Abstract
In eukaryotic cells, ribonucleoproteins (RNPs) form mesoscale condensates by liquid-liquid phase separation that play essential roles in subcellular dynamic compartmentalization. The formation and dissolution of many RNP condensates are finely dependent on the RNA-to-RNP ratio, giving rise to a window-like phase separation behavior. This is commonly referred to as reentrant liquid condensation (RLC). Here, using ribonucleoprotein-inspired polypeptides with low-complexity RNA-binding sequences as well as an archetypal disordered RNP, Fused in Sarcoma (FUS), as model systems, we investigate the molecular driving forces underlying this non-monotonous phase transition. We show that an interplay between short-range cation-π attractions and long-range electrostatic forces governs the heterotypic RLC behavior of RNP-RNA complexes. Short-range attractions, which can be encoded by both polypeptide chain primary sequence and nucleic acid base sequence, control the two-phase coexistence regime, regulate material properties of polypeptide-RNA condensates, and oppose condensate reentrant dissolution. In the presence of excess RNA, a competition between short-range attraction and long-range electrostatic repulsion drives the formation of a colloid-like cluster phase. With increasing short-range attraction, the fluid dynamics of the cluster phase is arrested, leading to the formation of a colloidal gel. Our results reveal that phase behavior, supramolecular organization, and material states of RNP-RNA assemblies are controlled by a dynamic interplay between molecular interactions at different length scales.
Keywords: Membraneless organelle, optical tweezer, complex coacervate, soft colloids, biomolecular condensate, sequence-encoded viscoelastic properties
Graphical Abstract
Introduction
Ribonucleoproteins (RNPs) form a diverse set of biomolecular condensates in eukaryotic cells by liquid-liquid phase separation (LLPS) 1. These condensates are non-membranous assemblies that can dynamically exchange their components with surrounding subcellular environment 2-5. From a physiologic point of view, RNP granules perform critical cellular functions that are conserved from yeast to humans 2, 6. From a pathological point of view, these granules enrich many disease-linked proteins such as FUS, hnRNPA1/A2, and TDP43 7-8, aggregation of which is often associated with multiple degenerative disease conditions 9.
Many nuclear RNPs spontaneously self-assemble into liquid droplets in vitro at their physiological concentrations 8. However, they predominantly exist in a homogeneous solution phase in cells 8, 10-11. This is attributed to the buffering effect of cellular RNAs 10. Recent in vitro studies have demonstrated that RNAs can inhibit RNP phase separation when present at high RNA-to-RNP ratios but promote RNP phase separation at low RNA-to-RNP ratios 12. The observed regulatory effect of RNAs is thought to be manifested via promiscuous RNA binding to evolutionarily conserved positively charged Arg/Gly-rich low-complexity domains (R/G-rich LCDs) 10, 12-13. R/G-rich LCDs represent a class of RNA interaction motifs that are ubiquitous in the eukaryotic RNPs 14-15. The RNA-dependent RNP condensation and subsequent decondensation can be explained by the reentrant liquid condensation (RLC) model for associative phase separation 12.
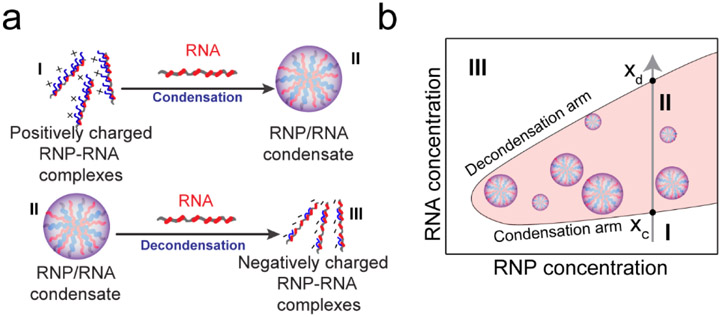
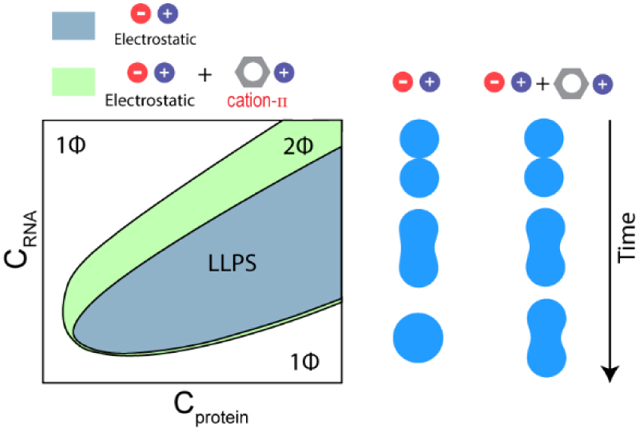
According to the RLC model, the condensation is driven by electrostatic attraction between positively charged R/G-rich LCDs and negatively charged RNA chains. The decondensation process, however, is best explained by a charge inversion on the surface of RNA or RNP chains. In the charge inverted state, the surface of a charged macromolecule is over-screened by oppositely charged ions, i.e., a greater number of counter-ions accumulate on the macromolecule than what is required to reach a charge-neutral state (Fig. 1a) 16. With excess bound counter-ions, charge inversion triggers an energetically unfavorable long-range coulomb repulsion, which drives the decondensation process. Consequently, a reentrant phase transition can be quantified using three distinct yet inter-linked parameters: (a) the condensation concentration (xc), (b) the decondensation concentration (xd), and (c) the mixture composition at which phase separation is most favorable (Fig. 1b) 16-18. When macromolecules interact only through screened coulomb forces, both condensation and decondensation boundaries are primarily determined by the linear charge densities and the number of charged residues in polycation (i.e., R/G-rich LCD) and polyanion (i.e., RNA) 17. Although this framework qualitatively recapitulates the reentrant nature of RNP-RNA condensation 12, its application to naturally occurring charged biological macromolecules remains limited, since the RLC model ignores intermolecular short-range forces which can be sequence-specific and are ubiquitously utilized by natural systems.
Figure 1: Reentrant liquid condensation (RLC) of RNP-RNA complexes.
a) A schematic representation of RLC showing distinct molecular species at three different RNP:RNA regimes. b) A corresponding phase diagram showing a window-like phase separation regime (shaded region-regime II). The condensation and decondensation arms are labeled. Grey arrow represents passage of the system through three distinct phases of peptide-RNA complexes when RNA concentration is progressively increased at a fixed peptide concentration. xc: condensation point; xd: decondensation point.
Short-range interactions are a critical segment of the biomolecular forces. In multiple recent reports, it was shown that short-range forces are vital in controlling the RNP-RNA complexation 7, 19-20. Given the regulatory roles of short-range interactions in driving RNP LLPS, here we ask how sequence-encoded short-range interactions, such as the cation-π attraction, impact the reentrant liquid condensation of RNP-RNA mixtures. By measuring phase diagrams of positively charged LCD-RNA systems and the fluid dynamics of their condensates using correlative fluorescence microscopy and optical tweezers, we show that combined intermolecular interactions on different length scales govern the phase behavior and material properties of RNP-RNA assemblies. Based on our experimental findings and theoretical modeling, we propose that long-range ionic interactions drive the overall reentrant phase behavior of RNP-RNA complexes, while sequence-specific short-range attractions dictate the width of the phase separation regime as well as the viscoelastic states of their condensates.
Results and Discussion
Arginine-rich and lysine-rich low-complexity domains (LCDs) produce distinct reentrant liquid condensation behavior
Polycationic R/G-rich LCDs are prevalent in eukaryotic RNA-binding proteome 14-15. R/G-rich LCDs promiscuously interact with diverse RNA sequences 13, 15, 21, but their role in RNP function remains relatively uncharacterized. We and others previously reported that the R/G-rich LCDs can act as a phase separation module via multivalent long-range electrostatic interactions with RNAs 12-13, 22. To investigate the roles of Arg-mediated short-range interactions in the phase separation of R/G-rich LCD and RNA mixtures, we designed two peptide sequences: [RGRGG]5 and [KGKGG]5 (Fig. 2a). Both polypeptides are estimated to carry the same overall charge (+10e at pH 7.5; Supplementary Table 1; Supplementary Fig. S2) and therefore, are expected to exhibit similar phase behavior with RNA, assuming inter-chain heterotypic ionic interactions are the only driving force.
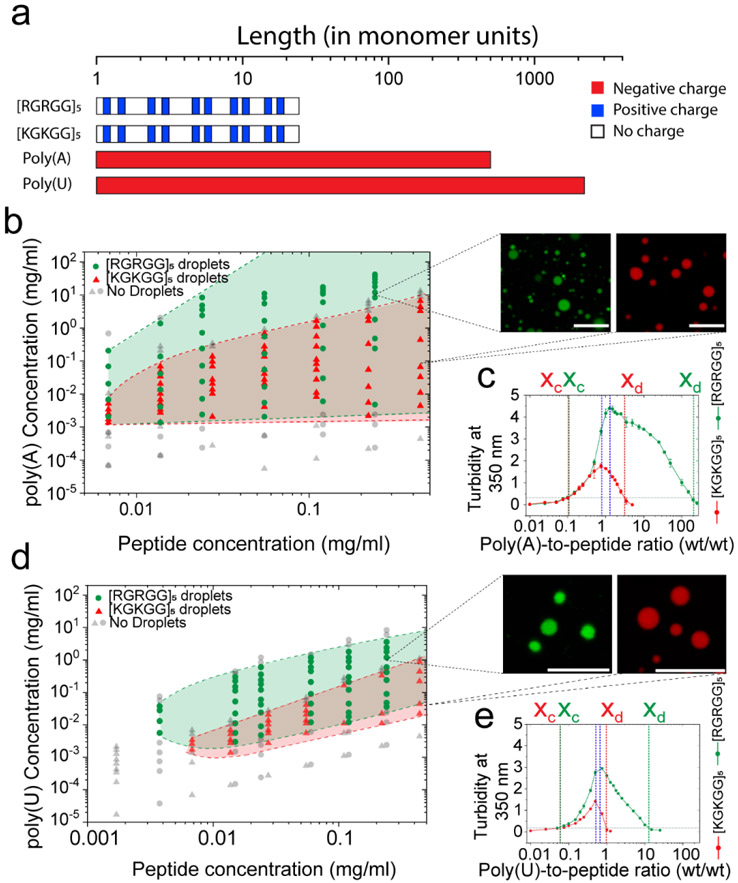
Figure 2: Liquid-liquid phase separation (LLPS) regimes of peptide-RNA complexes are dependent on polypeptide amino acid and RNA nucleobase sequences.
a) An approximate length of homopolymeric RNAs and peptides used in this study (length is provided as the number of monomers in respective biopolymers). b) Experimental phase diagrams of [RGRGG]5 and [KGKGG]5 with poly(A) RNA. The respective LLPS regimes are marked by shaded regions (green for [RGRGG]5; red for [KGKGG]5). Representative confocal fluorescence micrographs of peptide-RNA condensates are shown (pseudo colored) with scale bars of 10 μm. Dashed lines represent phase boundaries. c) Solution turbidity at 350 nm of [RGRGG]5 and [KGKGG]5 with poly(A) mixtures showing that the dissolution of [RGRGG]5 condensates requires ~50-fold higher poly(A) concentration (green) than [KGKGG]5 condensates (red). Blue lines represent the maximum phase separation point xo. d) Phase diagrams of [RGRGG]5 and [KGKGG]5 with poly(U) RNA. The respective LLPS regimes are marked by shaded regions (green for [RGRGG]5; red for [KGKGG]5). Representative confocal fluorescence images of peptide-RNA condensates are shown. Scale bars represent 10 μm. Dotted lines represent phase boundaries. e) Solution turbidity at 350 nm of [RGRGG]5 and [KGKGG]5 with poly(U) mixtures showing that [RGRGG]5 condensates are stable across a ~10-fold higher poly(U) concentration than [KGKGG]5 condensates. Blue lines represent the maximum phase separation point xo. Buffer used: 25 mM Tris-HCl, 20 mM DTT (pH 7.5).
Using a double-ringed homopolymeric RNA, poly(A), we determined phase diagrams of [RGRGG]5 and [KGKGG]5 peptides utilizing solution turbidity and optical microscopy measurements. Our phase diagrams clearly show that both [RGRGG]5 and [KGKGG]5 peptides display reentrant liquid condensation, i.e., peptide-RNA droplets are formed at a relatively low RNA-to-peptide ratio, whereas a homogeneous phase reemerges at higher RNA-to-peptide ratio (Fig. 2b&c). The condensation point (xc) and the isoelectric point (x0) were observed to be minimally altered by Lys-to-Arg substitutions (Fig. 2c). These results are in agreement with the charge inversion mechanism16, given the length and net charge per residue (NCPR) are the same for both peptides (Supplementary Fig. S2). However, the decondensation boundary (xd), which marks the re-appearance of the homogeneous phase, was more than an order of magnitude higher for [RGRGG]5 when compared with [KGKGG]5 (Figs. 2b&c). This results in a substantial widening of the liquid condensation regime (regime II) of the R/G-rich LCD as compared to the corresponding K/G-rich sequence. To probe for the charges of peptide-RNA assemblies across the reentrant phase boundary, we next measured their electrophoretic mobility by phase analysis light scattering (PALS). Electrophoretic light scattering measurements revealed similar charge inversion transitions for both peptide systems (Fig. 3a), suggesting charge inversion may not be sufficient to fully describe their phase behavior. Similar alteration in the decondensation boundary by Lys-to-Arg substitutions were also observed in the peptide-RNA phase diagrams with a single-ringed RNA homopolymer, poly(U). In this case, however, the difference was less pronounced as compared to poly(A) RNA (Figs. 2d&e). These data collectively suggest that despite carrying identical charges, R-rich LCDs and K-rich LCDs encode distinct reentrant phase behavior with RNA.
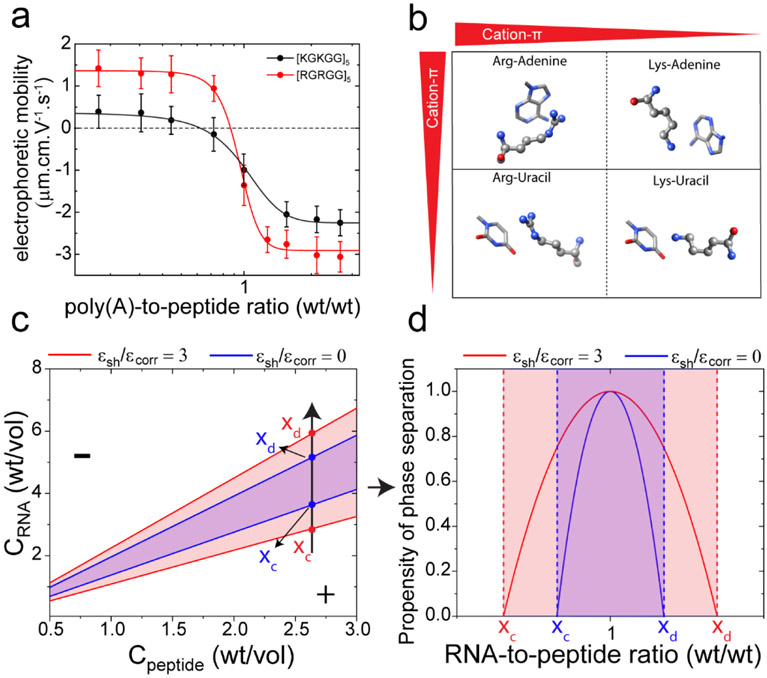
Figure 3: Short-range cation-π attraction widens the LLPS regime of peptide-RNA complexes.
a) Electrophoretic mobility of poly(A)-[RGRGG]5 and poly(A)-[KGKGG]5 complexes as a function of poly(A)-to-peptide ratio. Solid lines represent fits to a two-state model. b) Schematics showing cation-π interactions between Arg/Lys-nucleobases and their relative strengths. c) Simulated phase diagrams showing widening of the LLPS regimes (red vs. blue) due to short-range attraction. ϵcorr is the correlation energy that represents an enthalpic driving force of LLPS in RLC model and ϵsh is the short-range inter-complex attraction. (d) Propensity of phase separation calculated from equation-9 in SI-note 1. The two curves here represent the values of the RNA-to-peptide ratio at which the inter-complex attraction leading to phase separation is favorable. They correspond to the absolute value of the relative energy loss due to phase separation, which is maximal at RNA-to-peptide ratio of 1.0. In regions outside the boundary marked by xc and xd, phase separation is not favorable. Parameters used for generating (c) and (d) are: RNA charge Q=2200, peptide charge q = 10, correlation energy ϵcorr=1000. See SI Note-1 for further details.
Short-range cation-π attraction controls the two-phase coexistence regime of heterotypic peptide-RNA LLPS
Two key aspects of our experimentally determined phase diagrams (Fig. 2b-e) of heterotypic peptide-RNA systems are: (i) the R-rich LCDs encode a much wider regime of LLPS than the K-rich LCDs for a given RNA sequence, and (ii) the decondensation boundary is far more sensitively dependent on the polypeptide sequence than the condensation boundary for a given RNA sequence. In our first attempt to model the peptide-RNA phase behavior, we utilized the RLC theory that considers charge inversion (see SI Note-1 for further details), given both peptide systems show highly cooperative charge inversion transitions with RNA (Fig. 3a). In contrary to our experimental observations, RLC model suggests that both peptides should have identical LLPS regime in their phase diagrams since their charges are the same (Supplementary Fig S3a; SI Note-1). Therefore, we infer that a simple charge inversion model (that only considers ionic interactions 17) is inadequate to explain RNP-RNA condensation/decondensation completely.
To provide a physical basis for this apparent discrepancy between theory and experiments, we next consider whether additional forces are in play that can impact the peptide-RNA complexation. The potential of both arginine and lysine residues to engage in cation-π interactions with an RNA base is well documented in the literature 23-28. Ab initio calculations, crystal structure analyses, and molecular dynamics simulations previously established that the short-range Arg-RNA cation-π interactions are generally stronger than the Lys-RNA cation-π interactions 23-24, 27. Moreover, the delocalized π electrons of arginine guanidinium group can participate in π-π interactions with RNA bases 29-30. In addition to the electrostatic monopoles, arginine, but not lysine, contains higher order multipoles that can mediate weaker and shorter ranged directional interactions with RNA bases 30. Therefore, arginine side chains are capable of mediating a stronger short-range multi-modal interaction network with RNA, as compared to lysine (Fig. 3b) 30. To test this experimentally, we conducted phase separation measurements of [RGRGG]5-poly(U) mixture with increasing ionic strength of the buffer and compared the same with [KGKGG]5-poly(U) mixture (Supplementary Fig. S3b). For condensation mediated by electrostatic and polar interactions, increasing salt concentration should decrease the propensity for LLPS due to screening effect 7. Our salt titration experiments reveal that ~ 3-fold higher concentration of NaCl is required to suppress phase separation of [RGRGG]5 as compared to [KGKGG]5 with poly(U) RNA. This data supports the idea that R-rich LCD-RNA interactions are stronger as compared to K-rich LCD-RNA interactions.
We reasoned that if short-range cation-π interactions between peptide Arg and RNA nucleobases are primarily responsible for the observed differences between the reentrant phase behavior of R-rich and K-rich LCDs, then abolishing the cation-π interactions by removing nucleobases from the RNA chain should result in similar phase boundary curves for the two LCD types. To confirm this, we characterized the RLC behavior of R-rich and K-rich LCDs upon their interactions with polyphosphate (a model for the negatively charged nucleic acid backbone that lacks nucleobases). As expected, both [RGRGG]5 and [KGKGG]5 peptides displayed similar phase separation windows with polyphosphate (Supplementary Fig. S4), supporting the idea that cation-π interactions are indeed responsible for the observed differences between the cationic LCDs with respect to their phase behavior with RNA.
By applying a suitable modification to account for short-range attraction, we next examined the effect of cation-π forces on the phase diagrams for peptide-RNA mixtures using the RLC framework (SI Note-1). This is done by fixing the peptide/RNA charges and introducing a short-range attraction parameter ϵsh between peptide-RNA complexes. The resulting phase diagrams successfully recapitulate the widening of LLPS regime with increasing short-range force (Figs. 3c&d). However, the widening remains symmetric (i.e., both condensation and decondensation arms are symmetrically affected) while our experimental phase diagrams reveal an asymmetric broadening due to Lys-to-Arg substitutions (Fig. 2d-e). Therefore, a simple inclusion of inter-complex short-range attraction in the RLC model is not sufficient to account for the observed phase behavior of R-rich LCD-RNA system as compared with the analogous K-rich LCD-RNA system.
To explain the observed asymmetric effect of cation-π forces, next we consider the distinct microscopic nature of peptide-RNA complexes at either side of the charge neutral point. At peptide excess regime, RNA chains are overcharged with peptide molecules, and are likely to form a brush-like structure with an overall positive charge17 (Supplementary Fig. S5a). In this case, RNA bases are saturated with bound peptides and the possibility of inter-complex cation-π attraction is small. Contrastingly, at RNA excess regime, RNA chains are partially screened by the peptides, and the overall complex becomes negatively charged. In such a case, the exposed RNA bases are more likely to contribute to inter-complex cation-π bonding through multi-modal interactions involving R-rich LCDs. Building on these considerations, we further extend the RLC model (SI Note-1). Our extended model now produced theoretical phase diagrams (Supplementary Fig. S5b) that exhibited similar asymmetric effects as observed in our experiments, i.e., the condensation arm is minimally affected while the decondensation arm is significantly shifted due to peptide-RNA cation-π interactions.
Together, our experiments and phenomenological theory suggest that a competition between the long-range repulsion and short-range cation-π attraction can result in an effective widening of the phase separation window (Fig. 2). It should be noted that even though the exact order of cation-π interaction strengths in our peptide-RNA mixtures are yet to be determined experimentally, previous computational studies nevertheless can provide some key insights 23-28. Firstly, from the peptide sequence perspective, R-rich LCDs are expected to engage in a more pronounced short-range attraction as compared with K-rich LCDs. Secondly, from the ribonucleic acid sequence perspective, two-ringed purine bases are more efficient at forming cation-π bonds as compared with single-ringed pyrimidine bases 23-24, 27 (Fig. 3b). Therefore, [RGRGG]5-poly(A) cation-π interactions are expected to be the strongest and [KGKGG]5-poly(U) interactions will be the weakest amongst our four possible peptide-RNA combinations. Consistent with this argument, we observe that [RGRGG]5-poly(A) condensates resist the reentrant dissolution the most, whereas [KGKGG]5-poly(U) condensates are most easily dissolved by RNA (Supplementary Fig. S6).
Synergy between short-range and long-range forces controls the material properties of peptide-RNA condensates prior to a charge inversion
The material properties of complex fluids are collectively determined by the net strengths of respective intermolecular interaction networks 31-33. The combination of a tunable short-range force and a non-monotonic long-range electrostatic interaction between polypeptides and RNA chains (Figs. 2&3b) provides a unique potential for controlling the material properties of these condensates. We hypothesized that this can be achieved by a systematic variation of polypeptide and RNA sequences as well as their mixture compositions, which tunes short-range and long-range ionic forces, respectively. Before and at the charge neutral state (≤ x0), we expect that short-range cation-π interactions should act synergistically with electrostatic attraction. Assuming the long-range ion-ion interactions are similar in both K/G-rich and R/G-rich peptide systems with a given RNA sequence (Supplementary Fig. S2), we hypothesized that the material properties of peptide-RNA condensates will be determined by the net strengths of short-range attraction between respective polypeptide and RNA chains. Based on our experimentally determined phase diagrams (Fig. 2), we anticipate that R/G-poly(A) droplets will display slower fluid dynamics than K/G-poly(A) droplets. To test this idea, we employed two complementary techniques: Fluorescence Recovery After Photobleaching (FRAP) and controlled fusion of suspended peptide-RNA droplets using a dual-trap optical tweezer. Using FRAP experiments, we probed for the peptide diffusion in the condensed phase. For FRAP experiments, we used ~ 1% fluorescently labeled peptides (labeled by cys-maleimide chemistry-see methods for details) in a mixture with unlabeled peptides. Peptide diffusion was nearly arrested within R/G-poly(A) droplets, whereas peptides within K/G-poly(A) droplets had some degree of mobility (Fig. 4a; Supplementary Fig. S7). Furthermore, a greater degree of fluorescence recovery was observed for R/G-poly(U) droplets as compared to R/G-poly(A) droplets. These data are consistent with the idea that in-droplet peptide mobility at the nanoscale is partly dependent on the inter-molecular friction due to short-range interactions (cation-π forces in our case) with RNA, and with increasing strengths of such interactions, in-droplet diffusivity dynamics become slower 34. The observed trend in FRAP results is also in agreement with the rank order of the energetics of short-range interactions of amino acid-base combinations (Fig. 3b) 23-24, 27.
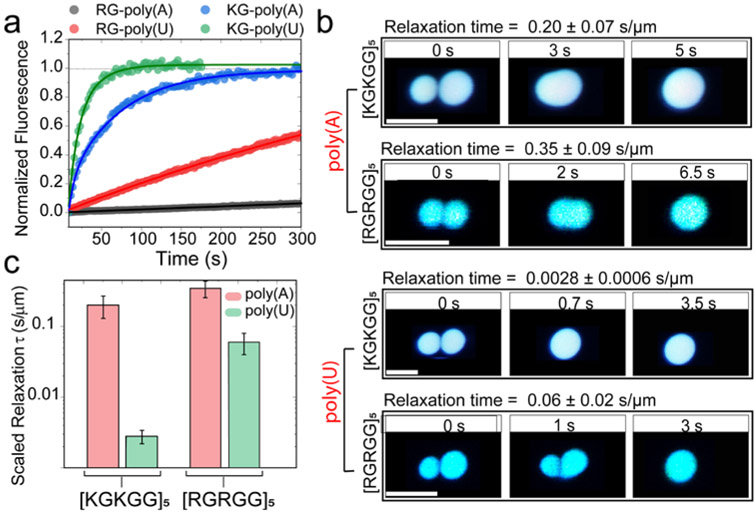
Figure 4: Short-range attraction tunes material states of RNA-peptide condensates.
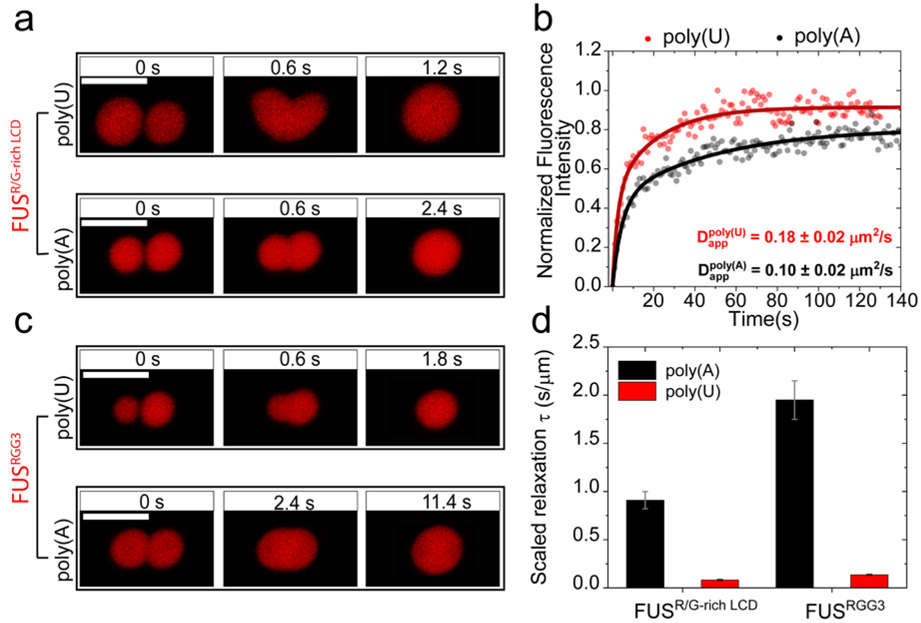
a) Normalized fluorescence recovery after photobleaching (FRAP) intensity time-traces for peptide-RNA systems. The mobile phase fraction (ratio of final intensity to pre-bleach intensity) and recovery time show a scaling with the strength of the short-range attraction between RNA and the peptide. R/G-poly(A) (black): no significant recovery observed with < 5% mobile fraction. KG-poly(U) (green): fastest recovery observed with ~100% mobile fraction. See Supplementary Fig. S7 for corresponding images. b) Time-lapse images of trap-induced fusion of RNA-[RGRGG]5 droplets showing slower fusion relaxation as compared to RNA-[KGKGG]5 droplets for both poly(U) and poly(A) RNAs. Corresponding movies are provided as Supplementary Movies 1-4. c) Relaxation times of the peptide-RNA condensates with poly(U) (green) and poly(A) (red). [KGKGG]5-poly(U) condensates display the fastest relaxation time, whereas [RGRGG]5-poly(A) condensates display the slowest relaxation time. Scale bars in b represent 5 μm. All samples were prepared at 1.0 mg/ml peptide concentration, 0.75 mg/ml RNA concentration (corresponding to the peak in their turbidity plots shown in Figs. 2c&e) in 25 mM Tris-HCl, 20 mM DTT and 25 mM NaCl (pH = 7.5).
Simultaneously, to probe for the dynamics of peptide-RNA condensates at the micron-scale, we performed force-induced droplet coalescence using an optical tweezer system (Figs. 4b&c). In these experiments, two droplets were optically trapped and brought slowly into contact to initiate droplet fusion. The fusion timescale, which provides direct insights into the material state of the condensates 35, is estimated from the force relaxation curves measured by the traps operating at ~12 μs time resolution. Similar to our observation in FRAP experiments, K/G-poly(U) droplets displayed the fastest relaxation time of coalescence (0.0028±0.0006 s/μm), which indicates a high degree of fluidity and low viscosity (Figs. 4a&b), whereas R/G-poly(A) droplets showed more than two orders of magnitude slower relaxation time (Fig. 4c). Together with the FRAP results, the fusion data confirms that a combination of short-range attraction and long-range forces controls the molecular diffusion and the mesoscale fluid dynamics of peptide-RNA condensates. We note that similar observations on the material properties of PR protein-RNA condensates were recently reported by Boeynaems et al.29, which are in agreement with our results reported here.
RNA sequence tunes material states of FUSR/G-rich LCD condensates
Our results using RNP-inspired polypeptide sequences indicate that the dynamics of the phase separated condensates formed by R/G-rich LCDs are finely dependent on RNA-base sequences. We next tested whether this effect is extended to naturally occurring RNA binding proteins with more complex sequence patterns. Using an RNA binding domain construct of FUS [FUSR/G-rich LCD; contains three disordered R/G-rich LCDs and one folded RNA recognition motif (RRM)] (Supplementary Fig. S2d), we employed FRAP and trap-induced fusion experiments to investigate the mesoscale RNP-RNA condensate dynamics (Fig. 5). In presence of poly(A) RNA, FUSR/G-rich LCD condensates showed almost an order of magnitude slower fusion kinetics than condensates formed by poly(U) RNA (Fig. 5a). Simultaneously, FRAP measurements revealed ~ 2-fold decrease in protein diffusion rate within poly(A)-FUSR/G-rich LCD condensates as compared to the poly(U)-FUSR/G-rich LCD droplets (Fig. 5b). Similar trend was also observed for the isolated third RGG box of FUS (FUSRGG3 (472−505); Supplementary Fig. S2c) with more pronounced difference in fusion relaxation timescales for FUSRGG3 condensates with poly(A) and poly(U) RNAs (Fig. 5c&d). These results suggest that RNA base sequence tunes the material states of RNP-RNA condensates through short-range cation-π interactions.
Figure 5: RNA base sequence tunes material states of FUSR/G-rich LCD condensates.
a) Trap-induced fusion time lapse images of poly(U)-FUSR/G-rich LCD droplets and poly(A)-FUSR/G-rich LCD droplets. b) Normalized Fluorescence recovery after photobleaching (FRAP) intensity time-trace for FUSR/G-rich LCD-RNA condensates. c) Trap-induced fusion time lapse images of droplets formed by poly(U) and poly(A) RNA with FUSRGG3, respectively. d) Relaxation times of the FUS-RNA condensates are approximately one order of magnitude higher for poly(A) RNA as compared to poly(U) RNA. Scale bars in a and c represent 5 μm. Corresponding movies for a and c are provided as Supplementary Movies 5 and 6.
Competition between short-range and long-range forces leads to the emergence of a cluster phase at a high RNA-to-RNP ratio
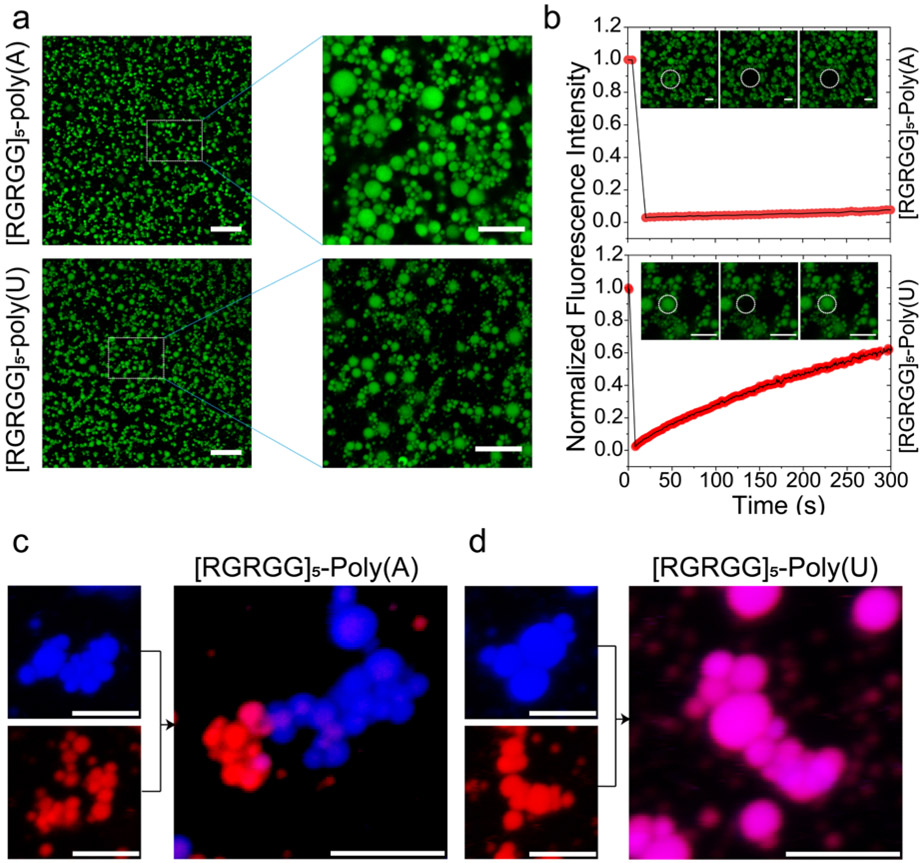
Reentrant liquid condensation of protein-RNA complexes is manifested by a highly non-monotonic electrostatic potential, which is sensitively dependent on the RNA-to-polypeptide ratio (Figs. 1&3a). So far, studies pertaining to electrostatically-driven biological phase separating systems are mostly limited to mixture compositions corresponding to the charge-neutral state (x0), and hence, little is known about the structure and material state of the condensates past the charge inversion point (i.e., at RNA-to-peptide ratios greater than respective x0’s) where a long-range repulsive force operates. In contrary to the charge neutral state for our peptide-RNA condensates, confocal microscopy of peptide-RNA mixtures past respective x0 points reveals the emergence of a cluster phase where small spherical peptide-RNA assemblies aggregated together to form a colloid-like solution that did not coalesce (Fig. 6a; Supplementary Movie 7). This behavior is more pronounced at lower buffer ionic strength (≤ 25 mM Na+) where electrostatic interactions exist over a longer range (i.e., higher Debye length). Using electrophoretic light scattering measurements, we confirmed that these condensates are strongly negatively charged (Fig. 3a; Supplementary Fig. S8), suggesting that the suspension of colloid-like condensates is kinetically stabilized by long-range electrostatic repulsions. This scenario is analogous to colloidal clusters formed by micro-particle suspensions where the colloidal particles interact via a mixed attractive and repulsive potential 36. Under such conditions, combining long-range repulsion with a weaker short-range attraction between colloidal particles in a suspension results in the formation of a dynamic cluster phase, while increasing inter-particle attraction leads to colloidal gel formation with cluster-like morphologies 36-38. Remarkably, we observed that while R/G-poly(U) droplet clusters displayed relatively fast diffusion dynamics (Fig. 6a), R/G-poly(A) droplet clusters showed no FRAP recovery under similar conditions (Fig. 6b). These data suggest a severely restricted in-droplet fluid dynamics for R/G-poly(A) clusters, which is consistent with the stronger Arg-adenine interactions as compared to the Arg-uracil pair.
Figure 6: Long-range ionic repulsion competes with short-range attraction at high RNA-to-peptide ratio leading to a colloid-like cluster phase:
a) Fluorescence micrographs showing cluster formation for [RGRGG]5-poly(A) (top) and [RGRGG]5-poly(U) condensates (bottom). Scale bar represents 20 μm (left) and 10 μm (right). b) RNA sequence tunes the fluid dynamics of [RGRGG]5 clusters showing arrested diffusion in poly(A) clusters and a more dynamic diffusion in poly(U) clusters as evidenced by the fluorescence recovery time traces. Scale bars represent 5 μm for poly(A) images and 4 μm for poly(U) images. Corresponding movies are provided as Supplementary Movies 8 and 9. c) [RGRGG]5-poly(A) condensates can form co-clusters that retain their composition in a complex mixture. d) The [RGRGG]5-poly(U) condensates exchange their content rapidly under similar conditions. Scale bar represents 5 μm for both (c) and (d).
To investigate whether R/G-poly(A) condensates exchange their components within a single cluster, we mixed independently prepared R/G-poly(A) clusters of different colors (using independently labeled peptides with Alexa488 and Alexa594 dyes, respectively). Two-color confocal microscopy imaging revealed that R/G-poly(A) clusters indeed can coexist in a complex mixture and retain the internal composition for an extended period of time (Fig. 6c). Contrastingly, R/G-poly(U) clusters exchange their materials rapidly under identical conditions (Fig. 6d). Weakening the electrostatic and polar interactions by increasing the ionic strength of the buffer, we observed that both R/G-poly(A) and R/G-poly(U) clusters transitions to larger spherical droplets by coalescence (Supplementary Fig. S9). Interestingly, cluster phase was not observed for K/G-poly(A) mixtures under similar conditions, and instead a relatively homogeneous suspension of negatively charged spherical condensates was visible (Supplementary Fig. S10). These results suggest that short-range forces are a critical determinant for the colloid-like cluster formation in peptide-RNA mixtures. Therefore, we conclude that a competition of sequence encoded short-range attraction and coulomb repulsion amongst peptide-RNA complexes can give rise to distinct supramolecular organization of peptide-RNA condensates at higher length-scales.
Conclusion
Electrostatic forces play a fundamental role in driving the formation of biomolecular condensates in the cell 39. Previously, we reported reentrant liquid condensation of R/G-rich LCDs with RNA 12, a phenomenon that can explain the enhanced solubility of prion-like RNA binding proteins in the nucleus (high RNA environment) 10. In this work, we explore how reentrant phase transition is controlled by RNA and polypeptide sequence variations at the molecular level. We demonstrate that intermolecular ionic interactions are important for the reentrant nature of peptide-RNA LLPS, whereas short-range forces stabilize the phase separated complexes against reentrant dissolution at higher RNA-to-polypeptide ratios. Sequence-encoded short-range forces, e.g. cation-π interactions, enable a fine control of the width of the phase separation window. Combined with the long-range ionic forces, these sequence-specific interactions can collectively determine the material states of polypeptide-RNA condensates. Although our current study focuses on the cationic LCD and RNA mixtures, the physical principles presented here are generally applicable to other biological systems (e.g., Alzheimer protein Tau 40) where phase separation is electrostatically driven 41. Additionally, we envision that posttranslational modifications that alter physicochemical properties of cationic amino acid residues can tune the phase behavior and fluid properties of cellular RNP condensates.
In a strict sense, our proposed extended RLC formalism is only applicable for equilibrium systems. However, our material state characterization data (FRAP and fusion kinetics data in Figs. 4&6) clearly show that R/G-poly(A) condensates form gel-like network at high RNA-to-peptide ratio, which may represent a kinetically arrested state. Thus, the onset of gelation may further complicate the application of a simple charge inversion framework. Similar complexities including gel-like network formation are also predicted in a recent theoretical work on reentrant phase behavior 42.
Unlike short-range attractions, electrostatic interactions (attractive vs. repulsive) are non-monotonic and are determined by the overall stoichiometry of respective complexes (Fig. 1a). This offers an intuitive means of tuning the structure and dynamics of the RNA-protein condensates by varying the heterotypic mixture composition. When complexes are neutral, the short-range and long-range forces act synergistically to determine the material properties of RNP-RNA condensates (Figs. 4&5). At higher RNA-to-peptide ratios (where complexes are negatively charged), a competition between short-range and long-range forces can lead to the formation of a colloid-like phase, which is marked by the formation of droplet clusters. Using peptide (R or K) and RNA sequence (purine or pyrimidine) variants, we show that the structure and dynamics of these clusters are also dependent on the strength of cation-π interactions. With relatively high short-range attraction between the R/G-rich peptide and poly(A) chains, these clusters behave analogously to a colloidal gel that can be formed via a mixed interaction potential 36, 43. These results indicate that the rules of sequence-specific molecular interactions are relevant not only to understand the reentrant liquid condensation behavior of RNP-RNA complexes, but also to potentially engineer new mesoscale materials where viscoelastic peptide-nucleic acid microspheres could be utilized as soft colloids for gelation. Unlike commonly used DNA and protein-coated polystyrene particles 44-45, the hardness of these peptide-RNA microspheres can be finely tuned by varying amino acid and nucleobase sequences of polypeptides and nucleic acids, respectively.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge UB north campus confocal imaging facility (supported by National Science Foundation MRI Grant: DBI 0923133) and its director, Mr. Alan Siegel for helpful assistance.
Funding
We gratefully acknowledge support for this work from University at Buffalo, SUNY, College of Arts and Sciences to P.R.B. P.R.B. is also supported by funding from the National Institute on Aging (NIA) of the National Institutes of Health (R21 AG064258).
Footnotes
Conflicts of Interest
The authors declare no conflict of interest.
References:
- 1.Alberti S, The wisdom of crowds: regulating cell function through condensed states of living matter. J Cell Sci 2017, 130 (17), 2789–2796. [DOI] [PubMed] [Google Scholar]
- 2.Banani SF; Lee HO; Hyman AA; Rosen MK, Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 2017, 18 (5), 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hyman AA; Weber CA; Julicher F, Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol 2014, 30, 39–58. [DOI] [PubMed] [Google Scholar]
- 4.Mitrea DM; Kriwacki RW, Phase separation in biology; functional organization of a higher order. Cell communication and signaling : CCS 2016, 14, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shin Y; Brangwynne CP, Liquid phase condensation in cell physiology and disease. Science 2017, 357 (6357). [DOI] [PubMed] [Google Scholar]
- 6.Riback JA; Katanski CD; Kear-Scott JL; Pilipenko EV; Rojek AE; Sosnick TR; Drummond DA, Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response. Cell 2017, 168 (6), 1028–1040 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brangwynne CP; Tompa P; Pappu RV, Polymer physics of intracellular phase transitions. Nat Phys 2015, 11 (11), 899–904. [Google Scholar]
- 8.Wang J; Choi JM; Holehouse AS; Lee HO; Zhang X; Jahnel M; Maharana S; Lemaitre R; Pozniakovsky A; Drechsel D; Poser I; Pappu RV; Alberti S; Hyman AA, A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018, 174 (3), 688–699 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gitler AD; Dhillon P; Shorter J, Neurodegenerative disease: models, mechanisms, and a new hope. Disease Models & Mechanisms 2017, 10 (5), 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maharana S; Wang J; Papadopoulos DK; Richter D; Pozniakovsky A; Poser I; Bickle M; Rizk S; Guillen-Boixet J; Franzmann TM; Jahnel M; Marrone L; Chang YT; Sterneckert J; Tomancak P; Hyman AA; Alberti S, RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 2018, 360 (6391), 918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mann JR; Gleixner AM; Mauna JC; Gomes E; DeChellis-Marks MR; Needham PG; Copley KE; Hurtle B; Portz B; Pyles NJ; Guo L; Calder CB; Wills ZP; Pandey UB; Kofler JK; Brodsky JL; Thathiah A; Shorter J; Donnelly CJ, RNA Binding Antagonizes Neurotoxic Phase Transitions of TDP-43. Neuron 2019, 102 (2), 321–338 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banerjee PR; Milin AN; Moosa MM; Onuchic PL; Deniz AA, Reentrant Phase Transition Drives Dynamic Substructure Formation in Ribonucleoprotein Droplets. Angew Chem Int Ed Engl 2017, 56 (38), 11354–11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boeynaems S; Bogaert E; Kovacs D; Konijnenberg A; Timmerman E; Volkov A; Guharoy M; De Decker M; Jaspers T; Ryan VH; Janke AM; Baatsen P; Vercruysse T; Kolaitis RM; Daelemans D; Taylor JP; Kedersha N; Anderson P; Impens F; Sobott F; Schymkowitz J; Rousseau F; Fawzi NL; Robberecht W; Van Damme P; Tompa P; Van Den Bosch L, Phase Separation of C9orf72 Dipeptide Repeats Perturbs Stress Granule Dynamics. Mol Cell 2017, 65 (6), 1044–1055 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Birney E; Kumar S; Krainer AR, Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Research 1993, 21 (25), 5803–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thandapani P; O’Connor TR; Bailey TL; Richard S, Defining the RGG/RG motif. Mol Cell 2013, 50 (5), 613–23. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen TT; Rouzina I; Shklovskii BI, Reentrant condensation of DNA induced by multivalent counterions. The Journal of chemical physics 2000, 112 (5), 2562–2568. [Google Scholar]
- 17.Zhang R; Shklovskii BI, Phase diagram of solution of oppositely charged polyelectrolytes. Physica A: Statistical Mechanics and its Applications 2005, 352 (1), 216–238. [Google Scholar]
- 18.Grosberg AY; Nguyen TT; Shklovskii BI, Colloquium: The physics of charge inversion in chemical and biological systems. Reviews of Modern Physics 2002, 74 (2), 329–345. [Google Scholar]
- 19.Alberti S, Phase separation in biology. Current Biology 2017, 27 (20), R1097–R1102. [DOI] [PubMed] [Google Scholar]
- 20.Boeynaems S; Alberti S; Fawzi NL; Mittag T; Polymenidou M; Rousseau F; Schymkowitz J; Shorter J; Wolozin B; Van Den Bosch L; Tompa P; Fuxreiter M, Protein Phase Separation: A New Phase in Cell Biology. Trends in cell biology 2018, 28 (6), 420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozdilek BA; Thompson VF; Ahmed NS; White CI; Batey RT; Schwartz JC, Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding. Nucleic Acids Res 2017, 45 (13), 7984–7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aumiller WM Jr.; Keating CD, Phosphorylation-mediated RNA/peptide complex coacervation as a model for intracellular liquid organelles. Nat Chem 2016, 8 (2), 129–37. [DOI] [PubMed] [Google Scholar]
- 23.Biot C; Buisine E; Kwasigroch JM; Wintjens R; Rooman M, Probing the energetic and structural role of amino acid/nucleobase cation-pi interactions in protein-ligand complexes. J Biol Chem 2002, 277 (43), 40816–22. [DOI] [PubMed] [Google Scholar]
- 24.Wintjens R; Lievin J; Rooman M; Buisine E, Contribution of cation-pi interactions to the stability of protein-DNA complexes. J Mol Biol 2000, 302 (2), 395–410. [DOI] [PubMed] [Google Scholar]
- 25.Mao L; Wang Y; Liu Y; Hu X, Multiple intermolecular interaction modes of positively charged residues with adenine in ATP-binding proteins. J Am Chem Soc 2003, 125 (47), 14216–7. [DOI] [PubMed] [Google Scholar]
- 26.Michael Gromiha M; Santhosh C; Suwa M, Influence of cation–π interactions in protein–DNA complexes. Polymer 2004, 45 (2), 633–639. [Google Scholar]
- 27.Zhang H; Li C; Yang F; Su J; Tan J; Zhang X; Wang C, Cation-pi interactions at non-redundant protein--RNA interfaces. Biochemistry (Mosc) 2014, 79 (7), 643–52. [DOI] [PubMed] [Google Scholar]
- 28.Vasilyev N; Polonskaia A; Darnell JC; Darnell RB; Patel DJ; Serganov A, Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP. Proc Natl Acad Sci U S A 2015, 112 (39), E5391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boeynaems S; Holehouse AS; Weinhardt V; Kovacs D; Van Lindt J; Larabell C; Van Den Bosch L; Das R; Tompa PS; Pappu RV; Gitler AD, Spontaneous driving forces give rise to protein-RNA condensates with coexisting phases and complex material properties. Proc Natl Acad Sci U S A 2019, 116 (16), 7889–7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burley SK; Petsko GA, Weakly polar interactions in proteins. Adv Protein Chem 1988, 39, 125–89. [DOI] [PubMed] [Google Scholar]
- 31.Rubinstein M; Semenov AN, Dynamics of entangled solutions of associating polymers. Macromolecules 2001, 34 (4), 1058–1068. [Google Scholar]
- 32.Semenov AN; Rubinstein M, Dynamics of entangled associating polymers with large aggregates. Macromolecules 2002, 35 (12), 4821–4837. [Google Scholar]
- 33.Semenov AN; Rubinstein M, Dynamics of strongly entangled polymer systems: activated reptation. Eur Phys J B 1998, 1 (1), 87–94. [Google Scholar]
- 34.Schavemaker PE; Boersma AJ; Poolman B, How Important Is Protein Diffusion in Prokaryotes? Front Mol Biosci 2018, 5 (93), 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elbaum-Garfinkle S; Kim Y; Szczepaniak K; Chen CC; Eckmann CR; Myong S; Brangwynne CP, The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci U S A 2015, 112 (23), 7189–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stradner A; Sedgwick H; Cardinaux F; Poon WC; Egelhaaf SU; Schurtenberger P, Equilibrium cluster formation in concentrated protein solutions and colloids. Nature 2004, 432 (7016), 492–5. [DOI] [PubMed] [Google Scholar]
- 37.Stradner A; Cardinaux F; Schurtenberger P, A small-angle scattering study on equilibrium clusters in lysozyme solutions. J Phys Chem B 2006, 110 (42), 21222–31. [DOI] [PubMed] [Google Scholar]
- 38.Stradner A; Thurston GM; Schurtenberger P, Tuning short-range attractions in protein solutions: from attractive glasses to equilibrium clusters. J Phys-Condens Mat 2005, 17 (31), S2805–S2816. [Google Scholar]
- 39.Zhou H-X; Pang X, Electrostatic Interactions in Protein Structure, Folding, Binding, and Condensation. Chemical reviews 2018, 118 (4), 1691–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X; Lin Y; Eschmann NA; Zhou H; Rauch JN; Hernandez I; Guzman E; Kosik KS; Han S, RNA stores tau reversibly in complex coacervates. PLoS Biol 2017, 15 (7), e2002183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin Y; McCarty J; Rauch JN; Delaney KT; Kosik KS; Fredrickson GH; Shea JE; Han S, Narrow equilibrium window for complex coacervation of tau and RNA under cellular conditions. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi J-M; Dar F; Pappu RV, LASSI: A lattice model for simulating phase transitions of multivalent proteins. bioRxiv 2019, 611095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sciortino F; Mossa S; Zaccarelli E; Tartaglia P, Equilibrium cluster phases and low-density arrested disordered states: the role of short-range attraction and long-range repulsion. Phys Rev Lett 2004, 93 (5), 055701. [DOI] [PubMed] [Google Scholar]
- 44.Obana M; Silverman BR; Tirrell DA, Protein-Mediated Colloidal Assembly. J Am Chem Soc 2017, 139 (40), 14251–14256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mirkin CA; Letsinger RL; Mucic RC; Storhoff JJ, A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 1996, 382 (6592), 607–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.