Abstract
Pancreatic ductal adenocarcinoma (PDAC) remains a major cause of cancer-associated mortality, with poor patient outcome. The present study aimed to identify key candidate genes and investigate the potential molecular mechanisms associated with the progression of PDAC. The GSE46234 dataset was downloaded from the Gene Expression Omnibus database, in order to identify the upregulated differentially expressed genes (DEGs) in PDAC. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed to determine the biological functions and pathways of the upregulated DEGs, and a protein-protein interaction (PPI) network was subsequently constructed to screen the hub genes. Subsequently, survival analyses of the hub genes were undertaken in patients with PDAC, using The Cancer Genome Atlas dataset. Reverse transcription-quantitative (RT-q)PCR analysis was performed to assess the mRNA expression levels of the hub genes associated with the prognosis of patients with PDAC. In the present study, 65 upregulated DEGs were identified. GO analysis suggested that the DEGs were enriched in response to hypoxia, calcium ion and negative regulation of catecholamine. KEGG analysis demonstrated that the DEGs were enriched in gastric acid secretion, the ECM-receptor interaction and the cGMP-PKG signaling pathway. Among the 18 hub genes determined by module screening of the PPI network, upregulation of three key genes, abnormal spindle-like microcephaly-associated protein (ASPM), mitotic checkpoint serine/threonine-protein kinase BUB1 β (BUB1B) and protein spindly (SPDL1), was significantly associated with worse overall survival and disease-free survival time in patients with PDAC. Furthermore, ASPM, BUB1B and SPDL1 were demonstrated to be associated with advanced tumor stage, and their upregulation in PDAC tumor tissues was validated using RT-qPCR analysis. Taken together, the results of the present study demonstrate that ASPM, BUB1B and SPDL1 may have the potential to function as prognostic markers and therapeutic targets for PDAC.
Keywords: pancreatic ductal adenocarcinoma, bioinformatics analysis, differentially expressed gene, prognostic markers
Introduction
Pancreatic ductal adenocarcinoma (PDAC) arises from the ductal cells of the exocrine pancreas and comprises the vast majority of all types of pancreatic cancer (1). PDAC is ranked the 7th leading cause of cancer-associated mortality worldwide, predominantly due to poor diagnosis (2) and the incidence of PDAC was 12.3/100,000 in the United States in 2011 (3). It has been reported that only 10–20% of patients are surgically resectable at the time of diagnosis, and PDAC has a global 5-year survival rate of <5% (4). Currently, only a limited number of chemotherapeutic agents have been demonstrated to be effective against PDAC, including gemcitabine and fluorouracil (5). Despite advancements in the treatment of pancreatic cancer, therapies for PDAC are inadequate and the prognosis of PDAC remains unoptimistic due to late detection, drug resistance, and high recurrence and metastatic rates (6,7). Thus, there is a critical need to improve survival rates by identifying novel treatment targets and prognostic factors for patients with PDAC.
With the development of modern biomedicine, growing evidence has demonstrated that multiple gene alterations, such as those in KRAS, cyclin-dependent kinase inhibitor 2A (CDKN2A) and TP53, may act as key determinants of tumor fate within the pancreas (8), and that several cellular signaling pathways are closely associated with the occurrence and progression of PDAC (9,10). Next-generation sequencing (NGS) technology has been widely applied as a notable tool in early cancer diagnosis, cancer grading and progression prediction (11). Furthermore, a substantial amount of biological and clinical data for PDAC cases has been generated with the development of NGS analysis, and these available datasets provide opportunities to systemically investigate and analyze a wide range of expression patterns and molecular signatures for PDAC (12).
The present study aimed to improve the understanding of the molecular basis of PDAC and to identify novel prognostic factors for PDAC. The GSE46234 dataset was downloaded from the Gene Expression Omnibus (GEO) database, in order to determine the upregulated differentially expressed genes (DEGs) in the tumor tissues of patients with PDAC. Subsequently, Gene Ontology (GO) functional enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed. Following identification of the hub genes via protein-protein interaction (PPI) network screening, their relevance with overall survival (OS) and disease-free survival (DFS) time in patients with PDAC was evaluated. Taken together, the results of the present study provide further insight into the potential candidate biomarkers for the progression and prognosis of PDAC at the molecular level.
Materials and methods
Study design and source of data
In the present study, GEO profiles with raw data of the CEL file type based on the GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array), with information on the probe ID, Gene Symbol and Entrez Gene ID were included. The gene expression profiles of the GSE46234 dataset were downloaded from the GEO database (ncbi.nlm.nih.gov/gds/), which comprised data from four healthy and four PDAC tissue samples.
Identification of DEGs
The GEO2R online tool (ncbi.nlm.nih.gov/geo/geo2r) within R software version 3.6.2 (13) was used to determine the DEGs between tumor tissues in patients with PDAC and normal tissues in healthy subjects. An unpaired Student's t-test was performed to identify the DEGs, and the DEGs were considered to be statistically significant according to the cut-off criteria of |logFC|≥2 and adjusted P<0.01 (14). Furthermore, visual hierarchical cluster analysis was applied to identify the upregulated DEGs using Morpheus online analysis software (software.broadinstitute.org/morpheus). A heat map was constructed to validate these results.
GO functional enrichment and KEGG pathway analyses of the DEGs
The Database for Annotation, Visualization and Integrated Discovery version 6.7 (DAVID) (david-d.ncifcrf.gov/) was used to perform GO functional enrichment and KEGG pathway analyses GO analysis is widely used to annotate specific genes and gene products for high-throughput genome and transcriptome data (14). In the present study, GO analysis was performed to predict the potential functions of the DEGs based on biological process (BP), molecular function (MF) and cellular component (CC). For both analyses, P<0.05 was considered to indicate statistical significance.
PPI network construction
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (15) was used to assess and integrate the PPIs, including direct (physical) and indirect (functional) associations among the DEGs. The PPI network for the DEGs was constructed using the Molecular Complex Detection plugin within Cytoscape software version 3.7.1 (16). PPI network modules were screened, and scores >3 and nodes >4 were used as the selection criteria. P<0.05 was considered to indicate a statistical significance.
Survival analysis and expression levels of the hub genes
The present study set out to identify candidate biomarkers for PDAC prognosis. Genes with the highest scores in the PPI network were selected as the hub genes, and the OS and DFS outcomes based on the hub gene expression levels were depicted according to the data downloaded from the Gene Expression Profiling Interactive Analysis database (17). The expression levels of the hub genes in patients at different tumor stages were downloaded from the interactive web resource, UALCAN (ualcan.path.uab.edu/index.html). The hub genes associated with both shorter OS and DFS were identified for further study, and their expression levels were compared in both PDAC tissues and normal adjacent tissues.
Patients and samples
The subsequent experiments were approved and authorized by The Ethics Committee of Taizhou Hospital of Zhejiang Province, and written informed consent was obtained from all patients prior to study commencement. PDAC tissues and normal adjacent tissues were provided by the Biobank of Taizhou Hospital of Zhejiang province (Taizhou, China). Samples obtained from six patients initially diagnosed by two pathologists of pathology department in Taizhou hospital were resected during surgery and immediately frozen in liquid nitrogen for 3 months. Clinical characteristics, including age, sex, tumor location, TNM stage (18) and differentiation are presented in Table I.
Table I.
Clinical characteristics of the six patients with pancreatic ductal adenocarcinoma.
| Sex | Age, years | Location of tumor | TNM stagea | Differentiation |
|---|---|---|---|---|
| Male | 60 | Head | T2N0M0 | Poor |
| Male | 57 | Body and tail | T2N1M0 | Poor |
| Female | 55 | Head | T3N1M0 | Moderate |
| Male | 70 | Head | T3N1M0 | Moderate |
| Male | 46 | Body and tail | T1N1M0 | Moderate |
| Female | 67 | Head | T2N0M0 | Well |
(18). TNM, Tumor-Node-Metastasis.
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from the six PDAC and normal adjacent tissue samples using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The purity and concentration of total RNA were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Inc.). A total of 1 µg total RNA was reverse transcribed into cDNA using M-MuLV Reverse Transcriptase (Toyobo Life Science) at a total volume of 20 µl, according to the manufacturer's protocol. The primer sequences (Genewiz, Inc.) used for qPCR are presented in Table II. The following thermocycling conditions were used for qPCR: Initial denaturation at 95°C for 3 min followed by 40 cycles of denaturation at 95°C for 10 sec and annealing and elongation at 60°C for 30 sec. The relative expression levels of abnormal spindle-like microcephaly-associated protein (ASPM), mitotic checkpoint serine/threonine-protein kinase BUB1 β (BUB1B) and protein spindly (SPDL1) were determined using the 2−ΔΔCq method (19) and normalized to the internal reference gene, GAPDH. The difference in mRNA expression level between the matched tumor and nontumor tissue samples was assessed using a paired Student's t-test within GraphPad Prism (version 7.0; GraphPad Software). P<0.05 was considered to indicate a statistically significant difference.
Table II.
Primer sequences used for reverse transcription-quantitative PCR analysis.
| Gene | Forward sequence (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| ASPM | GGCCCTAGACAACCCTAACGA | AGCTTGGTGTTTCAGAACATC |
| BUB1B | AAATGACCCTCTGGATGTTTGG | GCATAAACGCCCTAATTTAAGCC |
| SPDL1 | CGAGAGCTAGCTGAGCGAAT | CAGCCTCTTTGAGCCTGCAT |
| GAPDH | AAGCCTGCCGGTGACTAAC | GTTAAAAGCAGCCCTGGTGAC |
ASPM, abnormal spindle-like microcephaly-associated protein; BUB1B, mitotic checkpoint serine/threonine-protein kinase BUB1 β; SPDL1, protein spindly.
Results
Differential gene analysis at the mRNA level
Gene expression profiles from the GSE46234 dataset were downloaded from the GEO database; the significantly altered genes were screened to determine the DEGs in PDAC tissues. A total of 217 DEGs were identified via GEO2R analysis, including 65 upregulated genes and 152 downregulated genes (Fig. 1), and the top 10 most upregulated and downregulated DEGs are presented in Table III. The results were further validated using the Morpheus online tool, and the DEGs (including the top 50 most upregulated genes) are presented in a hierarchical clustering heat map (Fig. 2).
Figure 1.
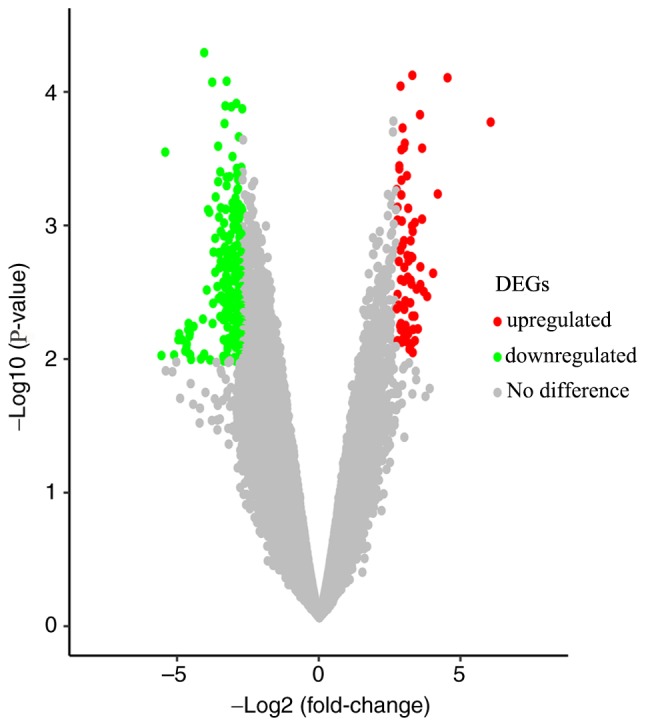
Volcano map of the DEGs in pancreatic ductal adenocarcinoma. Red dots represent upregulated genes, green dots represent downregulated genes and gray dots represent genes that are unchanged. DEGs, differentially expressed genes.
Table III.
Top 10 most upregulated and downregulated differentially expressed genes in the GSE46234 dataset.
| A, Upregulated | ||
|---|---|---|
| Gene symbol | Fold change | P-value |
| IAPP | 6.61 | 2.33×10−4,b |
| FFAR1 | 4.94 | 3.93×10−5,b |
| NUP210L | 4.78 | 6.57×10−4,b |
| CADPS | 4.71 | 6.87×10−4,b |
| LOC101928663 | 4.60 | 6.64×10−5,b |
| SMIM21 | 4.54 | 1.41×10−3,a |
| LINC00844 | 4.47 | 1.10×10−3,a |
| INS | 4.34 | 2.10×10−4,b |
| MAPT | 4.34 | 3.99×10−4,b |
| SLCO1A2 | 4.31 | 7.53×10−4,b |
| B, Downregulated | ||
| Gene symbol | Fold change | P-value |
| S100P | −7.35 | 1.37×10−4,b |
| DUOXA2 | −5.50 | 6.17×10−5,b |
| C19orf33 | −5.08 | 4.99×10−4,b |
| ST6GALNAC1 | −4.89 | 2.06×10−3,a |
| POSTN | −4.41 | 2.19×10−4,b |
| MUC5B | −4.40 | 7.80×10−4,b |
| DPCR1 | −4.33 | 1.84×10−3,a |
| BIK | −4.32 | 1.20×10−4,b |
| TMC7 | −4.09 | 8.32×10−4,b |
| IGK | −4.01 | 9.70×10−4,b |
P<0.01.
P<0.001.
Figure 2.
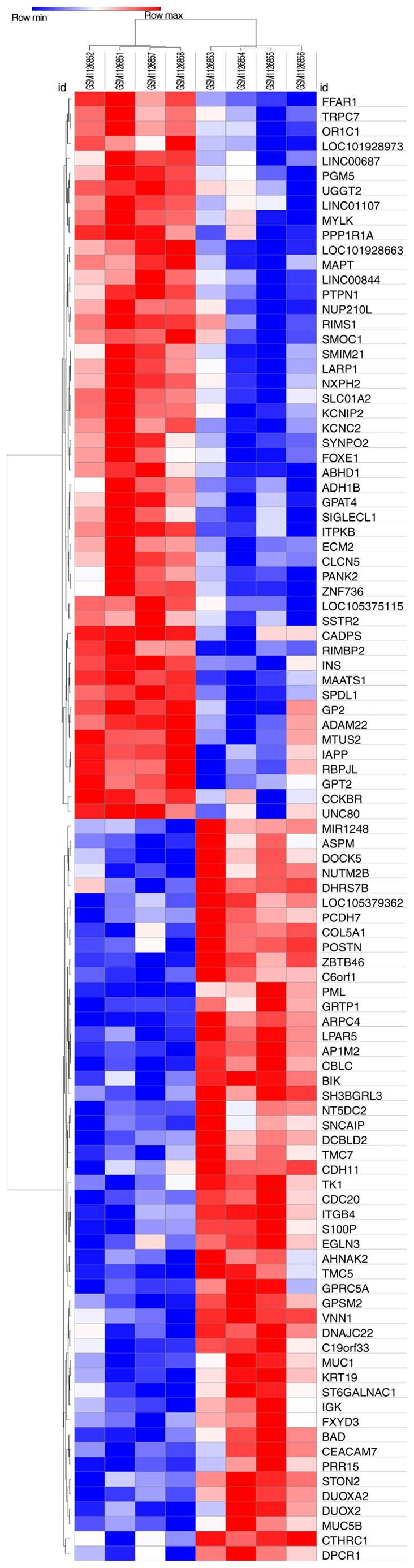
Heat map of the differentially expressed genes from all samples. Red represents upregulated genes and blue represents downregulated genes.
GO functional enrichment and KEGG pathway analyses
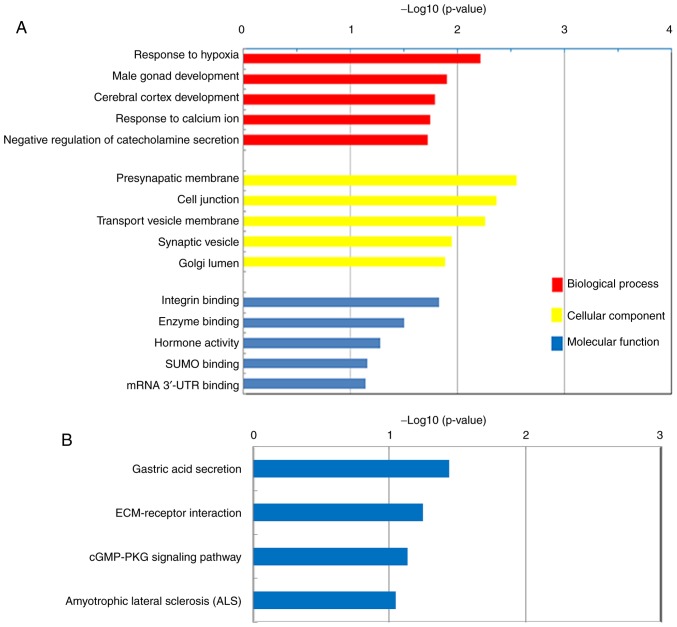
The top five BPs, CCs and MFs of the upregulated DEGs in PDAC, according to GO analysis, are presented in Fig. 3A. The majority of the DEGs were demonstrated to be significantly enriched in CCs, including the ‘Presynaptic membrane’, ‘Cell junction’, ‘Transport vesicle membrane’, ‘Synaptic vesicle’ and ‘Golgi lumen’. These DEGs were also demonstrated to be associated with BPs, including the ‘Response to hypoxia’, ‘Cerebral cortex development’, ‘Response to calcium ion’ and the ‘Negative regulation of catecholamine secretion’. Furthermore, the DEGs were indicated to exert MFs, including the regulation of ‘Integrin binding’, ‘Enzyme binding’ and ‘Hormone activity’. KEGG pathway analysis demonstrated that the upregulated DEGs were enriched in ‘Gastric acid secretion’, ‘ECM-receptor interaction’, ‘cGMP-PKG signaling pathway’ and ‘Amyotrophic lateral sclerosis’ (Fig. 3B).
Figure 3.
Gene Ontology enrichment and KEGG pathway analyses of the DEGs. (A) Top five biological processes, cellular components and molecular functions of the DEGs between patients with PDAC and healthy subjects. (B) KEGG pathway analysis of the DEGs between patients with PDAC and healthy subjects. KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes; PDAC, pancreatic ductal adenocarcinoma; UTR, untranslated region; ECM, extracellular matrix.
Module screening of the PPI network and hub gene identification
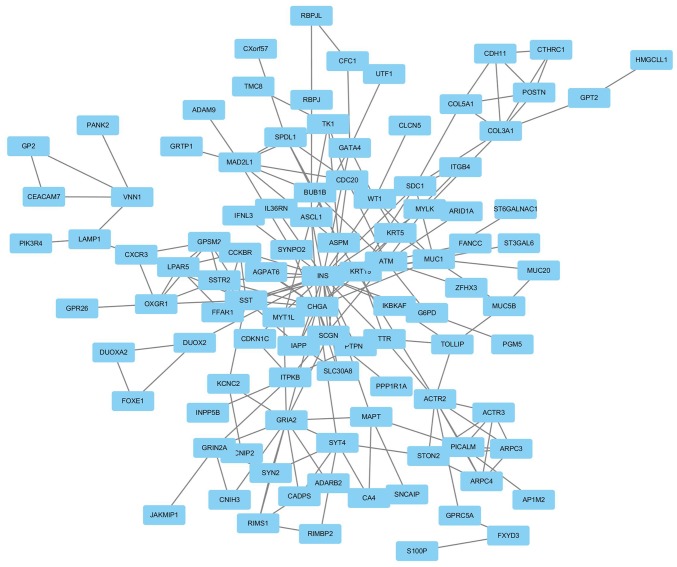
A PPI network was constructed using Cytoscape software based on the DEGs identified from the GSE46234 dataset. Genes with the highest scores were screened and considered hub genes, which were more likely to be associated with PDAC. The top 18 hub genes included ACTR2, ACTR3, ARPC3, ARPC4, ASPM, BUB1B, CDC20, CXCR3, GPSM2, LPAR5, MAD2L1, OXGR1, PICALM, SPDL1, SST, SSTR2, STON2 and TK1 (Fig. 4).
Figure 4.
Protein-protein interaction network of the differentially expressed genes in pancreatic ductal adenocarcinoma.
Upregulation of ASPM, BUB1B and SPDL1 predicts poor survival outcome in patients with PDAC
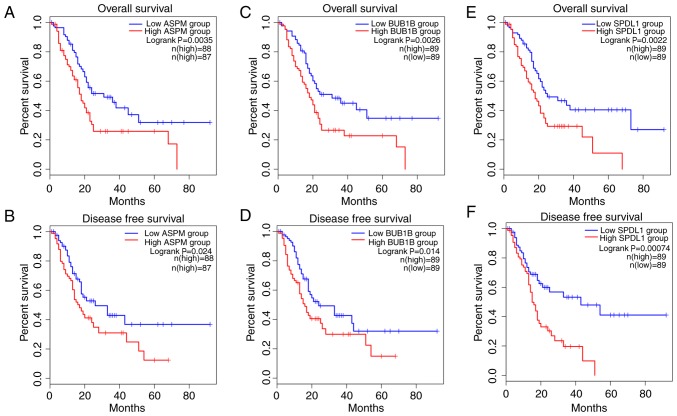
In order to identify candidate prognostic biomarkers, the top 18 hub genes identified in the PPI network were analyzed, and only the genes which were associated with both shorter OS and DFS were regarded as potential biomarkers for PDAC prognosis. Among the 18 hub genes, ASPM, BUB1B, CDC20, MAD2L1, SPDL1 and TK1 demonstrated association with prognosis. The results demonstrated that patients with PDAC with upregulated expression of ASPM, BUB1B, MAD2L1 and SPDL1 in tumors had significantly worse OS, while the upregulation of ASPM, BUB1B, CDC20 and SPDL1 was associated with significantly decreased DFS. Furthermore, upregulation of only ASPM, BUB1B and SPDL1 in tumors was significantly associated with both poor OS and DFS in patients with PDAC (all P<0.05; Fig. 5).
Figure 5.
Prognostic analysis of overall survival and disease-free survival for (A and B) ASPM, (C and D) BUB1B and (E and F) SPDL1.
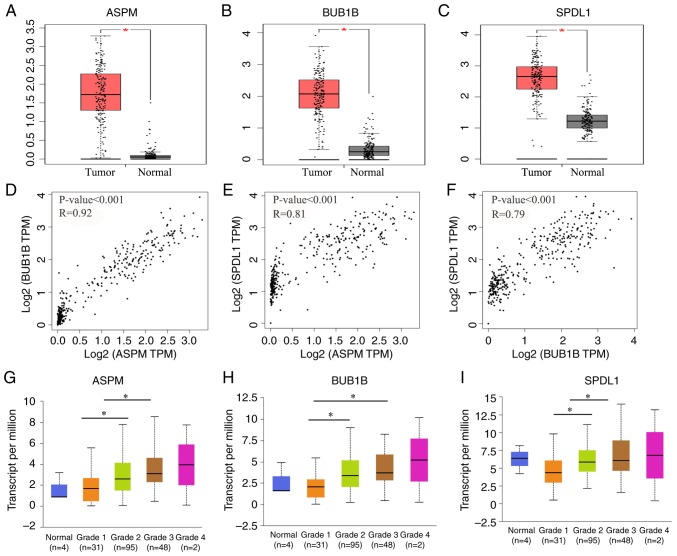
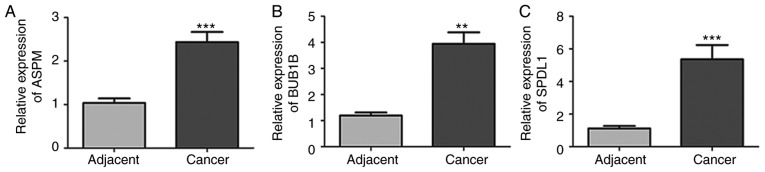
ASPM, BUB1B and SPDL1 were selected for further analysis. The expression of these three genes was demonstrated to be significantly higher in tumor tissues compared with normal adjacent tissues (all P<0.05; Fig. 6A-C). Furthermore, these three genes were also significantly upregulated in patients with grade G2 and G3 neoplasms compared with patients with G1 neoplasms (all P<0.05; Fig. 6G-I. Consistent with the results of the database analysis, mRNA expression of ASPM, BUB1B and SPDL1 was significantly higher in PDAC tissues compared with normal adjacent tissues (all P<0.01; Fig. 7).
Figure 6.
Increased expression levels of ASPM, BUB1B and SPDL1 in PDAC. Red represents tumor tissue and black represents normal tissue. (A) ASPM, (B) BUB1B and (C) SPDL1 were upregulated in the tumors of patients with PDAC and the associations between (D) ASPM, (E) BUB1B and (F) SPDL1 were analyzed via Pearson's correlation analysis. (G) ASPM, (H) BUB1B and (I) SPDL1 were also upregulated in the tumor tissues of patients with advanced PDAC. *P<0.05. ASPM, abnormal spindle-like microcephaly-associated protein; BUB1B, mitotic checkpoint serine/threonine-protein kinase BUB1 β; SPDL1, protein spindly; PDAC, pancreatic ductal adenocarcinoma; TPM, Transcripts Per Kilobase of exon model per Million mapped reads.
Figure 7.
Relative mRNA expression levels of ASPM, BUB1B and SPDL1 determined via reverse transcription-quantitative PCR analysis in the tumor and paired non-tumor tissue samples of six patients with PDAC. (A) ASPM, (B) BUB1B and (C) SPDL1 mRNA expression was increased in the tumor tissues of patients with PDAC compared with the matched non-tumor tissues. **P<0.01, ***P<0.001 vs. adjacent. PDAC, pancreatic ductal adenocarcinoma; ASPM, abnormal spindle-like microcephaly-associated protein; BUB1B, mitotic checkpoint serine/threonine-protein kinase BUB1 β; SPDL1, protein spindly.
Discussion
Compared with all other malignancies, PDAC has the poorest prognosis, with a 5% 5-year survival rate and a mean life expectancy of <6 months, which is predominantly due to resistance to standard therapy (20,21). Commonly used tumor markers, such as CA19-9 and CEA, are ineffective for the early detection of PDAC (22). Furthermore, they fail to provide information on the exact location of the cancer lesions, thus making it difficult to determine a therapeutic strategy (23,24). Therefore, identifying prognostic factors for patients with PDAC remains critical, with the aim to develop novel therapeutic approaches and select adequate treatment strategies.
In the present study, 217 DEGs were screened, including 65 upregulated and 152 downregulated genes, in the PDAC tissue samples of the GSE46234 dataset. GO analysis indicated that the 65 upregulated genes were significantly enriched in functions involving the regulation of ‘Integrin binding’, ‘Enzyme binding’ and ‘Hormone activity’. Previous studies have demonstrated that the interactions between ligands and integrins, such as αvβ6, could affect the proliferation, invasion, metastasis and angiogenesis of PDAC cells (5,25,26). Furthermore, hormone inhibitors, such as angiotensin system inhibitors were applied to improve the prognosis of PDAC (27). KEGG pathway analysis demonstrated that the upregulated genes were predominantly involved in ‘ECM-receptor interaction’ and the ‘cGMP-PKG signaling pathway’, which has been identified as a key signaling pathway in different types of cancer, including bladder cancer (28). A total of 18 hub genes were discovered following construction of the PPI network, in which three key genes (ASPM, BUB1B and SPDL1) were demonstrated to be enriched in the cell cycle biological processes/pathways, and were significantly associated with both shorter OS and DFS in patients with PDAC. The results of the present study were unable to be compared with the findings of the original study as it is unavailable. Thus, TCGA cohort analysis was performed; the results demonstrated high ASPM, BUB1B and SPDL1 expression levels in patients with PDAC, which was further validated by RT-qPCR analysis. Furthermore, upregulation of ASPM, BUB1B and SPDL1 was associated with the histological grade of the neoplasm. This suggests that high ASPM, BUB1B and SPDL1 expression levels may contribute to PDAC progression and a worse prognosis, indicating the prognostic value of ASPM, BUB1B and SPDL1 for patients with PDAC.
ASPM was originally identified as a centrosomal protein that regulates neurogenesis and brain size (29), and was reported to be extensively expressed in a variety of malignant tissues, such as gliomas, ovarian and hepatocellular cancer tissues (30–32). Bikeye et al (33) reported that ASPM is upregulated in recurrent tumors and demonstrates a positive association with the pathological grading of gliomas. Upregulation of ASPM has also been reported to be associated with the pathological grading and poor survival of patients with ovarian cancer and hepatocellular carcinoma (31,34). In the present study, ASPM expression was demonstrated to be higher in tumor tissues compared with normal adjacent tissues, and was associated with poor survival of patients with PDAC. These results validate the findings of previous studies, which suggest that ASPM may enhance PDAC tumor progression by promoting Wnt activity to regulate cancer stemness (32–35).
The results of the present study suggest that high BUB1B expression may play a key role in the tumorigenesis and progression of PDAC; this is consistent with the findings of previous bioinformatics analyses, which suggest that BUB1B demonstrates high connectivity degrees to PDAC, and may be useful as a therapeutic target (32,36,37). BUB1B upregulation is also reportedly associated with chromosomal instability in other malignancies, including bladder cancer, breast cancer and kidney carcinomas (38–40). Furthermore, BUB1B upregulation is also associated with the progression and recurrence of gastric cancer (37). SPDL1 is required for efficient chromosome congression and mitotic checkpoint regulation, and is also involved at the spindle checkpoint during mitosis (41). High SPDL1 expression is an independent prognostic indicator for cancer-specific survival and has been associated with increased cellular proliferation in oral cancer, highlighting its potential value in therapy (42). Hence, these data support the hypothesis that ASPM, BUB1B and SPDL1 may be candidate molecular markers for PDAC progression and prognosis, and thus may be useful as therapeutic targets.
In the present study, the DEGs between PDAC and normal tissue samples in the GSE46234 dataset were determined, and the hub genes among the DEGs were demonstrated to be associated with the prognosis of patients with PDAC patients. Furthermore, ASPM, BUB1B and SPDL1 were identified as potential candidate biomarkers for OS and DFS in patients with PDAC. High ASPM, BUB1B and SPDL1 mRNA expression levels were validated by TCGA cohort analysis and subsequent RT-qPCR analysis, which may preliminarily uncover the pathophysiological role of these hub genes in PDAC at the molecular level. However, the sample size for microarray analysis in the present study was small, thus further molecular and biological studies with larger sample sizes are required to validate these findings, and to confirm the functions of the key genes in PDAC.
Taken together, the results of the present study demonstrate that the upregulation of ASPM, BUB1B and SPDL1 in tumor tissues is closely associated with tumor development and poor OS and DFS in patients PDAC. This indicates their potential prognostic values and use as therapeutic targets.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Natural Science Foundation of Zhejiang Province, China (grant no. LQ19H200001).
Availability of data and materials
The datasets analyzed in the present study are available in the GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE46234) and TCGA (https://cancergenome.nih.gov) databases.
Authors' contributions
XT and NW participated in the study design, data collection and analysis. XT contributed to the collection of samples, RT-qPCR analysis and the writing of the article. NW critically reviewed the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by The Ethics Committee of Taizhou Hospital of Zhejiang Province (Taizhou, China) and written informed consent was obtained from all patients prior to study commencement.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Howlader N, Noone AM, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA, editors. National Cancer Institute; Bethesda, MD: 2019. SEER Cancer Statistics Review, 1975–2011. [Google Scholar]
- 4.Pozios I, Knösel T, Zhao Y, Assmann G, Pozios I, Müller MH, Bruns CJ, Kreis ME, Seeliger H. Expression of phosphorylated estrogen receptor beta is an independent negative prognostic factor for pancreatic ductal adenocarcinoma. J Cancer Res Clin Oncol. 2018;144:1887–1897. doi: 10.1007/s00432-018-2717-2. [DOI] [PubMed] [Google Scholar]
- 5.Kasuga A, Hamamoto Y, Takeuchi A, Kawasaki K, Suzuki T, Hirata K, Sukawa Y, Takaishi H, Kanai T. Positive relationship between subsequent chemotherapy and overall survival in pancreatic cancer: Meta-analysis of postprogression survival for first-line chemotherapy. Cancer Chemother Pharmacol. 2017;79:595–602. doi: 10.1007/s00280-017-3263-3. [DOI] [PubMed] [Google Scholar]
- 6.Karakas Y, Lacin S, Yalcin S. Recent advances in the management of pancreatic adenocarcinoma. Expert Rev Anticancer Ther. 2018;18:51–62. doi: 10.1080/14737140.2018.1403319. [DOI] [PubMed] [Google Scholar]
- 7.Ruckert MT, de Andrade PV, Santos VS, Silveira VS. Protein tyrosine phosphatases: Promising targets in pancreatic ductal adenocarcinoma. Cell Mol Life Sci. 2019;76:2571–2592. doi: 10.1007/s00018-019-03095-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heiser PW, Cano DA, Landsman L, Kim GE, Kench JG, Klimstra DS, Taketo MM, Biankin AV, Hebrok M. Stabilization of beta-catenin induces pancreas tumor formation. Gastroenterology. 2008;135:1288–1300. doi: 10.1053/j.gastro.2008.06.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Wu X, Wang G, Hu S, Zhang Y, Zhao S. CALD1, CNN1, and TAGLN identified as potential prognostic molecular markers of bladder cancer by bioinformatics analysis. Medicine (Baltimore) 2019;98:e13847. doi: 10.1097/MD.0000000000013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tell RW, Horvath CM. Bioinformatic analysis reveals a pattern of STAT3- associated gene expression specific to basal-like breast cancers in human tumors. Proc Natl Acad Sci USA. 2014;111:12787–12792. doi: 10.1073/pnas.1404881111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gentleman R, Ihaka R. R: A language and environment for statistical computing. Computing. 2011;1:12–21. [Google Scholar]
- 14.Saura M, Marquez S, Reventun P, Olea-Herrero N, Arenas MI, Moreno-Gómez-Toledano R, Gómez-Parrizas M, Muñóz-Moreno C, González-Santander M, Zaragoza C, Bosch RJ. Oral administration of bisphenol A induces high blood pressure through angiotensin II/CaMKII-dependent uncoupling of eNOS. FASEB J. 2014;28:4719–4728. doi: 10.1096/fj.14-252460. [DOI] [PubMed] [Google Scholar]
- 15.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen PJ, Kuk D, Castillo CF, Basturk O, Wolfgang CL, Cameron JL, Lillemoe KD, Ferrone CR, Morales-Oyarvide V, He J, et al. Multi-institutional validation study of the American Joint Commission on Cancer (8th Edition) changes for T and N staging in patients with pancreatic adenocarcinoma. Ann Surg. 2017;265:185–191. doi: 10.1097/SLA.0000000000001763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Tanemura M, Miyoshi E, Nagano H, Eguchi H, Matsunami K, Taniyama K, Hatanaka N, Akamatsu H, Mori M, Doki Y. Cancer immunotherapy for pancreatic cancer utilizing α-gal epitope/natural anti-Gal antibody reaction. World J Gastroenterol. 2015;21:11396–11410. doi: 10.3748/wjg.v21.i40.11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li N, Zhao X, You S. Identification of key regulators of pancreatic ductal adenocarcinoma using bioinformatics analysis of microarray data. Medicine (Baltimore) 2019;98:e14074. doi: 10.1097/MD.0000000000014074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakamoto T, Saito H, Amisaki M, Tokuyasu N, Honjo S, Fujiwara Y. Combined preoperative platelet-to-lymphocyte ratio and serum carbohydrate antigen 19-9 level as a prognostic factor in patients with resected pancreatic cancer. Hepatobiliary Pancreat Dis Int. 2019;18:278–284. doi: 10.1016/j.hbpd.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Piao J, Zhu L, Sun J, Li N, Dong B, Yang Y, Chen L. High expression of CDK1 and BUB1 predicts poor prognosis of pancreatic ductal adenocarcinoma. Gene. 2019;701:15–22. doi: 10.1016/j.gene.2019.02.081. [DOI] [PubMed] [Google Scholar]
- 24.Yin X, Wang M, Wang H, Deng H, He T, Tan Y, Zhu Z, Wu Z, Hu S, Li Z. Evaluation of neurotensin receptor 1 as a potential imaging target in pancreatic ductal adenocarcinoma. Amino Acids. 2017;49:1325–1335. doi: 10.1007/s00726-017-2430-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueda M, Fukushima T, Ogawa K, Kimura H, Ono M, Yamaguchi T, Ikehara Y, Saji H. Synthesis and evaluation of a radioiodinated peptide probe targeting αvβ6 integrin for the detection of pancreatic ductal adenocarcinoma. Biochem Biophys Res Commun. 2014;445:661–666. doi: 10.1016/j.bbrc.2014.02.086. [DOI] [PubMed] [Google Scholar]
- 26.Principe M, Borgoni S, Cascione M, Chattaragada MS, Ferri-Borgogno S, Capello M, Bulfamante S, Chapelle J, Di Modugno F, Defilippi P, et al. Alpha-enolase (ENO1) controls alpha v/beta 3 integrin expression and regulates pancreatic cancer adhesion, invasion, and metastasis. J Hematol Oncol. 2017;10:16. doi: 10.1186/s13045-016-0385-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, Naxerova K, Pinter M, Incio J, Lee H, Shigeta K, Ho WW, Crain JA, Jacobson A, Michelakos T, et al. Use of angiotensin system inhibitors is associated with immune activation and longer survival in nonmetastatic pancreatic ductal adenocarcinoma. Clinical Cancer Res. 2017;23:5959–5969. doi: 10.1158/1078-0432.CCR-17-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang F, He Z, Lei H, Chen Y, Lu Z, Zeng G, Wang H. Identification of differentially expressed genes and biological pathways in bladder cancer. Mol Med Rep. 2018;17:6425–6434. doi: 10.3892/mmr.2018.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kouprina N, Pavlicek A, Collins NK, Nakano M, Noskov VN, Ohzeki J, Mochida GH, Risinger JI, Goldsmith P, Gunsior M, et al. The microcephaly ASPM gene is expressed in proliferating tissues and encodes for a mitotic spindle protein. Hum Mol Genet. 2005;14:2155–2165. doi: 10.1093/hmg/ddi220. [DOI] [PubMed] [Google Scholar]
- 30.Raman P, Maddipati R, Lim KH, Tozeren A. Pancreatic cancer survival analysis defines a signature that predicts outcome. PLoS One. 2018;13:e0201751. doi: 10.1371/journal.pone.0201751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brüning-Richardson A, Bond J, Alsiary R, Richardson J, Cairns DA, McCormack L, Hutson R, Burns P, Wilkinson N, Hall GD, et al. ASPM and microcephalin expression in epithelial ovarian cancer correlates with tumor grade and survival. Br J Cancer. 2011;104:1602–1610. doi: 10.1038/bjc.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang WY, Hsu CC, Wang TY, Li CR, Hou YC, Chu JM, Lee CT, Liu MS, Su JJ, Jian KY, et al. A gene expression signature of epithelial tubulogenesis and a role for ASPM in pancreatic tumor progression. Gastroenterology. 2013;145:1110–1120. doi: 10.1053/j.gastro.2013.07.040. [DOI] [PubMed] [Google Scholar]
- 33.Bikeye SN, Colin C, Marie Y, Vampouile R, Ravassard P, Rousseau A, Boisselier B, Idbaih A, Calvo CF, Leuraud P, et al. ASPM-associated stem cell proliferation is involved in malignant progression of gliomas and constitutes an attractive therapeutic target. Cancer Cell Int. 2010;10:1. doi: 10.1186/1475-2867-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin SY, Pan HW, Liu SH, Jeng YM, Hu FC, Peng SY, Lai PL, Hsu HC. ASPM is a novel marker for vascular invasion, early recurrence, and poor prognosis of hepatocellular carcinoma. Clin Cancer Res. 2008;14:4814–4820. doi: 10.1158/1078-0432.CCR-07-5262. [DOI] [PubMed] [Google Scholar]
- 35.Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S, Ciciliano JC, Smas ME, Winokur D, Gilman AJ, et al. RNA sequencing of pancreatic circulating tumor cells implicates WNT signaling in metastasis. Nature. 2012;487:510–513. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long J, Zhang Z, Liu Z, Xu Y, Ge C. Identification of genes and pathways associated with pancreatic ductal adenocarcinoma by bioinformatics analyses. Oncol Lett. 2016;11:1391–1397. doi: 10.3892/ol.2015.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong S, Huang F, Zhang H, Chen Q. Overexpression of BUB1B, CCNA2, CDC20, and CDK1 in tumor tissues predicts poor survival in pancreatic ductal adenocarcinoma. Biosci Rep. 2019;39 doi: 10.1042/BSR20182306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinto M, Vieira J, Ribeiro FR, Soares MJ, Henrique R, Oliveira J, Jerónimo C, Teixeira MR. Overexpression of the mitotic checkpoint genes BUB1 and BUBR1 is associated with genomic complexity in clear cell kidney carcinomas. Cell Oncol. 2008;30:389–395. doi: 10.3233/CLO-2008-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scintu M, Vitale R, Prencipe M, Gallo AP, Bonghi L, Valori VM, Maiello E, Rinaldi M, Signori E, Rabitti C, et al. Genomic instability and increased expression of BUB1B and MAD2L1 genes in ductal breast carcinoma. Cancer Lett. 2007;254:298–307. doi: 10.1016/j.canlet.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto Y, Matsuyama H, Chochi Y, Okuda M, Kawauchi S, Inoue R, Furuya T, Oga A, Naito K, Sasaki K. Overexpression of BUBR1 is associated with chromosomal instability in bladder cancer. Cancer Genet Cytogenet. 2007;174:42–47. doi: 10.1016/j.cancergencyto.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 41.Barisic M, Sohm B, Mikolcevic P, Wandke C, Rauch V, Ringer T, Hess M, Bonn G, Geley S. Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation. Mol Biol Cell. 2010;21:1968–1981. doi: 10.1091/mbc.e09-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva PMA, Delgado ML, Ribeiro N, Florindo C, Tavares ÁA, Ribeiro D, Lopes C, do Amaral B, Bousbaa H, Monteiro LS. Spindly and Bub3 expression in oral cancer: Prognostic and therapeutic implications. Oral Dis. 2019;25:1291–1301. doi: 10.1111/odi.13089. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed in the present study are available in the GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE46234) and TCGA (https://cancergenome.nih.gov) databases.