Abstract
Purpose
Hepatocellular carcinoma (HCC) has a high incidence in China and exploring effective ways for early diagnosis is an important method to improve the prognosis of patients with HCC. Additional studies reported that. Some kinds of microRNA (miRNA) in plasma will change accordingly during HCC progress, and this change can be used to diagnose HCC, especially with miRNA-122, miRNA-21 and miRNA-96. We were aiming at investigating the values of the exosomal miRNAs in diagnosis and prognosis for HCC patients.
Patients and Methods
Blood samples from 50 patients with HCC and 50 patients with hepatic cirrhosis and 50 healthy volunteers were obtained. The diagnostic accuracy of the plasma and exosomal miRNAs and the comparisons among different groups were measured by the area under the curve (AUC) on receiver operating characteristic (ROC) curve analysis.
Results
Expression levels of miRNA-21 and miRNA-96 were significantly higher in patients with HCC and of miRNA-122 were significantly lower in HCC compared with cirrhotic patients in both exosomes and plasma. Among different groups, exosomal miRNA-122, miRNA-21 and miRNA-96 were significantly more accurate in diagnosing HCC than those miRNAs in plasma and the alpha-fetoprotein (AFP) level. The miRNA panel had high accuracy in discriminating HCC from the cirrhosis group (AUC 0.924; 95% CI; sensitivity 82%, specificity 92%) and healthy volunteers’ group. Exosomal miRNA-21 and miRNA-96 with low expression and miRNA-122 with high expression could be associated with a patient’s survival time. However, the miRNA panel could better predict the HCC patient’s survival time compared with each miRNA individually.
Conclusion
This study showed that the expression levels of miRNA-122, miRNA-21 and miRNA-96 in exosomes were more significantly changed than those miRNAs in plasma in patients with HCC compared with cirrhotic patients, and the exosomal miRNA panel containing miRNA-122, miRNA-21 and miRNA-96 could be defined as a diagnostic biomarker for patients with HCC. We also conclude that different expression of exosomal miRNAs, especially the miRNA panel, could predict the HCC patient’s prognosis.
Keywords: microRNA, diagnosis, HCC
Introduction
Hepatocellular carcinoma (HCC) is a frequently diagnosed cancer and increasingly occurring worldwide, especially in China.1,2 Several treatment options were administered and surgical resection remains the most commonly used curative therapy modality for HCC.3 Although the prognosis of patients with HCC has been improved recently, the overall survival is still unsatisfied and several factors are involved.4,5 The poor prognosis of patients with HCC is attributed to the lack of an effective means of early diagnosis. Only 30–40% of patients are candidates for potentially curative hepatectomy at the time of diagnosis.6 Discovery of an effective and reliable tool for early diagnosis of HCC would play a pivotal role in improving the prognosis of patients with HCC.
Exosomes are lipid bilayer cup-shaped nanovesicles 30–100 nm in diameter and released by almost all cell types.7,8 MicroRNAs (miRNAs) consist of 19–23 nucleotides which perform a significant function in biological stability and gene expression at a post-transcriptional level.9 Recently, miRNAs have been reported as a novel serum marker for diagnosing HCC, particularly serological detection, which provides a simple procedure for early diagnosis of HCC.10–12 Many studies have demonstrated a significant difference in miRNA, especially miRNA-96, miRNA-21 and miRNA-122, expression between hepatocarcinomatous and paracarcinoma tissues, and also affect tumor growth by regulating mRNA protein translation.13–15 Some studies believed that exosomes can be the best choice for non-invasive diagnosis. The reason is because the exosome is not degradable due to protection by cell membranes, so the large amounts of tumor cell information it carries, especially the miRNAs with tumor cell characteristics, are also preserved.16 Studies have shown that peripheral blood and exosome-mediated miRNA were crucial for human cancers.17 According to liver diseases, miRNAs derived from exosomes have already been used as a non-invasive diagnostic marker for hepatitis rating and classification.18 In the present study, we investigated the early diagnostic value of the panel of exosomal miRNA-96, miRNA-21 and miRNA-122 in patients with HCC from the peripheral blood, which can be easily available with a minimally invasive procedure. We also studied the influence of different exosomal miRNA expression on the prognosis of patients by follow-up patients’ survival time.
Patients and Methods
Patients
Blood samples from 50 patients with HCC and 50 patients with hepatic cirrhosis were obtained from the People’s Liberation Army (PLA) Rocket Force Characteristic Medical Center (Beijing, People's Republic of China) from September 2016 to November 2018, prior to definitive therapy. HCC diagnosis and the cell differentiation were based on criteria of the World Health Organization (WHO) and the 7th T(Tumor)N(Node)M(metastasis) staging classification was applied. Blood samples were also collected from 50 healthy volunteers with matching ages and genders to the patients. Clinical outcomes (PFS) were determined by chart review. Written consents were obtained from all subjects prior to recruitment. The study protocol was approved by the Institutional Review Board of Hospital Ethics Committee. All patients' and healthy volunteers' consents were written and informed, and this study was conducted in accordance with the Declaration of Helsinki. We did not include patients with a combination of other primary malignancies and conditions of autoimmune disease and infection into our clinical studies. Patients characteristics are shown in Table 1.
Table 1.
Patient and Tumor Characteristics (N=150)
| Variable | Control Group | Cirrhosis Group | HCC Group |
|---|---|---|---|
| Case, n | 50 | 50 | 50 |
| Age | 60.5±7.8 | 62.0±9.6 | 62.8±8.1 |
| Sex | |||
| Female | 19 | 19 | 17 |
| Male | 31 | 31 | 33 |
| HBsAg | |||
| Positive | 0 | 36 | 43 |
| Negative | 50 | 14 | 7 |
| HBeAg | |||
| Positive | 0 | 34 | 40 |
| Negative | 50 | 16 | 10 |
| Liver cirrhosis | |||
| Yes | 0 | 50 | 50 |
| No | 50 | 0 | 0 |
| TBL (µmol/l) | 12.5±8.3 | 15.1±7.3 | 16.1±8.2 |
| ALB (g/dl) | 39.4±6.6 | 38.9±6.5 | 37.9±4.6 |
| ALT (U/L) | 25.7±14.1 | 50.4±30.2 | 79.4±66.5 |
| AFP at diagnosis (ng/mL) | |||
| ≤400 | 50 | 42 | 32 |
| >400 | 0 | 8 | 18 |
| Tumor size (cm) | |||
| >5 cm | _ | _ | 23 |
| ≤5 cm | _ | _ | 27 |
| Microvascular invasion | |||
| Yes | _ | _ | 20 |
| No | _ | _ | 30 |
| TNM staging | |||
| I | _ | _ | 10 |
| II | _ | _ | 7 |
| III–IV | _ | _ | 33 |
| Metastases | |||
| Yes | _ | _ | 21 |
| No | _ | _ | 29 |
| Treatment | |||
| Target therapy | _ | _ | 30 |
| Chemotherapy | _ | _ | 8 |
| Partial hepatectomy | _ | _ | 6 |
| Anatomical liver resection | _ | _ | 6 |
Abbreviations: TBL, total bilirubin; ALB, albumin; ALT, alanine aminotransferase; AFP, alpha-fetoprotein; HBV, hepatitis B virus.
Extraction of Exosomes from Peripheral Blood
Exosomes were extracted from plasma depending on the manufacturer’s instructions for Exo-Quick exosome precipitation solution (System Biosciences, Mountain View, CA, USA). We extracted exosomes from peripheral whole blood, centrifuged at 3000 g for 15 min to remove cells or cell debris, placed the supernatant into a centrifuge tube, added 6 3 μL Exo-Quick per 250 μL, and stopped at 4 °C. The mixture was centrifuged at 1500 g for 30 min at 4 °C and precipitated at the bottom of the centrifuge tube. The supernatant was completely aspirated and centrifuged at 1500 g for 5 min at 4 °C. The supernatant was completely aspirated (the tube was not shaken), and the pellet was completely dissolved in PBS 200 μL and stored at −20 °C.
RNA Extraction from Exosomes
RNA was isolated from 200 µL exosome in PBS mixture using the RecoverALL Total Nucleic Acid Isolation Kit (Invitrogen/Life Technologies, USA) as per the recommended conditions. The mixture was placed in ice for 5 min. Then, an equal amount of acid-phenol:chloroform was added, the tube was shaken for 50 seconds, and the mixture was centrifuged at 10,000 g for 5 min. The supernatant was transferred to a fresh tube, the eluate was preheated, and the supernatant was added with 1.25 volumes of absolute ethanol. The mixture was placed in a filter cartridge, centrifuged for 15 seconds, and the lysate was discarded. We added 700 μL of miRNA washing solution 1 into the filter element, and after centrifugation at 10,000 g for 15 seconds, the lysate was discarded, 2/3 of the washing solution was added to 500 μL, centrifuged at 10,000 g for 15 seconds, and the lysate was discarded (repeated twice). The filter element was placed in a fresh tube, 35 μL of the 95 °C eluate was added, centrifuged at 10,000 g for 1 min, and the eluate was collected and stored at −70 °C. The purity and content of the extracted genomic RNA were determined by an ultraviolet–visible spectrophotometer.
Western Blot
Total proteins were extracted using RIPA lysis buffer (Beyotime Biotechnology, Shanghai, People's Republic of China) and then quantified using the BCA Protein Assay Kit (ThermoFisher, Shanghai, People's Republic of China). The protein samples were mixed with 5× loading buffer and separated by 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. Next, the proteins were transferred to a polyvinylidene fluoride (PVDF) membrane, and immersed in 5.0% non-fat milk for 45 min at 37 °C. The membrane was then incubated with CD63 (1:1000, ab134045; Abcam) at 4 °C overnight. Subsequently, the membrane was incubated with HRP-conjugated goat anti-rabbit IgG (1:10,000; Sigma, USA) for 1 h at room temperature.
RNA Extraction from Plasma
RNA was isolated from 200 µL plasma using the RecoverALL Total Nucleic Acid Isolation Kit (Invitrogen/Life Technologies, USA) as per the recommended conditions. The mixture was placed in ice for 5 min. Then, an equal amount of acid-phenol:chloroform was added, the tube was shaken for 50 seconds, and the mixture was centrifuged at 10,000 g for 5 min. The supernatant was transferred to a fresh tube, the eluate was preheated, and the supernatant was added with 1.25 volumes of absolute ethanol. The mixture was placed in a filter cartridge, centrifuged for 15 seconds, and the lysate was discarded. We added 700 μL of miRNA washing solution 1 into the filter element, and after centrifugation at 10,000 g for 15 seconds, the lysate was discarded, 2/3 of the washing solution was added to 500 μL, centrifuged at 10,000 g for 15 seconds, and the lysate was discarded (repeated twice). The filter element was placed in a fresh tube, 35 μL of the 95 °C eluate was added, centrifuged at 10,000 g for 1 min, and the eluate was collected and stored at −70 °C. The purity and content of the extracted genomic RNA were determined by an ultraviolet–visible spectrophotometer.
Quantification of miRNAs (miRNA-96, miRNA-21 and miRNA-122 Expressions) in Peripheral Bloods and Exosomes of Patients in Different Groups
For microRNA expression analysis, complementary DNA from 10 ng of total RNA was synthesized by the addition of a microRNA-specific 5× reverse transcription stem-loop primer and the TaqMan microRNA Reverse Transcription Kit, according to the manufacturer’s instructions. Real-time PCR was performed by diluting the complementary cDNA product in 2× TaqMan Universal Master Mix II (with UNG) and 20× TaqMan microRNA Expression. All four probes used in this study were synthesized by Life Technology, which were namely miRNA-122, item No.: 4427975, ID: 002130; miRNA-21, item No.: 4427975, ID: 000397; miRNA-96, item No.: 4427975, ID: 000186; and control group U6, item No.: 4427975, ID: 001973. Expression levels of miRNAs in plasma and exosomes were determined by the 2–∆∆Ct method, relative to the U6 expression.
Diagnosis and Treatment
HCC was diagnosed by tests in blood, chest X-ray, upper gastrointestinal endoscopy, abdominal ultrasound, contrast-enhanced computerized tomography (CT) and/or magnetic resonance imaging (MRI). A clinical diagnosis of HCC was based on the criteria of the American Association for the Study of Liver Diseases (AASLD).19 Most patients with stage 3 and stage 4 HCC receive sorafenib-targeted therapy and chemotherapy according to NCCN guidelines.
Statistical Analysis
Continuous variables were expressed as mean±SD (standard deviation) and compared using a two-tailed unpaired Student’s t test; categorical variables were compared using χ2 or Fisher analysis. Histopathological study of the resected specimens was carried out independently by three pathologists who came to a consensus by discussion if there was any discrepancy. The predictive performance of plasma and exosomal miRNAs was measured using the area under the ROC curve (AUC). AUCs were also used to compare plasma and exosomal miRNAs and AFP level using the Hanley and McNeil method.20 The miRNA panel was based on the logistic regression model for differentiation between the HCC group and the control group. Meanwhile, the calculation formula for the exosome miRNA panel was obtained: logit(P=HCC)=−2.275–2.258×miR-122–2.109×miR-21−1.796×miR-96. SPSS was used and P<0.05 was considered significant in all of the analyses.
Results
Characteristics of the Patients
The characteristics of patients with HCC, cirrhotic patients and the control group enrolled in this study are shown in Table 1.
Western Blot Quantity of Cryopreserved Exosome Extract
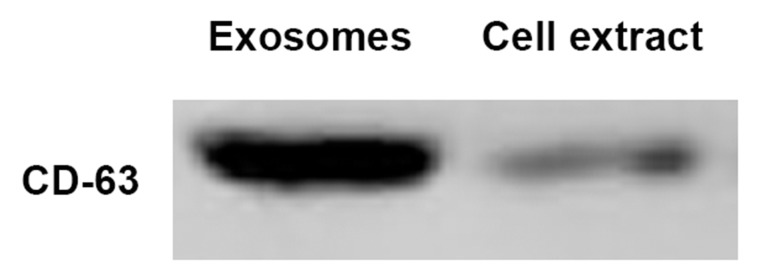
To ensure the quantity of the plasma exosome isolation, we characterized the western results of CD63 in the exosome solution and the supernatant remaining after extraction. Thisshowed that our extract contains a lot of exosomes (Figure 1).
Figure 1.
(A) CD63 in the exosome solution and the supernatant remaining after extraction. (B) Morphological characterization of exosomes isolated from serum samples by transmission electron microscopy. Bar, 100 nm.
Comparing Expression Levels of miRNA-122, miRNA-21 and miRNA-96 in Both Plasma and Exosomes Among the Three Groups
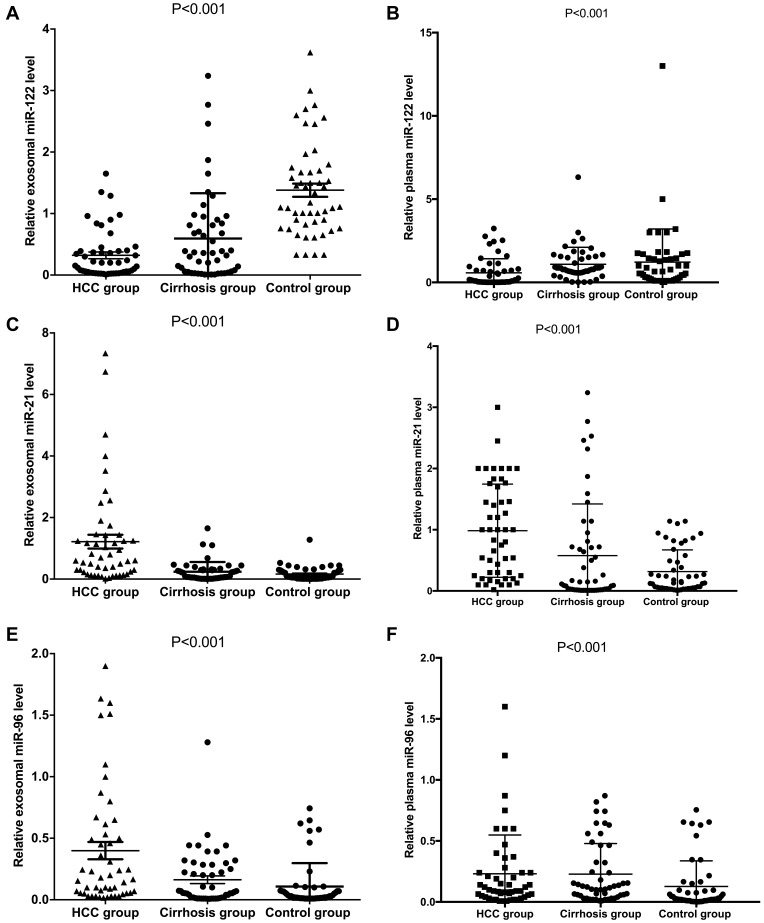
Expression of miRNA-122 was significantly lower in patients with HCC compared with cirrhotic patients and the control group in both exosomes and plasma (P<0.001, Figure 2A and B). Expression of miRNA-21 was significantly higher in patients with HCC compared with cirrhotic patients and the control group in both exosomes and plasma (P<0.001, Figure 2C and D). Expression of miRNA-96 was significantly higher in patients with HCC compared with cirrhotic patients and the control group in both exosomes and plasma (P<0.001, Figure 2E and F).
Figure 2.
Comparing expression levels of miRNA-122, miRNA-21 and miRNA-96 in both plasma and exosomes among the three groups. (A, C, E) miRNA levels in plasma and (B, D, F) exosomal levels.
Comparing Diagnostic Significance for Patients with HCC Among miRNAs in Exosome, Plasma and AFP Levels Between Different Groups
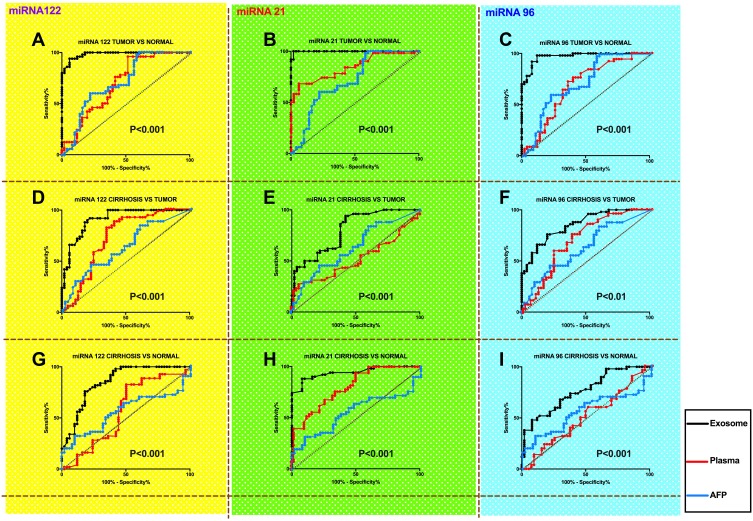
Between patients with HCC and normal groups, exosomal miRNA-122 was significantly more accurate in diagnosing HCC than plasma miRNA-122 and the AFP level; exosomal miRNA-21 was significantly more accurate in diagnosing HCC than plasma miRNA-21 and the AFP level; and exosomal miRNA-96 was significantly more accurate in diagnosing HCC than plasma miRNA-96 and the AFP level (P<0.001, Figure 3A–C). Between patients with HCC and the cirrhotic group, exosomal miRNA-122 was significantly more accurate in diagnosing HCC than plasma miRNA-122 and the AFP level; exosomal miRNA-21 was significantly more accurate in diagnosing HCC than plasma miRNA-21 and the AFP level; and exosomal miRNA-96 was significantly more accurate in diagnosing HCC than plasma miRNA-96 and the AFP level (P<0.001, Figure 3D and E, P<0.01, Figure 3F). Between the cirrhotic group and the control group, exosomal miRNA-122 was significantly more accurate in predicting cirrhosis than plasma miRNA-122 and the AFP level; exosomal miRNA-21 was significantly more accurate in predicting cirrhosis than plasma miRNA-21 and the AFP level; and exosomal miRNA-96 was significantly more accurate in predicting cirrhosis than plasma miRNA-96 and the AFP level (P<0.001, Figure 3G–I).
Figure 3.
Comparing diagnostic significance for patients with HCC among miRNAs in exosome, plasma and AFP levels between different groups (A, D, G: comparsion of miR-122; B, E, H: comparsion of miR-21; C, F, I: comparsion of miR-96). The black lines represent exosomal levels and the red lines represent plasma levels.
Multivariate Logistic Regression Analysis of the miRNA Panel Comprising Exosomal miRNA-122, miRNA-21 and miRNA-96 as a Diagnostic Marker for HCC
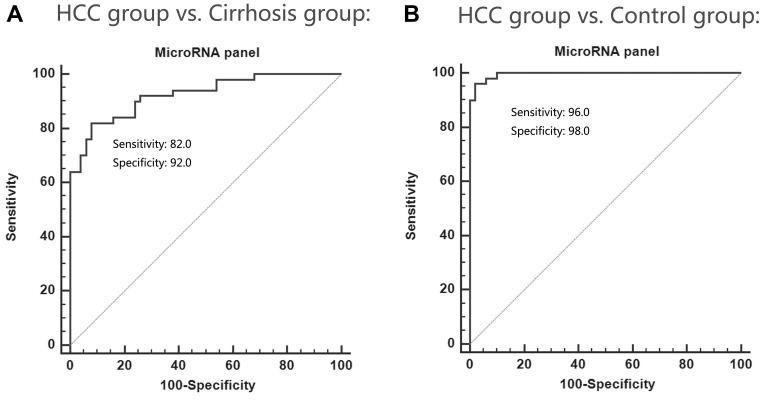
Considering that the exosomal expressions of miRNA-122, miRNA-21 and miRNA-96 were much better than the plasma expressions of the three miRNAs in terms of diagnostic specificity and sensitivity for HCC, we performed multivariate logistic regression analysis on the variables of miRNA-122, miRNA-21 and miRNA-96 and combining them as an miRNA panel to diagnose HCC (Table 2). The performance of the miRNA panel in differentiating the HCC group from the healthy and cirrhosis groups was also evaluated, respectively (Figure 4). The analysis demonstrated that the miRNA panel had high accuracy in discriminating the HCC group from the cirrhosis group (AUC 0.924; 95% CI; sensitivity 82%, specificity 92%) and the control group (AUC 0.996; 95% CI; sensitivity 96%, specificity 98%).
Table 2.
Logistic Analysis of Exosomal MicroRNA Profile and Diagnostic Performance of Patients with HCC Compared with Control Group
| Exosomal MicroRNA Group | AUC | Univariate | Multivariate | ||
|---|---|---|---|---|---|
| P value | HR | 95% CI | P value | ||
| miRNA-21 | 0.912 | 0.001 | 0.076 | 0.024–0.241 | 0.001 |
| miRNA-96 | 0.802 | 0.001 | 0.108 | 0.016–0.832 | 0.038 |
| miRNA-122 | 0.853 | 0.001 | 0.115 | 0.027–0.903 | 0.043 |
Notes: Control group includes healthy participants, patients with cirrhosis. logit(P=HCC)= −2.275–0.258×miR-122–2.109×miR-21−1.796×miR-96.
Abbreviations: CI, confidence interval; AUC, area under the receiver operating characteristic curve; HR, hazard risk; HCC, hepatocellular carcinoma.
Figure 4.
The performance of the miRNA panel in differentiating the HCC group from the healthy group (A) and the cirrhosis group (B).
Difference Expression Levels of Exosomal miRNA-122, miRNA-21, miRNA-96 Could Influence the Patients’ Survival Time, and the miRNA Panel Could Better Predict Patient Prognosis
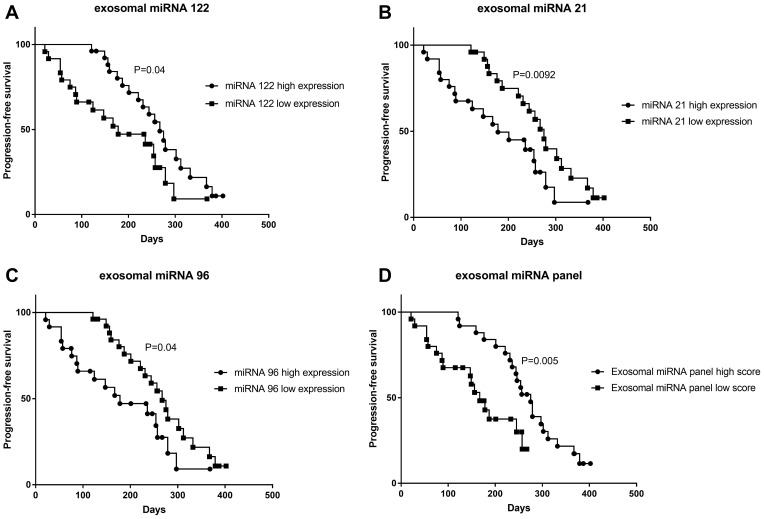
According to the above result, we had cleared that the exosomal miRNAs have a good diagnostic value. We further followed-up patients’ progression-free survival time to identify whether the miRNA panel could affect the prognosis of patients. First, we distinguish between high and low expression of miRNA-122, miRNA-21 and miRNA-96 respectively by the median value. Patients with overexpression of miRNA-21 (P=0.0092) and miRNA-96 (P=0.04) had a much poorer prognosis, while patients with overexpression of miRNA-122 (P=0.04) had a better prognosis (Figure 5).
Figure 5.
Differences in expression levels of exosomal miRNA-122 (A), miRNA-21 (B), miRNA-96 (C) and miRNA panel (D) could influence the patients’ progression-free survival time.
Discussion
Many studies showed that exosomes could be presented in the urine and pleuroperitoneal fluid, and the exosome has pleiotropic biological functions, including antigen-presenting, intracellular communication and transmission of signals and transfer of RNAs and miRNAs.21,22,23 Accumulating evidence has indicated that circulating miRNAs were involved in several pathophysiological processes and related to cancer.24,25 Numerous reports had illuminated the usefulness of circulating miRNAs as novel noninvasive biomarkers for cancers, such as colorectal cancer,26 breast cancer27,28 and renal cell carcinoma.29 Large amounts of exosomes can be secreted by tumor cells due to the influence of hypoxia, internal environmental changes and other factors. These exosomes are not easily degradable in either intercellular space or peripheral blood due to the protection by plasma membranes. With respect to patients with HCC, exosomal miR-21 has been reported as a biomarker for HCC diagnosis and with a key role in tumor progression.13 Therefore, this study aims to compare the expression levels of miRNAs derived from exosomes and plasmas among different groups and investigate the sensitivity and specificity of diagnostic and prognostic value for patients with HCC.
Increasing studies have shown that expression of miR-122 is dysregulated in many tumors and that it has specific functions in various tumors.14,30,31 To date, many oncogenic genes have been confirmed as targets of miR-122 that are involved in multiple biological pathways, including proliferation, invasion, differentiation, angiogenesis and energy metabolism.32 miRNA-122 plays a dual role in tumorigenesis, functioning as both a tumor suppressor and an oncogene. One article reported that the target genes of miRNA-122 may contribute to the composition of the nucleus and cytoplasm, and regulate a variety of biological processes, including cardiac muscle cell differentiation and glucose metabolic processes via protein biosynthesis, estrogen and glucagon-associated signaling pathways. These results revealed that miRNA-122 may be related to better prognosis of HCC patients.33
miRNA-21 is part of a microRNA family. This group of microRNAs has been extensively studied and has been shown to be downregulated in several cancers, such as gastric cancer, peripheral nerve sheath tumors, esophageal squamous cell carcinoma, melanoma, and breast cancer, among others.13,34 In these diseases, the increased expression of miR-21 leads to the up-regulation of its downstream targets that are involved in epigenetic modification, metastasis and cell proliferation.34
miRNA-96 could repress the cancer cell growth and migration respectively in a previous study. Furthermore, miRNA-96 expression was reduced in endometrioid adenocarcinoma, papillary thyroid carcinoma, ovarian cancer and hepatocellular carcinoma.15
In the present study, we first compared expression levels of miRNA-122, miRNA-21 and miRNA-96 in both plasma and exosomes among HCC, cirrhotic and control groups. We found that expression levels of miRNA-21 and miRNA-96 were significantly higher in patients with HCC and of miRNA-122 were significantly lower in HCC compared with cirrhotic patients in both exosomes and plasma. Secondly, we found that miRNAs in exosomes had better diagnostic significance for patients compared with plasma and AFP levels between different groups. We also combined exosomal miRNA-21, miRNA-122 and miRNA-96 as an miRNA panel to diagnose HCC. The performance of the exosomal miRNA panel had much better predictive value in diagnosing HCC. Finally, we compared the patients’ progression-free survival times in different expressions of miRNA-21, miRNA-122 and miRNA-96 respectively, and we found that varied quantitative miRNAs could influence the patient’s prognosis. However, this result could provide some help in assessing the patient’s condition.
At present, an increasing number of studies have been carried out to examine the role of long noncoding RNA (lncRNA), which regulates miRNA expression to regulate tumor invasion and proliferation.35,36 Our study discovers that miRNA partially affects patient prognosis, but most importantly, it is related to lncRNA regulation. To sum up, lncRNA is produced prior to miRNA production and affects tumor growth, as a result, the effect of miRNA on patient prognosis is closely correlated with lncRNA regulation. In the future, we will further investigate the role of lncRNA in regulating miRNA in the context of liver cancer.
There are limitations of this study: 1) the sample size is small in this study, and a further, larger sample study is needed to confirm the present experimental results; and 2) whether the three miRNAs have the optimal specificity and sensitivity for liver cancer diagnosis also needs future confirmation.
In conclusion, we found that expression of exosomal miRNA-122 was significantly lower in patients with HCC and exosomal miRNA-21 and miRNA-96 were significantly higher in patients with HCC compared with the cirrhotic and control groups, and we confirmed the exosomal miRNA panel containing miRNA-122, miRNA-21 and miRNA-96 could be defined as a diagnostic biomarker for patients with HCC. We also conclude that different expression of exosomal miRNAs in HCC patients could influence prognosis.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.v61:2 [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516 [DOI] [PubMed] [Google Scholar]
- 3.Arii S, Yamaoka Y, Futagawa S, et al. Results of surgical and nonsurgical treatment for small-sized hepatocellular carcinomas: a retrospective and nationwide survey in Japan. The Liver Cancer Study Group of Japan. Hepatology. 2000;32:1224–1229. [DOI] [PubMed] [Google Scholar]
- 4.Tateishi R, Yoshida H, Matsuyama Y, Mine N, Kondo Y, Omata M. Diagnostic accuracy of tumor markers for hepatocellular carcinoma: a systematic review. Hepatol Int. 2008;2:17–30. doi: 10.1007/s12072-007-9038-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nathan H, Schulick RD, Choti MA, Pawlik TM. Predictors of survival after resection of early hepatocellular carcinoma. Ann Surg. 2009;249:799–805. doi: 10.1097/SLA.0b013e3181a38eb5 [DOI] [PubMed] [Google Scholar]
- 6.Llovet JM, Di Bisceglie AM, Bruix J, et al. Design and endpoints of clinical trials in hepatocellular carcinoma. J Natl Cancer Inst. 2008;100:698–711. doi: 10.1093/jnci/djn134 [DOI] [PubMed] [Google Scholar]
- 7.Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7:e30679. doi: 10.1371/journal.pone.0030679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol. 2015;8:83. doi: 10.1186/s13045-015-0181-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naidu S, Magee P, Garofalo M. MiRNA-based therapeutic intervention of cancer. J Hematol Oncol. 2015;8:68. doi: 10.1186/s13045-015-0162-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017 [DOI] [PubMed] [Google Scholar]
- 11.Kharaziha P, Ceder S, Li Q, Panaretakis T. Tumor cell-derived exosomes: a message in a bottle. Biochim Biophys Acta. 2012;1826:103–111. doi: 10.1016/j.bbcan.2012.03.006 [DOI] [PubMed] [Google Scholar]
- 12.Lee TH, D’Asti E, Magnus N, Al-Nedawi K, Meehan B, Rak J. Microvesicles as mediators of intercellular communication in cancer–the emerging science of cellular ‘debris’. Semin Immunopathol. 2011;33:455–467. doi: 10.1007/s00281-011-0250-3 [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Hou L, Li A, Duan Y, Gao H, Song X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed Res Int. 2014;2014:864894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin Y, Wang J, Han J, Luo D, Sun Z. MiR-122 inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Snail1 and Snail2 and suppressing WNT/beta-cadherin signaling pathway. Exp Cell Res. 2017;360:210–217. doi: 10.1016/j.yexcr.2017.09.010 [DOI] [PubMed] [Google Scholar]
- 15.Yang N, Zhou J, Li Q, Han F, Yu Z. miR-96 exerts carcinogenic effect by activating AKT/GSK-3β/β-catenin signaling pathway through targeting inhibition of FOXO1 in hepatocellular carcinoma. Cancer Cell Int 2019;19:38. doi: 10.1186/s12935-019-0756-7. eCollection 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8:4083–4099. doi: 10.1002/pmic.200800109 [DOI] [PubMed] [Google Scholar]
- 17.Challagundla KB, Fanini F, Vannini I, et al. microRNAs in the tumor microenvironment: solving the riddle for a better diagnostics. Expert Rev Mol Diagn. 2014;14:565–574. doi: 10.1586/14737159.2014.922879 [DOI] [PubMed] [Google Scholar]
- 18.Murakami Y, Toyoda H, Tanahashi T, et al. Comprehensive miRNA expression analysis in peripheral blood can diagnose liver disease. PLoS One. 2012;7:e48366. doi: 10.1371/journal.pone.0048366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology. 2005;42:1208–1236. doi: 10.1002/hep.v42:5 [DOI] [PubMed] [Google Scholar]
- 20.Hanley JA. Receiver operating characteristic (ROC) methodology: the state of the art. Crit Rev Diagn Imaging. 1989;29:307–335. [PubMed] [Google Scholar]
- 21.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855 [DOI] [PubMed] [Google Scholar]
- 22.Raj DA, Fiume I, Capasso G, Pocsfalvi G. A multiplex quantitative proteomics strategy for protein biomarker studies in urinary exosomes. Kidney Int. 2012;81:1263–1272. doi: 10.1038/ki.2012.25 [DOI] [PubMed] [Google Scholar]
- 23.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267 [DOI] [PubMed] [Google Scholar]
- 24.Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nonaka R, Nishimura J, Kagawa Y, et al. Circulating miR-199a-3p as a novel serum biomarker for colorectal cancer. Oncol Rep. 2014;32:2354–2358. doi: 10.3892/or.2014.3515 [DOI] [PubMed] [Google Scholar]
- 27.Mar-Aguilar F, Mendoza-Ramirez JA, Malagon-Santiago I, et al. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis Markers. 2013;34:163–169. doi: 10.1155/2013/259454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Si H, Sun X, Chen Y, et al. Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J Cancer Res Clin Oncol. 2013;139:223–229. doi: 10.1007/s00432-012-1315-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redova M, Poprach A, Nekvindova J, et al. Circulating miR-378 and miR-451 in serum are potential biomarkers for renal cell carcinoma. J Transl Med. 2012;10:55. doi: 10.1186/1479-5876-10-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Q, Liu HO, Liu YD, et al. Decreased expression of hepatocyte nuclear factor 4alpha (Hnf4alpha)/microRNA-122 (miR-122) axis in hepatitis B virus-associated hepatocellular carcinoma enhances potential oncogenic GALNT10 protein activity. J Biol Chem. 2015;290:1170–1185. doi: 10.1074/jbc.M114.601203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Q, Zhang M, Tu J, Pang L, Cai W, Liu X. MicroRNA-122 affects cell aggressiveness and apoptosis by targeting PKM2 in human hepatocellular carcinoma. Oncol Rep. 2015;34:2054–2064. doi: 10.3892/or.2015.4175 [DOI] [PubMed] [Google Scholar]
- 32.Lin CJ, Gong HY, Tseng HC, Wang WL, Wu JL. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2008;375:315–320. doi: 10.1016/j.bbrc.2008.07.154 [DOI] [PubMed] [Google Scholar]
- 33.Dai M, Li L, Qin X. Clinical value of miRNA-122 in the diagnosis and prognosis of various types of cancer. Oncol Lett. 2019;17:3919–3929. doi: 10.3892/ol.2019.10024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083 [DOI] [PubMed] [Google Scholar]
- 35.Zhou X, Wang X, Zhou Y, Cheng L, Zhang Y, Zhang Y. Long noncoding RNA NEAT1 promotes cell proliferation and invasion and suppresses apoptosis in hepatocellular carcinoma by regulating miRNA-22-3p/akt2 in vitro and in vivo. Onco Targets Ther. 2019;12:8991–9004. doi: 10.2147/OTT.S224521 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Li S, Yang J, Xia Y, Fan Q, Yang KP. Long noncoding RNA NEAT1 promotes proliferation and invasion via targeting miR-181a-5p in non-small cell lung cancer. Oncol Res. 2018;26:289–296. doi: 10.3727/096504017X15009404458675 [DOI] [PMC free article] [PubMed] [Google Scholar]