Abstract
B-cell acute lymphoblastic leukemia (B-ALL) is a hematopoietic malignancy characterized by overproduction of immature B-lymphoblasts. B-ALL is the most common pediatric tumor and remains the leading cause of mortality in children and adolescents. Molecular and cytogenetic analyses of B-ALL revealed recurrent genetic and structural genomic alterations which are routinely applied for diagnosis, prognosis and choice of treatment regimen. The present case report describes a 4-year-old female diagnosed with B-ALL. GTG-banding at low resolution revealed an abnormal clone with 46,XX,?t(X;19)(q13;q13.3),der(9) besides normal cells. Molecular cytogenetics demonstrated a balanced translocation between chromosomes 16 and 19, and an unbalanced translocation involving chromosomes 5 and 9. A locus-specific probe additionally identified that the FUS gene in 16p11.2 was split and its 5′ region was translocated to subband 19q13.33, whereas the 3′ region of the FUS gene remained on the derivative chromosome 16. Overall, this complex karyotype included four different chromosomes and five break events. Further analyses, including array-comparative genomic hybridization, additionally revealed biallelic deletion of the tumor suppressor genes CDKN2A/B, and deletion of the NR3C1 and VPREB1 genes. The patient passed away under treatment due to sepsis.
Keywords: childhood B-cell acute lymphoblastic leukemia, FUS, NR3C1, VPREB1, CDKN2A/B, array-comparative genomic hybridization, complex karyotype
Introduction
B-cell acute lymphoplastic leukemia (B-ALL) is a heterogeneous disease, however from the genetic point of view it is the result of block differentiation and clonal proliferation of lymphoid B-cell progenitors in bone marrow, blood and extramedullary sites. B-ALL is the most common cancer in children, represents 80% of ALL cases and is the leading cause of cancer-related death in children (1,2).
A key role for B-ALL pathogenesis has been attributed to recurrent cytogenetic and molecular abnormalities which can be detected in ~60% of ALL cases. A diagnostic, treatment and prognostic significance is known for example for translocation t(12;21)(p13;q22) leading to the ETV6/RUNX1 gene-fusion, carriers of which are more likely to be cured than those with a translocation BCR/ABL1 or KMT2A/AFF1. Besides, high-hyperdiploidy (51–67 chromosomes), encompassing 25% of childhood B-ALL, has been associated with good survival and favorable outcome. Controversely, hypodiploidy (<44 chromosomes) has been linked to unfavorable prognosis (2,3). Complex karyotypes (CKs) are rare events in children with B-ALL and typically involve three to five or even more chromosomal abnormalities and usually result in activation of oncogenes or deletion of tumor suppression genes. CKs are found in ~5% of ALL cases and are also associated with an adverse outcome (2,4).
Accordingly it is not surprising that copy number alterations (CNAs) play a fundamental role in ALL development and progression, being present as deletion or duplication of genes involved in lymphoid development, cell cycle regulation and apoptosis, such as EBF1, PAX5, IKZF1 and VPREB1. Such genetic markers were already integrated into risk stratification systems, after their validation in large clinical cohorts (5–10).
Consequently, B-ALL is one of the genetically best characterized malignancies and the success of curing pediatric ALLs has increased dramatically reaching ~85%. Nonetheless, cryptic structural chromosomal abnormalities were and are still a challenge in cytogenetic diagnostics of B-ALL (2,10).
In the present study, a comprehensive analysis of a childhood B-ALL case was done using high resolution molecular approaches, revealing a cryptic complex karyotype with involvement of FUS, NR3C1 and VPREB1 genes.
Case report
Clinical description
A 4-year-old female was admitted to the Institute of Mother and Child of Serbia ‘Dr. Vukan Cupic’ because of bruisings and splenomegaly. At admission, the white blood cell (WBC) count was 229×109/l with of 92% lymphocytes, the hemoglobin was 103 g/l, the platelets were 52×109/l and red blood cells (RBC) count was 3.78×106/mm3. Serum uric acid (UA) value was 529 µmol/l (normal 150–450), and lactate-dehydrogenase (LDH) value was 6,579 IU/l (normal 120–250).
Bone marrow aspirate showed infiltration with more than 90% of lymphoblasts, FAB L1 60% and L2 30%. Periodic acid-Schiff stain (PAS) and Sudan Black B were negative. Flow cytometric (FCM) analysis characterized the expression of a variety of B-cell-specific antigens and were positive for CD10 89%, CD19 93%, CD45 74%, iCD79a 94%, iIgM 68%, sIgλ 18% and were negative for CD34, iCD3, sCD3, CD7, CD8, CD13, CD15, sIgκ and MPO. These findings were consistent with B-ALL.
Unfortunately, during induction therapy (ALL IC-BFM 2009), a diagnosis of sepsis was established with capillary leak syndrome and cardiopulmonary failure, resulting in a fatal outcome. So, it was not possible to perform a control bone marrow aspiration and determine whether a remission was achieved or not.
Chromosome analysis
Banding cytogenetic analyses was performed on unstimulated bone marrow aspirate according to standard procedures (11). A total of 20 metaphases were available for cytogenetic evaluation and analyzed on a banding level of 200 bands per haploid karyotype (12).
Molecular genetics
Total RNA was extracted from bone marrow cells, using phenol chloroform extraction protocol (13). cDNA was prepared from 1 µg of total RNA with the High Capacity Reverse Transcription Kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). Screening for BCR/ABL1 (p190, p210), MLL/AF4, ETV6/RUNX1 and PBX1/E2A fusion transcripts were performed according to BIOMED-1 Concerted Action program (BMH-CMT 94–1675), using Maxima Hot Start Taq DNA polymerase (Thermo Fisher Scientific, Inc.) (14).
Molecular cytogenetics
Fluorescence in situ hybridization (FISH) was done according to standard procedures and/or according to manufacturer's instructions. Homemade probes and probe sets were applied as follows: i) BAC (bacterial artificial chromosome) clones of interest were identified through the Human Genome Browser Database of the Genome Bioinformatics Group at the University of California at Santa Cruz (http://genome.ucsc.edu/) and Ensembl Genome Data Resources of the Sanger Institute Genome Database (http://www.ensembl.org/). DNA probes (Table I) obtained from Resources Center were labeled by PCR with SpectrumGreen, SpectrumOrange or TexasRed-dUTP and applied in two- or three-color FISH-approaches. ii) Chromosome specific high resolution array-proven multicolor-banding (aMCB) probes sets for #5, #9, #16, #19 and X were used. In combination with whole chromosome painting (WCP) for chromosomes 5, 6, 9, 16, 17, 18, 19, 20, 21, 22, X were applied as well (15,16).
Table I.
Locus specific probes used for FISH.
| Probe | Cytoband | Position [NCBI36/hg18] | Genes/locus | Result (signals on…) |
|---|---|---|---|---|
| RP11-434D11 | 5q23.2 | chr5:126,045,879-126,232,850 | n.d. | der(5) |
| RP11-114H7 | 5q23.3 | chr5:130,306,745-130,460,728 | n.d. | der(5) |
| LSI EGR1/D5S721, D5S23 (Vysis) | 5q31.2 | chr5:137,829,080-137,832,903 | EGR1 | der(5) |
| LSI PDGFRB DCBAP (Vysis) | 5q33.1 | chr5:149,473,595-149,515,615 | PDGFRB | der(9) |
| 5qTEL (Vysis) | 5q35.3 | chr5:180,510,748-180,711,420 | D5S2907 | der(9) |
| 9pTEL (Vysis) | 9p24.3 | chr9:131,486-331,767 | 305J7-T7 | der(5) |
| POSEIDON™ MLL/MLLT3 DCDFP (Kreatech) | 9p21.3 | chr9:20,331,667-20,612,514 | MLLT3 | der(5) |
| SPEC CDKN2A/CEN9 DCP (Zytovision) | 9p21.3 | chr9:21,957,751-21,965,038 | CDKN2A/B | biallelic deletion (9)(p21.3p21.3) |
| LSI BCR/ABL1 DCDFP (Vysis) | 9q34.12 | chr9:132,579,089-132,752,883 | ABL1 | der(9) |
| 9qTEL (Vysis) | 9q34.3 | chr9:140,000,171-140,200,260 | D9S325 | der(9) |
| SPEC BIRC3/MALT1 DCDFP (Zytovision) | 11q22.3 | chr11:101,693,404-101,713,675 | BIRC3 | #11 |
| RP11-142A12 | 16p12.1 | chr16:26,595,070-26,757,869 | n.d | der(16) |
| RP11-147I4 | 16p12.1 | chr16:27,071,343-27,211,332 | KDM8, NSMCE1 | der(16) |
| RP11-159J3 | 16p11.2 | chr16:28,012,684-28,171,002 | XPO6 | der(16) |
| SPEC FUS DCBAP (Zytovision) | 16p11.2 | chr16:31,098,954-31,110,600 | FUS | signal split on der(16) and der(19) |
| SPEC TP53/ATM DCP (Zytovision) | 17p13.1 | chr17:7,512,445-7,531,588 | TP53 | #17 |
| RP11-21J15 | 19q13.31 | chr19:49,726,602-49,900,222 | CEACAM22P, IGSF23, PVR, CEACAM19 | der(19) |
| RP11-84C16 | 19q13.31 | chr19:49,089,546-50,686,777 | BCL3 | der(19) |
| RP11-492P7 | 19q13.32 | chr19:51,987,986-52,169,539 | AP2S, ARHGAP35 | der(19) |
| SPEC 19q13/19p13 DCP (Zytovision) | 19q13.32 | chr19:52,477,388-53,038,398 | GLTSCR1, GLTSCR2, CRX | der(19) |
| RP11-3N16 | 19q13.32 | chr19:53,269,619-53,431,708 | PLA2G4C, LIG1, CARD8 | der(19) |
| RP11-264M8 | 19q13.33 | chr19:54,767,615-54,925,355 | PRRG2, PRRG2, IRF3, BCL2L12, PRMT1, CPTC1 | der(16) |
Locus specific probes used for FISH together with their location according to genome browser version NCBI36/hg18. This version was used as some applied FISH-probes are no longer available in newer genome browser versions. Results obtained are presented using standard gene abbreviations and such used according to the international system of cytogenomic nomenclature. der, derivative; FISH, fluorescence in situ hybridization.
Additionally, commercially available probes were applied: ZytoLight®SPEC CDKN2A/CEN9 (in 9p21.3 and 9p11q11 dual color probe), ZytoLight®SPEC BIRC3/MALT1 Dual Color Dual Fusion Probe (in 11q22.2 and 18q21.32), ZytoLight®SPEC FUS (16p11.2 Break Apart Probe) all from ZytoVision GmbH; LSI EGR1/D5S23, D5S721 (in 5q31 and 5p15.2), LSI PDGFRB (5q32-q33 Break apart probe), subtelomeric probe for 5qter (D5S2907), LSI ABL1/BCR (in 9q34.1 and 22q11.2 dual color probe), LSI p53/ATM (in 17p13.1 and 11q22.3 dual color probe) all from Vysis (Abbott GmbH & Company, KG) and POSEIDON MLL/MLLT3 (in 11q23 and 9p21 dual color probe, Kreatech Diagnostics).
A total of 10–15 metaphase spreads were analyzed and (where applicable) for interphase-FISH analysis, 200 interphase nuclei were examined, using a fluorescence microscope (AxioImager.Z1 mot; Zeiss) equipped with appropriate filter sets to discriminate between a maximum of five fluorochromes and the counterstain 4,6-diamidino-2-phenylindole (DAPI). Image capturing and processing were carried out using an ISIS imaging system (MetaSystems).
DNA isolation
Genomic DNA was extracted from bone marrow cells by Puregene DNA Purification Kit (Gentra Systems). DNA concentration was determined by a NanoDrop spectrophotometer. The quality of DNA was checked using agarose gel electrophoresis. DNA-samples extracted from fixed cells of 2 healthy males and 2 healthy females by the same method were used as reference samples.
High resolution array-comparative genomic (aCGH)
aCGH was performed using Agilent SurePrint G3 Human Genome microarray 180 K (Agilent Technologies), an oligonucleotide microarray containing approximately 180,000 probes 60-mer with a 17 kb average probe spacing. Genomic DNA of patient was co-hybridized with a male control DNA (Agilent Technologies). Labeling was performed using Agilent Genomic DNA enzymatic labeling kit (Agilent Technologies) according to the manufacturers' instructions. After hybridization, the aCGH slide was scanned on an Agilent scanner, processed with Feature Extraction software (v10.7) and results were analyzed using Cytogenomics (v2.9.1.3) using ADM2 as aberration algorithm.
Results
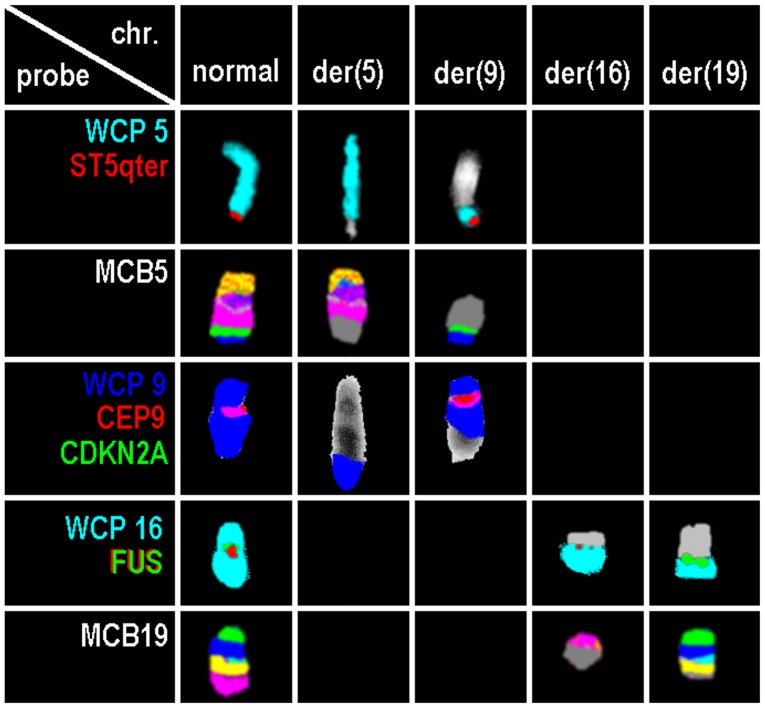
GTG-banding at low resolution revealed a female karyotype as 46,XX,t(X;19)(q13;q13.3),der(9)[10]/46,XX[10] (result not shown). Molecular analyses of RNA/cDNA did not detect any hint on presence of tested fusion genes BCR/ABL1 (p190, p210), MLL/AF4, ETV6/RUNX1 or PBX1/E2A (results not shown). Application of molecular cytogenetics approaches including WCPs, aMCB probe sets and locus specific probes revealed the karyotype 46, XX, der(5)t(5;9)(pter->q32::p21.3->pter), der(9)(9qter->9q33.3::9p21.3->9q33.3::5q32->5qter),t(16;19)(p11.2; q13.33) in the aberrant clone. Overall, the present case had genetic changes involving four chromosomes and five break events (Fig. 1).
Figure 1.
Results of normal and derivative chromosomes 5, 9, 16 and 19 for probes or probe sets. Probes or probe sets are indicated in the first column. chr., chromosome; der, derivative; WCP, whole chromosome probe; CDKN2A, cyclin-dependent kinase inhibitor 2A; CEP, centromeric probe; chr, chromosome; der, derivative chromosome; FUS, fused in sarcoma; MCB, multicolor banding; qter, long arm terminal; ST, subtelomeric probe.
The breakpoint for translocation t(16;19) covered FUS gene in 16p11.2 (Fig. 1). Using locus specific probes, the breakpoints at 5q32, 9p21.3, 9q33 and 19q13.33 could be narrowed down, as well (Table I). For the 19q13.33, the break was shown to be located between the positions: Chr19:53,431,708-54,767,615 (NCBI36/hg18); 48 OMIM genes are located there (Table II).
Table II.
Summary of CNAs detected by array-comparative genomic hybridization.
| Chromosome (alteration U or R) | Cytobands | NCBI36/hg18 | Size of imbalance (Mb) | Online Mendelian Inheritance in Man genes |
|---|---|---|---|---|
| 5 (U) | del(5)(q31.3q32) | chr5:142,130,476-145,871,262 | 3.74 | ARHGAP26, NR3C1, HMHB1, YIPF5, KCTD16, PRELID2, GRXCR2, SH3RF2, PLAC8L1, LARS, RBM27, POU4F3, TCERG1 |
| 9 (R) | del(9)(p21.3p21.3) | chr9:21,212,342-22,632,472 | 1.42 | IFNA17, IFNA14, IFNA22P, IFNA5, KLHL9, IFNA6, IFNA13, IFNA2, IFNA8, IFNA1, MIR31HG, IFNE, MIR31, MTAP, CDKN2A-DT, CDKN2A, CDKN2B-AS1, CDKN2B, DMRTA1 |
| 9 (U) | del(9)(q33.3q33.3) | chr9:124,813,871-125,060,804 | 0.24 | RABGAP1, GPR21, MIR600HG, STRBP |
| 22 (U) | del(22)(q11.22q11.22) | chr22:20,811,200-21,422,368 | 0.61 | VPREB1, BMS1P20, ZNF280B, ZNF280A, PRAME, LL22NC03-63E9.3, POM121L1P, GGTLC2 |
R and U acquired CNAs are correspondingly highlighted in the first column. Results obtained are presented using standard gene abbreviations and such used according to the international system of cytogenomic nomenclature. CNAs, copy number alterations; R, recurrent; U, unique.
Array-CGH revealed four additional small genomic imbalances: A loss of 1.42 Mb in the region of 9p21.3p21.3 between the positions 21,212,342 and 22,632,472 (NCBI36/hg18) with biallalic deletion of CDKN2A and CNKN2B and confirmed by locus specific FISH probe result (Fig. 1).
Additional genomic imbalances revealed by array-CGH were losses in: 5q31.3-q32 at positions 142,130,476-145,871,262 including loss of NR3C1; 9q33.2-q33.3 at positions 124,813,871 and 125,060,804 (no specific candidate gene identified), and 22q11.22 at positions 20,811,200 and 21,422,368 including loss of VPREB1 (Table II).
Discussion
In pediatric B-ALL, chromosomal rearrangements play a crucial role in leukemogenesis by contributing to inactivation of tumor suppressor genes and/or dysregulated expression of oncogenes and the generation of novel gene fusions. Thus, chromosomal break-events can be hints on genes with biologic function in the leukemogenesis. Here we showed involvement of Fused in Sarcoma (FUS) gene located in chromosome 16p11.2 in childhood B-ALL. Interestingly, very little is known about the oncogenic role of FUS gene in ALL (17). FUS gene was first identified as an oncogene in human myxoid liposarcomas, being activated by translocations (18). FUS gene encodes an RNA-binding protein (takes part in transcription initiation by binding RNA pol II and some transcription factors), the C-terminal end of which is involved in protein and RNA binding and which appears to be involved in transcriptional activation with its N-terminal end. FUS is a nucleoprotein that functions in DNA and RNA metabolism, including DNA repair, and the regulation of transcription, RNA splicing and export to the cytoplasm (19,20).
In acute leukemia, FUS gene rearrangements have been identified in both childhood and adults: The recurrent translocation t(16;21)(p11;q22)/FUS-ERG was yet seen in approximately 80 cases of acute myeloid leukemia (AML). It occurs in <0.5% of AML cases and is associated with poor prognosis and high risk of relapse (21,22). In ALL, the translocation t(16;21)(p11;q22)/FUS-ERG is very rare and only identified in 15 patients, including 10 adults and five children (17,22). In addition, the translocation t(9;16)(q34;p11.2) FUS/SET has been detected in one T-ALL patient (23).
To the best of our knowledge, the balanced translocation t(16;19)(p11.2;q13.3)?/FUS has not yet been reported according to the Mitelman database and the literature. This translocation seems to create a yet unknown fusion gene between FUS gene on 16p11.2 and a gene on 19q13.33. In 19q13.33 region 48 OMIM genes are located, including the EMP3 gene (epithelial membrane protein 3); the translocation t(11;19)(q23;q13)CEP164/EMP3 has been detected previously in only one T-ALL patient. However in gene-rich subband 19q13.33, it is difficult to determine which one might have provided a fusion with or strong promoter for FUS gene dysregulate expression. Overall, FUS gene at 16p11.2 is considered as one of the fusion products being critical for leukaemogenic process as a sole abnormality (2,17,22–25). On the other hand, an unbalanced translocation in this case involves three other breakpoints 5q32, 9p21.3 and 9q33. Participation of cancer-related oncogenes PDGFRB in 5q32, MLLT3 in 9p21.3, and ABL1 in 9q34.13 could be excluded (Table I), however, loss of two tumor suppressor genes in 9p21 was identified.
Interestingly, the unbalanced translocation involves simultaneously interstitial deletions and complex rearrangements. Tumor suppressor genes CDKN2A and CDKN2B at 9p21, which encode p14/ARF and p16/INK4A (both encoded by CDKN2A) and p15/INK4B (encoded by CDKN2B) proteins have a role in cell growth regulation and apoptosis. Thus, here observed homozygous deletion represents a marker of high risk of relapse and poor prognosis in children B-ALL as reported in previous studies (26–28).
NR3C1 gene in 5q31.3, encoding DNA- and hormone-binding domains of the human glucocorticoid receptor, can function both as a transcription factor that binds to glucocorticoid response elements in promoters of glucocorticoid responsive genes and activate their transcription, and as a regulator of other transcription factors. Interestingly, loss of NR3C1 gene is associated with glucocorticoid resistance, which is a key component of childhood ALL therapy. Besides, deletion of NR3C1 gene is rare in B-ALL and significantly associated with clinical high risk of relapse and an inferior outcome, where it correlated to high rate of induction failure and death (2,7,29).
Furthermore, VPREB1 gene deletion at 22q11.2 has been currently reported in association with poor prognosis and high risk of relapse in both children and adult B-ALL. VPREB1 gene encodes the iota polypeptide chains, which associate with the Ig-mu chain to form a molecular complex that expressed on the surface of pre-B cells. VPREB1 gene is located within the IGL@ locus among the variable immunoglobulin (Ig) segments, upstream from the VJ junction and is an essential gene in B-cell development and differentiation due to its role as part of the surrogate light chain in the pre-B cell receptor (pre-BCR). A recent report by Mangum et al (10) described a new mechanism of B-ALL development emerging by loss of VPREB1 gene during B-cell maturation. In this context, loss of VPREB1 gene contributes to leukemogenesis as a result of failure to form a viable surrogate light chain in the pre-BCR in both human and mice due to block the pro- to pre-B-cell transition in the bone marrow with a decrease in circulating mature B-cells. Besides, VPREB1 gene deletion was also identified in specific B-ALL subtypes including ETV6-RUNX1 and associate with higher WBC count as also in our case, BCR-ABL1 and TCF3-HLF (6,8,10,30,31).
In conclusion, further studies are required to establish whether such potentially poor outcome due to the combination of the here reported aberrations is linked directly to glucocorticoid resistance or whether it is part of a broader drug resistance profile. We suggest NR3C1 and VPREB1 genes as excellent candidates for further studying B-ALL leukemogenesis to improve treatment strategies in pediatric B-ALL. Overall, we provided evidence, that comprehensive molecular analyses are urgently necessary for the detection of cryptic rearrangements and identification of genes involved in B-ALL formation and progression.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
MAKO, BM and FAS performed the FISH studies. JBM, IMC and MAKO performed array comparative genomic hybridization analyses and interpretation. NL performed the reverse transcription PCR. MĐ, ZZ, DV and GS provided B-ALL-case, including clinical data, and performed banding cytogenetic data interpretation and made decisions on what tests should be performed in their lab. MAKO, RA and TL developed the study, made decisions on what tests should be performed, were involved in overall data interpretation and drafted the study. All authors read and approved the study.
Ethics approval and consent to participate
As the case was identified during routine diagnostics, ethical approval and consent to participate is not applicable, except for the agreement that diagnostics was performed to solve the clinical question.
Patient consent for publication
Informed consent was provided by the patient's parent.
Competing interests
BM works at Zytovison, the company providing one of the FISH-probes used. All other authors declare that they have no competing interests.
References
- 1.Malouf C, Ottersbach K. Molecular processes involved in B cell acute lymphoblastic leukaemia. Cell Mol Life Sci. 2018;75:417–446. doi: 10.1007/s00018-017-2620-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tasian SK, Hunger SP. Genomic characterization of paediatric acute lymphoblastic leukaemia: An opportunity for precision medicine therapeutics. Br J Haematol. 2017;176:867–882. doi: 10.1111/bjh.14474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moorman AV. New and emerging prognostic and predictive genetic biomarkers in B-cell precursor acute lymphoblastic leukemia. Haematologica. 2016;101:407–416. doi: 10.3324/haematol.2015.141101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moorman AV, Harrison CJ, Buck GA, Richards SM, SeckerWalker LM, Martineau M, Vance GH, Cherry AM, Higgins RR, Fielding AK, et al. Karyotype is an independent prognostic factor in adult acute lymphoblastic leukemia (ALL): Analysis of cytogenetic data from patients treated on the Medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) 2993 trial. Blood. 2007;109:3189–3197. doi: 10.1182/blood-2006-10-051912. [DOI] [PubMed] [Google Scholar]
- 5.Hamadeh L, Enshaei A, Schwab C, Alonso CN, Attarbaschi A, Barbany G, den Boer ML, Boer JM, Braun M, Dalla Pozza L, et al. Validation of the United Kingdom copy-number alteration classifier in 3239 children with B-cell precursor ALL. Blood Adv. 2019;3:148–157. doi: 10.1182/bloodadvances.2018025718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ribera J, Zamora L, Morgades M, Mallo M, Solanes N, Batlle M, Vives S, Granada I, Juncà J, Malinverni R, et al. Copy number profiling of adult relapsed B-cell precursor acute lymphoblastic leukemia reveals potential leukemia progression mechanisms. Genes Chromosomes Cancer. 2017;56:810–820. doi: 10.1002/gcc.22486. [DOI] [PubMed] [Google Scholar]
- 7.Irving JA, Enshaei A, Parker CA, Sutton R, Kuiper RP, Erhorn A, Minto L, Venn NC, Law T, Yu J, et al. Integration of genetic and clinical risk factors improves prognostication in relapsed childhood B-cell precursor acute lymphoblastic leukemia. Blood. 2016;128:911–922. doi: 10.1182/blood-2016-03-704973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Öfverholm I, Tran AN, Olsson L, Zachariadis V, Heyman M, Rudd E, Syk Lundberg E, Nordenskjöld M, Johansson B, Nordgren A, Barbany G. Detailed gene dose analysis reveals recurrent focal gene deletions in pediatric B-cell precursor acute lymphoblastic leukemia. Leuk Lymphoma. 2016;57:2161–2170. doi: 10.3109/10428194.2015.1136740. [DOI] [PubMed] [Google Scholar]
- 9.Othman MA, Melo JB, Carreira IM, Rincic M, Glaser A, Grygalewicz B, Gruhn B, Wilhelm K, Rittscher K, Meyer B, et al. High rates of submicroscopic aberrations in karyotypically normal acute lymphoblastic leukemia. Mol Cytogenet. 2015;8:45. doi: 10.1186/s13039-015-0153-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mangum DS, Downie J, Mason CC, Jahromi MS, Joshi D, Rodic V, Müschen M, Meeker N, Trede N, Frazer JK, et al. VPREB1 deletions occur independent of lambda light chain rearrangement in childhood acute lymphoblastic leukemia. Leukemia. 2014;28:216–220. doi: 10.1038/leu.2013.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claussen U, Michel S, Mühlig P, Westermann M, Grummt UW, Kromeyer-Hauschild K, Liehr T. Demystifying chromosome preparation and the implications for the concept of chromosome condensation during mitosis. Cytogenet Genome Res. 2002;98:136–146. doi: 10.1159/000069817. [DOI] [PubMed] [Google Scholar]
- 12.Shaffer LG, McGowan-Jordan J, Schmid M, editors. Karger; Basel: 2013. An International System for Human Cytogenetic Nomenclature; pp. 2108–2109. [Google Scholar]
- 13.Liu X, Harada S. RNA isolation from mammalian samples. Curr Protoc Mol Biol. 2013;Chapter 4 doi: 10.1002/0471142727.mb0416s103. Unit 4.16. [DOI] [PubMed] [Google Scholar]
- 14.van Dongen JJ, Macintyre EA, Gabert JA, Delabesse E, Rossi V, Saglio G, Gottardi E, Rambaldi A, Dotti G, Griesinger F, et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 Concerted Action: Investigation of minimal residual disease in acute leukemia. Leukemia. 1999;13:1901–1928. doi: 10.1038/sj.leu.2401592. [DOI] [PubMed] [Google Scholar]
- 15.Weise A, Mrasek K, Fickelscher I, Claussen U, Cheung SW, Cai WW, Liehr T, Kosyakova N. Molecular definition of high-resolution multicolor banding probes: First within the human DNA sequence anchored FISH banding probe set. J Histochem Cytochem. 2008;56:487–493. doi: 10.1369/jhc.2008.950550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liehr T, editor. Springer Verlag; Berlin: 2017. Fluorescence In Situ Hybridization (FISH)-Application Guide. [Google Scholar]
- 17.Coccé MC, Alonso CN, Rossi J, Felice MS, Gitter MR, Gallego MS. A case of pediatric ALL with t(16;21)(p11.2;q22) and FUS-ERG rearrangement. Blood Res. 2015;50:55–58. doi: 10.5045/br.2015.50.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aman P, Ron D, Mandahl N, Fioretos T, Heim S, Arheden K, Willén H, Rydholm A, Mitelman F. Rearrangement of the transcription factor gene CHOP in myxoid liposarcomas with t(12;16)(q13;p11) Genes Chromosomes Cancer. 1992;5:278–285. doi: 10.1002/gcc.2870050403. [DOI] [PubMed] [Google Scholar]
- 19.Naro C, Bielli P, Pagliarini V, Sette C. The interplay between DNA damage response and RNA processing: The unexpected role of splicing factors as gatekeepers of genome stability. Front Genet. 2015;6:142. doi: 10.3389/fgene.2015.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y, Liu S, Oztürk A, Hicks GG. FUS-regulated RNA metabolism and DNA damage repair: Implications for amyotrophic lateral sclerosis and frontotemporal dementia pathogenesis. Rare Dis. 2014;2:e29515. doi: 10.4161/rdis.29515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noort S, Zimmermann M, Reinhardt D, Cuccuini W, Pigazzi M, Smith J, Ries RE, Alonzo TA, Hirsch B, Tomizawa D, et al. Prognostic impact of t(16;21)(p11;q22) and t(16;21)(q24;q22) in pediatric AML: A retrospective study by the I-BFM Study Group. Blood. 2018;132:1584–1592. doi: 10.1182/blood-2018-05-849059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zerkalenkova E, Panfyorova A, Kazakova A, Baryshev P, Shelihova L, Kalinina I, Novichkova G, Maschan M, Maschan A, Olshanskaya Y. Molecular characteristic of acute leukemias with t(16;21)/FUS-ERG. Ann Hematol. 2018;97:977–988. doi: 10.1007/s00277-018-3267-z. [DOI] [PubMed] [Google Scholar]
- 23.Atak ZK, Gianfelici V, Hulselmans G, De Keersmaecker K, Devasia AG, Geerdens E, Mentens N, Chiaretti S, Durinck K, Uyttebroeck A, et al. Comprehensive analysis of transcriptome variation uncovers known and novel driver events in T-cell acute lymphoblastic leukemia. PLoS Genet. 2013;9:e1003997. doi: 10.1371/journal.pgen.1003997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitelman F, Johansson B, Mertens F. National Cancer Institute; Bethesda, MD: [Apr 18;2019 ]. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. [Google Scholar]
- 25.Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, McCastlain K, Edmonson M, Pounds SB, Shi L, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49:1211–1218. doi: 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kathiravan M, Singh M, Bhatia P, Trehan A, Varma N, Sachdeva MS, Bansal D, Jain R, Naseem S. Deletion of CDKN2A/B is associated with inferior relapse free survival in pediatric B cell acute lymphoblastic leukemia. Leuk Lymphoma. 2019;60:433–441. doi: 10.1080/10428194.2018.1482542. [DOI] [PubMed] [Google Scholar]
- 27.Agarwal M, Bakhshi S, Dwivedi SN, Kabra M, Shukla R, Seth R. Cyclin dependent kinase inhibitor 2A/B gene deletions are markers of poor prognosis in Indian children with acute lymphoblastic leukemia. Pediatr Blood Cancer. 2018;65:e27001. doi: 10.1002/pbc.27001. [DOI] [PubMed] [Google Scholar]
- 28.Braun M, Pastorczak A, Fendler W, Madzio J, Tomasik B, Taha J, Bielska M, Sedek L, Szczepanski T, Matysiak M, et al. Biallelic loss of CDKN2A is associated with poor response to treatment in pediatric acute lymphoblastic leukemia. Leuk Lymphoma. 2017;58:1162–1171. doi: 10.1080/10428194.2016.1228925. [DOI] [PubMed] [Google Scholar]
- 29.Donner KM, Hiltunen TP, Jänne OA, Sane T, Kontula K. Generalized glucocorticoid resistance caused by a novel two-nucleotide deletion in the hormone-binding domain of the glucocorticoid receptor gene NR3C1. Eur J Endocrinol. 2012;168:K9–K18. doi: 10.1530/EJE-12-0532. [DOI] [PubMed] [Google Scholar]
- 30.Eswaran J, Sinclair P, Heidenreich O, Irving J, Russell LJ, Hall A, Calado DP, Harrison CJ, Vormoor J. The pre-B-cell receptor checkpoint in acute lymphoblastic leukaemia. Leukemia. 2015;29:1623–1631. doi: 10.1038/leu.2015.113. [DOI] [PubMed] [Google Scholar]
- 31.Mårtensson IL, Ceredig R. Review article: Role of the surrogate light chain and the pre-B-cell receptor in mouse B-cell development. Immunology. 2000;101:435–441. doi: 10.1046/j.1365-2567.2000.00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.