Abstract
Background
Aedes aegypti is the principal mosquito vector of Zika, dengue, and yellow fever viruses. Two subspecies of Ae. aegypti exhibit phenotypic divergence with regard to habitat, host preference, and vectorial capacity. Chromosomal inversions have been shown to play a major role in adaptation and speciation in dipteran insects and would be of great utility for studies of Ae. aegypti. However, the large and highly repetitive genome of Ae. aegypti makes it difficult to detect inversions with paired-end short-read sequencing data, and polytene chromosome analysis does not provide sufficient resolution to detect chromosome banding patterns indicative of inversions.
Results
To characterize chromosomal diversity in this species, we have carried out deep Illumina sequencing of linked-read (10X Genomics) libraries in order to discover inversion loci as well as SNPs. We analyzed individuals from colonies representing the geographic limits of each subspecies, one contact zone between subspecies, and a closely related sister species. Despite genome-wide SNP divergence and abundant microinversions, we do not find any inversions occurring as fixed differences between subspecies. Many microinversions are found in regions that have introgressed and have captured genes that could impact behavior, such as a cluster of odorant-binding proteins that may play a role in host feeding preference.
Conclusions
Our study shows that inversions are abundant and widely shared among subspecies of Aedes aegypti and that introgression has occurred in regions of secondary contact. This library of 32 novel chromosomal inversions demonstrates the capacity for linked-read sequencing to identify previously intractable genomic rearrangements and provides a foundation for future population genetics studies in this species.
Keywords: Structural variation, Chromosomal inversions, Introgression, Aedes aegypti
Background
Since the early 1920s, chromosomal inversions have been a rich source of insight into speciation and adaptation. First discovering inversions as cytogenetic markers of divergence between related Drosophila species, Sturtevant correctly predicted that inversions would suppress meiotic recombination within their bounds [1], causing karyotypes to diverge even in the absence of selection. The selective pressures acting on inversions have been shown to include cases of divergent selection and balancing selection (summarized in Wellenreuther and Bernatchez [2]), with often conflicting selective pressures dependent on the alleles captured in an inverted region. What is apparent is that, by allowing the co-adaptation of linked genes, inversions can contribute to local adaptation [3]. For example, associations between inversions and climatic clines have independently developed on multiple continents in various Drosophila species [4]. Indeed, these linked blocks of co-adapted genes can enable rapid adaptation to novel environments by selecting from a stock of globally shared standing variation—as has been seen during colonization of independent freshwater habitats by marine stickleback populations [5]. These “supergenes” can also introgress between closely related species leading to the transfer of distinct phenotypes across species boundaries [6]. Moreover, inversions are commonly implicated in the speciation process [7, 8]. Although the precise mechanisms of speciation remain a matter of debate [9–12], fixed inversions are more commonly found between sympatric sister species than allopatric [9] and both underdominance of heterokaryotypes as well as inversion-associated assortative mating may play a role in speciation [13].
Inversions are commonly studied in malaria mosquito vectors. Mosquitoes are often found as complexes of closely related species that are distinguished by fixed inversions but maintain some degree of gene flow [14], while structured populations at the subspecies level have also been posited based on differing inversion polymorphisms [15]. Chromosomal inversions have also been directly associated with phenotypes that are important for vector competence: thermal tolerance underlying range expansion [16–18], feeding behavior [19, 20], oviposition site preferences [21, 22], insecticide resistance [23], and immune response to parasites [24, 25].
However, despite their utility in other dipterans, studies of chromosomal inversions have not yet been undertaken in the mosquito Aedes aegypti. This is perhaps a surprise since the population structure of this organism is not just of academic interest, but could be of key importance for global health. Ae. aegypti is the principal vector of many viral and parasitic diseases and is found throughout the tropics [26]. Along with this global distribution comes a similarly broad dispersal of Aedes-borne viruses. The most common, dengue, infects millions of people per year [27] and causes more than 20,000 deaths [28], while emergent Aedes-borne viruses such as Zika are capable of rapid global spread [28, 29]. While chromosomes have been directly observed in Aedine mosquitoes, resolution of chromosome banding patterns is relatively poor [30, 31]; low levels of replication in polytene chromosomes and a tendency to break rather than cleanly separating mean that as little as 4% of chromosome preparations show sufficient resolution to identify banding patterns [31] and no studies have so far been able to capture visual confirmation of chromosomal inversions via banding patterns for Ae. aegypti.
Detecting inversions in Ae. aegypti via conventional sequencing-based methods is also difficult. The Ae. aegypti genome is significantly larger than other well-studied dipterans (~ 1.25 Gb), being more than five times the size of Drosophila melanogaster (~ 180 Mb [32]) or Anopheles gambiae (~ 250 Mb [33]) despite having only around 20% more genes [34]. Much of this extra genome consists of transposable elements and repeats, which comprise 65% of the Ae. aegypti genome. The abundance of mobile elements is a significant problem for sequencing-based detection of chromosomal rearrangements: whole-genome resequencing experiments will effectively waste more than half of the sequence they produce on intractable repetitive sequence and the mobility of these elements can lead to spurious signals of structural variation. Paired-end methods of inversion detection generally rely upon accurate mapping of reads separated by 300–500 bp and are thus poorly adapted to finding inversion breakpoints where they are buried within repetitive sequence [35, 36]. For this reason, much of the current evidence for inversions in Ae. aegypti is indirect: inversions are inferred from decreases in recombination rate around the putative locus. This technique has been used to detect an inversion in the sex-determining “M locus” [37] and a number of large inversions on all three chromosomes in populations in Senegal [38]. While optical and physical mapping has shown capacity to confirm large inversions [34, 39], direct confirmation of the breakpoints, or mapping the coordinates of these inversions has not proven possible. A method of detecting chromosomal inversions via sequencing would therefore be a major advance, enabling the detection of inversions that are obscure to cytogenetic methods.
Despite the challenges of working with the Ae. aegypti genome, there have been a number of insights into Aedine population structure. While relatively homogeneous in most of the tropics, within sub-Saharan Africa, the species exhibits notable phenotypic diversity: in contrast to the anthropophily of the global Ae. aegypti aegypti (Aaa), across sub-Saharan Africa, the generalist Ae. aegypti formosus (Aaf) dominates. Isolated regions in which both Aaa and Aaf forms are present have been found along the coast of East Africa [40, 41] and possibly in northwest Senegal [42], in which the two subspecies display varying degrees of reproductive isolation, with more hybridization in urban settings where the two forms overlap [43]. These subspecies may be expected to segregate different complements of inversions.
In order to identify inversions within the Aedes genome, we have employed long-read (Pacific Biosciences and Oxford Nanopore) sequencing and linked-read (10X Genomics) sequencing, in which sequencing library constructs deriving from a common DNA molecule up to ~ 80 kb in length share a common barcode, enabling one to identify reads deriving from physically proximal sequences within the genome over a greater distance than is practical with standard paired-end sequencing libraries. Regions that are not proximal in the reference assembly, but share large numbers of barcodes, are strong candidates for structural rearrangement in the genome. This technology allows us to call both inversions and SNPs with the potential to link inversion polymorphisms to more tractable SNP markers. Cataloguing inversions in this manner not only is of utility for future studies of population structure in Ae. aegypti, but may also highlight regions that underlie the maintenance of reproductive isolation in the two subspecies. We applied this linked-read technology to 26 individual mosquitoes from 9 different colonies (Fig. 1), generating more than 500 candidate inversions; through rigorous validation of breakpoints using both sequencing types, we were able to confirm 32 of these in multiple independently analyzed samples. The majority of these inversions are “microinversions” (those below the cytological limit of detection—here considered to be below 500 kb). The microinversions we detected are widely shared between subspecies and some occur in regions exhibiting introgression with a sister species, Aedes mascarensis.
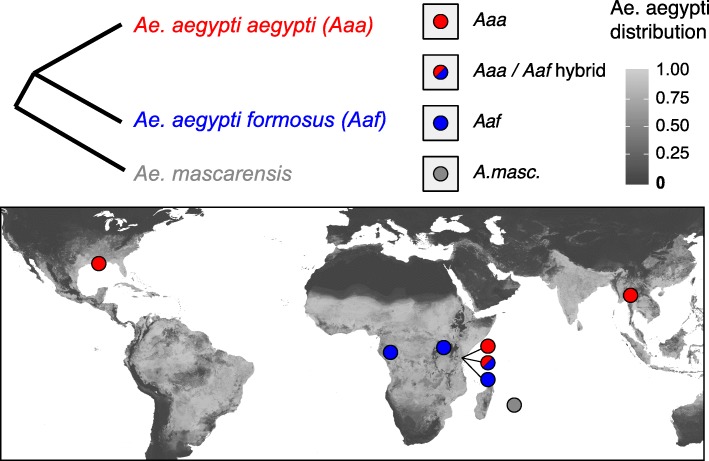
Fig. 1.
Colonies were selected to represent the extreme latitudes of both subspecies of Aedes aegypti: Ae. aegypti formosus (Aaf) found only in sub-Saharan Africa and Ae. aegypti aegypti (Aaa) found worldwide. Three colonies, consisting of pure Aaa, Aaf, and Aaa/Aaf hybrids, were founded from Rabai, Kenya: a zone of secondary contact between the two subspecies. An outgroup colony, Ae. mascarensis, was included from Mauritius
Results
More than 560 candidate inversions were called across all 28 samples (Fig S1); 465 were called by Long Ranger and 142 by GROC-SVs, with only 44 called by both. The size profile of candidates was also different between the two packages, with GROC-SVs calling larger inversions overall. There were 363 inversions called in only one sample (319/37/7 singletons called in Long Ranger/GROC-SVs/both, respectively).
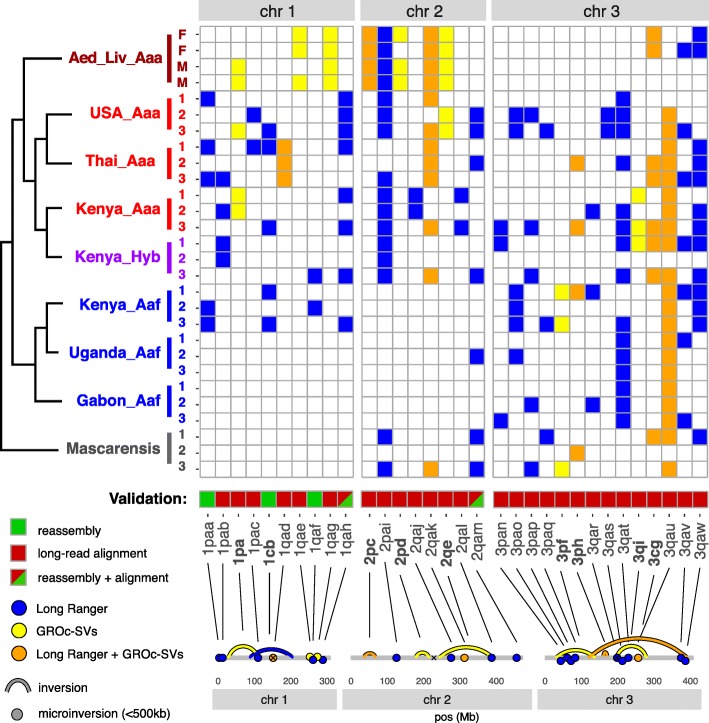
Due to the highly repetitive nature of this genome, and in particular the likelihood that any active transposons will generate false-positive inversion candidates, we took an aggressive approach to validation. Inversions were considered valid if we were able to reconstruct both inverted breakpoints—either via de novo reassembly of the breakpoint regions or by alignment of long-read sequence across the breakpoints. Reconstruction of both breakpoints was limited to 32 inversions (9 large inversions and 23 microinversions) (Fig. 2): 4 confirmed via reassembly, 24 by long-read alignment, and 4 by both methods; all nine large inversions were confirmed via long-read alignment. Microinversions are here defined as any inversions under 500 kb; median size was 43 kb.
Fig. 2.
Two structural variant callers were implemented on the same linked-read data, GROC-SVs and Long Ranger. Validation of candidate inversions was performed via long-read alignment across breakpoints, or de novo reassembly of breakpoint regions. Thirty-two inversions (9 inversions, 21 microinversions) could be confirmed in both breakpoints; extensive sharing of inversions between subspecies as well as between Ae. aegypti and Ae. mascarensis is seen for inversions on chromosomes 2 and 3. Notably, chromosome 1, the homomorphic chromosome that is the site of the sex-determining locus, shows no shared inversions between subspecies outside of the hybrid zone of Rabai
Phylogenetic results support divergence of A. aegypti aegypti from A. aegypti formosus
Divergence as measured by Fst was high between samples from all regions, with no difference whether we were comparing within or between subspecies (mean Fst within, 0.18; between, 0.19), likely representing the effect of colonization rather than allele fixation in wild populations (Fig S2). Consistent with its emergence from an Aaf ancestral population, diversity was significantly lower in Aaa colonies than in Aaf colonies, even after removing long runs of homozygosity (π Thai = 3.3e−3, USA = 4.0e−3, Uganda = 4.6e−3, Gabon = 5.7e−3; mean π Aaa 3.9e−3, Aaf 5.1e−3; Wilcoxon rank-sum test, P < 2.2e−16) (Fig S3).
A maximum parsimony phylogeny was generated from biallelic SNP data including all colony samples and two outgroup species (Ae. albopictus/Ae. bromeliae) (Fig S4). The phylogeny showed strong support for the separation of the Aaa/Aaf subgroups, with all bootstrap values between Aaa colonies above 40%, all Aaf colonies at 100%, and the branching of Aaa from Aaf or Ae. aegypti from Ae. mascarensis also supported by 100% of bootstrap replicates. All of these results are consistent with previous studies showing that Aaf is the ancestral population and founder effects have led to significantly reduced diversity of Aaa outside sub-Saharan Africa.
Pervasive introgression is found between subspecies and species
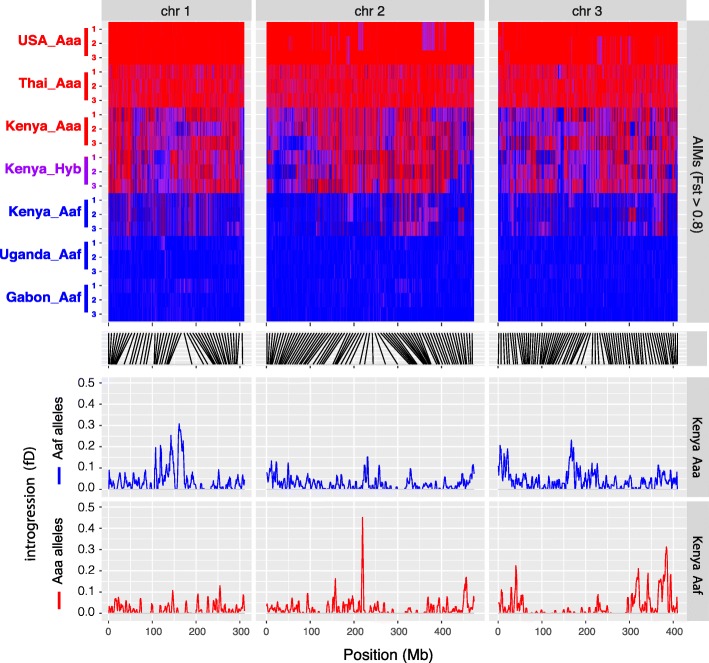
All three Kenyan colonies derive from mosquitoes collected in the Rabai region of Kenya. Both subspecies live in sympatry in this region and produce viable offspring after hybridization, creating an opportunity for genetic introgression between diverged populations. To test for this, we performed two tests of introgression: Patterson’s D for genome-wide introgression using block jackknifing to assess significance, and Martin’s ƒD to localize the genomic regions that have introgressed. At the subspecies level, tests were performed comparing Kenyan colonies to the “pure” colonies from other regions; significant introgression was detected between Aaf and Kenyan Aaa, and Aaa and Kenya Aaf—suggesting bidirectional gene flow between the two subspecies in Kenya (Fig S5). The Kenya_Aaa and hybrid colonies showed a similar distribution of “Aaf” ancestry-informative markers (AIMs); in a comparison of 10,000 AIMs, 31.57% of the AIM loci show a predominantly Aaf allele in the Kenya Aaa colony with 35.78% in the “hybrid” colony (Fig. 3), suggesting that gene flow is extensive in this region and “pure” Aaa is likely to be rare—consistent with a collapsing Aaa population at the time of sampling [C. McBride—personal communication]. Despite efforts to identify pure Aaa and hybrids, the two colonies appear to be sampling from the same population.
Fig. 3.
Introgression was assessed via Patterson’s D between global populations of Aaa/Aaf and colonies within Rabai, illustrating bidirectional gene flow between the two forms. Local peaks of introgression were identified using Martin’s ƒD. Ancestry-informative markers were selected as those with Fst > 0.8 between all pure Aaa/Aaf colonies and illustrate the assymetric nature of introgression in this region, with a higher proportion of Aaf alleles found within the Aaa and hybrid colonies than vice versa
Though gene flow is clearly bidirectional in Kenya, the degree of introgression into each subspecies is not equal—whether measured by ƒD or by examining AIMs, patterns of introgression in Aaa and Aaf colonies are markedly different (Fig. 3). Compared to more than 30% of Aaf markers in the Kenya_Aaa colony, only 12.90% of these loci show a majority Aaa allele in Kenya Aaf. The patterns of introgression are also different; in Kenya Aaa and Kenya Hyb colonies, introgression peaks and Aaf alleles are present across broad swaths of the genome, including the entire chromosome 1 centromere which encompasses the sex-determining locus. In contrast, Aaa alleles in the Aaf colony are limited to short haplotypes—potentially including genes under positive selection in Aaf. This pattern is consistent with asymmetric introgression; however, caution should be used when making inferences from a small number of founders for the Aaa and Hyb colonies (Table 1) as capture of Aaf individuals in the Aaa and Hyb founders could give similar results.
Table 1.
Colonies
| Colony | Spp./sub-spp. | Collection year | Collection location | Gens in colony | No. founders | Previously published |
|---|---|---|---|---|---|---|
| Liverpool_Aae | Ae. aegypti aegypti | – | – | Unknown* | – | Matthews et al. [34] |
| USA_Aae | Ae. aegypti aegypti | 2014 | New Orleans, USA | 7 | 4 females | Powell and Evans [44] |
| Thailand_Aae | Ae. aegypti aegypti | 2015 | Muang district, Thailand | 3 | > 500 larvae | |
| Kenya_Aae | Ae. aegypti aegypti | 2009 | Rabai, Kenya | 19 | 3 females | McBride et al. [40] |
| Kenya_Hyb | Aaa/Aaf hybrid | 2009 | Rabai, Kenya | 14 | 2 females | McBride et al. [40] |
| Kenya_Aaf | Ae. aegypti formosus | 2009 | Rabai, Kenya | 11 | 9 females | McBride et al. [40] |
| Uganda_Aaf | Ae. aegypti formosus | 2015/2016 | Zika, Uganda | 4 | ~ 100 inds, from 2 ovitraps | |
| Gabon_Aaf | Ae. aegypti formosus | 2016 | Franceville, Gabon | 2 | ~ 10 mated females from multiple sites | |
| Mascarensis | Ae. mascarensis | 2014 | Mauritius | 2 | 10 females |
*The Liverpool colony was founded circa 1935 and has undergone a large but unknown number of generations. The precise collection location and numbers of founders are not known. Our samples are the same generation used in Matthews et al. [34]
Interestingly, AIM analyses also showed a mixture of Aae and Aaf alleles in the Liverpool sequencing colony used for the genome assemblies [33, 34] with 44.63% of AIMs having a predominantly Aaf genotype in the Liverpool colony (Fig S6), consistent with a previously reported west African origin for Aae [42].
While introgression between sympatric populations of the same species in Kenya may not be surprising, gene flow is also detected between different species: comparing our two subspecies, and Ae. mascarensis with a more distant outgroup Ae. albopictus (Fig S5b), post-speciation gene flow was detected between our mascarensis colony and global Aaa samples. Much like the Kenya_Aaf samples, introgression into the Ae. mascarensis colony was limited to short haplotypes (Fig S5b); indeed, many of these short Aaa haplotypes appear to have introgressed into both Kenyan Aaf and Ae. mascarensis (Fig. 4).
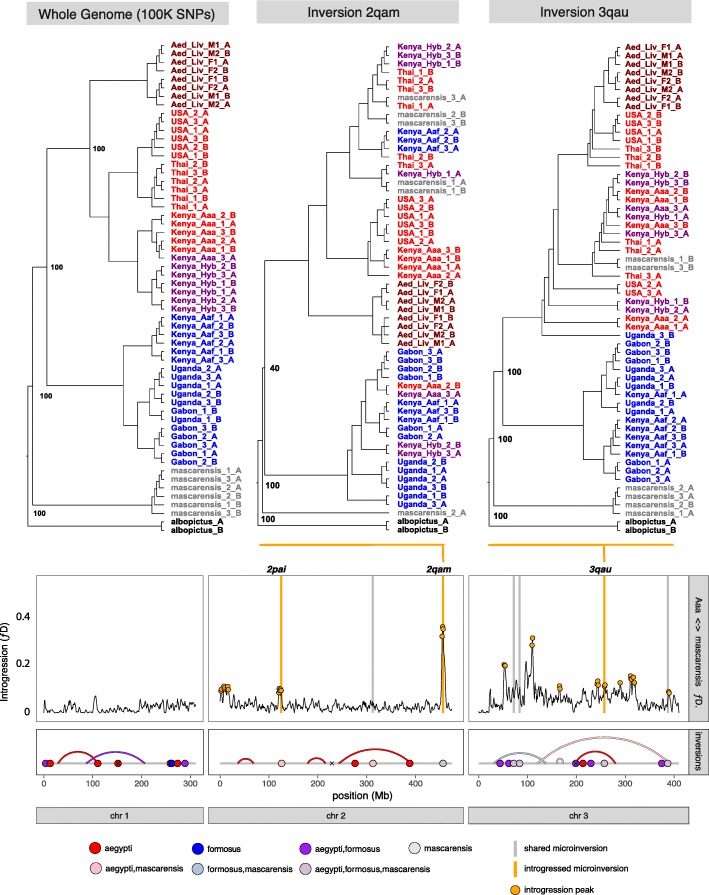
Fig. 4.
Evolutionary history of many microinversions differed from the consensus genome phylogeny; while genome-wide SNP panels showed uniform support for two separate Aaa/Aaf clades and Ae. mascarensis as an outgroup after 100 bootstrap replicates, phylogenies derived from the 1 MB around introgressed inversions (surrounding region showed ƒD > 1.5× IQ range and > 90% of local 200 Mb maximum) can illustrate introgression of haplotypes between diverged forms. Inversions 2qam and 3qau both show Ae. mascarensis haplotypes that cluster within the Aaa clade, indicating introgression from global Aaa populations into the local Ae. mascarensis populations
Inversions in Aedes aegypti do not appear to act as speciation islands
Though inversions have been proposed as speciation islands in other species [9, 11, 12], none of the 32 inversions we detected appear to be playing this role. No inversions were fixed between the Aaa and Aaf subspecies, and in most cases, Fst was not elevated within inversion regions (mean Aaa/Aaf Fst genome-wide, 0.077; inverted regions, 0.080). Microinversions with more than one gene exhibited increased Fst compared to uninverted regions, and four inversions individually showed elevated Fst between subspecies (1pab, 1pac, 1qag, 3qau) (Fig S7); however, two of these (1pac, 1qag) contain no genes; one (1pab) is present only on three haplotypes and only in Kenya; one (3qau) segregates widely across both subspecies. While we cannot rule out 1pab or 3qau being speciation islands, there is little evidence this could derive from inversion-related suppression of recombination.
Regions containing microinversions have introgressed between populations
Comparing the locations of microinversions with the peaks of ƒD, we frequently find that the two overlap. Of the 23 confirmed microinversions, 6 are found within peaks of ƒD indicating that microinversions themselves have introgressed between divergent populations.
The direction of that introgression can also be discerned by examining haplotypes within those inversions. Whereas the genome-wide phylogeny branches into clear Aaa/Aaf clades, phylogenies generated using only SNPs within or proximal to inversions frequently do not. One microinversion (2qam) shows haplotypes from Aaf individuals that are sited within an otherwise distinct Aaa clade; a clear indication of that a haplotype has introgressed from Aaa to Aaf in Rabai. Similarly, microinversion phylogenies indicate that a further 3 inversions (2qam, 3qas, and 3qau) have all introgressed from Aaa into the Ae. mascarensis colony in Mauritius (Fig S8). Introgression between Aaa and Ae. mascarensis exhibited significantly elevated fD within inverted regions (mean fD genome, 0.034; inversion, 0.043; Mann-Whitney P, 2.5e−4) (Table S2).
Though this study is not powered to detect associations of these inversions with specific phenotypes, the maintenance of introgressed inversions in multiple colonies could be because they confer a selective advantage. Examination of the genes within these inversions may therefore indicate potential functional consequences of particular inversion alleles.
We observed 11 microinversions containing no genes within the inverted regions, which are likely to be selectively neutral. A further six inversions contain single genes in which the increased linkage disequilibrium derived from the inversion itself is unlikely to provide any selective advantage (though one of these, 2qam, has introgressed from Aaa into both Aaf and Ae. mascarensis and would be an intriguing target for functional characterization). The remaining 15 inversions contained more than two genes and could facilitate co-adaptation of genes. Intriguingly, inversion 3qau contains ten genes, eight of which comprise a family of odorant-binding proteins, of which two (OBP11/OBP65) have previously been shown to be differentially expressed in zoophilic/anthropophilic colonies [40]. A full list of genes in inversions and proximal regions is in Table S2 and divergent non-synonymous markers in 3qau in Table S3.
Discussion
Using a combination of linked-read and long-read sequencing platforms, we have generated whole-genome sequences of 26 individual Aedes mosquitoes representing the extreme longitudes of the global distribution of the Aaa and Aaf subspecies. These technologies allowed us to detect both single-nucleotide and inversion polymorphisms from the same library and have allowed us to generate a catalogue of inversions based on fully independent detection of structural variants in each individual. This is the first large-scale survey for inversions in this species and a crucial first step in establishing the distribution of these variants in wild populations. It is important to note that the 23 novel microinversions that we have identified would likely not have been detectable by either traditional cytology or short-read sequencing approaches.
The phylogeny we generated based on SNP genotypes supports previous conclusions regarding the origins of the Aaa and Aaf subspecies and supports their genetic divergence [42, 43]; however, we did not detect any inversion that was fixed between Aaa and Aaf colonies. Most inversions segregated widely among colonies despite the large geographic distances separating the founders and elevated Fst was not found within most inversions. Tests for introgression showed that microinversions are commonly found within introgressed regions and that the distribution of microinversions in Kenya derives, at least in part, from introgression between subspecies.
Few of the more than 500 inversion candidates were validated by breakpoint reassembly or long-read sequence (Table 2). Indeed, few were reliably found in both technical replicates of the “Liverpool” samples, indicating that even with the advantage of long-range linking information, reliably calling inversions is challenging in Aedes spp. We cannot rule out the possibility that inversions are more plentiful but repetitive sequence flanking the breakpoints prevents us from identifying or validating them, or that breakpoints remain improperly assembled in the reference assembly as they are in anopheline genomes [45].
Table 2.
Inversion calls
| Inversion | Position | Size | Micro | Prog | Validation |
|---|---|---|---|---|---|
| 1paa | 1:3662097-3696373 | 34276 | * | Long Ranger | Assembly |
| 1pab | 1:12833450-12864379 | 30929 | * | Long Ranger | Long-read align |
| 1pa | 1:28526219-110482104 | 81955885 | GROC-SVs | Long-read align | |
| 1cb | 1:86573078-207047458 | 120474380 | Long Ranger | Long-read align | |
| 1pac | 1:110426504-110463452 | 36948 | * | Long Ranger | Assembly |
| 1qad | 1:151766620-151847533 | 80913 | * | Both | Long-read align |
| 1qae | 1:258805045-258838552 | 33507 | * | GROC-SVs | Long-read align |
| 1qaf | 1:261838607-261877216 | 38609 | * | Long Ranger | Assembly |
| 1qag | 1:273812946-273859121 | 46175 | * | GROC-SVs | Long-read align |
| 1qah | 1:288864540-288902671 | 38131 | * | Long Ranger | Both |
| 2pc | 2:36263327-68515898 | 32252571 | Both | Long-read align | |
| 2pai | 2:125768796-125807158 | 38362 | * | Long Ranger | Long-read align |
| 2pd | 2:178843909-215734436 | 36890527 | GROC-SVs | Long-read align | |
| 2qe | 2:242868946-392168275 | 149299329 | GROC-SVs | Long-read align | |
| 2qaj | 2:276157193-276249519 | 92326 | * | Long Ranger | Long-read align |
| 2qak | 2:312677653-312836936 | 159283 | * | Both | Long-read align |
| 2qal | 2:387870950-387930807 | 59857 | * | Long Ranger | Long-read align |
| 2qam | 2:456306790-456341898 | 35108 | * | Long Ranger | Both |
| 3pf | 3:30056138-138778486 | 108722348 | GROC-SVs | Long-read align | |
| 3pan | 3:43852147-43896847 | 44700 | * | Long Ranger | Long-read align |
| 3pao | 3:61571542-61611075 | 39533 | * | Long Ranger | Long-read align |
| 3pap | 3:72073163-72216512 | 143349 | * | Long Ranger | Long-read align |
| 3paq | 3:83753676-83794150 | 40474 | * | Long Ranger | Long-read align |
| 3cg | 3:119518403-392983952 | 273465549 | Both | Long-read align | |
| 3ph | 3:161230873-171356784 | 10125911 | Both | Long-read align | |
| 3qi | 3:196536178-280271694 | 83735516 | GROC-SVs | Long-read align | |
| 3qar | 3:198751700-198836215 | 84515 | * | Long Ranger | Long-read align |
| 3qas | 3:213245322-213670971 | 425649 | * | Long Ranger | Long-read align |
| 3qat | 3:229688967-229782178 | 93211 | * | Long Ranger | Both |
| 3qau | 3:257058020-257252866 | 194846 | * | Both | Both |
| 3qav | 3:374671141-374714093 | 42952 | * | Long Ranger | Assembly |
| 3qaw | 3:386773996-386844766 | 70770 | * | Long Ranger | Long-read align |
Nevertheless, the size and distribution of the inversions we called are unexpected. Prior studies had indicated large inversions around the centromere of chromosomes 1, as inferred due to the suppression of recombination seen between Aaa and Aaf [37, 38]; Dickson et al. also identified rearrangements of BAC clone markers indicating a pair of pericentromeric inversions on chromosome 3 and a large inversion on the 2p arm proximal to the chromosome 2 centromere and suggested that these rearrangements could be linked to a reduction in fecundity in Senegalese Aaf [39]. While we found inversions that may be consistent with the positions of these rearrangements (1cb, 2pd, 3cg), we did not detect any that were fixed or at high frequency in Aaa, none of these inversions showed elevated Aaa/Aaf divergence, and none was found in Aaf (fixation of these inversions in the reference Aaa strain would be detected as a polymorphism in the Aaf subspecies).
If the absence of large fixed inversions between Aaa and Aaf were confirmed, we would require an alternative explanation for the reduced recombination around the chromosome one centromere in Aaa/Aaf crosses identified by previous studies [38]. The extensive repeat structure could provide one possible mechanism for this reduced recombination. Aedes aegypti has long been characterized as a highly repetitive, short-period interspersion species [46], and differing complements of transposable elements or satellite lengths could act to reduce collinearity. The extent to which microinversions themselves suppress recombination is also not known; while it has been shown previously that large inversions act to suppress recombination up to 1.5 Mb outside of the inversion breakpoints [47], this work has not been applied to microinversions and the extent to which the lack of collinearity suppresses recombination is unknown.
The nature of introgression in Kenya is also instructive. Clear asymmetry is seen in the distribution of AIMS in these colonies with far more Aaf alleles are found in the Aaa colony than vice versa. Similar patterns have been seen in structured populations of both anopheline [48] and culicine [49] mosquitoes where this asymmetric pattern of backcrossing is thought to underlie divergence in the face of extensive hybridization [50]. Both the geographical and genomic distributions of inversions support this conclusion of hybridization between cosmopolitan Aaa and local populations in Kenya or closely related species in Mauritius. Phylogenetic analysis of inversion-linked SNPs further suggests that, although the predominant direction of gene flow in the Kenyan hybrid zone is from Aaf to Aaa, the predominant direction of gene flow within inversions is from synanthropic Aaa populations to Ae. mascarensis and sylvan Aaf populations. This may be indicative of adaptive introgression; however, caution should be taken when inferring that this is due to inversions—particularly as the strongest signal of introgression (2qam) contains only one gene and would derive little or no selective advantage from being sited within an inversion.
Indeed, caution must be taken when interpreting these population genetic signals from colony samples. The reduced haplotypic diversity in colonies relative to wild populations provided a valuable opportunity to validate the novel inversion calling approaches we employed through replicated calls. However, unlike samples taken directly from the field, these samples will be subject to allele loss deriving from the colonization process itself [51]. Signals such as the apparent asymmetric introgression could derive from stochastic loss of haplotypes or from neutral selection acting upon different population sizes [52] (Aaa was rare in this area when colony founders were collected (McBride pers. comm.)). Small founder numbers (Table 1) also require caution when inferring introgression signals within these colonies or the sampled founders are representative of the wider population. While signals such as the introgression of inversion 2qam into both Aaf and Ae. mascarensis populations (colonies that were collected more than 2000 km apart and maintained in different laboratories) are difficult to explain other than via true introgression, confirmation of all of these signals will ultimately require prospective population genetic projects in Aedes aegypti.
Nevertheless, adaptive introgression has been seen in other vector mosquitoes, where it was responsible for the transfer of advantageous traits between sister species. For example, in sympatric populations of Anopheles gambiae and Anopheles coluzzii, insecticide resistance alleles appear to have introgressed between An. gambiae and An. coluzzii [53, 54], and historical introgression of large inversions may also have occurred between more distantly related taxa [14, 55], with the potential to transfer adaptive traits underlying range expansion into xeric environments [17, 56]. Examining genes within introgressed inversions can therefore generate testable theories as to which phenotypes might be under positive selection. In many cases, our microinversions have not captured any genes within the inverted region itself, and are likely to be selectively neutral, or contain only single genes which would gain little advantage from suppression of recombination; however, some have captured more than one gene (Table S2).
The microinversion that has captured the most genes is 3qau, which encompasses 10 genes, eight of which are confirmed or putative odorant-binding proteins (OBPs). OBPs are short (typically < 20 KDA) proteins that are thought to bind and solubilize small hydrophobic molecules and to play some role in olfaction [57, 58]. The mechanism by which they do this is unclear; an OBP is not necessary to activate the odorant receptor complex [59]; instead, their binding of soluble odorants is thought to assist in transportation of odorants to the odorant receptor complex or to buffer olfactory stimuli enabling the olfactory response to function under varying levels of stimuli [60]. Though the specificity of OBPs to each class of odorants is yet to be determined, in two different mosquitoes, OBP1 (Ag/CqOBP1) has been shown to bind to compounds that are associated with oviposition sites [61, 62]. Whether through changes in conformation or expression, there is clear potential for these molecules to affect mosquito behavior and vectorial capacity.
Of the eight OBPs in inversion 3qau, two (OBP11/OBP65) have been demonstrated to have significantly different expression between zoophilic and anthropophilic colonies [40]. Of these two, one has an orthologue in An. gambiae (AaOBP11/AgOBP25) that has been shown to be expressed in mosquito antennae [63]. This inversion has introgressed from wild Aaa populations into the sampled Ae. mascarensis population after secondary contact. That this inversion is linked to increased anthropophily is a tantalizing possibility and one that bears further investigation.
Introgression of such an inversion between sympatric populations would be of more than academic interest. As has been seen within anopheline vectors, inversions can be associated with a wide variety of phenotypes important for vector competence. Even if partial reproductive isolation is maintained by asymmetric introgression, the transfer of inversions between divergent populations in secondary contact zones is likely to generate increased phenotypic plasticity within these regions and reduce our ability to reliably predict phenotypic profiles of vector species. Larval source management requires an understanding of where those larvae are oviposited: a key difference between Aaa and Aaf populations [64] and one that is linked to anthropophily [40]. Transference of anthropophilic biting tendencies to sylvan populations could impact larval control programs and lead to an increase in outbreaks of arboviral diseases. We have not detected any directional introgression from African Aaf into global Aaa populations, suggesting this may be a concern limited to regions of secondary contact between Aaa and Aaf populations. Yet regions of mixed Aaa/Aaf genotypes and increased phenotypic diversity include areas outside sub-Saharan Africa [43, 65] and importation of global Aaa is unlikely to be limited to east Africa and Mauritius, suggesting that this localized concern could become a global one.
Conclusions
We have genotyped chromosomal inversions in eight colonies of Ae. aegypti, representing the extreme latitudes of each subspecies, one location where they live in sympatry, and the outgroup species Aedes mascarensis. Applying a combination of linked-read and long-read methods, we have detected and validated 32 novel inversions. In contrast to anopheline mosquito species in which large, ancient (predating speciation) inversions predominate, we find large numbers of microinversions. Most inversion polymorphisms are shared between subspecies and post-divergence hybridization and genetic introgression has occurred between subspecies and with Ae. mascarensis. If repeated in other regions of the world, this introgression of inversions could affect phenotypic diversity in Ae. aegypti and has the potential to impact dengue control programs.
Methods
Colony sampling
To add to the previously generated linked-read data from the Liverpool colony [34], we selected 8 colonies of Aedes aegypti representing the extreme east and west of both Aaa and Aaf as well as a hybrid zone (Fig. 1). “Pure” Aaa colonies were established from North America (New Orleans, USA) and Asia (Chiang Mai, Thailand) and “pure” Aaf colonies from West Africa (Franceville, Gabon) and East Africa (Uganda). Mosquitoes were also sampled from an Aaa/Aaf hybrid zone in Rabai, Kenya, where the two subspecies are believed to live in sympatry but represent genetically distinct units [43]; three colonies were established from these samples—one Aaa, one Aaf, and one founded from hybrids. An outgroup colony was also founded from a closely related species Aedes mascarensis (Mauritius). Colony sampling dates, locations, number of generations, and founders are given in Table 1.
Library preparation/sequencing
High molecular weight DNA was extracted using the Qiagen MagAttract kit according to the manufacturer’s instructions with minor modifications (rapid vortexing was replaced by inversion and wide-bore pipette tips were used—both to prevent excessive shearing of DNA). DNA extracted from each individual pupa was loaded into a separate lane of the 10X Chromium for barcode tagging of the amplicons, then an Illumina library was prepared. Each resulting library was sequenced with a full lane of Illumina X10 sequencing (~ 120-Gb total output) for a target of 100-fold coverage.
Alignment/genotyping
Sequences were aligned to the reference using BWA via the LongRanger-Align function (longranger v. 2.1.3). Variants were called using GATK HaplotypeCaller (GATK version 3.5.0) and filtered for quality (QD > 5), strand bias (FS < 60), and read position (RankSum < 8). Only biallelic SNPs were used for subsequent analyses. Previously generated 10X library sequence was included from the reference Liverpool colony as described in Matthews et al. [34].
Additional sequence sets were obtained from Genbank for two further outgroup species to allow the determination of derived alleles in our colony samples. Aedes albopictus (SRA project: SRP064281) and Aedes bromeliae (SRA project: SRP092518) were both aligned and called in the same manner. Two resultant callsets were produced: the first with SNPs that could be reliably called across all 8 of our colonies (colony callset), and the second for SNPs that could be called in all colony samples and the two outgroup species (conserved callset).
Chromosomal inversion detection
The full Long Ranger “WGS” pipeline (Long Ranger v.2.1.5) was run on all samples, with memory overrides for both the SNP/INDEL phasing and SV calling stages required due to the high heterozygosity found in these samples. The pipeline was run with the pre-called VCF from the prior variant calling ensuring that the same sites were genotyped and phased in all samples. A second SV calling pipeline, GROC-SVs, was run on the BWA alignments generated for variant calling. Long Ranger was run with both repeatmasked and unmasked references to account for potential TE-associated false positives. Structural variants were compared between each individual and both methods and were merged if they showed a 95% pairwise overlap in position.
Phasing
10X-phased genotypes were also generated via the Long Ranger pipeline. Haplotypes generated by Long Ranger vary in size depending on the level of heterozygosity in the region, since variants that are significantly more distant than our typical molecule size cannot be efficiently phased. In regions where one sample showed a long pair of haplotypes, haplotypes in other samples within the same colony were compared in order of descending size; a haplotype was only considered novel if it showed more than 1 SNP difference per kilobase. Regions that showed less than this degree of divergence from a longer haplotype were also assumed to derive from that same founder haplotype.
After examination of the levels of hybridization within the Rabai colonies, this was also repeated across all nine samples from Rabai, enabling reconstruction of hybrid haplotypes and more detailed examination of the introgressed genotypes.
For the conserved callset, including A. albopictus/A. bromeliae, statistical phasing was performed via SHAPEIT (v2.837) allowing us to determine phylogenies for individual haplotypes.
Validation
Due to the large number of chromosomal inversion candidate regions detected, and the high probability of TE-related false positives, we took an aggressive approach to inversion validation in which independent confirmation of both breakpoints was required for validation. Breakpoint reconstruction took two forms: breakpoints could be reassembled de novo or long-read sequence could be aligned across the inverted breakpoint.
Long-read sequence for the Liverpool colony consisted of PacBio sequence generated during genome assembly [34], while all other colonies were sequenced using an Oxford Nanopore GridIon sequencer. Read N50 differed between the two sequencing runs (PacBio: N50 = 14,307, Nanopore N50 = 6789), most likely as a result of degradation in DNA stocks between PacBio and Nanopore sequencing runs. The longest reads (> 5 kb) were selected within each sequencing type, and competitive alignment was performed using Minimap2 (V2.11 using map-pb/map-ont presets) to a pan-genome sequence containing both the original reference breakpoints and artificially inverted breakpoints (along with the rest of the chromosome with the actual breakpoints masked). Reads were considered to align to the breakpoint if they extended at least 1 KB across the breakpoint on both sides. A second pan-genome was created consisting of 1000 artificial breakpoints that were generated to have a similar complement of repeats and transposable elements to our candidate regions; alignments to these artificial breakpoints were used to calculate the typical level of misalignment in an uninverted region with a false discovery rate of under 1% per breakpoint (1e−4 for both breakpoints): breakpoints with under 10X coverage or more than 2× the inter-quartile range were discarded and those that remained were considered valid if the alignment had more than 37% of long-read alignment to the inverted breakpoint.
Breakpoint assembly was performed using the linked-read-aware Supernova software (v1.1.4). In each case, all reads within 50 kb of the two candidate breakpoints were collected along with all reads linked to that region by at least one 10X barcode. This targeted sequence set was then aligned using Supernova, and “megabubble” sequences (i.e., phased supercontigs) greater than 10 kb in length were realigned to the reference using Minimap2 (v2.11). Those supercontigs aligning to either both upstream or both downstream regions of each breakpoint, with an alignment score above 60, were used to determine true inversions. False discovery rate was determined by running the reassembly script with our artificial inverted regions.
Heterozygosity/Fst/introgression/AIMs
Heterozygosity was calculated via VCFtools (v0.1.14) from the colony callset in 1-Mb windows. Long regions of homozygosity are assumed to be the result of inbreeding within colonies and were identified using the LROH function of VCFtools. Significance of heterozogosity differences between Aaa and Aaf samples was determined by Wilcoxon rank-sum test. Fst (divergence) was calculated between all pairs of colonies via the Fst function within VCFtools. Ancestry-informative markers (AIMs) for Aaa/Aaf were determined based on Fst between the unintrogressed colonies (i.e., Uganda + Gabon vs Thailand + USA); AIMs were chosen if they exhibited Fst > 0.8 and were callable in both colony and conserved datasets. Mann-Whitney tests were used to compare Fst between subspecies within our called inversions and within our artificial inverted regions, as well as between different classes of inversions (intergenic, monogenic, polygenic).
Genome-wide introgression was tested using Patterson’s D [66], and significance was determined by Z-score (2 or more considered significant) after calculation of standard deviation by block jackknifing. Where samples were found to have significant genome-wide “D,” localized introgressed regions were identified using Martin’s ƒD statistic, which controls for regions of low diversity [67]; peaks of introgression were defined as those with ƒD above 1.5× the IQ range and more than 90% of the local 200 Mb maxima. Mann-Whitney tests were used to compare ƒD within and outside microinversions.
Supplementary information
Additional file 1: Figure S1. All Unconfirmed Structural Variants. a) Over 500 inversion candidates were detected by linked-read analysis, of which 32 (9 inversions, 21 microinversions) could be confirmed by breakpoint reassembly or long-read alignment. b) a further 210 insertion and 404 deletion candidates were discovered using linked read analysis, though without a clear method for validation of candidates, these classes of structural variant were not further investigated.
Additional file 2: Figure S2.Fst Between Colonies. Fst values were calculated using vcftools based on the colony SNP set. Elevated Fst values were seen between all groups of samples following colonization, with the highest values between comparisons of USA or Ae. mascarensis colonies. Fst was uniformly lower on chromosome 3 (fig S3b) where the highest degrees of both introgression and inversion sharing were seen.
Additional file 3: Figure S3. Genome-wide Heterozygosity and Long Runs of Homozygosity. Genome-wide heterozygosity values are dominated large blocks of homoygosity due to inbreeding. After removal of these regions using VCFtools LRoH function, heterozygosity was seen to correlate weakly but significantly with time in colony (Pearson’s product-moment correlation r = − 0.19, P < 2.2e− 16) and was found to be lower in all pure Aaa colonies than Aaf (Wilcoxon rank-sum test, P < 2.2e− 16) consistent with the recent global emergence of this clade. While the Liverpool strain is shown on the figure, due to extreme inbreeding and an uncertain number of generations since its foundation in the mid 1930s, this colony was not included in any analyses.
Additional file 4: Figure S4. Bootstrapped Whole-Genome Phylogeny. A maximum parsimony phylogeny was derived from 10,000 genome-wide markers giving the background phylogeny to which we compared the inverted regions. Strong bootstrap support was shown for the separation between Aaa / Aaf, Ae. aegypti / Ae. mascarensis, and for the isolation of the Ae aegypti Liverpool colony.
Additional file 5: Figure S5. All Applied Introgression Tests. Patterson’s D was used to assess introgression between a potential introgressing clade and pair of putative sister clades, with an outgroup used to determine derived alleles for the other three clades; 5a) introgression into individual colonies was compared by applying Patterson’s D statistic to each colony as compared to the other two colonies of the same subspecies (e.g we examined Aaf introgression into Kenya_Aaa vs Thai_Aaa + USA_Aaa using Ae. mascarensis as the outgroup). Specific tests applied are given in figures i,iii,v, and the results for each shown in figs ii,iv,vi. Significance was tested by block jackknifing. 5b) The same test was applied to introgression between mascarensis and any individual population (i-iv), as well as between Ae mascarensis and all Aaf or Aaa populations (v-vi); all inter-specific tests used Ae albopictus as the outgroup. 5c) Significant introgression was detected in between Aaf and Kenya_Aaa, between Aaa and Kenya_Aaf, indicating bidirectional introgression between the two subspecies in Kenya. Martin’s fD statistic was applied to identify specific introgressed loci in these two populations and was also applied to testing for Aaf introgression into the ‘hybrid’ population in this region. 5d) significant introgression was also detected between Ae. mascarensis and global Aaa populations. Notably one fD peaks at the distal end of chromosome 2q appeared to have introgressed between Aaa and Kenya_Aaf and Aaa and Ae mascarensis.
Additional file 6: Figure S6. Aegypti / Formosus Ancestrally-Informative Markers, Liverpool colony. Contrary to expectation, the Ae. aegypti Liverpool strain used for sequencing was not a clear Aaa strain, but instead demonstrated evidence of both Aaa and Aaf alleles. This is consistent with a west African origin and prior evidence from Crawford et al. of forms ancestral to Aae/Aaf in this region [40].
Additional file 7: Figure S7. Elevation of Fst Within Inversions. Fst between pure Aaa and Aaf colonies was calculated in 1mb windows across the genome and windows that contained either an entire microinverisons or a larger inversion breakpoint were compared to a set of artificial inversions constructed of ‘breakpoints’ containing similar levels of TEs and repeats. Fst was compared between categories via Wilcoxon rank-sum test. A) Of the 32 inversions 4 (shown in red) showed significantly elevated levels of Fst indicating a higher degree of differentiation between subspecies; three on chromosome one, the fourth inversion showing elevated Fst is 3Qau, containing 10 genes, 8 of which are odorant binding proteins previously seen to be differentiated between the two subspecies. B) categories of inversions: macroinversions, microinversions without genes, with one gene, and with many genes, were compared to artificial inversions; while polygenic inversions did show elevated Fst compared to artificial inversions, there was no significant difference found when comparing 0 and 1-gene microinversions to polygenic microinversions.
Additional file 8: Figure S8. Within-Inversion Phylogenies. Maximum parsimony phylogenies were derived from the 1 M region surrounding each microinversion in order to establish the unique evolutionary history of these regions. In many cases haplotypes did not cluster into clean sub-species clades, but instead indicated extensive introgression of haplotypes from global Aaa populations into local sylvatic forms.
Additional file 9: Table S1. fD elevation in inverted regions.
Additional file 10: Table S2. Microinversion genes.
Additional file 11: Table S3. Non-synonymous variants: Inversion 3qau.
Acknowledgements
We thank Diego Ayala (IRD) and Le Centre International de Recherches Médicales de Franceville (CIRMF) for the supply of the colony material from Gabon. The authors would like to thank Jeff Powell (Yale) and Carolyn McBride (Princeton) for supplying vital colony material for this paper and for their valuable comments on the manuscript.
Authors’ contributions
DEN, SNR, IS, ZT, and MS designed the study; SNR analyzed and interpreted the data; MS and AS validated inversion candidates; SNR wrote the manuscript; DEN was a major contributor to the manuscript; ZT, IS, and MS provided comments and edits on the manuscript. The authors read and approved the final manuscript.
Funding
This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Grant Number U19AI110818 to the Broad Institute. SNR was further supported by a Broad Next10 Grant.
Availability of data and materials
All linked-read and long-read libraries generated for this study were deposited with SRA under project ID: PRJNA559933 [68]. Linked-read and PacBio libraries used for calling and validation of the “Liverpool” samples were previously deposited under project ID PRJNA318737 [69].
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12915-020-0757-y.
References
- 1.Sturtevant AH. A case of rearrangement of genes in Drosophila. Proc Natl Acad Sci U S A. 1921;7(8):235–237. doi: 10.1073/pnas.7.8.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wellenreuther M, Bernatchez L. Eco-evolutionary genomics of chromosomal inversions. Trends Ecol Evol. 2018;33(6):427–440. doi: 10.1016/j.tree.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Dobzhansky T, Sturtevant AH. Inversions in the chromosomes of Drosophila pseudoobscura. Genetics. 1938;23(1):28–64. doi: 10.1093/genetics/23.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krimbas KV, Powell JR. Drosophila inversion polymorphism. UK: CRC Press; 1992.
- 5.Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484(7392):55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson MJ, Jiggins CD. Supergenes and their role in evolution. Heredity (Edinb) 2014;113(1):1–8. doi: 10.1038/hdy.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White MJD. Modes of speciation. San Francisco: W. H. Freeman and Co.; 1978.
- 8.Dobzhansky T. Genetics of natural populations; a response of certain gene arrangements in the third chromosome of Drosophila pseudoobscura to natural selection. Genetics. 1947;32(2):142–160. doi: 10.1093/genetics/32.2.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noor MA, Grams KL, Bertucci LA, Reiland J. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci U S A. 2001;98(21):12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dagilis AJ, Kirkpatrick M. Prezygotic isolation, mating preferences, and the evolution of chromosomal inversions. Evolution (N Y) 2016;70(7):1465–1472. doi: 10.1111/evo.12954. [DOI] [PubMed] [Google Scholar]
- 11.Fuller ZL, Leonard CJ, Young RE, Schaeffer SW, Phadnis N. Ancestral polymorphisms explain the role of chromosomal inversions in speciation. PLoS Genet. 2018;14(7):e1007526. doi: 10.1371/journal.pgen.1007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruickshank TE, Hahn MW. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol. 2014;23(13):3133–3157. doi: 10.1111/mec.12796. [DOI] [PubMed] [Google Scholar]
- 13.Ayala D, Fontaine MC, Cohuet A, Fontenille D, Vitalis R, Simard F. Chromosomal inversions, natural selection and adaptation in the malaria vector Anopheles funestus. Mol Biol Evol. 2011;28(1):745–758. doi: 10.1093/molbev/msq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontaine MC, Pease JB, Steele A, Waterhouse RM, Neafsey DE, Sharakhov IV, et al. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science. 2014;347(6217):1258524. doi: 10.1126/science.1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coluzzi M, Sabatini A, della Torre A, Di Deco MA, Petrarca V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002;298(5597):1415–1418. doi: 10.1126/science.1077769. [DOI] [PubMed] [Google Scholar]
- 16.Simard F, Ayala D, Kamdem GC, Pombi M, Etouna J, Ose K, et al. Ecological niche partitioning between Anopheles gambiae molecular forms in Cameroon: the ecological side of speciation. BMC Ecol. 2009;9:17. 10.1186/1472-6785-9-17. [DOI] [PMC free article] [PubMed]
- 17.Fouet C, Gray E, Besansky NJ, Costantini C. Adaptation to aridity in the malaria mosquito Anopheles gambiae: chromosomal inversion polymorphism and body size influence resistance to desiccation. PLoS One. 2012;7(4):e34841. doi: 10.1371/journal.pone.0034841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng C, White BJ, Kamdem C, Mockaitis K, Costantini C, Hahn MW, et al. Ecological genomics of Anopheles gambiae along a latitudinal cline: a population-resequencing approach. Genetics. 2012;190(4):1417–1432. doi: 10.1534/genetics.111.137794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans R Soc Trop Med Hyg. 1979;73(5):483–497. doi: 10.1016/0035-9203(79)90036-1. [DOI] [PubMed] [Google Scholar]
- 20.Main BJ, Lee Y, Ferguson HM, Kreppel KS, Kihonda A, Govella NJ, et al. The genetic basis of host preference and resting behavior in the major African malaria vector, Anopheles arabiensis. PLoS Genet. 2016;12(9):e1006303. 10.1371/journal.pgen.1006303. [DOI] [PMC free article] [PubMed]
- 21.Manoukis NC, Powell JR, Toure MB, Sacko A, Edillo FE, Coulibaly MB, et al. A test of the chromosomal theory of ecotypic speciation in Anopheles gambiae. Proc Natl Acad Sci U S A. 2008;105(8):2940–2945. doi: 10.1073/pnas.0709806105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanford MR, Ramsay S, Cornel AJ, Marsden CD, Norris LC, Patchoke S, et al. A preliminary investigation of the relationship between water quality and Anopheles gambiae larval habitats in western Cameroon. Malar J. 2013;12(1):225. doi: 10.1186/1475-2875-12-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brooke BD, Hunt RH, Coetzee M. Resistance to dieldrin + fipronil assorts with chromosome inversion 2La in the malaria vector Anopheles gambiae. Med Vet Entomol. 2000;14(2):190–194. doi: 10.1046/j.1365-2915.2000.00222.x. [DOI] [PubMed] [Google Scholar]
- 24.Petrarca V, Beier JC. Intraspecific chromosomal polymorphism in the Anopheles gambiae complex as a factor affecting malaria transmission in the Kisumu area of Kenya. Am J Trop Med Hyg. 1992;46(2):229–237. doi: 10.4269/ajtmh.1992.46.229. [DOI] [PubMed] [Google Scholar]
- 25.Riehle MM, Bukhari T, Gneme A, Guelbeogo WM, Coulibaly B, Fofana A, et al. The Anopheles gambiae 2La chromosome inversion is associated with susceptibility to Plasmodium falciparum in Africa. Elife. 2017;23:6. doi: 10.7554/eLife.25813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraemer MUG, Sinka ME, Duda KA, Mylne A, Shearer FM, Barker CM, et al. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Elife. 2015;4:e08347. doi: 10.7554/eLife.08347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.W.H.O. Global Strategy for dengue prevention and control, 2012–2020. Switzerland: World Health Organization; 2012.
- 29.Faria NR, Azevedo RS d S, Kraemer MUG, Souza R, Cunha MS, Hill SC, et al. Zika virus in the Americas: early epidemiological and genetic findings. Science. 2016;352(6283):345–349. doi: 10.1126/science.aaf5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma GP, Mittal OP, Chaudhry S, Pal V. A preliminary map of the salivary gland chromosomes of Aedes (stegomyia) aegypti (Culicadae, Diptera) Cytobios. 1978;22(87–88):169–178. [PubMed] [Google Scholar]
- 31.Campos J, Andrade CFS, Recco-Pimentel SM. A technique for preparing polytene chromosomes from Aedes aegypti (Diptera, Culicinae) Mem Inst Oswaldo Cruz. 2003;98(3):387–390. doi: 10.1590/S0074-02762003000300017. [DOI] [PubMed] [Google Scholar]
- 32.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287(5461):2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 33.Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298(5591):129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 34.Matthews BJ, Dudchenko O, Kingan SB, Koren S, Antoshechkin I, Crawford JE, et al. Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature. 2018;563(7732):501–507. doi: 10.1038/s41586-018-0692-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medvedev P, Stanciu M, Brudno M. Computational methods for discovering structural variation with next-generation sequencing. Nat Methods. 2009;6(11):S13–S20. doi: 10.1038/nmeth.1374. [DOI] [PubMed] [Google Scholar]
- 36.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12(5):363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macdonald WW, Sheppard PM. Cross-over values in the sex chromosomes of the mosquito Aedes aegypti and evidence of the presence of inversions. Ann Trop Med Parasitol. 1965;59:74–87. doi: 10.1080/00034983.1965.11686285. [DOI] [PubMed] [Google Scholar]
- 38.Bernhardt SA, Blair C, Sylla M, Bosio C, Black WC, IV, Black WC. Evidence of multiple chromosomal inversions in Aedes aegypti formosus from Senegal. Insect Mol Biol. 2009;18(5):557–569. doi: 10.1111/j.1365-2583.2009.00895.x. [DOI] [PubMed] [Google Scholar]
- 39.Dickson LB, Sharakhova MV, Timoshevskiy VA, Fleming KL, Caspary A, Sylla M, et al. Reproductive incompatibility involving Senegalese Aedes aegypti (L) is associated with chromosome rearrangements. PLoS Negl Trop Dis. 2016;10(4):1–28. doi: 10.1371/journal.pntd.0004626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McBride CS, Baier F, Omondi AB, Spitzer SA, Lutomiah J, Sang R, et al. Evolution of mosquito preference for humans linked to an odorant receptor. Nature. 2014;515(7526):222–227. doi: 10.1038/nature13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tabachnick WJ. Evolutionary genetics and arthropod-borne disease: the yellow fever mosquito. Am Entomol. 1991;37(1):14–26. doi: 10.1093/ae/37.1.14. [DOI] [Google Scholar]
- 42.Crawford JE, Alves JM, Palmer WJ, Day JP, Sylla M, Ramasamy R, et al. Population genomics reveals that an anthropophilic population of Aedes aegypti mosquitoes in West Africa recently gave rise to American and Asian populations of this major disease vector. BMC Biol. 2017;15(1):16. doi: 10.1186/s12915-017-0351-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gloria-Soria A, Ayala D, Bheecarry A, Calderon-Arguedas O, Chadee DD, Chiappero M, et al. Global genetic diversity of Aedes aegypti. Mol Ecol. 2016;25(21):5377–5395. doi: 10.1111/mec.13866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Powell JR, Evans BR. How much does inbreeding reduce heterozygosity? Empirical results from aedes aegypti. Am J Trop Med Hyg. 2017;96:157–58. doi: 10.4269/ajtmh.16-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lobo NF, Sangaré DM, Regier AA, Reidenbach KR, Bretz DA, Sharakhova MV, et al. Breakpoint structure of the Anopheles gambiae 2Rb chromosomal inversion. Malar J. 2010;9(1):293. doi: 10.1186/1475-2875-9-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cockburn AF, Mitchell SE. Repetitive DNA interspersion patterns in Diptera. Arch Insect Biochem Physiol. 1989;10(2):105–113. doi: 10.1002/arch.940100202. [DOI] [Google Scholar]
- 47.Stevison LS, Hoehn KB, Noor MAF. Effects of inversions on within- and between-species recombination and divergence. Genome Biol Evol. 2011;3(0):830–841. doi: 10.1093/gbe/evr081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marsden CD, Lee Y, Nieman CC, Sanford MR, Dinis J, Martins C, et al. Asymmetric introgression between the M and S forms of the malaria vector, Anopheles gambiae, maintains divergence despite extensive hybridization. Mol Ecol. 2011;20(23):4983–4994. doi: 10.1111/j.1365-294X.2011.05339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gomes B, Sousa CA, Novo MT, Freitas FB, Alves R, Côrte-Real AR, et al. Asymmetric introgression between sympatric molestus and pipiens forms of Culex pipiens (Diptera: Culicidae) in the Comporta region, Portugal. BMC Evol Biol. 2009;9(1):262. doi: 10.1186/1471-2148-9-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee Y, Marsden CD, Norris LC, Collier TC, Main BJ, Fofana A, et al. Spatiotemporal dynamics of gene flow and hybrid fitness between the M and S forms of the malaria mosquito, Anopheles gambiae. Proc Natl Acad Sci U S A. 2013;110:19854–9. 10.1073/pnas.1316851110. [DOI] [PMC free article] [PubMed]
- 51.Gloria-Soria A, Soghigian J, Kellner D, Powell JR. Genetic diversity of laboratory strains and implications for research: the case of Aedes aegypti. PLoS Negl Trop Dis. 2019;13(12):e0007930. doi: 10.1371/journal.pntd.0007930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He C, Liang D, Zhang P. Asymmetric Distribution of Gene Trees Can Arise Under Purifying Selection if Differences in Population Size Exist. Gojobori J, editor. Mol Biol Evol. 2020;37:881–92. 10.1093/molbev/msz232. [DOI] [PubMed]
- 53.Miles A, Harding NJ, Bottà G, Clarkson CS, Antão T, Kozak K, et al. Genetic diversity of the African malaria vector Anopheles gambiae. Nature. 2017;552(7683):96. doi: 10.1038/nature24995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clarkson CS, Weetman D, Essandoh J, Yawson AE, Maslen G, Manske M, et al. Adaptive introgression between Anopheles sibling species eliminates a major genomic island but not reproductive isolation. Nat Commun. 2014;5:4248. doi: 10.1038/ncomms5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neafsey D, Waterhouse R, Abai M, Aganezov S, Alekseyev M, Allen J, et al. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science (80- ) 2015;347(6217):1258522. doi: 10.1126/science.1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White BJ, Cheng C, Sangaré D, Lobo NF, Collins FH, Besansky NJ. The population genomics of trans-specific inversion polymorphisms in Anopheles gambiae. Genetics. 2009;183(1):275–288. doi: 10.1534/genetics.109.105817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58(1):373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- 58.Brito NF, Moreira MF, Melo ACA. A look inside odorant-binding proteins in insect chemoreception. J Insect Physiol. 2016;95:51–65. doi: 10.1016/j.jinsphys.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 59.Gomez-Diaz C, Reina JH, Cambillau C, Benton R. Ligands for pheromone-sensing neurons are not conformationally activated odorant binding proteins. PLoS Biol. 2013;11(4):e1001546. doi: 10.1371/journal.pbio.1001546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Larter NK, Sun JS, Carlson JR. Organization and function of Drosophila odorant binding proteins. Elife. 2016;15:5. doi: 10.7554/eLife.20242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pelletier J, Guidolin A, Syed Z, Cornel AJ, Leal WS. Knockdown of a mosquito odorant-binding protein involved in the sensitive detection of oviposition attractants. J Chem Ecol. 2010;36(3):245–248. doi: 10.1007/s10886-010-9762-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Biessmann H, Andronopoulou E, Biessmann MR, Douris V, Dimitratos SD, Eliopoulos E, et al. The Anopheles gambiae odorant binding protein 1 (AgamOBP1) mediates indole recognition in the antennae of female mosquitoes. PLoS One. 2010;5(3):e9471. doi: 10.1371/journal.pone.0009471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mastrobuoni G, Qiao H, Iovinella I, Sagona S, Niccolini A, Boscaro F, et al. A proteomic investigation of soluble olfactory proteins in Anopheles gambiae. PLoS One. 2013;8(11):e75162. doi: 10.1371/journal.pone.0075162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lounibos LP. Habitat segregation among African treehole mosquitoes. Ecol Entomol. 1981;6(2):129–154. doi: 10.1111/j.1365-2311.1981.tb00601.x. [DOI] [Google Scholar]
- 65.Mangudo C, Aparicio JP, Gleiser RM. Tree holes as larval habitats for Aedes aegypti in urban, suburban and forest habitats in a dengue affected area. Bull Entomol Res. 2015;105(06):679–684. doi: 10.1017/S0007485315000590. [DOI] [PubMed] [Google Scholar]
- 66.Durand EY, Patterson N, Reich D, Slatkin M. Testing for ancient admixture between closely related populations. Mol Biol Evol. 2011;28(8):2239–2252. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin SH, Davey JW, Jiggins CD. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol Biol Evol. 2015;32(1):244–257. doi: 10.1093/molbev/msu269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Redmond SN, Sharma A, Sharakhov I, Tu Z, Sharakhova M, Neafsey DE, et al. Aedes aegypti global structural variant discovery. Short Read Archive [PRJNA55993]. Available from: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA559933/.
- 69.Matthews BJ, Dudchenko O, Kingan SB, Koren S, Antoshechkin I, Crawford JE, et al. Aedes aegypti Genome Working Group. Short Read Archive [PRJNA318737]. Available from: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA318737.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. All Unconfirmed Structural Variants. a) Over 500 inversion candidates were detected by linked-read analysis, of which 32 (9 inversions, 21 microinversions) could be confirmed by breakpoint reassembly or long-read alignment. b) a further 210 insertion and 404 deletion candidates were discovered using linked read analysis, though without a clear method for validation of candidates, these classes of structural variant were not further investigated.
Additional file 2: Figure S2.Fst Between Colonies. Fst values were calculated using vcftools based on the colony SNP set. Elevated Fst values were seen between all groups of samples following colonization, with the highest values between comparisons of USA or Ae. mascarensis colonies. Fst was uniformly lower on chromosome 3 (fig S3b) where the highest degrees of both introgression and inversion sharing were seen.
Additional file 3: Figure S3. Genome-wide Heterozygosity and Long Runs of Homozygosity. Genome-wide heterozygosity values are dominated large blocks of homoygosity due to inbreeding. After removal of these regions using VCFtools LRoH function, heterozygosity was seen to correlate weakly but significantly with time in colony (Pearson’s product-moment correlation r = − 0.19, P < 2.2e− 16) and was found to be lower in all pure Aaa colonies than Aaf (Wilcoxon rank-sum test, P < 2.2e− 16) consistent with the recent global emergence of this clade. While the Liverpool strain is shown on the figure, due to extreme inbreeding and an uncertain number of generations since its foundation in the mid 1930s, this colony was not included in any analyses.
Additional file 4: Figure S4. Bootstrapped Whole-Genome Phylogeny. A maximum parsimony phylogeny was derived from 10,000 genome-wide markers giving the background phylogeny to which we compared the inverted regions. Strong bootstrap support was shown for the separation between Aaa / Aaf, Ae. aegypti / Ae. mascarensis, and for the isolation of the Ae aegypti Liverpool colony.
Additional file 5: Figure S5. All Applied Introgression Tests. Patterson’s D was used to assess introgression between a potential introgressing clade and pair of putative sister clades, with an outgroup used to determine derived alleles for the other three clades; 5a) introgression into individual colonies was compared by applying Patterson’s D statistic to each colony as compared to the other two colonies of the same subspecies (e.g we examined Aaf introgression into Kenya_Aaa vs Thai_Aaa + USA_Aaa using Ae. mascarensis as the outgroup). Specific tests applied are given in figures i,iii,v, and the results for each shown in figs ii,iv,vi. Significance was tested by block jackknifing. 5b) The same test was applied to introgression between mascarensis and any individual population (i-iv), as well as between Ae mascarensis and all Aaf or Aaa populations (v-vi); all inter-specific tests used Ae albopictus as the outgroup. 5c) Significant introgression was detected in between Aaf and Kenya_Aaa, between Aaa and Kenya_Aaf, indicating bidirectional introgression between the two subspecies in Kenya. Martin’s fD statistic was applied to identify specific introgressed loci in these two populations and was also applied to testing for Aaf introgression into the ‘hybrid’ population in this region. 5d) significant introgression was also detected between Ae. mascarensis and global Aaa populations. Notably one fD peaks at the distal end of chromosome 2q appeared to have introgressed between Aaa and Kenya_Aaf and Aaa and Ae mascarensis.
Additional file 6: Figure S6. Aegypti / Formosus Ancestrally-Informative Markers, Liverpool colony. Contrary to expectation, the Ae. aegypti Liverpool strain used for sequencing was not a clear Aaa strain, but instead demonstrated evidence of both Aaa and Aaf alleles. This is consistent with a west African origin and prior evidence from Crawford et al. of forms ancestral to Aae/Aaf in this region [40].
Additional file 7: Figure S7. Elevation of Fst Within Inversions. Fst between pure Aaa and Aaf colonies was calculated in 1mb windows across the genome and windows that contained either an entire microinverisons or a larger inversion breakpoint were compared to a set of artificial inversions constructed of ‘breakpoints’ containing similar levels of TEs and repeats. Fst was compared between categories via Wilcoxon rank-sum test. A) Of the 32 inversions 4 (shown in red) showed significantly elevated levels of Fst indicating a higher degree of differentiation between subspecies; three on chromosome one, the fourth inversion showing elevated Fst is 3Qau, containing 10 genes, 8 of which are odorant binding proteins previously seen to be differentiated between the two subspecies. B) categories of inversions: macroinversions, microinversions without genes, with one gene, and with many genes, were compared to artificial inversions; while polygenic inversions did show elevated Fst compared to artificial inversions, there was no significant difference found when comparing 0 and 1-gene microinversions to polygenic microinversions.
Additional file 8: Figure S8. Within-Inversion Phylogenies. Maximum parsimony phylogenies were derived from the 1 M region surrounding each microinversion in order to establish the unique evolutionary history of these regions. In many cases haplotypes did not cluster into clean sub-species clades, but instead indicated extensive introgression of haplotypes from global Aaa populations into local sylvatic forms.
Additional file 9: Table S1. fD elevation in inverted regions.
Additional file 10: Table S2. Microinversion genes.
Additional file 11: Table S3. Non-synonymous variants: Inversion 3qau.
Data Availability Statement
All linked-read and long-read libraries generated for this study were deposited with SRA under project ID: PRJNA559933 [68]. Linked-read and PacBio libraries used for calling and validation of the “Liverpool” samples were previously deposited under project ID PRJNA318737 [69].