Abstract
Background
lncRNA-SNHG16 was identified as an oncogene in many cancers, but its involvement in prostate carcinoma is unknown.
Material and Method
Expression of lncRNA-SNHG16 and glucose transporter 1 (GLUT-1) in 52 prostate carcinoma tissues and 36 normal prostate tissues was analyzed by RT-qPCR. Transfections were performed to analyze gene interactions. Cell proliferation was analyzed by cell proliferation assay.
Results
Overexpression of lncRNA-SNHG16 effectively distinguished prostate carcinoma patients from normal ones. Expression levels of lncRNA-SNHG16 and GLUT-1 mRNA were significantly and positively correlated across prostate carcinoma tissues. In vitro cancer cell experiments revealed that lncRNA-SNHG16 siRNA silencing downregulated the expressions of GLUT-1 and reduced glucose uptake. lncRNA-SNHG16 siRNA silencing also significantly inhibited prostate carcinoma cell proliferation. However, lncRNA-SNHG16 siRNA silencing did not affect the normal prostate.
Conclusion
In conclusion, lncRNA-SNHG16 might be a possible treatment target for prostate cancer.
Keywords: prostate carcinoma, SNHG16, glucose transporter 1
Introduction
How to inhibit tumor growth is recognized as a major task for cancer treatment nowadays.1 Prostate carcinoma is a frequently diagnosed cancer among male patients and is also one of the major causes of cancer-related deaths across the world.2 Previous studies during the last several decades have proposed models and revealed some areas of the growth of prostate cancer.3,4 Alterations in some molecular pathways, such as PTEN, TP53 and AR signaling pathway, have proved to be critical players in the occurrence, development and progression of prostate cancer.3,4 However, the molecular mechanism of the growth of prostate carcinoma still has not been fully elucidated. Therefore, in-depth investigations of the molecular mechanism of prostate carcinoma growth might help its treatments.
Glucose uptake and consumption are critical for the survival of normal cell and cancer cell.5 Accelerated glucose uptake distinguishes cancer cells from normal healthy cells.6 In effect, inhibition of glucose has been proven as a potential therapeutic target for cancer treatment.6 GLUT-1 has the duty of membrane translocation of glucose and altered expression of GLUT-1 is involved in the development of many diseases.7,8 GLUT-1 achieves its biological roles under certain pathological conditions through the interactions with lncRNAs.9,10 LncRNA-SNHG16 is a newly identified RNA with oncogenic roles in several kinds of cancers such as bladder cancer11 and glioma.12 In bladder cancer, lncRNA-SNHG16 is overexpression and can promote cancer cell proliferation by epigenetically silencing p21.11 In glioma, lncRNA-SNHG16 is also overexpressed and can sponge miR-4518 to upregulate PRMT5, thereby playing oncogenic roles.12 Our preliminary RNA-seq analysis revealed the positive correlation between GLUT-1 mRNA and lncRNA-SNHG16 across prostate carcinoma patients (data not shown), indicating the potential interaction between them. In our study, we found that lncRNA-SNHG16 may interact with GLUT-1 to promote the growth of prostate carcinoma cells.
Materials and Methods
Patient Enrollment and Specimen Collection
From January 2014 to January 2018, 120 patients with prostate carcinoma were diagnosed in the first affiliated hospital of Anhui University. Among those patients, 52 were included in this study. The inclusion criteria are 1) patients diagnosed as prostate carcinoma by fine-needle prostate biopsies; 2) patients diagnosed and treated for the first time; 3) patients agreed to participant. And exclusion criteria: 1) patient complicated with other malignancies; 2) patient with other prostate lesions; 3) patient aged >80; 4) patients who were suffering from chronic diseases; 5) patients who had been treated. The 52 patients had an age range of 41–78 and a mean age of 59.1±5.2. During the same time period, a total of other 102 patients with suspected prostate lesions that also received prostate biopsies and prostate lesions were excluded in 56 cases of them. Prostate biopsies of those 36 out of those 56 cases (to math distribution of patient group) were control group. Control group had an age ranging from 40 to 79 and a mean age of 58.6±5.8. Ethics board of the first affiliated hospital of Anhui University approved this study (No. FAHAU20131213573). All enrolled subjects signed informed consent. The study was carried out in accordance with the principles of the Declaration of Helsinki.
RT-qPCR
Trizol (Invitrogen, USA) was utilized to extract total RNA from prostate biopsies with all operations performed following the manufacturer’s instructions. All RNA samples were digested with DNase I to remove genomic DNA. After cDNA synthesis through reverse transcriptions using Bio-Rad iScript cDNA Kit (Bio-Rad), PCR reactions were carried out using SYBR® Green Real-Time PCR Master Mixes (Thermo Fisher Scientific, USA). Thermal conditions were 95°C for 55s, and 40 cycles of 95°C for 22s and 60°C for 46s. Primers were: 5′-CCCAAGCTTGCGTTCTTTTC-3′ (sense) and 5′-CCGGAATTCTGACGGTAGTTTC-3′ (anti-sense) for lncRNA-SNHG16; 5′-AAGAAGCTGACGGGTCGCCTCATGC-3′ (sense) and 5′-TGAGAGGGACCAGAGCGTGGTG-3′ (anti-sense) for GLUT1; 5ʹ-GACCTCTATGCCAACACAGT-3ʹ (sense) and 5ʹ-AGTACTTGCGCTCAGGAGGA-3ʹ (anti-sense) for β-actin. 2−ΔΔCT was employed to calculate the expressions.
Cell Transfections
Human prostate carcinoma cell lines 22Rv1 and normal prostate epithelial cell line HPrEC were purchased from ATCC (USA). Cells were cultured in DMEM with 10% FBS (ATCC 30-2020) at 37°C, 5% CO2. LncRNA-SNHG16 siRNA and negative control (NC, target no genes in human genome) siRNA were from Genepharma (Shanghai, China). Lipofectamine 2000 reagent (Thermo Fisher Scientific, Inc.) was used to transfect 50 nM siRNA into 5×105 cells. Cells were cultured for 12 h before subsequent experiments. Cells without transfection were set as control (C) cells and cells transfected with empty vector were negative control (NC) cells.
Glucose Uptake Assay
After transfection, lncRNA-SNHG16 expression was detected by qRT-PCR and overexpression rate above 200% was reached. Cells were cultured in glucose-free DMEM for 16 h, followed by incubation with high-glucose DMEM for another 24 h. Levels of intracellular glucose were measured using a fluorescence-based glucose assay kit (BioVision) with all operations performed following the instructions in the kit.
Cell Counting Kit-8 (CCK-8) Assay
After transfection, lncRNA-SNHG16 expression was detected by qRT-PCR and an overexpression rate above 200% was reached. Cell growth was measured by CCK-8 assay. Cell suspensions with a cell density of 5 ×104 cells per mL were prepared and 0.1 mL cell suspension was added into each well of a 96-well plate. The plate was kept in an incubator (37°C, 5% CO2). CCK-8 solution (10 ul, Sigma-Aldrich) was added into each well 24, 48, 72 and 96 h later. Cells were cultured for additional 4h, followed by the measurement of OD values at 450 nm.
Western-Blot
Total protein was extracted using RIPA solution (Sigma-Aldrich) and protein concentration was measured by BCA assay (Sigma-Aldrich). Following protein denaturation in boiling water for 10 min, SDS-PAGE (10%) gel electrophoresis was carried out to separate proteins. PVDF membranes were used to transfer proteins and blocking was performed in PBS containing 5% non-fat milk for 2h at room temperature. After that, membranes were incubated with rabbit anti-human primary antibodies of GLUT-1 (1:1500, ab15309, Abcam) and GAPDH antibody (1:1500, ab9485, Abcam) overnight at 4°C, followed by incubation with anti-rabbit IgG-HRP (1:1000, 435,036, MyBioSource) for 1h at room temperature. Signals were then developed by adding ECL (Sigma-Aldrich, USA). Image J V1.34 software was taken for signal normalization to GAPDH endogenous control.
Statistical Analysis
ROC analysis was carried out using default parameters. All data were expressed as mean±SD. Unpaired t-test was taken for comparisons between 2 groups and one-way ANOVA followed by LSD test was taken for comparisons in different groups. Pearson correlation was taken for correlation analyses. p < 0.05 was considered to be statistically significant.
Results
LncRNA-SNHG16 and GLUT1 mRNA in Prostate Tissues Were Up-Regulated in Prostate Carcinoma Patient Than in Normal Ones
Expression of lncRNA-SNHG16 and GLUT1 mRNA in prostate tissues of prostate carcinoma patients and normal ones was detected by qRT-PCR. According to Figure 1, expression levels of lncRNA-SNHG16 (Figure 1A) and GLUT1 mRNA (Figure 1B) were significantly higher in prostate carcinoma patients than those in normal ones (p<0.05).
Figure 1.
LncRNA-SNHG16 and GLUT1 mRNA in prostate tissues were up-regulated in prostate carcinoma patients than in normal ones. Data here show the comparison of expression levels of lncRNA-SNHG16 (A) and GLUT1 mRNA (B) between prostate carcinoma patients and normal ones (*p<0.05).
Up-Regulation of lncRNA-SNHG16 Effectively Distinguished Prostate Carcinoma Patient from Normal Ones
Diagnostic value of lncRNA-SNHG16 for prostate carcinoma was evaluated by ROC curve analysis. According to Figure 2, area under the curve is 0.9031 (standard error: 0.03116; 95% confidence interval: 0.8420–0.9642). Therefore, up-regulation of lncRNA-SNHG16 effectively distinguished prostate carcinoma patients from normal ones.
Figure 2.
Up-regulation of lncRNA-SNHG16 effectively distinguished prostate carcinoma patients from normal ones.
LncRNA-SNHG16 and GLUT1 mRNA Expression Was Positively Correlated in Prostate Carcinoma Patient but Not in Normal Ones
Pearson correlation coefficient was taken for correlation analysis of lncRNA-SNHG16 and GLUT1 mRNA expression. According to Figure 3A, a significantly positive relation in lncRNA-SNHG16 and GLUT1 mRNA expression was observed in prostate carcinoma patient. However, no significant relations in lncRNA-SNHG16 and GLUT1 mRNA expression was found in normal ones (Figure 3B, p<0.05).
Figure 3.
LncRNA-SNHG16 and GLUT1 mRNA expression was positively correlated in prostate carcinoma patient but not in normal ones. This figure shows the Pearson results of lncRNA-SNHG16 and GLUT1 mRNA expression in prostate carcinoma patients (A) and normal ones (B).
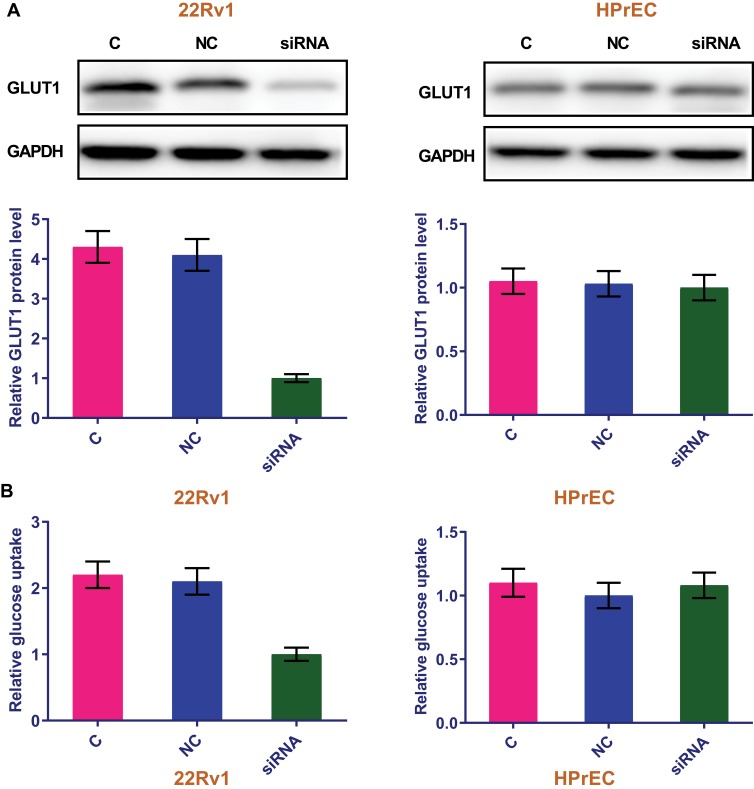
LncRNA-SNHG16 siRNA Silencing Inhibited GLUT1 Protein Expression and Reduced Glucose Uptake in Prostate Carcinoma Cells
To further investigate the relations in lncRNA-SNHG16 and GLUT1, cells of human prostate carcinoma cell lines 22Rv1 and normal prostate epithelial cell line HPrEC were transfected with lncRNA-SNHG16 siRNA and the effects on GLUT1 protein expression and GLUT1-related glucose uptake were detected by western and glucose uptake assay, respectively. According to Figure 4, in contrast to control cells (C, untransfected cells) and negative control cells (NC, cells transfected with NC siRNA), 22Rv1 cells with lncRNA-SNHG16 siRNA transfection showed significantly inhibited GLUT1 protein expression (Figure 4A) and reduced glucose uptake (Figure 4B) (p<0.05). However, lncRNA-SNHG16 siRNA transfection showed no significant effects on GLUT1 protein expression (Figure 4A) and glucose uptake (Figure 4B) in cells of normal prostate epithelial cell line HPrEC (p>0.05).
Figure 4.
LncRNA-SNHG16 siRNA silencing inhibited GLUT1 protein expression and reduced glucose uptake in prostate carcinoma cells. This figure shows the effects of lncRNA-SNHG16 siRNA silencing onGLUT1 protein expression (A) and glucose uptake (B) in cells of human prostate carcinoma cell lines 22Rv1 and normal prostate epithelial cell line HPrEC.
LncRNA-SNHG16 siRNA Silencing Inhibited Growth of Prostate Carcinoma Cells
CCK-8 assay was performed to explore the effects of lncRNA-SNHG16 siRNA silencing on the growth of prostate carcinoma cells. According to Figure 5, in contrast to control cells (C) and negative control cells (NC), 22Rv1 cells with lncRNA-SNHG16 siRNA transfection showed significantly inhibited cell growth (p<0.05). In contrast, lncRNA-SNHG16 siRNA silencing showed no significant effects on the growth of cells of normal prostate epithelial cell line HPrEC.
Figure 5.
LncRNA-SNHG16 siRNA silencing inhibited growth of prostate carcinoma cells including 22Rv1 (A) and HPrEC (B) (*p<0.05).
Discussions
The key finding of our study is that lncRNA-SNHG16 is also an oncogenic lncRNA in prostate carcinoma and the action of lncRNA-SNHG16 in prostate carcinoma is very likely to be achieved through the interaction with GLUT1.
The involvement of lncRNAs in the development and progression of prostate carcinoma has been extensively reported by previous studies. LncRNA HOTAIR was up-regulated in prostate carcinoma tissues and the overexpression of HOTAIR not only promotes the development of cancer but also reduces the sensitivity of cancer cells to chemotherapy.13 LncRNA ATB is also overexpressed in prostate carcinoma and promotes the growth of cancer cells.14 Up-regulation of lncRNA-SNHG16 has been observed in several types of cancers, such as bladder cancer11 and glioma.12 In our study we found significantly higher expression levels of lncRNA-SNHG16 in prostate carcinoma patients than those in normal ones, indicating the possible role of lncRNA-SNHG16 as an oncogenic lncRNA in this disease. It has been reported that altered expression of lncRNAs may provide guidance for the diagnosis of prostate carcinoma.15 In our study, we observed that up-regulation of lncRNA-SNHG16 effectively distinguished prostate carcinoma patient from normal ones. Therefore, up-regulation of lncRNA-SNHG16 may serve as a potential diagnostic marker for prostate carcinoma.
GLUT1 is a major player in glucose metabolism in cancer biology, which is usually up-regulated in different types of cancers.16 Overexpression of GLUT1 not only accelerated glucose metabolism but also protected cancer cells from glucose deprivation-induced oxidative stress.17 GLUT1 expression inhibition is considered to be a therapeutic target for cancer treatment.18 LncRNA has also been found to be involved in ovarian carcinoma.19 Being consistent with previous studies, our study observed significantly up-regulated expression of GLUT1 mRNA in prostate carcinoma patients than that in normal ones. It is known that GLUT-1 participates in certain pathological changes through the interactions with lncRNAs.9,10 Interestingly, our study observed a significantly positive relation in GLUT1 and lncRNA-SNHG16 expression in prostate carcinoma patient. In vitro experiment further confirmed that downregulation of GLUT1 leads to reduced expression levels of lncRNA-SNHG16 and decrease in glucose uptake. Therefore, lncRNA-SNHG16 is a positive regulator of GLUT1 in prostate carcinoma. However, this regulatory effect of lncRNA-SNHG16 on GLUT1 is very likely to be indirect because of less significant relations in lncRNA-SNHG16 and GLUT1 in normal ones. Certain pathological mediators should exit between lncRNA-SNHG16 and GLUT1.
It is worth to note that lncRNA-SNHG16 siRNA silencing showed no significant effects on glucose uptake and GLUT1 expression in normal prostate cells. In addition, lncRNA-SNHG16 siRNA silencing also did not significantly affect prostate carcinoma proliferation.
In conclusion, lncRNA-SNHG16 is overexpressed in prostate carcinoma. Inhibition of lncRNA-SNHG16 expression inhibits prostate carcinoma growth possibly by downregulating GLUT1.
Acknowledgments
The authors would like to thank the financial support from Shandong Natural Science Foundation (Grant No. ZR2016HL12).
Data Sharing Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Author Contributions
Jianan Zou supervised the whole study, had conception and design, analysis and interpretation of data, and revised the manuscript critically for important intellectual content. Mingfeng Shao, Ziqiang Yu had conception and design, acquisition of data, analysis and interpretation of data, and drafted the article. All authors had final approval of the version to be published; and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Disclosure
The authors declare that they have no competing interests in this work.
References
- 1.Goubran HA, Kotb RR, Stakiw J, et al. Regulation of tumor growth and metastasis: the role of tumor microenvironment. Cancer Growth Metastasis. 2014;7(CGM):S11285. doi: 10.4137/CGM.S11285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 3.Härmä V, Virtanen J, Mäkelä R, et al. A comprehensive panel of three-dimensional models for studies of prostate cancer growth, invasion and drug responses. PLoS One. 2010;5(5):e10431. doi: 10.1371/journal.pone.0010431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mulholland DJ, Tran LM, Li Y, et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell. 2011;19(6):792–804. doi: 10.1016/j.ccr.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vander Heiden MG, Plas DR, Rathmell JC, et al. Growth factors can influence cell growth and survival through effects on glucose metabolism. Mol Cell Biol. 2001;21(17):5899–5912. doi: 10.1128/MCB.21.17.5899-5912.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeBerardinis RJ, Sayed N, Ditsworth D, et al. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18(1):54–61. doi: 10.1016/j.gde.2008.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olsen JM, Sato M, Dallner OS, et al. Glucose uptake in brown fat cells is dependent on mTOR complex 2–promoted GLUT1 translocation. J Cell Biol. 2014;207(3):365–374. doi: 10.1083/jcb.201403080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meireles P, Sales‐Dias J, Andrade CM, et al. GLUT1-mediated glucose uptake plays a crucial role during plasmodium hepatic infection. Cell Microbiol. 2017;19(2):e12646. doi: 10.1111/cmi.12646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang Y, Zhu X, Xu Y, et al. Energy stress-induced lncRNA HAND2-AS1 represses HIF1α-mediated energy metabolism and inhibits osteosarcoma progression. Am J Cancer Res. 2018;8(3):526–537. [PMC free article] [PubMed] [Google Scholar]
- 10.Luo F, Liu X, Ling M, et al. The lncRNA MALAT1, acting through HIF-1α stabilization, enhances arsenite-induced glycolysis in human hepatic L-02 cells. Biochim Biophys Acta. 2016;1862(9):1685–1695. doi: 10.1016/j.bbadis.2016.06.004 [DOI] [PubMed] [Google Scholar]
- 11.Cao X, Xu J, Yue D. LncRNA-SNHG16 predicts poor prognosis and promotes tumor proliferation through epigenetically silencing p21 in bladder cancer. Cancer Gene Ther. 2018;25(1):10–17. doi: 10.1038/s41417-017-0006-x [DOI] [PubMed] [Google Scholar]
- 12.Y F L, Cai XL, Li ZZ, et al. LncRNA SNHG16 functions as an oncogene by sponging miR-4518 and up-regulating PRMT5 expression in glioma. Cell Physiol Biochem. 2018;45(5):1975–1985. doi: 10.1159/000487974 [DOI] [PubMed] [Google Scholar]
- 13.Zhang A, Zhao JC, Kim J, et al. LncRNA HOTAIR enhances the androgen-receptor-mediated transcriptional program and drives castration-resistant prostate cancer. Cell Rep. 2015;13(1):209–221. doi: 10.1016/j.celrep.2015.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu S, Yi XM, Tang CP, et al. Long non-coding RNA ATB promotes growth and epithelial-mesenchymal transition and predicts poor prognosis in human prostate carcinoma. Oncol Rep. 2016;36(1):10–22. doi: 10.3892/or.2016.4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee B, Mazar J, Aftab MN, et al. Long noncoding RNAs as putative biomarkers for prostate cancer detection. J Mol Diagn. 2014;16(6):615–626. doi: 10.1016/j.jmoldx.2014.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noguchi C, Kamitori K, Hossain A, et al. D-allose inhibits cancer cell growth by reducing GLUT1 expression. Tohoku J Exp Med. 2016;238(2):131–141. doi: 10.1620/tjem.238.131 [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez-Menendez P, Hevia D, Alonso-Arias R, et al. GLUT1 protects prostate cancer cells from glucose deprivation-induced oxidative stress. Redox Biol. 2018;17:112–127. doi: 10.1016/j.redox.2018.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shibuya K, Okada M, Suzuki S, et al. Targeting the facilitative glucose transporter GLUT1 inhibits the self-renewal and tumor-initiating capacity of cancer stem cells. Oncotarget. 2015;6(2):651–661. doi: 10.18632/oncotarget.v6i2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hang Y, Hu Y, Jin Z, et al. LncRNA snaR up-regulates GRB2-associated binding protein 2 and promotes proliferation of ovarian carcinoma cells. Biochem Biophys Res Commun. 2018;503(3):2028–2032. doi: 10.1016/j.bbrc.2018.07.152 [DOI] [PubMed] [Google Scholar]