Abstract
Context
It is important to identify patients at highest risk of fractures.
Objective
To compare the separate and combined performances of bone-related genetic risk scores (GRSs) for prediction of forearm, hip and vertebral fractures separately, as well as of trabecular and cortical bone microstructure parameters separately.
Design, Setting, and Participants
Using 1103 single nucleotide polymorphisms (SNPs) independently associated with estimated bone mineral density of the heel (eBMD), we developed a weighted GRS for eBMD and determined its contribution to fracture prediction beyond 2 previously developed GRSs for femur neck BMD (49 SNPs) and lumbar spine BMD (48 SNPs). Associations between these GRSs and forearm (ncases = 1020; ncontrols = 2838), hip (ncases = 1123; ncontrols = 2630) and vertebral (ncases = 288; ncontrols = 1187) fractures were evaluated in 3 Swedish cohorts. Associations between the GRSs and trabecular and cortical bone microstructure parameters (n = 426) were evaluated in the MrOS Sweden cohort.
Results
We found that eBMDGRS was the only significant independent predictor of forearm and vertebral fractures while both FN-BMDGRS and eBMDGRS were significant independent predictors of hip fractures. The eBMDGRS was the major GRS contributing to prediction of trabecular bone microstructure parameters while both FN-BMDGRS and eBMDGRS contributed information for prediction of cortical bone microstructure parameters.
Conclusions
The eBMDGRS independently predicts forearm and vertebral fractures while both FN-BMDGRS and eBMDGRS contribute independent information for prediction of hip fractures. We propose that eBMDGRS captures unique information about trabecular bone microstructure useful for prediction of forearm and vertebral fractures. These findings may facilitate personalized medicine to predict site-specific fractures as well as cortical and trabecular bone microstructure separately.
Keywords: bone mineral density, genetic risk scores, fractures, bone microstructure, trabecular, cortical
Osteoporosis is a disease characterized by low bone mass and micro-architectural deterioration of bone tissue, leading to increased risk of fragility fractures (1). Fracture risk is inversely associated with bone strength, which is dependent on bone mineral density (BMD) as well as bone quality parameters such as trabecular and cortical bone microstructure parameters (2, 3).
Ultrasound measures of the calcaneus (heel) predict fracture risk and this association partly remains after adjustment for hip BMD, suggesting that ultrasound captures unique bone property information of importance for fracture risk (4, 5). Detailed studies using three-dimensional high resolution peripheral quantitative computed tomography (HRpQCT) have revealed that both trabecular and cortical bone microstructure parameters also contribute to fracture prediction beyond two-dimensional dual-energy x-ray absorptiometry (DXA)-derived BMD (6–11). Recent data indicate that the different HRpQCT measures may predict fractures in a bone site–specific manner (12).
In the clinical setting, it is important to identify the patients at highest risk of fracture, who are most likely to benefit from osteoporosis treatment (3). Today, osteoporosis diagnosis and fracture risk prediction are based on imaging (particularly DXA) in combination with assessment of clinical risk factors; the latter are often incorporated in fracture risk prediction tools. It is likely that the genetic and environmental factors that determine fracture risk differ substantially between fractures at different bone sites with different proportions of trabecular and cortical bone. However, current available fracture risk prediction tools do not include information on trabecular or cortical bone microstructure parameters and are not designed to distinguish between differences in prediction of bone site–specific fractures (13, 14). It is possible that factors regulating trabecular bone microstructure are major determinants of vertebral fracture risk, while factors determining cortical bone parameters and risk of falls are major determinants of hip fracture risk. We propose that novel separate predictors for bone site–specific fractures as well as for cortical and trabecular bone microstructure parameters may be useful for the development of personalized medicine. A certain combination of predictors might then be used to identify high risk subjects for bone site–specific fractures and also to determine whether treatments mainly targeting trabecular or cortical bone mass should be considered.
Several bone parameters including femoral neck BMD (FN-BMD) and lumbar spine BMD (LS-BMD) measured by DXA, ultrasound measures at the calcaneus and HRpQCT-derived trabecular and cortical bone parameters are highly heritable (15–20). A meta-analysis of genome-wide association studies (GWAS) has identified 63 independent single nucleotide polymorphisms (SNPs) associated with either FN-BMD or LS-BMD (21), and a recent large scale GWAS in the UK-Biobank identified as many as 1103 independent SNPs associated with estimated BMD (eBMD) from ultrasound measures at the calcaneus (22). The identified SNPs can be used for the development of genetic risk scores (GRSs) that may be used to predict bone health-related phenotypes. Unfortunately, the number of SNPs identified by informative computed tomography (CT) measurements of trabecular and cortical bone parameters separately is low and the variance explained by these SNPs of the respective bone phenotype is marginal. Therefore, no powerful meaningful GRS for separate prediction of trabecular and cortical bone parameters can be developed (23–26).
Two previous studies have used available DXA-derived SNPs for the development of different GRSs to predict risk of fractures in independent data sets (3,27), demonstrating that these GRSs were modest predictors of fractures. Importantly, these studies did not determine the bone site-specific prediction of fracture risk. GRSs based on SNPs associated with ultrasound-derived eBMD (eBMDGRS) have neither been evaluated for fracture prediction in an independent data set nor for bone site-specific prediction of fracture risk (22,28).
In this study, we hypothesized that different GRSs or combination of GRSs, developed from recent well-powered GWAS on ultrasound-derived and DXA-derived relevant bone parameters, may be used for bone site–specific prediction of fracture as well as of separate prediction of trabecular and cortical bone microstructure parameters. In particular, we hypothesized that the eBMDGRS, developed from a high number of identified SNPs explaining a substantial part of the variance (20.3%) in eBMD at the calcaneus, may contribute information for bone site–specific fracture prediction beyond previously developed DXA-based GRSs (22). As the calcaneus is a bone with an exceptionally high trabecular bone content (29), we hypothesized that the eBMDGRS might capture unique information of trabecular bone microstructure, not identified by DXA-derived GRSs, useful for prediction of fracture risk at bone sites with a relatively high proportion of trabecular bone. The unique well-powered fracture cohorts from the Umeå Fracture and Osteoporosis (UFO) study, used in the present study for comparing bone site–specific fracture prediction of the different GRSs, did not have information on traditional clinical risk factors or FN-BMD. Therefore, the aim of the present explorative study was not to determine the clinical utility of the evaluated GRSs beyond current available fracture risk prediction tools.
Materials and Methods
Study participants
UFO cohort.
The Umeå Fracture and Osteoporosis (UFO) study is a population-based study designed to identify the genetic and environmental determinants of osteoporotic fractures. This cohort is sampled from the Northern Sweden Health and Disease Study (NSHDS), a longitudinal, population-based cohort study from Northern Sweden, consisting of blood samples, and lifestyle and dietary data from approximately 100 000 unique individuals from the county of Västerbotten (approximately 255 000 inhabitants as of Dec 31, 2007) (30, 31). The UFO-hip and UFO-forearm GWAS fracture cohorts (recruited in 2008) are subcohorts of the UFO cohort, currently including approximately 5000 fracture cases and approximately 5000 controls (30). Hip and forearm fracture cases were identified by merging the NSHDS cohort with medical records and radiographic reports. From 1993 to 2008, we identified 1086 subjects with low-trauma forearm fractures and 1086 subjects with hip fractures who were also represented with a DNA sample in the biobank. A low-trauma forearm fracture was defined as a fracture resulting from a fall from a standing position or lower. Thus, forearm fractures occurring after falls from >1 m or from traffic accidents were excluded. The UFO studies have been approved by local ethical committees and informed consent was obtained from all study participants.
The UFO-forearm fracture study is a nested case-control study in which each fracture case is compared with 1 control selected from the NSHDS cohort, matched for gender and age at recruitment, comprising a total of 2172 subjects of whom 2115 passed the quality check for genotyping. All patients in the UFO-forearm fracture study completed a survey about how the forearm fracture occurred. Only fractures caused by low energy trauma were included for forearm fractures. To refine the forearm fracture phenotype in this study, 36 subjects who had another known non-forearm fracture were excluded and 101 subjects were excluded for missing height and/or weight data, resulting in a final cohort of 1978 subjects (984 forearm cases and 994 controls). The inclusion criteria for cases in the UFO-forearm GWAS fracture cohort were an age of ≥ 30 years and having a low-trauma forearm fracture as defined by medical records and/or radiograph reports. The inclusion criteria for the forearm controls were an age of ≥ 30 years with no low-trauma fracture (until year 2008) and matching for gender and age at baseline (Table 1).
Table 1.
Characteristics of Study Participants
| UFO Hip-Fracture (n = 1873) | UFO Forearm-Fracture (n = 1978) | MrOS Sweden (n = 1880) | |
|---|---|---|---|
| Age (years) | 62.8 (11.7) | 61.0 (6.8) | 75.4 (3.2) |
| BMI (kg/m2) | 25.7 (4.3) | 25.5 (3.9) | 26.4 (3.5) |
| Height (cm) | 169.1 (9.1) | 165.7 (7.4) | 175.0 (6.5) |
| Weight (kg) | 73.7 (14.4) | 70.2 (12.5) | 80.7 (12.1) |
| Females (n,%) | 1069 (57.1) | 1677 (84.8) | 0 (0) |
| Hip fracture cases (n, %) | 994 (53.1) | 0 (0) | 129 (6.9) |
| Forearm fracture cases (n, %) | 0 (0) | 984 (49.7) | 36 (1.9) |
| Vertebral fractures cases (n, %) | 0 (0) | 0 (0) | 288 (19.5*) |
| eBMDGRS | 31.6 (0.6) | 31.6 (0.6) | 31.3 (0.6) |
| FN-BMDGRS | 2.33 (0.21) | 2.33 (0.21) | 2.35 (0.21) |
| LS-BMDGRS | 2.57 (0.22) | 2.58 (0.22) | 2.58 (0.23) |
Values are given as mean (SD) or n (%). *Percent of patients with validated vertebral fractures was calculated based on the 1475 participants with vertebral fracture information.
The UFO-hip fracture study is a case-cohort study in which the 1086 fracture cases are compared with a set of 934 controls, comprising a total of 2020 participants, of whom 1941 passed the quality check for genotyping. Fracture patients from the larger hospital in Umeå but not from the 2 smaller hospitals in Lycksele and Skelefteå completed a survey about how the hip fracture occurred. Only fractures caused by low energy trauma were included for hip fractures for the patients in Umeå. To refine the hip fracture phenotype in this study, 19 subjects who had another known non-hip fracture were excluded and 49 subjects were excluded for missing height and/or weight data, resulting in a final cohort of 1873 participants (994 hip cases and 879 controls). The inclusion criteria for the cases in the UFO-hip GWAS fracture cohort were an age of ≥ 20 years and a hip fracture defined by medical records and/or radiograph reports. Subjects with a previous known low-trauma forearm fracture were excluded. The UFO-hip GWAS fracture controls were drawn from the controls of a previous GWAS study of glioma (32) (Table 1).
MrOS Sweden
The Osteoporotic Fractures in Men (MrOS) study is a multicenter, prospective study including older men in Sweden, Hong Kong, and the United States. The MrOS Sweden cohort consists of 3 sub-cohorts from 3 Swedish cities (n = 1005 in Malmö, n = 1010 in Gothenburg, and n = 999 in Uppsala). Study participants (men aged 69 to 81 years) were randomly selected through national population registers, contacted, and asked to participate. A total of 45% of the subjects who were contacted participated in the study. To be eligible for the study, the participants had to be able to walk without assistance, provide self-reported data, and sign an informed consent (33). In this study, we included 1880 participants (953 from Gothenburg and 927 participants from Malmö) with available genetic data that passed the quality check for genotyping (Table 1). Participants in the MrOS Sweden cohort were followed for up to 10 years after the baseline examination. Fracture evaluation was done by searching digital x-ray archives and matching them with MrOS Sweden participants using unique personal identification number, which all Swedish citizens have. All reported fractures after baseline were confirmed by physician review of radiology report. Fractures with ICD10 code S72.0, S72.1, or S72.2 were classified as hip fractures while ICD10 code S52.5 and S52.6 were classified as forearm fractures (Table 1). Vertebral fractures were identified using thoracic spine x-ray at up to 3 occasions (at baseline, after 3 years and after 5 years). The radiographs were evaluated for radiographic vertebral fractures by an expert radiologist using a modified semiquantitative method developed by Genant and colleagues (34, 35). Individuals with a radiographic vertebral fracture at any of the occasions were classified as fracture cases while individuals with at least 1 x-ray and no vertebral fracture identified were classified as non-cases. Individuals without any thoracic spine x-ray were excluded from the analyses of radiographic vertebral fractures (Table 1). No information about the underlying cause of fracture is available for MrOS Sweden. The study was approved by the ethics committees at the Universities of Gothenburg, Lund, and Uppsala and informed consent was obtained from all study participants.
Genotyping
UFO cohort
UFO-forearm fracture cohort.
Genotyping of the forearm fracture cohort was performed using Illumina Omni express arrays. Genotypes were called using the BeadStudio calling algorithm. The sample quality control exclusion criteria were a sample call rate < 97.5%, gender discrepancy with genetic data from X-linked markers, excess autosomal heterozygosity > 0.33 (false discovery rate [FDR] < 0.1%), duplicates and/or first-degree relatives identified using identical-by-state (IBS) probabilities (>97%), ethnic outliers (3 standard deviations [SD] away from the population mean) using multidimensional scaling analysis with 4 principal components. Genotypes from 2115 individuals (1055 controls and 1060 cases) passed the sample quality control.
UFO hip fracture cohort.
Genotyping of the hip fracture cohort was performed using Illumina HumanHap660 arrays. Genotypes were called using the BeadStudio calling algorithm. The sample quality control exclusion criteria were a sample call rate < 97.5%, gender discrepancy with genetic data from X-linked markers, excess autosomal heterozygosity > 0.33 (~FDR < 0.1%), duplicates and/or first degree relatives identified using IBS probabilities (> 97%), ethnic outliers (3 SD away from the population mean) using multidimensional scaling analysis with 4 principal components. Genotypes from 1941 individuals (891 controls and 1050 cases) passed the sample quality control.
MrOS Sweden
Gothenburg part.
Genotyping, imputation, and quality controls were performed using the Illumina HumanOmni1_Quad_v1-0 B array. Genotypes were called using the Illumina’s BeadStudio calling algorithm. The sample quality control exclusion criteria were a sample call rate < 97%, excessive autosomal heterozygosity, first- and second-degree relatives, genotypic sex mismatch using X and Y chromosome probe intensities, and gross chromosome abnormalities. Genotyped SNPs with GenTrain scores < 0.6, cluster separation scores < 0.4, call rates < 97%, or minor allele frequency < 0.01 were excluded. Also, autosomal SNPs with Hardy-Weinberg Equilibrium P value < 10−4 were excluded and genotype clusters for SNPs on chrX, chrY, chrXY, and chrMT were reviewed manually. 714 543 autosomal SNPs passed quality control. Genotypes from 953 individuals passed the sample quality control.
Malmö part.
Genotyping and quality controls were performed using the HumanOmniExpress-12v1_B build 36. The sample quality control exclusion criteria were sample call rate < 97.5%, missing data, excessive autosomal heterozygosity, familiar relationship (1 sample excluded), genotypic sex mismatch, non-Caucasian ethnicity, and gross chromosome abnormalities. SNPs with call rates < 95% were excluded. 725 409 autosomal SNPs passed quality control. Genotypes from 927 individuals passed the sample quality control.
Imputation
For all cohorts, genotypes were imputed to the Haplotype Reference Consortium release 1.1 reference panel (36) yielding dosages for all SNPs, that is, continuous estimates of the number of risk alleles. The imputation was done separately for each cohort. Both pre-phasing and imputation were done using Sanger Imputation Service. SNPs with no variance or low imputation quality (< 0.3) were excluded. The HRC.r1 release consists of 64 940 haplotypes and a total of 40 405 505 variants of predominantly European ancestry.
Three different genetic risk scores
We defined 3 GRSs (FN-BMDGRS, LS-BMDGRS, and eBMDGRS) based on previously identified SNPs associated with the corresponding BMD measure in large-scale GWAS (21,22). A total of 1103 independent SNPs have been identified to associate with eBMD (22). In MrOS Sweden and the UFO cohorts, 1083 and 1085 SNPs, respectively, out of the 1103 SNPs had high-quality imputed dosage information available and were used for the calculation of the eBMDGRS. The GWAS results, including the effect sizes for the 1103 eBMD SNPs used in this study, are publicly available from Supplementary Table 2 in the original publication (22). Out of the 63 independent SNPs identified in the GWAS on FN-BMD and LS-BMD (21), 49 SNPs were associated with FN-BMD (P < 5 × 10–8) and included in the FN-BMDGRS while 48 of the 63 SNPs were associated with LS-BMD (P value < 5 × 10–8) and included in the LS-BMDGRS. All these 63 SNPs were imputed with high quality in UFO while 1 SNP had low imputation quality in MrOS Sweden and was excluded. The GWAS results, including the effect sizes for the 49 SNPs associated with FN-BMD and the 48 SNPs associated with LS-BMD are publicly available from Supplementary Tables 4A and 4B in the original publication (21). There is an overlap between the SNPs in the eBMDGRS and the 2 DXA-derived GRSs. A total of 27 of the SNPs in FN-BMDGRS and 28 of the SNPs in LS-BMDGRS are represented in the eBMDGRS by either the same SNP or a SNP with an R2 ≥ 0.8. For each individual, the GRSs were defined as the weighted sum of SNP dosages, where SNP effects from the corresponding BMD GWAS were used as weights. The GRSs were standardized to have a mean of zero and SD of 1.
Bone measures
Estimated BMD (eBMD) using ultrasound.
Speed of sound (SOS) and bone ultrasound attenuation (BUA) were measured by quantitative ultrasound (Hologic Sahara, Waltham, MA) at the left calcaneus in MrOS Sweden participants (37). The eBMD was then calculated as a linear combination of SOS and BUA using the following algorithm eBMD = 0.0025926 × (BUA + SOS) − 3.687) (38). Thus, eBMD is an indirectly measured BMD (Table 2). The eBMD is not used for osteoporosis diagnosis or fracture prediction in the clinical context, but some studies have shown that not only DXA-derived BMD but also ultrasound-derived eBMD is strongly associated with fracture risk (39, 40).
Table 2.
Bone Parameters in MrOS Sweden
| Ultrasound (n = 1880) | |
|---|---|
| Estimated BMD of Calcaneus (g/cm2) | 0.53 (0.15) |
| DXA (n = 1880) | |
| Femur Neck BMD (g/cm2) | 0.83 (0.13) |
| Lumbar Spine BMD (g/cm2) | 1.13 (0.20) |
| HR pQCT of Distal Radius (n = 426) | |
| Failure Load (N) | 3822 (827) |
| Trabecular parameters | |
| Trabecular vBMD (mg/cm3) | 165 (38) |
| Trabecular Number (1/mm) | 2.1 (0.3) |
| Trabecular Thickness (µm) | 67 (12) |
| Cortical prameters | |
| Cortical Area (mm2) | 53.2 (18.4) |
| Cortical vBMD (mg/cm3) | 886 (53) |
| Cortical Porosity (%) | 4.3 (1.7) |
Values are given as mean (SD).
Abbreviations: estimated BMD, estimated bone mineral density analysed by ultrasound; femur neck BMD, femoral neck bone mineral density analysed by dual energy absorptiometry (DXA); HRpQCT, high resolution peripheral quantitative computed tomography; lumbar spine BMD, lumbar spine bone mineral density analysed by DXA; vBMD, volumetric bone mineral density.
Areal BMDs using DXA.
Areal BMDs (g/cm2) of the femoral neck (FN-BMD) and lumbar spine (LS-BMD; L1–L4) were assessed using the Lunar Prodigy DXA (GE Lunar Corp., Madison, WI) for the subjects investigated in MrOS Sweden/Malmö, or the Hologic DXA Hologic QDR 4500/A‐Delphi (Hologic, Whaltman, MA) for subjects investigated in MrOS Sweden/Gothenburg. The coefficient of variations for the areal BMD measurements ranged from 0.5% to 3%, depending on application. To be able to use DXA measurements performed with equipment from the 2 different manufacturers, standardized BMD was calculated for these bone sites as previously described (33, 41–43) (Table 2).
High resolution peripheral quantitative computed tomography (HRpQCT).
Volumetric bone density (vBMD) and bone microarchitecture were assessed at the ultradistal radius with XtremeCT scanners (Scanco Medical, Switzerland), which were used by operators trained by the manufacturer (11). Scans were acquired with a nominal isotropic voxel size of 82 µm3. Scanning was done on the nondominant forearm. If a participant reported previous extremity fracture or had metal in the scan region, the contralateral extremity was examined. Anteroposterior scout views were used to place a reference line on the distal radial joint surface (44). The radial scan region was 9 mm in length (110 slices) and offset proximally to the reference line by 9.5 mm. Scanning of a quality control phantom limb containing rods of hydroxyapatite at densities of 0, 100, 200, 400, and 800 mg hydroxyapatite per cm3 was done daily to monitor long-term stability of the system. Scans were graded with a 5-point motion artefact scale (1 = none, 2 = minor, 3 = moderate, 4 = severe, and 5 = extreme) (45). For density measures, scans with movement artefacts graded 1 to 4 were retained, and for microarchitecture measures, those graded 1 to 3 were retained. A standard analysis program (Scanco software version 6.0) was used to assess total cross-sectional area, density, trabecular density, and trabecular microarchitecture, and a semi-automated cortical bone segmentation technique was used to assess cortical density and cortical microarchitecture (46). All bone measures were standardized to have a mean of zero and SD of 1 (Table 2).
Statistical analyses
Associations between the 3 GRSs and risk of forearm, hip, and vertebral fractures were first evaluated in separate logistic regression models, adjusted for age, sex, height, weight, and MrOS site when applicable. Differences between log odds for the separate GRSs were tested for significance using a z-test. Next, independent associations between the 3 GRSs and risk of forearm, hip, and vertebral fractures were evaluated in combined logistic regression models. The independently associated GRSs were selected by forward stepwise selection in logistic regression models starting from a fixed base model including age, sex, height, weight, and MrOS site. We then validated that the final models, including either 1 or 2 independently associated GRSs, also resulted in the lowest Akaike information criterion (AIC) for fracture prediction. The analyses were done separately for the UFO-forearm, the UFO-hip and the MrOS Sweden cohorts and the cohort–specific effects were then combined using inverse variance weighted meta-analysis. Association between the 3 GRSs and different bone parameters in MrOS Sweden were first evaluated in separate linear regression models, adjusted for age, sex, height, weight, and MrOS site. Differences between bone measure associations for the separate GRSs were tested for significance using a z-test. Next, independent association between the 3 GRSs and different bone parameters in MrOS Sweden were evaluated using stepwise selection in combined linear regression models, adjusted for age, sex, height, weight, and MrOS site. All analyses were performed using R version 3.4.2. Area under the receiver operating characteristic curve (AUC) and corresponding confidence intervals were calculated using the roc.test function in the pROC R-package (47).
Results
The ultrasound-based genetic risk score eBMDGRS displays modest correlations with the DXA-based genetic risk scores FN-BMDGRS and LS-BMDGRS
The means and SDs of the 3 bone-related GRSs, including the heel ultrasound-based eBMDGRS and the DXA-based FN-BMDGRS and LS-BMDGRS, were similar in the 3 evaluated Swedish cohorts, UFO hip fracture, UFO forearm fracture, and MrOS Sweden (Table 1). To determine if these 3 GRSs might have the potential to contribute independent information for prediction of bone health–related parameters, we first evaluated their intercorrelations in the 3 clinical cohorts used in the present study (Table 3). The FN-BMDGRS explained as much as 53.5% to 55% of the variance of the other DXA-based genetic risk score, LS-BMDGRS, in the 3 cohorts. In contrast, the variances in the 2 DXA-based risk scores explained by the newly-developed ultrasound-based eBMDGRS were rather modest: FN-BMDGRS (9.2% to 13.5%) and LS-BMDGRS (12.6% to 14.8%) (Table 3). Thus, the new eBMDGRS might contribute substantial independent information, beyond the 2 previously evaluated DXA-based GRSs, for prediction of different bone-related parameters. We next confirmed in the MrOS Sweden cohort that the different evaluated GRSs were associated with the underlying bone phenotype used for the development of the respective GRS (with a high GRS indicating a low value of the bone parameter). As expected, the eBMDGRS was significantly inversely associated with eBMD (variance explained R2 = 17.0%; P = 8.5 × 10−68), the FN-BMDGRS was significantly inversely associated with FN-BMD (R2 = 3.9%; P = 5.0 × 10−21) and the LS-BMDGRS was significantly inversely associated with LS-BMD (R2 = 4.6%; P = 1.2 × 10−23) (Table 4).
Table 3.
Cross Tab of Variance Explained (R2) for the 3 Genetic Risk Scores
| FN-BMDGRS | LS-BMDGRS | |
|---|---|---|
| UFO Hip-Fracture | ||
| eBMDGRS | 12.2% | 13.3% |
| FN-BMDGRS | 53.5% | |
| UFO Forearm-Fracture | ||
| eBMDGRS | 13.5% | 14.8% |
| FN-BMDGRS | 54.8% | |
| MrOS-Sweden | ||
| eBMDGRS | 9.2% | 12.6% |
| FN-BMDGRS | 55.0% |
Abbreviations: eBMDGRS, genetic risk score for estimated bone mineral density of the heel; FN-BMDGRS, genetic risk score for bone mineral density in femur neck; LS-BMDGRS, genetic risk score for bone mineral density in lumbar spine.
Table 4.
Association Between 3 Genetic Risk Scores (GRSs) and Different Bone Parameters in MrOS Sweden, Evaluated in Separate Linear Regression Models
| eBMDGRS | FN-BMDGRS | LS-BMDGRS | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Beta | SE | P | R 2 | Beta | SE | P | R 2 | Beta | SE | P | R 2 | |||
| Ultrasound Calcaneus | |||||||||||||||
| eBMD | 1567 | −0.41 | 0.02 | 8.5E-68 | 17.0% | −0.15 | 0.02 | 1.0E-09 | 2.3% | * | −0.16 | 0.02 | 1.0E-10 | 2.5% | * |
| DXA | |||||||||||||||
| FN-BMD | 1861 | −0.21 | 0.02 | 1.0E-24 | 4.6% | −0.20 | 0.02 | 5.0E-21 | 3.9% | −0.15 | 0.02 | 3.1E-12 | 2.2% | * | |
| LS-BMD | 1865 | −0.23 | 0.02 | 3.1E-27 | 5.4% | −0.17 | 0.02 | 2.5E-16 | 3.1% | −0.21 | 0.02 | 1.2E-23 | 4.6% | ||
| HRpQCT of Distal Radius | |||||||||||||||
| Failure Load | 354 | −0.38 | 0.05 | 7.0E-14 | 12.6% | −0.24 | 0.05 | 1.4E-06 | 5.5% | * | −0.22 | 0.05 | 3.1E-05 | 4.1% | * |
| Trabecular parameters | |||||||||||||||
| vBMD | 403 | −0.39 | 0.05 | 4.0E-15 | 13.8% | −0.24 | 0.05 | 1.2E-06 | 5.5% | * | −0.26 | 0.05 | 9.0E-07 | 5.6% | |
| Trabecular Number | 357 | −0.33 | 0.05 | 1.2E-09 | 9.2% | −0.20 | 0.05 | 9.2E-05 | 3.9% | −0.24 | 0.05 | 9.2E-06 | 5.0% | ||
| Trabecular Thickness | 357 | −0.31 | 0.05 | 9.7E-09 | 8.2% | −0.15 | 0.05 | 4.3E-03 | 2.1% | * | −0.14 | 0.05 | 1.2E-02 | 1.6% | * |
| Cortical parameters | |||||||||||||||
| Area | 426 | −0.24 | 0.05 | 3.8E-07 | 5.3% | −0.22 | 0.05 | 1.6E-06 | 4.8% | −0.23 | 0.05 | 2.2E-06 | 4.6% | ||
| vBMD | 397 | −0.20 | 0.05 | 3.9E-05 | 3.8% | −0.22 | 0.05 | 7.4E-06 | 4.5% | −0.20 | 0.05 | 1.3E-04 | 3.3% | ||
| Porosity | 352 | 0.04 | 0.06 | 4.7E-01 | 0.1% | 0.05 | 0.05 | 3.2E-01 | 0.3% | 0.05 | 0.06 | 4.1E-01 | 0.2% | ||
The models for ultrasound and dual energy absorptiometry (DXA) parameters, available both in the Gothenburg and the Malmö cohort of MrOS Sweden, are adjusted for age, height, weight, and MrOS site. The models for high resolution peripheral quantitative computed tomography (HRpQCT)-derived parameters in the distal radius, available only in the Gothenburg part of MrOS Sweden, are adjusted for age, height, and weight. Betas are expressed as SD change in bone parameter per SD increase in GRS.
Abbreviations: eBMD, estimated bone mineral density analysed by ultrasound; FN-BMD, femoral neck bone mineral density analysed by DXA; LS-BMD, lumbar spine bone mineral density analysed by DXA; vBMD, volumetric bone mineral density.
* P < 0.05 vs eBMDGRS
The eBMDGRS is an independent predictor of vertebral and forearm fractures, while both the FN-BMDGRS and the eBMDGRS contribute to hip fracture prediction
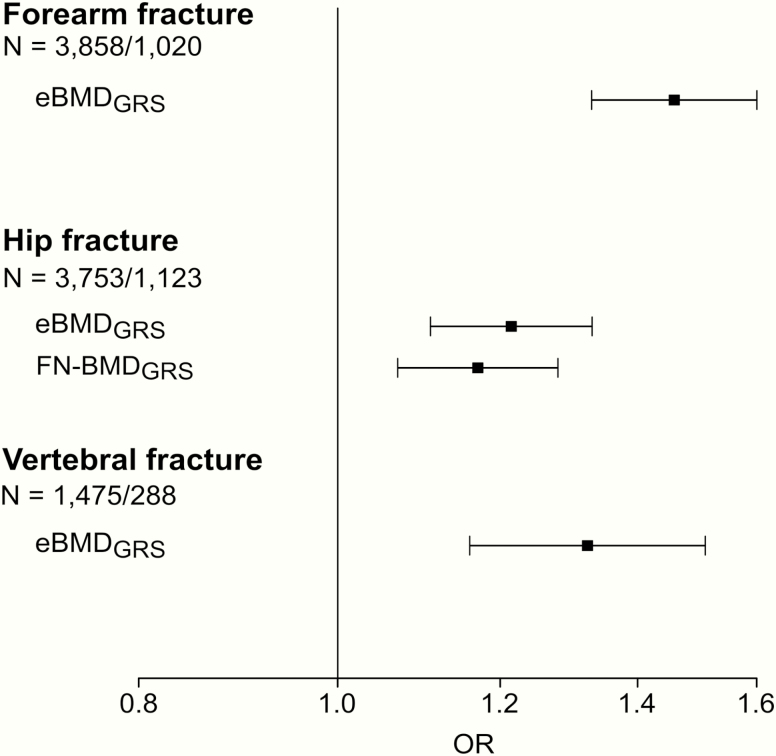
We next compared the separate and combined performances of the 3 bone-related GRSs for prediction of vertebral fractures, forearm, and hip fractures. Associations between the 3 GRSs and risk of forearm, hip, and vertebral fractures were first evaluated in separate logistic regression models. All 3 GRSs were significantly directly associated with fracture risk at all 3 bone sites (Fig 1). For forearm fractures and vertebral fractures, the effect sizes, expressed as odds ratio (OR) per SD increase in GRS, were more pronounced for the eBMDGRS compared with the 2 DXA-based GRSs, while for hip fractures the effect sizes were rather similar for the FN-BMDGRS and the eBMDGRS (Fig 1). AUCs for fracture discrimination for the GRSs in each of the included cohorts are given in Table 5. In general, these AUC data support the notion that the eBMDGRS is the most informative GRS for prediction of wrist and vertebral fractures, while the AUCs of the eBMDGRS and the FN-BMDGRS are of similar magnitude for hip fracture discrimination. As the GRSs were correlated with each other, we next evaluated the independent associations between the 3 GRSs and risk of fractures, using stepwise selection in combined logistic regression models (Fig 2). In these analyses, we observed that the eBMDGRS was the only independent predictor of forearm (OR [per SD increase] 1.46; 95% CI, 1.33-1.60) and vertebral fractures (OR 1.32; 95% CI, 1.16-1.51). In contrast, both the FN-BMDGRS (OR 1.17; 95% CI, 1.07-1.28) and the eBMDGRS (OR 1.21; 95% CI, 1.11-1.33) contributed independent information for prediction of hip fractures.
Figure 1.
Associations between 3 GRSs and risk of forearm, hip, and vertebral fractures, evaluated in separate logistic regression models. Models are adjusted for age, sex, height, weight, and MrOS site. Odds ratios (OR) and 95% confidence intervals given per SD increase of the genetic risk score (GRS) from inverse variance weighted meta-analysis of significant independent associations. The association between the eBMDGRS and forearm fractures was significantly stronger than the corresponding associations for the FN-BMDGRS (P = 0.002) and the LS-BMDGRS (P = 0.001). N = total number of subjects/fracture cases. Abbreviations: eBMD, estimated bone mineral density analysed by ultrasound; FN-BMD, femoral neck bone mineral density analysed by dual-energy absorptiometry; LS-BMD, lumbar spine bone mineral density analysed by dual-energy absorptiometry.
Table 5.
Area Under the ROC Curve (AUC) for 3 Genetic Risk Scores (GRSs) for Prediction of Risk of Forearm, Hip, and Vertebral Fractures, Evaluated in Separate Cohorts
| UFO Hip-Fracture N = 1873 | UFO Forearm Fracture N = 1978 | MrOS Sweden N = 1880* | |||||||
|---|---|---|---|---|---|---|---|---|---|
| AUC | 95% CI | N cases | AUC | 95% CI | N cases | AUC | 95% CI | N cases | |
| Hip fracture | 994 | 129 | |||||||
| eBMDGRS | 0.56 | (0.54, 0.59) | 0.56 | (0.51, 0.61) | |||||
| FN-BMDGRS | 0.56 | (0.53, 0.58) | 0.57 | (0.52, 0.62) | |||||
| LS-BMDGRS | 0.55 | (0.52, 0.58) | 0.56 | (0.50, 0.62) | |||||
| Forearm fracture | 984 | 36 | |||||||
| eBMDGRS | 0.60 | (0.57, 0.62) | 0.66 | (0.58, 0.75) | |||||
| FN-BMDGRS | 0.55 | (0.52, 0.57) | 0.60 | (0.50, 0.70) | |||||
| LS-BMDGRS | 0.55 | (0.53, 0.58) | 0.58 | (0.47, 0.69) | |||||
| Vertebral fracture | 288 | ||||||||
| eBMDGRS | 0.57 | (0.53, 0.61) | |||||||
| FN-BMDGRS | 0.55 | (0.51, 0.59) | |||||||
| LS-BMDGRS | 0.54 | (0.51, 0.58) | |||||||
Area under the ROC curve (AUC) and 95% confidence intervals (CI) are given for each GRS in separate unadjusted models. N = number of participants. N cases = number of fracture cases.
Abbreviations: eBMDGRS, genetic risk score for estimated bone mineral density of the heel; FN-BMDGRS, genetic risk score for bone mineral density in femur neck; LS-BMDGRS, genetic risk score for bone mineral density in lumbar spine.
*For vertebral fractures, only N = 1475 participants were included.
Figure 2.
GRSs independently associated with risk of forearm, hip and vertebral fractures. The independently associated genetic risk scores (GRSs) were selected by forward stepwise selection in logistic regression models starting from a fixed base model including age, sex, height, weight, and MrOS site. We then validated that the final models, including either 1 or 2 independently associated GRSs, also resulted in the lowest AIC. Odds ratios (OR) and 95% confidence intervals given per SD increase in GRS from inverse variance weighted meta-analysis of significant independent associations. N = total number of subjects/fracture cases.
Abbreviations: eBMD, estimated bone mineral density analysed by ultrasound; FN-BMD, femoral neck bone mineral density analysed by dual-energy absorptiometry.
We next performed an explorative subanalysis dividing up both the eBMDGRS and the FN-BMDGRS into 1 GRS that included only the common SNPs (27 SNPs) between the eBMDGRS and FN-BMDGRS and 1 GRS containing only the SNPs specific for either the FN-BMDGRS (22 SNPs) or the eBMDGRS (1076 SNPs). This results in 6 different GRSs (eBMDGRS, CommonSNP-eBMDGRS, SpecificSNP-eBMDGRS, FN-BMDGRS and CommonSNP-FNBMDGRS, SpecificSNP-FNBMDGRS) (Table 6) for comparison of their associations with fracture risk at different bone sites. The CommonSNP-eBMDGRS and the CommonSNP-FNBMDGRS, both including the same common 27 SNPs, displayed very similar patterns of association with fracture risk at all 3 bone sites (Table 6) and these associations were also very similar to the associations for the FN-BMDGRS for fractures at all 3 bone sites. In contrast, the eBMDGRS displayed stronger associations with forearm and vertebral fractures, but not with hip fractures, compared with the corresponding associations for the 2 Common SNP GRSs. Interestingly, the SpecificSNP-eBMDGRS demonstrated very similar fracture prediction as the eBMDGRS for fractures at all 3 bone sites while the SpecificSNP-FNBMDGRS was only associated with risk of hip fractures. These exploratory subanalyses support the notion that a substantial part of the better performance of the eBMDGRS compared with the FN-BMDGRS is dependent on the larger number of SNPs and variance explained by the eBMDGRS. Yet, the specific eBMD SNPs seem to add unique information not captured by the FN-BMDGRS for prediction of forearm and vertebral fractures.
Table 6.
Comparison of Fracture Risk Associations Using Different SNP Selections for Genetic Risk Scores (GRS)
| Forearm Fracture N = 3858/1020 | Hip Fracture N = 3753/1123 | Vertebral Fracture N = 1475/288 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Selection of SNPs | Number of SNPs | Variance (% explained of the underlying bone phenotype in MrOS Sweden) | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | |
| eBMDGRS | All SNPs | 1103 | 17.0% | 1.46 | (1.33, 1.60) | 1.2E-16 | 1.26 | (1.15, 1.37) | 1.4E-07 | 1.32 | (1.16, 1.51) | 4.5E-05 |
| FN-BMDGRS | All SNPs | 49 | 3.9% | 1.20 | (1.10, 1.30) | 5.2E-05 | 1.22 | (1.12, 1.33) | 3.3E-06 | 1.15 | (1.01, 1.31) | 3.1E-02 |
| CommonSNP-eBMDGRS | Common SNPs | 27 | 3.3% | 1.27 | (1.16, 1.38) | 1.0E-07 | 1.20 | (1.11, 1.31) | 6.2E-06 | 1.13 | (0.99, 1.29) | 6.0E-02 |
| CommonSNP-FNBMDGRS | Common SNPs | 27 | 1.8% | 1.24 | (1.14, 1.36) | 8.5E-07 | 1.20 | (1.11, 1.30) | 1.0E-05 | 1.14 | (1.00, 1.30) | 4.3E-02 |
| SpecificSNP-eBMDGRS | Specific SNPs | 1076 | 15.7% | 1.40 | (1.28, 1.53) | 9.4E-14 | 1.27 | (1.17, 1.38) | 3.8E-08 | 1.30 | (1.14, 1.49) | 9.3E-05 |
| SpecificSNP-FNBMDGRS | Specific SNPs | 22 | 2.2% | 1.02 | (0.94, 1.11) | 6.0E-01 | 1.23 | (1.13, 1.34) | 1.5E-06 | 1.07 | (0.94, 1.22) | 2.9E-01 |
Both eBMDGRS and the FN-BMDGRS were divided into 1 GRS including only the common SNPs (27 SNPs) between the eBMDGRS and FN-BMDGRS and 1 GRS including only SNPs specific for either the FN-BMDGRS (22 SNPs) or the eBMDGRS (1076 SNPs). This results in 6 different GRSs (eBMDGRS, CommonSNP-eBMDGRS, SpecificSNP-eBMDGRS, FN-BMDGRS and CommonSNP-FNBMDGRS, SpecificSNP-FNBMDGRS. The table displays odds ratios (OR) and 95% confidence intervals (CI) for fracture risk for the 6 different GRSs and the 3 different fracture types.
Abbreviations: eBMDGRS, genetic risk score for estimated bone mineral density of the heel; FN-BMDGRS, genetic risk score for bone mineral density in femur neck; SNP, single nucleotide polymorphism.
The eBMDGRS is the major predictor of trabecular bone microstructure parameters, while both the FN-BMDGRS and the eBMDGRS contribute independent information for prediction of cortical bone area and cortical density
eBMD and DXA-based BMD.
We first compared the associations between the 3 GRSs and the different underlying ultrasound and DXA-based bone parameters in MrOS Sweden, evaluated in separate models. We observed that for eBMD, the eBMDGRS explained substantially more of the variance than what the 2 DXA-based GRSs did (Table 4). In contrast, the FN-BMDGRS explained the variance of FN-BMD similarly well as the eBMDGRS did and the LS-BMDGRS explained the variance of LS-BMD similarly well as the eBMDGRS did. When evaluated using stepwise selection in combined linear regression models, only the eBMDGRS was independently associated with eBMD while both the FN-BMDGRS and the eBMDGRS were independently associated with FN-BMD, and both the LS-BMDGRS and the eBMDGRS were independently associated with LS-BMD (Table 7). Thus, the eBMDGRS contributes to the prediction of the DXA-based BMDs beyond the site-specific GRSs while the DXA-based GRSs do not contribute to the prediction of eBMD beyond the eBMDGRS.
Table 7.
Association Between 3 Genetic Risk Scores (GRSs) and Different Bone Parameters in MrOS Sweden, Evaluated Using Stepwise Selection in Combined Linear Regression Models
| eBMDGRS | FN-BMDGRS | LS-BMDGRS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | Beta | SE | P | Beta | SE | P | Beta | SE | P | |
| Ultrasound Calcaneus | ||||||||||
| eBMD | 1567 | −0.41 | 0.02 | 8.5E-68 | ||||||
| DXA | ||||||||||
| FN-BMD | 1861 | −0.17 | 0.02 | 3.0E-15 | −0.15 | 0.02 | 1.6E-11 | |||
| LS-BMD | 1865 | −0.17 | 0.02 | 3.4E-15 | −0.15 | 0.02 | 1.4E-11 | |||
| HRpQCT of Distal Radius | ||||||||||
| Failure Load | 354 | −0.34 | 0.05 | 3.9E-11 | −0.16 | 0.05 | 9.2E-04 | |||
| Trabecular parameters | ||||||||||
| vBMD | 403 | −0.35 | 0.05 | 4.3E-12 | −0.15 | 0.05 | 1.6E-03 | |||
| Trabecular Number | 357 | −0.29 | 0.05 | 1.1E-07 | −0.13 | 0.05 | 9.9E-03 | |||
| Trabecular Thickness | 357 | −0.31 | 0.05 | 9.7E-09 | ||||||
| Cortical parameters | ||||||||||
| Area | 426 | −0.19 | 0.05 | 4.9E-05 | −0.17 | 0.05 | 2.2E-04 | |||
| vBMD | 397 | −0.16 | 0.05 | 1.9E-03 | −0.18 | 0.05 | 3.5E-04 | |||
The models for ultrasound and dual energy absorptiometry (DXA) parameters, available both in the Gothenburg and the Malmö cohort of MrOS Sweden, are adjusted for age, height, weight, and MrOS site. The models for high resolution peripheral quantitative computed tomography (HRpQCT)-derived parameters in the distal radius, available only in the Gothenburg part of MrOS Sweden, are adjusted for age, height, and weight. Betas are expressed as SD change in bone parameter per SD increase in GRS of significant independent associations.
Abbreviations: eBMD, estimated bone mineral density analysed by ultrasound; FN-BMD, femoral neck bone mineral density analysed by DXA; LS-BMD, lumbar spine bone mineral density analysed by DXA; vBMD, volumetric bone mineral density.
Trabecular and cortical bone microstructure parameters.
We next hypothesized that the different GRSs might predict cortical and trabecular bone microstructure parameters differentially. The separate and combined associations between the 3 GRSs and bone microstructure parameters were evaluated using HRpQCT measurements at the distal radius (the bone site for forearm fractures) available in the MrOS Sweden cohort.
When comparing the separate association between the 3 GRSs and different bone microstructure parameters, we observed that the eBMDGRS was the main predictor of the trabecular bone parameters vBMD, trabecular number, and trabecular thickness, as well as of the overall bone strength-related parameter failure load. In contrast, the DXA-based FN-BMDGRS predicted the cortical bone parameters cortical area and cortical volumetric BMD similarly to the eBMDGRS (Table 4). Interestingly, none of the evaluated GRSs displayed any tendency of association with cortical porosity. When evaluated using stepwise selection in combined linear regression models, eBMDGRS was the only independent predictor of trabecular thickness (R2 = 8.2%) and the major predictor of trabecular vBMD (R2 = 13.8%), trabecular number (R2 = 9.2%), and failure load (R2 = 12.6%) (Table 7 and Fig 3). In contrast, both the FN-BMDGRS and the eBMDGRS contributed independently and approximately equally to the prediction of the cortical bone parameters cortical bone area and cortical vBMD (Table 7).
Figure 3.
The variance explained (R2) of different bone parameters in MrOS Sweden by eBMDGRS and FN-BMDGRS evaluated using stepwise selection in combined linear regression analyses. Black bars = variance explained independently by eBMDGRS. White bar = variance explained independently by FN-BMDGRS. Grey bar = variance explained shared by eBMDGRS and FN-BMDGRS. Trabecular thickness, cortical bone area and cortical volumetric BMD (cortical vBMD) were analysed by high resolution peripheral quantitative computed tomography. Abbreviations: eBMD, estimated bone mineral density analysed by ultrasound; FN-BMD, femoral neck bone mineral density analysed by dual-energy absorptiometry; GRS, genetic risk score.
Discussion
The differences in genetic contribution to bone site–specific fracture risk and to trabecular versus cortical bone microstructure parameters are poorly investigated. In this study, we hypothesized that different GRSs or combinations of GRSs, developed from recent well-powered GWAS on ultrasound-derived and DXA-derived relevant bone parameters, may be used for the separate prediction of fractures at different bone sites, including forearm, vertebral, and hip fractures, as well as for separate prediction of trabecular and cortical bone microstructure parameters. We demonstrate that the ultrasound-based calcaneus eBMDGRS captures unique information of trabecular bone microstructure parameters as well as of risk of vertebral and forearm fractures, while both the DXA-based FN-BMD and the eBMDGRS capture unique information of cortical bone mass and hip fracture risk (Fig 4).
Figure 4.
Summary of the separate and combined prediction of fracture types and trabecular and cortical bone microstructure by eBMDGRS and FN-BMDGRS. eBMDGRS but not FN-BMDGRS independently predicted trabecular bone thickness as well as vertebral and forearm fractures. In contrast, both FN-BMDGRS and eBMDGRS independently predicted cortical bone mass parameters (cortical area and cortical thickness) and hip fractures. We propose that eBMDGRS captures unique information of trabecular bone microstructure useful for the prediction of forearm and vertebral fractures. In contrast, both FN-BMDGRS and eBMDGRS capture unique information of cortical bone mass useful for the prediction of hip fractures. Abbreviations: eBMD, estimated bone mineral density analysed by ultrasound; FN-BMD, femoral neck bone mineral density analysed by dual-energy absorptiometry.
Two previous studies have shown that fracture risk prediction is modest when only using available DXA-derived GRSs (3, 27). The bone site-specific prediction of fracture risk was, however, not evaluated in these studies. In addition, no previous study has evaluated the impact of any bone-related GRS for cortical and trabecular bone microstructure parameters. In the present study we observed that the ultrasound-based eBMDGRS displayed modest correlations with the 2 DXA-based GRSs, demonstrating that the eBMDGRS might contribute with independent information beyond the information from DXA-derived GRSs for prediction of bone health–related parameters. Besides contributing unique bone property information not captured by the DXA-derived GRSs, the eBMDGRS explained approximately 5 times more of the variance of the underlying bone phenotype than the 2 evaluated DXA-based GRSs. The latter was most likely the result of the exceptional large UK-Biobank cohort used for the development of the eBMDGRS. This is most likely the reason why the eBMDGRS in general was a stronger predictor of fracture risk and bone microstructure parameters than the 2 DXA-based GRSs. However, importantly both for bone site-specific fracture prediction and for association with cortical versus trabecular bone microstructure parameters, there were clear differences in the patterns of the relative importance of the different GRSs. The eBMDGRS but not FN-BMDGRS independently predicted trabecular bone thickness as well as vertebral and forearm fractures. In contrast, both FN-BMDGRS and eBMDGRS independently predicted cortical bone mass parameters (cortical area and cortical thickness) and hip fractures (Fig 4). We propose that eBMDGRS captures unique information on trabecular bone microstructure useful for prediction of forearm and vertebral fractures. The strong prediction of trabecular bone parameters by eBMDGRS is most likely related to the exceptional high trabecular bone content in the calcaneus (90%) where ultrasound eBMD is measured (29). In contrast, both FN-BMDGRS and eBMDGRS capture unique information on cortical bone mass useful for prediction of hip fractures (Fig 4).
There are some clear advantages to using GRS for the prediction of different health outcomes. First, GRSs do not change over the lifetime, enabling both early identification of individuals at high risk to be considered for primary prevention strategies and for later risk assessment in patients considered for secondary prevention strategies. In addition, genome-wide array genotyping has a relatively low one-time cost and can be used to calculate GRSs not only for fracture risk prediction but also for prediction of several other complex diseases (48–50).
The main strength with the present study is the large number of x-ray–verified forearm, hip, and vertebral fractures, enabling separate fracture prediction by bone site. In addition, detailed HRpQCT analyses at the distal radius were available in a subset of MrOS Sweden, enabling us to evaluate the combined performances of the GRSs for trabecular and cortical bone microstructure parameters separately at the bone site for forearm fractures. Although we aimed to determine the predictive value of the GRSs for the separate prediction of fractures at different bone sites, as well as for trabecular and cortical bone microstructure, a limitation is that information on clinical risk factors and FN-BMD were not available in the 2 large fracture cohorts used in the present study. Further studies in large cohort with a high fracture prevalence and information on clinical risk factors and available FN-BMD are warranted to determine if the herein-identified combinations of GRSs predict bone site-specific fracture risk beyond clinical risk factors and FN-BMD. Nevertheless, current fracture models in clinical use are not designed to discriminate between risk for fractures at different bone sites (13, 14). It is a limitation with the present study that we only used stepwise regression and not more modern methods such as LASSO or Bayesian Model Averaging. Another limitation with the present study is that only individuals of white ethnic background were included, and because the genetic background varies in different populations, we cannot generalize our findings to other race/ethnic populations.
In conclusion, the eBMDGRS is the only independent GRS for prediction of forearm and vertebral fractures while both FN-BMDGRS and eBMDGRS contribute independent information for prediction of hip fractures. We propose that eBMDGRS captures unique information of trabecular bone microstructure useful for the prediction of forearm and vertebral fractures. The findings in the present study may facilitate personalized medicine to predict different fracture types as well as cortical and trabecular bone microstructure parameters separately. Personalized medicine has the potential to customize therapy with the best response and highest safety margin to ensure better patient care, by enabling each patient to receive earlier diagnoses, risk assessments, and optimal treatments.
Acknowledgments
UFO cohort
The UFO cohort is funded by the Swedish Research Council, the Swedish Foundation for Strategic Research, by grants from the Swedish state under the agreement between the Swedish government and the county councils, the ALF-agreement (ALFGBG-720331 and ALF-VLL 679611), the Lundberg Foundation, the Torsten and Ragnar Söderberg’s Foundation, the Novo Nordisk Foundation, and the European Commission grant HEALTH-F2-2008-201865-GEFOS, BBMRI.se, the Swedish Society of Medicine, the Kempe-Foundation (JCK-1021), the Medical Faculty of Umeå University, the county council of Västerbotten (Spjutspetsanslag VLL:159:33–2007). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
MrOS Sweden cohort
MrOS in Sweden is supported by the Swedish Research Council, the Swedish Foundation for Strategic Research, the ALF/LUA research grant in Gothenburg, the Lundberg Foundation, the Knut and Alice Wallenberg Foundation, the Torsten Soderberg Foundation, and the Novo Nordisk Foundation.
Financial Support: This work was supported by the Swedish Research Council, the Swedish Foundation for Strategic Research, by grants from the Swedish state under the agreement between the Swedish government and the county councils, the ALF research grants in Gothenburg and Västerbotten, the Lundberg Foundation, Knut and Alice Wallenberg Foundation, the Torsten and Ragnar Söderberg’s Foundation, the Novo Nordisk Foundation, the European Commission grant, BBMRI.se, the Swedish Society of Medicine, the Kempe-Foundation, the Medical Faculty of Umeå University and the county council of Västerbotten Foundation.
Glossary
Abbreviations
- AIC
Akaike information criterion
- AUC
area under the receiver operating characteristic curve
- BMD
bone mineral density
- BUA
bone ultrasound attenuation
- CT
computed tomography
- DXA
dual-energy x-ray absorptiometry
- eBMD
estimated bone mineral density of the calcaneus
- eBMDGRS
weighted genetic risk score for estimated bone mineral density of the calcaneus
- FDR
false discovery rate
- FN-BMD
femoral neck bone mineral density
- FN-BMDGRS
weighted genetic risk score for femoral neck bone mineral density
- GWAS
genome-wide association study
- HRpQCT
high resolution peripheral quantitative computed tomography
- IBS
identical-by-state
- LS-BMD
lumbar spine bone mineral density
- MrOS
Osteoporotic Fractures in Men
- NSHDS
Northern Sweden Health and Disease Study
- SNP
single nucleotide polymorphism
- SOS
speed of sound
- UFO
Umeå Fracture and Osteoporosis
- vBMD
volumetric bone mineral density
Additional Information
Disclosure Summary: The authors have nothing to disclose.
Data Availability: Restrictions apply to the availability of data generated or analyzed during this study to preserve patient confidentiality or because they were used under license. The corresponding author will on request detail the restrictions and any conditions under which access to some data may be provided.
References
- 1. Kanis JA, Melton LJ 3rd, Christiansen C, Johnston CC, Khaltaev N. The diagnosis of osteoporosis. J Bone Miner Res. 1994;9(8):1137–1141. [DOI] [PubMed] [Google Scholar]
- 2. Ohlsson C. Bone metabolism in 2012: Novel osteoporosis targets. Nat Rev Endocrinol. 2013;9(2):72–74. [DOI] [PubMed] [Google Scholar]
- 3. Eriksson J, Evans DS, Nielson CM, et al. Limited clinical utility of a genetic risk score for the prediction of fracture risk in elderly subjects. J Bone Miner Res. 2015;30(1):184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bauer DC, Ewing SK, Cauley JA, Ensrud KE, Cummings SR, Orwoll ES; Osteoporotic Fractures in Men (MrOS) Research Group Quantitative ultrasound predicts hip and non-spine fracture in men: the MrOS study. Osteoporos Int. 2007;18(6):771–777. [DOI] [PubMed] [Google Scholar]
- 5. Bauer DC, Glüer CC, Cauley JA, et al. Broadband ultrasound attenuation predicts fractures strongly and independently of densitometry in older women. A prospective study. Study of Osteoporotic Fractures Research Group. Arch Intern Med. 1997;157(6):629–634. [PubMed] [Google Scholar]
- 6. Langsetmo L, Peters KW, Burghardt AJ, et al. ; Osteoporotic Fractures in Men (MrOS) Study Research Group Volumetric bone mineral density and failure load of distal limbs predict incident clinical fracture independent HR-pQCT BMD and failure load predicts incident clinical fracture of FRAX and clinical risk factors among older men. J Bone Miner Res. 2018;33(7):1302–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ohlsson C, Sundh D, Wallerek A, et al. Cortical bone area predicts incident fractures independently of areal bone mineral density in older men. J Clin Endocrinol Metab. 2017;102(2):516–524. [DOI] [PubMed] [Google Scholar]
- 8. Szulc P, Boutroy S, Chapurlat R. Prediction of fractures in men using bone microarchitectural parameters assessed by high-resolution peripheral quantitative computed tomography-the prospective STRAMBO Study. J Bone Miner Res. 2018;33(8):1470–1479. [DOI] [PubMed] [Google Scholar]
- 9. Biver E, Durosier-Izart C, Chevalley T, van Rietbergen B, Rizzoli R, Ferrari S. Evaluation of radius microstructure and areal bone mineral density improves fracture prediction in postmenopausal women. J Bone Miner Res. 2018;33(2):328–337. [DOI] [PubMed] [Google Scholar]
- 10. Sornay-Rendu E, Boutroy S, Duboeuf F, Chapurlat RD. Bone Microarchitecture Assessed by HR-pQCT as Predictor of Fracture Risk in Postmenopausal Women: The OFELY Study. J Bone Miner Res. 2017;32(6):1243–1251. [DOI] [PubMed] [Google Scholar]
- 11. Samelson EJ, Broe KE, Xu H, et al. Cortical and trabecular bone microarchitecture as an independent predictor of incident fracture risk in older women and men in the Bone Microarchitecture International Consortium (BoMIC): a prospective study. Lancet Diabetes Endocrinol. 2019;7(1):34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Biver E, Samelson EJ, Xu H, et al. 2019 Site-specific prediction of fractures by BMD and bone microarchitecture in older women and men: the Bone Microarchitecture International Consortium (BoMIC). J Bone Miner Res. 34(Suppl 1). Abstracts from the 2019 Annual Meeting of the American Society for Bone and Mineral Research (ASBMR 2019). doi:10.1002/jbmr.3936 [Google Scholar]
- 13. Kanis JA, McCloskey EV, Johansson H, Oden A, Ström O, Borgström F. Development and use of FRAX in osteoporosis. Osteoporos Int. 2010;21(Suppl 2):S407–S413. [DOI] [PubMed] [Google Scholar]
- 14. Nguyen ND, Frost SA, Center JR, Eisman JA, Nguyen TV. Development of prognostic nomograms for individualizing 5-year and 10-year fracture risks. Osteoporos Int. 2008;19(10):1431–1444. [DOI] [PubMed] [Google Scholar]
- 15. Arden NK, Baker J, Hogg C, Baan K, Spector TD. The heritability of bone mineral density, ultrasound of the calcaneus and hip axis length: a study of postmenopausal twins. J Bone Miner Res. 1996;11(4):530–534. [DOI] [PubMed] [Google Scholar]
- 16. Howard GM, Nguyen TV, Harris M, Kelly PJ, Eisman JA. Genetic and environmental contributions to the association between quantitative ultrasound and bone mineral density measurements: a twin study. J Bone Miner Res. 1998;13(8):1318–1327. [DOI] [PubMed] [Google Scholar]
- 17. Karasik D, Demissie S, Zhou Y, et al. Heritability and genetic correlations for bone microarchitecture: the Framingham Study families. J Bone Miner Res. 2017;32(1):106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Karasik D, Myers RH, Hannan MT, et al. Mapping of quantitative ultrasound of the calcaneus bone to chromosome 1 by genome-wide linkage analysis. Osteoporos Int. 2002;13(10):796–802. [DOI] [PubMed] [Google Scholar]
- 19. Peacock M, Turner CH, Econs MJ, Foroud T. Genetics of osteoporosis. Endocr Rev. 2002;23(3):303–326. [DOI] [PubMed] [Google Scholar]
- 20. Ralston SH, Uitterlinden AG. Genetics of osteoporosis. Endocr Rev. 2010;31(5):629–662. [DOI] [PubMed] [Google Scholar]
- 21. Estrada K, Styrkarsdottir U, Evangelou E, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44(5):491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morris JA, Kemp JP, Youlten SE, et al. ; 23andMe Research Team An atlas of genetic influences on osteoporosis in humans and mice. Nat Genet. 2019;51(2):258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paternoster L, Lorentzon M, Lehtimäki T, et al. Genetic determinants of trabecular and cortical volumetric bone mineral densities and bone microstructure. Plos Genet. 2013;9(2):e1003247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paternoster L, Lorentzon M, Vandenput L, et al. Genome-wide association meta-analysis of cortical bone mineral density unravels allelic heterogeneity at the RANKL locus and potential pleiotropic effects on bone. Plos Genet. 2010;6(11):e1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zheng HF, Tobias JH, Duncan E, et al. WNT16 influences bone mineral density, cortical bone thickness, bone strength, and osteoporotic fracture risk. Plos Genet. 2012;8(7):e1002745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nielson CM, Liu CT, Smith AV, et al. Novel genetic variants associated with increased vertebral volumetric BMD, reduced vertebral fracture risk, and increased expression of SLC1A3 and EPHB2. J Bone Miner Res. 2016;31(12):2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ho-Le TP, Center JR, Eisman JA, Nguyen HT, Nguyen TV. Prediction of bone mineral density and fragility fracture by genetic profiling. J Bone Miner Res. 2017;32(2):285–293. [DOI] [PubMed] [Google Scholar]
- 28. Kim SK. Identification of 613 new loci associated with heel bone mineral density and a polygenic risk score for bone mineral density, osteoporosis and fracture. Plos One. 2018;13(7):e0200785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Langton CM. The 25th anniversary of BUA for the assessment of osteoporosis: time for a new paradigm? Proc Inst Mech Eng H. 2011;225(2):113–125. [DOI] [PubMed] [Google Scholar]
- 30. Englund U, Nordström P, Nilsson J, et al. Physical activity in middle-aged women and hip fracture risk: the UFO study. Osteoporos Int. 2011;22(2):499–505. [DOI] [PubMed] [Google Scholar]
- 31. Hallmans G, Agren A, Johansson G, et al. Cardiovascular disease and diabetes in the Northern Sweden Health and Disease Study Cohort - evaluation of risk factors and their interactions. Scand J Public Health Suppl. 2003;61:18–24. [DOI] [PubMed] [Google Scholar]
- 32. Rajaraman P, Melin BS, Wang Z, et al. Genome-wide association study of glioma and meta-analysis. Hum Genet. 2012;131(12):1877–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mellström D, Johnell O, Ljunggren O, et al. Free testosterone is an independent predictor of BMD and prevalent fractures in elderly men: MrOS Sweden. J Bone Miner Res. 2006;21(4):529–535. [DOI] [PubMed] [Google Scholar]
- 34. Genant HK, Wu CY, van Kuijk C, Nevitt MC. Vertebral fracture assessment using a semiquantitative technique. J Bone Miner Res. 1993;8(9):1137–1148. [DOI] [PubMed] [Google Scholar]
- 35. Kherad M, Rosengren BE, Hasserius R, et al. Low clinical relevance of a prevalent vertebral fracture in elderly men–the MrOs Sweden study. Spine J. 2015;15(2):281–289. [DOI] [PubMed] [Google Scholar]
- 36. McCarthy S, Das S, Kretzschmar W, et al. ; Haplotype Reference Consortium A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cronholm F, Rosengren BE, Nilsson JÅ, et al. The fracture predictive ability of a musculoskeletal composite score in old men - data from the MrOs Sweden study. BMC Geriatr. 2019;19(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kemp JP, Morris JA, Medina-Gomez C, et al. Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat Genet. 2017;49(10):1468–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johnell O, Kanis JA, Oden A, et al. Predictive value of BMD for hip and other fractures. J Bone Miner Res. 2005;20(7):1185–1194. [DOI] [PubMed] [Google Scholar]
- 40. McCloskey EV, Kanis JA, Odén A, et al. Predictive ability of heel quantitative ultrasound for incident fractures: an individual-level meta-analysis. Osteoporos Int. 2015;26(7):1979–1987. [DOI] [PubMed] [Google Scholar]
- 41. Lu Y, Fuerst T, Hui S, Genant HK. Standardization of bone mineral density at femoral neck, trochanter and Ward’s triangle. Osteoporos Int. 2001;12(6):438–444. [DOI] [PubMed] [Google Scholar]
- 42. Hanson J. Standardization of proximal femur BMD measurements. International Committee for Standards in Bone Measurement. Osteoporos Int. 1997;7(5):500–501. [DOI] [PubMed] [Google Scholar]
- 43. Genant HK, Grampp S, Glüer CC, et al. Universal standardization for dual x-ray absorptiometry: patient and phantom cross-calibration results. J Bone Miner Res. 1994;9(10):1503–1514. [DOI] [PubMed] [Google Scholar]
- 44. Boutroy S, Bouxsein ML, Munoz F, Delmas PD. In vivo assessment of trabecular bone microarchitecture by high-resolution peripheral quantitative computed tomography. J Clin Endocrinol Metab. 2005;90(12):6508–6515. [DOI] [PubMed] [Google Scholar]
- 45. Pialat JB, Burghardt AJ, Sode M, Link TM, Majumdar S. Visual grading of motion induced image degradation in high resolution peripheral computed tomography: impact of image quality on measures of bone density and micro-architecture. Bone. 2012;50(1):111–118. [DOI] [PubMed] [Google Scholar]
- 46. Buie HR, Campbell GM, Klinck RJ, MacNeil JA, Boyd SK. Automatic segmentation of cortical and trabecular compartments based on a dual threshold technique for in vivo micro-CT bone analysis. Bone. 2007;41(4):505–515. [DOI] [PubMed] [Google Scholar]
- 47. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Inouye M, Abraham G, Nelson CP, et al. ; UK Biobank CardioMetabolic Consortium CHD Working Group Genomic risk prediction of coronary artery disease in 480,000 adults: implications for primary prevention. J Am Coll Cardiol. 2018;72(16):1883–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lourida I, Hannon E, Littlejohns TJ, et al. Association of lifestyle and genetic risk with incidence of dementia. JAMA. 2019;322(5):430-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rutten-Jacobs LC, Larsson SC, Malik R, et al. ; MEGASTROKE consortium; International Stroke Genetics Consortium Genetic risk, incident stroke, and the benefits of adhering to a healthy lifestyle: cohort study of 306 473 UK Biobank participants. BMJ. 2018;363:k4168. doi:10.1136/bmj.k4168 [DOI] [PMC free article] [PubMed] [Google Scholar]