With the digitization of all records and processes, and prevalence of cloud‐driven services and Internet of Things, today's era can truly be considered as an era of data. Machine learning (ML) and artificial intelligence (AI) skills are among the most sought‐after skills today. McKinsey Global Institute research suggests that 45% of workplace activities in corporations could be automated with current technologies; 80% of that is attributable to existing ML capabilities, and breakthroughs in natural language processing could further the impact.1 Gartner forecasts that large‐scale data‐driven analytics could lead to huge benefits in health care; in the United States, where healthcare spending is 18% of gross domestic product, up to US$600 per person could be saved annually. Gartner also forecasts that data‐driven insights for demand‐supply matching could create an economic impact of $850 billion to $2.5 trillion.2 International Data Corporation forecasts that spending on AI and ML will grow to $79.2 billion by 2022, with a compound annual growth rate of 38% between the 2018 and 2022 period.3
Machine Learning
AI is defined as the study of intelligent agents, which can perceive the environment and intelligently act just as humans do.4 AI can philosophically be categorized as strong AI or weak AI.4 Machines that can act in a way as though intelligent (simulated thinking) are said to possess weak AI, and machines that are intelligent and can actually think are said to possess strong AI. In today's applications, most AI researchers are engaged in implementing weak AI to automate specific task(s).4 ML techniques are commonly used to learn from data and achieve weak AI. ML involves the scientific study of statistical models and algorithms that can progressively learn from data and achieve desired performance on a specific task. The knowledge/rules/findings inferred from the data using ML are expected to be nontrivial. Therefore, ML can be used in many tasks that need automation, and especially in scenarios where humans cannot manually develop a set of instructions to automate the desired tasks. Deep learning (DL) is a subfield of ML, which focuses on learning data representations with computational models composed of multiple processing layers.5 Figure 1 shows a commonly used diagram to illustrate the relationship between AI, ML, and DL.
Figure 1.
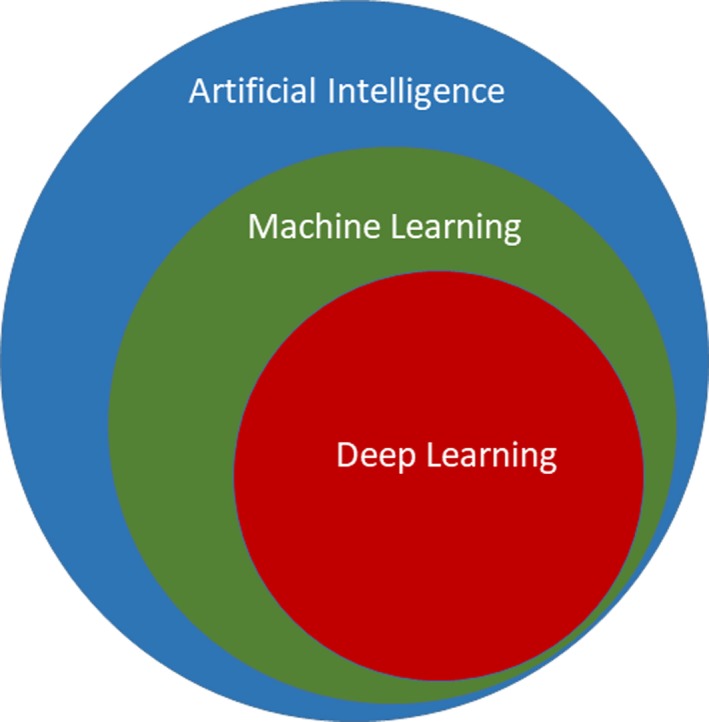
This figure illustrates the relationships between AI, ML, and DL. DL is a subfield of ML, while ML is a subfield of AI. AI indicates artificial intelligence; DL, deep learning; ML, machine learning.
ML can be broadly categorized into supervised learning, unsupervised learning, semisupervised learning, reinforcement learning, and active learning tasks.6 Supervised learning is the task of learning a function that maps input data to target labels. It is provided with a labeled training data set. These problems can be further categorized into problems of regression and classification. When target variables are continuous real number values, the supervised learning task(s) are known as regression problems, and when the target variables are categorical variables, the tasks are known as classification problems. Common supervised learning algorithms include linear regression,7 logistic regression (LR),8 decision tree,9 random forest (RF),10 support vector machine (SVM),11 k‐nearest neighbors (KNN) and artificial neural network (ANN). RF and SVM are among the most commonly used algorithms; they are illustrated and explained in Figures 2 and 3, respectively.
Figure 2.
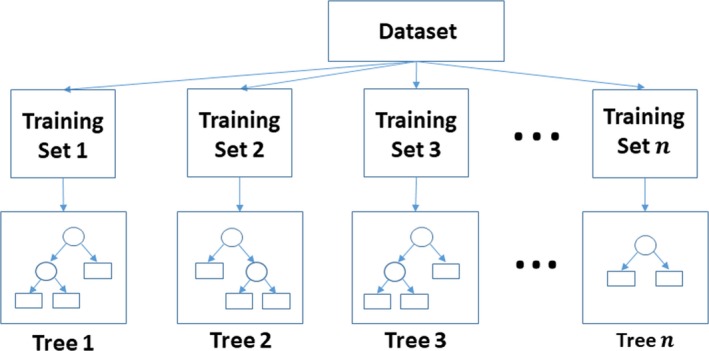
This figure illustrates the training on an RF classifier. RF is an ensemble machine learning algorithm. Let n be the number of trees in the random forest classifier; n different training sets are then generated using the bootstrapping technique, and for each training set, 1 decision tree is generated. The ovals in the trees represent the splits, while the rectangles represent the classes. While generating each tree, the most effective feature out of a random subset of features would be selected to create the splits. Gini's diversity index is a commonly used split criterion. During the phase of testing, features of new samples would be passed along all the trees. Each tree would vote for a decision, and the majority of the votes would represent the final decision. RF indicates random forest.
Figure 3.
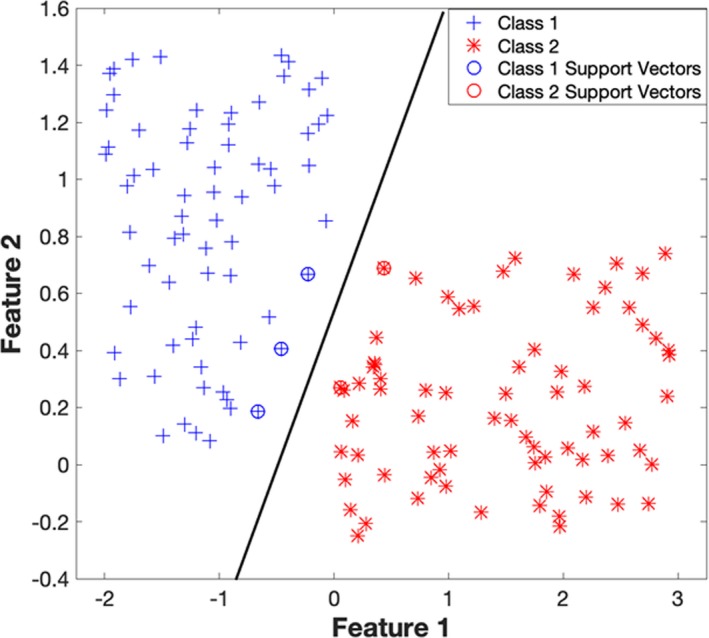
This figure illustrates the SVM binary classification algorithm, which has been trained over a sample data. Let class 1 refer to the samples belonging to the first class (on the left‐hand side) and class 2 refer to the samples belonging to the other class. The data points (both class 1 and class 2) which are encircled/starred, are the support vectors. The support vectors are those data points that the algorithm identifies to be hardest in getting correctly classified. The SVM algorithm picks an optimal hyperplane that maximizes the margins between itself and the support vectors. SVM indicates support vector machine.
Unsupervised learning is the task of discovering patterns from a data set consisting of input data without target labels. Examples of unsupervised learning tasks are (1) identifying the underlying distribution of data, (2) discovering natural grouping/clustering within data, and (3) dimensionality reduction and the like. Some commonly used unsupervised learning algorithms are k‐means clustering, hierarchical clustering, principal component analysis, auto‐encoders, and Parzen windows for density estimation. Figure 4 illustrates the use of unsupervised learning with a k‐means algorithm. Both semisupervised learning and active learning deal with situations in which data are partially labeled. With semisupervised learning, the objectives remain the same as those of supervised learning; however, the techniques in this category also use the unlabeled data, and they attempt to improve over the supervised learning performance, which uses only labeled data. Active learning methods are commonly used in situations where manual annotation of data is expensive. Active learning focuses on methods that best suggest which unlabeled data are to be labeled next, so as to attain the desired supervised learning task with minimum efforts of labeling. Reinforcement learning is concerned with how intelligent agents learn and perform actions to maximize a notion of cumulative rewards.
Figure 4.
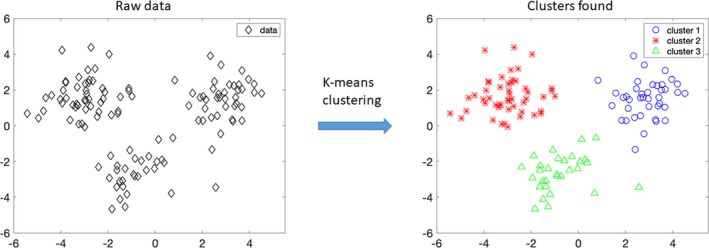
Clustering is a form of unsupervised learning. Clustering is a task of grouping unannotated data into distinct groups, such that samples of the same group are more similar to each other than those from the other groups. In this figure, unannotated data (data) on the left‐hand side are provided as input to the k‐means algorithm with k=3, and the algorithm groups the raw data into 3 distinct clusters, namely cluster 1, cluster 2, and cluster 3, as shown on the right‐hand side.
Typically, an ML pipeline consists of the following steps: (1) data acquisition, (2) data preprocessing, (3) feature extraction, (4) feature selection, and (5) supervised/unsupervised/reinforcement learning task. It is believed that for most objectives, creating an appropriate feature representation is among the most important steps in an ML workflow/pipeline.12 Before the introduction of deep learning techniques, feature representations were almost always handcrafted by subject matter experts. DL is useful because it can learn useful feature representations with multiple levels of abstraction. Many of the original ideas in DL were discovered years before they were used for practical applications because DL tasks can be computationally very expensive. Availability of cheaper computational resources have played a significant role in the rise of DL applications. Krizhevsky and colleagues’13 work stands out in this regard. They were the first to use graphics processing units for training a DL model, which in their case was a deep convolutional neural network (CNN). The significant findings taken from their work were that (1) graphics processing units can significantly speed up the learning process; (2) constructing deeper networks can significantly improve the overall performance; and (3) DL models are extremely powerful in learning feature representations, and in many tasks, can provide better performance than the state‐of‐the‐art custom‐built ML models (eg, computer vision,13 natural language processing14). This work significantly triggered the rise of DL applications and the rise in development of DL‐centric graphics processing units.
To obtain good generalization and desired performance, DL techniques must preferably be trained on large amounts of data, which may not always be possible to obtain. Most DL tasks have 2 steps: (1) pretraining and (2) fine‐tuning. In the pretraining step, DL models attempt to learn the underlying distribution of data and create feature representations in an unsupervised manner. In the fine‐tuning step, the feature representations are tuned for the specific task at hand such that maximum performance is achieved. Since the pretraining step is an unsupervised step (ie, which does not use annotations), it provides freedom for DL models to augment their performance by using data of the same modality but not pertaining to the relevant data set. The ability to store knowledge from other problems and apply it to a current problem is known as transfer learning. DL models are also notable because they provide convenient methods to perform transfer learning.
Deep neural network (DNN), CNN, recurrent neural network (RNN), and deep belief network are some notable classes of DL architectures.12 DNN is a feed‐forward ANN with multiple layers. CNN is a variation of DNN, and is designed to recognize visual patterns directly from pixel images with minimal preprocessing. CNNs are inspired from biological processes, such that the organization of neurons resembles that of the visual cortex of animals. CNNs are particularly effective in analyzing images as well as sequential data. RNNs are a class of ANNs in which the network connections form a directed graph along a temporal sequence, and the learning algorithm accounts for both the input data and the past internal states (of hidden nodes). RNNs work especially well in analyzing sequential data such as time series or natural text and can capture longer context (older information) than that of CNNs. Enhanced versions of RNNs, namely, long short‐term memory networks,12 memory networks,15 attention layers,16 and the like, have been designed, to enhance the memory retention capability (retaining older information) of RNNs. CNNs and RNNs are illustrated and explained in Figures 5 and 6, respectively. Deep belief network is a probabilistic generative graphical model, composed of multiple layers of latent variables. Unlike the other 3 architectures, a deep belief network learns probabilistic relationships between layers and how these can reconstruct the input. It should be noted that estimating the probabilities in a deep belief network is often not an easy task and can be computationally very expensive.12
Figure 5.
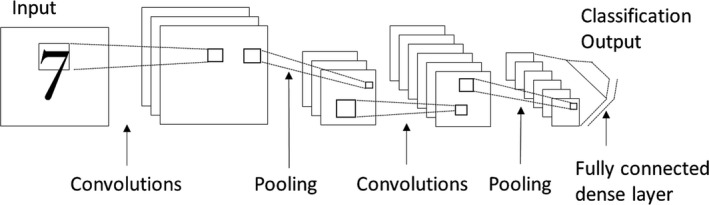
The figure shows a simple CNN meant for classifying images (in this case, images of digits). Most CNN architectures include (1) convolutional layers, (2) pooling layers, and (3) dense (fully connected) layers. A convolutional layer typically has multiple filters (similar to the image filters), wherein the filter weights are allowed to change and learn from the data. Each of these filters is moved across the length and breadth of the entire image as it is convolved with the image pixel values. It should be noted that these filters act like feature extractors, and the output (feature maps) obtained after performing the convolution operation is used as input to the next layer. The pooling layer provides an approach to down sample the feature maps while summarizing the presence of features, either locally or globally. Also, the pooling layer acts like a feature detector that helps identify important features and to a certain degree helps in providing rotational and translational invariance. The dense layer is a fully connected network wherein each neuron receives input from each neuron of the previous layer. Typically, the dense layer contributes to the greatest number of learnable parameters (weights and biases) and helps reduce the training error. The sharing of filters in convolutional layers helps the CNN to avoid overfitting. The network as a whole thus attempts to achieve low training error and high generalization ability. CNN indicates convolutional neural network.
Figure 6.
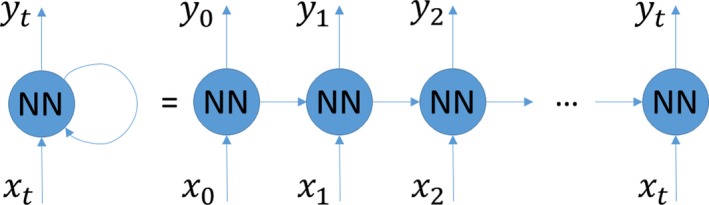
Illustration of an RNN. x i and y i are the input and output at the ith time step, respectively. In RNN, the output is dependent on (1) the current input, (2) the output from the previous time step, and (3) the network weights and biases. In other words, the RNN's output is dependent on the current and previous inputs together. This makes RNN suitable for analyzing sequential data. NN indicates neural network; RNN, recurrent neural network.
As will become apparent below, most studies employ supervised ML models. An important aspect to be considered in all supervised ML models is how well they generalize to the unseen data. It is therefore expected that studies have clear demarcations on what are the training data and what are the unseen test data, while the final ML models are assessed and compared only on the unseen test data. Some studies may have a third distinct group of data, known as the validation data. Validation data are generally used for tuning the hyperparameters of the ML model, before finally assessing it on the test data. When demarcations on the training, validation, and test data are missing (completely or in part), model validation techniques such as cross‐validation techniques are employed. In k‐fold cross validation, the entire data are sampled into k mutually exclusive groups. From the k groups, k−1 groups are selected for training the ML model, and the model's performance is assessed on data from the remaining 1 group; this procedure is repeated k times until each of the k groups is used at least once for model assessment. Leave‐one‐out cross validation is a special case, where the value of k equals the number of data samples; that is, only one sample is used for testing, and all other samples are used for training. Once the performances across the k test groups are found, their mean and standard deviation are commonly reported for model assessment and comparisons.
AI/ML/DL in Medicine
AI has been around since the 1950s, and it has already revolutionized industries such as finance,17 transportation,18 and advertising.19 Accordingly, many believe that AI can significantly and positively support the healthcare industry.20
AI can make health care more accessible and affordable for the general public, especially in the third‐world countries where there is shortage of physicians.21 With the ever‐increasing accumulation of multimodal and multidimensional data, the use of AI systems can help draw meaningful insights and ease the workloads of physicians.22 AI can also be used to provide second opinions and reduce human errors, as physicians can suffer from fatigue, inattention, or inexperience and thus provide wrong diagnoses.23, 24
AI has been used for diagnosing cancer25, 26 and for mining patient data such as electronic health records, vital signs, and genetics,27, 28, 29, 30, 31, 32, 33, 34, 35 with the intent to establish new patient stratification principles, reveal unknown disease correlations, and provide personalized medicine. The area of drug development has also benefited from the use of AI, as AI can help identify factors that are most predictive of the effectiveness of a drug.36, 37, 38, 39
Weber and Toyama have suggested that AI should develop intelligence into the existing systems/institutions, rather than starting from scratch, or hoping to replace the existing systems.40 Furthermore, AI/ML has many hopes in contributing to health care in resource‐poor settings.41 Gartner et al42 used ML techniques, namely, SVM and Bayesian models, for predicting diagnosis‐related groups and then used the predictions to allocate scarce hospital resources. Use of these methods led to improved use of operating rooms and beds. ML and natural language processing techniques have been used for surveillance and outbreak predictions using data from electronic health records and online/social media.41 AI‐based planning is also expected to help improve scheduling the travel of community health workers across multiple homes and communities.41
The main body of this article focuses on the state‐of‐the‐art ML methods used for improving cardiovascular outcomes in patients with cardiac disorders.
ML in the Cardiovascular System
There has been considerable research in using ML to improve cardiovascular outcomes in patients. We have reviewed and categorized the research into 5 application areas, namely, (1) imaging43, 44, 45, 46, 47, 48, 49, 50 (8 studies), (2) electrocardiography51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68 (18 studies), (3) in‐hospital monitoring69, 70, 71, 72, 73, 74, 75, 76, 77, 78 (10 studies), (4) mobile and wearable technology79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89 (11 studies), and (5) precision medicine90, 91, 92, 93, 94, 95, 96, 97, 98, 99 (10 studies). Each of these categories has been further divided into 2 subsections, namely, (1) diagnosis and disease classification, and (2) risk prediction and patient management.
Imaging
Diagnosis and disease classification
Narula et al43 developed an ensemble ML model of 3 classifiers (SVM, RF, ANN) to automate the discrimination of hypertrophic cardiomyopathy from physiological hypertrophy in athletes, using speckle‐tracking echocardiography data. The authors worked on a cohort of 139 male subjects with 77 verified athlete cases and 62 verified hypertrophic cardiomyopathy cases. Information gain was used for feature selection, and the volume, mid–left ventricular (LV) segmental longitudinal strain, average longitudinal strain, and mid‐LV segmental radial strain, were identified as the best predictors. The authors claimed that the ML model with speckle‐tracking echocardiography–based parameters demonstrated a diagnostic ability comparable to that of the conventional 2‐dimensional echocardiographic and Doppler‐derived parameters used in clinical practice.
Sengupta et al44 developed a cognitive ML classifier called associative memory classifier (AMC), which can integrate both the clinical data and the imaging data for use in discrimination of cardiac abnormalities. The AMC's use was validated on the problem of distinguishing patients with constrictive pericarditis from those with restrictive cardiomyopathy. AMC and other ML models were trained on data belonging to a cohort of 50 patients with constrictive pericarditis, and 44 patients with restrictive cardiomyopathy. The AMC with 15 speckle‐tracking echocardiography variables was able to make the discrimination with an area under the curve (AUC) value of 0.892, and this performance improved to 0.962 when 4 additional echocardiographic variables (e′, E/e′, interventricular septum, and posterior wall thickness of the left ventricle) were used by the AMC. The authors also found that the AMC performed better than other classifiers, namely, RF, SVM, KNN, and neural networks.
Recognizing the views in echocardiography is an essential first step for computer‐assisted interpretation. Madani et al45 employed DL to distinguish the standard views in echocardiography. The authors trained a CNN to simultaneously classify 15 standard views (12 video views and 3 still images) based on the annotated still images and videos from 267 transthoracic echocardiograms. The CNN achieved an overall test accuracy of 97.8% in classifying the 12 video views. On low‐resolution single still images from all 15 views, the DL model managed to correctly classify the views with 91.7% accuracy; in comparison, the board‐certified echocardiographers in the study could achieve accuracies of only 70.2% to 84.0% with the still images.
Attia et al46 used DL to identify asymptomatic LV dysfunction in patients. They used 12‐lead ECG and echocardiographic data (to compute LV ejection fraction) from 44 959 patients at the Mayo Clinic to train a CNN and identify patients with ventricular dysfunction using the ECG data alone. Patients with LV ejection fraction below 0.35 were identified to have asymptomatic LV dysfunction on an independent data set of 52 870 patients with an AUC, sensitivity, specificity, and accuracy of 0.93, 86.3%, 85.7%, and 85.7%, respectively.
DL has also been applied in cardiac magnetic resonance imaging. In a study published in 2016, Avendi et al used DL algorithms and deformable models together, to perform automatic segmentation of the left ventricle (LV) from cardiac magnetic resonance imaging data sets. The data of the Medical Image Computing and Computer Assisted Intervention Society 2009 challenge was used for validation and comparisons.47 CNNs were employed to automatically detect the LV chamber in the magnetic resonance imaging scan, and then stacked autoencoders were used to infer the LV shape. The inferred shape was then incorporated into/within deformable models, to further improve the accuracy and reliability of the segmentation. The combined DL and deformable model approach was found to outperform the prevalent state‐of‐the‐art methods.
Risk prediction and patient management
With respect to risk factors, a study by Motwani et al48 showed that ML methods gave higher AUC value in predicting all‐cause mortality in patients undergoing coronary computed tomographic angiography, as compared with the Framingham risk score or the coronary computed tomographic angiography severity scores.
In another study, Poplin et al49 trained DL models to make quantitative predictions of popular cardiovascular risk factors (CRFs) using retinal fundus photographs. The CRFs were age, sex, smoking status, systolic blood pressure (BP), and major adverse cardiac events. The authors trained the DL models over retinal fundus images from 48 101 patients of the UK Biobank and 236 234 patients of EyePACS, and tested the models on retinal fundus images from 12 026 patients of the UK Biobank and 999 patients of EyePACS. On testing with the UK Biobank test data, the proposed DL models were found to predict the (1) age with mean absolute error of 3.26 years (95% CI, 3.22–3.31) where CI refers to confidence interval, (2) sex with AUC of 0.97, (3) smoking status with AUC of 0.71, (4) systolic BP with mean absolute error within 11.23 mm Hg (95% CI, 11.18–11.51), and (5) major adverse cardiac events with AUC of 0.70.
In another study, Wang et al50 developed a coronary artery disease risk marker, wherein they used DL for detecting breast arterial calcifications from mammograms. They used a 12‐layer CNN to discriminate breast arterial calcification from non–breast arterial calcification, and a pixelwise, patch‐based procedure was applied for breast arterial calcification detection. The performance was evaluated using a set of 840 full‐field digital mammograms from 210 cases, using both free‐response receiver operating characteristics analysis and calcium mass quantification analysis. Application of CNN in free‐response receiver operating characteristics analysis provide a detection performance which was similar to that of the human experts.
Electrocardiography
Diagnosis and disease classification
Electrocardiography is a noninvasive way of measuring the electrical activity of the heart. Ever since the 2000s, there has been a fair amount of research to classify normal and abnormal heart rhythms in the ECG using ML algorithms.51, 52, 53, 54, 55, 56, 57, 58 The most common ML algorithms used here have been the linear discriminant analysis, ANN, and SVM.
For example, Li et al59 used SVM to detect life‐threatening arrhythmias ventricular fibrillation and ventricular tachycardia. In the study, 14 features such as complexity,60 leakage,61 kurtosis,62 and the like were extracted from the ECG signal, and different window lengths were tried while extracting the features. Features were then selected using a genetic algorithm‐based technique for optimal feature combinations. Three annotated public domain ECG databases, namely, the American Heart Association (AHA) Database, the Creighton University Ventricular Tachyarrhythmia Database, and the MIT‐BIH Malignant Ventricular Arrhythmia Database, were used as the training, test, and validation data sets. With 5‐fold cross validation on the out‐of‐sample validation data, the best performance values were achieved with a window length of 5 seconds, giving a test accuracy of 96.3±3.4%, sensitivity of 96.2±2.7%, and specificity of 96.2±4.6%.
There have been many studies on using DL for ECG beat classification. Zubair et al63 used CNN to classify the non–life‐threatening ECG beats into the 5 classes recommended by the Association for Advancement of Medical Instrumentation, namely: nonectopic, supraventricular ectopic, ventricular ectopic, fusion, and unknown. The authors used the MIT‐BIH database to evaluate the classification performance. The authors found that the proposed system achieved better accuracy and superior computational efficiency than most of the existing state‐of‐the art methods. Acharya et al64 conducted a similar study of classifying heartbeats from the MIT‐BIH Arrhythmia Database into 5 classes using a 9‐layer deep CNN. The data set was artificially augmented to balance the number of instances in 5 classes of heartbeats, and the signals were filtered to remove high‐frequency noise. When trained using the augmented data, the CNN achieved accuracy of 94.03% and 93.47% in the original and noise‐free ECGs, respectively. When the CNN was trained without the augmented data, the accuracy reduced to 89.07% and 89.3%, in noisy and noise‐free ECGs, respectively. This study showed that the proposed CNN model can be used as a tool for screening the ECG signal and quickly classify heartbeats.
Isin and Ozdalili65 were among the first to use transfer learning in ECG arrhythmia diagnostics. The authors used the AlexNet CNN13 trained on ImageNet image data, and then applied transfer learning to carry out ECG arrhythmia diagnostics. To train and test the ECG data, the R‐T segments of the preprocessed ECG beats, each one having 200 samples, were scaled as images of 256×256, and then reproduced as an RGB image, before being used as input to the CNN network. Features extracted from the sixth and seventh layers of the deep CNN were used independently, then principal component analysis was used to reduce the dimensionality, and a multilayered feed‐forward network was used for classification, which exhibited a recognition rate of 98.51% and testing accuracy of around 92%. The study showed that using transfer learning with CNNs could be useful in detecting cardiac arrhythmias.
CNNs have also been used for identifying the R‐wave peaks and P‐ and T‐waves of the ECG signal. Taking annotations from QT and the MIT‐BIH P‐Wave Database, Sodmann et al66 used a 9‐layered CNN to learn and generate features. An Extreme Gradient Boosted Tree classifier was then trained on the extracted features, to distinguish among the various waves in the ECG. The developed model identified the R‐wave peaks, P‐waves, and T‐waves with an accuracy of 98%, 92%, and 88%, respectively.
Deng et al67 developed a dynamic neural learning mechanism for ECG‐based cardiovascular disease classification. During the model training phase, cardiac dynamics within the ECG signals were extracted using the radial basis function neural networks through the deterministic learning mechanism. The authors claimed that such cardiac dynamics represented the beat‐to‐beat temporal change of ECG modifications and that it could provide a larger amount of discriminability than the original ECG signals. For the study, ECG signals from 52 healthy controls, 148 subjects with myocardial infarction, 18 subjects with heart failure (HF), 15 subjects with bundle branch block, 14 subjects with dysrhythmia, 7 subjects with myocardial hypertrophy, 6 subjects with valvular heart disease, and 4 subjects with myocarditis, were used. In the test phase, the cardiac dynamics of the test ECG pattern was compared with that of the training patterns, and the class of the test ECG pattern was recognized according to the smallest error principle. On leave‐one‐out cross validation, the model achieved accuracies between 86.6% and 100% for each disease.
Generative adversarial networks (GANs)100 were introduced in 2014. They are a class of ML (often DL) models where 2 models are trained simultaneously: a generative model G tries to capture the distribution of the training data, and a discriminative model D that, given a sample, estimates the sample's probability of whether it belongs to the training data or is generated by G. They are useful for unsupervised learning, semisupervised learning, and sometimes even for supervised learning. Recently, Zhu et al68 developed a generative adversarial network composed of bidirectional long short‐term memory as the generator and CNN as the discriminator, for generating synthetic ECG data, that had high morphological similarity to the real ECG recordings.
How the conventional ML models, which use handcrafted features, compare with the more recent DL models where features are learned and selected automatically, is an important discussion. Our observations on this topic are as follows. For some applications, DL models perform overwhelmingly better than the conventional ML ones, while for other applications they work either equally well or better only when the DL model is augmented with a conventional ML model. For example, in applications pertaining to QRS peak detection, and ECG beat delineation, multiple articles101, 102 have showcased that DL methods outperform the conventional ML and signal processing–based methods. In the application of heart rhythm classification using ECG, there has been an increasing trend of published research articles that show DL models performing better than the conventional ML models.81 On public domain databases like the Physionet 2017 challenge data, the top‐ranked performers83, 84 used a stacked classifier that contained a DL model and a conventional ML model in union. In more complex applications like disease classification/prediction, researchers typically use data from multiple modalities. Considering the trade‐off between the amount of available data and the number of features, it is common for researchers to represent the ECG signals, with handcrafted features.96, 102 In conclusion, given a large amount of data, the features learned and selected by DL in most cases can provide equal or better performance than the conventional features. However, with explainability being an important concern among healthcare practitioners, it is preferred that researchers be able to explain the significance/impact that each feature (either handcrafted or learned using DL) has on the concerned application.
In‐Hospital Monitoring
Critically ill patients are admitted to the intensive care unit (ICU) or the emergency department, and it is customary to have medical devices continuously monitor them. A large amount of data such as heart rate, BP, temperature, pulse oximetry, and respiratory rate are gathered from these patients. Such data provide major opportunities to use ML for improving patient outcomes.
Diagnosis and disease classification
In the ICU, the monitors raise alarm(s) whenever they believe patients to be at risk. Unfortunately, these monitors have been often found to generate false alarms.69 A study discovered that 88.8% of annotated arrhythmic alarms were false positives.69 Major causes of these false arrhythmic alarms were found to be noise and artifacts in the physiological signals, which again were probably caused by patient motion or loose electrodes. Too many false alarms can create a noisy environment and cause desensitization among caregivers to the extent that they may ignore the true alarms as well. There has been a large amount of research to reduce the number of false alarms in the ICU, some of which employs ML. In the “Reducing False Arrhythmia Alarms in the ICU: PhysioNet/Computing in Cardiology Challenge 2015,” Plesinger et al70 developed a method that was based on fuzzy logic, with promising results. ML‐based models using SVM71 and RF,72 have also been applied to tackle this problem of false ICU alarms.
Wiens et al73 used active learning to develop a clinically useful method for patient‐specific, adaptive heart beat classification. The proposed method, when tested on the MIT‐BIH data, was able to achieve excellent performance on the 2 main tasks suggested by the Association for Advancement of Medical Instrumentation, with over 90% less training data. The 2 tasks were (1) detecting ventricular ectopic beats and (2) detecting supraventricular ectopic beats. Furthermore, the authors used this method to develop a tool for cardiologists that produced excellent results.
Risk prediction and patient management
Ong et al74 proposed an ML‐based model that used features such as age, heart rate, variability parameters, and vital signs to predict cardiac arrest within 72 hours. An SVM classifier was used to classify patients at the time of presentation to the emergency department, on whether they would experience cardiac arrest within 72 hours. Additionally, a score for each patient was computed based on the Euclidean distances between the patient's data and two cluster centers, wherein one cluster consisted of patients having cardiac arrest or death as outcomes, and the other cluster consisted of patients without such outcomes. A score of 0 to 40 indicated low risk, 41 to 60 indicated intermediate risk, and 61 to 100 indicated high risk. The authors claimed that the ML‐based score was more accurate than the modified early warning score in predicting cardiac arrest within 72 hours in critically ill patients presented to the emergency department.
A multicenter study by Churpek et al75 found that ML methods predicted the clinical deterioration in patients on wards more accurately than the conventional regression methods. In the study, variables such as demographics, laboratory values, and vital signs were used in a discrete time survival analysis framework to predict the combined outcome of cardiac arrest, ICU transfer, or death. Among the examined models, the RF‐based model was found to be the most accurate, with an AUC of 0.80 (95% CI, 0.80–0.80). Furthermore, all tested ML algorithms, namely, gradient‐boosted machines, bagged trees, SVM, NN, LR, KNN, and decision trees, also gave better results than the modified early warning score. The authors then suggested that these techniques could be used for improved identification of critically ill patients on the wards.
Frizzell et al76 developed ML‐based approaches to predict the 30 day all‐cause readmission of patients discharged following a HF hospitalization. The variables used for developing the predictive models included information pertaining to demographics, socioeconomic status, medical history, characterization of HF (including admission symptoms), admission and discharge medications, vital signs, weight, selected laboratory treatments, and discharge interventions. The authors tried multiple predictive models, including a tree‐augmented naive Bayesian network, LR, gradient‐boosted models, and RF. Each model's performance was evaluated using the validation data set, and the C statistic was used to evaluate a model's ability to discriminate between a readmission or not. The authors found that all evaluated ML models showed only modest discrimination ability, with their C statistic varying between 0.59 and 0.62. Therefore, they concluded that ML models in this case did not provide a significant improvement in predicting HF readmissions, as compared with the traditional statistical models.
Sandu et al proposed a reinforcement learning–based model for BP regulation in post–cardiac surgery patients.77 The state‐of‐the‐art BP regulation technique uses a proportional–integral–derivative controller (based on classical feedback control loop) with predecided patient models. The authors in the study highlighted the advantage of reinforcement learning–based models, which do not have to use a predecided patient model; instead, they can directly use the clinical data for training. All experiments by the authors were done on simulated data. Reinforcement learning was also used by Rom et al78 while training a spiking neurons architecture for cardiac resynchronization therapy (CRT). The spiking neural network architecture allows the atrioventricular delay and interventricular interval parameters to adapt according to the information provided by the intracardiac electrograms and hemodynamic sensors. The adaptive cardiac resynchronization therapy prototype in simulations showed a 30% increase in cardiac output as compared with a nonadaptive cardiac resynchronization therapy device. The authors softly suggested that using an adaptive cardiac resynchronization therapy device can improve the quality of life for patients with congestive HF.
Mobile and Wearable Technology
The popular use of smartphones and wearable technology has caused an explosion of available biomedical data. Such data provide major opportunities for early diagnosis and prevention of cardiovascular diseases.79, 80
Diagnosis and disease classification
Hannun et al81 used an end‐to‐end DL approach for detecting multiple heart rhythm classes in patients who used a single‐lead ambulatory ECG monitoring device. The developed DNN model contained convolutional layer blocks and ResNet blocks. The authors trained the DNN on a huge annotated data set obtained from iRhythm Technologies, consisting of 91 232 single‐lead ECG records from 53 549 patients, that were divided into 12 rhythm classes. Testing across 328 records from 328 unique patients, which were annotated by a group of cardiologists, the DNN model showcased an average F1 score of 0.837, which was better than what individual cardiologists achieved, an average F1 score of 0.78.
Ortín et al82 proposed a fully automated, single‐lead, and real‐time ventricular heartbeat classifier. The proposed method used an echo state network to classify the ECG signals following the Association for Advancement of Medical Instrumentation recommendations with an interpatient scheme. The model was validated on the MIT‐BIH arrhythmia and the Incart data sets. On the MIT‐BIH arrhythmia data, the method respectively achieved a sensitivity and precision of 95.4% and 88.8% for lead II, and 90.9% and 89.2% for lead V1. The methodology was proven to be a competitive single‐lead ventricular heartbeat classifier and was comparable to the existing state‐of‐the‐art algorithms that used multiple leads.
Many single‐channel ECG signal classification algorithms were recently developed toward the 2017 PhysioNet/CinC Challenge. The objective of the CinC challenge was to encourage the development of ML and DL methods that could identify from a single, short ECG lead recording (30–60 seconds), whether it shows normal sinus rhythm, atrial fibrillation (AF), an alternative rhythm, or is too noisy to be classified. All recordings were collected using the AliveCor device. Teijeiro et al83 won the challenge by obtaining the highest F1 test score of 0.83. Teijeiro et al carefully crafted features, which included morphological and rhythm‐related features, and built 2 classifiers; one evaluated the ECG signal with aggregated feature values, and the other evaluated the ECG signal as a sequence of features extracted from each heartbeat using RNNs. The decisions of the classifiers were finally combined using a stacking technique. Another notable work was presented by Plesinger et al,84 who claimed to have achieved an overall test F1 score of 0.83 in the challenge. They used 2 ML approaches in parallel; the first approach used a 13‐layered CNN to process 6 seconds of ECG signal, while the second approach extracted 43 handcrafted features and used a bagged tree ensemble for classification. Of the 2 approaches, the CNN‐based approach was first applied to classify the ECG signal, and if the confidence of classification was not above a defined threshold, the second bagged tree ensemble–based approach was used for making the decision.
There have been multiple studies on identifying AF in patients using non‐ECG data and ML. Shashikumar et al85 used a DL approach to monitor and detect AF using wearable technology. For the study, photoplethysmographic data and triaxial accelerometry from 98 subjects (45 with AF and 53 with other rhythms) were gathered using a multichannel wrist‐worn device. A continuous wavelet transform was applied to the photoplethysmographic data to derive spectrograms, and then a CNN was trained on the spectrograms to project them to a 1‐dimensional feature vector. This feature vector, along with other features calculated on the basis of beat‐to‐beat variability and signal quality, were fed to an elastic net logistic classifier to classify each patient to AF class or non‐AF class. Leave‐one‐out cross validation resulted in an AUC value of 0.95 and accuracy of 91.8%. Lahdenoja et al86 used accelerometers and gyroscopes in smartphones to measure cardiogenic micromovements of the patients’ chests, and then used SVM, kernel SVM, and RF classifiers to distinguish patients with AF from healthy individuals. The study achieved a sensitivity of 93.8% and specificity of 100%.
The amount of data generated by biosensors in mobile and wearable technology is enormous, and it is not feasible to have physicians manually label all these data. To label biosensor data without access to the ground truth, Zhu et al87 proposed 2 Bayesian approaches for aggregating the labels from independent and potentially correlated annotators (algorithms and/or humans), to infer a more reliable label than each individual annotator. Results from applying the models on simulated data, and 2 publicly available biomedical data sets (2006 PhysioNet QT data set and Capnobase Respiratory Rate Database) showed that the proposed method could perform better than the existing approaches in literature.
A significant challenge while analyzing biosensor data is that they can be noisy. Baseline wandering, which is caused by the subjects’ motion, muscle artifacts, and the like, is among the most commons forms of noise. This is because the subjects having mobile/wearable devices are expected to move constantly and perform various tasks throughout the day. Effective noise detection is therefore an important research problem and is an active area of research. Satija et al88 proposed a novel unified framework for automatic detection, localization, and classification of single and combined ECG noise. In the framework, the ECG signals were first decomposed using the modified ensemble empirical mode decomposition algorithm for discriminating the ECG components from the noise and artifacts. Short‐term temporal features such as maximum absolute amplitude, number of zero crossings, and local maximum peak amplitude of the autocorrelation function, were then computed from the extracted high‐frequency and low‐frequency signals. Finally, a decision‐rule–based algorithm was used to detect the presence of noise and classify the type of noise. The proposed framework was evaluated on 5 benchmark ECG databases and on real‐time ECG signals. The proposed framework achieved an average sensitivity of 99.12% and specificity of 98.56% in detecting the presence of noise. With respect to classifying the different types of noise, it achieved an average sensitivity of 98.93%, positive predictive value of 98.39%, and classification accuracy of 97.38%. The authors claimed that their proposed framework performed better in noise detection than the existing state‐of‐the‐art methods, and localized short bursts of noise accurately with low–end‐point delineation errors. To assess the quality of ECG signals, Yaghmaie et al89 introduced a dynamic signal quality index. The index used a smoothed pseudo–Wigner‐Ville transform to derive the time‐frequency patterns of the ECG signal, and then based on a weighted cross‐correlation function, assigned a score between 0 and 1 to each ECG beat (identified by the Pan and Tompkins algorithm103) to indicate the signal quality. The index was validated by testing its effectiveness in noise detection. The index was used to discriminate noisy signals from normal ECG data and also noisy signals from the abnormal heart rhythms’ ECG data. On testing with the public databases on PhysioNet, the dynamic signal quality index achieved an AUC of 0.93 in discriminating normal versus noisy ECG data, and an AUC of 0.94 in discriminating abnormal heart rhythm versus noisy ECG data. The authors also claimed that these noise detection results were better than the previous state‐of‐the‐art metrics when used individually.
Precision Medicine
Precision medicine is defined as the approach to optimize the medical care provided to a patient by accounting for the patient's genes, environment, and lifestyle. Though precision medicine in cardiology is fairly new compared with that in oncology, there have been a considerable number of studies in this area.
Diagnosis and disease classification
RNNs have the ability to take temporal relations into account. A study by Choi et al90 found that using RNN in modeling the events in electronic health records could improve the performance in initial diagnosis of HF, as compared with that of conventional methods, which ignore temporality. Data were drawn from 3 884 HF incidents/cases and 28 903 controls. Events such as disease diagnosis, medication orders, and procedure orders were time stamped, and a 12‐ to 18‐month window of cases and controls were observed. It was found that RNNs performed better than the other baseline methods such as regularized LR, multilayer perceptron with 1 hidden layer, SVM and KNN, in both the 12‐month and the 18‐month observation windows. Also, the RNN using the 18‐month observation window performed better than the RNN using the 12‐month observation window.
In a study by Juhola et al,91 the authors showed that it was possible to separate different genetic cardiac diseases on the basis of Ca2+ transients using ML methods. The diseases studied were catecholaminergic polymorphic ventricular tachycardia, long QT syndrome, and hypertrophic cardiomyopathy. Classification accuracy of up to 87% was obtained for these diseases, and it indicated that Ca2+ transients are disease specific. Multiple classifiers, namely, KNN, RF, and least square SVM, were examined here, and RF was found to provide the highest accuracy.
Statistical methods and machine learning approaches have often been used in high throughput differential gene expression analyses from microarrays or RNA sequencing, with the intent to identify a list of genes that are altered in patients but not in controls. For example, in Kullo and colleagues’92 study, the single‐nucleotide polymorphism rs653178 in the ATXN2‐SH2B3 locus was found to be significantly associated with peripheral arterial disease. LR analysis adjusted for age and sex was used here to identify the single‐nucleotide polymorphisms associated with peripheral arterial disease case/control status in the discovery, replication, and combined sets.
In another study, Khera et al93 examined the relationship between familial hypercholesterolemia mutations and high polygenic score, to early‐onset myocardial infarction, using LR models that were adjusted for the first 4 principal components of ancestry. Of the patients scored, the top 5% were considered to have high polygenic scores. To obtain the polygenic score, the authors used a method that accounts the polygenic score to 6.6 million common DNA variants, and accordingly quantified the cumulative susceptibility conferred by these variants.104
Risk prediction and patient management
Weng et al showed that applying ML algorithms using routine clinical data significantly improved the accuracy in predicting the first cardiovascular event over 10 years, as compared with an established algorithm by American College of Cardiology (ACC) guidelines.94 RF, LR, gradient‐boosting machines, and neural networks were used in these studies, and all of them were found to give better results than the established algorithm of the American College of Cardiology.
In a recent study, ML models, namely, LR, RF, gradient‐boosted trees, CNN, and long short‐term memory, were applied on features extracted from longitudinal electronic health records, with and without the genotype information, to predict 10‐year cardiovascular disease events.95 The study was performed in a cohort of 9 824 cases and 99 666 controls and showed that ML models that employ longitudinal electronic health record features perform significantly better in prediction than the American College of Cardiology and the AHA pooled cohort risk equation, and also better than ML models that employ only American College of Cardiology/AHA features. A further improvement in predictive performance was also achieved when genotype data in the form of 204 single‐nucleotide polymorphisms were included as features.
Diller et al96 developed DL models to estimate prognosis and guide therapy in a cohort of 10 019 adult patients with adult congenital heart disease. The parameters/variables used as input to the DL algorithms were clinical and demographic data, ECG parameters, cardiopulmonary exercise testing, and few selected laboratory markers. On the test data, the authors were able to predict the patients’ need to discuss with the multidisciplinary teams about their management, surgical or catheter intervention, and device implantation, with 90.2% accuracy.
Bellot and Van der Schaar97 studied the problem of personalized survival estimates of patients in heterogeneous populations and proposed a novel ML‐based solution called the Hierarchical Bayesian Survival model. Their goals were (1) to make more accurate predictions by making these predictions personalized to a specific patient, (2) to better understand diseases and their risk factors, and (3) to provide model outputs that are interpretable by clinicians. The proposed Hierarchical Bayesian Survival model was a probabilistic survival model that captured individual traits through a hierarchical latent variable formulation. Survival paths were then estimated by jointly sampling the location and shape of the individual survival distribution. When compared with other baseline survival models, the Hierarchical Bayesian Survival was found to be computationally expensive, but it consistently achieved a higher time‐dependent concordance index105 and lower Brier score106 in predicting survival. The authors also introduced a personalized interpreter that could test the effect of covariates on each patient.
Shah et al98 studied 397 HF patients with preserved ejection fraction, with the goal of identifying phenotypically distinct HF categories with preserved ejection fraction. The authors performed detailed clinical, laboratory, electrocardiographic, and echocardiographic phenotyping of the patients. Different statistical learning algorithms, namely, unbiased hierarchical cluster analysis of phenotypic data and penalized model‐based clustering, were used to separate patients into mutually exclusive groups to comprise a novel classification of HF with preserved ejection fraction. Phenomapping analysis separated the patients into 3 distinct groups that had different clinical characteristics, cardiac structure/function, invasive hemodynamics, and outcomes. One of the phenomapping groups had an increased risk of HF hospitalization even after adjusting for traditional risk factors. All the phenomapping analyses were performed blinded to clinical outcomes. The authors were also able to replicate the HF with preserved ejection fraction phenomapping group classification on a validation cohort (n=107).
Okser et al99 developed a predictive modeling framework to predict the extreme classes of risk to atherosclerosis and their progression over a 6‐year period. The framework used CRFs and single‐nucleotide polymorphisms selected with ML techniques as features, and achieved AUCs of 0.84 and 0.76 for the risk prediction and disease progressions tasks, respectively. These performances were found to be significantly better than the performances achieved when using CRFs alone. The CRFs used were sex, age, BMI, waist circumference, systolic and diastolic BP, cholesterol levels, triglycerides, apolipoprotein A1, apolipoprotein B, and whether the patient smoked.
Limitations and Challenges in Applying ML
Although ML and DL seem to hold promise in medicine, they have limitations. DL requires vast amount of data. Unlike obtaining images of dogs and cats, it can be challenging and expensive to obtain a large amount of labeled data in medicine. First, to obtain data associated with a disease of interest, one has to find patients with the disease, and those patients need to be willing to share their data. Second, the data must be annotated by trained clinicians; the process of obtaining annotations for data can therefore be time consuming and expensive. ML algorithms are subject to the principle of garbage in–garbage out. Therefore, having high‐quality training data is essential to the performance of ML algorithms. The annotations provided by clinicians can be subjected to bias. Disagreements can often occur between clinicians regarding the diagnosis/annotation. To obtain high‐quality annotations, ideally a panel of highly trained clinicians would be required to provide the annotations, which understandably is an expensive procedure.
ML and DL algorithms are subject to bias. One type of bias is the sample bias, which occurs when the distribution of one's training data does not reflect the actual environment where the ML model will be applied. Consider a case where ML has been used to build a predictive model, which predicts sudden cardiac arrest using heart rate variability data from patients with HF. Such model(s) would be limited in application, since sudden cardiac arrests occur in a much broader population. Another type of bias is unconscious human bias. ML algorithms are trained with data gathered and labeled by humans. Humans can be subjective, and they are affected by their surroundings and upbringings. There have been studies that showed that most healthcare providers have an implicit bias in terms of positive attitudes toward whites and negative attitudes toward people of color.107, 108 Such human bias can easily get incorporated into the ML models trained for providing clinical decisions.
There are many stakeholders in the healthcare system. These include the patients, the physicians, the employers, the insurance companies, the pharmaceutical firms, and the government. All these stakeholders have competing interests. To deploy ML in medicine, all stakeholders have to be on board. For example, physicians may resist the deployment of ML in medicine, considering that their role/job could be replaced by ML‐based systems.
The issue of liability is another serious challenge when applying ML in medicine. In medicine, patients’ lives are at stake. Who should be blamed when an ML algorithm provides a wrong diagnosis or a bad recommendation for treatment and the patient dies because of that recommendation? Should the physician be blamed, or should the person who took responsibility in building the ML‐based recommendation/diagnoses model be blamed?
The issue of patient data privacy is another significant concern when applying ML in medicine. To obtain concrete insights, ML‐based systems need vast amounts of data to be trained upon. It is therefore required that patients be willing to share their personal data to fuel the growth of ML in medicine. Though efforts have been made to deidentify the medical records/data, there is still a risk of reidentifying the patients from such data.109
Many ML algorithms, for example, RF and especially the DL algorithms, work like a “black box.” In other words, their decision‐making processes are seldom understood in totality. If DL is employed to make recommendations on the patients’ treatment plan, the patients may want to know the reasons behind those recommendations. Also, when the algorithms work like a black box, it could be much harder for the physicians to trust and understand the working of ML‐based recommender systems and catch the incorrect recommendations, if any.
Future of ML‐Based Applications in the Cardiovascular System
The need for using ML in the cardiovascular systems will continue to grow. As wearable and mobile devices become more widely used, the amount of cardiovascular data made available for training ML‐based systems will explode. The use of ML in cardiovascular systems can enable high‐accuracy automated diagnosis and save expert clinicians a considerable amount of time. Wearable devices with sensors and software capable of analytics will be in high demand.
The cost of sequencing the human genome is rapidly decreasing with the development of high‐throughput sequencing technology. Such reduction in cost is fueling the growth of ML‐based research for precision medicine. The exponential growth of biomedical data is unquestionable. However, at the same time, the data can be dispersed and not well organized. Efforts to make integrated and curated data sets will enable ML efficacy, in the future. In fact, such efforts are already being made. For example, the AHA recently established the Precision Medicine Platform through the efforts of multiple AHA volunteers and a collaboration with Amazon Web Services.110 The goals of the platform are (1) to make the data available for researchers, (2) to make searching across orthogonal data sets easy, (3) to provide a secure workspace/facility for taking advantage of the power of cloud computing, and (4) to provide a place for users to share insights.
Continuous efforts are necessary to avoid bias in data. There are no quick and easy solutions to this problem. Solving this problem requires careful sampling of the data, and conscious efforts from the creators of the ML‐based systems to eliminate personal bias from the input data.
One of the future directions of research would be to build classifiers that are more interpretable. Interpretability in ML models would allow physicians and patients to trust the ML‐based systems. In a recent study, an interpretable ML model was built for accurate prediction of sepsis in ICU patients.111 Another example of an ML algorithm that is both accurate and interpretable is the optimal decision tree.112
While the appropriate use of ML algorithms can enable better health care, the limitations of ML algorithms need to be acknowledged. In the near future, ML algorithms can at most assist physicians, and obviously cannot replace them. Physicians should therefore not feel threatened by AI, but could rather embrace it as a tool for providing better health care to patients. That being said, once ML's use in medicine becomes prominent, there will be changes in how physicians work. Future physicians would not only be required to be well versed in traditional medicine but also would be required to use/understand the ML‐based systems effectively and have good knowledge in statistics and data analytics.
Conclusions
In this article, we have highlighted and summarized the state of the art in ML‐based applications for improving patient outcomes pertaining to the cardiovascular system. There has definitely been an explosion of cardiovascular data, as well as an explosion of interest in applying AI. Because ML is better suited to find structures within complex data sets than traditional statistical methods, their application can surely improve patient outcomes. ML has limitations: (1) being prone to bias and (2) being difficult to interpret. Additionally, there are challenges of accountability and competing interests of stakeholders, which may restrict the usability of ML‐based applications in medicine. Deployment of ML‐based models in medicine needs a well‐thought‐out plan. Once planned, their deployment could lead to lasting benefits for mankind.
Sources of Funding
The work was supported by a Grand‐in‐Aid (#15GRNT23070001) from the American Heart Association (AHA), the RICBAC Foundation, NIH grant 1 R01 HL135335‐01, 1 R21 HL137870‐01 and 1 R21EB026164‐01 and a Founders Affiliate Post‐doctoral Fellowship (#19POST34450149) from the AHA.
Disclosures
None.
J Am Heart Assoc. 2020;9:e013924 DOI: 10.1161/JAHA.119.013924.
References
- 1. Henke N, Bughin J, Chui M, Manyika J, Saleh T, Wiseman B, Sethupathy G. The age of analytics: Competing in a data‐driven world. McKinsey Global Institute; December 2016. Available at: https://www.mckinsey.com/business-functions/mckinsey-analytics/our-insights/the-age-of-analytics-competing-in-a-data-driven-world. [Google Scholar]
- 2. Research from Gartner—five ways data science and machine learning deliver business impacts. 2017.
- 3. Daquila M. Worldwide Artificial Intelligence Systems Spending 2018–2022 Forecast: Market Opportunity by Industry and Use Case. International Data Corporation; November 2018. Doc # US44430518. Available at: https://www.idc.com/getdoc.jsp?containerId=US44430518. [Google Scholar]
- 4. Russell SJ, Norvig P. Artificial Intelligence: A Modern Approach. Malaysia: Pearson Education Limited; 2016. [Google Scholar]
- 5. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436. [DOI] [PubMed] [Google Scholar]
- 6. Bishop CM. Pattern Recognition and Machine Learning. Berlin, Heidelberg: Springer‐Verlag; 2006. ISBN: 0387310738. [Google Scholar]
- 7. Seber GAF, Lee AJ. Linear Regression Analysis. 2nd ed Hoboken, NJ: John Wiley & Sons; 2003. ISBN: 978-0-471-41540-4 [Google Scholar]
- 8. Hosmer DW Jr, Lemeshow S, Sturdivant RX. Applied Logistic Regression. 3rd ed Hoboken, NJ: John Wiley & Sons; 2013. Print ISBN: 9780470582473. Online ISBN: 9781118548387. DOI: 10.1002/9781118548387 [DOI] [Google Scholar]
- 9. Quinlan JR. Induction of decision trees. Mach Learn. 1986;1:81–106. [Google Scholar]
- 10. Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 11. Boser BE, Guyon IM, Vapnik VN. A training algorithm for optimal margin classifiers. Proceedings of the fifth annual workshop on computational learning theory 1992:144–152.
- 12. Goodfellow I, Bengio Y, Courville A. Deep Learning. MIT Press; 2016. Available at: http://www.deeplearningbook.org. [Google Scholar]
- 13. Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Adv Neural Inf Process Syst. 2012;25:1097–1105. [Google Scholar]
- 14. Devlin J, Chang M‐W, Lee K, Toutanova K. Bert: pre‐training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:181004805 2018.
- 15. Weston J, Chopra S, Bordes A. Memory networks. arXiv preprint arXiv:14103916 2014.
- 16. Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I. Attention is all you need. Adv Neural Inf Process Syst. 2017;30:5998–6008. [Google Scholar]
- 17. Bahrammirzaee A. A comparative survey of artificial intelligence applications in finance: artificial neural networks, expert system and hybrid intelligent systems. Neural Comput Appl. 2010;19:1165–1195. [Google Scholar]
- 18. Janai J, Güney F, Behl A, Geiger A. Computer vision for autonomous vehicles: problems, datasets and state‐of‐the‐art. arXiv preprint arXiv:170405519 2017.
- 19. Perlich C, Dalessandro B, Raeder T, Stitelman O, Provost F. Machine learning for targeted display advertising: transfer learning in action. Mach Learn. 2014;95:103–127. [Google Scholar]
- 20. Chen M, Hao Y, Hwang K, Wang L, Wang L. Disease prediction by machine learning over big data from healthcare communities. IEEE Access. 2017;5:8869–8879. [Google Scholar]
- 21. Koch M. Artificial intelligence is becoming natural. Cell. 2018;173:531–533. [DOI] [PubMed] [Google Scholar]
- 22. Andreu‐Perez J, Poon CC, Merrifield RD, Wong ST, Yang G‐Z. Big data for health. IEEE J Biomed Health Inform. 2015;19:1193–1208. [DOI] [PubMed] [Google Scholar]
- 23. Seera M, Lim CP. A hybrid intelligent system for medical data classification. Expert Syst Appl. 2014;41:2239–2249. [Google Scholar]
- 24. Tucker PE, Cohen PA, Bulsara MK, Acton J. Fatigue and training of obstetrics and gynaecology trainees in Australia and New Zealand. Aust N Z J Obstet Gynaecol. 2017;57:502–507. [DOI] [PubMed] [Google Scholar]
- 25. Wang J, Yang X, Cai H, Tan W, Jin C, Li L. Discrimination of breast cancer with microcalcifications on mammography by deep learning. Sci Rep. 2016;6:27327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist‐level classification of skin cancer with deep neural networks. Nature. 2017;542:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bertsimas D, Kallus N, Weinstein AM, Zhuo YD. Personalized diabetes management using electronic medical records. Diabetes Care. 2017;40:210–217. [DOI] [PubMed] [Google Scholar]
- 28. Wang Z, Shah AD, Tate AR, Denaxas S, Shawe‐Taylor J, Hemingway H. Extracting diagnoses and investigation results from unstructured text in electronic health records by semi‐supervised machine learning. PLoS One. 2012;7:e30412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012;13:395. [DOI] [PubMed] [Google Scholar]
- 30. Miotto R, Li L, Kidd BA, Dudley JT. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep. 2016;6:26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rajkomar A, Oren E, Chen K, Dai AM, Hajaj N, Hardt M, Liu PJ, Liu X, Marcus J, Sun M. Scalable and accurate deep learning with electronic health records. NPJ Digit Med. 2018;1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnson AE, Ghassemi MM, Nemati S, Niehaus KE, Clifton DA, Clifford GD. Machine learning and decision support in critical care. Proc IEEE. 2016;104:444–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, Gaasenbeek M, Angelo M, Reich M, Pinkus GS. Diffuse large B‐cell lymphoma outcome prediction by gene‐expression profiling and supervised machine learning. Nat Med. 2002;8:68. [DOI] [PubMed] [Google Scholar]
- 34. Peterson TA, Doughty E, Kann MG. Towards precision medicine: advances in computational approaches for the analysis of human variants. J Mol Biol. 2013;425:4047–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Potti A, Dressman HK, Bild A, Riedel RF, Chan G, Sayer R, Cragun J, Cottrill H, Kelley MJ, Petersen R. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294. [DOI] [PubMed] [Google Scholar]
- 36. Lavecchia A. Machine‐learning approaches in drug discovery: methods and applications. Drug Discov Today. 2015;20:318–331. [DOI] [PubMed] [Google Scholar]
- 37. Gawehn E, Hiss JA, Schneider G. Deep learning in drug discovery. Mol Inform. 2016;35:3–14. [DOI] [PubMed] [Google Scholar]
- 38. Zhang L, Tan J, Han D, Zhu H. From machine learning to deep learning: progress in machine intelligence for rational drug discovery. Drug Discov Today. 2017;22:1680–1685. [DOI] [PubMed] [Google Scholar]
- 39. Chen H, Engkvist O, Wang Y, Olivecrona M, Blaschke T. The rise of deep learning in drug discovery. Drug Discov Today. 2018;23:1241–1250. [DOI] [PubMed] [Google Scholar]
- 40. Weber JS, Toyama K. Remembering the past for meaningful AI‐D. 2010 AAAI Spring Symposium Series 2010.
- 41. Wahl B, Cossy‐Gantner A, Germann S, Schwalbe NR. Artificial intelligence (AI) and global health: how can AI contribute to health in resource‐poor settings? BMJ Glob Health. 2018;3:e000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gartner D, Kolisch R, Neill DB, Padman R. Machine learning approaches for early DRG classification and resource allocation. INFORMS J Comput. 2015;27:718–734. [Google Scholar]
- 43. Narula S, Shameer K, Omar AMS, Dudley JT, Sengupta PP. Machine‐learning algorithms to automate morphological and functional assessments in 2D echocardiography. J Am Coll Cardiol. 2016;68:2287–2295. [DOI] [PubMed] [Google Scholar]
- 44. Sengupta PP, Huang Y‐M, Bansal M, Ashrafi A, Fisher M, Shameer K, Gall W, Dudley JT. A cognitive machine learning algorithm for cardiac imaging: a pilot study for differentiating constrictive pericarditis from restrictive cardiomyopathy. Circ Cardiovasc Imaging. 2016;9:e004330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Madani A, Arnaout R, Mofrad M, Arnaout R. Fast and accurate view classification of echocardiograms using deep learning. NPJ Digit Med. 2018;1:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Attia ZI, Kapa S, Lopez‐Jimenez F, McKie PM, Ladewig DJ, Satam G, Pellikka PA, Enriquez‐Sarano M, Noseworthy PA, Munger TM. Screening for cardiac contractile dysfunction using an artificial intelligence–enabled electrocardiogram. Nat Med. 2019;25:70. [DOI] [PubMed] [Google Scholar]
- 47. Avendi M, Kheradvar A, Jafarkhani H. A combined deep‐learning and deformable‐model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med Image Anal. 2016;30:108–119. [DOI] [PubMed] [Google Scholar]
- 48. Motwani M, Dey D, Berman DS, Germano G, Achenbach S, Al‐Mallah MH, Andreini D, Budoff MJ, Cademartiri F, Callister TQ. Machine learning for prediction of all‐cause mortality in patients with suspected coronary artery disease: a 5‐year multicentre prospective registry analysis. Eur Heart J. 2016;38:500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Poplin R, Varadarajan AV, Blumer K, Liu Y, McConnell MV, Corrado GS, Peng L, Webster DR. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng. 2018;2:158. [DOI] [PubMed] [Google Scholar]
- 50. Wang J, Ding H, Bidgoli FA, Zhou B, Iribarren C, Molloi S, Baldi P. Detecting cardiovascular disease from mammograms with deep learning. IEEE Trans Med Imaging. 2017;36:1172–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Colloca R, Johnson AE, Mainardi L, Clifford GD. A support vector machine approach for reliable detection of atrial fibrillation events. Computing in Cardiology Conference (CinC), 2013 2013:1047–1050.
- 52. Özçift A. Random forests ensemble classifier trained with data resampling strategy to improve cardiac arrhythmia diagnosis. Comput Biol Med. 2011;41:265–271. [DOI] [PubMed] [Google Scholar]
- 53. Asl BM, Setarehdan SK, Mohebbi M. Support vector machine‐based arrhythmia classification using reduced features of heart rate variability signal. Artif Intell Med. 2008;44:51–64. [DOI] [PubMed] [Google Scholar]
- 54. Ceylan R, Özbay Y. Comparison of FCM, PCA and WT techniques for classification ECG arrhythmias using artificial neural network. Expert Syst Appl. 2007;33:286–295. [Google Scholar]
- 55. Joshi AJ, Chandran S, Jayaraman VK, Kulkarni BD. Hybrid SVM for multiclass arrhythmia classification.BIBM'09 IEEE International Conference on Bioinformatics and Biomedicine 2009:287–290.
- 56. Yeh Y‐C, Wang W‐J, Chiou CW. Cardiac arrhythmia diagnosis method using linear discriminant analysis on ECG signals. Measurement. 2009;42:778–789. [Google Scholar]
- 57. Moavenian M, Khorrami H. A qualitative comparison of artificial neural networks and support vector machines in ECG arrhythmias classification. Expert Syst Appl. 2010;37:3088–3093. [Google Scholar]
- 58. Sree SV, Ghista DN, Ng K‐H. Cardiac arrhythmia diagnosis by HRV signal processing using principal component analysis. J Mech Med Biol. 2012;12:1240032. [Google Scholar]
- 59. Li Q, Rajagopalan C, Clifford GD. Ventricular fibrillation and tachycardia classification using a machine learning approach. IEEE Trans Biomed Eng. 2014;61:1607–1613. [DOI] [PubMed] [Google Scholar]
- 60. Zhang XS, Zhu YS, Thakor NV, Wang ZZ. Detecting ventricular tachycardia and fibrillation by complexity measure. IEEE Trans Biomed Eng. 1999;46:548–555. [DOI] [PubMed] [Google Scholar]
- 61. Kuo S. Computer detection of ventricular fibrillation. Proc of Computers in Cardiology, IEEE Computer Society. 1978:347–349.
- 62. Li Q, Mark RG, Clifford GD. Robust heart rate estimation from multiple asynchronous noisy sources using signal quality indices and a Kalman filter. Physiol Meas. 2007;29:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zubair M, Kim J, Yoon C. An automated ECG beat classification system using convolutional neural networks. 6th International Conference on IT Convergence and Security (ICITCS) 2016:1–5.
- 64. Acharya UR, Oh SL, Hagiwara Y, Tan JH, Adam M, Gertych A, San Tan R. A deep convolutional neural network model to classify heartbeats. Comput Biol Med. 2017;89:389–396. [DOI] [PubMed] [Google Scholar]
- 65. Isin A, Ozdalili S. Cardiac arrhythmia detection using deep learning. Procedia Comput Sci. 2017;120:268–275. [Google Scholar]
- 66. Sodmann P, Vollmer M, Nath N, Kaderali L. A convolutional neural network for ECG annotation as the basis for classification of cardiac rhythms. Physiol Meas. 2018;39:104005. [DOI] [PubMed] [Google Scholar]
- 67. Deng M, Wang C, Tang M, Zheng T. Extracting cardiac dynamics within ECG signal for human identification and cardiovascular diseases classification. Neural Netw. 2018;100:70–83. [DOI] [PubMed] [Google Scholar]
- 68. Zhu F, Ye F, Fu Y, Liu Q, Shen B. Electrocardiogram generation with a bidirectional LSTM‐CNN generative adversarial network. Sci Rep. 2019;9:6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Drew BJ, Harris P, Zègre‐Hemsey JK, Mammone T, Schindler D, Salas‐Boni R, Bai Y, Tinoco A, Ding Q, Hu X. Insights into the problem of alarm fatigue with physiologic monitor devices: a comprehensive observational study of consecutive intensive care unit patients. PLoS One. 2014;9:e110274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Plesinger F, Klimes P, Halamek J, Jurak P. Taming of the monitors: reducing false alarms in intensive care units. Physiol Meas. 2016;37:1313–1325. [DOI] [PubMed] [Google Scholar]
- 71. Kalidas V, Tamil LS. Cardiac arrhythmia classification using multi‐modal signal analysis. Physiol Meas. 2016;37:1253–1272. [DOI] [PubMed] [Google Scholar]
- 72. Eerikäinen LM, Vanschoren J, Rooijakkers MJ, Vullings R, Aarts RM. Reduction of false arrhythmia alarms using signal selection and machine learning. Physiol Meas. 2016;37:1204. [DOI] [PubMed] [Google Scholar]
- 73. Wiens J, Guttag JV. Active learning applied to patient‐adaptive heartbeat classification. Adv Neural Inf Process Syst. 2010;232442–2450. [Google Scholar]
- 74. Ong MEH, Ng CHL, Goh K, Liu N, Koh ZX, Shahidah N, Zhang TT, Fook‐Chong S, Lin Z. Prediction of cardiac arrest in critically ill patients presenting to the emergency department using a machine learning score incorporating heart rate variability compared with the modified early warning score. Crit Care. 2012;16:R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Churpek MM, Yuen TC, Winslow C, Meltzer DO, Kattan MW, Edelson DP. Multicenter comparison of machine learning methods and conventional regression for predicting clinical deterioration on the wards. Crit Care Med. 2016;44:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Frizzell JD, Liang L, Schulte PJ, Yancy CW, Heidenreich PA, Hernandez AF, Bhatt DL, Fonarow GC, Laskey WK. Prediction of 30‐day all‐cause readmissions in patients hospitalized for heart failure: comparison of machine learning and other statistical approaches. JAMA Cardiol. 2017;2:204–209. [DOI] [PubMed] [Google Scholar]
- 77. Sandu C, Popescu D, Popescu C. Post cardiac surgery recovery process with reinforcement learning. 19th International Conference on System Theory, Control and Computing (ICSTCC) 2015:658–661.
- 78. Rom R, Erel J, Glikson M, Lieberman RA, Rosenblum K, Binah O, Ginosar R, Hayes DL. Adaptive cardiac resynchronization therapy device based on spiking neurons architecture and reinforcement learning scheme. IEEE Trans Neural Netw. 2007;18:542–550. [DOI] [PubMed] [Google Scholar]
- 79. Burke LE, Ma J, Azar KM, Bennett GG, Peterson ED, Zheng Y, Riley W, Stephens J, Shah SH, Suffoletto B. Current science on consumer use of mobile health for cardiovascular disease prevention: a scientific statement from the American Heart Association. Circulation. 2015;132:1157–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Piette JD, List J, Rana GK, Townsend W, Striplin D, Heisler M. Mobile health devices as tools for worldwide cardiovascular risk reduction and disease management. Circulation. 2015;132:2012–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. >Hannun AY, Rajpurkar P, Haghpanahi M, Tison GH, Bourn C, Turakhia MP, Ng AY. Cardiologist‐level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med. 2019;25:65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ortín S, Soriano MC, Alfaras M, Mirasso CR. Automated real‐time method for ventricular heartbeat classification. Comput Methods Programs Biomed. 2019;169:1–8. [DOI] [PubMed] [Google Scholar]
- 83. Teijeiro T, García CA, Castro D, Félix P. Arrhythmia classification from the abductive interpretation of short single‐lead ECG records. Comput Cardiol. 2017;44:1–4. [Google Scholar]
- 84. Plesinger F, Nejedly P, Viscor I, Halamek J, Jurak P. Parallel use of a convolutional neural network and bagged tree ensemble for the classification of Holter ECG. Physiol Meas. 2018;39:094002. [DOI] [PubMed] [Google Scholar]
- 85. Shashikumar SP, Shah AJ, Li Q, Clifford GD, Nemati S. A deep learning approach to monitoring and detecting atrial fibrillation using wearable technology. IEEE EMBS International Conference on Biomedical & Health Informatics (BHI) 2017:141–144.
- 86. Lahdenoja O, Hurnanen T, Iftikhar Z, Nieminen S, Knuutila T, Saraste A, Kiviniemi T, Vasankari T, Airaksinen J, Pankaala M. Atrial fibrillation detection via accelerometer and gyroscope of a smartphone. IEEE J Biomed Health Inform. 2017;99:1–12. [DOI] [PubMed] [Google Scholar]
- 87. Zhu T, Pimentel MA, Clifford GD, Clifton DA. Unsupervised Bayesian inference to fuse biosignal sensory estimates for personalising care. IEEE J Biomed Health Inform. 2018;23:47–58. [DOI] [PubMed] [Google Scholar]
- 88. Satija U, Ramkumar B, Manikandan MS. Automated ECG noise detection and classification system for unsupervised healthcare monitoring. IEEE J Biomed Health Inform. 2018;22:722–732. [DOI] [PubMed] [Google Scholar]
- 89. Yaghmaie N, Maddah‐Ali MA, Jelinek HF, Mazrbanrad F. Dynamic signal quality index for electrocardiograms. Physiol Meas. 2018;39:105008. [DOI] [PubMed] [Google Scholar]
- 90. Choi E, Schuetz A, Stewart WF, Sun J. Using recurrent neural network models for early detection of heart failure onset. J Am Med Inform Assoc. 2016;24:361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Juhola M, Joutsijoki H, Penttinen K, Aalto‐Setälä K. Detection of genetic cardiac diseases by Ca 2+ transient profiles using machine learning methods. Sci Rep. 2018;8:9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kullo IJ, Shameer K, Jouni H, Lesnick TG, Pathak J, Chute CG, De Andrade M. The ATXN2‐SH2B3 locus is associated with peripheral arterial disease: an electronic medical record‐based genome‐wide association study. Front Genet. 2014;5:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Khera AV, Chaffin M, Zekavat SM, Collins RL, Roselli C, Natarajan P, Lichtman JH, D'Onofrio G, Mattera J, Dreyer R. Whole‐genome sequencing to characterize monogenic and polygenic contributions in patients hospitalized with early‐onset myocardial infarction. Circulation. 2019;139:1593–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Weng SF, Reps J, Kai J, Garibaldi JM, Qureshi N. Can machine‐learning improve cardiovascular risk prediction using routine clinical data? PLoS One. 2017;12:e0174944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zhao J, Feng Q, Wu P, Lupu RA, Wilke RA, Wells QS, Denny JC, Wei W‐Q. Learning from longitudinal data in electronic health record and genetic data to improve cardiovascular event prediction. Sci Rep. 2019;9:717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Diller G‐P, Kempny A, Babu‐Narayan SV, Henrichs M, Brida M, Uebing A, Lammers AE, Baumgartner H, Li W, Wort SJ. Machine learning algorithms estimating prognosis and guiding therapy in adult congenital heart disease: data from a single tertiary centre including 10 019 patients. Eur Heart J. 2019;40:1069–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bellot A, Van der Schaar M. A hierarchical Bayesian model for personalized survival predictions. IEEE J Biomed Health Inform. 2019;23:72–80. [DOI] [PubMed] [Google Scholar]
- 98. Shah SJ, Katz DH, Selvaraj S, Burke MA, Yancy CW, Gheorghiade M, Bonow RO, Huang C‐C, Deo RC. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation. 2015;131:269–279. DOI: 10.1161/CIRCULATIONAHA.114.010637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Okser S, Lehtimäki T, Elo LL, Mononen N, Peltonen N, Kähönen M, Juonala M, Fan Y‐M, Hernesniemi JA, Laitinen T. Genetic variants and their interactions in the prediction of increased pre‐clinical carotid atherosclerosis: the Cardiovascular Risk in Young Finns Study. PLoS Genet. 2010;6:e1001146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Goodfellow I, Pouget‐Abadie J, Mirza M, Xu B, Warde‐Farley D, Ozair S, Courville A, Bengio Y. Generative adversarial nets. Adv Neural Inf Process Syst. 2014;27:2672–2680. [Google Scholar]
- 101. Chandra BS, Sastry CS, Jana S. Robust heartbeat detection from multimodal data via CNN‐based generalizable information fusion. IEEE Trans Biomed Eng. 2018;66:710–717. [DOI] [PubMed] [Google Scholar]
- 102. Tison GH, Zhang J, Delling FN, Deo RC. Automated and interpretable patient ECG profiles for disease detection, tracking, and discovery. Circ Cardiovasc Qual Outcomes. 2019;12:e005289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Pan J, Tompkins WJ. A real‐time QRS detection algorithm. IEEE Trans Biomed Eng. 1985;32:230–236. [DOI] [PubMed] [Google Scholar]
- 104. Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT. Genome‐wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Gerds TA, Kattan MW, Schumacher M, Yu C. Estimating a time‐dependent concordance index for survival prediction models with covariate dependent censoring. Stat Med. 2013;32:2173–2184. [DOI] [PubMed] [Google Scholar]
- 106. Mogensen UB, Ishwaran H, Gerds TA. Evaluating random forests for survival analysis using prediction error curves. J Stat Softw. 2012;50:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Hall WJ, Chapman MV, Lee KM, Merino YM, Thomas TW, Payne BK, Eng E, Day SH, Coyne‐Beasley T. Implicit racial/ethnic bias among health care professionals and its influence on health care outcomes: a systematic review. Am J Public Health. 2015;105:e60–e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sabin DJA, Nosek DBA, Greenwald DAG, Rivara DFP. Physicians’ implicit and explicit attitudes about race by MD race, ethnicity, and gender. J Health Care Poor Underserved. 2009;20:896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. El Emam K, Dankar FK, Vaillancourt R, Roffey T, Lysyk M. Evaluating the risk of re‐identification of patients from hospital prescription records. Can J Hosp Pharm. 2009;62:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Kass‐Hout TA, Stevens LM, Hall JL. American Heart Association precision medicine platform. Circulation. 2018;137:647–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Nemati S, Holder A, Razmi F, Stanley MD, Clifford GD, Buchman TG. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit Care Med. 2018;46:547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Bertsimas D, Dunn J. Optimal classification trees. Mach Learn. 2017;106:1039–1082. [Google Scholar]


