Abstract
The endosperm cell walls of mature coffee seeds accumulate large amounts of mannan storage polysaccharides, which serve as nutrient reserve for embryo and contribute to beverage quality. Our study investigated the evolutionary patterns of key galactomannan (GM) biosynthesis genes using d N/d S ratio, synteny, and phylogenetic analysis and detected heterogeneity in rate of evolution among gene copies. Selection ratio index revealed evidence of positive selection in the branch editing gene Coffea canephora alpha (α) galactosidase (Cc‐alpha Gal) at Cc11_g15950 copy (ω = 1.12), whereas strong purifying selection on deleterious mutations was observed in the Coffea canephora uridine diphosphate (UDP)‐glucose 4′‐epimerase (Cc‐UG4E) and Coffea canephora mannose‐1P guanylytransferase (Cc‐MGT) genes controlling the crucial nucleotide carbon sugar building blocks flux in the pathway. Relatively low sequence diversity and strong syntenic linkages were detected in all GM pathway genes except in Cc‐alpha Gal, which suggests a correlation between selection pressure and nucleotide diversity or synteny analysis. In addition, phylogenetic analysis revealed independent evolution or expansion of GM pathway genes in different plant species, with no obvious inferable clustering patterns according to either gene family or congruent with evolutionary plants lineages tested due to high dynamic nature and specific biochemical cell wall modification requirements. Altogether, our study shows a significant high rate of evolutionary variation among GM pathway genes in the diploid C. canephora and demonstrates the inherent variation in evolution of gene copies and their potential role in understanding selection rates in a homogenously connected metabolic pathway.
Keywords: Coffea canephora, galactomannan, nucleotide diversity, positive selection, purifying selection, synonymous and nonsynonymous substitution
This study investigated the evolutionary patterns of key galactomannan biosynthesis genes using selection ratio index, synteny, and phylogenetic analysis and detected heterogeneity in rate of evolution among gene copies.

1. INTRODUCTION
Evolutionary studies have shown that plant genomes tend to have high rate of evolution as compared with animals due to polyploidization, local duplication events, and high retention rates of duplicate genes (Jeffrey, 2002). Gene duplication enables duplicated members to evolve under varied selective constraint (Panchy, Lehti‐Shiu, & Shiu, 2016), leading to evolutionary rate variation among copies as copies tend to undergo different selection pressure due to their contribution to fitness (Kimura, 1977; Rausher, Lu, & Meyer, 2008). Homogenously connected metabolic pathways are increasingly being used to clarify selection and evolutionary rate dynamics because genes operate as connected components based on their position, flux control, and downstream effects within the pathway (Rausher, Miller, & Tiffin, 1999). Pathway architecture has been shown to affect protein evolutionary rates (Greenberg, Stockwell, & Clark, 2008), for example, increased d N for downstream enzymes have previously been reported in carotenoid, gibberellin, and anthocyanin pathway genes (Livingstone & Anderson, 2009; Rausher et al., 1999; Yang, Zhang, & Ge, 2009). Thus, metabolic networks create the potential for predicting differential evolutionary rates as a function of the network properties. For example, low evolutionary rates in upstream genes have been reported due to their flux control properties (Vitkup, Kharchenko, & Wagner, 2006). In addition to pathway properties, rapid evolutionary rate pattern has been reported in regulatory genes than in structural genes (Purugganan & Wessler, 1994; Tucker & Lundrigan 1993), and in some plant lineages as a result of environmental factors (Pierce & Crawford, 1997).
Cell wall storage polysaccharides (CWSPs) are mainly composed of mannans, glucomannans, galactomannans, xyloglucans, and galactans. These polysaccharides structurally vary in backbone chain, branching residue and nucleotide sugars, and their biosynthesis requires different hydrolytic enzymes (Buckeridge & Reid, 1996). CWSPs of the mannan family are reserved in considerable amount as nutrient source for germinating embryo in endosperm of many albuminous monocot and dicot seed plants, such as date palm, guar, and coffee (Buckeridge, 2010). In addition, the mannan CWSPs enable seeds to retain living cellular endosperm at mature, confer seed firmness, and contribute to water buffering during drought (Buckeridge, 2010; Horbowicz & Obendorf, 1994). The galactomannans (GMs) structure consist of a linear mannose backbone chain with galactose side chains which vary in mannose: galactose ratio among species, for example, 1:1 and 20:1 in fenugreek and tobacco, respectively (Reid, Edwards, Dickson, Scott, & Gidley, 2003). The mannose/galactose ratio variation in legumes subfamilies has been shown to reflect systematics and evolutionary pattern in their adaptation to tropical and temperate environments. For example, tropical legumes exhibit high content of galactomannan as well as mannose/galactose ratio, which could be used as taxonomic markers (Buckeridge & Reid 1996; Polhill et al.,; Siol, Wright, & Barrett, 2010).
The endosperm of coffee seeds predominantly accumulates galactomannans, which account for over 50% of all CWSPs (Redgwell & Fischer, 2006). The biosynthetic pathway of galactomannans has been well characterized in plants (Dhugga, Barreiro, & Whitten, 2004; Edwards, Scott, Gidley, & Reid, 1992; Joersbo, Marcussen, & Brunstedt, 2001; Pré, Caillet, Sobilo, & McCarthy, 2008) and involves genes encoding mannose‐1P guanylytransferase (MGT), uridine diphosphate (UDP)‐glucose 4′‐epimerase (UG4E), mannan synthase (ManS), galactomannan galactosyltransferase (GMGT), and α‐galactosidase (α‐Gal) (Figure 1). MGT catalyzes the conversion of Mannose‐1P to GDP‐Mannose (GDP‐Man), which is subsequently converted to 1,4‐β‐Mannan by mannan synthase (ManS), whereas, UG4E catalyzes the conversion of UDP‐glucose to UDP‐galactose (UDP‐Gal). GMGT converts 1,4‐β‐Mannan to galactomannan through catalyzing the formation of galactose side branch from UDP‐Gal, and α‐Gal is responsible for the regulation of postdepositional substitution rate of galactose side branch.
Figure 1.
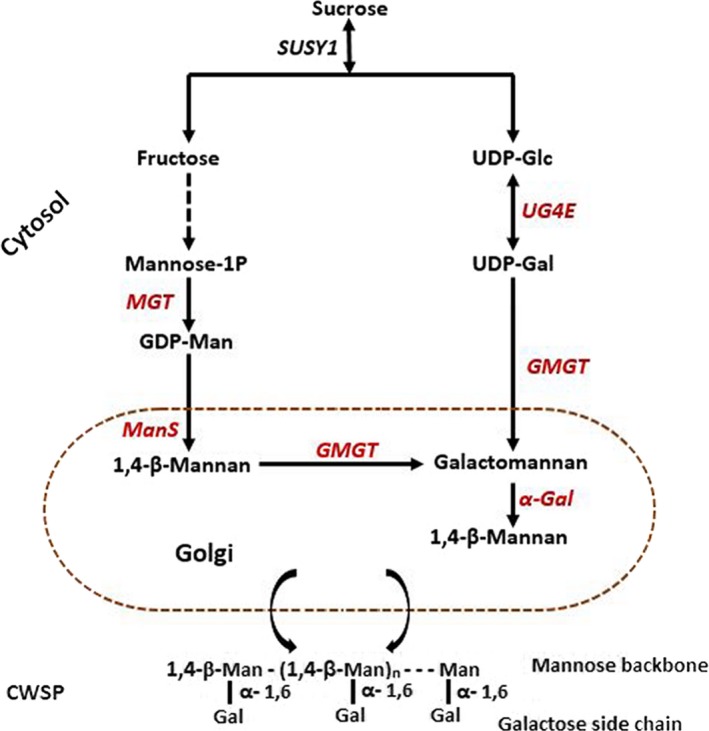
Simplified Galactomannan biosynthetic pathway in coffee seed
Coffee belongs to the Rubiaceae family, the genus Coffea with approximately 124 species. It is a tropical plant (Figure 2) with Coffea arabica and Coffea canephora as the two widely cultivated species. The enormous coffee endosperm which constitutes approximately 99% of total seeds mass along with its ability to accumulate large amount of galactomannans makes it a suitable model plant for investigating metabolism as well as the evolution of galactomannan pathway genes. The structure of the diploid C. canephora genome suggests that it has not undergone whole‐genome duplication (WGD) since triplication origin from core eudicots, and gene expansion is mainly attributed to local duplication (Denoeud et al., 2014). For example, tandem duplication in caffeine metabolic pathway genes has been implicated in convergent adaptive evolution in coffee. In this study, we investigated the evolutionary patterns of galactomannan biosynthesis genes in coffee using selection pressure, synteny, and phylogenetic analysis with an aim of demonstrating variation in evolution using multiple‐copy analysis in C. canephora.
Figure 2.

Coffee plant with mature cherries
2. MATERIALS AND METHODS
2.1. Plant material, DNA extraction, amplification, and sequencing
A total of 23 C. canephora including 18 cultivated and five wild accessions used in this study are maintained at Spice and Beverage Research Institute of Chinese Academy of Tropical Agricultural Sciences (Hainan, China). Cultivated accessions were selected based on our previously published polymorphic data (Ogutu et al., 2016; Yan et al., 2019). Seeds were randomly collected at mature stage, immediately frozen in liquid nitrogen, and then stored at −80°C until use. Total DNA was isolated from twenty‐three freshly frozen coffee seeds using our previously described modified protocol (Ogutu et al., 2016). PCR primers were designed for specific sequencing regions (Table S1). Amplification was carried out in a 50 μl reaction volume on a GeneAmp PCR System 9700 (ABI), containing a 5 μl 10 Ex‐PCR buffer (100 mM Tris‐HCl pH 8.8 at 25°C; 500 MmKCl, 0.8% [v/v] Nonidet), 4 μl 10 m MdNTPs, 1.5 μl 10 μM of each primer, 0.5 μl Ex‐Taq DNA polymerase (5 U/μl) (Sangon), 3 μl sample DNA, 34.5 μl ddH2O. PCR program was optimized as follows: an initial denaturing for 4 min at 95°C, followed by 40 cycles of 94°C for 30 s, 52–61°C Tm for 40 s, and 72°C for 2:30 min; and a final extension at 72°C for 10 min. PCR products were sequenced directly after separation on a 1% agarose gels stained with GoldView™ (ZomanBio) and purification using TIANgel Midi Purification kit (TIANGEN Biotech). All DNA sequences have been submitted to the GenBank databases with BankIt accession numbers:‐ BankIt2304042: MN960694–MN960808; BankIt2304536: MN960809–MN960900, BankIt2304544: MN960901–MN960969, BankIt2304547: MN960970–MN961015, BankIt2304548: MN961016–MN961107.
2.2. Sequence retrieval and alignment
Nonredundant NCBI blast search (Gish & States, 1993) was conducted to identify coding DNA sequences of C. canephora galactomannan gene copies using previously reported coding DNA sequences for Cc‐alpha, Cc‐Man‐S, and Cc‐alpha Gal as query sequences (Joët et al., 2014; Pré et al., 2008), except for Cc‐MGT and Cc‐UG4E whose query sequences were retrieved from Arabidopsis and rice, respectively (Conklin et al., 1999; Guevara, El‐Kereamy, Yaish, Mei‐Bi, & Rothstein, 2014). The partial Cc‐UG4E and Cc‐MGT sequences (Joët et al., 2014) were used to blast Arabidopsis and rice databases and the best hits results were selected and subsequently used to retrieve complete coding sequences from C. canephora genome after confirming their annotation.
2.3. Sequence diversity and codon usage bias test
The average pairwise nucleotide sequence diversity index, π, was calculated using Jukes and Cantor correction method in DnaSP v5.0 (Librado & Rozas, 2009). Effective number of codon (ENC) was tested in DnaSP and confirmed in CodonW (Peden, 1999). ENC is a codon usage bias test that can reflect the level of selective constraint on a particular gene and variation in synonymous substitution rate is related to codon usage (Sharp, 1991; Sharp, Tuohy, & Mosurski, 1986). Low ENC values represent greater codon bias, while high ENC represents low codon usage bias (Wright, 1990).
2.4. Variation in d N, d S, and ω among genes
The synonymous (d S) and nonsynonymous (d N) nucleotide substitution rates were calculated using CodeML feature in the phylogenetic analysis by maximum likelihood, PAML v4.3 (Yang, 1997) as described by (Jeffares, Tomiczek, Sojo, & Reis,). Briefly, coding DNA sequences were translated to protein sequences, and their orthology determined for tree construction and CodeML analysis in PALM. Overall, cumulative mutations across the whole gene region can effectively be used to estimate positive selection or selective pressure by calculating substitution ratio of nonsynonymous and synonymous mutations (d N/d S = ω). Under neutral selection, both nonsynonymous and synonymous are fixed at the same rate thus ω = 1. Purifying selection prevents fixation of deleterious mutation, thus ω < 1, due to excess synonymous over nonsynonymous substitutions. When nonsynonymous mutation offers selective advantage, is it fixed at a higher rate than synonymous mutation? Thus, ω > 1 (Yang & Nielsen, 2000). Due to complex demographic history of domesticated plants, parameters allowing for deviations from the standard neutral model were used to best reflect specific demographic events (Wright & Gaut, 2005). Evidence of positive selection was determined by calculating selection at each codon for all genes using likelihood‐ratio test (LRT) to compare M8 and M8a models (Swanson, Nielsen, & Yang, 2003), which allow for positive selection (beta, and ω > 1) and near neutral model M8a (ω = 1), respectively, for gene copies in PAML. Branch‐site analysis with model A was used to detect independent ω values of positive selection, and Bayes Empirical Bayes analysis (BEB) was performed to identify sequences under selection. For statistical significance, bivariate correlation between sequence length and mutation rates d S, d N, and ω was tested to confirm reliability of variation in the data.
2.5. Phylogenetic and synteny analysis of galactomannan pathway genes
All‐against‐all BlastP in different plant genome databases was conducted during the period of March 2018 to retrieve putative galactomannan pathway gene copies and their corresponding annotations using default parameters (Altschul, Madden, Schaffer, Zhang, & Zhang, 1997; Gu, Cavalcanti, Chen, Bouman, & Li, 2002). Homology was inferred for sequences with at least 40% identity and 70% align‐able region lengths and subsequently used for phylogenetic analysis. Protein sequence alignment was performed using ClustalW v1.83 (Thompson, Higgins, & Gibson, 1994) with the default parameters and corrected manually on MEGA v.6.0 (Tamura, Stecher, Peterson, Filipski, & Kumar, 2013). Prior to ClustalW alignment, redundant sequences were filtered using CD‐HIT program (Huang, Niu, Gao, Fu, & Li, 2010). Conserved sequences were trimmed and then used to guide CDS alignments in BioEdit 7.0.5.3 (Hall, 1999). Tree construction was conducted using bootstrap neighbor‐joining method in MEGA v6.0, with algae as outgroup, a Kimura 2‐parameters, and a bootstrap value of 1,000 to assess internal node stability.
Conserved synteny analysis of short chromosomal segments that included regions of GM pathway genes was conducted to visualize syntenic linkages in the C. canephora genome using genome comparison visualizer in Easyfig software (Sullivan, Petty, & Beatson, 2011). Alignment of chromosome segments that contained putative GM pathway gene copies and their flanking regions extending 100 kb in both forward and reverse was conducted in MAFFTv6.7 (Kazutaka & Daron, 2013), and two chromosome regions with best alignment hits were selected for synteny analysis.
3. RESULTS
3.1. Analysis of sequence polymorphism and codon usage bias
A total of 18 GM pathway gene copies were identified in C. canephora (Cc) genome and used for sequence variation test and evolutionary rates analysis. Evolutionary rate at silent (d S) and replacement (d N) mutation, nucleotide sequence polymorphism, and ω was compared among multiple gene copies of the GM pathway (Table 1). Overall, the average nucleotide sequence diversity (π) ranged from 0.03 in Cc‐MGT gene copies to 1.13 in Cc‐alpha Gal genes. Polymorphic variation within each gene copies varied from 0.02 to 0.24 for Cc08_g06540 and Cc10_g07720 in Cc‐ManS; from 0 to 0.18 for Cc07_g07210 and Cc07_g07220 in Cc‐GMGT and from 0.08 to 0.74 for Cc11_g00330 and Cc11_g15950 in Cc‐alpha Gal genes copies (Table 1). In contrast, a relatively low sequence polymorphism was observed in Cc‐UG4E and Cc‐MGT gene copies. Most codon usage bias was detected among Cc‐UG4E and Cc‐MGT gene copies based on lowest average ENC of 38.9 and 40.9, respectively, while Cc‐alpha Gal gene with the highest average d N/d S showed least codon usage bias with a mean average ENC of 57.8 (Table 2).
Table 1.
Sequence diversity and selection test in 23 sequenced genotypes
| Enzyme | Gene locus | π | d S | d N | d N /d S (ω) | LRT |
|---|---|---|---|---|---|---|
| Cc‐Man‐S | Cc06_g04240 | 0.09 | 0.14 | 0.02 | 0.14 | 0.00 |
| Cc01_g05870 | 0.47 | 0.31 | 0.11 | 0.35 | 0.00 | |
| Cc06_g05120 | 0.19 | 0.03 | 0.01 | 0.33 | 1.23NS | |
| Cc08_g06540 | 0.02 | 0.06 | 0.00 | 0.00 | 0.00 | |
| Cc10_g07720 | 0.24 | 0.98 | 0.72 | 0.73 | 0.00 | |
| Cc‐GMGT | Cc07_g07220 | 0.18 | 0.28 | 0.11 | 0.39 | 0.00 |
| Cc07_g07210 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Cc00_g25840 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Cc03_g01930 | 0.20 | 0.21 | 0.03 | 0.14 | 24.51 | |
| Cc‐alpha Gal | Cc02_g05490 | 0.16 | 0.41 | 0.29 | 0.71 | 7.94NS |
| Cc11_g15950 | 0.74 | 0.14 | 0.17 | 1.21 | 98.83 | |
| Cc04_g14280 | 0.15 | 1.07 | 0.58 | 0.54 | 0.00 | |
| Cc11_g00330 | 0.08 | 0.40 | 0.29 | 0.73 | 48.12 | |
| Cc‐UG4E | Cc11_g04810 | 0.04 | 0.34 | 0.02 | 0.06 | 0.00 |
| Cc07_g03170 | 0.02 | 0.24 | 0.03 | 0.13 | 0.00 | |
| Cc00_g06000 | 0.14 | 0.31 | 0.01 | 0.03 | 0.00 | |
| Cc‐MGT | Cc05_g00320 | 0.02 | 0.04 | 0.00 | 0.00 | 0.00 |
| Cc02_g07370 | 0.01 | 0.05 | 0.02 | 0.25 | 0.00 |
p < .001.
Abbreviations: d N, nonsynonymous substitution; d S, synonymous substitution; LRT, likelihood‐ratio test; NS, not significant (p < .51); π, nucleotide sequence diversity index; ω, selection index.
Table 2.
Information of the C. canephora GM genes used in this study
| Enzyme | Gene locus | Chr. Location | Length sequenced (bp) | Coding region | |||
|---|---|---|---|---|---|---|---|
| Total | CDS | Promoter | ENC | GC (GC3)a | |||
| Cc‐Man‐S | Cc06_g04240 | 6 | 2,274 | 1,593 | 681 | 41.24 | 0.36 (0.41) |
| Cc01_g05870 | 1 | 2,245 | 2,031 | 214 | 52.51 | 0.39 (0.43) | |
| Cc06_g05120 | 6 | 2,076 | 1,602 | 474 | 39.11 | 0.35 (0.31) | |
| Cc08_g06540 | 8 | 2,117 | 2,082 | 35 | 38.95 | 0.37 (0.30) | |
| Cc10_g07720 | 10 | 2,127 | 2,062 | 65 | 67.21 | 0.39 (0.32) | |
| Cc‐GMGT | Cc07_g07220 | 7 | 2,233 | 1,335 | 898 | 54.56 | 0.34 (0.40) |
| Cc07_g07210 | 7 | 2,218 | 1,347 | 871 | 37.71 | 0.28 (0.32) | |
| Cc00_g25840 | 0 | 2,120 | 1,311 | 809 | 39.93 | 0.30 (0.39) | |
| Cc03_g01930 | 3 | 1,823 | 1,383 | 440 | 50.13 | 0.31 (0.34) | |
| Cc‐alpha Gal | Cc02_g05490 | 2 | 2,157 | 1,275 | 882 | 58.24 | 0.41 (0.48) |
| Cc11_g15950 | 11 | 2,118 | 1,182 | 936 | 54.79 | 0.53 (0.52) | |
| Cc04_g14280 | 4 | 2,130 | 1,137 | 993 | 61.01 | 0.41 (0.37) | |
| Cc11_g00330 | 11 | 1,754 | 1,311 | 443 | 57.20 | 0.41 (0.49) | |
| Cc‐UG4E | Cc11_g04810 | 11 | 2,142 | 1,355 | 787 | 40.91 | 0.48 (0.57) |
| Cc07_g03170 | 7 | 2,177 | 2,116 | 61 | 43.42 | 0.40 (0.53) | |
| Cc00_g06000 | 0 | 1,928 | 1,350 | 578 | 38.33 | 0.38 (0.36) | |
| Cc‐MGT | Cc05_g00320 | 5 | 2,128 | 1,086 | 1,042 | 36.47 | 0.44 (0.58) |
| Cc02_g07370 | 2 | 1,736 | 1,086 | 650 | 41.31 | 0.37 (0.43) | |
Abbreviation: ENC, effective number of codon.
Average for 23 C. canephora accessions, GC3, average GC content at the third‐codon positions of coding genes for all accessions
3.2. Comparing d N, d S, and ω among gene copies
The average d S and d N values are shown in Figure 3. Interestingly, variation pattern suggesting a slow evolutionary rate for the most upstream pathway genes as compared with the most downstream genes was observed. For example, low average d S and d N values were observed for two upstream genes, Cc‐UG4E (d N = 0.02; d S = 0.30; p < .001) and Cc‐MGT (d N = 0.01; d S = 0.05; p < .001), while significantly higher average d S and d N values were detected for the downstream Cc‐alpha Gal gene (d N = 0.33; d S = 0.51; p < .001) (Figure 3; Table 1). The highest d S and d N values were observed in Cc‐alpha Gal at Cc04_g14280 (d S = 1.07) and in Cc‐Man‐S at Cc10_g07720 (d N = 0.72), respectively. Test for positive selection using d N/d S variation among gene copies revealed an excess of replacement substitution in Cc‐alpha Gal at Cc11_g15950 (Table 1). This result was confirmed by maximum likelihood test of ω in PAML, which revealed strong evidence of significant positive selection sites based on LRT values in Cc‐alpha Gal gene at Cc11_g15950; Cc11_g00330 with LRT of 98.83 (p < .001); 48.12 (p < .001), respectively, and in Cc‐GMGT gene at Cc03_g01930 with LRT of 24.51 (p < .001). It is noteworthy that high LRT of 1.23 and 7.94 for Cc‐ManS at locus Cc06_g05120 and Cc‐alpha Gal at locus Cc02_g05490, respectively, were observed. However, the values were not significant which could be due to the effects of either sequence length, sequence divergence, and the strength of positive selection which have been shown to affect the power of LRT (Maria, Joseph, & Ziheng, 2001).
Figure 3.
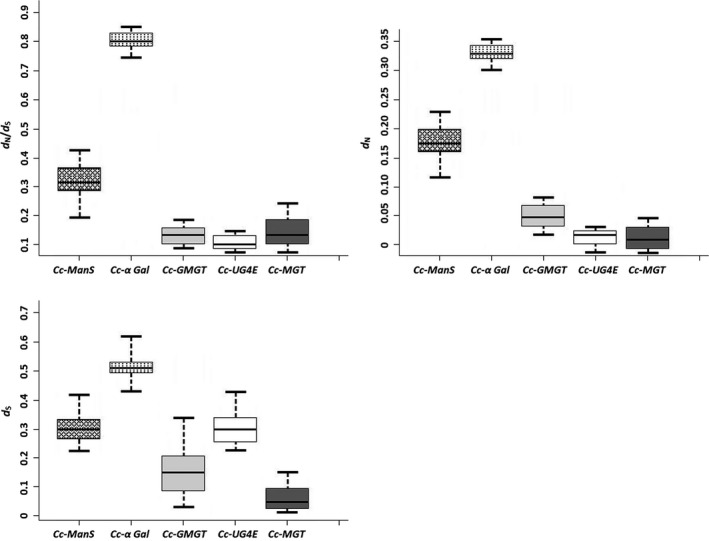
Evolutionary rates at silent and replacement sites of the GM pathway genes. Average values of d S, d N, and d N/d S are represented by horizontal bars within the boxes at 95% confidence intervals
In addition, increased nonsynonymous substitution was detected in Cc‐Man‐S gene at Cc10_g07720 (ω = 0.73) and in Cc‐alpha Gal gene at Cc02_g05490 (ω = 0.71), Cc04_g14280 (ω = 0.54) and Cc11_g00330 (ω = 0.73). However, the LRT test was not significant for Cc02_g05490. Overall, high d N/d S ratio variation among and within gene copies was observed, which demonstrates a significantly diverse evolutionary rate in GM pathway genes. These results suggested different rates of selection events on GM pathway gene copies in C. canephora. More selection events appeared to have occurred in Cc‐alpha Gal gene, while Cc‐UG4E and Cc‐MGT copies showed evidence of strong selective constraint. However, interpreting whether the observed difference in selection rates across the pathway is actually more than expected at random requires conjunction of this data with results from population genetics models that include recombination between selected sites and nearby neutral marker (Charlesworth, 2006; Schmid, Ramos‐Onsins, Ringys‐Beckstein, Weisshaar, & Mitchell‐Olds, 2005).
3.3. Synteny and phylogenetic analysis
Phylogenetic and synteny analysis was conducted to get insight into evolutionary history of GM pathway gene copies. Extensive synteny was observed for most sequence segments with strong conservation of tandemly linked regions spanning ~60% in length for duplicate copies in Cc‐Man‐S, Cc‐GMGT, Cc‐UG4E, and Cc‐MGT (Figure 4; Figure S1). In contrast, a relatively weak synteny was detected at Cc‐alpha Gal, which might be due to more selection events accompanying its duplication in C. canephora.
Figure 4.
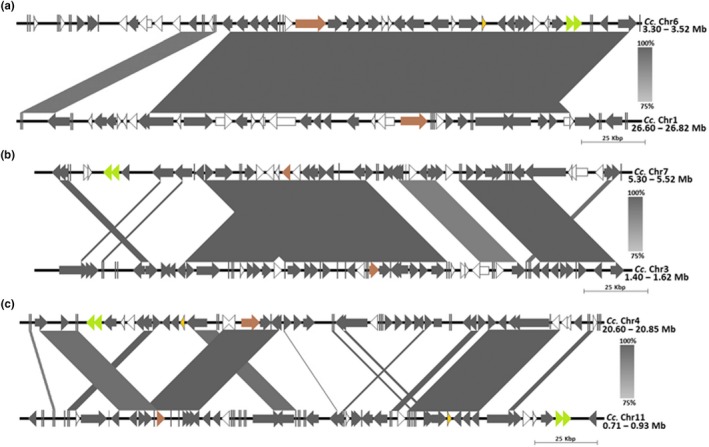
Synteny analysis of tandem chromosomal regions for Man‐S (a), Cc‐GMGT (b), showing extensive alignments, and in Cc‐alpha Gal (c), with less conserved (<60%) syntenic alignment of the two chromosome segments. Arrow heads represent different segment features, and target gene position is shown in dark brown color. Color intensity of gray diagonal blocks represents rate of conserved sequence sites on chromosomes
The phylogenetic analysis, however, failed to support nucleotide diversity in the dataset with paralogs in most species tested forming distinct subclusters. In addition, no obvious orthologous clustering congruent with species evolution could be inferred. Each gene family appeared to have unique expansion patterns or have undergone independent evolutionary events in different plant lineages analyzed in this study. In addition, analysis of GMGT and UG4E phylogenies indicated independent subclades that corresponded with monocots and dicots groups (Figure 5; Figures S2 and S3).
Figure 5.
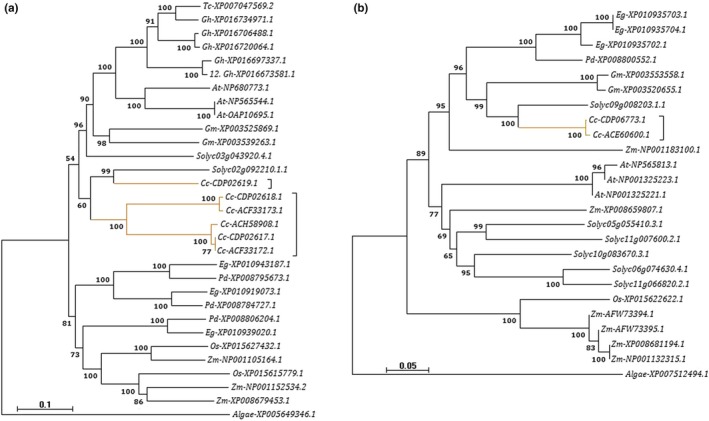
Consensus phylogenetic trees of (a) GMGT and (b) Man‐S gene family members from different plant lineages. Italicized abbreviations represents species name: Tc—Theobroma cocoa, Gh—Gossypium hirsutum, At—Arabidopsis thaliana, Cc—Coffea canephora, Gm—Glycine max, Eg—Elaeis guineensis, Pd—Phoenix dactylifera, Os—Oryza sativa, Zm—Zea mays, Solyc—Solanum lycopersicum. Gene accession numbers are shown in‐front of species name, and bootstrap values indicating confidence levels ≥50 for clustering are shown at the nodes. Tree branches for C. canephora copies are highlighted in brown color
4. DISCUSSION
Our study examined the evolutionary rate analysis of the galactomannan pathway genes based on multiple copies analysis in the diploid C. canephora this is because multiple‐copy encoding enzymes can provide direct evidence of gene evolution patterns in metabolic pathways (Chu, Wang, Cheng, Yang, & Yu, 2014). Five wild and eighteen accessions representing independent, divergent species (Kryazhimskiy & Plotkin, 2008; Nielsen & Yang, 2003) were sequenced to analyze evolutionary patterns in 18 GM pathway gene copies. Differential selection rates on genes cause effect on patterns of nucleotide sequence diversity (Ma et al.,2015, Ramsay, Rieseberg, & Ritland, 2009). In this study, we detected a correlation between different selection pressure and patterns of sequence diversity, which was consistent with the observed level of syntenic linkage analysis. The Cc‐alpha Gal gene showing evidence of positive and/or relaxed selection pressure had the highest sequence diversity index and displayed weak synteny. In contrast, Cc‐UG4E and Cc‐MGT under intense purifying selection had the lowest sequence diversity and displayed the strongest syntenic linkage. Cc‐UG4E and Cc‐MGT control the crucial carbon flux step from sucrose to galactomannan through synthesis of UDP‐galactose and GDP‐mannose nucleotide sugar building blocks, respectively. Thus, a strong negative selection is needed to optimize and remove deleterious mutations (Lu & Rausher, 2003) to ensure their novel evolutionary function remains fixed in the population. In addition, previous studies reveal that high gene expression is correlated to their fitness and significant role in survival (Blanc & Wolfe, 2004; Popescu, Borza, Bielawski, & Lee, 2006). Fitness genes exhibit strong codon bias due to intense purifying selection for translation efficiency and their expression levels are usually higher (Blanc & Wolfe, 2004; Brown & Kelly, 2018; Lavin, Herendeen, & Wojciechowski, 2005). More recently, expression of core GM pathway genes, including Cc‐UG4E and Cc‐MGT, was reported to be positively correlated with the amount of stored cell wall storage polysaccharides (Joët et al., 2014). Altogether, these reports are in agreement with the strong negative selection and greater codon bias observed for Cc‐UG4E and Cc‐MGT in our study.
Frequency of positive selection and selective constraints are most likely factors to account for evolutionary variation among gene copies in a metabolic pathway (Braverman, Hamilton, & Johnson, 2016; Gaut, Yang, Takuno, & Eguiarte, 2011; Yang et al., 2009). High ratio of synonymous (silent) to nonsynonymous (replacement) variation among and within gene copies observed in this study demonstrates a significant rate of evolutionary heterogeneity in GM pathway genes, which is consistent with high degree of endogenous heterogeneity reported in several cell wall storage polysaccharides (Gruppen, Kormelink, & Voragen, 1993; Toole et al., 2012). The d N/d S ratio and codon‐based analysis detected signature of positive selection in Cc‐alpha Gal gene copy. Interestingly, different isoforms of α‐Gal gene have been reported to exist in plant tissues (Chrost & Schmitz, 2000; Dey & Del Campillo 1984) which could contribute to high rate gene mutation (Lynch & Conery, 2000). In addition, enzyme position in a metabolic pathway has been linked to evolutionary rate variation with upstream genes evolving more slowly than the downstream genes (Clotault, Peltier, Soufflet‐Freslon, Briard, & Geoffriau, 2012). This is because upstream genes have greater pleiotropic effects on a number of pathway end products (Cork & Purugganan, 2004; Pal, Papp, & Lercher, 2006; Rausher et al., 1999). The Cc‐alpha Gal is a debranching enzyme catalyzing the postdepositional degree of galactose side branch to determine the mannose to galactose ratio, and thus, it is possible that its position in the GM biosynthesis pathway and occurrence in different isoforms could have played a role in its accelerated rate of accumulating adaptive mutations in C. canephora. No positive selection was observed in Cc‐GMGT or Cc‐Man‐S, which together catalyze the polymerization of core galactomannan structure, despite the latter showing relatively high average d N/d S ratio in some copies, suggesting that it might be under relaxed selective constraint.
In contrast, phylogenetic analysis failed to support nucleotide diversity with most paralogs in species tested forming distinct subclusters. In addition, no obvious orthologous clustering congruent with species phylogeny could be inferred. Similar results have also been observed in other plant cell wall genes (Ahn et al., 2007; Figueiredo, Lashermes, & Aragão, 2011). This may be due to the highly variable and dynamic nature of plant cell walls throughout growth and development process (Campbell & Braam, 1999). Trees revealed that GM pathway genes might have independently evolved or expanded in distinct patterns in different plant species in order to cope with specific biochemical cell wall modification requirements. However, despite the observed high amino acid sequence diversity, the core biochemically active motifs of specific genes were conserved in different plant lineages, allowing them to retain their primary functional properties. Phylogenies of GMGT and UG4E gene families suggested that they are evolutionary more distant between monocots and dicots which supports previous hypothesis that Nucleotide‐diphospho‐sugar interconversion enzymes (NSEs) families such as Epimerases are highly divergent in monocots and dicots plants (Yin, Huang, Gu, Bar‐Peled, & Xu, 2011).
In summary, this study investigated the evolution patterns of five core structural genes of the galactomannan pathway using multi‐copy based analysis and identified significant variation in evolutionary rate among genes. Positive selection detected in the GM branch editing gene Cc‐alpha Gal is likely to explain the reason for varying mannose to galactose ratio in coffee seeds. Highly conserved Cc‐UG4E and Cc‐MGT suggest a strong negative selection, which purge changes that cause deleterious effects on the fitness due to their crucial role in sucrose carbon flux for galactomannan accumulation in coffee seeds. We also observed lineage‐specific gene family expansion for MGT, alpha Gal, and Man‐S, while independent expansion of GMGT and UG4E gene families in monocots and dicots. Our results provide insights into the evolutionary rate variations in endosperm cell wall storage polysaccharides genes in the diploid C. canephora.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
AUTHORS CONTRIBUTION
Mr. Collins Ogutu conceived the idea, designed, and conducted the experiments. Ms. Sylvia Cherono and Ms. Charmaine Ntini helped with the experiments and manuscript revision. Mr. Md. Dulal Ali Mollah, Mr. Lei Zhao, and Mr. Mohammad A. Belal helped with data analysis.
CONSENT FOR PUBLICATION
Not applicable.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All methods used to collect observational data were noninvasive.
Supporting information
ACKNOWLEDGMENTS
This project was supported by funds received from the Overseas Construction Plan for Science and Education Base, China‐Africa Center for Research and Education, Chinese Academy of Sciences (grant no. SAJC201327).
Ogutu C, Cherono S, Ntini C, et al. Evolutionary rate variation among genes involved in galactomannan biosynthesis in Coffea canephora . Ecol Evol. 2020;10:2559–2569. 10.1002/ece3.6084
DATA AVAILABILITY STATEMENT
All DNA sequences have been submitted to the GenBank databases, and accession numbers are provided in the methods section.
REFERENCES
- Ahn, Y. O. , Zheng, M. , Bevan, D. R. , Esen, A. , Shiu, S.‐H. , Benson, J. , … Poulton, J. E. (2007). Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 35. Phytochemistry, 68, 1510–1520. 10.1016/j.phytochem.2007.03.021 [DOI] [PubMed] [Google Scholar]
- Altschul, S. F. , Madden, T. L. , Schaffer, A. A. , Zhang, J. , Zhang, Z. , Miller, W. , & Lipman, D. J. (1997). Gapped BLAST and PSI‐BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25, 3389 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, G. , & Wolfe, K. H. (2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. The Plant Cell, 16, 1679–1691. 10.1105/tpc.021410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman, J. M. , Hamilton, M. B. , & Johnson, B. A. (2016). Patterns of substitution rate variation at many nuclear loci in two species trios in the Brassicaceae partitioned with ANOVA. Journal of Molecular Evolution, 83, 97–109. 10.1007/s00239-016-9752-x [DOI] [PubMed] [Google Scholar]
- Brown, K. E. , & Kelly, J. K. (2018). Antagonistic pleiotropy can maintain fitness variation in annual plants. Journal of Evolutionary Biology, 31, 46–56. 10.1111/jeb.13192 [DOI] [PubMed] [Google Scholar]
- Buckeridge, M. (2010). Seed cell wall storage polysaccharides: Models to understand cell wall biosynthesis and degradation. Plant Physiology, 154, 1017–1023. 10.1104/pp.110.158642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckeridge, M. , & Reid, J. (1996). Major cell wall polysaccharides in legume seeds: Structure, catabolism and biological functions. Ciência e Cultura, 48, 153–162. [Google Scholar]
- Campbell, P. , & Braam, J. (1999). Xyloglucan endotransglycosylases: Diversity of genes, enzymes and potential wall‐modifying functions. Trends in Plant Science, 4, 361–366. 10.1016/S1360-1385(99)01468-5 [DOI] [PubMed] [Google Scholar]
- Charlesworth, D. (2006). Balancing selection and its effects on sequences in nearby genome regions. PLoS Genetics, 2, 64 10.1371/journal.pgen.0020064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrost, B. , & Schmitz, K. (2000). Purification and characterization of multiple forms of α‐galactosidases in Cucumis melo plants. Journal of Plant Physiology, 156, 483–491. [Google Scholar]
- Chu, S. S. , Wang, J. , Cheng, H. , Yang, Q. , & Yu, D. Y. (2014). Evolutionary study of the isoflavonoid pathway based on multiple copies analysis in soybean. BMC Genetics, 15, 76 10.1186/1471-2156-15-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotault, J. , Peltier, D. , Soufflet‐Freslon, V. , Briard, M. , & Geoffriau, E. (2012). Differential selection on carotenoid biosynthesis genes as a function of gene position in the metabolic pathway: A study on the carrot and dicots. PLoS ONE, 7, e38724 10.1371/journal.pone.0038724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin, P. L. , Norris, S. R. , Wheeler, G. L. , Williams, E. H. , Smirnoff, N. , & Last, R. L. (1999). Genetic evidence for the role of GDP mannose in plant ascorbic acid (vitamin C) biosynthesis. Proceedings of the National Academy of Sciences of the United States of America, 96, 4198–4203. 10.1073/pnas.96.7.4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cork, J. M. , & Purugganan, M. D. (2004). The evolution of molecular genetic pathways and networks. BioEssays, 26, 479–484. 10.1002/bies.20026 [DOI] [PubMed] [Google Scholar]
- Denoeud, F. , Carretero‐Paulet, L. , Dereeper, A. , Droc, G. , Guyot, R. , Pietrella, M. , … Lashermes, P. (2014). The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science, 345, 1181–1184. 10.1126/science.1255274 [DOI] [PubMed] [Google Scholar]
- Dey, P. M. , & Del Campillo, E. (1984). Biochemistry of multiple forms of glycosidases in plants. Advances in Enzymology and Related Areas of Molecular Biology, 56, 141–249. [DOI] [PubMed] [Google Scholar]
- Dhugga, K. , Barreiro, R. , Whitten, B. , Stecca, K. , Hazebroek, J. , Randhawa, G. S. , … Anderson, P. (2004). Guar seed betamannan synthase is a member of the cellulose synthase super gene family. Science, 303, 363–366. [DOI] [PubMed] [Google Scholar]
- Edwards, M. , Scott, C. , Gidley, M. J. , & Reid, J. S. G. (1992). Control of mannose galactose ratio during galactomannan formation in developing legume seeds. Planta, 187, 67–74. 10.1007/BF00201625 [DOI] [PubMed] [Google Scholar]
- Figueiredo, S. A. , Lashermes, P. , & Aragão, F. J. (2011). Molecular characterization and functional analysis of the b‐galactosidase gene during Coffea arabica (L.) fruit development. Journal of Experimental Botany, 62, 2691–2703. [DOI] [PubMed] [Google Scholar]
- Gaut, B. , Yang, L. , Takuno, S. , & Eguiarte, L. E. (2011). The patterns and causes of variation in plant nucleotide substitution rates. Annual Review of Ecology, Evolution, and Systematics, 42, 245–266. 10.1146/annurev-ecolsys-102710-145119 [DOI] [Google Scholar]
- Gish, W. , & States, D. J. (1993). Identification of protein coding regions by database similarity search. Nature Genetics, 3, 266 10.1038/ng0393-266 [DOI] [PubMed] [Google Scholar]
- Greenberg, A. J. , Stockwell, S. R. , & Clark, A. G. (2008). Evolutionary constraint and adaptation in the metabolic network of Drosophila. Molecular Biology and Evolution, 25, 2537–2546. 10.1093/molbev/msn205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruppen, H. , Kormelink, F. , & Voragen, A. (1993). Water‐unextractable cell wall material from wheat flour. 3. A structural model for arabinoxylans. Journal of Cereal Science., 18, 111–128. 10.1006/jcrs.1993.1040 [DOI] [Google Scholar]
- Gu, Z. , Cavalcanti, A. , Chen, F. , Bouman, P. , & Li, W. (2002). Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Molecular Biology and Evolution, 19, 256–262. 10.1093/oxfordjournals.molbev.a004079 [DOI] [PubMed] [Google Scholar]
- Guevara, D. R. , El‐Kereamy, A. , Yaish, M. W. , Mei‐Bi, Y. , & Rothstein, S. J. (2014). Functional characterization of the rice UDP‐glucose 4‐epimerase 1, OsUGE1: A potential role in cell wall carbohydrate partitioning during limiting nitrogen conditions. PLoS ONE, 9, e96158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, T. A. (1999). BioEdit: A user‐friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98. [Google Scholar]
- Horbowicz, M. , & Obendorf, R. L. (1994). Seed desiccation tolerance and storability: Dependence on flatulence‐producing oligosaccharides and cyclitols—Review and survey. Seed Science Research, 4, 385–405. 10.1017/S0960258500002440 [DOI] [Google Scholar]
- Huang, Y. , Niu, B. , Gao, Y. , Fu, L. , & Li, W. (2010). CD‐HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics, 26, 680–682. 10.1093/bioinformatics/btq003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffares, D. C. , Tomiczek, B. , Sojo, V. , & dos Reis, M. (2015). A beginners guide to estimating the nonsynonymous to synonymous rate ratio of all protein-coding genes in a genome. Methods in Molecular Biology., 1201, 65–90. [DOI] [PubMed] [Google Scholar]
- Jeffrey, L. (2002). Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica, 115, 29–36. [DOI] [PubMed] [Google Scholar]
- Joersbo, M. , Marcussen, J. , & Brunstedt, J. (2001). In vivo modification of the cell wall polysaccharide galactomannan of guar transformed with an alpha‐galactosidase gene cloned from senna. Molecular Breeding, 7, 211–219. [Google Scholar]
- Joët, T. , Laffargue, A. , Salmona, J. , Doulbeau, S. , Descroix, F. , Bertrand, B. , … Dussert, S. (2014). Regulation of galactomannan biosynthesis in coffee seeds. Journal of Experimental Botany, 65, 323–337. 10.1093/jxb/ert380 [DOI] [PubMed] [Google Scholar]
- Kazutaka, K. , & Daron, M. S. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M. (1977). Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature, 267, 275–276. 10.1038/267275a0 [DOI] [PubMed] [Google Scholar]
- Kryazhimskiy, S. , & Plotkin, J. B. (2008). The population genetics of dN/dS. PLoS Genetics, 4(12), e1000304 10.1371/journal.pgen.1000304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin, M. , Herendeen, P. S. , & Wojciechowski, M. F. (2005). Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Systematic Biology, 54, 575–594. 10.1080/10635150590947131 [DOI] [PubMed] [Google Scholar]
- Librado, P. , & Rozas, J. (2009). DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- Livingstone, K. , & Anderson, S. (2009). Patterns of variation in the evolution of carotenoid biosynthetic pathway enzymes of higher plants. Journal of Heredity, 100, 754–761. 10.1093/jhered/esp026 [DOI] [PubMed] [Google Scholar]
- Lu, Y. , & Rausher, M. D. (2003). Evolutionary rate variation in anthocyanin pathway genes. Molecular Biology and Evolution, 20, 1844–1853. 10.1093/molbev/msg197 [DOI] [PubMed] [Google Scholar]
- Lynch, M. , & Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science, 290, 1151–1155. 10.1126/science.290.5494.1151 [DOI] [PubMed] [Google Scholar]
- Ma, Y. , Wang, J. , Zhong, Y. , Geng, F. , Cramer, G. R. , & Cheng, Z. M. (2015). Subfunctionalization of cation/proton antiporter 1 genes in grapevine in response to salt stress in different organs. Horticulture Research, 2, 15031 10.1038/hortres.2015.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maria, A. , Joseph, P. , & Ziheng, Y. (2001). Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Molecular Biology and Evolution, 18, 1585–1592. [DOI] [PubMed] [Google Scholar]
- Nielsen, R. , & Yang, Z. (2003). Estimating the distribution of selection coefficients from phylogenetic data with applications to mitochondrial and viral DNA. Molecular Biology and Evolution, 20, 1231–1239. 10.1093/molbev/msg147 [DOI] [PubMed] [Google Scholar]
- Ogutu, C. , Fang, T. , Yan, L. , Wang, L. , Huang, L. , Wang, X. , … Han, Y. (2016). Characterization and utilization of microsatellites in the Coffea canephora genome to assess genetic association between wild species in Kenya and cultivated coffee. Tree Genetics and Genomes, 12, 54 10.1007/s11295-016-1014-y [DOI] [Google Scholar]
- Pal, C. , Papp, B. , & Lercher, M. J. (2006). An integrated view of protein evolution. Nature Reviews Genetics, 7, 337–348. 10.1038/nrg1838 [DOI] [PubMed] [Google Scholar]
- Panchy, N. , Lehti‐Shiu, M. , & Shiu, S. H. (2016). Evolution of gene duplication in plants. Plant Physiology, 171, 2294–2316. 10.1104/pp.16.00523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden, J. F. (1999). CodonW: Analysis of codon usage. PhD thesis, University of Nottingham, UK. [Google Scholar]
- Pierce, V. , & Crawford, D. (1997). Phylogenetic analysis of glycolytic enzyme expression. Science, 276, 256–259. 10.1126/science.276.5310.256 [DOI] [PubMed] [Google Scholar]
- Polhill, R. M. , Raven, P. H. , & Stirto, C. H. (1981). Evolution and Systematics of the Leguminosae In Polhill R. M., & Raven P. H. (Eds.), Advances in Legume Systematics (pp. 1–26). Kew: Part I, Royal Botanic Garden. [Google Scholar]
- Popescu, C. E. , Borza, T. , Bielawski, J. P. , & Lee, R. W. (2006). Evolutionary rates and expression level in Chlamydomonas. Genetics, 172, 1567–1576. 10.1534/genetics.105.047399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pré, M. , Caillet, V. , Sobilo, J. , & McCarthy, J. (2008). Characterization and expression analysis of genes directing galactomannan synthesis in coffee. Annals of Botany, 102, 207–220. 10.1093/aob/mcn076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan, M. , & Wessler, S. (1994). Molecular evolution of the plant R regulatory gene family. Genetics, 138, 849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay, H. , Rieseberg, L. H. , & Ritland, K. (2009). The correlation of evolutionary rate with pathway position in plant terpenoid biosynthesis. Molecular Biology and Evolution, 26, 1045–1053. 10.1093/molbev/msp021 [DOI] [PubMed] [Google Scholar]
- Rausher, M. , Lu, Y. , & Meyer, K. (2008). Variation in constraint versus positive selection as an explanation for evolutionary rate variation among anthocyanin genes. Journal of Molecular Evolution, 67, 137–144. 10.1007/s00239-008-9105-5 [DOI] [PubMed] [Google Scholar]
- Rausher, M. , Miller, R. , & Tiffin, P. (1999). Patterns of evolutionary rate variation among genes of the anthocyanin biosynthetic pathway. Molecular Biology and Evolution, 16, 266–274. 10.1093/oxfordjournals.molbev.a026108 [DOI] [PubMed] [Google Scholar]
- Redgwell, R. , & Fischer, M. (2006). Coffee carbohydrates. Brazilian Journal of Plant Physiology, 18, 165–174. 10.1590/S1677-04202006000100012 [DOI] [Google Scholar]
- Reid, J. , Edwards, M. , Dickson, C. , Scott, C. , & Gidley, M. (2003). Tobacco transgenic lines that Express fenugreek galactomannan galactosyltransferase constitutively have structurally altered galactomannans in their seed endosperm cell walls. Plant Physiology, 131, 1487–1495. 10.1104/pp.102.016840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, K. J. , Ramos‐Onsins, S. , Ringys‐Beckstein, H. , Weisshaar, B. , & Mitchell‐Olds, T. (2005). A multilocus sequence survey in Arabidopsis thaliana reveals a genome‐wide departure from a neutral model of DNA sequence polymorphism. Genetics, 169, 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, P. (1991). Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurium: Codon usage, map position, and concerted evolution. Journal of Molecular Evolution, 33, 23–33. 10.1007/BF02100192 [DOI] [PubMed] [Google Scholar]
- Sharp, P. , Tuohy, T. , & Mosurski, K. (1986). Codon usage in yeast: Cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Research, 14, 5125–5143. 10.1093/nar/14.13.5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siol, M. , Wright, S. I. , & Barrett, S. C. (2010). The population genomics of plant adaptation. New Phytologist, 188, 313–332. 10.1111/j.1469-8137.2010.03401.x [DOI] [PubMed] [Google Scholar]
- Sullivan, M. J. , Petty, N. K. , & Beatson, S. A. (2011). Easyfig: A genome comparison visualizer. Bioinformatics, 27, 1009–1010. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, W. J. , Nielsen, R. , & Yang, Q. (2003). Pervasive adaptive evolution in mammalian fertilization proteins. Molecular Biology and Evolution, 20, 18–20. 10.1093/oxfordjournals.molbev.a004233 [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. , & Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D. , Higgins, D. G. , & Gibson, T. J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toole, G. A. , Le Gall, G. , Colquhoun, I. J. , Drea, S. , Opanowicz, M. , Bedő, Z. , … Mills, E. (2012). Spectroscopic analysis of diversity in the spatial distribution of arabinoxylan structures in endosperm cell walls of cereal species in the HEALTHGRAIN diversity collection. Journal of Cereal Science, 56, 134–141. 10.1016/j.jcs.2012.02.016 [DOI] [Google Scholar]
- Tucker, P. , & Lundrigan, B. (1993). Rapid evolution of the sex‐determining loci in Old World mice and rats. Nature, 364, 715–717. [DOI] [PubMed] [Google Scholar]
- Vitkup, D. , Kharchenko, P. , & Wagner, A. (2006). Influence of metabolic network structure and function on enzyme evolution. Genome Biology, 7, R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, F. (1990). The “effective number of codons” used in a gene. Gene, 87, 23–29. 10.1016/0378-1119(90)90491-9 [DOI] [PubMed] [Google Scholar]
- Wright, S. I. , & Gaut, B. S. (2005). Molecular population genetics and the search for adaptive evolution in plants. Molecular Biology and Evolution, 22, 506–519. 10.1093/molbev/msi035 [DOI] [PubMed] [Google Scholar]
- Yan, L. , Ogutu, C. , Huang, L. , Wang, X. , Zhou, H. , Lv, Y. , … Han, Y. (2019). Genetic diversity and population structure of coffee germplasm collections in China revealed by ISSR markers. Plant Molecular Biology Reporter, 37, 204–213. 10.1007/s11105-019-01148-3 [DOI] [Google Scholar]
- Yang, Y. H. , Zhang, F. M. , & Ge, S. (2009). Evolutionary rate patterns of the gibberellin pathway genes. BMC Evolutionary Biology, 9, 206 10.1186/1471-2148-9-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. (1997). PAML: A program package for phylogenetic analysis by maximum likelihood. Computer Applications in the Biosciences, 13, 555–556. 10.1093/bioinformatics/13.5.555 [DOI] [PubMed] [Google Scholar]
- Yang, Z. , & Nielsen, R. (2000). Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Molecular Biology and Evolution, 17, 32–43. 10.1093/oxfordjournals.molbev.a026236 [DOI] [PubMed] [Google Scholar]
- Yin, Y. , Huang, J. , Gu, X. , Bar‐Peled, M. , & Xu, Y. (2011). Evolution of plant nucleotide sugar interconversion enzymes. PLoS ONE, 6, e27995 10.1371/journal.pone.0027995 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All DNA sequences have been submitted to the GenBank databases, and accession numbers are provided in the methods section.


