Abstract
Long non-coding RNA (lncRNA) prostate cancer-associated transcript 1 (PCAT-1) is a long non-coding RNA involved in the development of various cancer types. Tongue squamous cell carcinoma (TSCC) is the most frequently diagnosed type of oral cancer worldwide. The present study aimed to explore the role of lncRNA PCAT-1 in TSCC, and to provide a preliminary assessment of the downstream pathways involved. The expression levels of lncRNA PCAT-1 in TSCC specimens were first assessed. Subsequently, lncRNA PCAT-1 was knocked down in TSCC cell lines and the resulting effect on the proliferation and apoptosis, as well as the metastatic and invasive potential of TSCC cells was investigated. To assess the downstream signaling resulting from lncRNA PCAT-1 inhibition, the expression of p21 was also detected. The results indicated that lncRNA PCAT-1 was upregulated in clinical TSCC compared with peri-tumor tissues. Inhibition of lncRNA PCAT-1 resulted in reduced proliferation and apoptosis of TSCC cells, in addition to the suppression of migration and invasion. Furthermore, the expression of p21 was induced following lncRNA PCAT-1 inhibition. Collectively, the present study demonstrated an oncogenic role for lncRNA PCAT-1 in TSCC; inhibition of lncRNA PCAT-1 reduced the growth, metastasis and invasion of TSCC, at least in part via the induction of p21 signaling.
Keywords: metastasis, prostate cancer-associated transcript 1, p21, tongue squamous cell carcinoma
Introduction
In recent decades, non-coding RNAs (ncRNAs) were assumed to be transcripts of junk DNA that had no involvement in biological processes. However, in emerging studies focusing on the functions of ncRNAs, they were revealed to be important regulators of multiple pathways. ncRNAs were also revealed to be involved in the regular development of organisms, in addition to the progression of various diseases (1,2). Furthermore, the development and progression of high-throughput genomic technologies have allowed scientists to analyze complex cellular transcriptomes, leading to the discovery of a myriad of novel ncRNAs (3,4). A significant class of ncRNAs is the long ncRNAs (lncRNAs), which are defined as transcripts of >200 nucleotides with no protein-coding potential (5). The regulatory roles of lncRNAs have been demonstrated in a large number of distinct biological processes (6–8), and multiple lncRNAs have proven their potential as therapeutic candidates in cancer (9–11). Given that cancer is largely caused by genetic alterations distributed in non-coding regions of the genome, the roles of lncRNAs in the progression of cancer have been increasingly emphasized in the last few decades (12–14).
lncRNA prostate cancer (PCa)-associated transcript 1 (PCAT-1) has been reported as an oncogenic factor in PCa (15,16). Upregulation of lncRNA PCAT-1 promoted the proliferation of PCa cells and was associated with poor prognosis in patients with PCa (17). Regarding its role in other cancer types, the upregulation of lncRNA PCAT-1 was also indicated in esophageal squamous carcinoma (18). Additionally, in a study by Qiao et al (19), the inhibition of lncRNA PCAT-1 suppressed the multidrug resistance and aggressiveness of colorectal cancer cells.
In spite of the widely accepted oncogenic role of lncRNA PCAT-1, the underlying mechanisms of this role remain to be fully elucidated. A comprehensive investigation of the downstream signaling of lncRNA PCAT-1 in different cancer types and stages may lead to the development of lncRNA PCAT-1-based anti-tumor therapies. Huang et al (20) reported that lncRNA PCAT-1 acted as an oncogene in osteosarcoma by reducing p21 levels; given that p21 is involved in the anti-tumor effects of multiple agents (20–22), lncRNA PCAT-1 inhibition may represent a potential treatment strategy by restoring the levels of p21 in numerous cancer types.
Tongue squamous cell carcinoma (TSCC) is the most prevalent malignancy of the oral cavity, accounting for ~30% of all oral cancer cases worldwide (23). TSCC is a rapid-growth tumor type with a high risk of regional and distant metastasis (24), thus, early prediction and diagnosis are key to the successful management of TSCC (24). According to a study by Gao et al (25), multiple lncRNAs, including lnc-PPP2R4-5, SPRR2D-1, MAN1A2-1 and FAM46A-1 are dysregulated in TSCC, indicating a close interaction between lncRNAs and the oncogenesis of tongue cells. Regarding the role of lncRNA PCAT-1 in the onset and progression of TSCC, initial clinical investigations in the present study identified the upregulation of lncRNA PCAT-1 in TSCC tissues. It was therefore hypothesized that the inhibition of lncRNA PCAT-1 may impair the proliferation and metastasis of TSCC associated with 21 upregulation.
To verify this hypothesis, lncRNA PCAT-1 was knocked down in TSCC cell lines and the subsequent effects on the proliferation, apoptosis, motility and p12 were assessed. The results indicated that depletion of lncRNA PCAT-1 impaired the growth, increased the apoptotic rate and reduced the metastatic and invasive potential of TSCC cells, accompanied by an increase in the expression levels of p21.
Materials and methods
TSCC specimen collection
A total of 23 pairs of TSCC and corresponding peri-tumor samples were obtained from volunteers at the People's Hospital of Tongliang District Chongqing City (Chongqing, China) between January and December 2015. The cohort included 14 males (60.9%) and 9 females (39.1%), with an average patient age of 51.5 years (range, 23–75 years). The patients were included based on the following criteria: i) Patients were diagnosed with primary tongue squamous cell carcinoma according to American Joint Committee on Cancer; ii) no prior history of chemotherapy or radiotherapy; and iii) patients underwent radical tumor resection. The peri-tumor tissues were collected from regions 1.5 cm from the tumor. Following dissection, the samples were stored at −80°C prior to analysis using reverse transcription-quantitative PCR (RT-qPCR). All patients provided written informed consent for the use of their tissues and the study was approved by the ethics committee of the People's Hospital of Tongliang District Chongqing City (Chongqing, China); all procedures were performed in accordance with the Declaration of Helsinki (https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/).
Cell culture
The TSCC cell line CAL27 (cat. no. ZQ0606) was obtained from OriGene Technologies, Inc. and the human TSCC cell line Tca-8113 (cat. no. TCHu 77) was purchased from the cell bank of the Type Culture Collection of the Chinese Academy of Sciences. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (both from Thermo Fisher Scientific, Inc.) at 37°C and 5% CO2.
Construction of lncRNA PCAT-1 small hairpin (sh)RNA vectors and transfection
shRNAs targeting PCAT-1 (shRNA-1, 5′-GCTCACGCCTGTAATCTCA-3′; and shRNA-2, 5′-GAACCTAACTGGACTTTAA-3′) were synthesized by Sangon Biotech Co., Ltd. and inserted into the pRNA-H1.1 plasmid (between BamIII and HindIII) to construct 2 PCAT-1 suppression vectors (shRNA-1 and shRNA-2, respectively). A non-targeting shRNA was employed as a negative control (NC; 5′-GATCCCCTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTT-3′). Transfections were performed using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol, using 2 µg plasmid. Cells with stable lncRNA PCAT-1 knockdown were selected using 500 µg/ml G418 (Invitrogen; Thermo Fisher Scientific Inc.). Subsequent experiments were conducted 48 h following transfection.
MTT assay
The viability of PCAT-1-knockdown TSCC cells was determined using an MTT assay. Briefly, cells were seeded into a 96-well plate and cultured for 96 h at 37°C; at 24-h intervals, 5 mg/ml MTT was added to 3 wells from each group, and the cells were incubated for an additional 4 h at room temperature, leading to the formation of a colored precipitate. DMSO was added to the wells to dissolve the purple formazan crystals. The optical density at 490 nm (OD490) was detected using a microplate reader (ELX-800; Biotek Instruments, Inc.).
Flow cytometry detection of apoptosis
The effects of PCAT-1 knockdown on TSCC cell apoptosis were detected using an Apoptosis Detection kit (cat. no. KGA106; Nanjing KeyGen Biotech Co., Ltd.), using 1×105 cells/well, according to the manufacturer's protocol. The apoptotic cells were detected using a FACScan flow cytometer (BD Biosciences) and analyzed using the FlowJo 7.6.1 software (Tree Star, Inc.) The total apoptotic rate was determined as the sum of the late and the early apoptotic rates.
Wound healing assay
The effects of lncRNA PCAT-1 inhibition on cell motility was determined using a wound-healing assay. TSCC cells (2×104 cells/well) were seeded in a 24-well plate and reference points were recorded to ensure the acquisition of the identical area for imaging. After culturing for 2 days at 37°C (5% CO2), the cell monolayers were scratched to generate a cell-free wound and rinsed with PBS to remove cell debris from the wound edges. The cells were incubated in DMEM once more, and the migration distances (as the percentage of gap closure) were measured at 0-, 12- and 24-h time points.
Transwell assay
The effect of lncRNA PCAT-1 inhibition on cell invasion potential was detected using a Transwell assay. TSCC cells (1×105 cells/well) suspended in serum-free DMEM were added to the upper chamber of the Transwell inserts (membranes were pre-coated with 40 µl Matrigel for 2 h at 37°C) and incubated for 2 h at 37°C; the cells were allowed to penetrate through the porous membrane to the lower chamber [supplemented with 30% FBS (HyClone; Thermo Fisher Scientific Inc.)] for 4 h. After removal of the cells on the upper surface, the cells in the lower chamber were stained with 0.5% (w/v) crystal violet for 5 min at room temperature. Images were captured (magnification, ×200) under an inverted light microscope (AE31; Motic) and the number of invaded cells was calculated using Image-Pro Plus software version 6.0 (Media Cybernetics, Inc.).
RT-qPCR
The total RNA of TSCC cells was extracted using the RNApure High-purity Total RNA Rapid Extraction kit (cat. no. RP1201; BioTeke Corporation) according to the manufacturer's instructions. The cDNA templates were synthesized using Super M-MLV (cat. no. PR6502, BioTeke Corporation) according to the manufacturer's instruction; cDNA synthesis was conducted at 70°C for 5 min with 1 µl oligo(dT)15, 1 µl random primers and 2 µl dNTPs (2.5 mM). The PCR system contained 10 µl SYBR® Green master mix (Beijing Solarbio Science & Technology Co., Ltd.), 0.5 µl of each primer (PCAT-1 forward, 5′-ACAGGCTGAGGCAGGAGAAT-3′; PCAT-1 reverse; 5′-CTTTGGGAAGTGCTTTGGAG-3′; β-actin forward, 5′-CTTAGTTGCGTTACACCCTTTCTTG-3′; and β-actin reverse, 5′-CTGTCACCTTCACCGTTCCAGTTT-3′), 1 µl cDNA template and 8 µl double-distilled H2O. The amplification conditions were as follows: Denaturation at 95°C for 10 min, followed by 40 cycles of amplification at 95°C for 10 sec, 60°C for 20 sec and 72°C for 30 sec. The reaction was terminated at 25°C for 5 min. The relative expression levels of lncRNA PCAT-1 were determined using a Real-time PCR system (Exicycler 96; Bioneer Corporation) according to the 2−∆∆Cq method (26), and β-actin was used as the internal reference.
Western blot analysis
Total protein was extracted from TSCC cells using RIPA lysis buffer (cat. no. P0013B; Beyotime Institute of Biotechnology) and collected by centrifugation at 10,000 × g for 10 min at 4°C. SDS-PAGE (using a 12% gel) was performed with 40 µg protein/sample. The proteins were transferred to PVDF membranes that were subsequently blocked for 1 h at room temperature using 5% skimmed milk powder. The membranes were then incubated with primary antibodies against p21 (1:500 dilution; cat. no. D153319; Sangon Biotech Co., Ltd.) and β-actin (1:1,000 dilution; cat. no. sc-47778; Santa Cruz Biotechnology, Inc.) at 4°C overnight, followed by incubation with secondary horseradish peroxidase-conjugated IgG antibodies (1:5,000 dilution; cat. no. A0216 and A0208; Beyotime Institute of Biotechnology) at 37°C for 45 min. The membranes were developed using ECL Plus reagent (cat. no. P0018; Beyotime Institute of Biotechnology) and the relative expression levels were quantified using the Gel-Pro-Analyzer software version 4.0 (Media Cybernetics, Inc.) with β-actin as the internal reference.
Statistical analysis
The data are presented as the mean ± standard deviation of three replicates. Statistical analyses were performed using SPSS version 19.0 (IBM Corp.). One-way analysis of variance and Duncan's multiple range test were conducted for comparisons between >3 variables. The differences in lncRNA PCAT-1 expression levels between TSCC and peri-tumor tissue groups were analyzed using paired Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
lncRNA PCAT-1 is upregulated in TSCC specimens
The expression levels of lncRNA PCAT-1 were detected in 23 pairs of TSCC specimens and corresponding peri-tumor tissues using RT-qPCR. As presented in Fig. 1, the expression levels of lncRNA PCAT-1 were significantly upregulated in tumor tissues compared with those in the peri-tumor tissues. The results demonstrated a possible positive association between lncRNA PCAT-1 expression levels and TSCC progression.
Figure 1.
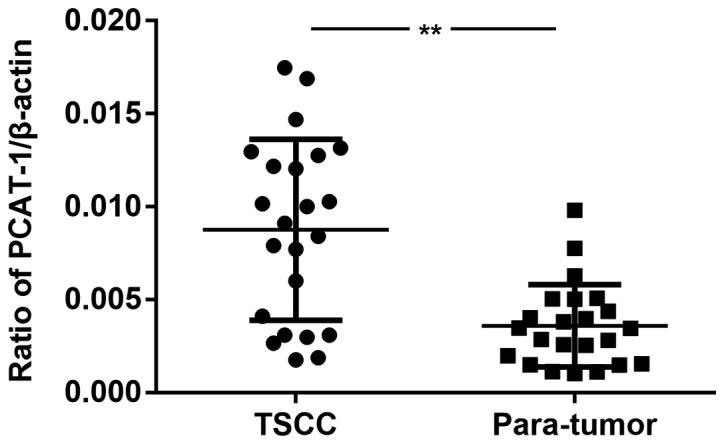
Expression of lncRNA PCAT-1 is upregulated in TSCC samples. The expression levels of lncRNA PCAT-1 in the tumor and peri-tumor tissues of 23 patients with TSCC were detected using reverse transcription-quantitative PCR. **P<0.01. lncRNA PCAT-1, long non-coding RNA prostate cancer-associated transcript-1; TSCC, tongue squamous cell carcinoma.
Inhibition of lncRNA PCAT-1 suppresses proliferation and induces apoptosis in TSCC cells
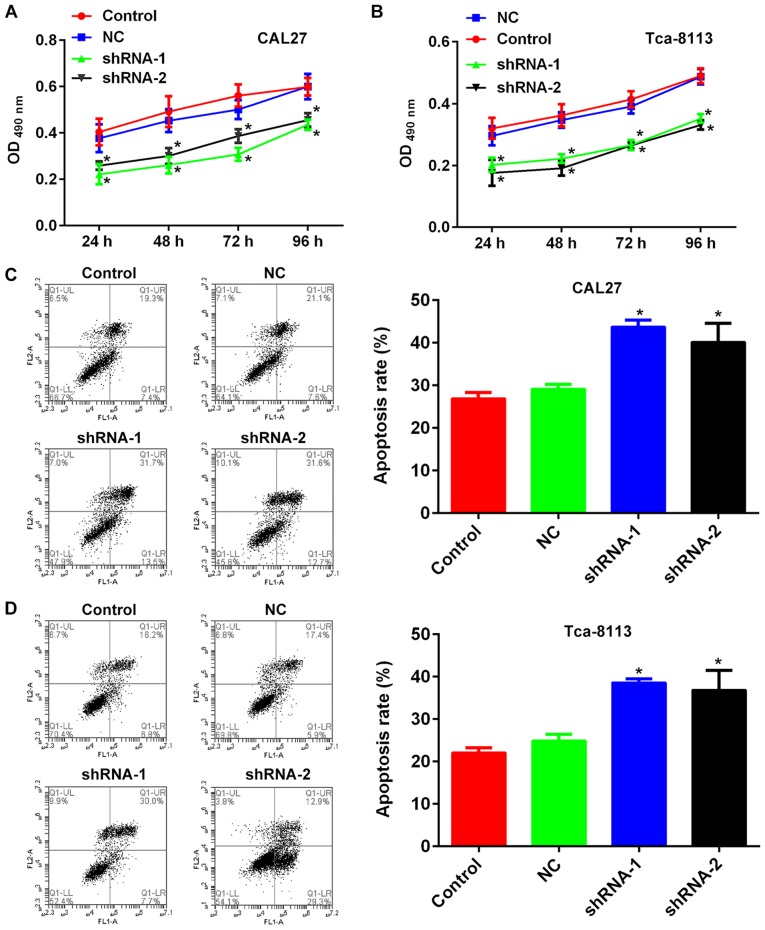
The TSCC cell lines CAL27 and Tca-8113 were transfected with lncRNA PCAT-1-shRNA vectors and the knockdown was confirmed using RT-qPCR (Fig. 2). MTT and flow cytometry assays were subsequently performed to establish the influence of lncRNA PCAT-1 inhibition on cell proliferation and apoptosis. For each cell line, the OD490 values of the knockdown groups were significantly lower than those of the NC-transfected groups (Fig. 3A and B). Furthermore, lncRNA PCAT-1 knockdown resulted in a marked increase in apoptotic rate compared with the NC group (Fig. 3C and D). Taken together, these results confirmed that lncRNA PCAT-1 inhibition impaired proliferation and induced apoptosis in TSCC cells.
Figure 2.
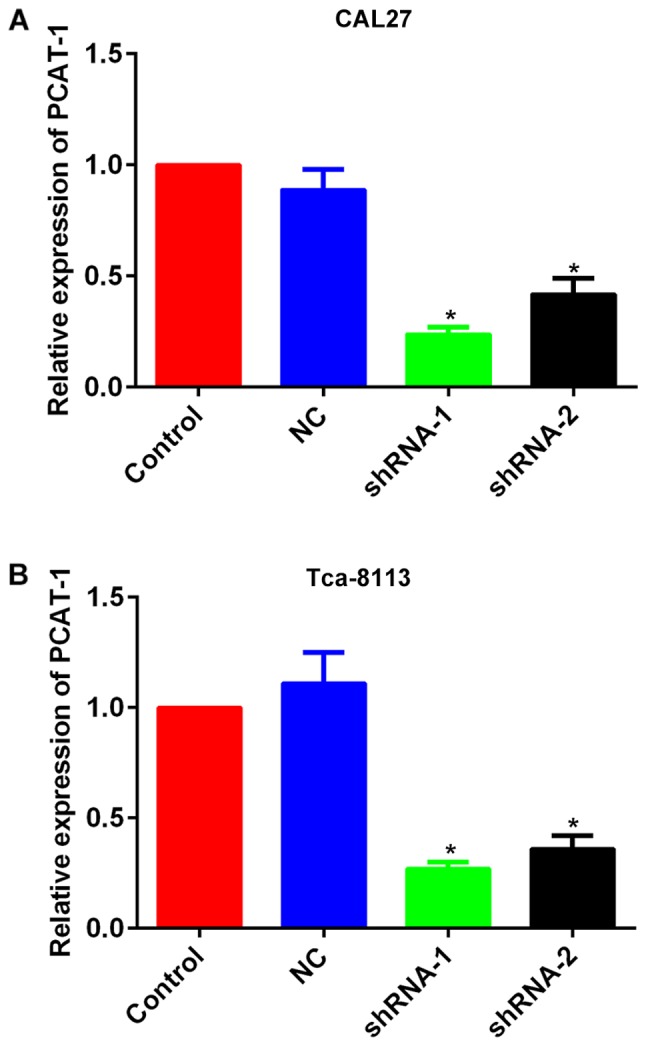
Expression of PCAT-1 was inhibited in tongue squamous cell carcinoma cell lines by transfection of specific shRNAs. Quantitative analysis using reverse transcription-quantitative PCR detection of the expression levels of lncRNA PCAT-1 in (A) CAL27 and (B) Tca-8113 cells. *P<0.05 vs. NC group. Each assay was performed in triplicate. lncRNA PCAT-1, long non-coding RNA prostate cancer-associated transcript-1; NC, negative control; shRNA, small hairpin RNA.
Figure 3.
Inhibition of lncRNA PCAT-1 impairs growth, while inducing apoptosis in tongue squamous cell carcinoma cell lines. Quantitative analysis results of MTT cell proliferation assays with (A) CAL27 and (B) Tca-8113 cells. Representative images and quantitative analysis of flow cytometry detection of apoptosis in (C) CAL27 and (D) Tca-8113 cells. *P<0.05 vs. NC group. Each assay was performed in triplicate. lncRNA PCAT-1, long non-coding RNA prostate cancer-associated transcript-1; NC, negative control; shRNA, small hairpin RNA.
Inhibition of lncRNA PCAT-1 reduces the metastatic potential of TSCC cells
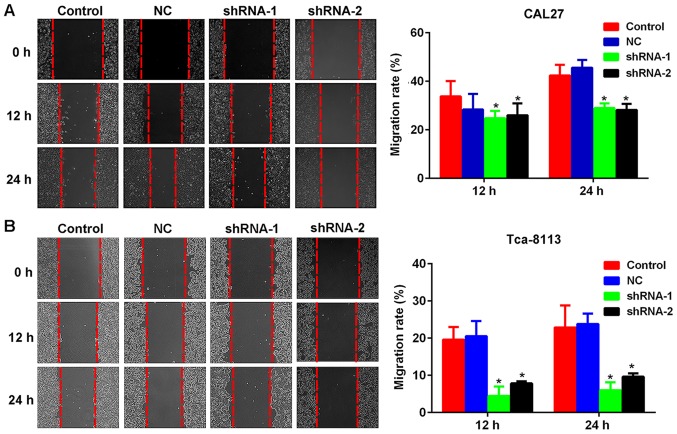
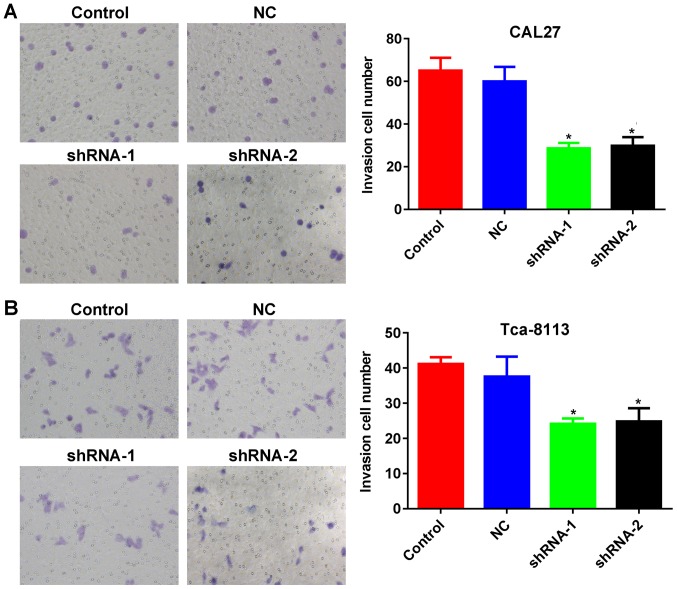
As presented in Fig. 4A and B, lncRNA PCAT-1 inhibition reduced the closure of scratch wounds compared with that in the Control and NC groups (Fig. 4A and B). The differences in the wound-healing rate between lncRNA PCAT-1-knockdown and NC cells were statistically significant (P<0.05; Fig. 4A and B). The results suggested that lncRNA PCAT-1 inhibition significantly suppressed TSCC cell migration. In addition, Transwell assays indicated that the number of TSCC cells penetrating through the membrane were significantly lower in the knockdown groups compared with those in the control groups (P<0.05; Fig. 5A and B), demonstrating inhibited invasive potential as a result of lncRNA PCAT-1 inhibition.
Figure 4.
Inhibition of lncRNA PCAT-1 reduces the migration ability of tongue squamous cell carcinoma cell lines. Representative images and quantitative analysis of wound healing assays using (A) CAL27 and (B) Tca-8113 cells. Magnification, ×200. *P<0.05 vs. NC group. Each assay was performed in triplicate. lncRNA PCAT-1, long non-coding RNA prostate cancer-associated transcript-1; NC, negative control; shRNA, small hairpin RNA.
Figure 5.
Inhibition of lncRNA PCAT-1 reduces the invasiveness of tongue squamous cell carcinoma cell lines. Representative images and quantitative analysis of Transwell assays with (A) CAL27 and (B) Tca-8113 cells. Magnification, ×200. *P<0.05 vs. NC group. Each assay was performed in triplicate. lncRNA PCAT-1, long non-coding RNA prostate cancer-associated transcript-1; NC, negative control; shRNA, small hairpin RNA.
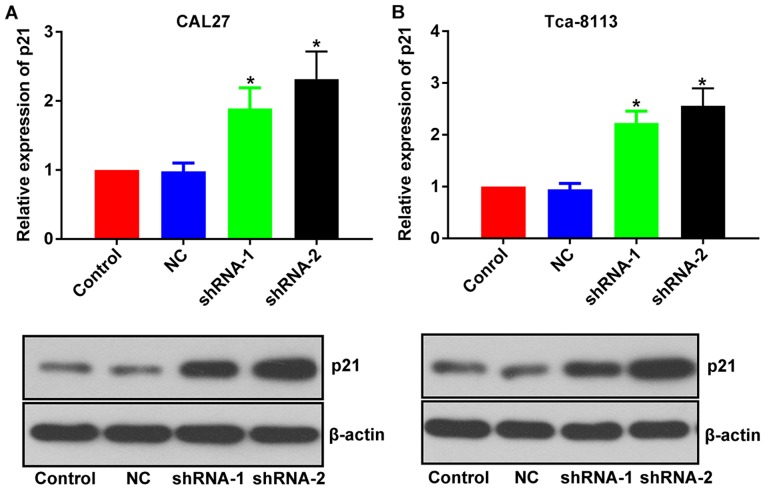
Inhibitory effect of lncRNA PCAT-1 knockdown in TSCC cells is associated with the upregulation of p21
The mechanisms associated with the inhibitory effect of lncRNA PCAT-1-knockdown were further investigated by focusing on its influence on p21 expression level. Following the inhibition of lncRNA PCAT-1, the expression level of p21 was upregulated in both TSCC cell lines (Fig. 6). Given the well-documented anti-tumor effect of p21 (20–22), the anti-TSCC effect of lncRNA PCAT-1-knockdown may be associated with the activation of p21 signaling.
Figure 6.
Inhibition of lncRNA PCAT-1 increases the expression of p21 in tongue squamous cell carcinoma cell lines. Representative images and quantitative analysis of p21 western blotting in (A) CAL27 and (B) Tca-8113 cells. *P<0.05 vs. NC group. Each assay was performed in triplicate. lncRNA PCAT-1, long non-coding RNA prostate cancer-associated transcript-1; NC, negative control; shRNA, small hairpin RNA.
Discussion
In the past 5 years, TSCC has become the most common type of oral cancer (23). The incidence and mortality rates associated with TSCC have been steadily increasing, and patients with TSCC currently account for one third of all oral cancer cases worldwide. Furthermore, TSCC is one of the most aggressive subtypes of oral cancer and in spite of the rapid progression in diagnostic and therapeutic strategies, the 5-year survival rate has remained unchanged (27). Therefore, it is necessary to explore novel targets for the development of additional anti-TSCC therapies.
Advancements in sequencing techniques have resulted in the identification of multiple cancer-associated lncRNAs in TSCC (25), and lncRNA-PPP2R4-5, SPRR2D-1, MAN1A2-1 and FAM46A-1 have been reported to be dysregulated in TSCC (25). In light of previous findings, the present study explored the role of lncRNA PCAT-1 on the growth and metastatic potential of TSCC. Furthermore, given the fact that lncRNA PCAT-1 is known to inhibit expression of the anti-tumor protein p21, the present study also explored the downstream pathway mediating its function, focusing on its interaction with p21. The results demonstrated that the expression of lncRNA PCAT-1 was upregulated in clinical TSCC samples, and that the inhibition of this lncRNA in TSCC cell lines impaired proliferation, induced apoptosis and suppressed metastatic and invasive potential. Regarding the downstream mechanisms at the molecular level, knockdown of PCAT-1 resulted in the upregulation of p21, indicating that these inhibitory effects on TSCC cells are, at least in part associated with the activation of p21 and downstream pathways.
The gene encoding lncRNA PCAT-1 is located on chromosome 8q24, and it was originally identified as a biomarker for prostate cancer (15). Subsequently, altered expression levels of lncRNA PCAT-1 were also reported in other cancer types; Shi et al (18) reported that the upregulation of lncRNA PCAT-1 was associated with the development of esophageal squamous cell carcinoma. A study by Qiao et al (19) reported that inhibition of lncRNA PCAT-1 impaired the multidrug resistance and aggressiveness of colorectal cancer cells. Given the involvement of lncRNA PCAT-1 in the genesis of various different cancer types, and the results of current clinical investigations with TSCC samples, the present study hypothesized that lncRNA PCAT-1 may also contribute to the onset and progression of TSCC. The results of the present study have supported this hypothesis, where inhibition of lncRNA PCAT-1 not only reduced cell growth and induced apoptosis in TSCC cells, but also suppressed the metastatic and invasive potential of these cells. The effects of lncRNA PCAT-1 inhibition demonstrated the critical function of this lncRNA in maintaining the normal biological characteristics of TSCC cells. They also inferred that this inhibition may represent a promising strategy for the development of anti-TSCC therapies.
Apart from determining the role of lncRNA PCAT-1 in the progression of TSCC, the present study also attempted to elucidate its mechanism. Therefore, the expression levels of p21 in TSCC PCAT-1-knockdown cells were examined. It was revealed that the expression levels of p21 were significantly upregulated following lncRNA PCAT-1 inhibition. The results were consistent with the previously reported effect of lncRNA PCAT-1, acting as an oncogene by reducing p21 expression levels (20). p21 is one of the most important cyclin-dependent kinases and regulates cell cycle transition from the G1 to the S phase (28). A previous study by Zhang et al (29) demonstrated that downregulation of p21 was closely associated with poor prognosis in patients with TSCC. Collectively, this indicates that the inhibitory effect of lncRNA PCAT-1 knockdown in TSCC is, at least in part associated with the induction of p21 expression.
The results of the present study indicated that lncRNA PCAT-1 is overexpressed in TSCC specimens, and that its inhibition impairs the growth, metastasis and invasiveness of TSCC, whilst inducing apoptosis. Furthermore, the inhibitory effect of lncRNA PCAT-1 knockdown on TSCC cells was associated with the upregulation of p21, indicating an interaction between lncRNA PCAT-1 and p21 signaling during the progression of TSCC. However, the present results only provide a preliminary evaluation of the mechanism by which lncRNA PCAT-1 acts in the genesis and progression of TSCC. Due to the original experimental design, the clinicopathological information of some patients was not collected, thus analyzing the potential of lncRNA PCAT-1 in predicating the progression of TSCC was not possible. To fully explain the role of lncRNA PCAT-1 in the oncogenesis of tongue tissues, more comprehensive studies with complete patient information and modulation of the downstream effectors of lncRNA PCAT-1 are required, which may aid the development of lncRNA PCAT-1-based anti-TSCC therapies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed are available from the corresponding author on reasonable request.
Authors' contributions
MY designed the experiments, performed the data collection and drafted the manuscript. TZ designed the experiments and drafted the manuscript. LF performed the data collection and analyzed the data. RT performed the data collection and analyzed the data. DL analyzed the data and YD analyzed the data and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of The People's Hospital of Tongliang District, Chongqing City. All patients provided written informed consent for the use of their tissues.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109:2093–2100. doi: 10.1111/cas.13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun H, Huang Z, Sheng W, Xu M. Emerging roles of long non-coding RNAs in tumor metabolism. J Hematol Oncol. 2018;11:106. doi: 10.1186/s13045-018-0648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huarte M, Rinn JL. Large non-coding RNAs: Missing links in cancer? Hum Molr Genet. 2010;19:152–161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ewan AG, Carolyn JB, Wan LL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.John RP, Arul MC. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough H, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by non-coding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabory A, Jammes H, Dandolo L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32:473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 11.Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, Zatloukal K. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 12.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 14.Rasool M, Malik A, Zahid S, Basit Ashraf MA, Qazi MH, Asif M, Zaheer A, Arshad M, Raza A, Jamal MS. Non-coding RNAs in cancer diagnosis and therapy. Noncoding RNA Res. 2016;1:69–76. doi: 10.1016/j.ncrna.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani I, Grasso CS, Kominsky HD, et al. Transcriptome sequencing identifies PCAT-1, a novel lincRNA implicated in prostate cancer progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prensner JR, Wei C, Sumin H, Iyer MK, Cao Q, Kothari V, Evans JR, Knudsen KE, Paulsen MT, Ljungman M, et al. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia. 2014;16:900–908. doi: 10.1016/j.neo.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi WH, Wu QQ, Li SQ, Yang TX, Liu ZH, Tong YS, Tuo L, Wang S, Cao XF. Upregulation of the long noncoding RNA PCAT-1 correlates with advanced clinical stage and poor prognosis in esophageal squamous carcinoma. Tumour Biol. 2015;36:2501–2507. doi: 10.1007/s13277-014-2863-3. [DOI] [PubMed] [Google Scholar]
- 19.Qiao L, Liu X, Tang Y, Zhao Z, Zhang J, Liu H. Knockdown of long non-coding RNA prostate cancer-associated ncRNA transcript 1 inhibits multidrug resistance and c-Myc-dependent aggressiveness in colorectal cancer Caco-2 and HT-29 cells. Mol Cell Biochem. 2018;441:99–108. doi: 10.1007/s11010-017-3177-8. [DOI] [PubMed] [Google Scholar]
- 20.Huang J, Deng G, Liu T, Chen W, Zhou Y. Long noncoding RNA PCAT-1 acts as an oncogene in osteosarcoma by reducing p21 levels. Biochem Biophys Res Commun. 2018;495:2622–2629. doi: 10.1016/j.bbrc.2017.12.157. [DOI] [PubMed] [Google Scholar]
- 21.Fang C, He W, Xu TY, Dai J, Xu L, Sun F. Upregulation of lncRNA DGCR5 correlates with better prognosis and inhibits bladder cancer progression via transcriptionally facilitating P21 expression. J Cell Physiol. 2019;234:6254–6262. doi: 10.1002/jcp.27356. [DOI] [PubMed] [Google Scholar]
- 22.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 23.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 24.Okuyemi OT, Piccirillo JF, Spitznagel E. TNM staging compared with a new clinicopathological model in predicting oral tongue squamous cell carcinoma survival. Head Neck. 2014;36:1481–1489. doi: 10.1002/hed.23486. [DOI] [PubMed] [Google Scholar]
- 25.Gao W, Chan YW, Wong TS. Long non-coding RNA deregulation in tongue squamous cell carcinoma. Biomed Res Int. 2014;2014:405860. doi: 10.1155/2014/405860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 28.Tedeschi A, Wutz G, Huet S, Jaritz M, Wuensche A, Schirghuber E, Davidson IF, Tang W, Cisneros DA, Bhaskara V, et al. Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature. 2013;501:564–568. doi: 10.1038/nature12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Chen W, Fu X, Su X, Yang A. CBX3 promotes tumor proliferation by regulating G1/S phase via p21 downregulation and associates with poor prognosis in tongue squamous cell carcinoma. Gene. 2018;654:49–56. doi: 10.1016/j.gene.2018.02.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed are available from the corresponding author on reasonable request.