Abstract
Sexual differentiation of the brain is a crucial developmental process that enables the lifelong expression of sexually dimorphic behaviors, including those necessary for successful reproduction. During a perinatal sensitive period, gonadal hormones defeminize and masculinize the male brain, and a lack of gonadal steroids allows for feminization in the female. This hormonally-induced differentiation permanently alters neural structures, creating highly dimorphic brain regions; however, the mechanism by which hormones exert their long-lasting effects are still largely unknown. Epigenetic processes such as DNA methylation and histone modifications serve as an interface for environmental stimuli to exert control over the genome. These modifications have the capacity to activate or repress gene expression, thereby shaping the developmental outcomes of cells, circuits, and structures in the brain. Sex differences in methylation, methyl-binding proteins, and chromatin modifications indicate that epigenetic mechanism may be important for sexual differentiation of the brain. The data outlined in this review provide evidence that gonadal hormones in the neonatal brain influence epigenetic processes such as DNA methylation and histone acetylation, but also emphasize the primitive status of our current understanding of epigenetics and sexual differentiation and the brain.
Key Words: Sexual differentiation, Sexual dimorphism, Epigenetics, DNA methylation, Histone acetylation
Sexual Differentiation of the Brain
Sexual differentiation of the brain is a hormonally-driven process involving steroids synthesized by the recently differentiated gonads in the developing embryo. Sexual differentiation is distinguishable by sex differences, which may occur in the adult and be the product of sex-specific gonadal steroids, or the result of genes on the X or Y chromosome [1,2]. Adult sex differences established by hormones should disappear following gonadectomey, whereas those endpoints that are truly sexually differentiated will endure. Whether genetics contributes only to sex differences or also impacts sexual differentiation remains to be determined. Hormonally-mediated sexual differentiation consists of two active processes: defeminization and masculinization. Defeminization involves the permanent elimination of the female brain phenotype controlling sexual behavior and therefore a loss of the ability to express female sexual behavior. Masculinization is the active development of the male-typical neural anatomy and the capacity to express male sexual behavior in adulthood. Together, these two processes differentiate the male brain from the female, the development of which is defined as feminization [3].
In the rodent there is a carefully defined critical period for brain sexual differentiation that occurs between embryonic day 18 (E18) and approximately postnatal day 10 (PN10) [4,5]. The onset of this critical period coincides with the beginning of testosterone production by the testes in genetic males, while the offset is defined as the point after which females are no longer masculinized by exogenous steroid treatment. In male fetuses high levels of testosterone synthesized by the testis reach the brain where a portion is converted to estradiol by the enzyme p450 aromatase [6,7]. Estradiol acts via its nuclear receptors, estrogen receptors α (ERα) and β (ERβ) to initiate defeminization and masculinization [8,9]. When bound by ligand, both ER isoforms can act as transcription factors, thereby influencing gene expression [10]. Administering testosterone or estradiol to a genetic female during the critical period for sexual differentiation of the brain leads to a defeminized and masculinized neural phenotype, rendering this female anovulatory and therefore sterile [11], and capable of male-typical behavior in adulthood if treated with testosterone [12]. Conversely, blocking aromatization of testosterone to estradiol or antagonizing ER binding during the critical period impedes organization of the male brain [13,14]. Importantly, these manipulations have no impact on the determination of brain sex if made before or after the critical period, indicating a highly regulated and tightly orchestrated timeframe for these crucial organizational events. If one considers the importance of matching brain and body sex for successful reproduction and species propagation, it seems logical to have a highly controlled critical period for sexual differentiation of the brain. However, the question of how such fleeting hormone exposure can have enduring, lifelong consequences on the nervous system, and ultimately reproductive fitness, remains to be determined.
Epigenetic Processes
How does brief hormone exposure during early development permanently organize sex differences in the brain? Emerging interest in the field of epigenetics has provided potential answers to this question. The term epigenetics, literally meaning ‘above genetics’, refers to modifications made to the genome that can impact gene expression without affecting the underlying DNA sequence. Epigenetic processes are important for coordinating the impact of environmental factors during developmental sensitive periods [reviewed in [15]]. Steroid hormones are a component of the internal environment that vary between males and females, and steroid receptors associate directly with DNA and enzymes that mediate some forms of epigenetic changes, making them ideal candidates to exert a lasting effect on the epigenome, or the overall epigenetic state of a cell.
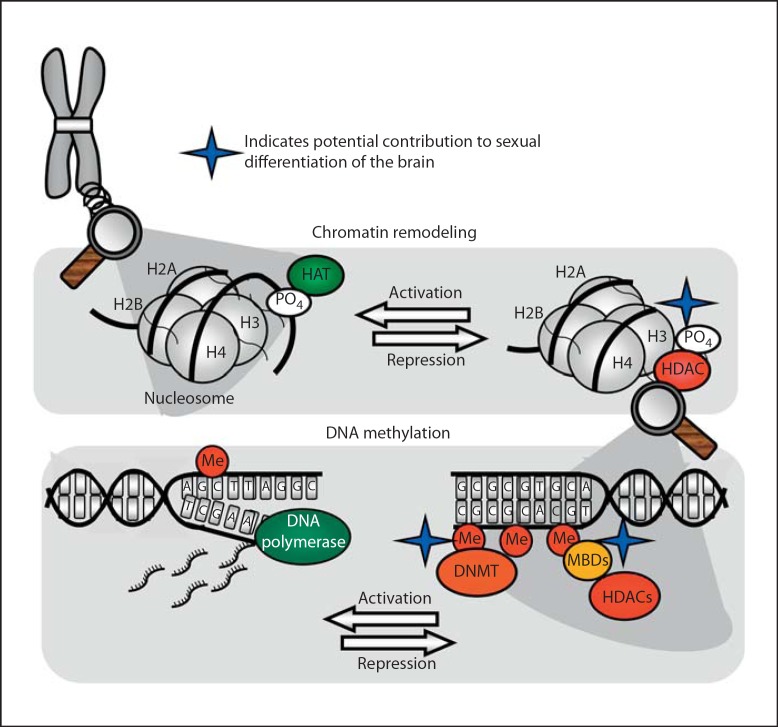
The genome is contained within chromosomes, which are comprised of a highly organized mix of DNA and histone proteins called chromatin. Chromatin's specialized structure ensures that lengthy stretches of DNA are efficiently packaged within the confines of a cell's nucleus in addition to aiding mitosis, meiosis, and regulating gene expression [16]. Two copies of each core histone protein (H2A, H2B, H3, H4) form a histone octamer around which 147 DNA base pairs wrap, forming a nucleosome, the basic building block of chromatin structure [17]. Chromatin in its loosened state, referred to as euchromatin, is often associated with active gene transcription. Genes are typically thought to be in a silent state when chromatin is in its tightly condensed heterochromatin state. The N-terminal tail of each histone protein protrudes from the nucleosome allowing for posttranslational modifications to alter chromatin structure and ultimately gene expression (fig. 1). Modifications made to histones affect gene transcription in two ways: (1) by affecting electrostatic links between DNA and histone proteins causing chromatin loosening and thereby enhancing access of transcriptional machinery to the DNA, and (2) by changing histone-histone interactions [18,19], which impacts chromatin reassembly following transcription [20]. Such modifications include histone acetylation, methylation, phosphorylation, ubiquitination, and sumoylation.
Fig. 1.
Epigenetic control of gene expression. Chromosomes have complex higher order structures, the basic unit of which is called the nucleosome. Nucleosomes consist of segments of DNA wrapped around core histone proteins with protruding N-terminal tails. Histone acetyltransferase enzymes (HATs) acetylate histone protein tails, opening chromatin structure and activating transcription. Removal of acetyl groups by histone deacetylase enzymes (HDACs) results in chromatin condensing and transcriptional repression. Interference with HDAC activity can interfere with masculinization of the neonatal brain. Chromatin in a repressed state is usually highly methylated by DNA methyltransferase enzymes (DNMTs). Methylation at CpG dinucleotides attracts methyl-binding proteins (MBDs), which recruit chromatin remodeling proteins such as HDACs. Sex differences in DNA methylation and MBD expression in sexually dimorphic brain regions during the critical period suggest an involvement of DNA methylation in brain organization. Promoters with low levels of DNA methylation are typically associated with active transcription, but many empirical findings challenge this dogma.
The impact of posttranslational modifications on gene expression depends on which modification is made, as well as which histone tail and specific amino acid residue is modified [21]. Because of its location within the nucleosome structure, histone H3 is the most highly modified histone protein and has received the most focus for its role in epigenetic control of gene expression [22]. Histone phosphorylation and acetylation release DNA from histones enhancing gene transcription [23,24]. In the case of acetylation, histone acetyltransferase (HAT) enzymes covalently attach a negatively charged acetyl group to a histone tail, causing repulsion of the negatively charged phosphate backbone of DNA. In this state, acetylation allows for DNA to separate from the nucleosome and to be accessed by transcriptional machinery. Removal of the acetyl group by a histone deacetylase (HDAC) enzyme restores the nucleosome's tightly wound structure [25] (fig. 1). Other modifications, such as histone methylation and ubiquitination, which do not impact electromagnetic charges within the nucleosome, have various outcomes based on their titer and placement. For instance, methylation of lysine 9 on histone H3 is associated with active gene transcription, whereas di- or trimethylation of the same residue causes chromatin condensing [26]. Reversible, protein mono-ubiquitination functions as a signaling mechanism at a given substrate [27], and can be coupled to other histone modifications such as methylation [28]. Although less is known about the function of histone ubiquitination, it has been shown that ubiquitination of histone H2A results in gene repression, whereas ubiquitination of H2B can cause transcriptional activation or silencing [29]. Sumoylation is a negative regulator of transcriptional activity, which has been shown to block chromatin remodeling and gene expression induced by acetylation and ubiquitination [30]. Histone alterations induced by small ubiquitin-related modifier (SUMO) are associated with heterochromatin, and SUMO has been shown to interact with HDACs and other members of repressor complexes [31].
In addition to histone protein modifications, direct DNA methylation often has a profound epigenetic impact. DNA methyltransferase (DNMT) enzymes covalently affix a methyl group to the cytosine residue within a CpG dinucleotide (referred to as a CpG site). CpG sites are commonly methylated throughout the genome, however CpG sites are typically unmethylated within so-called CpG islands, which are regions containing large quantities of CpGs in the 5′ regulatory region, or promoter, of a gene [32,33,34]. Low baseline methylation levels in regulatory regions enable small changes in methylation at specific CpG sites within a gene's promoter to have a significant effect on gene expression.
The prevailing dogma is that methylation of cytosine residues is permanent, irreversible, and consistently results in transcriptional repression. This view has proved true for some biological systems, although recently large differences in DNA methylation were found in embryonic and fetal cells during cellular differentiation, revealing that cytosine methylation is a highly dynamic process [35]. Within the central nervous system, DNA methylation can be transient and is not necessarily associated with decreased gene expression. DNA methylation impacts gene expression and chromatin structure in several ways. Most notably, methylated cytosines can be used as docking sites for methyl-binding proteins which then recruit chromatin remodeling complexes, typically containing an HDAC enzyme, resulting in tightening of the chromatin structure. Proteins containing methyl-binding domains (MBD) such as MeCP2 and MBD1, 2, 3, and 4 all bind to methylated CpG sites but exert distinct outcomes [36]. MeCP2 is critical for neurological development and is typically associated with gene silencing through its recruitment of HDACs [37,38]. However, recent evidence indicates MeCP2 actually enhances expression of 85% of its effector genes [39]. In fact, the majority of MeCP2-binding sites are outside of genes, associated with active transcription, and only a small percentage of these sites are methylated cytosines [39], suggesting MeCP2 plays opposing roles in regulating gene transcription in addition to having other functions. Other methyl-binding proteins such as MBD2 and MBD4 are primarily associated with transcriptional silencing, yet both have been linked to hypomethylation [40], and may be involved in demethylation [41] and DNA repair [42].
In addition to recruiting methyl-binding proteins, DNA methylation can directly block transcriptional machinery from accessing binding sites within a gene's promoter [43]. In genes with multiple promoters, DNA methylation can dictate promoter utilization [44]. As our knowledge of epigenetic processes expands and the dogma regarding DNA methylation unravels, we will likely learn that DNA methylation serves other as yet unknown functions.
Epigenetic Consequences of Early Life Experiences
Much of our knowledge of epigenetic processes stems from research on cell fate determination during embryogenesis and tumorigenesis. More recently, epigenetic modifications have been observed in the brain in response to various exogenous stimuli such as maternal behavior, drugs, and learning [45,46,47]. The work that fueled an interest in epigenetics among neuroendocrinologists comes from Michael Meaney and colleagues who discovered that a seminal link between maternal care and adult hypothalamic-pituitary-adrenal-axis (HPAA) function involves epigenetic alterations of the glucocorticoid receptor [48]. Meany's model revealed that rat pups reared by less attentive, low licking/grooming (low LG) dams have lower levels of acetylation at lysine 9 of histone H3 (a site synonymous with active chromatin [49]), increased methylation of a critical region of the glucocorticoid receptor promoter, and decreased transcription factor binding. Importantly, changes in epigenetic control of the glucocorticoid receptor are established neonatally but persist into adulthood, wherein the animals exhibit impaired HPAA function. Moreover, reduced maternal care increases methylation of the ERα-1b promoter resulting in decreased ERα expression in the female preoptic area (POA) [49,50]. Conversely, reduced maternal care increased ERα expression in other regions, such as the anteroventral periventricular nucleus of the hypothalamus (AVPV) and ventromedial nucleus of the hypothalamus (VMN), two areas critical for the control of gonadatropin release, estrous cyclicity, and female sexual behavior [51].
Sex-Specific DNA Methylation during Development and Beyond
DNA methylation directs gonadal sex determination by controlling the timing and spatial expression of the sry gene in genetic males [52] and may influence offspring sex ratio [53]. Similarly, DNA methylation is emerging as a critical component of steroid hormone-mediated organization of the brain. During sexual differentiation of the brain, estradiol derived from testosterone causes permanent structural and behavioral masculinization and defeminization via activation of ERα and ERβ, respectively [54,55]. Because these receptors are critical in organizing sex differences in the brain, their promoters have been thoroughly examined as potential targets of epigenetic control via DNA methylation.
In the rat, five alternative promoters control ERα gene expression. A system of untranslated exons and regulatory elements determine ERα transcription initiation [56,57,58]. Estrogens are known to regulate the selective usage of these promoter systems [59] and although it has not been explicitly tested, sex differences in ERα promoter methylation may be related to alternative promoter selection and thereby involved in sexual differentiation of the brain. The POA is a highly sexually dimorphic region necessary for male sex behavior [60,61]. An analysis of ERα alternative promoter usage in the POA following exposure to endocrine disrupting chemicals revealed that sex and exposure to estrogenic compounds determines the methylation status and usage of alternative promoters. These differences are dependent on both developmental timepoint and compound dosage [62].
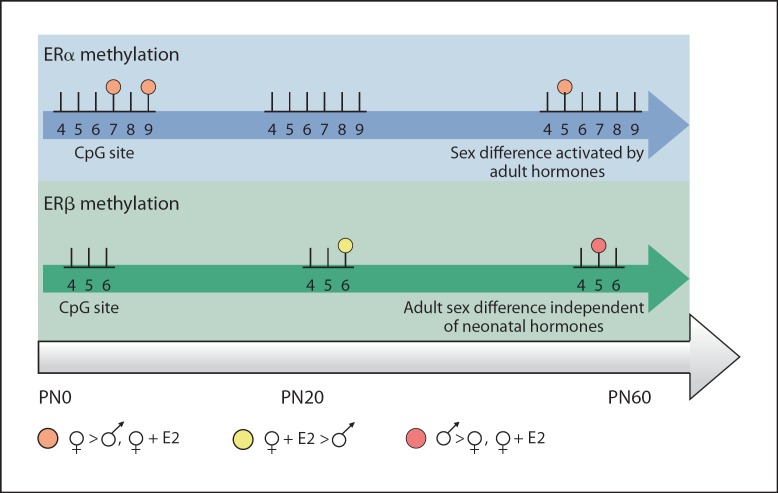
More recent studies have examined the impact of physiologically relevant masculinizing doses of estradiol. At the close of the sensitive period (PN10), males have higher levels of methylation at two CpG sites on the noncoding exon 1b of the ERα promoter within the POA compared to females. Treatment of female pups with estradiol to mimic the hormonal milieu of a male results in increased methylation at one site [63]. However, just upstream of exon 1b, in intron 1, a region containing high sequence homology to the intron 1 enhancer region in humans [64,65], the methylation pattern is markedly different. Newborn females have significantly higher levels of methylation at two CpG cites compared to males, and again, treating females with estradiol decreases methylation levels at these sites to that of males [66]. Interestingly, these differences are no longer present by around 3 weeks of age, a timepoint after the close of the sensitive period but before the onset of adult levels of circulating hormone. Even more surprising, a sex difference in CpG methylation reappears in the POA in adulthood in gonadally intact animals. A different pattern of methylation emerges in a CpG island located just upstream of ERβ exon 2. There are no sex differences present at birth but instead they appear at 3 weeks and 60 days of age (fig. 2). Thus within the POA the methylation status of ER regulatory regions can be organized with neonatal hormones and these differences may become apparent only in adulthood when hormones are present, a classic example of how early hormones organize the ability for adult hormones to activate sex differences in the brain. Within the medial basal hypothalamus, another region differentiated by neonatal estradiol exposure, newborn females have higher methylation of ERα intron 1 compared to males at two different CpG sites. This sex difference appears to be a transient developmental difference as methylation levels are equal between the sexes at later timepoints [66].
Fig. 2.
Dynamic methylation of ER regulatory regions. Schematic representation of the methylation patterns seen on ERα intron 1, and ERβ exon 2 within the POA across the lifespan following neonatal estradiol treatment (♀+E2). CpG methylation on both promoters is transient and affected by both neonatal hormonal milieu and adult hormone status. In the neonatal POA, females have higher methylation at two sites on ERα intron 1 compared to males and estradiol-treated females; however, this difference is not permanently organized because it disappears by PN20. With the activation by adult circulating hormones at PN60, the same trend (females having higher methylation compared to males and estradiol-treated females) reemerges. Conversely, the early hormonal milieu does not appear to play a role in the methylation patterns seen on ERβ exon 2, since treating females with a masculinizing dose of estradiol during development does not significantly alter methylation levels at any timepoint compared to vehicle-treated females.
In addition to sexually differentiating the brain, estrogens are important for normal development of many regions not typically considered highly sexually dimorphic [67]. ERα is highly expressed in the neonatal cortex, but its expression declines by around 10 days of life and is nonexistent in the adult [68,69]. The suppression of ERα expression in the adult cortex is the result of increased DNA methylation at specific areas of the ERα 5′ untranslated region, and coincides with higher expression of DNMT1, the methyltransferase isoform responsible for maintenance of DNA methylation [70].
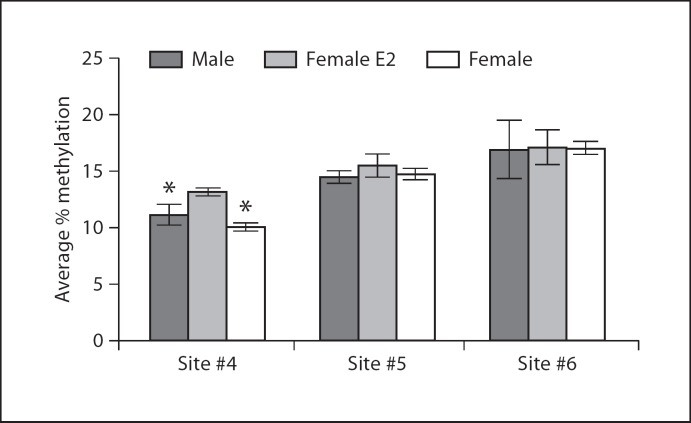
ERβ is highly expressed in the hippocampus [71] and has been implicated in controlling stress and learned-helplessness-related behaviors associated with depression [72,73,74]. In newborn and 3-week-old rats, sex and neonatal estradiol treatment have no impact on methylation status of the ERβ exon 2 promoter, whereas neonatal estradiol treatment causes a significant increase in methylation at one CpG site in adults (for detailed methods see Schwarz et al. [66]; fig. 3). This finding supports the idea that early hormone exposure can impact an animal's epigenome across its lifespan. In adulthood, acute estradiol is known to enhance memory consolidation [75]. Infusing estradiol into the dorsal hippocampus alters both DNMT and HDAC protein levels, and inhibition of DNMT activity blocks estradiol's enhancement of memory, indicating that estradiol works through epigenetic mechanisms to boost memory consolidation in adult animals [76].
Fig. 3.
ERβ promoter methylation in the hippocampus. Female rat pups were treated with a masculinizing dose of estradiol on the day of birth, and levels of CpG methylation were assessed in males, females, and estradiol-treated females on P1, PN20, and PN60. There was no developmental impact of hormone treatment on CpG methylation of ERβ in the hippocampus (data not shown), but a significant difference in methylation at CpG site 4 of ERβ intron 1 emerged in adulthood (ANOVA F2,13 = 6.4, p < 0.05; * p < 0.05 compared to estradiol-treated female).
The progesterone receptor (PR) is an additional target for neonatal hormones that may be important in sexual differentiation of the brain. Estradiol induces PR expression in the neonatal brain [77], and newborn males have significantly higher levels of PR in subregions of the POA [78]. While the functional relevance of this sex difference is unknown, blocking PR activity attenuates estrogen-induced increases in POA subnucleus volume [79]. Although estradiol is known to enhance PR expression in the POA, estradiol treatment at birth significantly increases methylation within exon 1 of the PR promoter in the neonatal POA [66]. This unexpected finding provides another example of how gene methylation does not always correspond to gene expression and further supports the view of new roles for DNA methylation in brain organization.
A sex difference in levels of MeCP2 provides further evidence for a role of DNA methylation in sexual differentiation of the brain. Females have more MeCP2 in the amygdala and hypothalamus during the critical period than do males, but expression levels are equal between the sexes by day 10 [80]. Females also express higher levels of nuclear receptor corepressor (NCoR) in the developing amygdala, and estradiol treatment reduces NCoR levels to that of males [81]. NCoR interacts with ERα [82] as well as various methyl-binding proteins, such as MeCP2, and forms corepressor complexes with HDAC enzymes [83].
Although fewer studies of sex differences in DNA methylation have been reported in the adult brain, a study profiling changes in the human epigenome associated with major psychosis revealed sex-specific methylation patterns in genes associated with bipolar disorder and schizophrenia [84], two diseases with sex differences in prevalence, symptomology, and age of onset [85,86]. It remains to be seen if the differential gene expression found in this study is the result of hormonal or genetic sex difference and whether developmental factors are at play.
Hormonal Modulation of Chromatin Structure
Within the perinatal cortex and hippocampus, males have higher levels of acetylated and trimethylated histone H3 than females. The sex difference in H3 aceylation appears to be hormonally dependent as testosterone treatment administered to rat dams during their final days of gestation raises H3 acetylation in the cortex and hippocampus of neonatal females [87]. However, testosterone treatment fails to alter methylation of histone H3, suggesting this sex difference is not organized by early hormone exposure and may be the result of other genomic differences.
The principle nucleus of the bed nucleus of the stria terminalis (BNSTp) is a forebrain structure organized by perinatal hormone exposure that is larger and contains more neurons in males than females [88]. During the critical period for sexual differentiation of the brain, Bax-dependent apoptotic cell death kills cells in the female BNSTp, while gonadal steroids protect males and hormone-treated females from cell loss in this region [89,90]. Interestingly, the protective effect of hormones is not evident until about 5 days after hormone treatment, leading Forger, de Vries and colleagues [91] to investigate epigenetic mechanisms controlling this cellular memory. They found that inhibition of histone deacetylation with the anticonvulsant drug, valproic acid (VPA), significantly impairs masculinization of BNSTp volume and neuron number in genetic males and testosterone-treated females, suggesting hormones influence chromatin conformation during sexual differentiation.
Histone modifications also appear to play a role in sexual differentiation of the POA. During the sensitive period males have higher levels of acetylation of histone H3 and H4 around the ERα 1b and aromatase promoters compared to females, suggesting males may have higher levels of transcriptional activity at these genes during the critical period [92]. HDAC protein levels are equal in both sexes neonatally, but males have greater levels of HDAC2 and HDAC4 bound to the ERα 1b and aromatase promoters. These sex differences appear to be critical for neonatal masculinization of the brain because inhibiting HDAC activity with TSA or antisense oligonucleotides directed against HDAC2 and HDAC4 impairs male sexual behavior performance in adulthood.
Conclusions
As interest in epigenetic processes grows, the importance of these genomic modifications in normal and pathological biological systems is becoming apparent. The currently unknown mechanisms for permanent sexual differentiation of the neonatal brain are beginning to be addressed through the study of epigenetics. As outlined above, recent findings suggest that epigenetic processes such as DNA methylation and histone acetylation may play an important role in the creation and maintenance of sexual dimorphisms in the brain. Sex differences in the epigenome are prevalent in the developing brain in regions classically considered sexually dimorphic as well as other brain regions known to exhibit sex differences but not necessarily of the same magnitude or obvious functional significance. Importantly, many of the observed epigenetic changes are mediated by hormones and are only apparent during the critical period for sexual differentiation of the brain, lending credence to the hypothesis that hormone exposure during the critical period organizes the bipotential brain via epigenomic alterations. It is well established that epigenetic processes are integral to determining cell fate in the developing brain [93]. Likewise, gonadal steroids exert ubiquitous actions on the developing brain to determine cell fate and neuronal anatomy, the question remains as to how, when, and how much hormonally-induced epigenetic mechanisms contribute. For instance, do estradiol-induced patterns of gene methylation determine sex difference in the distribution of different cell types by controlling gene expression, or does estradiol differentiate cellular fate which in turn causes sex differences in epigenetic markers? The study of the involvement of epigenetic mechanisms in sexual differentiation of the brain is still in its infancy. A better understanding of the mechanisms by which hormones alter the epigenome and a more comprehensive analysis of the genes impacted by epigenetic regulation during sexual differentiation is necessary to fully understand the importance of these processes in the establishment of the sexually dimorphic brain.
References
- 1.McCarthy MM. Estradiol and the developing brain. Physiol Rev. 2008;1:91–124. doi: 10.1152/physrev.00010.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold AP, Chen X. What does the ‘four core genotypes’ mouse model tell us about sex differences in the brain and other tissues? Front Neuroendocrinol. 2009;1:1–9. doi: 10.1016/j.yfrne.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCarthy M. How it's made: organizational effects of hormones on the developing brain. J Neuroendocrinol. 2010;7:736–742. doi: 10.1111/j.1365-2826.2010.02021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhees RW, Shryne JE, Gorski RA. Onset of the hormone-sensitive perinatal period for sexual differentiation of the sexually dimorphic nucleus of the preoptic area in female rats. J Neurobiol. 1990;5:781–786. doi: 10.1002/neu.480210511. [DOI] [PubMed] [Google Scholar]
- 5.Arnold AP, Gorski RA. Gonadal steroid induction of structural sex differences in the central nervous system. Annu Rev Neurosci. 1984;1:413–442. doi: 10.1146/annurev.ne.07.030184.002213. [DOI] [PubMed] [Google Scholar]
- 6.Weisz J, Ward IL. Plasma testosterone and progesterone titers of pregnant rats, their male and female fetuses, and neonatal offspring. Endocrinology. 1980;1:306–316. doi: 10.1210/endo-106-1-306. [DOI] [PubMed] [Google Scholar]
- 7.McEwen BS, Lieberburg I, Chaptal C, Krey LC. Aromatization: important for sexual differentiation of the neonatal rat brain. Horm Behav. 1977;3:249–263. doi: 10.1016/0018-506x(77)90060-5. [DOI] [PubMed] [Google Scholar]
- 8.Lephart ED. A review of brain aromatase cytochrome P450. Brain Res Rev. 1996;1:1–26. [PubMed] [Google Scholar]
- 9.Naftolin F, Ryan KJ, Davies IJ, Reddy VV, Flores F, Petro Z, Kuhn M, White RJ, Takaoka Y, Wolin L. The formation of estrogens by central neuroendocrine tissues. Recent Prog Horm Res. 1975:295–319. doi: 10.1016/b978-0-12-571131-9.50012-8. [DOI] [PubMed] [Google Scholar]
- 10.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;4854:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barraclough CA. Production of anovulatory, sterile rats by single injections of testosterone propionate. Endocrinology. 1961;1:62–67. doi: 10.1210/endo-68-1-62. [DOI] [PubMed] [Google Scholar]
- 12.Baum MJ. Differentiation of coital behavior in mammals: a comparative analysis. Neurosci Biobehav Rev. 1979;4:265–284. doi: 10.1016/0149-7634(79)90013-7. [DOI] [PubMed] [Google Scholar]
- 13.Vreeburg J, van der Vaart PDM, Van Der Schoot P. Prevention of central defeminization but not masculinization in male rats by inhibition neonatally of oestrogen biosynthesis. J Endocrinol. 1977;3:375. doi: 10.1677/joe.0.0740375. [DOI] [PubMed] [Google Scholar]
- 14.Bakker J, Honda S, Harada N, Balthazart J. Sexual partner preference requires a functional aromatase (cyp19) gene in male mice. Horm Behav. 2002;2:158–171. doi: 10.1006/hbeh.2002.1805. [DOI] [PubMed] [Google Scholar]
- 15.Roth TL, David Sweatt J. Annual Research Review: Epigenetic mechanisms and environmental shaping of the brain during sensitive periods of development. J Child Psychol Psychol. doi: 10.1111/j.1469-7610.2010.02282.x. (DOI 10.1111/j.1469–7610.2010. 02282.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Widom J. Structure, dynamics, and function of chromatin in vitro. Annu Rev Biophys Biomol Struct. 1998;1:285–327. doi: 10.1146/annurev.biophys.27.1.285. [DOI] [PubMed] [Google Scholar]
- 17.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, review fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 18.Hansen JC, Tse C, Wolffes AP. Structure and function of the core histone N-termini: more than meets the eye. Biochemistry. 1998;51:17637–17641. doi: 10.1021/bi982409v. [DOI] [PubMed] [Google Scholar]
- 19.Wolffe AP, Hayes JJ. Chromatin disruption and modification. Nucleic Acids Res. 1999;3:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye J, Ai X, Eugeni EE, Zhang L, Carpenter LR, Jelinek MA, Freitas MA, Parthun MR. Histone H4 lysine 91 acetylation: a core domain modification associated with chromatin assembly. Mol Cell. 2005;1:123–130. doi: 10.1016/j.molcel.2005.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;6765:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 22.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;6921:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 23.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;6649:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 24.Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;1:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 25.Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;8:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 26.Rosenfeld JA, Wang Z, Schones DE, Zhao K, DeSalle R, Zhang MQ. Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genomics. 2009;10:143. doi: 10.1186/1471-2164-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sigismund S, Polo S, Di Fiore PP. Signaling through monoubiquitination. Curr Top Microbiol Immunol. 2004;286:149–185. doi: 10.1007/978-3-540-69494-6_6. [DOI] [PubMed] [Google Scholar]
- 28.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;2:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 29.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;6:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 30.Nathan D, Ingvarsdottir K, Sterner DE, Bylebyl GR, Dokmanovic M, Dorsey JA, Whelan KA, Krsmanovic M, Lane WS, Meluh PB. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 2006;8:966–976. doi: 10.1101/gad.1404206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia-Dominguez M, Reyes JC. SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim Biophys Acta. 2009;6–8:451–459. doi: 10.1016/j.bbagrm.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 33.Illingworth R, Kerr A, DeSousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;1:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caiafa P, Zampieri M. DNA methylation and chromatin structure: the puzzling CpG islands. J Cell Biochem. 2005;2:257–265. doi: 10.1002/jcb.20325. [DOI] [PubMed] [Google Scholar]
- 35.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bogdanović O, Veenstra GJC. DNA methylation and methyl-CpG-binding proteins: developmental requirements and function. Chromosoma. 2009;5:549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samaco RC, Nagarajan RP, Braunschweig D, LaSalle JM. Multiple pathways regulate MeCP2 expression in normal brain development and exhibit defects in autism-spectrum disorders. Hum Mol Genet. 2004;6:629–639. doi: 10.1093/hmg/ddh063. [DOI] [PubMed] [Google Scholar]
- 38.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;6683:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 39.Yasui DH, Peddada S, Bieda MC, Vallero RO, Hogart A, Nagarajan RP, Thatcher KN, Farnham PJ, LaSalle JM. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci USA. 2007;49:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balada E, Ordi-Ros J, Serrano-Acedo S, Martinez-Lostao L, Vilardell-Tarres M. Transcript overexpression of the MBD2 and MBD4 genes in CD4 T cells from systemic lupus erythematosus patients. J Leukoc Biol. 2007;6:1609–1616. doi: 10.1189/jlb.0107064. [DOI] [PubMed] [Google Scholar]
- 41.Detich N, Theberge J, Szyf M. Promoter-specific activation and demethylation by MBD2/demethylase. J Biol Chem. 2002;39:35791–35794. doi: 10.1074/jbc.C200408200. [DOI] [PubMed] [Google Scholar]
- 42.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;6750:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 43.Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Dev. 1993;2:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 44.Sasaki M, Dharia A, Oh BR, Tanaka Y, Fujimoto S, Dahiya R. Progesterone receptor B gene inactivation and CpG hypermethylation in human uterine endometrial cancer. Cancer Res. 2001;1:97–102. [PubMed] [Google Scholar]
- 45.Fagiolini M, Jensen CL, Champagne FA. Epigenetic influences on brain development and plasticity. Curr Opin Neurobiol. 2009;2:207–212. doi: 10.1016/j.conb.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Renthal W, Nestler EJ. Epigenetic mechanisms in drug addiction. Trends Mol Med. 2008;8:341–350. doi: 10.1016/j.molmed.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;2:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- 48.Weaver ICG, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;8:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 49.Kadonaga JT. Eukaryotic transcription: an interlaced review network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 50.Champagne FA, Weaver ICG, Diorio J, Dymov S, Szyf M, Meaney MJ. Maternal care associated with methylation of the estrogen receptor-α1b promoter and estrogen receptor-α expression in the medial preoptic area of female offspring. Endocrinology. 2006;6:2909–2915. doi: 10.1210/en.2005-1119. [DOI] [PubMed] [Google Scholar]
- 51.Cameron N, Shahrokh D, Del Corpo A, Dhir S, Szyf M, Champagne F, Meaney M. Epigenetic programming of phenotypic variations in reproductive strategies in the rat through maternal care. J Neuroendocrinol. 2008;6:795–801. doi: 10.1111/j.1365-2826.2008.01725.x. [DOI] [PubMed] [Google Scholar]
- 52.Nishino K, Hattori N, Tanaka S, Shiota K. DNA methylation-mediated control of Sry gene expression in mouse gonadal development. J Biol Chem. 2004;21:22306–22313. doi: 10.1074/jbc.M309513200. [DOI] [PubMed] [Google Scholar]
- 53.Cisneros FJ, Branch S. Transplacental exposure to the DNA demethylating agent, 5-AZA-CdR, affects the sexual behavior of CD-1 male mice. Neurotoxicology. 2004;3:411–417. doi: 10.1016/j.neuro.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 54.Kudwa A, Michopoulos V, Gatewood J, Rissman E. Roles of estrogen receptors α and β in differentiation of mouse sexual behavior. Neuroscience. 2006;3:921–928. doi: 10.1016/j.neuroscience.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 55.Kudwa AE, Bodo C, Gustafsson JÅ, Rissman EF. A previously uncharacterized role for estrogen receptor β: defeminization of male brain and behavior. Proc Natl Acad Sci USA. 2005;12:4608. doi: 10.1073/pnas.0500752102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koike S, Sakai M, Muramatsu M. Molecular cloning and characterization of rat estrogen receptor cDNA. Nucleic Acids Res. 1987;6:2499–2513. doi: 10.1093/nar/15.6.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirata S, Koh T, Yamada-Mouri N, Hoshi K, Kato J. The untranslated first exon ‘exon OS’ of the rat estrogen receptor gene. FEBS Lett. 1996;3:371–373. doi: 10.1016/0014-5793(96)00987-8. [DOI] [PubMed] [Google Scholar]
- 58.Hirata S, Koh T, Yamada-Mouri N, Kato J. The novel untranslated first exon ‘exon 0N’ of the rat estrogen receptor gene. Biochem Biophys Res Commun. 1996;2:653. doi: 10.1006/bbrc.1996.1262. [DOI] [PubMed] [Google Scholar]
- 59.Donaghue C, Westley B, May F. Selective promoter usage of the human estrogen receptor-α gene and its regulation by estrogen. Mol Endocrinol. 1999;11:1934–1950. doi: 10.1210/mend.13.11.0366. [DOI] [PubMed] [Google Scholar]
- 60.De Jonge F, Louwerse A, Ooms M, Evers P, Endert E, Van de Poll N. Lesions of the SDN-POA inhibit sexual behavior of male Wistar rats. Brain Res Bull. 1989;6:483–492. doi: 10.1016/0361-9230(89)90194-9. [DOI] [PubMed] [Google Scholar]
- 61.Gorski RA, Gordon J, Shryne J, Southam A. Evidence for a morphological sex difference within the medial preoptic area of the rat brain. Brain Res. 1978;2:333–346. doi: 10.1016/0006-8993(78)90723-0. [DOI] [PubMed] [Google Scholar]
- 62.Monje L, Varayoud J, Luque EH, Ramos JG. Neonatal exposure to bisphenol A modifies the abundance of estrogen receptor-α transcripts with alternative 5′-untranslated regions in the female rat preoptic area. J Endocrinol. 2007;1:201–212. doi: 10.1677/JOE-07-0014. [DOI] [PubMed] [Google Scholar]
- 63.Kurian JR, Olesen KM, Auger AP. Sex differences in epigenetic regulation of the estrogen receptor-α promoter within the developing preoptic area. Endocrinology. 2010;5:2297–2305. doi: 10.1210/en.2009-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Freyschuss B, Grandien K. The 5′-flank of the rat estrogen receptor gene: structural characterization and evidence for tissue- and species-specific promoter utilization. J Mol Endocrinol. 1996;3:197–206. doi: 10.1677/jme.0.0170197. [DOI] [PubMed] [Google Scholar]
- 65.Maruyama H, Toji H, Harrington CR, Sasaki K, Izumi Y, Ohnuma T, Arai H, Yasuda M, Tanaka C, Emson PC. Lack of an association of estrogen receptor-α gene polymorphisms and transcriptional activity with Alzheimer disease. Arch Neurol. 2000;2:236–240. doi: 10.1001/archneur.57.2.236. [DOI] [PubMed] [Google Scholar]
- 66.Schwarz JM, Nugent BM, McCarthy MM. Developmental and hormone-induced epigenetic changes to estrogen and progesterone receptor genes in brain are dynamic across the life span. Endocrinology. 2010;151:4871–4881. doi: 10.1210/en.2010-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McEwen B. Steroid hormones: effect on brain development and function. Horm Res. 1992;3:1–10. doi: 10.1159/000182393. [DOI] [PubMed] [Google Scholar]
- 68.Prewitt AK, Wilson ME. Changes in estrogen receptor-α mRNA in the mouse cortex during development. Brain Res. 2007;1134:62–69. doi: 10.1016/j.brainres.2006.11.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miranda RC, Toran-Allerand CD. Developmental expression of estrogen receptor mRNA in the rat cerebral cortex: a nonisotopic in situ hybridization histochemistry study. Cereb Cortex. 1992;1:1–15. doi: 10.1093/cercor/2.1.1. [DOI] [PubMed] [Google Scholar]
- 70.Westberry JM, Trout AL, Wilson ME. Epigenetic regulation of estrogen receptor-α gene expression in the mouse cortex during early postnatal development. Endocrinology. 2010;2:731–740. doi: 10.1210/en.2009-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shughrue PJ, Lane MV, Merchenthaler I. Comparative distribution of estrogen receptor-α and -β mRNA in the rat central nervous system. J Comp Neurol. 1997;4:507–525. doi: 10.1002/(sici)1096-9861(19971201)388:4<507::aid-cne1>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 72.Krezel W, Dupont S, Krust A, Chambon P, Chapman PF. Increased anxiety and synaptic plasticity in estrogen receptor β-deficient mice. Proc Natl Acad Sci USA. 2001;21:12278–12282. doi: 10.1073/pnas.221451898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Imwalle DB, Gustafsson JÅ, Rissman EF. Lack of functional estrogen receptor-β influences anxiety behavior and serotonin content in female mice. Physiol Behav. 2005;1:157–163. doi: 10.1016/j.physbeh.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 74.Rocha BA, Fleischer R, Schaeffer JM, Rohrer SP, Hickey GJ. 17β-Estradiol-induced antidepressant-like effect in the forced swim test is absent in estrogen receptor-β knockout (BERKO) mice. Psychopharmacology. 2005;3:637–643. doi: 10.1007/s00213-004-2078-1. [DOI] [PubMed] [Google Scholar]
- 75.Fernandez SM, Lewis MC, Pechenino AS, Harburger LL, Orr PT, Gresack JE, Schafe GE, Frick KM. Estradiol-induced enhancement of object memory consolidation involves hippocampal extracellular signal-regulated kinase activation and membrane-bound estrogen receptors. J Neurosci. 2008;35:8660–8667. doi: 10.1523/JNEUROSCI.1968-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao Z, Fan L, Frick K. Epigenetic alterations regulate estradiol-induced enhancement of memory consolidation. Proc Natl Acad Sci USA. 2010;12:5605–5610. doi: 10.1073/pnas.0910578107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wagner CK, Pfau JL, De Vries GJ, Merchenthaler IJ. Sex differences in progesterone receptor immunoreactivity in neonatal mouse brain depend on estrogen receptor-α expression. J Neurobiol. 2001;3:176–182. doi: 10.1002/neu.1025. [DOI] [PubMed] [Google Scholar]
- 78.Quadros PS, Pfau JL, Goldstein AYN, De Vries GJ, Wagner CK. Sex differences in progesterone receptor expression: a potential mechanism for estradiol-mediated sexual differentiation. Endocrinology. 2002;10:3727–3739. doi: 10.1210/en.2002-211438. [DOI] [PubMed] [Google Scholar]
- 79.Quadros PS, Lopez V, De Vries GJ, Chung WCJ, Wagner CK. Progesterone receptors and the sexual differentiation of the medial preoptic nucleus. J Neurobiol. 2002;1:24–32. doi: 10.1002/neu.10040. [DOI] [PubMed] [Google Scholar]
- 80.Kurian JR, Forbes-Lorman RM, Auger AP. Sex difference in mecp2 expression during a critical period of rat brain development. Epigenetics. 2007;3:173–178. doi: 10.4161/epi.2.3.4841. [DOI] [PubMed] [Google Scholar]
- 81.Jessen HM, Kolodkin MH, Bychowski ME, Auger CJ, Auger AP. The nuclear receptor corepressor has organizational effects within the developing amygdala on juvenile social play and anxiety-like behavior. Endocrinology. 2010;3:1212–1220. doi: 10.1210/en.2009-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang HJ, Norris JD, McDonnell DP. Identification of a negative regulatory surface within estrogen receptor-α provides evidence in support of a role for corepressors in regulating cellular responses to agonists and antagonists. Mol Endocrinol. 2002;8:1778–1792. doi: 10.1210/me.2002-0089. [DOI] [PubMed] [Google Scholar]
- 83.Wen YD, Perissi V, Staszewski LM, Yang WM, Krones A, Glass CK, Rosenfeld MG, Seto E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci USA. 2000;13:7202–7207. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, Jia P, Assadzadeh A, Flanagan J, Schumacher A. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;3:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Arnold LM. Gender differences in bipolar disorder. Psychiatr Clin North Am. 2003;3:595–620. doi: 10.1016/s0193-953x(03)00036-4. [DOI] [PubMed] [Google Scholar]
- 86.Häfner H. Gender differences in schizophrenia. Psychoneuroendocrinology. 2003;28:17–54. doi: 10.1016/s0306-4530(02)00125-7. [DOI] [PubMed] [Google Scholar]
- 87.Tsai HW, Grant PA, Rissman EF. Sex differences in histone modifications in the neonatal mouse brain. Epigenetics. 2009;1:47–53. doi: 10.4161/epi.4.1.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hines M, Allen LS, Gorski RA. Sex differences in subregions of the medial nucleus of the amygdala and the bed nucleus of the stria terminalis of the rat. Brain Res. 1992;2:321–326. doi: 10.1016/0006-8993(92)90068-k. [DOI] [PubMed] [Google Scholar]
- 89.Forger NG, Rosen GJ, Waters EM, Jacob D, Simerly RB, De Vries GJ. Deletion of Bax eliminates sex differences in the mouse forebrain. Proc Natl Acad Sci. 2004;37:13666–13671. doi: 10.1073/pnas.0404644101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chung WCJ, Swaab DF, De Vries GJ. Apoptosis during sexual differentiation of the bed nucleus of the stria terminalis in the rat brain. J Neurobiol. 2000;3:234–243. [PubMed] [Google Scholar]
- 91.Murray EK, Hien A, de Vries GJ, Forger NG. Epigenetic control of sexual differentiation of the bed nucleus of the stria terminalis. Endocrinology. 2009;9:4241–4247. doi: 10.1210/en.2009-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Matsuda KI, Mori H, Nugent BM, Pfaff DW, McCarthy MM, Kawata M. Histone deacetylation during masculinization of the brain. J Neurosci (under review) doi: 10.1210/en.2011-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hsieh J, Gage FH. Epigenetic control of neural stem cell fate. Curr Opin Genet Dev. 2004;5:461–469. doi: 10.1016/j.gde.2004.07.006. [DOI] [PubMed] [Google Scholar]