Abstract
Over the last 5 years, multiple stakeholders in the field of spinal cord injury (SCI) research have initiated efforts to promote publications standards and enable sharing of experimental data. In 2016, the National Institutes of Health/National Institute of Neurological Disorders and Stroke hosted representatives from the SCI community to streamline these efforts and discuss the future of data sharing in the field according to the FAIR (Findable, Accessible, Interoperable and Reusable) data stewardship principles. As a next step, a multi-stakeholder group hosted a 2017 symposium in Washington, DC entitled “FAIR SCI Ahead: the Evolution of the Open Data Commons for Spinal Cord Injury research.” The goal of this meeting was to receive feedback from the community regarding infrastructure, policies, and organization of a community-governed Open Data Commons (ODC) for pre-clinical SCI research. Here, we summarize the policy outcomes of this meeting and report on progress implementing these policies in the form of a digital ecosystem: the Open Data Commons for Spinal Cord Injury (ODC-SCI.org). ODC-SCI enables data management, harmonization, and controlled sharing of data in a manner consistent with the well-established norms of scholarly publication. Specifically, ODC-SCI is organized around virtual “laboratories” with the ability to share data within each of three distinct data-sharing spaces: within the laboratory, across verified laboratories, or publicly under a creative commons license (CC-BY 4.0) with a digital object identifier that enables data citation. The ODC-SCI implements FAIR data sharing and enables pooled data-driven discovery while crediting the generators of valuable SCI data.
Keywords: community-based repository, data sharing, FAIR data, ODC-SCI, spinal cord injury
Introduction and History
Published research articles generally include methods, results, and supplementary data. Data shared in publications about pre-clinical research, however, represent only a fraction of the data produced. The full results of the majority of research studies are actually never made public. For example, surgical data or data that do not fit the “story” of a manuscript are often not included in published articles.2 Further, experiments with so-called negative outcomes will likely not be published in a journal at all.3–5 Thus, a large amount of data are not accessible. These “dark data” are estimated to make up 85% of all data collected in biomedical research,3 equating to around $200 billion of the ∼$240 billion global research expenditure.3,6 The inability to discover and access all data dramatically impedes research and translational efforts by creating positive publication bias that overinflates the reported effect sizes.7,8 Further, the lack of transparency (i.e., accessibility of all data) does not allow funders to verify that data are reusable beyond the funded project they supported, journals are unable to deliver the entire picture of the experiments, and researchers repeat studies that have been performed by others, incorporating incorrect protocols and flawed scientific premises without ever knowing it.
This causes a significant waste of time and resources and also triggers ethical concerns regarding animal use and human subjects who receive experimental therapies. Another issue is that the current publication model does not facilitate replications, independent validations, or bench-to-bedside translation.9,10 Last, collecting a large body of data would allow more sophisticated meta-analysis based on individual subject-level raw data, thereby facilitating new discoveries and translation.11 Indeed, the “father” of meta-analysis, Gene Glass, stated: “Meta-analysis was created out of the need to extract useful information from the cryptic records of inferential data analyses in the abbreviated reports of research in journals and other printed sources….”12,13
With the advent of digital data-sharing technologies, the full vision of meta-analysis based on shared pools of data is within reach. This is well evidenced in physics and genomics/transcriptomics where the culture of sharing pre-publication findings through public data repositories and pre-print servers (e.g., arXiv.org; biorxiv.org) and the rapid and fruitful evolution of new approaches for managing and analyzing “big data” have driven discoveries.1,14 Potential reasons for the genomics/transcriptomics community being much further ahead in data sharing include the fact that their data are less complex (four bases and highly structured measures) than data derived from spinal cord injury (SCI) or stroke studies for instance. Highly complex manipulations on diverse tissue in the central nervous system and their complex impact on function/behavior is harder characterize through individual, independent data components. Further, the genomics field benefited from early data-sharing mandates and federally funded data infrastructure efforts around 30 years ahead of other fields.
Researchers, funding institutions, and publishers are all increasingly aware of and concerned about the lack of data sharing and transparency, yet changes in practice and policy are slow, and with few easy opportunities for sharing, most data remain inaccessible. The heterogeneity of pre-clinical research, particularly SCI research, makes sharing data even more essential to reduce waste and identify meaningful outcomes.1 As a consequence, biomedical research is experiencing a cultural shift in approaches to collection of big data and data sharing. In 2011, a meeting of international leaders in data science known as “The Future of Research Communications and e-Scholarship,” or FORCE 11, ultimately published recommendations for data sharing (https://www.force11.org/). One product was the development of “FAIR Data Principles,”15 which describe a set of key principles that will ensure data's value to the research community, that is, that the data are Findable (with sufficient explicit metadata), Accessible (open and available to other researchers), Interoperable (using standard definitions and common data elements [CDEs]), and Reusable (meeting community standards and sufficiently documented). The Office of Data Science at the National Institutes of Health (NIH) endorsed the FAIR Data Principles and NIH plans to incorporate these standards in future data sharing recommendations and programs.15
Also, the SCI field has embraced the challenge of FAIR data sharing, and since 2013, five projects have added data resources and tools for the SCI pre-clinical research community: 1) the VISION-SCI data repository with source data contributed by multiple research laboratories16,17; 2) a consensus guideline of minimal reporting standards for pre-clinical SCI research (MIASCI)18; 3) a knowledge base and ontology for integration of SCI research (RegenBase)19; 4) the Preclinical Spinal Cord Injury Knowledge Base (PSINK by HW Müller and colleagues20); and 5) the recently developed Open Data Commons for SCI research (ODC-SCI).1 These efforts are not competitive, but rather address different goals. Similar efforts can be observed in clinical research with the European Multicenter Study about Spinal Cord Injury (EMSCI), the Rick Hansen registry, and the Neurorecovery Network (NRN), the North American Clinical Trials Network® (NACTN), and in veterinarian SCI research with the Canine Spinal Cord Injury Consortium (CANSORT-SCI) data registery,21 among others.
The present work focuses on the ODC-SCI, which at the time of writing is in public beta release, with a full production version planned for late 2019. The ODC-SCI is a combined repository of raw study data, as opposed to other repositories of research metadata only or some journal storage of non-integrated study data. It evolved from previous efforts by the Ferguson SCI data science research group. A precursor was VISION-SCI, a collection and curation of data starting with 14 SCI labs representing ∼$60 million worth of previous NIH-funded data collection. Analysis of these legacy data has resulted in four publications that serve as a proof of the feasibility of the ODC-SCI.16,17,22,23 The process of establishing a community-driven database in pre-clinical SCI research is progressing with a steering committee that coordinates development, guidelines, and the outreach to the broader community, including researchers, funders, and journals.
A milestone in this development was the recent workshop “Developing a FAIR Share Community.”1 This was followed by the 2017 “FAIR-SCI Ahead” workshop held in November 2017 in conjunction with the Society for Neuroscience (SFN) meeting in Washington, DC. The workshop was organized by a steering committee (co-chairs: K.F. and A.R.F., members: the primary authors of this article) and sponsored by Wings for Life, the Craig Neilsen Foundation, the Rick Hansen Institute, and International Spinal Research Trust. The workshop focused on: 1) when and how data should be shared; 2) who it should be shared with; and 3) discussion of polices surrounding governance and how outreach to the community should be addressed. To gain a diverse perspective and broad stakeholder input, the meeting participants consisted of biomedical SCI researchers, bioinformatics/data scientists, funding, publishing and industry representatives, governmental agencies (NIH, Department of Defense, and Department of Veterans Affairs), university administrators, and individuals with SCIs. A full list is noted in the “FAIR-SCI Ahead Workshop Participants” author block in the Acknowledgments.
Major Points of Discussion
The discussions and outcomes of the FAIR-SCI Ahead meeting are summarized below.
What data should be shared?
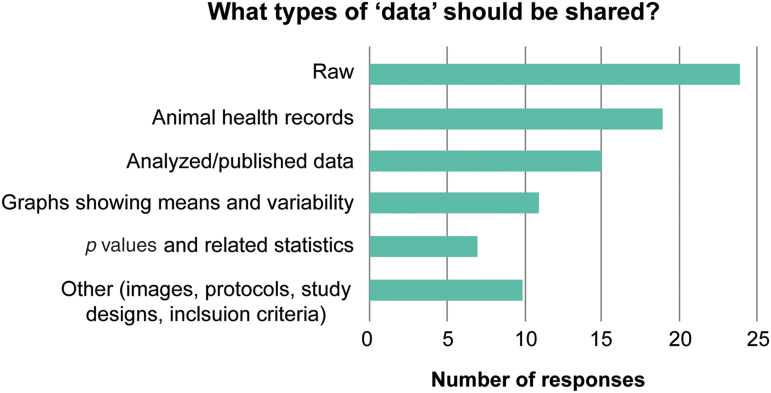
Experimental data are very broad, ranging from animal health records to statistical values calculated from the raw data. Currently, raw data are considered to be, in the case of ODC-SCI, the endpoint tabular data represented by numbers and categories rather than the original raw format in which data were collected (images, or electrophysiological traces, transcript counts, etc.). Even if published in a journal, it is likely that not all of these data will be accessible or published. Consequently, the amount of data to be shared will not only determine the required workload needed to upload all these different data, but could also introduce privacy issues, where users might feel uncomfortable sharing data. To explore this topic, the first question for the workshop participants was: What data should be shared in the ODC-SCI? Participants were asked to select all answers they thought applied from the following list: A) raw individual-level data; B) animal health records; C) analyzed/published data; D) graphs showing means and variability (standard deviation, standard error of the mean, and confidence interval); E) p values and related statistics (effect sizes, test statistics, etc.)? or F) other. Responses are shown in Figure 1. Nearly all (24 of 26) respondents indicated that raw, individual-level data should be shared, and most (19 of 25) thought that animal health records should also be included. Of the minority of respondents that indicated summary statistics should be included, most specified these were only useful in an abstract describing the underlying data set. Suggestions for “other” data to be shared included images or imaging data, and inclusion/exclusion criteria and/or protocols.
FiG. 1.
Responses to the question “what is ‘data’ that should be shared?” (n = 26). Color image is available online.
One concern voiced by several researchers was that publication of individual animal health records could be problematic because of universities' restrictions and because publicizing the total number of animals used could cause investigators to be targeted by animal rights organizations. Others, including editors from the journal Lab Animal, countered that increasing transparency is necessary for the public to understand the actual investments required for translational advances; moreover, sharing data from animals that were not included in a publication helps ensure that their deaths are not wasted.
Who has access to the data and who gives permission for that access?
A somewhat contentious topic is “public” access to data once they are entered into the ODC-SCI. Traditionally, researchers are protective of their data, especially given that they might consider use of these data as the basis for their future studies. At the FAIR-SCI Ahead workshop, three models of data access were presented and discussed: 1) time embargoed, where the access of data by others is granted after a set period; 2) content based, where the type of data dictates the level of access (e.g., lab principal investigator [PI] decides who may access specific data based on the sensitivity of the content and/or readiness to be shared and made accessible); and 3) “prep-kitchen,” where data go through levels of “semishared” curation and only the final, highly curated data sets are “published,” once this is approved by the owner of the data. The second and third approaches were extensively discussed, with major themes being the quality and utility of the data, as well as the sensitivity of the data (e.g., researchers being targeted by animal rights activists or being scooped by a competing investigator/industry). Although no consensus was reached, the discussion clarified the need for a flexible sharing policy that could incorporate different sharing concerns and goals.
Setting standards and quality control
One of the most challenging tasks for any database is quality control. Ensuring that data are properly vetted, cleaned, and well described is labor- and time-intensive, particularly with retrospective studies that may not have been collected with data standards in mind. This process is, however, essential given that data quality is a concern of both funders and users. Some automatic quality checks can be integrated into the upload platform for quality control, including completeness of data and checking that values are within range and that appropriate units are provided. However, some standard quality checks require expert knowledge and curation to determine appropriate ranges (e.g., discrepancies between outcome measures and injury type) or decide whether outcome measures are applicable to the uploaded data set. Our current process involves so-called data wranglers that can assist with data upload and curation. Levels of data standardization were described as a “FAIR onion,” with layers of metadata starting with the core data descriptors (e.g., who, what, where, when), community vocabularies (Neurolex/Interlex, MIASCI, RegenBase ontology, and PSINK ontology),18–20,24 domain-specific metadata (impact parameters, SCI-specific endpoints like Basso, Beattie, Bresnahan Locomotor Rating Scale/Basso Mouse Scale for Locomotion score, epicenter sparing, and regeneration), and highly specific metadata (e.g., experiment-specific treatments). Creating these terms requires ongoing expert discussions given that standards and tools are currently being developed and are best curated by the research community.
Part of identifying the appropriate level of curation and quality is determining how the ODC-SCI repository will be used. Specifically, a balance has to be reached between data quality/curation versus ease of use/low barrier for entry to data submission. Therefore, how the data are to be used is critical to determine what level of curation is needed. Further, adding layers of curation will also directly increase in cost of the data upload process. There was agreement that the ODC-SCI should be flexible to accommodate multiple research goals. However, determining the specific curation levels required by the ODC-SCI community remains a topic for a future workshop.
Why sharing?
The goals of sharing data within the ODC-SCI were classified as 1) discovery, 2) retrospective analysis, and 3) prospective collection. Sharing data for discovery science has the lowest bar for entry given that its primary utility is to provide raw data for exploratory usage. Participants envisioned a text searchable catalog to help identify whether others have already tested an intervention, used a specific reagent, or worked with a specific method. The value of a discovery-centered repository would be to reduce repeated dead-end experiments or find collaborators and/or advisors, but should be subjected to minimal curation to facilitate sharing and speed discovery. In contrast, the retrospective data usage would consist of complete experiments or projects, which may combine both published and unpublished datasets. The goal of accessing these data sets is to mine them using advanced analytical tools to identify potential interventions or variables that may contribute to outcomes. These types of data are also the hardest to curate and upload given that they likely require extensive cleaning and expert review to align with a data dictionary (where experimental terms are defined), or evaluate the quality of the research approach. These data are also most likely to warrant restrictions, given that they can be misused by competitive laboratories, or because the data donor may hope to use them in the future.
Use of the data most likely will require investigator approval and/or collaboration. Finally, prospective translational studies are the most purpose curated, but should also be publicly accessible. These would include new studies specifically testing an intervention with the intention of translation and designed for sharing. These data sets could come from studies pre-registered with a journal that agrees to publish it regardless of outcome, or a grant that requires data sharing as part of its funding requirement. Advantages of the prospective data set include the idea that the data dictionary and data collection forms can be set up before data are collected and can use the best available standards. Data quality can also be checked during data collection and can improve experiment quality. Each of these goals represents important ways to utilize otherwise dark data, and it may be possible to develop a database and governance structure that has processes and features that can achieve these different data-use goals. The ODC-SCI infrastructure was therefore designed with all three goals in mind.
Copyright and authorship
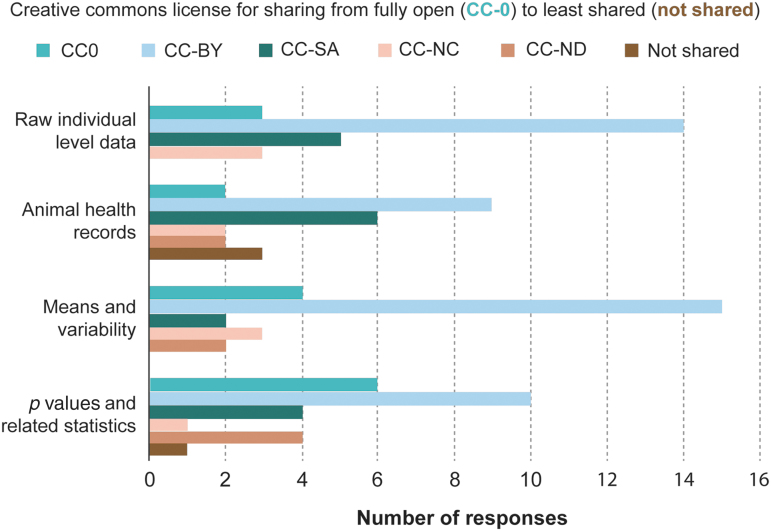
Pre-clinical data sets generally do not have strict copyright standards; nevertheless, universities normally own intellectual property associated with investigators' research. Therefore, it is the responsibility of the investigators to find out what restrictions may apply to their data. In pre-clinical SCI research as in other fields of discovery research, most data are not considered proprietary, and investigators are unlikely to have many restrictions, but it is important to discuss this with a university representative before making potentially sensitive data publicly available. There are various examples and levels of licenses available from the Open Definition Organization (see opendefinition.org/licenses), which were introduced at the meeting. Workshop participants were asked to indicate what level of restriction they felt was appropriate for different types of data. Most indicated fairly unrestricted levels of access, with the redistribution and reuse on the condition that the creator is appropriately credited (“CC-BY”; Fig. 2). Digital object identifiers (DOIs) are commonly applied to data sets in many fields and organizations (e.g., NASA) and data contributors are cited accordingly. Many communities, including funding agencies, have an interest in increasing data sharing by rewarding investigators who produce valuable data sets and recognizing those data sets from their sharing index (similar to an “H-index” for article citations). However, most institutions have yet to implement any reward structure for data sharing.
FIG. 2.
Responses to the level of license considered appropriate for various types of data (n = 26; *four respondents chose more than one answer, only the most conservative/restricted was counted). CC, Creative Commons. Color image is available online.
Pre-empting potential conflict must also be addressed in any data-sharing environment. Suggestions for mitigating or resolving potential conflicts included setting expectations early (ability to know or decide what level of sharing you are participating in) and what the authorship expectations are (Can you cite this data set or do you need a co-authorship?). Other suggestions included setting tiered levels of authorship (the researcher who did the experiment vs. the one who uploaded and cleaned the data). Most conflicts can be resolved through explicit license agreements, a governance committee, and/or a university ombudsman, if necessary.
Sustainability
To be successful, the ODC-SCI must become self-sustainable. This means moving away from reliance on grants and toward a subscription, pay per use, or other sustainability model. The question that was immediately raised by the participants was: Who should pay this fee and how much would it amount to? During the discussion, most participants felt it would be unfair to expect investigators to spend funds as well as put time and effort into sharing. Others felt that if investigators pay to publish an article, why shouldn't they also pay to publish a data set? However, budgeting is not easy for the entire process of data submission, and requiring data contributors to publication fees on top for the submission might create an additional barrier. Although the NIH and other funders are encouraging investigators to incorporate data-sharing costs into their budgets, or request supplements to share valuable data, this is not yet in general practice. However, it is generally viewed as a potential model for the future that sharing costs will be eligible or even a required part of any operating grant budget.
Other sustainability models include universities and/or publishing companies having a low-cost subscription fee for their investigators/authors to use a database, similar to libraries paying fees for journal subscriptions. Considering that more and more journals request data sharing without offering a viable option, this seems to be a reasonable model for the future. There is also an evolution of rewards regarding data sharing from institutions (e.g., the QUEST Open Data research award from the Berlin Institute of Health) and funders (e.g., NIH Big Data to Knowledge initiative). Last, a successful data commons would offer itself as a powerful resource for industry, which then could be charged for access to restricted data sets, if applicable. These ideas have to be further explored for the ODC-SCI to evolve into a community-based repository.
Outreach
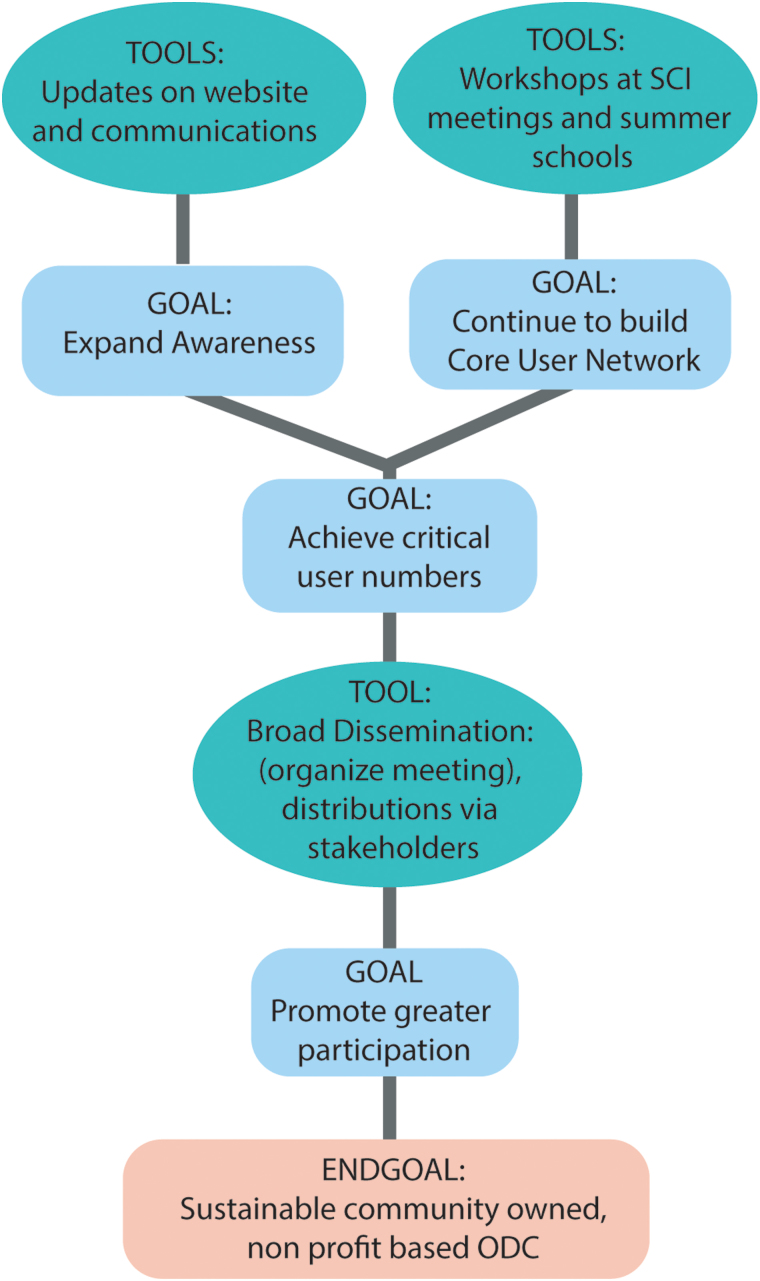
Data-sharing efforts are increasing across all fields, and most funders and publishers encourage or require data sharing. Developing a SCI community repository will take continuous community engagement and incentives. The challenges for such outreach include the need for funding and the need to find an approach that appeals to a broad body of researchers. The question is who is organizing the outreach considering that a data commons should be community driven/owned in the future. Various suggestions were brought forward on how to perform outreach most effectively. The most appreciated approach was to continue to organize an open annual meeting/workshop in parallel to a SCI-specific meeting or SFN. The second-highest-rated approach was to organize workshops at meeting of foundations for higher frequency and broader dissemination. To add a discussion board on the ODC-SCI website was ranked third. In contrast, mass advertisement, e-mail lists with newsletters, and the organization of smaller workshops were rated unfavorably. In addition to these proposed approaches, the participants recommended several areas for engagement (Fig. 3). The key approach was to continue to build a core user network to achieve a critical mass. This can be achieved by continuing to present the progress of the ODC-SCI at SCI-specific meetings and summer courses. Once a greater awareness of the ODC-SCI has been achieved, and its potential for the field has been communicated, a broader dissemination could be accomplished by hosting a larger meeting.
FIG. 3.
Outreach suggestions from meeting participants. ODC, Open Data Commons; SCI, spinal cord injury. Color image is available online.
Engineering FAIR-SCI Goals into ODC-SCI.org
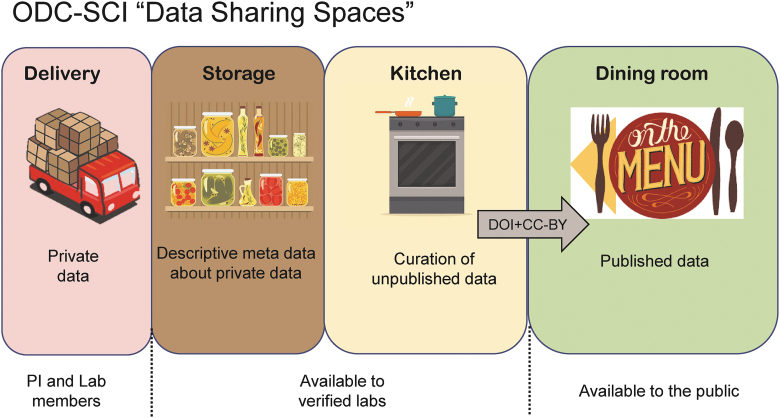
Based on the community requirements articulated above, the ODC-SCI was developed and publicly beta-released on the web at (http://ODC-SCI.org), with a full production release planned for late 2019. At the time of writing, ODC-SCI contains data from 52 laboratories and 173 data sets and is starting to show signs of steady expansion. The ODC-SCI architecture, implemented using the generalized SciCrunch platform,25 fulfills SCI community data-sharing priorities gleaned from the 2016 FAIR-Share meeting in Bethesda, Maryland (previously reported1), as well as the policy and governance feedback from the 2017 FAIR-SCI Ahead meeting in Washington, DC reported in the present article. The ODC-SCI core engineering team has worked with the steering committee to build a staged ecosystem to help manage the data-sharing life cycle from private data management to public release through an accessible DOI citation numbers issued under a creative commons-BY (CC-BY) license (Fig. 4).
FIG. 4.
Data-sharing spaces within the ODC-SCI. Based on community input from the 2016 (Bethesda, MD) and 2017 (Washington, DC) workshops, we have built the ODC-SCI architecture around distinct data-sharing spaces using a restaurant metaphor (from delivery, storage, processing in the kitchen and serving it in a public space, i.e., the dining room). The spaces range from the most closed, private, and secure space (left) to the most open and public space (right). The “lab” is the organizational unit of the ODC-SCI, and within the lab private space, the PI has total control over data-sharing permissions. Once data are uploaded and curated within the lab, the PI can submit the data to the semiprivate space of the ODC-SCI that is shared among verified laboratories. This will allow pooled research and data curation. Once data have been fully vetted through the processes described in this proposal, data can be shared publicly, will be given a permanent digital object identifier (DOI) and released using a creative commons BY (CC-BY) license, the same publication license used by open-access publishers. This will make data searchable using web search engines and will allow data citation and linkage of source data to their associated published articles. CC, Creative Commons; ODC-SCI, Open Data Commons/spinal cord injury; PI, principal investigator. Color image is available online.
The ODC-SCI architecture is built around the concept of digital laboratory spaces that control the level of access to data uploaded to the system. This intentionally provides multiple levels of access ranging from private to fully public, at the discretion of a responsible data steward, referred to as the “principal investigator” within the ODC-SCI system. Any individual with a verified institutional e-mail address can register and join the ODC-SCI ecosystem as a user and gain access to publicly accessible data sets. However, to unlock the full data management, data curation, and collaborative data-sharing capabilities, users must join a verified laboratory and be granted access from that laboratory's PI (or PI designee). Once ODC-SCI users have joined a verified laboratory, they gain the ability to upload data to the laboratory and access to private data that are in the process of being curated within the laboratory (Fig. 4, “Delivery” space). As part of the curation process, the ODC-SCI system encourages laboratory members to register their data elements to Common Data Elements, which are stored in the back end of the ODC-SCI system in a standard vocabulary of interlex terms (.ilx) using Neuroscience Information Framework core web standards and ontologies.24 This capability will make data interoperable with other data sets at the time of data upload/curation.
In addition, members of verified laboratories have access to data that are shared in a pre-public data commons available only to verified laboratories within the ODC-SCI ecosystem (Fig. 4, “Storage” and “Kitchen” spaces). This shared colaboratory space is designed to enable collaboration and data sharing and promote peer review of data for quality assurance and quality control before public release. This pre-public data space is equivalent to a pre-print server for scientific articles, enabling controlled community feedback before “publication.” Once data are vetted either through the pre-review space or through other mechanisms (e.g., manuscripts associated with ODC-SCI data are submitted for peer-reviewed publications), the laboratory PI has the ability to make the data set “public” and will be given a permanent DOI number that can be used to find and access the data using a standard web browser in much the same way that published articles receive DOIs.26 The DOI will enable data reuse through an open-access license (CC-BY) and derivative works as long as the data creator is credited (cited) upon reuse. For example, the first of these publicly available data sets can be found by typing the DOI citation number (http://doi.org/10.7295/W9T72FMZ) into a standard web browser. In addition, reuse will be promoted by the ODC-SCI architecture itself because the system has a built-in application programming interface that enables the development of new functionalities using ODC-SCI data resources.
In this sense, the ODC-SCI will function like an open-source cloud-based “operating system” so that data sharing can evolve over time, gaining new features and add-ons, as new open source applications for data sharing, data analysis, and data visualization are developed. In summary, the core proof-of-concept technology of the ODC-SCI implements many of the community goals of making SCI data FAIR.
Future Steps
It will be important to continue to get the word out in order to engage investigators, funders, journals, and the entire SCI community. Various presentations and workshops at SCI-specific meetings and SFN have piqued interest in a process that needs to be continued. At the same time, for success, it will be critical to streamline the data upload and find ways of rewarding data contributors. Further, during the next year, the steering committee will need to undergo a change into a “curation board” with appropriate leadership with terms of reference designed to make the ODC-SCI a truly community-run repository. Similarly, governance and financial independence need to evolve in the near future, to ensure survival of the ODC-SCI and enable a future with less dark data and data bias, but transparency and repeatability and with successful translation of pre-clinical experiments.
FAIR-SCI Ahead Workshop Participants
The participants of the FAIR-SCI Ahead Workshop are: Sabina Alam, PhD, Faculty of 1000, now Taylor and Francis Group; Mark Bacon, PhD, Spinal Research/ISRT; Linda Bambrick, PhD, Department of Defense; Michele Basso, EdD, PT, Ohio State University; Michael Beattie, PhD, University of California San Francisco; John Bixby, PhD, University of Miami; Jacqueline Bresnahan, PhD, University of California San Francisco; Alison Callahan, PhD, Stanford University; Adam Ferguson, PhD, University of California San Francisco; Karim Fouad, PhD, University of Alberta; John Gensel, PhD, University of Kentucky; Dustin Graham, PhD, Nature Lab Animal; Jeff Grethe, PhD, University of California San Diego; J. Russell Huie, PhD, University of California San Francisco; Lyn Jakeman, PhD, NIH/NINDS; Linda Jones, PT, MS, Craig H. Neilsen Foundation; Patricia Kabitzke, PhD, Cohen Veterans Bioscience; Naomi Kleitman, PhD, Craig H. Neilsen Foundation; Audrey Kusiak, PhD, U.S. Department of Veteran's Affairs; Brian Kwon, MD, PhD, University of British Columbia; Rosi Lederer, MD, Wings for Life; Vance Lemmon, PhD, University of Miami; Malcom MacLeod, PhD, University of Edinburgh; David Magnuson, PhD, University of Louisville; Maryann Martone, PhD, University of California San Diego; Verena May, PhD, Wings for Life; Ellen Neff, MS, Nature Lab Animal; Jessica Nielson, PhD, University of Minnesota; Sasha Rabchevsky, PhD, University of Kentucky; Jan Schwab, MD, PhD, Ohio State University; Carol Taylor-Burds, PhD, NIH/NINDS; Wolfram Tetzlaff, MD, PhD, University of British Columbia; and Abel Torres-Espin, PhD, University of Alberta, now University of California San Francisco.
Contributor Information
Collaborators: the FAIR-SCI Ahead Workshop Participants, Sabina Alam, Mark Bacon, Linda Bambrick, Michele Basso, Michael Beattie, Jacqueline Bresnahan, John Gensel, Dustin Graham, Jeff Grethe, J. Russell Huie, Linda Jones, Patricia Kabitzke, Naomi Kleitman, Audrey Kusiak, Brian Kwon, Rosi Lederer, Malcom MacLeod, Verena May, Ellen Neff, and Sasha Rabchevsky
Funding Information
The meeting to host the FAIR share meeting was supported by Wings for Life, International Spinal Research Trust, Craig H. Neilsen Foundation, with material support from NIH/NINDS.
Author Disclosure Statement
Lyn Jakeman, PhD, and Carol Taylor-Burds, PhD, are employees of the National Institute of Neurological Disorders and Stroke (NINDS) at the National Institutes of Health (NIH). The views expressed here are those of the authors and do not represent those of the NIH, the NINDS, or the U.S. government, nor do they imply endorsement by the U.S. government of any products of this work.
References
- 1. Callahan A., Anderson K.D., Beattie M.S., Bixby J.L., Ferguson A.R., Fouad K., Jakeman L.B., Nielson J.L., Popovich P.G., Schwab J.M., and Lemmon V.P. (2017). Developing a data sharing community for spinal cord injury research. Exp. Neurol. 295, 135–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Steward O., Popovich P.G., Dietrich W.D., and Kleitman N. (2012). Replication and reproducibility in spinal cord injury research. Exp. Neurol. 233, 597–605 [DOI] [PubMed] [Google Scholar]
- 3. Macleod M.R., Michie S., Roberts I., Dirnagl U., Chalmers I., Ioannidis J.P.A., Salman R.A.-S., Chan A.-W., and Glasziou P. (2014). Biomedical research: increasing value, reducing waste. Lancet 1509, 101–104 [DOI] [PubMed] [Google Scholar]
- 4. Ioannidis J.P.A. (2014). How to make more published research true. PLoS Med. 11, e1001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ioannidis J.P.A. (2005). Why most published research findings are false. PLoS Med. 2, e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chalmers I., and Glasziou P. (2009). Avoidable waste in the production and reporting of research evidence. Lancet 374, 86–89 [DOI] [PubMed] [Google Scholar]
- 7. Watzlawick R., Rind J., Sena E.S., Brommer B., Zhang T., Kopp M.A., Dirnagl U., Macleod M.R., Howells D.W., and Schwab J.M. (2016). Olfactory ensheathing cell transplantation in experimental spinal cord injury: effect size and reporting bias of 62 experimental treatments: a systematic review and meta-analysis. PLoS Biol. 14, e1002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Watzlawick R., Antonic A., Sena E.S., Kopp M.A., Rind J., Dirnagl U., Macleod M., Howells D.W., and Schwab J.M. (2019). Outcome heterogeneity and bias in acute experimental spinal cord injury: a meta-analysis. Neurology 93, e40–e51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Begley C.G., and Ioannidis J.P.A. (2015). Reproducibility in science: Improving the standard for basic and preclinical research. Circ. Res. 116, 116–126 [DOI] [PubMed] [Google Scholar]
- 10. Schulz J., Cookson M., and Hausmann L. (2016). The impact of fraudulent and irreproducible data to the translational research crisis—solutions and implementation. Neurochemistry 139, 253–270 [DOI] [PubMed] [Google Scholar]
- 11. Ferguson A.R., Nielson J.L., Cragin M.H., Bandrowski A.E., and Martone M.E. (2014). Big data from small data: data-sharing in the “long tail” of neuroscience. Nat..Neurosci. 17, 1442–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Glass G.V. (1976). Primary, secondary, and meta-analysis of research. Educ. Res. 5, 3–8 [Google Scholar]
- 13. Glass G.V.(2000). Meta-Analysis at 25. www.gvglass.info/papers/meta25.html (last accessed May24, 2019)
- 14. Kelley K.W., Nakao-inoue H., Molofsky A. V, and Oldham M.C. (2018). Variation among intact tissue samples reveals the core transcriptional features of human CNS cell classes. Nat. Neurosci. 21, 1171–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wilkinson M.D., Dumontier M., Aalbersberg Ij.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., Santos L.B. da S., Bourne P.E., Bouwman J., Brookes A.J., Clark T., Crosas M., Ingrid Dillo, Dumon O., Edmunds S., Evelo C.T., Finkers R., Gonzalez-Beltran A., Gray A.J.G., Groth P., Goble C., Grethe J.S., Heringa J., Hoen P.A.C. ’t Hooft, R., Kuhn T., Kok R., Kok J., Lusher S.J., Martone M.E., Mons A., Packer A.L., Persson B., Rocca-Serra P., Roos M., Schaik R. van, Sansone S.-A., Schultes E., Sengstag T., Slater T., Strawn G., Swertz M.A., Thompson M., Lei J. van der Mulligen, E. van Velterop, J., Waagmeester A., Wittenburg P., Wolstencroft K., Zhao J., and Mons B. (2016). The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nielson J.L., Guandique C.F., Liu A.W., Burke D.A., Lash A.T., Moseanko R., Hawbecker S., Strand S.C., Zdunowski S., Irvine K.-A., Brock J.H., Nout-Lomas Y.S., Gensel J.C., Anderson K.D., Segal M.R., Rosenzweig E.S., Magnuson D.S.K., Whittemore S.R., McTigue D.M., Popovich P.G., Rabchevsky A.G., Scheff S.W., Steward O., Courtine G., Edgerton V.R., Tuszynski M.H., Beattie M.S., Bresnahan J.C., and Ferguson A.R. (2014). Development of a database for translational spinal cord injury research. J. Neurotrauma 31, 1789–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nielson J.L., Paquette J., Liu A.W., Guandique C.F., Tovar C.A., Inoue T., Irvine K.A., Gensel J.C., Kloke J., Petrossian T.C., Lum P.Y., Carlsson G.E., Manley G.T., Young W., Beattie M.S., Bresnahan J.C., and Ferguson A.R. (2015). Topological data analysis for discovery in preclinical spinal cord injury and traumatic brain injury. Nat. Commun. 6, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lemmon V.P., Ferguson A.R., Popovich P.G., Xu X.-M., Snow D.M., Igarashi M., Beattie C.E., and Bixby J.L. (2014). Minimum information about a spinal cord injury experiment: a proposed reporting standard for spinal cord injury experiments. J. Neurotrauma 31, 1354–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Callahan A., Abeyruwan S.W., Al-Ali H., Sakurai K., Ferguson A.R., Popovich P.G., Shah N.H., Visser U., Bixby J.L., and Lemmon V.P. (2016). RegenBase: a knowledge base of spinal cord injury biology for translational research. Database 2016, baw040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brazda N., Kruse M., Kruse F., Kirchhoffer T., Klinger R., and Müller H. (2013). The CNR preclinical database for knowledge management in spinal cord injury research. Abstracts of the Society of Neurosciences 148 [Google Scholar]
- 21. Moore S.A., Zidan N., Spitzbarth I., Granger Y.S.N.N., Ronaldo C., Jonathan C., Nick M.L., Veronika D.J., Andrea M.S., and Olby N.J. (2018). Development of an International Canine Spinal Cord Injury observational registry: a collaborative data-sharing network to optimize translational studies of SCI. Spinal Cord 56, 656–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nielson J.L., Haefeli J., Salegio E.A., Liu A.W., Guandique C.F., Stück E.D., Hawbecker S., Moseanko R., Strand S.C., Zdunowski S., Brock J.H., Roy R.R., Rosenzweig E.S., Nout-Lomas Y.S., Courtine G., Havton L.A., Steward O., Reggie Edgerton V., Tuszynski M.H., Beattie M.S., Bresnahan J.C., and Ferguson A.R. (2015). Leveraging biomedical informatics for assessing plasticity and repair in primate spinal cord injury. Brain Res. 1619, 124–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ferguson A.R., Irvine K.-A., Gensel J.C., Nielson J.L., Lin A., Ly J., Segal M.R., Ratan R.R., Bresnahan J.C., and Beattie M.S. (2013). Derivation of multivariate syndromic outcome metrics for consistent testing across multiple models of cervical spinal cord injury in rats. PLoS One 8, e59712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Larson S.D., and Martone M.E. (2013). NeuroLex.org: an online framework for neuroscience knowledge. Front. Neuroinform. 7, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grethe J.S., Bandrowski A., Banks D.E., Condit C., Gupta A., Larson S.D., Li Y., Ozyurt I.B., Stagg A.M., Whetzel P.L., Marenco L., Miller P., Wang R., Shepherd G.M., and Martone M.E. (2014). SciCrunch: a cooperative and collaborative data and resource discovery platform for scientific communities. Front. Neuroinform. Conference Abstract: Neuroinformatics 2014. doi: 10.3389/conf.fninf.2014.18.00069 [DOI] [Google Scholar]
- 26. Bierer B.E., Crosas M., and Pierce H.H. (2017). Data authorship as an incentive to data sharing. N. Engl. J. Med. 376, 1684–1687 [DOI] [PubMed] [Google Scholar]