Abstract
Purpose
Synchronous colorectal liver metastasis (SCLM) had limited availability of tools to predict survival and tumor recurrence. LncRNA CRNDE and lncRNA SNHG7 have been proven to be closely related to cancer progression. However, the predictive value of lncRNA CRNDE and lncRNA SNHG7 in cancer prognosis is still unclear. The purpose of this study was to investigate whether lncRNA CRNDE and lncRNA SNHG7 could be used as promising biomarkers for prognosis prediction of SCLM patients who underwent hepatectomy.
Methods
The expression profile of lncRNA CRNDE and lncRNA SNHG7 in serum of SCLM patients was examined by qRT-PCR. The relationship between lncRNA expression and clinicopathological characteristics was analyzed. The Cox proportional-hazards regression model and Kaplan-Meier analysis were performed to analyze the association between lncRNA expression and overall survival (OS) and tumor recurrence of SCLM patients.
Results
Levels of lncRNA CRNDE and lncRNA SNHG7 in patients who underwent recurrence or death were significantly higher than that of patients with recurrence-free or survival (P<0.01). Both lncRNA CRNDE high level and lncRNA SNHG7 high level showed a significant correlation with differentiation of primary tumor, invasion depth of primary focus, lymph node metastases, number of liver metastases, and liver metastasis grade. High levels of lncRNA CRNDE or lncRNA SNHG7 predicted shorter recurrence time, shorter OS time, higher recurrence rate and lower OS rate. Furthermore, lncRNA CRNDE and lncRNA SNHG7 were independent risk factors for high recurrence and poor OS in SCLM underwent hepatectomy.
Conclusion
Taken together, lncRNA CRNDE and lncRNA SNHG7 could be promising biomarkers for prediction of OS and tumor recurrence in SCLM underwent hepatectomy.
Keywords: lncRNA CRNDE, lncRNA SNHG7, prognosis, recurrence, synchronous colorectal liver metastasis, SCLM
Introduction
Colorectal cancer is one of the most common digestive tract cancers which account for much of the global morbidity and mortality.1 Clinical research showed that the death of colorectal cancer usually results from uncontrolled metastatic disease.2,3 The liver is the single most frequent organ of distant metastasis in patients with colorectal cancer.4 According to the Chinese Guidelines for Diagnosis and Comprehensive Treatment of Liver Metastases from Colorectal Cancer, 15–25% of patients with colorectal cancer have liver metastasis at the time of diagnosis, while another 15–25% of patients will have liver metastasis after radical resection of primary colorectal cancer.5 For synchronous colorectal liver metastasis (SCLM), in many situations, a hepatectomy is recommended because it is currently the most effective therapy alone. Whereas for only 20% to 30% of SCLM patients is the initial hepatectomy a curative treatment, 50% to 75% of SCLM patients, clinical research reported, experience tumor recurrence after the initial hepatectomy.6–8 The identification of independent preoperative factors of prognosis for SCLM patients underwent hepatectomy is of great significance, because it may help to stratify subgroups of SCLM patients and establish individual therapeutic regimens.
Recently, it is increasingly recognized that long-noncoding RNAs (lncRNAs) constitute an important component in the regulation of cellular development, differentiation, and many other biological processes.9–11 LncRNAs are dysregulated in various cancers, indicating both tumor-suppressive and oncogenic roles, thus suggesting that aberrant expression of lncRNAs may contribute to cancer development.12,13 Drawing on a review of published studies,14–20 it enlightens us that lncRNA CRNDE and lncRNA SNHG7, two oncogenes, may be promising biomarkers of SCLM prognosis. For biological function, several studies have shown that lncRNA CRNDE levels were upregulated in numbers of solid tumors, including hepatic carcinoma and colorectal cancer.14, 15 The study of Jiang et al demonstrated that lncRNA CRNDE accelerates cell proliferation and migration in colorectal carcinoma via activating Ras/MAPK signaling pathways.16 Furthermore, lncRNA SNHG7 was found to be involved in cell proliferation, invasion and migration of various tumors, including colorectal cancer and lung cancer.17,18 As showed in Shan et al’s study, lncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulated GALNT1.19 These studies indicated that lncRNA CRNDE and lncRNA SNHG7 are involved in the development and metastasis of liver cancer and colorectal cancer. The biological function of lncRNA CRNDE and lncRNA SNHG7 provides convincing theoretical basis that these lncRNAs are promising biomarkers for prognosis prediction in SCLM. However, the prognostic role of lncRNA CRNDE and lncRNA SNHG7 in SCLM has not been reported. Recently, the prognostic role of lncRNA CRNDE and lncRNA SNHG7 is reported in gastrointestinal cancer. For example, Shan et al provided evidence supporting that high expression of lncRNA SNHG7 was associated with poor overall survival of colorectal cancer.19 In addition, Xia et al have demonstrated that overexpression of lncRNA CRNDE correlates with poor prognosis in intrahepatic cholangiocarcinoma.20 According to the biological function as well as the prognostic role in other tumors, we believe that lncRNA CRNDE and lncRNA SNHG7 are promising biomarkers for prognosis of SCLM.
In this study, we investigated the prognostic ability of serum lncRNA CRNDE and lncRNA SNHG7 in SCLM patients who had undergone colorectal and liver surgery. These observations suggest that lncRNA CRNDE and lncRNA SNHG7 might represent promising biomarkers of poor prognosis in SCLM and could be potential therapeutic targets for gene therapy.
Patients and Methods
Patients and Clinical Samples
Clinical and pathological data for synchronous colorectal liver metastasis (SCLM) patients who underwent hepatectomy at Yan’an Hospital of Kunming City & Yan’an Hospital Affiliated to Kunming Medical University between August 2013 and January 2015 were retrospectively reviewed. Colorectal cancer and SCLM were diagnosed via ultrasound, pathological examinations, X-rays, adequate radiological imaging, or cytological examination. Further imaging examination including magnetic resonance imaging or computed tomography scans were executed if clinically indicated. Patient demographics as well as therapy protocol, imaging data, laboratory data, and follow-up records were collected. Routine tests, such as serum tumor markers, serum biochemical indices and complete blood counts were conducted within the week before hepatectomy. The exclusion criteria were as follows: (1) unresectable primary tumor, (2) presence of extrahepatic metastasis, (3) overt double primary malignancy, (4) patients with familial adenomatous polyposis and hereditary nonpolyposis colorectal cancer patients, (5) loss of follow-up after surgery, (6) presence of prior history of liver-directed treatment, (7) patients with other liver disease, such as liver cirrhosis, (8) liver metastasis arose after surgical treatment. The study was received the ethical approval from the Clinical Research Ethics Committee of Yan’an Hospital of Kunming City & Yan’an Hospital Affiliated to Kunming Medical University. Written informed consents were provided from all SCLM patients. Before treatment, venous blood samples were collected from each SCLM patients and then initially isolated into serum by centrifugation (3000 rpm, 15 min) at 4°C. The supernatant was collected and followed by further centrifugation (12,000 rpm, 5 min) to obtain the serum. All serum samples were stored at −80°C for further processing. Follow-up was performed and ended on March 31, 2019. Death and dates of recurrence were verified by hospital records or by phone contact with the patients or their relatives. Recurrence time and overall survival (OS) time were defined according to the time after hepatectomy. All SCLM patients were followed up regularly every 1 month after therapy through telephone and outpatient follow-up.
Cancer Treatment
The treatment strategies for individual SCLM patients were discussed within a multi-disciplinary team (MDT) of experts including hepatobiliary specialist, colorectal surgeon, medical oncologists, radiation oncologist, interventional ultrasound physician, and diagnostic radiologist. Then, the treatment strategy would be explained to SCLM patients and their relatives and Written informed consents would be obtained. SCLM patients received fluorouracil-based preoperative neoadjuvant chemotherapy regimens as a first-line or second-line treatment. For patients with wild-type of Kras status, the patients further treated with cetuximab; For patients with Kras mutation, patients further received bevacizumab therapy. After that, all SCLM patients treated with hepatectomy.
RNA Extraction and qRT-PCR
Total RNA extractor kit (Sangon Biotech (Shanghai) Co., Ltd., Shanghai, China) was used to extract total RNA from patient serum. Then, total RNA from serum was eluted by RNase-free water (50 μL, Invitrogen). A260/A280 ratio was used to assess the quality of total RNA. According to manufacturer’s instructions of UEIris II RT-PCR System for First-Strand cDNA Synthesis System (US Everbright Inc., Suzhou, Jiangsu, China), total RNAs were reversely transcribed into complementary DNA (cDNA). qRT-PCR was completed using and a ProFlex qRT-PCR system (Thermo Fisher Scientifc, Inc., Waltham, MA, USA) and Power SYBR Green PCR Master Mix Kit (Thermo Fisher Scientifc, Inc., Waltham, MA, USA). 5 μL cDNA were prepared as templates for PCR reaction. The thermal cycling conditions were as follows: 95°C at 600 s for a hot start, then 45 loops at 93°C for 15 s, 54°C for 0.5 min, and 72°C for 0.5 min. qRT-PCR was done in triplicate. GAPDH was used as control. The −ΔCt method was used to calculate the relative level of lncRNA. The sequences of lncRNA-SNHG7 primers were: 5′-GTT GGG GTG TTG GCA TTC TTG TT-3′ (sense) and 5′-TGG TCA GCC TGG TCA CTC TGG-3′ (antisense). The sequences of lncRNA-CRNDE primers were 5ʹ-TGA AGG AAG GAA GTG GTG CA-3ʹ (sense) and 5ʹ-TCC AGT GGC ATC CTA CAA GA-3ʹ (antisense); The sequences of GAPDH primers were 5ʹ-GTC AGC CGC ATC TTC TTT TG-3ʹ (sense) and 5ʹ-GCG CCC AAT ACG ACC AAA TC-3ʹ (antisense).15,18
Statistical Analysis
All the statistical analyses were performed using SPSS 22.0 software (IBM Corp., Armonk, NY, USA). Fisher exact test and Mann–Whitney U-tests were used to examine whether the protein correlated with the clinical and pathological parameters. The Kaplan–Meier method was used to analyze the survival rates and recurrence rates, and the log-rank test was performed to compare the differences. A multivariate analysis of all the variables that were found to be significantly correlated in the univariate analysis was performed using a Cox proportional-hazards regression model. A two-tailed P <0.05 was considered to indicate a statistically significant relationship.
Result
Demographic Features and Clinicopathological Data
A total of 96 SCLM patients with 174 metastatic lesions were involved in this study. Demographic features and clinicopathological data are shown in Table 1. The present study cohort included 76 men and 20 women with a median age of 53.81±7.68 years and a median liver tumor size of 24.39±6.41 mm. The primary tumors were poorly differentiated in 43.75% of SCLM patients and 53.13% in invasive depth of primary focus at T3-T4. Type of hepatectomy was minor in 15.63% of SCLM patients and major in 84.37%. Lymph node metastases with N2-N3 appeared in 53.13% SCLM patients. Liver metastasis grade with H2-H3 occurred in 56.25% SCLM patients. Of the 96 patients, 69 (71.88%) received neoadjuvant chemotherapy including non-targeted therapy (14, 14.58%), combination therapy with bevacizumab (29, 30.21%) and combination therapy with cetuximab (26, 37.68%). For SCLM patients treated with cetuximab, the Kras status was confirmed as wild-type before cetuximab administration. Thirty-five patients with Kras mutation were confirmed, including 6 patients received non-targeted therapy and 29 patients received combination therapy with bevacizumab.
Table 1.
Baseline Characteristics in 96 Patients with SCLM
| Characteristics | No. | |
|---|---|---|
| Age (years) | <55 | 56 |
| ≥55 | 40 | |
| Gender | Male | 76 |
| Female | 20 | |
| Location of primary tumor | Colon | 54 |
| Rectum | 42 | |
| Invasion depth of primary focus | T1-T2 | 45 |
| T3-T4 | 51 | |
| Differentiation of primary tumor | Well-/moderately differentiation | 54 |
| Poorly differentiation | 42 | |
| Lymph node metastases | N1 | 45 |
| N2-3 | 51 | |
| Liver tumor size (mm) | >20 | 53 |
| ≤20 | 43 | |
| Number of liver metastases | Single | 46 |
| Multiple | 50 | |
| Liver metastasis grade | H1 | 42 |
| H2-H3 | 54 | |
| Resection status of liver metastases | R0 | 70 |
| R1-2 | 26 | |
| Type of hepatectomy | Minor | 15 |
| Major | 81 | |
| CEA (ng/mL) | 98.72±20.67 | |
| CA19-9 (U/mL) | 137.44±43.21 | |
| Neoadjuvant chemotherapy | Short duration | 39 |
| Long duration | 30 | |
| Postoperative chemotherapy | Yes | 82 |
| No | 14 | |
| Recurrence after hepatectomy | Intrahepatic rec. | 58 |
| Extrahepatic rec. | 13 | |
| Intra and extrahepatic | 8 | |
| Rec. adjuvant chemotherapy | Present | 54 |
| Absent | 16 | |
| Repeat hepatectomy | Present | 50 |
| Absent | 8 | |
The median follow-up period was 44 (range: 39–48) months. Of the 96 SCLM patients, the recurrence rate was 73.95% (71/96) including 60.42% patients with intrahepatic recurrence, 13.54% patients with extrahepatic recurrence and 8.33% patients with intra & extrahepatic recurrence. 56.25% of patients received recurrent adjuvant chemotherapy and 52.08% of patients received repeat hepatectomy. The 3-year OS rate of SCLM patients was 67.71% (65/96).
The Expression Profile of lncRNA CRNDE and lncRNA SNHG7 in SCLM Patients
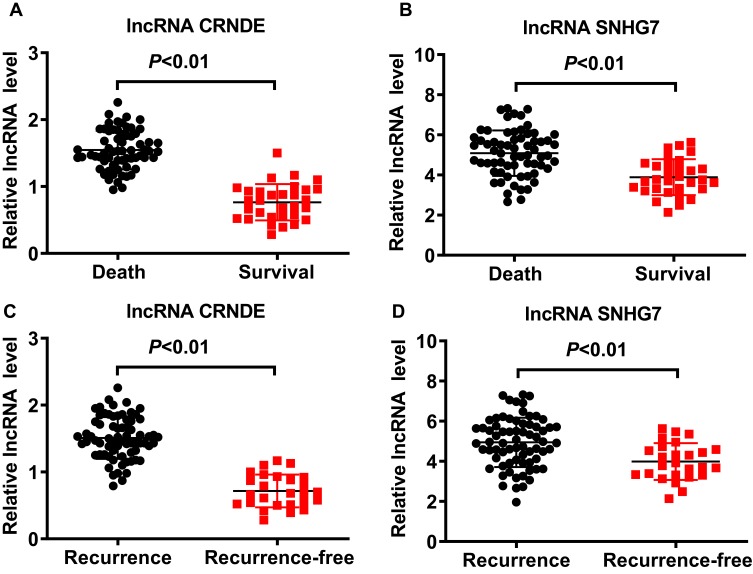
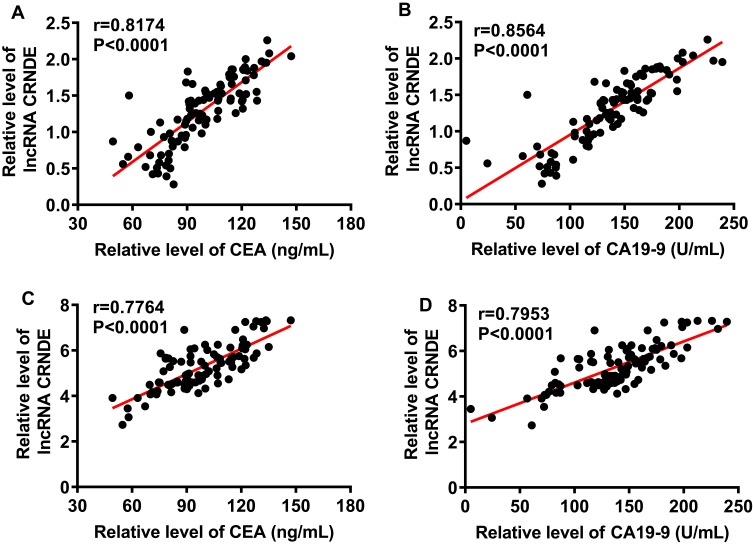
Expression profiles of lncRNA CRNDE and lncRNA SNHG7 in serum were investigated before treatment. As shown in Figure 1A and B, the levels of lncRNA CRNDE and lncRNA SNHG7 in patients with recurrence were significantly higher than that of patients with recurrence-free (P< 0.01). We also found that the levels of lncRNA CRNDE and lncRNA SNHG7 in patients underwent death were significantly higher than that of surviving patients (P< 0.01), shown in Figure 1C and D. The associations of the clinicopathological factors with lncRNA CRNDE expression or lncRNA SNHG7 expression are shown in Table 2. According to the median of lncRNA CRNDE, 96 SCLM patients were divided into two groups: low lncRNA CRNDE group (<1.395, n=48) and high lncRNA CRNDE group (>1.395, n=48). lncRNA CRNDE high expression showed a significant correlation with differentiation of primary tumor (P=0.003), invasion depth of primary focus (P=0.002), lymph node metastases (P=0.004), number of liver metastases (P<0.001) and liver metastasis grade (P<0.001). However, no association was observed between lncRNA CRNDE and age, gender, location of primary tumor, liver tumor size and type of hepatectomy (P>0.05). According to the median of lncRNA SNHG7, 96 SCLM patients were also divided into two groups: low lncRNA SNHG7 group (<4.605, n=48) and high lncRNA SNHG7 group (>4.605, n=48). lncRNA SNHG7 was significantly associated with differentiation of primary tumor (P=0.014), invasion depth of primary focus (P<0.001), lymph node metastases (P=0.013), number of liver metastases (P<0.001) and liver metastasis grade (P<0.014). Whereas no association was observed between lncRNA SNHG7 and age, gender, location of primary tumor, liver tumor size and type of hepatectomy (P>0.05). Subsequently, we evaluated the relationship between lncRNA levels and CEA or CA19-9 levels through pearson correlation analysis. As shown in Figure 2A, there was an obvious positive correlation between lncRNA CRNDE levels and CEA levels (r = 0.8174, P < 0.0001); Additionally, an obvious positive correlation was also observed between lncRNA CRNDE levels and CA19-9 levels (r =0.8564, P < 0.0001, Figure 2B). Furthermore, pearson correlation analysis revealed an obvious positive correlation between lncRNA SNHG7 levels and CEA levels (r =0.7764, P < 0.0001), as well as between lncRNA SNHG7 levels and CA19-9 levels (r =0.7953, P < 0.0001), shown in Figure 2C and D. Taken together, these data suggested that lncRNA CRNDE and lncRNA SNHG7 may be promising factors for poor prognosis in SCLM patients.
Figure 1.
The levels of lncRNA CRNDE and lncRNA SNHG7 in the serum of SCLM patients. (A) The levels of lncRNA CRNDE in the serum of SCLM patients. (B) The levels of lncRNA SNHG7 in the serum of SCLM patients. (C) The levels of lncRNA CRNDE in the serum of SCLM patients with or with recurrence. (D) The levels of lncRNA SNHG7 in the serum of SCLM patients with or with recurrence.
Table 2.
Clinical and Pathological Characteristics Among 96 SCLM Patients with Different lncRNA Levels
| Characteristics | lncRNA CRNDE | lncRNA SNHG7 | ||||
|---|---|---|---|---|---|---|
| Low (n=48) | High (n=48) | P value | Low (n=48) | High (n=48) | P value | |
| Age (years) | 0.679 | 0.408 | ||||
| <55 | 27 | 29 | 30 | 26 | ||
| ≥55 | 21 | 19 | 18 | 22 | ||
| Gender | 0.615 | 0.132 | ||||
| Male | 37 | 39 | 41 | 35 | ||
| Female | 11 | 9 | 7 | 13 | ||
| Location of primary tumor | 0.100 | 0.681 | ||||
| Colon | 31 | 23 | 28 | 26 | ||
| Rectum | 17 | 25 | 20 | 22 | ||
| Differentiation of primary tumor | 0.003 | 0.014 | ||||
| Well-/moderately differentiation | 36 | 18 | 33 | 21 | ||
| Poorly differentiation | 12 | 30 | 15 | 27 | ||
| Invasion depth of primary focus | 0.002 | 0.000 | ||||
| T1-T2 | 30 | 15 | 33 | 12 | ||
| T3-T4 | 18 | 33 | 15 | 36 | ||
| Lymph node metastases | 0.004 | 0.013 | ||||
| N1 | 27 | 13 | 26 | 14 | ||
| N2-3 | 21 | 35 | 22 | 34 | ||
| Liver tumor size (mm) | 0.151 | 0.538 | ||||
| >20 | 23 | 30 | 25 | 28 | ||
| ≤20 | 25 | 18 | 23 | 20 | ||
| Type of hepatectomy | 0.745 | 0.160 | ||||
| Minor | 8 | 7 | 5 | 10 | ||
| Major | 39 | 41 | 43 | 38 | ||
| Number of liver metastases | 0.000 | 0.000 | ||||
| Single | 36 | 10 | 34 | 12 | ||
| Multiple | 12 | 38 | 14 | 36 | ||
| Liver metastasis grade | 0.000 | 0.014 | ||||
| H1 | 30 | 12 | 27 | 15 | ||
| H2-H3 | 18 | 36 | 21 | 33 | ||
Figure 2.
The relationship between lncRNA levels and CEA or CA19-9 levels. (A) Pearson correlation analysis between lncRNA CRNDE levels and CEA levels. (B) Pearson correlation analysis between lncRNA CRNDE levels and CA19-9 levels. (C) Pearson correlation analysis between lncRNA SNHG7 levels and CEA levels. (D) Pearson correlation analysis between lncRNA SNHG7 levels and CA19-9 levels.
The Prognostic Values of lncRNA CRNDE and lncRNA SNHG7 in OS of SCLM Patients
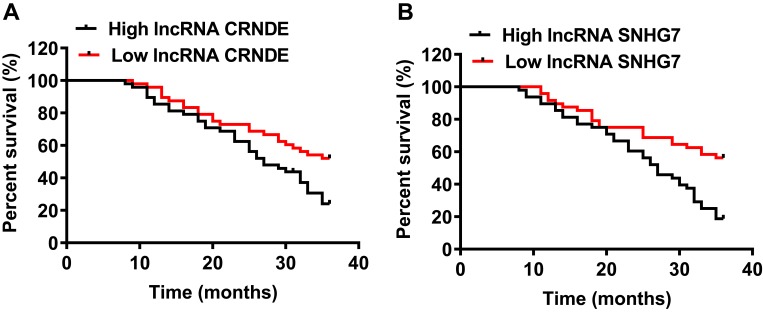
To compare OS rate and median OS time of SCLM patients in different lncRNA expression group, Kaplan-Meier survival analysis and log-rank test were performed. The results of survival analysis in Figure 3A showed that the median OS time was 27 months in the high lncRNA CRNDE group, which was significantly lower than that in the low lncRNA CRNDE group (>36 months, P<0.01). The OS rate was 22.92% (11/48) in the high lncRNA CRNDE group, which was also significantly lower than patients with low lncRNA CRNDE expression (52.08%, 25/48; P<0.01). Furthermore, the results in Figure 3B revealed that the SCLM patients with high lncRNA SNHG7 expression also had 27 months of median OS time, which was significantly lower than that in the low lncRNA SNHG7 group (>36 months, P<0.01). The OS rate of SCLM patients with low lncRNA SNHG7 expression was 56.25% (27/48) in relative to patients with high lncRNA SNHG7 expression (18.75%, 9/48; P<0.01). These results revealed that high levels of lncRNA CRNDE or lncRNA SNHG7 indicated high death risk in SCLM patients.
Figure 3.
Prognostic value of lncRNA CRNDE and lncRNA SNHG7 in each SCLM patient. (A) Kaplan-Meier survival curves for OS of SCLM patients according to lncRNA CRNDE levels. (B) Kaplan-Meier survival curves for OS of SCLM patients according to lncRNA SNHG7 levels.
As showed as in Table 3, univariate Cox regression analysis identified 7 prognosis factors for OS: invasion depth of primary focus (P<0.001), differentiation of primary tumor (P=0.036), lymph node metastases (P<0.001), number of liver metastases (P=0.018), liver metastasis grade (P=0.004), lncRNA CRNDE expression (P<0.001) and lncRNA SNHG7 expression (P<0.001). Next, multivariate Cox regression analysis was performed to determine if these variables were independently related to OS. Multivariate Cox regression analysis revealed that lncRNA CRNDE and lncRNA SNHG7 could be regarded as independent prognostic factors for OS in SCLM patients. Taken together, these data demonstrated that high expression of lncRNA CRNDE and lncRNA SNHG7 predicted a poor prognosis and could be regarded as independent prognostic factors for OS of SCLM.
Table 3.
Univariate and Multivariate Cox Regression Analysis for Overall Survival
| Characteristics | HR | P value | 95% CI | |
|---|---|---|---|---|
| Lower | Upper | |||
| Univariate | ||||
| Age (<55vs ≥55 years) | 0.99 | 0.968 | 0.603 | 1.627 |
| Gender (male vs female) | 0.918 | 0.777 | 0.508 | 1.659 |
| Location of primary tumor (colon vs rectum) | 1.024 | 0.926 | 0.627 | 1.67 |
| Invasion depth of primary focus (T1-T2 vs T3-T4) | 2.489 | <0.001 | 1.512 | 4.098 |
| Differentiation of primary tumor (well-/moderately vs poorly differentiation) | 1.696 | 0.036 | 1.034 | 2.782 |
| Lymph node metastases (N1 vs N2-3) | 2.439 | <0.001 | 1.488 | 3.996 |
| Liver tumor size (>20 vs ≤20 mm) | 0.97 | 0.904 | 0.595 | 1.583 |
| Number of liver metastases (single vs multiple) | 1.806 | 0.018 | 1.108 | 2.945 |
| Liver metastasis grade (H1 vs H2-H3) | 2.055 | 0.004 | 1.259 | 3.354 |
| Type of hepatectomy (minor vs major) | 0.989 | 0.974 | 0.504 | 1.94 |
| lncRNA CRNDE (low expression vs high expression) | 5.482 | <0.001 | 3.208 | 9.37 |
| lncRNA SNHG7 (low expression vs high expression) | 1.637 | <0.001 | 1.317 | 2.035 |
| Multivariate | ||||
| Invasion depth of primary focus (T1-T2 vs T3-T4) | 2.398 | 0.003 | 1.343 | 4.282 |
| Differentiation of primary tumor (well-/moderately vs poorly differentiation) | 1.813 | 0.032 | 1.052 | 3.124 |
| Lymph node metastases (N1 vs N2-3) | 1.843 | 0.036 | 1.039 | 3.267 |
| Number of liver metastases (single vs multiple) | 1.811 | 0.022 | 1.089 | 3.012 |
| Liver metastasis grade (H1 vs H2-H3) | 2.905 | 0.001 | 1.591 | 5.301 |
| lncRNA CRNDE (low expression vs high expression) | 7.527 | <0.001 | 3.81 | 14.873 |
| lncRNA SNHG7 (low expression vs high expression) | 1.344 | 0.016 | 1.058 | 1.709 |
Abbreviations: HR, hazard ratio; CI, confidence interval.
The Predictive Values of lncRNA CRNDE and lncRNA SNHG7 in Recurrence of SCLM Patients
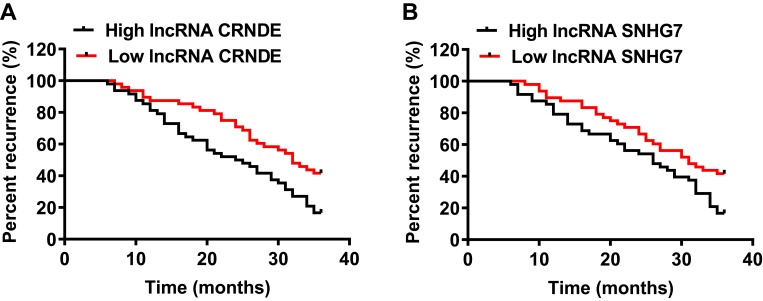
Kaplan-Meier survival analysis and log-rank test were also performed to investigate these factors in predicting tumor recurrence. The results in Figure 4A showed that SCLM patients with high lncRNA CRNDE level showed significantly shorter recurrence time compared with patients in the low lncRNA CRNDE group (24.5 months vs 32 months, P<0.01); meanwhile, we also found that the recurrence time of patients with high lncRNA SNHG7 was shorter than patients with low lncRNA SNHG7 expression (26 months vs 31 months, P<0.01), shown in Figure 4B. The recurrence rate of SCLM patients was 58.33% (28/48) in the low lncRNA CRNDE group and 83.33% (40/48) in high lncRNA CRNDE level group (P<0.01, Figure 4A). Meanwhile, the patients had a significantly higher recurrence rate in the high lncRNA SNHG7 group (72.92%, 29/48) than that in the low lncRNA SNHG7 group (60.42%, 35/48; P<0.01, Figure 4B).
Figure 4.
Prognostic value of lncRNA CRNDE and lncRNA SNHG7 in each SCLM patient. (A) Kaplan-Meier survival curves for tumor recurrence of SCLM patients according to lncRNA CRNDE levels. (B) Kaplan-Meier survival curves for tumor recurrence of SCLM patients according to lncRNA SNHG7 levels.
From univariate Cox regression analysis, we found that invasion depth of primary focus (P=0.002), differentiation of primary tumor (P=0.122), lymph node metastases (P=0.001), number of liver metastases (P=0.085), liver metastasis grade (P=0.025), lncRNA CRNDE expression (P<0.001) and lncRNA SNHG7 expression (P<0.001) were significantly associated with increased risk of recurrence (Table 4). The Cox multivariate analysis was also used and showed that lncRNA CRNDE and lncRNA SNHG7 were independent risk factors for recurrence. We conclude from these data that lncRNA CRNDE and lncRNA SNHG7 could be regarded as independent markers for predicting recurrence.
Table 4.
Univariate and Multivariate Cox Regression Analysis for Recurrence
| Characteristics | HR | P value | 95% CI | |
|---|---|---|---|---|
| Lower | Upper | |||
| Univariate | ||||
| Age (<55vs≥55 years) | 1.031 | 0.901 | 0.64 | 1.66 |
| Gender (male vs female) | 1.032 | 0.915 | 0.575 | 1.853 |
| Location of primary tumor (colon vs rectum) | 0.992 | 0.972 | 0.621 | 1.584 |
| Invasion depth of primary focus (T1-T2 vs T3-T4) | 2.144 | 0.002 | 1.336 | 3.44 |
| Differentiation of primary tumor (well-/moderately vs poorly differentiation) | 1.447 | 0.122 | 0.906 | 2.314 |
| Lymph node metastases (N1 vs N2-3) | 2.152 | 0.001 | 1.345 | 3.444 |
| Liver tumor size (>20 vs ≤20 mm) | 1.015 | 0.95 | 0.635 | 1.623 |
| Number of liver metastases (single vs multiple) | 1.507 | 0.085 | 0.945 | 2.402 |
| Liver metastasis grade (H1 vs H2-H3) | 1.714 | 0.025 | 1.07 | 2.744 |
| Type of hepatectomy (minor vs major) | 0.991 | 0.977 | 0.521 | 1.884 |
| lncRNA CRNDE (low expression vs high expression) | 4.803 | <0.001 | 2.863 | 8.06 |
| lncRNA SNHG7 (low expression vs high expression) | 1.45 | <0.001 | 1.179 | 1.784 |
| Multivariate | ||||
| Invasion depth of primary focus (T1-T2 vs T3-T4) | 1.929 | 0.014 | 1.143 | 3.254 |
| Differentiation of primary tumor (well-/moderately vs poorly differentiation) | 1.86 | 0.023 | 1.09 | 3.174 |
| Lymph node metastases (N1 vs N2-3) | 2.236 | 0.009 | 1.221 | 4.096 |
| Number of liver metastases (single vs multiple) | 1.94 | 0.011 | 1.167 | 3.226 |
| Liver metastasis grade (H1 vs H2-H3) | 2.731 | 0.002 | 1.466 | 5.09 |
| lncRNA CRNDE (low expression vs high expression) | 10.597 | <0.001 | 5.303 | 21.175 |
| lncRNA SNHG7 (low expression vs high expression) | 1.331 | 0.017 | 1.053 | 1.682 |
Abbreviations: HR, hazard ratio; CI, confidence interval.
Discussion
SCLM still has a clinical challenge, mainly because of the limited availability of tools that use promising prognostic markers to guide treatment.21,22 Thus, there is an urgent requirement to find reliable predictors for prediction of survival and tumor recurrence in SCLM patients underwent hepatectomy. At present, mounting evidence indicates the role of serum-circulating lncRNAs as novel serum biomarkers for diagnosis and prognosis prediction of cancer.23,24 For example, Gong et al25 evaluated the diagnostic efficacy and prognostic value of serum lncRNA-HIF1A-AS1 in patients with colorectal carcinoma and found that elevated serum lncRNA-HIF1A-AS1 could be clinically functioned as a potential biomarker for colorectal carcinoma diagnoses and prognosis. Importantly, Ren et al26 reported that lncRNAs in serum or plasma are are quite stable and are readily detected by RT-qPCR. Therefore, examining serum-circulating lncRNAs is feasible and helpful for SCLM prognosis evaluation. The most important contribution of our study is to evaluate the prognostic significance of lncRNA CRNDE and lncRNA SNHG7 for SCLM patients who underwent hepatectomy. In this retrospective cohort study, we found that levels of both lncRNA CRNDE and lncRNA SNHG7 were significantly increased in recurrence or death patients, and was significantly associated with various clinicopathological parameters of SCLM. From a clinical perspective, lncRNA CRNDE and lncRNA SNHG7 were independent prognostic factors for prediction of survival and tumor recurrence in SCLM underwent hepatectomy. According to our knowledge, this is the first study to elucidate the relationship of lncRNA CRNDE and lncRNA SNHG7 with the prognosis of SCLM.
LncRNA CRNDE expression is significantly increased in various malignant tumors, including colorectal cancer, hepatocellular carcinoma and glioma.14,15,27 Some previous studies have suggested that lncRNA CRNDE is associated with tumorigenesis and metastasis.28,29 In the present study, serum levels of lncRNA CRNDE and lncRNA SNHG7 in patients underwent recurrence was significantly higher than that of patients with recurrence-free. Furthermore, serum levels of lncRNA CRNDE and lncRNA SNHG7 in patients who underwent death was also significantly higher than that of surviving patients. Similar patterns were also observed in previous studies that the expression levels of the lncRNA CRNDE and lncRNA SNHG7 were upregulated in clinical tumor tissue samples.29,30 Several studies indicated that lncRNA CRNDE and lncRNA SNHG7 as oncogene contribute to develop the proliferation, invasion and migration and other biological behavior of breast cancer, lung cancer and other tumors.28,29,31,32 Interestingly, we found that lncRNA CRNDE high expression showed a significant correlation with differentiation of primary tumor, invasion depth of primary focus, lymph node metastases, number of liver metastases and liver metastasis grade. A similar pattern was also observed for lncRNA SNHG7. Furthermore, obvious positive correlation was found between lncRNA CRNDE levels and CEA levels and between lncRNA CRNDE levels and CA19-9 levels, as well as between lncRNA SNHG7 levels and CEA levels and between lncRNA SNHG7 levels and CA19-9 levels. These clinicopathological parameters were reported to be closely related to poor prognosis of SCLM. The relationship reflects the functions of lncRNA CRNDE and lncRNA SNHG7 that they accelerate cell proliferation, cycle progression, migration and invasion by regulating CRNDE, FAIM2, and other proteins.18,29 Therefore, it is conceivable to speculate that lncRNA CRNDE and lncRNA SNHG7 might be independent factors for prediction of survival and tumor recurrence in SCLM underwent therapy.
Clinical studies showed that the 5- and 10-y survival rates of SCLM patients after hepatectomy are approximately 40% and 25%, respectively.33,34 Furthermore, the recurrence rate of SCLM patient underwent radical resection can be as high as 57%-78%, while approximately 50% of the recurrence sites are liver, which is the main cause of treatment failure and patient death.35,36 The unsatisfactory outcome of prognosis reflects the absence of precise criteria that previously developed prognostic scoring systems that use clinicopathologic variables to select patients for hepatectomy.37 This underscores the need for a new prognostic model that uses biologic and clinicopathologic factors to guide treatment of the SCLM patients. Recent clinical trials have explored whether there is any prognostic role of lncRNA in colorectal cancer and hepatocellular carcinoma, lacking of hepatic metastasis from a primary colorectal cancer. In the current study, our results show that lncRNA CRNDE and lncRNA SNHG7 from serum possess independent prognostic significance for prediction of survival and tumor recurrence in SCLM underwent hepatectomy. This notion is supported by the fact that, in our present data, SCLM patients with high expression of lncRNA CRNDE and lncRNA SNHG7 possess significantly lower OS rate and higher recurrence rate, which are similar to previous studies. For example, as Jing et al described, cancer patients with higher expression of lncRNA CRNDE showed worse overall survival compared with patients with lower expression of lncRNA CRND.38 What’s more, Ren et al evaluated patient survival rates relative to SNHG7 expression levels and found that higher expression of SNHG7 in glioblastoma patients correlated with poorer prognosis.31 It is possible that lncRNA CRNDE and lncRNA SNHG7 become clinically useful markers to predict OS and recurrence of SCLM patients. At present, the function and molecular mechanism of lncRNA CRNDE and lncRNA SNHG7 have been investigated.16,39 Increasing evidence indicates that lncRNA CRNDE promotes colorectal cancer cell proliferation, metastasis and chemoresistance via epigenetically silencing DUSP5/CDKN1A expression,40 miR-181a-5p-mediated regulation of Wnt/β-catenin signaling,14 and activating Ras/MAPK signaling pathways.16 Furthermore, mounting evidence indicates that lncRNA SNHG7 facilitates the proliferation and liver metastasis of colorectal cancer through miR-216b-mediated upregulation of GALNT1 and activating PI3K/Akt/mTOR pathway.17,19 These previous studies provide guidance for lncRNA CRNDE and lncRNA SNHG7 in the prognosis evaluation of SCLM.
There were still some limitations in this study that need to be solved in the future. The retrospective nature potentially introduces the risk of selection bias. Therefore, the well-defined inclusion criteria and relatively large number of subjects may have mitigated this problem. Furthermore, we need to extend the follow-up time to further clarify the prognosis value of lncRNA CRNDE and lncRNA SNHG7 on SCLM patients. Finally, the prognosis value of lncRNA CRNDE and lncRNA SNHG7 was not validated in an independent SCLM patient group. Prospective validation in an independent population would need for their widespread clinical use.
Conclusion
In conclusion, we reported for the first time that high lncRNA CRNDE and lncRNA SNHG7 expression were significantly correlated with worse survival and higher recurrence rate in SCLM patients, predicting the survival and recurrence of patients with SCLM. Examination of lncRNA CRNDE and lncRNA SNHG7 may help to stratify subgroups of SCLM patients and establish individual therapeutic regimens.
Ethics Approval and Consent to Participate
The present study obtained the informed consent of all patients and was approved by the Clinical Research Ethics Committee of Yan’an Hospital of Kunming City & Yan’an Hospital Affiliated to Kunming Medical University. The study was carried out in accordance with the principles of the Declaration of Helsinki.
Patient Consent for Publication
The present study obtained the informed consent of all patients.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Troeger C, Forouzanfar M, Rao PC, et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the Global Burden of Disease Study. Lancet Infect Dis. 2017;17(11):1133–1161. doi: 10.1016/S1473-3099(17)30396-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canova C, Richiardi L, Merletti F, et al. Alcohol, tobacco and genetic susceptibility in relation to cancers of the upper aerodigestive tract in northern Italy. Tumori. 2010;96(1):1–10. doi: 10.1177/030089161009600101 [DOI] [PubMed] [Google Scholar]
- 3.Reding D, Pestalozzi BC, Breitenstein S, et al. Treatment strategies and outcome of surgery for synchronous colorectal liver metastases. Swiss Med Wkly. 2017;147:w14486. doi: 10.4414/smw.2017.14486 [DOI] [PubMed] [Google Scholar]
- 4.Shady W, Petre EN, Gonen M, et al. Percutaneous radiofrequency ablation of colorectal cancer liver metastases: factors affecting outcomes-A 10-year experience at a single center. Radiology. 2016;278(2):601–611. doi: 10.1148/radiol.2015142489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu J, Qin X, Wang J, et al. Chinese guidelines for the diagnosis and comprehensive treatment of hepatic metastasis of colorectal cancer. J Cancer Res Clin Oncol. 2011;137(9):1379–1396. doi: 10.1007/s00432-011-0999-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung U, Gönen M, Allen PJ, et al. Colorectal cancer liver metastases and concurrent extrahepatic disease treated with resection. Ann Surg. 2017;265(1):158–165. doi: 10.1097/SLA.0000000000001624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janowski E, Timofeeva O, Chasovskikh S, et al. Yttrium-90 radioembolization for colorectal cancer liver metastases in KRAS wild-type and mutant patients: clinical and ccfDNA studies. Oncol Rep. 2017;37(1):57–65. doi: 10.3892/or.2016.5284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cercek A, Boucher TM, Gluskin JS, et al. Response rates of hepatic arterial infusion pump therapy in patients with metastatic colorectal cancer liver metastases refractory to all standard chemotherapies. J Surg Oncol. 2016;114(6):655–663. doi: 10.1002/jso.24399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li K, Tang J, Hou Y. LncRNA GATA6-AS inhibits cancer cell migration and invasion in gallbladder cancer by downregulating miR-421. Onco Targets Ther. 2019;12:8047–8053. doi: 10.2147/OTT.S212231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu J, Ni G, Mao L, et al. LINC00565 promotes proliferation and inhibits apoptosis of gastric cancer by targeting miR-665/AKT3 axis. Onco Targets Ther. 2019;12:7865–7875. doi: 10.2147/OTT.S212231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang FW, Cao CH, Han K, et al. APC-activated long noncoding RNA inhibits colorectal carcinoma pathogenesis through reduction of exosome production. J Clin Invest. 2019;129(2):727–743. doi: 10.1172/JCI122478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong H, Ni Z, He J, et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36(25):3528–3540. doi: 10.1038/onc.2016.521 [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, Yuan W, Song J, et al. LncRna CPS1-IT1 suppresses cell proliferation, invasion and metastasis in colorectal cancer. Cell Physiol Biochem. 2017;44(2):567–580. doi: 10.1159/000485091 [DOI] [PubMed] [Google Scholar]
- 14.Han P, Li JW, Zhang BM, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer. 2017;16(1):9. doi: 10.1186/s12943-017-0583-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z, Yu C, Zhan L, et al. LncRNA CRNDE promotes hepatic carcinoma cell proliferation, migration and invasion by suppressing miR-384. Am J Cancer Res. 2016;6(10):2299–2309. [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang H, Wang Y, Ai M, et al. Long noncoding RNA CRNDE stabilized by hnRNPUL2 accelerates cell proliferation and migration in colorectal carcinoma via activating Ras/MAPK signaling pathways. Cell Death Dis. 2017;8(6):e2862. doi: 10.1038/cddis.2017.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Zeng C, Hu J, et al. Long non-coding RNA-SNHG7 acts as a target of miR-34a to increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in colorectal cancer progression. J Hematol Oncol. 2018;11(1):89. doi: 10.1186/s13045-018-0632-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.She K, Huang J, Zhou H, et al. lncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep. 2016;36(5):2673–2680. doi: 10.3892/or.2016.5105 [DOI] [PubMed] [Google Scholar]
- 19.Shan Y, Ma J, Pan Y, et al. LncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Dis. 2018;9(7):722. doi: 10.1038/s41419-018-0759-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia XL, Xue D, Xiang TH, et al. Overexpression of long non-coding RNA CRNDE facilitates epithelial-mesenchymal transition and correlates with poor prognosis in intrahepatic cholangiocarcinoma. Oncol Lett. 2018;15(4):4105–4112. doi: 10.3892/ol.2018.7815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Illemann M, Eefsen RH, Bird NC, et al. Tissue inhibitor of matrix metalloproteinase-1 expression in colorectal cancer liver metastases is associated with vascular structures. Mol Carcinog. 2016;55(2):193–208. doi: 10.1002/mc.22269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu ZH, Peng JH, Wang FL, et al. Bevacizumab with preoperative chemotherapy versus preoperative chemotherapy alone for colorectal cancer liver metastases: a retrospective cohort study. Medicine. 2016;95(35):e4767. doi: 10.1097/MD.0000000000004767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y, Wang YQ, Weng WW, et al. A serum-circulating long noncoding RNA signature can discriminate between patients with clear cell renal cell carcinoma and healthy controls. Oncogenesis. 2016;5:e192. doi: 10.1038/oncsis.2015.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang K, Shi H, Xi H, et al. Genome-wide lncRNA microarray profiling identifies novel circulating lncRNAs for detection of gastric cancer. Theranostics. 2017;7(1):213–227. doi: 10.7150/thno.16044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong W, Tian M, Qiu H, et al. Elevated serum level of lncRNA-HIF1A-AS1 as a novel diagnostic predictor for worse prognosis in colorectal carcinoma. Cancer Biomark. 2017;20(4):417–424. doi: 10.3233/CBM-170179 [DOI] [PubMed] [Google Scholar]
- 26.Ren S, Wang F, Shen J, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. 2013;49(13):2949–2959. doi: 10.1016/j.ejca.2013.04.026 [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Wang Y, Li J, et al. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015;367(2):122–128. doi: 10.1016/j.canlet.2015.03.027 [DOI] [PubMed] [Google Scholar]
- 28.Meng YB, He X, Huang YF, et al. Long noncoding RNA CRNDE promotes multiple myeloma cell growth by suppressing miR-451. Oncol Res. 2017;25(7):1207–1214. doi: 10.3727/096504017x14886679715637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Tang Y, Xing W, et al. LncRNA, CRNDE promotes osteosarcoma cell proliferation, invasion and migration by regulating Notch1 signaling and epithelial-mesenchymal transition. Exp Mol Pathol. 2018;104(1):19–25. doi: 10.1016/j.yexmp.2017.12.002 [DOI] [PubMed] [Google Scholar]
- 30.Qi H, Wen B, Wu Q, et al. Long noncoding RNA SNHG7 accelerates prostate cancer proliferation and cycle progression through cyclin D1 by sponging miR-503. Biomed Pharmacother. 2018;102:326–332. doi: 10.1016/j.biopha.2018.03.011 [DOI] [PubMed] [Google Scholar]
- 31.Ren J, Yang Y, Xue J, et al. Long noncoding RNA SNHG7 promotes the progression and growth of glioblastoma via inhibition of miR-5095. Biochem Biophys Res Commun. 2018;496(2):712–718. doi: 10.1016/j.bbrc.2018.01.109 [DOI] [PubMed] [Google Scholar]
- 32.Xu LJ, Yu XJ, Wei B, et al. LncRNA SNHG7 promotes the proliferation of esophageal cancer cells and inhibits its apoptosis. Eur Rev Med Pharmacol Sci. 2018;22(9):2653–2661. doi: 10.26355/eurrev_201805_14961 [DOI] [PubMed] [Google Scholar]
- 33.Cavnar MJ, Turcotte S, Katz SC, et al. Tumor-associated macrophage infiltration in colorectal cancer liver metastases is associated with better outcome. Ann Surg Oncol. 2017;24(7):1835–1842. doi: 10.1245/s10434-017-5812-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alzahrani N, Ung L, Valle SJ, et al. Synchronous liver resection with cytoreductive surgery for the treatment of liver and peritoneal metastases from colon cancer: results from an Australian centre. ANZ J Surg. 2017;87(11):E167–E172. doi: 10.1111/ans.13231 [DOI] [PubMed] [Google Scholar]
- 35.Reddy SK, Parker RJ, Leach JW, et al. Tumor histopathology predicts outcomes after resection of colorectal cancer liver metastases treated with and without pre‐operative chemotherapy. J Surg Oncol. 2016;113(4):456–462. doi: 10.1002/jso.24144 [DOI] [PubMed] [Google Scholar]
- 36.Bradatsch A, Kornprat P, Bacher H, et al. The value of lymph node dissection in the surgery of colorectal cancer liver metastases. Anticancer Res. 2016;36(6):2993. [PubMed] [Google Scholar]
- 37.Fleten KG, Bakke KM, Mælandsmo GM, et al. Use of non-invasive imaging to monitor response to aflibercept treatment in murine models of colorectal cancer liver metastases. Clin Exp Metastasis. 2017;34(1):51. doi: 10.1007/s10585-016-9829-3 [DOI] [PubMed] [Google Scholar]
- 38.Jing SY, Lu YY, Yang JK, et al. Expression of long non-coding RNA CRNDE in glioma and its correlation with tumor progression and patient survival. Eur Rev Med Pharmacol Sci. 2016;20(19):3992–3996. [PubMed] [Google Scholar]
- 39.She K, Yan H, Huang J, et al. miR-193b availability is antagonized by LncRNA-SNHG7 for FAIM2-induced tumour progression in non-small cell lung cancer. Cell Prolif. 2018;51(1):e12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding J, Li J, Wang H, et al. Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis. 2017;8(8):e2997. [DOI] [PMC free article] [PubMed] [Google Scholar]