Abstract
Probiotics play a key role in the modulation of the gut immune system in health and disease and their action is mediated by molecules exposed on the microorganism surface or secreted probiotic-derived factors. In particular, Lactobacillus gasseri OLL2809, a probiotic microorganism isolated from human feces, has the potential to modulate various immune responses. The dendritic cells (DCs) are considered the main players in orchestrating the immune response, and their contact with intestinal microbiota is crucial for the development and homeostasis of gut immunity. To gain a perspective on the molecular mechanisms involved in the maturation process of DCs and investigate factors that could modulate these processes, a differential proteomic analysis was performed on the secretome of immature DCs, mature DCs (mDCs, induced by lipopolysaccharide (LPS)), and immature DCs challenged with L. gasseri OLL2809 before treatment with LPS (LGmDCs). The maturation process of DCs was associated to profound changes in the protein secretome and probiotic pre-treatment led to a dramatic modulation of several secreted proteins of mDC, not only classical immune mediators (i.e., cytokines, complement factors, T cell Receptor ligands) but also proteins involved in the contractile and desmosome machineries. The latter data highlight a novel mechanism by which L. gasseri can modulate the maturation process of DCs, reinforcing the concept of a protective anti-inflammatory role ascribed to this probiotic strain.
Keywords: dendritic cells, immunomodulation, probiotics, proteomics, secreted proteins, mass spectrometry, Lactobacillus gasseri
1. Introduction
Probiotics play a key role in the modulation of the gut immune system in health and disease and their action is mediated by molecules exposed on the microorganism surface or secreted probiotic-derived factors [1]. Lactobacillus gasseri OLL2809, a probiotic microorganism isolated from human feces, has the potential to modulate various immune responses [2]. In particular, it is able to enhance the oral tolerance induction and to act as an immune-modulator for the prevention of or early remission from food allergy [3]. The beneficial activity of this strain on mucosal inflammation has also been shown in mice, where the administration of OLL2809 was effective in reducing endometriotic lesions [4]. The immune properties of this microorganism were also exhibited by heat-killed cells that stimulated a high IL-12 (p70) production and a reduction in antigen-specific IgE levels in animal models of allergy [5]. In addition, heat-killed OLL2809 cells had suppressive effects on inflammatory conditions through the inhibition of CD4+ T-cell proliferation [6]. We previously found that γ-irradiated L. gasseri OLL2809 possessed the ability to modulate the cytokine profile in murine bone-marrow dendritic cells (DCs) [7]. DCs are considered to be major players in orchestrating the immune response, and their contact with intestinal microbiota is crucial for the development and homeostasis of gut immunity. However, the precise mechanisms by which intestinal microbiota can influence the development and function of DCs remain to be further elucidated.
Proteomics have been widely applied to elucidate differences in whole-cell protein patterns of immature DCs (iDCs) and mature DCs (mDCs), and to study exosome proteome, and, recently, phagosomal proteome [8,9,10,11,12,13]. However, only a few proteomic studies have characterized the protein pattern secreted (secretome) by iDCs and mDCs [14,15,16]. Therefore, the secretome analysis could represent an interesting area to exploit to gain a perspective on the molecular mechanisms involved in the maturation process of DCs and investigate factors that could modulate these processes.
Herein, a quantitative proteomic study was performed on the secretome of iDCs, lipopolysaccharide (LPS)-induced mDCs, and mDCs that were obtained from iDCs challenged with L. gasseri OLL2809 before inducing the maturation process with LPS (LGmDCs). A label-free quantitative approach based on spectral counting (normalized spectral abundance factor, NSAF) [17] was applied to gain insights into the immunomodulatory properties of L. gasseri OLL2809 and thoroughly investigate the cross-talk between dendritic cells and this probiotic microorganism.
2. Materials and Methods
2.1. Preparation of L. gasseri
The probiotic strain Lactobacillus gasseri OLL28099 was isolated from the human intestine (Patent Microorganisms Depositary, National Institute of Technology and Evaluation, Japan, Accession n. NITE BP-72) [5]. Working cultures were grown in deMan Rogosa Sharpe (MRS) broth (Difco, Detroit, MI, USA) for 24 h at 37 °C under microaerobic conditions. For co-culture experiments, bacterial strains were harvested in the late log phase and irradiated with 2800 Gy (Gray) γ-irradiation (MDS Nordion γ-cell 1000, Nordion, Ottawa, ON, Canada). This treatment completely inactivated the L. gasseri preparations.
2.2. Dendritic Cell Preparation and Co-Culture
BALB/c mice were maintained in pathogen-free conditions at our animal facility (accreditation no. DM.161/99). Animal studies were approved by the review committee of Health Ministry, General Division of Animal Health and of Veterinary Medicine (2016) and performed according to European regulations (EU Directive 2010/63/EU). Murine DCs were generated according to a previously published method [18]. In brief, bone marrow cells were collected in 10 mL RPMI 1640 medium (Sigma, St. Louis, MO, USA) containing antibiotics (penicillin 100 IU/mL; streptomycin 100 IU/mL, Sigma), 10% fetal calf serum (Sigma), 1% non-essential amino acids (EuroClone, Milan, Italy), and 20 ng/mL granulocyte–macrophage colony-stimulating factor (GM-CSF; PharMingen, San Diego, CA, USA) (complete medium). On day 3, 10 mL complete medium was added; on day 7, the 10 mL medium was replaced with fresh medium. This procedure eliminated all other bone marrow cells.
To obtain a comprehensive picture of the protein secretome from murine bone marrow DCs, protein contribution from cell culture medium was drastically reduced by incubating cells under serum-free conditions [18]. Specifically, on day 9, non-adherent immature DCs (iDCs) were harvested, placed in 24-well plates (1 × 106 cells/mL) and transferred in X-Vivo 15 medium (Lonza, Basel, Switzerland). iDCs were then incubated for 24 h with irradiated bacteria re-suspended in X-Vivo 15 medium at a 30:1 (bacteria: eukaryotic cell ratio). Finally, 1 μg/mL lipopolysaccharide (LPS) was added to the culture for 6 h to induce the maturation of DCs. Following incubation, cell viability was microscopically evaluated by a dye-exclusion test using Nigrosin (1% solution) and found ≥ 90% live cells in all experiments. To estimate the residual percentage of lysed cells in culture supernatants, Lactate Dehydrogenase (LDH) assay was used [19]. Results were expressed as a percentage of total LDH release from control cultures treated with 1% (w/v) NP-40 and calculated as follows:
| [(experimental value − blank value)/(total lysis − blank value) × 100] | (1) |
2.3. Analysis of Cytokine Production
Supernatants from DCs cultures were analyzed for IL-12 and IL-10 protein levels using an in-house sandwich ELISA. One hundred microliters of capture antibody solution (BioLegend, San Diego, CA, USA) were plated into each well of a 96-well plate (NuncMaxisorb, eBioscience Inc., San Diego, CA, USA) and incubated overnight at 4 °C. After antibody removal, 100 μL of PBS supplemented with 1% BSA (blocking buffer) were added to each well and incubated for 2 h at room temperature (RT). Wells were then washed with 0.05% Tween-20 in PBS. Next, cytokine standards (BioLegend, San Diego, CA, USA) or diluted samples were added to the wells and incubated for 2 h at RT. After the washing steps, 100 μL aliquots of biotinylated antibody solution were added and incubated for 2 h at RT. Streptavidin–horseradish peroxidase conjugate solution (1:2000 dilution; BioLegend) was then added to the wells and incubated for 1 h at RT. Finally, after washing, reaction was developed by adding 100 μL of 63 mM Na2HPO4, 29 mM citric acid (pH 6.0) containing 0.66 mg/mL o-phenylenediamine/HCl and 0.05% hydrogen peroxide. The cytokine concentration was calculated by reading absorbance at 415 nm. Values were expressed as pg/mL.
2.4. Fluorescence-activated Cell Sorting (FACS) Analysis
DCs were stained with phycoerythrin (PE)- or fluorescein isothiocyanate (FITC)-conjugated Abs (BioLegend) toward mouse CD80 and CD86 surface markers. Cell staining was analyzed using a CyFlow Space flow cytometer (Partec, Munster, Germany) and FlowJo software (Tree Star Inc., Ashland, OR, USA). For each Ab, an isotype control of the appropriate subclass was used.
2.5. Preparation of Samples for Proteomic Analyses
Secreted proteins were extracted from supernatants of iDCs, mDCs, and LGmDCs samples by using a trichloroacetic acid (TCA) precipitation protocol. Four biological replicates were prepared for each sample. TCA and sodium deoxycholate were added to supernatants to 7.5% and 0.1% w/v final concentrations, respectively, and precipitation was carried out for 2 h at 4 °C. Protein pellets, collected by centrifugation (10,000 × g for 10 min at 4 °C), were washed twice with 0.5 mL of ice-cold acetone, recovered by centrifugation (10,000× g for 10 min at 4 °C) and dried under nitrogen. Pellets were solubilized in 20 mM Tris-HCL, 65 mM DTT, 8 M urea, 2% CHAPS, pH 8.5, and protein concentration was measured by a Bradford assay (BioRad, Hercules, CA, USA). One hundred and twenty microgram aliquots of each sample were loaded on 12% Mini-PROTEAN® TGX™ Precast Protein Gels (BioRad). Gels were run at 120 V using the Mini-PROTEAN Tetra Cell (BioRad). Protein profiles were visualized by staining with Coomassie Brilliant Blue R-250. Each lane was cut in 10 slices that were destained with 50% acetonitrile in 50 mM ammonium bicarbonate. Proteins contained in each slice were submitted to in-gel reduction with 10 mM dithiotreitol in 50 mM ammonium bicarbonate pH 8.0 for 1 h at 56 °C, alkylation with 50 mM iodacetamide in the same buffer for 30 min at RT in the dark and digestion with 0.2 µg modified trypsin (Promega, Madison, WI, USA) in 25 mM ammonium bicarbonate pH 8.4 overnight at 37 °C. Peptide extraction was carried out with 5% formic acid and 50% acetonitrile. Peptides mixtures were dried in a Speed-Vac centrifuge (Savant) and solubilized in 30 µL 0.1% formic acid.
2.6. LC-MS/MS Analysis, Protein Identification, and Functional Classification
Tryptic peptide mixtures were analyzed using a Q-ExactiveTM mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) interfaced with an UltiMate 3000 RSLCnano LC system (Thermo Fisher Scientific). Peptide mixtures were concentrated and desalted on a trapping pre-column (Acclaim PepMap C18, 300 μm × 5 mm nanoViper, 5 μm, 100 Å, Thermo Fisher Scientific), using 0.05% formic acid and 2% acetonitrile at a flow rate of 10 μL/min. The peptide separation was performed at 35 °C using a C18 column (Acclaim Easy Spray PepMap RSLC C18, 75 μm × 15 cm nanoViper, 3 μm, 100 Å, Thermo Fisher Scientific), using as eluent A 0.1% formic acid and as eluent B 80% acetonitrile in 0.08% formic acid at a flow rate of 0.3 μL/min and a linear gradient from 4% to 40% B over 45 min, hold for 10 min, from 40% to 90% B over 1 min, hold for 10 min before column re-equilibration to 4% B.
Mass spectra were acquired in the m/z range 350–1600. Data acquisition was performed in a data-dependent mode Full MS/ddMS2, enabling the acquisition of MS/MS spectra for the ten most intense precursor ions (top ten) and dynamic exclusion of 10 s. The resolution was set to 70,000 for MS spectra acquisition and 17,500 for MS/MS spectra acquisition.
MS data processing for protein identification was performed using ProteomeDiscoverer™ platform (version 2.1.0.81; Thermo Fisher Scientific), interfaced with the Sequest HT Search Engine server (Washburn et al., 2001). The parameters used for the database searches were as follows: Mus musculus protein database (taxon 10,090, downloaded from UniProtKB on June 2018, https://www.uniprot.org/) and a contaminant protein database (PD_Contaminants_2015_5.fasta, provided by the manufacturer), trypsin as proteolytic enzyme, up to two missed cleavages, carbamidomethyl as fixed modification for cysteine residues, oxidation of methionine residues, and formation of pyro-glutamic acid of N-terminal glutamine residues as dynamic modifications, 20 ppm mass tolerance for precursor ions and 0.02 Da mass tolerance for MS/MS fragments. Protein identification based on at least two peptides (FDR < 0.05) was considered reliable. Proteins identified in three out of four biological replicates were considered reproducibly present in one sample.
Quantitative proteomic analysis was carried out using the spectral counting approach to compare protein abundance levels in the secretomes of mDCs vs. iDCs, and LGmDCs vs. mDCs. To calculate each protein normalized spectral abundance factor (NSAF), the number of spectral counts (SpCs; i.e., peptide spectra matches, PSM) was divided by the protein length (thus obtaining the SAF value) and normalized to the sum of SAFs in each sample [17].
t-test values were calculated from Log2(NSAF) values of the four biological replicates of the compared samples (mDCs vs. iDCs and LGmDCs vs. mDCs). A p-value ≤ 0.05 indicated statistically significant differences in protein abundance. Changes in protein abundance (fold change, FC) were calculated as the ratio between the average NSAF of each protein in the two samples. Proteins showing fold changes ≥ ±2 were assumed to be present in different amounts in one of the compared samples. However, fold changes ≥ ±1.5 and p-values ≤ 0.1 in these proteins were assumed to give biologically relevant information (Table S1 (quantitation analysis) and Table S2 in Supplementary Materials).
Identified proteins were analyzed using the SignalP 5.0 server (http://www.cbs.dtu.dk/services/SignalP) that predicts the presence and location of signal peptide cleavage sites in amino acid sequences for translocation across cell membranes [20]. Cellular localization of proteins carrying a signal peptide was postulated based on sequence information using DeepLoc 1.0 server (http://www.cbs.dtu.dk/services/DeepLoc-1.0/index.php) [21] and on Uniprot annotation for subcellular localization reported in the UniProtKb database (https://www.uniprot.org). The prediction of non-classically secreted proteins (such as moonlighting proteins) was achieved by the SecretomeP 2.0 server (http://www.cbs.dtu.dk/services/SecretomeP) [22].
Identified proteins were functionally classified using the Reactome Pathway Database (https://reactome.org) that statistically determines whether certain pathways are enriched in the data. This analysis produces a probability score, which is corrected for false discovery rate (FDR) using the Benjamani–Hochberg method [23].
Protein–protein interactions were explored using the web resource STRING v.9.1 (Search Tool for the Retrieval of Interacting Genes/Proteins, http://string-db.org/). Active interaction sources used in our analysis were a neighborhood, co-expression, experiments, gene fusions, gene co-occurrence, databases, and text mining, using high confidence value (0.700) [24].
Gene names for some of the proteins included in functional analyses were deduced using BLASTP (https://blast.ncbi.nlm.nih.gov) (Table S1 (Blast Results) in Supplementary Materials).
2.7. Statistical Analysis
Differences among the various treatment groups were determined by the ANOVA test followed by Tukey’s multiple comparisons test. A cut-off of p < 0.05 was selected to denote a significant difference.
3. Results and Discussion
3.1. Phenotypic Characterization and Secretome Analysis of Dendritic Cells
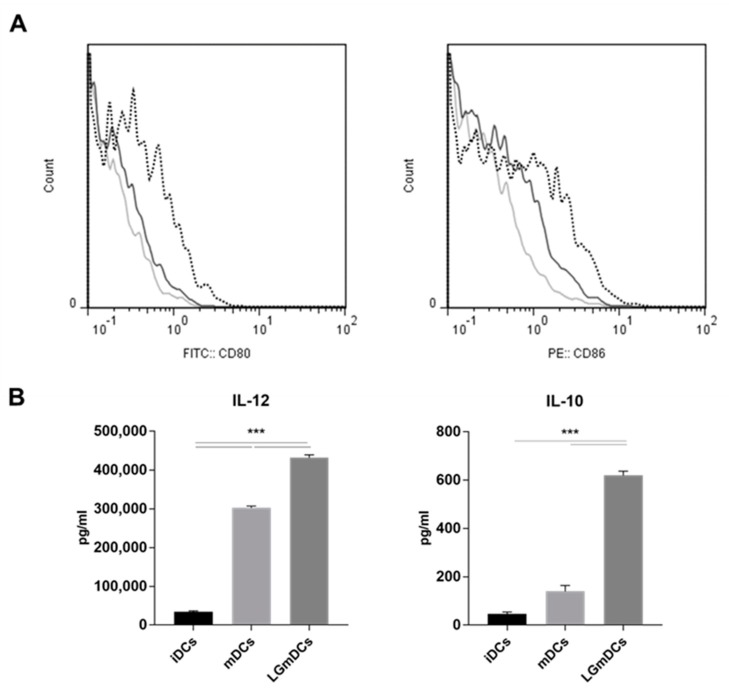
A quantitative proteomic study was designed aimed at investigating both the maturation process in DCs and the effects of a L. gasseri challenge on this process. The study focused on secreted proteins, as these proteins (cytokines, chemokines, proteases, protease inhibitors, growth factors, etc.) play a major role in both innate and adaptive responses driven by DCs [15]. To obtain a more inclusive picture of the secretome, unspecific protein contribution from cell culture was drastically reduced by incubating iDCs in a serum-free medium. Specifically, in vitro generated iDCs were pre-incubated for 24 h in X-Vivo 15 medium [25]. In this medium, we found ≥ 90% live cells. Cell integrity was assessed by measuring the percentage LDH release in the spent medium and this assay showed that DC maturation was not associated with any decrease in cell integrity. In fact, iDCs and mDCs showed comparable values %LDH release (11.1 ± 0.8 vs. 10.6 ± 0.6; mDCs vs. iDCs; mean ± SD; P = 0.71) and LGmDCs showed a non-significant increase (14.6 + 2.7; P = 0.08). The increased expression of both CD80 and CD86 surface markers testified the effective maturation of DCs (Figure 1A). We also estimated the induced phenotype by evaluating the production of interleukin (IL)-12 and IL-10, a pro- and an anti-inflammatory cytokine, respectively. As reported in Figure 1B, IL-12 significantly increased in mDCs, whereas changes in IL-10 levels were not relevant. Therefore, induced mDCs had a pro-inflammatory signature. Interestingly, pre-incubation with L. gasseri further induced the expression of surface markers (Figure 1A) as well as increased the levels of IL-12 and IL-10 (Figure 1B); the latter findings confirmed the ability of this probiotic strain to modulate the phenotype of mDCs.
Figure 1.
Phenotype of dendritic cells (DCs) cultured in X-Vivo 15 medium and challenged with L. gasseri. Immature dendritic cells (iDCs) were challenged or not with irradiated L. gasseri and then subjected to a 6-h lipopolysaccharide (LPS) pulse to induce DC maturation. (A) Fluorescence-activated cell sorting (FACS) analysis of DCs double-stained for CD80 and CD86; light grey line, iDCs; dark grey line, mDCs; dotted line, L. gasseri OLL2809 challenged iDCs (LGmDCs). (B) Cytokine analysis (Interleukin-10 (IL-10) and Interleukin-12 (IL-12))of iDCs, mature dendritic cells (mDCs), and LGmDCs. Data were collected from ungated cells and are representative of three independent experiments. *** p < 0.01 (ANOVA test).
Moreover, the modulation of secretome profiles was investigated by proteomics analyzing the secretomes of iDCs, mDCs, and LGmDCs. This study led to the identification of 156 proteins included in the dataset that were reproducibly present in three biological replicates (out of four replicates analyzed in the study) of at least one sample (Table S1 (All Proteins) in Supplementary Materials).
Bioinformatics processing of the proteins included in the dataset using the SignalP tool highlighted that 33 proteins were predicted to contain a signal peptide for translocation across cell membranes. DeepLoc analysis predicted extracellular localization for 21 of these proteins, and for the other five proteins (Ctss, Il1rn, Ctsb, Pla2g7, Lgals1), the extracellular localization was deduced from Uniprot annotation. An additional 21 proteins were predicted to be non-classically secreted proteins by SecretomeP analysis. Therefore, 30% of the identified proteins were extracellularly located based on bioinformatics processing. However, it cannot be ruled out that other proteins identified in this study could be extracellularly located. For example, it has been reported that cathepsins, generally located in the lysosomes and endosomes, could be secreted into the extracellular milieu as proenzymes and have also been found in cell-cultured medium and in the circulation of both experimental animals and human beings under physiological conditions [26,27]. Similarly, peptidyl-prolyl cis-trans isomerase has been observed in the conditioned medium of lipopolysaccharide-stimulated murine macrophages [28,29]. In addition, due to partial cell lysis occurring during cell growth or sample preparation, minor cytoplasmic protein contamination should also be taken into consideration.
The functional classification of these proteins, performed using the Reactome Pathway Database, highlighted that the immune system, signal transduction, cellular responses to external stimuli, and vesicle-mediated transport were among the most enriched Reactome pathways. These data are in tight agreement with the physiology of DCs and their modification following stimuli (Table S3 in Supplementary Materials).
3.2. Secretome Analysis of the Immature DCs
A first overall picture of the secretome of iDCs was gained through proteomics leading to the identification of 61 proteins, and 21 of these (34%) were predicted to be extracellularly localized (Table S1 (iDCs) in Supplementary Materials). It is worth noting, all the proteins identified in the iDCs secretome were also present in the secretome of mDCs (Table S1 (mDCs) in Supplementary Materials). Similarly to data reported by Gundacker and colleagues [15], iDCs secreted several proteases and protease inhibitors (Cystatin-C (Cst3), Cathepsin B (Ctsb), Cathepsin D (Ctsd), Cathepsin L (Ctsl), Cathepsin S (Ctss), Cathepsin Z (Ctsz)) involved in antigen processing and Galectin 3 (Lgals3) involved in inflammatory responses including neutrophil activation and adhesion, as well as proteins involved in redox regulation (Prdx1 and Prdx5, Sod1) and in protein folding (Hsp90ab1, Hspa8, Ppia). In addition, actin, MMP12 (probably involved in cell trafficking), and peroxiredoxin 1 were already detected in the secretome of iDCs by Villiers and colleagues [16].
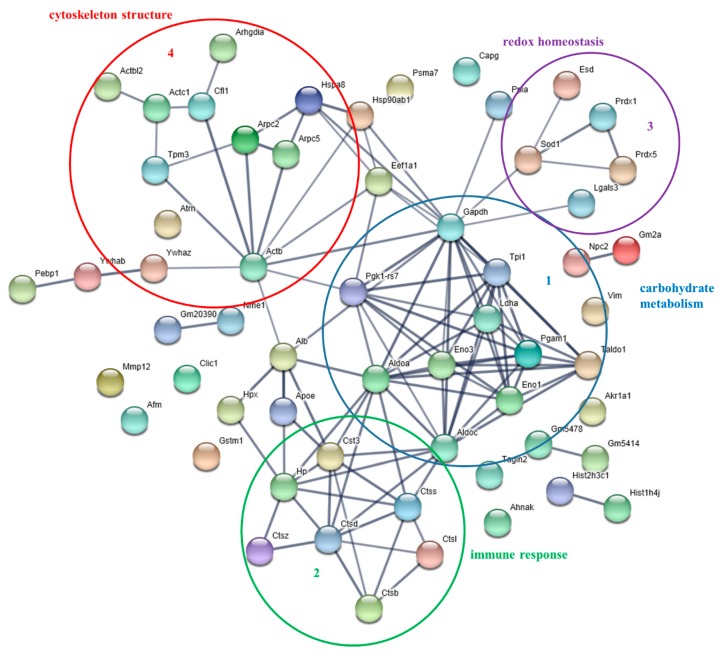
Protein–protein interactions were analyzed, processing data reported in Table S1 (iDCs), by means of the web resource STRING (Figure 2). Notably, STRING analysis highlighted highly interconnected clusters of proteins. The main clusters included proteins involved in carbohydrate catabolic processes (Cluster 1: Eno1, Eno2, Ldha, Aldoa, Aldoc, etc.), in immune response (Cluster 2: several cathepsins acting in protein degradation and turnover), in redox homeostasis (Cluster 3: Prdx1 and Prdx5, Sod1 and Esd) and finally, in cytoskeleton structure (Cluster 4: mainly joining actins and tropomyosin). More interestingly, coherent with the immunological role of iDCs, the Reactome Pathway analysis showed that the most enriched pathways of this dataset were “Innate Immune System” (MMU-168249, 23 proteins, fold enrichment + 8.37, FDR 6.6E-13), and in particular “Neutrophil degranulation” (including Cst3, Gm2a, Aldoa, Aldoc, Arpc5, Gm20390, Hp, Hsp90ab1, Npc2, Pgam1, Lgals3), and “Adaptive Innate Immune System” (MMU-1280218, 4 proteins, fold enrichment + 12.6, FDR 2.4E-2) and in particular “MHC class II antigen presentation”, including proteins of the cathepsin family (Ctss, Ctsd, Ctsb, Ctsl). (Table S3 (iDCs)).
Figure 2.
Protein–protein interaction network of proteins identified in the secretome of immature DCs. The Gene Name according to UniProtKB are reported. The network was obtained using the EMBL STRING with a confidence cut-off of 0.700 [24]. The four main functional modules are circled and numbered.
3.3. Modulation of DCs Secretome Induced by the Maturation Process
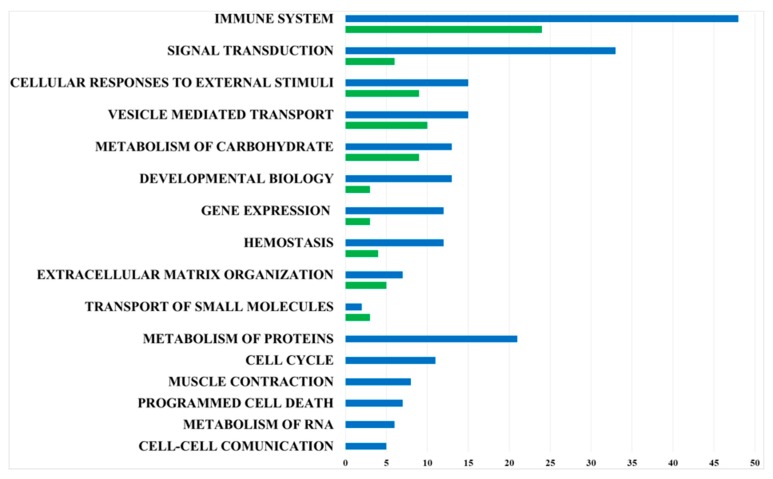
The proteomic analysis of mDCs secretome led to the identification of 130 proteins present in at least three biological replicates. Twenty proteins were predicted to be extracellularly located, and 18 proteins were secreted through non-classical pathways (29% extracellular proteins) (Table S1 (mDCs) in Supplementary Materials). The functional classification confirmed that the maturation process further enriched the secretome profile with proteins involved in immunity. In fact, 46 proteins were classified as belonging to “Innate Immune System” (MMU-168249, fold enrichment + 7.71, FDR 6.55E-25), including C3, Lcn2, Rock1, that plays a functional role in the dynamic reorganization of cytoskeletal proteins, while, for instance, only 23 proteins of the iDCs secretome were involved in “Innate Immune System”. Similarly, the maturation process led to the expression of a larger number of proteins involved in Signal Transduction (Figure 3, Table S3 (mDCs) in Supplementary Materials).
Figure 3.
Bar diagram summarizing the functional classification of proteins present in the mDCs secretome (blue bars) and in the secretome of iDCs (green bars) achieved by Reactome Pathway Database analysis.
A quantitative proteomic analysis led to the identification of 37 proteins exhibiting a statistically significant variation in abundance when comparing the secretome of mDCs vs. iDCs. The maturation process triggered the abundance of 32 proteins and reduced the amount of five proteins in mDCs (Table 1; Table S2 in Supplementary Materials)).
Table 1.
Results of the differential proteomic study: proteins whose abundance was affected by the maturation process of DCs challenged or not with L. gasseri and functional classification obtained by the Reactome Pathway Database.
| ACCESSION UNIPROTKB |
DESCRIPTION | GENE NAME | FUNCTIONAL CLASSIFICATION | FOLD CHANGE | |
|---|---|---|---|---|---|
| mDCs/iDCs | LGmDCs/mDCs | ||||
| IMMUNE SYSTEM | |||||
| Q542G9 | Annexin 2 | Anxa2 | Innate Immune System Neutrophil degranulation |
32.54 | |
| P01027 | Complement C3 | C3 | Innate Immune System Neutrophil degranulation |
12.29 | −16.16 |
| Q3U6A3 | Monocyte differentiation antigen CD14 | Cd14 | Innate Immune System Neutrophil degranulation |
8.91 | |
| Q3UD32 | Uncharacterized protein | Ctss | Innate Immune System Neutrophil degranulation |
−2.42 | |
| Q9ES94 | Cathepsin Z | Ctsz | Innate Immune System Neutrophil degranulation |
1.97 | 1.53a |
| E9Q557 | Desmoplakin | Dsp | Innate Immune System Neutrophil degranulation |
14.13 | −5.86 |
| Q3UBK2 | Uncharacterized protein | Hmgb1 | Innate Immune System Neutrophil degranulation |
−3.80 | 3.85 |
| Q02257 | Junction plakoglobin | Jup | Innate Immune System Neutrophil degranulation |
8.07 | −5.13 |
| P08905 | Lysozyme C-2 | Lyz2 | Innate Immune System Neutrophil degranulation |
−2.97 | |
| Q3TFQ8 | Alpha-1,4 glucan phosphorylase | Pygb | Innate Immune System Neutrophil degranulation |
25.10 | −4.13 |
| Q4FJQ0 | MCG130610 | Rab7 | Innate Immune System Neutrophil degranulation |
3.80 | −3.52 |
| Q60842 | Chromosome 24p3 (Fragment) | Lcn2 | Innate Immune System Neutrophil degranulation |
18.34 | −24.10 |
| Q5FWA0 | Ribonuclease T2B | Rnaset2b | Innate Immune System Neutrophil degranulation |
17.80 | −6.96 |
| B8JJN0 | Uncharacterized protein | Cfb | Innate Immune System | 10.86 | |
| P43432 | Interleukin-12 subunit beta | Il12b | Cytokine Signaling in Immune System | 19.36 | 1.89a |
| P25085 | Interleukin-1 receptor antagonist protein | Il1rn | Cytokine Signaling in Immune System | 3.65 | |
| P29391 | Ferritin light chain 1 | Ftl1 | IMMUNE SYSTEM | 3.63 | −2.28 |
| P01921 | H-2 class II histocompatibility antigen, A-D beta chain | H2-Ab1 | IMMUNE SYSTEM | 22.45 | |
| P06343 | H-2 class II histocompatibility antigen, A-K beta chain | H2-Ab1 | IMMUNE SYSTEM | 8.33 | |
| SIGNAL TRANSDUCTION | |||||
| Q64426 | Histone H2A (Fragment) | Hist2h2aa1 | Signaling by Nuclear Receptors | 12.83 | |
| Q6P8R3 | C-X-C motif chemokine | Pf4 | Signaling by GPCR | 5.72 | −1.98a |
| Q3US43 | Annexin 1 | Anxa1 | Signaling by GPCR | 61.76 | −22.14 |
| P60766 | Cell division control protein 42 homolog | Cdc42 | Signaling by Rho GTPases | -2.88 | |
| Q3THE2 | Myosin regulatory light chain 12B | Myl12b | Signaling by Rho GTPases /MUSCLE CONTRACTION Smooth Muscle Contraction | 3.80 | |
| MUSCLE CONTRACTION | |||||
| Q91VH3 | Tpm2 protein | Tmp2 | Striated Muscle Contraction Smooth Muscle Contraction |
4.71 | −2.20 |
| F8WID5 | Tropomyosin alpha-1 chain | Tpm1 | Striated Muscle Contraction Smooth Muscle Contraction |
3.55 | |
| B2RQQ1 | MCG133649, isoform CRA_a | Myh6 | Striated Muscle Contraction | 13.33 | −9.83 |
| B2RWW8 | Myosin, heavy polypeptide 8 | Myh8 | Striated Muscle Contraction | 35.12 | −25.91 |
| B2RWX0 | Myosin, heavy polypeptide 1 | Myh1 | Striated Muscle Contraction | 37.49 | −27.66 |
| B1AR69 | Myosin, heavy polypeptide 13 | Myh13 | Striated Muscle Contraction | 11.44 | −8.44 |
| Q5SX39 | Myosin-4 | Myh4 | Striated Muscle Contraction | 37.62 | −27.75 |
| HEMOSTASIS | |||||
| Q8BTM8 | Filamin-A | Flna | Platelet activation, signaling, and aggregation | 14.59 | −18.85 |
| P11276 | Fibronectin | Fn1 | Platelet activation, signaling, and aggregation | −74.75 | |
| Q9WVA4 | Transgelin-2 | Tagln2 | Platelet activation, signaling, and aggregation | 2.30 | |
| EXTRACELLULAR MATRIX ORGANIZATION | |||||
| P34960 | Macrophage metalloelastase | Mmp12 | Degradation of extracellular matrix | −1.80a | 4.47 |
| P24369 | Peptidyl-prolyl cis-trans isomerase B | Ppib | Collagen Formation | 3.44 | |
| METABOLISM | |||||
| H3BKH6 | S-formylglutathione hydrolase | Esd | Biological Oxidations | 2.08 | |
| P08226 | Apolipoprotein E | Apoe | Metabolism of vitamins and cofactors | −3.64 | |
| Q9WUB3 | Glycogen phosphorylase | Pygm | Metabolism of carbohydrate | 51.35 | −10.93 |
| P63242 | Eukaryotic translation initiation factor 5A-1 | Eif5a | Metabolism of proteins Post-translational protein modification |
13.51 | |
| A2AB79 | Histone H2A | Hist3h2a | Metabolism of proteins Post-translational protein modification |
11.72 | |
| UNCLASSIFIED | |||||
| Q3U1N0 | Coronin | Coro1a | 2.52 | ||
| Q20BD0 | Heterogeneous nuclear ribonucleoprotein A/B, isoform CRA_a | Hnrnpa | 7.24 | ||
| P04918 | Serum amyloid A-3 protein | Saa3 | 14.27 | ||
| Q99LB4 | Capping protein (Actin filament), gelsolin-like, isoform CRA_a | Capg | −2.33 | 1.84a | |
| B9EIU3 | Attractin | Atrn | −2.10 | 2.61 | |
a proteins with fold change values in the range ± 1.5–2.0.
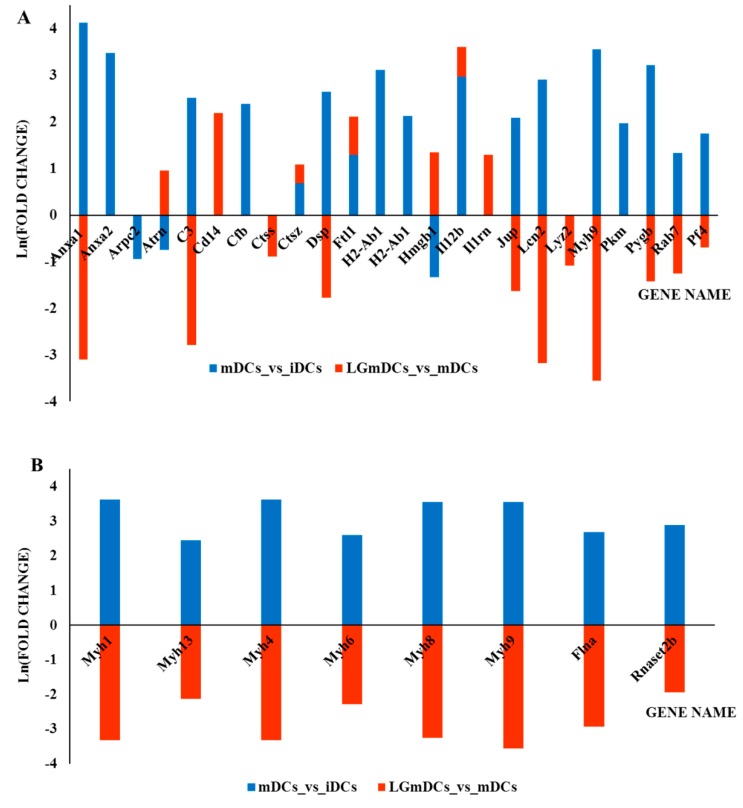
Functional classification of these proteins highlighted that proteins related to “Innate Immune System” (MMU-168249, 13 proteins, fold enrichment + 7.75 FDR 7.1E-06), such as Anxa2, Arpc2, C3, Ctsz, Dsp, Jup, Myh9, Pkm, Pygb, Rab7, were significantly more abundant in mDCs. In addition, the “Signaling by RHO GTPase” pathway was significantly enriched (MMU-194315, 5 proteins, fold enrichment + 12.24 FDR 1.5E-02) (Figure 4A, Table 1, Table S3 (mDCs vs. iDCs) in Supplementary Materials).
Figure 4.
Bar diagram summarizing the most relevant results of the quantitative proteomic analyses and reporting the natural logarithm (Ln) of fold change values obtained comparing mDCs vs. iDCs and LGmDCs vs. mDCs. Fold change values are reported in Table 1. Positive values of Ln (fold change) indicate an increase in protein amount, while negative values of Ln (fold change) indicate a decrease in protein amount. (A) Proteins mainly involved in the immune response. (B) Proteins mainly involved in contractile machinery. These proteins showed a significant increase in fold changes when comparing mDCs vs. iDCs (blue bars) and a dramatic decrease when comparing LGmDCs vs. mDCs (red bars).
Proteins involved in intracellular transport, cell–cell communication, and cytoskeletal proteins, in particular myosin isoforms, tropomyosin and filamin, were highly abundant in mDCs compared to iDCs, thus indicating that DCs are preparing for migration, which requires profound alterations in cell morphology and motility (Figure 4B).
IL-12 subunit beta was present in the mDCs secretome and absent in the iDCs one, while IL-10 was not detected in this analysis, probably due to the instrumental detection limit. These results are in tight agreement with the ELISA data (Figure 1B). Interestingly, we detected an increased abundance of canonical extracellular proteins with general anti-microbial activity, such as Complement 3 (C3), Complement factor b (Cfb), and Lipocalin-2 (Chromosome 24p3, Lcn2). We also detected an increased presence of proteins with more specific roles, such as Platelet aggregating factor 4 (Pf4) and Serum amyloid A3 protein (Saa3). The latter one stimulates neutrophils to secrete IL-22 for protecting the integrity of the epithelial barrier [30]. The pleiotropic role of mDCs in the immunological synapse was confirmed by the massive presence of sMHC II (H2-Ab1), Annexins, and extracellular released Histone 2, known to engage the T cell receptor or toll-like receptors. Through these molecules, both the innate and adaptive arms of the immune response could be activated (Table 1). Furthermore, the increased abundance of contractile machinery proteins (myosin polypeptide, filamin-A, tropomyiosin, Tpm2) and desmosome proteins (desmoplakin, junction plakoglobin) could testify an increased migration/cell interaction activity of mDCs. Unexpectedly, we also registered an increased abundance of eukaryotic initiation factor 5A (eIF-5A). In DCs, eIF-5A is mainly involved in the intracellular transport pathway of transcript encoding for CD83, a maturation marker [31]. Noteworthy, in cardiac myocytes, eIF5A undergoes tyrosine sulfation in the trans-Golgi and is rapidly secreted in response to hypoxia, inducing apoptosis via an autocrine mechanism [32]. Therefore, our findings suggested that secreted eIF5A could work as a pro-apoptotic ligand in different cell types. Intriguingly, the analysis also revealed the presence of enzymes generally found intracellularly in the secretome of mDCs (Pkm, Ppib, Pyg). Several non-canonical functions have already been attributed to such enzymes. In particular, neutrophils at tissue damaged sites could release pyruvate kinase m1/2 to promote angiogenesis and wound healing [33]. Furthermore, extracellular cyclophilins (peptidylprolyl isomerase) have been shown to modulate the levels of TNFα [29]. The increased presence of RNase T2 could also be expected, as it degrades RNAs in the extracellular space, thus playing a role in innate host defense. On the contrary, the secretion of glycogen phosphorylases (Pyg), classical intracellular enzymes, remains to be clarified.
3.4. Modulation of the Secretome of mDCs by L. gasseri Challenge
To further elucidate the modulatory effects induced by this probiotic strain on the maturation process, the secretomes of LGmDCs and mDCs were compared. The microbial treatment affected the abundance level of 37 proteins, increasing the level of 13 proteins and decreasing that of 24 proteins. These proteins were functionally classified in “Immune System” (fold enrichment + 4.77, FDR 2.3E-04, 14 proteins), in “Platelet activation, signaling, and aggregation” (MMU-76002, fold enrichment + 17.22, FDR 3.0E-02, four proteins: Pf4, Flna, Fn1, Tagln2) and “Extracellular matrix organization” (MMU-1474244, fold enrichment + 9.50, FDR 4.2E-02, five proteins: Ctss, Ppib, Mmp12, Fn1, Dsp) (Figure 4, Table S3 (LGmDCs vs. mDCs) in Supplementary Materials).
Most importantly, L. gasseri pre-treatment was associated with a dramatic decrease of 19 proteins that mainly featured the mDC secretome, thanks to their relatively high abundance (as from LGmDCs vs. mDCs and mDCs vs. iDCs comparisons). In particular, we found that not only proteins closely related to the immune function (Annexins, C3, Lcn2) but also contractile machinery proteins (Myosins, Filamin-A) and desmosome proteins were drastically reduced (Figure 4). The abundance of Il12b and Ctsz was instead increased in both the comparisons (Table 1, Figure 4).
In addition, seven proteins (Tagln2, Ppib, Coro1a, Il1rn, Esd, Hnrnpa, CD14) were specifically involved in the maturation process mediated by L. gasseri (Table1), increasing their amount. In particular, the increased abundance of Interleukin-1 receptor antagonist protein, an early inhibitory cytokine that suppresses pro-inflammatory cytokines and Monocyte differentiation antigen CD14, a co-receptor for the detection of LPS and other pathogen-associated molecular patterns (i.e., lipoteichoic acid), could suggest a possible protective role of LG treatment against inflammation. Recently, Transgelin-2 (Tagln2) has been recognized as an important immune regulator in the context of immune cell-cell interaction [34] and is suggested to be also involved in the stability of the immunological synapse in B-cells [35]. Coronin (Coro1a, a component of the cytoskeleton of highly motile cells) plays a key role in modulating the activity of lymphocytes, neutrophils, and macrophages [36]. Finally, the preventive bacterial challenge caused a strong reduction of fibronectin, classically involved in different cell adhesion and migration processes (Table 1).
4. Conclusions
The present proteomic study established that the maturation process of DCs was associated with profound changes in the protein secretome. These changes went beyond the increased expression of classical immune mediators (i.e., cytokines, complement factors, TcR ligands), but involved some unexpected new mediators (i.e., eIF5A), as well as the contractile and desmosome machineries. The latter findings will help to explain the role of DCs in the current cell migration model, where integrin adhesions are built upon initial cell contact with the matrix, followed by the generation of traction forces on the adhesions through local actomyosin-based contractile units. L. gasseri pre-treatment was associated with a dramatic modulation of several proteins secreted by mDC, not only belonging to the immune synapse. In fact, L. gasseri appeared to reduce junction formation and the secretion of actomyosin-based contractile units. It is known that L. gasseri drives a physiological activity for regulating cell adherence and integrating/activating or controlling signals in dendritic and T/B cell networks. Therefore, these findings suggest a novel mechanism by which L. gasseri could modulate the maturation process of DCs, thus confirming that this probiotic strain exerts a protective anti-inflammatory role. Further studies are needed to investigate whether these findings reflect the general properties of probiotic strains.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6643/12/2/555/s1, Table S1: Proteomic data gathered in this study. List of the proteins identified in this study. The accession number (UniProtKB), the gene name and cell localization are reported together with the number of matched peptides and peptide mass spectra matches (PSM) for each protein in each analyzed sample (All proteins sheet); List of the proteins identified in the secretome of iDCs (iDCs sheet); List of the proteins identified in the secretome of mDCs (mDCs sheet); Quantitative label-free analysis performed on the proteomic data applying the spectral counting approach (Quantitation_analysis sheet); Gene names for some of the proteins included in functional analyses as deduced using BLASTP (https://blast.ncbi.nlm.nih.gov) (Blast results sheet); Table S2: Results of the differential proteomic study: proteins whose abundance was affected by the maturation process of DCs challenged or not with L. gasseri; Table S3: Functional analysis of the proteins identified in the proteomic study. Reactome Pathway analysis of proteins identified in the secretome of iDCs (iDCs sheet); Reactome Pathway analysis of proteins identified in the secretome of mDCs (mDCs sheet); Reactome Pathway analysis of proteins involved in the maturation process (mDCs vs. iDCs sheet); Reactome Pathway analysis of proteins involved in the maturation process modulated by L. gasseri (LGmDCs vs. mDCs sheet).
Author Contributions
Conceptualization, M.F.M., D.L., T.S., M.R., and R.A.S.; Funding acquisition, M.R. and R.A.S.; Investigation, M.F.M., D.L., T.S., M.R., and R.A.S.; Methodology, M.F.M., D.L., M.R., and R.A.S.; Software, M.F.M.; Writing—original draft, M.F.M., M.R., and R.A.S.; Writing—review & editing, M.F.M., D.L., T.S., M.R., and R.A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the CNR grant under the Agreement of Scientific Cooperation CNR-JSPS 2010-11 and by CNR-ISA 2018 grant.
Conflicts of Interest
The authors state no conflict of interest.
References
- 1.Bron P.A., van Baarlen P., Kleerebezem M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat. Rev. Microbiol. 2011;10:66–78. doi: 10.1038/nrmicro2690. [DOI] [PubMed] [Google Scholar]
- 2.Selle K., Klaenhammer T.R. Genomic and phenotypic evidence for probiotic influences of Lactobacillus gasseri on human health. FEMS Microbiol. Rev. 2013;37:915–935. doi: 10.1111/1574-6976.12021. [DOI] [PubMed] [Google Scholar]
- 3.Aoki-Yoshida A., Yamada K., Hachimura S., Sashihara T., Ikegami S., Shimizu M., Totsuka M. Enhancement of oral tolerance induction in DO11.10 mice by Lactobacillus gasseri OLL2809 via increase of effector regulatory T cells. PLoS ONE. 2016;11:e0158643. doi: 10.1371/journal.pone.0158643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itoh H., Sashihara T., Hosono A., Kaminogawa S., Uchida M. Lactobacillus gasseri OLL2809 inhibits development of ectopic endometrial cell in peritoneal cavity via activation of NK cells in a murine endometriosis model. Cytotechnology. 2011;63:205–210. doi: 10.1007/s10616-011-9343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sashihara T., Sueki N., Ikegami S. An analysis of the effectiveness of heat-killed lactic acid bacteria in alleviating allergic diseases. J. Dairy Sci. 2006;89:2846–2855. doi: 10.3168/jds.S0022-0302(06)72557-7. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida A., Yamada K., Yamazaki Y., Sashihara T., Ikegami S., Shimizu M., Totsuka M. Lactobacillus gasseri OLL2809identifying and its RNA suppress proliferation of CD4(+) T cells through a MyD88-dependent signalling pathway. Immunology. 2011;133:442–451. doi: 10.1111/j.1365-2567.2011.03455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luongo D., Miyamoto J., Bergamo P., Nazzaro F., Baruzzi F., Sashihara T., Tanabe S., Rossi M. Differential modulation of innate immunity in vitro by probiotic strains of Lactobacillus gasseri. BMC Microbiol. 2013;13:298. doi: 10.1186/1471-2180-13-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira G.B., Mathieu C., Overbergh L. Understanding dendritic cell biology and its role in immunological disorders through proteomic profiling. Proteom. Clin. Appl. 2010;4:190–203. doi: 10.1002/prca.200900162. [DOI] [PubMed] [Google Scholar]
- 9.Ferret-Bernard S., Castro-Borges W., Dowle A.A., Sanin D.E., Cook P.C., Turner J.D., MacDonald A.S., Thomas J.R., Mountford A.P. Plasma membrane proteomes of differentially matured dendritic cells identified by LC-MS/MS combined with iTRAQ labelling. J. Proteom. 2012;75:938–948. doi: 10.1016/j.jprot.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hussaarts L., Kaisar M.M.M., Guler A.T., Dalebout H., Everts B., Deelder A.M., Palmblad M., Yazdanbakhsh M. Human dendritic cells with Th2-polarizing capacity: Analysis using label-free quantitative proteomics. Int. Arch. Allergy Immunol. 2017;174:170–182. doi: 10.1159/000479512. [DOI] [PubMed] [Google Scholar]
- 11.Korkmaz A.G., Popov T., Peisl L., Codrea M.C., Nahnsen S., Steimle A., Velic A., Macek B., von Bergen M., Bernhardt J., et al. Proteome and phosphoproteome analysis of commensally induced dendritic cell maturation states. J. Proteom. 2018;180:11–24. doi: 10.1016/j.jprot.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Arya S., Wiatrek-Moumoulidis D., Synowsky S.A., Shirran S.L., Botting C.H., Powis S.J., Stewart A.J. Quantitative proteomic changes in LPS-activated monocyte-derived dendritic cells: A SWATH-MS study. Sci. Rep. 2019;9:4343. doi: 10.1038/s41598-019-40773-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pauwels A.M., Härtlova A., Peltier J., Driege Y., Baudelet G., Brodin P., Trost M., Beyaert R., Hoffmann E. Spatiotemporal changes of the phagosomal proteome in dendritic cells in response to LPS stimulation. Mol. Cell. Proteom. 2019;18:909–922. doi: 10.1074/mcp.RA119.001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chevallet M., Diemer H., Van Dorssealer A., Villiers C., Rabilloud T. Toward a better analysis of secreted proteins: The example of the myeloid cells secretome. Proteomics. 2007;7:1757–1770. doi: 10.1002/pmic.200601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gundacker N.C., Haudek V.J., Wimmer H., Slany A., Griss J., Bochkov V., Zielinski C., Wagner O., Stöckl J., Gerner C. Cytoplasmic proteome and secretome profiles of differently stimulated human dendritic cells. J. Proteome Res. 2009;8:2799–2811. doi: 10.1021/pr8011039. [DOI] [PubMed] [Google Scholar]
- 16.Villiers C., Chevallet M., Diemer H., Couderc R., Freitas H., Van Dorsselaer A., Marche P.N., Rabilloud T. From secretome analysis to immunology: Chitosan induces major alterations in the activation of dendritic cells via a TLR4-dependent mechanism. Mol. Cell. Proteom. 2009;8:1252–1264. doi: 10.1074/mcp.M800589-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zybailov B., Mosley A.L., Sardiu M.E., Coleman M.K., Florens L., Washburn M.P. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 18.Lutz M.B., Kukutsch N., Ogilvie A.L., Rossner S., Koch F., Romani N., Schuler G. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J. Immunol. Methods. 1999;13:77–92. doi: 10.1016/S0022-1759(98)00204-X. [DOI] [PubMed] [Google Scholar]
- 19.Luongo D., Severino L., Bergamo P., De Luna R., Lucisano A., Rossi M. Interactive effects of fumonisin B1 and alpha-zearalenol on proliferation and cytokine expression in Jurkat T cells. Toxicol. Vitro. 2006;20:1403–1410. doi: 10.1016/j.tiv.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Almagro Armenteros J.J., Tsirigos K.D., Sønderby C.K., Petersen T.N., Winther O., Brunak S., von Heijne G., Nielsen H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 21.Almagro Armenteros J.J., Sønderby C.K., Sønderby S.K., Nielsen H., Winther O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics. 2017;33:3387–3395. doi: 10.1093/bioinformatics/btx431. [DOI] [PubMed] [Google Scholar]
- 22.Bendtsen J.D., Kiemer L., Fausbøll A., Brunak S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005;5:58. doi: 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B., et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen L.J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sisto A., Luongo D., Treppiccione L., De Bellis P., Di Venere D., Lavermicocca P., Rossi M. Effect of Lactobacillus paracasei culture filtrates and artichoke polyphenols on cytokine production by dendritic cells. Nutrients. 2016;8:E635. doi: 10.3390/nu8100635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu H., Du Q., Dai Q., Ge J., Cheng X. Cysteine protease cathepsins in atherosclerotic cardiovascular diseases. J. Atheroscler. Thromb. 2018;25:111–123. doi: 10.5551/jat.RV17016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reiser J., Adair B., Reinheckel T. Specialized roles for cysteine cathepsins in health and disease. J. Clin. Investig. 2010;120:3421–3431. doi: 10.1172/JCI42918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherry B., Yarlett N., Strupp A., Cerami A. Identification of cyclophilin as a proinflammatory secretory product of lipopolysaccharide-activated macrophages. Proc. Natl. Acad. Sci. USA. 1992;89:3511–3515. doi: 10.1073/pnas.89.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ditiatkovski M., Neelisetti V.N., Cui H.L., Malesevic M., Fischer G., Bukrinsky M., Sviridov D. Inhibition of extracellular cyclophilins with cyclosporine analog and development of atherosclerosis in apolipoprotein E-deficient mice. J. Pharmacol. Exp. Ther. 2015;353:490–495. doi: 10.1124/jpet.115.223420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang G., Liu J., Wu L., Fan Y., Sun L., Qian F., Chen D., Ye R.D. Elevated expression of Serum Amyloid A 3 protects colon epithelium against acute injury through TLR2-dependent induction of neutrophil IL-22 expression in a mouse model of colitis. Front. Immunol. 2018;9:1503. doi: 10.3389/fimmu.2018.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lechmann M., Berchtold S., Hauber J., Steinkasserer A. CD83 on dendritic cells: More than just a marker for maturation. Trends Immunol. 2002;23:273–275. doi: 10.1016/S1471-4906(02)02214-7. [DOI] [PubMed] [Google Scholar]
- 32.Seko Y., Fujimura T., Yao T., Taka H., Mineki R., Okumura K., Murayama K. Secreted tyrosine sulfated-eIF5A mediates oxidative stress-induced apoptosis. Sci. Rep. 2015;5:13737. doi: 10.1038/srep13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y., Li L., Liu Y., Liu Z.R. PKM2 released by neutrophils at wound site facilitates early wound healing by promoting angiogenesis. Wound Repair Regen. 2016;24:328–336. doi: 10.1111/wrr.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jo S., Kim H.R., Mun Y., Jun C.D. Transgelin-2 in immunity: Its implication in cell therapy. J. Leukoc. Biol. 2018;104:903–910. doi: 10.1002/JLB.MR1117-470R. [DOI] [PubMed] [Google Scholar]
- 35.Na B.R., Kwon M.S., Chae M.W., Kim H.R., Kim C.H., Jun C.D., Park Z.Y. Transgelin-2 in B-Cells controls T-cell activation by stabilizing T cell-B cell conjugates. PLoS ONE. 2016;11:e0156429. doi: 10.1371/journal.pone.0156429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tokarz-Deptuła B., Malinowska M., Adamiak M., Deptuła W. Coronins and their role in immunological phenomena. Cent. Eur. J. Immunol. 2016;41:435–441. doi: 10.5114/ceji.2016.65143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.