Abstract
Background and Purpose
Animal toxins have contributed significantly to our understanding of the neurobiology of receptors and ion channels. We studied the venom of the coral snake Micrurus fulvius fulvius and identified and characterized the structure and pharmacology of a new homodimeric neurotoxin, fulditoxin, that exhibited novel pharmacology at nicotinic ACh receptors (nAChRs).
Experimental Approach
Fulditoxin was isolated by chromatography, chemically synthesized, its structure determined by X‐ray crystallography, and its pharmacological actions on nAChRs characterized by organ bath assays and two‐electrode voltage clamp electrophysiology.
Key Results
Fulditoxin's distinct 1.95‐Å quaternary structure revealed two short‐chain three‐finger α‐neurotoxins (α‐3FNTxs) non‐covalently bound by hydrophobic interactions and an ability to bind metal and form tetrameric complexes, not reported previously for three‐finger proteins. Although fulditoxin lacked all conserved amino acids canonically important for inhibiting nAChRs, it produced postsynaptic neuromuscular blockade of chick muscle at nanomolar concentrations, comparable to the prototypical α‐bungarotoxin. This neuromuscular blockade was completely reversible, which is unusual for snake α‐3FNTxs. Fulditoxin, therefore, interacts with nAChRs by utilizing a different pharmacophore. Unlike short‐chain α‐3FNTxs that bind only to muscle nAChRs, fulditoxin utilizes dimerization to expand its pharmacological targets to include human neuronal α4β2, α7, and α3β2 nAChRs which it blocked with IC50 values of 1.8, 7, and 12 μM respectively.
Conclusions and Implications
Based on its distinct quaternary structure and unusual pharmacology, we named this new class of dimeric Micrurus neurotoxins represented by fulditoxin as Σ‐neurotoxins, which offers greater insight into understanding the interactions between nAChRs and peptide antagonists.
Abbreviations
- 3FTx
three‐finger toxin
- AA
amino acid
- Boc
tertbutoxycarbonyl
- CCh
carbachol
- CBCM
chick biventer cervicis muscle
- EbTx‐a
erabutoxin‐a
- EbTx‐b
erabutoxin‐b
- EC50
half‐maximal effective concentration
- ESI‐MS
electrospray ionization MS
- Fmoc
9‐fluorenylmethoxycarbonyl
- IC50
half‐maximal inhibitory concentration
- MFV
Micrurus fulvius fulvius venom
- nFulditoxin
native fulditoxin
- RP‐HPLC
reversed‐phase HPLC
- sFulditoxin
synthetic fulditoxin
- TEVC
two‐electrode voltage clamp
- α‐3FNTx
three‐finger α‐neurotoxin
- α‐BgTx
α‐bungarotoxin
- α‐CbTx
α‐cobratoxin
What is already known
Snake three‐finger α‐neurotoxins competitively and selectively inhibit muscle and neuronal nAChRs.
Their selectivity for nAChR subtypes is defined by conserved functionally invariant amino acid residues.
What does this study add
Fulditoxin represents a novel dimeric three‐finger α‐neurotoxin family which lacks canonical functionally invariant residues.
Unlike monomeric short‐chain α‐neurotoxins, fulditoxin inhibits α7, α3β2, α4β2, and muscle nAChRs.
What is the clinical significance
This novel neurotoxin class offers new insights into interactions between peptide antagonists and nAChRs.
1. INTRODUCTION
The identification and characterization of the first neurotransmitter receptor protein, the https://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=76 (nAChR), was a significant milestone in molecular pharmacology, which was possible only with the concurrent discovery of https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=3964 (α‐BgTx), the prototypical snake neurotoxin antagonist of the receptor (Changeux, 2012). The nAChR is a heteropentameric, allosteric receptor formed by homologous subunits around a central transmembrane cation channel, with extracellular https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=294 binding sites at subunit interfaces. The combinatorial assembly of a number of different subunits (https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=462&familyId=76&familyType=IC to https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=470&familyId=76&familyType=IC, https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=471&familyId=76&familyType=IC to https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=474&familyId=76&familyType=IC, https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=476&familyId=76&familyType=IC, γ, or ε) generates a diversity of receptor subtypes with distinct physiological, pharmacological, and clinical significance. At the neuromuscular junction, the postsynaptic nAChR is of the (α1)2β1γδ (fetal) or (α1)2β1δε (adult) subunit stoichiometry. Neuronal nAChRs are composed of various combinations of https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=463&familyId=76&familyType=IC to α10 and https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=472&familyId=76&familyType=IC to β4 subunits and play significant physiological roles in the CNS including cognition, memory, and addiction and are involved in cellular signalling pathways at extraneuronal locations (see Bouzat & Sine, 2018; Wonnacott, Bermudez, Millar, & Tzartos, 2018).
The snake three‐finger α‐neurotoxins (α‐3FNTxs) constitute one of the largest groups within the three‐finger toxin (3FTx) family, characterized by three finger‐like β‐stranded loops converging at a disulfide‐rich hydrophobic core (Kessler, Marchot, Silva, & Servent, 2017; Tsetlin, 2015). Despite this conserved protein fold, 3FTxs have markedly different biological targets, illustrating functional divergence in evolution (Kini & Doley, 2010; Tsetlin, 2015). α‐3FNTxs are classified into short‐chain (Type I; 60‐62 amino acid [AA] residues with four disulfide bonds) and long‐chain (Type II; 66‐75 AA residues with five disulfide bonds) α‐3FNTxs. Both subfamilies bind with high affinity (K D 10−10–10−11 M) to muscle nAChRs, whereas long‐chain, but not short‐chain, α‐3FNTxs bind with high affinity (K D 10−8–10−9 M) to neuronal https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=468&familyId=76&familyType=IC, https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=469&familyId=76&familyType=IC, and α9/α10 nAChRs (McIntosh, Absalom, Chebib, Elgoyhen, & Vincler, 2009). Furthermore, some long‐chain α‐3FNTxs also inhibit the structurally homologous https://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=72 receptors (K D 5–20−6 M; Kudryavtsev et al., 2015). Although most α‐3FNTxs bind irreversibly to nAChRs, novel α‐3FNTxs that produced rapid and completely reversible blockade of muscle nAChRs have also been described (Harvey, Hider, Hodges, & Joubert, 1984; Nirthanan, Charpantier, et al., 2003).
The short‐chain α‐3FNTx, erabutoxin‐a (EbTx‐a), and long‐chain α‐3FNTxs, α‐cobratoxin and α‐BgTx use a common core of canonical AA residues in loops I and II, in addition to specific combinations of conserved AA residues for binding to muscle and neuronal α7 nAChRs (Antil‐Delbeke et al., 2000; Teixeira‐Clerc, Menez, & Kessler, 2002; see Figure S1). Interestingly, in Type III α‐3FNTxs (Jackson et al., 2013) and Ω‐neurotoxins (Hassan‐Puttaswamy, Adams, & Kini, 2015), two unrelated classes of nAChR antagonists, almost all conserved functional AA residues of α‐3FNTxs are replaced, revealing that these toxins may utilize alternative functional sites to bind to a common molecular target within nAChRs.
Although most 3FTxs are monomers, a small number exist as dimers. κ‐Bungarotoxin (κ‐BgTx) is a long‐chain α‐3FNTx homodimer, which binds with high affinity to https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=464&familyId=76&familyType=ICβ2 and weakly to α7 and https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=465&familyId=76&familyType=ICβ2 nAChRs, but not to muscle nAChRs (Dewan, Grant, & Sacchettini, 1994). We described haditoxin (Ophiophagus hannah), a short‐chain α‐3FNTx homodimer that inhibits muscle α1β1γδ and neuronal α7, α3β2, and α4β2 nAChRs (Roy et al., 2010) and the first covalently linked heterodimeric α‐3FNTx, irditoxin (Boiga irregularis), that exhibits taxa‐specific neurotoxicity for avian α1β1γδ nAChRs (Pawlak et al., 2009). A covalently bound α‐cobratoxin homodimer (Naja kaouthia) retains the monomeric α‐cobratoxin's ability to inhibit α1β1γδ and α7 nAChRs, but additionally, inhibits α3β2 nAChRs (Osipov et al., 2008; Osipov et al., 2012). All these dimers share functionally invariant AA residues with short‐ and long‐chain α‐3FNTxs. Thus, dimeric α‐3FNTxs constitute a structurally heterogeneous group of toxins exhibiting novel structural conformations which enables them to diversify their receptor specificity (Tsetlin, 2015).
Human envenomation by Micrurus coral snakes causes high mortality due to neurotoxicity attributed to α‐3FNTxs and https://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=275 β‐neurotoxins (Kitchens & Van Mierop, 1987). Micrurus 3FTxs exhibit significant sequence diversity as observed in transcriptomic analyses of their venom glands (Correa‐Netto et al., 2011; Guerrero‐Garzon et al., 2018; Margres, Aronow, Loyacano, & Rokyta, 2013); and they lack the conserved functional AA residues from previously characterized clades of 3FTxs, providing an impetus to investigate novel Micrurus toxins (Dashevsky & Fry, 2018).
Here, we describe the identification, and pharmacological and structural characterization of a dimeric α‐3FNTx, fulditoxin, from Micrurus fulvius fulvius venom. The 1.95‐Å crystal structure of fulditoxin revealed a homodimer of short‐chain α‐3FNTxs non‐covalently bound by hydrophobic interactions, with the ability to form a tetrameric complex in the presence of https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=566 (Zn2+), exhibiting a unique metal‐binding capability not previously reported for 3FTxs. Interestingly, despite lacking all the functionally important AA residues critical for α‐3FNTx interaction with nAChRs, fulditoxin produced potent, but completely reversible, postsynaptic neuromuscular blockade, as well as broad spectrum inhibition of α1β1δε, α4β2, α7, and α3β2 nAChRs.
2. METHODS
2.1. Animal experimentation protocols
Domestic chicks (Gallus gallus domesticus) were purchased from Chew's Agricultural Pte Ltd (Singapore; ISO9001 certified) and delivered on the day of experimentation and killed by exposure to 100% carbon dioxide. All experiments were conducted in accordance with Protocol 103/08A approved by the Institutional Animal Care and Use Committee (IACUC) of the National University of Singapore which conforms to the World Health Organization's International Guiding Principles for Animal Research (Howard‐Jones, 1985), adapted by the Council for International Organizations of Medical Sciences in 1985.
For electrophysiological studies using the Xenopus oocyte expression system, all experiments on Xenopus laevis frogs conformed to the Geneva Canton Rules on Animal Experimentation (Accreditation # G171/3551) and the RMIT University Animal Ethics committee (Protocol # 1222, 1223). Female X. laevis frogs were sourced from the Centre de Resources Biologiques (Rennes, Cedex, France) and maintained in an aquarium as approved by the Ethics Committee for Animal Experimentation (Swiss Academy of Medical Sciences) and from Nasco (Fort Atkinson, WI, USA) and housed in the RMIT Aquatic Facility. A maximum of three frogs were kept in purpose‐built 10‐L tanks at 20–25°C with 12‐hr light/dark cycle within. Electrophysiological experiments were performed using oocytes obtained from three ~5‐year‐old frogs. Frogs were anaesthetized with 1.7 mg·ml−1 of ethyl 3‐aminobenzoate methanesulfonate (pH 7.4 with NaHCO3), and for post‐surgery recovery, animals were placed in fresh water at a level below the nostrils. Frogs were left to recover for a minimum of 4 months between surgeries. Terminal anaesthesia with 5.0 mg·ml−1 of ethyl 3‐aminobenzoate methanesulfonate (pH 7.4 with NaHCO3) was performed on frogs at the sixth surgery.
Animal studies are reported in compliance with the ARRIVE guidelines (Kilkenny, Browne, Cuthill, Emerson, & Altman, 2010; McGrath, Drummond, McLachlan, Kilkenny, & Wainwright, 2010) and with the recommendations made by the British Journal of Pharmacology.
2.2. Chromatographic purification of proteins from Micrurus fulvius venom
The isolation and purification of fulditoxin from crude Micrurus fulvius venom (MFV) was carried out using established protocols in our laboratory (Hassan‐Puttaswamy, Adams, & Kini, 2015; Pawlak et al., 2009; Roy et al., 2010). Crude MFV (50 mg dissolved in 1‐ml MilliQ water and filtered using a 0.45‐μm syringe filter) was loaded onto a Superdex 30 Hiload size‐exclusion chromatography column, equilibrated with 50‐mM Tris‐hydrochloric acid (Tris–HCl) buffer (pH 7.4), and proteins were eluted with the same buffer using an ÄKTA purifier system (GE Healthcare, Little Chalfont, UK). Fractions containing the protein of interest were further purified by reversed phase‐HPLC (RP‐HPLC) using a Jupiter C18 preparative column that was equilibrated with 0.1% (v/v) TFA. Proteins were eluted with a linear gradient of 80% (v/v) MeCN in 0.1% (v/v) TFA. Elution of proteins was monitored at 280 and 215 nm.
2.3. Determination of molecular mass and homogeneity of fulditoxin
2.3.1. Electrospray ionization‐MS
The molecular mass and homogeneity of the purified protein were determined by injecting the protein samples into an API‐300 LC‐tandem MS system (PerkinElmer Life Sciences, Massachusetts, USA) as described previously (Roy et al., 2010). Ion spray, orifice, and ring voltages were set at 4600, 50, and 350 V respectively. Nitrogen was used as the nebulizer and curtain gas. A Shimadzu LC‐10 AD pump was used for solvent delivery [50% (v/v) MeCN in 0.1% (v/v) formic acid] at a flow rate of 40 μl·min−1. Analyst software (PerkinElmer Life Sciences, MA, USA) was used to analyse and deconvolute the raw MS data. Fractions that showed the expected molecular mass of fulditoxin were pooled and lyophilized.
2.3.2. Capillary zone electrophoresis
The homogeneity of purified fulditoxin was assessed by capillary zone electrophoresis which was performed on a BioFocus3000 system (Bio‐Rad, Foster City, CA, USA). The native protein was dissolved in MilliQ water (1.9 μg·μl−1) and injected into a 25 μm × 17 cm coated capillary under a pressure mode (5 ψ·s−1) and run in 0.1‐M phosphate buffer (pH 2.5) under 18 kV at 20°C for 7 min. Migration of the protein sample was monitored at 200 nm.
2.4. N‐terminal AA sequencing of fulditoxin
Amino terminal sequencing of the native fulditoxin (nFulditoxin) and peptides derived from chemical digestion was performed by automated Edman degradation using a Procise Model 494 pulsed liquid‐phase sequencer (PerkinElmer Life Sciences, Massachusetts, USA) with an online 785 A phenylthiohydantoin derivative analyser. The phenylthiohydantoin AAs were subsequently identified by mapping the respective separation profiles with the standard chromatogram.
2.4.1. Protein reduction and alkylation
Lyophilized protein (1.04 mg) was dissolved in 520 μl of denaturing buffer (0.13 mol·L−1 of Tris–HCl, 1 mmol·L−1 of EDTA, 6 mol·L−1 of guanidine‐HCl, pH 8.5). β‐Mercaptoethanol (1.05 μl) was added, and the solution was incubated under nitrogen at room temperature for 2 hr. The alkylating agent, 4‐vinylpyridine (4.82 μl), was subsequently added and the solution was incubated under nitrogen for another 3 hr at room temperature. After the reaction, the S‐pyridylethylated protein was separated from the reaction mixture by RP‐HPLC on a Jupiter C18 semi‐preparative column using a linear gradient of 80% MeCN in 0.1% TFA at a flow rate of 2 ml·min−1. The molecular mass of the S‐pyridylethylated protein was determined by electrospray ionization‐MS (ESI‐MS).
2.4.2. Chemical cleavage
The S‐pyridylethylated protein was subjected to chemical cleavage at the C‐terminus of methionine residues with cyanogen bromide (CNBr). The S‐pyridylethylated protein (500 μg) was dissolved in 250 μl of 70% TFA to which 5 μl of β‐mercaptoethanol was added. A 200‐fold molar excess (over methionine residues) of CNBr in 70% TFA was added to make a final protein concentration of 1 mg·ml−1. After 24 hr of incubation in the dark at room temperature, 10 volumes of MilliQ water was added to the reaction mixture and lyophilized. Lyophilized sample containing cleaved peptides were reconstituted in 0.1% TFA and separated by RP‐HPLC on a Jupiter C18 analytical column using a linear gradient of 80% MeCN in 0.1% TFA at a flow rate of 1 ml·min−1. Molecular masses of the separated peptide fragments were determined by ESI‐MS and their AA sequences determined by N‐terminal sequencing. The complete AA sequence of fulditoxin was determined by alignment of the sequences of the CNBr‐cleaved peptides.
2.5. Size‐exclusion chromatography for determination of oligomeric states of fulditoxin
The oligomeric states of fulditoxin (0.15–1.5 μM) were determined by analytical size‐exclusion chromatography using a Superdex 75 column (1 × 30 cm) equilibrated with 50 mM of Tris–HCl buffer, pH 7.4, in the absence and presence of 8‐M urea. Size‐exclusion chromatography was carried out on an ÄKTA purifier system at a flow rate of 0.6 ml·min−1. Columns were calibrated with BSA (66 kDa), carbonic anhydrase (29 kDa), cytochrome C (12.9 kDa), aprotinin (6.5 kDa), and blue dextran (200 kDa) as molecular mass markers. Native proteins as well as protein samples in 8‐M urea were loaded separately onto the column.
2.6. Chemical synthesis of fulditoxin
Using established protocols (Dawson, Muir, Clark‐Lewis, & Kent, 1994; Schnolzer, Alewood, Jones, Alewood, & Kent, 1992), the C‐terminal 42‐63 fragment (sequence CHEGAYNVCCSTDLCNKSSTSG) of fulditoxin was assembled on a Symphony (Protein Technologies Inc.) automated peptide synthesizer on an Fmoc (9‐fluorenylmethoxycarbonyl)–Gly–Wang polystyrene resin (loading 0.37 mmol·g−1) on a 0.1‐mmol scale. Fmoc deprotections were achieved using 30% piperidine/N,N‐dimethylformamide (DMF; 1 × 1.5 min, then 1 × 4 min). Couplings were performed in DMF using 5 equivalents of Fmoc‐AA/2‐(1H‐benzotriazol‐1‐yl)‐1,1,3,3‐tetramethyluronium hexafluorophosphate (HBTU)/(N,N‐diisopropylethylamine) DIEA (1:1:1) relative to resin loading for 2 × 20 min. AA side chains were protected as Asn (Trityl [Trt]), Asp (t‐Butyl ester [OtBu]), Cys (Trt), Glu (OtBu), His (Trt), Lys (tert‐butoxycarbonyl [Boc]), Ser (tertbutyl [tBu]), Thr (tBu), and Tyr (tBu). Cleavage from the resin and removal of side‐chain protecting groups was achieved by treatment with 95% TFA/2.5% triisopropylsilane (TIPS)/2.5% water at room temperature for 2 hr. After most of the cleavage solution was evaporated under a stream of nitrogen gas, the product was precipitated and washed with cold diethyl ether (Et2O) and lyophilized from 50% MeCN/0.1% TFA/water. The crude product was purified by preparative HPLC. Mass of the purified product was determined by ESI‐MS (described below).
The N‐terminal 1‐41‐thioester fragment of fulditoxin (sequence LKCYSSRTETMTCPEGEDKCEKYAVGLMHGSFFFIYTCTSK‐COSR) was assembled using manual Boc protocols on a 0.25‐mmol scale. Boc deprotections were achieved using TFA (2 × 1 min). Couplings were performed in DMF using four equivalents of Boc‐AA/HBTU/DIEA (8:8:11) relative to resin loading for a minimum of 10 min. AA side chains were protected as Asp (Cyclohexyl ester [OcHxl]), Arg (4‐toluenesulfonyl [Tos]), Cys (4‐methylbenzyl [Meb]), Glu (OcHxl), His (Benzyloxymethyl [Bom]), Lys (2‐chlorobenzyloxycarbonyl [2‐ClZ]), Ser (benzyl [Bzl]), Thr (Bzl), and Tyr (2‐bromobenzyloxycarbonyl [2‐BrZ]). S‐trityl‐3‐mercaptopropionic acid was coupled to Arg (Tos)‐4‐(oxymethyl) phenylacetamidomethyl (OCH2‐PAM) polystyrene resin (loading 0.56 mmol·g−1) then the S‐trityl group removed with 2 × 1 min treatments of 5% TIPS/TFA. The first AA of the sequence (Lys) was coupled to the resultant free thiol to form the thioester. Cleavage from the resin and removal of side‐chain protecting groups was achieved by treatment with anhydrous hydrogen fluoride (HF)/p‐cresol/H‐Cys‐OEt at 0°C for 1 hr. After evaporation of HF, the product was precipitated and washed with cold Et2O and lyophilized from 50% MeCN/0.1% TFA/water. The crude product was purified by preparative HPLC and its mass determined by ESI‐MS (described below).
Both fragments of fulditoxin were purified by RP‐HPLC using solvents, 0.05% TFA/water as solution A and 90% MeCN/0.043% TFA/water as solution B. Analytical HPLC was performed on a Shimadzu LC20AT system using a Thermo Hypersil GOLD 2.1 × 100 mm C18 column heated at 40°C with flow rate of 0.3 ml·min−1. A gradient of 10 to 55% B over 30 min was used, with detection at 214 nm. Preparative HPLC was performed on a Vydac 218TP1022 column (22 × 250 mm) running at a flow rate of 16 ml·min−1 using a gradient of 10 to 50% B over 40 min. MS was performed on an API2000 (ABI Sciex) mass spectrometer in positive ion mode.
2.6.1. Native chemical ligation
Purified 1‐41‐thioester (22 mg) and 42‐63 (11 mg) fragments were combined and stirred in a solution containing 6‐M guanidine hydrochloride (GnHCl)/0.2‐M sodium phosphate/20‐mM tris (2‐carboxyethyl) phosphine (TCEP)/50‐mM MPAA (pH 7.0, 2.5 ml) for 16 hr at room temperature under an argon atmosphere; 1‐63 octathiol fulditoxin was isolated by preparative HPLC (8 mg) and its mass determined by ESI‐MS.
2.6.2. Oxidative folding
Purified reduced peptide (8 mg), reduced GSH (100 equivalent), and oxidized GSH (10 equivalent) were dissolved in 6‐M GnHCl (8 ml) then added to a solution of 1.55‐M GnHCl/0.37‐M ammonium acetate (pH 8.0, 71 ml) and stirred at 4°C with exposure to air for 48 hr. The predominant product was isolated by preparative HPLC (2 mg) and its mass determined by ESI‐MS. Analytical HPLC was carried out to compare the synthetic fulditoxin (sFulditoxin) with venom‐purified native protein, nFulditoxin.
2.7. Screening of fulditoxin action on isolated tissues in organ bath studies
Isolated tissue experiments were conducted as described previously (Nirthanan, Gao, et al., 2002; Nirthanan, Charpantier, et al., 2003; Pawlak et al., 2009; Roy et al., 2010) using a conventional organ bath (6 ml) containing physiological Krebs–Henseleit buffer of the following composition (in mM): 118‐mM NaCl, 4.8 KCl, 1.2 KH2PO4, 2.5 CaCl2, 2.4 MgSO4, 25 NaHCO3, and 11 D‐(+) glucose), pH 7.4. Organ bath chambers were continuously aerated with carbogen (5% carbon dioxide in oxygen) and maintained at 37°C throughout the experiment. The resting tension of the isolated tissues was maintained between 1‐ and 2‐g tension, and the tissues were allowed to equilibrate for 30–45 min before the start of an experiment. Electrical field stimulation was carried out through platinum ring electrodes using a Grass stimulator S88 (Grass instruments, West Warwick, Rhode Island, USA). The magnitude of the contractile responses was measured in gram tension. Data were continuously recorded on a PowerLab LabChart 6 data acquisition system using a force displacement transducer (Model MLT0201; ADInstruments, Bella Vista, NSW, Australia).
2.7.1. Chick biventer cervicis muscle preparation
The chick biventer cervicis muscle (CBCM) nerve‐skeletal muscle preparation (Ginsborg & Warriner, 1960) was isolated from 3‐ to 5‐day‐old chicks and mounted in the organ bath chamber under similar experimental conditions as described above. Motor responses of the muscle were evoked by stimulating the motor nerve supramaximally by electrical field stimulation (7–10 V, 0.1 ms, 0.2 Hz). Submaximal contractures to exogenously applied ACh (200 μM for 30 s), https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=298 (CCh; 20 μM for 90 s), and KCl (30 mM for 60 s) were obtained in the absence of electrical field stimulation, prior to the addition of the toxin, and after complete blockade of nerve‐evoked twitch responses in the muscle. The effect of crude MFV (1–100 μg·ml−1) and fulditoxin (5–500 nM) on nerve‐evoked twitch responses of the CBCM was studied. The neuromuscular blockade produced by nFulditoxin was compared with that produced by EbTx‐b (0.01–1 μM) and α‐BgTx (0.001–1 μM). The neuromuscular blockade was expressed as a percentage of the original twitch height, after 30 min of exposure of the CBCM to either crude MFV or toxins. The recovery of the CBCM from complete neuromuscular blockade produced by crude MFV or toxins was assessed by washing out the toxin by bath overflow (16 ml·min−1) for the first 5 min followed by a slow drip‐wash at a flow rate of ∼8 ml·min−1 with fresh Krebs solution until maximal recovery. Three independent experiments were conducted for each concentration of toxin tested.
2.8. Electrophysiological studies on native and sFulditoxin
2.8.1. Electrophysiological studies on nFulditoxin
Functional characterization of nFulditoxin was carried out via electrophysiological recordings on nAChRs expressed in Xenopus oocytes using an automated two‐electrode voltage clamp (TEVC) system (GeneClamp Amplifier, Axon Instruments, Foster City, CA, USA) as described previously (Nirthanan, Charpantier, et al., 2002; Roy et al., 2010). The isolation, preparation, and injection of Xenopus oocytes were carried out as described previously and all experiments conformed to the Geneva canton rules on animal experimentation (accreditation number G171/3551; Hogg, Bandelier, Benoit, Dosch, & Bertrand, 2008). Oocytes were injected intra‐nuclearly with expression vectors containing the various cDNAs (2 ng) that encode for the different nAChR subunits using a Roboinject Automatic Injection System (Multi Channel Systems, Reutlingen, Germany) and maintained in 96‐well plate at 18°C in filtered Barth solution containing NaCl (88 mM), KCl (1 mM), NaHCO3 (2.4 mM), HEPES (10 mM, pH 7.5), MgSO4·7H2O (0.82 mM), Ca (NO3)2·4H2O (0.33 mM), CaCl2·6H2O (0.41 mM), at pH 7.4, and supplemented with 20 μg·ml−1 of kanamycin, 100 U·ml−1 of penicillin, and 100 μg·ml−1 of streptomycin. TEVC recordings were made 2 to 3 days after injection. The oocytes were clamped at −100 mV and perfused with OR2 (oocyte Ringer) medium containing (in mM) NaCl (82.5), KCl (2.5), MgCl2 (1.0), CaCl2 (2.5), HEPES (5.0), and 20 μg·ml−1 of BSA, at pH 7.4, during recordings. https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=320 (0.5 μM) was added to all solutions to block all activity attributed to muscarinic AChRs. ACh and nFulditoxin were prepared fresh in OR2 solution and applied to the bath by gravity‐driven perfusion. Control responses were recorded before toxin exposure and toxin incubation were carried out in the recording chamber between 3 and 5 min with agitation. All recordings were performed with a TEVC automated robot system. The data were digitized and analysed offline using MATLAB (Mathworks, Natick, MA, USA; RRID:SCR_001622).
2.8.2. Electrophysiological studies on sFulditoxin
Independent of the experiments described above, functional characterization of sFulditoxin at receptor level was carried out using TEVC electrophysiology as described previously (Hassan‐Puttaswamy, Adams, & Kini, 2015). Stage V–VI oocytes were harvested from mature female Xenopus laevis under anaesthesia with 0.1% Tricaine as approved by the RMIT Animal Ethics Committee (Protocol # 1222, 1223). cRNA encoding for nAChR subunits were prepared and microinjected into oocytes for nAChR expression. Briefly, cDNA encoding the human α3, α4, α9, α10, α7, β2, and β4 subunits, and rodent α1, β1, γ, δ, and ε subunits of nAChRs were sub‐cloned into the oocyte expression vector pT7Ts and used for cRNA preparation using the mMESSAGE mMACHINE kit (Ambion®, Life Technologies Australia Pty. Ltd., Mulgrave, VIC, Australia); 25‐ng cRNA was microinjected into oocytes, at α:β ratio of 1:1 for the human heteromeric neuronal nAChR subtypes and α1:β1:δ:ε ratio of 2:1:1:1 for the rat muscle receptor. The oocytes were kept at 18°C in ND96 buffer (96‐mM NaCl, 2‐mM KCl, 1‐mM MgCl2, and 5‐mM HEPES, pH 7.4) supplemented with 50 mg·L−1 of gentamicin and 100 units/μg·ml−1 of penicillin–streptomycin for 2 to 5 days before recordings were undertaken. The oocytes were voltage clamped (virtual ground circuit) using a GeneClamp 500B amplifier (Molecular Devices, Sunnyvale, CA, USA). Voltage recording and current‐injecting electrodes were pulled from borosilicate glass capillaries (GC150T‐7.5, Harvard Apparatus Ltd., Holliston, MA, USA) and filled with 3‐M KCl to have resistances of 0.3–1.5 MΩ.
All TEVC recordings were done manually, at room temperature (20–23°C), using a bath solution of ND96. The oocytes were continuously perfused at a flow rate of 2 ml·min−1 with ND96 buffer during recordings, with toxin incubated for 5 min before ACh was added. The concentration of ACh, calculated to be the EC50 concentration for the respective nAChR subtype, was applied for 2 s at 2 ml·min−1, with 5‐min washout periods between applications. Oocytes were voltage clamped at a holding potential of −80 mV for nAChRs. Data were filtered at 100 Hz and sampled at 500 Hz.
2.9. Data analyses
In screening of fulditoxin on various nAChR subtypes, concentration response curves for antagonists were fitted by unweighted non‐linear regression to the logistic equation:
Y = bottom+(top – bottom)/(1+10 ^ ((Log IC50 – X) * Hill Slope)), where Y is the normalized response, X is the antagonist concentration, top and bottom are maximal and minimal normalized responses, respectively, and IC50 is the antagonist concentration giving 50% inhibition of the maximal response.
EC50 shift of concentration–response plots were used to determine the competitive or non‐competitive nature of the antagonist. The EC50 shift concentration–response plot (Arunlakshana & Schild, 1959) was plotted using the equation:
Y = bottom+(top – bottom)/(1+10 ^ ((Log EC – X) * Hill Slope)), where EC50 is the concentration of agonist that gives half maximal response; pA2 is the negative logarithm of the concentration of antagonist needed to shift the concentration–response curve by a factor of 2; Hill slope describes the steepness of the family of curves; Schild slope quantifies how well the shifts correspond to the prediction of competitive interaction and if the action is competitive, the Schild slope will equal 1.0, and top and bottom are plateaus in the units of the Y axis. Each data point represents the average ± SEM of three independent experiments. Computation was done using GraphPad Prism 6.03 (GraphPad software, Inc., La Jolla, CA, USA; RRID:SCR_002798).
The data and statistical analysis in this study comply with the recommendations of the British Journal of Pharmacology on experimental design and analysis in pharmacology (Curtis et al., 2018). Given the limitations in the availability of fulditoxin due to its ultra‐low yield in venom (<0.2%), and complexity in its chemical synthesis, statistical analyses have not been performed because fewer than five independent experiments were performed during the course of our studies. The pharmacology studies were not blinded as there was no expected outcome of screening fulditoxin across nAChR subtypes for activity and determining the concentration–response relationships.
2.10. Crystal structure determination of nFulditoxin
Crystallization screens were performed using Hampton Research Crystal Screen I and II by hanging‐drop vapour diffusion method in the mosquito crystallization robot. Initial hits were obtained from the condition containing 0.2‐M zinc acetate dihydrate, 0.1‐M sodium cacodylate trihydrate, pH 6.5, 18% w/v polyethylene glycol 8000. The identified initial condition was further optimized using grid screen, which was further optimized by grid screening. The complete diffraction data from nFulditoxin crystals were collected at Beamline X6A of the National Synchrotron Light source. The crystals diffracted up to 1.95‐Å resolution (Table 1). The data sets were processed and scaled using the program HKL2000 (Otwinowski & Minor, 1997). The Matthews coefficient was calculated as 2.18 Å3/Da (Matthews, 1968) corresponding to a solvent content of 44%. The structure was solved by the molecular replacement method using the program Phaser from the Phenix suite (McCoy et al., 2007; Zwart et al., 2008; RRID:SCR_014224) with β‐cardiotoxin (PDB ID 3PLC) as the search model. The initial model was refined using Phenix‐refine followed by model building using the Phenix AutoBuild program (Adams et al., 2004). The missing residues were manually built using the Coot program (Emsley & Cowtan, 2004; RRID:SCR_014222), followed by refinement with Phenix‐refine (Adams et al., 2002). No non‐crystallographic symmetry restrain was used in the final refinement cycle. Finally, 141 well‐defined water molecules were added, and the R value converged to 20.0% (R free = 23.7%) with good stereochemical parameters (Table 1).
Table 1.
Crystallographic data and refinement statistics for fulditoxin
| Data collection | |
|---|---|
| Unit cell parameters (Å) | a = b = 58.8, c = 70.57 |
| Space group | P4212 |
| Resolution range (Å) | 50–1.95 (1.98–1.95) |
| Wavelength (Å) | 0.97640 |
| Observed reflections | 130267 |
| Unique reflections | 9570 |
| Completeness (%) | 99.9 (100) |
| Overall I/σ(I) | 10.8 |
| Redundancy | 13.6 (13.7) |
| R Sym (%)a | 0.08 (0.42) |
| Refinement and quality | |
| Resolution range (Å) | 30–1.95 |
| R work (%)b | 20.1 |
| R free (%)c | 23.7 |
| RMSD bond length (Å) | 0.007 |
| RMSD bond angles (°) | 1.244 |
| Average B‐factors (Å2) | |
| Main chain | 18.89 |
| Side chain and waters | 23.37 |
| Ramachandran plot | |
| Most favoured regions (%) | 85.3 |
| Additionally allowed regions (%) | 14.7 |
| Generously allowed regions (%) | 0 |
| Disallowed regions (%) | 0 |
Rsym = ∑|Ii − <I>|/∑|Ii|, where Ii is the intensity of the ith measurement and <I> is the mean intensity for that reflection.
Rwork = ∑|Fobs − Fcalc|/∑|Fobs|, where Fcalc and Fobs are the calculated and observed structure factor amplitudes respectively.
Rfree = as for Rwork, but for 10.0% of the total reflections chosen at random and omitted from refinement.
2.11. Dynamic light scattering
The apparent hydrodynamic radii of fulditoxin (5 mg·ml−1) in the absence and presence of Zn2+ (3–30 μM) were examined by Protein Solutions DynaPro‐99‐E‐50 Dynamic Light Scattering Module (Wyatt Technology, Santa Barbara, CA, USA).
2.12. Materials
Lyophilized MFV was purchased from Medtoxin Venom Laboratories (DeLand, FL, USA). α‐BgTx and EbTx‐b were purchased from Latoxan (Valence, France). Superdex 30 Hiload (16/60), Superdex 75 (1 × 30 cm), Jupiter C18 preparative (5 μm, 300 Å, 21.2 × 250 mm), semi‐preparative (5 μm, 300 Å, 10 × 250 mm), and analytical (5 μm, 300 Å, 4.6 × 250 mm) columns were purchased from GE Healthcare (Little Chalfont, UK). The following reagents and chemicals used for purification of MFV were from the sources indicated: reagents for N‐terminal sequencing by Edman degradation (Applied Biosystems, Foster City, California, USA), potassium chloride (KCl), acetonitrile (MeCN), and trifluoroacetic acid (TFA; Merck KGaA, Darmstadt, Germany). Crystal screening solution and accessories were purchased from Hampton Research (Aliso Viejo, California, USA). All other chemicals were purchased from Sigma‐Aldrich (St Louis, MO, USA). All the reagents used were of the highest purity grade. Water was purified using a MilliQ system (MilliPore, Billerica, MA, USA).
2.13. Data deposition
The coordinates and structure factors of fulditoxin are deposited in the Protein Data Bank (http://www.pdb.org; RRID:SCR_006555) with the code 4RUD.
2.14. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20 (Alexander et al., 2019).
3. RESULTS
3.1. Bioassay‐guided purification of fulditoxin from Micrurus fulvius venom
Organ bath experiments with the isolated chick biventer cervicis nerve‐skeletal muscle preparation (CBCM; Ginsborg & Warriner, 1960) were used as a screening bioassay to undertake two‐step HPLC chromatography of MFV to isolate and purify the neurotoxic components. Crude MFV was first fractionated based on MW, by size‐exclusion chromatography (Figure 1a), and fractions which elicited neuromuscular blocking activity (indicated by horizontal black bar) were pooled and sub‐fractionated by RP‐HPLC (Figure 1b). Peak 3c (indicated by an arrow), which elicited potent neuromuscular blockade, was further purified to homogeneity by RP‐HPLC using a shallow gradient (Figure 1c). The molecular mass and homogeneity of fulditoxin was determined by ESI‐MS to be 6947.4 ± 0.6 Da (Figure 1d). Capillary electrophoresis confirmed the homogeneity of purified fulditoxin, revealing a single protein peak in the electropherogram indicating the absence of contaminants (Figure 1e).
Figure 1.
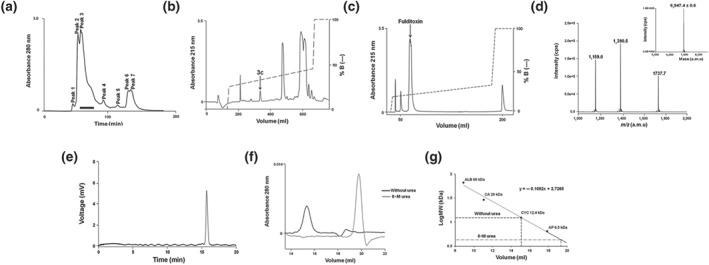
Isolation and purification of fulditoxin from crude Micrurus fulvius fulvius venom. Multi‐step HPLC purification of M. fulvius venom, determination of fulditoxin's MW by MS, and determination of its purity by capillary electrophoresis. (a) Size‐exclusion chromatography of crude MFV (50 mg·ml−1) on a Superdex 30 HiLoad (16/60) column. The column was pre‐equilibrated with Tris–HCl buffer (50 mM), pH 7.4. Proteins were eluted at a flow rate of 1.0 ml·min−1 using the same buffer. Protein elution was monitored at 280 nm. The fractions indicated by a horizontal black bar corresponding to peak 3 were pooled and subjected to RP‐HPLC. (b) RP‐HPLC profile of size‐exclusion chromatography peak 3 on a Jupiter C18 preparative column (5 μm, 300 Å, 21.2 × 250 mm) using a linear gradient (25–50% over 108 min; dotted line) of buffer B (80% ACN in 0.1% TFA) at a flow rate of 5 ml·min−1. Protein elution was monitored at 215 nm. The peak indicated by an arrow corresponding to peak 3c was rechromatographed on a shallower gradient. (c) Rechromatogram of peak 3c obtained by RP‐HPLC on a semi‐preparative column (5 μm, 300 Å, 10 × 250 mm) using a shallow gradient (25–45% over 80 min; dotted line) of buffer B (80% ACN in 0.1% TFA) at a flow rate of 2 ml·min−1. Protein elution was monitored at 215 nm. The resulting protein peak indicated by the arrow was named fulditoxin. (d) ESI‐MS profile of fulditoxin. The spectrum shows a series of multiply charged ions with mass/charge (m/z) ratios ranging from +4 to +6 charges. Inset, reconstructed mass spectrum of fulditoxin corresponding to a single homogeneous protein with MW of 6947.4 Da; CPS = counts s−1; a.m.u = atomic mass units. (e) Capillary electrophoresis of fulditoxin shows a single peak supporting homogeneity. (f) Analytical size‐exclusion chromatography profile of fulditoxin; 1.5‐μM fulditoxin was loaded onto a Superdex 75 column (1 × 30 cm) equilibrated with 50‐mM Tris–HCl buffer, pH 7.4 at a flow rate of 0.6 ml·min−1 in the presence and absence of 8‐M urea respectively. (G) MW determination on size‐exclusion column. Column calibration was done using BSA (ALB; 66 kDa), carbonic anhydrase (CA; 29 kDa), cytochrome c (CYC; 12.9 kDa), aprotinin (AP; 6.5 kDa), and blue dextran (200 kDa) as molecular mass markers
3.2. Fulditoxin forms a non‐covalent dimer in solution
During size‐exclusion chromatography, it was observed that fulditoxin was contained in peak 3 (Figure 1a), in contrast to most 3FTxs which eluted later, as observed in our previous studies (Nirthanan, Charpantier, et al., 2002; Rajagopalan et al., 2007). This suggested that fulditoxin may exist in dimeric form. In analytical size‐exclusion chromatography, fulditoxin eluted as a single peak corresponding to a relative molecular mass of 3.88 kDa in the presence of 8‐M urea, compared to as a single peak corresponding to a mass of 11.56 kDa in the absence of urea (Figure 1f). Hence, this purified toxin was named “fulditoxin”—Micrurus fulvius fulvius dimeric neurotoxin.
3.3. Amino acid sequence of fulditoxin
N‐terminal sequencing of fulditoxin was carried out by automated Edman degradation. The first 40 AA residues of the toxin were identified by direct sequencing of the native toxin, and the remaining residues were determined by sequencing of overlapping fragments of chemically‐cleaved fulditoxin (see Figure S2). The calculated mass of 6946.85 Da of the full‐length fulditoxin sequence agreed with its experimentally determined molecular mass (6947.4 ± 0.6 Da). Basic Local Alignment Sequence Tool (BLAST) search (Altschul et al., 1997; RRID:SCR_004870) showed that fulditoxin shares the highest identity (68–99%) with sequences of Micrurus 3FTxs identified by transcriptomic analyses (Correa‐Netto et al., 2011; Margres, Aronow, Loyacano, & Rokyta, 2013; Rey‐Suarez et al., 2012; Vergara et al., 2014; Figure 2a).
Figure 2.
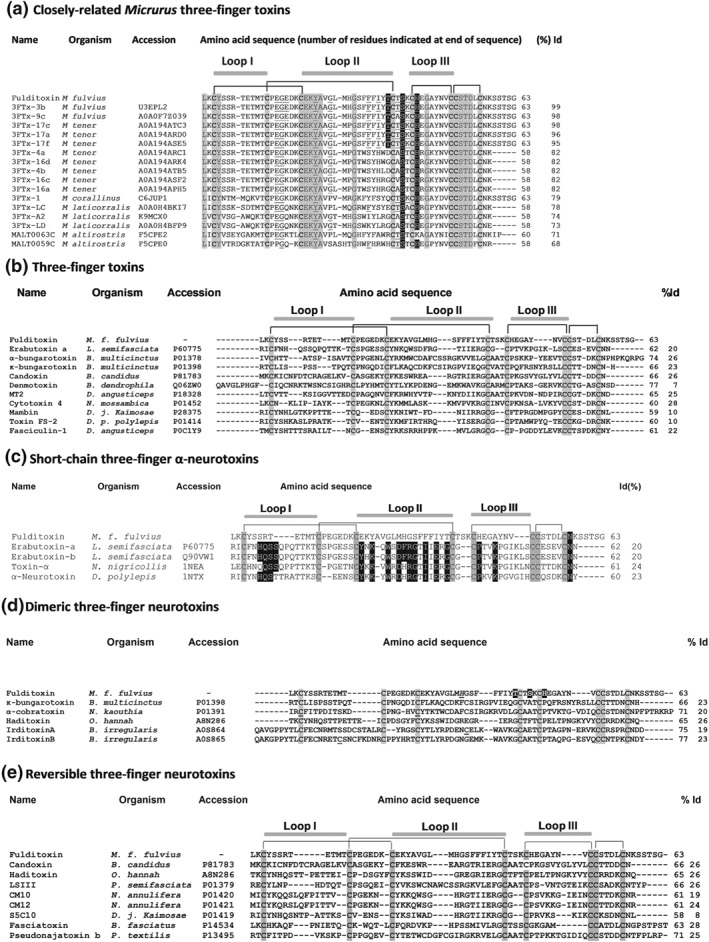
Comparison of the amino acid sequence of fulditoxin with sequences other snake three‐finger toxins. In all panels, the accession numbers and the source organism are indicated. Only the conserved cysteine residues within each group are shaded in grey. The number of amino acid (AA) residues and the percentage identity (% Id) of the respective toxins with fulditoxin is indicated at the end. The disulfide linkages and segments contributing to the three loops are also shown. (a) The AA sequence of fulditoxin was subjected to Protein BLAST® search for sequence homology with protein databases. The top 16 protein sequences that shared the highest sequence homology with fulditoxin are shown. Identical AA residues across all 17 sequences are shaded in grey. The AA residues that contribute to hydrogen bond formation (shaded in black) and hydrophobic interactions (underlined) in the dimeric interface of fulditoxin are highlighted across the sequences. (b) The sequence of fulditoxin as compared with the sequences of other 3FTxs, each representing a different subfamily: erabutoxin‐a, short‐chain α‐neurotoxin; α‐bungarotoxin, long‐chain α‐neurotoxin; κ‐bungarotoxin, κ‐neurotoxins; candoxin, elapid non‐conventional three‐finger α‐neurotoxin; denmotoxin, colubrid non‐conventional three‐finger α‐neurotoxins; MT2, muscarinic three‐finger toxin; cytotoxin 4, cardiotoxin; mambin, antagonist of cell‐adhesion processes; Toxin FS‐2, L‐type calcium channel antagonist; and fasciculin‐1, AChE inhibitor. (c) Comparison of the AA sequence of fulditoxin with the sequences of short‐chain 3F‐αNTxs. The conserved AA residues in short‐chain 3F‐αNTxs deemed critical for the recognition and binding to muscle‐type nAChRs are highlighted in black. (d) Comparison of the AA sequence of fulditoxin with the sequences of dimeric 3F‐NTxs. The AA residues in fulditoxin contributing to dimerization are highlighted in black; and His29 involved in Zn2+ binding is italicized and underlined. The cysteine residues involved in the formation of intermolecular disulfide linkages in the α‐cobratoxin dimer and irditoxin A–irditoxin B dimer are underlined. (e) Comparison of the AA sequence of fulditoxin with the sequences of reversible neurotoxins
Multiple sequence alignment of fulditoxin with other classes of 3FTxs, including α‐neurotoxins, cardiotoxins, cytotoxins, and AChE inhibitors, showed little sequence similarities (Figure 2b). Fulditoxin shared only ~20% to 30% sequence identity with other monomeric short‐chain α‐3FNTxs (Figure 2c), dimeric α‐3FNTxs (Figure 2d), and α‐3FNTxs which are reversible in their neuromuscular blocking action (Figure 2e), a pharmacological characteristic shared by fulditoxin. Notably, none of the conserved functional AA residues experimentally shown to be critical for α‐3FNTxs to inhibit nAChRs are present in the sequence of fulditoxin.
3.4. Chemical synthesis of fulditoxin
Solid‐phase assembly of full‐length fulditoxin (1‐63) was initially attempted using automated Fmoc (9‐fluorenylmethoxycarbonyl) chemistry. While the correct mass was observed in the crude product, it was not possible to efficiently separate it from accompanying impurities. It was therefore decided to synthesize the peptide fragments that could be subsequently assembled using native chemical ligation (Dawson, Muir, Clark‐Lewis, & Kent, 1994). The 1‐41‐thioester segment was synthesized as a single chain using Boc (tert‐butyloxycarbonyl) chemistry and subsequently solubilized and purified (Alewood et al., 1997). ESI‐MS of the purified product revealed a mass of 4937.2 Da. Likewise, the C‐terminal 42‐63 fragment was synthesized and solubilized, and the purified product showed a mass of 2278.8 Da. Native chemical ligation of the 1‐41‐thioester and 42‐63 fragments then proceeded efficiently to yield full‐length linear fulditoxin 1‐63 (Figure S3A–C). ESI‐MS of the purified 1‐63 octathiol fulditoxin revealed a mass of 6956.8 Da. Oxidative folding of the reduced peptide produced a single predominant isomer with expected mass of 6946.8 Da (Figure S3D,E). The isolated major product (sFulditoxin) was indistinguishable from nFulditoxin by analytical RP‐HPLC (Figure S3F).
3.5. Fulditoxin is a reversible postsynaptic neuromuscular blocker
The effects of purified nFulditoxin (5–500 nM) on neuromuscular transmission were studied in ex vivo organ bath experiments using the CBCM. nFulditoxin produced time‐ and concentration‐dependent blockade of nerve‐evoked twitch responses in indirectly stimulated CBCM (IC50 = 27.8 nM; 95% CI [17.7, 43.1]; Figure 3a–c), based on the percentage of neuromuscular blockade observed after 30‐min incubation with the toxin. The contractile responses of the CBCM to exogenously applied agonists ACh and CCh were also largely abolished in the presence of fulditoxin, whereas the KCl‐induced contraction was unaffected, indicating a blockade of postsynaptic muscle nAChRs, with no pharmacological evidence of myotoxicity (Figures 3a and S4B). This was consistent with preliminary screening of crude MFV that also showed no evidence of myotoxicity in the CBCM (Figure S4A). The neurotoxic potency of nFulditoxin was comparable to α‐BgTx (IC50 = 11.4 nM; 95% CI [7.9, 16.2]) and EbTx‐b (IC50 = 31.7 nM; 95% CI [21.8, 45.8]; Figure 3c). Thus, fulditoxin is among the most potent antagonists of ACh transmission in the CBCM (avian nAChR). In contrast, candoxin (Bungarus candidus) from the non‐conventional class of α‐3FNTxs (Nirthanan, Charpantier, et al., 2002; Nirthanan, Charpantier, et al., 2003) showed weaker inhibition of the CBCM (IC50 = 1.6 μM; 95% CI [1.3, 2.0]; Figure S4C).
Figure 3.
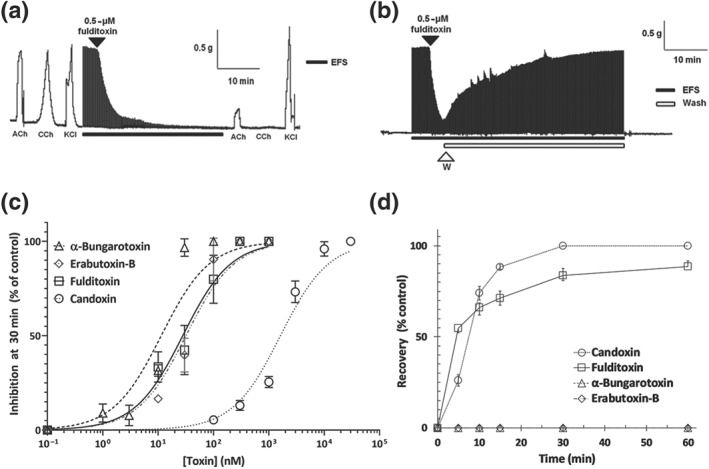
Reversible postsynaptic neuromuscular blockade produced by fulditoxin in the avian neuromuscular junction. Pharmacological characterization of native fulditoxin (nFulditoxin) on isolated nerve‐muscle models in organ bath experiments. (a) Segment of tracing showing the blockade of nerve‐evoked twitches of the chick biventer cervicis muscle (CBCM) produced by fulditoxin (0.5 μM). Contractions produced by exogenous ACh (200 μM), carbachol (CCh; 20 μM), and KCl (30 mM) before and after incubation with nFulditoxin are also shown. (b) Segment of tracing showing the reversible blockade of nerve‐evoked twitches of the CBCM produced by nFulditoxin (0.5 μM). Upon 80% inhibition of the nerve‐evoked twitch responses, the muscle was washed with fresh Krebs solution (washes lasting 30 s, at intervals of 1 min, for a period of 30 min) to remove the toxin from the bath chamber. In panels (a) and (b), the black horizontal bar indicates EFS (0.2 Hz, 0.1 ms and 7–10 V) and the unfilled bar indicates the duration of the wash. (c) Concentration–response curves for the neuromuscular blockade produced by nFulditoxin, α‐bungarotoxin and erabutoxin‐b and candoxin. Neuromuscular blockade was calculated as the twitch height of the muscle after 30‐min exposure to the respective toxin, expressed as a percentage of the control twitch responses of the muscle to supramaximal nerve stimulation. The IC50 values of nFulditoxin, α‐bungarotoxin, erabutoxin‐b, and candoxin on the CBCM are 27.8 nM (95% CI [17.7, 43.1]), 11.4 nM (95% CI [7.9, 16.2]), and 31.75 nM (95% CI [21.8, 45.8]) respectively. Each data point represents the mean ± SEM of three independent experiments. (d) Comparison of reversibility of neuromuscular blockade showing the time course of reversal of the neuromuscular blockade produced by nFulditoxin, α‐bungarotoxin, erabutoxin‐b, and candoxin. The recovery is calculated as a percentage of the control twitch responses. Each data point represents the mean ± SEM of three independent experiments
The reversibility of the neuromuscular blockade produced by fulditoxin was evaluated through intermittent washing of the CBCM with fresh Krebs solution. Over 90% recovery of nerve‐evoked twitch responses was attained within 30 min of tissue washing (Figure 3b), in contrast to α‐BgTx and EbTx‐b which are poorly reversible α‐3FNTxs (Figure 3d). The reversibility of neuromuscular blockade produced by fulditoxin was also compared with candoxin (Figure 3d); and the time taken for 50% recovery from neuromuscular blockade was similar (fulditoxin = 5.3 min; candoxin = 5.1 min).
3.6. Selectivity of fulditoxin for muscle and neuronal nAChR subtypes
As nFulditoxin produced potent neuromuscular blockade in isolated nerve‐muscle preparations, it was screened for its effects on human muscle (hα1β1δε) nAChRs expressed in Xenopus oocytes using TEVC electrophysiology. nFulditoxin produced partially (~65%) reversible, concentration‐dependent inhibition of hα1β1δε nAChRs (IC50 = 2.56 μM; see Figure S5A,B). Thus, the affinity towards hα1β1δε nAChRs was ~100 times lower than that for avian muscle nAChRs. Additionally, nFulditoxin also produced reversible blockade of human neuronal α7 (hα7) nAChRs (IC50 = 6.57 μM; Figure S5C,D). Due to the scarcity of nFulditoxin, the highest concentration tested was 15 μM, which produced only 45% inhibition of ACh‐evoked currents in hα7 nAChRs but was adequate to completely block hα1β1δε nAChRs.
We then characterized the selectivity of sFulditoxin for seven different nAChR subtypes, namely, rodent adult muscle (rα1β1δε) and human neuronal hα7, hα9α10, hα4β2, hα4β4, hα3β2, and hα3β4 nAChRs (Figure 4a). sFulditoxin was selective for rα1β1δε (IC50 = 2.6 μM; 95% CI [2.2, 3.1]) as well as hα4β2 (IC50 = 1.8 μM; 95% CI [1.6, 2.1]), hα7 (IC50 = 7.0 μM; 95% [CI 5.8, 8.4]), and hα3β2 (IC50 = 12.6 μM; 95% CI [11.3, 13.9]) subtypes. It also weakly inhibited hα4β4 (IC50 not determined), but not the hα9α10 and hα3β4 nAChRs, even at 30 μM (Figure 4b,c). The n H of fulditoxin binding to rα1β1δε, hα4β2, hα7, and hα3β2 receptors were −1.5, −1.1, −1.7, and −1.2 respectively, suggesting non‐cooperative ligand–receptor interaction (Prinz, 2010). Importantly, it was observed that sFulditoxin blocked rα1β1δε (IC50 = 2.6 μM) and hα7 (IC50 = 7.0 μM) subtypes with IC50 values similar to those for hα1β1δε (IC50 = 2.56 μM) and hα7 (IC50 = 6.57 μM) nAChRs, noting that the experiments with sFulditoxin and nFulditoxin were independently carried out in different laboratories. Therefore, we conclude that sFulditoxin and nFulditoxin are structurally and functionally equivalent.
Figure 4.
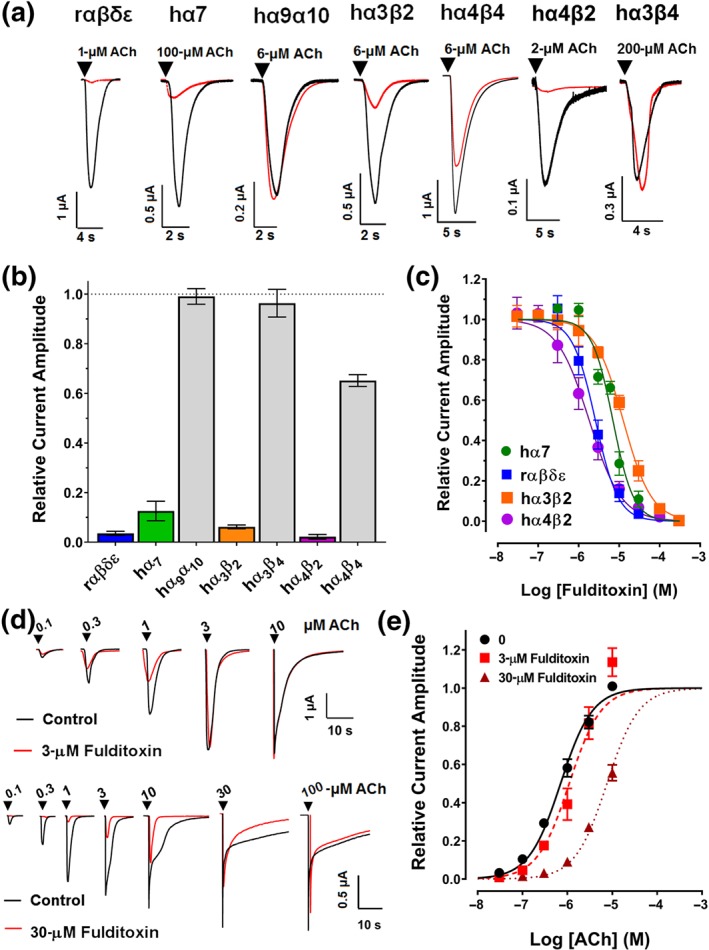
Activity of fulditoxin at nAChR subtypes expressed in Xenopus oocytes. TEVC electrophysiological characterization of synthetic fulditoxin (sFulditoxin) on nAChR subtypes expressed in Xenopus oocytes. (a) Superimposed representative ACh‐evoked currents from Xenopus oocytes expressing rαβδε, hα7, hα9α10, hα3β2, hα4β4, hα4β2, and hα3β4 nAChRs in the absence (black trace) and presence of 30‐μM sFulditoxin (red trace). For the nAChR subtypes indicated, r and h represent the species, rodent and human respectively. Whole‐cell nAChR‐mediated currents were activated by ACh at a concentration that represented the EC50 of ACh for the respective subtype expressed in the oocyte. (b) Histogram showing the effects of 30‐μM sFulditoxin on relative ACh‐evoked current amplitude mediated by rαβδε (n = 3), hα7 (n = 3), hα9α10 (n = 3), hα3β2 (n = 4), hα3β4 (n = 3), hα4β2 (n = 3), and hα4β4 (n = 4) nAChRs. (c) Concentration–response curves of sFulditoxin inhibition of selective nAChR subtypes revealed the following IC50 values for the nAChR subtypes tested: hα7 = 7.0 μM (95% CI [5.8, 8.4]; n = 3), rαβδε = 2.6 μM (95% CI [2.2, 3.1]; n = 3), hα3β2 = 12.6 μM (95% CI [11.3, 13.9]; n = 4), and hα4β2 = 1.8 μM (95% CI [1.6, 2.1; n = 4). (d) TEVC electrophysiological characterization of sFulditoxin on rodent muscle αβδε nAChRs expressed in Xenopus oocytes showing competitive antagonism of ACh binding. Representative superimposed traces of responses to varying concentrations of ACh, with or without 3‐μM sFulditoxin (top) or 30‐μM sFulditoxin (bottom). (e) Concentration–response curves obtained from the traces of ACh‐evoked responses, with or without sFulditoxin at 3 μM and 30 μM. Each data point is the mean ± SEM of three independent experiments
3.7. Fulditoxin competitively inhibits ACh binding to the ACh‐binding pocket of muscle nAChRs
To determine whether fulditoxin's low sequence identity with other α‐3FNTxs (including the absence of canonical functional AA residues) would allow it to potentially bind at a different site at the nAChR (non‐competitive allosteric antagonism) or exhibit a unique mode of interaction (competitive orthosteric antagonism), we analysed sFulditoxin's effects at rα1β1δε nAChRs activated by serial concentrations of ACh (Figure 4d). At concentrations of 3 and 30 μM, fulditoxin shifted the agonist concentration–response curve to the right, without any change in the apparent maximum response (Figure 4e). Thus, fulditoxin acts as a competitive antagonist of ACh, with a pA2 value of −0.68 (95% CI [−0.89, −0.46]).
3.8. Fulditoxin is a unique non‐covalent dimer
The crystal structure of fulditoxin was determined to 1.95‐Å resolution (Figure 5; Table 1). There were two molecules, each consisting of AA residues Leu1 to Lys58 (Figure 5a), forming a tight dimer in the asymmetric unit. Both monomers were well defined in the electron density map except for five C‐terminal residues (Figure 5b). Each monomer of the asymmetric unit has the characteristic three‐finger protein scaffold consisting of three β‐sheeted loops extending from a central core which is stabilized by four highly conserved disulfide bridges (Cys3‐Cys20, Cys13‐Cys38, Cys42‐Cys50, and Cys51‐56). Loop I is formed by two anti‐parallel β‐strands βA (Lys2‐Tyr4) and βB (Thr10‐Thr12), whereas loops II and III formed a three‐stranded β‐sheet, βC (Lys19 ‐ Met28), βD (Ser31‐Thr39), and βE (Val49 ‐ Cys51; Figure 5a). Both monomers were related by a twofold symmetry, and their superimposition yielded an RMSD of 0.623 Å for 58 Cα atoms (Figure 5d).
Figure 5.
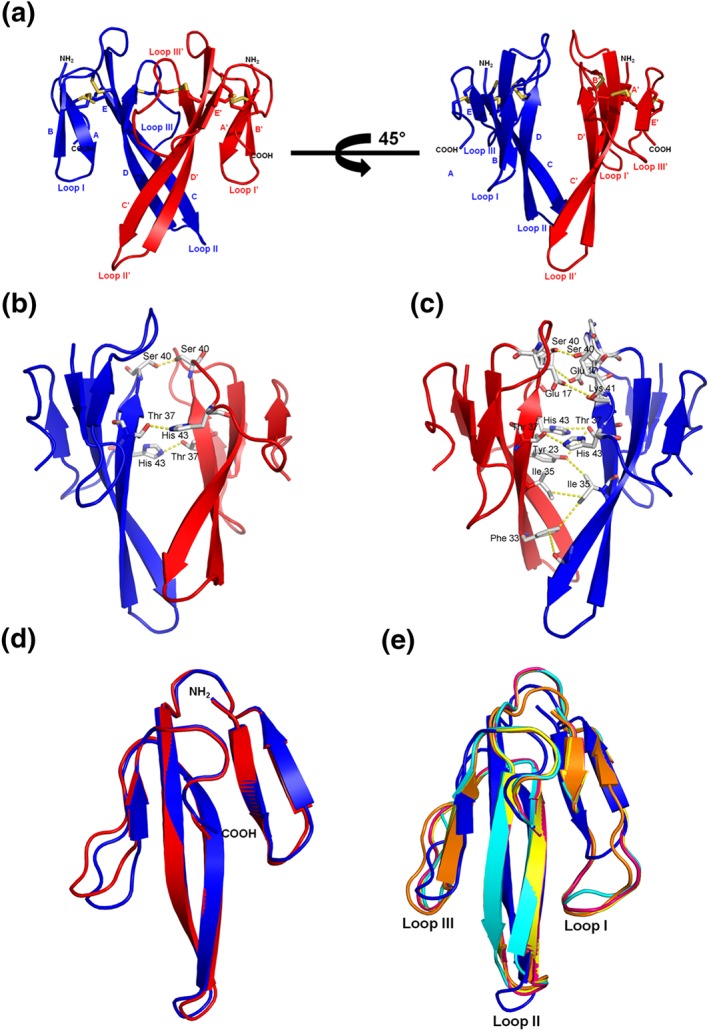
Overall structure of fulditoxin (PDB # 4RUD). X‐ray crystallographic structure of fulditoxin. (a) Fulditoxin dimer. Monomer A and monomer B of fulditoxin are shown in blue and red respectively. The N‐ and C‐termini are labelled as NH2 and COOH respectively. β‐strands and loops I–III (I′′–III') are labelled. Disulfide linkages are shown in yellow. (b) Dimeric interface of fulditoxin showing the hydrogen bonding interactions. Residues involved in the hydrogen bonding are labelled. All the hydrogen bonds are side chain‐mediated hydrogen bonds. (c) Hydrophobic interactions in the dimeric interface of fulditoxin. Residues involved in the hydrophobic interactions are labelled. (d) Superimposition of fulditoxin monomers [(a) in blue and (b) in red]. Both the monomers were related by a twofold symmetry and their superimposition yielded an RMSD of 0.6233 Å for 58 Cα atoms. The N‐ and C‐termini are labelled as NH2 and COOH respectively. (e) Superimposition of fulditoxin monomer A with short‐chain three‐finger α‐neurotoxins. Fulditoxin monomer A is shown in blue, erabutoxin‐a in yellow (5EBX; RMSD 1.86 Å for 53 Cα atoms), erabutoxin‐b in pink (3EBX; RMSD 1.87 Å for 53 Cα atoms), toxin‐α in orange (1IQ9: RMSD 1.88 Å for 53 Cα atoms), and neurotoxin‐β in cyan (1NXB; RMSD 1.98 Å for 51 Cα atoms). Loops I to III are indicated. The PDB codes are given in parentheses
A search for topologically similar proteins within the PDB database was performed with the program DALI (Holm & Sander, 1998), which showed similarity with several 3FTxs including neurotoxins and cardiotoxins (Table S1). Each monomer of fulditoxin is structurally similar to short‐chain α‐3FNTxs such as EbTx‐a (PDB code 5EBX) and Naja nigricollis toxin‐α (PDB code 1IQ9; Figure 5e), but loops I and III in fulditoxin are shorter, and loop II is orientated in the opposite direction when compared to the corresponding loops in other short‐chain α‐3FNTxs. Fulditoxin also showed overall structural similarity with long neurotoxin‐1 (PDB code 1YI5), but significant differences were observed in the length of loop I and orientation of loop II between long neurotoxin‐1 and short‐chain α‐3FNTxs, EbTx‐b (PDB code 6EBX), and fulditoxin (Figure S6).
3.9. Dimer interface comprised primarily of hydrophobic interactions
Approximately 514‐Å2 (~12% of the total) surface areas were involved in the dimerization as calculated by the Protein Interfaces, Surfaces and Assemblies server analysis (Krissinel & Henrick, 2007). There are 20 AA residues from both monomers involved in the dimerization; with close contacts between the monomers maintained by 29 hydrophobic interactions and three hydrogen bonds (<3.2 Å; see Tables S2A,B and Figure 5b). These three hydrogen bonds were all side chain–side chain contacts, with two of them observed between Thr37 and His43 from both monomers. The hydrogen bonding contacts were observed between βE of monomer A and βD of monomer B and vice versa. The third hydrogen bond was observed between Ser40 of both monomers located in the loop which connects βD and βE (Table S2A; Figure 5b). These observations strongly suggest the existence of fulditoxin as a non‐covalently linked homodimer, which is consistent with analytical size‐exclusion chromatography observations (Figure 1f).
3.10. Zinc‐binding enables fulditoxin to form a tetrameric complex
The crystal structure of fulditoxin, determined under conditions containing Zn2+, revealed two Zn2+ ions, each bound to four fulditoxin monomers arranged in a tetrahedral coordination geometry, resulting in the formation of a tetramer of dimers (Figure S7A). The tetrameric complex is formed by four dimeric fulditoxin molecules held together by two Zn2+, with each Zn2+ interacting with His29 of four different monomers from the four fulditoxin molecules (Figure S7B). Thus, Zn2+ appears to induce the oligomerization of fulditoxin molecules. Dynamic light scattering experiments confirmed that fulditoxin exists as a dimer (relative molecular mass of 16.1 kDa) in the absence of Zn2+ and as a tetramer of dimers (relative molecular mass of 34.3 kDa) in the presence of 3‐mM Zn2+. Furthermore, in the presence of 8‐M urea, reduction of the fulditoxin dimer to the monomer species (relative molecular mass of 6.18 kDa) was observed (Table S3).
4. DISCUSSION
In the five decades since the discovery of α‐BgTx, the scope of using animal toxins as molecular probes for localization and characterization of membrane receptors and ion channels and as therapeutic leads for drug discovery has broadened substantially (King, 2011; Lewis & Garcia, 2003). Notwithstanding the extensive structure–function analyses done on typical snake α‐3FNTxs such as α‐BgTx, the discovery of α‐3FNTxs with novel structural characteristics (dimeric α‐3FNTxs [Pawlak et al., 2009; Roy et al., 2010]; non‐conventional α‐3FNTxs [Nirthanan, Gopalakrishnakone, Gwee, Khoo, & Kini, 2003]; or toxins with unique sequences [Ω‐neurotoxins]; Hassan‐Puttaswamy, Adams, & Kini, 2015) suggests that some α‐3FNTxs may interact with nAChRs via different functional sites and display selectivity for nAChR subtypes not described previously. Here, we have described the discovery, chemical synthesis, and pharmacological and X‐ray crystallographic characterization of fulditoxin, a structurally unique, homodimeric neurotoxin from coral snake (Micrurus fulvius fulvius) venom, which showed high selectivity towards avian muscle nAChRs, and broad selectivity for rodent and human muscle and neuronal nAChRs while lacking all AA residues critical for binding to nAChRs. It is also the first example of a snake dimeric neurotoxin that produced reversible neuromuscular blockade and the first 3FTx to show metal binding that enables the formation of a tetrameric complex.
4.1. Fulditoxin is the first dimeric neurotoxin to produce reversible neuromuscular blockade
Fulditoxin produced neuromuscular blockade in avian muscle with an IC50 of ~27 nM, comparable to potent α‐3FNTxs, α‐BgTx, and EbTx‐b. However, unlike α‐BgTx and EbTx‐b, which produced irreversible ex vivo inhibition of muscle nAChRs, the neuromuscular blockade produced by fulditoxin was almost completely and rapidly reversible. This atypical reversible neuromuscular blockade has been reported with a few monomeric α‐3FNTxs (Harvey, Hider, Hodges, & Joubert, 1984), notably from the non‐conventional α‐3FNTx subfamily which includes candoxin (Bungarus candidus; Nirthanan, Charpantier, et al., 2003).
The mechanism of reversible interaction of snake α‐3FNTxs with muscle nAChRs is complex and not well understood. Studies on conventional monomeric long‐chain α‐3FNTxs have found their off‐rates from nAChRs to be extremely slow, sometimes lasting days (Young, Herbette, & Skita, 2003), with a half‐time for dissociation of the [3H]α‐toxin‐Torpedo nAChR‐rich membrane complex to be ~60 hr (Weber & Changeux, 1974). Therefore, our experimental design using miniaturized organ baths to estimate reversal of α‐3FNTx‐induced neuromuscular blockade was limited, given the spontaneous decline in contractility of isolated CBCM observed after ~300 min.
However, reversibility could not simply be attributed to weak or strong binding affinities of the toxin to the receptor. For instance, fulditoxin is as potent as α‐BgTx and EbTx‐b in producing neuromuscular blockade in avian muscle but is almost completely reversible in its action in contrast to α‐BgTx and EbTx‐b. Likewise, candoxin is reversible in its action at hα1β1δε, but not at hα7 receptors, while binding with nanomolar affinity to both subtypes (Nirthanan, Charpantier, et al., 2003). The reversibility of α‐3FNTx action has also been attributed to a specific site of interaction on the toxin, distinct from its functional site, based on multiple sequence analyses which revealed that readily reversible α‐3FNTxs lacked the conserved Asp31, postulating a role for this residue in determining reversibility (Harvey, Hider, Hodges, & Joubert, 1984; Nirthanan, Charpantier, et al., 2003). As fulditoxin binds to the ACh‐binding pocket (described below) with a yet‐to‐be‐identified, distinct functional site, the mechanism of fulditoxin's reversible neuromuscular blockade is presently unknown.
4.2. Fulditoxin shows broad selectivity for nAChR subtypes but lacks all functionally important AA residues for nAChR recognition
Fulditoxin (native and synthetic) produced comparable blockade of rα1β1δε or hα1β1δε (IC50 = 2.5 μM) and hα7 (IC50 = 7 μM) nAChRs in electrophysiological experiments. It also blocked neuronal hα4β2 (IC50 = 1.8 μM) and hα3β2 (IC50 = 12 μM) receptors, revealing a breadth of nAChR selectivity, which is unusual for short‐chain α‐3FNTxs, that are highly selective for muscle nAChRs (Servent et al., 1997). Pairwise alignment of mature protein sequences of human β2 and β4 subunits showed 70% homology in the extracellular domain. Homology modelling and overlay of their inter‐subunit interfaces revealed side‐chain differences between the non‐conserved residues of the β2 and β4 subunits (Cuny, Kompella, Tae, Yu, & Adams, 2016). These differences may account for fulditoxin's weak interaction with hα4β4 and hα3β4 subtypes in contrast to β2‐containing nAChRs. Interestingly, fulditoxin shares only ~20% identity (including the eight conserved cysteines) with other short‐chain α‐3FNTxs, as well as with other classes of snake α‐3FNTxs, emphasizing its structural and functional distinctiveness.
The AA residues critical for short‐chain α‐3FNTxs to bind to muscle nAChRs were identified as Lys27, Trp29, Asp31, Phe32, Arg33, and Lys47; in addition to residues (His6, Gln7, Ser8, Ser9, Gln10, Tyr25, Gly34, Ile36, and Glu38) that play a supporting role (Figure S1; Teixeira‐Clerc, Menez, & Kessler, 2002). Although fulditoxin competitively inhibits the chick muscle nAChR by binding to the ACh‐binding site at nanomolar concentrations (Figure 4e), comparable to prototypical snake α‐3FNTxs, none of the previously delineated functionally invariant AAs are present in fulditoxin (Figure 2c). Thus, fulditoxin must interact with avian nAChRs by utilizing alternative combinations of AA residues to account for its potent neuromuscular blockade.
Differences in assay methods notwithstanding, the affinity of fulditoxin for human muscle nAChRs was ~100 times lower compared to avian muscle nAChRs. Mammals are not the usual prey for Micrurus spp. which feeds primarily on reptiles including other snakes and lizards; and coral snakes themselves are prey for predatory birds (Jackson & Franz, 1981). Therefore, fulditoxin is likely to be optimized to target its natural prey (offence) and predators (defence) (Margres, Aronow, Loyacano, & Rokyta, 2013).
4.3. Fulditoxin displays structural plasticity in its three‐finger fold
The highly conserved “three‐finger” scaffold is held together by eight conserved cysteines forming four disulfide linkages at its hydrophobic core, as well as other key AA residues such as Tyr25, Gly40, Pro44, Pro48, Arg39, and Glu58, integral to its structural stability (Kini & Doley, 2010; Ricciardi et al., 2000; Tsetlin, 2015). Fulditoxin, however, retains just the eight conserved cysteines, lacking all the other structurally invariant AA residues, suggesting that the four disulfide bonds are sufficient to retain its three‐finger structure. Furthermore, structural plasticity in the loop regions of 3FTxs permits the adoption of a variety of functional conformations in order to interact with different pharmacological targets (Ricciardi et al., 2000). In this context, fulditoxin has a longer loop III (Figure 2c) and shows conformational differences at the tip of loop II (Figure 5e), suggesting that these fundamental structural differences could have important functional implications.
4.4. Fulditoxin forms a distinct dimer primarily through hydrophobic interactions
The elution on size‐exclusion chromatography (Figure 1a), dynamic light scattering (Figure S7C), and crystal structure (Figure 5a–c) of fulditoxin indicated that it exists as a non‐covalent homodimer of two short‐chain α‐3FNTx monomers. This dimer formation is distinct from other non‐covalent α‐3FNTx homodimers, such as κ‐neurotoxins and haditoxin composed of long‐chain and short‐chain α‐3FNTx monomers respectively. κ‐Neurotoxins and haditoxin share a similar quaternary structure, with their two monomeric units held in an antiparallel orientation through extensive hydrogen bonding between their loop III β‐strands (Figure 6; Dewan, Grant, & Sacchettini, 1994; Roy et al., 2010). In contrast, the fulditoxin dimer is held together primarily by 29 hydrophobic interactions between AA residues from loop II; and only three side chain–side chain hydrogen bonding contacts.
Figure 6.
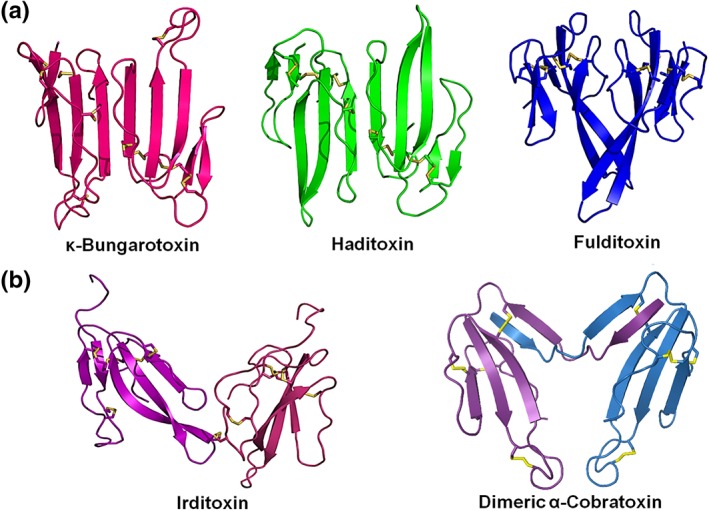
Structures of dimeric three‐finger neurotoxins. (a) Non‐covalently linked homodimers ҡ‐bungarotoxin (Bungarus multicinctus; Elapidae; 1KBA), haditoxin (Opiophagus hannah; Elapidae; 3HH7), and fulditoxin (Micrurus fulvius; Elapidae). (b) Covalently linked heterodimer irditoxin (Boiga irregularis; Colubridae; 2H7Z) and covalently linked homodimeric α‐cobratoxin (Naja kaouthia; Elapidae; 4AEA). The figures were generated using the respective PDB structures (accession number indicated in parenthesis) of each toxin. The species and family of the source are indicated. Disulfide bonds are shown in yellow
A BLAST search revealed that the AA residues involved in dimerization of fulditoxin are conserved among other Micrurus 3FTxs (Figure 2a). Hence, we propose that these Micrurus 3FTxs are also likely to form non‐covalent dimers, and fulditoxin is the first member of this new subfamily of dimeric α‐3FNTxs.
4.5. Effects of dimerization on the pharmacology of fulditoxin
Typical monomeric short‐chain α‐3FNTxs like EbTx‐a and EbTx‐b have been shown to be selective and potent blockers of muscle nAChRs while being ineffective at neuronal nAChRs (Kessler, Marchot, Silva, & Servent, 2017; Servent et al., 1997). In contrast, haditoxin, a homodimer of short‐chain α‐3FNTxs, inhibits both muscle α1β1γδ as well as neuronal α7, α3β2, and α4β2 nAChRs (Roy et al., 2010). Fulditoxin, also a homodimer of short‐chain α‐3FNTxs, albeit with a significantly different quaternary structure, exhibits nanomolar and micromolar affinity for chick and mammalian muscle nAChRs, respectively, in addition to inhibiting several neuronal nAChR subtypes. Thus, dimerization appears to expand the selectivity of short‐chain α‐3FNTxs for nAChRs.
Besides non‐covalent α‐3FNTx dimers like haditoxin and fulditoxin, covalent dimers where α‐3FNTx monomers are linked by intermolecular disulfide bonds have also been reported (Figure 2d; Table 2; Osipov et al., 2012; Pawlak et al., 2009). Dimeric α‐cobratoxin (Naja kaouthia) developed the additional capacity to block neuronal α3β2 nAChRs, unlike monomeric α‐cobratoxin that is restricted to targeting only muscle and α7 nAChRs (Osipov et al., 2012). The covalently linked heterodimeric α‐3FNTx, irditoxin (Boiga irregularis), exhibits taxa‐specific neurotoxicity for avian α1β1γδ nAChRs representing the usual prey of this arboreal species (Pawlak et al., 2009). Therefore, dimeric α‐3FNTxs represent a structurally heterogeneous group of unique snake toxins (Figure 6) exhibiting different receptor specificities from that expected for their monomeric components. Hence, we propose that dimerization is likely to present novel structural conformations for α‐3FNTxs to enable interactions with new targets, diversifying the biological activity of snake toxins.
Table 2.
Selected dimeric snake three‐finger neurotoxins that interact with nicotinic ACh receptors
| Dimeric toxin | Dimeric linkage | Monomeric subunit | nAChR activity | Source | References |
|---|---|---|---|---|---|
| Non‐covalent dimers | |||||
| Fulditoxin |
Non‐covalent 3 hydrogen bonds 29 hydrophobic interactions |
Short‐chain α‐3FNTx |
Muscle (α1β1δε)(IC50 = 2.6 μM) α3β2 (IC50 = 12.6 μM) α7 (IC50 = 7 μM) α4β2 (1.8 μM) |
Eastern coral snake Micrurus fulvius fulvius (Elapidae) |
This paper |
| κ‐Bungarotoxin |
Non‐covalent 9 hydrogen bonds |
Long‐chain α‐3FNTx |
α3β2 (IC50 = 3 nM) α7 (weak inhibition) α4β2 (weak inhibition) |
Taiwanese multi‐banded krait Bungarus multicinctus (Elapidae) |
Osipov et al., 2008 |
| Haditoxin |
Non‐covalent 14 hydrogen bonds |
Short‐chain α‐3FNTx |
α7 (IC50 = 0.2 μM) Muscle (α1β1δγ) (IC50 = 0.5 μM) α3β2 (IC50 = 0.50 μM) α4β2‐ (IC50 = 2.60 μM) |
King cobra Ophiophagus hannah (Elapidae) |
Roy et al., 2010 |
| Covalent dimers | |||||
| Irditoxin |
Covalent one inter‐molecular disulfide bridge |
Non‐conventional α‐3FNTx |
Avian muscle (α1β1δγ) (IC50 = 10 nM) Rodent muscle (α1β1γδ) (IC50 > 10 μM) |
Brown tree snake Boiga irregularis (Colubridae) |
Pawlak et al., 2009 |
| α‐Cobratoxin dimer |
Covalent two inter‐molecular disulfide bridges |
Long‐chain α‐3FNTx (or long‐chain α‐3FNTx and cytotoxin) |
Torpedo (α1β1δγ) (IC50 = 10 nM) α7 (IC50 = 0.2 μM) α3β2 (IC50 = 0.15 μM) |
Monocellate cobra Naja kaouthia (Elapidae) |
Osipov et al., 2012 |
4.6. Fulditoxin is the first metal‐binding 3FTx
Four dimeric fulditoxin molecules coalesced and formed a tetrameric complex around two Zn2+ atoms, with each Zn2+ atom coordinated with four His29 residues at the tip of loop II (Figure S7B). Metal‐binding proteins in snake venom are predominantly enzymes whose catalytic activity involves zinc. Metals can also stabilize the conformation of proteins and contribute to their function (Moroz et al., 2009). Fulditoxin is the first 3FTx to show metal‐binding capability. Interestingly, His29 is conserved in some 3FTxs from M. fulvius (Figure 2a), and a histidine is also present at the tip of loop II of Ω‐neurotoxins (Hassan‐Puttaswamy, Adams, & Kini, 2015), suggesting that these 3FTXs may also undergo Zn2+‐coordinated tetramerization.
Analytical size‐exclusion chromatography of fulditoxin at pharmacologically relevant concentrations in the presence of Zn2+ showed that it existed in the native dimeric species, suggesting that zinc‐induced oligomerization is likely to be concentration‐dependent and may occur only at fulditoxin concentrations above 5 mg·ml−1, which is functionally inconsequential since fulditoxin produced neuromuscular blockade at nanomolar concentrations (IC50 values about 10 nM; 69 ng·ml‐1).
4.7. Fulditoxin constitutes a new class of nAChR‐targeting neurotoxins
Altogether, fulditoxin is structurally and functionally distinct from other nAChR‐targeting snake neurotoxins. We propose that fulditoxin and other Micrurus toxins identified through transcriptomic analysis of venom glands (Correa‐Netto et al., 2011; Guerrero‐Garzon et al., 2018; Margres, Aronow, Loyacano, & Rokyta, 2013; Figure 2a) belong to a new class of nAChR‐targeting neurotoxins which we named Σ‐neurotoxins.
Snake 3FTxs utilize a common protein scaffold to target a variety of molecular targets and exhibit diverse biological effects, underpinning functional evolutionary divergence (Kini, 2011; Kini & Doley, 2010). Interestingly, structurally distinct protein scaffolds can also exhibit functional similarity through convergent evolution (Tsetlin, 2015), including animal toxins such as monomeric/dimeric α‐3FNTxs, waglerins, PLA2, azemiopsin, Ω‐neurotoxins, and α‐conotoxins, which have significantly diverse structures but bind to the ACh‐binding pocket of nAChRs (Kini, 2011; Kini & Doley, 2010). Hence, Σ‐neurotoxins represented by fulditoxin, while retaining the protein scaffold of α‐3FNTxs, use a yet‐to‐be determined pharmacophore to bind to the ACh‐binding pocket of the nAChR in another example of evolutionary functional convergence. Σ‐neurotoxins offer new tools for studying nAChR subtypes, as well as greater insight into interactions between peptide antagonists and nAChRs.
In conclusion, fulditoxin is a novel homodimeric short‐chain α‐3FNTx held together primarily by hydrophobic interactions and able to form a tetrameric complex by binding Zn2+, the first report of metal‐binding capability for the 3FTx family. Fulditoxin exhibited broad spectrum inhibition of α1β1δε, α4β2, α7, and α3β2 nAChRs and produced postsynaptic neuromuscular blockade at nanomolar concentrations, which was completely reversible in contrast to typical α‐3FNTxs. Furthermore, fulditoxin competitively antagonized muscle nAChRs at the ACh‐binding site despite lacking all canonical α‐3FNTx functional AA residues, suggesting that this toxin used a novel and different pharmacophore for its interaction with nAChRs. This is an example of unusual functional convergence in evolution, in contrast to the characteristic functional divergence seen within the 3FTx family.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
This work was conceptualized by S.N. and R.M.K. The experiments were performed, analysed, and interpreted as follows: C‐S.F. and R.M.K. (protein chemistry); C‐S.F. and S.N. (pharmacology); Z.D. and P.F.A. (chemical synthesis); D.B. (nFulditoxin electrophysiology); V.H‐P., H‐S.T., and D.J.A. (sFulditoxin electrophysiology); and C.J. and J.S. (x‐ray crystallography). All authors contributed to the writing of the manuscript and reviewed, revised, and approved the final paper.
DECLARATION OF TRANSPARENCY AND SCIENTIFIC RIGOUR
This Declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research as stated in the BJP guidelines for https://bpspubs.onlinelibrary.wiley.com/doi/full/10.1111/bph.14207, and https://bpspubs.onlinelibrary.wiley.com/doi/full/10.1111/bph.14206, and as recommended by funding agencies, publishers and other organisations engaged with supporting research.
Supporting information
Figure S1. (A, B) Critical amino acid residues in erabutoxin‐a and α‐cobratoxin for the Torpedo (or muscle) nicotinic acetylcholine receptor.
Figure S2. Amino acid sequence determination of fulditoxin.
Figure S3. Chemical synthesis of fulditoxin.
Figure S4. Pharmacological characterization of Micrurus fulvius venom and fulditoxin on the chick biventer cervicis muscle.
Figure S5. Effect of native fulditoxin (nFulditoxin) on human nAChRs expressed in Xenopus oocytes.
Figure S6. Superposition of fulditoxin monomer A with short‐chain (fulditoxin; erabutoxin‐b) and long‐chain (long neurotoxin‐1) three‐finger α‐neurotoxins.
Figure S7. Zinc‐dependent tetramerization of fulditoxin.
Table S1. Structural homologs of fulditoxin (results from DALI server).
Table S2. Hydrogen bonding and hydrophobic interactions in the dimeric interface of fulditoxin.
Table S3. Zinc‐dependent tetramerization of fulditoxin.
ACKNOWLEDGEMENTS
This research was supported by the following sources: Australian Research Council Discovery Project Grant (DP150103990; D.J.A); National Health and Medical Research Council (Australia) Fellowship and Program Grant (APP1072113; P.F.A; D.J.A); Ministry of Education Academic Research Grant from the National University of Singapore (R154000A72‐114; partially supported J.S); Griffith Health Institute Project Grants (HFC2132940 and GRC1009) (S.N); and National Medical Research Council, Singapore Grant (NMRC/CBRG/0025/2012; R.M.K).
We acknowledge Norman Benton for the photograph of Micrurus fulvius (CC BY‐SA 3.0) used in the graphical abstract.
Foo CS, Jobichen C, Hassan‐Puttaswamy V, et al. Fulditoxin, representing a new class of dimeric snake toxins, defines novel pharmacology at nicotinic ACh receptors. Br J Pharmacol. 2020;177:1822–1840. 10.1111/bph.14954
Present Address: Chun Shin Foo, School of Applied Science, Temasek Polytechnic, 21 Tampines Avenue 1, 529757 Singapore. Varuna Hassan‐Puttaswamy, Institute for Stem Cell Biology and Regenerative Medicine, Bellary Road, Bangalore 560065, India.
Contributor Information
Selvanayagam Nirthanan, Email: niru.nirthanan@griffith.edu.au.
R. Manjunatha Kini, Email: dbskinim@nus.edu.sg.
REFERENCES
- Adams, P. D. , Gopal, K. , Grosse‐Kunstleve, R. W. , Hung, L. W. , Ioerger, T. R. , McCoy, A. J. , … Terwilliger, T. C. (2004). Recent developments in the PHENIX software for automated crystallographic structure determination. Journal of Synchrotron Radiation, 11(Pt 1), 53–55. 10.1107/s0909049503024130 [DOI] [PubMed] [Google Scholar]
- Adams, P. D. , Grosse‐Kunstleve, R. W. , Hung, L. W. , Ioerger, T. R. , McCoy, A. J. , Moriarty, N. W. , … Terwilliger, T. C. (2002). PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallographica Section D: Biological Crystallography, 58(Pt 11), 1948–1954. [DOI] [PubMed] [Google Scholar]
- Alewood, P. , Alewood, D. , Miranda, L. , Love, S. , Meutermans, W. , & Wilson, D. (1997). Rapid in situ neutralization protocols for Boc and Fmoc solid‐phase chemistries. Methods in Enzymology, 289, 14–29. 10.1016/s0076-6879(97)89041-6 [DOI] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Mathie, A. , Peters, J. A. , Veale, E. L. , Striessnig, J. , Kelly, E. , … Collaborators, C. (2019). The Concise Guide to PHARMACOLOGY 2019/20: Ion channels. British Journal of Pharmacology, 176(Suppl 1), S142–S228. 10.1111/bph.14749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F. , Madden, T. L. , Schaffer, A. A. , Zhang, J. , Zhang, Z. , Miller, W. , & Lipman, D. J. (1997). Gapped BLAST and PSI‐BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25(17), 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antil‐Delbeke, S. , Gaillard, C. , Tamiya, T. , Corringer, P. J. , Changeux, J. P. , Servent, D. , & Menez, A. (2000). Molecular determinants by which a long chain toxin from snake venom interacts with the neuronal α‐nicotinic acetylcholine receptor. Journal of Biological Chemistry, 275(38), 29594–29601. 10.1074/jbc.M909746199 [DOI] [PubMed] [Google Scholar]
- Arunlakshana, O. , & Schild, H. O. (1959). Some quantitative uses of drug antagonists. British Journal of Pharmacology and Chemotherapy, 14(1), 48–58. 10.1111/j.1476-5381.1959.tb00928.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzat, C. , & Sine, S. M. (2018). Nicotinic acetylcholine receptors at the single‐channel level. British Journal of Pharmacology, 175(11), 1789–1804. 10.1111/bph.13770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changeux, J. P. (2012). The nicotinic acetylcholine receptor: The founding father of the pentameric ligand‐gated ion channel superfamily. Journal of Biological Chemistry, 287(48), 40207–40215. 10.1074/jbc.R112.407668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa‐Netto, C. , Junqueira‐de‐Azevedo Ide, L. , Silva, D. A. , Ho, P. L. , Leitao‐de‐Araujo, M. , Alves, M. L. , … Calvete, J. J. (2011). Snake venomics and venom gland transcriptomic analysis of Brazilian coral snakes, Micrurus altirostris and M. corallinus. Journal of Proteomics, 74(9), 1795–1809. 10.1016/j.jprot.2011.04.003 [DOI] [PubMed] [Google Scholar]
- Cuny, H. , Kompella, S. N. , Tae, H. S. , Yu, R. , & Adams, D. J. (2016). Key structural determinants in the agonist binding loops of human β2 and β4 nicotinic acetylcholine receptor subunits contribute to a3b4 subtype selectivity of α‐conotoxins. Journal of Biological Chemistry, 291(45), 23779–23792. 10.1074/jbc.M116.730804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M. J. , Alexander, S. , Cirino, G. , Docherty, J. R. , George, C. H. , Giembycz, M. A. , … Ahluwalia, A. (2018). Experimental design and analysis and their reporting II: updated and simplified guidance for authors and peer reviewers. British Journal of Pharmacology, 175, 987–993. 10.1111/bph.14153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashevsky, D. , & Fry, B. G. (2018). Ancient diversification of three‐finger toxins in Micrurus coral snakes. Journal of Molecular Evolution, 86(1), 58–67. 10.1007/s00239-017-9825-5 [DOI] [PubMed] [Google Scholar]
- Dawson, P. E. , Muir, T. W. , Clark‐Lewis, I. , & Kent, S. B. (1994). Synthesis of proteins by native chemical ligation. Science, 266(5186), 776–779. [DOI] [PubMed] [Google Scholar]
- Dewan, J. C. , Grant, G. A. , & Sacchettini, J. C. (1994). Crystal structure of k‐bungarotoxin at 2.3‐Å resolution. Biochemistry, 33(44), 13147–13154. 10.1021/bi00248a026 [DOI] [PubMed] [Google Scholar]
- Emsley, P. , & Cowtan, K. (2004). Coot: Model‐building tools for molecular graphics. Acta Crystallographica . Section D: Biological Crystallography, 60(Pt 12 Pt 1), 2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Ginsborg, B. L. , & Warriner, J. (1960). The isolated chick biventer cervicis nerve‐muscle preparation. British Journal of Pharmacology and Chemotherapy, 15, 410–411. 10.1111/j.1476-5381.1960.tb01264.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero‐Garzon, J. F. , Benard‐Valle, M. , Restano‐Cassulini, R. , Zamudio, F. , Corzo, G. , Alagon, A. , & Olvera‐Rodriguez, A. (2018). Cloning and sequencing of three‐finger toxins from the venom glands of four Micrurus species from Mexico and heterologous expression of an α‐neurotoxin from Micrurus diastema . Biochimie, 147, 114–121. 10.1016/j.biochi.2018.01.006 [DOI] [PubMed] [Google Scholar]
- Harding, S. D. , Sharman, J. L. , Faccenda, E. , Southan, C. , Pawson, A. J. , Ireland, S. , … Nc‐Iuphar (2018). The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: Updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Research, 46(D1), D1091–D1106. 10.1093/nar/gkx1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, A. L. , Hider, R. C. , Hodges, S. J. , & Joubert, F. J. (1984). Structure‐activity studies of homologues of short chain neurotoxins from Elapid snake venoms. British Journal of Pharmacology, 82(3), 709–716. 10.1111/j.1476-5381.1984.tb10810.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan‐Puttaswamy, V. , Adams, D. J. , & Kini, R. M. (2015). A distinct functional site in O‐neurotoxins: Novel antagonists of nicotinic acetylcholine receptors from snake venom. ACS Chemical Biology, 10(12), 2805–2815. 10.1021/acschembio.5b00492 [DOI] [PubMed] [Google Scholar]
- Hogg, R. C. , Bandelier, F. , Benoit, A. , Dosch, R. , & Bertrand, D. (2008). An automated system for intracellular and intranuclear injection. Journal of Neuroscience Methods, 169(1), 65–75. 10.1016/j.jneumeth.2007.11.028 [DOI] [PubMed] [Google Scholar]
- Holm, L. , & Sander, C. (1998). Touring protein fold space with Dali/FSSP. Nucleic Acids Research, 26(1), 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard‐Jones, N. (1985). A CIOMS ethical code for animal experimentation. World Health Organisation Chronicles, 39(2), 51–56. [PubMed] [Google Scholar]
- Jackson, D. R. , & Franz, R. (1981). Ecology of the Eastern coral snake (Micrurus fulvius) in Northern Peninsular Florida. Herpetologica, 37(4), 213–228. [Google Scholar]
- Jackson, T. N. , Sunagar, K. , Undheim, E. A. , Koludarov, I. , Chan, A. H. , Sanders, K. , … Fry, B. G. (2013). Venom down under: Dynamic evolution of Australian elapid snake toxins. Toxins, 5(12), 2621–2655. 10.3390/toxins5122621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, P. , Marchot, P. , Silva, M. , & Servent, D. (2017). The three‐finger toxin fold: a multifunctional structural scaffold able to modulate cholinergic functions. Journal of Neurochemistry, 142(Suppl 2), 7–18. 10.1111/jnc.13975 [DOI] [PubMed] [Google Scholar]
- Kilkenny, C. , Browne, W. , Cuthill, I. C. , Emerson, M. , & Altman, D. G. (2010). Animal research: Reporting in vivo experiments: the ARRIVE guidelines. British Journal of Pharmacology, 160, 1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, G. F. (2011). Venoms as a platform for human drugs: Translating toxins into therapeutics. Expert Opinion on Biological Therapy, 11(11), 1469–1484. 10.1517/14712598.2011.621940 [DOI] [PubMed] [Google Scholar]
- Kini, R. M. (2011). Evolution of three‐finger toxins—A versatile mini protein scaffold. Acta Chimica Slovenica, 58, 693–701. [PubMed] [Google Scholar]
- Kini, R. M. , & Doley, R. (2010). Structure, function and evolution of three‐finger toxins: Mini proteins with multiple targets. Toxicon, 56(6), 855–867. 10.1016/j.toxicon.2010.07.010 [DOI] [PubMed] [Google Scholar]
- Kitchens, C. S. , & Van Mierop, L. H. (1987). Envenomation by the Eastern coral snake (Micrurus fulvius fulvius). A study of 39 victims. Journal of the American Medical Association, 258(12), 1615–1618. [PubMed] [Google Scholar]
- Krissinel, E. , & Henrick, K. (2007). Inference of macromolecular assemblies from crystalline state. Journal of Molecular Biology, 372(3), 774–797. 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- Kudryavtsev, D. S. , Shelukhina, I. V. , Son, L. V. , Ojomoko, L. O. , Kryukova, E. V. , Lyukmanova, E. N. , … Utkin, Y. N. (2015). Neurotoxins from snake venoms and α‐conotoxin ImI inhibit functionally active ionotropic g‐aminobutyric acid (GABA) receptors. Journal of Biological Chemistry, 290(37), 22747–22758. 10.1074/jbc.M115.648824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, R. J. , & Garcia, M. L. (2003). Therapeutic potential of venom peptides. Nature Reviews. Drug Discovery, 2(10), 790–802. 10.1038/nrd1197 [DOI] [PubMed] [Google Scholar]
- Margres, M. J. , Aronow, K. , Loyacano, J. , & Rokyta, D. R. (2013). The venom‐gland transcriptome of the eastern coral snake (Micrurus fulvius) reveals high venom complexity in the intragenomic evolution of venoms. BMC Genomics, 14, 531 10.1186/1471-2164-14-531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). Solvent content of protein crystals. Journal of Molecular Biology, 33(2), 491–497. 10.1016/0022-2836(68)90205-2 [DOI] [PubMed] [Google Scholar]
- McCoy, A. J. , Grosse‐Kunstleve, R. W. , Adams, P. D. , Winn, M. D. , Storoni, L. C. , & Read, R. J. (2007). Phaser crystallographic software. Journal of Applied Crystallography, 40(Pt 4), 658–674. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath, J. C. , Drummond, G. B. , McLachlan, E. M. , Kilkenny, C. , & Wainwright, C. L. (2010). Guidelines for reporting experiments involving animals: The ARRIVE guidelines. British Journal of Pharmacology, 160(7), 1573–1576. 10.1111/j.1476-5381.2010.00873.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh, J. M. , Absalom, N. , Chebib, M. , Elgoyhen, A. B. , & Vincler, M. (2009). a9 Nicotinic acetylcholine receptors and the treatment of pain. Biochemical Pharmacology, 78(7), 693–702. 10.1016/j.bcp.2009.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz, O. V. , Burkitt, W. , Wittkowski, H. , He, W. , Ianoul, A. , Novitskaya, V. , … Bronstein, I. B. (2009). Both Ca2+ and Zn2+ are essential for S100A12 protein oligomerization and function. BMC Biochemistry, 10, 11 10.1186/1471-2091-10-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirthanan, S. , Charpantier, E. , Gopalakrishnakone, P. , Gwee, M. , Khoo, H. , Cheah, L. , … Bertrand, D. (2003). Neuromuscular effects of candoxin, a novel toxin from the venom of the Malayan krait (Bungarus candidus). British Journal of Pharmacology, 139(4), 832–844. 10.1038/sj.bjp.0705299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirthanan, S. , Charpantier, E. , Gopalakrishnakone, P. , Gwee, M. C.‐E. , Khoo, H.‐E. , Cheah, L.‐S. , … Kini, R. M. (2002). Candoxin, a novel toxin from Bungarus candidus, is a reversible antagonist of muscle (αβγδ) but a poorly reversible antagonist of neuronal α7 nicotinic acetylcholine receptors. Journal of Biological Chemistry, 277(20), 17811–17820. 10.1074/jbc.M111152200 [DOI] [PubMed] [Google Scholar]
- Nirthanan, S. , Gao, R. , Gopalakrishnakone, P. , Gwee, M. , Khoo, H. , Cheah, L. , & Kini, R. M. (2002). Pharmacological characterization of mikatoxin, an α‐neurotoxin isolated from the venom of the New‐Guinean small‐eyed snake Micropechis ikaheka . Toxicon, 40(7), 863–871. 10.1016/s0041-0101(01)00268-9 [DOI] [PubMed] [Google Scholar]
- Nirthanan, S. , Gopalakrishnakone, P. , Gwee, M. C. , Khoo, H. E. , & Kini, R. M. (2003). Non‐conventional toxins from Elapid venoms. Toxicon, 41(4), 397–407. 10.1016/s0041-0101(02)00388-4 [DOI] [PubMed] [Google Scholar]
- Osipov, A. V. , Kasheverov, I. E. , Makarova, Y. V. , Starkov, V. G. , Vorontsova, O. V. , Ziganshin, R. , … Utkin, Y. N. (2008). Naturally occurring disulfide‐bound dimers of three‐fingered toxins: A paradigm for biological activity diversification. Journal of Biological Chemistry, 283(21), 14571–14580. 10.1074/jbc.M802085200 [DOI] [PubMed] [Google Scholar]
- Osipov, A. V. , Rucktooa, P. , Kasheverov, I. E. , Filkin, S. Y. , Starkov, V. G. , Andreeva, T. V. , … Tsetlin, V. I. (2012). Dimeric α‐cobratoxin X‐ray structure: Localization of intermolecular disulfides and possible mode of binding to nicotinic acetylcholine receptors. Journal of Biological Chemistry, 287(9), 6725–6734. 10.1074/jbc.M111.322313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. , & Minor, W. (1997). Processing of X‐ray diffraction data collected in oscillation mode In Methods in enzymology (Vol. 276) (pp. 307–326). Cambridge, MA, USA: Academic Press. [DOI] [PubMed] [Google Scholar]
- Pawlak, J. , Mackessy, S. P. , Sixberry, N. M. , Stura, E. A. , Le Du, M. H. , Ménez, R. , … Kini, R. M. (2009). Irditoxin, a novel covalently linked heterodimeric three‐finger toxin with high taxon‐specific neurotoxicity. FASEB Journal, 23(2), 534–545. 10.1096/fj.08-113555 [DOI] [PubMed] [Google Scholar]
- Prinz, H. (2010). Hill coefficients, dose‐response curves and allosteric mechanisms. Journal of Chemical Biology, 3(1), 37–44. 10.1007/s12154-009-0029-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan, N. , Pung, Y. F. , Zhu, Y. Z. , Wong, P. T. , Kumar, P. P. , & Kini, R. M. (2007). β‐Cardiotoxin: A new three‐finger toxin from Ophiophagus hannah (king cobra) venom with β‐blocker activity. FASEB Journal, 21(13), 3685–3695. 10.1096/fj.07-8658com [DOI] [PubMed] [Google Scholar]
- Rey‐Suarez, P. , Floriano, R. S. , Rostelato‐Ferreira, S. , Saldarriaga‐Cordoba, M. , Nunez, V. , Rodrigues‐Simioni, L. , & Lomonte, B. (2012). Mipartoxin‐I, a novel three‐finger toxin, is the major neurotoxic component in the venom of the redtail coral snake Micrurus mipartitus (Elapidae). Toxicon, 60(5), 851–863. 10.1016/j.toxicon.2012.05.023 [DOI] [PubMed] [Google Scholar]
- Ricciardi, A. , le Du, M. H. , Khayati, M. , Dajas, F. , Boulain, J. C. , Menez, A. , & Ducancel, F. (2000). Do structural deviations between toxins adopting the same fold reflect functional differences? Journal of Biological Chemistry, 275(24), 18302–18310. 10.1074/jbc.275.24.18302 [DOI] [PubMed] [Google Scholar]
- Roy, A. , Zhou, X. , Chong, M. Z. , D'Hoedt, D. , Foo, C. S. , Rajagopalan, N. , … Kini, R. M. (2010). Structural and functional characterization of a novel homodimeric three‐finger neurotoxin from the venom of Ophiophagus hannah (king cobra). Journal of Biological Chemistry, 285(11), 8302–8315. 10.1074/jbc.M109.074161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnolzer, M. , Alewood, P. , Jones, A. , Alewood, D. , & Kent, S. B. (1992). In situ neutralization in Boc‐chemistry solid phase peptide synthesis. Rapid, high yield assembly of difficult sequences. International Journal of Peptide and Protein Research, 40(3–4), 180–193. 10.1111/j.1399-3011.1992.tb00291.x [DOI] [PubMed] [Google Scholar]
- Servent, D. , Winckler‐Dietrich, V. , Hu, H. Y. , Kessler, P. , Drevet, P. , Bertrand, D. , & Menez, A. (1997). Only snake curaremimetic toxins with a fifth disulfide bond have high affinity for the neuronal a7 nicotinic receptor. Journal of Biological Chemistry, 272(39), 24279–24286. 10.1074/jbc.272.39.24279 [DOI] [PubMed] [Google Scholar]
- Teixeira‐Clerc, F. , Menez, A. , & Kessler, P. (2002). How do short neurotoxins bind to a muscular‐type nicotinic acetylcholine receptor? Journal of Biological Chemistry, 277(28), 25741–25747. 10.1074/jbc.M200534200 [DOI] [PubMed] [Google Scholar]
- Tsetlin, V. I. (2015). Three‐finger snake neurotoxins and Ly6 proteins targeting nicotinic acetylcholine receptors: Pharmacological tools and endogenous modulators. Trends in Pharmacological Sciences, 36(2), 109–123. 10.1016/j.tips.2014.11.003 [DOI] [PubMed] [Google Scholar]
- Vergara, I. , Pedraza‐Escalona, M. , Paniagua, D. , Restano‐Cassulini, R. , Zamudio, F. , Batista, C. V. , … Alagon, A. (2014). Eastern coral snake Micrurus fulvius venom toxicity in mice is mainly determined by neurotoxic phospholipases A2. Journal of Proteomics, 105, 295–306. 10.1016/j.jprot.2014.02.027 [DOI] [PubMed] [Google Scholar]
- Weber, M. , & Changeux, J. P. (1974). Binding of Naja nigricollis (3H)α‐toxin to membrane fragments from Electrophorus and Torpedo electric organs I. Binding of the Tritiated α‐Neurotoxin in the Absence of Effector. Molecular Pharmacology, 10(1), 1–14. [PubMed] [Google Scholar]
- Wonnacott, S. , Bermudez, I. , Millar, N. S. , & Tzartos, S. J. (2018). Nicotinic acetylcholine receptors. British Journal of Pharmacology, 175(11), 1785–1788. 10.1111/bph.14209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, H. S. , Herbette, L. G. , & Skita, V. (2003). α‐Bungarotoxin binding to acetylcholine receptor membranes studied by low angle X‐ray diffraction. Biophysical Journal, 85(2), 943–953. 10.1016/S0006-3495(03)74533-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart, P. H. , Afonine, P. V. , Grosse‐Kunstleve, R. W. , Hung, L. W. , Ioerger, T. R. , McCoy, A. J. , … Adams, P. D. (2008). Automated structure solution with the PHENIX suite. Methods in Molecular Biology, 426, 419–435. 10.1007/978-1-60327-058-8_28 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. (A, B) Critical amino acid residues in erabutoxin‐a and α‐cobratoxin for the Torpedo (or muscle) nicotinic acetylcholine receptor.
Figure S2. Amino acid sequence determination of fulditoxin.
Figure S3. Chemical synthesis of fulditoxin.
Figure S4. Pharmacological characterization of Micrurus fulvius venom and fulditoxin on the chick biventer cervicis muscle.
Figure S5. Effect of native fulditoxin (nFulditoxin) on human nAChRs expressed in Xenopus oocytes.
Figure S6. Superposition of fulditoxin monomer A with short‐chain (fulditoxin; erabutoxin‐b) and long‐chain (long neurotoxin‐1) three‐finger α‐neurotoxins.
Figure S7. Zinc‐dependent tetramerization of fulditoxin.
Table S1. Structural homologs of fulditoxin (results from DALI server).
Table S2. Hydrogen bonding and hydrophobic interactions in the dimeric interface of fulditoxin.
Table S3. Zinc‐dependent tetramerization of fulditoxin.


