 A screen of 37 compounds identified four inhibitors that exhibited dual on-target activity against Mycobacterium tuberculosis aminoacyl-tRNA synthetases.
A screen of 37 compounds identified four inhibitors that exhibited dual on-target activity against Mycobacterium tuberculosis aminoacyl-tRNA synthetases.
Abstract
Effective treatment of tuberculosis is challenged by the rapid development of Mycobacterium tuberculosis (Mtb) multidrug resistance that presumably could be overcome with novel multi-target drugs. Aminoacyl-tRNA synthetases (AARSs) are an essential part of protein biosynthesis machinery and attractive targets for drug discovery. Here, we experimentally verify a hypothesis of simultaneous targeting of structurally related AARSs by a single inhibitor. We previously identified a new class of mycobacterial leucyl-tRNA synthetase inhibitors, N-benzylidene-N′-thiazol-2-yl-hydrazines. Molecular docking of a library of novel N-benzylidene-N′-thiazol-2-yl-hydrazine derivatives into active sites of M. tuberculosis LeuRS (MtbLeuRS) and MetRS (MtbMetRS) resulted in a panel of the best ranking compounds, which were then evaluated for enzymatic potency. Screening data revealed 11 compounds active against MtbLeuRS and 28 compounds active against MtbMetRS. The hit compounds display dual inhibitory potency as demonstrated by IC50 values for both enzymes. Compound 3 is active against Mtb H37Rv cells in in vitro bioassays.
Introduction
Tuberculosis is one of the deadliest human diseases and public problems throughout the world. The main problem of tuberculosis treatment is the rapid development of Mycobacterium tuberculosis (Mtb) multidrug resistance that stimulates the search for antibiotics with novel mechanisms of action as well as identification of novel targets for anti-tuberculosis treatment. The current outline of promising drugs and treatment opportunities has been recently reviewed.1–3 In the last decade, multi-target drugs have attracted much attention for the treatment of diseases associated with drug resistance.4 According to the strategy “one target–one drug”, which is the commonly favored approach in drug development, the application of highly specific drugs diminishes the risk of side effects due to off-target effects. However, biological systems demonstrate resilience to single-point perturbations.5 Multi-target drugs are considered to reduce resistance emergence, a considerable challenge in drug discovery.
Aminoacyl-tRNA synthetases (AARSs) play a pivotal role in living cells and hence represent promising drug targets for antibiotic development.6–10 They catalyse the coupling of amino acids to their cognate tRNAs, providing aminoacyl-tRNA substrates for protein biosynthesis. The aminoacylation reaction starts with amino acid activation to generate an aminoacyl-adenylate intermediate (AA-AMP), followed by transfer of the amino acid moiety to the 3′ end of its cognate tRNA, resulting in aminoacyl-tRNA formation. Most prokaryotic cells possess 20 different AARSs, one for each cognate amino acid–tRNA pair. All known AARSs are divided into two structural classes. Within each class, enzymes share a catalytic domain with a common fold that binds substrates for the aminoacylation reaction: amino acid, ATP and 3′-terminus of tRNA.11 Simultaneous targeting of AARS enzymes by a cocktail of two AARS inhibitors reportedly overcame the considerable resistance liabilities of Staphylococcus aureus associated with inhibitors acting against a single AARS.12 Multi-targeting of structurally related AARSs by a single inhibitor is assumed,6,10,12 but experimental data is missing.
Specificity of the AA-AMP formation in the first step consisting of the aminoacylation reaction strictly depends on proper binding of the cognate amino acid within the active site of an AARS enzyme. Competitive inhibitors, mimicking the AA-AMP intermediate, usually display strong inhibitory potency against the target AARS [reviewed in ref. 10]. In contrast, potential dual-target inhibitors would be compatible with the active sites of two AARSs. The binding affinity of such an inhibitor for a particular enzyme is expected to be compromised, compared to cognate aminoacyl-adenylate. Nevertheless, we hypothesized that the exclusion of the amino acid-binding pocket from the design of a dual-target inhibitor would yield compounds exhibiting too broad specificity. Such inhibitors would likely target several ATP-binding proteins, not only aminoacyl-tRNA synthetases, which could result in unpredictable and significant off-target effects. For this reason, binding in the amino acid pocket of the active sites would enhance the on-target selectivity of a universal inhibitor.
Here, we report a strategy that yielded small molecular inhibitors capable of targeting two M. tuberculosis AARSs, leucyl-tRNA synthetase (MtbLeuRS) and methionyl-tRNA synthetase (MtbMetRS), simultaneously. Leucyl-, isoleucyl-, valyl-, and methionyl-tRNA synthetases share a well conserved overall structure and belong to the Ia subclass.13 They are known to effectively activate near-cognate proteinogenic and nonproteinogenic amino acids, demonstrating insufficient discrimination capability toward structurally related amino acids.14–18 Docking and molecular dynamics results displayed preferential binding of noncognate homocysteinyl-adenylate in similar positions in all four Ia subclass AARSs, suggesting marked structural resemblance between their active sites.19 Hence, this enzyme group is a promising potential target for the design of dual-target inhibitors.
Derivatives of N-benzylidene-N′-thiazol-2-yl-hydrazine have been previously identified as a chemical class of mycobacterial leucyl-tRNA synthetase (MtbLeuRS) inhibitors.20 We conjectured that M. tuberculosis LeuRS inhibitors could potentially bind to and inhibit at least one of another Ia subclass AARS. In this respect, methionyl-tRNA synthetase (MetRS) is of particular interest because it catalyses not only the charging of elongator tRNAMet but also of initiator tRNAFMet. MetRS is a validated molecular target for drug design against protozoa, staphylococci, Giardia intestinalis, Clostridium difficile, and Pseudomonas aeruginosa.21–27 Recently, the crystal structure of the mycobacterial MetRS (MtbMetRS) has been reported making this enzyme of great interest in the structure-based design of antitubercular drugs.28,29
Results and discussion
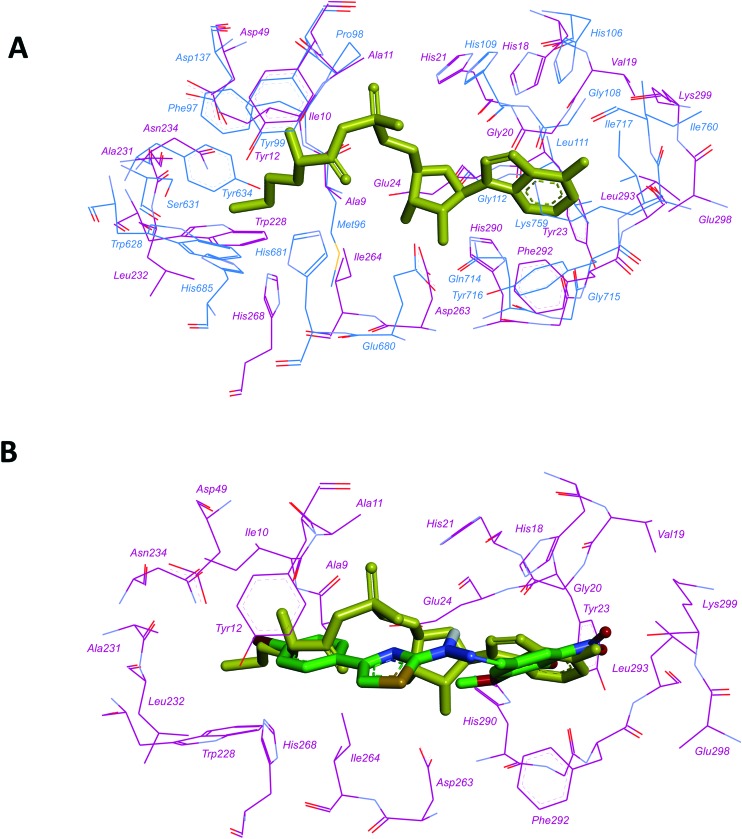
We compared the aminoacyl-adenylate binding pockets of MtbMetRS (PDB accession code: ; 6AX8) and MtbLeuRS (homology model, obtained by us earlier20) and revealed their striking structural resemblance (Fig. 1A). Molecular docking calculations indicated similar binding modes of 4-{[4-(4-bromophenyl)-thiazol-2-yl]hydrazonomethyl}-2-methoxy-6-nitrophenol (compound 1)20 to both active sites: 2-methoxy-6-nitrophenyl interacted with the adenine-binding region and the 4-substituted phenyl ring occupied the amino acid binding pocket (Fig. 1B and ESI† Fig. S1-1), suggesting that MtbMetRS could be a probable target for N-benzylidene-N′-thiazol-2-yl-hydrazine derivatives.
Fig. 1. Aminoacyl-adenylate binding pockets of M. tuberculosis LeuRS and MetRS. A. Superposition of the aminoacyl-adenylate binding pockets of MtbLeuRS (homology model)20and MtbMetRS (PDB: ; 6AX8). Carbon atoms of MtbLeuRS are coloured blue, carbon atoms of MtbMetRS are coloured dark magenta, methionyl-adenylate is coloured yellow); B. docked pose of compound 1 in the active site of MtbMetRS. Colour scheme is the same as that in A. Carbon atoms of compound 1 are coloured green.
We performed molecular docking of a preselected library of N-benzylidene-N′-thiazol-2-yl-hydrazine derivatives (334 compounds) into the active sites of MtbLeuRS and MtbMetRS using the DOCK program as described previously.20 According to the docking results (score values and bonding interactions) and visual analysis of the bound compounds (complementarity between ligand and receptor surfaces, the correctness of torsion angles, stacking interactions, etc.), a panel of the 35 best ranking novel compounds was evaluated for enzymatic potency against MtbLeuRS and MtbMetRS. Additionally, compounds 1 and 2, previously identified as the most potent inhibitors of MtbLeuRS, were subjected to evaluation of their dual-targeting capability.
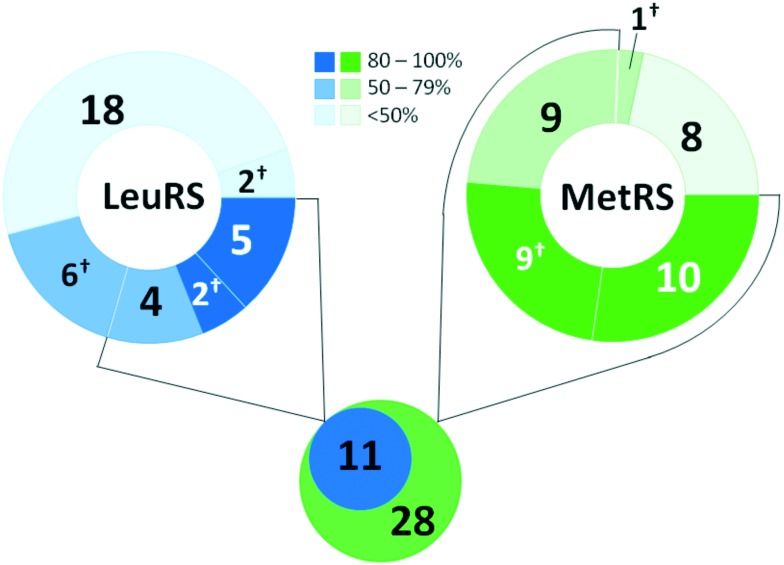
MtbLeuRS (gene ID: 887040) and MtbMetRS (gene ID: 886050) were recombinantly expressed in E. coli cells and purified. The prepared enzymes were >90% pure as judged by SDS-PAGE followed by staining with Coomassie Brilliant Blue (shown in Fig. S3-1†). The N-benzylidene-N′-thiazol-2-yl-hydrazine derivatives were synthesized according to the general procedure previously reported by our group.20 The representative structures of the compounds were confirmed with spectral data (1H NMR and 13C NMR), and their purity was checked by chromatographic techniques (HPLC-MS). All selected compounds were dissolved in DMSO and screened for inhibitory activity against MtbLeuRS and MtbMetRS in aminoacylation assays (Table 1). The aminoacyl-tRNA synthetase activity was determined by monitoring the level of inorganic pyrophosphate released in the aminoacylation reaction. A coupled enzyme, inorganic pyrophosphatase, was present in the reaction mixture to convert the pyrophosphate molecule into two inorganic phosphate molecules. The phosphate concentration was measured using the BIOMOL® GREEN reagent (Enzo Life Sciences). Control incubations revealed that 10 compounds affected the steps downstream of the aminoacylation reaction mainly by attenuating the BIOMOL® GREEN signal (up to 35%) (ESI† Table S1). These compounds were eliminated from the investigation based on this assay interference. Nevertheless, all these compounds, except compound 8, demonstrated substantial inhibition of MtbMetRS (over 80%), and two of them, compounds 2 and 25, effectively inhibited the MtbLeuRS activity, suggesting that these compounds could be considered to show over 50% inhibition in screening assays. Excluding these compounds, there were nine potential inhibitors for MtbLeuRS and 18 for MtbMetRS with an inhibition cut-off taken as 50%. Taken together, screening data revealed 11 compounds active against MtbLeuRS and 28 compounds active against MtbMetRS (Fig. 2), suggesting that MtbMetRS is as relevant a target for N-benzylidene-N′-thiazol-2-yl-hydrazines as MtbLeuRS.
Table 1. In vitro structure–activity relationships of N-benzylidene-N′-thiazol-2-yl-hydrazines.

| ||||||||
| Compound number | R1 | R2 | R3 | R4 | R5 | R6 | Inhibition (%) |
|
| MtbLeuRS | MtbMetRS | |||||||
| 1 a | C6H4-4-Br | H | H | NO2 | OH | OCH3 | 113 ± 14 (5) | 110 ± 6 (3) |
| 2 | C6H4-4-NO2 | H | H | NO2 | OH | OCH3 | 103 ± 9 (5) | 94 ± 23 (4) |
| 3 | C6H4-4-F | H | H | Br | OCH3 | H | 109 ± 3 (2) | 121 ± 8 (2) |
| 4 | C6H3-2,4-dCl | H | H | H | C(O)OH | H | 101 ± 4 (2) | 92 ± 4 (2) |
| 5 | C6H4-3-NO2 | H | Br | H | H | H | 80 ± 12 (2) | 93 ± 10 (2) |
| 6 | C6H4-3-NO2 | H | H | OC6H5 | H | H | 11 ± 15 (2) | 65 ± 15 (2) |
| 7 | C6H4-3-NO2 | H | H | H | C2H5 | H | 3 ± 23 (2) | 27 ± 9 (2) |
| 8 a | C6H5 | H | H | NO2 | OH | H | 53 ± 10 (2) | 72 ± 16 (2) |
| 9 a | C6H5 | CH3 | H | OH | H | H | 65 ± 18 (2) | 112 ± 13 (2) |
| 10 a | C6H5 | H | H | H | N(CH3)CH3 | H | 65 ± 18 (2) | 122.7 ± 0.3 (2) |
| 11 | C6H5 | H | OH | H | H | H | 7 ± 8 (2) | 51 ± 4 (2) |
| 12 | 3-Pyridyl | H | OCH3 | H | H | OCH3 | 74 ± 4 (2) | 115 ± 13 (2) |
| 13 | CH2C(O)OC2H5 | H | H | H | C(O)OH | H | 12 ± 4 (2) | 22 ± 6 (2) |
| 14 | CH3 | C(O)OC2H5 | CF3 | H | H | H | 34 ± 3 (2) | 45 ± 4 (2) |
| 15 | CH3 | C(O)OC2H5 | H | Br | OH | Br | 55 ± 4 (2) | 51 ± 17 (4) |
| 16 | CH3 | C(O)OC2H5 | H | Br | OH | OCH3 | 54 ± 9 (2) | 94 ± 5 (2) |
| 17 a | CH3 | C(O)OC2H5 | H | H | Br | H | 39 ± 8 (2) | 111 ± 6 (2) |
| 18 | CH3 | C(O)OC2H5 | Br | H | H | H | 67 ± 15 (2) | 99 ± 13 (2) |
| 19 | CH3 | C(O)OC2H5 | H | H | C(O)OH | H | –7 ± 11 (2) | 6 ± 8 (2) |
| 20 | CH3 | C(O)OC2H5 | H | OH | H | H | 1 ± 2 (2) | 71 ± 11 (2) |
| 21 | CH3 | C(O)OC2H5 | H | H | H | H | 19 ± 1 (2) | 73 ± 13 (2) |
| 22 a | CH3 | C(O)OCH3 | H | H | Br | H | 46 ± 1 (2) | 88 ± 18 (2) |
| 23 | CH3 | C(O)OCH3 | H | H | C(O)OH | H | 12 ± 1 (2) | –11 ± 3 (2) |
| 24 | CH3 | C(O)OCH3 | H | H | NO2 | H | 88 ± 1 (2) | 75 ± 12 (3) |
| 25 a | CH3 | C(O)CH3 | H | OCH3 | OCH2C6H4–2Cl | H | 87 ± 21 (5) | 113 ± 3 (2) |
| 26 | CH3 | C(O)CH3 | H | H | OC2H5 | H | 20 ± 11 (2) | 53 ± 0 (2) |
| 27 | CH3 | CH3 | H | H | OCH3 | H | 1 ± 10 (2) | 1 ± 8 (2) |
| 28 | CH3 | H | OCH3 | OCH3 | H | H | 17 ± 8 (3) | 25 ± 12 (2) |
| 29 | CH3 | H | H | H | C(O)OH | H | –4 ± 17 (2) | –22 ± 2 (2) |
| 30 | CH3 | H | H | H | NO2 | H | 20 ± 6 (2) | 92 ± 3 (2) |
| 31 a | H | H | H | OCH3 | OC(O)C6H3–2,4-dCl | H | 73 ± 16 (2) | 142 ± 2 (2) |
| 32 a | H | H | H | Br | OCH2C6H5 | OCH3 | 55 ± 6 (2) | 106 ± 1 (2) |
| 33 a | H | H | H | OCH3 | OCH2C6H4–2-Cl | H | 64 ± 1 (2) | 102 ± 2 (2) |
| 34 | H | H | H | Br | OCH3 | H | 29 ± 11 (2) | 90 ± 23 (4) |
| 35 | CH3 | H | OH | H | H | Br | 39 ± 3 (2) | 113 ± 11 (2) |
| 36 | H | H | H | OCH3 | OC6H13 | H | 47 ± 6 (2) | 62 ± 5 (2) |
| 37 | H | H | NO2 | H | H | H | 45 ± 35 (4) | 51 ± 25 (4) |
aCompounds that affected the steps downstream of the aminoacylation reaction in the assays (inorganic pyrophosphatase activity and/or development of BIOMOL® GREEN signal).
Fig. 2. Summary of screening. Ranges of inhibition of M. tuberculosis LeuRS and MetRS activity, the number of compounds within the scope and the total number of compounds with inhibition activity above 50% are shown. †The number of compounds affected the steps downstream of the aminoacylation reaction in the assays.
Analysis of the compounds' chemical structures indicated that the phenyl group was preferentially required as the R1 substituent to improve efficiency against MtbLeuRS, whereas the methyl group as the R1 substituent was also commonly found in compounds showing significant inhibition of MtbMetRS activity. Notably, all MtbLeuRS inhibitors are capable of reducing the MtbMetRS activity, demonstrating that they exhibit dual-targeted potency.
The compounds found to result in at least 80% inhibition of activity of both MtbLeuRS and MtbMetRS were further analysed to determine IC50 values (Table 2). All compounds demonstrated no remarkable enzyme-dependent difference in inhibition activity, while they varied from one to another in potency by 2–5-fold. Compounds 2 and 5 inhibited both enzymes with IC50 values below 10 μM. The potency of compound 3 was slightly lower (IC50 values of 12.5 and 10.7 μM for MtbLeuRS and MtbMetRS, respectively). Compound 4 demonstrated markedly increased IC50 values, suggesting the requirement of at least a single substitution at R3 or R4 with a bromine atom or methoxy group for improved inhibitory activity against both enzymes.
Table 2. IC50 values of dual-target compounds against M. tuberculosis LeuRS and MetRS.
| Compound number | R1 | IC50 (μM) |
|
| MtbLeuRS | MtbMetRS | ||
| 2 | C6H4-4-NO2 | 5.8 ± 4.1 (3) | 4.2 ± 1.3 (3) |
| 3 | C6H4-4-F | 12.5 ± 4.7 (3) | 10.7 ± 8.4 (4) |
| 3a | CH3 | 95 ± 10 (2) | ND |
| 4 | C6H3-2,4-dCl | 27.3 ± 1.3 (2) | 22.7 ± 7.2 (3) |
| 5 | C6H4-3-NO2 | 8.9 ± 1.7 (2) | 6.5 ± 0.4 (2) |
| 5a | C6H5 | 24.1 ± 5.5 (3) | 37.5 ± 5.3 (3) |
| 5b | CH3 | 38.4 ± 9.2 (3) | 123 ± 15 |
To get some insight into the role of the phenyl ring as an R1 substituent, we evaluated the inhibition potency of compounds 3a, 5a and 5b, which differ from the corresponding compounds 3 and 5 only in their R1 substituent. According to the docking results, these compounds were ranked lower from the best ranking analogues. Substitution of the 4-fluoro-phenyl group (compound 3) with the methyl group (compound 3a) led to a severe decrease in the targeting capacity toward both AARSs. Compound 29, which differs from compound 4 in terms of bearing the methyl group instead of 2,4-dichlorophenyl, did not show any inhibition of either MtbLeuRS or MtbMetRS in the primary screen (Table 1). Compounds 5a and 5b with an R1 phenyl and methyl group, respectively, demonstrated markedly increased IC50 values when compared to compound 5, especially against MtbMetRS. Both derivatives, though exhibit robust inhibitory activity, suggesting that the 3-nitro-phenyl moiety does not play a crucial role as an R1 substituent. This notion is supported by the marginal to absent activity of compounds 6 and 7 in the primary screen (Table 1). Combined with IC50 data for previously reported derivatives,20 our findings suggest that the phenyl group with a halogen atom or NO2 group in the para-position as an R1 substituent provides a substantial improvement in the ability of N-benzylidene-N′-thiazol-2-yl-hydrazine derivatives to inhibit both MtbLeuRS and MtbMetRS.
The potency of compounds 2–5 was tested against Mtb cultures with the H37Rv laboratory strain to determine the minimum inhibitory concentration (MIC). Cell cultures were incubated for a total of 14 days in parallel under four different growth conditions consisting of Middlebrook 7H9-based media supplemented with either glucose or cholesterol/dipalmitoyl phosphatidylcholine (DPPC) as a carbon source in the presence or absence of 5% bovine serum albumin fraction V (BSA). All growth media contained 0.05% Tyloxapol as a surfactant with BSA-free media containing 0.3% Bacto casitone. MIC1 and MIC2 values were generated after incubation for 7 and 14 days, respectively (Table 3). Compounds 2, 4 and 5 exhibited no significant inhibitory activity against Mtb at concentrations below 50 μM. The level of inhibition needed for both MtbLeuRS and MtbMetRS that results in growth cessation is not known. This lack of whole cell activity is perhaps not surprising given that the IC50 values for enzymatic inhibition are in the range of 4–22 μM. The cytosolic concentrations of these compounds would depend on the balance of uptake and efflux as well as compound metabolism by the pathogen. In contrast, compound 3 was found to exert potent inhibition of cells, exhibiting whole cell growth inhibition as observed from its MIC values which were noticeably lower than the IC50 values for on-target inhibition, suggesting that compound 3 could inhibit additional targets. This suggestion is supported by the carbon source selectivity of compound 3 as demonstrated by the more than 10-fold increased potency against the pathogen growing in media containing glucose as opposed to cholesterol/DPPC as the major carbon source. Thus, careful validation of intracellular targets of compound 3 is required.
Table 3. Inhibitory activity of the compounds against M. tuberculosis H37Rv cell growth.
| Compound number | MIC (μM) |
|||||||
| Medium A |
Medium B |
Medium C |
Medium D |
|||||
| MIC1 | MIC2 | MIC1 | MIC2 | MIC1 | MIC2 | MIC1 | MIC2 | |
| 2 | 50 | ≥50 | 50 | >50 | >50 | >50 | ≥50 | >50 |
| 3 | 1.56 | 2.3 | 19 | 37 | 0.78 | 2.3 | 9.4 | 25 |
| 4 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| 5 | >50 | >50 | ≥50 | >50 | 37 | >50 | 37 | 50 |
| Isoniazid | 0.1 | 0.3 | 0.6 | 0.6 | 0.07 | 0.1 | 0.39 | 0.39 |
We subsequently examined compound 3 for toxicity against human cells. The in vitro cytotoxic activity of the compound was evaluated by the MTT assay against the human embryonic kidney 293 cell line (HEK239) and the human hepatocellular carcinoma cell line (HepG2) following procedures described previously.30 Compound 3, tested in the concentration range of 6.25–100 μM, demonstrated CC50 (concentration to cause 50% cytotoxicity) values greater than 100 μM, exhibiting low toxicities to human cell lines (ESI† Table S2). This indicates the mycobacteria-selective inhibitory potency of the compound.
Experimental
The general procedure of N-benzylidene-N′-thiazol-2-yl-hydrazine derivative synthesis and 1H NMR, 13C NMR and HPLC-MS data for compounds 3–37, 3a, 5a, and 5b are provided in the ESI.†
Molecular docking
The receptor–ligand flexible docking was performed using the DOCK program.31–34 As a receptor, we used the homology model of M. tuberculosis LeuRS, which was obtained by us earlier,20 and crystal structure of M. tuberculosis MetRS (PDB ID: ; 6AX8). Ligand geometry was calculated using the YFF force field.35 Partial atomic charges of the ligands were added with the Kirchhoff method.36
Docking parameters have been set as described previously,37 except for several parameters. In our experiments, the active site atoms included the atoms of amino acid residues selected within 8 Å from the reference ligand. The active site spheres for the ligand docking were calculated with the DOCK sphgen software. The spheres which were outside of the active site were deleted manually. Grid maps were calculated using the program Grid, with a grid spacing of 0.3 Å. Proteins were represented by an all atom model. In our virtual screening experiments, the ‘multiple anchors’ parameter was chosen, the minimum of heavy atoms in the anchor was set to 6, and the maximum number of orientations was 1000.
The superposition of the active sites of M. tuberculosis LeuRS and MetRS was analyzed using Discovery Studio Visualizer.38 The amino acids of the MetRS active site were used as a template during molecular overlay. The parameters for superposition were set as the following: align by field: 50% steric, 50% electrostatic. The RMSD of superposition was 0.73.
Docking data for compounds 1–37 could be found in the ESI.†
Cloning of M. tuberculosis MetRS
The gene encoding MetRS was isolated from the genomic DNA of M. tuberculosis H37Rv (National Institute of Phthisiology and Pulmonology named after F.G. Yanovsky of the NAMS of Ukraine, Kyiv). For the His6-MetRS construct, primers MetRSMt-N (5′-CCA TGG AGC CCT ATT ACG TCA CCA CCG CG) and MetRSMt-C (5′-AAG CTT GCC TTC GGG TGG TTG CGG CGG CTG G) were designed according to the NCBI reference sequence NC_000962.3. Appropriate sites for NcoI and HindIII restriction enzymes were included into MetRSMt-N and MetRSMt-C, respectively. Cloning into a final expression vector was done by isolating the NcoI–HindIII fragment and ligating it into NcoI–HindIII digested E. coli expression plasmid pET28b. Resulting plasmid pET28b/MetRS was transformed into E. coli BL21 (DE3) Star cells (Invitrogen).
Purification of M. tuberculosis MetRS and LeuRS
For the production of C-terminally tagged His6 derivative of M. tuberculosis MetRS (MtbMetRS), the cells were grown in LB medium supplemented with 3% v/v ethanol, and the expression was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside for 5 hours at 16 °C. The cells from 1 L culture were harvested and sonicated in 8–10 mL of a lysis buffer (50 mM Tris-HCl, pH 8.0 at 25 °C, 300 mM NaCl, 10% v/v glycerol, 0.5 mM phenylmethylsulfonyl fluoride, 0.5% v/v Triton X-100) containing protease inhibitors (cOmplete, EDTA-free protease inhibitor cocktail tablet; Roche Applied Science). The cell debris was removed by centrifugation at 10 000g for 25 min. His6-MetRS was purified by a standard procedure employing affinity chromatography on Ni2+–nitroacetic acid (Ni–NTA) Sepharose Fast Flow resin (GE Healthcare). Briefly, the cell-free extract was supplemented with imidazole to a final concentration of 15 mM and combined with 2 ml Ni–NTA Sepharose Fast Flow resin equilibrated in the lysis buffer supplemented with 15 mM imidazole. After extensive washing with the same buffer, His6-MetRS was eluted with a lysis buffer containing 250 mM imidazole. The fractions containing His6-MetRS (judged by Coomassie Brilliant Blue staining after SDS-PAGE) were pooled and further purified by size-exclusion chromatography on a Hi-Load 16/60 Superdex 200 (150 mL, Pharmacia Biotech) column, equilibrated with 50 mM HEPES–NaOH at pH 7.0, 10 mM β-mercaptoethanol, 100 mM NaCl, 10% v/v glycerol, and 0.003% w/v NaN3, with a flow rate of 1 mL min–1. The purified protein was concentrated to 3.75 mg mL–1 and stored at –80 °C.
Mycobacterium tuberculosis LeuRS cloned into the pET28a vector was a generous gift from Stephen Cusack and Andres Palencia. The enzyme was expressed in E. coli and purified as described previously.20
The final purity of proteins was monitored by SDS-PAGE and UV spectroscopy. Protein concentrations were determined by the Bradford assay39 using Roti® Quant (Roth).
MtbLeuRS and MtbMetRS inhibition studies
All the tested compounds but one were dissolved in 100% DMSO (Merck) to a concentration of 10 mM. The stock solution of compound 25 was at a concentration of 5 mM.
Assays were conducted at 25 °C in clear, flat bottom, polystyrene, 96-well plates.
Each assay was performed in a 50 μL reaction volume consisting of 100 mM HEPES–NaOH, pH 7.5, 10 mM MgCl2, 15 mM KCl, 1 mM DTT, 50 μg mL–1 BSA (for MtbLeuRS) or 50 mM HEPES–NaOH, pH 7.5, 15 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 10 μg mL–1 BSA (MtbMetRS standard reaction buffer) supplemented with 200 μM ATP (Sigma), 1 mg mL–1 total E. coli MRE 600 tRNA (Boehringer Mannheim), 50 nM MtbLeuRS or MtbMetRS, cognate amino acid (1 mM leucine (Sigma) or 0.5 mM methionine (Pierce), respectively) and 0.25 U mL–1 yeast inorganic pyrophosphatase (iPPase; Sigma). Each compound at the desired concentrations (2.5 μL in 100% DMSO) was incubated with one of the enzymes in 25 μL of appropriate reaction buffer for 30 min before the aminoacylation reaction was initiated by the addition of a 25 μL mix of ATP, cognate amino acid, tRNA and iPPase in the same reaction buffer. After a 10 min incubation, the reaction was stopped by the addition of 100 μL BIOMOL® GREEN reagent (Enzo Life Science). The signal of BIOMOL® GREEN reagent was developed for 30 min at 25 °C and the absorbance was read at 630 nm using a Biotek ELx800™ (Biotek) plate reader.
Data points were normalized to a percentage: The blank and control wells contained 2.5 μL 100% DMSO instead of a compound. The blank wells also lacked an aminoacyl-tRNA synthetase. The IC50 values were calculated using nonlinear regression (sigmoidal logistic curve) from Origin 6.0. The processed data may be found in the ESI.†
The blank and control wells contained 2.5 μL 100% DMSO instead of a compound. The blank wells also lacked an aminoacyl-tRNA synthetase. The IC50 values were calculated using nonlinear regression (sigmoidal logistic curve) from Origin 6.0. The processed data may be found in the ESI.†
The impact of compounds on reactions downstream of the aminoacylation reaction was monitored in two sets of control assays.
Set 1. Assays of iPPase activity were performed in 50 μL MtbMetRS standard reaction buffer supplemented with 5 μM inorganic pyrophosphate and 50 μM inhibitor. Inorganic PPase was present at a concentration of 0.25 U mL–1. The reaction was initiated by the addition of enzyme, allowed to proceed for 10 min and stopped by the addition of 100 μL BIOMOL® GREEN reagent, followed by processing and analysis as described above. Control and blank incubations were conducted with DMSO only. The blank wells lacked enzymes.
Set 2. To measure the compounds' impact on the development of BIOMOL® GREEN signal, 100 μL reagent was added to 50 μL MtbMetRS standard reaction buffer supplemented with 10 μM phosphate standard (Enzo Life Sciences) and 50 μM inhibitor. After 30 min, the incubation absorbance was read at 630 nm and analysed. The blank and control wells contained DMSO only. In the blank wells, the phosphate standard was absent.
Determination of minimum inhibitory concentrations (MICs)
Minimum inhibitor concentrations (MICs) were determined in four Middlebrook 7H9-based media: 7H9/glucose/BSA/Tyloxapol (4.7 g L–1 7H9 broth base, 5 g L–1 BSA, 4 g L–1 glucose, 0.81 g L–1 NaCl, 0.5 ml L–1 Tyloxapol); 7H9/DPPC/cholesterol/BSA/Tyloxapol (4.7 g L–1 7H9 broth base, 5 g L–1 BSA, 5 mg L–1 DPPC, 0.062 mM cholesterol, 0.81 g L–1 NaCl, 0.5 ml L–1 Tyloxapol); 7H9/glucose/casitone/Tyloxapol (4.7 g L–1 7H9 broth base, 0.3 g L–1 Bacto casitone, 4 g L–1 glucose, 0.81 g L–1 NaCl, 0.5 ml L–1 Tyloxapol); 7H9/DPPC/casitone/Tyloxapol (4.7 g L–1 7H9 broth base, 0.3 g L–1 Bacto casitone, 14 mg L–1 DPPC, 0.81 g L–1 NaCl, 0.5 ml L–1 Tyloxapol). The compounds were serially diluted in required medium two-fold across wells in 96-well clear round-bottom plates, plated in duplicates. Isolated Mtb cells (ATCC 27294) were grown to an OD600 of 0.2–0.3 in the required medium and diluted 103-fold in the same medium. Then, 50 μL was added to each well of the compound-containing plates, approximating 104 cells per well in a total volume of 100 μL per well giving a final compound concentration range of 0.049–50 μM. Isoniazid and DMSO (drug-free) were used as positive and negative controls, respectively. All plates were incubated for a total of 14 days at 37 °C in zip-lock bags. At 7 and 14 days, cell growth was estimated using an inverted enlarging mirror plate reader with the MIC taken as the lowest compound concentration that inhibited all visible growth. At 14 days, 10 μL Alamar Blue was added to each well in plates with the Middlebrook 7H9 regular medium and those with the cholesterol medium. The plates were incubated at 37 °C for 24 hours and read using visual scoring to distinguish cholesterol precipitation from growths which occur if the media cooled down during the MIC set-up.
In vitro cytotoxicity assay
The human embryonic kidney 293 cell line (HEK239) was obtained from the Russian Cell Culture Collection (Institute of Cytology of the Russian Academy of Science, St. Petersburg, Russia). The human hepatocellular carcinoma cell line (HepG2) was obtained from the Bank of Cell Lines from human and animal tissue (R. E. Kavetsky Institute of Experimental Pathology, Oncology and Radiobiology, NAS of Ukraine, Kyiv, Ukraine). HEK293 and HepG2 cell viability was examined by MTT assay as described previously.30 Viable cells were dispensed into a 96-well tissue plate at ∼5000 cells per well (HEK293) or ∼104 cells per well (HepG2), incubated for 24 hours, and then treated with a compound at a final concentration range of 6.25–100 μM. After a 72 hour treatment, the cells were processed with MTT (3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyl-tetrazolium bromide; Sigma). The cell viability was expressed as a percentage relative to the untreated control cells.
Conclusions
In this work, we provide the first attempt to achieve simultaneous targeting of two AARSs by a single small molecular inhibitor. Considering the structural resemblance in aminoacyl-adenylate binding sites of the two enzymes, M. tuberculosis LeuRS and MetRS, and using the chemical scaffold of known MtbLeuRS inhibitors, N-benzylidene-N′-thiazol-2-yl-hydrazines, we identified four compounds that exhibited dual on-target activity as demonstrated by their IC50 values in enzymatic activity assays. The hit compound 3 showed anti-tubercular whole cell activity against Mtb H37Rv in in vitro bioassays as well as low cytotoxicity to HEK293 and HepG2 human cell lines. Our findings suggest that the compound is valuable for further biological evaluation, including the determination of its on- and potentially off-target effects in Mtb, and the frequency of resistance of Mtb H37Rv against the compound as well as the activity of the compound against drug-resistant clinical isolates of Mtb.
Conflicts of interest
The authors declare no conflicts of interest from a financial or commercial standpoint.
Supplementary Material
Acknowledgments
This work was supported by the Science and Technology Center in Ukraine (contract No. 6258) and by the Intramural Research Program of NIH, NIAID (AI000693-25). The authors are grateful to Dr. Stephen Cusack and Dr. Andres Palencia (EMBL Grenoble Outstation, France) for the gift of plasmid encoding M. tuberculosis LeuRS. We also thank Prof. Vasyl Mel'nyk (National Institute of Phthisiology and Pulmonology named after F.G. Yanovsky of the NAMS of Ukraine, Kyiv, Ukraine) for providing the genomic DNA of M. tuberculosis H37Rv. We are indebted to Anna Iatsyshyna, Institute of Molecular Biology and Genetics NAS Ukraine, for helping with cytotoxicity tests. We are grateful to Dr. Clifton Barry III (National Institute of Allergy and Infectious Disease, National Institute of Health, Maryland, USA) for his continued interest and support.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c9md00347a
References
- Libardo M. D. J., Boshoff H. I., Barry 3rd C. E. Curr. Opin. Pharmacol. 2018;42:81. doi: 10.1016/j.coph.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. F. K., Laughon B. E., McHugh T. D., Lipman M. Curr. Opin. Pulm. Med. 2019;25:271. doi: 10.1097/MCP.0000000000000570. [DOI] [PubMed] [Google Scholar]
- Torfs E., Piller T., Cos P., Cappoen D. Int. J. Mol. Sci. 2019;20:2868. doi: 10.3390/ijms20122868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Shurig-Briccio L. A., Feng X., Upadhyay A., Pujari V., Lechartier B., Fontes F. L., Yang H., Rao G., Zhu W., Gulati A., No J. H., Cintra G., Bogue S., Liu Y. L., Molohon K., Orlean P., Mitchell D. A., Freitas-Junior L., Ren F., Sun H., Jiang T., Li Y., Guo R. T., Cole S. T., Gennis R. B., Crick D. C., Oldfield E. J. Med. Chem. 2014;57:3126. doi: 10.1021/jm500131s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins A. L. Nat. Chem. Biol. 2008;4:682. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- Vondenhoff G. H. M., Van Aerschot A. Eur. J. Med. Chem. 2011;46:5227. doi: 10.1016/j.ejmech.2011.08.049. [DOI] [PubMed] [Google Scholar]
- Pang Y. L., Poruri K., Martinis S. A. Wiley Interdiscip. Rev.: RNA. 2014;5:461. doi: 10.1002/wrna.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewan V., Reader J., Forsyth K. M. Top. Curr. Chem. 2014;344:293. doi: 10.1007/128_2013_425. [DOI] [PubMed] [Google Scholar]
- Ho J. M., Bakkalbasi E., Söll D., Miller C. A. RNA Biol. 2018;15:667. doi: 10.1080/15476286.2018.1429879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francklyn C. S., Mullen P. J. Biol. Chem. 2019;294:5365. doi: 10.1074/jbc.REV118.002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Nature. 1990;347:203. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Randall C. P., Rasina D., Jirgensons A., O'Neill A. J. Antimicrob. Agents Chemother. 2016;60:6359. doi: 10.1128/AAC.00674-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donoghue P., Luthey-Schulten Z. Microbiol. Mol. Biol. Rev. 2003;67:550. doi: 10.1128/MMBR.67.4.550-573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English S., English U., von der Haar F., Cramer F. Nucleic Acids Res. 1986;14:7529. doi: 10.1093/nar/14.19.7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincecum T. L., Martinis S. A. SAAS Bull. Biochem. Biotechnol. 2000;13:25. [Google Scholar]
- Jakubowski H. Acta Biochim. Pol. 2011;58:149. [PubMed] [Google Scholar]
- Cvetesic N., Palencia A., Halasz I., Cusack S., Gruic-Sovulj I. EMBO J. 2014;33:1639. doi: 10.15252/embj.201488199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilus M., Semanjski M., Mocibob M., Zivkovic I., Cvetesic N., Tawfik D. S., Toth-Petroczy A., Macek B., Gruic-Sovulj I. J. Mol. Biol. 2019;431:1284. doi: 10.1016/j.jmb.2019.01.029. [DOI] [PubMed] [Google Scholar]
- Fortowsky G. B., Simard D. J., Aboelnga M. M., Gauld J. W. Biochemistry. 2015;54:5757. doi: 10.1021/acs.biochem.5b00588. [DOI] [PubMed] [Google Scholar]
- Gudzera O. I. Bioorg. Med. Chem. Lett. 2016;24:1023. doi: 10.1016/j.bmc.2016.01.028. [DOI] [PubMed] [Google Scholar]
- Huang W. Bioorg. Med. Chem. Lett. 2017;27:2702. doi: 10.1016/j.bmcl.2017.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain T., Yogavel M., Sharma A. Antimicrob. Agents Chemother. 2015;59:1856. doi: 10.1128/AAC.02220-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranade R. M., Zhang Z., Gillespie J. R., Shibata S., Verlinde C. L., Hol W. G., Fan E., Buckner F. S. Antimicrob. Agents Chemother. 2015;59:7128. doi: 10.1128/AAC.01573-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrie L. S., Brand S., Robinson D. A., Ko E. J., Stojanovski L., Simeons F. R. C., Wyllie S., Thomas J., Ellis L., Osuna-Cabello M., Epemolu O., Nühs A., Riley J., MacLean L., Manthri S., Read K. D., Gilbert I. H., Fairlamb A. H., De Rycker M. ACS Infect. Dis. 2017;3:718. doi: 10.1021/acsinfecdis.7b00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvest R. L., Berge J. M., Berry V., Boyd H. F., Brown M. J., Elder J. S., Forrest A. K., Fosberry A. P., Gentry D. R., Hibbs M. J., Jaworski D. D., O'Hanlon P. J., Pope A. J., Rittenhouse S., Sheppard R. J., Slater-Radosti C., Worby A. J. Med. Chem. 2002;45:1959. doi: 10.1021/jm025502x. [DOI] [PubMed] [Google Scholar]
- Eissa A. G., Blaxland J. A., Williams R. O., Metwally K. A., El-Adl S. M., Lashine El-S. M., Baillie L. W., Simons C. J. Enzyme Inhib. Med. Chem. 2016;31:1694. doi: 10.3109/14756366.2016.1140754. [DOI] [PubMed] [Google Scholar]
- Robles S., Hu Y., Resto T., Dean F., Bullard J. M. Curr. Drug Discovery Technol. 2017;14:156. doi: 10.2174/1570163814666170330100238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros-Álvarez X., Turley S., Ranade R. M., Gillespie J. R., Duster N. A., Verlinde C. L. M. J., Fan E., Buckner F. S., Hol W. G. J. Acta Crystallogr., Sect. F: Struct. Biol. Commun. 2018;74:245. doi: 10.1107/S2053230X18003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Qin B., Wojdyla J. A., Wang M., Gao X., Cui S. IUCrJ. 2018;5:478. doi: 10.1107/S2052252518008217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volynets G., Lukashov S., Borysenko I., Gryshchenko A., Starosyla S., Bdzhola V., Ruban T., Iatsyshyna A., Lukash L., Bilokin Ya., Yarmoluk S. Monatsh. Chem. 2019 doi: 10.1007/s00706-019-02493-5. [DOI] [Google Scholar]
- Stoichet B. K., Stroud R. M., Santi D. V., Kuntz I. D., Perry K. M. Science. 1993;259:1445. doi: 10.1126/science.8451640. [DOI] [PubMed] [Google Scholar]
- Bodian D. L., Yamasaki R. B., Buswell R. L., Stearns J. F., White J. M., Kuntz I. D. Biochemistry. 1993;32:2967. doi: 10.1021/bi00063a007. [DOI] [PubMed] [Google Scholar]
- Ring C. S., Sun E., McKerrow J. H., Lee G. K., Rosenthal P. J., Kuntz I. D., Cohen F. E. Proc. Natl. Acad. Sci. U. S. A. 1993;90:3583. doi: 10.1073/pnas.90.8.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing T. J., Makino S., Skillman A. G., Kuntz I. D. J. Comput.-Aided Mol. Des. 2001;15:411. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- Yakovenko O. Ya., Oliferenko A. A., Golub A. G., Bdzhola V. G., Yarmoluk S. M. Ukr. Bioorg. Acta. 2007;1:52. [Google Scholar]
- Yakovenko O. Ya., Oliferenko A. A., Bdzhola V. G., Palyulin V. A., Zefirov N. S. J. Comput. Chem. 2008;29:1332. doi: 10.1002/jcc.20892. [DOI] [PubMed] [Google Scholar]
- Bursulaya B. D., Totrov M., Abagyan R., Brooks III C. L. J. Comput.-Aided Mol. Des. 2003;17:755. doi: 10.1023/b:jcam.0000017496.76572.6f. [DOI] [PubMed] [Google Scholar]
- Discovery Studio Visualizer 4.0. https://www.3dsbiovia.com/products/collaborative-science/biovia-discovery-studio/visualization-download.php, (accessed May 2019).
- Bradford M. M. Anal. Biochem. 1976;72:248. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.