Abstract
Introduction
Due to the increasing resistance to nucleot(s)ide analogs in patients with chronic hepatitis B, development of new antiviral drugs to eradicate hepatitis B virus is still urgently needed.
Material and methods
To date, most studies on evaluating anti-HBV drugs have been performed using cell lines where the HBV genomic DNA is chromosomally integrated, e.g. Hep2.2.15 in HBV-infected livers of the viral episomal genome replicates in the nucleus and covalently closed circular DNA (cccDNA) serves as a transcriptional template. Another option involves the use of HBV-infected cells of HepaRG or NTCP-overexpressing cells. However, the development of the infection system is expensive and laborious, and its HBV expression level remained low.
Results
Compared to HuH7 cells, the established stable cell lines based on episomal-type pEB-Multi vectors can been expressed HBV wild-type by qRT-PCR and immunoblotting (p < 0.05). These two vectors are also sensitive to Entecavir and against nucleoside analog Lamivudine in mutants cellines.
Conclusions
It is worth demonstrating how useful the established cell system is for evaluating antiviral agents and their mechanisms of action.
Keywords: hepatitis B virus, stable cell lines; hepatitis B virus genome, episomal replication; antiviral drug evaluation
Introduction
Hepatitis B virus (HBV) is a member of the hepadnavirus family, which comprises unique DNA viruses that initiate reverse transcription during replication. About 350–400 million people are chronically infected with HBV and HBV-related liver diseases such as cirrhosis, liver failure and hepatocellular carcinoma, resulting in one million deaths annually worldwide [1]. Nucleoside analog inhibitors of HBV DNA polymerase are the current treatment options for chronic hepatitis B that has resistance to antiviral drugs; particularly lamivudine resistance is seen in 80% of patients treated for 5 years, and has a cumulative annual incidence of 14–32% [2, 3]. Thus, screening new antiviral drugs remains essential for the cure of clinical patients.
The HepG2.2.15 cells containing two integrated head-to-tail copies of the genotype D and stably replicating the HBV genome [4] are currently used to evaluate the effect of antiviral compounds [5–7]. However, in HBV-infected liver cells, the viral episomal genome cccDNA serves as a transcriptional template, but does not integrate the formation. Other options such as recombinant baculovirus infection and a transient transfection system are used for evaluating the drug resistance of a given mutant archive and cross-resistance profile of HBV mutants [8] due to their capability and convenience to efficiently initiate viral DNA replication. However, detailed investigations based on reliable cell-based assays are required for variant mechanisms of actions of viral mutants resistant to lamivudine and high throughput screening (HTS) of compounds that inhibit virus mutants resistance to lamivudine HBV infection [9]. To address this issue, several cell lines with stably replicating HBV virus mutants resistant to lamivudine have been reported [10–14]. However, these cell lines promote less persistence of viral particle-DNA copies in the supernatant and evaluation efficiency of antiviral agents in the cell lines harboring wild-type HBV genome and virus mutants resistant to lamivudine.
Hence, in the current study, we established a stable cell system based on the episomal-type pEB-Multi vector that can stably replicate HBV genome of genotype Ce. Furthermore, we constructed virus mutants with the stable transfection of lamivudine mutation into hepatoma HuH7 cells. This cell line can persistently produce hepatitis B surface antigen (HBsAg) over 21 days, pregenomic RNA over 60 days, and particle-DNA with high stable expression in at least 7 days. Therefore, this cell line is considered to be suitable for screening new antiviral agents against HBV mutant lamivudine resistance.
Material and methods
Plasmid construction
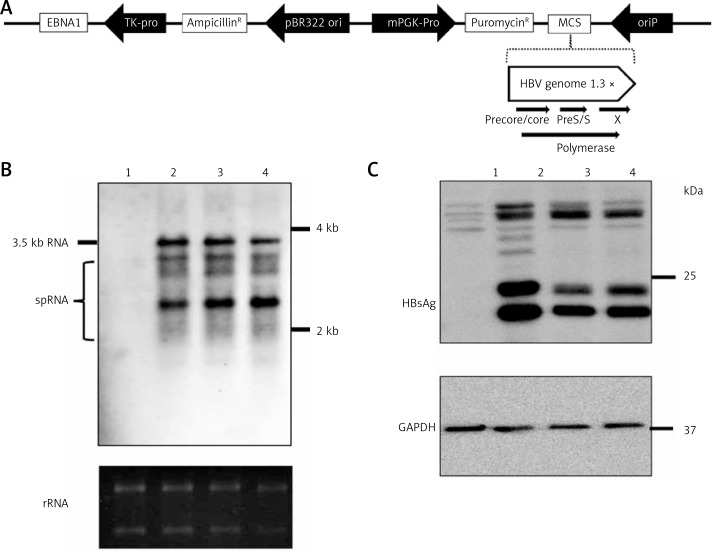
The mutant plasmid pEB-HBV-puromycin (L180M + M204V) was constructed by using the HBV genome (genotype C, subtype adw, GenBank Accession No. AY066028) isolated from FH4. The oriP-EBNA1 system and the puromycin resistance gene are contained in the PEB-multicarrier. A 1.3 × unit length HBV genome was inserted downstream of the puromycin genome (Figure 1 A). The pEB-HBV-puromycin (L180M + M204V) plasmid contains the L180M + M204V HBV genome and its pregenomic RNA expression under the control of the base core promoter element was derived from pEB-HBCe-puromycin (wild-type) by site-directed mutagenesis.
Figure 1.
Construction of HBV wild-type and mutant plasmids. A – Genomic organization of vector pEB-puro carrying HBV genome 1.3 times the length of the fully functional HBV genome. The backbone vector pEB-puro contains the oriP-EBNA1 system and puromycin resistance gene. A 1.3 × unit-length HBV genome was inserted into the multiple cloning site. The overlength of a HBV genome can generate authentic pgRNA (B) and HBs (C) protein under the control of a viral promoter. 1 pUC19-vector, 2 pUC19-HBCe, 3 pEB-HBCe, 4 pEB-HBCe (L180M + M204V)
Cell culture, transfection and selection of stable cell lines
HuH7 liver cancer cells of human origin were maintained in 10% fetal bovine serum supplemented with Dulbecco’s Modified Eagle medium (DMEM). Cells (2 × 106 cells/well in 10 cm plates) were transiently transfected with 5 μg of plasmid DNA mixed with Lipofectamine LTX purchased from Thermo Fisher Scientific. After 24-hour culture, medium containing 5 μg/ml puromycin was changed and after a further 7 days of culture with puromycin treated cells were prepared for assay.
Quantification of HBV DNA and RNA
Quantification of HBV DNA and RNA was performed as previously described [15]. The HBV DNA in the culture supernatant collected from the transfected cells was treated with PNE solution (8.45% PEG, 0.445 mole NaCl and 13 mmol EDTA) for 1 h on ice. The pellets were incubated with DNase I (TAKARA, Shiga, Japan) and RNase (TaKaRa) for 1 h at 37°C. The pellets were then treated with proteinase K for 12 h at 56°C, and HBV DNA was separated by phenol/chloroform extraction and ethanol precipitation. HBV DNA copies were determined by qPCR. For quantification of HBV 3.5 kb pgRNA, total RNA was extracted from HBV-transfected cells using TRI reagent (Molecular Research Center, Cincinnati, OH, USA). After treatment with DNase I and RNase inhibitor, cDNA templates were synthesized and HBV RNAs were quantified by qPCR using the SYBR qPCR Mix kit (Toyobo, Osaka, Japan) using 5′-TCCCTCGCCTCGCAGACG-3′ and 5′-GTTTCCCACCTTATGAGTC-3′ for unspliced 3.5 kb RNA, β-actin mRNA primers (5′- TTCTACAATGAGCTGCGTGTG-3′ and 5′-GGGGTGTTGAAGGTCTCAAA-3′). For semi-quantitative RT-PCR, cDNA templates were amplified with primers as previously reported [15].
Immunoblotting
Immunoblotting was performed as previously described [16]. Briefly, cell lysates were separated by SDS-PAGE and transferred onto PVDF membranes. The membrane was blocked after 1 h, and an anti-HBc antibody was produced by the HBc protein rabbit, HBs antibody (Immunology Institute, Tokyo, Japan), and GAPDH (Santa Cruz Biotechnology) which were immunologically and biologically expressed. After washing, the membrane was incubated with HRP-conjugated secondary antibody (manufactured by Cell Signaling Technology, Danvers, MA) for 0.5–1 h. Antigen-antibody complexes were measured using the ChemiDoc Imaging System (Bio-Rad Laboratories, Tokyo, Japan).
Results
Construction of HBV wild-type and mutant plasmids
Firstly, we constructed episomal-type HBV wild-type and mutant plasmids pEB-HBCe (L180M + M204V) and pEB-HBCe. After that, we evaluated whether they were successfully constructed and their replication ability in hepatocarcinoma cells. Northern and western blotting under the control of basal core promoter of the virus after transfection into HuH7 cells were performed. The results revealed that the HBV expression plasmids pEB-HBV-puromycin (L180M + M204V) and pEB-HBCe-puromycin could produce authentic HBV RNA (Figure 1 B) and HBs protein in cells (Figure 1 C). These results confirmed the successful construction of HBV wild-type and mutant plasmids.
Establishment of pEB-Multi plasmid based cell lines by stably replicating HBV wild-type and virus mutant resistant to antiviral drugs
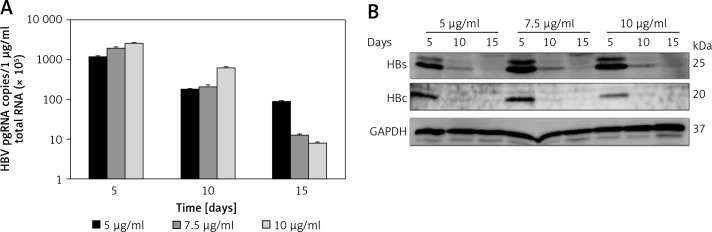
To confirm HBV replication activity in pEB-Multi plasmid based cell lines, HBV pregenomic RNA (pgRNA) and protein production were determined after transfection of HuH7 cells with pEB-HBV-puromycin (L180M + M204V) and pEB-HBCe-puromycin under increasing concentrations of puromycin. The results showed that 5 μg/ml puromycin was more suitable for cell growth and higher HBV pgRNA expression and protein production (Figures 2 A, B). These results indicated that the cell lines could stably express wild-type HBV and mutant plasmids, and the mutant type was resistant to antiviral drugs.
Figure 2.
Establishment of pEB-Multi plasmid based cell lines. HuH7 cells were transfected with pEB-HBV-puromycin (L180M + M204V). After 24-hour culture, transfected cells were transferred to fresh medium containing increasing concentrations of 5 μg/ml, 7.5 μg/ml and 10 μg/ml puromycin. After further culture for 5, 10, and 15 days, (A) HBV pgRNA level and (B) protein production in cells were determined by RT-qPCR and western blot, respectively
Values were represented as means ± SD from three independent samples.
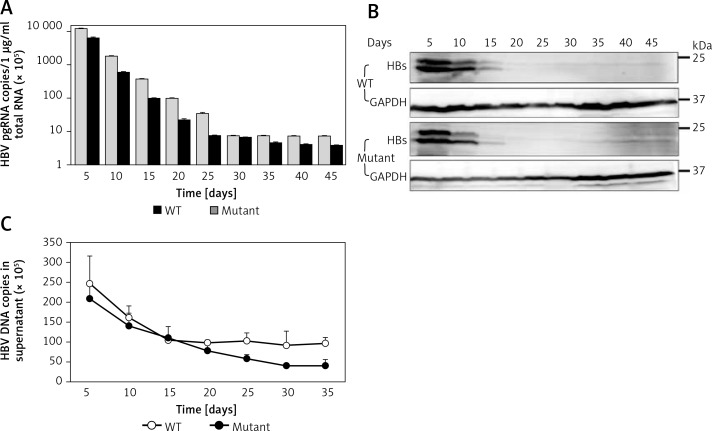
Time course of expression of viral antigens, viral RNAs and DNA formed in the supernatant
To determine the time course of expression of HBV wild-type and mutant, we measured the production of viral DNAs, RNAs, and antigens after culturing the novel stable cell line. Time-dependent expression of HBV RNAs (Figure 3 A) and antigens (Figure 3 B) was observed in cell lines stably replicating HBV wild-type and virus mutants resistant to lamivudine after 45 days in cell culture containing 5 μg/ml puromycin. We also observed time-dependent expression of HBV DNAs (Figure 3 C) in the supernatant. These results demonstrated that pEB-based cell lines can persistently replicate with HBV and virus mutants.
Figure 3.
Time course of expression of viral production. Under 5 μg/ml puromycin maintenance, HBV stably replicating cells and supernatant were harvested every 5 days. HBV production derived from wild-type (WT) and lamivudine resistant vector (Mutant) was measured by RT-qPCR (A), western blotting (B), and DNA quantification (C)
Values were represented as means ± SD from three independent samples.
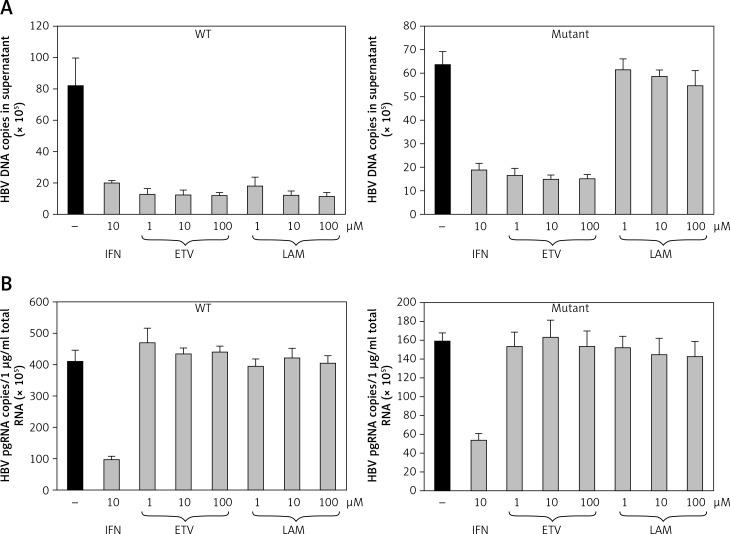
Effect of antiviral drugs on HBV replication of wild-type and virus mutants resistant to lamivudine
The antiviral activities in the presence of increasing concentrations of lamivudine and entecavir were screened to validate whether this cell line would be appropriate for screening antiviral agents. HuH7 cells transfected by puromycin at 7 days were selected, and cell lines stably replicating HBV wild-type and mutants were treated with lamivudine and entecavir in a dose-dependent manner. Cells were harvested after 7 days and further cultured using nucleoside analog treatment. HBV DNA (Figure 4 A) and viral pgRNA (Figure 4 B) levels were detected by real-time PCR in supernatant and cells. The effective concentrations of entecavir were 0.1, 1, and 10 μM and those of lamivudine were 1, 10, and 100 μM for HBV DNA and pgRNA against the wild-type and lamivudine resistant cell lines, respectively. We found that HBV DNA levels were suppressed by entecavir as well as lamivudine in the wild-type cell line, but were decreased only by entecavir treatment in the lamivudine resistant cell line. No effect on pgRNA was observed in both cell lines treated with lamivudine and entecavir. These results indicated that the stable cell lines based on episomal-type pEB-Multi vectors were resistant to lamivudine, but sensitive to interferon or entecavir. This remained useful for evaluating antiviral agents and investigating their mechanisms of action.
Figure 4.
Drug sensitivity assay of pEB-HBV cell line. pEB-HBV cells derived from wild-type (WT) and lamivudine resistant vector (Mutant) were cultured for drug assay and grown in the presence of interferon, lamivudine, or entecavir, respectively. Real-time PCR analysis of supernatant viral replication (A) and RT-qPCR of intracellular HBV pgRNA (B) demonstrated that this cell line was resistant to lamivudine, but sensitive to interferon and entecavir
Discussion
Large containers of drug-resistant viral variants of nucleoside analogs for the extensive clinical use of chronic HBV therapy, especially for lamivudine and adefovir. The resistance to these old drugs has seriously affected the efficacy of the newer drugs. HepG2.2.15 cells, which are still the most widely used for cell HBV life cycle analysis and antiviral studies, are genotype D of HBV stable production cell lines regardless of significant limitations, such as no viral replication from cccDNA (This unclear sentence is duplicated below) [10]. However, different methods have been used to study HBV antiviral drugs for sensitivity in vitro. Stable transfected HBV cell lines have become an important tool for HBV and anti-HBV drug research [4].
In our study, we established stable cell lines based on episomal-type pEB-Multi vectors stably replicating HBV wild-type and virus mutants resistant to lamivudine that can stably replicate HBV for over 1 month. HBV production and particle-associated HBV DNA in culture supernatants (Figure 3 C) that are sensitive to entecavir and against lamivudine (Figure 4 A) in mutants cell line were determined. Unlike techniques that artificially introduce a few particular mutations within a wild-type HBV background, the full length amplification and transfection method allows the determination of antiviral efficacy on samples and is more useful to screen antiviral agents. Lamivudine was approved for use in 1998, followed by adefovir, telbivudine, entecavir and tenofovir in 2003, 2005, 2006 and 2008, respectively. Since the number of HBV antiviral drugs is limited, transmission of mutant virus is of particular importance for HBV infection as mutations that confer cross-reactivity can leave patients with few therapeutic options. Indeed, there are recent reports of lamivudine- and adefovir-related mutations in acute HBV infection in both China and Japan [17].
Increased use of antiviral drugs for chronic hepatitis B has led to increased antiviral resistance. Because of this, transmission of resistant HBV is a growing concern. A similar scenario has already been witnessed in acute HIV infection. Novel cell lines are not only useful for assessing new antiviral inhibitors, but also for investigating their mechanisms of actions. On the other hand, cell lines are convenient to determine the roles of host proteins that have DNA- and RNA-binding properties in HBV replication. There are a variety of liver-enriched and ubiquitous transcription factors that target the promoter and enhancer regions to regulate viral transcription and replication [18–21]. A number of host cytokines that have been identified to interact with the ENII/BCP region to modulate HBV transcriptional activity are mostly active activators that stimulate cis-acting elements. For example, liver-enriched or ubiquitous transcription factors such as PPAR, HNF4, HNF3, C/EBP, RXR, FTF/LRH-1, TBP, FXR, PGC-1, SIRT1 and SP1 [22, 23] bind to ENII, contributing to the up-regulation of the core promoter activity.
Regarding the transcriptional repression mechanism of HBV gene expression, it is known that a negative regulatory element (NRE) located immediately upstream of ENII participates in down-regulation of core promoter activity in a direction-independent manner. In addition, ENII has not been widely reported, but it involves negative regulation of activities. Prox 1, called FTF/LRH-1 co-repressor, inhibits FTF/LRH-1 mediated ENII activation [24]. Down-regulation of IL-4 expression by C/EBP may inhibit core promoter activity [25]. It also indicates that TRIM protein and COUP-TF1 potentially contribute to the inhibitory activity of ENII [26, 27]. LUC7L3, a member of the SR protein family, is a novel ENII-mediated negative regulator of HBV replication [16, 28, 29].
In conclusion, we constructed novel stable HBV-producing cell lines with HBV wild-type and lamivudine resistant. These novel stable HBV-producing cell lines can serve as valuable tools for screening antiviral agents and analyzing virus-host interaction in vitro.
Acknowledgments
Suofeng Sun and Yuan Li equally contributed to this research and should be considered as co-first authors.
This work was supported by the Research Program on Hepatitis from the Japan Agency for Medical Research and Development and the Japanese Ministry of Education, Culture, Sports, Science, and Technology and the Dr. Start-up Fund of Henan Provincial People’s Hospital.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Dienstag JL. Hepatitis B virus infection. N Engl J Med. 2008;359:1486–500. doi: 10.1056/NEJMra0801644. [DOI] [PubMed] [Google Scholar]
- 2.Lai CL, Dienstag J, Schiff E, et al. Prevalence and clinical correlates of YMDD variants during lamivudine therapy for patients with chronic hepatitis B. Clin Infect Dis. 2003;36:687–96. doi: 10.1086/368083. [DOI] [PubMed] [Google Scholar]
- 3.Lok AS, Lai CL, Leung N, et al. Long-term safety of lamivudine treatment in patients with chronic hepatitis B. Gastroenterology. 2003;125:1714–22. doi: 10.1053/j.gastro.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 4.Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–9. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deres K, Schroder CH, Paessens A, et al. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science. 2003;299:893–6. doi: 10.1126/science.1077215. [DOI] [PubMed] [Google Scholar]
- 6.Dougherty AM, Guo H, Westby G, et al. A substituted tetrahydro-tetrazolo-pyrimidine is a specific and novel inhibitor of hepatitis B virus surface antigen secretion. Antimicrob Agents Chemother. 2007;51:4427–37. doi: 10.1128/AAC.00541-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aziz H, Raza A, Ali K, Khattak JZ, Irfan J, Gill ML. Polymorphism of the IL28B gene (rs8099917, rs12979860) and virological response of Pakistani hepatitis C virus genotype 3 patients to pegylated interferon therapy. Int J Infect Dis. 2015;30:91–7. doi: 10.1016/j.ijid.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 8.Durantel D. Fitness and infectivity of drug-resistant and cross-resistant hepatitis B virus mutants: why and how is it studied? Antivir Ther. 2010;15:521–7. doi: 10.3851/IMP1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zoulim F. In vitro models for studying hepatitis B virus drug resistance. Semin Liver Dis. 2006;26:171–80. doi: 10.1055/s-2006-939759. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Zhang Y, Kang Y, et al. Generation of a human hepatoma cell line supporting efficient replication of a lamivudine resistant hepatitis B virus. J Virol Methods. 2014;201:51–6. doi: 10.1016/j.jviromet.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Westland CE, Yang H, Delaney WE, et al. Activity of adefovir dipivoxil against all patterns of lamivudine-resistant hepatitis B viruses in patients. J Viral Hepat. 2005;12:67–73. doi: 10.1111/j.1365-2893.2005.00578.x. [DOI] [PubMed] [Google Scholar]
- 12.Yang H, Qi X, Sabogal A, Miller M, Xiong S, Delaney WE. Cross-resistance testing of next-generation nucleoside and nucleotide analogues against lamivudine-resistant HBV. Antivir Ther. 2005;10:625–33. [PubMed] [Google Scholar]
- 13.Fu L, Cheng YC. Characterization of novel human hepatoma cell lines with stable hepatitis B virus secretion for evaluating new compounds against lamivudine- and penciclovir-resistant virus. Antimicrob Agents Chemother. 2000;44:3402–7. doi: 10.1128/aac.44.12.3402-3407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ladner SK, Miller TJ, Otto MJ, King RW. The hepatitis B virus M539V polymerase variation responsible for 3TC resistance also confers cross-resistance to other nucleoside analogues. Antivir Chem Chemother. 1998;9:65–72. [PubMed] [Google Scholar]
- 15.Sun S, Nakashima K, Ito M, et al. Involvement of PUF60 in transcriptional and post-transcriptional regulation of hepatitis Bvirus pregenomic RNA expression. Sci Rep. 2017;7:12874. doi: 10.1038/s41598-017-12497-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Ito M, Sun S, Chida T, Nakashima K, Suzuki T. LUC7L3/CROP inhibits replication of hepatitis B virus via suppressing enhancer II/basal core promoter activity. Sci Rep. 2016;6:36741. doi: 10.1038/srep36741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi K, Katano Y, Ishigami M, et al. Prevalence and clinical characterization of patients with acute hepatitis B induced by lamivudine-resistant strains. J Gastroenterol Hepatol. 2010;25:745–9. doi: 10.1111/j.1440-1746.2009.06118.x. [DOI] [PubMed] [Google Scholar]
- 18.Cheng Y, Seet BL, Ong CS, et al. Are in vitro hepatitis B core promoter mutations important for clinical alterations in viral load? Antiviral Res. 2006;69:142–51. doi: 10.1016/j.antiviral.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Nan Y, Zhang YJ. Molecular biology and infection of hepatitis E virus. Front Microbiol. 2016;7:1419. doi: 10.3389/fmicb.2016.01419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology. 2015;479-480:672–86. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ploss A, Dubuisson J. New advances in the molecular biology of hepatitis C virus infection: towards the identification of new treatment targets. Gut. 2012;61(Suppl 1):i25–35. doi: 10.1136/gutjnl-2012-302048. [DOI] [PubMed] [Google Scholar]
- 22.Moolla N, Kew M, Arbuthnot P. Regulatory elements of hepatitis B virus transcription. J Viral Hepat. 2002;9:323–31. doi: 10.1046/j.1365-2893.2002.00381.x. [DOI] [PubMed] [Google Scholar]
- 23.Quarleri J. Core promoter: a critical region where the hepatitis B virus makes decisions. World J Gastroenterol. 2014;20:425–35. doi: 10.3748/wjg.v20.i2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin J, Zhai J, Hong R, et al. Prospero-related homeobox protein (Prox1) inhibits hepatitis B virus replication through repressing multiple cis regulatory elements. J Gen Virol. 2009;90:1246–55. doi: 10.1099/vir.0.006007-0. [DOI] [PubMed] [Google Scholar]
- 25.Lin SJ, Shu PY, Chang C, Ng AK, Hu CP. IL-4 suppresses the expression and the replication of hepatitis B virus in the hepatocellular carcinoma cell line Hep3B. J Immunol. 2003;171:4708–16. doi: 10.4049/jimmunol.171.9.4708. [DOI] [PubMed] [Google Scholar]
- 26.Fischer SF, Schmidt K, Fiedler N, et al. Genotype-dependent activation or repression of HBV enhancer II by transcription factor COUP-TF1. World J Gastroenterol. 2006;12:6054–8. doi: 10.3748/wjg.v12.i37.6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang S, Guo JT, Wu JZ, Yang G. Identification and characterization of multiple TRIM proteins that inhibit hepatitis B virus transcription. PLoS One. 2013;8:e70001. doi: 10.1371/journal.pone.0070001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao G, Dudley SC., Jr RBM25/LUC7L3 function in cardiac sodium channel splicing regulation of human heart failure. Trends Cardiovasc Med. 2013;23:5–8. doi: 10.1016/j.tcm.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao G, Xie A, Huang SC, et al. Role of RBM25/LUC7L3 in abnormal cardiac sodium channel splicing regulation in human heart failure. Circulation. 2011;124:1124–31. doi: 10.1161/CIRCULATIONAHA.111.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]