Abstract
Cellular signal transduction is predominantly based on protein interactions and their posttranslational modifications, which enable a fast response to input signals. Due to difficulties in designing new unique protein–protein interactions, designed cellular logic has focused on transcriptional regulation; however, this has a substantially slower response requiring transcription and translation. Here, we present a de novo design of modular, scalable signaling pathways based on proteolysis and designed coiled-coils (CC) implemented in mammalian cells. A set of split proteases with highly specific orthogonal cleavage motifs was constructed and combined with strategically positioned cleavage sites and designed orthogonal CC dimerizing domains of tunable affinity for competitive displacement after proteolytic cleavage. This enabled implementation of Boolean logic functions and signaling cascades in mammalian cells. Designed split protease-cleavable orthogonal CC-based logic (SPOC logic) circuits enable response to chemical or biological signals within minutes rather than hours, useful for diverse medical and nonmedical applications.
Introduction
Responsiveness to external and internal signals is a key feature of living cells, allowing appropriate response to environmental conditions, intracellular communication and many other functions. Physicochemical signals are typically sensed by diverse protein receptors that relay and transduce the signal to trigger an appropriate cellular response. Signaling in both prokaryotes and eukaryotes is predominantly achieved through protein–protein interactions and their posttranslational modifications, such as phosphorylation or proteolytic cleavage and degradation. Although many proteins in signaling pathways are composed of related modular domains 1, protein-protein interactions have been optimized during evolution for the high specificity and orthogonality of pathways operating in parallel. Several physiological responses such as secretion of insulin upon increased glucose concentration, blood vessel dilation or response to noxious agents, need to occur rapidly, within minutes. Since de novo biosynthesis of signal mediators through transcription and translation is time consuming, fast signaling responses are often accomplished by rapid processing of premade mediators. The design and introduction of new signaling pathways based on protein modification rather than transcription regulation may therefore enable therapeutic or biotechnological benefits and contribute to elucidation of the principles of signaling through naturally evolved pathways.
Signaling pathways have already been rewired and transferred between organisms, for example, in yeast and mammalian cells 2–6. However, in order to introduce specific, adjustable, and scalable regulation, the information-processing pathways should preferably be designed de novo, as this minimizes the unwanted interactions with the cellular chassis and makes the pathways highly programmable for implementing designed cellular logic. Up to now, most designed cell circuits have been based on transcriptional regulation, drawing on the modular DNA recognition and transcriptional effector domains 7–9. Transcriptional regulation–based cell logic is however inherently slower than protein interaction and modification-based systems.
In contrast to the transcriptional regulation–based response, the protein interaction/modification-based response occurs in cells within minutes and typically combines specific protein–protein interactions and catalytic steps arranged in several interconnected layers, which can combine multiple input signals, mediators, modifiers and information processing steps (Fig. 1a). Several natural pathways utilize proteolysis, either for proteasome-mediated degradation of selected proteins that generate or expose a degradation-targeting motif (e.g., IκB in the inflammatory signaling cascade) or through cleavage at defined sites (e.g., Notch signaling, apoptosis, or coagulation cascade) 10–12. Proteolytic regulation had already been engineered into mammalian cells, for example, through degrons 4,13 and cleavage of transcription factors 14 and their translocation triggered by proteolytic cleavage 15,16. In addition, in vitro systems for detection of proteolytic cleavage have been designed for a limited number of logic functions 17,18. However, the design of a fast, modular, scalable protein modification–based signaling platform for the construction of logic functions in mammalian cells remained a challenge, especially in terms of achieving the cellular response at the sub-hour time scale. Use of proteolysis for logic circuit design imposes several prerequisites, specifically 1) availability of a sufficient number of orthogonal proteases, each specific to its own substrate without interfering with other components or processes within the circuit or the cellular chassis, 2) a mechanism to activate proteases by selected internal or external signals, 3) a mechanism to convert the proteolytic processing into an output activity in a functionally complete way, allowing the design of diverse logic functions, and 4) a mechanism to render further information processing (protease cleavage) dependent on the output of the upstream logic function, allowing coupling of functional layers in a modular way (scalability).
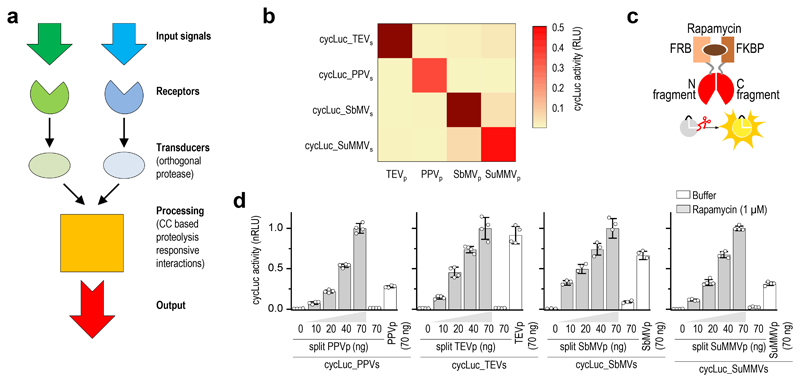
Figure 1. Design of the proteolysis-based signaling pathways and orthogonal proteases.
(a) Scheme of components of the proteolysis-based signaling pathways. (b) Heat map showing orthogonality of the four potyviral protease homologues tested in HEK293T cells, detected by the cycLuc reporter with a matching protease cleavage site. (c,d) Chemically inducible reconstitution of the split proteases 24h after induction with rapamycin (three-dimensional homology models of orthogonal split proteases are shown in Supplementary Fig. 1e). Values in (d) are the mean of four cell cultures ± (s.d.) and are representative of two independent experiments. Transfection plasmid mixtures are listed in Supplementary Table 1.
Here, we present a platform for the design of a proteolysis-based signaling pathway, called split protease and orthogonal coiled-coil logic (SPOC logic). We show the design of a set of orthogonal split proteases as signal transducers, and a set of orthogonal proteolysis-responsive coiled-coil (CC)-based modules as designable and scalable information-processing modules. Proteolytic cleavage induces the conformational rearrangement within and between the CC modules leading to the reconstitution of downstream functional protein domains. We present construction of a full set of Boolean logic gates. Mammalian cells with designed logic functions are responsive to small molecule inducers and respond within minutes after the chemical signal, and the scalability of the modular system is validated by construction of multi-level cascades. Finally, the applicability of the SPOC logic system is demonstrated as a viral protease detection system with a built in chemically regulated safety switch.
Results
Design of a highly specific orthogonal split protease set
Protease-based signaling pathways require proteases with high target specificity that minimally interfere with the existing cellular chassis. Protease orthogonality is required to establish logic functions based on the proteolytic cleavage of signaling mediators. Initially we sought to construct a toolbox of orthogonal split proteases based on mutants and orthologues of the tobacco etch virus protease (TEVp), one of the most widely used proteases in biotechnology, which recognizes a seven–amino acid residue sequence and is nontoxic to mammalian cells 19. Two previously designed TEVp mutants with modified substrate specificity 20 displayed a high preference for cleavage of their respective substrate variant and were mutually orthogonal; however they also maintained a propensity for cleavage of the wild type TEVp substrate, making the TEVp mutants less appropriate for our purpose of robust construction of an orthogonal protease set (Supplementary Fig. 1a,b). In contrast, three other proteases of the potyviral family—plum pox virus protease (PPVp) 19, soybean mosaic virus protease (SbMVp) 21, and sunflower mild mosaic virus protease (SuMMVp) 14, displayed excellent substrate cleavage orthogonality in human embryonic kidney (HEK) 293T cells when tested on cyclic luciferase reporters with designed cleavage sites 22 (Fig. 1b; Supplementary Fig. 1c,d). Furthermore, to make the selected proteolytic activity inducible by chemical input signals or protein–protein interactions required along the signaling pathway, the proteases should be designed as split enzymes. The homology of the potyviral orthologues with TEVp allowed us to design split proteases using three-dimensional modeling of the orthologues based on the tertiary structure of TEVp 23 alongside its split design24 (Supplementary Fig. 1e). Complementary split fragments of each protease, fused to either FKBP and FRB (whose heterodimerization is inducible with rapamycin) or ABI and PYL1 (whose heterodimerization is inducible with abscisic acid, ABA) 25 domain pairs, gained proteolytic activity upon addition of the appropriate chemical inducer, with undetectable leakage in its absence while maintaining the orthogonality (Fig. 1c-d, Supplementary Fig. 2a-c, Supplementary Fig. 3).
Proteolysis-dependent coiled-coil mediated rearrangement
The second component of the designed proteolysis-based signaling pathway demanded scalable implementation of orthogonal protein–protein interaction modules responsive to proteolysis. Coiled-coil (CC) domains are well understood mediators of protein-protein interaction that can be designed de novo 26–29 and have been used for various designed protein assemblies, from CC protein origami 30,31 to in vitro protease sensors 17. In canonical coiled-coils, two or more helices fold in a left-handed supercoiled assembly with each coil characterized by a repeat of seven residues per two turns of the helix 32. This seven amino acid pattern is denoted with the string abcdefg, also referred to as a heptad repeat 33. To guide information flow and implementation of logic functions in vivo, several orthogonal CC sets, fused to split-output proteins and functional in the complex environment of the mammalian cell cytosol, are needed. Moreover, variants of orthogonal CC pairs with different affinities and orientations (parallel vs. antiparallel) are required for designing autoinhibitor and displacer peptides, such that a displacer peptide fused to a functional split protein can effectively displace an autoinhibitory peptide fused to a target segment partner upon proteolytic cleavage of the linker between the autoinhibitor and target CC partner (Fig. 2a, b).
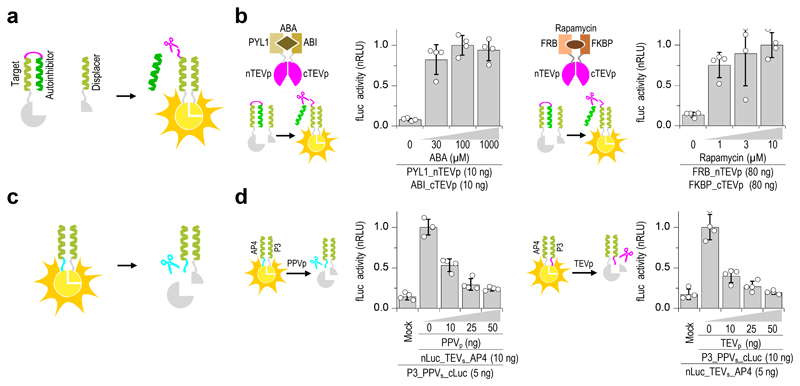
Figure 2. Design of proteolytic cleavage-responsive coiled-coil (CC) interaction modules.
(a) Design of the proteolytic cleavage-responsive CC rearrangement reconstituting the functional split protein. Upon linker cleavage, an autoinhibitory coil is replaced by a displacer segment with higher binding affinity to reconstitute the split effector/reporter. (b) Abscisic acid (ABA) and rapamycin inducible activation was demonstrated in HEK293T cells measured 30 min after induction with indicated concentration of ABA or rapamycin. (c) Design of a proteolytic cleavage-inactivated module (logical negation). A protease cleavage site is introduced between the CC-forming segments and split effector/reporter domain. After cleavage of the linker, split luciferase dissociates. (d) Decrease in luciferase activity upon co-transfection of logical negation functions with plasmids coding for specific proteases. Values in (b), and (d) are the mean of four cell cultures ± (s.d.) and representative of two independent experiments. Transfection plasmid mixtures are listed in Supplementary Table 1.
To allow for an easy implementation with various split output proteins fused to the coiled-coils, we designed our system such that an antiparallel coiled-coil pair is used for the autoinhibition, but once the linker between the autoinhibitory and target coils is cleaved by a protease, either a parallel or an antiparallel displacer peptide can interact with the target, depending on what is needed for the reconstitution of the fused split protein. Since the available characterized CC heterodimers are predominantly parallel 26,34, we designed and tested new heterodimeric antiparallel CC pairs (Supplementary Fig. 3a-c). To ensure pairing in either a parallel or an antiparallel orientation and to retain the orthogonality between different coiled-coil couples, we based our designed CC peptides on a previously published parallel CC set 35,36. In order to change the parallel orientation of helices in heterodimers P3/P4 and P9/P10 into antiparallel, the sequences of P4 and P10 were reversed, and the resulting peptides were named AP4 and AP10 (Supplementary Fig. 3a, b). Reversing the sequence of these coiled-coil segments generated the contacts necessary for association in an antiparallel orientation 37,38(detailed description in the online methods- Design of coiled-coil target-autoinhibitor-displacer sequence sets). The new designed antiparallel pairs P3/AP4 and P9/AP10 fused to split luciferase fragments were experimentally tested and demonstrated to reconstitute luciferase activity in HEK 293T. Those peptide sets were found to be orthogonal to each other and the concentration range in which the pair functions has been determined (Supplementary Fig. 3c, d).
The antiparallel pairs where further modified for use as autoinhibitory segments (Supplementary Fig. 4a-c). Displacement of such an autoinhibitory segment, occurring after the proteolytic cleavage of the linker in the hairpin, should be promoted by the formation of a more stable pair between the target and displacer peptide in a concentration dependent fashion. We sought to favor the formation of a coiled-coil dimer between the displacer coil and the target coil by decreasing the stability of the interaction between the autoinhibitory and target peptides. The defined register of coiled-coil dimers allows for predictable site specific modifications that can be used to tune the coiled-coil dimer stability. In particular positions b, c and f, which are not involved in protein-protein interactions, can be substituted in order to alter the stability of dimers without affecting their specificity of binding 39. Since the autoinhibitory displacement system is based on the concentration dependent displacement, there is a stability window in which the interaction of the target coil with the autoinhibitory segment is stable enough to prevent binding in the absence of the signal, yet sufficiently weak to allow effective displacement after proteolysis of the linker between the two coils. Previously, we destabilized coiled-coil dimers by mutations at noncontact positions, specifically by replacing high helical propensity alanine residues at positions b and c in the heptad repeats of P3 and P4 by less helical polar residues serine and glutamine in P3mS and P4mS 40. Therefore the autoinhibitory antiparallel coils were designed using the same principle (Supplementary Fig. 4a-c). These mutations were found to be effective at facilitating the displacement of autoinhibitory peptides by displacer peptides and structural re-arrangement upon chemical signal-triggered proteolysis while providing sufficient inhibition of split protein reconstitution without proteolytic cleavage. Indeed, very little leakage in target protein reconstitution was observed even in excess of a displacer peptide, which resulted in a wide range of autoinhibited target to displacer ratios effectively responding to a proteolytic signal (Supplementary Fig. 5a-c). On the other hand, additional Leu or Ile to Ala replacements at core positions a and d excessively destabilized the CC pair, thus promoting displacement even in the absence of proteolytic activity and high leakage (peptides P3mS1A2A, P3mS2A2 and P3mS1A22A2, Supplementary Fig. 6a-c).
Construction of SPOC logic functions
A cleavage site in the antiparallel hairpin linker, as used before 17, enables implementation of only a limited set of logic functions (e.g., OR, AND) but not the functions comprising a logical negation (NOT, NOR, NAND, etc.). To overcome this limitation, an additional cleavage site type between the CC forming segments and an effector domain (i.e., at the site of fusion between the split luciferase or split protease) was introduced, which enabled functional split enzyme inactivation by proteolysis (Fig. 2a-d, Supplementary Fig.7).
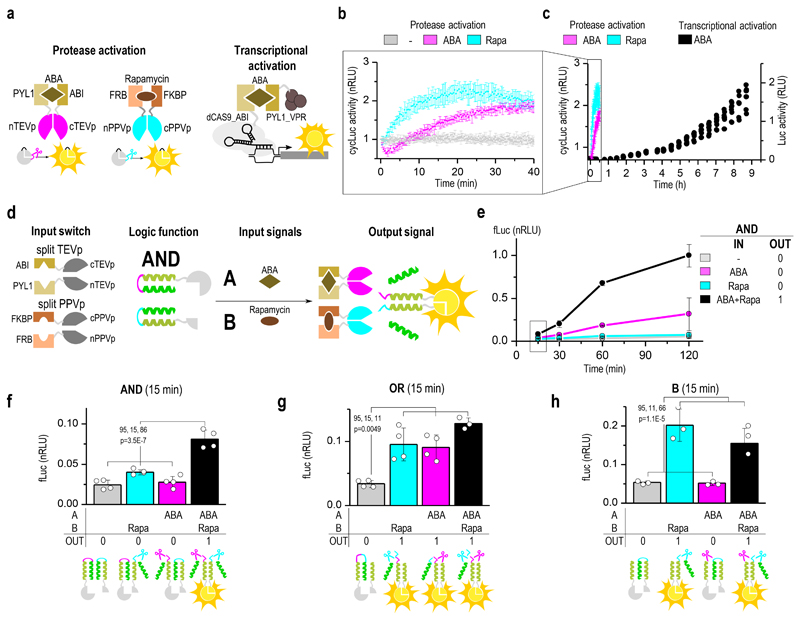
These designed interaction modules, encompassing cleavage sites for selected proteases at different positions within the CC modules, represent a sufficient set to construct all binary Boolean logic functions. Altogether, 13 variants of protease-responsive interaction modules were constructed, combining CC modules with effector protein (sub)domains and cleavage sites at up to two different positions (Supplementary Fig. 8a-c). Additional protease cleavage sites could be introduced into each of these positions in order to extend the logic functions towards multiple inputs. Single input functions were constructed when only one of the building blocks was cleavable (NOT A, NOT B, A, B, depending on the position of the cleavage site; (Fig. 3a, Supplementary Fig. 9a-c). An AND function was formed when both reporter segments are activated by a cleavage between the target and an autoinhibitory segment (Fig. 3c). Analysis of the concentration range of both applied inputs demonstrated the expected symmetric response (Supplementary Fig. 10). A NOR function is constructed when both reporter modules are deactivated by cleavage between the CC-forming domain and split reporter fragment (Fig. 3b). Introducing a linker comprising cleavage sites for both input proteases within the single building block, an OR function was obtained (Fig. 3d). When one of the building blocks is cleavage-activated by one input protease and the other building block is cleavage-deactivated by the other input protease, the system results in NIMPLY functions (Fig. 3e, Supplementary Fig. 9d).
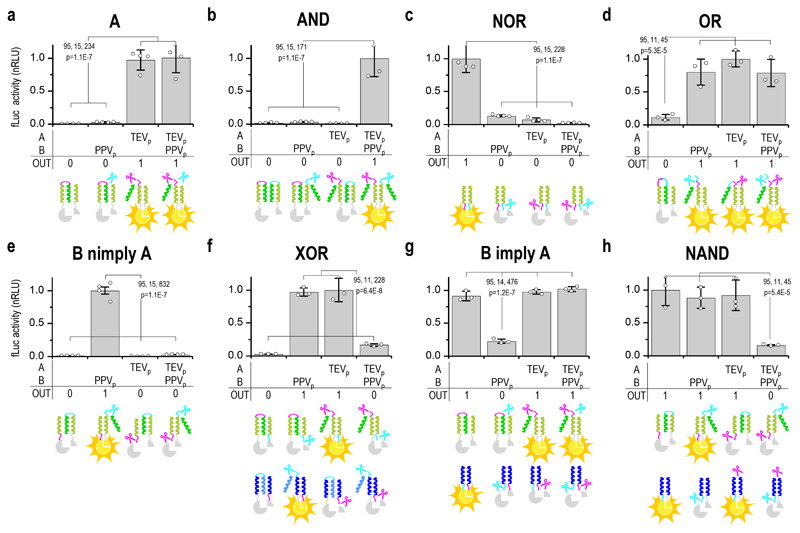
Figure 3. Design of Boolean logic functions implemented by split protease-cleavable orthogonal coiled-coil (CC)-based logic (SPOC logic).
Experimental analysis of SPOC logic function designs in HEK293T cells with introduced genetic circuits. The design and expected response to all input combinations is schematically shown below the graphs with experimental luciferase based results. Input signals are combinations of two orthogonal proteases TEVp and PPVp, and the output signal is split luciferase activity. The remaining Boolean SPOC logic circuits (B, NOT B, NOT A, A nimply B, A imply B, XNOR, NAND) are shown in Supplementary Fig. 9. Transfection plasmid mixtures are listed in Supplementary Table 2. Values are the mean of three (d-h) and four (a-c,e) cell cultures ± (s.d.) and are representative of two independent experiments. Significance was tested by 1-way ANOVA with Tukey’s comparison (values CI, df, F and p are indicated on graphs).
The functions obtained by combining the output of two split reporter fragments on two orthogonal CC sets complete the full set of nontrivial Boolean functions with XOR (A nimply B in conjunction with orthogonal B nimply A, Fig. 3g), A imply B (AND in conjunction with orthogonal NOT A, Supplementary Fig. 9e), XNOR (AND in conjunction with orthogonal NOR, Supplementary Fig. 9f), B imply A (A in conjunction with orthogonal NOR, Fig. 3f) and NAND (B nimply A in conjunction with orthogonal NOT B, Fig. 3h). A robust and significant difference between the active and inactive states was observed for all the designed Boolean functions (p < 0.001).
Multilayered signaling
The modular design of SPOC logic supports scalability by extension to multiple layers. A reconstituted split protease acting as an output, can serve as the input of the next protease-regulated logic layer. This design allows the construction of multilayer circuits, such as inverter cascades.
When a split protein is reconstituted through a CC linker cleavage, the total interaction energy difference between the ON and OFF state consists of the contribution of both the coiled-coil peptides as well as the interaction of the split protein fragments. Split proteins domain pairs with a large interaction surface and a strong binding affinity might therefore result in a leaky activation, where the blocking the CC interaction with an autoinhibitory coil may not be sufficient to prevent reconstitution driven by the split protein domain interaction. We indeed observed this effect in case of reconstitution of TEVp with high undesired activity in the OFF state (Supplementary Fig. 11). To solve this issue, a catalytically inactive split domain, fused to the autoinhibitory segment was introduced. This equalized the interaction energy contribution from the split protein reconstitution in the autoinhibited and active (displaced) states, resulting in compensation and suppression of leakage in the uninduced state. The above was achieved by fusing a fragment of TEVp with a mutation in the active site, previously shown to be catalytically inactive 41, to the autoinhibitory coil. Upon cleavage of the linker, the inactive TEVp fragment is displaced along with the autoinhibitory coil by an active TEVp fragment fused to the displacer coil. This modification resulted in an excellent performance of the proteolytic cascade, with a decreased background activity and substantially improved fold activation (Fig. 4a).
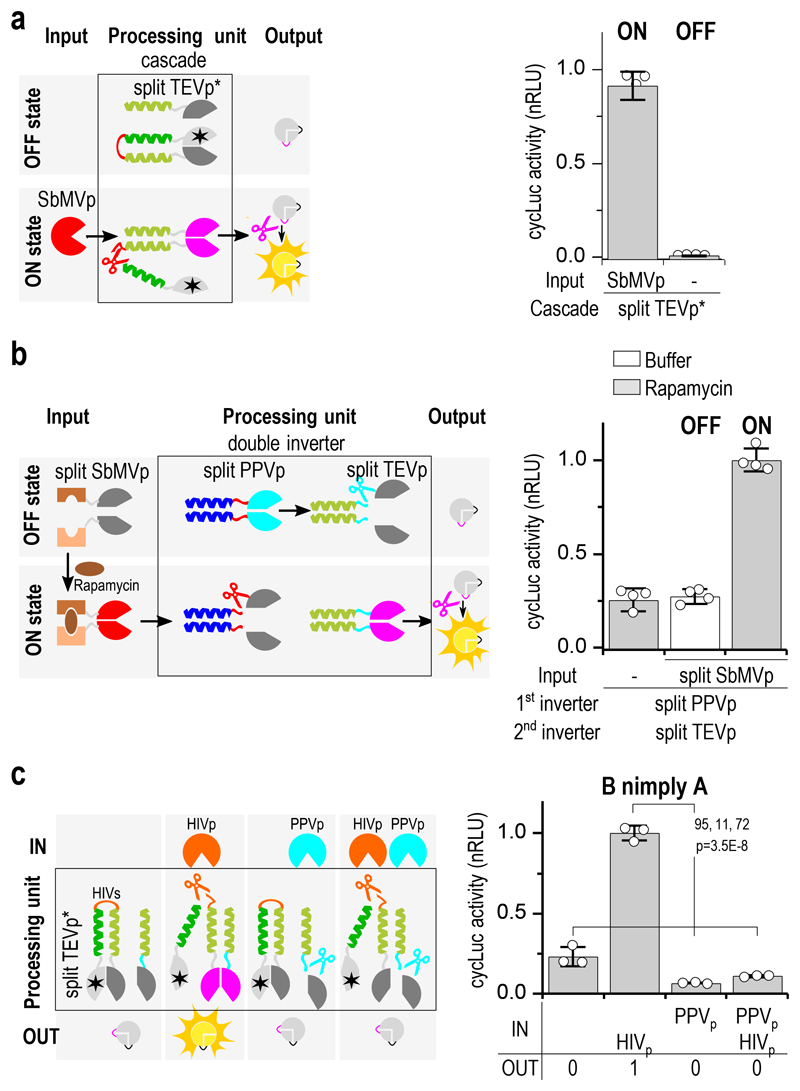
Figure 4. Multilayer design of proteolysis-based signaling pathways.
(a) Two-layer protease-cascade function with a catalytically inactive split tobacco etch virus protease (TEVp*) domain fused to the autoinhibitory CC shows decreased leakage and higher fold activation (see also Supplementary Fig. 11). (b) Double inverter consisting of a split TEVp regulated by split plum pox virus protease (PPVp), where PPVp is regulated by the rapamycin-induced split soybean mosaic virus protease (SbMVp). (c) B nimply A logic function combining human immunodeficiency virus-1 (HIV-1p) and PPVp as input signals. “SQVSQNYPIVQNLQ” recognition sequence for HIV-1 protease was used. Transfection plasmid mixtures are listed in Supplementary Table 1, 4. Values are the mean of three (c) and four (a,b) cell cultures ± (s.d.) and are representative of at least two independent experiments, significance tested by 1-way ANOVA with Tukey’s comparison between the indicated ON and OFF states (CI=95%, df=11, F=72 (c)).
To demonstrate building a cascade based on SPOC logic, a chemically inducible double inverter was constructed utilizing chemical reconstitution of PPVp that can inactivate CC-linked split TEVp, which in turn inactivates a reporter. Addition of rapamycin resulted in response that was at least comparable to the activation of the cascade by constitutive PPVp (Supplementary Fig. 12a). An alternative double negation cascade was constructed with three proteases, combining a cycLuc reporter with a PPVp-inactivated TEVp and a layer consisting of SbMVp-inactivated split PPVp (Supplementary Fig. 12b). In the default state, the final protease in this system, TEVp, is inactive, since the reconstituted split PPVp cleaves off the split TEVp fragments from the coupled CC in the absence of an input signal. On the other hand, in the presence of an active SbMVp, the split PPVp fragments are cleaved off, enabling pairing of the split TEVp in the second layer, resulting in an active output. This type of a double inverter was also shown to be inducible with rapamycin (Fig. 4b).
To demonstrate that the SPOC logic platform performs independently of other cellular components and that it could be adaptable to diverse chassis, the AND function was constructed in NIH3T3, CHO and Neuro 2a cell lines and was shown to be highly efficient (Supplementary Fig. 13a-d). Furthermore, in addition to potyviral proteases, SPOC logic can also be adapted to respond to any biological signals that could be coupled to a specific proteolytic activity. As an example, for use as a diagnostic or theranostic tool, the cleavage sequences in the SPOC logic constructs can be replaced by sequences cleaved by pathogen-specific proteases. Such a system could be controlled by multiple inputs, combined either into an AND function recognizing multiple physiological signals or into a NIMPLY function, providing a safety switch operated by an external signal (molecule) that can regulate arming of the pathogen sensor. The feature of combining the detection of viral protease with an additional signal was demonstrated by the NIMPLY function triggered in the presence of human immunodeficiency virus-1 (HIV-1) protease activity, but only in the case when the small molecule-regulated PPVp has not been activated (Fig. 4c).
Kinetics of the SPOC logic signaling pathway
One of the most important advantages of protein signaling in comparison to transcriptional regulation is the possibility of a substantially faster response. We demonstrated this feature of the designed signaling pathway by comparing the kinetics of the response of inducible split proteases fused to either the rapamycin- or ABA-responsive protein domains (Fig. 5a) with the transcription regulation-based modules fused to the same type of input-responsive domains. A significant response could be detected already in less than 5 minutes after the addition of chemical inducers to the cells carrying constructs coding for split proteases with cyclic luciferase reporters (Fig. 5b, 5c). Presumably due to different kinetics of rapamycin and ABA diffusion through cell membranes and the differences in the kinetics of the binding induced by these small molecules 42, a higher amount of rapamycin-inducible proteases was used to achieve a comparable short-term response from both induction systems by compensating for the slower induction of binding with rapamycin as compared to ABA (Supplementary Fig. 2). Nevertheless, a significant and specific response of both one- and two-input functions was observed in less than 15 minutes after the addition of the inducers (Fig. 5d-h, Supplementary Fig. 14a-d). No changes in protease or luciferase fragment levels were observed in this timeframe, confirming that the change in output is in fact due to the cleavage and re-arrangement of coiled-coil coupled luciferase fragments (Supplementary Fig. 15) rather than differences in transcription/translation. In comparison, the same ligands used to trigger transcriptional activation required 2 hours for a detectable increase of the reporter activity in cells, demonstrating a speeding-up of the information processing in the cells by close to an order of magnitude.
Figure 5. Fast kinetics of the proteolysis-mediated signaling pathway.
(a) Scheme of the reconstitution of the split tobacco etch virus protease (TEVp) by abscisic acid (ABA) and split plum pox virus protease (PPVp) by rapamycin measured using cyclic luciferase reporter in HEK293 cells. A CRISPR/dCas9-based activator was used as comparison for the kinetics of the transcriptional control of luciferase activation. (b) Continuous online monitoring of chemically induced luciferase activity in HEK293T cells and comparison with the kinetics of transcriptional activation (c). (d) Schematic presentation of building blocks for inducible split protease-cleavable orthogonal CC-based logic (SPOC logic) functions. (e) Kinetics of chemically regulated AND function where both segments of the split luciferase are activated by proteolytic cleavage. Significant response was detected in less than 15 minutes after the addition of chemical inducers. (f-h) SPOC logic functions AND, OR, and B regulated by rapamycin and ABA 15 minutes after induction. Transfection mixtures are listed in Supplementary Table 5, 6. Values are the mean of three (f) and four (b, c, e-g) cell cultures ± (s.d.) and are representative of two independent experiments, significance tested by 1-way ANOVA with Tukey’s comparison between the indicated ON and OFF states (values CI, df, F and p are indicated on graphs (f-h)).
Discussion
All components of SPOC logic presented here rely solely on protein–protein interactions and modifications: the sensory input modules (split proteases) depend on small molecule binding and could also be regulated by light or other signals; the information mediating modules (coiled-coils) rely on specific protein-protein interactions unleashed by proteolytic cleavage; and the output modules (split enzymes) result in the reconstitution of enzymatic activity or other functions. The concepts of modularity and scalability built into the platform, provide a significant freedom in designing the functions best suited to any specific application. SPOC proteolytic processing could be wired to enable a wide range of downstream functions, such as secretion of hormones, antimicrobial compounds, enzymes or other signaling molecules. It could be made to interact with native intracellular protein-interaction-based signaling pathways, to destabilize or activate transcription factors 14, with potential applications from therapy to diagnostics and biotechnology. While proteolysis coupled to transcriptional regulation has been described before 13,14 it could not exploit the full potential of the protein processing-based signaling logic due to its reliance on a transcriptional and translational step. Kinetics of the SPOC system is governed by the interaction affinity and concentration of the components, and is independent from the slower processes of transcription and translation or cellular recycling machinery. With a response time of a few minutes, SPOC logic mimics natural signaling pathways in their speed.
Furthermore, this platform can support multiple inputs and advanced combinations of input signals from external or internal processes. Induction of proteolytic cleavage by rapamycin and ABA has been demonstrated here; however, it would be easy to adapt SPOC logic to respond to other chemical inducers of dimerization or even to intracellular processes, by fusing to dimerization domains dependent on the presence of selected biological molecules. While orthogonality of four proteases was demonstrated here, tens of additional potyviral protease homologues are known that could be used to expand the toolbox 43. In addition, other natural process- or pathogen-specific proteases, such as caspases and viral proteases (e.g., from HIV-1 or HCV) can be implemented as an input. The demonstrated cascade using HIV-1 protease provides an example of an application in which the system is employed to detect a disease-specific signal, but also contains a built-in safety switch that can regulate arming of the sensor. SPOC logic is thus particularly useful for enhancing the cell’s response to infection or physiological states that require a rapid response, such as secretion of insulin as a response to a change in glucose concentration, where transcriptional regulation is clearly too slow, or as a rapid sensing and response to pathogens, that may multiply before the transcriptional response provides the defense molecules. The coagulation cascade and the complement system, that provide a response to injury and infection, are natural extracellular examples of proteolytic cascades matched in speed by the SPOC logic and, when dysregulated by disease or mutations, might be rescued by implementation of fast artificial signaling pathways.
While this paper was under review, a report was published based on utilizing viral protease-based protein logic called CHOMP, which also described construction of Boolean logic functions in mammalian cells 44. That approach is based on the protease-induced regulation of the activity of degrons fused to the output reporters, which either leads to targeting of proteins for degradation when the degron is exposed by cleavage or to stabilization of proteins that would otherwise be directed to proteasomal degradation, if the degron is removed by cleavage. In comparison to CHOMP, SPOC logic seems to be substantially faster as the proteolytic cleavage directly (in)activates the effector protein and does not rely on the cellular degradation machinery. Additionally, since it is based on direct tunable protein-protein interactions and protease inactivation, SPOC is largely independent of the chassis (in contrast to cell-specific degron and proteasome efficiency) and is expected to work in most cell types in addition to the five cell lines demonstrated here. The availability of a set of designable orthogonal coiled-coils enables scalability of information processing logic. Since the degron- as well as coiled-coil based modules described in both contributions are based on orthogonal split proteases, they could potentially be combined into even more powerful platforms for posttranscriptional modification based logic circuits.
Online Methods
Recombinant DNA construct
Plasmids were constructed using the standard procedures of molecular cloning or Gibson assembly45. The amino acid sequences of all constructs are provided in Supplementary Table 8. All protease coding sequences were codon optimized for expression in human cells, and the DNA was synthesized by IDT (PPVp, SbMVp, and SuMMVp) or Life Technologies (TEVp). Split N- and C-fragments of proteases in fusion with CCs or dimerization partners FKBP/FRB and ABI/PYL1 were PCR amplified and inserted into the pcDNA3 vector. The FKBP and FRB domains were obtained from the pC4-RHE and pC4EN-F1 plasmids. ABI and PYL1 were obtained from pSLQ2816 pPB: CAG-PYL1-VPR-p2A-GID1-ABI-WPRE PGK-GAI-tagBFP-SpdCas9 (Addgene: 84261)46. The HIV-1 protease gene was obtained from plasmid pNL4-3.HSA.R-.E- (from Dr. N. Landau, Division of AIDS, NIAID). The firefly luciferase gene was obtained from pGL4.16 (Promega). The cleavable firefly luciferase (fLuc) inverse reporter was constructed by the addition of a protease cleavage site between the N- and C-parts of the Firefly luciferase at position 490 and 491, which inactivates the luciferase by cleavage. The cyclic luciferase reporter was constructed by circularly permuting firefly luciferase, i.e. placing the amino acid residues 234-544 N terminally, followed by a short protease-specific cleavable linker and then residues 234-544 as described in Kanno et al22. The IntN and IntC domains of the NpuDnaE intein obtained from the pSKDuet01 (Addgene: 12172) and pSKBAD2 (Addgene: 15335)47 were fused to the C- and N-ends of the circularly permutated firefly luciferase, respectively. phRL-TK (Promega) was used as transfection control in the dual luciferase assay. A set of plasmids for SPOC logic, comprising the split orthogonal proteases, cyclic luciferase reporters and CC-building modules, was deposited to Addgene (Addgene ID: 119866-118970; 119182; 119207-119214; 119299-119303).
Cell culture
The human embryonic kidney (HEK) 293T cell line was cultured in DMEM medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS; BioWhittaker, Walkersville, MD, USA) at 37°C in a 5% CO2 environment. NIH3T3 and CHO cell line was cultured in DMEM-F12 medium (Invitrogen) supplemented with 10% FBS and Neuro 2a cell line was cultured in OptiMEM medium (Invitrogen) supplemented with 10% FBS at 37°C in a 5% CO2 environment.
Chemical inducers
Rapamycin (Sigma-Aldrich) and abscisic acid (ABA, Sigma-Aldrich) were each dissolved in dimethyl sulfoxide (DMSO) at concentrations 1 mM and 50 mM, respectively. Before stimulation, stock concentrations of rapamycin and ABA were diluted in DMEM at 30 μM (Rapamycin) and 1mM (ABA) and 10 μL of the diluted inducer molecules were added to each well in 96-well plates for final concentration of 3 μM and 100 μM, respectively.
Transfection and dual luciferase assay
HEK293T cells were seeded in white 96-well plates (CoStar, Corning) at 2 × 104 cells per well. At a confluence of 50–70%, cells were transfected with a mixture of DNA and PEI. For every 1 μg of DNA transfected, 8 μl PEI at stock concentration of 0.324 mg/ml, pH 7.5, was diluted in 150 mM NaCl and mixed at a 1:1 ratio with the appropriate DNA also diluted in 150 mM NaCl. This was incubated at room temperature for 15 minutes and added to the cell media in 96-well plates. Detailed information about the amounts of plasmids in each experiment is provided in Supplementary Tables 2-8. Forty-eight hours after transfection, the cells were harvested and lysed with 30 μl of 1× passive lysis buffer (Promega). For experiments with split proteases 48 hours after the transfection, cells were stimulated with rapamycin and/or ABA as described above. At indicated time points after induction, cells were harvested and lysed with 30 μl of 1× passive lysis buffer (Promega).
NIH3T3, CHO and Neuro 2a cells were seeded in 96-well plates (CoStar, Corning) at 2,5 × 104 cells per well. At a confluence of 30-40%, cells were transfected with Lipofectamine 2000 (Invitrogen). For every 1 μg of DNA transfected, 4 μl Lipofectamine 2000, was diluted in Opti-MEM medium and mixed at a 1:1 ratio with the appropriate DNA also diluted in Opti-MEM medium. This was incubated at room temperature for 5 minutes and added to the cell media in 96-well plates. 24h after transfection, medium was replaced with fresh medium. Detailed information about the amounts of plasmids in each experiment is provided in Supplementary Table 3. Forty-eight hours after transfection, the cells were harvested and lysed with 30 μl of 1× passive lysis buffer (Promega).
Firefly luciferase and Renilla luciferase expressions were measured using a dual luciferase assay (Promega) and an Orion II microplate reader (Berthold Technologies). Luciferase activity presented as relative light units (RLU) was calculated by dividing each sample’s firefly luciferase activity by the constitutive Renilla luciferase activity determined in the same sample and then normalizing to the highest value obtained in each experiment (nRLU).
Design of coiled-coil target-autoinhibitor-displacer peptide sequence sets
Sequences of antiparallel CC pairs (target-autoinhibitor) were designed to maintain the charge and hydrophobic/hydrophilic residue complementarity in agreement with the canonical contact interactions in the antiparallel orientation (Supplementary Fig. 3a, Supplementary Fig. 4a). Target dimeric coiled coil pairs P3/P4 and P9/P10, used previously for the design of the orthogonal parallel CC set26 were chosen and used as template for antiparallel (autoinhibitory and displacer) designs. The design of antiparallel pairs was based on reversing the sequence of P4 and P10, to obtain AP4 and AP10 respectively, able to bind P3 and P9 in the antiparallel orientation. In contrast to parallel pairing, where amino acids in e and g’ positions form salt bridges, antiparallel dimers are stabilized by salt bridges between e and e’and g and g’ positions (with e and g representing residues in one helix and e’ and g’ representing the corresponding residues in the other helix). In P4 and P10, e and g positions in each heptad are occupied by the same residues, which allowed the reversed sequence to retain a correct pattern of interaction in the opposite orientation. On the other hand, positions a and d of the heptad form a tightly packed interface, also called knobs-into-holes 48,49, characterized by beta-branched hydrophobic amino acids intercalating in layers. Between the two different orientations the layers change from a-a’ and d-d’ in parallel coils to a-d’ and d-a’ in antiparallel coils 50. Reversing the sequence of P4 to AP4 relocates the leucine residues from positions d of P4 to positions a in AP4. The resulting heterodimeric peptides are therefore likely to bind in the antiparallel orientation in order to retain the favorable knobs-in-holes interactions (Supplementary Fig. 3a, b). Destabilized variants of CC peptides with maintained binding specificity and orthogonality, but expected decreased helical propensity, were designed as described before31 by replacing hydrophobic residues with high helical propensity at positions b and c with more hydrophilic residues (Supplementary Fig. 4, Supplementary Fig. 6). The helical wheels projections were drawn in DrawCoil 1.051. Per residue helical propensity was calculated with AGADIR52.
In situ measurements of split protease reconstitution
HEK293T cells seeded in a 96-well white plate at 3 × 104 cells per well (100 μl) were transfected with PEI reagent (6 μl PEI/500 ng DNA) at 40-50% confluency. At 40–48 h post transfection, at near 100% confluency, the growth medium was removed and replaced with 100 μl of assay medium (DMEM supplemented with 10% FBS; 2 mM ATP; 0.54 mM D-Luciferin). Detailed information about the amounts of plasmids in each experiment is provided in Supplementary Table 6. Cells were incubated in assay medium for 1 h at a controlled room temperature of 21°C. Measurements were obtained with the Orion II Microplate Luminometer (Berthold Detection Systems) with 2 s acquisition times of emitted light. Selected wells were continuously measured in cycles of 23 s for 5 min to obtain a base light emission in the absence of inducers. After baseline acquisition, the measurement was briefly paused (<1 min) for manual addition of the inducer rapamycin (3 μM, final concentration) or ABA (100 μM, final concentration in assay medium). Measurements resumed for at least 30 min to 1 h post induction. The firefly luciferase units were normalized in terms of activity of constitutively expressed Renilla luciferase (presented as RLU), which was further normalized on the background RLU values in the experiments (nRLU).
In situ measurements of transcriptional activation
HEK293T cells seeded in a 96-well white plate at 2 × 104 cells per well (100 μl) were transfected with PEI reagent (6 μl PEI/500 ng DNA) at 20–30% confluency. Detailed information about the amounts of plasmids in each experiment is provided in Supplementary Table 6. Measurements were performed 24 h post transfection on the Orion II Microplate Luminometer (Berthold Detection Systems) with 1-s acquisition times of emitted light. Before the addition of the inducer, 90 μl of growth medium was replaced with 50 μl of fresh DMEM with 10% FBS and 20 μl of assay medium (DMEM supplemented with 10% FBS, 27 mM coenzyme A, 2 M DTT, 0.53 mM ATP, 14 mM D-Luciferin) and background values of firefly luciferase were recorder in cycles of 23 s for 5 min. Transcription was induced by the addition of ABA (100 μM, final concentration in assay medium) and measured continuously for 23 min. The plate was then returned to the incubator with a controlled atmosphere of 37°C and 5% CO2, and it was measured again every 20 min for three sequential measurements.
Immunoblotting
The HEK293T cells (5-7 × 105) were seeded in 6-well plates (Techno Plastic Products). The next day, at a confluence of 50–70% cells were transiently transfected with a mixture of DNA and PEI (8 μl PEI/1,000 ng DNA). Detailed information about the amounts of plasmids in each experiment is provided in Supplementary Table 7. 48h post transfection the cells were washed with 1 mL PBS and lysed in 100 μL of lysis buffer (40 mM Tris-HCl (pH 8.0); 4 mM EDTA; 2% Triton X-100; 274 mM NaCl) containing a cocktail of protease inhibitors (Roche). Cells were lysed for 20 min on ice and centrifuged for 15 min at 17 400 rpm to remove cell debris. The total protein concentration in supernatant was determined using BCA assay. Proteins from the supernatant were separated on 12% SDS-PAGE gels (120 V, 60 min) and transferred to a nitrocellulose membrane (350 mA, 90 min). Membrane blocking, antibody binding and membrane washing were performed using iBind Flex Western device (ThermoFisher) according to the manufacturer’s protocol. The primary antibodies were rabbit-anti Myc (Sigma C3956; diluted 1:2000), rabbit-anti HA (Sigma H6908; diluted 1:2000) and mouse β-aktin (Cell Signaling techn., 3700; diluted 1:2000). The secondary antibodies were HRP-conjugated goat anti rabbit IgG, diluted 1:3000 (Abcam ab6721) and HRP-conjugated goat anti mose IgG (Santa Cruz, sc2005; diluted 1:3000). The secondary antibodies were detected with ECL Western blotting detection reagent (Super Signal West Femto; ThermoFisher) according to the manufacturer’s protocol.
Software and statistics
Graphs were prepared with Origin 8.1 (http://www.originlab.com/), and GraphPad Prism 5 (http://www.graphpad.com/) was used for statistical purposes. Values are the means of at least three experimental replicates (transfections off cell culture in individual wells) ± standard deviation (s.d.) and are representative of at least two independent experiments. An unpaired two-tailed t-test (equal variance was assessed with the F-test assuming normal data distribution) or one way ANOVA with post-hoc Tukey test was used for the statistical comparison of the data Confidence interval (CI), degree of freedom (df), and F value (F) are indicated as (CI, df, F).
Supplementary Material
Acknowledgments
The idea and proof of principle for this work was conceived as the part of the Slovenian iGEM 2016 project, and members of the team that are not among the authors Maja Meško, Miha Mraz, Miha Moškon, Dejan Križaj, Rok Krese, Nik Franko, Lidija Magdevska, Miha Gradišek, Žiga Pušnik, Samo Roškar and Kosta Cerović are acknowledged for their contribution.
Funding
The project was funded by the Slovenian Research Agency (P4-0176 and J3-7034) and an ERC project MaCChines (to RJ).
Footnotes
Data availability
The authors declare that the data supporting the findings of this study are available in the paper and its supplementary information files. The raw data are available from the corresponding author upon reasonable request.
Author contributions: TF, JL, AP, TP, KL, NJ, EM performed the experiments and analyzed the results; TF, JL, TL designed SPOC logic gates; ŽS and FL designed antiparallel CCs; MB, TF, JL, RJ wrote and edited the manuscript, MB supervised the experimental work, RJ conceptualized the study and acquired funding.
Competing financial interests: The authors declare no competing interests.
References
- 1.Cohen GB, Ren R, Baltimore D. Modular binding domains in signal transduction proteins. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 2.Nandagopal N, Elowitz MB. Synthetic biology: integrated gene circuits. Science. 2011;333:1244–8. doi: 10.1126/science.1207084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Good MC, Zalatan JG, Lim WA. Scaffold proteins: Hubs for controlling the flow of cellular information. Science (80-. ) 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordley RM, et al. Engineering dynamical control of cell fate switching using synthetic phospho-regulons. Proc Natl Acad Sci. 2016;113:13528–13533. doi: 10.1073/pnas.1610973113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dueber JE, Yeh BJ, Chak K, Lim WA. Reprogramming control of an allosteric signaling switch through modular recombination. Science. 2003;301:1904–8. doi: 10.1126/science.1085945. [DOI] [PubMed] [Google Scholar]
- 6.Howard PL, Chia MC, Del Rizzo S, Liu F-F, Pawson T. Redirecting tyrosine kinase signaling to an apoptotic caspase pathway through chimeric adaptor proteins. Proc Natl Acad Sci U S A. 2003;100:11267–72. doi: 10.1073/pnas.1934711100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lonzaric J, Fink T, Jerala R. Design and applications of synthetic information processing circuits in mammalian cells. Synthetic Biology: Volume 2. 2017:1–34. doi: 10.1039/9781782622789-00001. [DOI] [Google Scholar]
- 8.Haellman V, Fussenegger M. Synthetic biology – Engineering cell-based biomedical devices. Curr Opin Biomed Eng. 2017;4:50–56. [Google Scholar]
- 9.Kitada T, DiAndreth B, Teague B, Weiss R. Programming gene and engineered-cell therapies with synthetic biology. Science. 2018;359 doi: 10.1126/science.aad1067. eaad1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neurath H, Walsh KA. Role of proteolytic enzymes in biological regulation (a review) Proc Natl Acad Sci U S A. 1976;73:3825–32. doi: 10.1073/pnas.73.11.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strasser A, O’Connor L, Dixit VM. Apoptosis Signaling. Annu Rev Biochem. 2000;69:217–245. doi: 10.1146/annurev.biochem.69.1.217. [DOI] [PubMed] [Google Scholar]
- 12.Kovall Rhett A, Gebelein Brian, Sprinzak David, K R. The Canonical Notch Signaling Pathway: Structural and Biochemical Insights into Shape, Sugar, and Force. Dev Cell. 2017;41:228–241. doi: 10.1016/j.devcel.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Rodriguez J, Voigt CA. Post-translational control of genetic circuits using Potyvirus proteases. Nucleic Acids Res. 2016;44:6493–6502. doi: 10.1093/nar/gkw537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Majerle A, Gaber R, Benčina M, Jerala R. Function-based mutation-resistant synthetic signaling device activated by HIV-1 proteolysis. ACS Synth Biol. 2015;4:667–72. doi: 10.1021/sb5002483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarz KA, Daringer NM, Dolberg TB, Leonard JN. Rewiring human cellular input-output using modular extracellular sensors. Nat Chem Biol. 2017;13:202–209. doi: 10.1038/nchembio.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shekhawat SS, Porter JR, Sriprasad A, Ghosh I. An Autoinhibited Coiled-Coil Design Strategy for Split-Protein Protease Sensors. J Am Chem Soc. 2009;131:15284–90. doi: 10.1021/ja9050857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stein V, Alexandrov K. Protease-based synthetic sensing and signal amplification. Proc Natl Acad Sci U S A. 2014;111:15934–9. doi: 10.1073/pnas.1405220111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng N, et al. Specific and efficient cleavage of fusion proteins by recombinant plum pox virus NIa protease. Protein Expr Purif. 2008;57:153–162. doi: 10.1016/j.pep.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yi L, et al. Engineering of TEV protease variants by yeast ER sequestration screening ( YESS ) of combinatorial libraries. 2013;110 doi: 10.1073/pnas.1215994110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo J-K, Choi H-S, Kim K-H. Engineering of soybean mosaic virus as a versatile tool for studying protein–protein interactions in soybean. Sci Rep. 2016;6 doi: 10.1038/srep22436. 22436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanno A, Yamanaka Y, Hirano H, Umezawa Y, O T. Cyclic Luciferase for Real Time Sensing of Caspase 3 Activities in Living Mammals. Angew Chemie - Int Ed. 2007;46:7595–7599. doi: 10.1002/anie.200700538. [DOI] [PubMed] [Google Scholar]
- 23.Zdanov AS, Phan J, Evdokimov AG, Tropea JE, Peters HK, III, Kapust RB, Li M, Wlodawer A, Waugh DS. Tobacco Etch Virus Protease: Crystal Structure of the Active Enzyme and Its Inactive Mutant. Russ J Bioorganic Chem. 2003;29:415–418. [PubMed] [Google Scholar]
- 24.Wehr MC, et al. Monitoring regulated protein-protein interactions using split TEV. Nat Methods. 2006;3:985–93. doi: 10.1038/nmeth967. [DOI] [PubMed] [Google Scholar]
- 25.Liang F-S, Ho WQ, Crabtree GR. Engineering the ABA Plant Stress Pathway for Regulation of Induced Proximity. Sci Signal. 2011;4:18. doi: 10.1126/scisignal.2001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gradišar H, Jerala R. De novo design of orthogonal peptide pairs forming parallel coiled-coil heterodimers. J Pept Sci. 2011;17:100–6. doi: 10.1002/psc.1331. [DOI] [PubMed] [Google Scholar]
- 27.Woolfson DN. The design of coiled-coil structures and assemblies. Adv Protein Chem. 2005;70:79–112. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- 28.Grigoryan G, Keating AE. Structural specificity in coiled-coil interactions. Curr Opin Struct Biol. 2008;18:477–483. doi: 10.1016/j.sbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinke AW, Grant RA, Keating AE. A synthetic coiled-coil interactome provides heterospecific modules for molecular engineering. J Am Chem Soc. 2010;132:6025–31. doi: 10.1021/ja907617a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gradišar H, et al. Design of a single-chain polypeptide tetrahedron assembled from coiled-coil segments. Nat Chem Biol. 2013;9:362–6. doi: 10.1038/nchembio.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ljubetič A, et al. Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo. Nat Biotechnol. 2017;35:1094–1101. doi: 10.1038/nbt.3994. [DOI] [PubMed] [Google Scholar]
- 32.Crick FHC. Is α-keratin a coiled coil? Nature. 1952;170:882–883. doi: 10.1038/170882b0. [DOI] [PubMed] [Google Scholar]
- 33.Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 34.Berrington WR, Smith KD, Skerrett SJ, Hawn TR. Nucleotide-binding oligomerization domain containing-like receptor family, caspase recruitment domain (CARD) containing 4 (NLRC4) regulates intrapulmonary replication of aerosolized Legionella pneumophila. BMC Infect Dis. 2013;13:371. doi: 10.1186/1471-2334-13-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gradišar H, Jerala R. De novo design of orthogonal peptide pairs forming parallel coiled-coil heterodimers. J Pept Sci. 2011;17:100–106. doi: 10.1002/psc.1331. [DOI] [PubMed] [Google Scholar]
- 36.Ljubetič A, et al. Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo. Nat Biotechnol. 2017;35:1094–1101. doi: 10.1038/nbt.3994. [DOI] [PubMed] [Google Scholar]
- 37.Oakley MG, Hollenbeck JJ. The design of antiparallel coiled coils. Curr Opin Struct Biol. 2001;11:450–457. doi: 10.1016/s0959-440x(00)00232-3. [DOI] [PubMed] [Google Scholar]
- 38.Mittl PRE, et al. The retro-GCN4 leucine zipper sequence forms a stable three-dimensional structure. Proc Natl Acad Sci. 2000;97:2562–2566. doi: 10.1073/pnas.97.6.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drobnak I, Gradišar H, Ljubetič A, Merljak E, Jerala R. Modulation of Coiled-Coil Dimer Stability through Surface Residues while Preserving Pairing Specificity. J Am Chem Soc. 2017;139:8229–8236. doi: 10.1021/jacs.7b01690. [DOI] [PubMed] [Google Scholar]
- 40.Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys J. 1998;75:422–7. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phan J, et al. Structural Basis for the Substrate Specificity of Tobacco Etch Virus Protease *. 2002;277:50564–50572. doi: 10.1074/jbc.M207224200. [DOI] [PubMed] [Google Scholar]
- 42.Liang Fu-Sen, Ho Wen Qi, C GR. Engineering the ABA Plant Stress Pathway for Regulation of Induced Proximity. Sci Signal. 2011;4 doi: 10.1126/scisignal.2001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riechmann JL, Lain S, Garcia JA. Highlights and prospects of potyvirus molecular biology. J Gen Virol. 1992;73:1–16. doi: 10.1099/0022-1317-73-1-1. [DOI] [PubMed] [Google Scholar]
- 44.Gao XJ, Chong LS, Kim MS, Elowitz MB. Programmable protein circuits in living cells. Science (80-. ) 2018;361:1252–1258. doi: 10.1126/science.aat5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibson Daniel G, Young Lei, Chuang Ray-Yuan, Venter J Craig, HI CA, S HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 46.Gao Y, et al. Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat Methods. 2016;13:1043–1049. doi: 10.1038/nmeth.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iwai H, Züger S, Jin J, Tam P-H. Highly efficient protein trans -splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett. 2006;580:1853–1858. doi: 10.1016/j.febslet.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 48.Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 49.CRICK FHC. Is α-Keratin a Coiled Coil? Nature. 1952;170:882–883. doi: 10.1038/170882b0. [DOI] [PubMed] [Google Scholar]
- 50.Oakley MG, H J. The design of antiparallel coiled coils. Curr Opin Struct Biol. 2001;11:450–457. doi: 10.1016/s0959-440x(00)00232-3. [DOI] [PubMed] [Google Scholar]
- 51.A GGK. Structural specificity in coiled-coil interactions. Curr Opin Struct Biol. 2008;18:477–483. doi: 10.1016/j.sbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lacroix E, Viguera AR, S L. Elucidating the folding problem of alpha-helices: local motifs, long-range electrostatics, ionic-strength dependence and prediction of NMR parameters. J Mol Biol. 1998;284:173–191. doi: 10.1006/jmbi.1998.2145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.