Abstract
The establishment of separated pulmonary and systemic circulation in vertebrates, via cardiac outflow tract (OFT) septation, is a sensitive developmental process accounting for 10% of all congenital anomalies. Neural Crest Cells (NCC) colonising the heart condensate along the primitive endocardial tube and force its scission into two tubes. Here, we show that NCC aggregation progressively decreases along the OFT distal-proximal axis following a BMP signalling gradient. Dullard, a nuclear phosphatase, tunes the BMP gradient amplitude and prevents NCC premature condensation. Dullard maintains transcriptional programs providing NCC with mesenchymal traits. It attenuates the expression of the aggregation factor Sema3c and conversely promotes that of the epithelial-mesenchymal transition driver Twist1. Altogether, Dullard-mediated fine-tuning of BMP signalling ensures the timed and progressive zipper-like closure of the OFT by the NCC and prevents the formation of a heart carrying the congenital abnormalities defining the tetralogy of Fallot.
Research organism: Mouse
Introduction
The heart outflow tract (OFT) is an embryonic structure which ensures the connection between the muscular heart chambers and the embryonic vascular network. Initially, forming a solitary tube called truncus arteriosus, it gets progressively remodelled into two tubes which give rise to the aortic (Ao) and pulmonary (Pa) arteries (Brickner et al., 2000; Figure 1A). This remodelling stands as one of the most sensitive processes during heart morphogenesis. As such, faulty septation of the OFT represents 30% of all congenital heart diseases, with poor clinical prognosis due to improper mixing of oxygenated and deoxygenated blood. This thus calls for a better understanding of the cellular and molecular cues by which the OFT gets septated during development.
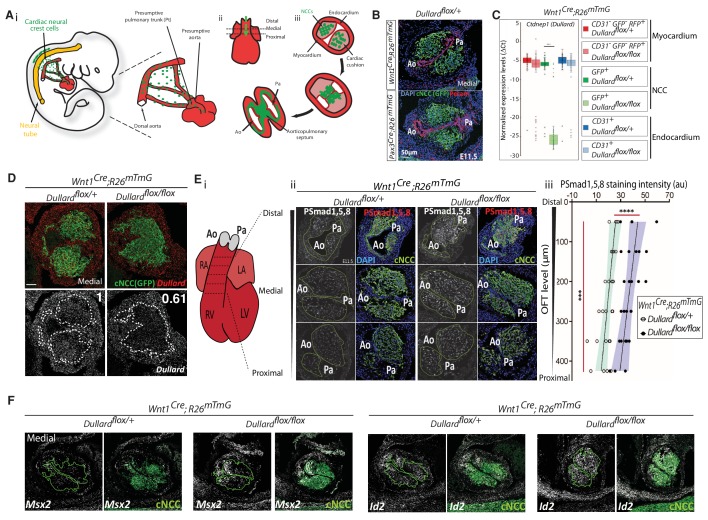
Figure 1. Dullard acts as a Smad1/5/8 activity inhibitor in cardiac NCC.
(A) Ai. Schematic representation of the migration routes the cardiac NCC (green) have taken to reach the heart region (red) in a E10.5 mouse embryo. Aii. Schematics of the embryonic heart at E11.5 showing the distal-proximal axis of the OFT. Aiii. Schematic representation of transverse sections through the OFT showing discrete stages of NCC condensation and endocardium septation along the OFT distal-proximal axis. (B) Pecam and GFP immunolabelling and DAPI staining on transverse sections throughout the medial OFT of E11.5 Wnt1Cre or Pax3Cre; Dullardflox/+; Rosa26mTmG embryos. (C) Normalized expression levels of Dullard assayed by q-RT-PCR on single cells isolated after immuno-marking endothelial CD31+ cells from E11.5 Wnt1Cre; Dullardflox/+ and Wnt1Cre; Dullardflox/flox; Rosa26mTmG hearts (dots: value for a single cell; boxplot: mean ± s.e.m.). The primers used to amplify Dullard specifically binds to exons 2 and 3, which are excised by the Cre recombinase. (D) Dullard mRNA distribution detected using RNAscope probes, in transverse sections of E11.5 control and mutant OFTs, assessed by RNAscope. Dullard mRNA levels were significantly reduced in mutant cardiac cushions compared to controls; however, mRNA signals were still detected given the binding of Z pair probes to non-recombined exons 5 to 8 and UTR region. (E) Ei. Schematics of E11.5 heart showing the position of the transverse sections used to quantify the levels of the phosphorylated forms of Smad1/5/8 in iii. Eii. Immunolabelling for P-Smad1/5/8 and GFP, and DAPI staining on transverse sections across the OFT at three distinct distal-proximal levels in E11.5 embryos with the indicated genotype. Eiii. Quantification of P-Smad1/5/8 levels in cardiac NCC along the OFT distal-proximal axis of E11.5 embryos with the indicated genotype (dots: values obtained on a given section; n > 4 embryos per genotype recovered from at least three liters; the black line is the linear regression, the coloured areas delineate the 95% confidence intervals, ***: p-value<0001 for a two-way Anova statistical test). (F) Msx2 and Id2 mRNA distribution detected using RNAscope probes (grey) and immunostaining of GFP (green) in transverse sections of E11.5 control and mutant OFTs (n = 2 embryos). On all A-F panels: green dotted lines delineate the area colonised by cardiac NCC. Ao: aortic artery, Pa: pulmonary artery.
Figure 1—figure supplement 1. Dullard phosphatase is a negative regulator of BMP signalling in several mammalian cells.
Morphogenesis of the OFT is orchestrated in time and space by cross-interaction between several cell types including the myocardial progenitors of the second heart field (SHF), the endocardial cells (EC) delineating the OFT lumen, and the cardiac neural crest cells (cardiac NCC) (Kelly, 2012; Keyte and Hutson, 2012; Figure 1A). Various genetic manipulations or ablation models have highlighted the predominant role of cardiac NCC in initiating and controlling OFT septation (Bockman et al., 1987; Phillips et al., 2013). Originally, cardiac NCC delaminate from the dorsal neural tube and migrate through the pharyngeal mesoderm to reach the developing OFT (Figure 1A). There, they invade the two cardiac cushions, condense toward the endocardium and trigger its rupture, thereby inducing cardiac cushions fusion and creating the two great arteries (Plein et al., 2015; Waldo et al., 1998). The rupture of the endocardium is first detected in the regions of the OFT which are the most distal from the heart chambers. In mouse embryos this rupture initiates around 11.5 days of embryonic development (E11.5; Figure 1A) and then expands progressively to more proximal levels. In parallel to these morphogenetic events, NCC differentiate into the vascular smooth muscles of the aortic arch (Keyte and Hutson, 2012) and also contribute to the arterial valves (Odelin et al., 2018).
Intense investigations to identify the molecular cues controlling the stereotyped behaviour and differentiation of cardiac NCC in the OFT have established the importance of the Bone Morphogenic Proteins (BMP), secreted by the outlying myocardium cells from E8.75 onwards (Danesh et al., 2009; Jiao et al., 2003; Liu et al., 2004; McCulley et al., 2008). Indeed, ablation of the BMP receptor Bmpr1a, ablation of the key downstream transcriptional effector Smad4, or forced expression of the BMP signalling antagonist Smad7 within the NCC lineage all lead to the formation of hypoplastic cushions, a shorter and non-septated OFT, thus phenocopying cardiac NCC ablation experiments (Jia et al., 2007; Stottmann et al., 2004; Tang et al., 2010). Knock-out of the ligand BMP4 from the myocardium similarly prevents OFT septation (Liu et al., 2004). However, little is known about the cardiac NCC behaviour and molecular cascades triggered by BMP signalling and responsible for the cardiac NCC mediated OFT septation.
To gain insights into these molecular cascades, we decided to dissect the role of Dullard (Ctdnep1), a perinuclear phosphatase that functions as a negative intracellular BMP inhibitor, during OFT morphogenesis (Sakaguchi et al., 2013; Urrutia et al., 2016; Sardi et al., 2019). In the canonical BMP signalling cascade, binding of BMP ligands to their transmembrane receptors leads to the phosphorylation of the transcription factors Smad1/5/8 which translocate to the nucleus and modify the transcriptional landscape of targeted cells (Bruce and Sapkota, 2012). Dullard stands out as one of the few cytoplasmic modulators of this phosphorylation step, which also includes PP1A, PP2B, the inhibitory Smads 6 and 7 and the Ubiquitin degradation pathway (Bruce and Sapkota, 2012). The Dullard protein is evolutionary conserved from yeast to mammals and expressed in many embryonic tissues, including the developing neural tube and neural crest cells (Sakaguchi et al., 2013; Satow et al., 2006; Tanaka et al., 2013; Urrutia et al., 2016; Figure 1D). Several pieces of evidence from Drosophila, xenopus, and mouse embryos indicate that this enzyme dampens Smad1/5/8 phosphorylation levels upon BMP stimulation (Sakaguchi et al., 2013; Satow et al., 2006; Urrutia et al., 2016). However, this activity is likely to be tissue specific, as depleting Dullard in gastrulating mouse embryos or later in the limb bud mesenchyme did not impair BMP signalling, while its depletion in the mouse embryonic kidney led to an elevated BMP response (Sakaguchi et al., 2013; Tanaka et al., 2013; Hayata et al., 2015). Regardless of the effect on BMP signalling, Dullard appears as a key regulator of various morphogenetic events regulating the elaboration of embryonic tissues. Early in development, it is required for the expansion of extraembryonic tissues, and later on, it prevents cell death by apoptosis in kidney nephrons or favours the ossification of limb bones (Sakaguchi et al., 2013; Tanaka et al., 2013; Hayata et al., 2015).
We showed here that deletion of Dullard in the cardiac NCC increases Smad1/5/8 activity, leading to premature and asymmetric septation of the OFT and pulmonary artery closure. BMP overactivation in the cardiac NCC occurs concurrently with the downregulation of mesenchymal markers (Snai2, Twist1, Rac1, Mmp14 and Cdh2) and upregulation of Sema3c, which is associated with premature cardiac NCC condensation to the endocardium. Our data converge to a model whereby graded BMP activity, Sema3c expression and cardiac NCC condensation along the OFT axis set the tempo of OFT septation from its distal to its proximal regions. Hence, our findings reveal that fine tuning of BMP signalling levels in cardiac NCC orchestrates OFT septation in time and space.
Results
Dullard deletion triggers hyperactivation of BMP intracellular signalling in cardiac NCC
In order to ablate Dullard in cardiac NCC, we crossed mice carrying floxed alleles of Dullard with mice expressing the Cre recombinase from the Pax3 locus or thanks to Wnt1 enhancer (Danielian et al., 1998; Engleka et al., 2005; Sakaguchi et al., 2013). Cell lineage tracing was achieved by using a ubiquitous double-fluorescent Cre reporter allele, Rosa26mTmG, in which Cre-mediated recombination labels the cells with membrane-targeted GFP (Muzumdar et al., 2007). The pattern of cell recombination in the cardiac cushions of E11.5 control embryos carrying either Cre driver matched with the pattern of colonising cardiac NCC described by previous lineage analyses (Figure 1B; Brown et al., 2001; Jiang et al., 2000). RT-qPCR on single cells isolated by Fluorescence-activated cell sorting (FACS) from dissected OFT and RNAscope in situ hybridization on histological sections were used to monitor Dullard expression and validate its deletion on GFP+ cells upon Cre recombination of Dullard flox alleles (Figure 1C,D). At E11.5, Dullard was ubiquitously expressed in all OFT layers of control embryos. In recombined Wnt1Cre; Dullardflox/flox; Rosa26mTmG embryos, the cardiac NCC displayed a strong reduction in Dullard levels compared to control littermates, while the surrounding tissues remained Dullard positive. Strikingly, in these mutants, the NCC formed a unique mass at the distal part of the OFT, while two distinct NCC cushions were present in the control embryos (Figure 1D), indicating that Dullard regulates the spatial organization of NCC in the OFT (see below).
We next assessed the relationship between Dullard and the activity of the intracellular effectors of BMP signalling, that is the Smad1/5/8 transcription factors, in mammalian cells. As previously shown (Satow et al., 2006), Dullard overexpression in the myogenic cell line C2C12 strongly decreased the levels of phosphorylated Smad1/5/8 induced by BMP2 treatment (Figure 1—figure supplement 1A). In addition, by generating a version of Dullard carrying a phosphatase dead domain, we showed that the role of Dullard as negative modulator of BMP signaling relied on its phosphatase activity (Figure 1—figure supplement 1A). Accordingly, Dullard deletion in cardiac NCC was sufficient to double the levels of P-Smad1/5/8 within the NCC whatever their position along the distal-proximal OFT axis of E11.5 hearts (Figure 1E, Figure 1—figure supplement 1B). To confirm this result, we also looked at the distribution and/or the levels of expression of several well-established BMP signalling pathway downstream targets, namely Id1, Id2, Msx1 and Msx2 (Figure 1F, Figure 1—figure supplement 1C; Miyazono et al., 2005). Quantitative RT-qPCR on isolated E11.5 NCC showed that the levels of Id1 transcripts were significantly elevated by Dullard loss and that Msx1 and Msx2 expression range was shifted towards higher values in mutants compared to control embryos (Figure 1—figure supplement 1C). In agreement, the amount of Msx2 transcripts detected by in situ hybridisation within the NCC in the vicinity of the aorta was greater in mutants compared to control embryos, so was that of Id2 transcripts present throughout all cardiac NCC (Figure 1F). This further indicates that in the cardiac NCC lineage, Dullard acts as a BMP intracellular signalling inhibitor. It is worth mentioning that Dullard was also required to dampen the levels of activated Smad1/5/8 in other cell types, including myogenic cells (Figure 1—figure supplement 1D). Furthermore, while Dullard has also been shown to decrease the levels of phosphorylation of Smads acting downstream of the TGF𝞫 in bone precursors (Hayata et al., 2015), the modulation of Dullard activity did not alter the phosphorylation state of one of these Smads, namely Smad2, in cardiac NCC (Figure 1—figure supplement 1E).
Remarkably, in both control and mutant contexts, P-Smad1/5/8 levels were more elevated distally than proximally (Figure 1Eiii, Figure 1—figure supplement 1Bii), indicating that BMP signalling elicits a graded response in cardiac NCC, which declines as they colonise more proximal OFT areas. Altogether, our results show that Dullard is required in cardiac NCC to dampen the magnitude of the BMP signalling gradient along the proximo-distal axis of the OFT, but is not required for its establishment, which is still observed in mutant embryos.
Dullard deletion in cardiac NCC leads to the emergence of heart abnormalities present in Fallot’s tetralogy
Given that cardiac NCC control OFT septation (Bockman et al., 1987; Phillips et al., 2013; Plein et al., 2015), we next sought to examine the morphology of the OFT in control and Dullard mutants. No gross morphological defects were detected in E10.5 Dullard mutant OFTs (Figure 2—figure supplement 1A). Notably, the endocardium morphology, the thickness and position of the myocardium, the NCC distribution along the outflow tract, were comparable in control and mutant embryos (Figure 2—figure supplement 1A). Strikingly, 24 hr later, severe and penetrant morphological OFT defects were observed in Dullard mutant embryos (Figure 2A,B, Figure 2—figure supplement 1B). To characterize these defects, we first analyzed E11.5–12 hearts labelled for the arterial marker Pecam using 3D lightsheet and confocal microscopy (Figure 2A,B, Figure 2—figure supplement 1B, Videos 1 and 2). At distal levels, the OFT of control embryos displayed symmetrical septation with two great arteries of similar size (Figure 2Ai,Bi, Figure 2—figure supplement 1Bi - Video 1). In contrast, Pax3Cre or Wnt1Cre; Dullardflox/flox embryos exhibited an asymmetric breakdown of the endocardium on the pulmonary side with obstruction of the pulmonary artery (Pa) (Figure 2Aii,Biv, Figure 2—figure supplement 1Biv - Video 2). At more medial levels in control embryos, the pulmonary pole of the endocardium was still connected to its aortic pole (Figure 2Bii, Figure 2—figure supplement 1Bii). This pole was also attached to the presumptive pulmonary valve intercalated-cushion (PV-IC), a cell cluster recognisable by faint levels of endothelial markers such as Pecam (Mifflin et al., 2018). Conversely, the aortic and pulmonary poles of the endocardium were prematurely septated and the NCC cushions were fused in the medial portion of the mutant OFT (Figure 2Bv, Figure 1—figure supplement 1Bv). Similarly to the observations made at distal levels, pulmonary endocardium cells were aggregated together failing to delineate a lumen. Furthermore, NCC often intervened between the residual pulmonary endocardium cells and the presumptive pulmonary valve intercalated-cushion (PV-IC) (Figure 2Bv). At proximal regions, the shape of the OFT endocardium and the surrounding NCC cushions were similar in both mutant and control embryos (Figure 2A,Biii,vi, Figure 2—figure supplement 1Biii,vi). We also checked for the state of the peripheral sheet of myocardium composing E11.5 control and Dullard mutant OFTs by immunolabelling the myosin heavy chain or the transcription factor Islet1/2 (Isl1/2) (Figure 2—figure supplement 1C,D). No significant differences could be detected between the control and mutant OFTs.
Figure 2. Dullard deletion in cardiac NCC causes asymmetric and premature OFT septation similar to Fallot’s tetralogy.
(A) Three-dimensional rendering of the Pecam+ endocardium of E12 Pax3Cre; Dullardflox/+ and Pax3Cre; Dullardflox/flox embryos after 3Disco clearing and lightsheet acquisition (n = 3 per genotype). The fine oblique white line marks the Pa width. The OFT levels along its distal-proximal axis analyzed in B are also indicated. (B) Immunolabelling for Pecam (red), GFP (green) and DAPI (blue) on transverse sections along the distal-proximal axis of the OFT in E11.5 embryos with the indicated genotypes (n > 10 embryos collected from more than three liters). Brackets in i and iv highlight the symmetric and asymmetric Ao and Pa poles in control and mutant embryos, respectively. Arrowheads in ii and v point at the unruptured and ruptured endocardium in control and mutant embryos, respectively. (C) Percentage of living Dullard mutant embryos before E12.5 and after E12.5, carrying the indicated Cre driver. (D) Immunolabelling for Pecam, GFP and DAPI staining on sections through the hearts of E14.5 (i,ii,v,vi) and E18.5 (iii,iv) embryos with the indicated genotypes (n = 2 embryos per genotype). Arrowheads in ii and arrow in iv point at a septation defect, the star in vi indicates the lack of Pa. (E) Whole dissected E18.5 hearts coming from embryos with the indicated genotype (n = 2 per genotype). (F) Two cycles of blood flow measured at the level of the abdominal artery of E11.75 control embryos and indication of the parameters analysed. VTI: velocity time integral. Parameters (i-iii) of the blood flow velocity measured in the abdominal artery of E11.75 control (turquoise dots) and Wnt1Cre; Dullardflox/flox embryos (purple squares)(dots and squares: mean of two to five measures obtained on a single embryo, bars: mean ± s.e.m; differences evaluated using a Mann-Whitney test: N.S. non-significant, *: p<0.05, **: p<0.01, ***: p<0.001). i. peak systolic velocity, ii. end-diastolic velocity, iii. mean of three velocity time integrals (n = 10 mutants and n = 32 controls). Ao: aortic artery, Pa: pulmonary artery, PV IC: pulmonary valve intercalated-cushion.
Figure 2—figure supplement 1. Morphological defects in Dullard mutants become striking from E11.5 onwards.
Video 1. Three-dimensional rendering of cardiac NCC (green) over Pecam (white) after BABB clearing and Lightsheet acquisition of Pax3Cre; Dullardflox/+; Rosa26mTmG and Pax3Cre; Dullardflox/flox; Rosa26mTmG E12 embryos.
No defect in the OFT colonisation of mutant cardiac NCC is observed.
Video 2. Three-dimensional rendering of cardiac NCC (green) over Pecam (white) after BABB clearing and Lightsheet acquisition of Pax3Cre; Dullardflox/+; Rosa26mTmG and Pax3Cre; Dullardflox/flox; Rosa26mTmG E12 embryos.
No defect in the OFT colonisation of mutant cardiac NCC is observed.
In order to evaluate the physiological impact of these OFT defects on cardiac function, we first attempted to harvest older embryos to conduct a histological characterisation of their hearts. The premature death of Pax3Cre or Wnt1Cre; Dullardflox/flox embryos after E12.5 complicated this task (Figure 2C). Nevertheless, in the handful of Dullard mutant embryos collected alive at E14.5 or E18.5, we consistently observed hearts with an interventricular septum (compare Figure 2Dii,iv with 2Di,iii), a hypertrophy of their right ventricle compared to their left one (compare Figure 2Div with 2Diii), a wide aorta connected to both ventricles (Figure 2Dvi, E) and pulmonary stenosis (compare Figure 2Dvi with 2Dv, asterisk in E). These are the four major heart morphological traits defining the tetralogy of Fallot (TOF)’s condition (Neeb et al., 2013). Second, we imaged the blood flow passing through the abdominal artery of E11.75 Wnt1Cre; Dullardflox/flox and Wnt1Cre; Dullardflox/+ embryos using doppler ultrasound (Figure 2F; see Material and methods section; Nomura-Kitabayashi et al., 2009). Several hemodynamic parameters were affected in Dullard mutants compared to controls. Notably, the systolic velocity peaked at a lower level in mutants compared to controls, suggesting a compromised blood ejection from the heart (Figure 2Fi). Conversely, the heart relaxation phase was less affected as no differences in the diastolic velocity were detected (Figure 2Fii). Overall, the blood flow was weaker in the mutants than in controls, as indicated by a decrease in the mean of the velocity time integral (VTI) (Figure 2Fiii).
Dullard prevents the premature condensation of cardiac NCC
We next wanted to further investigate the cellular mechanisms by which Dullard in NCC ensures OFT septation and started evaluating the migrative, proliferation and death status of cardiac NCC (Figure 3A, Figure 3—figure supplement 1, Videos 1–4). Whole mount immunostaining and 3D-reconstructions revealed that GFP+ cardiac NCC reached similar OFT levels in E11.5 control and Dullard mutants, showing that Dullard is not required for NCC colonisation of the OFT (Figure 3A, Videos 1–4). Similarly, quantification of cell proliferation and apoptosis on tissue sections, using antibodies raised against the phosphorylated form of histone H3 and the cleaved version of Caspase three respectively, indicated that Dullard does not control the proliferation nor the survival of cardiac NCC (Figure 3—figure supplement 1Ai-iv). In agreement with these observations, the total number of GFP+ cells colonising the OFT in mutant embryos was not significantly different from that found in controls (Figure 3—figure supplement 1Av, vi).
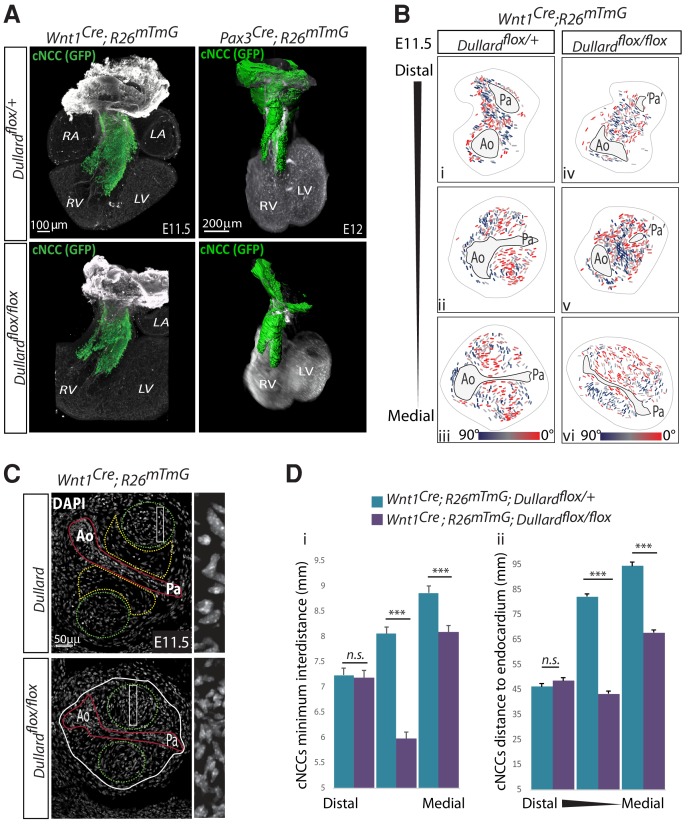
Figure 3. Dullard does not affect NCC migration, but prevents NCC premature condensation.
(A) Three-dimensional rendering of cardiac NCC (green) over Pecam (white) after BABB clearing and confocal acquisition (Wnt1cre samples) or 3disco clearing and lightsheet microscopy (Pax3cre samples) of whole E11.5 hearts isolated from embryos with the indicated genotype (n = 2 per genotype). (B) Coloured coded orientation of the major axis of NCC cells relative to Ao-Pa axis colour-coded as indicated in the section shown in Figure 2Bi–vi. (C) DAPI staining on transverse sections through the medial part of the OFT of E11.5 embryos with the indicated genotype. Magnified regions on the right are indicated by white rectangles in NCC cushions. The entire OFT is circled with a while line. The endocardium is delineated in red, the condensed and round NCC in green, the loose and elongated NCC in yellow. (D) Minimum distances between NCCs (Di) and distances between NCCs and the endocardium (Dii) quantified along the distal-proximal axis of the OFT in E11.5 embryos with the indicated genotypes (n = 3 embryos from distinct liters were analyzed for each genotype and OFT level, bars: mean ±s.d.; ***: p-value<0.0001 for Student statistical t-test). Ao: Aorta; Pa: pulmonary artery; Lv: left ventricle; PV-IC: Pulmonary valve intercalated-cushion; Rv: right ventricle.
Figure 3—figure supplement 1. Morphogenetic defects in Dullard NCC mutants.
Video 3. Three-dimensional rendering of cardiac NCC (green isosurface) over Pecam (red isosurface) and Dapi (white) of Wnt1Cre; Dullardflox/+; Rosa26mTmG and Wnt1Cre; Dullardflox/flox; Rosa26mTmG E11.5 embryos.
No defect in the OFT colonisation of mutant cardiac NCC is observed, and reduction of the pulmonary artery is visible in the mutant.
Video 4. Three-dimensional rendering of cardiac NCC (green isosurface) over Pecam (red isosurface) and Dapi (white) of Wnt1Cre; Dullardflox/+; Rosa26mTmG and Wnt1Cre; Dullardflox/flox; Rosa26mTmG E11.5 embryos.
No defect in the OFT colonisation of mutant cardiac NCC is observed, and reduction of the pulmonary artery is visible in the mutant.
Finally, we wondered whether the morphogenetic defects of the mutant OFT could stem from differences in cell-cell arrangements, looking at the position and orientation of NCC and endocardial cell nuclei (Figure 3B–D, Figure 3—figure supplement 1B–C). The orientation of the cardiac NCC nuclei relative to the endocardium appeared spatially regulated along the proximal-distal axis of the OFT, in both mutant and control hearts (Figure 3B,C, Figure 3—figure supplement 1B). In controls, NCC perpendicular to the endocardium could be found at distal levels, while at proximal levels no orientation preference could be assigned (blue dashes in Figure 3B, Figure 3—figure supplement 1B). Strikingly, in Wnt1Cre; Dullardflox/flox OFTs the perpendicular orientation was more widely observed at medial levels than in control OFT (blue dashes in Figure 3B, Figure 3—figure supplement 1B). Moreover, quantification of the shortest distance between adjacent cardiac NCC nuclei indicated that in E11.5 control hearts NCC condensation was also variable along the distal-proximal axis of the OFT (Figure 3C,Di). Cells were closer to each other at distal levels than in proximal regions. This progression of NCC condensation along the OFT axis was impaired in Dullard mutants, whereby mutant NCC prematurely condensed within the medial region of the OFT (Figure 3C,Di). Finally, the position of NCC to the endocardium was variable along the OFT axis of control embryos with NCC being closer to this epithelium at distal levels than at proximal levels (Figure 3C,Dii). In the mutants, NCC were in a closer vicinity of the endocardium than control cells, so that in medial levels they displayed traits of cells normally found at distal levels in control hearts (Figure 3C,Dii). In agreement with these data, the OFT area was reduced in mutants and remained more constant along the distal to proximal axis (Figure 3—figure supplement 1C).
Taken together Figures 2 and 3 data demonstrate that Dullard stands as a key modulator of NCC behaviour dynamics in the heart and hence of OFT septation. It precipitates NCC condensation, and thereby leads to the premature breakage of the endocardium and obstruction of the pulmonary artery. This weakens the embryonic hemodynamics and compromises the living of Dullard mutant embryos (see discussion). Our data also brings further support to the idea that morphogenetic defects in the NCC-derived cushions stand as one possible cause of Fallot's tetralogy (Neeb et al., 2013).
Dullard deletion in NCC mainly affects the transcriptional state of NCC
To decipher the molecular basis of the defective OFT remodeling observed in mutants, we micro-dissected Wnt1Cre; Rosa26mTmG E11.5 control and Dullard mutant heart OFTs and sorted the cardiac NCC (GFP+) and endocardial cells (CD31+; RFP+) from the other OFT cell-types (CD31-; RFP+) (Figure 4A). We then performed single-cell RT-qPCR for 44 genes implicated in epithelial-mesenchymal transition (EMT), migration and/or specification of the different OFT progenitor subtypes (Supplementary file 1), and their expression levels were normalised to GAPDH and ActB (Figure 4B).
Figure 4. Single-cell transcriptional analyses of all OFT cells at E11.5.
(A) Experimental steps performed to profile gene expression in OFT single-cells sorted from five Wnt1Cre; Dullardflox/+; Rosa26mTmG and five Wnt1Cre; Dullardflox/flox; Rosa26mTmG E11.5 embryos. At least 70 cells were isolated per gate and genotype (GFP+, CD31+, RFP+). (B) Graph showing the distribution of all cells analysed (dots) as a function of normalised expression values of the house keeping genes Actb and Gapdh and a linear regression (red line). (C) t-SNE plot showing the distribution of 44-genes-based transcriptomes of 433 OFT cells expressing the indicated markers isolated from both Wnt1Cre; Dullard+/flox; Rosa26mTmG and Wnt1Cre; Dullardflox/flox; Rosa26mTmG E11.5 embryos. (D) Unsupervised clustering heatmap of the 433 OFT isolated cells from Wnt1Cre; Dullard+/flox; Rosa26mTmG and Wnt1Cre; Dullardflox/flox; Rosa26mTmG based on the gene expression level of the 44 genes included in the panel (Supplementary file 1). Six different groups of cells can be discriminated, among which the endocardial (Group 1) cells expressing high levels of Flt1, Kdr, Nfatc1 and Tek, and the epicardial (Group 5) cells expressing high levels of Wt1, Tcf21..
T-statistic Stochastic Neighbour Embedding (t-SNE) was first used to plot the distances existing between the 44 gene-based-transcriptomes of individual cells (Figure 4C). It revealed that the 44 chosen genes were sufficient to segregate the three isolated cell subtypes, GFP+ NCC, the RFP+;CD31+endocardial cells and the other RFP+;CD31- OFT cells, validating our approach. Unsupervised hierarchical clustering analysis of all cells refined this segregation and identified six distinct groups of OFT cells (Figure 4D). Importantly, some of these groups contained both control and Dullard mutant cells (Groups 1, 3, 5) meaning that their 44 gene-based-transcriptome was not drastically dependent on Dullard. Instead, the three other groups were enriched for cells with a given genotype (Groups 2, 4, 6), hence harboured a Dullard dependent transcriptional state. Importantly, most GFP+ NCC were contained in the Groups 2, 4, 6, while the other groups were enriched for other cell types. For instance, the RFP+;CD31+ (Flt1+;Kdr+;Nfatc+;Tek+) endocardial cells and the RFP+;CD31- (Tcf21+;Wt1+) epicardial cells defined the Groups 1 and 5, respectively. Group 3 contained Isl1+ and Six2+ cells coming from GFP+ (NCC) and RFP+;CD31- (including myocardial) lineages. Overall, it suggests that Dullard deletion in NCC mainly alters the transcriptional states cell-autonomously and modulates to a much lesser extent the surrounding cell types present in the developing E11.5 heart.
Transcriptomic heterogeneity in non NCC-derived populations upon Dullard deficiency
We next focused on the transcriptomic variations operating in the distinct OFT specific cell populations, starting with the RFP+;CD31+endocardial cells and RFP+;CD31- OFT cells (Figure 5Ai–ii’, Figure 5—figure supplement 1). Hierarchical clustering and two-dimensional visualisation of cells on diffusion maps indicated that in both cell types, some transcriptomic heterogeneity was found and distinct subpopulations could be isolated (Figure 5Ai–ii’, Figure 5—figure supplement 1A). All 5 RFP+;CD31- subpopulations identified contained both control and mutant cells (Figure 5Ai,i’), sustaining the idea that Dullard loss in cardiac NCC does not impair the differentiation of the SHF-derived myocardium and smooth muscle, nor the differentiation of the epicardium (see also Figure 2—figure supplement 1C). Similarly, the vast majority of RFP+;CD31+ cells (four out of six subpopulations (sub-pops 2 to 5)) presented both mutant and control cells (Figure 5Aii,ii’, Figure 5—figure supplement 1A). The transcriptomic heterogeneity between these RFP+;CD31+ subpopulations was mild and these cells were all Kdr+, Foxc1+, Nfatchigh, Flt1+, Nrp1+, Plxnd1 High, as expected for the endocardium (blue rectangles in Figure 5—figure supplement 1A). However, the subpopulation 1 of RFP+;CD31+ cells was enriched in control cells while the subpopulation six in mutant cells (Figure 5Aii,ii’). From the genes that drive the segregation of these CD31+ subpopulations (Figure 5—figure supplement 1B), very few of them were specifically induced or repressed in the subpopulation six or the subpopulation one compared to the others subpopulation (Figure 5—figure supplement 1C). Out of them stood Twist1 and Sox9, which were enriched in the subpopulation 1. Accordingly, while few Twist1+ cells could be immunolabelled within the endocardium of control embryos, these were almost undetectable in mutant embryos (Figure 6A). Twist1 being one of the epithelial-mesenchymal transition (EMT) drivers, it suggests that endocardial EMT is affected by the absence of Dullard in the NCC (see discussion).
Figure 5. Impact of Dullard deletion in NCC on the transcriptomic variations within the distinct cellular subtypes of E11.5 hearts.
(A) Projected position of 44 genes-based transcriptomes assessed in mutant and control CD31+; RFP+ and CD31-; RFP+ OFT cells on diffusion maps made using the first two Diffusion Component 1 (DC1) and 2 (DC2) (Figure 5—figure supplement 1B). Sub-populations defined with hierarchical clustering are presented in i and ii, while the genotype of cells is illustrated in i’ and ii’. (B, C) Projected position of 44 genes-based transcriptomes assessed in mutant and control cardiac GFP+ NCC on diffusion maps made using the first two Diffusion Component 1 (DC1) and 2 (DC2) (see Figure 5—figure supplement 2B). The five subpopulations defined in Figure 3A are highlighted. (D) Percentage of cardiac NCC in each subpopulation. (E) Heatmaps showing the levels of expression of selected genes in all GFP+ NCC in the five subpopulations identified using unsupervised hierarchical clustering (Figure 5—figure supplement 2A) coming from control (blue) or mutant (orange) E11.5 embryos. (F) Boxplot representation of the expression levels of genes differentially expressed between the five NCC subpopulations (Sub-Pop 1: 44 cells, Sub-Pop 2: 29 cells, Sub-Pop3: 20 cells, Sub-Pop 4: 34 cells, Sub-Pop 5: 24 cells) (mean ±s.d.).
Figure 5—figure supplement 1. Transcriptional profiling of CD31+; RFP+ OFT cells.
Figure 5—figure supplement 2. Single-cell transcriptional profiling of GFP+ NCC at E11.5.
Figure 6. Dullard prevents NCC from acquiring epithelial-like traits and prolongs the expression of mesenchymal drivers.
(E) Twist1 immunolabelling and RFP signal (red; grey), Snai2 mRNA distribution assessed by RNAscope (red; grey), GFP (green) immunolabelling and DAPI staining on transverse sections through the medial OFT of E11.5 Wnt1Cre; Dullardflox/+; Rosa26mTmG and Wnt1Cre; Dullardflox/flox; Rosa26mTmG embryos. The red lines mark the endocardium, green arrowheads point at NCC, while the white ones indicate the endocardium. The white lines delineate the cardiac NCC cushions. RFP and GFP mark cell membrane whereas Twist1 is cytoplasmic or nuclear. (B) Sema3c expression assessed by ISH on transverse sections from distal to medial OFT levels of E11.5 Wnt1Cre; Dullardflox/+ and Wnt1Cre; Dullardflox/flox embryos. The endocardium is delineated with a red line, the cardiac NCC areas with a green line and the myocardium with grey lines. (C) Normalized expression levels of Sema3c assayed by q-RT-PCR on single cells isolated after immuno-marking endothelial CD31+ cells from E11.5 Wnt1Cre; Dullardflox/+ and Wnt1Cre; Dullardflox/flox; Rosa26mTmG hearts (dots: value for a single cell; boxplot: mean ± s.e.m.). (D) Model for the molecular and cellular cues controlling OFT septation. Upper panel: Morphogenesis of the single truncus arteriosus at E10.5 into fully formed great arteries at E14.0. Middle panel: Shape of P-Smad1/5/8 and Sema3c gradients along the OFT distal-proximal axis in a control situation. NCC in the cardiac cushions condense toward the endocardium between the aorta and the pulmonary trunk. Lower panel: Premature condensation of NCC at the pulmonary trunk in absence of Dullard in NCC, is associated with increased levels of Sema3c and BMP signalling.
Figure 6—figure supplement 1. Dullard loss in cardiac NCC alters the expression of Sema3c specifically in NCC.
Dullard controls the mesenchymal transcriptional state of cardiac NCC
Given the key defects observed in NCC condensation in Dullard mutants, we then focused on the gene expression signature of GFP+ NCC. With similar approaches as above we could identify five GFP+ NCC subpopulations based on their gene expression signature (Sub Pops 1 to 5) (Figure 5B, Figure 5—figure supplement 2A), each of them containing an unbalanced ratio of mutant versus control cells (Figure 5C,D), suggesting that Dullard influences the fate of all NCC subtypes. The NCC subpopulation 1 was characterised by the expression of Nfatc1, Tcf21, Postn (Periostin) (Figure 5—figure supplement 2A–C), which are all expressed in the heart valves (Acharya et al., 2011; Norris et al., 2008; Wu et al., 2011), a heart structure that is also colonised by NCC (Odelin et al., 2018). The slight enrichment for control cells in this subpopulation raised the possibility that Dullard is required to favour the contribution of NCC to this structure (Figure 5C). The second subpopulation was defined by the predominant expression of cardiac progenitor markers Tbx1, Six2, Gja1, Isl1, and contained both control and mutant cardiac NCC (Figure 5—figure supplement 2A–C), indicating that Dullard is not required for the entry of NCC into the smooth muscle lineage (Zhou et al., 2017). This is in agreement with the distribution of Isl1 we observed in control and mutant embryos (Figure 2—figure supplement 1C). Conversely, the emergence of the subpopulation 3 was strictly dependent on Dullard, as this subpopulation barely contained mutant cells (Figure 5C,D, Figure 5—figure supplement 2A). Subpopulation 3 corresponded to NCC further differentiated toward the smooth lineage as defined by expression of Myh11 and Acta2 (Huang et al., 2008; Figure 5D,E). The subpopulations 4 and 5 were enriched for mutant cardiac NCC (Figure 5C–E, Figure 5—figure supplement 2A). The transcriptomic state of the cells in these two subpopulations diverged from that of the subpopulation 3 cells (Figure 5C,E,D). This divergence was greater for the subpopulation 5 than for the Ssubpopulation 4. Yet, the position of cells within the diffusion maps suggests a close relationship between these two subsets of cells which might represent two states of the differentiation path of Dullard deleted cardiac NCC (Figure 5C).
Strikingly, genes that were differentially expressed between subpopulation 3 and the subpopulation 4/5 encode for known regulators of cell adhesion and epithelial-mesenchymal transition. On the one hand, the levels of mesenchymal markers (Snai2, Twist1, Cdh2, Mmp14, Rac1) in both subpopulations 4 and 5 were lower than in the subpopulation 3. In control embryos, NCC expressing these markers were found near the endocardium, as demonstrated by an ISH for Snai2 or immunolabelling for Twist1 (Figure 6A). In mutants, these pro-epithelial-mesenchymal transition factors were barely detected in NCC. These data are in agreement with the NCC over-condensation phenotype observed in Dullard mutants nearby the endocardium. On the other hand, cells in subpopulations 4 and 5 displayed higher levels of the epithelial markers Cdh5 and of Sema3c compared to subpopulation 3 cells. In situ hybridization of Sema3c further confirmed these results with cellular resolution (Figure 6B, Figure 6—figure supplement 1A,B), showing that at distal OFT levels, Sema3C was expressed in a scattered fashion in NCC in cushions of control embryos, while it was found in almost all NCC in mutants. Importantly, we observed a graded decrease of Sema3c expression along the OFT axis in both control and mutant embryos. This was reminiscent of the BMP response and condensation gradients described previously (Figures 1E and 3D) and in agreement with the established role of Sema3c in the regulation of cohesive/metastatic balance in several cancers (Tamagnone, 2012).
Altogether, our results show that Dullard prevents the establishment of an epithelial-like state and promotes the expression of pro-mesenchymal genes in NCC. In addition, it may also impair the epithelial mesenchymal transition of the endothelial cells (see discussion).
Discussion
Our analysis of the functions of the Dullard phosphatase in cardiac NCC brings us to propose a model whereby a BMP-dependent gradient of cardiac NCC condensation would set the timing of cardiac cushions fusion and thereby the septation of the OFT into the aorta and pulmonary artery. In addition, the analogy between the phenotype of Dullard mutant mice and patients suffering from Fallot’s tetralogy condition calls for a discussion on the cellular and molecular aetiology of this complex congenital heart disease.
First, we showed that a gradient of BMP activation exists in the cardiac NCC along the OFT axis and matches with a gradient of NCC condensation. The magnitude of this BMP gradient is under the control of the phosphatase Dullard. Yet, the establishment of this gradient is dependent on other mechanisms, as the gradient is still observed in absence of Dullard in the cardiac NCC. The establishment of this gradient is unlikely to result from a corresponding gradient of ligand that would diffuse from a localized distal source, as described in other contexts (Bier and De Robertis, 2015). In fact, BMP4 is homogeneously expressed in the myocardium throughout the entire length of the OFT and not solely at the distal level (Jia et al., 2007; Jiao et al., 2003; Jones et al., 1991; Zhang et al., 2010). Rather, intracellular signalling inhibitors could allow a temporal adaptation of cardiac NCC to BMP signals along with their migration towards proximal levels of the OFT. Prominent expression of Smad6, a BMP negative feedback effector, and of the diffusible inhibitor Noggin, are indeed observed in the OFT from E10.5 throughout the great arteries formation and thus represent promising candidates (Choi et al., 2007; Galvin et al., 2000).
Secondly, several lines of evidence support the idea that the elevation of BMP signalling in Dullard mutants underpins the premature condensation of cardiac NCC and the strong pulmonary artery atrophy. While in other biological systems, Dullard loss has been associated with decreased Wnt-𝞫-Catenin signalling (Tanaka et al., 2013) or enhanced TGF𝞫-Smads2/3 signalling (Hayata et al., 2015), these signalling pathways are probably not affected upon Dullard deletion in the cardiac NCC. On the one hand, we showed that the phosphorylation state of Smad2 in Dullard mutant and control cardiac NCC was comparable (Figure 1—figure supplement 1E). On the other hand, loss of Wnt signalling in cardiac NCC leads to persistent truncus arteriosus, which is associated with reduced NCC-mediated cushion fusion (Brade et al., 2006). Furthermore, gain- and loss-of-function experiments argue that BMP levels stand as a sufficient parameter to tune the degree of condensation of cardiac NCC towards the endocardium. Lowering BMP levels was shown to prevent fusion of the cardiac cushions and leads to persistent truncus arteriosus (Jia et al., 2007; Stottmann et al., 2004). Instead, we showed that increasing downstream BMP signalling due to Dullard loss triggers premature cardiac NCC condensation to the endocardium and accentuated OFT septation (Figure 2). This phenotype is reminiscent of the misplaced septation and narrowing of one of the OFT tubes observed in mouse mutants for the BMP and TGF𝞫 signalling antagonist Smad6 (Galvin et al., 2000). As such, the BMP signalling gradient can be seen as a means that allow septation of the OFT to proceed in a zipper-like fashion. This is further supported by the fact that altering the ability of cardiac NCC to contract, migrate and adhere, hence to condense, prevents the correct formation and positioning of the aorticopulmonary septum (Luo et al., 2006; Phillips et al., 2013; Plein et al., 2015).
Thirdly, we have uncovered part of the cellular and molecular mechanism by which Dullard controls cardiac NCC behaviour. Dullard deletion triggers a downregulation of mesenchymal markers reminiscent of a cardiac NCC transition towards epithelial-like states, with a loss of migratory freedom and increased cohesiveness between cells (Kim et al., 2017). Furthermore, out of the transcriptional changes induced by the loss of Dullard, several lines of evidence support the idea that the up-regulation of Sema3c is likely to play a predominant role in the increased condensation of the cardiac NCC. In fact, the expression of Sema3c in the cardiac NCC is required for their convergence to the endocardium and was also shown to promote the aggregation of cardiac NCC in primary cultures as well as in cancer cells in vivo (Delloye-Bourgeois et al., 2017; Feiner et al., 2001; Kodo et al., 2017; Plein et al., 2015; Toyofuku et al., 2008). In addition, the Sema3C-Nrp1 signalling has been proposed as a triggering signal for the EMT of the endocardium which accompanies its rupture (Plein et al., 2015). Intriguingly, our data show that the elevation of Sema3c in Dullard mutants is associated with variations in the expression of EMT factors in the endocardium (Figure 5—figure supplement 1), that would indicate that either the endocardium EMT is inhibited or has taken place prematurely. Testing the first possibility would require a fine quantification over time of rare EMT figures within the endocardium of control and Dullard mutants at several developmental stages. Yet, in the light of the premature septation of the endocardium we observed in Dullard mutants, we favour the second possibility. The elevation of Sema3c in Dullard mutants raises also the question about the nature of the regulatory relationship existing between BMP signalling and Sema3c expression. Embryos from the Pax3Cre driver line harbour myogenic recombined cells which, as the cardiac NCC, show a striking increase in phosphorylation of Smad1/5/8 and Sema3c expression when Dullard is deleted (Figure 1—figure supplement 1E). This suggests that the regulatory influence of BMP signaling on Sema3c expression is not restricted to the context of cardiac NCC but also to other cell types. However, it remains unclear if Smads act directly on Sema3c expression or indirectly via an intermediate transcription factor. In fact, Gata6, a member of the zinc finger family of transcription factors, has been described as an activator of Sema3c expression in the cardiac NCC (Kodo et al., 2017; Lepore et al., 2006). Yet, we did not observe any significant difference of Gata6 expression in Dullard deleted cardiac NCC (data not shown).
Finally, our data indicates that mice in which Dullard is deleted in the NCC harbours the traits of patients affected by Fallot’s tetralogy condition, and thus represents one of the rare animal models for this pathology (Neeb et al., 2013). Interestingly, while the few well-characterized genetic causal factors identified in humans (such as Jag1, Nkx2.5, Tbx5) and the analysis of few mouse mutants would primarily incriminate defects at the level of the SHF, our data suggest that this sheet of cells is unaffected in Dullard mutants (Neeb et al., 2013; Morgenthau and Frishman, 2018). Instead, our data provide further evidence for the NCC as one of the OFT cell lineages whose development defects can lead to the formation of hearts with an aorta overriding the interventricular septum (High et al., 2007). Further explorations would be required to assess whether the malformations led by defects in the NCC or the SHF are comparable and whether, in both cases, the molecular and cellular defects lead to more pressure on the endocardium and forces its rupture.
Materials and methods
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | C57BL/6JRj | Janvier Labs | ||
| Genetic reagent (Mus musculus) | Wnt1Cre | PMID: 9843687 | MGI:2386570 | Dr A Pierani (Imagine Institute) |
| Genetic reagent (Mus musculus) | Pax3Cre | PMID: 15882581 | MGI: 3573783 | Dr F Relaix (Créteil University) |
| Genetic reagent (Mus musculus) | Rosa26mTmG | PMID: 17868096 | MGI: 3716464 | Dr F Relaix (Créteil University) |
| Genetic reagent (Mus musculus) | DullardFlox | PMID: 23360989 | other | Dr R Nishinakamura (Kumamoto University) |
| Antibody | Anti-GFP (Chicken) | Aves Labs | GFP-1020 RRID:AB_10000240 |
IF(1:500) |
| Antibody | anti-PECAM (Rat monoclonal) | Santa-Cruz Biotechnology | Sc-18916 RRID:AB_627028 |
IF(1:200) |
| Antibody | Anti-phosphoSmad1/5/8 (Rabbit monoclonal) | Cell Signalling Technology | 13820S RRID:AB_2493181 |
IF (1:500) |
| Antibody | Anti-Myosin Heavy Chain (mouse monoclonal) | DSHB | MF20 RRID:AB_2147781 |
IF (1:300) |
| Antibody | Anti-phospho-histone H3 (rabbit polyclonal) | Cell Signalling Technology | 9701 RRID:AB_331535 |
IF (1:500) |
| Antibody | Anti-cleaved Caspase 3 (Rabbit monoclonal) | Cell Signalling Technology | 9664 RRID:AB_2070042 |
IF (1:500) |
| Antibody | Anti-Isl1 (Rabbit polyclonal) | Abcam | Ab20670 RRID:AB_881306 |
IF (1:500) |
| Antibody | AF 488 donkey IgG anti-chicken IGG (H+L) | Interchim | 703-545-155 RRID:AB_2340375 |
IF (1:500) |
| Antibody | Alexa Fluor 555 Goat Anti-Rabbit IgG (H+L), highly cross-adsorbed | Life Technologies | A-21429 RRID:AB_2535850 |
IF (1:500) |
| Antibody | AF 647 Donkey IgG Anti Rabbit IGG (H+L) | Interchim | 711-605-152 RRID:AB_2492288 |
IF (1:500) |
| Antibody | Alexa Fluor 647 Goat Anti-Rat IgG (H+L) | Life Technologies | A21247 RRID:AB_141778 |
IF (1:500) |
| Antibody | Alexa Fluor 647 Goat Anti-Mouse IgG (H+L) Antibody, highly cross-adsorbed | Life Technologies | A-21236 RRID:AB_2535805 |
IF (1:500) |
| Antibody | Alexa Fluor 488 Goat Anti-Mouse IgG (H+L) Antibody, highly cross-adsorbed | Life Technologies | A-11029 RRID:AB_2534088 |
IF (1:500) |
| Antibody | Alexa Fluor 488 Goat Anti-Rabbit IgG (H+L) Antibody | Life Technologies | A-11008 RRID:AB_143165 |
IF (1:500) |
| Antibody | Anti-Phospho Smad 2/3 (Rabbit monoclonal) | Ozyme | 8828 s RRID:AB_2631089 |
IF (1:200) |
| Sequence-based reagent | Sema3C | PMID: 16397144 | other | RNA probe from Dr S Zaffran (Aix Marseille University) |
| Sequence-based reagent | Dullard | Advanced Cell Diagnostics | #456911 | RNA probe |
| Sequence-based reagent | Sema3C | Advanced Cell Diagnostics | #441441 | RNA probe |
| Sequence-based reagent | Twist1 | Advanced Cell Diagnostics | #414701 | RNA probe |
| Sequence-based reagent | Snai2 | Advanced Cell Diagnostics | #451191 | RNA probe |
| Sequence-based reagent | Msx2 | Advanced Cell Diagnostics | #421851 | RNA probe |
| Sequence-based reagent | Id2 | Advanced Cell Diagnostics | #445871 | RNA probe |
| Antibody | Anti-Ter119 Pe-Cy7 (Mouse monoclonal) | Sony | 1181110 | Facs (1:300) |
| Antibody | Anti-CD45 APC-Cy7 (Mouse monoclonal) | BD Pharmingen | 557659 RRID:AB_396774 |
Facs (1:300) |
| Antibody | Anti-CD31 APC (Mouse monoclonal) | BD Pharmingen | Clone MEC 13.3 Catalog No. 561814 RRID:AB_10893351 |
Facs (1:300) |
| Commercial assay or kit | 7AAD PE-Cy7 | BD Pharmingen | 559925 | Facs (1:800) |
| Commercial assay, kit | CellsDirect One-Step qRTPCR Kit | Invitrogen | 11753100 | |
| Commercial assay, kit | 48.48 Sample/Loading Kit— 10 IFCs | Fluidigm Corporation | BMK-M10- 48.48 | |
| Cell Line (Mus musculus) | C2C12 | ATCC, PMID: 28966089 | CRL-1772, RRID:CVCL_0188 | |
| Peptide, recombinant protein | Human BMP-2 | Thermo Fisher Scientific | #PHC7145 | 50 ng/ml |
| Software, algorithm | R, pHeatmap (v1.0.10) | R foundation | R package (v3.2.2), RRID:SCR_016418 | |
| Software, algorithm | R, phenograph (v0.99.1) | R foundation | R package (v3.2.2), RRID:SCR_016919 | |
| Software, algorithm | R, ggplot2 (v3.1.0) | R foundation | R package (v3.2.2), RRID:SCR_014601 | |
| Software, algorithm | R, Destiny (v2.6.1) | R foundation | R package (v3.6) | https://github.com/theislab/destiny |
| Recombinant DNA reagent | pEGFP-Dullard (plasmid) | This paper | GFP-Dullard expression plasmid | |
| Recombinant DNA reagent | pEGFP-Dullard D67E (plasmid) | This paper | GFP-Dullard D67E expression plasmid | |
| Recombinant DNA reagent | pEGFP-N1 | Clontech | 6085–1 | |
| Commercial assay or kit | Gateway LR Clonase | ThermoFisher | 11791100 | |
| Commercial assay or kit | Taq Polymerase, Superscript III | Thermofisher | 11732020 | |
| Sequence-based reagent | Dull_FL_C1_FW | This paper | PCR primers | GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGATGCGGACGCAGTGT |
| Sequence-based reagent | Dull_FL_C1_Rev | This paper | PCR primers | GGGGACCACTTTGTACAAGAAAGTGGGTCTCACCAGAGCCTATGTTGGTG |
| Sequence-based reagent | Dull_D67E_FW | This paper | PCR primers | GATCCTGGTGCTGGAACTGGACGAAACCCTG |
| Sequence-based reagent | Dull_D67E_Rev | This paper | PCR primers | CAGGGTTTCGTCCAGTTCCAGCACCAGGATC |
Animals
All animal experiments were approved by the Animal Ethics Committee of Sorbonne University. We used the mouse strains described in the following papers and MGI IDs: Dullardflox/flox (Sakaguchi et al., 2013; in these mice exons 2 to 4 are floxed), Pax3Cre (Engleka et al., 2005, MGI: 3573783), Wnt1Cre (Danielian et al., 1998, MGI:2386570), Rosa26mTmG (Muzumdar et al., 2007, MGI: 3716464), and C57BL/6JRj (Janvier Labs).
Immunohistochemistry and imaging
Mouse embryos were collected at E11.5 and dissected in cold PBS, incubated 5 min in 200 mM KCl to stop heart beating and fixed for 2–3 hr in 4% PFA (Electron Microscopy Science, #15710S) at 4°C.
For immunostaining on cryosections, embryos were cryoprotected in 20% sucrose overnight at 4°C, embedded in OCT, and cryosectioned at 12 µm thickness. Sections were permeabilized 10 min in PBS/0.5% Triton, incubated for 1 hr in blocking buffer (5% goat serum in PBS) and overnight in primary antibody solution (in 1% BSA in PBS). After thorough washing in PBS, they were incubated 1 hr in secondary antibody solution (in 1% BSA in PBS), washed in PBS and mounted in Fluoromount-G (Clinisciences, 0100–01). Immunostainings were acquired using a Nikon Ti2 microscope, driven by Metamorph (Molecular Devices), equipped with a motorized stage and a Yokogawa CSU-W1 spinning disk head coupled with a Prime 95 sCMOS camera (Photometrics), then assembled and analyzed on Fiji (Schindelin et al., 2012).
For wholemount staining, we followed the 3Disco protocol (Belle et al., 2017) to immunostain, clear and image with a lightsheet ultramicroscope (LaVision BioTec). Alternatively, micro-dissected hearts immunostained as described in Belle et al. (2017), were clarified with BABB and imaged using a LSM700 confocal microscope (Carl Zeiss) (Gopalakrishnan et al., 2015). 3D renderings were generated using the Imaris software.
The primary antibodies used were raised against: GFP (chicken, Aves Labs, GFP-1020, 1/500), Pecam (rat monoclonal, Santa-Cruz Biotechnology, sc-18916, 1/200), Phospho-Smad1/5/8 (rabbit monoclonal, Cell Signalling Technology, 13820S, 1/500), myosin heavy chain, MyHC (mouse monoclonal, DSHB, MF20, 1/300), Phospho-Histone H3 (rabbit, Cell Signalling Technology, 9701, 1/500), Cleaved Caspase-3 (rabbit monoclonal, Cell Signalling Technology, Asp175, 1/500), Isl-1 (Abcam, ab20670), Phospho-Smad2,3 (rabbit, Ozyme, 8828 s, 1:200). Secondary antibodies were bought from Life technologies or Interchim and were Donkey or Goat Igg coupled to Alexa fluorophores.
In situ hybridisation
The Sema3c probe was provided by the lab of S. Zaffran (Bajolle et al., 2006). In situ hybridization on cryosections were processed following the protocol described in Chotteau-Lelièvre et al. (2006).Fluorescent in situ hybridization probes for Dullard (#456911), Sema3c (#441441), Twist1 (#414701), Snai2 (#451191), Msx2 (#421851) and Id2 (#445871) were obtained from Advanced Cell Diagnostics, Inc In situ hybridization was performed using the RNAscope V2-fluorescent kit according to the manufacturer's instructions. For sample pre-treatments: H2O2 treatment was performed during 10 min at RT, retrieval 2 min at 98°C and slides were digested with Protease Plus reagent for 15 min at 40°C. After the probe detection steps immunostaining was performed as described above with fluorescent secondary antibodies. Sections were imaged using a 40x objective on a LSM700 microscope (Zeiss) or Nikon Ti2 microscope.
Image analyses, quantification, statistical analysis
Mean levels of P-Smad1/5/8 in the cardiac NCC were quantified using Image J thanks to a mask established on the GFP channel. Distances between cardiac NCC and between cardiac NCC and the endocardium, as well as the angle of the major axis of NCC to an axis linking the Ao and Pa poles of the endocardium were measured using Metamorph (Molecular Devices) and a home-made algorithm on Excel. Statistical analysis was performed with the Student’s t-test or Mann-Whitney test depending on normality. The analysis was performed using Prism Software (GraphPad). Statistical significance is represented as follows: ***p<0.001. All results are shown as mean ± standard deviation.
Plasmids
Dullard was directly cloned from an NIH 3T3 mRNA library using the SuperScript III One‐Step RT‐PCR System (Life Technologies) using primers Dull_FL_C1_FW 5 ´‐GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGATGCGGACGCAGTGT‐3’ and Dull_FL_C1_Rev 5 ´‐GGGGACCACTTTGTACAAGAAAGTGGGTCTCACCAGAGCCTATGTTGGTG‐3’ for N‐terminal tag destination vectors (pDONR221). For phosphatase‐null point mutant Dullard D67E, site‐directed mutagenesis was performed by PCR amplification of pDONR 221 Dullard full‐length vector using primers Dull_D67E_FW 5 ´‐GATCCTGGTGCTGGAACTGGACGAAACCCTG‐3’ and Dull_D67E_Rev 5 ´‐CAGGGTTTCGTCCAGTTCCAGCACCAGGATC‐3’ followed by DpnI endonuclease mediated digestion of the parent (methylated) DNA chain. After sequence confirmation, entry vectors were recombined with pEGFP GW C1 for N‐terminal (Life Technologies) GFP‐tag fusion proteins using the Gateway system (Thermofisher).
Cell culture and transfection
C2C12 cells were cultivated at 37°C/5% CO2, in growth medium (DMEM, 4.5 g/L, D-glucose, 4 mM L-glutamine, 1 mM sodium pyruvate, 10% fetal calf serum). Plasmid transfection was performed using Lipofectamine 2000 (Life Technologies). 24 hr after transfection, BMP2 recombinant human protein (Thermo Fisher Scientific, #PHC7145) was applied for 1 hr on cells. Cells were fixed with 4% PFA and immunostained with the phospho-Smad1/5/8 antibody. Alternatively, cells were washed in PBS, collected with PBS 1% SDS and passed through Qiashredder columns (Qiagen) to disrupt nucleic acids. Proteins extracts were then processed for western blotting using pre-cast gels (Life Technologies) and transferred on nitrocellulose by semi-dry transfer (Bio-Rad). C2C12 (ATCC CRL-1772) is a murine myogenic cell line. Its myogenic profile is regularly checked by placing them in low serum to trigger the formation of myotubes through cell-cell fusion. Their mycoplasma contamination status resulted negative.
Tissue dissociation and FACS sorting
Mouse embryos were collected at E11.5 and placed in HBSS/1% FBS (HBSS +/+, Invitrogen) during genotyping. OFT were micro-dissected and dissociated by 15 min incubation in collagenase (0.1 mg/ml in HBSS, C2139 Sigma) and thorough pipetting. HBSS (10% FBS) was added to the cells medium to stop the enzymatic reaction. OFT cell suspensions were centrifuged and resuspended in HBSS (1% FBS) before immunostaining. The panel of conjugated antibodies used for FACS included Ter119 Pe-Cy7 (Erythroid Cells, anti-mouse, Sony, Catalog No. 1181110, 1/300), CD45 APC-Cy7 (Rat anti-mouse, BD Pharmingen, Catalog No. 557659, 1/300), CD31 APC (Rat anti-mouse, BD Pharmingen, Clone MEC 13.3 Catalog No. 561814, 1/300) diluted in HBSS (1% FBS). Cells were centrifuged and resuspended in the antibody solution for a 25 min incubation period (4°C, dark), washed three times, filtered (Fisher cell strainer, 70 μm mesh) and 7AAD PE-Cy7 (1/800) was added in the cells suspension to exclude dead cells. Cells were sorted in a BD FACSAria III into 96-well plates loaded with RT-STA reaction mix (CellsDirect One-Step qRTPCR Kit, Invitrogen) and 0.2x specific TaqMan Assay mix (see Supplementary file 1 for assays list).
Single-cell gene expression
We proceeded as described in Valente et al. (2019). Cells were sorted in RT-STA reaction mix from the CellsDirect One-Step qRT-PCR Kit (Life Technologies), reverse transcribed and specific target pre-amplified (20 cycles), according to the manufacturer’s procedures. Pre-amplified samples were diluted 5x with low EDTA TE buffer prior to qPCR analysis using 48.48 Dynamic Array IFCs and the BioMark TM HD System (Fluidigm). The same TaqMan gene expression assays (20x, Life Technologies) were individually diluted 1:1 with 2x assay loading reagent (Fluidigm). Pre-amplified samples were combined with TaqMan Universal Master Mix (Life Technologies) and 20x GE sample loading reagent (Fluidigm). Loading of the 48.48 Dynamic Array TM IFCs and qPCR cycling conditions followed the Fluidigm procedure for TaqMan gene expression assays.
Bioinformatic analysis
The analytic framework used followed the one described in Perchet et al. (2018); Valente et al. (2019). It included notably a normalization of all the cycle threshold (Ct) values extracted from the Biomark chips using the mean value of Actb and Gapdh housekeeping genes (Figure 4B). For visualisation of the single-cell multiplex qPCR, done on 44 genes, we generated a heatmap using the pHeatmap (v1.0.10) R package (v3.2.2). For unsupervised clustering, we used PhenoGraph that takes as input a matrix of N single-cell measurements and partitions them into subpopulations by clustering a graph that represents their phenotypic similarity. PhenoGraph builds this graph in two steps. Firstly, it finds the k nearest neighbors for each cell (using Euclidean distance), resulting in N sets of k-neighborhoods. Secondly, it operates on these sets to build a weighted graph such that the weight between nodes scales with the number of neighbors they share. The Louvain community detection method is then used to find a partition of the graph that maximizes modularity. Given a dataset of N d-dimensional vectors, M distinct classes, and a vector providing the class labels for the first L samples, the PhenoGraph classifier assigns labels to the remaining N_L unlabeled vectors. Firstly, a graph is constructed as described above. The classification problem then corresponds to the probability that a random walk originating at unlabeled node x will first reach a labeled node from each of the M classes. This defines an M-dimensional probability distribution for each node x that records its affinity for each class. PhenoGraph is implemented as Rphenograph (v0.99.1) R package (v3.2.3).
We used boxplot for gene expressions of clusters obtained with PhenoGraph algorithm, from package ggplot2 (v3.1.0) R package (v3.2.2). For visualisation and pathway analyses (tree), Destiny (v2.6.1) on R package (v3.6) generates the diffusion maps. Destiny calculates cell-to-cell transition probabilities based on a Gaussian kernel with a width σ to create a sparse transition probability matrix M. For σ, Destiny employs an estimation heuristic to derive this parameter. Destiny allows for the visualization of hundreds of thousands of cells by only using distances to the k nearest neighbors of each cell for the estimation of M. An eigen decomposition is performed on M after density normalisation, considering only transition probabilities between different cells. The resulting data-structure contains the eigenvectors with decreasing eigenvalues as numbered diffusion components (DC), the input parameters and a reference to the data. These DC are pseudotimes identifying differentiation dynamics from our sc-qPCR data.
Impact of gene expressions creating the different dimensions is represented as horizontal box plots, showing cells up- or down-regulating indicated genes in the plot.
Ultrasound imaging
Pregnant mice were anaesthetised (3% isoflurane in air and maintained at 1.5%), installed on a heating pad and monitored for respiration frequency, ECG and temperature (Vevo Imaging Station, Visualsonics). Pregnant mice were intraperitoneally injected with Metacam (1mg/kg body weight; Boehringer Ingelheim). A laparotomy was then performed and the uterine horns were gently exteriorized to allow direct visualisation of embryos using a high-resolution ultrasound imaging scanner (VEVO2100, Visualsonics) equipped with a 60 MHz probe (MS-700). To ensure contact between the ultrasound probe and the embryos, a warm sterile gel (Aquasonic) was used. Long axis view of the heart was performed on each embryo to measure left and right ventricle dimensions. PW Doppler measurements were achieved positioning the caliper on the descending abdominal aorta with embryo in sagittal view. From Abdominal Aorta the following measurements were performed: Abdominal Aorta Peak Systolic Velocity (AA PSV, mm/s), End Diastolic Velocity (AA EDV, mm/s), Velocity Time Integral of mean velocities (AA VTI, ms). From these measurements, the following parameters were calculated: Mean Velocity (AA VTI Mean Vel), Abdominal Aorta Pulsatility Index (AA PI), Abdominal Aorta Resistive Index (AA RI).
Acknowledgements
We thank the Cadot, Bitoun and Ribes Laboratories for discussions, Sigolène Meilhac for her help on understanding heart development concepts, Edgar Gomes laboratories, Isabelle Le Roux and Pascale Gilardi-Hebenstreit for useful comments, Stéphane Zaffran for the Sema3c ISH probe and Morgane Belle for Lightsheet microscopy. The Pax3Cre and Rosa26mTmG transgenic lines were kindly provided by F Relaix, and the Wnt1Cre line by A Pierani. This work was supported by Agence Nationale pour la Recherche (ANR-14-CE09-0006-04) to BC; Association Institut de Myologie to BC. VR is an INSERM researcher, and work in her lab is supported by CNRS/INSERM ATIP-AVENIR program, as well as by a Ligue Nationale Contre le Cancer grant (PREAC2016.LCC). MV has a postdoctoral fellowship from the Laboratoire d'Excellence Revive (Investissement d'Avenir; ANR-10-LABX-73). In vivo imaging was performed at the Life Imaging Facility of Paris Descartes University (Plateforme Imageries du Vivant – PIV) and is supported by France Life Imaging (grant ANR-11-INBS-0006).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Vanessa Ribes, Email: vanessa.ribes@ijm.fr.
Bruno Cadot, Email: cadotbruno@gmail.com.
Richard P Harvey, Victor Chang Cardiac Research Institute, Australia.
Marianne E Bronner, California Institute of Technology, United States.
Funding Information
This paper was supported by the following grants:
Agence Nationale de la Recherche ANR-14-CE09-0006-04 to Bruno Cadot.
Ligue Contre le Cancer PREAC2016.LCC to Vanessa Ribes.
Agence Nationale de la Recherche ANR-10-LABX-73 to Mariana Valente.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Data curation, Formal analysis, Investigation, Visualization, Methodology.
Data curation, Investigation, Visualization, Methodology.
Data curation, Formal analysis, Validation, Investigation, Visualization.
Investigation.
Formal analysis, Validation, Investigation.
Resources.
Resources.
Investigation.
Investigation.
Conceptualization, Data curation, Supervision, Validation, Investigation, Visualization, Methodology, Project administration.
Conceptualization, Resources, Data curation, Supervision, Funding acquisition, Validation, Investigation, Visualization, Methodology, Project administration.
Ethics
Animal experimentation: All animal experiments (APAFIS#4163-2016042809186990) were approved by the Animal Ethics Committee of Sorbonne University (Permit Number: A751320).
Additional files
Data availability
All data generated or analysed during this study are included in the manuscript and supporting files.
References
- Acharya A, Baek ST, Banfi S, Eskiocak B, Tallquist MD. Efficient inducible Cre-mediated recombination in Tcf21 cell lineages in the heart and kidney. Genesis. 2011;49:870–877. doi: 10.1002/dvg.20750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajolle F, Zaffran S, Kelly RG, Hadchouel J, Bonnet D, Brown NA, Buckingham ME. Rotation of the myocardial wall of the outflow tract is implicated in the normal positioning of the great arteries. Circulation Research. 2006;98:421–428. doi: 10.1161/01.RES.0000202800.85341.6e. [DOI] [PubMed] [Google Scholar]
- Belle M, Godefroy D, Couly G, Malone SA, Collier F, Giacobini P, Chédotal A. Tridimensional visualization and analysis of early human development. Cell. 2017;169:161–173. doi: 10.1016/j.cell.2017.03.008. [DOI] [PubMed] [Google Scholar]
- Bier E, De Robertis EM. EMBRYO DEVELOPMENT. BMP gradients: a paradigm for morphogen-mediated developmental patterning. Science. 2015;348:aaa5838. doi: 10.1126/science.aaa5838. [DOI] [PubMed] [Google Scholar]
- Bockman DE, Redmond ME, Waldo K, Davis H, Kirby ML. Effect of neural crest ablation on development of the heart and arch arteries in the chick. American Journal of Anatomy. 1987;180:332–341. doi: 10.1002/aja.1001800403. [DOI] [PubMed] [Google Scholar]
- Brade T, Männer J, Kühl M. The role of wnt signalling in cardiac development and tissue remodelling in the mature heart. Cardiovascular Research. 2006;72:198–209. doi: 10.1016/j.cardiores.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Brickner ME, Hillis LD, Lange RA. Congenital heart disease in adults. New England Journal of Medicine. 2000;342:256–263. doi: 10.1056/NEJM200001273420407. [DOI] [PubMed] [Google Scholar]
- Brown CB, Feiner L, Lu MM, Li J, Ma X, Webber AL, Jia L, Raper JA, Epstein JA. PlexinA2 and semaphorin signaling during cardiac neural crest development. Development. 2001;128:3071–3080. doi: 10.1242/dev.128.16.3071. [DOI] [PubMed] [Google Scholar]
- Bruce DL, Sapkota GP. Phosphatases in SMAD regulation. FEBS Letters. 2012;586:1897–1905. doi: 10.1016/j.febslet.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Choi M, Stottmann RW, Yang YP, Meyers EN, Klingensmith J. The bone morphogenetic protein antagonist noggin regulates mammalian cardiac morphogenesis. Circulation Research. 2007;100:220–228. doi: 10.1161/01.RES.0000257780.60484.6a. [DOI] [PubMed] [Google Scholar]
- Chotteau-Lelièvre A, Dollé P, Gofflot F. Expression analysis of murine genes using in situ hybridization with radioactive and nonradioactively labeled RNA probes. Methods Mol Biol. . 2006;326:61–87. doi: 10.1385/1-59745-007-3:61. [DOI] [PubMed] [Google Scholar]
- Danesh SM, Villasenor A, Chong D, Soukup C, Cleaver O. BMP and BMP receptor expression during murine organogenesis. Gene Expression Patterns. 2009;9:255–265. doi: 10.1016/j.gep.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of cre recombinase. Current Biology. 1998;8:1323–1326. doi: 10.1016/S0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Delloye-Bourgeois C, Bertin L, Thoinet K, Jarrosson L, Kindbeiter K, Buffet T, Tauszig-Delamasure S, Bozon M, Marabelle A, Combaret V, Bergeron C, Derrington E, Castellani V. Microenvironment-Driven shift of cohesion/Detachment balance within tumors induces a switch toward metastasis in neuroblastoma. Cancer Cell. 2017;32:427–443. doi: 10.1016/j.ccell.2017.09.006. [DOI] [PubMed] [Google Scholar]
- Engleka KA, Gitler AD, Zhang M, Zhou DD, High FA, Epstein JA. Insertion of cre into the Pax3 locus creates a new allele of splotch and identifies unexpected Pax3 derivatives. Developmental Biology. 2005;280:396–406. doi: 10.1016/j.ydbio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Feiner L, Webber AL, Brown CB, Lu MM, Jia L, Feinstein P, Mombaerts P, Epstein JA, Raper JA. Targeted disruption of semaphorin 3C leads to persistent truncus arteriosus and aortic arch interruption. Development. 2001;128:3061–3070. doi: 10.1242/dev.128.16.3061. [DOI] [PubMed] [Google Scholar]
- Galvin KM, Donovan MJ, Lynch CA, Meyer RI, Paul RJ, Lorenz JN, Fairchild-Huntress V, Dixon KL, Dunmore JH, Gimbrone MA, Falb D, Huszar D. A role for Smad6 in development and homeostasis of the cardiovascular system. Nature Genetics. 2000;24:171–174. doi: 10.1038/72835. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan S, Comai G, Sambasivan R, Francou A, Kelly RG, Tajbakhsh S. A cranial mesoderm origin for esophagus striated muscles. Developmental Cell. 2015;34:694–704. doi: 10.1016/j.devcel.2015.07.003. [DOI] [PubMed] [Google Scholar]
- Hayata T, Ezura Y, Yoichi E, Asashima M, Nishinakamura R, Noda M. Dullard/Ctdnep1 regulates endochondral ossification via suppression of TGF-β signaling. Journal of Bone and Mineral Research. 2015;30:318–329. doi: 10.1002/jbmr.2343. [DOI] [PubMed] [Google Scholar]
- High FA, Zhang M, Proweller A, Tu L, Parmacek MS, Pear WS, Epstein JA. An essential role for notch in neural crest during cardiovascular development and smooth muscle differentiation. Journal of Clinical Investigation. 2007;117:353–363. doi: 10.1172/JCI30070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Cheng L, Li J, Chen M, Zhou D, Lu MM, Proweller A, Epstein JA, Parmacek MS. Myocardin regulates expression of contractile genes in smooth muscle cells and is required for closure of the ductus arteriosus in mice. Journal of Clinical Investigation. 2008;118:515–525. doi: 10.1172/JCI33304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Q, McDill BW, Li SZ, Deng C, Chang CP, Chen F. Smad signaling in the neural crest regulates cardiac outflow tract remodeling through cell autonomous and non-cell autonomous effects. Developmental Biology. 2007;311:172–184. doi: 10.1016/j.ydbio.2007.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Jiao K, Kulessa H, Tompkins K, Zhou Y, Batts L, Baldwin HS, Hogan BL. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes & Development. 2003;17:2362–2367. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Lyons KM, Hogan BL. Involvement of bone morphogenetic Protein-4 (BMP-4) and Vgr-1 in morphogenesis and neurogenesis in the mouse. Development. 1991;111:531–542. doi: 10.1242/dev.111.2.531. [DOI] [PubMed] [Google Scholar]
- Kelly RG. The second heart field. Current Topics in Developmental Biology. 2012;100:33–65. doi: 10.1016/B978-0-12-387786-4.00002-6. [DOI] [PubMed] [Google Scholar]
- Keyte A, Hutson MR. The neural crest in cardiac congenital anomalies. Differentiation. 2012;84:25–40. doi: 10.1016/j.diff.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Jackson TR, Davidson LA. On the role of mechanics in driving mesenchymal-to-epithelial transitions. Seminars in Cell & Developmental Biology. 2017;67:113–122. doi: 10.1016/j.semcdb.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodo K, Shibata S, Miyagawa-Tomita S, Ong SG, Takahashi H, Kume T, Okano H, Matsuoka R, Yamagishi H. Regulation of Sema3c and the interaction between cardiac neural crest and second heart field during outflow tract development. Scientific Reports. 2017;7:6771. doi: 10.1038/s41598-017-06964-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepore JJ, Mericko PA, Cheng L, Lu MM, Morrisey EE, Parmacek MS. GATA-6 regulates semaphorin 3C and is required in cardiac neural crest for cardiovascular morphogenesis. Journal of Clinical Investigation. 2006;116:929–939. doi: 10.1172/JCI27363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Selever J, Wang D, Lu MF, Moses KA, Schwartz RJ, Martin JF. Bmp4 signaling is required for outflow-tract septation and branchial-arch artery remodeling. PNAS. 2004;101:4489–4494. doi: 10.1073/pnas.0308466101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, High FA, Epstein JA, Radice GL. N-cadherin is required for neural crest remodeling of the cardiac outflow tract. Developmental Biology. 2006;299:517–528. doi: 10.1016/j.ydbio.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulley DJ, Kang JO, Martin JF, Black BL. BMP4 is required in the anterior heart field and its derivatives for endocardial cushion remodeling, outflow tract septation, and semilunar valve development. Developmental Dynamics. 2008;237:3200–3209. doi: 10.1002/dvdy.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifflin JJ, Dupuis LE, Alcala NE, Russell LG, Kern CB. Intercalated cushion cells within the cardiac outflow tract are derived from the myocardial troponin T type 2 (Tnnt2) Cre lineage. Developmental Dynamics. 2018;247:1005–1017. doi: 10.1002/dvdy.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Maeda S, Imamura T. BMP receptor signaling: transcriptional targets, regulation of signals, and signaling cross-talk. Cytokine & Growth Factor Reviews. 2005;16:251–263. doi: 10.1016/j.cytogfr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Morgenthau A, Frishman WH. Genetic origins of tetralogy of fallot. Cardiology in Review. 2018;26:86–92. doi: 10.1097/CRD.0000000000000170. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Neeb Z, Lajiness JD, Bolanis E, Conway SJ. Cardiac outflow tract anomalies. Wiley Interdisciplinary Reviews: Developmental Biology. 2013;2:499–530. doi: 10.1002/wdev.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura-Kitabayashi A, Phoon CK, Kishigami S, Rosenthal J, Yamauchi Y, Abe K, Yamamura K, Samtani R, Lo CW, Mishina Y. Outflow tract cushions perform a critical valve-like function in the early embryonic heart requiring BMPRIA-mediated signaling in cardiac neural crest. American Journal of Physiology-Heart and Circulatory Physiology. 2009;297:H1617–H1628. doi: 10.1152/ajpheart.00304.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris RA, Moreno-Rodriguez RA, Sugi Y, Hoffman S, Amos J, Hart MM, Potts JD, Goodwin RL, Markwald RR. Periostin regulates atrioventricular valve maturation. Developmental Biology. 2008;316:200–213. doi: 10.1016/j.ydbio.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odelin G, Faure E, Coulpier F, Di Bonito M, Bajolle F, Studer M, Avierinos J-F, Charnay P, Topilko P, Zaffran S. Krox20 defines a subpopulation of cardiac neural crest cells contributing to arterial valves and bicuspid aortic valve. Development. 2018;145:dev151944. doi: 10.1242/dev.151944. [DOI] [PubMed] [Google Scholar]
- Perchet T, Petit M, Banchi EG, Meunier S, Cumano A, Golub R. The notch signaling pathway is balancing type 1 innate lymphoid cell immune functions. Frontiers in Immunology. 2018;9:1252. doi: 10.3389/fimmu.2018.01252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips HM, Mahendran P, Singh E, Anderson RH, Chaudhry B, Henderson DJ. Neural crest cells are required for correct positioning of the developing outflow cushions and pattern the arterial valve leaflets. Cardiovascular Research. 2013;99:452–460. doi: 10.1093/cvr/cvt132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plein A, Fantin A, Ruhrberg C. Current Topics in Developmental Biology. Trainor PA: Academic Press; 2015. Chapter Six - Neural Crest Cells in CardiovascularDevelopment; pp. 183–200. [DOI] [PubMed] [Google Scholar]
- Sakaguchi M, Sharmin S, Taguchi A, Ohmori T, Fujimura S, Abe T, Kiyonari H, Komatsu Y, Mishina Y, Asashima M, Araki E, Nishinakamura R. The phosphatase dullard negatively regulates BMP signalling and is essential for nephron maintenance after birth. Nature Communications. 2013;4:1398. doi: 10.1038/ncomms2408. [DOI] [PubMed] [Google Scholar]
- Sardi J, Bener MB, Simao T, Descoteaux AE, Slepchenko BM, Inaba M. Mad dephosphorylation at the nuclear envelope is essential for asymmetric stem cell division. bioRxiv. 2019 doi: 10.1101/798116. [DOI] [PMC free article] [PubMed]
- Satow R, Kurisaki A, Chan TC, Hamazaki TS, Asashima M. Dullard promotes degradation and dephosphorylation of BMP receptors and is required for neural induction. Developmental Cell. 2006;11:763–774. doi: 10.1016/j.devcel.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stottmann RW, Choi M, Mishina Y, Meyers EN, Klingensmith J. BMP receptor IA is required in mammalian neural crest cells for development of the cardiac outflow tract and ventricular myocardium. Development. 2004;131:2205–2218. doi: 10.1242/dev.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagnone L. Emerging role of semaphorins as major regulatory signals and potential therapeutic targets in cancer. Cancer Cell. 2012;22:145–152. doi: 10.1016/j.ccr.2012.06.031. [DOI] [PubMed] [Google Scholar]
- Tanaka SS, Nakane A, Yamaguchi YL, Terabayashi T, Abe T, Nakao K, Asashima M, Steiner KA, Tam PP, Nishinakamura R. Dullard/Ctdnep1 modulates WNT signalling activity for the formation of primordial germ cells in the mouse embryo. PLOS ONE. 2013;8:e57428. doi: 10.1371/journal.pone.0057428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Snider P, Firulli AB, Conway SJ. Trigenic neural crest-restricted Smad7 over-expression results in congenital craniofacial and cardiovascular defects. Developmental Biology. 2010;344:233–247. doi: 10.1016/j.ydbio.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku T, Yoshida J, Sugimoto T, Yamamoto M, Makino N, Takamatsu H, Takegahara N, Suto F, Hori M, Fujisawa H, Kumanogoh A, Kikutani H. Repulsive and attractive semaphorins cooperate to direct the navigation of cardiac neural crest cells. Developmental Biology. 2008;321:251–262. doi: 10.1016/j.ydbio.2008.06.028. [DOI] [PubMed] [Google Scholar]
- Urrutia H, Aleman A, Eivers E. Drosophila dullard functions as a mad phosphatase to terminate BMP signaling. Scientific Reports. 2016;6:32269. doi: 10.1038/srep32269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente M, Resende TP, Nascimento DS, Burlen-Defranoux O, Soares-da-Silva F, Dupont B, Cumano A, Pinto-do-Ó P. Mouse HSA+ immature cardiomyocytes persist in the adult heart and expand after ischemic injury. PLOS Biology. 2019;17:e3000335. doi: 10.1371/journal.pbio.3000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldo K, Miyagawa-Tomita S, Kumiski D, Kirby ML. Cardiac neural crest cells provide new insight into septation of the cardiac outflow tract: aortic sac to ventricular septal closure. Developmental Biology. 1998;196:129–144. doi: 10.1006/dbio.1998.8860. [DOI] [PubMed] [Google Scholar]
- Wu X, Northcott PA, Croul S, Taylor MD. Mouse models of medulloblastoma. Chinese Journal of Cancer. 2011;30:442–449. doi: 10.5732/cjc.011.10040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chang JY, Huang Y, Lin X, Luo Y, Schwartz RJ, Martin JF, Wang F. The FGF-BMP signaling axis regulates outflow tract valve primordium formation by promoting cushion neural crest cell differentiation. Circulation Research. 2010;107:1209–1219. doi: 10.1161/CIRCRESAHA.110.225318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Wang J, Guo C, Chang W, Zhuang J, Zhu P, Li X. Temporally distinct Six2-Positive second heart field progenitors regulate mammalian heart development and disease. Cell Reports. 2017;18:1019–1032. doi: 10.1016/j.celrep.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]