Abstract
Wild animals have an ecological function and can serve as sentinels to identify infectious agents and as indicators of environmental health. Among the zoonotic pathogens, Salmonella spp. deserve special attention due to their high worldwide prevalence and their ubiquity of hosts. With the aim of investigating the presence of Salmonella spp. in wild birds from the Atlantic Forest in southern Bahia, Brazil, we collected 114 fecal samples of wild birds (14 families) between 2016 and 2017. Fecal samples were collected by means of cloacal swab and subjected to microbiological culture to isolate and serotype Salmonella spp. specifically. Antibiotic susceptibility was determined using the disk diffusion test protocol. Only one bird, Ceratopipra rubrocapilla, tested positive for Salmonella enterica subsp. enterica serotype Agona, which is the first record for this bird species. This isolate exhibited intermediate sensitivity to amikacin and gentamicin and sensitivity to the other 13 antibiotics tested. Results may indicate environmental preservation since the studied areas had minimal human activity and good sanitary quality. Despite the low prevalence, it is necessary to monitor wildlife and establish disease control and surveillance systems, especially for zoonotic diseases.
1. Introduction
The Atlantic Forest is the second largest forest in South America. Despite its extremely high levels of endemism (up to 40% for plants and 60% for amphibians), this biodiversity hotspot is also the most endangered biome of Brazil, as only 8% of its original area remains. The Brazilian population has been historically concentrated along the Southeastern coast, which resulted in more than 60% of the country's population (more than 100 million inhabitants) occupying the areas originally covered by the forest. This proximity to the effervescent development of the country's major capitals of the country leads to the extended cycles of land exploitation for the intensive production of commodity exports, particularly wood, sugarcane, and coffee. Additionally, the expansion of urban limits and country houses, illegal logging, subsidized soybean plantation, pine and eucalyptus production, palm heart extraction, wildlife poaching, and hydroelectric dams were additional factors that lead to the profound transformation of this landscape. The unprofitable areas of mountain ranges, marshes, and mangroves were the only areas spared from the devastation, alongside with a few small protected areas [1–3].
Currently, this biome accounts for less than 10% of the original native forest of the south of Bahia (Brazil) [3]. Even with deforestation, these remaining forest fragments are considered the centers of endemism of this ecosystem [4]. Human actions, such as irregular forest occupation and deforestation, may alter and influence the epidemiology of zoonosis by creating conditions for the dissemination of infectious agents and even the resurgence of emerging diseases [5]. Consequently, these ecosystems should be monitored regarding the presence of pathogens that are important for public health, to ensure the early implementation of control strategies.
Wild animals have an ecological function and can serve as sentinels to identify infectious agents and as indicators of environmental health [6]. Among the zoonotic pathogens, Salmonella spp. deserve special attention due to their high worldwide prevalence and their ubiquity of hosts. This bacterium is often associated with outbreaks and food infections and it is one of the world's three main food-borne diseases [7] with high morbidity and mortality rates [8]. Salmonella spp. are usually found in the intestinal tract of humans and animals, and they are transmitted through feces that eliminate the agent and contaminate water and food [9].
Studies indicate that Salmonella Typhimurium is the most prevalent serovar in wild birds [10, 11] with reports of outbreaks of septicemia and deaths in wild birds in the United Kingdom, Canada, Sweden, Switzerland, and Norway [12–16]. Wild birds affected by Salmonella spp. can present specific symptoms, such as pneumonia, anorexia, diarrheal stools, and neurological disorders or even sudden death [17, 18]. On the other hand, one of the main characteristics of Salmonella is the latent carriers. Latency corresponds to a condition in which the individual does not present clinical symptoms but remains intermittently eliminating the agent in the feces [19]. However, there is a period in which the etiological agent remains hidden in an intracellular compartment and is not eliminated and therefore may mask laboratory results. Thus, these latent carriers become natural reservoirs and therefore maintain the pathogen in both the food chain and nature [20].
Studies on the health of wildlife, especially on the role of species as carriers of Salmonella spp., can help identify the elements involved in the dissemination of the bacterium in the environment and the association with health factors and epidemiological variables. Additionally, such studies can help detect species that are more vulnerable to the agent and correlate the presence of the bacterium to anthropic factors such as deforestation and human occupation of environmental areas.
Thus, the aim of this study was to investigate the presence of Salmonella spp. in wild birds of the Atlantic Forest biome, in the mesoregion of southern Bahia, and characterize it phenotypically regarding serotype and antimicrobial susceptibility. These data can complement information on epidemiology, control, and prevention of this agent and, consequently, help maintain the quality of public health.
2. Material and Methods
2.1. Study Area
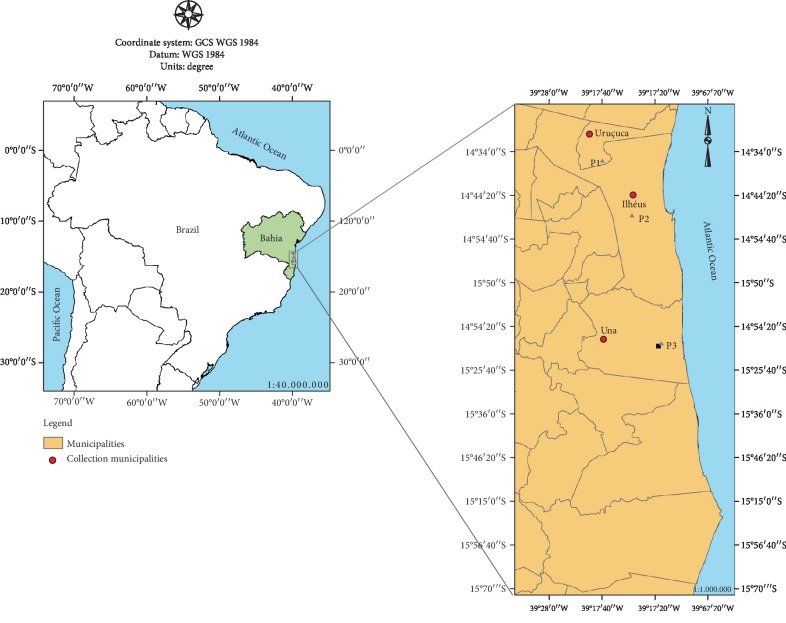
The animals were sampled in three collection points in three municipalities of southern Bahia (Ilhéus, Una, and Uruçuca) (Figure 1), in areas of the Atlantic Forest biome. Climate in this region is typically humid tropical with an annual average temperature of 24°C and an annual rainfall of around 1300 mm [21].
Figure 1.
Bird collection sites in southern Bahia, Brazil.
The collection points (P1, P2, and P3) have the following geographic coordinates: P1 (municipality of Uruçuca in the Fazenda Matinha Area: 14°35′58.3″S, 39°16′33.7″W), P2 (municipality of Ilhéus, at the UESC: 14°47′52.06″S, 39°10′33.366″W), and P3 (municipality of Una at the RPPN Nova Angélica Estate: 15°14′59.0″S, 39°04′41.0″W). Collection point P1 is a fragment of the Cabruca agroforestry system, which is a reforested area with cocoa plantations. The other collection points (P2 and P3) are fragments of native Atlantic Forest.
2.2. Sampling of Animals
In all, 114 birds were caught using fog nets for about 10 hours per day in each area. In the municipality of Uruçuca (P1), samples were collected from May to June 2017 in a single sampling. The nets were inspected every 40 minutes for 4 consecutive days, applying the sample effort of 32400 h·m2. The site in Ilhéus (P2) was sampled in November 2016 and April and June 2017. In Una (P3), samples were collected in November 2016 and March and August 2017. In P2 and P3, nets were inspected every 40 minutes for 5 consecutive days, applying a sampling effort of 40500 h·m2 in each collection period. In P1, P2, and P3, the nets were controlled 15 times a day with the interval of 40 minutes. The number of leaks and losses was 17, and there were 8 deaths in total.
For the acquisition of fecal samples by means of cloacal swab, the birds were kept in paper boxes for about 1 hour. Sterile swab samples were collected from the cloacal region. After this procedure, the samples were identified and placed in sterile microtube containing 1 mL of Buffered Peptone Water (APT) culture medium under temperature conditions (15-25°C), to be transported to the facilities of the Santa Cruz State University Veterinary Hospital, where they were analyzed for the presence of Salmonella spp.
Once captured, the specimens were removed from the nets, identified, and classified by order and family [22]. Then, they were numbered in increasing order, photographed, marked temporarily with non-toxic ink, and released in the sampled site. Bird capture was carried out under authorization No. 53000-1, issued by the Chico Mendes Institute for Biodiversity Conservation (ICMBio), and approval No. 014/2014 of the Ethics Committee on Animal Use of the State University of Santa Cruz (UESC).
2.3. Isolation and Identification of Salmonella spp.
Cloacal swab was incubated in 1 mL of Buffered Peptone Water–APT (Liofilchem) at 37°C/24 hours, for pre-enrichment. Then, selective enrichment was performed in 1 mL of Rappaport-Vassiliadis (RV) broth (Oxoid Ltd.) at 43°C/18-24 hours. On the third day, isolation was performed in xylose-lysine-deoxycholate-XLD agar (Neogen Corporation-Acumedia) and Hektoen-HE enteric agar (Becton, Dickinson and Company Sparks), both incubated in an incubator at 37°C/18-24 hours. The suspected colonies (red colonies with or without black center on XLD Agar and green to bluish-green colonies with or without black center in HE agar) were inoculated in 1 mL of Tryptone Soya Broth (TSB; HiMedia Laboratories Pvt. Ltd.) and incubated at 37°C/18-24 hours. Presumptive biochemical identification was performed using Triple Sugar Iron Agar (TSI; HiMedia Laboratories Pvt. Ltd.), Lysine Iron Agar (LIA; HiMedia Laboratories Pvt. Ltd.), and a urease test (Neogen Corporation-Acumedia).
The presumptive colonies of Salmonella spp. identified in the biochemical tests were confirmed through polymerase chain reaction (PCR), based on a previous study [23]. Each 25 μL of the reaction contained 1X PCR (Invitrogen, Carlsbad, CA, USA); 1.25 mM MgCl2 (Invitrogen); 200 μM dNTP; 10 pmol of each initiator ST11 (5′-AGCCAACCATTGCTAAATTGGCGCA-3′) and ST15 (5′-TTTGCGACTATCAGGTTACCGTGG-3′), specific for the genus Salmonella [24]; 1.25 U of Taq DNA polymerase (Invitrogen); and a suspicious colony of the pathogen. The reaction volume was completed to 25 μL with sterile water free of nucleases. Salmonella enteritidis PT1 was used for positive control, the colony being omitted from the reaction for the negative control. Reactions were performed in a ProFlex PCR thermal cycler (Applied Biosystems, Life Technologies, Carlsbad, USA). The amplification cycles consisted of 5 min at 94°C for initial denaturation, followed by 35 cycles (30 sec at 94°C, 30 sec at 62°C, and 1 min at 72o C), and a final extension step of 7 min at 72°C. The PCR product was visualized in 1% agarose gel stained with SYBER-Green (Invitrogen) and examined under UV light.
Positive PCR samples were cultivated in tryptic soy agar (TSA; HiMedia Laboratories Pvt. Ltd.) and sent to the Enterobacteria Laboratory of the Oswaldo Cruz Foundation (FIOCRUZ), Rio de Janeiro, for serotyping using serogroup- and serotype-specific antisera.
2.4. Antimicrobial Susceptibility Test
Antibiotic susceptibility was determined using the disk diffusion procedure in Mueller-Hinton agar (HiMedia Laboratories Pvt. Ltd.), with bacterial suspension adjusted to a turbidity of 0.5 McFarland standard, according to the Kirby-Bauer method [25] and following the guidelines by the Clinical Laboratory Standards Institute [26]. The disks used in the test were amikacin (30 μg), amoxicillin/clavulanic acid (20/10 μg), ampicillin + sulbactam (10/10 μg), cefepime (30 μg), cefoxitin (30 μg), ciprofloxacin (5 μg), chloramphenicol (30 μg), gentamicin (10 μg), imipenem (10 μg), lomefloxacin (10 μg), norfloxacin (10 μg), piperacillin-tazobactam (100/10 piperacillin-tazobactam), sulphazotrim (trimethoprim/sulfamethoxazole 25 μg), tobramycin (10 μg), and trimethoprim (5 μg) (LABORCLIN—Produtos para Laboratórios Ltda, Pinhais—Paraná, Brazil). The culture of Escherichia coli ATCC 25922 was used to control test quality.
2.5. Statistical Data Analysis
The signal test, which is a nonparametric statistical method, was applied to verify the agreement of the methodologies tested in the identification of Salmonella spp.
3. Results
A total of 114 samples of wild birds were studied: 77 being of undefined sex (13 juveniles and 64 adults), 19 males (7 juveniles and 12 adults), and 18 females (5 juveniles and 13 adults). The samples were of 32 different species, 4 of which were not identified (Table 1). None of the animals showed clinical signs or gross lesions that could be associated with salmonellosis (e.g., diarrhea, anorexia, neurological disorders, polyuria/polydipsia syndrome, and respiratory problems, such as dyspnea and pneumonia) [17, 18].
Table 1.
Taxonomic and geographic identification of wild birds from the Atlantic Forest of southern Bahia, Brazil.
| Order | Family | Species | Collection points (number of individuals) |
|---|---|---|---|
| Apodiformes | Trochilidae | Anthracothorax nigricollis | P2 (1) |
| Glaucis dohrnii | P2 (1) | ||
|
| |||
| Columbiformes | Columbidae | Geotrygon montana | P3 (1) |
| Leptotila rufaxilla | P2 (5) | ||
|
| |||
| Passeriformes | Dendrocolaptidae | Campylorhamphus trochilirostris | P3 (1) |
| Dendrocincla turdina | P1 (3), P2 (3), P3 (1) | ||
| Dendrocolaptes platyrostris | P2 (2) | ||
| Unidentified species | P2 (1) | ||
| Glyphorynchus spirurus | P3 (4) | ||
| Sittasomus griseicapillus | P1 (1) | ||
| Xiphorhynchus fuscus | P1 (2), P2 (2), P3 (4) | ||
| Fringillidae | Euphonia chlorotica | P2 (1) | |
| Furnariidae | Automolus leucophthalmus | P3 (1) | |
| Grallariidae | Unidentified species | P3 (1) | |
| Onychorhynchidae | Myiobius barbatus | P2 (1) | |
| Passerellidae | Arremon taciturnus | P2 (1) | |
| Pipridae | Ceratopipra rubrocapilla | P2 (7∗), P3 (5) | |
| Dixiphia pipra | P3 (9) | ||
| Machaeropterus regulus | P1 (1), P3 (5) | ||
| Manacus manacus | P2 (3), P3 (4) | ||
| Rhynchocyclidae | Unidentified species | P3 (1) | |
| Thraupidae | Coereba flaveola | P2 (1) | |
| Saltator maximus | P2 (1) | ||
| Tangara palmarum | P2 (2) | ||
| Tangara seledon | P2 (1) | ||
| Turdidae | Turdus amaurochalinus | P2 (1) | |
| Turdus leucomelas | P1 (1), P2 (9), P3 (8) | ||
| Turdus rufiventris | P1 (7), P2 (5), P3 (1) | ||
| Tyrannidae | Attila rufus | P1 (2) | |
| Unidentified species | P3 (1) | ||
| Rhytipterna simplex | P2 (1) | ||
|
| |||
| Piciformes | Picidae | Celeus flavescens | P3 (1) |
∗One sample was positive for Salmonella enterica subsp. enterica serotype Agona.
After biochemical tests, the PCR confirmed only one positive sample for Salmonella spp., in a bird of the species Ceratopipra rubrocapila (Figure 2). This specimen was an adult female found in the Cabruca system of the UESC site (P2).
Figure 2.

Exemplary of the Ceratopipra rubrocapilla species positive for Salmonella Agona. Photo: Josiane Moreira Rocha, 2017.
In serotyping, this isolate was identified as Salmonella enterica subsp. enterica serotype Agona (S. Agona). This is to the authors' knowledge the first isolation of Salmonella spp. in this bird species in Brazil. This sample was subjected to the antimicrobial susceptibility test and exhibited intermediate resistance to amikacin and gentamicin and sensitivity to the other antimicrobial agents tested.
4. Discussion
Our study showed a low prevalence of Salmonella spp. in wild birds caught in the forest in southern Bahia, probably because they are individuals from areas of fragments of native Atlantic Forest, with little human activity. In fact, the only positive specimen for Salmonella spp. was found in a fragment close to a urban area (P2). Bird species is usually found in the Atlantic Forest and in the Amazon region, in the south of river Amazonas. It is also found in Bolivia and Peru, particularly in wetlands and capoeira areas [27]. It is a bird of the Passeriformes order of the family Pipridae [22] and, in ecological terms, plays an important role in preserving the forest [28] because it usually feeds on small fruits and, consequently, helps scatter the seeds of different species of flora [29].
The low prevalence of Salmonella spp. in wild free-living birds has also been reported in other studies that found few positive results for this bacteria in free-living birds [11, 30]. The research conducted with 62 wild birds, namely 30 Columba livia, 19 Pterodroma baraui, 10 Puffinus lherminieri, and three Phaethon lepturus, from industrial sites and rescue centers, and found that only one specimen of two species each tested positive for Salmonella spp. [31]. The study with 364 Passeriformes and Piciformes found no positive isolate for Salmonella spp. [32]. However, birds from urban areas may exhibit a higher prevalence, as found in a study with 126 Columbiformes of the species Columbia livia in an urban area, in which 7.96% tested positive for bacteria of the genus Salmonella: 8 were Salmonella Typhimurium, one was Salmonella enterica subsp. enterica serotype 4,12,i and one was Salmonella enterica subsp. enterica serotype 4,12 [33]. This higher prevalence may be due to birds' access to areas of environmental contamination, e.g., sewage and human waste sites [34].
The prevalence of pathogens is higher when studying captive animals, since infectious agents in the captive environment can be more easily transmitted due to the greater contact between individuals and the plurality of their health status [35, 36]. It analyzed fecal samples and cloacal swabs of 30 Passeriformes of the species Paroaria dominicana and 19 of Paroaria coronata apprehended in the fight against the illegal wildlife traffic in the state of São Paulo, Brazil. Although the highest prevalence was for Escherichia coli (85.7%) and Klebsiella pneumoniae (57.1%), two specimens of Paroaria dominican were positive for Salmonella spp. (4%). The possible cause for the health status of these cardinals was attributed to the stress arising from the conditions of trafficking [37].
By associating the frequency of enterobacteria with the taxonomic order of birds, Suphoronski et al. [38] found that Salmonella spp. was more prevalent in Passeriformes, while Escherichia coli was more prevalent in Columbiformes. Moreover, it was observed that E. coli occurred in 82.33% of Columbiformes and Salmonella spp. occurred in 46.67% of the Passeriformes.
Salmonella Agona has been isolated from several species of animals. It can affect reptiles, domestic birds (anseriformes and galliformes), non-domestic birds, and mammals. Furthermore, it is one of the most prevalent serotypes isolated from cattle with clinical signs of salmonellosis; however, in pigs, it may occur in sick and asymptomatic animals [39]. This serotype was also isolated from seabirds [40], both in seagulls leaving in the proximity of garbage deposits [34] and in Magellan penguins from the Chilean Patagonia [41].
From an epidemiological perspective, the presence of this serotype in wild birds should not be ignored, despite the low prevalence, since in recent years the number of worldwide outbreaks determined by S. Agona has been considerable. From 2014 to July 2018, 147 cases were reported in the United Kingdom, Finland, Denmark, Germany, and Ireland. Of these 147 cases, the United Kingdom was responsible for 129 occurrences. Between 2007 and 2016, in countries of the European Union, S. Agona caused 13 outbreaks with 636 human cases and 12 hospitalizations, none of which resulted in deaths. In these countries, between 2004 and 2015, 4144 animals of different species tested positive for Salmonella Agona, especially chicken (n = 3236), cattle (n = 322), pigs (n = 271), ewes (n = 183), and turkeys (n = 61). In the same period, S. Agona was isolated from several food sources, mainly meat and its derivatives, especially pork (n = 512) and chicken meat (n = 422). To a lesser extent, this serotype can be isolated from other food categories, such as eggs and derivatives, fish products, fruits and vegetables, and food for animal consumption [42].
Studies have reported that S. Agona is resistant to sulfonamide, tetracycline [24], gentamicin, and chloramphenicol [43]. However, in our study, of the 15 tested antimicrobials, S. Agona was intermediately susceptible only to amikacin and gentamicin and sensitive to the others [44]. In our region (southern Bahia, Brazil), sulfonamide-resistant S. Agona was reported as the second most prevalent serotype in a study that analyzed 30 giant tegus (Tupinambis merianae) born in captivity and was isolated in 27% of asymptomatic adult individuals [24]. Increased growth of multidrug-resistant bacteria poses a potential risk to human and animal health. In wild animals, the prevalence of antibiotic-resistant strains reaches levels that are similar to those observed in humans and in domestic animals. This is explained by the presence of resistance genes, which are expressed in environments in which the selection pressure is a consequence of antimicrobial residues which contaminate the environment and harm the wildlife [44]. Environmental contamination caused by antimicrobials in forest areas may be more evident in perirural areas, because some property owners use these substances to treat and prevent animal diseases or apply subtherapeutic dosages in feed to foster growth [45]. The indiscriminate use of antimicrobials increases the dissemination of resistant agents in zoonosis and may indicate failures in treatment and misconceptions in the disease control system [46]. All these conditions, isolated or associated with each other, may cause resistance of the pathogens to the most diverse antimicrobial agents to increase or multiply [45]. The identification of these causes, as well as their subsequent control and neutralization, is critical to prevent the development of new resistance profiles [46].
5. Conclusion
The results of this study suggest a low prevalence of Salmonella spp. in wild birds from the Atlantic Forest in southern Bahia. However, considering the environmental heterogeneity of the studied sites, the multiplicity of species and niches of the Atlantic Forest, and the increasing anthropization of this biome, it is necessary to periodically monitor forest areas to identify pathogens of importance to public health and evaluate the effectiveness of the already implemented control measures. Some mitigating actions include reducing deforestation, controlling human occupation, and eliminating waste and wastewater in forests, in addition to implementing and maintaining environmental protection areas in rural properties. In parallel, the recorded data should be evaluated together with epidemiological elements and sanitary and environmental variables. This type of investigation is necessary because wild animals play a critical ecological role and may act as a reservoir of Salmonella spp., which means they can serve as sentinel species to identify infectious agents, indicate the quality of human and environmental health, and reveal the results of human action in forest areas.
Acknowledgments
This work was supported by Fundação de Amparo à Pesquisa do Estado da Bahia, Brasil (FAPESB; Grant PNED 001/2014) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; Grant 306308/2015-0). This study was also financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brasil (CAPES), Finance Code 001, for the scholarship to Eliege Jullia Eudoxia dos Santos.
Data Availability
The analyse data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
References
- 1.Dean W. A Ferro e Fogo: História e a Devastação da Mata Atlântica. São Paulo: Companhia das Letras; 1995. [Google Scholar]
- 2.Galindo-Leal C., Jacobsen T. R., Langhammer P. F., Olivieri S. State of the hotspots: the dynamics of biodiversity loss. The Atlantic Forest of South America: biodiversity status, threats, and outlook. 2003:12–23. [Google Scholar]
- 3.Tabarelli M., Pinto L. P., Silva J. M. C., Hirota M. M., Bedê L. C. Desafios e oportunidades para a conservação da biodiversidade na Mata Atlântica brasileira. Megadiversidade. 2005;1(1):132–138. [Google Scholar]
- 4.Araujo M., Alger K., Rocha R., Mesquita C. Mata Atlântica do sul da Bahia: situação atual, ações e perspectivas. 1998;8 [Google Scholar]
- 5.Hassell J. M., Begon M., Ward M. J., Fèvre E. M. Urbanization and disease emergence: dynamics at the wildlife–livestock–human interface. Trends in Ecology & Evolution. 2017;32(1):55–67. doi: 10.1016/j.tree.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foti M., Siclari A., Mascetti A., Fisichella V. Study of the spread of antimicrobial-resistant Enterobacteriaceae from wild mammals in the National Park of Aspromonte (Calabria, Italy) Environmental Toxicology and Pharmacology. 2018;63:69–73. doi: 10.1016/j.etap.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Wuyts V., Denayer S., Roosens N. H. C., et al. Whole genome sequence analysis of Salmonella enteritidis PT4 outbreaks from a National Reference Laboratory’s viewpoint. PLoS Currents. 2015;7 doi: 10.1371/currents.outbreaks.aa5372d90826e6cb0136ff66bb7a62fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arya G., Holtslander R., Robertson J., et al. Epidemiology, pathogenesis, genoserotyping, antimicrobial resistance, and prevention and control of non-typhoidal Salmonella serovars. Current Clinical Microbiology Reports. 2017;4(1):43–53. doi: 10.1007/s40588-017-0057-7. [DOI] [Google Scholar]
- 9.Kallapura G., Morgan M. J., Pumford N. R., et al. Evaluation of the respiratory route as a viable portal of entry for Salmonella in poultry via intratracheal challenge of Salmonella Enteritidis and Salmonella Typhimurium. Poultry Science. 2014;93(2):340–346. doi: 10.3382/ps.2013-03602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamer S. A., Lehrer E., Magle S. B. Wild birds as sentinels for multiple zoonotic pathogens along an urban to rural gradient in greater Chicago, Illinois. Zoonoses and Public Health. 2012;59(5):355–364. doi: 10.1111/j.1863-2378.2012.01462.x. [DOI] [PubMed] [Google Scholar]
- 11.Krawiec M., Kuczkowski M., Kruszewicz A., Wieliczko A. Prevalence and genetic characteristics of Salmonella in free-living birds in Poland. BMC Veterinary Research. 2015;11(1):p. 15. doi: 10.1186/s12917-015-0332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pennycott T. W., Mather H. A., Bennett G., Foster G. Salmonellosis in garden birds in Scotland, 1995 to 2008: geographic region, Salmonella enterica phage type and bird species. Veterinary Record. 2010;166(14):419–421. doi: 10.1136/vr.b4761. [DOI] [PubMed] [Google Scholar]
- 13.Daoust P. Y., Busby D. G., Ferns L., et al. Salmonellosis in songbirds in the Canadian Atlantic provinces during winter-summer 1997-98. The Canadian Veterinary Journal. 2000;41(1):54–59. [PMC free article] [PubMed] [Google Scholar]
- 14.Tauni M. A., Österlund A. Outbreak of Salmonella Typhimurium in cats and humans associated with infection in wild birds. Journal of Small Animal Practice. 2000;41(8):339–341. doi: 10.1111/j.1748-5827.2000.tb03214.x. [DOI] [PubMed] [Google Scholar]
- 15.Giovannini S., Pewsner M., Hüssy D., et al. Epidemic of salmonellosis in passerine birds in Switzerland with spillover to domestic cats. Veterinary Pathology. 2013;50(4):597–606. doi: 10.1177/0300985812465328. [DOI] [PubMed] [Google Scholar]
- 16.Refsum T., Vikøren T., Handeland K., Kapperud G., Holstad G. Epidemiologic and pathologic aspects of Salmonella Typhimurium infection in passerine birds in Norway. Journal of Wildlife Diseases. 2003;39(1):64–72. doi: 10.7589/0090-3558-39.1.64. [DOI] [PubMed] [Google Scholar]
- 17.Godoy S. N. Psittaciformes (arara, papagaio, periquito) In: Cubas Z. S., Silva J. C. R., Catão-Dias J. L., editors. Tratado de Animais Selvagens. Roca: São Paulo: 2007. pp. 222–251. [Google Scholar]
- 18.Harcourt-Brown N. H., Tully T. N., Jr., Dorrestein G. M., Jones A. K. Clínica de Aves. 2nd. Rio de Janeiro: Elsevier; 2010. Aves psitaciformes; pp. 122–123. [Google Scholar]
- 19.Tischler A. D., Mckinney J. D. Contrasting persistence strategies in _Salmonella_ and _Mycobacterium_. Current Opinion in Microbiology. 2010;13(1):93–99. doi: 10.1016/j.mib.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng S.-X., Cheng A.-C., Wang M.-S., Cao P. Gastrointestinal tract distribution of Salmonella enteritidis in orally infected mice with a species-specific fluorescent quantitative polymerase chain reaction. World Journal of Gastroenterology. 2007;13(48):6568–6574. doi: 10.3748/wjg.v13.i48.6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas W. W. Corredor de Biodiversidade da Mata Atlântica do Sul da Bahia. Publicação em CD-ROM, Ilhéus, IESB/CI/CABS/UFMG/UNICAMP; 2003. Natural vegetation types in southern Bahia; pp. 1–4. [Google Scholar]
- 22.Sigrist T. Guia de Campo Avis Brasilis: Avifauna Brasileira. Avisbrasilis Editora; 2013. [Google Scholar]
- 23.Maciel B. M., Argôlo Filho R. C., Nogueira S. S. C., Dias J. C. T., Rezende R. P. High prevalence of Salmonella in tegu lizards (Tupinambis merianae), and susceptibility of the serotypes to antibiotics. Zoonoses and Public Health. 2010;57(7-8):e26–e32. doi: 10.1111/j.1863-2378.2009.01283.x. [DOI] [PubMed] [Google Scholar]
- 24.Aabo S., Rasmussen O. F., Rossen L., Sorensen P. D., Olsen J. E. Salmonella identification by the polymerase chain reaction. Molecular and Cellular Probes. 1993;7(3):171–178. doi: 10.1006/mcpr.1993.1026. [DOI] [PubMed] [Google Scholar]
- 25.Bauer A. W., Kirby W. M. M., Sherris J. C., Turck M. Antibiotic susceptibility testing by a standardized single disk method. American Journal of Clinical Pathology. 1966;45(4_ts):493–496. doi: 10.1093/ajcp/45.4_ts.493. [DOI] [PubMed] [Google Scholar]
- 26.Wayne P. A. Clinical and Laboratory Standards Institute: performance standards for antimicrobial susceptibility testing: twenty-fourth informational supplement, M100-S24. 1. Vol. 34. Clinical and Laboratory Standards Institute (CLSI); 2014. [Google Scholar]
- 27.Snow D. Red-headed manakin (Ceratopipra rubrocapilla) In: Hoyo J., Elliott A., Sargatal J., Christie D. A., Juana E., editors. Handbook of the Birds of the World Alive. Barcelona: Lynx Edicions; 2018. https://www.hbw.com/node/57066. [Google Scholar]
- 28.Succo P. A., Vecchi M. B., Henud K. F., Ribeiro E. A., Alves M. A. S. Dieta de Ceratopipra rubrocapilla (aves: pipridae) na reserva biológica união. In: Monteiro L. R., Nascimento M. T., Júnior W. C., editors. I Encontro de Pesquisadores da Reserva Biológica União. Rio de Janeiro: 2016. pp. 53–55. (Org.) [Google Scholar]
- 29.Dario F. R. Diversity of frugivorous and omnivorous birds in different stages of ecological succession in Amazon Rainforest fragments. World News of Natural Sciences. 2017;15:37–48. [Google Scholar]
- 30.Gaukler S. M., Linz G. M., Sherwood J. S., et al. Escherichia coli, Salmonella and Mycobacterium avium subsp. paratuberculosis in wild European starlings at a Kansas cattle feedlot. Avian Diseases. 2009;53(4):544–551. doi: 10.1637/8920-050809-Reg.1. [DOI] [PubMed] [Google Scholar]
- 31.Tessier C., Atiana L. P., Lagadec E., Le Minter G., Denis M., Cardinale E. Wild fauna as a carrier of Salmonella in Reunion Island: Impact on pig farms. Acta Tropica. 2016;158:6–12. doi: 10.1016/j.actatropica.2016.01.027. [DOI] [PubMed] [Google Scholar]
- 32.Brittingham M. C., Temple S. A., Duncan R. M. A survey of the prevalence of selected bacteria in wild birds. Journal of Wildlife Diseases. 1988;24(2):299–307. doi: 10.7589/0090-3558-24.2.299. [DOI] [PubMed] [Google Scholar]
- 33.de Sousa E., Júnior A. B., Pinto A. A., et al. Prevalence of Salmonella spp. antibodies to Toxoplasma gondii, and Newcastle disease virus in feral pigeons (Columba livia) in the city of Jaboticabal, Brazil. Journal of Zoo and Wildlife Medicine. 2010;41(4):603–607. doi: 10.1638/2008-0166.1. [DOI] [PubMed] [Google Scholar]
- 34.Wahlstrom H., Tysen E., Engvall E. O., et al. Survey of Campylobacter species, VTEC O157 and Salmonella species in Swedish wildlife. Veterinary Record. 2003;153(3):74–80. doi: 10.1136/vr.153.3.74. [DOI] [PubMed] [Google Scholar]
- 35.Alves R. R. N., Lima J. R. D. F., Araujo H. F. P. The live bird trade in Brazil and its conservation implications: an overview. Bird Conservation International. 2013;23(1):53–65. doi: 10.1017/S095927091200010X. [DOI] [Google Scholar]
- 36.Fernandes-Ferreira H., Mendonça S. V., Albano C., Ferreira F. S., Alves R. R. N. Hunting, use and conservation of birds in Northeast Brazil. Biodiversity and Conservation. 2012;21(1):221–244. doi: 10.1007/s10531-011-0179-9. [DOI] [Google Scholar]
- 37.Cunha M. P. V., Guimarães M. B., Davies Y. M., Milanelo L., Knöbl T. Bactérias Gram-negativas em cardeais (Paroaria coronata e Paroaria dominicana) apreendidos do tráfico de animais silvestres. Brazilian Journal of Veterinary Research and Animal Science. 2016;53(1):107–111. doi: 10.11606/issn.1678-4456.v53i1p107-111. [DOI] [Google Scholar]
- 38.Suelen A. S., Tania F. R., Nadia C. W., Meire C. S., Adriano O. T. C. Occurrence of Salmonella sp. and Escherichia coli in free-living and captive wild birds from 2010-2013 in Guarapuava, Paran, Brazil. African Journal of Microbiology Research. 2015;9(29):1778–1782. doi: 10.5897/ajmr2015.7593. [DOI] [Google Scholar]
- 39.Hoelzer K., Switt A. M., Wiedmann M. Animal contact as a source of human non-typhoidal salmonellosis. Veterinary Research. 2011;42(1):p. 34. doi: 10.1186/1297-9716-42-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fresno M., Barrera V., Gornall V., et al. Identification of diverse Salmonella serotypes, virulotypes, and antimicrobial resistance phenotypes in waterfowl from Chile. Vector-Borne and Zoonotic Diseases. 2013;13(12):884–887. doi: 10.1089/vbz.2013.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dougnac C., Pardo C., Meza K., et al. Detection of Salmonella enterica in Magellanic penguins (Spheniscus magellanicus) of Chilean Patagonia: evidences of inter-species transmission. Epidemiology & Infection. 2015;143(6):1187–1193. doi: 10.1017/S0950268814002052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.EFSA - European Food Safety Authority and European Centre for Disease Prevention and Control. Multi-country outbreak of Salmonella Agona infections possibly linked to ready-to-eat food. 7, article 1465E. Vol. 15. EFSA Supporting Publications; 2018. [Google Scholar]
- 43.Donahue J. M. Emergence of antibiotic-resistant Salmonella Agona in horses in Kentucky. Journal of the American Veterinary Medical Association. 1986;188(6):592–594. [PubMed] [Google Scholar]
- 44.Gilliver M. A., Bennett M., Begon M., Hazel S. M., Hart C. A. Antibiotic resistance found in wild rodents. Nature. 1999;401(6750):233–234. doi: 10.1038/45724. [DOI] [PubMed] [Google Scholar]
- 45.Graziani C., Galetta P., Busani L., et al. Le Infezioni da Salmonella: Diagnostica, Epidemiologia e Sorveglianza-Salmonella Infections: Diagnosis, Epidemiology and Surveillance. 2005.
- 46.Botti V., Navillod F. V., Domenis L., et al. Salmonella spp. and antibiotic-resistant strains in wild mammals and birds in north-western Italy from 2002 to 2010. Veterinaria Italiana. 2013;49(2):195–202. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyse data used to support the findings of this study are included within the article.