Abstract
Introduction
Klebsiella pneumoniae with OXA-48-like enzymes were introduced into Tshwane Tertiary Hospital (TTH) (Pretoria, South Africa) during September 2015, causing nosocomial outbreaks.
Methods
PCR methodologies and WGS were used to characterize K. pneumoniae with carbapenemases (n = 124) from TTH (July 2015–December 2016).
Results
PCR was used to track K. pneumoniae ST307 with OXA-181 among 60% of carbapenemase-positive isolates in different wards/units over time and showed the transmission of IncX3 plasmids to other K. pneumoniae clones. WGS identified different ST307 clades: 307_OXA181 (consisting of two lineages, A and B) with OXA-181 on IncX3 plasmids (named p72_X3_OXA181) and clade 307_VIM with VIM-1 on IncFII plasmids. Clade 307_OXA181 lineage B was responsible for the rapid increase and transmission of OXA-181 K. pneumoniae in various wards/units throughout TTH, while the numbers of clade 307_OXA181 lineage A and clade 307_VIM remained low. Separate outbreaks were due to K. pneumoniae ST17 and ST29 with p72_X3_OXA181 plasmids.
Conclusions
The high-risk clone K. pneumoniae ST307 with OXA-181 rapidly spread to different wards/units despite infection and prevention measures. ST307 clades and lineages seemingly acted differently in outbreak situations. This study also highlighted the threat of promiscuous plasmids such as p72_X3_OXA181.
Introduction
Carbapenemases are important causes of carbapenem resistance because carbapenemase genes are typically part of mobile genetic elements (MGEs) that can move between different Gram-negative species, such as the Enterobacterales.1 The most common carbapenemases among Enterobacterales are the KPC (Amber class A), NDM (Ambler class B) and OXA-48-like (Ambler class D) enzymes.2
Genomic surveillance studies have shown that OXA-48-like carbapenemases from Klebsiella pneumoniae and Escherichia coli are the most common carbapenemases among Enterobacterales in certain regions (e.g. Middle East, North Africa and countries such as Belgium and Spain).3 OXA-48, OXA-181, OXA-232, OXA-204, OXA-162 and OXA-244 are the most common enzymes identified among the OXA-48-like carbapenemases.3
K. pneumoniae with OXA-48-like enzymes were introduced into Tshwane Tertiary Hospital (TTH) (Pretoria, South Africa) during September 2015 after a patient was transferred from a private Pretoria hospital. TTH then experienced a significant increase in the numbers of K. pneumoniae with OXA-48-like enzymes. Several South African private hospitals were part of province-wide outbreaks due to the high-risk clone K. pneumoniae ST307 with blaOXA-181.4 A genomic study was designed to establish whether ST307 with OXA-181 was responsible for the increase in OXA-48-like K. pneumoniae.
Methods
Ethics
Ethics approval was obtained from the Conjoint Health Research Ethics Board at the University of Calgary (REB17-1010) and from the Research Ethics Committee (REC), Faculty of Health Sciences, University of Pretoria (protocol numbers: 240/2016 and 104/2017).
Setting
Please refer to the Supplementary data available at JAC Online for details of TTH.
Bacterial isolates, identification, susceptibility and carbapenemase testing
During July 2015–December 2016, 148 carbapenem-non-susceptible (i.e. ertapenem and/or meropenem non-susceptible) non-repeat K. pneumoniae were isolated at the TTH microbiology laboratory from clinical and surveillance cultures; 124 tested phenotypically positive for carbapenemases and were referred to the National Institute for Communicable Diseases for PCR confirmation. CLSI criteria for broth dilution were used for ertapenem and meropenem.5 For details of identification, susceptibility and carbapenemase testing please refer to the Supplementary data.
PCR methodologies
PCR primers for the detection of OXA-181,6 ST307, IncX3 plasmids and the IS3000–OXA junction4 were used to screen K. pneumoniae with OXA-48-like enzymes. K. pneumoniae with other carbapenemases were screened for ST307.
WGS and data analysis
K. pneumoniae (n = 106), E. coli (n = 4) and Enterobacter sp. (n = 1) were sequenced with the NextSeq (Illumina, San Diego, CA, USA) platform. Libraries were prepared with the Nextera XT Kit to produce paired-end reads of 150 bp for a predicted minimum coverage of 75×. ST307 genomes were mapped to the reference genome I72 (accession CP034281) and recombination-free SNP phylogenetic trees were generated as previously described.4 Genomic data from this study were submitted to NCBI under the Bioproject PRJNA488070.
The blaOXA181-harbouring IncX3 plasmid named p72_X3_OXA181 was previously sequenced using the RSII platform (Pacific Biosciences)4 and identification was conducted by de novo assembly using plasmidSPAdes,7 Bandage assembly graph viewer8 and BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) as well as S1-PFGE in combination with Southern blotting. The genome sequences were de novo assembled using plasmidSPAdes,7 followed by extraction of the blaOXA181-harbouring contigs, using blastdbcmd. The blaOXA-181 contigs were ‘BLASTed’ against the PlasmidFinder9 database. The results showed that blaOXA-181 co-existed on the same contig as a truncated ColKp3 replicon gene, similar to that of p72_OXA181_X3. The identity of p72_OXA181_X3 was determined using Bandage assembly graph viewer8 and BLASTn. The plasmid region was identified by BLASTn searching for the p72_OXA181_X3 ‘core’ genes, excluding ISs or repeated sequences.
Infection prevention and control (IP&C) measures taken at TTH
Routine auditing of hospital-acquired infections formed part of the continuous antimicrobial-resistant organism (ARO) surveillance at TTH. AROs isolated at the microbiology laboratory were communicated to the treating physicians and IP&C practitioners on a daily basis. ARO weekly reports were generated and reviewed by IP&C as part of standard surveillance practice. Monthly reports were communicated to the TTH Chief Executive Officer.
A case definition of carbapenem-resistant Enterobacterales (CRE) was adopted in September 2015 when an isolate showed imipenem and/or meropenem MICs of ≥1 mg/L and/or an ertapenem MIC of ≥0.5 mg/L or an isolate tested positive for carbapenemases. A CRE outbreak was declared in November 2015 and the CDC (Atlanta, GA, USA) guidelines for CRE IP&C were implemented by the hospital management.10 Hospital-acquired cases were classified as patients that developed infections >48 h after admission to TTH.
Outbreak response teams were created by hospital management and IP&C and outbreak management measures (i.e. cohorting of case patients, barrier precautions and active surveillance) were applied. IP&C practitioners held weekly information sessions with all the wards and monitored hand hygiene practices. Contact precaution training and education on donning and removal of gloves and aprons were monitored. Environmental cleaning was actively carried out.
Case patients that met the case definition and were admitted to ICUs or the high-risk unit (HRU) were cohorted in each unit. Non-ICU patients that met the case definition were cohorted in two outbreak-isolation wards. Strict cohorting of staff was also implemented. Strict access control to outbreak-isolation wards for staff and visitors was applied. Case patients were bathed with 2% chlorhexidine daily and when transferred out of TTH.
Surveillance cultures (i.e. rectal, wounds, ostomy, endotracheal suctions) were used to identify all epidemiologically linked patients who may have become colonized. This was directed by IP&C and formed part of contact tracing and unit-wide prevalence screening. The patients included inpatients on the same wards/units as the case patient and subsequent patients.
Surveillance cultures were undertaken weekly from all patients on the affected units until negative screening cultures were obtained for two consecutive weeks. Roommates and unit contacts of case patients also had surveillance specimens collected before discharge. Screening swabs were placed into Copan M40 transport system tubes containing Amies gel transport medium. Stool specimens and endotracheal suctions were submitted in sterile containers without transport medium. The CDC protocol was used to screen for carbapenemase-producing Gram-negative bacteria.10 Environmental cultures taken throughout the hospital were negative for K. pneumoniae with carbapenemases. Surveillance cultures from staff were not obtained.
Results
Carbapenemases
TTH detected 124 PCR-positive carbapenemase-producing K. pneumoniae from July 2015 to December 2016 that included NDM (n = 18), VIM (n = 7) and OXA-48-like (n = 99) enzymes. Before July 2015, 12 cases of NDMs were obtained in 2013 and 2014. The first OXA-48-like isolate at TTH was obtained in September 2015 and the numbers of isolates with these enzymes increased substantially, especially during October–December 2015 (Figure 1). VIM- and NDM-positive isolates were detected in July 2015 and the numbers remained low (Figure 1).
Figure 1.
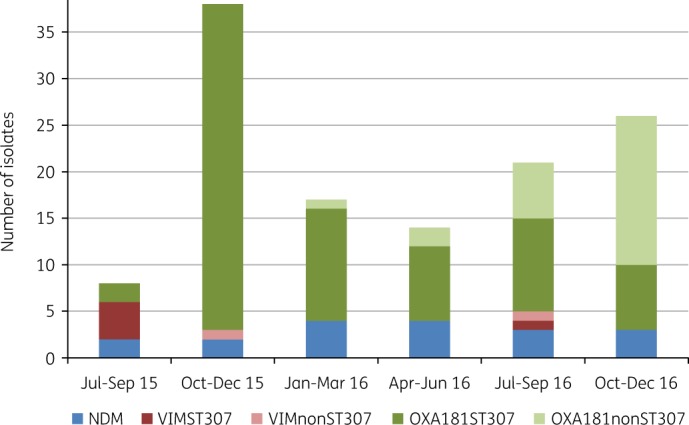
PCR results of K. pneumoniae with carbapenemases from TTH, July 2015–December 2016. VIMST307, PCR positive for VIM and ST307; VIMnonST307, PCR positive for VIM and negative for ST307; OXA181ST307, PCR positive for OXA-181 and ST307; OXA181nonST307, PCR positive for OXA-181 and negative for ST307. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
PCR screening showed high prevalence of K. pneumoniae ST307 with blaOXA-181
PCR screening of OXA-48-like K. pneumoniae showed that 74/99 (75%) were positive for OXA-181, ST307, IncX3 and IS3000–OXA (Figure 1) (referred to as OXA181ST307). K. pneumoniae with blaVIM (5/7) also tested positive for ST307 (referred to as VIMST307), while the NDM-positive isolates were negative for ST307 (Figure 1). OXA181ST307 was introduced into TTH during September 2015, via a patient that was transferred from a private hospital, and then increased exponentially during November–December 2015. VIMST307 was detected in July 2015 (Figure 1).
K. pneumoniae with OXA-48-like enzymes that tested negative by PCR for ST307, but positive for OXA-181, IncX3 and IS3000–OXA [referred to as OXA181nonST307 (n = 25)], appeared in March 2016 and increased, especially during October–December 2016 (Figure 1). Additionally, E. coli (n = 4) and Enterobacter sp. (n = 1) with OXA-48-like enzymes emerged during October–December 2016 and were positive for OXA-181, IncX3 and IS3000–OXA. These PCR data suggested that IncX3 plasmids with OXA-181 were introduced by ST307 into TTH and then transferred to non-ST307 K. pneumoniae, as well as other Enterobacterales.
K. pneumoniae ST307 from TTH belonged to different clades and lineages
WGS confirmed that 73/74 OXA181ST307 belonged to ST307 and showed that the OXA181nonST307 (n = 25) belonged to ST15 (n = 1), ST17 (n = 15), ST29 (n = 6), ST45 (n = 2) and ST336 (n = 1). The OXA-181 PCR results correlated with WGS results.
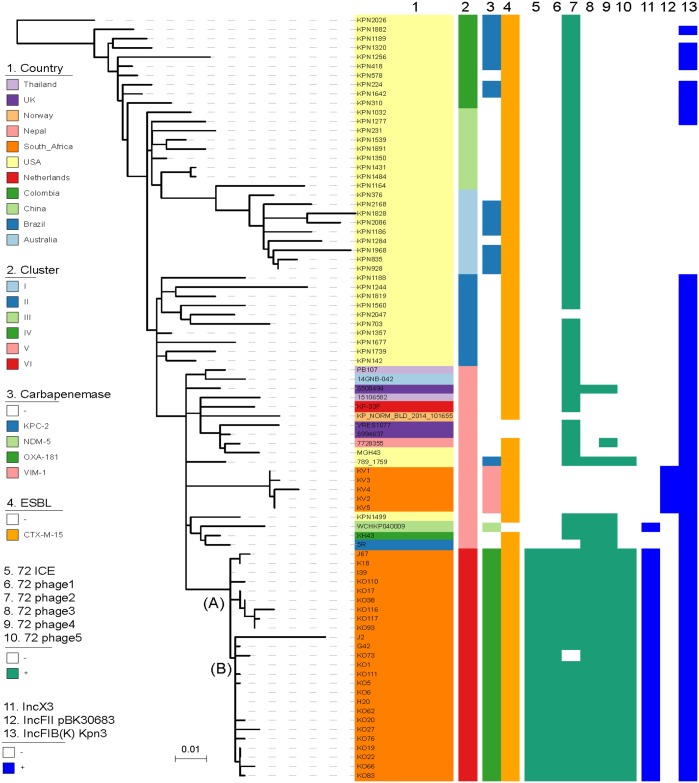
The K. pneumoniae ST307 with OXA-181 from TTH (n = 73) belonged to the previously described clade VI that is unique to South Africa4 (Figure 2). Genomic analysis identified two distinct lineages: A (n = 7) and B (n = 66) among ST307 with OXA-181 (Figure 2). The p72_X3_OXA181 plasmid was identified in the ST307 with OXA-181.
Figure 2.
Phylogenetic analysis of K. pneumoniae ST307 from TTH including some representative global ST307 isolates to highlight different clades. Isolates from TTH (n = 24), isolates from private Pretoria hospitals (n = 6) and global isolates (n = 53) are included. The global genomes were obtained from the NCBI WGS database. Clades and lineages were defined using previously published criteria.4 OXA-181 ST307 isolates belonged to a distinct South African clade, namely clade VI. Analysis identified two distinct lineages among this clade, namely ST307_181A (A) and ST307_181B (B), as illustrated at the nodes. Isolates J67, K18, I39, J2, G42 and H20 were obtained from a previous study involving private South African hospitals.4 Isolates KO110, KO17, KO38, KO116, KO117, KO93, KO73, KO1, KO111, KO5, KO6, KO62, KO20, KO27, KO76, KO19, KO22, KO66 and KO83 were from this study. VIMST307 (KV1–KV5 from this study) belonged to a different ST307 clade (cluster V) that clustered with other international ST307. ST307 clades I–IV are limited to the USA. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
VIMST307 (n = 5) belonged to a different ST307 clade and contained IncFII-like plasmids that harboured blaVIM-1 (Figure 2). The VIMnonST307 (n = 2) belonged to ST231.
The characteristics shared by ST307 with OXA-181 and ST307 with VIM-1 included the presence of the capsular locus wzi-173, capsule 2, the π-fimbrial cluster, type IV secretion system, ParC-80I and GyrA-83I mutations and blaCTX-M-15. ST307 with OXA-181 contained five prophages (namely 72_phage 1 to 5) and one novel integrative conjugative element (72_ICE) that were absent in ST307 with blaVIM-1 (Figure 2).
WGS confirmed clonal outbreaks of K. pneumoniae ST307 with OXA-181 across TTH
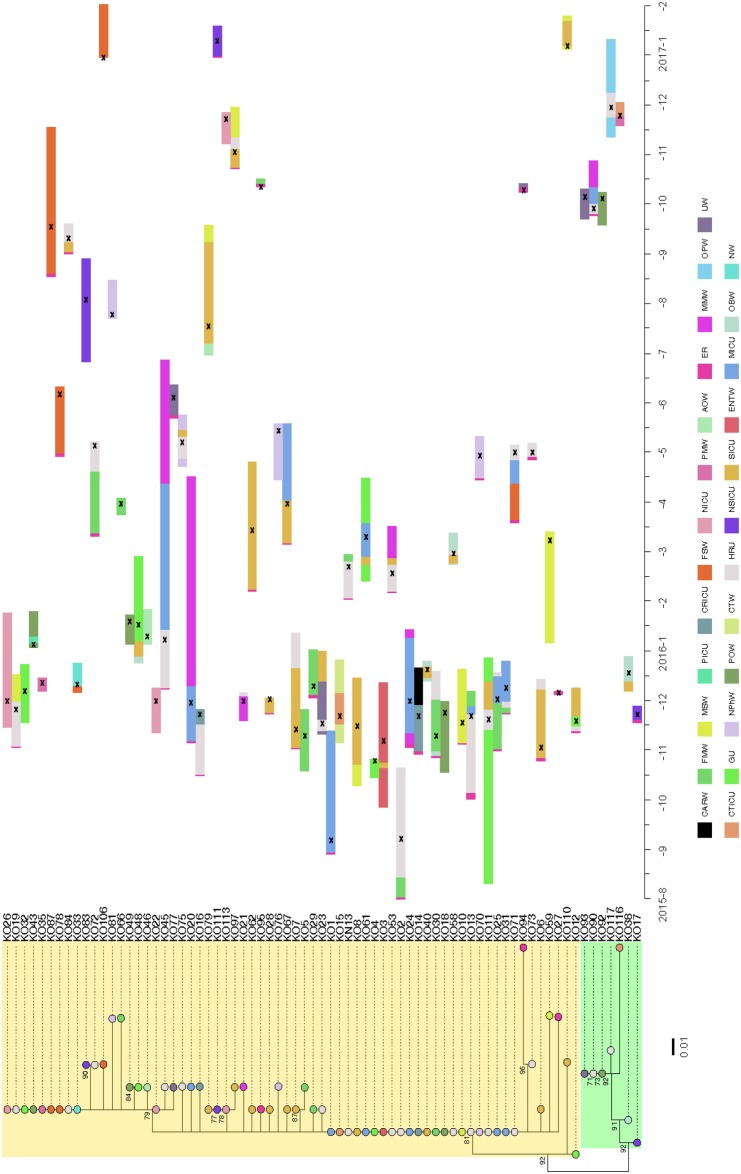
Genomic relationships of ST307 with blaOXA-181 were combined with clinical epidemiology data from patients admitted to TTH to identify transmission events (Figure 3).
Figure 3.
Genomic relationships of K. pneumoniae ST307 with OXA-181 combined with the epidemiology data from TTH, July 2015–December 2016. A maximum likelihood phylogenetic tree is shown on the left, labelled with major genomic lineages (lineage B is highlighted in yellow and lineage A is highlighted in light green) and supporting bootstrap values (70%). Nodes of the tree are coloured by the wards/units from which each isolate was obtained from. Coloured horizontal bars on the right indicate admission and transfer dates over time. X indicates the detection of the ST307 isolate. Lineages were defined using previously published criteria.4 Ward/Unit abbreviations: ER, Emergency Room; FMW, Female Medical Ward; HRU, High-Risk Unit; ENTW, Ear Nose Throat Ward; MSW, Male Surgery Ward; GU, Gynaecology Unit; MICU, Medical ICU; CTW, Cardiothoracic Ward; CTICU, Cardiothoracic ICU; SICU, Surgical ICU; CRICU, Coronary ICU; POW, Paediatric Oncology Ward; NICU, Neonatal ICU; MMW, Male Medical Ward; UW, Urology Ward; OBW, Obstetrics Ward; FSW, Female Surgery Ward; PMW, Paediatric Medical Ward; PICU, Paediatric ICU; AOW, Adult Oncology Ward; NSICU, Neurosurgical ICU; NPhW, Nephrology Ward; OPW, Orthopaedic Ward; NW, Neurology Ward; and CARW, Cardiology Ward. K. pneumoniae ST307 lineage B (n = 66) (referred to as ST307_181B) was introduced into TTH in September 2015 (i.e. KO1 from a patient with a documented transfer from a private hospital) to the medical ICU via the ER. Isolate KO2, identical to KO1, was obtained a day later, from a patient admitted to the HRU. Identical or highly related ST307_181B isolates then spread to patients within the medical ICU, HRU and surgical ICU. The main transmission events seemingly occurred in the medical ICU, medical wards, HRU and surgical ICU. Continuous transmission events with ST307_181B continued during 2016, especially in the HRU and surgical ICU. The majority of patients infected/colonized with ST307_181B during 2016 had previous contact with the medical wards, HRU and surgical ICU. K. pneumoniae ST307 lineage A (n = 7) (referred to as ST307_181A) was obtained from a patient (i.e. KO17) with a documented transfer from a private hospital during November 2015 and admitted to the neurosurgical ICU via the ER. Transmission events with ST307_181A occurred in September 2016 (i.e. KO90, KO92 and KO93). No obvious association with a specific ward/unit was evident. Patients at TTH were often moved between wards for reasons of clinical care. The two outbreak isolation wards were the male medical ward and the female medical ward. The critical cohorting units consisted of the medical ICU and surgical ICU. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
K. pneumoniae ST307 lineage B (n = 66) (referred to as ST307_181B) was introduced into TTH in September 2015 (i.e. isolate KO1 from a patient with a documented transfer from a private hospital) to the medical ICU via the emergency room (ER) (Figure 3). Isolate KO2, identical to KO1, was obtained a day later from a patient admitted to the HRU. This patient was admitted to TTH during July 2015 via the ER, transferred to the medical female ward and then to the HRU during October 2015 (Figure 3). ST307_181B isolates, identical or highly related to KO1, were then obtained from patients admitted to the gynaecology unit (i.e. KO4), the surgical ICU (i.e. KO6) and the ear/nose/throat ward (i.e. KO3) during October 2015 (Figure 3). These patients did not have obvious contacts with the medical ICU or the HRU.
Identical or highly related ST307_181B isolates then spread to patients within the medical ICU (n = 4), HRU (n = 5) and surgical ICU (n = 4), as well as various wards/units during November and December 2015 (Figure 3). The main transmission events seemingly occurred in the medical ICU, medical wards, HRU and surgical ICU (Figure 3). Continuous transmission events with ST307_181B continued during 2016 especially in the HRU (n = 8) and surgical ICU (n = 5). The majority of patients infected/colonized with ST307_181B during 2016 had previous contact with the medical wards, HRU and surgical ICU (Figure 3).
K. pneumoniae ST307 lineage A (n = 7) (referred to as ST307_181A) was obtained from a patient (i.e. KO17) with a documented transfer from a private hospital during November 2015 and admitted to the neurosurgical ICU via the ER (Figure 3). Transmission events with ST307_181A occurred in September 2016 (i.e. KO90, KO92 and KO93) (Figure 3). No obvious association with a specific ward/unit was evident.
Plasmid analysis confirmed the transmission of p72_X3_OXA181 to other members of the Enterobacterales
Plasmid p72_X3_OXA181 was identified among OXA181nonST307 K. pneumoniae (n = 25), E. coli (n = 4) and Enterobacter sp. (n = 1). K. pneumoniae ST17 with p72_X3_OXA181 (n = 15) first appeared in the medical ICU during July 2016 and the numbers increased, especially during November–December 2016 (n = 9), mainly from HRU patients. K. pneumoniae ST29 with p72_X3_OXA181 (n = 6) first appeared in the paediatric oncology unit during September 2016 and the numbers increased during December 2016 (n = 3) from HRU patients.
K. pneumoniae ST15, ST45 and ST336 with p72_X3_OXA181 were identified during 2016 from the gynaecology unit, neurosurgical ICU and the paediatric medical ward, respectively. The E. coli (n = 4) and Enterobacter sp. (n = 1) with p72_X3_OXA181 were obtained during September–December 2016 from the HRU and surgical and medical ICUs.
Discussion
Low- and middle-income countries (LMICs) bear a considerable burden of the widespread prevalence of antimicrobial-resistant (AMR) bacteria, but lack adequate genomic diagnostic tools and surveillance systems to curb the spread of such bacteria.11 LMICs urgently need on-site rapid, accurate and cost-effective genomic technologies to identify country-specific AMR determinants. The genomic methodologies should be simple, user-friendly and accordant with local economic and social constraints.11 We described such an approach that was utilized in South Africa, a middle-income country, to rapidly identify nosocomial outbreaks over a 3–4 week period at a large tertiary hospital due to K. pneumoniae ST307 with OXA-181. PCR methodologies were used to identify OXA-181 and then track ST307 and MGEs associated with OXA-181 in different wards/units over time. This approach was accurate, rapid, simple and cost-effective and can easily be utilized by LMICs to identify potential nosocomial outbreaks, especially in settings with high prevalences of OXA-181 and K. pneumoniae ST307. The PCR also identified ST307 with VIMs and demonstrated that plasmids with OXA-181 were transferred to non-ST307 K. pneumoniae, E. coli and Enterobacter sp.
OXA-181 is the second most common global OXA-48-like derivative and was first described in Indian hospitals during 2006–07.12 Currently, Enterobacterales with blaOXA-181 are endemic on the Indian subcontinent and several global nosocomial outbreaks have been described.3 OXA-181 genes are present on several types of plasmids (i.e. ColE2-type, IncT and IncN), but IncX3 plasmids are mainly responsible for the global dissemination of blaOXA-181.3 Clonal outbreaks (e.g. K. pneumoniae ST147, K. pneumoniae ST307 and E. coli ST410) have also contributed to the international distribution of blaOXA-181.3 This study described a rapidly evolving nosocomial outbreak over 18 months of OXA-181-producing K. pneumoniae that contained IncX3 plasmids (i.e. p72_X3_OXA181) identical to other plasmids with blaOXA-181 previously reported from China,13 Angola14 and Portugal,15 highlighting the global distribution of OXA-181 IncX3 plasmids. The p72_X3_OXA181 plasmid was introduced into TTH by K. pneumoniae ST307 and was transferred to non-ST307 K. pneumoniae STs, E. coli and Enterobacter sp. The non-ST307 K. pneumoniae were responsible for separate nosocomial outbreaks, illustrating the threat of promiscuous plasmids such as p72_X3_OXA181. IP&C measures, in general, were designed and developed to specifically curb and control clonal spread during outbreak situations. The results of this study suggested that such measures are not necessarily that effective against the horizontal spread of plasmids.
K. pneumoniae ST307 is an emerging AMR high-risk clone that emerged during the mid-1990s and has been responsible for several worldwide nosocomial outbreaks.16 Certain characteristics of ST307 may lead to increased fitness, persistence and adaptation to the hospital environment.17K. pneumoniae ST307 belongs to six distinct clades, namely the USA clades I–IV, an international clade V and a uniquely South African clade VI.4K. pneumoniae ST307 clade VI with blaOXA-181 was responsible for countrywide nosocomial outbreaks within six South African provinces.4 Clade VI emerged around March 2013 and evolved during 2014 into two distinct lineages, namely ST307_181A and ST307_181B.4
This study identified two distinct clades among K. pneumoniae ST307 obtained from TTH. ST307 with blaOXA-181 belonged to clade VI with the same lineages as before.4 Lineage ST307_181B was introduced into TTH in September 2015 and was mostly responsible for the rapid increase and movement of ST307 clade VI across various wards/units. The HRU and surgical and medical ICUs were identified as the ‘hotspots’ for continuous transmission of this lineage. Lineage ST307_181A was introduced during November 2015, but the numbers remained low and it failed to gain the same success as lineage ST307_181B. The use of IP&C procedures implemented during November–December 2015 decreased the numbers of ST307_181B during 2016. However, TTH experienced another increase in K. pneumoniae with OXA-181 during October–December 2016 that were mainly due to K. pneumoniae ST17 and ST29 with p72_X3_OXA181 obtained from the HRU. K. pneumoniae ST17 and ST29 are rarely reported AMR STs.18,19
The ST307 clade with blaVIM-1 clustered within the international ST307 clade V (Figure 2) and was isolated from the HRU and surgical ICU during July 2015. Even though these units were identified as ‘hotspots’ for transmission and spread of the ST307_181B lineage, the numbers of the VIM clade remained low and it failed to become a dominant clone at TTH, suggesting that ST307 clades behaved differently during outbreaks. K. pneumoniae ST307 with blaVIM-1 was recently described in Italy.20
To conclude, K. pneumoniae ST307 clade VI with IncX3 plasmids containing blaOXA-181 was introduced into a South African tertiary public healthcare institution and spread rapidly to different wards/units. Cost-effective PCR methodologies (as compared with sequencing, which is time-consuming and expensive) were able to track this clone over an 18 month period and showed the transmission of plasmids with blaOXA-181 to other K. pneumoniae clones that were then responsible for separate outbreaks. WGS showed that ST307 clades and lineages acted differently in outbreak situations. Limitations of this study include that we could not rule out continuous introduction of different lineages from other hospitals and comprehensive screening of all patients was not performed.
Future studies are urgently required to investigate the reasons for the high transmission rates of ST307 clade VI. Such projects will serve as models to predict what can possibly happen in the future with the continuing emergence of successful clones among clinically relevant bacteria.1
Supplementary Material
Acknowledgements
We would like to thank the National Health Laboratory Service for the use of their facilities and E. Shashkina, P. Dhawan and R. Donnelly (New Jersey Medical School) as well as the TTH infection control department (i.e. Elsie Lewis and Helga Daniels) for their contributions.
Funding
This work was supported in part by research grants from Calgary Laboratory Services (10015169; J.D.D.P.), the National Institutes of Health (R01AI090155; B.N.K.), the NHLS Research Trust (M.M.K.) and RESCOM, University of Pretoria (M.M.K.). The funders of the study had no role in the study design, data collection, data analysis, interpretation or writing of the report.
Transparency declarations
J.D.D.P. has received research funds from Merck and AstraZeneca. All other authors: none to declare.
The corresponding author had full access to all the data and takes final responsibility for this publication.
Disclaimer
Opinions expressed and conclusions arrived at are those of the authors and are not necessarily to be attributed to the National Institutes of Health.
References
- 1. Mathers AJ, Peirano G, Pitout JD.. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 2015; 28: 565–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pitout JD, Nordmann P, Poirel L.. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother 2015; 59: 5873–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pitout JDD, Peirano G, Kock MM. et al. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev 2019; 33: e00102-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lowe M, Kock MM, Coetzee J. et al. Klebsiella pneumoniae ST307 with blaOXA-181, South Africa, 2014-2016. Emerg Infect Dis 2019; 25: 739–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CLSI. Performance Standards for Antimicrobial Susceptibility Testing—Twenty-Sixth Edition: M100.2016.
- 6. Peirano G, Matsumura Y, Nobrega D. et al. A cost-effective method for identifying Enterobacterales with OXA-181. J Clin Microbiol 2019; 57: e01281-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Antipov D, Hartwick N, Shen M. et al. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 2016; 32: 3380–7. [DOI] [PubMed] [Google Scholar]
- 8. Wick RR, Schultz MB, Zobel J. et al. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 2015; 31: 3350–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carattoli A, Zankari E, Garcia-Fernandez A. et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 2014; 58: 3895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CDC. Guidance for the Control of Carbapenem-Resitant Enterobacteriaceae (CRE) 2012 Toolkit.2012.
- 11. Seale AC, Gordon NC, Islam J. et al. AMR surveillance in low and middle-income settings - a roadmap for participation in the Global Antimicrobial Surveillance System (GLASS). Wellcome Open Res 2017; 2: 92.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Castanheira M, Deshpande LM, Mathai D. et al. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006-2007. Antimicrob Agents Chemother 2011; 55: 1274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu Y, Feng Y, Wu W. et al. First report of OXA-181-producing Escherichia coli in China and characterization of the isolate using whole-genome sequencing. Antimicrob Agents Chemother 2015; 59: 5022–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kieffer N, Nordmann P, Aires-de-Sousa M. et al. High prevalence of carbapenemase-producing Enterobacteriaceae among hospitalized children in Luanda, Angola. Antimicrob Agents Chemother 2016; 60: 6189–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aires-de-Sousa M, Ortiz de la Rosa JM, Goncalves ML. et al. Epidemiology of carbapenemase-producing Klebsiella pneumoniae in a hospital, Portugal. Emerging Infect Dis . Emerg Infect Dis 2019; 25: 1632–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wyres KL, Hawkey J, Hetland MAK. et al. Emergence and rapid global dissemination of CTX-M-15-associated Klebsiella pneumoniae strain ST307. J Antimicrob Chemother 2019; 74: 577–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Villa L, Feudi C, Fortini D. et al. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb Genom 2017; 3: e000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jure MA, Castillo M, Musa HE. et al. Novel patterns in the molecular epidemiology of KPC-producing Klebsiella pneumoniae in Tucuman, Argentina . J Glob Antimicrob Resist 2019; 19: 183–7. [DOI] [PubMed] [Google Scholar]
- 19. Markovska R, Stoeva T, Boyanova L. et al. Dissemination of successful international clone ST15 and clonal complex 17 among Bulgarian CTX-M-15 producing K. pneumoniae isolates. Diagn Microbiol Infect Dis 2017; 89: 310–3. [DOI] [PubMed] [Google Scholar]
- 20. Piazza A, Comandatore F, Romeri F. et al. Identification of blaVIM-1 gene in ST307 and ST661 Klebsiella pneumoniae clones in Italy: old acquaintances for new combinations. Microb Drug Resist 2019; 25: 787–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.