Abstract
Tropomodulin-1 (TMOD1) is a key regulator of actin dynamics, which caps the pointed end of actin filaments. TMOD1 has been reported to be involved in several cellular processes, including neurite outgrowth, spine formation and cell migration. Increasing evidence demonstrates that TMOD1 is implicated in several aspects of cancer development. The present study aimed to investigate the role of TMOD1 in cervical cancer. HeLa and CaSki cell lines, derived from human cervical cancer, were used to evaluate the function of TMOD1. Cell motility was measured via a wound-healing assay, with the TMOD1 short hairpin (sh)RNAs transfected cells. Subsequently, cell proliferation was assessed using low serum cell culture condition, while cell cycle distribution was analyzed via flow cytometry. The results demonstrated that downregulated TMOD1 promoted cell motility and proliferation, which is attributed to promotion of G1/S phase transition in HeLa and CaSki cells. Furthermore, it was indicated that co-expression of shRNA resistant TMOD1 rescued these phenomena. The clinical data demonstrated that high TMOD1 expression is associated with good pathological status in patients with cervical cancer. Overall, the results of the present study indicated that TMOD1 may act as a tumor suppressor in cervical cancer, whereby its downregulated expression was demonstrated to have direct effects on cell motility and cell proliferation. These results provide new evidence for the prognostic prediction of cervical cancer, which may serve as a promising therapeutic strategy for patients with cervical cancer.
Keywords: cell cycle, cell proliferation, cell motility, cervical cancer, tropomodulin-1
Introduction
Cervical cancer is the fourth most common cancer of woman worldwide (1). In 2012, the incidence rate for cervical cancer was ~7.5/100,000 women worldwide (2). Currently, human papillomavirus is observed in nearly all reported cases of cervical cancer (3), whereby persistent infection with human papillomavirus is associated with an increased risk of developing cervical cancer (3). Surgical removal, chemotherapy, radiotherapy or concurrent chemoradiation are the common methods used to treat the different subtypes of cervical cancer (4,5). Although cisplatin-based chemoradiation yields overall survival rates of 66%, cisplatin is considered to cause kidney damage following long-term usage (6). Thus, the development of novel targets for the treatment of cervical cancer remains critical.
Tropomodulins (TMOD) are a family of proteins that cap the pointed end of actin filaments and regulate their depolymerization (7). There are currently four confirmed TMODs: TMOD1, 2, 3 and 4 that are involved in a variety of cellular functions, such as myofibril alignment (8), neurite extension (9) and endothelial cell migration (10). TMOD1 and TMOD3 demonstrate broad expression, while TMOD2 and TMOD4 demonstrate specific expression in the nervous system and skeletal muscle, respectively (7). Due to the role of TMODs in regulating actin dynamics, TMODs were observed to be associated with the negative regulation of neurite outgrowth (9) and cell migration (10). As a key member of the TMOD family, TMOD1 has been extensively researched thus far (7). Despite its involvement in the nervous system and in cardiac muscle, TMOD1 has been implicated in cancer development through its function in actin dynamics regulation (11). High TMOD1 expression has been reported to restore thymosin β-10 mediated apoptosis in SKOV-3 ovarian cancer cells (12). Furthermore, TMOD1 has been demonstrated to enhance hematopoietic reconstitution (13). It has also been reported that TMOD1 may serve as a target gene for NF-κB and promote the development of breast cancer via the NF-κB-TMOD1-β-catenin-MMP13 axis (14). Furthermore, overexpression of TMOD1 has been demonstrated to enhance regional lymph node metastasis in human oral cancer (15). Recently, the TMOD1 loci was screened as a potential risk loci in patients with esophageal adenocarcinoma using a high-density single nucleotide polymorphism array (16).
TMOD1 has been implicated in several types of cancer; however, its role in cervical cancer remains unknown. The present study investigated TMOD1 function in two cervical cancer cell lines, HeLa and CaSki, assessing its effects on cell motility, cell proliferation and cell cycle by downregulating or overexpressing TMOD1 in cervical cancer cells. Furthermore, clinical data obtained from patients with cervical cancer was assessed. The results indicate an association between the TMOD1 expression and the development of cervical cancer, which provides novel evidence on the ambiguous role of TMOD1 in cervical cancer.
Materials and methods
Plasmids
Non-targeting short hairpin (sh)RNA and specific shRNA for human TMOD1 were purchased from Sigma-Aldrich (Merck KGaA). The targeting sequence of TMOD1 shRNA-1 localizes in the untranslated region of the TMOD1 gene. The TMOD1 overexpression vector was constructed with the empty vector, CSII-CMV-MCS (RIKEN BioResource Center) and amplified according to the coding sequences of the TMOD1 gene. CSII-CMV-MCS was used as a control vector in overexpression experiments. The TMOD1 overexpression vector was used as a shRNA resistant TMOD1 expression vector. The sequences of shRNAs are presented in Table I.
Table I.
Sequences of shRNAs.
| shRNA clone | Antisense sequence |
|---|---|
| Human TMOD1 shRNA-1 | 5′-ATTTGGCAACTTTAATCGAGG-3′ |
| Human TMOD1 shRNA-2 | 5′-ATATTCCGGATATTATTGAGG-3′ |
TMOD1, tropomodulin-1; sh, short hairpin.
Cell culture and transfection
The HeLa and CaSki cervical cancer cell lines were obtained from the American Type Culture Collection. Cells were cultured in DMEM supplemented with 10% heat inactivated FBS at 37°C in a 5% CO2 incubator. Cells were plated into 6-well plates at a density of 1×105 cells/ml/plate and transfected with 6 µl of Fugene HD (Promega Corporation), followed by 1.5 µg of TMOD1 shRNAs (1 µg/µl) with or without 1.5 µg of TMOD1 overexpression vector (1 µg/µl), thus the final volume of transfection mixture was 100 µl. After 24 h, cells were treated with puromycin (Thermo Fisher Scientific, Inc.) at 37°C, for 2 consecutive days.
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from the cultured cells using ISOGEN lysis buffer (Nippon Gene Co., Ltd.), according to the manufacturer's protocol. Total RNA was reverse transcribed into cDNA using the ReverTra Ace® qPCR RT kit (Toyobo Life Science), according to the manufacturer's protocol. qPCR was subsequently performed using the SYBR® Green Real-time PCR master mix (Toyobo Life Science), as previously described (17). The method of quantification was performed according to a previous report (18). The primer sequences of TMOD1, Twist, Snail and GAPDH are presented in Table II.
Table II.
Primer sequences for PCR experiments.
| Primer name | Forward primer sequence (5′-3′) | Reverse primer sequence (5′-3′) |
|---|---|---|
| Human TMOD1 | GTGGAAATGGAGATTGTGAGC | TCATGCTGCCAGCATTTTGC |
| Human GAPDH | ATCATCCCTGCCTCTACTGG | CCCTCCGACGCCTGCTTCAC |
| Human Snail | TCGGAAGCCTAACTACAGCGA | AGATGAGCATTGGCAGCGAG |
| Human Twist | GTCCGCAGTCTTACGAGGAG | GCTTGAGGGTCTGAATCTTGCT |
TMOD1, tropomodulin-1.
Western blot analysis
Total protein was extracted from HeLa or CaSki cells using RIPA lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP-40, 1% sodium dexycholate, 0.1% SDS), and protein concentration was measured using a Pierce bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Inc). A total of 15 µg protein/lane was separated via SDS-PAGE on a 12% gel. The separated proteins were subsequently transferred onto a nitrocellulose membrane (GE Healthcare) and blocked with 5% skimmed milk, supplemented in PBS with 0.1% Tween-20 for 1 h at room temperature. The membranes were incubated with primary antibodies against TMOD1 (1:1,000; cat. no. H00007111-B01P; Novus Biologicals, LCC) and β-actin (1:1,000; cat. no. A5441; Sigma-Aldrich; Merck KGaA) overnight at 4°C. Membranes were washed three times with 0.1% Tween-20 in PBS. Following the primary antibody incubation, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (1:5,000; cat. no. 115-035-003; Jackson ImmunoResearch Laboratories, Inc.) for 1 h at room temperature. Membranes were washed three times again with 0.1% Tween-20 in PBS, and protein bands were visualized using the ECL western blotting substrate (GE Healthcare), according to the manufacturer's protocol.
In vitro wound-healing assay
The wound-healing assay was performed as described by Zhang et al (19), with a modification on the culture time following wound generation. A total of 8 ×105 cells/ml HeLa cells or 1.2× 106 cells/ml CaSki cells were seeded into a 6-well plate and incubated at 37°C with DMEM supplied with 10% heat-inactivated FBS, until ~100% confluence was achieved. The straight cell-free wound was created using a 100 µl pipette tip in the center of the plate. Cells were subsequently washed twice with PBS to remove any debris and incubated with serum-free media at 37°C for 24 h. Image was captured using a light microscope (magnification, ×100). The distance migrated by the cell monolayer was measured during this period, in order to close the wounded area. The results were presented as a relative migration ratio as follows: Distance migrated by TMOD1 shRNAs or shRNA-resistant TMOD1, with TMOD1 shRNA-1 treated cells are relative to the distance migrated by control shRNAs treated cells. The distance was determined using ImageJ software [version 1.8.0; (20)].
Cell proliferation assay
The transfected cells were re-seeded into 96 well plates at a density of 1×104 cells/ml of HeLa cells or 3×104 cells/ml of CaSki cells and allowed to adhere overnight. At day 0, the culture medium was changed from 10% FBS to DMEM supplemented with 2% heat inactivated FBS. At day 4, cell number was measured using the Cell Counting Kit-8 (Dojindo Molecular Technologies, Inc.) at a wavelength of 450 nm, according to the manufacturer's protocol.
Flow cytometry
Cell cycle analysis was performed as described by Lu et al (17). A total of 1×106 cells/ml HeLa cells or CaSki cells were collected following treatment with trypsin at 37°C for 5 min, and washed once with 5 ml of cold PBS prior to fixation with absolute ethanol overnight at −20°C. Fixed cells were centrifuged at 167.7 × g/min at 4°C for 5 min and re-suspended in 1 ml of PBS. Subsequently, cells were treated with 10 µl of RNase A (10 mg/ml) for 30 min at 37°C, and supplemented with 50 µl of propidium iodide (1 mg/ml) for 30 min at 4°C. Cell cycle analysis was performed using a FACS Canto flow cytometer (BD Biosciences) and FlowJo software (version 7.6; (TOMY Digital Biology Co., Ltd.).
Terminal deoxynucleotidyl-transferase-mediated dUTP nick end labeling (TUNEL) assay
Following transfection with TMOD1 shRNA or control shRNA, HeLa or CaSki cells were re-seeded into 8-well plates (BD Biosciences) at a density of 3×104 cells/chamber, after 3 days. After 24 h, the TUNEL assay was performed, according to the manufacturer's protocol (21). Briefly, cells were washed three times with PBS and subsequently fixed with 4% formaldehyde for 15 min at 4°C. Cells were incubated with 0.5% Tween 20 in PBS supplemented with 0.2% BSA at room temperature for 15 min and re-washed three times with PBS, prior to incubation in 100 µl of TdT mixture (90 µl of TdT buffer, 5 µl of FITC-dUTP and 5 µl of TdT; Medical & Biological Laboratories, Co., Ltd.) at 37°C for 1 h. Cell nuclei were counterstained with DAPI (final concentration: 2 µg/ml) for 10 min at room temperature and mounted with mounting medium (Abcam). TUNEL-positive cells were observed in three randomly-selected fields under a fluorescent microscope (magnification, ×100).
Matrigel invasion assay
Transwell membranes (pore size, 8 mm; BD Biosciences) were precoated with 100 µl of 5 mg/ml Matrigel (BD Biosciences) at 37°C for 6 h. A total of 5×104 HeLa or CaSki cells were harvested 48 h post-transfection and plated in the upper chambers of Transwell plates in % BSA. Simultaneously, 10% FBS was plated in the lower chambers. Following incubation at 37°C for 24 h, the non-migratory cells in the upper chambers were removed and the migratory cells were stained with DAPI for 10 min at room temperature. Stained cells were counted in four randomly-selected fields under a fluorescent microscope (magnification, ×100).
Clinical patient data
The raw TMOD1 gene expression data and corresponding clinical parameters were downloaded from the publicly available R2 database (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi). The GSE2109 cervical cancer dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2109) employed in R2 was downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). The GEO database includes 36 clinical samples, of which 25 cervical cancer specimens where selected in the present study, which exhibited detectable TMOD1 expression. All samples were hybridized to Affymetrix human genome U133 plus 2.0 microarrays. TMOD1 expression levels of 25 individuals are presented in Table III.
Table III.
TMOD1 expression levels in the clinical database.
| Stage | Sample name | Relative TMOD1 expression level | Histology | Patient age, years |
|---|---|---|---|---|
| I (n=12) | gsm117626 | 4.27 | Endometrioid carcinoma | >40 |
| gsm152580 | 4.68 | Squamous cell carcinoma, spindle cell type | <40 | |
| gsm152635 | 6.12 | Squamous cell carcinoma | <40 | |
| gsm152667 | 5.09 | Adenocarcinoma | <40 | |
| gsm152719 | 4.47 | Adenocarcinoma | <40 | |
| gsm179853 | 7.02 | Adenosquamous carcinoma | <40 | |
| gsm179907 | 4.49 | Squamous cell carcinoma, non-keratinizing | >40 | |
| gsm179956 | 4.49 | Squamous cell carcinoma, non-keratinizing | <40 | |
| gsm203742 | 4.69 | Squamous cell carcinoma, non-keratinizing | >40 | |
| gsm325835 | 8.46 | Mucinous Adenocarcinoma, endocervical type | <40 | |
| gsm46919 | 5.46 | Squamous cell carcinoma, verrucous | >40 | |
| gsm46942 | 6.01 | Squamous cell carcinoma, non-keratinizing | <40 | |
| II (n=1) | gsm203799 | 4.79 | Squamous cell carcinoma, non-keratinizing | >40 |
| III (n=9) | gsm117576 | 2.00 | Squamous cell carcinoma, keratinizing | <40 |
| gsm152587 | 2.68 | Squamous cell carcinoma | >40 | |
| gsm152723 | 3.66 | Squamous cell carcinoma, keratinizing | >40 | |
| gsm152751 | 0.58 | Squamous cell carcinoma, keratinizing | >40 | |
| gsm152788 | 2.94 | Squamous cell carcinoma | >40 | |
| gsm102527 | 1.49 | Squamous cell carcinoma | >40 | |
| gsm203622 | 5.35 | Adenocarcinoma | >40 | |
| gsm76614 | 1.32 | Squamous cell carcinoma, non-keratinizing | >40 | |
| gsm89050 | 5.30 | Squamous cell carcinoma, non-keratinizing | <40 | |
| IV (n=3) | gsm102481 | 4.83 | Squamous cell carcinoma, non-keratinizing | <40 |
| gsm152787 | 2.83 | Squamous cell carcinoma, non-keratinizing | <40 | |
| gsm152800 | 1.77 | Squamous cell carcinoma | >40 |
TMOD1, tropomodulin-1; gsm: Gene Expression Omnibus database sample.
Statistical analysis
Statistical analysis was performed using GraphPad (version 5; GraphPad Software, Inc.). Data are presented as the mean ± standard deviation. Statistical analysis was determined using one-way analysis of variance followed by Tukey's post hoc test. All experiments were performed in triplicate. P<0.05 was considered to indicate a statistically significant difference.
Results
Downregulation of TMOD1 promotes cell motility in HeLa and CaSki cells
Given the close association of TMOD1 with filamentary actin dynamics, the present study determined to investigate the involvement of TMOD1 in cell motility.
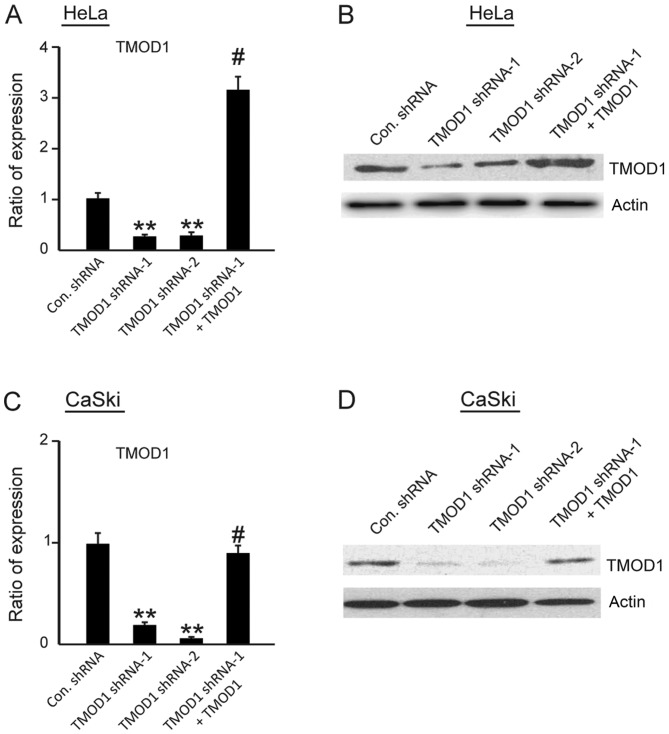
First, HeLa and CaSki cells were transfected with control non-targeting shRNA, TMOD1 shRNA-1, TMOD1 shRNA-2, or shRNA-resistant TMOD1 plus TMOD1 shRNA-1, respectively. After 24 h, puromycin (final concentration: 2 µg/ml) was added to the cell culture medium, in order to obtain the shRNA positive cells. Following transfection after 3 days, total RNA and protein were extracted, and RT-qPCR and western blot analysis were performed, respectively, to determine knockdown efficiency. The results demonstrated that both TMOD1 shRNA-1 and TMOD1 shRNA-2 significantly suppressed TMOD1 expression in HeLa and CaSki cells (all P<0.001; Fig. 1A-D).
Figure 1.
TMOD1 shRNAs suppresses TMOD1 expression in HeLa and CaSki cells. Total RNA and whole cell lysate were extracted from stable HeLa and CaSki cells transfected with control shRNA, TMOD1 shRNA-1, TMOD1 shRNA-2 or hRNA-resistant TMOD1 with TMOD1 shRNA-1 and subjected to (A and C) reverse transcription-quantitative PCR and (B and D) western blot analysis, respectively. The data are presented as the mean ± standard deviation (n=3). **P<0.001 vs. control shRNA group; #P<0.001 vs. TMOD1 shRNA-1 group. TMOD1, tropomodulin-1; Con, control; sh, short hairpin.
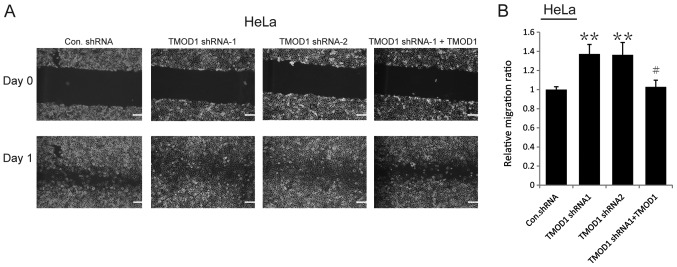
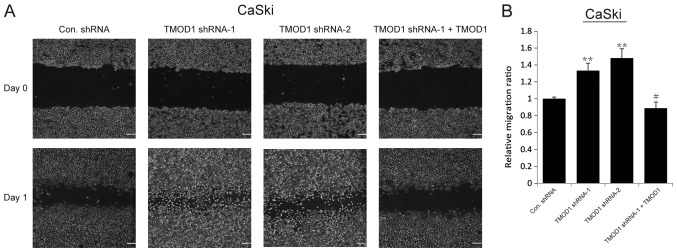
The results of the wound-healing assay demonstrated that the motility of TMOD1 shRNAs-treated cells was more notable compared with the control cells (all P<0.001; Figs. 2 and 3). In order to confirm this phenomenon, rescue experiments were performed in which a shRNA-1-resistant TMOD1 expressing vector was used to transfect TMOD1 shRNA-1 treated cells. The results indicated that TMOD1 expression significantly rescued the phenotype caused by TMOD1 shRNA-1 (all P<0.01; Figs. 2 and 3). A Matrigel assay was performed to investigate the role of TOMOD1 in cervical cancer cell invasion. The results demonstrated that the tendency of cell invasion was promoted in both Hela and CaSki cells following TMOD1 knockdown, and differences between the control and TMOD1 shRNAs treated groups were not significant (data not shown). The expression of two epithelial-mesenchymal transition (EMT) markers, Twist and Snail, was also analyzed via RT-qPCR to determine whether or not TMOD1 modulates cell migration by EMT, and no significant difference was demonstrated (data not shown).
Figure 2.
Downregulation of TMOD1 enhances cell motility. The stably transfected HeLa cells were seeded in 6-well plates and scratched the next day. (A) The images were captured at 0 24 h, respectively, and (B) the corresponding quantitative results are demonstrated. The data are presented as the mean ± standard deviation (n=3). Scale bar, 150 µm. **P<0.001 vs. control shRNA group; #P<0.01 vs. TMOD1 shRNA-1 group. Scale bar, 100 µm. TMOD1, tropomodulin-1; Con, control; sh, short hairpin.
Figure 3.
Downregulation of TMOD1 enhances cell motility. The stably transfected CaSki cells were seeded in 6-well plates and scratched the next day. (A) The images were captured at 0 and 24 h, respectively, and (B) the corresponding quantitative results are demonstrated. The data are presented as the mean ± standard deviation (n=3). Scale bar, 150 µm. **P<0.001 vs. control shRNA group; #P<0.01 vs. TMOD1 shRNA-1 group. Scale bar, 100 µm. TMOD1, tropomodulin-1; Con, control; sh, short hairpin.
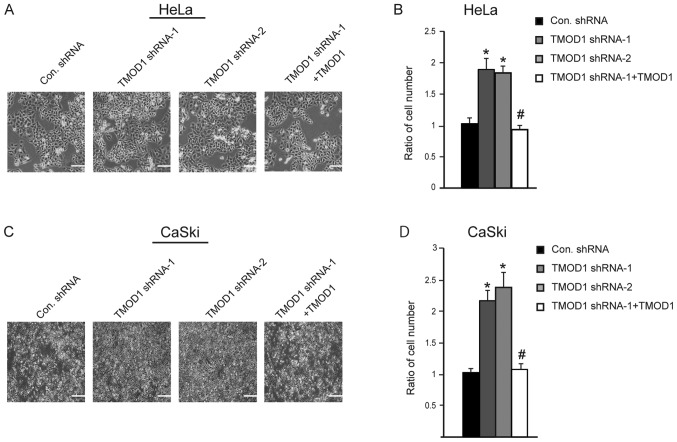
Downregulation of TMOD1 promotes cell proliferation and G1/S transition in HeLa and CaSki cells
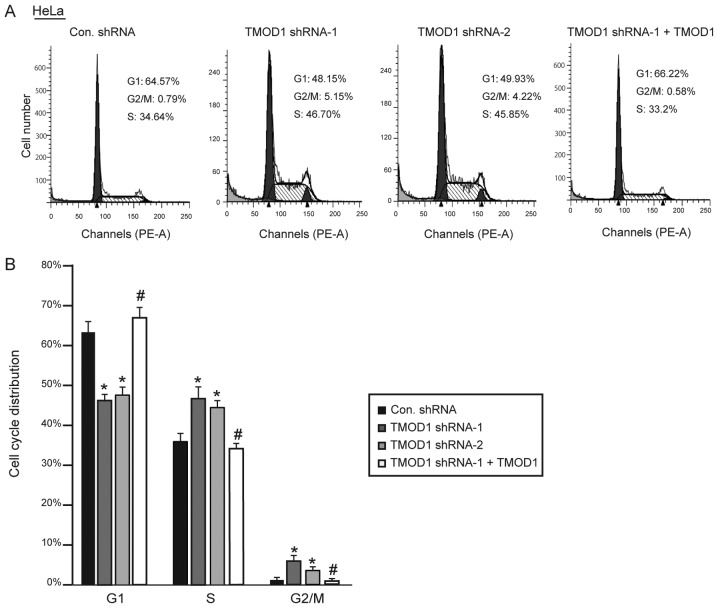
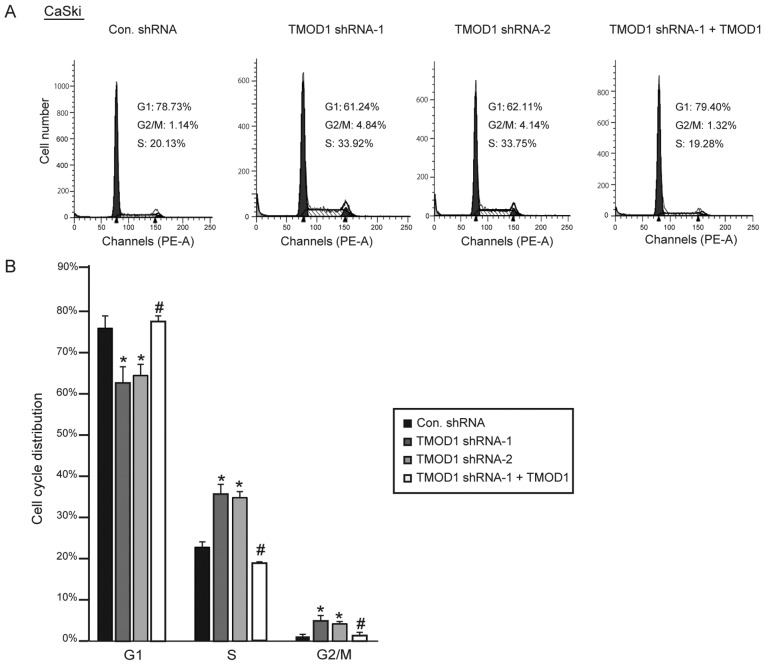
In order to improve the understanding of the role of TMOD1 in cervical cancer, its expression on HeLa and CaSki cell proliferation was subsequently assessed. The stable shRNA transfected cells were seeded and cultured in DMEM supplied with 2% heat inactivated FBS, and cell number was counted at day 4. The results indicated that the cells transfected with TMOD1 shRNAs proliferated at a faster rate compared with control cells (all P<0.01; Fig. 4A-D). Simultaneously, the level of apoptotic cells was detected via the TUNEL assay at day 4. No significant differences in cell death were demonstrated between the four groups (data not shown). Cell cycle analysis was performed via flow cytometry, in order to determine whether TMOD1 regulates cell proliferation by altering the cell cycle. The results demonstrated that downregulation of TMOD1 promoted G1/S phase transition in HeLa and CaSki cells, and the percentage of cells in the G2/M phase was significantly increased, while shRNA-resistant TMOD1 expression in TMOD1 shRNA treated cells was demonstrated to rescue the phenomenon induced by TMOD1 shRNA-1 (all P<0.05; Figs. 5 and 6).
Figure 4.
Downregulation of TMOD1 promotes HeLa and CaSki cell proliferation. The stably transfected HeLa and CaSki cells were cultured in DMEM supplied with 2% FBS. Phase-contrast images were captured at day 4 of (A) HeLa and (C) CaSki cells. Scale bar, 100 µm. Cell numbers were counted, and the data quantified for (B) HeLa and (D) CaSki cells. The data are presented as the mean ± standard deviation (n=3). *P<0.01 vs. control shRNA group; #P<0.01 vs. TMOD1 shRNA-1 group. Scale bar, 100 µm. TMOD1, tropomodulin-1; Con, control; sh, short hairpin.
Figure 5.
TMOD1 regulates the cell cycle in HeLa cells. The stably transfected HeLa cells were cultured in DMEM supplied with 2% FBS. (A) Cell cycle distribution was detected via flow cytometry at day 4. (B) Cell cycle distribution was measured and quantified. The data are presented as the mean ± standard deviation (n=3). *P<0.01 vs. control shRNA group; #P<0.05 vs. TMOD1 shRNA-1 group. TMOD1, tropomodulin-1; Con, control; sh, short hairpin.
Figure 6.
TMOD1 regulates cell cycle in CaSki cells. The stably transfected CaSki cells were cultured in DMEM supplied with 2% FBS. (A) Cell cycle distribution was detected via flow cytometry at day 4. (B) Cell cycle distribution was measured. The data are presented as the mean ± standard deviation (n=3). *P<0.01 vs. control shRNA group; #P<0.05 vs. TMOD1 shRNA-1 group. TMOD1, tropomodulin-1; Con, control; sh, short hairpin.
Low TMOD1 expression is associated with worse pathological status
Whether or not the in vitro data were relevant to the clinical features of human cervical cancer was subsequently determined. TMOD1 expression was analyzed in different pathological stages using the R2 database (http://r2.amc.nl), which is a free, and publicly accessible web-based genomics analysis and visualization platform that allows researchers to integrate, analyze and visualize clinical and genomics data. High TMOD1 expression was observed in patients in the early pathological stages, while low TMOD1 expression was presented in patients in the late stages (P<0.05; Fig. 7). Patients within the Stage II group was excluded due to lack of an adequate sample size required for statistical analysis. In all, these results were consistent with the in vitro data, further suggesting that TMOD1 may act as a tumor suppressor gene in human cervical cancer.
Figure 7.
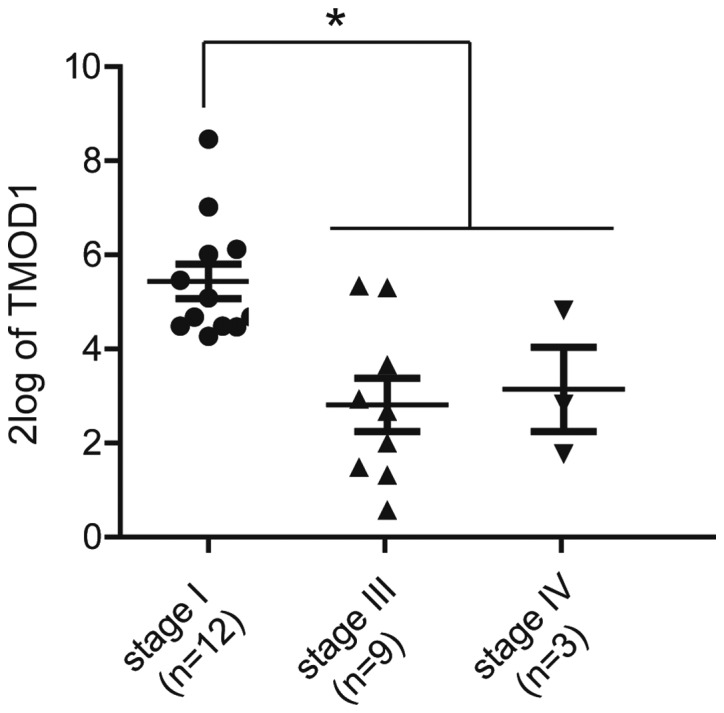
TMOD1 is associated with poor pathological status. TMOD1 expression was analyzed in three pathological subgroups of 25 patients with cervical cancer, from the clinical databases, GEO (GSE2019) and R2 (tumor cervix-Expo MAS5.0). Patients within the stage II group were excluded due to lack of an adequate sample size required for statistical analysis. TMOD1 expression levels for individual samples were analyzed using Affymetrix Human Genome U133 Plus 2.0 Array. *P<0.05. TMOD1, tropomodulin-1; GEO, Gene Expression Omnibus database.
Discussion
Actin is one of the multi-functional proteins in eukaryotic cells, which can polymerize into filamentary actin and also depolymerize into monomer actin (22). Depending on the dynamics of filamentary actin, actin regulates several physiological activities, such as regulating the subcellular localization of p53 (22) and regulating gene transcription (23). Filamentary actin has two ends, the barded end and the pointed end. Thus far, TMODs are the only known molecules that have the ability to cap the pointed end of filamentary actin. Capping by TMOD1 inhibits the release of free monomer actin from the pointed end, therefore prevents filamentary actin renewal and elongation (24). The tight association between actin and TMOD1 led researchers to focus on the function of TMOD1 on cell morphology (25,26), motility (10) and metastasis (15) or neuronal growth (9). It has been reported that TMOD1 knockdown in N2a cells increases the number of neuritis/cell and decreases the mean primary neurite length (9).
Despite its extensive roles within the nervous system, a number of previous reports have demonstrated that TMOD1 promotes cancer cell proliferation and metastasis by regulating filamentary actin, which indicates that TMOD1 may act as an oncogene in certain types of cancer (13–15,27). However, the results of the present study indicated that downregulation of TMOD1 promoted cell migration and proliferation in both HeLa and CaSki cell lines. Furthermore, the results from the clinical database suggested that low TMOD1 expression is associated with relative higher pathological grading and staging, while high expression is associated with relative lower pathological grading and staging in patients with human cervical cancer. Taken together, these results indicated that TMOD1 may act as a tumor suppressor in human cervical cancer, which seems to vary in its role across different types of cancer.
Cell cycle analysis demonstrated that downregulation of TMOD1 increased the percentage of cells in the S phase; however, it decreased the number of cells in the G1 phase. Despite the lack of current data on the association between TMOD1 and cell cycle progression, the notable roles of actin dynamics in cell cycle progress have been broadly researched (28,29). Disruption of filamentary actin dynamics with drugs, including cytochalasin D (inhibits actin monomer assembly by binding to the barbed ends of microfilaments), latrunculin B (prevents actin polymerization by binding to actin monomers near their ATP-binding site) and Jasplakinolide (stabilizes and promotes actin polymerization by binding to F-actin), has been reported to result in cell cycle arrest (30–32). Actin and several actin binding proteins, including Myosin II and septin have been demonstrated to interact with cyclins and cyclin-dependent kinases in different phases of the cell cycle (28). Furthermore, Sui et al (33) demonstrated that TMOD3 depletion increases the number of cells in the S phase, while fewer cells were observed in the G0/G1 phase in TMOD3−/− fetal liver tissue. Among the four TMODs, the amino acid sequences of their two major function domains (the TM-Cap and the LRR-Cap domains) have a 70–80% and 80–90% similarity, respectively (34). Considering the similar molecular structure and biological functions between TMOD1 and TMOD3, TMOD1 may also be involved in cell cycle regulation; however, its underlying molecular mechanism remains unclear.
The wound-healing assay results demonstrated that cell migration was notably enhanced following TMOD1 depletion. Conversely, the role of TMOD1 in cell invasion and EMT was assessed via a Matrigel invasion assay, and analysis of Twist and Snail expression levels, respectively. However, no significant differences were demonstrated between control cells and TMOD1 shRNA-treated cells in these experiments. Overall, TMOD1 expression was demonstrated to inhibit cell migration, but not invasion.
Increasing evidence suggests that the cyclins which modulate G1/S transition also promote cell migration. For instance, cyclin D1 has been reported to promote cell migration by stimulating the activity of Rac1 and Ral GTPases (35), and cyclin E2 has been demonstrated to enhance cell proliferation and migration of non-small cell lung cancer (36). The results of the present study demonstrated that downregulation of TMOD1 promoted G1/S transition, which suggests that TMOD1 may regulate the cell cycle by affecting the expression levels of G1 cyclins. Future studies will focus on investigating the associations between TMOD1, G1 cyclins and cell migration.
Acknowledgements
The authors of the present study would like to thank Dr. Peng Zou from Shenyang Medical College (Shenyang, China) for his general support.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
FL and PM designed the present study. FL, DC and LZ acquired the data, while FL and BM analyzed and interpreted the data. FL and PM drafted the initial manuscript and made substantial revisions. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Pimple S, Mishra G, Shastri S. Global strategies for cervical cancer prevention. Curr Opin Obstet Gynecol. 2016;28:4–10. doi: 10.1097/GCO.0000000000000241. [DOI] [PubMed] [Google Scholar]
- 2.GLOBOCAN 2012, corp-author. Estimated cancer incidence, mortality and prevalence worldwide in 2012. International Agency for Research on Cancer, World Health Organization [Google Scholar]
- 3.Small W, Jr, Bacon MA, Bajaj A, Chuang LT, Fisher BJ, Harkenrider MM, Jhingran A, Kitchener HC, Mileshkin LR, Viswanathan AN, Gaffney DK. Cervical cancer: A global health crisis. Cancer. 2017;123:2404–2412. doi: 10.1002/cncr.30667. [DOI] [PubMed] [Google Scholar]
- 4.Lippman SM, Hawk ET. Cancer prevention: From 1727 to milestones of the past 100 years. Cancer Res. 2009;69:5269–5284. doi: 10.1158/0008-5472.CAN-09-1750. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Yu J, Qian L, Zhang H, Ma J. Extended field intensity-modulated radiotherapy plus concurrent nedaplatin treatment in cervical cancer. Oncol Lett. 2016;11:3421–3427. doi: 10.3892/ol.2016.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duenas-Gonzaleza A, Cetina L, Coronel J, Gonzalez-Fierro A. The safety of drug treatments for cervical cancer. Expert Opin Drug Saf. 2016;15:169–180. doi: 10.1517/14740338.2016.1130127. [DOI] [PubMed] [Google Scholar]
- 7.Fischer RS, Fowler VM. Tropomodulins: Llife at the slow end. Trends Cell Biol. 2003;13:593–601. doi: 10.1016/j.tcb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Gokhin DS, Fowler VM. Cytoplasmic gamma-actin and tropomodulin isoforms link to the sarcoplasmic reticulum in skeletal muscle fibers. J Cell Biol. 2011;194:105–120. doi: 10.1083/jcb.201011128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fath T, Fischer RS, Dehmelt L, Halpain S, Fowler VM. Tropomodulins are negative regulators of neurite outgrowth. Eur J Cell Biol. 2011;90:291–300. doi: 10.1016/j.ejcb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer RS, Fritz-Six KL, Fowler VM. Pointed-end capping by tropomodulin3 negatively regulates endothelial cell motility. J Cell Biol. 2003;161:371–380. doi: 10.1083/jcb.200209057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nowak RB, Papoin J, Gokhin DS, Casu C, Rivella S, Lipton JM, Blanc L, Fowler VM. Tropomodulin 1 controls erythroblast enucleation via regulation of F-actin in the enucleosome. Blood. 2017;130:1144–1155. doi: 10.1182/blood-2017-05-787051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rho SB, Chun T, Lee SH, Park K, Lee JH. The interaction between E-tropomodulin and thymosin beta-10 rescues tumor cells from thymosin beta-10 mediated apoptosis by restoring actin architecture. FEBS Lett. 2004;557:57–63. doi: 10.1016/S0014-5793(03)01438-8. [DOI] [PubMed] [Google Scholar]
- 13.Ting SB, Deneault E, Hope K, Cellot S, Chagraoui J, Mayotte N, Dorn JF, Laverdure JP, Harvey M, Hawkins ED, et al. Asymmetric segregation and self-renewal of hematopoietic stem and progenitor cells with endocytic Ap2a2. Blood. 2012;119:2510–2522. doi: 10.1182/blood-2011-11-393272. [DOI] [PubMed] [Google Scholar]
- 14.Ito-Kureha T, Koshikawa N, Yamamoto M, Semba K, Yamaguchi N, Yamamoto T, Seiki M, Inoue J. Tropomodulin 1 expression driven by NF-κB enhances breast cancer growth. Cancer Res. 2015;75:62–72. doi: 10.1158/0008-5472.CAN-13-3455. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki T, Kasamatsu A, Miyamoto I, Saito T, Higo M, Endo-Sakamoto Y, Shiiba M, Tanzawa H, Uzawa K. Overexpression of TMOD1 is associated with enhanced regional lymph node metastasis in human oral cancer. Int J Oncol. 2016;48:607–612. doi: 10.3892/ijo.2015.3305. [DOI] [PubMed] [Google Scholar]
- 16.Gharahkhani P, Fitzgerald RC, Vaughan TL, Palles C, Gockel I, Tomlinson I, Buas MF, May A, Gerges C, Anders M, et al. Genome-wide association studies in oesophageal adenocarcinoma and Barrett's oesophagus: A large-scale meta-analysis. Lancet Oncol. 2016;17:1363–1373. doi: 10.1016/S1470-2045(16)30240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu F, Zheng Y, Donkor PO, Zou P, Mu P. Downregulation of CREB promotes cell proliferation by mediating G1/S phase transition in hodgkin lymphoma. Oncol Res. 2016;24:171–179. doi: 10.3727/096504016X14634208142987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Zhang CG, Huang JC, Liu T, Li XY. Anticancer effects of bishydroxycoumarin are mediated through apoptosis induction, cell migration inhibition and cell cycle arrest in human glioma cells. J BUON. 2015;20:1592–1600. [PubMed] [Google Scholar]
- 20.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mu P, Nagahara S, Makita N, Tarumi Y, Kadomatsu K, Takei Y. Systemic delivery of siRNA specific to tumor mediated by atelocollagen: Combined therapy using siRNA targeting Bcl-xL and cisplatin against prostate cancer. Int J Cancer. 2009;125:2978–2990. doi: 10.1002/ijc.24382. [DOI] [PubMed] [Google Scholar]
- 22.Saha T, Guha D, Manna A, Panda AK, Bhat J, Chatterjee S, Sa G. G-actin guides p53 nuclear transport: Potential contribution of monomeric actin in altered localization of mutant p53. Sci Rep. 2016;6:32626. doi: 10.1038/srep32626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadhukhan S, Sarkar K, Taylor M, Candotti F, Vyas YM. Nuclear role of WASp in gene transcription is uncoupled from its ARP2/3-dependent cytoplasmic role in actin polymerization. J Immunol. 2014;193:150–160. doi: 10.4049/jimmunol.1302923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber A, Pennise CR, Babcock GG, Fowler VM. Tropomodulin caps the pointed ends of actin filaments. J Cell Biol. 1994;127:1627–1635. doi: 10.1083/jcb.127.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moyer JD, Nowak RB, Kim NE, Larkin SK, Peters LL, Hartwig J, Kuypers FA, Fowle VM. Tropomodulin 1-null mice have a mild spherocytic elliptocytosis with appearance of tropomodulin 3 in red blood cells and disruption of the membrane skeleton. Blood. 2010;116:2590–2599. doi: 10.1182/blood-2010-02-268458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nowak RB, Fischer RS, Zoltoski RK, Kuszak JR, Fowler VM. Tropomodulin1 is required for membrane skeleton organization and hexagonal geometry of fiber cells in the mouse lens. J Cell Biol. 2009;186:915–928. doi: 10.1083/jcb.200905065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agnelli L, Mereu E, Pellegrino E, Limongi T, Kwee I, Bergaggio E, Ponzoni M, Zamò A, Iqbal J, Piccaluga PP, et al. Identification of a 3-gene model as a powerful diagnostic tool for the recognition of ALK-negative anaplastic large-cell lymphoma. Blood. 2012;120:1274–1281. doi: 10.1182/blood-2012-01-405555. [DOI] [PubMed] [Google Scholar]
- 28.Heng YW, Koh CG. Actin cytoskeleton dynamics and the cell division cycle. Int J Biochem Cell Biol. 2010;42:1622–1633. doi: 10.1016/j.biocel.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 29.Lee YJ, Keng PC. Studying the effects of actin cytoskeletal destabilization on cell cycle by cofilin overexpression. Mol Biotechnol. 2005;31:1–10. doi: 10.1385/MB:31:1:001. [DOI] [PubMed] [Google Scholar]
- 30.Moulding DA, Blundell MP, Spiller DG, White MR, Cory GO, Calle Y, Kempski H, Sinclair J, Ancliff PJ, Kinnon C, et al. Unregulated actin polymerization by WASp causes defects of mitosis and cytokinesis in X-linked neutropenia. J Exp Med. 2007;204:2213–2224. doi: 10.1084/jem.20062324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee K, Song K. Actin dysfunction activates ERK1/2 and delays entry into mitosis in mammalian cells. Cell Cycle. 2007;6:1487–1495. doi: 10.4161/cc.6.12.4303. [DOI] [PubMed] [Google Scholar]
- 32.Gachet Y, Tournier S, Millar JB, Hyams JS. A MAP kinase-dependent actin checkpoint ensures proper spindle orientation in fission yeast. Nature. 2001;412:352–355. doi: 10.1038/35085604. [DOI] [PubMed] [Google Scholar]
- 33.Sui Z, Nowak RB, Bacconi A, Kim NE, Liu H, Li J, Wickrema A, An XL, Fowler VM. Tropomodulin3-null mice are embryonic lethal with anemia due to impaired erythroid terminal differentiation in the fetal liver. Blood. 2014;123:758–767. doi: 10.1182/blood-2013-03-492710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamashiro S, Gokhin DS, Kimura S, Nowak RB, Fowler VM. Tropomodulins: Pointed-end capping proteins that regulate actin filament architecture in diverse cell types. Cytoskeleton (Hoboken) 2012;69:337–370. doi: 10.1002/cm.21031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedraza N, Cemeli T, Monserrat MV, Garí E, Ferrezuelo F. Regulation of small GTPase activity by G1 cyclins. Small GTPases. 2019;10:47–53. doi: 10.1080/21541248.2016.1268665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vannini I, Wise PM, Challagundla KB, Plousiou M, Raffini M, Bandini E, Fanini F, Paliaga G, Crawford M, Ferracin M, et al. Transcribed ultraconserved region 339 promotes carcinogenesis by modulating tumor suppressor microRNAs. Nat Commun. 2017;8:1801. doi: 10.1038/s41467-017-01562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.