SPTLC3, GALC, PLPP3, ASAH1 | 4 | 1.17E-02 |
| hsa04360 | Axon guidance | NRP1, LIMK2, SEMA6D, FYN, GNAI1, PLXNA2, SEMA3D, UNC5C, CXCL12, EPHA3 | 3 | 1.22E-02 |
| hsa04141 | Protein processing in endoplasmic reticulum | HSPA1L, HSP90B1, ERO1B, UBE4B, PDIA6, HSPA1A, HSPA1B, HSPA5, CALR, MAN1C1, SEC62 | 2 | 2.68E-02 |
| hsa04510 | Focal adhesion | ITGA9, CAV2, CAV1, CCND2, FYN, PGF, JUN, SOS2, ITGB3, LAMB1, KDR, MYL9 | 2 | 3.95E-02 |
(b)
| Symbol | Description | Feature |
|---|---|---|
| EGFR | The epidermal growth factor receptor | Hub protein |
| KIT | KIT proto-oncogene receptor tyrosine kinase | Hub protein |
| IRS1 | Insulin receptor substrate 1 | Hub protein |
| KDR | kinase insert domain receptor | Hub protein |
| BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B | Hub protein |
| CDH1 | Cadherin-1 | Hub protein |
| BUB1 | BUB1 mitotic checkpoint serine/threonine kinase | Hub protein |
| TEK | TEK receptor tyrosine kinase | Hub protein |
| TPM2 | tropomyosin 2 | Hub protein |
| NR4A2 | nuclear receptor subfamily 4 group A member 2 | Hub protein |
| FOXC1 | Forkhead Box C1 | Reporter Transcription Factor |
| GATA2 | GATA Binding Protein 2 | Reporter Transcription Factor |
| YY1 | YY1 Transcription Factor | Reporter Transcription Factor |
| FOXL1 | Forkhead Box L1 | Reporter Transcription Factor |
| SPTLC1 | Serine Palmitoyltransferase Long Chain Base Subunit 1 | Reporter Transcription Factor |
| E2F1 | E2F Transcription Factor 1 | Reporter Transcription Factor |
| CD36 | cluster of differentiation 36 | Reporter Transcription Factor |
| SRF | Serum Response Factor | Reporter Transcription Factor |
| POU2F2 | POU class 2 homeobox 2 | Reporter Transcription Factor |
| EGR2 | Early growth response 2 | Reporter Transcription Factor |
| hsa-mir-548c-3p | MicroRNA 548c | Reporter microRNA |
| hsa-mir-335-5p | MicroRNA 335 | Reporter microRNA |
| hsa-mir-26b-5p | MicroRNA 26b | Reporter microRNA |
| hsa-mir-155-5p | MicroRNA 155 | Reporter microRNA |
| hsa-mir-124-3p | MicroRNA 124 | Reporter microRNA |
| hsa-mir-1-3p | MicroRNA 1 | Reporter microRNA |
| hsa-mir-16-5p | MicroRNA 16 | Reporter microRNA |
| hsa-mir-1-1 | MicroRNA 1 | Reporter microRNA |
| hsa-mir-192-5p | MicroRNA 192 | Reporter microRNA |
| hsa-mir-215-5p | MicroRNA 215 | Reporter microRNA |
It was also observed that eight pathways were significantly over-represented. Mineral absorption, thyroid hormone synthesis, pathways in cancer, oestrogen signalling, sphingolipid metabolism, protein processing in the endoplasmic reticulum, axon guidance, and focal adhesion pathways (Table 1 (a)) were identified. These are associated with the DEGs expressed at lower levels in FTC lesions compared to FTA. We then performed GO analysis using DAVID to obtain further insight into the molecular roles and biological function of DEGs identified in this study. From this analysis, nine GO groups were associated with DEGs that were more highly expressed in FTC (see Table 2 (a)). Reflecting their greater number, the genes with lower expression in FTC compared to FTA were associated with 80 GO groups; the 10 most significant GO groups are presented in Table 2 (b). Fold enrichment, adjusted p-values, and genes associated with these ontologies are also indicated.
Table 2.
(a) Gene ontologies associated with genes with higher expression in FTC. (b) Gene ontologies associated with genes with lower expression in FTC compared to FTA. The 10 most significant biological functional ontologies are indicated and are those associated with these significant genes. Fold enrichment, adjusted p-values and genes associated with these ontologies are presented in these tables.
(a)
| Go Term | Pathway | Genes in the Pathway | Fold Enrichment | Adjusted p-Value |
|---|---|---|---|---|
| GO:0006636 | unsaturated fatty acid biosynthetic process | ELOVL4, SCD, ELOVL7 | 45 | 1.88E-03 |
| GO:0007094 | mitotic spindle assembly checkpoint | MAD2L1, BUB1, BUB1B | 33 | 3.39E-03 |
| GO:0045773 | positive regulation of axon extension | TNFRSF12A, MAP1B, FN1 | 24 | 6.83E-03 |
| GO:0044772 | mitotic cell cycle phase transition | CDK1, CKS2 | 67 | 2.92E-02 |
| GO:0046602 | regulation of mitotic centrosome separation | KIF11, NEK2 | 67 | 2.92E-02 |
| GO:0021930 | cerebellar granule cell precursor proliferation | ATF5, RORA | 45 | 4.35E-02 |
| GO:0035999 | tetrahydrofolate interconversion | TYMS, SHMT2 | 45 | 4.35E-02 |
| GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process | ELOVL4, ELOVL7 | 45 | 4.35E-02 |
| GO:0046777 | protein autophosphorylation | EGFR, NEK2, STK26, MELK | 5 | 4.78E-02 |
(b)
| Go Term | Pathway | Genes in the Pathway | Fold Enrichment | Adjusted p-Value |
|---|---|---|---|---|
| GO:0045926 | negative regulation of growth | ING5, MT1M, MT2A, MT1E, MT1H, MT1X, MT1G, MT1F | 18 | 1.37E-07 |
| GO:0071294 | cellular response to zinc ion | MT1M, MT2A, MT1E, MT1H, MT1X, MT1G, MT1F | 16 | 3.17E-06 |
| GO:0006590 | thyroid hormone generation | DIO2, CPQ, FOXE1, TPO, DIO1, IYD | 21 | 4.56E-06 |
| GO:0001525 | angiogenesis | FGFR2, EMCN, CAV1, NRP1, ACVRL1, EPAS1, PGF, MMP2, KDR, PKNOX1, CTGF, ID1, JUN, MAPK14, TEK, SCR, RHOB, CALCRL | 3 | 1.89E-05 |
| GO:0001938 | positive regulation of endothelial cell proliferation | CAV2, NRP1, ACVRL1, PGF, F3, JUN, TEK, ITGB3, CXCL12, KDR | 6 | 2.97E-05 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | FGFR2, GLIS3, TSHZ3, ZMYND11, CAV1, TFCP2L1, ZNF366, PRDM16, CALR, CBX7, CITED2, MINA, TCF4, BHLHE41, NFIL3, NR2F1, TXNIP, EGR1, NR4A2, TLE1, FOSB, SNAI2, FOXP2, CD36, BTG2, ID1, SALL1, FOXE1, LRP8, ID3, PRDM1, SMARCA2, KLF4, NFIA, RERE, PEG3 | 2 | 3.16E-05 |
| GO:0010628 | positive regulation of gene expression | CAV1, EPHX2, GJA1, TLE1, CDK6, KIT, CALR, CITED2, ANK2, GSN, CTGF, ID1, MAPK14, CYP26B1, NTRK2, RGCC, PRDM1, NFIL3, KLF4 | 3 | 4.33E-05 |
| GO:0030198 | extracellular matrix organization | B4GALT1, FBN1, OLFML2A, CDH1, ITGB3, KDR, CSGALNACT1, SMOC2, ITGA9, TNFRSF11B, COL9A3, ERO1B, FBLN5, JAM2, LAMB1, CYR61 | 4 | 5.55E-05 |
| GO:0043434 | response to peptide hormone | BTG2, GNAI1, CTGF, TEK, ANXA1, GJA1, CXCL12, IRS1 | 8 | 6.31E-05 |
| GO:0071276 | cellular response to cadmium ion | MT1E, MT1H, MT1X, MT1G, MT1F | 13 | 5.36E-04 |
3.2.1. Proteomic Signatures: Hub Target Proteins from PPI Analysis
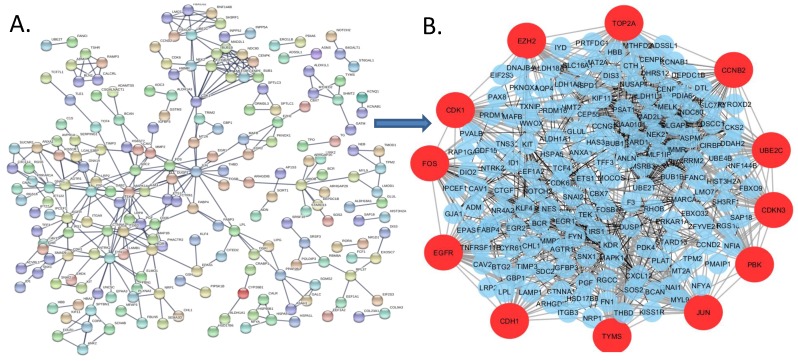
A PPI network was constructed using the DEGs identified in this study (Figure 2A) by using the STRING package. Topological analyses using STRING and further analysis by Cytoscape’s Cyto-Hubba plugin identified the most significant hub proteins, which were identified as gene products of TOP2A, JUN, EGFR, CDK1, FOS, CDKN3, EZH2, TYMS, PBK, CDH1, UBE2C, and CCNB2. The simplified PPI network for the most significant hub genes was constructed by using Cyto-Hubba plugin and is shown in Figure 2B.
Figure 2.
(A) The protein-protein interaction network constructed using the DEGs identified in the FTC/FTA comparison. (B) The protein-protein interaction(PPI) network identified in FTC/FTA dataset DEGs showing the most significant hub genes on the periphery (red).
3.2.2. Enrichment Analysis of the Modules Found in PPI of DEG
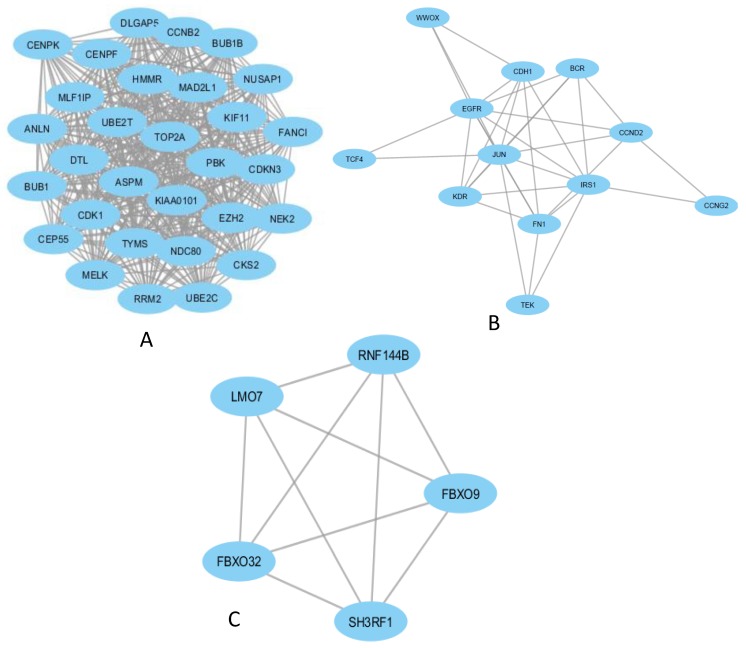
The PPI network was analysed by using MCODE plug-in in Cytoscape (version 3.5.1), and top three modules were selected (Figure 3).
Figure 3.
Top three modules in the protein-protein interaction network of the DEGs in TC. The nodes indicate the DEGs and the edges indicate the interactions between two genes. (A): Module-1; (B): Module-2; (C): Module-3.
The enrichment analysis of the top three modules was analysed by using DAVID. Module 1 represented a set of biological process significantly enriched for ATP binding, cell division, and cell proliferation; the associated enriched cellular components were the kinetochore, the condensed chromosome kinetochore (both of which relate to cell division), and nucleoplasm and cytoplasm, which are very general categories. Molecular functions significantly enriched in Module 1 included chromatin binding and protein kinase activity (Table 3 (a)). Module 2 showed significantly enriched biological process positive regulation of protein phosphorylation and protein autophosphorylation; these significantly enriched cellular components included membrane raft, endosome, apical plasma membrane, and focal adhesion. Molecular functions of Module 2 were enriched in protein tyrosine kinase activity, enzyme binding, protein binding, and integrin binding (Table 3 (b)).
Table 3.
(a) The enrichment analysis of the subnetwork module-1 (b) The enrichment analysis of the subnetworks module-2 and module-3.
(a) Subnetwork Module 1
| Biological Process of Subnetwork Module 1 | ||
|---|---|---|
| Term | p Values | Genes |
| ATP binding | 1.00E-13 | CDK1, KIF11, CCNB2, NEK2, BUB1, CENPF, BUB1B, NDC80, ANLN, PBK, CEP55, ASPM |
| Cell division | 1.28E-10 | CDK1, KIF11, MAD2L1, CCNB2, NEK2, CKS2, BUB1, CENPF, BUB1B, NDC80, UBE2C |
| Sister chromatidcohesion | 6.92E-07 | MAD2L1, BUB1, CENPF, BUB1B, NDC80, CENPK |
| Cell proliferation | 1.76E-06 | CDK1, TYMS, DLGAP5, CKS2, BUB1, CENPF, BUB1B, MELK |
| mitotic spindle assembly checkpoint | 4.64E-06 | MAD2L1, BUB1, CENPF, BUB1B |
| chromosome segregation | 4.68E-06 | KIF11, NEK2, CENPF, NDC80, TOP2A |
| mitotic sister chromatidsegregation | 9.32E-06 | MAD2L1, NEK2, NUSAP1, NDC80 |
| G2/M transition of mitotic cell cycle | 7.46E-05 | CDK1, CCNB2, NEK2, MELK, HMMR |
| negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 2.20E-04 | CDK1, MAD2L1, BUB1B, UBE2C |
| positive regulation of ubiquitin-protein ligase activity involved in regulation of mitotic cell cycle transition | 2.69E-04 | CDK1, MAD2L1, BUB1B, UBE2C |
| Cellular Component of Subnetwork Module 1 | ||
| Kinetochore | 1.39E-07 | MAD2L1, NEK2, BUB1, CENPF, BUB1B, NDC80 |
| Condensed chromosome kinetochore | 1.99E-07 | MAD2L1, NEK2, BUB1, BUB1B, NDC80, CENPK |
| Nucleoplasm | 1.74E-05 | CDK1, DTL, EZH2, KIAA0101, CENPF, ANLN, UBE2C, CENPK, TYMS, CCNB2, FANCI, RRM2, BUB1, TOP2A, UBE2T |
| Nucleus | 3.38E-05 | CDK1, DTL, NEK2, DLGAP5, EZH2, KIAA0101, NUSAP1, CENPF, NDC80, PBK, CDKN3, CENPK, TYMS, CCNB2, MAD2L1, RRM2, TOP2A, UBE2T, ASPM, MELK |
| Midbody | 4.30E-05 | CDK1, NEK2, CENPF, CEP55, ASPM |
| Cytoplasm | 9.43E-05 | CDK1, KIF11, DTL, NEK2, DLGAP5, EZH2, KIAA0101, NUSAP1, CENPF, UBE2C, CDKN3, TYMS, FANCI, RRM2, BUB1, BUB1B, TOP2A, UBE2T, ASPM |
| Centrosome | 4.30E-04 | CDK1, CCNB2, DTL, NEK2, CENPF, CEP55 |
| Cytosol | 5.65E-04 | CDK1, KIF11, NEK2, CENPF, NDC80, UBE2C, CENPK, HMMR, TYMS, CCNB2, MAD2L1, RRM2, BUB1, BUB1B |
| Spindle pole | 6.11E-04 | KIF11, MAD2L1, NEK2, CENPF |
| Spindle microtubule | 0.0020693 | CDK1, KIF11, NUSAP1 |
| Molecular Function of Subnetwork Module 1 | ||
| Chromatin binding | 3.63E-05 | CDK1, EZH2, CKS2, KIAA0101, CENPF, TOP2A, UBE2T |
| Protein kinaseactivity | 2.78E-04 | CDK1, NEK2, BUB1, BUB1B, PBK, MELK |
| protein serine/threoninekinaseactivity | 3.44E-04 | CDK1, NEK2, BUB1, BUB1B, PBK, MELK |
| ATP binding | 4.78E-04 | CDK1, KIF11, NEK2, BUB1, BUB1B, PBK, UBE2C, TOP2A, UBE2T, MELK |
| Protein binding | 9.40E-04 | CDK1, DTL, NEK2, DLGAP5, EZH2, KIAA0101, NUSAP1, CENPF, NDC80, PBK, CEP55, UBE2C, CENPK, CDKN3, HMMR, MAD2L1, CCNB2, FANCI, RRM2, BUB1, CKS2, BUB1B, TOP2A, MELK |
| Ubiquitinconjugating enzyme activity | 4.70E-2 | UBE2C, UBE2T |
(b) Module 2 and Module 3
| Biological Process of Module-2 | ||
|---|---|---|
| Term | p Values | Genes |
| positive regulation of protein phosphorylation | 6.67E-05 | EGFR, CCND2, TEK, KDR |
| peptidyl-tyrosine phosphorylation | 1.16E-04 | EGFR, BCR, TEK, KDR |
| protein autophosphorylation | 1.64E-04 | EGFR, BCR, TEK, KDR |
| positive regulation of cell proliferation | 1.65E-04 | EGFR, CCND2, IRS1, KDR, FN1 |
| positive regulation of ERK1 and ERK2 cascade | 1.73E-04 | EGFR, JUN, TEK, KDR |
| positive regulation of GTPase activity | 3.47E-04 | EGFR, BCR, JUN, TEK, IRS1 |
| Angiogenesis | 3.52E-04 | JUN, TEK, KDR, FN1 |
| positive regulation of fibroblast proliferation | 5.48E-04 | EGFR, JUN, FN1 |
| positive regulation of endothelial cell proliferation | 8.94E-04 | JUN, TEK, KDR |
| transmembrane receptor protein tyrosine kinase signaling pathway | 1.72E-03 | EGFR, TEK, KDR |
| Molecular Function of Subnetwork Module-2 | ||
| protein tyrosine kinase activity | 5.51E-05 | EGFR, BCR, TEK, KDR |
| transmembrane receptor protein tyrosine kinase activity | 2.20E-04 | EGFR, TEK, KDR |
| enzyme binding | 8.24E-04 | EGFR, BCR, JUN, WWOX |
| protein binding | 1.45E-03 | EGFR, BCR, CCND2, JUN, TEK, CDH1, TCF4, IRS1, WWOX, KDR, FN1 |
| integrin binding | 1.67E-03 | EGFR, KDR, FN1 |
| Ras guanyl-nucleotide exchange factor activity | 2.00E-03 | EGFR, TEK, IRS1 |
| receptor signaling protein tyrosine kinase activity | 5.91E-03 | EGFR, KDR |
| identical protein binding | 8.26E-03 | EGFR, JUN, TCF4, FN1 |
| growth factor binding | 1.59E-02 | TEK, KDR |
| chromatin binding | 2.13E-02 | EGFR, JUN, TCF4 |
| Cellular Component of Subnetwork Module-2 | ||
| membrane raft | 6.54E-03 | EGFR, TEK, KDR |
| endosome | 7.76E-03 | EGFR, CDH1, KDR |
| apical plasma membrane | 1.27E-02 | EGFR, TEK, FN1 |
| focal adhesion | 2.22E-02 | EGFR, TEK, CDH1 |
| cell junction | 3.00E-02 | BCR, CDH1, KDR |
| microvillus | 3.39E-02 | TEK, WWOX |
| Biological Process of Subnetwork Module-3 | ||
| protein ubiquitination | 3.82E-05 | SH3RF1, LMO7, FBXO32, FBXO9 |
| Molecular Function of Subnetwork Module-3 | ||
| ubiquitin ligase complex | 2.07E-04 | RNF144B, LMO7, FBXO9 |
| SCF ubiquitin ligase complex | 1.09E-02 | FBXO32, FBXO9 |
| Cellular Component of Subnetwork Module-3 | ||
| ubiquitin-protein transferase activity | 2.89E-05 | RNF144B, LMO7, FBXO32, FBXO9 |
| zinc ion binding | 2.62E-02 | SH3RF1, RNF144B, LMO7 |
Module 3’s gene ontology annotation showed a significant enrichment for protein ubiquitination, with significantly enriched cellular components being ubiquitin ligase complex and SCF ubiquitin ligase complex, which have involvement in proteosomal and autophagy functions. Module 3’s molecular functions showed significantly enrichment in ubiquitin-protein transferase activity and zinc ion binding (Table 3 (b)). It is important to note that pathway enrichment analyses found Modules 1 and 2 were significantly enriched in protein binding pathways, which was not seen in Module 3 (Table 3 (b)).
3.3. Regulatory Signatures: TFs and miRNAs Affecting DEG Transcript Levels
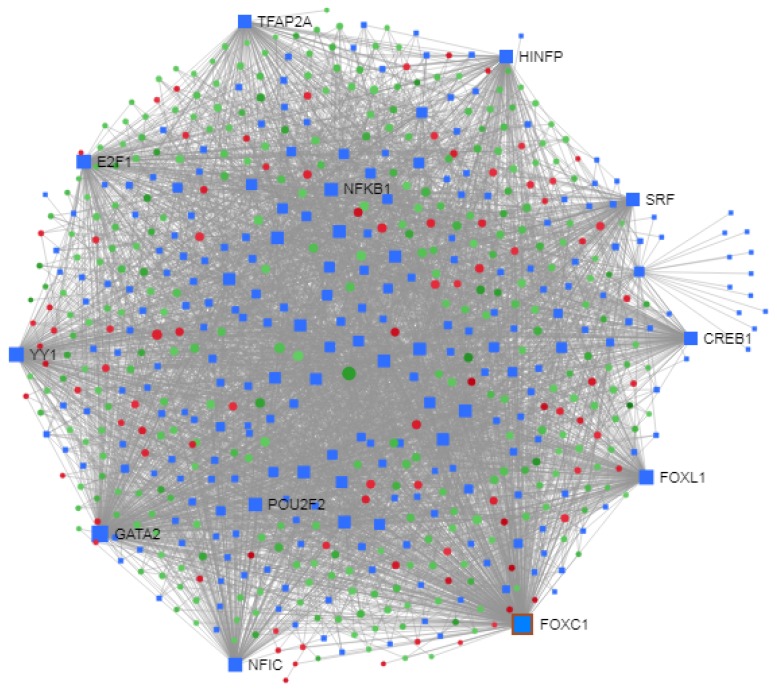
3.3.1. Transcriptional Regulatory Network Construction and TF Enrichment Analysis
The construction of reporter TFs and DEGs interaction network by NetworkAnalyst revealed a number of potentially important TFs selected by the topological analysis using a dual metric approach involving degree and betweenness (Figure 4). The top 10 ranked TFs with the highest degree and betweenness centrality included FOXC1, GATA2, YY1, FOXL1, E2F1, NFIC, SRF, TFAP2A, HINFP, and CREB1. Pathway enrichment analysis of the TFs that differ between FTC and FTA and DEGs mainly identified as statistically significant, a number of relevant pathways which included transcriptional misregulation in cancer, acute myeloid leukaemia, pathways in cancer, thyroid cancer, cell cycle, dorso-ventral axis formation, arrhythmogenic right ventricular cardiomyopathy (ARVC), cocaine addiction, and prostate cancer pathways (Table 4 (b)).
Figure 4.
Construction of regulatory networks of TF-DEG interactions. Red nodes indicate up-regulation and green nodes represent down-regulated DEGs.
Table 4.
(a) Top 10 KEGG pathway in genes of micro-RNAs and differentially expressed genes interaction networks (b) Top 10 KEGG pathway in genes of TFs and differentially expressed genes interaction networks
(a)
| Pathway | Total | Expected | Hits | p-Value |
|---|---|---|---|---|
| Endocytosis | 101 | 0.906 | 4 | 1.22E-02 |
| PPAR signaling pathway | 64 | 0.574 | 3 | 1.92E-02 |
| Hypertrophic cardiomyopathy(HCM) | 25 | 0.224 | 2 | 2.07E-02 |
| Complement and coagulation cascades | 67 | 0.601 | 3 | 2.16E-02 |
| Cytokine-cytokine receptor interaction | 253 | 2.27 | 6 | 2.40E-02 |
| Adherens junction | 70 | 0.628 | 3 | 2.43E-02 |
| Thyroid cancer | 28 | 0.251 | 2 | 2.56E-02 |
| Glycine, serine and threonine metabolism | 33 | 0.296 | 2 | 3.48E-02 |
| Tyrosine metabolism | 33 | 0.296 | 2 | 3.48E-02 |
| Pathways in cancer | 310 | 2.78 | 6 | 5.62E-02 |
(b)
| Pathway | Total | Expected | Hits | p-Value |
|---|---|---|---|---|
| Pathways in cancer | 310 | 5.37 | 20 | 1.66E-07 |
| Chronic myeloid leukemia | 73 | 1.26 | 8 | 3.07E-05 |
| MAPK signaling pathway | 265 | 4.59 | 15 | 3.67E-05 |
| Maturity onset diabetes of the young | 24 | 0.415 | 5 | 4.50E-05 |
| Pertussis | 52 | 0.9 | 6 | 2.41E-04 |
| Transcriptional misregulation in cancer | 19 | 0.329 | 4 | 2.65E-04 |
| Acute myeloid leukemia | 57 | 0.987 | 6 | 4.00E-04 |
| HTLV-I infection | 199 | 3.44 | 11 | 5.53E-04 |
| Prostate cancer | 87 | 1.51 | 7 | 6.87E-04 |
| Thyroid cancer | 28 | 0.485 | 4 | 1.24E-03 |
3.3.2. miRNA Regulatory Network Construction and Enrichment Analysis of miRNA
We constructed TC DEG-miRNA interactions networks using NetworkAnalyst (Figure 5). By topological analysis, the top 10 ranked miRNAs by highest degree and betweenness centrality were selected, which included hsa-mir-335-5p, hsa-mir-26b-5p, hsa-mir-124-3p, hsa-mir-16-5p, hsa-mir-192-5p, hsa-mir-1-3p, hsa-mir-17-5p, hsa-mir-92a-3p, hsa-mir-215-5p, and hsa-mir-20a-5p. The analysis indicates that these are miRNAs having the highest potential to regulate levels of the DEG transcripts differently in FTC and FTA. The pathway enrichment analysis of the miRNA associated DEGs networks identified the p53 signalling pathway, dorso-ventral axis formation, pertussis, adherens junction, thyroid cancer, pathways in cancer, focal adhesion, sphingolipid metabolism, PPAR signalling pathway, and the ECM-receptor interaction signalling system as statistically significant (Table 4 (a))
Figure 5.
Construction of regulatory networks of the DEGs-miRNAs interaction. Red nodes indicate up-regulation and green nodes represent down-regulated DEGs.
4. Discussion
To clarify the molecular mechanisms underlying the pathological features (especially malignancy potential) that distinguish malignant and benign thyroid lesions, we examined differential gene expression between FTC and FTA. Such a comparison is likely to be more informative than making such a comparison of malignant and normal tissues, which would contain many more differences that relate to neoplastic cell development. In addition, the lesions contain significant non-neoplastic cell components (such as mesenchymally-derived and endothelial cells) that are also likely to differ markedly from those of normal tissue.
The DEGs identified from the FTC/FTA comparison were used to identify possible regulatory patterns, key molecular pathways, and protein-protein interactions among these pathway gene products. Such an analytical approach can be used to find molecular signatures that may serve either as potential therapeutic targets or as biomarkers to differentially diagnose or identify FTC. As did the original study by Wojtas et al. [8], we identified significantly altered genes that may be candidate biomarkers that distinguish FTC. However, another important use of such data (and our main focus here) is to identify and characterise biological functions associated with these genes that may give insights into the biology and behaviour of FTC lesions themselves. Wang et al. [36] examined on normal tissue vs. FTC and normal tissue vs. FTA in the microarray dataset accession number GSE27155. They identified some important pathways that included drug metabolism cytochrome p450, viral carcinogenesis, tyrosine metabolism, cytokine-cytokine receptor interaction, and cocaine Addition as significant pathways distinguishing normal from FTC tissue. Our analysis of the FTA vs. FTC tissue data found several important pathways, as described in the above results section. However, there were no pathways that we identified (in our FTC/FCA comparison) which were also seen in the studies comparing FTC to normal tissues. This almost certainly reflects the fact that a tumour has very different cellular profiles compared to normal tissue (e.g., infiltrating leukocytes and stromal cells) so in this circumstance, where we want to better understand the behaviour of the tumour, it is be better to compare with a benign or non-aggressive tumour such as FTA. Our study thus identified a number of important pathways that were differentially expressed in FTC, some of which would be expected by the malignant profile of FTC compared to FTA. For example, the p53 signalling pathway plays an important role in cancer cell apoptosis and DNA repair by causing cycle arrest in response to DNA damage [37], and has been previously implicated in thyroid cancers [38]. Cell cycle progression is regulated and facilitated by cyclin-dependent kinases that are activated by cyclins such as cyclin D1. Indeed, cyclin D1 levels can influence tumour progression, and may have prognostic significance in FTC [39]. Related to this, it has also been reported that the one-carbon metabolism pathway is actively involved in cancer progression, probably due to its involvement in nucleotide synthesis [40]. We have summarised the significant GO groups associated with the DEGs, as shown in Table 2.
PPI network reconstruction and the analysis of reconstructed PPI network represent a powerful approach for understanding disease mechanisms, so to construct a PPI network for the DEGs in our study, we combined results of statistical analyses with the protein interactome network. To identify potential hub proteins, topological analysis strategies were employed. This method identified 12 hub genes (Table 1 (b)), including a number of genes commonly associated with cancers (including thyroid cancers), such as EGRF, JUN, FOS, CDK1, and other cyclin pathway genes, E-cadherin (CDH1) and E2F1. The hub protein EGFR is linked to growth in many types of cancer, and cardiovascular diseases, and has been previously linked to thyroid cancers [41]. JUN and FOS are best characterised as subunits of the AP-1 transcription factor that has a crucial role in many types of cell differentiation and inflammation processes [42]. GO databases also delineated particular roles for JUN in angiogenesis and the regulation of endothelial cell proliferation, both of which have central roles in metastasis and invasion (Table 2 (b)). CDK1 is an important cell cycle regulator and is involved in breast, lung, and ovary carcinomas [43]. CDK1 overexpression has been documented in lung cancer, lymphoma, and advanced melanoma, while loss of cytoplasmic CDK1 predicts poor patient survival and may confer chemotherapeutic resistance in the latter [44]. Cyclin B1 (in the same family as CDK1) overexpression and/or mislocalisation has been described in several primary cancers, including thyroid carcinoma and colon, gastric, prostate, breast, and non small-cell lung cancers [44]. It is also actively involved in mitotic cell cycle phase transition (Table 2 (a)). CDKN3 encodes a cyclin inhibitor (regulating cell cycle) and has been described as being overexpressed in lung adenocarcinoma (ADC), squamous cell carcinoma (SCC), hepatocellular carcinoma, cervical cancer, and epithelial ovarian cancer [45]. Over-expression of EZH2 is frequently observed in many cancer types [46].
TYMS expression is also associated with the risk of development of epithelial cancers and lymphoma cancer [47]. In addition, in hereditary diffuse gastric cancer (HDGC), the hub gene CDH1 mutations are connected with an increased incidence of lobular carcinoma of the breast, and possibly, prostate cancer and colorectal carcinoma [48]. This gene is also involved in extracellular matrix organisation (Table 2 (b)). Hub gene CCNB2 is known to be underexpressed in thyroid cell tumours [49]. We found that CDK1 and CCNB2 hubs genes were actively associated with the p53 signalling, progesterone-mediated oocyte maturation, and cell cycle pathways. The hub gene TYMS is also involved in one carbon pool by folate pathway; FOS, JUN, and CDH1 genes are involved in pathways in cancer; and JUN and FOS genes are involved in the oestrogen signalling pathway. A number of other hub genes have been described as associated with malignant thyroid cancers, including EXH2, UBCH10,TFAP2A, TOP2A, and SRF [50,51,52,53,54]. By considering the possible roles of these hub proteins in pathogenesis of FTC and other related diseases, new roles for these proteins may be identified. In addition to the above-mentioned hub genes, we also identified a number of factors not previously noted as having a role in FTC or thyroid cancers, including UBA52, GATA2, FOXL1, and NFIC.
As the regulation of gene expression is controlled by TFs and miRNAs at post-transcriptional and/or transcriptional levels, changes in these molecules may provide crucial information regarding the dysregulation of gene expression in FTC. Thus, we investigated TFs and their relationship to DEGs in these tumours, including FOXC1. One prior study found that FOXC1 is strongly associated with thyroid cancers [55]. The overexpression of YY1 in differentiated thyroid cancers has also been noted [56]. The pathway analysis of these TFs showed statistically significance in FTC. From the DEGs, a miRNA interaction network of miRNAs (Table 1 (b)) was also identified and analysed. Our pathway analysis of these miRNA also showed statistically significant relationships with FTC. Notably, the target DEGs of these miRNAs and TFs included the p53 signalling pathway, PPAR signalling pathway, dorso-ventral axis formation pathway, focal adhesion pathway (Table 4 (a)), pathways in cancer, transcriptional misregulation in cancer, and thyroid cancer pathway. The miRNA species we identified that have previously been shown to have a role in thyroid cancer, including hsa-mir-335-5p, hsa-124-3p, hsa-mir-17-5p, and hsa-mir-20a-5p [57,58,59,60]. However, six other miRNAs we identified have not previously been associated with thyroid lesions. miRNA species have wide ranging effects on gene expression which are indirect, so further analysis for these are needed.
We analysed the dataset originally generated by Wojtas et al., who used combined micro-array methods and literature meta-analysis to achieve this to find markers to distinguish FTC and FTA. The gene markers they identified included TFF3, CPQ (previously characterised FTC markers), PLVAP, and ACVRL1, all genes expressed in the thyroid, although ACVR1 is an activin receptor with wide expression. These latter genes were not identified by our analytical approach, but it should be noted that our study used very different analytic tools, focussing on identifying pathways and on finding evidence for gene functions using PPI and other resources. Our analytical approach was somewhat similar to that of Wang et al. [36] who examined an older thyroid cancer dataset (GSE27155) containing 10 FTA, 14 FTC, and 4 normal thyroid tissue samples plus those of other types of thyroid lesions. While we employed a number of additional analytical tools (some developed previously by us), there were a number of significant genes and pathways that were also found in the work by Wang et al. [36] where they compared FTC and FTA. These notably included the AP-1 TF and its components (e.g., JUN and FOS), BCL2, factors relating to DNA damage, and cyclin-related factors that regulate the cell cycle. As noted above, We identified no genes that Wang et al. identified when comparing FTC and normal tissues. Recent studies by Shang et al. and by Liang and Sun [61] examined datasets generated from papillary thyroid cancer lesions that were compared with normal tissues. DEGs identified in these studies were analysed by methods related to our approach [35]. However, unlike the Wang et al. study cited above, very few of the hub genes these studies identified were found in our analysis, BCL2 being the main exception. This lack of concordance with the FTC studies may reflect the different tumour types (and lower malignancy rates) represented by papillary thyroid cancer, and it might be useful to compare all these datasets in further detail to determine how these types of tumour differ. We also note that work by Pfeifer et al. [62] developed a classification using gene expression data that considered only five genes, and this distinguished FTC from FTA with good accuracy. They identified five genes (ELMO1, EMCN, ITIH5, KCNAB1, and SLCO2A1) as biomarkers. However, it should be emphasised that the focus of our work and methodology is different (since we studied pathway differences not gene classifiers), so not surprisingly, there were no significant genes identified in common between our work and Pfeifer et al.
5. Conclusions
Our data-driven approach has uncovered a number of significant molecular mechanisms that may underlie the pathogenic differences seen between FTC and FTA. In this study we used integrated bioinformatic analyses to study gene expression profiles in FTC and FTA, and used this information to identify candidate hub proteins that could play significant roles in FTC development. Our results included a number candidate genes and pathways that indicate the directions for future experimental work needed to clarify the roles of these cellular factors. This study identified gene networks that advance our understanding of FTC pathogenesis and indicates new avenues to develop therapies for FTC.
Abbreviations
The following abbreviations are used in this manuscript:
| ADC | adenocarcinoma |
| DEG | differencially expressed genes |
| FC | fold change |
| FTC | follicular thyroid carcinomas |
| FTA | follicular thyroid adenoma |
| GO | Gene Ontology |
| GEO | Gene Expression Omnibus |
| HDGC | hereditary diffuse gastric cancer |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| miRNA | micro-RNA |
| PPI | protein-protein Interactions |
| SCC | squamous cell carcinoma |
| TF | transcription factor |
Author Contributions
Conceptualisation, M.A.H., M.A.M., and T.A.A.; methodology, M.A.H. and T.A.A.; software, M.A.H.; validation, M.A.H., T.A.A., and M.A.M.; formal analysis, M.A.H.; investigation, M.A.H.; resources, M.A.H.; data curation, M.A.H. and T.A.A.; writing—original draft preparation, M.A.H.; writing—review and editing, M.A.H., M.M.R., J.M.W.Q., A.A.M., and M.A.M.; visualisation, M.A.H. and T.A.A.; supervision, M.A.M.; project administration, S.U., A.A.M., and M.A.M.; funding acquisition, S.U. and M.A.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
We have No conflict of interest.
References
- 1.Jung K.W., Won Y.J., Kong H.J., Oh C.M., Seo H.G., Lee J.S. Cancer statistics in Korea: Incidence, mortality, survival and prevalence in 2010. Cancer Res. Treat. 2013;45:1. doi: 10.4143/crt.2013.45.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thyroid Cancer: Statistics—Cancer Net. [(accessed on 6 October 2018)];2017 Available online: https://www.cancer.net/cancer-types/thyroid-cancer/statistics.
- 3.Bansal A., Ramana J. TCGDB: A compendium of molecular signatures of thyroid cancer and disorders. J. Cancer Sci. Ther. 2015;7:198–201. [Google Scholar]
- 4.Nguyen Q.T., Lee E.J., Huang M.G., Park Y.I., Khullar A., Plodkowski R.A. Diagnosis and treatment of patients with thyroid cancer. Am. Health Drug Benefits. 2015;8:30. [PMC free article] [PubMed] [Google Scholar]
- 5.Hossain M.A., Asa T.A., Quinn J.M., Rahman M.M., Huq F., Moni M.A. Network-based genetic profiling, and therapeutic target identification of Thyroid Cancer. bioRxiv. 2018 doi: 10.1101/480632. [DOI] [Google Scholar]
- 6.Hossain M.A., Asa T.A., Huq F., Quinn J.M., Moni M.A. Proceedings of International Joint Conference on Computational Intelligence. Springer; Singapore: 2020. A network-based approach to identify molecular signatures and comorbidities of thyroid cancer; pp. 235–246. [Google Scholar]
- 7.Rahman M., Islam T., Gov E., Turanli B., Gulfidan G., Shahjaman M., Banu N.A., Mollah M., Haque N., Arga K.Y., et al. Identification of Prognostic Biomarker Signatures and Candidate Drugs in Colorectal Cancer: Insights from Systems Biology Analysis. Medicina. 2019;55:20. doi: 10.3390/medicina55010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wojtas B., Pfeifer A., Oczko-Wojciechowska M., Krajewska J., Czarniecka A., Kukulska A., Eszlinger M., Musholt T., Stokowy T., Swierniak M., et al. Gene expression (mRNA) markers for differentiating between malignant and benign follicular thyroid tumours. Int. J. Mol. Sci. 2017;18:1184. doi: 10.3390/ijms18061184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moni M.A., Liò P. comoR: A software for disease comorbidity risk assessment. J. Clin. Bioinform. 2014;4:1. doi: 10.1186/2043-9113-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hossain M.A., Islam S.M.S., Quinn J., Huq F., Moni M.A. Identification of Ovarian Cancer Gene Expression Patterns Associated with Disease Progression and Mortality. bioRxiv. 2018 doi: 10.1101/473165. [DOI] [Google Scholar]
- 11.Hossain M.A., Islam S.M.S., Quinn J.M., Huq F., Moni M.A. Machine learning and bioinformatics models to identify gene expression patterns of ovarian cancer associated with disease progression and mortality. J. Biomed. Inform. 2019;100:103313. doi: 10.1016/j.jbi.2019.103313. [DOI] [PubMed] [Google Scholar]
- 12.Moni M.A., Liò P. How to build personalized multi-omics comorbidity profiles. Front. Cell Dev. Biol. 2015;3:28. doi: 10.3389/fcell.2015.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahman M.F., Rahman M.R., Islam T., Zaman T., Shuvo M.A.H., Hossain M.T., Islam M.R., Karim M.R., Moni M.A. A bioinformatics approach to decode core genes and molecular pathways shared by breast cancer and endometrial cancer. Inform. Med. Unlocked. 2019;17:100274. doi: 10.1016/j.imu.2019.100274. [DOI] [Google Scholar]
- 14.Hossain M.A., Asa T.A., Rahman M.R., Moni M.A. Network-based approach to identify key candidate genes and pathways shared by thyroid cancer and chronic kidney disease. Inform. Med. Unlocked. 2019;16:100240. doi: 10.1016/j.imu.2019.100240. [DOI] [Google Scholar]
- 15.Ria R., Simeon V., Melaccio A., Di Meo G., Trino S., Mazzoccoli C., Saltarella I., Lamanuzzi A., Morano A., Gurrado A., et al. Gene expression profiling of normal thyroid tissue from patients with thyroid carcinoma. Oncotarget. 2016;7:29677. doi: 10.18632/oncotarget.8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y., Prasad M., Lemon W.J., Hampel H., Wright F.A., Kornacker K., LiVolsi V., Frankel W., Kloos R.T., Eng C., et al. Gene expression in papillary thyroid carcinoma reveals highly consistent profiles. Proc. Natl. Acad. Sci. USA. 2001;98:15044–15049. doi: 10.1073/pnas.251547398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahman M.R., Islam T., Turanli B., Zaman T., Faruquee H.M., Rahman M.M., Mollah M.N.H., Nanda R.K., Arga K.Y., Gov E., et al. Network-based approach to identify molecular signatures and therapeutic agents in Alzheimer’s disease. Comput. Biol. Chem. 2019;78:431–439. doi: 10.1016/j.compbiolchem.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 18.Moni M.A., Liò P. Network-based analysis of comorbidities risk during an infection: SARS and HIV case studies. BMC Bioinform. 2014;15:1. doi: 10.1186/1471-2105-15-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rana H.K., Akhtar M.R., Ahmed M.B., Lio P., Quinn J.M., Huq F., Moni M.A. Genetic effects of welding fumes on the progression of neurodegenerative diseases. Neurotoxicology. 2019;71:93–101. doi: 10.1016/j.neuro.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Rahman M.R., Islam T., Al-Mamun M.A., Zaman T., Karim M.R., Moni M.A. The influence of depression on ovarian cancer: Discovering molecular pathways that identify novel biomarkers and therapeutic targets. Inform. Med. Unlocked. 2019;16:100207. doi: 10.1016/j.imu.2019.100207. [DOI] [Google Scholar]
- 21.Kori M., Gov E., Arga K.Y. Molecular signatures of ovarian diseases: Insights from network medicine perspective. Syst. Biol. Reprod. Med. 2016;62:266–282. doi: 10.1080/19396368.2016.1197982. [DOI] [PubMed] [Google Scholar]
- 22.Barrett T., Troup D.B., Wilhite S.E., Ledoux P., Rudnev D., Evangelista C., Kim I.F., Soboleva A., Tomashevsky M., Edgar R. NCBI GEO: Mining tens of millions of expression profiles—Database and tools update. Nucleic Acids Res. 2006;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smyth G.K., Michaud J., Scott H.S. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 24.Xu H., Moni M.A., Liò P. Network regularised Cox regression and multiplex network models to predict disease comorbidities and survival of cancer. Comput. Biol. Chem. 2015;59:15–31. doi: 10.1016/j.compbiolchem.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Moni M.A., Lio’ P. Genetic profiling and comorbidities of Zika infection. J. Infect. Dis. 2017;216:703–712. doi: 10.1093/infdis/jix327. [DOI] [PubMed] [Google Scholar]
- 26.Smyth G., Ritchie M., Thorne N. Linear Models for Microarray Data User’s Guide. Bioinformatics. 2011;20:3705–3706. [Google Scholar]
- 27.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2010;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moni M.A., Xu H., Lio P. CytoCom: A Cytoscape app to visualize, query and analyse disease comorbidity networks. Bioinformatics. 2014;31:969–971. doi: 10.1093/bioinformatics/btu731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen S.H., Chin C.H., Wu H.H., Ho C.W., Ko M.T., Lin C.Y. cyto-Hubba: A Cytoscape plug-in for hub object analysis in network biology; Proceedings of the 20th International Conference on Genome Informatics; Pacifico Yokohama, Japan. 14–16 December 2009. [Google Scholar]
- 31.Xia J., Gill E.E., Hancock R.E. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 2015;10:823. doi: 10.1038/nprot.2015.052. [DOI] [PubMed] [Google Scholar]
- 32.Khan A., Fornes O., Stigliani A., Gheorghe M., Castro-Mondragon J.A., van der Lee R., Bessy A., Cheneby J., Kulkarni S.R., Tan G., et al. JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2017;46:D260–D266. doi: 10.1093/nar/gkx1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sethupathy P., Corda B., Hatzigeorgiou A.G. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu S.D., Lin F.M., Wu W.Y., Liang C., Huang W.C., Chan W.L., Tsai W.T., Chen G.Z., Lee C.J., Chiu C.M., et al. miRTarBase: A database curates experimentally validated microRNA—Target interactions. Nucleic Acids Res. 2010;39:D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shang J., Ding Q., Yuan S., Liu J.X., Li F., Zhang H. Network Analyses of Integrated Differentially Expressed Genes in Papillary Thyroid Carcinoma to Identify Characteristic Genes. Genes. 2019;10:45. doi: 10.3390/genes10010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q., Shen Y., Ye B., Hu H., Fan C., Wang T., Zheng Y., Lv J., Ma Y., Xiang M. Gene expression differences between thyroid carcinoma, thyroid adenoma and normal thyroid tissue. Oncol. Rep. 2018;40:3359–3369. doi: 10.3892/or.2018.6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu W., Hu H., Qiu P., Yan G. Triptolide induces apoptosis in human anaplastic thyroid carcinoma cells by a p53-independent but NF-κB-related mechanism. Oncol. Rep. 2009;22:1397–1401. doi: 10.3892/or_00000580. [DOI] [PubMed] [Google Scholar]
- 38.He W., Qi B., Zhou Q., Lu C., Huang Q., Xian L., Chen M. Key genes and pathways in thyroid cancer based on gene set enrichment analysis. Oncol. Rep. 2013;30:1391–1397. doi: 10.3892/or.2013.2557. [DOI] [PubMed] [Google Scholar]
- 39.Wang S., Lloyd R.V., Hutzler M.J., Safran M.S., Patwardhan N.A., Khan A. The role of cell cycle regulatory protein, cyclin D1, in the progression of thyroid cancer. Mod. Pathol. 2000;13:882. doi: 10.1038/modpathol.3880157. [DOI] [PubMed] [Google Scholar]
- 40.Newman A.C., Maddocks O.D. One-carbon metabolism in cancer. Br. J. Cancer. 2017;116:1499. doi: 10.1038/bjc.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarajlić A., Janjić V., Stojković N., Radak D., Pržulj N. Network topology reveals key cardiovascular disease genes. PLoS ONE. 2013;8:e71537. doi: 10.1371/journal.pone.0071537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang C., Zhang C., Zhao X., Kuang X., Tang H., Xiao X. Differential regulation of mitogen-activated protein kinase signalling pathways in human with different types of mitral valvular disease. J. Surg. Res. 2013;181:49–59. doi: 10.1016/j.jss.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 43.Liu Z., Liang G., Tan L., Su A., Jiang W., Gong C. High-efficient Screening Method for Identification of Key Genes in Breast Cancer Through Microarray and Bioinformatics. Anticancer. Res. 2017;37:4329–4335. doi: 10.21873/anticanres.11826. [DOI] [PubMed] [Google Scholar]
- 44.Peyressatre M., Prével C., Pellerano M., Morris M.C. Targeting cyclin-dependent kinases in human cancers: From small molecules to peptide inhibitors. Cancers. 2015;7:179–237. doi: 10.3390/cancers7010179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zang X., Chen M., Zhou Y., Xiao G., Xie Y., Wang X. Identifying CDKN3 gene expression as a prognostic biomarker in lung adenocarcinoma via meta-analysis. Cancer Inform. 2015;14:CIN-S17287. doi: 10.4137/CIN.S17287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamaguchi H., Hung M.C. Regulation and Role of EZH2 in Cancer. Cancer Res. Treat. 2014;46:209. doi: 10.4143/crt.2014.46.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Junior J.d.S.N., de Lima Marson F.A., Bertuzzo C.S. Thymidylate synthase gene (TYMS) polymorphisms in sporadic and hereditary breast cancer. BMC Res. Notes. 2012;5:676. doi: 10.1186/1756-0500-5-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Norton J.A., Ham C.M., Jacques Van Dam R., Longacre T.A., Huntsman D.G., Chun N., Kurian A.W., Ford J.M. CDH1 truncating mutations in the E-cadherin gene: An indication for total gastrectomy to treat hereditary diffuse gastric cancer. Ann. Surg. 2007;245:873. doi: 10.1097/01.sla.0000254370.29893.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hossain M.A., Asa T.A., Islam S.M., Hussain M.S., Moni M.A. Identification of genetic association of Thyroid Cancer with Parkinsons disease, Osteoporosis, chronic heart failure, Chronic kidney disease, Type 1 diabetes and Type 2 diabetes; Proceedings of the 2019 5th International Conference on Advances in Electrical Engineering (ICAEE); Dhaka, Bangladesh. 26–28 September 2019; pp. 832–837. [Google Scholar]
- 50.Masudo K., Suganuma N., Nakayama H., Oshima T., Rino Y., Iwasaki H., Matsuzu K., Sugino K., Ito K., Kondo T., et al. EZH2 Overexpression as a Useful Prognostic Marker for Aggressive Behaviour in Thyroid Cancer. In Vivo. 2018;32:25–31. doi: 10.21873/invivo.11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee J.J., Au A.Y., Foukakis T., Barbaro M., Kiss N., Clifton-Bligh R., Staaf J., Borg Å., Delbridge L., Robinson B.G., et al. Array-CGH identifies cyclin D1 and UBCH10 amplicons in anaplastic thyroid carcinoma. Endocr.-Relat. Cancer. 2008;15:801–815. doi: 10.1677/ERC-08-0018. [DOI] [PubMed] [Google Scholar]
- 52.De Andrade J.P., Lorenzen A.W., Wu V.T., Bogachek M.V., Park J.M., Gu V.W., Sevenich C.M., Cassady V.C., Beck A.C., Kulak M.V., et al. Targeting the S.U.MO pathway as a novel treatment for anaplastic thyroid cancer. Oncotarget. 2017;8:114801. doi: 10.18632/oncotarget.21954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rossing M. Classification of follicular cell-derived thyroid cancer by global RNA profiling. J. Mol. Endocrinol. 2013;50:R39–R51. doi: 10.1530/JME-12-0170. [DOI] [PubMed] [Google Scholar]
- 54.Kim H.J., Kim K.R., Park H.S., Jang K.Y., Chung M.J., Shong M., Moon W.S. The expression and role of serum response factor in papillary carcinoma of the thyroid. Int. J. Oncol. 2009;35:49–55. [PubMed] [Google Scholar]
- 55.Bullock M., Lim G., Li C., Choi I.H., Kochhar S., Liddle C., Zhang L., Clifton-Bligh R.J. Thyroid transcription factor FOXE1 interacts with ETS factor ELK1 to co-regulate TERT. Oncotarget. 2016;7:85948. doi: 10.18632/oncotarget.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arribas J., Castellví J., Marcos R., Zafón C., Velázquez A. Expression of YY1 in differentiated thyroid cancer. Endocr. Pathol. 2015;26:111–118. doi: 10.1007/s12022-015-9359-6. [DOI] [PubMed] [Google Scholar]
- 57.Geraldo M., Yamashita A., Kimura E. MicroRNA miR-146b-5p regulates signal transduction of TGF-β by repressing SMAD4 in thyroid cancer. Oncogene. 2012;31:1910. doi: 10.1038/onc.2011.381. [DOI] [PubMed] [Google Scholar]
- 58.Yu S., Liu X., Zhang Y., Li J., Chen S., Zheng H., Reng R., Zhang C., Chen J., Chen L. Circulating microRNA124-3p, microRNA9-3p and microRNA196b-5p may be potential signatures for differential diagnosis of thyroid nodules. Oncotarget. 2016;7:84165. doi: 10.18632/oncotarget.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu L., Yang J., Zhu X., Li D., Lv Z., Zhang X. Long noncoding RNA H19 competitively binds miR-17-5p to regulate YES 1 expression in thyroid cancer. Febs J. 2016;283:2326–2339. doi: 10.1111/febs.13741. [DOI] [PubMed] [Google Scholar]
- 60.Jahanbani I., Al-Abdallah A., Ali R.H., Al-Brahim N., Mojiminiyi O. Discriminatory miRNAs for the management of papillary thyroid carcinoma and noninvasive follicular thyroid neoplasms with papillary-like nuclear features. Thyroid. 2018;28:319–327. doi: 10.1089/thy.2017.0127. [DOI] [PubMed] [Google Scholar]
- 61.Liang W., Sun F. Identification of key genes of papillary thyroid cancer using integrated bioinformatics analysis. J. Endocrinol. Investig. 2018;41:1237–1245. doi: 10.1007/s40618-018-0859-3. [DOI] [PubMed] [Google Scholar]
- 62.Pfeifer A., Wojtas B., Oczko-Wojciechowska M., Kukulska A., Czarniecka A., Eszlinger M., Musholt T., Stokowy T., Swierniak M., Stobiecka E., et al. Molecular differential diagnosis of follicular thyroid carcinoma and adenoma based on gene expression profiling by using formalin-fixed paraffin-embedded tissues. BMC Med. Genom. 2013;6:38. doi: 10.1186/1755-8794-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]