Abstract
Objectives
Azole resistance among Aspergillus fumigatus isolates is a growing concern worldwide. Induction of mutations during azole therapy, environment-acquired mutations caused by azole fungicides and intrinsic resistance of cryptic Fumigati species all contribute to the burden of resistance. However, there is a lack of data in Canada on this emerging threat.
Methods
To gain insights into the magnitude and mechanisms of resistance, a 14 year collection of Aspergillus section Fumigati comprising 999 isolates from 807 patients at a Montreal hospital was screened for azole resistance, and resistance mechanisms were investigated with the combined use of genome sequencing, 3D modelling and phenotypic efflux pump assays.
Results
Overall azole resistance was low (4/807 patients; 0.5%). A single azole-resistant A. fumigatus sensu stricto strain, isolated from a patient with pulmonary aspergillosis, displayed efflux-pump-mediated resistance. Three patients were colonized or infected with azole-resistant cryptic Fumigati species (one Aspergillus thermomutatus, one Aspergillus lentulus and one Aspergillus turcosus). Evidence is presented that azole resistance is efflux-pump-mediated in the A. turcosus isolate, but not in the A. lentulus and A. thermomutatus isolates.
Conclusions
Azole resistance is rare in our geographic area and currently driven by cryptic Fumigati species. Continued surveillance of emergence of resistance is warranted.
Introduction
Aspergillus species can produce a wide spectrum of fungal diseases, including life-threatening invasive aspergillosis (IA) in the most vulnerable hosts.1,2 Many different species may cause aspergillosis, among which the ubiquitous species Aspergillus fumigatus is the leading agent.3–5 Antifungal triazoles are the mainstay of prophylaxis and treatment of IA.6 However, a growing number of cases of aspergillosis implicating azole-resistant strains are reported worldwide,7 raising significant public health concerns.
Azole resistance in A. fumigatus is associated with three distinct mechanisms. First, wild-type (WT) isolates can acquire resistance by mutating in the tissues of infected patients exposed to prolonged azole therapy.8 Second, azole-naive patients may be infected by isolates that have already acquired resistance in the environment. The majority of these resistant isolates, although not clonal, harbour identical mutations in the cyp51a gene (TR34/L98H and TR46/Y121F/T289A), likely acquired through environmental exposure to azole fungicides.9,10 These mutations have now been identified worldwide, including in the USA,11 but not in Canada. Finally, ‘cryptic’ species within the section Fumigati also contribute to the burden of resistance. The Aspergillus section Fumigati complex comprises >30 species.12,13 So-called cryptic species are phenotypically very close to A. fumigatus sensu stricto and are often misidentified as A. fumigatus by clinical laboratories.14,15 Several of the cryptic species have been shown to be intrinsically resistant to azole antifungals and are recognized as agents of aspergillosis.16,17 Yet, the true prevalence of these cryptic species and their relative contribution to azole resistance remain unclear. The molecular mechanisms that govern azole resistance in these species are also unexplored,18,19 except for Aspergillus lentulus.20,21
We aimed to provide a comprehensive retrospective survey of azole resistance within the A. fumigatus species complex at a tertiary care Canadian hospital. Specifically, we sought to define the relative contribution of intrinsically resistant cryptic species to the burden of resistance, and to establish the presence of environment-associated mutations. We screened a collection of Aspergillus section Fumigati clinical isolates collected at our centre from 2000 to 2013. We determined the prevalence and clinical significance of a culture positive for any cryptic species or resistant A. fumigatus. We also characterized efflux pump- and cyp51a-mediated resistance mechanisms in resistant isolates, including previously unexplored cryptic species.
Materials and methods
Ethics
This study was approved by the hospital ethics committee (reference number 2015-598-14034) and conducted in accordance with national standards aligned with the Declaration of Helsinki. Patient consent was not deemed necessary.
Hospital description and clinical isolates
Maisonneuve-Rosemont is a 520 bed university-affiliated teaching hospital, serving patients at risk of aspergillosis including transplant recipients. From 2000 onwards, all clinical isolates routinely morphotyped as A. fumigatus at the hospital clinical mycology laboratory were archived by suspending colony fragments in 15% glycerol, stored at −80°C. In this retrospective study, all isolates collected from 2000 to 2013 were included.
Screening for cryptic species and azole-resistant isolates
Frozen isolates were thawed and cultured on Sabouraud dextrose agar (Oxoid, Netean, ON, Canada) at 37°C for 48–72 h. Upon regrowth, viable organisms were first re-identified by morphotyping and those compatible with A. fumigatus were retained for further study. Isolates were then screened for cryptic species using phenotypic characteristics (thermotolerance and sporulation rate).12 All atypical isolates were submitted to sequence-based identification, along with a subset of typical isolates. Additional details on cryptic species screening and confirmation can be found in the Supplementary data (available at JAC Online). Confirmed cryptic species were subjected to CLSI M38-compliant antimicrobial susceptibility testing (AST) for MIC determination with eight antifungal agents.22A. fumigatus sensu stricto isolates were screened for azole resistance using a modified broth microdilution (BMD) assay (Figure S1). All potentially resistant (screening-positive) isolates, as well as a subset of screening-negative isolates, underwent standard AST. Because CLSI clinical breakpoints are not available, both cryptic and A. fumigatus sensu stricto isolates displaying azole MICs above the epidemiological cut-off values (ECVs) for A. fumigatus23,24 were considered resistant, regardless of species. The term ‘resistant’ was favoured over ‘non-susceptible’ or ‘non-WT’ for simplicity and consistency across the manuscript, recognizing that it does not represent clinical resistance.
For details regarding conidia collection and genomic DNA extraction, control strains, reference sequences and accession numbers, please see the Supplementary data.
Efflux pump inhibitor assay
A 96-well microplate chequerboard assay was designed to quantify the effect of an efflux pump inhibitor on susceptibility to voriconazole of cryptic species isolates. The assay contained, each in duplicate, voriconazole concentrations ranging from 0 to 64 mg/L (2-fold dilutions) and the broad-spectrum efflux pump inhibitor MC-207,110 (l-Phe-l-Arg-β-naphthylamide)25 at concentrations of 0, 32, 64 and 128 mg/L. Inoculum, growth conditions and interpretation were the same as the CLSI BMD method. After incubation at 35°C for 48 h, MICs of voriconazole were determined for each concentration of the inhibitor.
Nile red efflux pump assay
A previously described microplate Nile red assay26 was adapted to the requirements of a filamentous fungus. Detailed methodology can be found in the Supplementary data.
WGS
Three cryptic species isolates underwent WGS: one azole-resistant Aspergillus turcosus (HMR-AF-1038), one azole-susceptible A. turcosus (HMR-AF-23; randomly selected out of three) and one azole-resistant Aspergillus thermomutatus (HMR-AF-39; one of seven isolates from a single patient). Detailed methodology of genome sequencing and annotation of these isolates was previously reported.27,28 In addition, two A. fumigatus sensu stricto isolates were sequenced in a similar fashion: one azole-resistant isolate (HMR-AF-270) and one randomly selected susceptible isolate (HMR-AF-706).
Sequencing of A. lentulus cyp51a gene
The cyp51a gene of azole-resistant A. lentulus isolate HMR-AF-1185, which did not undergo WGS, was amplified by PCR (see Supplementary data) and sequenced using the Sanger method at the Génome Québec Innovation Centre using a 3730xl DNA Analyzer (Applied Biosystems).
Efflux pump and metabolic gene identification
BLAST was used to identify efflux pumps and metabolic genes implicated in antifungal azole resistance, from genomic sequences determined in this study or reported elsewhere.29 Genes previously reported in A. fumigatus strain AF29330–33 or Candida albicans strain SC531426,34–36 were used as references. Identity percentages were calculated using BLAST.
CYP51A homology modelling
Multiple sequence alignments were performed using the default settings of Clustal Omega.37 3D models of CYP51A were constructed by homology modelling using MODELLER 9.18,38 following the same multiple template procedure. The A. fumigatus CYP51A sequence was 3D-modelled from the structural alignment performed using three homologous crystal structure templates: (i) the voriconazole-bound 14-α demethylase (CYP51B) from A. fumigatus (PDB 4UYM); (ii) the lanosterol-bound lanosterol 14-α demethylase from Saccharomyces cerevisiae (PDB 4LXJ); and (iii) the ketoconazole-bound lanosterol 14-α demethylase from Homo sapiens (PDB 3LD6). These templates respectively share 65%, 51% and 38% sequence identity with CYP51A from A. fumigatus. From the five homology models generated, the one presenting the best DOPE score was selected and further validated using PROCHECK.39 Cavity volumes for the different CYP51A models were computed using CAVER Analyst.40
Medical chart review
Medical charts of patients with cryptic and/or resistant isolates were reviewed to collect demographic and clinical data. Aspergillus-associated disease was classified according to the standard definitions.1 IA was further categorized following EORTC/MSG definitions.41
Results
Description of isolates included in the study
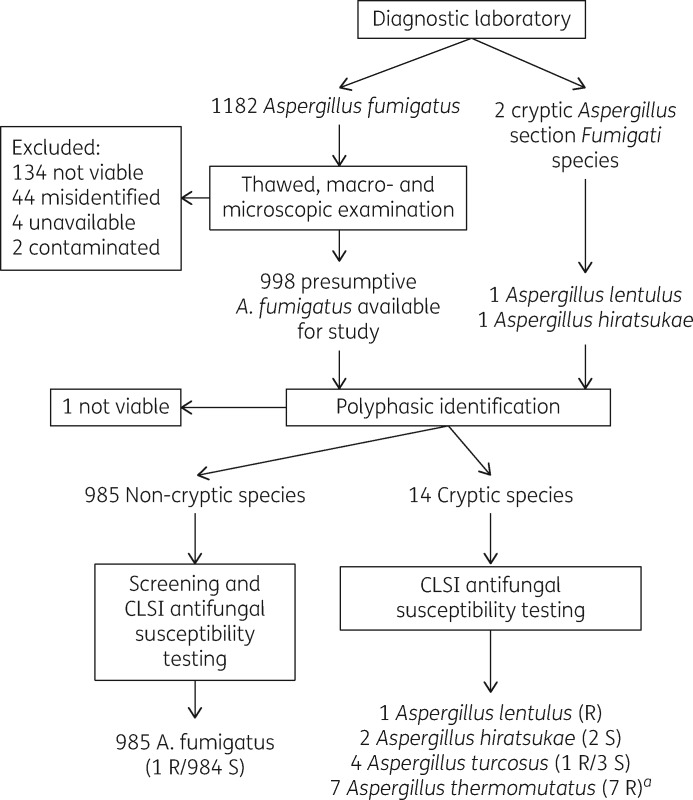
From 2000 to 2013, 1182 clinical isolates were identified as presumptive A. fumigatus by morphotyping in the diagnostic laboratory (Figure 1). Upon regrowth, 184 were excluded because of loss of viability, misidentification, unavailability or contamination, leaving 998 presumptive A. fumigatus isolates. In addition, two isolates (one each of A. lentulus and Aspergillus hiratsukae) that had been sent to a reference laboratory as part of clinical care were included, for a total of 1000 isolates. One isolate lost viability after initial thawing, leaving 999 isolates for further identification and susceptibility screening (Figure 1). These 999 isolates were obtained from 807 patients.
Figure 1.
Identification and susceptibility testing algorithm of 1184 Aspergillus section Fumigati isolates. Numbers in parentheses represent isolates susceptible (S) or resistant (R) to any triazole (itraconazole, posaconazole or voriconazole). aAll seven A. thermomutatus isolates were collected from a single patient.
Prevalence of cryptic species
We first established the prevalence of cryptic species. Upon phenotypic screening, 22 of 999 isolates displayed atypical characteristics (decreased thermotolerance and/or slow sporulation). Of these, sequence-based identification revealed 8 A. fumigatus sensu stricto, while 14 isolates were confirmed as cryptic species: A. lentulus (n = 1), A. hiratsukae (n = 2), A. turcosus (n = 4)42 and A. thermomutatus (n = 7). To assess the sensitivity of our phenotypic screening, 82 randomly selected typical A. fumigatus isolates underwent sequence-based identification and all were confirmed as A. fumigatus sensu stricto. All A. thermomutatus isolates were collected from a single patient, while all other cryptic isolates were recovered from separate patients. Overall, the per-sample and per-patient prevalence of cryptic species in this collection of Aspergillus section Fumigati isolates were 1.4% (14/999) and 0.99% (8/807), respectively.
Antifungal susceptibility
We then screened the entire collection for azole resistance. Isolates deemed to be A. fumigatus sensu stricto (n = 985) were submitted to microplate antifungal susceptibility screening (Figure S1), which revealed 12 potentially resistant isolates (itraconazole, n = 3; voriconazole, n = 9), all of which were further tested using the CLSI BMD method. Of these, only one A. fumigatus isolate (HMR-AF-270) was confirmed as resistant, displaying mildly elevated MICs of itraconazole, posaconazole and voriconazole (Table 1 and Table S1). CLSI BMD testing was also performed on 32 randomly selected resistance-screening-negative A. fumigatus sensu stricto isolates and all displayed WT MICs for triazoles. This confirmed that the screening method reliably detected azole resistance. Finally, antifungal susceptibility testing (CLSI BMD) of cryptic Aspergillus section Fumigati isolates (n = 14) showed that all seven A. thermomutatus isolates were resistant to itraconazole, posaconazole and voriconazole, one A. turcosus isolate was resistant to itraconazole and voriconazole and the single A. lentulus isolate was resistant to both itraconazole and voriconazole (Table 1). The resistance level was also low in the cryptic isolates. Susceptibility testing results for other antifungal agents are provided in Table S1. Overall, 1.0% (10/999) of all the Aspergillus section Fumigati isolates were azole resistant. Per-patient prevalence of resistance was 0.5% (4/807), including one patient with A. fumigatus sensu stricto and three patients with cryptic species.
Table 1.
Underlying diseases and Aspergillus-associated conditions of patients with an azole-susceptible or -resistant cryptic Aspergillus section Fumigati isolate or an azole-resistant A. fumigatus isolate
| Patient | Sex/ age (years) | Comorbidities | Aspergillus-associated clinical conditiona | Specimen | Collection date (dd/mm/yyyy) | Antifungals received prior to collection date | Isolate | Identification | MIC (mg/L)b |
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ITC | PSC | VRC | |||||||||
| 1 | M/62 | CLL; COPD; community-acquired pneumonia | colonization | BAL | 23/05/2000 | none | HMR AF 23 | A. turcosus | 0.25 | 0.12 | 0.25 |
| 2 | M/52 | allogeneic stem cell transplantation | non-invasive sinus aspergillosis and nasal septum abscess | nasal sinus abscess | 31/07/2000 | FLC | HMR AF 39c | A. thermomutatus | 2 | 1 | 2 |
| 3 | M/55 | probable old TB cavity | chronic cavitary pulmonary aspergillosis | expectorated sputum | 18/12/2002 | ITC | HMR AF 270 | A. fumigatus | 2 | 1 | 2 |
| 4 | F/70 | lymphoma (cutaneous); COPD | chronic cavitary pulmonary aspergillosis | BAL | 26/06/2009 | none | HMR AF 832 | A. hiratsukae | 0.25 | 0.12 | 1 |
| 5 | M/51 | COPD; SAHS; AECB | colonization | BAL | 14/10/2010 | none | HMR AF 1185 | A. lentulus | 2 | 0.5 | 2 |
| 6 | M/71 | congestive heart failure; SAHS; idiopathic haemoptysis | colonization | BAL | 08/12/2011 | none | HMR AF 1186 | A. hiratsukae | 0.5 | 0.25 | 0.5 |
| 7 | M/70 | diabetes mellitus; COPD; pulmonary Mycobacterium avium disease | colonization | BAL | 16/01/2012 | none | HMR AF 1038 | A. turcosus | 2 | 1 | 2 |
| 8 | M/68 | COPD; AECB | colonization | expectorated sputum | 15/02/2012 | none | HMR AF 1041 | A. turcosus | 0.25 | 0.12 | 0.5 |
| 9 | F/57 | kidney transplantation; PCP | colonization | BAL | 23/01/2013 | none | HMR AF 1120 | A. turcosus | 0.25 | 0.06 | 0.25 |
Abbreviations: M, male; F, female; SAHS, sleep apnoea/hyponoea syndrome; AECB, acute exacerbation of chronic bronchitis; PCP, Pneumocystis jirovecii pneumonia; BAL, bronchoalveolar lavage; FLC, fluconazole; ITC, itraconazole; PSC, posaconazole; VRC, voriconazole.
Conditions shown in bold are considered Aspergillus-related diseases.
MIC values shown in bold are considered non-WT (indicating resistance).
Six other A. thermomutatus isolates were recovered from the same patient.
Clinical findings
Clinical significance of resistant/cryptic isolates was assessed by chart review (Table 1). Three patients had an Aspergillus-associated clinical condition: one patient had nasal sinus aspergillosis with nasal septum abscess (without evidence of necrosis or extension across bony barriers), while two others had chronic cavitary pulmonary aspergillosis. Two patients had received antifungal therapy prior to collection of the resistant isolate. One of these (Patient 3) was the only patient infected with a resistant A. fumigatus sensu stricto and had been treated with itraconazole for 8 months prior to the recovery of the resistant isolate. He was experiencing clinical and radiological failure at that time. Interestingly, two prior isolates recovered from the same patient were fully susceptible, suggesting acquired resistance within the host.
Resistance mechanisms
Finally, we investigated the mechanisms of azole resistance in the resistant isolates, with a focus on the unexplored cryptic species A. thermomutatus and A. turcosus. Based on known mechanisms in A. fumigatus, A. lentulus and C. albicans,21,43 we specifically explored CYP51A- and efflux-pump-mediated mechanisms.
CYP51A-mediated mechanisms
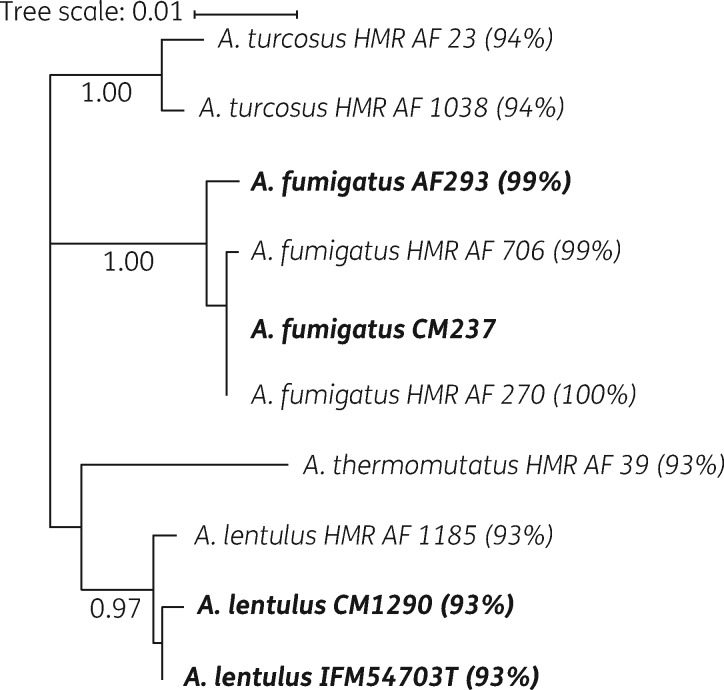
Tandem repeats were not found within the promoter of the azole-resistant HMR-AF-270 A. fumigatus isolate. Upon alignment of the CYP51A amino acid sequences, no mutation was identified for this azole-resistant A. fumigatus isolate, compared with a reference sequence (CM237) (Figure S2) and the promoter region of this isolate was also WT. The azole-susceptible A. fumigatus HMR-AF-706 isolate exhibited an amino acid substitution at position 9 that has been previously shown to be silent.44 Numerous differences in CYP51A amino acid sequences were found between species (Figure S2). A phylogenetic tree of the CYP51A amino acid sequences illustrates relative conservation of the enzyme across species (Figure 2). Interestingly, differences in CYP51A amino acid sequences were also found between azole-susceptible and -resistant A. turcosus isolates.
Figure 2.
Molecular phylogenetic analysis of the CYP51A amino acid sequences of Aspergillus strains, using the maximum likelihood method based on the JTT matrix-based model. Reference strains are in bold, bootstrap results greater than 0.90 are on each branch and percentage identities compared with strain CM237 are indicated in parentheses.
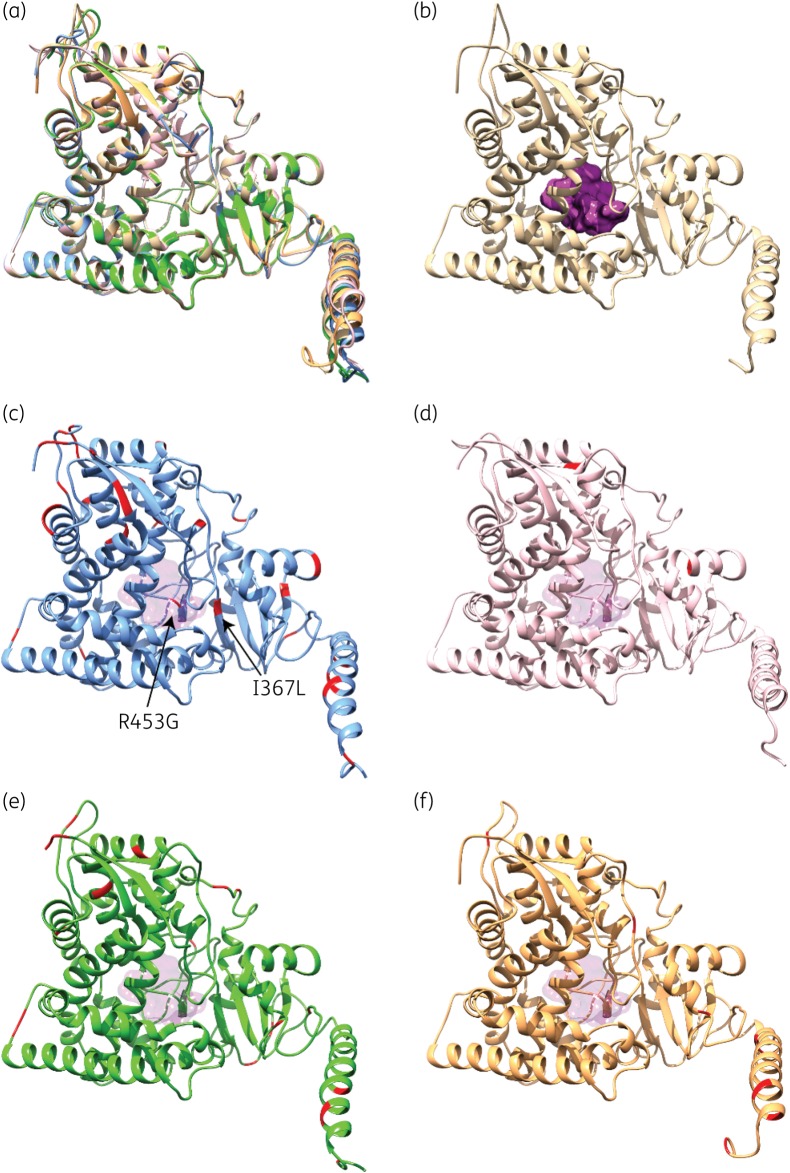
To clarify the significance of such differences, we undertook structural investigation of CYP51A homologues through in silico computational analyses. We constructed 3D homology models of CYP51A from WGS isolates and A. lentulus. As shown in Figure 3(a) and Tables S2 and S3, the overall CYP51A fold is highly conserved among structural homologues, exhibiting small differences primarily in unstructured regions (i.e. surface loops). The active site of CYP51A is buried, forming a hydrophobic pocket inside which the haem cofactor is located (Figure 3b). Most mutations observed in our enzyme variants are structurally localized away from the active site of CYP51A (Figure 3c–e). Consistently, the geometry and volume of the active-site cavity in both the azole-susceptible and azole-resistant CYP51A variants is highly similar (Tables S2 and S3). This indicates that resistance amongst our isolates is not the consequence of a reduction in azole recognition and stabilization resulting from a significant reshaping of the active-site cavity.
Figure 3.
Structural homology models of the five CYP51A variants investigated in this study. (a) Structural overlay of CYP51A models from HMR-AF-270 (beige; azole-resistant A. fumigatus with WT CYP51A), HMR-AF-23 (blue; azole-susceptible A. turcosus), HMR-AF-1038 (pink; azole-resistant A. turcosus), HMR-AF-39 (green; azole-resistant A. thermomutatus) and HMR-AF-1185 (orange; azole-resistant A. lentulus). (b) 3D homology model of WT CYP51A from HMR-AF-270, highlighting the buried active-site cavity (purple surface). (c) 3D homology model of CYP51A from HMR-AF-23, highlighting silent amino acid replacements diverging from the WT A. fumigatus sequence (in red). (d–f) 3D homology models of CYP51A from HMR-AF-1038 (d, pink), HMR-AF-39 (e, green) and HMR-AF-1185 (f, orange), highlighting amino acid replacements diverging from the WT A. fumigatus sequence (in red). For positional comparison, active-site cavity surface transparency is shown in all CYP51A variants.
CYP51A sequence comparison between the azole-susceptible A. turcosus HMR-AF-23 and a WT A. fumigatus strain (CM237) displays 30 amino acid replacements (Figure S2). Two of these mutations (I367L and R453G) are close to the active site (Figure 3c). However, since HMR-AF-23 is azole susceptible, these amino acid replacements do not appear to alter CYP51A structure or function. CYP51A variants from A. turcosus strains HMR-AF-23 and HMR-AF-1038 (azole resistant) almost share the same mutational pattern, except for F61L and F478L, both only present in HMR-AF-1038. Assuming that all other CYP51A amino acid replacements between WT A. fumigatus and HMR-AF-23 are silent, these two remaining mutations were of high interest. However, these mutations are located outside the active-site cavity and far from the substrate channels (Figure 3d).
Azole-resistant strains HMR-AF-39 (A. thermomutatus) and HMR-AF-1185 (A. lentulus) show 36 and 31 amino acid replacements relative to the WT A. fumigatus strain (CM237) (Figure S2), of which 21 and 18 replacements are located in silent positions, respectively. They also each exhibit 15 and 13 amino acid replacements lying outside the range of CYP51A entry channels or the active site (Figure 3e and f).
Efflux pumps
We next explored the possible contribution of efflux pumps to azole resistance. As a first step, we probed the genomes of cryptic species, looking for orthologues of known efflux pump genes in A. fumigatus and C. albicans (Table S4). Many orthologues were found in A. turcosus, A. thermomutatus and A. lentulus, with some sharing high identity, including ABC transporter genes previously associated with azole resistance (e.g. cdr1B, AfuMDR1–4, atrF). These observations confirmed that efflux-pump-mediated resistance was a biologically plausible mechanism for our cryptic species isolates.
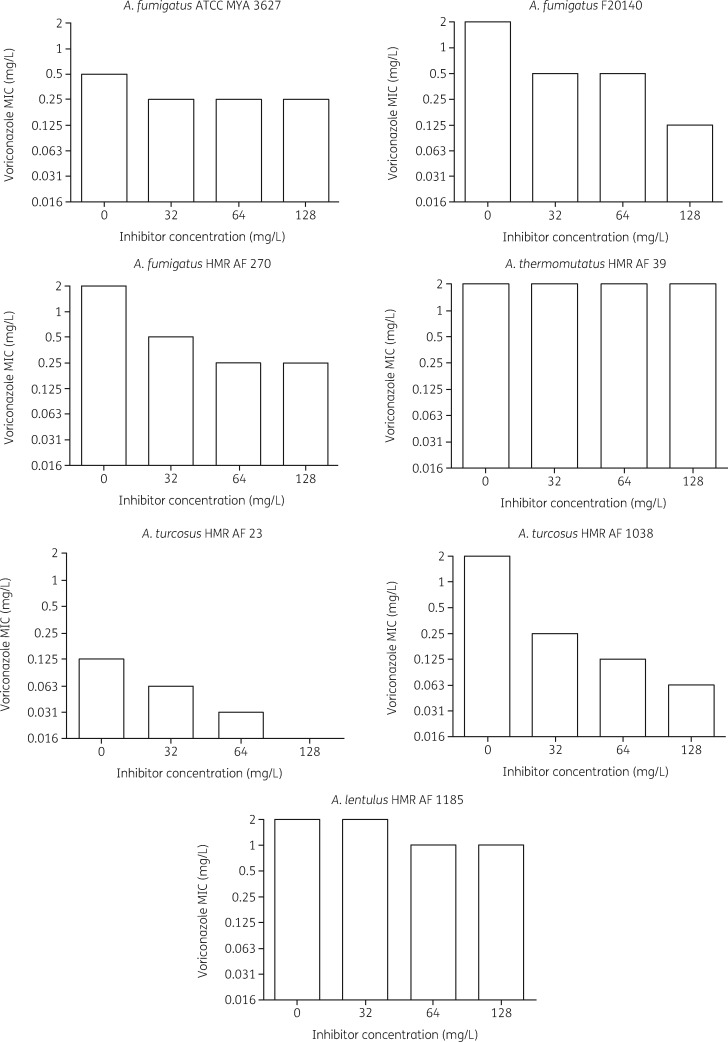
We devised a phenotypic assay, based on voriconazole susceptibility in the presence of the broad-spectrum efflux pump inhibitor MC-207,110 (Figure 4). At all tested inhibitor concentrations, the voriconazole MICs for the azole-susceptible A. fumigatus control strain ATCC MYA-3627 only decreased 2-fold. Conversely, the azole-resistant A. fumigatus control strain F20140, known to overexpress efflux pump genes and carrying WT cyp51a, showed a 4-fold lower MIC, at inhibitor concentrations of 32 and 64 mg/L, and a 16-fold lower MIC at a concentration of 128 mg/L. The resistant A. fumigatus isolate HMR-AF-270 displayed an 8-fold MIC reduction in the presence of the inhibitor. Azole-resistant A. turcosus strain HMR-AF-1038 also displayed a strong decrease (32-fold) in MIC at the highest inhibitor concentration. Interestingly, A. turcosus strain HMR-AF-23, despite being susceptible to voriconazole, showed an inhibitor concentration-dependent 8-fold decrease in MIC, from 0.125 to 0.016 mg/L, suggesting an effect of the inhibitor on basal efflux pump activity. In contrast, the efflux pump inhibitor had little or no effect on the MICs for A. thermomutatus HMR-AF-39 and A. lentulus HMR-AF-1185.
Figure 4.
Efflux pump inhibitor assay. MICs of voriconazole for Aspergillus strains at three concentrations of MC-207,110 efflux pump inhibitor. A second experiment produced comparable results (data not shown).
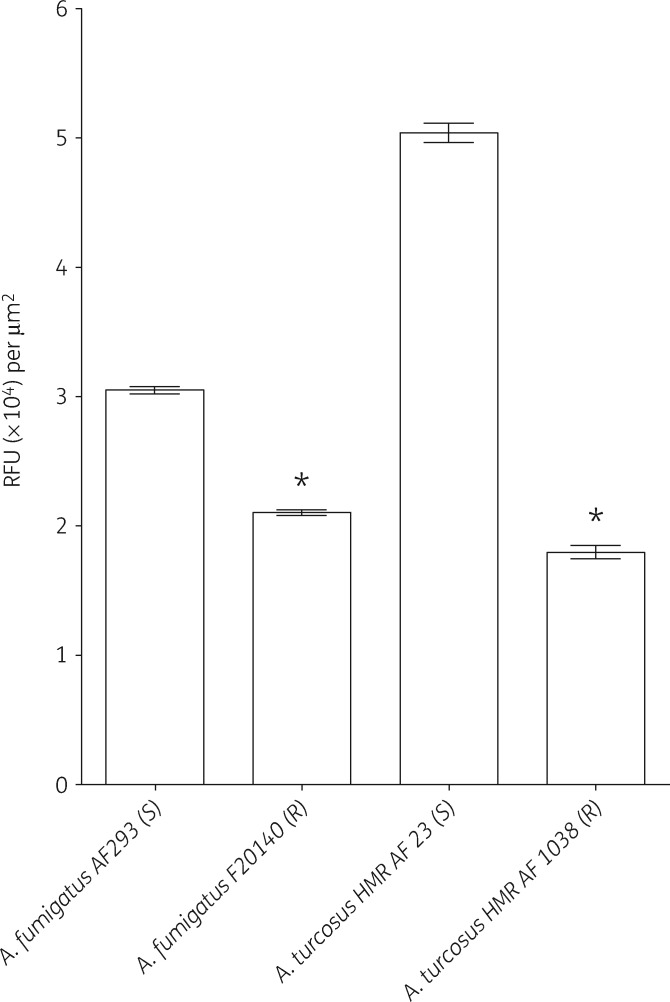
Since resistance and its underlying mechanisms have not been previously reported in A. turcosus, we sought to corroborate these findings with a second method. We designed another phenotypic assay, utilizing the fluorescent efflux pump substrate Nile red. The assay quantifies Nile red accumulation in germ tubes of tested isolates as an indirect measurement of efflux pump activity. As predicted, the control azole-resistant A. fumigatus strain F20140, overexpressing efflux pumps, accumulated significantly less Nile red than the control azole-susceptible strain AF293 (Figure 5). Two other control azole-resistant A. fumigatus strains (F17727 and F18304) with overexpression of efflux pumps produced comparable results (data not shown). Azole-resistant A. turcosus strain HMR-AF-1038 accumulated significantly less Nile red than azole-susceptible A. turcosus strain HMR-AF-23, again consistent with enhanced efflux pump activity.
Figure 5.
Accumulation of Nile red in azole-susceptible (S) and -resistant (R) Aspergillus isolates. Data represent mean ± SEM of a single experiment. A second experiment produced comparable results (data not shown). An asterisk indicates P < 0.001 compared with the susceptible strain of the same species, Student’s t-test. RFU, relative fluorescence units.
Discussion
This first (to the best of our knowledge) hospital-based survey of Aspergillus section Fumigati isolates in Canada reveals a low prevalence of azole resistance. Overall, 4 of 807 patients (0.5%) yielded resistant isolates, including only one A. fumigatus sensu stricto and three cryptic species (A. thermomutatus, A. lentulus and A. turcosus). The level of resistance was low in all the isolates, with MICs close to the reported ECV for A. fumigatus.23,24 Our findings suggest that resistance in the single resistant A. fumigatus isolate was efflux-pump mediated and that it was acquired in vivo during prolonged azole therapy (host-derived), although typing of serially collected isolates was not performed. In contrast to many other countries, cyp51a mutations associated with environment-derived resistance were not observed in these Canadian A. fumigatus isolates.
The percentage of cryptic Aspergillus species isolates in this study (8/807; 1%) falls within the range of previous surveys conducted in other countries (0.7%–5.4%).15,45–50 This finding adds to the body of evidence supporting that cryptic Aspergillus section Fumigati species share the geographic ubiquity of A. fumigatus sensu stricto, while they consistently differ by their far less frequent isolation from clinical specimens. Low prevalence in clinical samples could be a result of low environmental abundance or reduced virulence, but this remains speculative. The study of the relative pathogenicity of cryptic species has been limited to Aspergillus udagawae, which exhibited reduced virulence compared with A. fumigatus.51 With the exception of A. turcosus,42 the cryptic Aspergillus section Fumigati species isolated in the present survey (A. thermomutatus, A. hiratsukae and A. lentulus) had all been previously identified as agents of aspergillosis.15,18,19,46,48,50,52,53 This study underlines the pathogenic potential of these species by providing two additional cases of significant infections. Our recovery from respiratory samples of four A. turcosus isolates from four individual patients documents for the first time the isolation of this cryptic Aspergillus species from human clinical material.
We investigated the underlying mechanisms of azole resistance in three cryptic Aspergillus species. Our 3D model of CYP51A homologues provided important insights on possible molecular mechanisms of azole resistance. Much like other cytochrome P450 superfamily members, we observed that the structural fold adopted by CYP51A enzymes is highly tolerant of amino acid substitutions, displaying high structural plasticity. Consequently, mutations distant from the active site are unlikely to cause significant reshaping of its buried cavity. This is in accordance with previous findings in A. fumigatus showing that the main codons involved in resistance (G54, G138 and M220) are located near the opening of a ligand access channel.54 In the newly described A. turcosus specifically, comparison between azole-susceptible and -resistant isolates revealed CYP51A amino acid variations that did not translate into meaningful structural differences. On the other hand, two distinct phenotypic assays consistently showed the role of efflux pumps, suggesting that this mechanism is associated with azole resistance, at least in this particular isolate. Whether this finding extends to other representatives of this species remains to be determined. Quantitative efflux pump gene expression studies will be required to identify the specific genes responsible for this phenotype. In A. lentulus, the contribution of altered CYP51A structural characteristics to intrinsic azole resistance is supported by previous in silico and transformation studies.20,21 It has been deduced from docking studies that BC loop positioning differences observed in A. lentulus are responsible for weaker voriconazole stabilization at its putative binding site. Consistent with the report of Alcazar-Fuoli et al.,20 no significant difference could be deduced from CYP51A structural comparisons between our A. lentulus isolate and A. fumigatus. Furthermore, our isolate displayed very high CYP51A sequence identity compared with the strain reported by Alcazar-Fuoli et al.,20 suggesting that a similar mechanism could be at play. Importantly, the present work further emphasized the contribution of target alteration in azole resistance in A. lentulus, by demonstrating the lack of effect of the efflux pump inhibitor, a mechanism that was not investigated in previous work on A. lentulus. Finally, no contribution of efflux pumps could be demonstrated for A. thermomutatus. As with A. lentulus, this species displayed CYP51A differences located away from the active site or binding channels. Hypothetically, long-range mutations could still explain decreased voriconazole binding through a potential allosteric mechanism, similar to that observed in A. lentulus, especially considering the high sequence and structural identity between the two enzymes. This warrants further studies.
This work has several limitations. First, although the sensitivity and specificity of our identification and azole susceptibility screening methods were assessed, we cannot formally rule out that some cryptic and/or resistant isolates were missed. Also, the spectrum of the efflux pump inhibitor MC-207,110 is not precisely known, and hence the potential contribution of uninhibited efflux pumps to the resistance phenotype may have been overlooked. Finally, only known resistance mechanisms were investigated, based on current knowledge derived from A. fumigatus studies. The contribution of transcription factors relevant to cyp51a and/or efflux pump expression (e.g. HapE, AtrR, SbrA) was not assessed in the present work and will require further investigation.
In conclusion, overall azole resistance was rare in this large survey of Aspergillus section Fumigati clinical isolates and mainly driven by cryptic species. A. turcosus is now recognized as a human colonizer and this cryptic species may exhibit azole resistance, likely through efflux pump activity. Surveillance of the emergence of cryptic species is warranted and further studies are required to uncover resistance mechanisms.
Supplementary Material
Acknowledgements
We thank Dr Michel Laverdière for having set up the Aspergillus clinical isolate collection in 2000. We also thank Martine Raymond for helpful discussions on efflux pump assays and Monique Vasseur for assistance with fluorescence microscopy. We are also grateful to the platform personnel at Génome Québec Innovation Center for performing the Sanger sequencing. The information contained in this manuscript was presented in part at ASM Microbe 2016 in Boston, MA, USA (Poster FRIDAY-306).
Funding
This work was supported in part by a start-up grant from the Hôpital Maisonneuve-Rosemont Microbiology Research Fund (to S.F.D.). This work was also supported in part by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH) (under award number R01GM105978, to N.D.). N.D. holds a Fonds de Recherche Québec – Santé (FRQS) Research Scholar Junior 2 Career Award. M.P.-M. is the recipient of an award from the Gabriel-Marquis Scholarship Fund.
Transparency declarations
S.F.D. has received funds for advisory boards (Merck Canada Inc., Avir Pharma Inc.) and investigator-initiated research grants (bioMérieux Canada Inc.). All other authors: none to declare.
References
- 1. Hope WW, Walsh TJ, Denning DW.. The invasive and saprophytic syndromes due to Aspergillus spp. Med Mycol 2005; 43: S207–38. [DOI] [PubMed] [Google Scholar]
- 2. Kosmidis C, Denning DW.. The clinical spectrum of pulmonary aspergillosis. Thorax 2015; 70: 270–7. [DOI] [PubMed] [Google Scholar]
- 3. Latge JP. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev 1999; 12: 310–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kontoyiannis DP, Marr KA, Park BJ. et al. Prospective surveillance for invasive fungal infections in hematopoietic stem cell transplant recipients, 2001-2006: overview of the Transplant-Associated Infection Surveillance Network (TRANSNET) database. Clin Infect Dis 2010; 50: 1091–100. [DOI] [PubMed] [Google Scholar]
- 5. Steinbach WJ, Marr KA, Anaissie EJ. et al. Clinical epidemiology of 960 patients with invasive aspergillosis from the PATH Alliance registry. J Infect 2012; 65: 453–64. [DOI] [PubMed] [Google Scholar]
- 6. Patterson TF, Thompson GR 3rd, Denning DW. et al. Practice guidelines for the diagnosis and management of aspergillosis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 2016; 63: e1–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hagiwara D, Watanabe A, Kamei K. et al. Epidemiological and genomic landscape of azole resistance mechanisms in Aspergillus fungi. Frontiers Microbiol 2016; 7: 1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Howard SJ, Cerar D, Anderson MJ. et al. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg Infect Dis 2009; 15: 1068–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chowdhary A, Kathuria S, Xu J. et al. Emergence of azole-resistant Aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog 2013; 9: e1003633.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Snelders E, Camps SM, Karawajczyk A. et al. Triazole fungicides can induce cross-resistance to medical triazoles in Aspergillus fumigatus. PLoS One 2012; 7: e31801.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wiederhold NP, Gil VG, Gutierrez F. et al. First detection of TR34 L98H and TR46 Y121F T289A Cyp51 mutations in Aspergillus fumigatus isolates in the United States. J Clin Microbiol 2016; 54: 168–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Samson RA, Hong S, Peterson SW. et al. Polyphasic taxonomy of Aspergillus section Fumigati and its teleomorph Neosartorya. Stud Mycol 2007; 59: 147–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Samson RA, Visagie CM, Houbraken J. et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol 2014; 78: 141–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Balajee SA, Gribskov J, Brandt M. et al. Mistaken identity: Neosartorya pseudofischeri and its anamorph masquerading as Aspergillus fumigatus. J Clin Microbiol 2005; 43: 5996–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Balajee SA, Kano R, Baddley JW. et al. Molecular identification of Aspergillus species collected for the Transplant-Associated Infection Surveillance Network. J Clin Microbiol 2009; 47: 3138–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khare R, Gupta S, Arif S. et al. Misidentification of Neosartorya pseudofischeri as Aspergillus fumigatus in a lung transplant patient. J Clin Microbiol 2014; 52: 2722–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alcazar-Fuoli L, Mellado E, Alastruey-Izquierdo A. et al. Aspergillus section Fumigati: antifungal susceptibility patterns and sequence-based identification. Antimicrob Agents Chemother 2008; 52: 1244–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Howard SJ. Multi-resistant aspergillosis due to cryptic species. Mycopathologia 2014; 178: 435–9. [DOI] [PubMed] [Google Scholar]
- 19. Lamoth F. Aspergillus fumigatus-related species in clinical practice. Frontiers Microbiol 2016; 7: 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alcazar-Fuoli L, Cuesta I, Rodriguez-Tudela JL. et al. Three-dimensional models of 14α-sterol demethylase (Cyp51A) from Aspergillus lentulus and Aspergillus fumigatus: an insight into differences in voriconazole interaction. Int J Antimicrob Agents 2011; 38: 426–34. [DOI] [PubMed] [Google Scholar]
- 21. Mellado E, Alcazar-Fuoli L, Cuenca-Estrella M. et al. Role of Aspergillus lentulus 14-α sterol demethylase (Cyp51A) in azole drug susceptibility. Antimicrob Agents Chemother 2011; 55: 5459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi—Second Edition: M38 2008.
- 23.CLSI. Epidemiological Cutoff Values for Antifungal Susceptibility Testing—Second Edition: M59 2018.
- 24. Espinel-Ingroff A, Diekema DJ, Fothergill A. et al. Wild-type MIC distributions and epidemiological cutoff values for the triazoles and six Aspergillus spp. for the CLSI broth microdilution method (M38-A2 document). J Clin Microbiol 2010; 48: 3251–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rajendran R, Mowat E, McCulloch E. et al. Azole resistance of Aspergillus fumigatus biofilms is partly associated with efflux pump activity. Antimicrob Agents Chemother 2011; 55: 2092–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsao S, Weber S, Cameron C. et al. Positive regulation of the Candida albicans multidrug efflux pump Cdr1p function by phosphorylation of its N-terminal extension. J Antimicrob Chemother 2016; 71: 3125–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parent-Michaud M, Dufresne PJ, Fournier É. et al. Draft genome sequences of azole-resistant and azole-susceptible Aspergillus turcosus clinical isolates recovered from bronchoalveolar lavage fluid samples. Microbiol Resourc Announce 2019; 8: e01446-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Parent-Michaud M, Dufresne PJ, Fournier É. et al. Draft genome sequence of azole-resistant Aspergillus thermomutatus (Neosartorya pseudofischeri) strain HMR-AF-39, isolated from a human nasal septum abscess aspirate. Microbiol Res Announce 2019; 8: e01444-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kusuya Y, Sakai K, Kamei K. et al. Draft genome sequence of the pathogenic filamentous fungus Aspergillus lentulus IFM 54703T. Genome Announce 2016; 4: e01568-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu M, Zeng R, Zhang L. et al. Multiple cyp51A-based mechanisms identified in azole-resistant isolates of Aspergillus fumigatus from China. Antimicrob Agents Chemother 2015; 59: 4321–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Losada L, Sugui JA, Eckhaus MA. et al. Genetic analysis using an isogenic mating pair of Aspergillus fumigatus identifies azole resistance genes and lack of MAT locus’s role in virulence. PLoS Pathog 2015; 11: e1004834.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fraczek MG, Bromley M, Buied A. et al. The cdr1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus. J Antimicrob Chemother 2013; 68: 1486–96. [DOI] [PubMed] [Google Scholar]
- 33. Mellado E, Diaz-Guerra TM, Cuenca-Estrella M. et al. Identification of two different 14-α sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J Clin Microbiol 2001; 39: 2431–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schubert S, Barker KS, Znaidi S. et al. Regulation of efflux pump expression and drug resistance by the transcription factors Mrr1, Upc2, and Cap1 in Candida albicans. Antimicrob Agents Chemother 2011; 55: 2212–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kanafani ZA, Perfect JR.. Antimicrobial resistance: resistance to antifungal agents: mechanisms and clinical impact. Clin Infect Dis 2008; 46: 120–8. [DOI] [PubMed] [Google Scholar]
- 36. Tsao S, Rahkhoodaee F, Raymond M.. Relative contributions of the Candida albicans ABC transporters Cdr1p and Cdr2p to clinical azole resistance. Antimicrob Agents Chemother 2009; 53: 1344–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goujon M, McWilliam H, Li W. et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucl Acids Res 2010; 38: W695–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sali A, Blundell TL.. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 1993; 234: 779–815. [DOI] [PubMed] [Google Scholar]
- 39. Laskowski RA, Rullmann JA, MacArthur MW. et al. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 1996; 8: 477–86. [DOI] [PubMed] [Google Scholar]
- 40. Kozlikova B, Sebestova E, Sustr V. et al. CAVER Analyst 1.0: graphic tool for interactive visualization and analysis of tunnels and channels in protein structures. Bioinformatics 2014; 30: 2684–5. [DOI] [PubMed] [Google Scholar]
- 41. De Pauw B, Walsh TJ, Donnelly JP. et al. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis 2008; 46: 1813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hong SB, Shin HD, Hong J. et al. New taxa of Neosartorya and Aspergillus in Aspergillus section Fumigati. Antonie Van Leeuwenhoek 2008; 93: 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cowen LE, Sanglard D, Howard SJ. et al. Mechanisms of antifungal drug resistance. Cold Spring Harb Perspect Med 2014; 5: a019752.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Camps SM, van der Linden JW, Li Y. et al. Rapid induction of multiple resistance mechanisms in Aspergillus fumigatus during azole therapy: a case study and review of the literature. Antimicrob Agents Chemother 2012; 56: 10–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alastruey-Izquierdo A, Mellado E, Pelaez T. et al. Population-based survey of filamentous fungi and antifungal resistance in Spain (FILPOP Study). Antimicrob Agents Chemother 2013; 57: 3380–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Escribano P, Pelaez T, Munoz P. et al. Is azole resistance in Aspergillus fumigatus a problem in Spain? Antimicrob Agents Chemother 2013; 57: 2815–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sabino R, Verissimo C, Parada H. et al. Molecular screening of 246 Portuguese Aspergillus isolates among different clinical and environmental sources. Med Mycol 2014; 52: 519–29. [DOI] [PubMed] [Google Scholar]
- 48. Negri CE, Goncalves SS, Xafranski H. et al. Cryptic and rare Aspergillus species in Brazil: prevalence in clinical samples and in vitro susceptibility to triazoles. J Clin Microbiol 2014; 52: 3633–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mortensen KL, Johansen HK, Fuursted K. et al. A prospective survey of Aspergillus spp. in respiratory tract samples: prevalence, clinical impact and antifungal susceptibility. Eur J Clin Microbiol Infect Dis 2011; 30: 1355–63. [DOI] [PubMed] [Google Scholar]
- 50. Hubka V, Kubatova A, Mallatova N. et al. Rare and new etiological agents revealed among 178 clinical Aspergillus strains obtained from Czech patients and characterized by molecular sequencing. Med Mycol 2012; 50: 601–10. [DOI] [PubMed] [Google Scholar]
- 51. Sugui JA, Vinh DC, Nardone G. et al. Neosartorya udagawae (Aspergillus udagawae), an emerging agent of aspergillosis: how different is it from Aspergillus fumigatus? J Clin Microbiol 2010; 48: 220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Alastruey-Izquierdo A, Alcazar-Fuoli L, Cuenca-Estrella M.. Antifungal susceptibility profile of cryptic species of Aspergillus. Mycopathologia 2014; 178: 427–33. [DOI] [PubMed] [Google Scholar]
- 53. Van Der Linden JW, Warris A, Verweij PE.. Aspergillus species intrinsically resistant to antifungal agents. Med Mycol 2011; 49: S82–9. [DOI] [PubMed] [Google Scholar]
- 54. Snelders E, Karawajczyk A, Schaftenaar G. et al. Azole resistance profile of amino acid changes in Aspergillus fumigatus CYP51A based on protein homology modeling. Antimicrob Agents Chemother 2010; 54: 2425–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.